Abstract
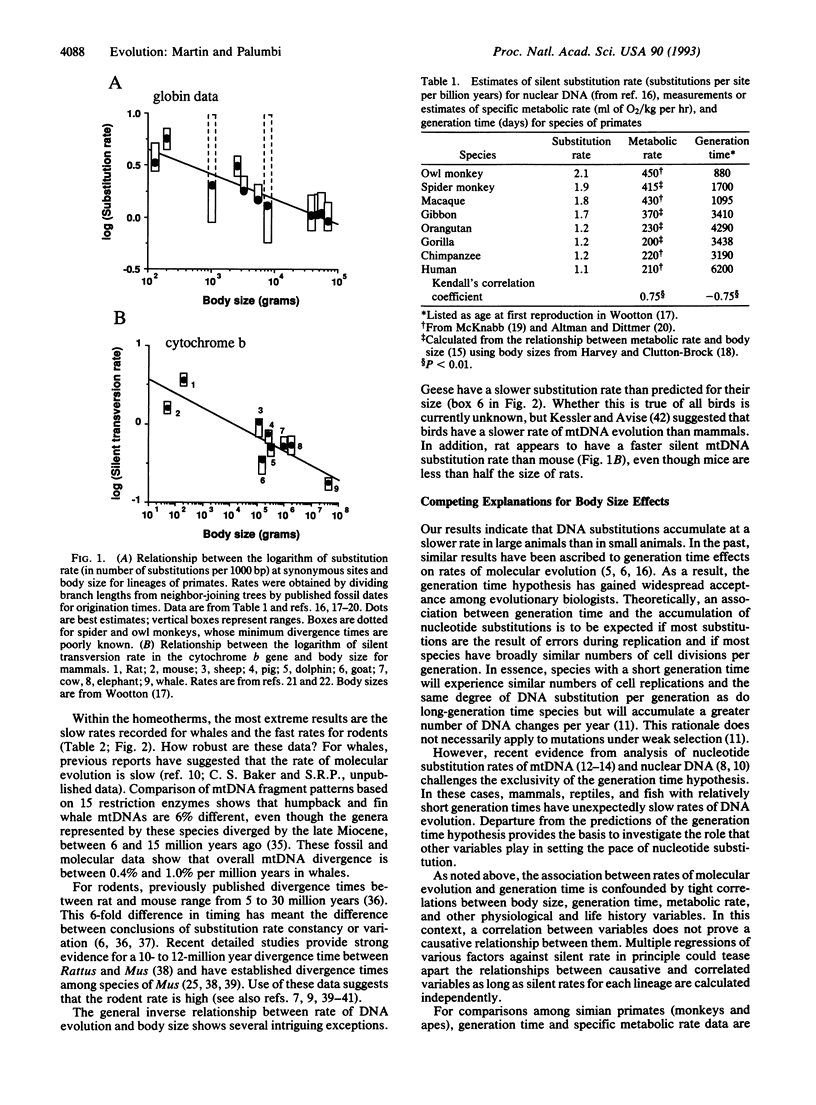
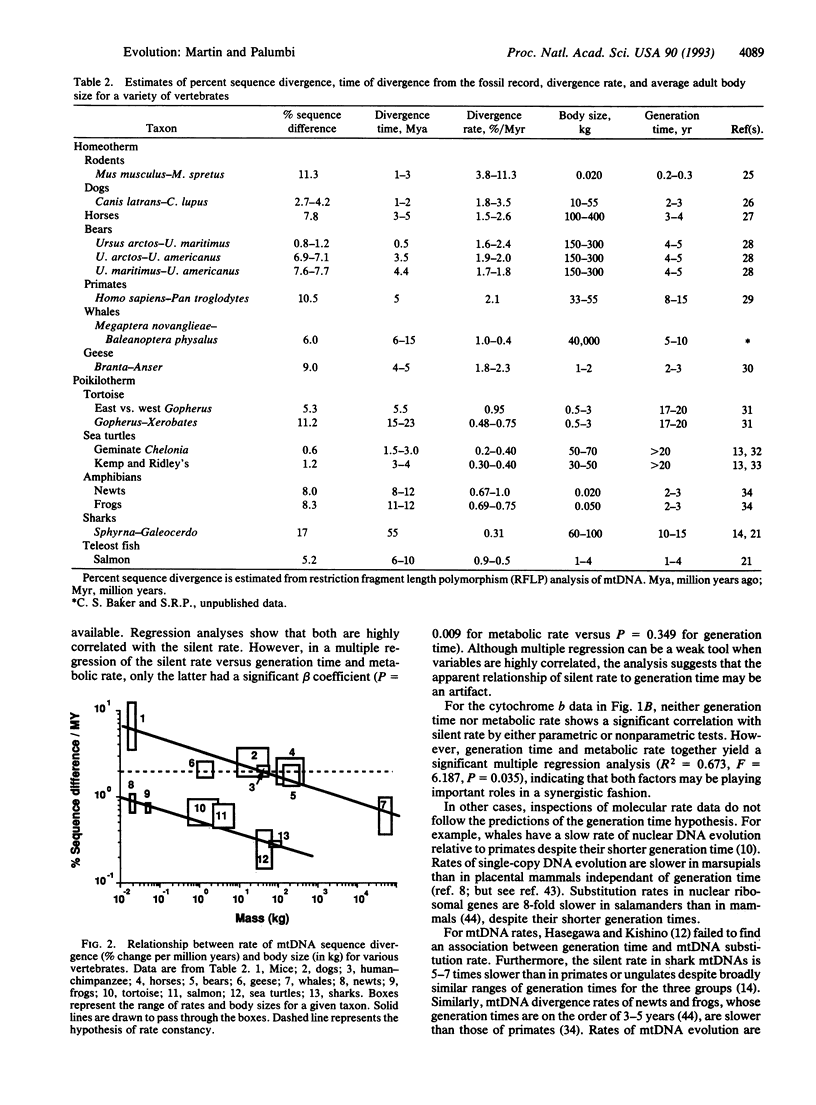
There is increasing evidence for variation in rates of nucleotide substitution among divergent taxonomic groups. Here, we summarize published rate data and show a strong relationship between substitution rate and body size. For instance, rates of nuclear and mtDNA evolution are slow in whales, intermediate in primates, and fast in rodents. A similar relationship exists for poikilothermic vertebrates. However, these taxa have slower mtDNA substitution rates overall than do homeotherms of similar size. A number of physiological and life history variables are highly correlated with body size. Of these, generation time and metabolic rate explain some patterns of rate heterogeneity equally well. In many cases, however, differences in metabolic rate explain important exceptions to the generation time model. Correlation between metabolic rate and nucleotide substitution may be mediated by (i) the mutagenic effects of oxygen radicals that are abundant by-products of aerobic respiration, and (ii) increased rates of DNA synthesis and nucleotide replacement in organisms with higher metabolic rates. Both of these factors increase mutation rate by decreasing the "nucleotide generation time," the average length of time before a nucleotide is copied either through replication or repair. Reconsideration of the generation time hypothesis to include physiological effects such as metabolic rate improves the theoretical underpinnings of molecular evolution.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Avise J. C., Bowen B. W., Lamb T., Meylan A. B., Bermingham E. Mitochondrial DNA evolution at a turtle's pace: evidence for low genetic variability and reduced microevolutionary rate in the Testudines. Mol Biol Evol. 1992 May;9(3):457–473. doi: 10.1093/oxfordjournals.molbev.a040735. [DOI] [PubMed] [Google Scholar]
- Bailey W. J., Fitch D. H., Tagle D. A., Czelusniak J., Slightom J. L., Goodman M. Molecular evolution of the psi eta-globin gene locus: gibbon phylogeny and the hominoid slowdown. Mol Biol Evol. 1991 Mar;8(2):155–184. doi: 10.1093/oxfordjournals.molbev.a040641. [DOI] [PubMed] [Google Scholar]
- Bowen B. W., Meylan A. B., Avise J. C. Evolutionary distinctiveness of the endangered Kemp's ridley sea turtle. Nature. 1991 Aug 22;352(6337):709–711. doi: 10.1038/352709a0. [DOI] [PubMed] [Google Scholar]
