Graphical abstract
Keywords: Chagas disease, Trypanosoma cruzi, Genetic exchange, Recombination, Cryptic sexuality, Mitochondrial introgression, Clonality
Highlights
-
•
The principal reproductive mode of Trypanosoma cruzi is controversial.
-
•
Field studies indicate recombination is frequent, non-obligatory and idiosyncratic.
-
•
These observations challenge the paradigm of clonal evolution in T. cruzi.
Abstract
Many eukaryotic pathogenic microorganisms that were previously assumed to propagate clonally have retained cryptic sexual cycles. The principal reproductive mode of Trypanosoma cruzi, the aetiological agent of Chagas disease, remains a controversial topic. Despite the existence of two recent natural hybrid lineages, a pervasive view is that recombination has been restrained at an evolutionary scale and is of little epidemiological relevance to contemporary parasite populations. This article reviews the growing number of field studies which indicate that natural hybridization in T. cruzi may be frequent, non-obligatory and idiosyncratic; potentially involving independent exchange of kinetoplast and nuclear genetic material as well as canonical meiotic mechanisms. Together these observations now challenge the traditional paradigm of preponderate clonal evolution in T. cruzi and highlight the need for additional, intensive and appropriately sampled field surveys, complemented by high resolution, combined nuclear and mitochondrial population genetics analyses.
1. Introduction
The principal reproductive mode of a number of parasitic protozoan species is the subject of an enduring debate (Smith et al., 1993, Tibayrenc et al., 1986, Tibayrenc et al., 1990, Tibayrenc and Ayala, 1991, Tibayrenc and Ayala, 2012, Tibayrenc and Ayala, 2013, Tibayrenc and Ayala, 2014a, Tibayrenc and Ayala, 2014b, Ramírez and Llewellyn, 2014, Tomasini et al., 2014a, Tomasini et al., 2014b). At the two extremes are the preponderate clonal evolution (PCE) model, which suggests that genetic exchange is too infrequent to break the predominant pattern of clonality, such that only ‘restrained recombination’ occurs at an evolutionary scale (Tibayrenc and Ayala, 2012, Tibayrenc and Ayala, 2013, Tibayrenc and Ayala, 2014a, Tibayrenc and Ayala, 2014b), and the counter-proposition that hybridization is in fact pervasive, albeit challenging to detect, among some natural disease foci (Ramírez and Llewellyn, 2014).
Trypanosoma cruzi, the aetiological agent of Chagas disease, often fulfils some key assumptions of PCE, which have been cited as compelling evidence that it is essentially a clonal organism, namely strong linkage disequilibrium, deviations from Hardy–Weinberg allele frequencies and structuring of populations into stable, distinct genetic clades, or discrete typing units (DTUs) (Tibayrenc and Ayala, 2012, Tibayrenc and Ayala, 2013). T. cruzi isolates display remarkable genetic diversity, which is widely believed to contribute to the considerable biological, epidemiological and clinical variation observed among Chagas disease foci (Miles et al., 2009). Current international consensus recognizes a minimum of six genetic lineages or DTUs (TcI–TcVI), with distributions loosely defined by geography, ecology and transmission cycle (Zingales et al., 2009). Genotyping using an array of markers indicate DTUs TcI–TcIV form monophyletic clades and TcV and TcVI are recent, natural inter-lineage hybrids of TcII and TcIII (Machado and Ayala, 2001, Brisse et al., 2003, Lewis et al., 2011, Yeo et al., 2011). Molecular dating indicates that these hybrid lineages evolved recently, within the last 60,000 years (Lewis et al., 2011), possibly from human disruption of sylvatic transmission cycles in the Southern Cone of South America, suggesting there may still be a risk of genetic exchange driving the emergence of novel recombinants (Flores-López and Machado, 2011, Lewis et al., 2011). These hybrid DTUs circulate almost exclusively in domestic transmission cycles and are sympatric with severe clinical sequelae in southern endemic areas. However, the frequency of recombination occurring among natural T. cruzi populations, the precise cytological mechanisms underlying genetic exchange events and the effect of hybridization on parasite phenotype remain largely undefined.
A clear understanding of the impact of genetic exchange on the ecological and geographical distributions and clinical characteristics of T. cruzi strains is crucial to establish the epidemiological risk associated with recombinant genotypes and to reconcile the implications parasite hybridization has at both the generational and evolutionary scales. However, detecting genetic exchange among natural populations is inherently complicated by choice of samples and marker resolution, given that strains most likely to be recombining may be closely related and potentially indistinguishable.
2. Genetic exchange among T. cruzi field populations
With improved sampling strategies and the development of higher resolution nuclear and mitochondrial genotyping techniques (Llewellyn et al., 2009, Messenger et al., 2012, Messenger et al., 2015, Ramírez et al., 2012), a growing number of field studies now indicate that natural recombination in T. cruzi may be frequent, non-obligatory and idiosyncratic; potentially involving independent exchange of kinetoplast and nuclear genetic material as well as canonical meiotic mechanisms (Table 1).
Table 1.
