Abstract
Purpose
We investigated the effect of basal protein expression on trastuzamab response in patients with Her2+ breast cancer who received trastuzamab and in Her2+ breast cancer cell lines.
Methods
Expression of CK5/6, CK14, and EGFR was evaluated after immunohistochemical staining in paraffin-embedded tissue of 97 patients with Stage 1-3 Her2+ breast cancer treated with chemotherapy/trastuzamab. Groups with and without basal protein expression were compared with respect to clinicopathologic parameters and survival. We treated 4 cell lines (2 basal-Her2(HCC1569, HCC1954) and 2 non-basal-Her2(BT474, SKBR3)) each with vehicle, trastuzamab (T), Paclitaxel (P), and T+P. Cell viability was assessed and Her2 pathway suppression was compared between groups using immunoblotting. Mammosphere formation was used to assess BCSC properties.
Results
EFGR expression was significant associated with cancer-specific survival (CSS) (p=0.05). CK5/6 expression strongly correlated with overall (OS), disease-free survival (DFS), and CSS (p=0.03, p=0.04, and p=0.03, respectively). Statistical significance was maintained for EGFR and CK5/6 after adjusting for covariates. CK14 was not associated with survival. All cell lines expressed similar levels of Her2. Both T and P alone inhibited proliferation of non-basal cell lines; T+P had an additive cytotoxic effect. Basal cells were resistant to T, P inhibited proliferation, but T+P had no additive cytotoxic effect on cell growth in basal cells. Immunoblotting showed a significant decrease in p-Akt levels after treatment with T or T+P in non-basal cells but not in basal cells. Akt blockade suppressed growth of basal and non-basal Her2+ cells. Furthermore, basal Her2 cell lines had increased mammosphere formation suggesting increased stem cell properties compared to non-basal Her2 cell lines.
Conclusions
CK5/6 and EGFR expression are predictive of worse prognosis in Her2+ breast cancer patients treated with trastuzamab. Basal-Her2 breast cancer cell lines are resistant to trastuzamab which is mediated through the Akt pathway; AKT inhibition abrogates this resistance. Basal Her2 cell lines also have increased stem cell properties which may play a role in the resistance pathway
Keywords: basal breast cancer, Her2 overexpression, trastuzamab resistance
Introduction
Human epidermal growth factor receptor 2-overexpressing (Her2+) breast cancer represents 20-25% of breast cancer and is associated with high relapse rates and poor prognosis.[1-3] Trastuzamab is a monoclonal antibody that targets the Her2 extracellular domain of the Her2 gene and inhibits downstream signaling of intracellular transduction cascades that control cell proliferation, survival, and differentiation. [4-8] While the exact mechanism of anti-tumor activity of trastuzamab in Her2+ breast cancer is unknown,[9-22] its introduction significantly impacted the treatment of Her2+ breast cancer with reduction in relapse rates of up to 50%.[23-26] However, some patients with Her2+ tumors have de novo resistance to trastuzamab, and 60-85% of patients with Her2+ metastatic breast cancer that initially respond to trastuzamab acquire resistance within a year.[23, 24, 27, 28]
Multiple targeted therapies have been developed to treat trastuzamab-resistant Her2+ breast cancer. All of these therapies target various downstream components of the pathway associated with Her2 signaling. However, acquired resistance may continue to be a challenge. This raises the question whether mechanisms outside of Her2 signaling should be investigated to target Her2+ breast cancer. In addition, there is evidence suggesting that there is heterogeneity of Her2 overexpression within Her2+ tumors [29-31] and there may be specific biologic features that predict which tumors exhibit more aggressive behavior.
We hypothesized that the basal phenotype, defined by expression of basal proteins, is a distinct biologic property associated with increased risk of recurrence and resistance to trastuzamab in Her2+ breast cancer. This subset of Her2+ breast cancer (basal-Her2) has been shown to carry a worse prognosis, [32-35] but little is known about how those with the basal-Her2 subtype respond to trastuzamab. The purpose of our study was to investigate the effect of basal protein expression on prognosis and trastuzamab response in both patients with Her2+ breast cancer and Her2+ breast cancer cell lines.
Methods
Patient and tumor specimen selection
Patients were identified from the Cedars-Sinai Medical Center (CSMC) Cancer Registry from January, 2005 through December, 2011 with Stage I-III Her2+ breast cancer who had surgery followed by chemotherapy and trastuzamab and were followed at CSMC. Patients who presented with Stage 4 disease, whose tumor tissue was not available for marker evaluation, who did not receive follow-up at CSMC, and who did not receive chemotherapy and trastuzamab were excluded. The following clinicopathologic data was obtained from review of medical records: age at diagnosis, tumor size, grade, histology, nodal status, estrogen and progesterone receptor (ER and PR) status, Her2 status, type of surgery, chemotherapy, radiation therapy, hormonal therapy, date and status of last follow-up.
Tumor specimen analysis
Archived paraffin-embedded tissue blocks from the primary tumor of patients who met the selection criteria were retrieved from the CSMC Department of Pathology. Pathologic review of slides was performed by a breast pathologist (SB) blinded to the results to confirm Her2+ status according to published guidelines [36, 37]. All 97 cases were confirmed to be amplified by fluorescence in situ hybridization (FISH). Positivity for HER2 amplification was defined by ratio of Her2 to CEP17 of greater than 2.2. All slides were reviewed to identify sections for further analysis. Selected tumor blocks were cut and stained according to standard protocol. Standard IHC techniques were used to stain tumors for CK5/6, CK14, and EGFR antibodies as described in our previously published work[32, 55]. Semi-quantitative analysis was performed and degree of immunoreactivity was independently scored by 2 investigators (SB, AC) blinded to clinical data based on percentage of positively stained slides and intensity of positive staining (0, 1+, 2+, or 3+). For EGFR, intensity score was based on the following scoring system: 1+ for faint or partial membranous staining, 2+ for complete weak membranous staining, and 3+ for strong diffuse membranous staining (Figure 1). Scores were estimated visually. Normal tissue was also selected as a control for all staining procedures. A final basal marker expression score was calculated by multiplying percentage of positively stained cells by intensity score. This score was used to determine EGFR expression, CK14 expression and CK5/6 expression for each of the respective markers. Statistical analysis was conducted to determine whether the expression score of each marker was independently associated with disease-free survival (DFS), cancer-specific survival (CSS) and/or overall survival (OS). Groups with and without significant basal marker expression were compared with respect to clinicopathologic parameters and survival.
Fig 1.
Representative slides of weak versus moderate versus strong staining for basal markers: a) CK5/6, b) CK14, and c) EGFR
Statistical Analysis
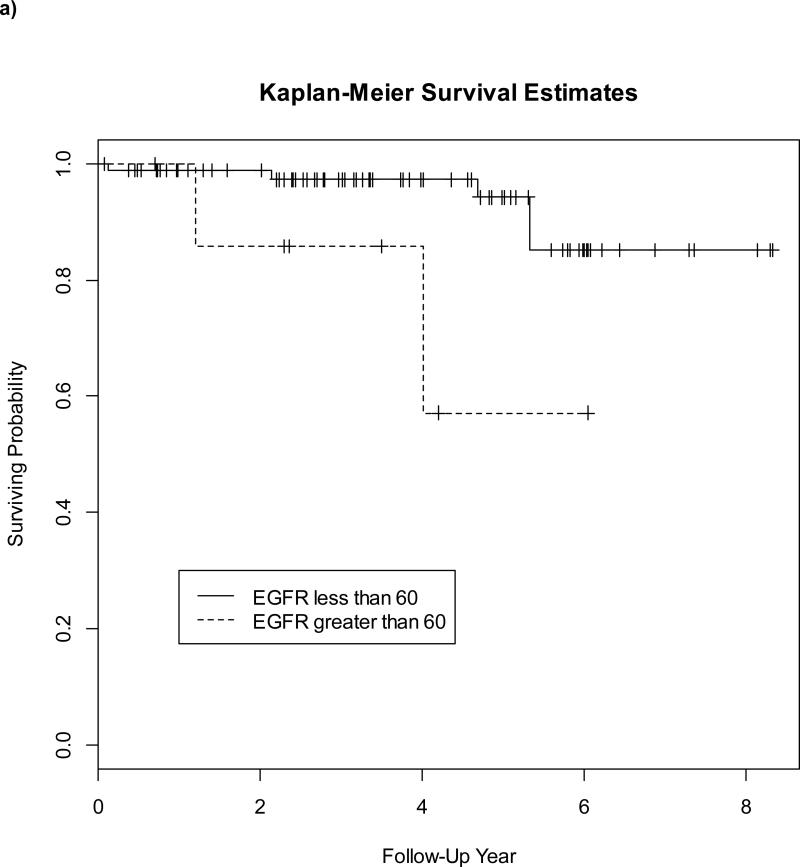
We used spearman correlations to test the association between the EGFR expression score and age at diagnosis and between tumor characteristics and the CK5/6 expression score. We used Wilcoxon rank sum test to test the association between the EGFR expression score and ER and PR status. Chi-square tests were conducted to test correlations between two categorical data points and Fisher exact tests were considered whenever there were one or more spare cells in the contingency table of two categorical variables. Survival analysis was performed using Cox regression models. The markers found to have a significant correlation with survival after univariable analysis were used to perform multivariable analysis adjusting for the following covariates: age at diagnosis, grade, tumor size, number of positive nodes, ER status and PR status. Kaplan–Meier curves for CSS and DFS were drawn for EGFR with the cut-point at 60 and the log-rank test was used for testing the significance between those above 60 versus below 60.
