An Introduction to Autophagy
Autophagy is an evolutionarily conserved recycling pathway that maintains protein and organelle quality control in systems ranging from unicellular organisms such as yeast to complex multicellular systems i.e., flies, worms, and mammals [28]. In essence, autophagic pathways entail the recognition, sequestration, and delivery of cytosolic cargo to lysosomes for degradation [28]. In mammalian systems, three distinct forms of autophagy have been described, macroautophagy [28], microautophagy [61], and chaperone-mediated autophagy (CMA) [1, 14], that differ from each other in mechanism of delivery and the type of cargo delivered to the lysosome. Macroautophagy is an in-bulk degradative pathway that turns over redundant or damaged organelles and protein aggregates as well as soluble proteins (Fig. 3.1). Macroautophagy requires the de novo formation of double membrane structures termed autophagosomes, which sequester cargo and then fuse with lysosomes or late endosomes to form autophago-lysosomes and amphisomes, respectively. These fusion events result in the exposure of sequestered cargo to lysosomal acid-sensitive hydrolases that lead to cargo degradation (Fig. 3.1). In contrast, CMA selectively degrades single soluble proteins containing a specific amino acid signature, the KFERQ motif, which is recognized by cytosolic Hsc70 followed by substrate delivery to lysosomes by the CMA receptor, lysosome-associated membrane protein (LAMP)-2A (Fig. 3.2) [1]. In essence, autophagy pathways are considered as protective pathways, and a significant body of evidence now supports this notion wherein cell/tissue-specific loss of autophagy has been shown to give rise to neurodegenerative disorders, metabolic defects, and cancers, to mention just a few. Macroautophagy and CMA have also been shown to decrease with age [9, 12], underscoring the possibility that compromised autophagy activity with age contributes to development of age-related diseases, for instance, neurodegeneration [39] and metabolic defects [66]. Macroautophagy and CMA are better characterized out of the autophagy pathways, and thus the discussion on the roles of these autophagy pathways on development of age-associated conditions will remain the focus of this chapter.
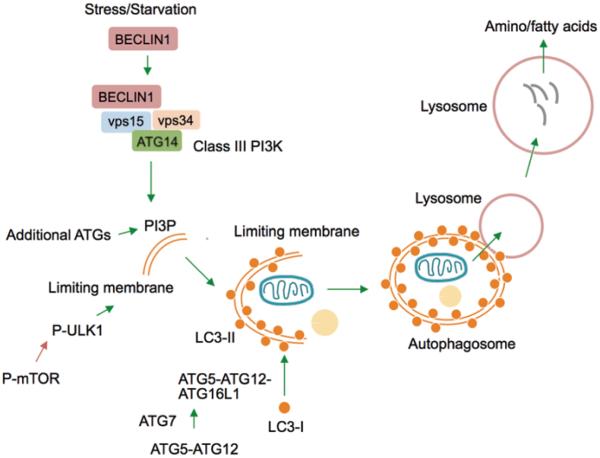
Fig. 3.1.
Steps in the macroautophagy pathway. Macroautophagy is a cellular quality control mechanism that requires greater than 30 Atg gene products to deliver cytosolic cargo to lysosomes for their degradation. Starvation or stressors activate macroautophagy by increasing the interaction of Beclin1 with additional proteins to generate the Class III phosphoinositide 3-kinase (PI3K) complex, which gives rise to PI3Ps to recruit ATG proteins (e.g., ULK1) for autophagosome formation. Activation of ATG7 (E1-like ligase) forms the ATG5-ATG12 conjugate, which binds to ATG16L1 to form the ATG5-ATG12-ATG16L1 complex. ATG5-ATG12-ATG16L1 facilitates lipidation of cytosolic LC3-I into autophagosome membrane-bound LC3-II. LC3-II-positive autophagosomes engulf cargo and target it to lysosomes, wherein a battery of hydrolases degrades cargo into amino- and fatty acids. A tight regulatory crosstalk between the nutrient sensor mTOR and ULK1 orchestrates autophagy. Green arrows indicate activating steps and red arrows indicate inhibitory steps
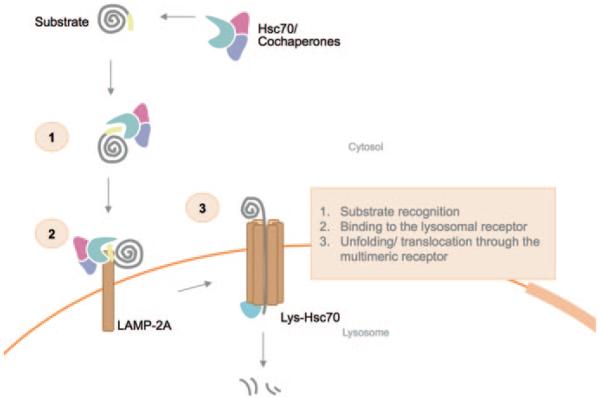
Fig. 3.2.
Regulation of chaperone-mediated autophagy. Chaperone-mediated autophagy ensues by (1) recognition of substrate by the Hsc70/Co-chaperone complex followed by (2) binding of the substrate to the cytosolic portion of the LAMP-2A receptor. (3) Binding is followed by substrate unfolding, and along with additional regulatory proteins and lysosomal resident Hsc70 (Lys-Hsc70) the unfolded substrate is translocated across the lysosomal membrane substrate via LAMP-2A multimers for their degradation
The Macroautophagy Apparatus
Yeast genetic studies have identified over 30 autophagy gene (ATG) proteins that regulate tight orchestration of macroautophagy during basal conditions and its activation during stress or starvation [26, 62, 73]. In mammals, induction of macroautophagy requires Beclin-1 to interact with additional ATG proteins and vacuole protein sorting (vps) proteins that together constitute class III PI3K activity (Fig. 3.1) [74]. Activation of class III PI3K generates PI3Ps that recruit additional ATG proteins for autophagic limiting membrane formation [28]. Two ubiquitin-like conjugation cascades allow the expansion of the limiting membranes into autophagosomes [22]. An identity of some of the critical ATG proteins involved in autophagosome biogenesis is as follows: ATG7 displays a unique ubiquitin E1-like ligase activity that initiates binding of ATG12 with ATG5 (Fig. 3.1) [54, 70]. In subsequent steps, ATG12-ATG5 interacts with ATG16L1 to give rise to ATG12-ATG5-ATG16L1 [59]. ATG12-ATG5-ATG16L1 trimers, in turn, catalyze the conjugation microtubule-associated protein 1 light chain 3 (LC3) to phosphatidylethanolamine on the autophagic membrane (Fig. 3.1) resulting in membrane formation [23]. Lipidated LC3, i.e. LC3-II, is a molecular signature of the autophagosome [71], and is structurally and functionally essential for autophagosome formation and cargo recognition due to its ability to bind to a cargo adaptor p62 [52]. Autophagosomes traffic along microtubules to fuse with lysosomes in a SNARE protein-dependent manner resulting in cargo degradation [30].