- Britten R. J. Rates of DNA sequence evolution differ between taxonomic groups. Science. 1986 Mar 21;231(4744):1393–1398. doi: 10.1126/science.3082006. [DOI] [PubMed] [Google Scholar]
- Brown W. M., George M., Jr, Wilson A. C. Rapid evolution of animal mitochondrial DNA. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1967–1971. doi: 10.1073/pnas.76.4.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown W. M., Prager E. M., Wang A., Wilson A. C. Mitochondrial DNA sequences of primates: tempo and mode of evolution. J Mol Evol. 1982;18(4):225–239. doi: 10.1007/BF01734101. [DOI] [PubMed] [Google Scholar]
- Bulmer M., Wolfe K. H., Sharp P. M. Synonymous nucleotide substitution rates in mammalian genes: implications for the molecular clock and the relationship of mammalian orders. Proc Natl Acad Sci U S A. 1991 Jul 15;88(14):5974–5978. doi: 10.1073/pnas.88.14.5974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng K. C., Cahill D. S., Kasai H., Nishimura S., Loeb L. A. 8-Hydroxyguanine, an abundant form of oxidative DNA damage, causes G----T and A----C substitutions. J Biol Chem. 1992 Jan 5;267(1):166–172. [PubMed] [Google Scholar]
- Cockburn A., Mansergh I. M., Broome L. S., Ward S. Molecular clocks and generation time in burramyid marsupials. Mol Biol Evol. 1990 May;7(3):283–285. doi: 10.1093/oxfordjournals.molbev.a040603. [DOI] [PubMed] [Google Scholar]
- Crozier R. H., Crozier Y. C., Mackinlay A. G. The CO-I and CO-II region of honeybee mitochondrial DNA: evidence for variation in insect mitochondrial evolutionary rates. Mol Biol Evol. 1989 Jul;6(4):399–411. doi: 10.1093/oxfordjournals.molbev.a040553. [DOI] [PubMed] [Google Scholar]
- Dutton D. R., Bowden G. T. Indirect induction of a clastogenic effect in epidermal cells by a tumor promoter. Carcinogenesis. 1985 Sep;6(9):1279–1284. doi: 10.1093/carcin/6.9.1279. [DOI] [PubMed] [Google Scholar]
- Easteal S. The relative rate of DNA evolution in primates. Mol Biol Evol. 1991 Jan;8(1):115–127. doi: 10.1093/oxfordjournals.molbev.a040632. [DOI] [PubMed] [Google Scholar]
- George M., Jr, Ryder O. A. Mitochondrial DNA evolution in the genus Equus. Mol Biol Evol. 1986 Nov;3(6):535–546. doi: 10.1093/oxfordjournals.molbev.a040414. [DOI] [PubMed] [Google Scholar]
- Gross N. J., Getz G. S., Rabinowitz M. Apparent turnover of mitochondrial deoxyribonucleic acid and mitochondrial phospholipids in the tissues of the rat. J Biol Chem. 1969 Mar 25;244(6):1552–1562. [PubMed] [Google Scholar]
- Hasegawa M., Kishino H. Heterogeneity of tempo and mode of mitochondrial DNA evolution among mammalian orders. Jpn J Genet. 1989 Aug;64(4):243–258. doi: 10.1266/jjg.64.243. [DOI] [PubMed] [Google Scholar]
