ABSTRACT
Smallpox was declared eradicated in 1980 after an intensive vaccination program using different strains of vaccinia virus (VACV; Poxviridae). VACV strain IOC (VACV-IOC) was the seed strain of the smallpox vaccine manufactured by the major vaccine producer in Brazil during the smallpox eradication program. However, little is known about the biological and immunological features as well as the phylogenetic relationships of this first-generation vaccine. In this work, we present a comprehensive characterization of two clones of VACV-IOC. Both clones had low virulence in infected mice and induced a protective immune response against a lethal infection comparable to the response of the licensed vaccine ACAM2000 and the parental strain VACV-IOC. Full-genome sequencing revealed the presence of several fragmented virulence genes that probably are nonfunctional, e.g., F1L, B13R, C10L, K3L, and C3L. Most notably, phylogenetic inference supported by the structural analysis of the genome ends provides evidence of a novel, independent cluster in VACV phylogeny formed by VACV-IOC, the Brazilian field strains Cantagalo (CTGV) and Serro 2 viruses, and horsepox virus, a VACV-like virus supposedly related to an ancestor of the VACV lineage. Our data strongly support the hypothesis that CTGV-like viruses represent feral VACV that evolved in parallel with VACV-IOC after splitting from a most recent common ancestor, probably an ancient smallpox vaccine strain related to horsepox virus. Our data, together with an interesting historical investigation, revisit the origins of VACV and propose new evolutionary relationships between ancient and extant VACV strains, mainly horsepox virus, VACV-IOC/CTGV-like viruses, and Dryvax strain.
IMPORTANCE First-generation vaccines used to eradicate smallpox had rates of adverse effects that are not acceptable by current health care standards. Moreover, these vaccines are genetically heterogeneous and consist of a pool of quasispecies of VACV. Therefore, the search for new-generation smallpox vaccines that combine low pathogenicity, immune protection, and genetic homogeneity is extremely important. In addition, the phylogenetic relationships and origins of VACV strains are quite nebulous. We show the characterization of two clones of VACV-IOC, a unique smallpox vaccine strain that contributed to smallpox eradication in Brazil. The immunogenicity and reduced virulence make the IOC clones good options for alternative second-generation smallpox vaccines. More importantly, this study reveals the phylogenetic relationship between VACV-IOC, feral VACV established in nature, and the ancestor-like horsepox virus. Our data expand the discussion on the origins and evolutionary connections of VACV lineages.
INTRODUCTION
Smallpox, caused by variola virus (genus Orthopoxvirus, family Poxviridae), accounted for millions of deaths throughout human history. The disease was declared eradicated in 1980 after a worldwide program implemented by the WHO involving strict surveillance and intensive vaccination using vaccinia virus (VACV; Poxviridae). After disease eradication, smallpox vaccination was discontinued on all continents (1, 2), and variola virus stocks were transferred to two maximum security laboratories at the CDC, Atlanta, GA, and Vector, Koltsovo, Novosibirsk region, Russia (3).
Since the 1990s, and mainly after the terrorist attacks on the United States in 2001, there has been a general concern that smallpox could reemerge as a biological weapon (3). Therefore, different countries and the WHO have increased their stockpile of smallpox vaccine over the years (4, 5). However, the first-generation vaccines used to eradicate smallpox had rates of adverse effects that are not acceptable by current health care standards (6, 7). Therefore, the search for new-generation vaccines that combine low pathogenicity and immune protection is important.
Recent studies using different sequencing technologies demonstrate that first-generation vaccines are genetically heterogeneous and consist of a pool of quasispecies related to the uncontrolled passage in calf/sheep skin or embryonated chicken eggs for several decades. Viral clones isolated from these vaccines may differ in virulence, efficacy of immune protection, and genetic content (8–12). In fact, the second-generation smallpox vaccine currently produced in the United States, ACAM2000, is a clone of the first-generation vaccine used in the country, Dryvax. ACAM2000 induces immune responses and protection similar to that of the parental strain and was selected in parallel to the neurovirulent clone 3 (CL-3), also isolated from the Dryvax vaccine (4, 9). Work involving additional clonal selection further confirmed the genotypic diversity of Dryvax (11, 13).
In addition to the search for safer vaccines, recent genetic studies of clonal variants of first-generation vaccines have improved our understanding of the phylogenetic relationships and obscure origins of different VACV strains (8, 11, 12). Nevertheless, numerous questions remain unanswered.
The major smallpox vaccine producer in Brazil, among three others, was Instituto Oswaldo Cruz in Rio de Janeiro (1, 14). VACV strain IOC (VACV-IOC) was exclusively used by the Institute to manufacture the vaccine that was widely distributed in Brazil during the eradication campaign (1). The vaccine was produced in calf skin or chicken eggs (14) and usually had take rates of >98% (1) and low rates of adverse effects (H. Schatzmayr, personal communication). Unfortunately, poor documentation resulted in scant existing information about the origins of VACV-IOC. However, it is suggested that the VACV-IOC used in the 1970s originated from the Beaugency strain (J. A. Espmark to D. A. Henderson, correspondence on March 31, 1969, file 88-001-10, Sanofi Pasteur Archives [Connaught Campus]) that probably constituted the first animal-based vaccine samples imported to Brazil in 1887 from Chambon Institute, France (15, 16).
Damaso et al. (17) named the vaccine VACV-IOC, after the Instituto Oswaldo Cruz, during comparative studies with Cantagalo virus (CTGV). This virus is a VACV strain that infects dairy cattle and milkers in Brazil, and it constitutes a rare example of the occurrence of VACV in nature. Studies have suggested that VACV-IOC is phylogenetically related to CTGV and to other field variants related to CTGV (CTGV-like viruses), which were isolated in subsequent outbreaks after 1999 (18–21). Nevertheless, the analyses were based on a few sequenced genes and deletion patterns (17, 22). Hence, full-genome sequencing would be necessary to place VACV-IOC correctly in relation to other VACV strains, particularly Cantagalo virus.
In addition to the lack of genetic information regarding VACV-IOC, little is known about the biological and immunological features of this vaccine strain. Taking into account the genetic diversity of first-generation vaccines, we isolated two viral clones at random to characterize genetically homogenous populations of VACV-IOC. In this work, we show that the selected clones combined low virulence to mice with effective immune protection from lethal challenge and shared a common fragmentation pattern of several virulence genes. More importantly, based on the full-genome sequencing, we provide evidence that VACV-IOC branches as a novel independent cluster in VACV phylogeny together with the Brazilian field strains CTGV and Serro 2 virus as well as horsepox virus (HSPV). Our data strongly support the hypothesis that CTGV represents a feral VACV that evolved in parallel with the VACV-IOC used in the 1970s after splitting from a most recent common ancestor, probably an ancient smallpox vaccine sample related to horsepox virus. This work brings new insights into the origins and evolutionary relationships of VACV lineages.
MATERIALS AND METHODS
Cells and viruses.
BSC-40 and HEp-2 cells were grown at 37°C in Dulbecco modified Eagle's medium (DMEM; Invitrogen, Carlsbad, CA) supplemented with 7% heat-inactivated fetal bovine serum (FBS; Invitrogen), 500 U/ml penicillin, 100 μg/ml streptomycin, and 2.5 μg/ml amphotericin B (Fungizone).
Ampoules of freeze-dried smallpox vaccine (calf lymph material) containing VACV strain IOC (VACV-IOC) were manufactured by the Instituto Oswaldo Cruz, Rio de Janeiro, Brazil, and were kindly provided to our laboratory by Herman Schatzmayr (Instituto Oswaldo Cruz) in 1981. An ampoule was reconstituted in phosphate-buffered saline (PBS), passaged once in BSC-40 cells, and stored at −80°C (17). Serial dilutions of VACV-IOC crude stock were inoculated onto BSC-40 cells incubated under semisolid medium. Isolated viral plaques were selected and two viral clones (B141 and B388) were obtained after two additional rounds of plaque purification (23). VACV strains WR, ACAM2000, and IOC (original stock and clones) were propagated and titrated by plaque assay in BSC-40 cells, as described previously (17). For DNA isolation, clones B141 and B388 were propagated in the human HEp-2 cell line. Intracellular virus particles were purified from cell lysates by high-speed centrifugation through a 36% sucrose cushion, followed by sedimentation in a 25% to 45% potassium tartrate gradient, as described previously (24). Virus yields were determined by plaque assay in BSC-40 cells as described previously (17).