Signaling Pathways and Regulatory Steps of Macroautophagy
Induction of macroautophagy is tightly regulated by multiple signaling pathways that allow appropriate fine-tuning of this cascade in response to diverse environmental cues [28]. An important negative regulator of macroautophagy is the key nutrient sensor, mammalian target of rapamycin (mTOR) [56]. Nutrient availability and growth factors activate mTOR, which decreases macroautophagy activity by phosphorylating and inhibiting a key upstream kinase that activates macroautophagy, Ulk1 [36]. In contrast, nutrient depletion decreases cellular ATP and activates adenosine mono-phosphate-activated protein kinase (AMPK), a kinase known to initiate a cellular response following depletion of nutrients [25]. Activated AMPK induces macroautophagy by phosphorylating Ulk1 at residues [20] distinct from those phosphorylated by mTOR. The activation of Ulk1 facilitates trafficking of select ATG proteins to sites of de novo autophagic membranogenesis [78] through direct phosphorylation of these ATGs [53]. It had long been considered that macroautophagy activity did not require induction of ATG or lysosomal gene expression; however, recent studies from Ballabio and colleagues have identified a gene regulatory network that enhances macroautophagy by increasing lysosomal biogenesis [63]. Indeed, using systems biology, Ballabio and colleagues identified that a basic HLH-leucine zipper, transcription factor EB (TFEB), is a master regulator that positively controls expression of lysosomal and Atg genes by binding to their promoters [63]. In consistency with the known role of macroautophagy in quality control, follow up studies have now demonstrated that TFEB-induced activation of macroautophagy protects against α-synuclein toxicity in midbrain dopaminergic neurons, and in this way prevents progression of Parkinson’s disease [16]. Similarly, overexpression of TFEB alleviates glycogen buildup in muscles as a result of Pompe disease [68], a severe form of metabolic myopathy occurring as a consequence of an absence of acid α-glucosidase. Increasing TFEB availability led to activation of macroautophagy followed by the exocytosis of autophagolysosomes containing glycogen cargo.
TFEB is regulated by its nuclear exclusion, and studies exploring regulation of TFEB activity identified two kinases—extracellular signal-regulated kinase (ERK2) and mTOR that control macroautophagy by modulating TFEB phosphorylation and hence its cellular localization [63, 64]. It has been shown that nutrient/ growth factor-stimulated ERK2 activation results in TFEB phosphorylation, its cytoplasmic accumulation and macroautophagy blockage [63]. Work from the same group has also elucidated a novel lysosome-to-nucleus signaling by TFEB that involves mTOR [64]. In this regulatory axis, the availability of nutrients recruits mTOR to the lysosomal surface [64]. At the lysosomal surface, active mTOR interacts and phosphorylates TFEB at Serine-142, which retains it in the cytosol and reduces macroautophagy. In contrast, fasting releases mTOR from lysosomes and decreases TFEB phosphorylation, which allows it to traverse to the nucleus and express macroautophagy genes. Recent work has also elucidated a master repressor of macroautophagy genes, ZKSCAN3, a zinc finger DNA-binding protein [6]. Indeed, knocking down ZKSCAN3 increased lysosomal biogenesis and activated macroautophagy [6]. Whether TFEB and ZKSCAN3 regulatory cascades are perturbed with aging remain unclear. Given the intricate control of macroautophagy at several steps by a plethora of regulatory proteins, it is conceivable that disruptions in any of the regulatory steps would contribute to age-related decline sin macroautophagy and cell function.
Molecular Players in Chaperone-Mediated Autophagy
A second form of autophagy, chaperone-mediated autophagy (CMA), functions in a manner quite distinct from macroautophagy. CMA utilizes a cytosolic chaperone, heat shock-cognate protein of 70 KDa (hsc70) for recognizing substrates, and the lysosomal-associated membrane protein 2A (LAMP-2A) receptor for internalizing these substrates into the lysosome [1]. The steps in the CMA pathway include (i) substrate recognition and delivery to lysosomes, (ii) protein binding and unfolding, and (iii) protein translocation into the lysosome followed by (iv) degradation in the lysosomal lumen (Fig. 3.2) [1]. The recognition motif for proteins selectively targeted for degradation via CMA lies in the primary structure of the protein itself, i.e., in the amino acid sequence KFERQ of the substrate protein (Fig. 3.2) [1]. The interaction of the substrate with cytosolic hsc70 allows its targeting to the lysosomal surface where it binds to the CMA receptor, lysosome membrane-spanning protein LAMP-2A (Fig. 3.2) [10]. The binding of substrate to monomeric form of LAMP-2A is followed by substrate unfolding possibly assisted by hsc70 and its co-chaperones [10]. It has also been shown that LAMP-2A multimerization at the lysosomal surface plays a critical role in substrate internalization [2]. The complete internalization of a substrate protein across the lysosome membrane also requires the lysosome-resident form of hsc70 (lys-hsc70) [1]. Lys-hsc70 together with lysosomal hsp90 also contributes to the assembly and disassembly of LAMP-2A at the lysosome membrane [2]. In addition, two interacting proteins, GFAP and EF1α, have been described to regulate CMA activity by modulating LAMP-2A multimerization, and its mobilization to lipid microdomains [3]. It is thus possible that changes in activity or levels of any of these proteins can compromise CMA activity with age.
Basal CMA activity has been described in multiple cell types as a quality control system to maintain functionality of the proteome [1]. Macroautophagy is the first line of defense during cellular stress, for instance, during starvation or oxidative stress; however, when the insult persists beyond six to eight hours, the induction of CMA ensures cellular homeostasis through selective removal of damaged proteins. [1]. However the response to starvation appears to occur in a cell- and tissue-specific manner. Starvation-induced CMA is accompanied by changes in the distribution and properties of the lysosomal compartment [13]. The percentage of CMA-active lysosomes amplifies to accommodate cellular requirements for enhanced degradation. Such an expansion of the CMA-competent lysosomal system is paralleled by an increase in the levels of LAMP-2A present at the lysosomal membrane [13]. The elevated amount of LAMP-2A in these conditions is attained not by de novo synthesis but by decreased degradation of this CMA receptor at lysosomes [15]. The intrinsic selectivity of CMA is also well suited for the removal of proteins damaged during stress without perturbing nearby normally functioning forms of the same protein, by way of differential accessibility to the Hsc70 chaperone, of the CMA-targeting motif in its folded and unfolded state.