Cell Culture
The non-basal Her2+ human breast cancer cell lines, SKBR3 and BT474 from the American Type Culture Collection (ATCC, Rockville, MD), and the basal-Her2+ cell lines, HCC1569, HCC1954 (ATCC, Rockville, MD), and JIMT-1 (http://www.dsmz.de/), were used. These cell lines are well established as basal or non-basal based on gene expression microarray data[38, 39]. Numerous studies have shown that nonbasal-Her2 cells such as SKBR3 and BT474 are sensitive to Herceptin treatment.[14, 40-43] Cancer cell lines were cultured in DMEM or RPMI containing 10% fetal bovine serum (FBS) in 5% CO2 at 37°C. Cells were plated at 2,000 per well in 96-well microtiter plates and treated with vehicle control, trastuzamab (Herceptin) 20 ug/mL (T), Paclitaxel 0.01 uM (P), and trastuzamab (Herceptin) 20 ug/mL + Paclitaxel 0.01 uM (T+P). Cell proliferation was assessed using the Celltiter-Glo Luminescent Cell Viability kit from Promega Corporation (Madison, WI) according to the manufacturer's instructions.
Immunoblotting
Basal-Her2 and non-basal-Her2 cell lines were then treated for 2 hours with the same treatment regimen as described in the previous paragraph. Proteins were extracted from human breast cancer cells using RIPA lysis buffer (Sigma-Aldrich) and protein concentration was determined by the BCA Protein Assay Kit (Thermo). Proteins (20 μg) were separated by SDS–PAGE on 4-20% gradient gels and transferred onto PVDF membrane using Trans-Blot Turbo transfer buffer (Bio-Rad) and the Trans-Blot Turbo transfer system (Bio-Rad). Membranes were blocked in Odyssey blocking buffer (LI-COR) and incubated with primary antibodies (anti-human HER2, p-AKT, AKT, p-ERK, ERK and beta-actin) overnight at 4°C, and then incubated with IRDye 800CW secondary antibodies (LI-COR) for 1 hour at room temperature. The membranes were scanned using the Odyssey infrared imaging system (LICOR).
Immunofluorescence (IF)
Cells were placed into chamber slides at 70-80% confluence. Cells were fixed with 4% paraformaldehyde, permeabilized with 0.5% Triton X-100, blocked with 5% BSA, incubated with the same primary antibodies as used for immunoblotting overnight at 4°C and secondary antibodies for 1 hour at room temperature. The nuclei were stained with DAPI (4',6-diamidino-2-phenylindole). Images were acquired with Olympus microscope.
Mammosphere Culture
Complete MammoCult™ Medium was prepared by adding 50 mL of thawed MammoCult™ Proliferation Supplements to 450 mL of MammoCult™ Basal Medium (Human) supplemented with 4μg/ml Heparin and 0.48μg/ml Hydrocortisone per the manufacturer's instruction (StemCell Technologies). Single cells of all cell lines were plated on 6-well ultra-low attachment plates (Corning) at a density of 2000 viable cells/mL in Complete MammoCult™ Medium and incubated for 7 days. Fresh medium was added on day 3. The number of spheres greater than 50 μm in diameter were counted per well and this was repeated in triplicate. Mean number of mammospheres ± SEM were calculated for each cell line.
Results
Patient and tumor specimen analysis
We identified 97 patients with Stage 1-3 Her2+ invasive breast cancer treated with chemotherapy and trastuzamab who had adequate archived paraffin-embedded tissue available for analysis. Mean age at diagnosis was 53 (range 25-82). Table 1 lists the clinicopathologic characteristics (including the number of missing data points) of the 97 patients. Among the 97 patients for whom data was available, there was no significant difference in the number of patients with tumors that were T1 versus T2 or T3, no difference in the number of patients with PR-positive versus PR-negative tumors, nor was there any difference in the number of patients who received hormonal therapy versus those who did not receive hormonal therapy. More tumors were node-negative compared to node-positive (p<0.01), more tumors were high grade compared to low or intermediate grade (p<0.01), more tumors that were ER-positive tumors than ER-negative (p<0.01), more tumors were of ductal histology compared to tumors of other histology (p<0.01); more patients had lumpectomy compared to mastectomy (p<0.01), and more patients had radiation versus no radiation (p<0.01). Expression of at least one basal marker was demonstrated in 36/97 (37.1%): 15/97 (15.4%) had expression of CK 5/6, 8/97 (8.2%) expressed CK14, and 33/97 (34.0%) expressed EGFR. Sixty-one (62.9%) patients did not express any basal markers and only 6 (6.2%) patients had expression of all 3 markers. One patient (1.0%) had expression of only CK5/6; one (1.0%) had expression of CK14 alone; and 20 (20.6%) had only EGFR expression; 14 (14.4%) expressed at least 2 basal markers.
Table 1.
Tumor characteristics of patient cohort (n=97) using Chis-squared analysis.
