Abstract
Congenital pancreatic triglyceride lipase (PNLIP) deficiency is a rare disorder with uncertain genetic background as most cases were described before gene sequencing was readily available. Recently, two brothers with PNLIP deficiency were found to carry a homozygous missense mutation, c.662C > T (p.T221M) in the PNLIP gene (J. Lipid Res. 2014. 55:307–312). Molecular modeling suggested the substitution would change the orientation of residues in the catalytic site and disrupt the function of p.T221M PNLIP. To test the effect of the p.T221M mutation on PNLIP function, we expressed wild-type and p.T221M PNLIP in human embryonic kidney (HEK) 293A cells and dexamethasone-differentiated AR42J rat acinar cells. In both cellular models, wild-type PNLIP was secreted into the conditioned medium where it was readily detectable by protein staining, immunoblot or lipase activity assays. In contrast, mutant p.T221M was not secreted into the medium, but it was present in cell lysates where it accumulated in the insoluble fraction. Intracellular retention of mutant p.T221M resulted in endoplasmic reticulum (ER) stress as measured by elevated XBP1 splicing and increased levels of ER chaperones. Our results demonstrate that the presence of methionine at position 221 in the PNLIP protein sequence causes misfolding and aggregation of the p.T221M mutant inside the cell. The consequent loss of enzyme secretion adequately explains the clinical phenotype of PNLIP deficiency reported for homozygous carriers of p.T221M. Furthermore, the ability of mutant p.T221M to induce ER stress suggests that this form of PNLIP deficiency might cause acinar cell damage as well.
Keywords: Lipase, Fat digestion, Protein misfolding, Endoplasmic reticulum stress response
1. Introduction
Although congenital pancreatic triglyceride lipase (PNLIP) deficiency is discussed in most textbooks of pediatric gastroenterology, there are only a few reports of patients with evidence of this rare disorder [1–10]. In each case, the PNLIP deficiency was demonstrated by enzymatic assay of pancreatic secretions. In no case, was a mutation in the protein or gene reported. A recent effort to identify mutations in the PNLIP gene of four patients with absent lipase activity in their pancreatic fluid failed to find any nonsense or missense mutations that could explain the deficiency [11]. Thus, it has never been clear if the absence of PNLIP activity in the pancreatic secretion of the reported patients resulted from a gene mutation or another reason such as the presence of a lipase inhibitor or technical problems with the sample collection or with the lipase assay.
Recently, Behar et al. reported two brothers from a consanguineous marriage who had clinical PNLIP deficiency with steatorrhea and a novel homozygous missense mutation in the PNLIP gene [12]. The heterozygous carrier parents were clinically unaffected. A single base substitution in exon 6 (c.662C > T) changed the amino acid at position 221 (p.T221M). Thr221 is conserved in all known PNLIP sequences. It is located in the β9 loop, which contributes to the active site of PNLIP [13]. In the crystal structure of human PNLIP, Thr221 forms a hydrogen bond with Asp193, a residue in the Ser-His-Asp catalytic triad [14,15]. Molecular modeling suggests the substitution of Thr221 with the larger amino acid, methionine, destabilizes the active site of PNLIP [12]. The authors speculated that the brothers’ steatorrhea resulted from decreased activity of p.T221M PNLIP.
To test the hypothesis that the p.T221M mutation alters PNLIP function, we expressed Thr221 PNLIP and Met221 PNLIP in HEK 293A and AR42J cells. We then determined the effect of the p.T221M mutation on PNLIP expression, secretion and activity. Our results demonstrate that the presence of methionine at position 221 in the protein sequence causes misfolding of p.T221M PNLIP. The misfolded lipase accumulates in the cell, is inactive and is not secreted into the medium.
2. Materials and methods
2.1. Nomenclature
Nucleotide numbering reflects coding DNA numbering with +1 corresponding to the A of the ATG translation initiation codon in PNLIP. Amino acid residues are numbered starting with the initiator methionine of the primary translation product for human PNLIP.
2.2. Expression plasmids, mutagenesis and adenovirus
The cDNA encoding human PNLIP was amplified by PCR using our previously obtained full length human PNLIP cDNA clone as template and subsequently subcloned into the mammalian protein expression vector pcDNA3 (Invitrogen, Carlsbad, CA) [16]. The c.662C > T (p.T221M) mutation was introduced by overlap extension PCR. The sequence of all plasmid DNA constructs was verified by dideoxynucleotide sequencing. Recombinant adenovirus carrying a His-tagged form of wild-type PNLIP or mutant p.T221M was generated by Viraquest, Inc. (North Liberty, IA). Control adenovirus was purchased from Viraquest.
2.3. Culture and transfection of mammalian cells
HEK 293A cells were purchased from ATCC (Manassas, VA) and maintained in DMEM supplemented with 10% fetal bovine serum, 100 units/ml penicillin and 100 μg/ml streptomycin (#15140-122 GIBCO, Life Technologies, Grand Island, NY) at 37 °C in a 5% CO2-humidified incubator. Sixteen hours prior to transfection, cells (2 ml/ well for 6-well plates and 10 ml/dish for 10-cm culture dishes) were seeded at 75% confluence. Transfections were carried out using FuGENE 6 (Roche Diagnostics, Indianapolis, IN). Briefly, in 100 μl of Opti-MEM I reduced serum medium (Invitrogen), 5 μl of FuGENE 6 was mixed with 1.65 μg of plasmid DNA (pcDNA3, pcDNA3/PNLIP, or pcDNA3/ PNLIP p.T221M). The mixture was then added to the cells in each well of 6-well plates. For RNA extraction, a mixture of 25 μl of FuGENE 6 and 10 μg of each plasmid DNA in 500 μl of Opti-MEM I reduced serum medium was added for each 10-cm dish.