Analysis of virus progeny production in cell culture.
For one-step growth curves, BSC-40 cells were grown in 35-mm dishes and infected with the IOC clones or VACV-WR at a multiplicity of infection (MOI) of 5. After adsorption for 30 min, the monolayers were washed with PBS and received fresh culture medium. Cells were harvested at the time points indicated in the figure legend and the infectious particles were titrated by plaque assay, as described elsewhere (17). A similar procedure was used for multistep growth analyses, except for an MOI of 0.0005 and 2 h of adsorption. To determine the extracellular virus (EV) yield, supernatants of infected cells first were incubated with anti-L1 neutralizing antibody (kindly provided by Chwang Hong Foo, Gary Cohen, and Andreas Nitsche) for 1 h at 37°C to eliminate contaminant intracellular mature viruses (MVs) (25). The mixture then was titrated by plaque assay (17). Comet tail assays were performed essentially as described elsewhere (25).
Analysis of actin tails by immunofluorescence assay.
BSC-40 cells were seeded on 13-mm round glass coverslips and were either mock infected or infected with clones B141, B388, or VACV-WR at an MOI of 10 for 14 h. Cells were fixed with 4% paraformaldehyde-PBS and permeabilized with 0.5% Triton X-100 (23). Virus particles were stained with anti-D8 antibody (kindly provided by Geoffrey Smith), followed by incubation with Alexa 488-conjugated anti-mouse IgG (Jackson ImmunoResearch, West Grove, PA). Actin filaments were stained with Texas Red X-phalloidin (Molecular Probes/Invitrogen), and DNA was stained with 4,6-diamidino-2-phenylindole (DAPI). Cells were visualized using a Zeiss Axio Observer Z1 inverted microscope, and virus-tipped actin tails were counted manually during random image acquisition. Actin tails were measured using ImageJ v. 1.46r software.
Infection assays in mice.
Four- to 6-week-old female BALB/c mice were purchased from CECAL (Fundação Oswaldo Cruz, Rio de Janeiro, Brazil) or FIOTEC-Unicamp (Campinas, São Paulo, Brazil). All experiments were conducted according to the NIH Guide for the Care and Use of Laboratory Animals (26) and the protocols were approved by the Ethics Committee of the Laboratory Animal Use of the Centro de Ciências da Saúde, Universidade Federal do Rio de Janeiro (CEUA-CCS; protocols IBCCF 092 and 184/13). The number of animals used per group is indicated in the figure legends.
(i) Intranasal infection.
Mice were anesthetized with 100 mg/kg of body weight ketamine and were either mock infected (PBS) or infected intranasally with 1 × 107 PFU of purified VACV-IOC (original stock and clones) or ACAM2000 in 10 μl of PBS. For VACV-WR, 100% morbidity was achieved using 5 × 105 PFU in 10 μl of PBS; therefore, this dose was set as a positive control of weight loss. Mice were weighed daily for 14 days, and the following clinical signs were scored: ruffled fur, arched back, breathing difficulty, and reduced mobility. Mice that lost 25% of the initial weight were euthanized. To detect infectious virus particles in organs, the animals were euthanized 3 days postinfection and trachea, lungs, spleen, and liver were removed. After maceration in PBS, protein concentration was determined in cleared supernatants followed by virus titration by plaque assay (25).
(ii) Immunization via tail scarification and protection assays.
Mice were anesthetized with 120 mg/kg ketamine and 8 mg/kg xylazine and were either mock inoculated (PBS) or inoculated with 1 × 106 PFU of purified VACV-IOC (original stock), the IOC clones, or ACAM2000 in 10 μl of PBS via tail scarification, as previously described (25). To measure antibody-mediated immune responses, sera were obtained from blood samples of animals euthanized 21 days postimmunization. To evaluate cell-mediated immune responses, spleens of euthanized mice were removed 21 days postimmunization and were processed as described later. For protection assays, mice were immunized as described above, and 4 weeks postimmunization the animals were either mock challenged or challenged by intranasal infection with 100 50% lethal doses (LD50) of VACV-WR (1 × 107 PFU). Mice were weighed daily for 14 days, and those that lost 25% of the initial weight were euthanized.
Evaluation of antibody-mediated immune response. (i) Anti-VACV IgG detection by enzyme-linked immunosorbent assay (ELISA).
Purified VACV-WR particles were UV inactivated (10 μg/ml) and used as the antigen to coat 96-well Nunc-MaxiSorp plates for 16 h at 4°C. Wells were washed with PBS–0.05% Tween 20 and were incubated with PBS–10% FBS for 2 h at 37°C. Serial dilutions of heat-inactivated serum samples in PBS–10% FBS were incubated in the coated plates for 16 h at 4°C. After washing, the wells received AP-conjugated anti-mouse IgG (Sigma-Aldrich, St. Louis, MO), 1:2,000, for 2 h at 37°C. After extensive washing, 1 μg/ml paranitrophenylphosphate was added to the wells for 30 min at room temperature in Tris-HCl-MgCl2, pH 9.8, and absorbance was measured at 405 nm (27). IgG endpoint titers were defined as the reciprocal of the serum dilution that yielded absorbance values corresponding to the mean absorbance values of negative-control sera plus 2 times the standard deviations (SD) of those controls.
(ii) PRNT.
The plaque reduction neutralization test (PRNT) was performed as previously described (28). Briefly, 150 PFU of purified VACV-WR were preincubated with serially diluted, heat-inactivated serum samples for 1 h at 37°C. The mixtures next were inoculated onto BSC-40 cells grown in 24-well plates, and infection proceeded for 24 h. Monolayers were fixed and stained with crystal violet solution, and viral plaques were counted manually. PRNT50 titers were defined as the reciprocal of the serum dilution that reduced the number of viral plaques by 50%.
(iii) Comet tail inhibition assay.
Comet tail assay was performed essentially as described previously (25), except that after virus adsorption for 2 h, the inocula were removed and either fresh medium or a 1:50 dilution of pooled, heat-inactivated sera of immunized or control mice was added onto the cells.
Evaluation of cell-mediated immune response. (i) Detection of gamma interferon (IFN-γ) secretion by ELISA.
Splenocytes were prepared by homogenization of spleens removed from mice 21 days postimmunization. Red blood cells were lysed with ammonium-chloride-potassium (ACK) solution, and splenocytes were resuspended in RPMI 1640 supplemented with 10% FBS and 2 mM l-glutamine. Spleen cells from individual mice were seeded in 96-well plates (5 × 105 cells/well) and were stimulated with UV-inactivated VACV-WR at an MOI of 10 for 72 h at 37°C. Alternatively, spleen cells from animals of the same group were pooled and plated as described above. Supernatants were collected and IFN-γ concentration was determined using the Ready-SET-Go! ELISA set (eBiosciences, San Diego, CA) according to the manufacturer's instructions (29).
(ii) Intracellular cytokine staining.
Splenocytes prepared as described above were stimulated with UV-inactivated VACV-WR for 24 h at 37°C. Brefeldin A (3 μg/ml) was added to the cells for the last 10 h, followed by washing with PBS–1% FBS and fixation with 4% paraformaldehyde for 30 min at 4°C. After blocking with PBS–1% FBS–2% mouse serum, the splenocytes were stained with allophycocyanin (APC)-conjugated anti-CD4 (1:400) and phycoerythrin (PE)-Cy5-conjugated anti-CD8 (1:800) (eBioscience) for 30 min at room temperature. The cells were washed, permeabilized with 0.1% saponin, and subsequently stained with fluorescein isothiocyanate (FITC)-conjugated anti-IFN-γ (1:200) and PE-conjugated anti-interleukin-2 (IL-2) (1:400) or PE-conjugated anti-tumor necrosis factor alpha (TNF-α) (1:400) antibodies (eBiosciences) for 1 h at room temperature (29). Samples were acquired using a FACSAria II flow cytometer (BD Biosciences, San Jose, CA), which recorded 5 × 105 events within an forward scatter-side scatter gate relative to lymphocytes. Data were analyzed using FlowJo v. 10 software.
Genome sequencing, assembly, and annotation.