| Tumor Characteristic | Number of patients (%) | Number Missing (%) | P-value |
|---|---|---|---|
| Tumor Size | 3 (3.1%) | 0.06* | |
| T1 | 38 (39.2%) | ||
| T2 | 47 (48.5%) | ||
| T3 | 9 (9.3%) | ||
| Nodal Status | 10 (10.3%) | <0.01* | |
| N0 | 51 (52.5%) | ||
| N1 | 20 (20.6%) | ||
| N2 | 11 (11.3%) | ||
| N3 | 5 (5.2%) | ||
| Tumor Grade | 2 (2.1%) | ||
| Low | 0 | <0.01 | |
| Intermediate | 18 (18.6%) | ||
| High | 77 (79.4%) | ||
| Tumor Histology | 0 | <0.01* | |
| Ductal | 86 (88.7%) | ||
| Lobular | 2 (2.1%) | ||
| Mixed Ductal-Lobular | 5 (5.2%) | ||
| Other | 4 (4.1%) | ||
| ER1 status | 1 (1%) | <0.01 | |
| Positive | 62 (63.9%) | ||
| Negative | 34 (35.1%) | ||
| PR2 status | 3 (3.1%) | 1.00 | |
| Positive | 47 (48.5%) | ||
| Negative | 47 (48.5%) | ||
| Primary Surgery Type | 4 (4.1%) | <0.01 | |
| Lumpectomy | 64 (66.0%) | ||
| Mastectomy | 29 (29.9%) | ||
| Radiation Therapy | 6 (6.2%) | <0.01 | |
| Yes | 67 (69.1%) | ||
| No | 24 (24.7%) | ||
| Hormonal Therapy | 1 (1%) | 0.22 | |
| Yes | 43 (44.3%) | ||
| No | 53 (54.6%) |
ER=estrogen receptor
PR=progesterone receptor
groups were combined for Chi-squared analysis (T2 and T3 for Tumor Size; N1, N2, N3 for Nodal Status; Lobular and Mixed Ductal-Lobular for Tumor Histology)
For each marker, we conducted survival analysis. EFGR expression had a significant association with CSS (p=0.049 using Cox regression analysis), and there was a trend towards significance for OS (p=0.056). Statistical significance was maintained after adjusting for covariates (p=0.03). There was no correlation between EGFR expression and DFS. CK5/6 expression had a strong correlation with CSS, DFS and OS (p=0.03, p=0.04, and p=0.03, respectively) . Statistical significance was maintained for CSS, DFS and OS after adjusting for covariates (p=0.03, p=0.01, and p=0.02, respectively). There was no significant correlation between CK14 expression and survival. Five-year CSS, DFS and OS of patients with CK5/6 expression compared to survival in those without basal marker expression was approximately 70% vs. 90.4% (p=0.39), 75% vs. 92.3% (p=0.33) and 70% vs. 90.6% (p=0.43), respectively. Additionally, statistical analysis identified a final basal marker expression score cutoff of 60 for which EGFR was significantly associated with worse CSS and there was a trend towards a significant correlation for this cutoff with DFS (p=0.06) but no significant threshold was identified for CK5/6 expression. Figure 2 demonstrates the Kaplan-Meier curves comparing CSS (A) and DFS (B) in patients with EFGR expression > 60 versus those < 60. Five-year CSS, DFS and OS of patients with EGFR expression > 60 compared to survival in those <60 was approximately 57% vs. approximately 90% (p=0.02), 67% vs. 90% (p=0.04) and 58.3% vs. 93.7% (p=0.07), respectively. Since a significant threshold for CK5/6 expression could not be identified, a similar Kaplan-Meier curve was not generated. Tumor factors associated with EGFR score > 60 versus < 60 included ER (p=0.001 by Fisher's exact test) and PR status (p=0.03 by Fisher's exact test). Factors associated with EGFR expression included younger age at diagnosis (p=0.035 by Spearman correlation), ER and PR expression (=<0.001 and p=0.0002, respectively, Wilcoxon rank sum). Tumor factors associated with CK5/6 expression included only tumor size (p=0.04 by Spearman correlation). Grade, histology, and nodal status were not associated with EGFR or CK5/6 expression. The spearman correlation between CK5/6 and EGFR was 0.49 (p-value < 0.01), demonstrating that CK5/6 and EGFR expression are highly correlated with one another. This strong correlation suggests that the statistical modeling must be performed individually for each basal marker. Therefore, analyses assessing various combinations of basal marker expression were not performed.
Fig 2.
Comparison of cancer-specific survival (a) and disease-free survival (b) for EGFR expression > 60 (dashed line) versus EGFR expression < 60 (solid line)