Macroautophagy and Aging
Decreased macroautophagy with age has been reported extensively in a variety of systems [18, 60]. The earliest studies to identify macroautophagy genes in the aging process were conducted in model organisms. An unbiased screen for chronological aging factors in yeast Saccharomyces cerevisiae identified multiple short-lived mutants with defects in macroautophagy [49]. Decreased lifespan in the nematode Caenorhabditis elegans ensued in mutants with loss-of-function of Atg1 (Unc-51), Atg7, Atg18 and Beclin-1 [72]. Similar findings in the fruit fly Drosophila melanogaster also revealed reduced lifespan in mutants with reduced expression of Atg1 and Atg8 [65]. The generation of conditional mice models to selectively delete Atg7 [38], Atg5 [24], Beclin [44] or Atg16l1 [5] was a significant step in our understanding of the consequences of loss of each of these genes in a tissue-specific manner. Indeed, whole body knockout of essential Atg genes leads to early postnatal death in murine models underscoring the crucial role of these genes in quality control, organogenesis, and differentiation. In contrast, conditional tissue-specific knockouts of Atg7 or Atg5 revealed milder phenotypes whereby younger knockout mice mimicked multiple age-associated phenomena such as aggregation of intracellular inclusion bodies and neurodegeneration [39], accumulation of lysosomes filled with the age-related pigment lipofuscin [69], disordered and defective mitochondria [35], accumulation of lipid droplets [67], increased protein oxidation [51], decreased muscle mass [48] as observed during sarcopenia of aging, and reduced differentiation of myogenic progenitors into muscle or fat [46]. A unifying theme emerging from all of these studies was the central role of macroautophagy in maintenance of quality control, and these studies suggest that a major determinant of the aging process is progressive loss of quality control. Consequently, it is not difficult to conceive that accumulation of toxic insults over a period of time as macroautophagy function wanes off results in age-associated pathologies, which include reduced muscle mass, neurodegeneration, cardiac malfunction, lipid accumulation and insulin insensitivity.
In addition to changes in quality control, secondary factors may come into play to determine loss of tissue function with age. For instance, a recent study reveals that acutely depleting Atg7 during adulthood was sufficient to dedifferentiate brown adipose tissue (BAT) and promote lipid accumulation [46]. Indeed, Atg7-depleted BAT displayed reduced expression of typical BAT markers, for instance the mitochondrial uncoupling protein-1. It is thus possible that lack of macroautophagy with age may allow highly specialized tissues, for instance muscle or brown fat, to dedifferentiate and lose their molecular and morphological signatures, which in turn would disrupt tissue function. A third layer of compromise added onto aged tissues is defective genome maintenance occurring as a result of the age-associated loss of macroautophagy. While it is well established that generation of oxidative stress and cumulative toxic insults with age associate with DNA damage, whether macroautophagy plays a role in genome maintenance remained unclear. To that end, two recent studies have now identified roles of macroautophagy gene products in regulation of genomic maintenance. In the first study, authors revealed that absence of macroautophagy results in a persistent delay in cell cycle progression that associates with aneuploidy due to insufficient cell growth and defective nuclear division [50]. In the second study, the authors highlight that UVRAG, a protein associated with macroautophagy, interacts with DNA repair enzymes to promote DNA doublestrand break repairs, and that the absence of UVRAG results in heightened sensitivity of cells to genomic instability due to exposure to irradiation. In addition, the authors reveal that UVRAG interacts with centrosomes to enhance their stability, and interfering with this function gives rise to aneuploidy [80]. Given these recent developments, it is easy to place ATG proteins on a functional framework to consider how their loss could accelerate aging through effects on quality control, tissue differentiation and aberrant genomic stability.
How macroautophagy decreases with age remains unclear. Given the complexities in the orchestration of macroautophagy, and the stochastic nature of the aging process, it is likely that the mechanisms contributing to inhibition of macroautophagy are complex and multifactorial. Transcriptional down-regulation of key macroautophagy genes such as Atg5 and Atg7 was detected by genome-wide analysis in human brain during normal aging [45]. Age-associated decreases in autophagic proteolysis have been detected in vivo in rodent models and also in vitro in isolated hepatocytes from these animals [4, 19]. Our work also revealed age-associated decreases in ATG7 protein levels and macroautophagy activity in hypothalamic proopiomelanocortin (POMC) neurons, which regulate food intake and energy balance [33]. The precise mechanisms leading to decreases in ATG levels remain unclear, and whether reduced ATG protein levels with age are due to a general failure to maintain Atg gene expression or reduced upstream signaling remains to be seen.
To that end, mechanisms disrupting signaling pathways that converge on the macroautophagy pathway could also disrupt quality control and accelerate aging. The aging process also associates with differential effects on the regulation of macroautophagy activity by hormones; for instance, during aging, glucagon’s stimulatory effect on macroautophagy is blunted while insulin’s inhibitory effect remains largely intact. Thus, it is possible that early changes in the regulation of macroautophagy may be secondary to age-related perturbations in metabolism and aberrant hormonal responses to starvation [17–19]. Perhaps the best-described regulatory mechanism contributing to suppression of macroautophagy with age is mTOR signaling. As detailed in the previous sections, the nutrient sensor mTOR exerts an inhibitory effect on macroautophagy at multiple steps [56, 64], and aging and obesity are both associated with hyperphosphorylation, and hence hyperactivation, of mTOR [8, 31, 75]. It is thus plausible that increased mTOR signaling plays a predominant role in age-associated suppression of macroautophagy and in many of the previously reported metabolic and neurodegenerative disorders occurring with age. In support of this notion, studies have demonstrated that dietary incorporation of rapamycin, an agent that activates macroautophagy by suppressing mTOR, increases longevity in male and female rodents [27], although the direct contribution of macroautophagy activation to longevity in this model has remained untested. Furthermore, a recent study has revealed that intracerebroventricular administration of rapamycin reduced age-related defects in energy balance and obesity, at least in part, from restoring the function of hypothalamic POMC neurons [77].