Summary of field evidence of genetic exchange in T. cruzi.
| T. cruzi population(s) | Transmission cycle, location | Type of genetic exchange | Genetic markers examined | Evidence of genetic exchange | Putative mechanism | Reference |
|---|---|---|---|---|---|---|
| TcI | Arboreal sylvatic, Bolivia | Intra-lineage | MLMT | - Mitochondrial introgression with no detectable nuclear involvement | Asymmetric mitochondrial introgression | Messenger et al. (2015) |
| mtMLST | - Dissimilar heterozygosity estimates | |||||
| TcI | Bolivia | Inter-lineage | GPIa | - Mitochondrial introgression between TcI and TcIII/IV/V/VI with no detectable nuclear involvement | - Asymmetric mitochondrial introgression | Barnabé and Breniere (2012) |
| ND1b | ||||||
| TcI | Bolivia | Intra-lineage | MLMT | - H-W allele frequencies | - Meiotic | Barnabé et al. (2013) |
| - Linkage equilibrium between loci | ||||||
| - Lack of repeated MLGs | ||||||
| TcI | Arboreal sylvatic, Brazil | Intra-lineage | MLEE | - Putative homozygous parents and heterozygous progeny | - Meiotic | Carrasco et al. (1996) |
| RAPD | - H–W phenotype frequencies | |||||
| TcII | Domestic, Brazil | Intra-lineage | MLMT | - H–W allele frequencies among local populations | - Meiotic | Baptista et al. (2014) |
| ND4b, ND7b | - Linkage equilibrium between loci | - Asymmetric mitochondrial inheritance | ||||
| - Independent inheritance of mitochondrial and nuclear genes | ||||||
| TcI | Domestic, peridomestic, sylvatic, Colombia | Intra-lineage | MLMT | - Mitochondrial introgression with no detectable nuclear involvement | -Asymmetric mitochondrial introgression | Ramírez et al. (2012) |
| mtMLST | - Recombinant mitochondrial genotype | - Biparental mitochondrial inheritance | ||||
| TcI | Domestic, peridomestic, sylvatic, Colombia | Intra-lineage | nMLST | - Linkage equilibrium between loci | - Meiotic | Ramírez et al. (2013) |
| - Putative recombination breakpoints | ||||||
| TcI | Domestic, Ecuador | Intra-lineage | MLMT | - H–W allele frequencies | - Meiotic | Ocaña-Mayorga et al. (2010) |
| - Linkage equilibrium between loci | ||||||
| - Lack of repeated MLGs | ||||||
| TcI and TcIII/IV | North America, Brazil, Bolivia | Inter-lineage | GPIa | - Mitochondrial introgression between TcIII and TcIV with no detectable nuclear involvement | - Asymmetric mitochondrial introgression | Lewis et al. (2011) |
| COII-ND1b | ||||||
| MLMT | ||||||
| TcI and TcIII/IV | North America, Venezuela, Argentina, Bolivia and Brazil | Inter- and intra-lineage | MLMT | - Mitochondrial introgression between TcI and TcIII/IV | - Asymmetric mitochondrial introgression | Messenger et al. (2012) |
| mtMLST | - Intra-TcI mitochondrial introgression | |||||
| - No detectable nuclear involvement | ||||||
| TcI and TcIV | North America | Inter-lineage | 24Sα rRNAa, 18S rRNAa, TcMSH2a, Tc55a, DHFR-TSa, COII-ND1b | - Mitochondrial introgression between TcI and TcIV | - Asymmetric mitochondrial introgression | Roellig et al. (2013) |
H–W: Hardy–Weinberg; MLEE: multilocus enzyme electrophoresis; MLG: multilocus genotype; MLMT: multilocus microsatellite typing; mtMLST: maxicircle multilocus sequence typing; nMLST: nuclear multilocus sequence typing; RAPD: random amplification of polymorphic DNA.
Nuclear gene.
Mitochondrial gene.
At the inter-lineage level, DTUs TcV and TcVI are unequivocal hybrids, which resemble diploid, heterozygous Mendelian F1 progeny, sharing intact alleles from their parental progenitors (TcII and TcIII) (Machado and Ayala, 2001, Brisse et al., 2003, Barnabé et al., 2011, Lewis et al., 2009, Lewis et al., 2011, Yeo et al., 2011). The origin(s) of these hybrid DTUs is presently unresolved; it is unclear whether they arose from two independent genetic exchange events (De Freitas et al., 2006, Lewis et al., 2011) or a single incidence of hybridization followed by clonal divergence (Westenberger et al., 2005, Sturm and Campbell, 2010, Flores-López and Machado, 2011). The status of TcIII and TcIV as ancient recombinants of TcI and TcII (Westenberger et al., 2005), or sister groups of TcI (Tomasini and Diosque, 2015), is more contentious, and varies based on the array of nuclear loci (Westenberger et al., 2005, Tomasini and Diosque, 2015) or mitochondrial haplotypes examined (Lewis et al., 2011, Messenger et al., 2012). The evidence for any contemporary recombination between major DTUs is more limited and the genetic identity of each lineage appears largely preserved. However, it is not known to what extent this is maintained by genetic reproductive barriers between DTUs or ecological isolation, considering, for example, historical parents TcII and TcIII now circulate in almost completely separate transmission cycles.
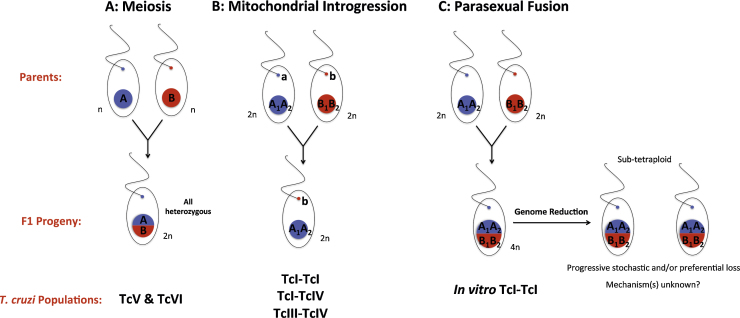
At the intra-lineage level, genetic exchange is increasingly reported, particularly among TcI populations. It is unclear whether this is due to the examination of representatives from intensely sampled populations that are minimally-subdivided spatially and temporally, and therefore more likely to undergo hybridization, or if it truly reflects the analysis of strains that are more permissive to recombination (Prugnolle and De Meeus, 2010, Ramírez and Llewellyn, 2014). The underlying cytological mechanisms of natural intra-TcI recombination vary between studies and genetic markers used (Table 1 and Fig. 1).