Effects of trastuzamab on cell viability in different Her2 cell lines
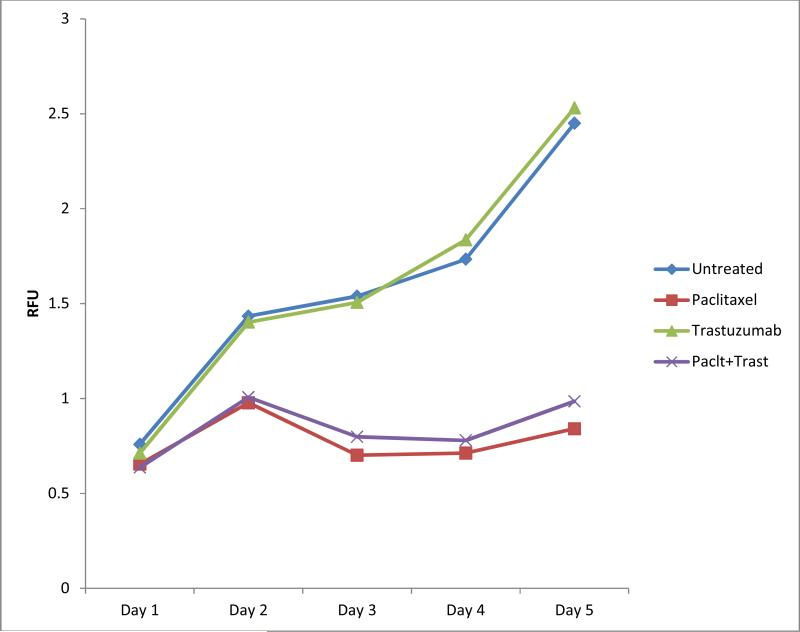
To explore the effects of basal protein expression on cellular responses to trastuzamab, we selected 4 breast cancer cell lines: 2 non-basal Her2 (BT474, SKBR3) and 2 basal Her2 (HCC1569, HCC1954) subtypes. All cell lines expressed similar levels of Her2 (Figure A1a). Her2 expression was further confirmed using IF. Basal and non-basal properties were confirmed by IF of basal cytokeratin CK5 and luminal cytokeratin CK18, respectively. Using cell viability assays, we found that H and P each inhibited cell proliferation and H+P enhanced the cytotoxic effect in non-basal-Her2 cell lines (Figure 3a and b). However, HCC1569 basal-Her2 cells were resistant to H when used alone or in combination with P (Figure 3c). Basal-Her2 HCC1954 (Figure 3d) and JIMT-1 (Figure A2) showed some effect with P alone and H+P.
Fig 3.
Non-basal Her2 cell lines BT474 (a) and SKBR3 (b) and basal Her2 cell lines HCC1569 (c) and HCC1954 (d) were treated with vehicle control (C), Trastuzamab 20 ug/mL (T), Paclitaxel 0.01 uM (P), or Trastuzamab 20 ug/mL + Paclitaxel 0.01 uM (T+P) over a 5 day course and cell proliferation was measured using CellTiter-Glo luminescent cell viability assay
Effects of trastuzamab on Akt signaling
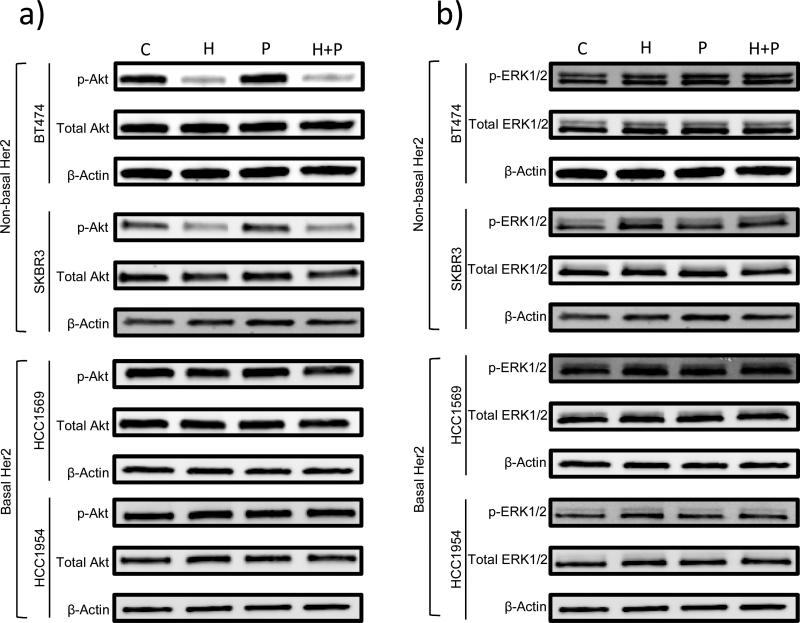
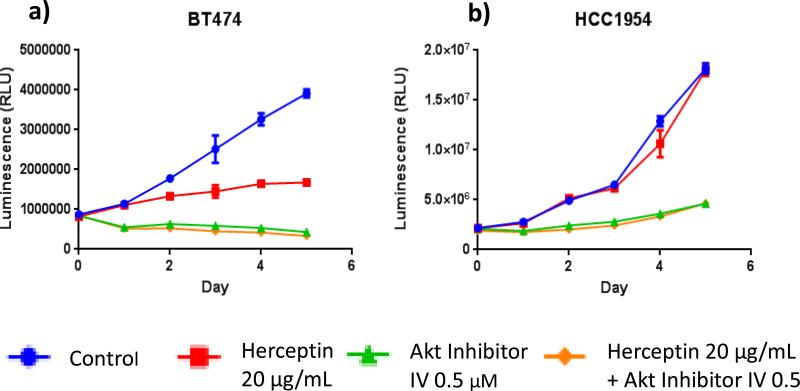
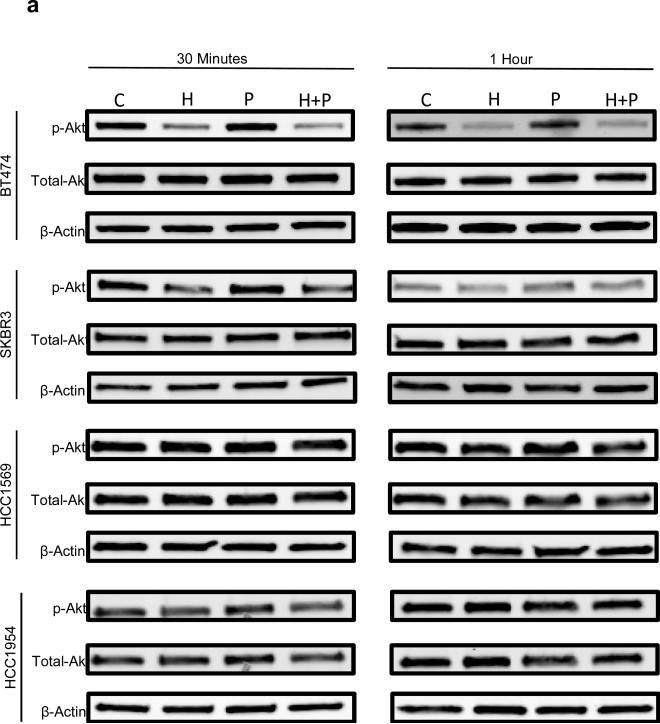
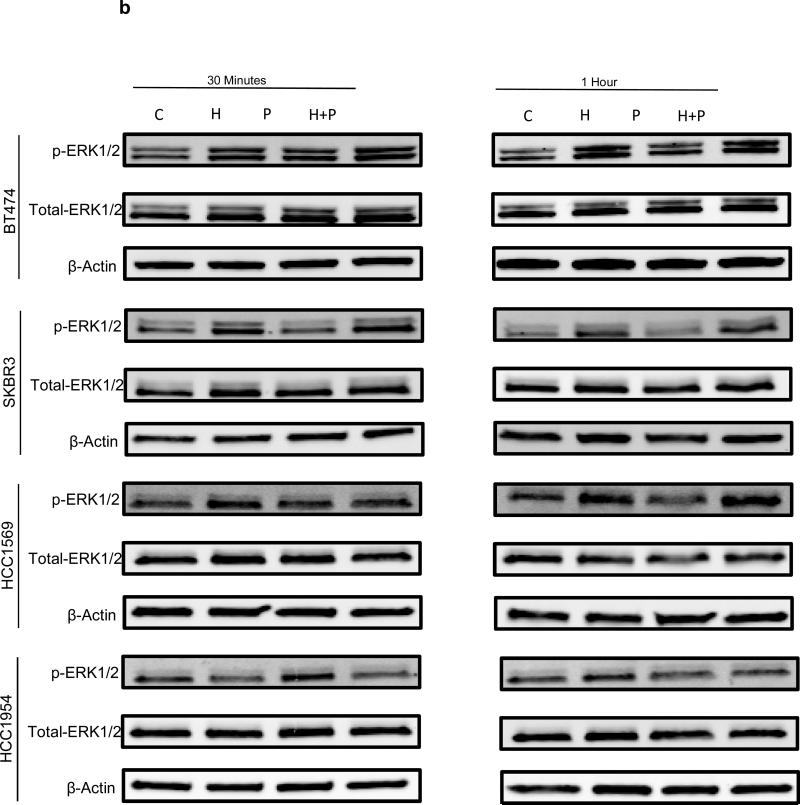
To investigate the molecular events underlying distinct responses to trastuzamab in different Her2 breast cancer cells, we studied the effects of H, P, or H+P on the Akt and Erk signaling pathways. Western blot analysis demonstrated a significant decrease in phospho-Akt levels after treatment with H or H+P in the non-basal-Her2 cell lines but not in the basal-Her2 cell lines (Figure 4a and A3a). In contrast, there were no detectable differences in the phospho-Erk levels in all tested cell lines under the same treatment (Figure 4b and A3b). Therefore, persistent and constitutively active Akt may be involved in resistance to trastuzamab and may be critical for Her2+ breast cancer cell growth. This was confirmed by a specific Akt inhibitor AI-IV. Akt blockade suppressed the growth of both basal and non-basal Her2+ cells (Figure 5). Taken together, these results suggest that trastuzamab resistance in basal-Her2 breast cancers may be mediated by the PI3K/Akt cell signaling pathway.
Fig 4.
Immunoblotting of non-basal Her2 (BT474, SKBR3) and basal Her2 (HCC1569, HCC1954) cell lines using a) p-Akt, total Akt and β-Actin antibodies and b) p-ERK 1/2, total ERK 1/2, and β-Actin antibodies after 2 hours of treatment with vehicle control (C), Trastuzamab 20 ug/mL (T), Paclitaxel 0.01 uM (P), or Trastuzamab 20 ug/mL + Paclitaxel 0.01 uM (T+P)
Fig 5.
Akt inhibition results in decreased cell proliferation in basal Her2 cell lines. Non-basal Her2 cell line BT474 (a) and basal Her2 cell line HCC1954 (b) were treated with vehicle control (blue), Trastuzamab 20 ug/mL (red), Akt Inhibitor IV 0.5 uM (green), and Trastuzamab 20 ug/mL + Akt Inhibitor IV 0.5 uM (orange) over a 5 day course and cell proliferation was measured using CellTiter-Glo luminescent cell viability assay
Breast cancer stem cell properties
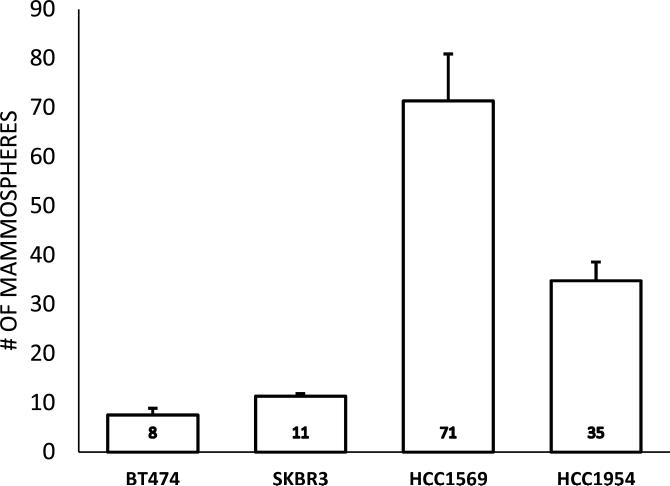
Cancer stem cells have been shown to be enriched in basal-like breast cancer. To explore whether basal Her2 breast cancer cells exhibit cancer stem cell properties, we examined the mammosphere growth capacity, which is a commonly used marker for BCSC, in the 4 breast cancer cell lines. Compared with non-basal Her2 cells, the mammosphere formation capacity was significantly higher in basal Her2 cells (Figure 6). These data demonstrate that basal Her2 cells have more cancer stem cell properties.
Fig 6.