Genomic DNA was isolated from purified B141 and B388 particles using a Wizard Genomic DNA purification kit (Promega, Madison, WI) and submitted to Fasteris SA (Plan-les-Ouates, Switzerland) for a 2 × 100 + 7 paired-end sequencing run using an Illumina HiSeq 2500 platform. The resulting raw reads were checked for quality control and removal of adaptors by Fasteris SA. Additionally, 1 μg of B141 was used to generate a library with the Titanium rapid library preparation kit (Roche Applied Sciences, Branford, CT). The library was amplified by emulsion PCR before sequencing based on the 454 FLX+ pyrosequencing chemistry (Roche Applied Sciences), and base calling was performed by the instrument's software. Illumina raw reads for the B141 genome were mapped to the final contig obtained from 454 sequencing using BWA v. 0.7.12 (30). Mapping was visualized using Tablet v. 1.13.07_21, and homopolymeric errors were corrected by manual inspection of conflicting bases indicated by Illumina reads or by Sanger sequencing. Illumina raw reads for the B388 genome were used for de novo assembly using Velvet v. 1.2.10 with a kmer of 95, average paired-read insert size of 250, and minimum contig size of 150 (31). Sanger sequencing was used to fill the gaps between contigs, to correct homopolymeric errors, and to extend the genome terminal regions. SeqMan (DNAStar package; Lasergene Inc.) was used for final assembly of next-generation sequencing (NGS) contigs with Sanger reads resulting from sequencing of the genome terminal regions. Genomes were annotated based on VACV-Cop sequence using the GATU software (32), followed by manual editing using CLC Main Workbench v. 6.9.1 to call open reading frames (ORFs) absent from the VACV-Cop genome. We annotated only ORFs longer than 30 amino acids starting with ATG, with less than 35% overlap with other ORFs, and with similarity to Orthopoxvirus ORFs (BLASTP; http://blast.ncbi.nlm.nih.gov/Blast.cgi). Genome annotations for B141 and B388 were merged into a pan-IOC annotation file that contained 255 putative ORFs numbered sequentially. This pan-IOC annotation file was used to reannotate B141 and B388 genomes so that orthologs had the same gene number despite the order of appearance in the genomes.
Phylogenetic inference and analysis of genome ends.
Multiple nucleotide sequences spanning the region between orthologs of VACV-Cop F9L and A24R were aligned using MAFFT v. 7 (33), followed by visual inspection. The alignment was used to reconstruct phylogenetic trees using MEGA v. 6.06 (34) by two methods: neighbor-joining, opting for the Kimura 2-p substitution model with 1,000 bootstrap replicates, and maximum likelihood, opting for the Tamura-Nei substitution model, neighbor-joining input tree, five-category discrete gamma model, and 1,000 bootstrap replicates. Additionally, Bayesian phylogenetic inference was conducted using BEAST v. 1.8.1 (35), opting for the GTR model plus a gamma correction for site rate, with default prior settings, 4 chains of 10 million generations of Markov chain Monte Carlo (MCMC) with tree sampling every 1,000th generation, and a burn-in of 1,500,000 states. The final Bayesian tree was constructed as a maximum clade credibility tree and was visualized using FigTree v. 1.4.2.
To analyze the structure of the genome terminal regions, the nucleotide sequences of multiple genomes were trimmed to select approximately 20 kb of the left or right ends. For the 5′ end, the sequences were aligned using MAFFT v. 7 (33), and the alignment was visualized using CLC Main Workbench v. 6.9.1. Minor deletions (<250 nucleotides [nt]) were omitted in order to highlight major deletion patterns. To analyze the 3′ end, a genome blast atlas was generated using GView Server based on a synthetic reference genome that contained all possible genes represented in the 3′ end of the genomes used in the analysis. A scheme was drawn based on the resulting blast atlas using Adobe Photoshop CS6. The intergenic regions and minor deletions (<250 nt) were removed, and the ORFs were represented contiguously in order to highlight major differences between viruses.
The following genome sequences (GenBank accession numbers) were used for phylogenetic inferences and analyses of the terminal regions: orthopoxviruses camelpox virus strain CMS (AY009089); taterapox virus (NC_008291); variola virus strains BSH95 (DQ437581) and BRZ66 (DQ441419); cowpox virus strain GRI-90 (X94355); horsepox virus strain MNR76 (DQ792504); VACV strains WR (AY243312), Copenhagen (M35027), LC16m8 (AY678275), Serro 2 virus (KF179385), Cantagalo virus isolate CM-01 (KT013210), and rabbitpox virus–Utrecht (AY484669); IHD-W (KJ125439); Lister 107 (DQ121394); 3737 (DQ377495); Duke (DQ439815); modified vaccinia Ankara (MVA) (DQ983238); chorioallantois vaccinia virus Ankara (CVA) (AM501482); Dryvax clones DPP15 (JN654981), DPP20 (JN654985), DPP13 (JN654980), DPP17 (JN654983), DPP21 (JN654986), ACAM2000 (AY313847), and CL-3 (AY313848); TianTan clones TT9 (JX489136), TP3 (KC207810), and TP5 (KC207811); and Tashkent clones TKT3 (KM044309) and TKT4 (KM044310).
Statistical analyses.
Statistical analyses were performed using GraphPad Prism v. 5.01 (Graph-Pad Software, Inc.) by nonparametric Kruskal-Wallis test or one-way analysis of variance (ANOVA) followed by Tukey's multiple comparison test, as indicated in the figure legends. P values of <0.05 were considered statistically significant.
Nucleotide sequence accession numbers.
The genome sequences of the IOC clones B141 and B388 and Cantagalo virus isolate CM-01 have been deposited in GenBank under accession numbers KT184690, KT184691, and KT013210, respectively.
RESULTS
Isolation of clones B141 and B388 from stocks of VACV-IOC: plaque phenotype, virus production, and spread in cell culture.
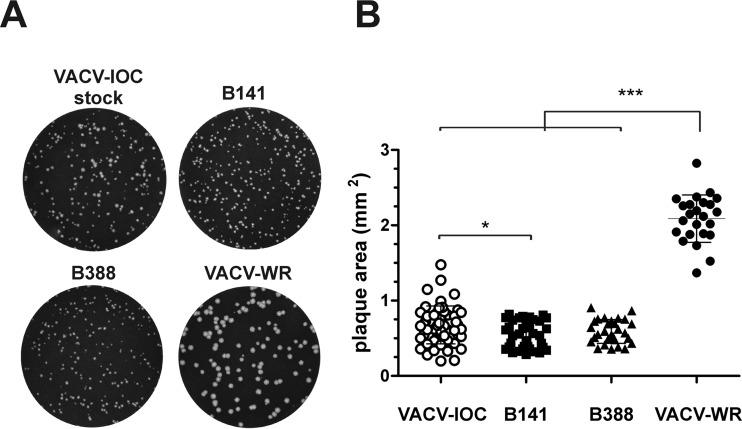
To characterize the VACV-IOC smallpox vaccine, we first selected two clones from the reconstituted stock of VACV-IOC after three rounds of limiting dilution and plaque purification on BSC-40 cells. Clones B141 and B388 produced viral plaques of uniform size compared with the heterogeneity of plaque sizes produced by the VACV-IOC stock (Fig. 1A). Plaque size varied similarly for clones B141 and B388 and was comparable to the mean size of VACV-IOC plaques (0.67 ± 0.25 mm2). In addition, plaques produced by both clones were significantly smaller than the plaques produced by VACV-WR (2.1 ± 0.31 mm2) (Fig. 1B).
FIG 1.
Plaque phenotypes produced by the IOC clones B141 and B388. BSC-40 cells were infected with 150 PFU of the original stock of VACV-IOC, clone B141, B388, or VACV-WR and were incubated for 48 h under semisolid medium. (A) Images of crystal violet-stained cells are representative of 2 independent experiments. (B) Random viral plaques (n > 24) were photographed at ×4 magnification, and the individual areas were measured. *, P < 0.05; ***, P < 0.001 (one-way ANOVA followed by Tukey's posttest).