- Hayasaka K., Gojobori T., Horai S. Molecular phylogeny and evolution of primate mitochondrial DNA. Mol Biol Evol. 1988 Nov;5(6):626–644. doi: 10.1093/oxfordjournals.molbev.a040524. [DOI] [PubMed] [Google Scholar]
- Holmes E. C. Different rates of substitution may produce different phylogenies of the eutherian mammals. J Mol Evol. 1991 Sep;33(3):209–215. doi: 10.1007/BF02100671. [DOI] [PubMed] [Google Scholar]
- Hruszkewycz A. M. Evidence for mitochondrial DNA damage by lipid peroxidation. Biochem Biophys Res Commun. 1988 May 31;153(1):191–197. doi: 10.1016/s0006-291x(88)81207-5. [DOI] [PubMed] [Google Scholar]
- Irwin D. M., Kocher T. D., Wilson A. C. Evolution of the cytochrome b gene of mammals. J Mol Evol. 1991 Feb;32(2):128–144. doi: 10.1007/BF02515385. [DOI] [PubMed] [Google Scholar]
- Joenje H. Genetic toxicology of oxygen. Mutat Res. 1989 Jul;219(4):193–208. doi: 10.1016/0921-8734(89)90001-5. [DOI] [PubMed] [Google Scholar]
- Kessler L. G., Avise J. C. A comparative description of mitochondrial DNA differentiation in selected avian and other vertebrate genera. Mol Biol Evol. 1985 Mar;2(2):109–125. doi: 10.1093/oxfordjournals.molbev.a040339. [DOI] [PubMed] [Google Scholar]
- Kimura M. Model of effectively neutral mutations in which selective constraint is incorporated. Proc Natl Acad Sci U S A. 1979 Jul;76(7):3440–3444. doi: 10.1073/pnas.76.7.3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanave C., Preparata G., Saccone C., Serio G. A new method for calculating evolutionary substitution rates. J Mol Evol. 1984;20(1):86–93. doi: 10.1007/BF02101990. [DOI] [PubMed] [Google Scholar]
- Larson A., Wilson A. C. Patterns of ribosomal RNA evolution in salamanders. Mol Biol Evol. 1989 Mar;6(2):131–154. doi: 10.1093/oxfordjournals.molbev.a040539. [DOI] [PubMed] [Google Scholar]
- Li W. H., Gouy M., Sharp P. M., O'hUigin C., Yang Y. W. Molecular phylogeny of Rodentia, Lagomorpha, Primates, Artiodactyla, and Carnivora and molecular clocks. Proc Natl Acad Sci U S A. 1990 Sep;87(17):6703–6707. doi: 10.1073/pnas.87.17.6703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W. H., Tanimura M., Sharp P. M. An evaluation of the molecular clock hypothesis using mammalian DNA sequences. J Mol Evol. 1987;25(4):330–342. doi: 10.1007/BF02603118. [DOI] [PubMed] [Google Scholar]