Breast Cancer Stem Cell (BCSC) Activity: Non-basal Her2 cell lines BT474 (a) and SKBR3 (b) as well as basal Her2 cell lines HCC1569 (c) and HCC1954 (d) were cultured at low density (2000 cells per 9.5 cm2) in MammoCult™ Media for 7 days forming mammospheres according to previously published protocols. Mammospheres greater than 50 μm in diameter were counted per well and this was repeated in triplicate. Mean number of mammospheres ± SEM were calculated for each cell line
Discussion
Trastuzamab resistance remains a significant problem in the management of Her2+ breast cancer, and heterogeneity among Her2+ tumors may contribute to challenges in treating this disease. The basal-Her2 subtype appears to be a distinct entity that is associated with worse prognosis. Our clinical findings not only corroborate data reported in the literature associating the basal-Her2 subtype with worse survival, [44-46] but also provide evidence that basal-Her2 tumors are more likely to be resistant to trastuzamab.
The incidence of basal marker expression in Her2+ breast cancer is variable among studies. Our patient population had a higher rate of basal marker expression than reported in other studies when evaluating expression of any of the 3 markers. However, the rate of expression of the individual markers was lower: 15% for CK5/6 expression, 8% for CK14 expression and 34% for EGFR. In our previous work we identified 9% of Her2+ cases with expression of either CK5/6 or CK14.[32] Liu et al found the basal Her2 phenotype in 8.2% of cases.[34] Basal tumors are known to be characterized by expression of several markers, including CK5, CK5/6, CK14, CK17 and/or EGFR.[47-50] However, there is inconsistency in the expression of these markers and it is not clear which markers are most accurate in defining basal-like breast cancer either alone or in combination. We found CK5/6 and EGFR to be better predictors of outcome than CK14. We also found that CK5/6 expression was highly correlated with EGFR expression. CK 14 was only expressed in 8% of our cases, which may account for the lack of correlation with survival. However, Alshareeda et al also identified CK14 to be inferior to CK5/6 as a prognostic factor. They evaluated IHC-staining of CK5, CK5/6, CK14, CK17, in 995 invasive breast tumors with long-term follow-up and found that CK14 was expressed in 50% fewer cases compared to CK5/6 [50] and CK5/6 expression significantly correlated with survival but CK14 expression did not. Fulford et al evaluated CK14 expression in 443 invasive tumors and found that long-term survival was better in those with CK14 expression who did not develop distant metastases.[51]
We found CK5/6 and EGFR expression to be significantly associated with CSS, and there was a significant correlation between CK5/6 expression and OS. The association between EGFR expression and OS approached statistical significance. These findings confirm work previously reported in the literature. Bagaria and colleagues found 5-year OS of patients with Her2+ breast cancer and CK5/6 or CK14 expression to be 65% compared to 94% in those without basal marker expression.[32] Blows et al evaluated over 10,000 breast cancer specimens assessed by IHC for CK5/6 or EGFR expression [33] and found that the nonluminal-Her2+ cases had a poorer prognosis compared to luminal-Her2+ cases. Liu and colleagues performed tissue microarray and IHC to characterize 713 ER-negative breast cancers and demonstrated that the basal-Her2 subtype had the worst OS among all of the other subtypes.[34] Complete information on trastuzamab treatment was not available in these studies; therefore, conclusions regarding response to trastuzamab could not be made.
In this study we included only patients that received adjuvant chemotherapy and trastuzamab, in attempt to control for adjuvant treatment as a confounding variable. The patients that recurred in the face of trastuzamab had greater basal marker expression suggesting that these tumors were resistant to systemic therapy. Molecular profiling has been used to predict response to various chemotherapy regimens, and investigators have found the basal subtype to be associated with stronger response to chemotherapy compared to the other molecular subtypes.[52, 53] Therefore, the higher recurrence rates in patients with basal-Her2 tumors was less likely due to resistance to chemotherapy, rather, we believe the tumors were resistant to trastuzamab. Harris et al evaluated pathologic response of 48 patients with Her2+ breast cancer to neoadjuvant vinorelbine and trastuzamab and found that large tumors expressing basal markers demonstrated intrinsic resistance to this drug regimen.[54]
In addition to the clinical evidence that is growing to support our hypothesis, we have also presented preclinical evidence that the basal phenotype may play a role in trastuzamab resistance. Our cell line work confirms that trastuzamab enhances the effect of chemotherapy by causing a greater decrease in cell survival, and this was clearly demonstrated in the nonbasal cell lines. However, there was a significant increase in the viability of basal cells when treated with trastuzamab compared to nonbasal cells. This change was most pronounced in HCC1569 cells indicating that basal cells do not respond to trastuzamab. Wang et al. treated a panel of 13 Her2+ cell lines with trastuzamab and lapatinib and identified SKBR3 and BT474 cells to be responsive to both drugs and HCC1569 to have minimal growth inhibition, suggesting de novo resistance to both of these drugs in vitro.[55] There have been similar findings in the literature using the JIMT-1 cell line, which was also confirmed by our laboratory (Figure A2). JIMT-1 cells were originally isolated from the pleural effusion of a patient with trastuzamab-resistant Her2+ metastatic breast cancer.[56] This cell line has been used as a model to investigate potential mechanisms for drug resistance in Her2+ breast cancer.
There has been a rising interest in cancer stem cells as a potential biologic cause of resistance to cytotoxic therapy. Several studies have reported enrichment of breast cancer stem cells (BCSC) in breast tumors that are resistant to therapy.[57-62] In our stem cell work, we demonstrated that HCC1569 and HCC1954 cells were associated with a higher level of mammosphere formation compared to SKBR3 and BT474, indicating higher stem cell activity in the basal cells. Mammosphere-forming cells have been shown to be useful as a tool to predict CSC-like cells.[63, 64] Croker and colleagues demonstrated that high ALDH expression and CD44+/CD24− expression were associated with increased colony growth, adhesion, migration and invasion in cell lines and enhanced tumorigenicity and metastasis in mouse models.[65] In a later study the investigators found that inhibition of ALDH activity reduces chemotherapy and radiation resistance of stem-like ALDHhiCD44+ human breast cancer cells.[66] Assessment of ALDH or CD44+/24− activity in the 4 cell lines would be strong confirmatory tests for stem cell activity and is a plan for future experimentation in our laboratory.
Trastuzamab blocks Her1/Her3/PI3K complex formation which inhibits Akt.[67] We explored ERK and Akt, 2 major downstream signaling pathways, as potential pathways involved in trastuzamab resistance. Our immunoblotting results suggest that Akt signaling may play a role in trastuzamab resistance of the basal cells, but ERK does not. Our Akt inhibition findings suggest that trastuzamab sensitivity is restored by Akt inhibition. In a previous study of Akt inhibition in basal breast cancer, our laboratory found Akt to play a role in transcription of FOXC1, a critical mediator of EGFR function in basal breast cancer, and that EGFR activation induces FOXC1 through ERK and Akt pathways.[68] Chandarlapaty et al [69] evaluated specimens from patients with Her2+ trastuzamab-resistant breast cancer and found high rates of PI3K-AKT activation. O'Brien and colleagues [70] evaluated response to trastuzumab and lapatinib in 18 Her2+ cell lines innately resistant to these drugs and found that increased activation of the PI3K/AKT pathway correlated with resistance to trastuzumab, which could be overcome by lapatinib. Basal cell lines were among three of the 18 cell lines evaluated in this study. Fabi et al [71] investigated the clinical impact of the expression of PTEN, p-Akt and PI3K in Her2+ metastatic breast cancer patients treated with trastuzumab and found that patients co-expressing PTEN and p-Akt had a statistically significant longer PFS as compared to the rest of patients (p = 0.01). The literature supports involvement of the Akt pathway in resistance to trastuzamab, but many of the studies do not provide information on the molecular subtype of the tumor or cells in question. Our work demonstrates a link between basal marker expression, stem cell marker expression, Akt signaling, and resistance to trastuzamab.
Conclusions
Trastuzamab resistance is a complex and challenging problem in management of Her2+ breast cancer. As new targeted therapies continue to be developed and molecular subtyping becomes further refined, multiple avenues of overcoming drug resistance will likely evolve. Our data support the basal phenotype, defined by basal and stem cell marker expression, and its link to Akt signaling as one clinically relevant pathway of trastuzamab resistance. Further studies are needed to characterize the role of Akt in this phenomenon with the hope that additional therapy may be tailored to improve outcome in patients with this aggressive subtype of breast cancer.