AR42J rat pancreatic acinar cells (ATCC #CRL-1492) were maintained in DMEM supplemented with 20% fetal bovine serum, 4 mM glutamine and 100 units/ml penicillin and 100 μg/ml streptomycin at 37°C. Prior to transfection, cells were plated in 6-well plates at a density of 106 cells per well and were grown in the presence of 100 nM dexamethasone for 48 h. Transduction with adenovirus were performed in 1 ml OptiMEM containing 100 nM dexamethasone using the indicated viral concentrations.
2.4. Collection of culture media and cells
Seventy-two hours after transfection, conditioned media from HEK 293A cells were withdrawn and subjected to 200 g × 5 min centrifugation to obtain cell free conditioned media. Cells were washed twice gently with ice-cold PBS and washed off the plates in 1.5 ml of PBS and centrifuged at 200 g × 5 min. For whole cell lysates, the resultant cell pellets were resuspended in 300 μl of 1 × Laemmli buffer, followed by 3 × 15 s sonication. Otherwise, cells were resuspended in 300 μl of RIPA buffer (150 mM NaCl, 1.0% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, and 50 mM Tris, pH 8.0) supplemented with protease and phosphatase inhibitors and incubated on ice for 10 min, followed by 3 × 5 s sonication. The cell lysates were clarified by centrifugation at 16,000 g × 20 min at 4 °C, and the supernatants were designated as soluble lysate fraction. The pellets were washed twice with ice-cold PBS and followed by 16,000 g × 5 min. The final pellets were resuspended in 100 μl of 1 × Laemmli buffer and sonicated until no particle was visible. This part was designated as insoluble lysate fraction.
Conditioned media from AR42J cells were harvested 24 h after transduction. Cells were washed twice with PBS, scraped from the tissue culture plates in 1 ml PBS and centrifuged for 10 min at 850 g. Cells were resuspended in 200 μl Reporter Lysis Buffer (Promega, Madison, WI) and 2 μl Halt Protease and Phosphatase Inhibitor Single-Use Cocktail (#78442, Pierce — Thermo Scientific, Rockford, IL) was added. Cells were incubated on ice for 15 min and the lysate was cleared by centrifugation for 10 min at 16,000 g. For all cell lysate samples, the protein concentration was measured with the Micro BCA Protein Assay Kit (Pierce — Thermo Scientific).
2.5. Measurement of PNLIP protein secretion from AR42J cells
Proteins in the conditioned media (40 μl from AR42J cells) were precipitated with 10% trichloroacetic acid (final concentration), resuspended in 20 μl Laemmli sample buffer containing 100 mM dithiothreitol, heat-denatured at 95 °C for 5 min and electrophoresed on 15% SDS-polyacrylamide gels. Gels were stained with Coomassie Blue.
2.6. Lipase activity assay
Lipase activity of human PNLIP was determined at 25 °C by using the standard 5-min pH-stat method with the emulsion of tributyrin (114 mM) in a total volume of 15 ml. The reactions were carried out in the presence or absence of over 5-fold molar excess of colipase at 4 mM sodium taurodeoxycholate (NaTDC) concentration. Fifty microliters of conditioned medium from HEK 293A cells or 100 μl of conditioned medium from AR42J cells were added to the reaction. For the assay of the soluble cell lysate from HEK 293A cells, 100 μl (total volume of 300 μl) was added to the reaction. Lipase activities are expressed as μmols of fatty acid released per min.
2.7. Protein immunoblot
Conditioned media and cell lysates from HEK 293A cells were treated with 2 × Laemmli sample buffer supplemented with 2-mercaptoethanol. After being heated to 95 °C for 5 min, the prepared conditioned medium (20 μl) and cell lysate samples (10–30 μg for whole cell lysate and soluble fraction and 5–10 μg for insoluble fraction) were resolved on 4–15% Mini-Protean TGX gels (Bio-Rad, Hercules, CA) and transferred onto an Immobilon-P membrane (Millipore, Bedford, MA). For AR42J cells, conditioned media (0.1 μl) and cell lysates (0.25 μg total protein for His-tag detection, 2.5 μg total protein for α-tubulin detection or 5 μg total protein for PNLIP detection) were treated as above and electrophoresed on 15% Tris-glycine minigels and transferred onto an Immobilon-P membrane. After blocking with 5% non-fat milk in PBS supplemented with 0.1% Tween 20, the membrane was incubated with primary antibody at 4° C overnight and then with secondary antibody for 1 h at room temperature. The bands were detected using the SuperSignal West Femto Maximum Sensitivity Substrate or the SuperSignal West Pico Chemiluminescent Substrate (Pierce — Thermo Scientific, Rockford, IL).
Antibodies used in this study were as following: rabbit polyclonal antibody against human PNLIP (1:5,000) was produced in our laboratory [17]; rabbit anti-BiP (3177; 1:1,000), GRP94 (2104; 1:1,000), calnexin (2679; 1:2,000), calreticulin (12238; 1:2,000), and rabbit anti-β tubulin (2128; 1:5,000) and GAPDH (5174; 1:5,000) were purchased from Cell Signaling (Danvers, MA); mouse monoclonal anti-α tubulin antibody (T5168; 1:2,000) was from Sigma-Aldrich (St. Louis, MO), HRP-conjugated goat polyclonal anti-mouse IgG (ab6798; 1:2,500) was from Abcam (Cambridge, MA) and horse-radish peroxidase (HRP)-conjugated mouse monoclonal penta-His antibody (34460; 1:2,000) was purchased from Qiagen (Louisville, KY). The goat anti-rabbit IgG conjugated with horseradish peroxidase (31460; 1:50,000) was purchased from Pierce —Thermo Scientific.