- Martin A. P., Naylor G. J., Palumbi S. R. Rates of mitochondrial DNA evolution in sharks are slow compared with mammals. Nature. 1992 May 14;357(6374):153–155. doi: 10.1038/357153a0. [DOI] [PubMed] [Google Scholar]
- McNab B. K. Complications inherent in scaling the basal rate of metabolism in mammals. Q Rev Biol. 1988 Mar;63(1):25–54. doi: 10.1086/415715. [DOI] [PubMed] [Google Scholar]
- Richter C., Park J. W., Ames B. N. Normal oxidative damage to mitochondrial and nuclear DNA is extensive. Proc Natl Acad Sci U S A. 1988 Sep;85(17):6465–6467. doi: 10.1073/pnas.85.17.6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlötterer C., Amos B., Tautz D. Conservation of polymorphic simple sequence loci in cetacean species. Nature. 1991 Nov 7;354(6348):63–65. doi: 10.1038/354063a0. [DOI] [PubMed] [Google Scholar]
- Shields G. F., Wilson A. C. Calibration of mitochondrial DNA evolution in geese. J Mol Evol. 1987;24(3):212–217. doi: 10.1007/BF02111234. [DOI] [PubMed] [Google Scholar]
- Shigenaga M. K., Gimeno C. J., Ames B. N. Urinary 8-hydroxy-2'-deoxyguanosine as a biological marker of in vivo oxidative DNA damage. Proc Natl Acad Sci U S A. 1989 Dec;86(24):9697–9701. doi: 10.1073/pnas.86.24.9697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohal R. S., Svensson I., Brunk U. T. Hydrogen peroxide production by liver mitochondria in different species. Mech Ageing Dev. 1990 Apr 30;53(3):209–215. doi: 10.1016/0047-6374(90)90039-i. [DOI] [PubMed] [Google Scholar]
- Springer M. S., Kirsch J. A. Rates of single-copy DNA evolution in phalangeriform marsupials. Mol Biol Evol. 1989 Jul;6(4):331–341. doi: 10.1093/oxfordjournals.molbev.a040557. [DOI] [PubMed] [Google Scholar]
- Suarez R. K., Lighton J. R., Brown G. S., Mathieu-Costello O. Mitochondrial respiration in hummingbird flight muscles. Proc Natl Acad Sci U S A. 1991 Jun 1;88(11):4870–4873. doi: 10.1073/pnas.88.11.4870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vawter L., Brown W. M. Nuclear and mitochondrial DNA comparisons reveal extreme rate variation in the molecular clock. Science. 1986 Oct 10;234(4773):194–196. doi: 10.1126/science.3018931. [DOI] [PubMed] [Google Scholar]
- Wolfe K. H., Sharp P. M., Li W. H. Mutation rates differ among regions of the mammalian genome. Nature. 1989 Jan 19;337(6204):283–285. doi: 10.1038/337283a0. [DOI] [PubMed] [Google Scholar]
- Wu C. I., Li W. H. Evidence for higher rates of nucleotide substitution in rodents than in man. Proc Natl Acad Sci U S A. 1985 Mar;82(6):1741–1745. doi: 10.1073/pnas.82.6.1741. [DOI] [PMC free article] [PubMed] [Google Scholar]