Fig. 1.
Putative cytological mechanisms and patterns of allele inheritance observed among natural field populations of T. cruzi (A and B) and during in vitro genetic exchange experiments (C).
In general, genetic exchange at the nuclear level has been demonstrated by Hardy–Weinberg allele frequencies, linkage equilibrium between loci, a lack of repeated multilocus genotypes (MLGs) (Barnabé et al., 2013, Baptista et al., 2014, Ocaña-Mayorga et al., 2010, Ramírez et al., 2013) and more rarely, excess heterozygosity (Messenger et al., 2015), all consistent with meiotic allele inheritance. In those studies that also examine kinetoplast DNA, mitochondrial introgression, evidenced by phylogenetic incongruence between nuclear and mitochondrial loci, is emerging as a common feature of natural transmission cycles especially within TcI populations (Lima et al., 2014, Messenger et al., 2012, Messenger et al., 2015, Ramírez et al., 2012, Zumaya-Estrada et al., 2012) but also historically, between major lineages (Lewis et al., 2011, Messenger et al., 2012, Barnabé and Breniere, 2012, Roellig et al., 2013) (Table 1 and Fig. 1). One explanation, given their crucial role in growth, development and metabolism, is that asymmetric mitochondrial introgression, may satisfy the elevated necessity to escape Muller's ratchet (the irreversible accumulation of deleterious mutations resulting from clonal reproduction) compared to the nuclear genome (Messenger et al., 2015, Neiman and Taylor, 2009, Ramírez and Llewellyn, 2014); others have attributed these observations to gross differences in evolutionary pressures and molecular clocks between non-coding microsatellites and coding maxicircle genes (Tibayrenc and Ayala, 2013). However, it is highly improbable that mutation rate variation could account for the observation of nearly identical nuclear genotypes with radically divergent mitochondrial genomes, particularly when putative donors and recipients are identified within the same population (Messenger et al., 2012, Ramírez et al., 2012).
Reciprocal nuclear recombination among parasite strains undergoing mitochondrial introgression has yet to be explicitly detected, which may support an asymmetric, cryptic hybridization mechanism, or perhaps more likely, reflect the minor amount of nuclear genetic information sampled (20% of the mitochondrial genome vs. <0.1% of the nuclear genome); without whole nuclear genome sequences for introgression hybrids and parental isolates, it is impossible to distinguish between these two hypotheses. However, by analogy to other medically important trypanosome species, the presence of alternate, covert sexual mechanisms within the same species is not entirely unexpected (Rougeron et al., 2009, Rougeron et al., 2011, Duffy et al., 2013, Hickman et al., 2013, Rogers et al., 2014, Ramírez and Llewellyn, 2014).
Interestingly, a recent study from Colombia identified biparental mitochondrial inheritance as a putative consequence of genetic exchange events (Ramírez et al., 2012). A mosaic maxicircle sequence was detected in a human TcI isolate and the presence of a recombination breakpoint confirmed by allele-specific PCR. Such a sequence is expected to arise following inter-molecular maxicircle recombination, which necessitates the inheritance of mixed mitochondrial complements. Uniparental inheritance of highly heteroplasmic maxicircles might present an indistinguishable scenario (if strains were characterized using basic dye-terminator sequencing) but reported mitochondrial heteroplasmy in T. cruzi, (examined using high coverage Illumina sequencing reads), is thus far low (Messenger et al., 2012). Parallel observations from experimental crosses of Trypanosoma brucei brucei (Gibson and Garside, 1990, Gibson et al., 2008), suggest that biparental mitochondrial inheritance might be a fundamental, as yet, uncharacterized, biological phenomenon in trypanosomatids.
3. Reconciling mechanisms of T. cruzi recombination in vitro and among natural populations
The generation of intra-TcI hybrids in vitro demonstrates that at least some T. cruzi strains have an extant capacity for genetic exchange (Gaunt et al., 2003). Putative parental isolates identified by Carrasco et al. (1996) were transformed with episomal recombinant plasmids containing either hygromycin B or neomycin resistance genes and co-passaged through in vitro (mammalian cell cultures) and in vivo (mice and triatomine bugs) cycles (Gaunt et al., 2003). Isolation of six clones by double drug selection from in vitro axenic cultures, and subsequent genetic characterization by MLEE, karyotyping, microsatellite genotyping and nucleotide sequencing of housekeeping genes, indicated that these intra-lineage recombinants inherited all parental alleles at most loci and one parental maxicircle genotype (Gaunt et al., 2003).
By analogy with Candida albicans (Bennett and Johnson, 2003, Forche et al., 2008), it was proposed that nuclear fusion had created a tetraploid intermediate, followed by homologous recombination, gradual genome erosion and reversion to aneuploidy (Fig. 1). FACs analysis of hybrid isolates reported a stable DNA content, on average, ∼69% higher than parental strains (Lewis et al., 2009); natural T. cruzi isolates are minimally diploid but overall genome size can vary by up to 48% between different DTUs (Lewis et al., 2009). Subsequent prolonged maintenance of the experimental hybrids in axenic cultures demonstrated a gradual, progressive decline in DNA content, with no evidence of any true meiotic reductive division; to date these strains remain sub-tetraploid (Lewis et al., 2010).