Studies have identified several steps in macroautophagy that could be affected so as to disrupt cargo degradation. For instance, “cargo recognition failure”, wherein the macroautophagy apparatus fails to recognize typical degradation signatures associated with aged or damaged organelles result in accumulation of unwanted substrates [47]. Amongst others, these mechanisms contribute to the development of neurological disorders, for example, Huntington’s disease and Familial Parkinson’s disease. In the latter condition, mutations in proteins, parkin and PINK1 that give rise to the “eat me” signal to activate mitophagy, result in accumulation of leaky, reactive oxygen species (ROS)-generating mitochondria [21], which in turn compromises cellular function. A crucial protein p62 serves as an adaptor that allows cargo to be recognized and sequestered by autophagosomes [52]. It is thus expected that mutations in p62/SQSTM1 would result in the failure to recognize cargo. Indeed, p62 mutations associate with Paget’s disease of the bone and the severely debilitating neurological condition, amyotrophic lateral sclerosis [57], while mutations in Huntingtin, which alters its binding to p62, has been shown to give rise to a generic cargo recognition failure [47]. In addition, post-translational changes to p62, for instance phosphorylation of p62, have been linked to an antioxidant response via its ability to regulate the Keap1-Nrf2 antioxidant response pathway [29], and thus in principle, age-associated changes to p62 phosphorylation could modify macroautophagy and resistance to oxidative stress. Mutations disrupting early events in macroautophagy, for example autophagosome formation and membrane elongation, have also been linked to a number of disorders. While loss of a single allele of Beclin1/Atg6 (Class III PI3K component) disrupts macroautophagy and results in increased incidence of cancers [44], mutations in Atg16l1, which determines autophagosome membrane elongation, has been associated with Crohn’s disease and inflammation of the bowel [5]. Autophagosome-lysosome fusion events are also amenable to disruptions that would lead to accumulation of cargo-laden autophagosomes, which in turn would compromise quality control. Recent work from the Cuervo laboratory [37] has identified that the previously reported inhibition of macroautophagy in rodent obesity models [67] occurs, at least in part, due to changes in the lipid composition of autophagosome and lysosomal membranes [37], which decreases their abilities to fuse with each other. It is thus conceivable that age-associated alterations in lipid metabolism and lipid accumulation could trigger a vicious cycle, which would further increase lipid accumulation and promote metabolic compromise by blocking autophago-lysosomal fusion.
Since it appears from the above observations that aging is associated with insufficient macroautophagy, the question arose whether increasing macroautophagy would delay aging and extend lifespan. In most animals studied to date, caloric restriction, which is reduced food intake without malnutrition, is the key anti-aging intervention [7] and is the most potent physiological inducer of macroautophagy [40, 43]. Caloric restriction not only extends lifespan but reduces the incidence of diabetes, cardiovascular disease, cancer, and brain-related diseases [7]. The effects of caloric restriction on macroautophagy induction are believed to be mediated by the activation of either of two energy sensors, AMPK and Sirtuin 1 (SIRT1) or by the inhibition of the insulin/insulin-like growth factor signaling pathway that ultimately leads to inhibition of mTOR signaling.
While inhibition of mTOR, either pharmacologically with rapamycin or genetically, extends lifespan in yeast, C. elegans, and D. melanogaster, the addition of caloric restriction does not further promote longevity. This suggests that there is a common mechanism that mediates the anti-aging effects of both these interventions, and macroautophagy may be the mediator as it has been shown that knockdown of essential Atg genes abrogates all life-extending effects of rapamycin in all species investigated. While rapamycin potently induces macroautophagy, it may also promote anti-aging effects via its anti-inflammatory properties. Likewise, inhibition of mTOR has major effects on protein translation and thus it is unclear whether mTOR inhibition extends lifespan primarily through induction of macroautophagy or whether the effects are mediated by macroautophagy-independent changes to the proteome. While transgenic overexpression of SIRT1 clearly delays aging and promotes lifespan, how SIRT1 triggers macroautophagy remains incompletely understood. Although it has been shown that SIRT1 forms complexes with essential macroautophagy proteins, ATG7, ATG5 and ATG8 and maintains them in the deacetylated state [42], further studies would be necessary to reveal the precise mechanism that is behind the requirement of SIRT1 in activation of macroautophagy.
A direct role of macroautophagy in promoting life-span, or perhaps more critically increasing healthspan, comes from studies showing that liver-specific overexpression of ATG7 prevents hepatic endoplasmic reticulum stress, increases hepatic insulin action, and improves glucose tolerance in a model of diet-induced obesity [76]. Studies in C. elegans have revealed that overexpression of the TFEB orthologue hlh-30 extends lifespan that associates with increased nuclear translocation of this transcription factor indicating that longevity was a result of increased TFEB-driven expression of macroautophagy-related genes [41]. Interestingly, livers of dietary-restricted mice also revealed increased nuclear TFEB content [41]. On similar lines, overexpression of Atg5 in mice has been shown to extend lifespan by ~ 17 %, increase leanness, and improve glucose clearance and motor function [55]. Furthermore, fibroblasts from transgenic ATG5 overexpressors have been shown to resist oxidative stress-induced cell death [55]. These restorative studies are a testament of the important role played by macroautophagy in extending lifespan and improving healthspan across multiple model systems. Given the function of macroautophagy in maintaining “clean” cells, it is likely that upholding macroautophagy function during aging will reduce the amount of toxic protein aggregates, reduce cell death, and improve cell function, all of which would, in principle, improve cellular function. Although, the effects of aging on the second form of autophagy, microautophagy [61], remain unknown, it is likely that microautophagy activity will decrease with age. However, further studies would be necessary to elucidate the consequences of aging on microautophagy.