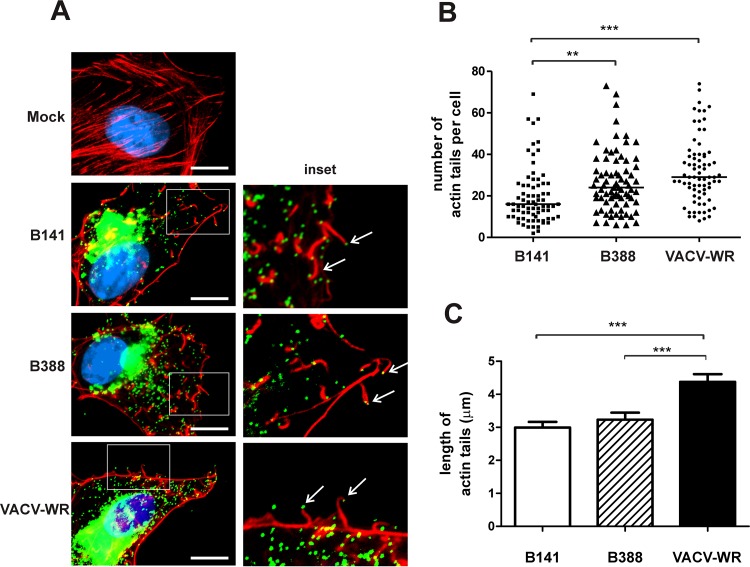
The size of viral plaques mainly results from cell-to-cell spread of infection via extracellular VACV on the tip of actin tails (36). Because B141 and B388 formed small plaques during infection, we investigated the formation of actin tails in BSC-40 cells infected for 16 h with each IOC clone. Figure 2A shows that clones B141 and B388 induced the formation of actin tails of regular morphology compared with those formed by VACV-WR. Nevertheless, clone B141 produced 1.6 and 1.9 times fewer virus-tipped actin tails per cell than clone B388 and VACV-WR, respectively (Fig. 2B). In addition, both clones induced the formation of actin tails of comparable length, but they were significantly shorter than those induced by VACV-WR infection (Fig. 2C). Together, these results suggest that clones B141 and B388 may not spread efficiently in cell culture.
FIG 2.
Formation of actin tails induced by B141 and B388 in BSC-40 cells. BSC-40 cells were either mock infected or infected with clone B141, B388, or VACV-WR and were processed for immunofluorescence assay 14 h postinfection. (A, left) Intact actin stress fibers (red) in mock-infected cells (Mock) and actin tails (red) with virus particles (green) on the tip (arrows) in virus-infected cells. DNA was stained with DAPI (blue). (Right) Closeup of the boxed area from the panel on the left. Representative images of 3 independent experiments are shown. Bars, 10 μm. (B) The number of actin tails (n = 75 cells) was counted in 3 independent experiments. Bars represent median values from each data set. **, P < 0.01; ***, P < 0.001 (nonparametric Kruskal-Wallis test). (C) Mean length of actin tails (n = 200) induced by B141, B388, or VACV-WR was measured (±SD) in 3 independent experiments. ***, P < 0.001 (one-way ANOVA followed by Tukey's posttest).
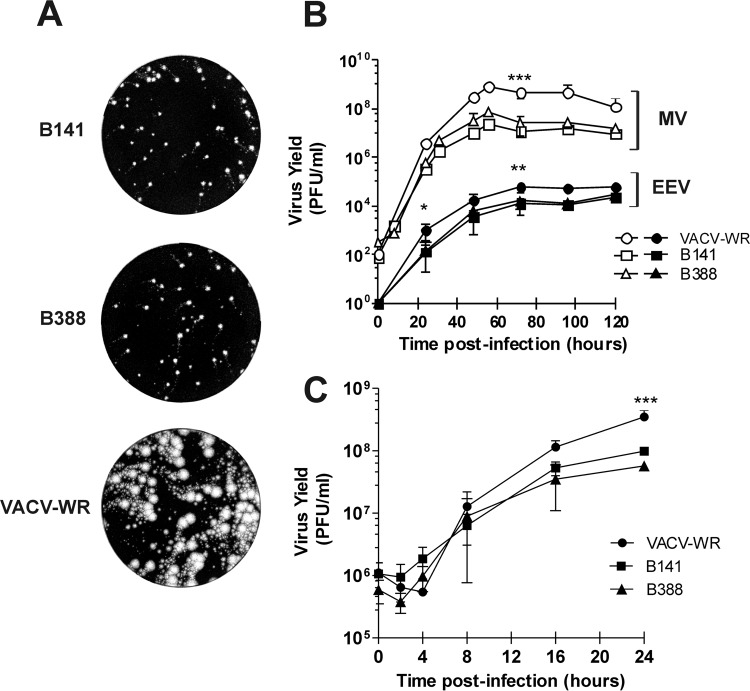
To investigate virus spread, we analyzed the formation of comet tails in BSC-40 cells infected with clones B141 and B388. Comet tails result from the release of extracellular virus (EEV) in the cell medium and subsequent infection of distant sites away from the primary viral plaque. Figure 3A shows that both clones formed comet tails of similar phenotype, but they were significantly smaller than those formed by VACV-WR 72 h postinfection. The reduced size of comet tails indicated a deficiency of virus spread, which was confirmed by the titration of infectious particles in a multistep growth curve. Clones B141 and B388 produced significantly less EEV and intracellular mature virus (MV) than VACV-WR (Fig. 3B). Similarly, in a one-step growth curve the yield of MV by clones B141 and B388 was consistently lower after the first 8 h of infection than those by VACV-WR (Fig. 3C).
FIG 3.
Progeny production and spread of B141 and B388 in BSC-40 cells. (A) Comet tail formation by B141, B388, or VACV-WR 72 h postinfection. Representative images of 2 independent experiments are shown. (B) Multistep growth curve of clones B141 and B388. Cells were infected with B141, B388, or VACV-WR, and at the indicated time points, the monolayers (open symbols) and supernatants (closed symbols) were harvested for virus titration. (C) One-step growth curve of B141 and B388. Cells were infected with the indicated viruses and were harvested for virus titration at 0, 2, 4, 8, 16, and 24 h postinfection. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (one-way ANOVA followed by Tukey's posttest for time points in which 3 experiments were conducted).
Immune response following immunization of mice with IOC clones B141 and B388.
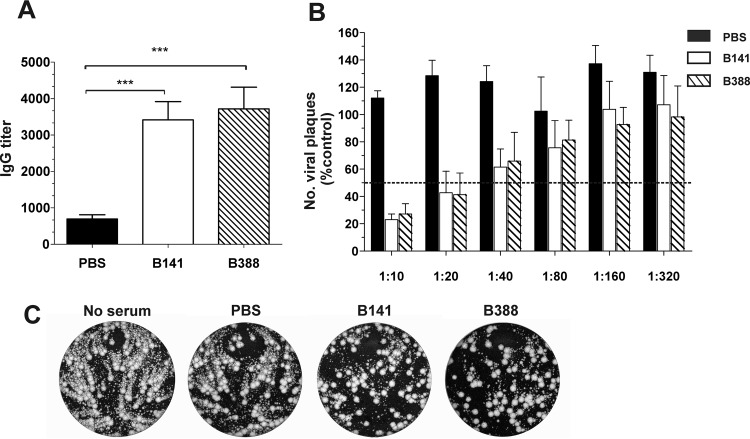
To further characterize B141 and B388 as clones of the Brazilian smallpox vaccine, we investigated the immune response elicited by these clones. BALB/c mice were immunized with 106 PFU of each clone by tail scarification. Sera of immunized mice were collected 21 days postimmunization, and the IgG titers were determined by ELISA using inactivated VACV-WR particles as antigens. Mice immunized with either B141 or B388 produced high levels of VACV-specific IgG, with mean titers of 3,415 ± 501.3 and 3,717 ± 597.4, respectively (Fig. 4A). To test the ability of sera to neutralize virus infection, purified VACV-WR MVs were incubated with serially diluted sera of immunized mice, and titers of neutralizing antibodies were determined by PRNT. In contrast to mock immunization, B141 and B388 immunization induced the production of MV-neutralizing antibodies at comparable levels, with mean PRNT50 titers of 28.76 ± 9.14 and 28.92 ± 14.3, respectively (Fig. 4B). Sera of mice immunized with B141 and B388 also neutralized extracellular virus particles (EEVs), as assessed by comet tail inhibition assays (Fig. 4C).
FIG 4.