While this parasexual mechanism of genetic exchange has a precedent in fungal species, it is challenging to reconcile with both the predominant patterns of allele inheritance observed among natural T. cruzi populations (Table 1 and Fig. 1) as well as the conservation of meiosis-specific orthologues within the T. cruzi genome (Ramesh et al., 2005). A similar paradox exists in T. b. brucei where canonical meiotic recombination (Peacock et al., 2011), including the formation of haploid life cycle stages (Peacock et al., 2014), has been explicitly described, but is not the exclusive or obligate reproductive mechanism reported from transmission cycles (Balmer et al., 2011, Duffy et al., 2013). Likewise, experimental hybridization in Leishmania resembles meiosis with recurrent triploidy (Akopyants et al., 2009, Inbar et al., 2013), but inbreeding also appears frequent in nature (Calvo-Álvarez et al., 2014, Rougeron et al., 2009, Rougeron et al., 2011, Sterkers et al., 2011, Sterkers et al., 2014, Rogers et al., 2014).
4. Problems of detecting recombination among field populations
It is clear that to detect natural recombination, sample strategy, population allocation and marker choice are crucial for study design. Grouping of divergent non-recombining subgroups (in the case of T. cruzi, major DTUs) can inflate genetic linkage statistics and mask recombination events occurring between more closely related individuals (Smith et al., 1993). Recent observations of the Wahlund effect obscuring Hardy–Weinberg allele frequencies and linkage equilibrium within Brazilian TcII strains, caution the interpretation of statistics derived from inappropriately assigned parasite populations (Baptista et al., 2014). The use of multiple, different types of molecular markers (nuclear and mitochondrial, coding and non-coding) are required, in combination with targeted investigation of potential ‘hybridization’ zones, i.e. areas where recently diverged, genetically distinguishable subpopulations regularly interact (Messenger et al., 2015, Ramírez and Llewellyn, 2014). The value of such high-density sampling has already been demonstrated in defining the population structures of other trypanosomatid species, e.g. Trypanosoma brucei gambiense (Koffi et al., 2009), Trypanosoma congolense (Morrison et al., 2009), Leishmania braziliensis (Rougeron et al., 2009) and Leishmania guyanensis (Rougeron et al., 2011), including establishing putative levels of genetic exchange. However, the low circulating parasitaemia that defines chronic Chagas disease patients often prohibits parasite isolation, and thus many studies are overly reliant on historical collections of references isolates assembled over many years.
5. Implications of hybridization for parasite phenotype
Importantly, the effects of genetic exchange on T. cruzi phenotype are unknown. Hybrid vigour (heterosis) is a well-documented phenomenon among parasitic protozoa (Detwiler and Criscione, 2010). Observations of natural Leishmania hybrids indicate that genetic exchange can impact vector permissibility (Volf et al., 2007), increase (Akopyants et al., 2009) or alter (Cortes et al., 2012, Calvo-Álvarez et al., 2014) virulence, including the ability to disseminate and colonize visceral organs (Romano et al., 2014), as well as generate recombinant progeny that are capable of widespread clonal propagation (Schwenkenbecher et al., 2006, Nolder et al., 2007); a scenario reminiscent of the successful establishment of TcV and TcVI among domestic transmission cycles in the Southern Cone. Similarly, if mitochondrial introgression is exploitable as a putative mechanism of host range extension, hybrids might be expected to present higher mammalian infectivity and growth rates, especially in vectors (Messenger et al., 2015). The pathological implications of recombinant genotypes in human infections with regards to virulence, transmissibility and drug susceptibility warrant further investigation in conjunction with improved methods of identification and isolation of natural hybrids strains.
6. Conclusions
The majority of field studies now indicate that natural genetic exchange in T. cruzi is both contemporary and historical, responsible for shaping current parasite population structures, as well as the evolution of distinct T. cruzi DTUs. Together these observations challenge the traditional paradigm of PCE in T. cruzi and highlight the need for additional, intensive and appropriately sampled field surveys, complemented by high resolution, combined nuclear and mitochondrial population genetics analyses.
The precedent in experimental design established by such studies may represent the most promising intermediary in T. cruzi population genetics until imminently superseded by comparative population genomics.
Acknowledgements
This review was largely based on research funded by the Wellcome Trust and the European Commission Framework Programme Project “Comparative epidemiology of genetic lineages of Trypanosoma cruzi” ChagasEpiNet (contract #223034). LAM was supported by a BBSRC Doctoral Training Grant. The authors thank Dr. Martin Llewellyn for critical reading of this manuscript.