Clinical Practice Points.
Tumor expression of basal proteins has been shown to be associated with worse prognosis and can be expressed in Her2+ breast cancer.
Our research provides clinical and laboratory evidence that basal protein expression in Her2+ tumors is associated with resistance to anti-Her2 therapy, and that Akt signaling may play a significant role in the mechanism of resistance.
As the mechanisms of drug resistance by basal tumors becomes further elucidated, it may be important to consider alternative therapies in patients with the basal subtype of Her2+ breast cancer.
Aknowledgments & Funding
This work was supported by UCLA CTSI Clinical Scholars Program (NIH/National Center for Advancing Translational Science UCLA Clinical and Translational Science Institute Grant No. UL1TR000124), the Cedars-Sinai Medical Center Clinical Scholars Program (CSMC CTSI Whiting Eigler Grant), National Institutes of Health (CA151610, CA129822, CA140995), the Avon Foundation for Women (02-2014-063), and David Salomon Translational Breast Cancer Research Fund to Xiaojiang Cui, the Fashion Footwear Charitable Foundation of New York, Inc., Associates for Breast and Prostate Cancer Studies, the Margie and Robert E. Petersen Foundation, and the Linda and Jim Lippman Research Fund to Armando Giuliano.
Appendix
Fig A1.
a Immunoblotting of non-basal Her2 (BT474, SKBR3) and basal Her2 (HCC1569, HCC1954) cell lines with HER2 antibody
Fig A1b Non-basal Her2 and basal Her2 cell lines were co-stained for CK5 (basal cytokeratin) and CK18 (luminal cytokeratin) antibodies. BT474 and SKBR3 were positive for CK18 but not CK5 whereas HCC1569 and HCC1954 were positive for CK5 but not CK18 confirming the molecular phenotypes of all four cell lines
Fig A2.
Cell proliferation was assessed using the CellTiter-Glo luminescent assay after treating basal Her2 cell line JIMT-1 with vehicle control (C), Trastuzamab 20 ug/mL (T), Paclitaxel 0.01 uM (P), and Trastuzamab 20 ug/mL + Paclitaxel 0.01 uM (T+P) over a 5 days. Similar to the other basal Her2 cell lines (HCC1569, HCC1954), JIMT-1 cell proliferation was not inhibited by trastuzamab (grey) relative to the control (blue). Paclitaxel alone (green) did inhibit cell proliferation in JIMT-1 cells but the combination of Trastuzamab and Paclitaxel did not have an additive cytotoxic effect
Fig A3a.
Immunoblotting of non-basal Her2 (BT474, SKBR3) and basal Her2 (HCC1569, HCC1954) cell lines with p-Akt, total Akt and β-Actin antibodies after 30 minutes and 1 hour of treatment with vehicle control (C), Trastuzamab 20 ug/mL (T), Paclitaxel 0.01 uM (P), or Trastuzamab 20 ug/mL + Paclitaxel 0.01 uM (T+P)
Fig A3b.
Immunoblotting of non-basal Her2 (BT474, SKBR3) and basal Her2 (HCC1569, HCC1954) cell lines with p-ERK 1/2, total ERK 1/2 and β-Actin antibodies after 30 minutes and 1 hour of treatment with vehicle control (C), Trastuzamab 20 ug/mL (T), Paclitaxel 0.01 uM (P), or Trastuzamab 20 ug/mL + Paclitaxel 0.01 uM (T+P)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of Interest
The authors have no conflicts of interest to disclose.
References
- 1.Slamon DJ, Clark GM, Wong SG, et al. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987;235:177–182. doi: 10.1126/science.3798106. [DOI] [PubMed] [Google Scholar]
- 2.Sjogren S, Inganas M, Lindgren A, et al. Prognostic and predictive value of c-erbB-2 overexpression in primary breast cancer, alone and in combination with other prognostic markers. J Clin Oncol. 1998;16:462–469. doi: 10.1200/JCO.1998.16.2.462. [DOI] [PubMed] [Google Scholar]
- 3.Owens MA, Horten BC, Da Silva MM. HER2 amplification ratios by fluorescence in situ hybridization and correlation with immunohistochemistry in a cohort of 6556 breast cancer tissues. Clin Breast Cancer. 2004;5:63–69. doi: 10.3816/cbc.2004.n.011. [DOI] [PubMed] [Google Scholar]
- 4.Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nat Rev Mol Cell Biol. 2001;2:127–137. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- 5.Park JW, Neve RM, Szollosi J, Benz CC. Unraveling the biologic and clinical complexities of HER2. Clin Breast Cancer. 2008;8:392–401. doi: 10.3816/CBC.2008.n.047. [DOI] [PubMed] [Google Scholar]
- 6.Lin NU, Winer EP. Brain metastases: the HER2 paradigm. Clin Cancer Res. 2007;13:1648–1655. doi: 10.1158/1078-0432.CCR-06-2478. [DOI] [PubMed] [Google Scholar]
- 7.Fendly BM, Kotts C, Vetterlein D, et al. The extracellular domain of HER2/neu is a potential immunogen for active specific immunotherapy of breast cancer. J Biol Response Mod. 1990;9:449–455. [PubMed] [Google Scholar]
- 8.Lewis GD, Figari I, Fendly B, et al. Differential responses of human tumor cell lines to anti-p185HER2 monoclonal antibodies. Cancer Immunol Immunother. 1993;37:255–263. doi: 10.1007/BF01518520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lazar GA, Dang W, Karki S, et al. Engineered antibody Fc variants with enhanced effector function. Proc Natl Acad Sci U S A. 2006;103:4005–4010. doi: 10.1073/pnas.0508123103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clynes RA, Towers TL, Presta LG, Ravetch JV. Inhibitory Fc receptors modulate in vivo cytotoxicity against tumor targets. Nat Med. 2000;6:443–446. doi: 10.1038/74704. [DOI] [PubMed] [Google Scholar]
- 11.Cooley S, Burns LJ, Repka T, Miller JS. Natural killer cell cytotoxicity of breast cancer targets is enhanced by two distinct mechanisms of antibody-dependent cellular cytotoxicity against LFA-3 and HER2/neu. Exp Hematol. 1999;27:1533–1541. doi: 10.1016/s0301-472x(99)00089-2. [DOI] [PubMed] [Google Scholar]
- 12.Molina MA, Codony-Servat J, Albanell J, et al. Trastuzumab (herceptin), a humanized anti-Her2 receptor monoclonal antibody, inhibits basal and activated Her2 ectodomain cleavage in breast cancer cells. Cancer Res. 2001;61:4744–4749. [PubMed] [Google Scholar]
- 13.Anido J, Scaltriti M, Bech Serra JJ, et al. Biosynthesis of tumorigenic HER2 C-terminal fragments by alternative initiation of translation. EMBO J. 2006;25:3234–3244. doi: 10.1038/sj.emboj.7601191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yakes FM, Chinratanalab W, Ritter CA, et al. Herceptin-induced inhibition of phosphatidylinositol-3 kinase and Akt Is required for antibody-mediated effects on p27, cyclin D1, and antitumor action. Cancer Res. 2002;62:4132–4141. [PubMed] [Google Scholar]
- 15.Asanuma H, Torigoe T, Kamiguchi K, et al. Survivin expression is regulated by coexpression of human epidermal growth factor receptor 2 and epidermal growth factor receptor via phosphatidylinositol 3-kinase/AKT signaling pathway in breast cancer cells. Cancer Res. 2005;65:11018–11025. doi: 10.1158/0008-5472.CAN-05-0491. [DOI] [PubMed] [Google Scholar]
- 16.Le XF, Lammayot A, Gold D, et al. Genes affecting the cell cycle, growth, maintenance, and drug sensitivity are preferentially regulated by anti-HER2 antibody through phosphatidylinositol 3-kinase-AKT signaling. J Biol Chem. 2005;280:2092–2104. doi: 10.1074/jbc.M403080200. [DOI] [PubMed] [Google Scholar]