Antibody-mediated immune response induced by clones B141 and B388 in immunized mice. BALB/c mice were either mock immunized (PBS) or immunized with 106 PFU of B141 or B388 via tail scarification, and sera were collected 21 days postimmunization. (A) Anti-VACV IgG titers in sera from individual mice were determined by ELISA. Data are presented as mean IgG titers (± standard errors of the means [SEM]) from 2 independent experiments (n = 3 mice per group per experiment). ***, P < 0.001 (one-way ANOVA followed by Tukey's posttest). (B) Detection of MV-neutralizing antibodies by PRNT. Serum dilutions were incubated with VACV-WR followed by infection of BSC-40 cells. Data from 2 independent experiments (n = 3 mice per group per experiment) are presented as the mean percentages (±SEM) of viral plaques relative to control infections in the absence of serum and considered 100%. (C) Detection of EEV-neutralizing antibodies by comet tail inhibition assay. BSC-40 cells were infected with VACV-WR and incubated for 72 h either in the absence or in the presence of pooled sera (1:50) from mock-immunized or immunized mice. Representative images are shown.
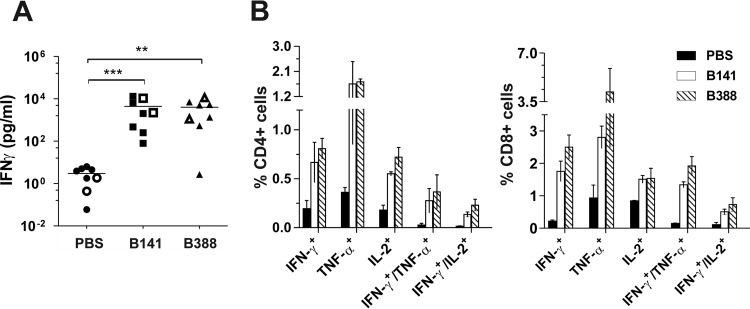
To investigate the T-cell-mediated response, splenocytes of mice immunized with B141 and B388 were harvested 21 days postimmunization and were stimulated in vitro with inactivated VACV-WR. Cells obtained from mice immunized with both clones produced increased levels of IFN-γ when cultured with VACV-WR compared to those of cells obtained from mock-immunized mice. The levels of secreted IFN-γ were similar between B141 and B388, with mean values of 4,403 ± 1,811 pg/ml and 3,996 ± 1,410 pg/ml, respectively (Fig. 5A). Furthermore, the phenotype of responsive splenocytes, determined by intracellular cytokine staining, revealed that immunization with B141 and B388 effectively primed CD4+ and CD8+ T cells that produced IFN-γ, TNF-α, or IL-2 (Fig. 5B). It is also worth noting that a subset of primed CD4+ and CD8+ cells exhibited a polyfunctional phenotype, with the concurrent expression of IFN-γ and TNF-α or IFN-γ and IL-2 (Fig. 5B). Taken together, these results show that B141 and B388 induced humoral and cellular immune responses to a similar extent and in a very similar pattern.
FIG 5.
T-cell-mediated immune response elicited by clones B141 and B388 in immunized mice. BALB/c mice were either mock immunized or immunized with 1 × 106 PFU of B141 or B388 via tail scarification, and spleens were removed 21 days postimmunization. (A) Detection of secreted IFN-γ by ELISA. Splenocytes from individual mice were plated separately (closed symbol) or were pooled by group before plating (open symbols). Data are presented as the mean IFN-γ concentration from 4 independent experiments (n = 3 mice per group per experiment). A lognormal distribution was assumed for all data sets. **, P < 0.01; ***, P < 0.001 (one-way ANOVA followed by Tukey's posttest). (B) Immunophenotyping of mouse splenocytes. Splenocytes were pooled, stimulated in vitro, and then stained with the indicated fluorochrome-conjugated antibodies, followed by flow cytometry analysis. Data from 2 independent experiments are presented as the mean percentages (±SEM) from CD4 (left) or CD8 cells (right) that express the indicated combinations of cytokines.
B141 and B388 differences in virulence in vivo.
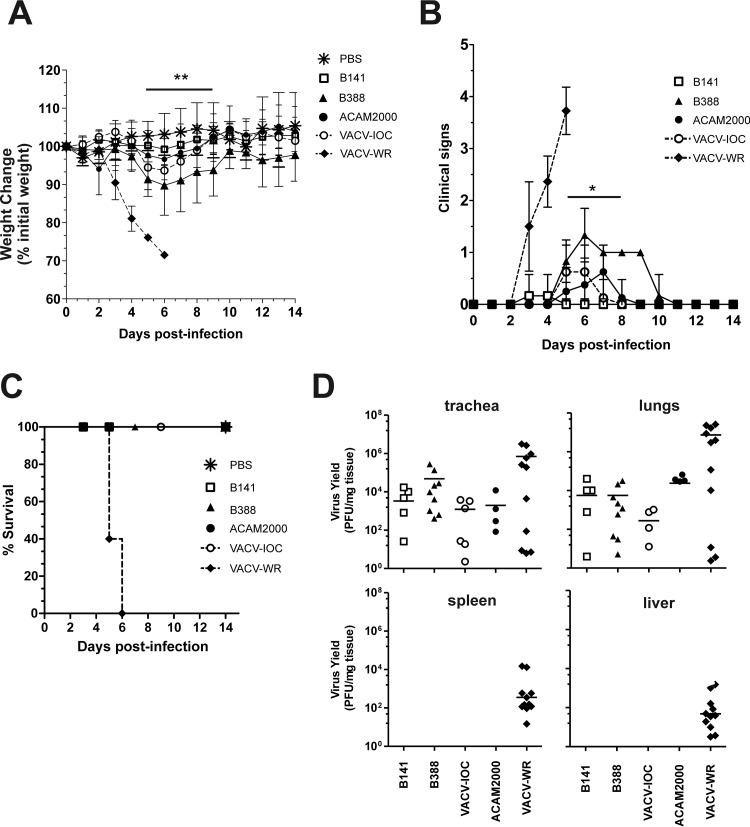
B141 and B388 did not show significant differences in virus spread in vitro and in the profile of the induced immune response in the mouse model. We next assessed whether these similarities reflected other biological properties, such as virulence and virus dissemination in vivo. Mice were intranasally infected with 1 × 107 PFU of B141 or B388, and clinical signs of infection were monitored for 14 days. We also included the licensed vaccine ACAM2000 (1 × 107 PFU) and the virulent strain VACV-WR (5 × 105 PFU) as references of low- and high-virulence strains, respectively.
Infection of mice with VACV-WR caused severe weight loss beginning on day 3 postinfection (Fig. 6A), when mice also showed increasing signs of illness (ruffled fur, arched back, breathing difficulty, and reduced mobility) (Fig. 6B). Mice infected with B141 did not lose weight (Fig. 6A) and developed no signs of illness (Fig. 6B), whereas B388 caused a transient weight loss of up to 10% of the initial weight until day 6, with full recovery 10 days postinfection (Fig. 6A). In the meantime, all mice infected with B388 developed at least one sign of illness, which was mostly ruffled fur, recovering on day 10 postinfection (Fig. 6B). The difference in virulence between clones B141 and B388 prompted us to investigate the pathogenicity of the original vaccine stock of VACV-IOC. Interestingly, infection with VACV-IOC (1 × 107 PFU) showed an intermediate pattern between B141 and B388, causing less weight loss, fewer signs of illness, and faster recovery than the infection with B388 (Fig. 6A and B). Similar to B141, ACAM2000 did not cause significant weight loss (Fig. 6A). However, some of the ACAM2000-infected mice developed at most one sign of illness between days 5 and 7 postinfection (Fig. 6B). Despite the differences observed in virulence, all mice survived the infection with all vaccine strains tested, in contrast to mice infected with the virulent strain VACV-WR, which succumbed in a maximum of 6 days postinfection (Fig. 6C). These animals either died or lost 25% of their initial weight, in which case they were euthanized.
FIG 6.
Low virulence of B141 and B388 in murine intranasal infection model. BALB/c mice were infected intranasally with 1 × 107 PFU of B141, B388, VACV-IOC, or ACAM2000 or 5 × 105 PFU of VACV-WR. Weight changes (A), clinical signs (B), and survival (C) were monitored daily for 14 days. (A) Symbols represent the mean weight percentage (±SD) relative to day zero postinfection in 2 independent experiments (n = 4 mice per group per experiment). (B) Data points represent mean scores, and error bars represent ±SD. Mock-infected animals (PBS) did not show any clinical signs, and symbols were omitted for the sake of clarity. In panels A and B, horizontal bars above the symbols indicate the time points in which differences between B141- and B388-infected mice were statistically significant. *, P < 0.05; **, P < 0.01 (one-way ANOVA followed by Tukey's posttest). (D) Virus replication in primary and secondary sites of infection. Mice were euthanized 3 days postinfection when trachea, lungs, spleen, and liver were removed for virus titration as described in Materials and Methods. Each data point represents the virus titer of individual mice normalized by tissue protein concentration. Bars represent mean virus titers from 2 independent experiments (n = 5 mice per group per experiment).