References
- Akopyants N.S., Kimblin N., Secundino N., Patrick R., Peters N., Lawyer P., Dobson D.E., Beverley S.M., Sacks D.L. Demonstration of genetic exchange during cyclical development of Leishmania in the sand fly vector. Science. 2009;324:265–268. doi: 10.1126/science.1169464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balmer O., Beadell J.S., Gibson W., Caccone A. Phylogeography and taxonomy of Trypanosoma brucei. PLoS Negl. Trop. Dis. 2011;5:e961. doi: 10.1371/journal.pntd.0000961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baptista Rde P., D’Avila D.A., Segatto M., do Valle I.F., Franco G.R., Valadares H.M., Gontijo E.D., Galvão L.M., Pena S.D., Chiari E., Machado C.R., Macedo A.M. Evidence of substantial recombination among Trypanosoma cruzi II strains from Minas Gerais. Infect. Genet. Evol. 2014;22:183–191. doi: 10.1016/j.meegid.2013.11.021. [DOI] [PubMed] [Google Scholar]
- Barnabé C., De Meeus T., Noireau F., Bosseno M.F., Monje E.M., Renaud F., Breniere S.F. Trypanosoma cruzi discrete typing units (DTUs): microsatellite loci and population genetics of DTUs TcV and TcI in Bolivia and Peru. Infect. Genet. Evol. 2011;11:1752–1760. doi: 10.1016/j.meegid.2011.07.011. [DOI] [PubMed] [Google Scholar]
- Barnabé C., Breniere S.F. Scarce events of mitochondrial introgression in Trypanosoma cruzi: new case with a Bolivian strain. Infect. Genet. Evol. 2012;12:1879–1883. doi: 10.1016/j.meegid.2012.08.018. [DOI] [PubMed] [Google Scholar]
- Barnabé C., Buitrago R., Bremond P., Aliaga C., Salas R., Vidaurre P., Herrera C., Cerqueira F., Bosseno M.F., Waleckx E., Breniere S.F. Putative panmixia in restricted populations of Trypanosoma cruzi isolated from wild Triatoma infestans in Bolivia. PLOS ONE. 2013;8:e82269. doi: 10.1371/journal.pone.0082269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett R.J., Johnson A.D. Completion of a parasexual cycle in Candida albicans by induced chromosome loss in tetraploid strains. EMBO J. 2003;22:2505–2515. doi: 10.1093/emboj/cdg235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brisse S., Henriksson J., Barnabé C., Douzery E.J., Berkvens D., Serrano M., De Carvalho M.R., Buck G.A., Dujardin J.C., Tibayrenc M. Evidence for genetic exchange and hybridization in Trypanosoma cruzi based on nucleotide sequences and molecular karyotype. Infect. Genet. Evol. 2003;2:173–183. doi: 10.1016/s1567-1348(02)00097-7. [DOI] [PubMed] [Google Scholar]
- Calvo-Álvarez E., Alvarez-Velilla R., Jiménez M., Molina R., Pérez-Pertejo Y., Balaña-Fouce R., Reguera R.M. First evidence of intraclonal genetic exchange in trypanosomatids using two Leishmania infantum fluorescent transgenic clones. PLoS Negl. Trop. Dis. 2014;8:e3075. doi: 10.1371/journal.pntd.0003075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrasco H.J., Frame I.A., Valente S.A., Miles M.A. Genetic exchange as a possible source of genomic diversity in sylvatic populations of Trypanosoma cruzi. Am. J. Trop. Med. Hyg. 1996;54:418–424. doi: 10.4269/ajtmh.1996.54.418. [DOI] [PubMed] [Google Scholar]
- Cortes S., Esteves C., Mauricio I., Maia C., Cristovao J.M., Miles M., Campino L. In vitro and in vivo behavior of sympatric Leishmania (V.) braziliensis L. (V.) peruviana and their hybrids. Parasitology. 2012;139:191–199. doi: 10.1017/S0031182011001909. [DOI] [PubMed] [Google Scholar]
- De Freitas J.M., Augusto-Pinto L., Pimenta J.R., Bastos-Rodrigues L., Goncalves V.F., Teixeira S.M.R., Chiari E., Junqueira A.C.V., Fernandes C., Macedo A.M., Machado C.R., Pena S.D.J. Ancestral genomes, sex and the population structure of Trypanosoma cruzi. PLoS Pathog. 2006;2:e24. doi: 10.1371/journal.ppat.0020024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detwiler J.T., Criscione C.D. An infection topic in reticulate evolution: introgression and hybridization in animal parasites. Genes. 2010;1:102–123. doi: 10.3390/genes1010102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy C.W., MacLean L., Sweeney L., Cooper A., Turner C.M., Tait A., Sternberg J., Morrison L.J., MacLeod A. Population genetics of Trypanosoma brucei rhodesiense: clonality and diversity within and between foci. PLoS Negl. Trop. Dis. 2013;7:e2526. doi: 10.1371/journal.pntd.0002526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores-López C.A., Machado C.A. Analyses of 32 loci clarify phylogenetic relationships among Trypanosoma cruzi lineages and support a single hybridization prior to human contact. PLoS Negl. Trop. Dis. 2011;5:e1272. doi: 10.1371/journal.pntd.0001272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forche A., Alby K., Schaefer D., Johnson A.D., Berman J., Bennett R.J. The parasexual cycle in Candida albicans provides an alternative pathway to meiosis for the formation of recombinant strains. PLoS Biol. 2008;6:e110. doi: 10.1371/journal.pbio.0060110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaunt M.W., Yeo M., Frame I.A., Stothard J.R., Carrasco H.J., Taylor M.C., Mena S.S., Veazey P., Miles G.A., Acosta N., de Arias A.R., Miles M.A. Mechanism of genetic exchange in American trypanosomes. Nature. 2003;421:936–939. doi: 10.1038/nature01438. [DOI] [PubMed] [Google Scholar]