2.8. RT-PCR and real-time PCR analysis
Total RNA was isolated from HEK 293A cells transfected with given plasmid DNAs using TRIzol (Invitrogen), and 1.5 μg of RNA was then reverse-transcribed with Moloney murine leukemia virus reverse transcription kit from Ambion (Austin, TX) at 44 °C for 1 h in a 20-μl reaction volume. Semiquantitative measurements were performed by PCR of the X-box binding protein-1 (XBP1) cDNA and its spliced form, using the following primers that amplify both forms as follows: XBP1 forward primer, 5′-CCT TGT AGT TGA GAA CCA GG-3′, and XBP1 reverse primer, 5′-GGG CTT GGT ATA TAT GTG G-3′. The XBP1 primers amplify 441- and 415-bp amplicons from the unspliced and spliced cDNAs, respectively. As an endogenous control, a 261-bp fragment of GAPDH was also amplified, using the following primers: GAPDH sense primer, 5′-GTC CAC TGG CGT CTT CAC CA-3′, and GAPDH antisense primer, 5′-GTG GCA GTG ATG GCA TGG AC-3′. PCR products were run on 2% agarose gels, and the bands were visualized by ethidium bromide staining.
Total RNA was extracted from AR42J cell lysates using the RNeasy Plus Mini kit (Qiagen). RNA was reverse-transcribed using High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Grand Island, NY). XBP1 splicing was studied by PCR using a primer set forward: 5′-GCT TGT GAT TGA GAA CCA GG-3′, reverse: 5′-AGG CTT GGT GTA TAC ATG G-3′ that flanked the spliced region and amplified both spliced (421 bp) and unspliced (447 bp) forms. The PCR products were resolved on 2% agarose gels and stained with ethidium bromide. Expression of mRNA for BiP and calreticulin was measured by real time PCR (7500 Real Time PCR System, Applied Biosystems) using TaqMan primers with TaqMan Universal PCR Mastermix (Applied Biosystems). Gene expression was quantitated using the comparative CT method (ΔΔCT method). Threshold cycle (CT) values were determined with the 7500 System Sequence Detection Software 1.3. Expression levels of target genes were first normalized to the GAPDH internal control gene (ΔCT) and then to expression levels measured in cells infected with 2 × 107 pfu/ml control (vector only) adenovirus (ΔΔCT). Results were expressed as fold changes calculated with the formula 2−ΔΔCT.
3. Results
3.1. Secretion and folding of p.T221M PNLIP
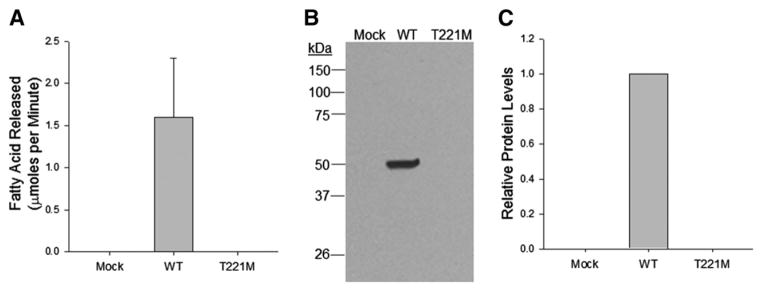
To determine if the p.T221M substitution affects the folding or function of PNLIP, we first transfected HEK 293A cells with empty vector, vector containing PNLIP or vector containing p.T221M PNLIP. These cells are relatively easy to culture and can be reproducibly transfected with plasmids at high efficiency. Additionally, they have been used to study secretion of a large variety of proteins and represent a good model to study protein secretion. When we assayed conditioned medium from the transfected cells for lipase activity, only the cells transfected with PNLIP had detectable activity (Fig. 1A). The medium from cells transfected p.T221M PNLIP had no lipase activity. We next determined if inactive p.T221M PNLIP was present in the conditioned medium by immunoblot analysis with our rabbit anti-human PNLIP antibody (Fig. 1B and C). Whereas PNLIP was detectable in conditioned medium, p.T221M PNLIP was undetectable suggesting the mutant lipase was either poorly expressed in HEK 293T cells or not secreted by these cells.
Fig. 1.
Secretion of PNLIP and p.T221M PNLIP from transfected HEK 293A cells. The media from transfected cells was sampled 72 h after transfection. (A) The lipase activity against tributyrin in 50 μl of medium was measured in the standard 5 minute pH-stat assay. The mean and SD of 3 separate determinations is plotted. (B) The presence of PNLIP was determined by SDS PAGE and protein immunoblot with the rabbit anti-human PNLIP antisera followed by detection with the HRP conjugated goat anti-rabbit IgG antisera. (C) The relative amounts of lipase in the samples were determined by densitometry of 3 separate experiments.