Chaperone-Mediated Autophagy and Aging
In parallel to the inhibition of macroautophagy with age, CMA has also been reported to decrease with age [11]. Studies in livers from young and old rats showed a reduction in levels of the CMA receptor LAMP-2A in lysosomal membranes from aged livers [11]. These decreases in LAMP-2A levels did not associate with any changes in levels of cytosolic Hsc70, in fact a compensatory increase in lysosomal Hsc70 levels was observed in livers from aged rats [11]. Age-associated reduction in LAMP-2A levels correlated with decreased substrate binding to lysosomes and reduced substrate uptake by lysosomes, which would in all likelihood impact proteolysis rates and cellular quality control. Substrate binding/uptake assays in aged livers revealed a greater compromise in substrate uptake by lysosomes than in the ability of substrates to bind to lysosomes. These functional studies provided the first direct evidence that CMA decreases with age [11]. Subsequent work from the same group revealed that decreased LAMP-2A levels in aged livers were not due to a decrease in transcriptional regulation of LAMP-2A but due to increases in its degradation rates [34]. Kiffin et. al. used early passage fibroblasts and senescent late passage fibroblasts that display decreased LAMP-2A levels and CMA activity as observed in aged animals, revealed reduced half-life and faster degradation rates of LAMP-2A in late passage fibroblasts [34]. Indeed metabolic labeling and immunoprecipitation of LAMP-2A followed by quantification of radioactivity revealed a significant reduction in the half-life of LAMP-2A from 38 h (h) in early passage fibroblasts to 26 h in late passage senescent fibroblasts under basal conditions [34]. Although the mechanism for decreased lysosomal LAMP-2A levels with age remains unclear, it has been suggested that alteration in lysosomal membrane cholesterol content with age could contribute to changes in levels of LAMP-2A in cholesterol-rich lysosomal lipid microdomains [34] that are typically enriched in LAMP-2A [32]. Interestingly, feeding rodents a high fat diet or a diet enriched in cholesterol resulted in decreased lysosomal LAMP-2A levels [58]. This reduction in LAMP-2A mimicked those observed in livers from aged mice [11] and occurred due to mobilization of LAMP-2A to specific lipid regions on the lysosomal membrane where the receptor was unstable and underwent rapid degradation. Furthermore, changes in lysosomal membrane lipid composition following dietary challenges were similar to those observed in aged mice levels [58]. These results demonstrate that as observed with macroautophagy, chronic overnutrition plays a suppressive role of autophagy pathways. It thus remains possible that chronic dietary stress could contribute, in part, to decreased macroautophagy and CMA observed with age.
The final evidence of the benefit of increasing LAMP-2A levels to upregulate CMA and combat age-related conditions comes from a study demonstrating that supplying an extra copy of this receptor in the liver reduces damaged protein content [79]. Transgenic LAMP-2A overexpressors demonstrate improved cellular homeostasis, reduced oxidized protein load, and improved liver function. Employing zoxazolamine-induced paralysis time assay wherein recovery times are noted as a measure of hepatic function, it was shown that LAMP-2A-overexpressing aged mice were able to metabolize zoxazolamine in their livers at comparable rates to young wild-type controls [79]. This work demonstrates a direct role of restoring CMA-mediated quality control on improving organ function and builds a case for the activation of autophagy as an attractive strategy in the fight against age-related disorders.
Concluding Remarks
Autophagic pathways play crucial roles in cellular quality control, and a plethora of evidence has linked compromised autophagy to development of age-related disorders. While accumulation of damaged proteins, organelles, and aggregates were major considerations for age-associated loss of cellular function; the recently elucidated roles for autophagy in mobilization of lipid droplets and glycogen granules, tumor development, and microbial pathogenesis has expanded the list of conditions likely to be associated with loss of autophagy during aging. Although multiple mechanisms have been attributed to the loss of autophagy during aging, restoration studies with rapamycin or with upregulation of critical regulatory genes have demonstrated that longevity and health-span can potentially be improved by activating these pathways. Consequently, understanding the mechanisms contributing to reduction of autophagy with age, and exploring new ways to activate autophagy remain critical in fighting age-related disorders and improving healthspan in the aging population.
Acknowledgements
We thankfully acknowledge NIH grants, DK087776 and AG043517, and an Ellison Medical Foundation award to RS. DA is supported by training grants 5T32GM728837 and T32GM007288. We thank Dr. Susmita Kaushik for assisting with designs of the models in Figs. 3.1 and 3.2.
Contributor Information
Nuria Martinez-Lopez, Department of Medicine and Department of Molecular Pharmacology, Albert Einstein College of Medicine, Bronx, NY 10461, USA.
Diana Athonvarangkul, Department of Medicine and Department of Molecular Pharmacology, Albert Einstein College of Medicine, Bronx, NY 10461, USA.
Rajat Singh, Department of Medicine (Endocrinology), and Molecular Pharmacology, Diabetes Research Center, Albert Einstein College of Medicine, 1300 Morris Park Avenue, Forchheimer Building, Room 505D, Bronx, NY 10461, USA.
References
- 1.Arias E, Cuervo AM. Chaperone-mediated autophagy in protein quality control. Curr Opin Cell Biol. 2011;23(2):184–189. doi: 10.1016/j.ceb.2010.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bandyopadhyay U, Kaushik S, Varticovski L, Cuervo AM. The chaperone-mediated autophagy receptor organizes in dynamic protein complexes at the lysosomal membrane. Mol Cell Biol. 2008;28(18):5747–5763. doi: 10.1128/MCB.02070-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bandyopadhyay U, Sridhar S, Kaushik S, Kiffin R, Cuervo AM. Identification of regulators of chaperone-mediated autophagy. Mol Cell. 2010;39(4):535–547. doi: 10.1016/j.molcel.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bergamini E, Del Roso A, Fierabracci V, Gori Z, Masiello P, Masini M, Pollera M. A new method for the investigation of endocrine-regulated autophagy and protein degradation in rat liver. Exp Mol Pathol. 1993;59(1):13–26. doi: 10.1006/exmp.1993.1023. [DOI] [PubMed] [Google Scholar]