To test whether differences in weight loss and in other clinical signs were due to differences in virus dissemination to organs, infectious virus particles were titrated in trachea, lungs, spleen, and liver of infected mice on day 3 postinfection. At this time point, all vaccine strains replicated in trachea and lungs but did not spread to spleen or liver, in contrast with the virulent strain VACV-WR (Fig. 6D). On day 5 postinfection, no infectious virus particle of B141 was detected in trachea and lungs, whereas B388 still was detected in these tissues (data not shown). On the other hand and more importantly, B388 did not spread to spleen and liver even at this time point (data not shown). Overall, despite showing differences in virulence patterns, B141 and B388 presented low virulence in the mouse model.
Protection from lethal challenge by immunization with B141 and B388.
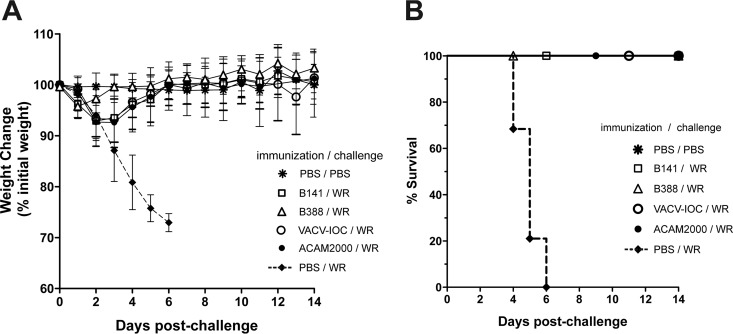
Infection with B141 and B388 caused mild effects in mice, similar to the effects caused by the infection with the licensed vaccine ACAM2000. We next investigated whether immune responses induced by B141 and B388 would be sufficient to confer protection following a lethal challenge. For that, mice were immunized with B141 or B388 by tail scarification and challenged after 28 days with 1 × 107 PFU of VACV-WR by the intranasal route. For comparison, mice also were immunized with parental VACV-IOC or ACAM2000.
Mice immunized with all vaccine strains survived the lethal challenge and did not lose weight through the entire observation period, despite showing a transient weight loss (<10%) on the first days postchallenge (Fig. 7A and B). On the other hand, mice that were challenged with no prior immunization (PBS/WR) began to lose weight on day 3 postinfection (Fig. 7A) and had died or were euthanized by day 6 postinfection. In conclusion, immunization with B141 and B388 efficiently protected mice from a lethal challenge, and this profile was comparable with the protection conferred by ACAM2000 and by parental VACV-IOC.
FIG 7.
B141 and B388 confer immune protection against a lethal challenge. Mice were either mock immunized or immunized with 1 × 106 PFU of B141, B388, VACV-IOC, and ACAM2000. Challenge was conducted 4 weeks postimmunization by intranasal infection with PBS (mock-challenged control) or 100 LD50 of VACV-WR. (A) Weight changes were monitored daily for 14 days. Symbols represent mean weight percentages (±SD) relative to that at day zero postchallenge in 2 independent experiments (n = 6 mice per group per experiment). (B) The survival percentage is presented.
Full-genome analysis and phylogeny inference of clones B141 and B388.
To gain a better understanding of the differences in virulence between the IOC clones and analyze their evolutionary relationships with other VACV strains, we determined the full genome sequences of B141 and B388. For that, DNA was isolated from purified B141 and B388 particles and sequenced using one or both NGS platforms, the 454 FLX+ system and/or Illumina HiSeq 2500, as described in Materials and Methods. Sanger sequencing was used to correct possible errors in homopolymeric regions, to fill the gaps between contigs, and to extend the ends of the genome. A summary of the sequencing data and statistics is presented in Table S1 in the supplemental material. The assembly of NGS-derived contigs together with Sanger reads generated a final contiguous sequence of 192,017 bp for clone B141 (1,256× genome coverage) and 186,980 bp for clone B388 (603× genome coverage). Nucleotide 1 was arbitrarily defined as the leftmost nucleotide in the 5′ end of the assembled genomes. The genome of B141 has a central region of 170,071 bp flanked by inverted terminal repeats (ITRs) of 10,973 bp at both ends. On the other hand, the B388 genome has shorter ITRs of 6,623 bp and a central region of 173,734 bp.
For the annotation of B141 and B388 genomes, we considered ORFs encoding ≥30 amino acids with less than 35% overlap sharing similarity with Orthopoxvirus ORFs. The complete list of ORFs encoded by clones B141 and B388 is shown in Table S2 in the supplemental material. Most ORFs were intact genes, but several fragmented genes were observed, which were annotated as truncated genes or pseudogenes. The B141 genome has 253 putative ORFs, of which 180 are intact genes and 73 are gene fragments. Clone B388 encodes 244 putative ORFs, of which 179 are full-length genes and 65 are fragmented genes.
Table 1 lists major differences in ORF length between B141 and B388 genomes as well as differences between both clones and other VACV strains. The pairwise alignment of B141 and B388 genomes revealed a nearly 4,300-bp deletion in the 3′ ITR of the B388 genome immediately downstream of gene IOC_234 (ortholog of VACV-Cop B19R) (Table 1). The deletion was confirmed by long-range PCR of the 3′ end and Sanger sequencing (data not shown). The 4.3-kb deletion contained the VACV-Cop orthologs B20R (IOC_235), C10L (IOC_236/237), C11R (IOC_238), and C12L (IOC_239) (Table 1). Of these, only IOC_238 and IOC_239 are full-length genes, whereas ORFs IOC_235 and IOC_236/237 correspond to fragmented genes. These ORFs, except for IOC_235, are present in the 5′ ITR of both B388 and B141 genomes; therefore, only the B141 clone is diploid for them.
TABLE 1.