To help distinguish between these possibilities, we next determined if p.T221M PNLIP was present inside the transfected cells. Protein immunoblot analysis of the whole cell lysate revealed the presence of both PNLIP and p.T221M PNLIP in the cells (Fig. 2A). The amount of the mutant lipase was significantly higher (P ≤ 0.001). We then separated the intracellular lipase into a soluble and detergent-insoluble fraction by detergent extraction and centrifugation. Both lipases were present in the soluble fraction (Fig. 2B). The amount of soluble p.T221M PNLIP was significantly higher than the amount of PNLIP (P ≤ 0.001). Lipase assay 100 μl of the PNLIP soluble intracellular fraction using tributyrin as substrate revealed an activity of 1.8 ± 0.097 μmol fatty acid released per min and no detectable activity in extracts from mock transfected or p.T221M PNLIP (n = 3) suggesting the soluble p.T221M was in an inactive conformation. Importantly, the amount of p.T221M PNLIP in the insoluble fraction was about 7-fold higher than the amount of PNLIP (P ≤ 0.001) (Fig. 2C). In both the soluble fraction and the insoluble fractions we detected higher molecular weight antibody positive proteins migrating above 100 kDa in the p.T221M samples. The bands were not present in the PNLIP samples suggesting they did not result from non-specific binding of the antibodies. The nature of these bands was not further investigated.
Fig. 2.
The intracellular accumulation of PNLIP and p.T221M PNLIP in transfected HEK 293A cells. Seventy-two hours after transfection the cells were harvested and processed for total, soluble and insoluble fractions as described in Materials and methods. The presence of PNLIP was determined by SDS PAGE and protein immunoblot with the rabbit anti-human PNLIP antisera followed by detection with the HRP conjugated goat anti-rabbit IgG antisera. The relative amounts of lipase in the samples were determined by densitometry. The mean and SD of 3 separate determinations are plotted. (A) The total amount of PNLIP and p.T221M PNLIP are shown. GAPDH serves as the loading control. (B) The amount of each lipase in the soluble intracellular fraction is shown. (C) The amount of each lipase in the insoluble intracellular fraction is shown. ***P ≤ 0.001.
We repeated these experiments in a rat acinar cell line, AR42J, transduced with recombinant adenovirus. The amount of PNLIP in the medium of transduced AR42J cells increased with increasing amounts of adenovirus as determined by lipase assay, by SDS-PAGE and protein staining and by protein immunoblot (Fig. 3A). As found in the HEK 293A cells, lipase activity was present in the conditioned medium from cells transduced with PNLIP adenovirus but not in cells transduced with p.T221M PNLIP adenovirus. Similarly, PNLIP protein was only detected in the conditioned medium from cells transduced with PNLIP adenovirus by protein staining or immunoblot. p.T221M PNLIP was not detected in conditioned medium by either protein staining or immunoblot.
Fig. 3.
Expression of PNLIP and p.T221M PNLIP in adenovirus transduced AR42J cells. The cells were transduced with three different titers of adenovirus. (A) Aliquots of conditioned medium were assayed by SDS PAGE followed by Coomassie Blue staining (upper panel) or by protein immunoblot with the HRP-conjugated mouse monoclonal penta-His antibody (middle panel) or were assayed for lipase activity against tributyrin in 100 μl of medium in the standard 5 minute pH-stat assay (bottom panel). The mean and SD of 3 separate determinations are plotted. (B) Aliquots of total cell lysate were analyzed by SDS PAGE and protein immunoblot with the HRP-conjugated mouse monoclonal penta-His antibody. α-Tubulin was used as a loading control. (C) AR42J cells were transfected with 108 pfu adenovirus and the total, soluble and insoluble fractions were analyzed by protein immunoblot. ***P ≤ 0.001.
Examination of the AR42J cellular fraction revealed the presence of p.T221M PNLIP in the cells as seen in the HEK 293A cells (Fig. 3B). The amount of PNLIP and p.T221M PNLIP in the cells varied proportionately with increasing viral titers. The amount of intracellular lipase was higher in the cells transduced with p.T221M PNLIP. The intracellular recombinant lipases were separated into a soluble and an insoluble fraction by centrifugation (Fig. 3C). As observed in the HEK 293A cells, both PNLIP and p.T221M PNLIP were present in the soluble fraction or supernatant. Only a minor portion of PNLIP appeared in the insoluble fraction. In contrast, a significant fraction of the intracellular p.T221M PNLIP was in the insoluble fraction.
3.2. Activation of the ER stress response
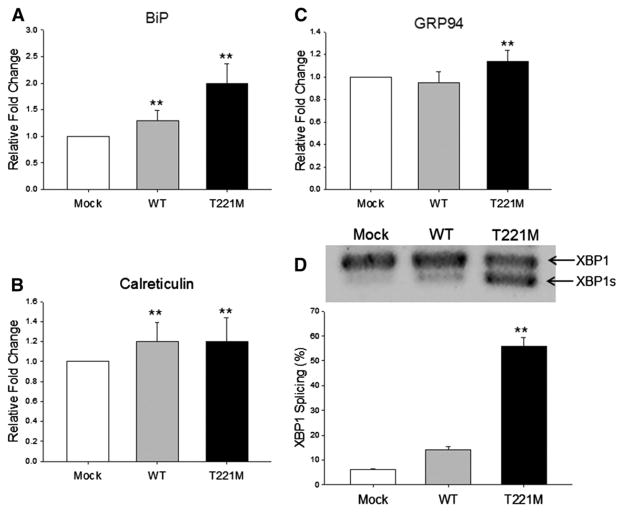
The accumulation of intracellular proteins in the detergent-insoluble fraction suggests the protein has misfolded and formed insoluble aggregates. If this is the case, an ER stress response will be activated. To determine if p.T221M PNLIP activates the UPR and ER stress response, we measured the protein levels of several major ER chaperones, BiP, calreticulin and GRP94 and the splicing of the mRNA encoding XBP1 in transfected HEK 293A cells. Overexpression of PNLIP and p.T221M PNLIP resulted in upregulation of BiP protein levels compared to the vector control (P ≤ 0.01) (Fig. 4A). Furthermore, levels of BiP in the cells transfected with p.T221M PNLIP were significantly higher than the BiP levels in cells transfected with PNLIP (P ≤ 0.01). The levels of calreticulin were elevated above controls in HEK 293A cells expressing PNLIP or p.T221M PNLIP (P ≤ 0.01) but the levels were not different between cells expressing the two recombinant lipases (Fig. 4B). Levels of GRP94 were significantly increased in cells transfected with p.T221M PNLIP compared to the levels in control and PNLIP transfected cells (P ≤ 0.01) (Fig. 4C). The GRP94 levels in cells expressing PNLIP did not differ from the GRP94 levels in control cells.