- Gibson W., Garside L. Kinetoplast DNA minicircles are inherited from both parents in genetic hybrids of Trypanosoma brucei. Mol. Biochem. Parasitol. 1990;42:45–53. doi: 10.1016/0166-6851(90)90111-x. [DOI] [PubMed] [Google Scholar]
- Gibson W., Peacock L., Ferris V., Williams K., Bailey M. The use of yellow fluorescent hybrids to indicate mating in Trypanosoma brucei. Parasit. Vectors. 2008;1:4. doi: 10.1186/1756-3305-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman M.A., Zeng G., Forche A., Hirakawa M.P., Abbey D., Harrison B.D., Wang Y.M., Su C.H., Bennet R.J., Wang Y., Berman J. The ‘obligate diploid’ Candida albicans forms mating-competent haploids. Nature. 2013;494:55–59. doi: 10.1038/nature11865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inbar E., Akopyants N.S., Charmoy M., Romano A., Lawyer P., Elnaiem D.E., Kauffmann F., Barhoumi M., Grigg M., Owens K., Fay M., Dobson D.E., Shalik J., Beverley S.M., Sacks D. The mating competence of geographically diverse Leishmania major strains in their natural and unnatural sand fly vectors. PLoS Genet. 2013;9:e1003672. doi: 10.1371/journal.pgen.1003672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koffi M., De Meeus T., Bucheton B., Solano P., Camara M., Kaba D., Cuny G., Ayala F.J., Jamonneau V. Population genetics of Trypanosoma brucei gambiense, the agent of sleeping sickness in Western Africa. Proc. Natl. Acad. Sci. U. S. A. 2009;106:209–214. doi: 10.1073/pnas.0811080106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis M.D., Llewellyn M.S., Gaunt M.W., Yeo M., Carrasco H.J., Miles M.A. Flow cytometric analysis and microsatellite genotyping reveal extensive DNA content variation in Trypanosoma cruzi populations and expose contrasts between natural and experimental hybrids. Int. J. Parasitol. 2009;39:1305–1317. doi: 10.1016/j.ijpara.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis M.D., Llewellyn M.S., Yeo M., Miles M.A. Experimental and natural recombination in Trypanosoma cruzi. In: Telleria J., Tibayrenc M., editors. American Trypanosomiasis Chagas Disease: One Hundred Years of Research. Elsevier; London: 2010. pp. 459–474. [Google Scholar]
- Lewis M.D., Llewellyn M.S., Yeo M., Acosta N., Gaunt M.W., Miles M.A. Recent, independent and anthropogenic origins of Trypanosoma cruzi hybrids. PLoS Negl. Trop. Dis. 2011;5:e1363. doi: 10.1371/journal.pntd.0001363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima V., Jansen A.M., Messenger L.A., Rocha F., Miles M.A., Llewellyn M.S. Wild Trypanosoma cruzi I genetic diversity in Brazil suggests admixture and disturbance in parasite populations from the Atlantic Forest regions. Parasit. Vectors. 2014;7:263. doi: 10.1186/1756-3305-7-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llewellyn M.S., Miles M.A., Carrasco H.J., Lewis M.D., Yeo M., Vargas J., Torrico F., Diosque P., Valente V., Valente S.A., Gaunt M.W. Genome-scale multilocus microsatellite typing of Trypanosoma cruzi discrete typing unit I reveals phylogeographic structure and specific genotypes linked to human infection. PLoS Pathog. 2009;5:e1000410. doi: 10.1371/journal.ppat.1000410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado C., Ayala F.J. Nucleotide sequences provide evidence of genetic exchange among distantly related lineages of Trypanosoma cruzi. Proc. Natl. Acad. Sci. U. S. A. 2001;98:7396–7401. doi: 10.1073/pnas.121187198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messenger L.A., Llewellyn M.S., Bhattacharyya T., Franzén O., Lewis M.D., Ramírez J.D., Carrasco H.J., Andersson B., Miles M.A. Multiple mitochondrial introgression events and heteroplasmy in Trypanosoma cruzi revealed by maxicircle MLST and next generation sequencing. PLoS Negl. Trop. Dis. 2012;6:e1584. doi: 10.1371/journal.pntd.0001584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messenger L.A., Garcia L., Vanhove M., Huaranca C., Bustamante M., Torrico M., Torrico F., Miles M.A., Llewellyn M.S. Ecological host fitting of Trypanosoma cruzi TcI in Bolivia: mosaic population structure, hybridization and a role for humans in Andean parasite dispersal. Mol. Ecol. 2015;24:2406–2422. doi: 10.1111/mec.13186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miles M.A., Llewellyn M.S., Lewis M.D., Yeo M., Baleela R., Fitzpatrick S., Gaunt M.W., Mauricio I. The molecular epidemiology and phylogeography of Trypanosoma cruzi and parallel research on Leishmania: looking back and to the future. Parasitology. 2009;136:1509–1528. doi: 10.1017/S0031182009990977. [DOI] [PubMed] [Google Scholar]