Major differences in ORF sizes between VACV-IOC clonesa
| Gene or gene fragmentb | Description/putative function | Ortholog(s)c | Length of genes or gene fragments (bp) |
||||
|---|---|---|---|---|---|---|---|
| B141 | B388 | VACV-WR | VACV-Cop | Other OPVd | |||
| IOC_001/255 | Hypothetical protein | VARV-Gar_1966-209/HSPV-001 | 255 | 429 (VARV)/219 (HSPV) | |||
| IOC_002/254 | Hypothetical protein | HSPV-001 | 147 | 147 | 219 (HSPV) | ||
| IOC_003/253 | Chemokine-binding protein | Cop-C23L/WR-001 | 681 | 711 | 735 | 735 | 747 (HSPV) |
| IOC_004/252 | TNF-α receptor | WR-002 | 192 | 186 | 1,050 (HSPV) | ||
| IOC_005/251 | TNF-α receptor | WR-002 | 105 | 105 | 186 | ||
| IOC_006/250 | TNF-α receptor | Cop-C22L/WR-004 | 360 | 342 | 369 | 369 | 1,050 (HSPV) |
| IOC_008/248 | Ankyrin-like protein | Cop-C21L/WR-006 | 201 | 201 | 195 | 342 | 1,764 (HSPV) |
| IOC_009/247 | Ankyrin-like protein | Cop-C21L/WR-006 | 141 | 141 | |||
| IOC_010/246 | Ankyrin-like protein | Cop-C18L/WR-007 | 186 | 186 | 330 | 453 | 2,019 (CPXV-GRI) |
| IOC_011/245 | Ankyrin-like protein | Cop-C17L | 318 | 318 | 1,161 | 1,194 (HSPV) | |
| IOC_012/244 | Ankyrin-like protein | Cop-C17L | 192 | 192 | |||
| IOC_013/243 | Ankyrin-like protein | Cop-C17L | 702 | 702 | |||
| IOC_014/242 | Predicted Bcl-2 protein | Cop-C16L | 531 | 522 | 546 | 462 (HSPV) | |
| IOC_015 | Hypothetical protein | HSPV-228 | 123 | 123 | 204 (HSPV) | ||
| IOC_016 | Hypothetical protein | Cop-C15L | 258 | 258 | 276 | ||
| IOC_019 | IL-1 receptor antagonist; binds to (PHD)2 protein; DNA-PK binding protein | Cop-C10L/WR-010 | 279 | 279 | 996 | 996 | 996 (HSPV) |
| IOC_020 | IL-1 receptor antagonist; binds to (PHD)2 protein; DNA-PK binding protein | Cop-C10L/WR-010 | 573 | 573 | |||
| IOC_021 | Ubiquitin ligase | WR-011 | 252 | 252 | 546 | 729 (CPX-GRI) | |
| IOC_023 | Interleukin-18-binding protein | WR-013 | 363 | 381 | 381 | 381 (HSPV) | |
| IOC_024 | Ankyrin-like protein | WR-014 | 273 | 273 | 714 | 2,007 (CPXV-GRI) | |
| IOC_025 | Ankyrin-like protein | WR-014 | 525 | 414 | |||
| IOC_027 | Ankyrin-like protein | WR-016 and WR-017 | 747 | 384 | 234 and 216 | 1,179 (VARV) | |
| IOC_028 | Ankyrin-like protein | Cop-C9L/WR-019 | 924 | 924 | 1,905 | 1,905 | 1,905 (HSPV) |
| IOC_029 | Ankyrin-like protein | Cop-C9L/WR-019 | 153 | 153 | |||
| IOC_030 | Ankyrin-like protein | Cop-C9L/WR-019 | 207 | ||||
| IOC_031 | Ankyrin-like protein | Cop-C9L/WR-019 | 684 | 495 | |||
| IOC_035 | Predicted Bcl-2 protein; IRF3 inhibitor | Cop-C6L/WR-022 | 270 | 261 | 456 | 456 | 468 (HSPV) |
| IOC_036 | Predicted Bcl-2 protein; IRF3 inhibitor | Cop-C6L/WR-022 | 126 | 126 | |||
| IOC_038 | Putative IL-1 receptor antagonist; NF-kB inhibitor | Cop-C4L/WR-024 | 189 | 189 | 951 | 951 | 951 (VARV) |
| IOC_039 | Putative IL-1 receptor antagonist; NF-kB inhibitor | Cop-C4L/WR-024 | 231 | 231 | |||
| IOC_040 | Putative IL-1 receptor antagonist; NF-kB inhibitor | Cop-C4L/WR-024 | 123 | 123 | |||
| IOC_041 | Putative IL-1 receptor antagonist; NF-kB inhibitor | Cop-C4L/WR-024 | 120 | 120 | |||
| IOC_042 | Secreted complement C3b/C4b-binding protein | Cop-C3L/WR-025 | 393 | 792 | 792 | 792 (HSPV) | |
| IOC_043 | Secreted complement C3b/C4b-binding protein | Cop-C3L/WR-025 | 189 | 786 | |||
| IOC_044 | POZ/BTB kelch domain protein | Cop-C2L/WR-026 | 105 | 105 | 1,539 | 1,539 | 1,539 (HSPV) |
| IOC_045 | POZ/BTB kelch domain protein | Cop-C2L/WR-026 | 534 | 534 | |||
| IOC_046 | POZ/BTB kelch domain protein | Cop-C2L/WR-026 | 813 | 813 | |||
| IOC_049 | Predicted Bcl-2 protein; IRF3 inhibitor | Cop-N2L/WR-029 | 513 | 513 | 528 | 528 | 534 (VARV) |
| IOC_050 | Ankyrin-like protein | Cop-M1L/WR-030 | 198 | 198 | 1,419 | 1,419 | 1,341 (VARV) |
| IOC_051 | Ankyrin-like protein | Cop-M1L/WR-030 | 645 | 645 | |||
| IOC_052 | Ankyrin-like protein | Cop-M1L/WR-030 | 474 | 474 | |||
| IOC_055 | Serine protease inhibitor-3 (SPI-3) | Cop-K2L/WR-033 | 1,095 | 1,095 | 1,110 | 1,110 | 1,122 (HSPV) |
| IOC_056 | eIF2 alpha-like PKR inhibitor | Cop-K3L/WR-034 | 183 | 267 | 267 | 267 (HSPV) | |
| IOC_057 | eIF2 alpha-like PKR inhibitor | Cop-K3L/WR-034 | 183 | 267 | |||
| IOC_063 | Predicted Bcl-2 protein; apoptosis inhibitor | Cop-F1L/WR-040 | 462 | 462 | 681 | 681 | 765 (HSPV) |
| IOC_064 | Predicted Bcl-2 protein; apoptosis inhibitor | Cop-F1L/WR-040 | 207 | 204 | |||
| IOC_093 | Involved in sustained ERK1/2 activation | Cop-O1L/WR-068 | 1,992 | 1,992 | 2,001 | 2,001 | 2,001 (HSPV) |
| IOC_134 | Virion core protein | Cop-D3R/WR-108 | 702 | 702 | 714 | 714 | 714 (HSPV) |
| IOC_149 | 39-kDa core protein | Cop-A4L/WR-123 | 840 | 822 | 846 | 846 | 852 (CPXV-GRI) |
| Cowpox A-type inclusion protein | Cop A25L/WR-145 | 198 | 198 | 213 (HSPV) | |||
| IOC_171 | Cowpox A-type inclusion protein | WR-146 | 180 | 180 | 465 | 612 (HSPV) | |
| IOC_185 | WV transmembrane phosphoprotein | Cop-A36R/WR-159 | 639 | 630 | 666 | 666 | 672 (HSPV) |
| IOC_189 | Semaphorin-like protein | Cop-A39R/WR-163 | 681 | 642 | 888 | 1,212 | 981 (HSPV) |
| IOC_190 | Semaphorin-like protein | Cop-A39R/WR-164 | 258 | 258 | 429 | ||
| IOC_191 | Semaphorin-like protein | Cop-A39R/WR-164 | 123 | 123 | |||
| IOC_193 | Chemokine binding protein | Cop-A41L/WR-166 | 639 | 639 | 660 | 660 | 657 (VARV) |
| IOC_204 | associates with and stabilizes ubiquitin | Cop-A51R/WR-177 | 210 | 210 | 1,005 | 1,005 | 1,005 (VARV) |
| IOC_205 | Associates with and stabilizes ubiquitin | Cop-A51R/WR-177 | 540 | 540 | |||
| IOC_208 | Secreted TNF-receptor-like protein; CrmC | Cop-A53R/WR-179 | 303 | 303 | 312 | 312 | 969 (CPX-GRI) |
| IOC_209 | kelch-like protein | Cop-A55R/WR-180 | 255 | 255 | 1,695 | 1,695 | 1,695 (HSPV) |
| IOC_210 | kelch-like protein | Cop-A55R/WR-180 | 1,416 | 1,416 | |||
| IOC_211 | Hemagglutinin | Cop-A56R/WR-181 | 927 | 927 | 945 | 948 | 945 (CPXV-GRI) |
| IOC_216 | schlafen-like protein | Cop-B3R/WR-185 | 183 | 183 | 504 | 375 | 375 (HSPV) |
| IOC_227 | Serine protease inhibitor (SPI-2/CrmA) | Cop-B13R/WR-195 | 351 | 351 | 1,038 | 351 | 1,038 (HSPV) |
| IOC_228 | Serine protease inhibitor (SPI-2/CrmA) | Cop-B14R/WR-195 | 144 | 144 | 669 | ||
| IOC_229 | Serine protease inhibitor (SPI-2/CrmA) | Cop-B14R/WR-195 | 534 | 534 | |||
| IOC_235 | Ankyrin-like protein | Cop-B20R/WR-202/WR-203 | 162 | 162 | 384 | 2,376 (HSPV) | |
| 930 | |||||||
| IOC_236 | IL-1 receptor antagonist; DNA-PK binding protein; binds to (PHD)2 protein | Cop-C10L/WR-209 | 573 | 996 | 996 | 996 (HSPV) | |
| IOC_237 | IL-1 receptor antagonist; DNA-PK binding protein; binds to (PHD)2 protein | Cop-C10L/WR-209 | 279 | ||||
| IOC_238 | Secreted epidermal growth factor-like | Cop-C11R/WR-009; WR-210 | 423 | 423 | 429 | 423 (HSPV) | |
| IOC_239 | Serine protease inhibitor (Serpin)-1 (SPI-1) | Cop-C12L/WR-205 | 1,062 | 1,062 | 1,074 (HSPV) | ||
The table lists only major differences in ORF length among VACV-IOC clones B141, B388, and other orthopoxviruses. The complete list of ORFs annotated in the genomes of B141 and B388 is shown in Table S2 in the supplemental material.