Fig. 4.
Effect of expression of p.T221M PNLIP in HEK 293A cells on ER stress. Cellular lysates of transfected HEK 293A cells were prepared as described in Materials and methods. At least three measurements from separate experiments were done in each case. (A) The relative amounts of BiP determined by protein immunoblot and quantitated by densitometry. (B) The relative amounts of calreticulin determined by protein immunoblot and quantitated by densitometry. (C) The relative amounts of GRP94 determined by protein immunoblot and quantitated by densitometry. (D) Total RNA was isolated from transfected HEK 293A cells. The amount of XBP1 mRNA splicing was measured by RT-PCR, separation by agarose gel electrophoresis, ethidium bromide staining and densitometry. The upper panel is a representative agarose gel of the RT-PCR products. The bottom panel is the amount of XBP1 mRNA splicing quantitated by densitometry. **P ≤ 0.01.
Because BiP protein levels were elevated in cells transfected with p.T221M PNLIP, we next determined if there was a downstream effect in the UPR. During ER stress, IRE1 is activated, which catalyzes the cytoplasmic splicing of XBP1 mRNA to generate a shorter XBP1 mRNA variant. This variant encodes a transcriptional activator critical for the UPR. We performed RT-PCR analysis of XBP1 mRNA in mock, PNLIP and p.T221M PNLIP transfected HEK 293A cells (Fig. 4D). Splicing of XBP1 mRNA was increased about 10-fold in cells transfected with p.T221M PNLIP compared to the control cells (P ≤ 0.01). XBP1 mRNA splicing was not increased in cells transfected with PNLIP.
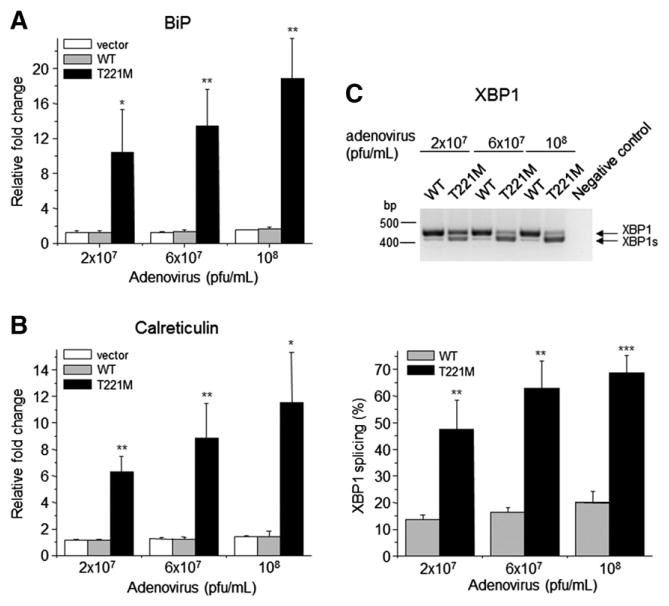
We next assessed the levels of BiP and calreticulin mRNA and splicing of XBP1 mRNA in AR42J cells transduced with adenovirus constructs at various titers. In these cells there was a robust upregulation of BiP in cells transduced with p.T221M PNLIP adenovirus compared to cells transduced with control or PNLIP adenovirus (Fig. 5A). The increase in BiP mRNA was about 18-fold above the levels in control cells at the highest viral titer tested (P ≤ 0.01). In contrast to the findings in HEK 293A cells, there was an 11-fold upregulation of calreticulin mRNA in the cells transduced with the highest titers of p.T221M PNLIP adenovirus (P ≤ 0.05) (Fig. 5B). Like the HEK 293A cells, the expression of p.T221M PNLIP in AR42J cells caused an increase in XBP1 mRNA splicing compared to the levels in cells expressing PNLIP (P ≤ 0.001) (Fig. 5C).
Fig. 5.
Effect of expression of p.T221M PNLIP in AR42J cells on ER stress. The cells were transduced with three different titers of each adenovirus construct. Total RNA was isolated from the cells as described. The amount of mRNA encoding BiP and calreticulin was determined by real-time RT-PCR and the amount of XBP1 mRNA splicing determined by RT-PCR, separation by agarose gel electrophoresis, ethidium bromide staining and densitometry. (A) Mean and SD of the relative BiP levels. (B) Mean and SD of the relative calreticulin levels. (C) Amount of XBP1 mRNA splicing is given. The upper panel shows a representative agarose gel separation of the RT-PCR products. The bottom panel presents the mean and SD of 3 experiments. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001.