- Morrison L.J., Tweedie A., Black A., Pinchbeck G.L., Christley R.M., Schoenefeld A., Hertz-Fowler C., MacLeod A., Turner C.M., Tait A. Discovery of mating in the major African livestock pathogen Trypanosoma congolense. PLoS ONE. 2009;4:e5564. doi: 10.1371/journal.pone.0005564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neiman M., Taylor D.R. The causes of mutation accumulation in mitochondrial genomes. Proc. R. Soc. Biol. Sci. 2009;276:1201–1209. doi: 10.1098/rspb.2008.1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolder D., Roncal N., Davies C.R., Llanos-Cuentas A., Miles M.A. Multiple hybrid genotypes of Leishmania (Viannia) in a focus of mucocutaneous leishmaniasis. Am. J. Trop. Med. Hyg. 2007;76:573–578. [PubMed] [Google Scholar]
- Ocaña-Mayorga S., Llewellyn M.S., Costales J.A., Miles M.A., Grijalva M.J. Sex, subdivision, and domestic dispersal of Trypanosoma cruzi lineage I in Southern Ecuador. PLoS Negl. Trop. Dis. 2010;4:e915. doi: 10.1371/journal.pntd.0000915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peacock L., Ferris V., Sharma R., Sunter J., Bailey M., Carrington M., Gibson W. Identification of the meiotic life cycle stage of Trypanosoma brucei in the tsetse fly. Proc. Natl. Acad. Sci. U. S. A. 2011;108:3671–3676. doi: 10.1073/pnas.1019423108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peacock L., Bailey M., Carrington M., Gibson W. Meiosis and haploid gametes in the pathogen Trypanosoma brucei. Curr. Biol. 2014;24:181–186. doi: 10.1016/j.cub.2013.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prugnolle F., De Meeus T. Apparent high recombination rates in clonal parasitic organisms due to inappropriate sampling design. Heredity. 2010;104:135–140. doi: 10.1038/hdy.2009.128. [DOI] [PubMed] [Google Scholar]
- Ramesh M.A., Malik S.B., Logsdon J.M., Jr. A phylogenomic inventory of meiotic genes; evidence for sex in Giardia and an early eukaryotic origin of meiosis. Curr. Biol. 2005;15:185–191. doi: 10.1016/j.cub.2005.01.003. [DOI] [PubMed] [Google Scholar]
- Ramírez J.D., Guhl F., Messenger L.A., Lewis M.D., Montilla M., Cucunuba Z., Miles M.A., Llewellyn M.S. Contemporary cryptic sexuality in Trypanosoma cruzi. Mol. Ecol. 2012;21:4216–4226. doi: 10.1111/j.1365-294X.2012.05699.x. [DOI] [PubMed] [Google Scholar]
- Ramírez J.D., Tapia-Calle G., Guhl F. Genetic structure of Trypanosoma cruzi in Colombia revealed by a high-throughput nuclear multilocus sequence typing (nMLST) approach. BMC Genet. 2013;14:96. doi: 10.1186/1471-2156-14-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez J.D., Llewellyn M.S. Reproductive clonality in protozoan pathogens – truth or artefact? Mol. Ecol. 2014;23:4195–4202. doi: 10.1111/mec.12872. [DOI] [PubMed] [Google Scholar]
- Roellig D.M., Savage M.Y., Fujita A.W., Barnabé C., Tibayrenc M., Steurer F.J., Yabsley M.J. Genetic variation and exchange in Trypanosoma cruzi isolates from the United States. PLOS ONE. 2013;8:e56198. doi: 10.1371/journal.pone.0056198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers M.B., Downing T., Smith B.A., Imamura H., Sanders M., Svobodova M., Volf P., Berriman M., Cotton J.A., Smith D.F. Genomic confirmation of hybridisation and recent inbreeding in a vector-isolated Leishmania population. PLoS Genet. 2014;10:e1004092. doi: 10.1371/journal.pgen.1004092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano A., Inbar E., Debrabant A., Charmoy M., Lawyer P., Ribeiro-Gomes F., Barhoumi M., Grigg M., Shaik J., Dobson D., Beverley S.M., Sacks D.L. Cross-species genetic exchange between visceral and cutaneous strains of Leishmania in the sand fly vector. Proc. Natl. Acad. Sci. U. S. A. 2014;111:16808–16813. doi: 10.1073/pnas.1415109111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rougeron V., De Meeus T., Hide M., Waleckx E., Bermudez H., Arevalo J., Llanos-Cuentas A., Dujardin J.C., De Doncker S., Le Ray D., Ayala F.J., Banuls A.L. Extreme inbreeding in Leishmania braziliensis. Proc. Natl. Acad. Sci. U. S. A. 2009;106:10224–10229. doi: 10.1073/pnas.0904420106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rougeron V., Banuls A.L., Carme B., Simon S., Couppie P., Nacher M., Hide M., De Meeus T. Reproductive strategies and population structure in Leishmania: substantial amount of sex in Leishmania (Viannia) guyanensis. Mol. Ecol. 2011;20:3116–3127. doi: 10.1111/j.1365-294X.2011.05162.x. [DOI] [PubMed] [Google Scholar]
- Schwenkenbecher J.M., Wirth T., Schnur L.F., Jaffe C.L., Schallig H., Al-Jawabreh A., Hamarsheh O., Azmi K., Pratlong F., Schonian G. Microsatellite analysis reveals genetic structure of Leishmania tropica. Int. J. Parasitol. 2006;36:237–246. doi: 10.1016/j.ijpara.2005.09.010. [DOI] [PubMed] [Google Scholar]
- Smith J.M., Smith N.H., O’Rourke M., Spratt B.G. How clonal are bacteria? Proc. Natl. Acad. Sci. U. S. A. 1993;90:4384–4388. doi: 10.1073/pnas.90.10.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterkers Y., Lachaud L., Crobu L., Bastien P., Pages M. FISH analysis reveals aneuploidy and continual generation of chromosomal mosaicism in Leishmania major. Cell. Microbiol. 2011;13:274–283. doi: 10.1111/j.1462-5822.2010.01534.x. [DOI] [PubMed] [Google Scholar]