- 5.Cadwell K, Liu JY, Brown SL, Miyoshi H, Loh J, Lennerz JK, Kishi C, Kc W, Carrero JA, Hunt S, Stone CD, Brunt EM, Xavier RJ, Sleckman BP, Li E, Mizushima N, Stappenbeck TS, Virgin HW., 4th A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature. 2008;456(7219):259–263. doi: 10.1038/nature07416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chauhan S, Goodwin JG, Chauhan S, Manyam G, Wang J, Kamat AM, Boyd DD. ZKSCAN3 is a master transcriptional repressor of autophagy. Mol Cell. 2013;50(1):16–28. doi: 10.1016/j.molcel.2013.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Colman RJ, Anderson RM, Johnson SC, Kastman EK, Kosmatka KJ, Beasley TM, Allison DB, Cruzen C, Simmons HA, Kemnitz JW, Weindruch R. Caloric restriction delays disease onset and mortality in rhesus monkeys. Science. 2009;325(5937):201–204. doi: 10.1126/science.1173635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cornu M, Albert V, Hall MN. mTOR in aging, metabolism, and cancer. Curr Opin Genet Dev. 2013;23(1):53–62. doi: 10.1016/j.gde.2012.12.005. [DOI] [PubMed] [Google Scholar]
- 9.Cuervo AM. Autophagy and aging: keeping that old broom working. Trends Genet. 2008;24(12):604–612. doi: 10.1016/j.tig.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cuervo AM. Chaperone-mediated autophagy: selectivity pays off. Trends Endocrinol Metab. 2010;21(3):142–150. doi: 10.1016/j.tem.2009.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cuervo AM, Dice JF. Age-related decline in chaperone-mediated autophagy. J Biol Chem. 2000;275(40):31505–31513. doi: 10.1074/jbc.M002102200. [DOI] [PubMed] [Google Scholar]
- 12.Cuervo AM, Wong E. Chaperone-mediated autophagy: roles in disease and aging. Cell Res. 2014;24(1):92–104. doi: 10.1038/cr.2013.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cuervo AM, Knecht E, Terlecky SR, Dice JF. Activation of a selective pathway of lysosomal proteolysis in rat liver by prolonged starvation. Am J Physiol. 1995;269(5 Pt 1):C1200–1208. doi: 10.1152/ajpcell.1995.269.5.C1200. [DOI] [PubMed] [Google Scholar]
- 14.Cuervo AM, Dice JF, Knecht E. A population of rat liver lysosomes responsible for the selective uptake and degradation of cytosolic proteins. J Biol Chem. 1997;272(9):5606–5615. doi: 10.1074/jbc.272.9.5606. [DOI] [PubMed] [Google Scholar]
- 15.Cuervo AM, Mann L, Bonten EJ, d’Azzo A, Dice JF. Cathepsin A regulates chaper-one-mediated autophagy through cleavage of the lysosomal receptor. EMBO J. 2003;22(1):47–59. doi: 10.1093/emboj/cdg002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Decressac M, Mattsson B, Weikop P, Lundblad M, Jakobsson J, Bjorklund A. TFEB-mediated autophagy rescues midbrain dopamine neurons from alpha-synuclein toxicity. Proc Natl Acad Sci U S A. 2013;110(19):E1817–1826. doi: 10.1073/pnas.1305623110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Del Roso A, Bombara M, Fierabracci V, Masini M, Masiello P, Pollera M, Bergamini E. Effect of dietary restriction on the age-related changes in hormone-regulated protein breakdown. Aging (Milano) 1991;3(4):407–408. doi: 10.1007/BF03324052. [DOI] [PubMed] [Google Scholar]
- 18.Del Roso A, Vittorini S, Cavallini G, Donati A, Gori Z, Masini M, Pollera M, Bergamini E. Ageing-related changes in the in vivo function of rat liver macroautophagy and proteolysis. Exp Gerontol. 2003;38(5):519–527. doi: 10.1016/s0531-5565(03)00002-0. [DOI] [PubMed] [Google Scholar]
- 19.Donati A, Cavallini G, Paradiso C, Vittorini S, Pollera M, Gori Z, Bergamini E. Age-related changes in the autophagic proteolysis of rat isolated liver cells: effects of antiaging dietary restrictions. J Gerontol A Biol Sci Med Sci. 2001;56(9):B375–383. doi: 10.1093/gerona/56.9.b375. [DOI] [PubMed] [Google Scholar]
- 20.Egan DF, Shackelford DB, Mihaylova MM, Gelino S, Kohnz RA, Mair W, Vasquez DS, Joshi A, Gwinn DM, Taylor R, Asara JM, Fitzpatrick J, Dillin A, Viollet B, Kundu M, Hansen M, Shaw RJ. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science. 2011;331(6016):456–461. doi: 10.1126/science.1196371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Geisler S, Holmstrom KM, Skujat D, Fiesel FC, Rothfuss OC, Kahle PJ, Springer W. “PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol. 2010;12(2):119–131. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- 22.Geng J, Klionsky DJ. The Atg8 and Atg12 ubiquitin-like conjugation systems in macroautophagy. ‘Protein modifications: beyond the usual suspects’ review series. EMBO Rep. 2008;9(9):859–864. doi: 10.1038/embor.2008.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hanada T, Noda NN, Satomi Y, Ichimura Y, Fujioka Y, Takao T, Inagaki F, Ohsumi Y. The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J Biol Chem. 2007;282(52):37298–37302. doi: 10.1074/jbc.C700195200. [DOI] [PubMed] [Google Scholar]
- 24.Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, Yokoyama M, Mishima K, Saito I, Okano H, Mizushima N. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441(7095):885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 25.Hardie DG, Carling D, Halford N. Roles of the Snf1/Rkin1/AMP-activated protein kinase family in the response to environmental and nutritional stress. Semin Cell Biol. 1994;5(6):409–416. doi: 10.1006/scel.1994.1048. [DOI] [PubMed] [Google Scholar]
- 26.Harding TM, Hefner-Gravink A, Thumm M, Klionsky DJ. Genetic and phenotypic overlap between autophagy and the cytoplasm to vacuole protein targeting pathway. J Biol Chem. 1996;271(30):17621–17624. doi: 10.1074/jbc.271.30.17621. [DOI] [PubMed] [Google Scholar]