4. Discussion
4.1. Misfolding of p.T221M PNLIP
In this study, we determined the effect of the p.T221M PNLIP substitution mutation on the function of PNLIP. The mutation was recently identified in subjects with clinical PNLIP deficiency. On the basis of structural modeling it was suggested that p.T221M should impair enzymatic activity of PNLIP by destabilizing the region around the active site [12]. In addition, Thr221 flanks the β9-loop, which contributes to an acyl chain binding site [13,18]. Disruption of either structure could result in a decrease or loss of activity and would explain the clinical phenotype of PNLIP deficiency. Our results demonstrated absent lipase activity and a marked secretion defect of p.T221M PNLIP expressed in two cell culture models. The secretion defect is a result of misfolding and pathological accumulation of p.T221M PNLIP inside the cells. The presence of p.T221M in an insoluble intracellular fraction suggests the misfolded mutant protein forms aggregates, most likely in the ER. This result could not be accurately predicted. The reported analysis of protein stability using the Eris server suggests the p.T221M change decreases stability of the protein, but does not address effects on folding [12]. Folding RaCe (http://www.iitm.ac.in/bioinfo/proteinfolding/Heading.html) predicted that the substitution of Met for Thr221 resulted in a logarithmic increase in the folding rate of 0.89/s, but this finding also does not address the fidelity of folding [19]. Still, Thr221 is conserved in all known PNLIP and PNLIPRP2 sequences and does not participate in the active site directly or in acyl chain binding suggesting it is critical for maintaining structure.
By extrapolation, it is unlikely that pancreatic acinar cells secrete p.T221M PNLIP to any significant extent. A secretory defect would explain the steatorrhea reported in the two brothers carrying p.T221M PNLIP in the homozygous state [12]. Unfortunately, lipase activity levels in the pancreatic secretions of the brothers were not reported. The report documented low serum lipase levels in the two brothers and this finding in concert with steatorrhea was interpreted as evidence for a functional defect in p.T221M PNLIP activity. The interpretation of the low serum lipase levels is complicated. Although the method for measuring lipase activity is not reported, they were likely done in a clinical chemistry laboratory. The commercially available systems for measuring serum lipase activity are not specific for PNLIP and the low activity might be from other lipases such as carboxyl ester lipase or pancreatic lipase related protein 2 (PNLIPRP2) rather than from low levels of p.T221M PNLIP or normal protein levels of p.T221M PNLIP with low specific activity.
4.2. Comparison of PNLIP deficiency in humans and mice
The clinical course of the two brothers resembles that reported for other patients with probable PNLIP deficiency [1–10]. The major clinical symptom was greasy, voluminous stools. Poor growth was not present in the brothers or in the previously described patients. In fact, the brothers’ compliance with pancreatic enzyme replacement therapy suffered because they were willing to cope with large, greasy stools after meals in exchange for an “inability to gain weight despite very high caloric intake [12].” That is, the brothers were resistant to developing obesity. The inability to gain weight differentiated the two brothers from their unaffected family members.
Preserved weight gain and resistance to obesity is mimicked in PNLIP-deficient mice [20,21]. Early in life, PNLIP-deficient mice gain weight normally [21]. But, when placed on a high-fat, high-cholesterol diet and followed over a longer time, the PNLIP-deficient mice are resistant to the obesity that develops in wild-type mice [12]. The PNLIP-deficient mice weigh less and have less fat body mass than the wild-type mice fed the same diet.
The presence of significant steatorrhea in the humans with PNLIP deficiency differs from the phenotype of PNLIP-deficient mice. The mice have minimal to no steatorrhea depending on the method used to measure fat absorption [20,21]. The explanation for this difference is unclear and is further confounded by results in colipase-deficient mice. Adult colipase-deficient mice have significant steatorrhea on a high-fat, high cholesterol diet [22]. Since mouse PNLIP is dependent on colipase for activity in the presence of micellar concentrations of bile salts and PNLIP is the predominant colipase-dependent lipase secreted by the pancreas, the effect of colipase deficiency was attributed to deficient PNLIP activity [23–25].
4.3. The role of PNLIPRP2 and CEL in compensating for PNLIP deficiency in humans and mice
The combination of steatorrhea in colipase-deficient mice but not in PNLIP-deficient mice suggests other lipases contribute to fat digestion in adult mice. To this point, mice deficient in both PNLIP and carboxyl ester lipase had steatorrhea although they still absorbed about 60% of the fat in the high-fat, high-cholesterol diet [20]. The authors suggested that PNLIPRP2 might account for some of the remaining fat digestion. PNLIPRP2 activity is stimulated by colipase and it accounts for all of the colipase-dependent lipase activity in the pancreas of a newborn mouse, an age when PNLIP is not expressed [23,26]. It is plausible that carboxyl ester lipase and PNLIPRP2 compensate for the loss of PNLIP in the PNLIP-deficient mice.
In the case of these brothers, it appears that PNLIPRP2 and CEL cannot compensate for the loss of PNLIP and the result is steatorrhea. The reason for this may lay in the marked differences in enzyme kinetics between the mouse and human PNLIPRP2 lipases [23,27]. In particular the specific activity of mouse PNLIPRP2 against long-chain triglycerides, the major dietary fat, is about 7-fold greater than the activity of human PNLIPRP2 even under optimal conditions [23,28]. In fact, the specific activity of mouse PNLIPRP2 against long chain triglycerides is only slightly lower than the specific activity of mouse PNLIP. Furthermore, mouse PNLIPRP2 has 10-fold higher specific activity against short and medium chain triglycerides than human PNLIPRP2. The higher specific activity may allow mouse PNLIPRP2 to adequately compensate for the absence of PNLIP in PNLIP-deficient mice.