- Sterkers Y., Crobu L., Lachaud L., Pages M., Bastien P. Parasexuality and mosaic aneuploidy in Leishmania: alternative genetics. Trends Parasitol. 2014;30:429–435. doi: 10.1016/j.pt.2014.07.002. [DOI] [PubMed] [Google Scholar]
- Sturm N.R., Campbell D.A. Alternative lifestyles: the population structure of Trypanosoma cruzi. Acta Trop. 2010;115:35–43. doi: 10.1016/j.actatropica.2009.08.018. [DOI] [PubMed] [Google Scholar]
- Tibayrenc M., Ward P., Moya A., Ayala F.J. Natural populations of Trypanosoma cruzi, the agent of Chagas disease, have a complex multiclonal structure. Proc. Natl. Acad. Sci. U. S. A. 1986;83:115–119. doi: 10.1073/pnas.83.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibayrenc M., Kjellberg F., Ayala F.J. A clonal theory of parasitic protozoa: the population structures of Entamoeba, Giardia, Leishmania, Naegleria, Plasmodium, Trichomonas and Trypanosoma and their medical and taxonomical consequences. Proc. Natl. Acad. Sci. U. S. A. 1990;87:2414–2418. doi: 10.1073/pnas.87.7.2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibayrenc M., Ayala F.J. Towards a population genetics of microorganisms: the clonal theory of parasitic protozoa. Parasitol. Today. 1991;7:228–232. doi: 10.1016/0169-4758(91)90234-f. [DOI] [PubMed] [Google Scholar]
- Tibayrenc M., Ayala F.J. Reproductive clonality of pathogens: a perspective on pathogenic viruses, bacteria, fungi and parasitic protozoa. Proc. Natl. Acad. Sci. U. S. A. 2012;109:e3305–e3313. doi: 10.1073/pnas.1212452109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibayrenc M., Ayala F.J. How clonal are Trypanosoma and Leishmania? Trends Parasitol. 2013;29:264–269. doi: 10.1016/j.pt.2013.03.007. [DOI] [PubMed] [Google Scholar]
- Tibayrenc M., Ayala F.J. Cryptosporidium, Giardia, Cryptococcus, Pneumocystis genetic variability: cryptic biological species or clonal near-clades? PLoS Pathog. 2014;10:e1003908. doi: 10.1371/journal.ppat.1003908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibayrenc M., Ayala F.J. New insights into clonality and panmixia in Plasmodium and Toxoplasma. Adv. Parasitol. 2014;84:253–268. doi: 10.1016/B978-0-12-800099-1.00005-3. [DOI] [PubMed] [Google Scholar]
- Tomasini N., Lauthier J.J., Monje Rumi M.M., Ragone P.G., Alberti D’Amato A.M., Brandán C.P., Basombrío M.A., Diosque P. Preponderate clonal evolution of Trypanosoma cruzi I from Argentinean Chaco revealed by Multilocus Sequence Typing (MLST) Infect. Genet. Evol. 2014;27:348–354. doi: 10.1016/j.meegid.2014.08.003. [DOI] [PubMed] [Google Scholar]
- Tomasini N., Lauthier J.J., Ayala F.J., Tibayrenc M., Diosque P. How often do they have sex? A comparative analysis of the population structure of seven eukaryotic microbial pathogens. PLOS ONE. 2014;9:e103131. doi: 10.1371/journal.pone.0103131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasini N., Diosque P. Evolution of Trypanosoma cruzi: clarifying hybridisations, mitochondrial introgressions and phylogenetic relationships between major lineages. Mem. Inst. Oswaldo Cruz. 2015;110:403–413. doi: 10.1590/0074-02760140401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volf P., Benkova I., Myskova J., Sadlova J., Campino L., Ravel C. Increased transmission potential of Leishmania major/Leishmania infantum hybrids. Int. J. Parasitol. 2007;37:589–593. doi: 10.1016/j.ijpara.2007.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westenberger S.J., Barnabé C., Campbell D.A., Sturm N.R. Two hybridization events define the population structure of Trypanosoma cruzi. Genetics. 2005;171:527–543. doi: 10.1534/genetics.104.038745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo M., Mauricio I.L., Messenger L.A., Lewis M.D., Llewellyn M.S., Acosta N., Bhattacharyya T., Diosque P., Carrasco H.J., Miles M.A. Multilocus Sequence Typing (MLST) for lineage assignment and high resolution diversity studies in Trypanosoma cruzi. PLoS Negl. Trop. Dis. 2011;5:e1049. doi: 10.1371/journal.pntd.0001049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zingales B., Andrade S.G., Briones M.R., Campbell D.A., Chiari E., Fernandes O., Guhl F., Lages-Silva E., Macedo A.M., Machado C.R., Miles M.A., Romanha A.J., Sturm N.R., Tibayrenc M., Schijman A.G., Second Satellite Meeting A new consensus for Trypanosoma cruzi intraspecific nomenclature: second revision meeting recommends TcI to TcVI. Mem. Inst. Oswaldo Cruz. 2009;104:1051–1054. doi: 10.1590/s0074-02762009000700021. [DOI] [PubMed] [Google Scholar]
- Zumaya-Estrada F.A., Messenger L.A., Lopez-Ordonez T., Lewis M.D., Flores-Lopez C.A., Martínez-Ibarra A.J., Pennington P.M., Cordon-Rosales C., Carrasco H.J., Segovia M., Miles M.A., Llewellyn M.S. North American import? Charting the origins of an enigmatic Trypanosoma cruzi domestic genotype. Parasit. Vectors. 2012;5:226. doi: 10.1186/1756-3305-5-226. [DOI] [PMC free article] [PubMed] [Google Scholar]