- 27.Harrison DE, Strong R, Sharp ZD, Nelson JF, Astle CM, Flurkey K, Nadon NL, Wilkinson JE, Frenkel K, Carter CS, Pahor M, Javors MA, Fernandez E, Miller RA. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature. 2009;460(7253):392–395. doi: 10.1038/nature08221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ichimura Y, Waguri S, Sou YS, Kageyama S, Hasegawa J, Ishimura R, Saito T, Yang Y, Kouno T, Fukutomi T, Hoshii T, Hirao A, Takagi K, Mizushima T, Motohashi H, Lee MS, Yoshimori T, Tanaka K, Yamamoto M, Komatsu M. Phosphorylation of p62 activates the Keap1-Nrf2 pathway during selective autophagy. Mol Cell. 2013;51(5):618–631. doi: 10.1016/j.molcel.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 30.Itakura E, Kishi-Itakura C, Mizushima N. The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell. 2012;151(6):1256–1269. doi: 10.1016/j.cell.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 31.Johnson SC, Rabinovitch PS, Kaeberlein M. mTOR is a key modulator of ageing and age-related disease. Nature. 2013;493(7432):338–345. doi: 10.1038/nature11861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaushik S, Massey AC, Cuervo AM. Lysosome membrane lipid microdomains: novel regulators of chaperone-mediated autophagy. EMBO J. 2006;25(17):3921–3933. doi: 10.1038/sj.emboj.7601283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaushik S, Arias E, Kwon H, Lopez NM, Athonvarangkul D, Sahu S, Schwartz GJ, Pessin JE, Singh R. Loss of autophagy in hypothalamic POMC neurons impairs lipolysis. EMBO Rep. 2012;13(3):258–265. doi: 10.1038/embor.2011.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kiffin R, Kaushik S, Zeng M, Bandyopadhyay U, Zhang C, Massey AC, Martinez-Vicente M, Cuervo AM. Altered dynamics of the lysosomal receptor for chaperone-mediated autophagy with age. J Cell Sci. 2007;120(Pt 5):782–791. doi: 10.1242/jcs.001073. [DOI] [PubMed] [Google Scholar]
- 35.Kim I, Rodriguez-Enriquez S, Lemasters JJ. Selective degradation of mitochondria by mitophagy. Arch Biochem Biophys. 2007;462(2):245–253. doi: 10.1016/j.abb.2007.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13(2):132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koga H, Kaushik S, Cuervo AM. Altered lipid content inhibits autophagic vesicular fusion. FASEB J. 2010;24(8):3052–3065. doi: 10.1096/fj.09-144519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, Ezaki J, Mizushima N, Ohsumi Y, Uchiyama Y, Kominami E, Tanaka K, Chiba T. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169(3):425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, Ueno T, Koike M, Uchiyama Y, Kominami E, Tanaka K. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441(7095):880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 40.Kroemer G, Levine B. Autophagic cell death: the story of a misnomer. Nat Rev Mol Cell Biol. 2008;9(12):1004–1010. doi: 10.1038/nrm2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lapierre LR, De Magalhaes Filho CD, Quary PR, Chu CC, Visvikis O, Chang JT, Gelino S, Ong B, Davis AE, Irazoqui JE, Dillin A, Hansen M. The TFEB orthologue HLH-30 regulates autophagy and modulates longevity in Caenorhabditis elegans. Nat Commun. 2013;4:2267. doi: 10.1038/ncomms3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee IH, Cao L, Mostoslavsky R, Lombard DB, Liu J, Bruns NE, Tsokos M, Alt FW, Finkel T. A role for the NAD-dependent deacetylase Sirt1 in the regulation of autophagy. Proc Natl Acad Sci U S A. 2008;105(9):3374–3379. doi: 10.1073/pnas.0712145105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132(1):27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, Levine B. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402(6762):672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- 45.Lipinski MM, Zheng B, Lu T, Yan Z, Py BF, Ng A, Xavier RJ, Li C, Yankner BA, Scherzer CR, Yuan J. Genome-wide analysis reveals mechanisms modulating autophagy in normal brain aging and in Alzheimer’s disease. Proc Natl Acad Sci U S A. 2010;107(32):14164–14169. doi: 10.1073/pnas.1009485107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martinez-Lopez N, Athonvarangkul D, Sahu S, Coletto L, Zong H, Bastie CC, Pessin JE, Schwartz GJ, Singh R. Autophagy in Myf5 + progenitors regulates energy and glucose homeostasis through control of brown fat and skeletal muscle development. EMBO Rep. 2013;14(9):795–803. doi: 10.1038/embor.2013.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Martinez-Vicente M, Talloczy Z, Wong E, Tang G, Koga H, Kaushik S, de Vries R, Arias E, Harris S, Sulzer D, Cuervo AM. Cargo recognition failure is responsible for inefficient autophagy in Huntington’s disease. Nat Neurosci. 2010;13(5):567–576. doi: 10.1038/nn.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Masiero E, Agatea L, Mammucari C, Blaauw B, Loro E, Komatsu M, Metzger D, Reggiani C, Schiaffino S, Sandri M. Autophagy is required to maintain muscle mass. Cell Metab. 2009;10(6):507–515. doi: 10.1016/j.cmet.2009.10.008. [DOI] [PubMed] [Google Scholar]
- 49.Matecic M, Smith DL, Pan X, Maqani N, Bekiranov S, Boeke JD, Smith JS. A micro-array-based genetic screen for yeast chronological aging factors. PLoS Genet. 2010;6(4):e1000921. doi: 10.1371/journal.pgen.1000921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Matsui A, Kamada Y, Matsuura A. The role of autophagy in genome stability through suppression of abnormal mitosis under starvation. PLoS Genet. 2013;9(1):e1003245. doi: 10.1371/journal.pgen.1003245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nezis IP, Stenmark H. p62 at the interface of autophagy, oxidative stress signaling, and cancer. Antioxid Redox Signal. 2012;17(5):786–793. doi: 10.1089/ars.2011.4394. [DOI] [PubMed] [Google Scholar]
- 52.Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Overvatn A, Bjorkoy G, Johansen T. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282(33):24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 53.Papinski D, Kraft C. Atg1 kinase organizes autophagosome formation by phosphorylating Atg9. Autophagy. 2014;10(7):1338–1340. doi: 10.4161/auto.28971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Phillips AR, Suttangkakul A, Vierstra RD. The ATG12-conjugating enzyme ATG10 Is essential for autophagic vesicle formation in Arabidopsis thaliana. Genetics. 2008;178(3):1339–1353. doi: 10.1534/genetics.107.086199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pyo JO, Yoo SM, Ahn HH, Nah J, Hong SH, Kam TI, Jung S, Jung YK. Overexpression of Atg5 in mice activates autophagy and extends lifespan. Nat Commun. 2013;4:2300. doi: 10.1038/ncomms3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ravikumar B, Vacher C, Berger Z, Davies JE, Luo S, Oroz LG, Scaravilli F, Easton DF, Duden R, O’Kane CJ, Rubinsztein DC. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat Genet. 2004;36(6):585–595. doi: 10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- 57.Rea SL, Majcher V, Searle MS, Layfield R. SQSTM1 mutations-bridging Paget disease of bone and ALS/FTLD. Exp Cell Res. 2014;325(1):27–37. doi: 10.1016/j.yexcr.2014.01.020. [DOI] [PubMed] [Google Scholar]