4.4. ER stress response
The second important finding of our study is the up-regulation of the ER stress response in cells expressing p.T221M PNLIP. A robust ER stress response can activate cell-death pathways, which could potentially damage the pancreas. This mechanism has been suggested for other misfolding variants of PRSS1, CTRC and CPA1 [29–31]. Each of these variants increases ER stress in transfected cells and are risk factors for chronic pancreatitis in humans. The accumulation of misfolded p.T221M PNLIP could also increase the risk for chronic pancreatitis. In the report of the two brothers, there are suggestions that they had pancreatic insufficiency and not just PNLIP deficiency. In addition to steatorrhea, low serum carnitine and low serum vitamin E levels, both brothers had low fecal elastase levels and the proband had an abnormal pancreolauryl test. Neither brother had evidence for an intestinal mucosal defect that might impair fatty acid absorption. No radiographic imaging of the pancreas was reported so it is not known if the brothers’ pancreas had anatomic abnormalities consistent with chronic pancreatitis. Even so, the available data suggests that the p.T221M PNLIP may have broader affects not limited to fat digestion. Although the p.T221M PNLIP mutation has only been reported in a single consanguineous family, the possibility that other mutations in PNLIP increase the risk for pancreatic disease should be considered.
Acknowledgments
This work was supported by National Institutes of Health Grants R01DK097241 and R01DK080820 to M.E.L. and R01DK082412, R01DK058088, and R01DK095753 to M.S-T.
Abbreviations
- BiP
immunoglobulin binding protein
- ER
endoplasmic reticulum
- GRP94
glucose-regulated protein-94
- IRE1
inositol-requiring enzyme 1
- PNLIP
pancreatic triglyceride lipase
- PNLIPRP2
pancreatic lipase related protein 2
- XBP1
X-box binding protein-1
Footnotes
Transparency document
The Transparency document associated with this article can be found, in the online version.
References
- 1.Figarella C, De Caro A, Leupold D, Poley JR. Congenital pancreatic lipase deficiency. J Pediatr. 1980;96:412–416. doi: 10.1016/s0022-3476(80)80683-4. [DOI] [PubMed] [Google Scholar]
- 2.Figarella C, Negri GA, Sarles H. Presence of colipase in a congenital pancreatic lipase deficiency. Biochim Biophys Acta. 1972;280:205–211. doi: 10.1016/0005-2760(72)90227-5. [DOI] [PubMed] [Google Scholar]
- 3.Ligumsky M, Granot E, Branski D, Stankiewicz H, Goldstein R. Isolated lipase and colipase deficiency in two brothers. Gut. 1990;31:1416–1418. doi: 10.1136/gut.31.12.1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Larbre F, Hartemann E, Cotton JB, Mathieu M, Charrat A, Moreau P. Chronic diarrhea due to pancreatic lipase deficiency. Pediatrie. 1969;24:807–813. [PubMed] [Google Scholar]
- 5.Ghishan FK, Moran JR, Durie PR, Greene HL. Isolated congenital lipase–colipase deficiency. Gastroenterology. 1984;86:1580–1582. [PubMed] [Google Scholar]
- 6.Sheldon W. Congenital pancreatic lipase deficiency. Arch Dis Child. 1964;39:268–271. doi: 10.1136/adc.39.205.268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rey J, Frezal J, Royer P, Lamy M. Congenital absence of pancreatic lipase. Arch Fr Pediatr. 1966;23:5–14. [PubMed] [Google Scholar]
- 8.Verger P, Babin R, Guillard JM, Babin JP, Cixous P, Laigle JL. Chronic steatorrhea in children due to a congenital insufficiency of pancreatic lipase. Arch Fr Pediatr. 1971;28:992. [PubMed] [Google Scholar]
- 9.Muller DP, McCollum JP, Trompeter RS, Harries JT. Proceedings: studies on the mechanism of fat absorption in congenital isolated lipase deficiency. Gut. 1975;16:838. [PubMed] [Google Scholar]
- 10.Balzer E. Angeborener Mangel an Pankreaslipase. Z Gastroenterol. 1967;5:239–246. [Google Scholar]
- 11.Hegele RA, Ramdath DD, Ban MR, Carruthers MN, Carrington CV, Cao H. Polymorphisms in PNLIP, encoding pancreatic lipase, and associations with metabolic traits. J Hum Genet. 2001;46:320–324. doi: 10.1007/s100380170066. [DOI] [PubMed] [Google Scholar]
- 12.Behar DM, Basel-Vanagaite L, Glaser F, Kaplan M, Tzur S, Magal N, Eidlitz-Markus T, Haimi-Cohen Y, Sarig G, Bormans C, Shohat M, Zeharia A. Identification of a novel mutation in the PNLIP gene in two brothers with congenital pancreatic lipase deficiency. J Lipid Res. 2014;55:307–312. doi: 10.1194/jlr.P041103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carriere F, Withers-Martinez C, van Tilbeurgh H, Roussel A, Cambillau C, Verger R. Structural basis for the substrate selectivity of pancreatic lipases and some related proteins. Biochim Biophys Acta. 1998;1376:417–432. doi: 10.1016/s0304-4157(98)00016-1. [DOI] [PubMed] [Google Scholar]
- 14.van Tilbeurgh H, Gargouri Y, Dezan C, Egloff MP, Nesa MP, Ruganie N, Sarda L, Verger R, Cambillau C. Crystallization of pancreatic procolipase and of its complex with pancreatic lipase. J Mol Biol. 1993;229:552–554. doi: 10.1006/jmbi.1993.1054. [DOI] [PubMed] [Google Scholar]
- 15.Winkler FK, D’Arcy A, Hunziker W. Structure of human pancreatic lipase. Nature. 1990;343:771–774. doi: 10.1038/343771a0. [DOI] [PubMed] [Google Scholar]