- 17.Zhou W, Carpenter G. Heregulin-dependent translocation and hyperphosphorylation of ErbB-2. Oncogene. 2001;20:3918–3920. doi: 10.1038/sj.onc.1204517. [DOI] [PubMed] [Google Scholar]
- 18.Sabbatini P, McCormick F. Phosphoinositide 3-OH kinase (PI3K) and PKB/Akt delay the onset of p53-mediated, transcriptionally dependent apoptosis. J Biol Chem. 1999;274:24263–24269. doi: 10.1074/jbc.274.34.24263. [DOI] [PubMed] [Google Scholar]
- 19.Le XF, Claret FX, Lammayot A, et al. The role of cyclin-dependent kinase inhibitor p27Kip1 in anti-HER2 antibody-induced G1 cell cycle arrest and tumor growth inhibition. J Biol Chem. 2003;278:23441–23450. doi: 10.1074/jbc.M300848200. [DOI] [PubMed] [Google Scholar]
- 20.Le XF, Pruefer F, Bast RC., Jr. HER2-targeting antibodies modulate the cyclin-dependent kinase inhibitor p27Kip1 via multiple signaling pathways. Cell Cycle. 2005;4:87–95. doi: 10.4161/cc.4.1.1360. [DOI] [PubMed] [Google Scholar]
- 21.Lane HA, Motoyama AB, Beuvink I, Hynes NE. Modulation of p27/Cdk2 complex formation through 4D5-mediated inhibition of HER2 receptor signaling. Ann Oncol. 2001;12(Suppl 1):S21–22. doi: 10.1093/annonc/12.suppl_1.s21. [DOI] [PubMed] [Google Scholar]
- 22.Izumi Y, Xu L, di Tomaso E, et al. Tumour biology: herceptin acts as an anti-angiogenic cocktail. Nature. 2002;416:279–280. doi: 10.1038/416279b. [DOI] [PubMed] [Google Scholar]
- 23.Slamon DJ, Leyland-Jones B, Shak S, et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med. 2001;344:783–792. doi: 10.1056/NEJM200103153441101. [DOI] [PubMed] [Google Scholar]
- 24.Cobleigh MA, Vogel CL, Tripathy D, et al. Multinational study of the efficacy and safety of humanized anti-HER2 monoclonal antibody in women who have HER2-overexpressing metastatic breast cancer that has progressed after chemotherapy for metastatic disease. J Clin Oncol. 1999;17:2639–2648. doi: 10.1200/JCO.1999.17.9.2639. [DOI] [PubMed] [Google Scholar]
- 25.Romond EH, Perez EA, Bryant J, et al. Trastuzumab plus adjuvant chemotherapy for operable HER2-positive breast cancer. N Engl J Med. 2005;353:1673–1684. doi: 10.1056/NEJMoa052122. [DOI] [PubMed] [Google Scholar]
- 26.Piccart-Gebhart MJ, Procter M, Leyland-Jones B, et al. Trastuzumab after adjuvant chemotherapy in HER2-positive breast cancer. N Engl J Med. 2005;353:1659–1672. doi: 10.1056/NEJMoa052306. [DOI] [PubMed] [Google Scholar]
- 27.Baselga J, Tripathy D, Mendelsohn J, et al. Phase II study of weekly intravenous recombinant humanized anti-p185HER2 monoclonal antibody in patients with HER2/neu-overexpressing metastatic breast cancer. J Clin Oncol. 1996;14:737–744. doi: 10.1200/JCO.1996.14.3.737. [DOI] [PubMed] [Google Scholar]
- 28.Vogel CL, Cobleigh MA, Tripathy D, et al. Efficacy and safety of trastuzumab as a single agent in first-line treatment of HER2-overexpressing metastatic breast cancer. J Clin Oncol. 2002;20:719–726. doi: 10.1200/JCO.2002.20.3.719. [DOI] [PubMed] [Google Scholar]
- 29.Pectasides D, Gaglia A, Arapantoni-Dadioti P, et al. HER-2/neu status of primary breast cancer and corresponding metastatic sites in patients with advanced breast cancer treated with trastuzumab-based therapy. Anticancer Res. 2006;26:647–653. [PubMed] [Google Scholar]
- 30.Mittendorf EA, Wu Y, Scaltriti M, et al. Loss of HER2 amplification following trastuzumab-based neoadjuvant systemic therapy and survival outcomes. Clin Cancer Res. 2009;15:7381–7388. doi: 10.1158/1078-0432.CCR-09-1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hurley J, Doliny P, Reis I, et al. Docetaxel, cisplatin, and trastuzumab as primary systemic therapy for human epidermal growth factor receptor 2-positive locally advanced breast cancer. J Clin Oncol. 2006;24:1831–1838. doi: 10.1200/JCO.2005.02.8886. [DOI] [PubMed] [Google Scholar]
- 32.Bagaria SP, Ray PS, Wang J, et al. Prognostic Value of Basal Phenotype in HER2-overexpressing Breast Cancer. Ann Surg Oncol. 2011 doi: 10.1245/s10434-011-2032-5. [DOI] [PubMed] [Google Scholar]
- 33.Blows FM, Driver KE, Schmidt MK, et al. Subtyping of breast cancer by immunohistochemistry to investigate a relationship between subtype and short and long term survival: a collaborative analysis of data for 10,159 cases from 12 studies. PLoS Med. 2010;7:e1000279. doi: 10.1371/journal.pmed.1000279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu H, Fan Q, Zhang Z, et al. Basal-HER2 phenotype shows poorer survival than basal-like phenotype in hormone receptor-negative invasive breast cancers. Hum Pathol. 2008;39:167–174. doi: 10.1016/j.humpath.2007.06.012. [DOI] [PubMed] [Google Scholar]
- 35.Oliveras-Ferraros C, Vazquez-Martin A, Martin-Castillo B, et al. Pathway-focused proteomic signatures in HER2-overexpressing breast cancer with a basal-like phenotype: new insights into de novo resistance to trastuzumab (Herceptin). Int J Oncol. 2010;37:669–678. doi: 10.3892/ijo_00000716. [DOI] [PubMed] [Google Scholar]
- 36.Pathologists' Guideline Recommendations for Immunohistochemical Testing of Estrogen and Progesterone Receptors in Breast Cancer. Breast Care (Basel) 2010;5:185–187. doi: 10.1159/000315039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gong Y, Sweet W, Duh YJ, et al. Chromogenic in situ hybridization is a reliable method for detecting HER2 gene status in breast cancer: a multicenter study using conventional scoring criteria and the new ASCO/CAP recommendations. Am J Clin Pathol. 2009;131:490–497. doi: 10.1309/AJCPI00TVGIGYXAA. [DOI] [PubMed] [Google Scholar]
- 38.Neve RM, Chin K, Fridlyand J, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–527. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kao J, Salari K, Bocanegra M, et al. Molecular profiling of breast cancer cell lines defines relevant tumor models and provides a resource for cancer gene discovery. PLoS One. 2009;4:e6146. doi: 10.1371/journal.pone.0006146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O'Neill F, Madden SF, Clynes M, et al. A gene expression profile indicative of early stage HER2 targeted therapy response. Mol Cancer. 2013;12:69. doi: 10.1186/1476-4598-12-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brockhoff G, Heckel B, Schmidt-Bruecken E, et al. Differential impact of Cetuximab, Pertuzumab and Trastuzumab on BT474 and SK-BR-3 breast cancer cell proliferation. Cell Prolif. 2007;40:488–507. doi: 10.1111/j.1365-2184.2007.00449.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Diermeier S, Horvath G, Knuechel-Clarke R, et al. Epidermal growth factor receptor coexpression modulates susceptibility to Herceptin in HER2/neu overexpressing breast cancer cells via specific erbB-receptor interaction and activation. Exp Cell Res. 2005;304:604–619. doi: 10.1016/j.yexcr.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 43.Diermeier-Daucher S, Breindl S, Buchholz S, et al. Modular anti-EGFR and anti-Her2 targeting of SK-BR-3 and BT474 breast cancer cell lines in the presence of ErbB receptor-specific growth factors. Cytometry A. 2011;79:684–693. doi: 10.1002/cyto.a.21107. [DOI] [PubMed] [Google Scholar]