- 58.Rodriguez-Navarro JA, Kaushik S, Koga H, Dall’Armi C, Shui G, Wenk MR, Di Paolo G, Cuervo AM. Inhibitory effect of dietary lipids on chaperone-mediated autophagy. Proc Natl Acad Sci U S A. 2012;109(12):E705–714. doi: 10.1073/pnas.1113036109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Romanov J, Walczak M, Ibiricu I, Schuchner S, Ogris E, Kraft C, Martens S. Mechanism and functions of membrane binding by the Atg5-Atg12/Atg16 complex during autophagosome formation. EMBO J. 2012;31(22):4304–4317. doi: 10.1038/emboj.2012.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rubinsztein DC, Marino G, Kroemer G. Autophagy and aging. Cell. 2011;146(5):682–695. doi: 10.1016/j.cell.2011.07.030. [DOI] [PubMed] [Google Scholar]
- 61.Sahu R, Kaushik S, Clement CC, Cannizzo ES, Scharf B, Follenzi A, Potolicchio I, Nieves E, Cuervo AM, Santambrogio L. Microautophagy of cytosolic proteins by late endosomes. Dev Cell. 2011;20(1):131–139. doi: 10.1016/j.devcel.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schlumpberger M, Schaeffeler E, Straub M, Bredschneider M, Wolf DH, Thumm M. AUT1, a gene essential for autophagocytosis in the yeast Saccharomyces cerevisiae. J Bacteriol. 1997;179(4):1068–1076. doi: 10.1128/jb.179.4.1068-1076.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Settembre C, Di Malta C, Polito VA, Garcia Arencibia M, Vetrini F, Erdin S, Erdin SU, Huynh T, Medina D, Colella P, Sardiello M, Rubinsztein DC, Ballabio A. TFEB links autophagy to lysosomal biogenesis. Science. 2011;332(6036):1429–1433. doi: 10.1126/science.1204592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Settembre C, Zoncu R, Medina DL, Vetrini F, Erdin S, Erdin S, Huynh T, Ferron M, Karsenty G, Vellard MC, Facchinetti V, Sabatini DM, Ballabio A. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J. 2012;31(5):1095–1108. doi: 10.1038/emboj.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Simonsen A, Cumming RC, Brech A, Isakson P, Schubert DR, Finley KD. Promoting basal levels of autophagy in the nervous system enhances longevity and oxidant resistance in adult Drosophila. Autophagy. 2008;4(2):176–184. doi: 10.4161/auto.5269. [DOI] [PubMed] [Google Scholar]
- 66.Singh R, Cuervo AM. Autophagy in the cellular energetic balance. Cell Metab. 2011;13(5):495–504. doi: 10.1016/j.cmet.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, Tanaka K, Cuervo AM, Czaja MJ. Autophagy regulates lipid metabolism. Nature. 2009;458(7242):1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Spampanato C, Feeney E, Li L, Cardone M, Lim JA, Annunziata F, Zare H, Polishchuk R, Puertollano R, Parenti G, Ballabio A, Raben N. Transcription factor EB (TFEB) is a new therapeutic target for Pompe disease. EMBO Mol Med. 2013;5(5):691–706. doi: 10.1002/emmm.201202176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stroikin Y, Dalen H, Loof S, Terman A. Inhibition of autophagy with 3-methyladenine results in impaired turnover of lysosomes and accumulation of lipofuscin-like material. Eur J Cell Biol. 2004;83(10):583–590. doi: 10.1078/0171-9335-00433. [DOI] [PubMed] [Google Scholar]
- 70.Tanida I, Tanida-Miyake E, Ueno T, Kominami E. The human homolog of Saccharomyces cerevisiae Apg7p is a protein-activating enzyme for multiple substrates including human Apg12p, GATE-16, GABARAP, and MAP-LC3. J Biol Chem. 2001;276(3):1701–1706. doi: 10.1074/jbc.C000752200. [DOI] [PubMed] [Google Scholar]
- 71.Tanida I, Ueno T, Kominami E. LC3 conjugation system in mammalian autophagy. Int J Biochem Cell Biol. 2004;36(12):2503–2518. doi: 10.1016/j.biocel.2004.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Toth ML, Sigmond T, Borsos E, Barna J, Erdelyi P, Takacs-Vellai K, Orosz L, Kovacs AL, Csikos G, Sass M, Vellai T. Longevity pathways converge on autophagy genes to regulate life span in Caenorhabditis elegans. Autophagy. 2008;4(3):330–338. doi: 10.4161/auto.5618. [DOI] [PubMed] [Google Scholar]
- 73.Tsukada M, Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 1993;333(1–2):169–174. doi: 10.1016/0014-5793(93)80398-e. [DOI] [PubMed] [Google Scholar]
- 74.Wei Y, Pattingre S, Sinha S, Bassik M, Levine B. JNK1-mediated phosphorylation of Bcl-2 regulates starvation-induced autophagy. Mol Cell. 2008;30(6):678–688. doi: 10.1016/j.molcel.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yang Z, Ming XF. mTOR signalling: the molecular interface connecting metabolic stress, aging and cardiovascular diseases. Obes Rev. 2012;13(Suppl 2):58–68. doi: 10.1111/j.1467-789X.2012.01038.x. [DOI] [PubMed] [Google Scholar]
- 76.Yang L, Li P, Fu S, Calay ES, Hotamisligil GS. Defective hepatic autophagy in obesity promotes ER stress and causes insulin resistance. Cell Metab. 2010;11(6):467–478. doi: 10.1016/j.cmet.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yang SB, Tien AC, Boddupalli G, Xu AW, Jan YN, Jan LY. Rapamycin ameliorates age-dependent obesity associated with increased mTOR signaling in hypothalamic POMC neurons. Neuron. 2012;75(3):425–436. doi: 10.1016/j.neuron.2012.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Young AR, Chan EY, Hu XW, Kochl R, Crawshaw SG, High S, Hailey DW, Lippincott-Schwartz J, Tooze SA. Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. J Cell Sci. 2006;119(Pt 18):3888–3900. doi: 10.1242/jcs.03172. [DOI] [PubMed] [Google Scholar]
- 79.Zhang C, Cuervo AM. Restoration of chaperone-mediated autophagy in aging liver improves cellular maintenance and hepatic function. Nat Med. 2008;14(9):959–965. doi: 10.1038/nm.1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhao Z, Oh S, Li D, Ni D, Pirooz SD, Lee JH, Yang S, Lee JY, Ghozalli I, Costanzo V, Stark JM, Liang C. A dual role for UVRAG in maintaining chromosomal stability independent of autophagy. Dev Cell. 2012;22(5):1001–1016. doi: 10.1016/j.devcel.2011.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]