- 16.Jennens ML, Lowe ME. The C-terminal domain of human pancreatic lipase is required for stability and maximal activity but not colipase reactivation. J Lipid Res. 1995;36:1029–1036. [PubMed] [Google Scholar]
- 17.Yang Y, Lowe ME. Human pancreatic triglyceride lipase expressed in yeast cells: purification and characterization. Protein Expr Purif. 1998;13:36–40. doi: 10.1006/prep.1998.0874. [DOI] [PubMed] [Google Scholar]
- 18.Dridi K, Amara S, Bezzine S, Rodriguez JA, Carriere F, Gaussier H. Partial deletion of beta9 loop in pancreatic lipase-related protein 2 reduces enzyme activity with a larger effect on long acyl chain substrates. Biochim Biophys Acta. 2013;1831:1293–1301. doi: 10.1016/j.bbalip.2013.04.010. [DOI] [PubMed] [Google Scholar]
- 19.Chaudhary P, Naganathan AN, Gromiha MM. Folding RaCe: a robust method for predicting changes in protein folding rates upon point mutations. Bioinformatics. 2015 doi: 10.1093/bioinformatics/btv091. ePub Feb 16. [DOI] [PubMed] [Google Scholar]
- 20.Gilham D, Labonte ED, Rojas JC, Jandacek RJ, Howles PN, Hui DY. Carboxyl ester lipase deficiency exacerbates dietary lipid absorption abnormalities and resistance to diet-induced obesity in pancreatic triglyceride lipase knockout mice. J Biol Chem. 2007;282:24642–24649. doi: 10.1074/jbc.M702530200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huggins KW, Camarota LM, Howles PN, Hui DY. Pancreatic triglyceride lipase deficiency minimally affects dietary fat absorption but dramatically decreases dietary cholesterol absorption in mice. J Biol Chem. 2003;278:42899–42905. doi: 10.1074/jbc.M303422200. [DOI] [PubMed] [Google Scholar]
- 22.D’Agostino D, Cordle RA, Kullman J, Erlanson-Albertsson C, Muglia LJ, Lowe ME. Decreased postnatal survival and altered body weight regulation in procolipase deficient mice. J Biol Chem. 2002;277:7170–7177. doi: 10.1074/jbc.M108328200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xiao X, Ross LE, Miller RA, Lowe ME. Kinetic properties of mouse pancreatic lipase-related protein-2 suggest the mouse may not model human fat digestion. J Lipid Res. 2011;52:982–990. doi: 10.1194/jlr.M014290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li X, Lindquist S, Lowe M, Noppa L, Hernell O. Bile salt-stimulated lipase and pancreatic lipase-related protein 2 are the dominating lipases in neonatal fat digestion in mice and rats. Pediatr Res. 2007;62:537–541. doi: 10.1203/PDR.0b013e3181559e75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lowe ME, Kaplan MH, Jackson-Grusby L, D’Agostino D, Grusby MJ. Decreased neonatal dietary fat absorption and T cell cytotoxicity in pancreatic lipase-related protein 2-deficient mice. J Biol Chem. 1998;273:31215–31221. doi: 10.1074/jbc.273.47.31215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.D’Agostino D, Lowe ME. Pancreatic lipase-related protein 2 is the major colipase-dependent pancreatic lipase in suckling mice. J Nutr. 2004;134:132–134. doi: 10.1093/jn/134.1.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Amara S, Lafont D, Fiorentino B, Boullanger P, Carriere F, De Caro A. Continuous measurement of galactolipid hydrolysis by pancreatic lipolytic enzymes using the pH-stat technique and a medium chain monogalactosyl diglyceride as substrate. Biochim Biophys Acta. 2009;1791:983–990. doi: 10.1016/j.bbalip.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 28.Xiao X, Mukherjee A, Ross LE, Lowe ME. Pancreatic lipase-related protein-2 (PLRP2) can contribute to dietary fat digestion in human newborns. J Biol Chem. 2011;286:26353–26363. doi: 10.1074/jbc.M111.249813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kereszturi E, Szmola R, Kukor Z, Simon P, Weiss FU, Lerch MM, Sahin-Toth M. Hereditary pancreatitis caused by mutation-induced misfolding of human cationic trypsinogen: a novel disease mechanism. Hum Mutat. 2009;30:575–582. doi: 10.1002/humu.20853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Szmola R, Sahin-Toth M. Pancreatitis-associated chymotrypsinogen C (CTRC) mutant elicits endoplasmic reticulum stress in pancreatic acinar cells. Gut. 2010;59:365–372. doi: 10.1136/gut.2009.198903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Witt H, Beer S, Rosendahl J, Chen JM, Chandak GR, Masamune A, Bence M, Szmola R, Oracz G, Macek M, Jr, Bhatia E, Steigenberger S, Lasher D, Buhler F, Delaporte C, Tebbing J, Ludwig M, Pilsak C, Saum K, Bugert P, Masson E, Paliwal S, Bhaskar S, Sobczynska-Tomaszewska A, Bak D, Balascak I, Choudhuri G, Reddy DN, Rao GV, Thomas V, Kume K, Nakano E, Kakuta Y, Shimosegawa T, Durko L, Szabo A, Schnur A, Hegyi P, Rakonczay Z, Jr, Pfutzer R, Schneider A, Groneberg DA, Braun M, Schmidt H, Witt U, Friess H, Algul H, Landt O, Schuelke M, Kruger R, Wiedenmann B, Schmidt F, Zimmer KP, Kovacs P, Stumvoll M, Bluher M, Muller T, Janecke A, Teich N, Grutzmann R, Schulz HU, Mossner J, Keim V, Lohr M, Ferec C, Sahin-Toth M. Variants in CPA1 are strongly associated with early onset chronic pancreatitis. Nat Genet. 2013;45:1216–1220. doi: 10.1038/ng.2730. [DOI] [PMC free article] [PubMed] [Google Scholar]