- 44.Ribelles N, Perez-Villa L, Jerez JM, et al. Pattern of recurrence of early breast cancer is different according to intrinsic subtype and proliferation index. Breast Cancer Res. 2013;15:R98. doi: 10.1186/bcr3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Voduc KD, Cheang MC, Tyldesley S, et al. Breast cancer subtypes and the risk of local and regional relapse. J Clin Oncol. 2010;28:1684–1691. doi: 10.1200/JCO.2009.24.9284. [DOI] [PubMed] [Google Scholar]
- 46.Rakha EA, El-Sayed ME, Green AR, et al. Prognostic markers in triple-negative breast cancer. Cancer. 2007;109:25–32. doi: 10.1002/cncr.22381. [DOI] [PubMed] [Google Scholar]
- 47.Cheang MC, Voduc D, Bajdik C, et al. Basal-like breast cancer defined by five biomarkers has superior prognostic value than triple-negative phenotype. Clin Cancer Res. 2008;14:1368–1376. doi: 10.1158/1078-0432.CCR-07-1658. [DOI] [PubMed] [Google Scholar]
- 48.Nielsen TO, Hsu FD, Jensen K, et al. Immunohistochemical and clinical characterization of the basal-like subtype of invasive breast carcinoma. Clin Cancer Res. 2004;10:5367–5374. doi: 10.1158/1078-0432.CCR-04-0220. [DOI] [PubMed] [Google Scholar]
- 49.Livasy CA, Karaca G, Nanda R, et al. Phenotypic evaluation of the basal-like subtype of invasive breast carcinoma. Mod Pathol. 2006;19:264–271. doi: 10.1038/modpathol.3800528. [DOI] [PubMed] [Google Scholar]
- 50.Alshareeda AT, Soria D, Garibaldi JM, et al. Characteristics of basal cytokeratin expression in breast cancer. Breast Cancer Res Treat. 2013;139:23–37. doi: 10.1007/s10549-013-2518-x. [DOI] [PubMed] [Google Scholar]
- 51.Fulford LG, Reis-Filho JS, Ryder K, et al. Basal-like grade III invasive ductal carcinoma of the breast: patterns of metastasis and long-term survival. Breast Cancer Res. 2007;9:R4. doi: 10.1186/bcr1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rouzier R, Perou CM, Symmans WF, et al. Breast cancer molecular subtypes respond differently to preoperative chemotherapy. Clin Cancer Res. 2005;11:5678–5685. doi: 10.1158/1078-0432.CCR-04-2421. [DOI] [PubMed] [Google Scholar]
- 53.Carey LA, Dees EC, Sawyer L, et al. The triple negative paradox: primary tumor chemosensitivity of breast cancer subtypes. Clin Cancer Res. 2007;13:2329–2334. doi: 10.1158/1078-0432.CCR-06-1109. [DOI] [PubMed] [Google Scholar]
- 54.Harris LN, You F, Schnitt SJ, et al. Predictors of resistance to preoperative trastuzumab and vinorelbine for HER2-positive early breast cancer. Clin Cancer Res. 2007;13:1198–1207. doi: 10.1158/1078-0432.CCR-06-1304. [DOI] [PubMed] [Google Scholar]
- 55.Wang YC, Morrison G, Gillihan R, et al. Different mechanisms for resistance to trastuzumab versus lapatinib in HER2-positive breast cancers--role of estrogen receptor and HER2 reactivation. Breast Cancer Res. 2011;13:R121. doi: 10.1186/bcr3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nagy P, Friedlander E, Tanner M, et al. Decreased accessibility and lack of activation of ErbB2 in JIMT-1, a herceptin-resistant, MUC4-expressing breast cancer cell line. Cancer Res. 2005;65:473–482. [PubMed] [Google Scholar]
- 57.Phillips TM, McBride WH, Pajonk F. The response of CD24(−/low)/CD44+ breast cancer-initiating cells to radiation. J Natl Cancer Inst. 2006;98:1777–1785. doi: 10.1093/jnci/djj495. [DOI] [PubMed] [Google Scholar]
- 58.Woodward WA, Chen MS, Behbod F, et al. WNT/beta-catenin mediates radiation resistance of mouse mammary progenitor cells. Proc Natl Acad Sci U S A. 2007;104:618–623. doi: 10.1073/pnas.0606599104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Diehn M, Cho RW, Lobo NA, et al. Association of reactive oxygen species levels and radioresistance in cancer stem cells. Nature. 2009;458:780–783. doi: 10.1038/nature07733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Duru N, Fan M, Candas D, et al. HER2-associated radioresistance of breast cancer stem cells isolated from HER2-negative breast cancer cells. Clin Cancer Res. 2012;18:6634–6647. doi: 10.1158/1078-0432.CCR-12-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shafee N, Smith CR, Wei S, et al. Cancer stem cells contribute to cisplatin resistance in Brca1/p53-mediated mouse mammary tumors. Cancer Res. 2008;68:3243–3250. doi: 10.1158/0008-5472.CAN-07-5480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Opyrchal M, Salisbury JL, Iankov I, et al. Inhibition of Cdk2 kinase activity selectively targets the CD44+/CD24−/Low stem-like subpopulation and restores chemosensitivity of SUM149PT triple-negative breast cancer cells. Int J Oncol. 2014;45:1193–1199. doi: 10.3892/ijo.2014.2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cioce M, Gherardi S, Viglietto G, et al. Mammosphere-forming cells from breast cancer cell lines as a tool for the identification of CSC-like- and early progenitor-targeting drugs. Cell Cycle. 2010;9:2878–2887. [PubMed] [Google Scholar]
- 64.Rappa G, Lorico A. Phenotypic characterization of mammosphere-forming cells from the human MA-11 breast carcinoma cell line. Exp Cell Res. 2010;316:1576–1586. doi: 10.1016/j.yexcr.2010.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Croker AK, Goodale D, Chu J, et al. High aldehyde dehydrogenase and expression of cancer stem cell markers selects for breast cancer cells with enhanced malignant and metastatic ability. J Cell Mol Med. 2009;13:2236–2252. doi: 10.1111/j.1582-4934.2008.00455.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Croker AK, Allan AL. Inhibition of aldehyde dehydrogenase (ALDH) activity reduces chemotherapy and radiation resistance of stem-like ALDHhiCD44(+) human breast cancer cells. Breast Cancer Res Treat. 2012;133:75–87. doi: 10.1007/s10549-011-1692-y. [DOI] [PubMed] [Google Scholar]
- 67.Junttila TT, Akita RW, Parsons K, et al. Ligand-independent HER2/HER3/PI3K complex is disrupted by trastuzumab and is effectively inhibited by the PI3K inhibitor GDC-0941. Cancer Cell. 2009;15:429–440. doi: 10.1016/j.ccr.2009.03.020. [DOI] [PubMed] [Google Scholar]
- 68.Jin Y, Han B, Chen J, et al. FOXC1 is a Critical Mediator of EGFR Function in Human Basal-like Breast Cancer. Ann Surg Oncol. 2014 doi: 10.1245/s10434-014-3980-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chandarlapaty S, Sakr RA, Giri D, et al. Frequent mutational activation of the PI3K-AKT pathway in trastuzumab-resistant breast cancer. Clin Cancer Res. 2012;18:6784–6791. doi: 10.1158/1078-0432.CCR-12-1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.O'Brien NA, Browne BC, Chow L, et al. Activated phosphoinositide 3-kinase/AKT signaling confers resistance to trastuzumab but not lapatinib. Mol Cancer Ther. 2010;9:1489–1502. doi: 10.1158/1535-7163.MCT-09-1171. [DOI] [PubMed] [Google Scholar]
- 71.Fabi A, Metro G, Di Benedetto A, et al. Clinical significance of PTEN and p-Akt co-expression in HER2-positive metastatic breast cancer patients treated with trastuzumab-based therapies. Oncology. 2010;78:141–149. doi: 10.1159/000312656. [DOI] [PubMed] [Google Scholar]