ORFs located in both ITRs (ITR paralogs) are indicated after a slash. ORFs in boldface are full-length or near-full-length genes in either one or both IOC clones. All other ORFs correspond to truncated genes or gene fragments.
Orthologs in VACV-Cop and VACV-WR are indicated. In the absence of those, the best matching sequence is indicated.
ORF size in other Orthopoxvirus (OPV) members is indicated for comparison with full-length (or near-full-length) genes. VARV, variola virus; CPXV-GRI, cowpox virus strain GRI.
It is interesting that, in addition to the presence of the 4.3-kb deletion, the B388 genome differs from the B141 genome in that it harbors full-size copies of two virulence genes that are fragmented in the B141 genome: gene IOC_043 (VACV-Cop C3L) encodes a secreted complement C3b/C4b-binding protein, and gene IOC_057 (VACV-Cop K3L) encodes an eIF2 alpha-like protein. These results highlight important differences in virulence genes between clones B141 and B388. Despite these differences, the IOC clones share a panel of virulence genes that are fragmented and probably nonfunctional in both genomes (Table 1). Of particular interest, B141 and B388 encode truncated forms of ankyrin-like proteins (orthologs of VACV-Cop C9L and M1L, among others), POZ/BTB kelch-domain proteins (VACV-Cop C2L and A55R), IRF-3 and IRF-7 inhibitor (VACV-Cop C6L), NF-κB inhibitor (VACV-Cop C4L), apoptosis inhibitors (VACV-Cop F1L and B13R/B14R), and DNA-PK binding protein (VACV-Cop C10L) (Table 1).
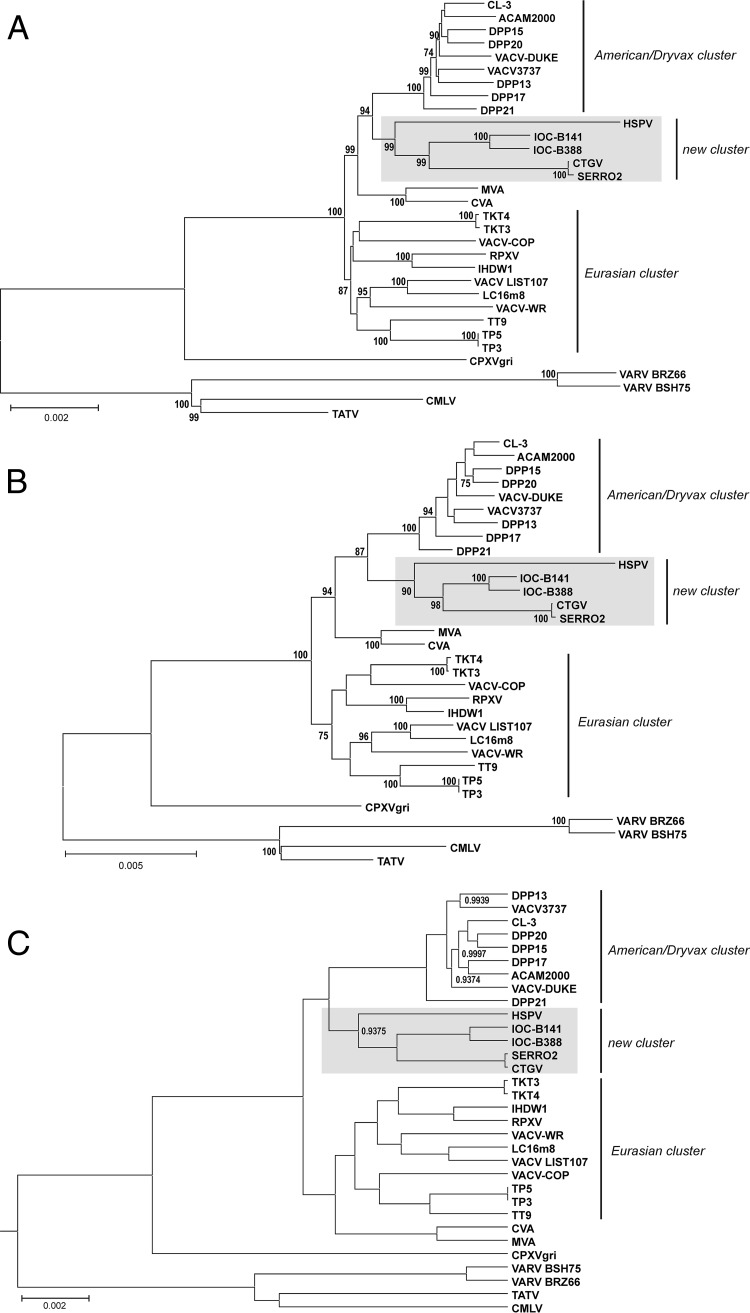
The phylogeny of clones B388 and B141 was inferred based on the multialignment of a conserved region (∼99,000 bp) spanning IOC_072 to IOC_170 orthologs (VACV-Cop F9L to A24R) in different VACV strains and other Orthopoxvirus members. Figure 8 shows that clones B141 and B388 branched as a new cluster within VACV, grouping with horsepox virus (HSPV), Cantagalo virus (CTGV), and Serro 2 virus. HSPV is a VACV-like virus that has the largest genome of the VACV group (212,633 bp) and is suggested to be closely related to an ancestor of the VACV lineage (37, 38). CTGV and Serro 2 virus, collectively called CTGV-like viruses, are field strains of VACV isolated in Brazil during outbreaks of VACV infection in cattle and milkers, respectively (17, 21). CTGV previously has been suggested to be related to VACV-IOC based on the presence of conserved deletions and the sequence of a few genes (17, 22).
FIG 8.
B141 and B388 branch as a novel cluster in VACV phylogeny. The multialignment of a conserved region from 31 Orthopoxvirus genomes (orthologs of VACV-Cop F9L through A24R) was used for phylogenetic inference. (A) Neighbor-joining (NJ) tree based on Kimura 2-parameter substitution model with 1,000 bootstrap replicates. (B) Maximum likelihood (ML) tree based on the Tamura-Nei substitution model, a five-category discrete gamma model with 1,000 bootstrap replicates. NJ and ML trees were constructed using MEGA 6.06, and bootstrap values of >70% are shown next to branch nodes. (C) Maximum clade credibility tree constructed by Bayesian inference (BEAST v. 1.8.1.) with default prior settings, 4 chains of 10 million generations of Markov chain Monte Carlo (MCMC) with tree sampling every 1,000th generation, and a burn-in of 1,500,000 states. Nodes showing posterior probabilities of <1.00 are labeled. (A, B, and C) The scale bars indicate the number of substitutions per site. The VACV clusters are indicated on the right. Gray boxes highlight the new VACV cluster grouping IOC-B141, IOC-B388, CTGV, Serro 2 virus, and HSPV.
The analysis of this novel cluster indicated that the IOC clones and the CTGV-like viruses shared a most recent common ancestor and diverged as separate subgroups after this point of splitting (Fig. 8). This coevolution path after splitting from a common ancestor is compatible with the observation that CTGV-like viruses contain sequences missing from the IOC clones and vice versa. Nonetheless, the genomes of both CTGV and Serro 2 virus (181,774 bp and 184,572 bp, respectively) have an overall greater loss of genetic information than the IOC genomes.
The IOC/CTGV-like branch formed a separate lineage from that of HSPV that mapped as an independent, long-branched subgroup within this new cluster (Fig. 8). Interestingly, maximum-likelihood analysis of whole genomes revealed a smaller estimated distance between IOC clones and HSPV (mean value of 0.01835) than between CTGV-like viruses and HSPV (mean value of 0.02382). Accordingly, a greater number of IOC ORFs than CTGV ORFs had the highest identity scores with HSPV ORFs, as indicated in Table S2 in the supplemental material. In fact, some sequences of the IOC clones revealed a striking similarity to HSPV sequences. For example, a 168-nt region of ORF IOC_003 from B141, but not B388, is virtually identical to the sequence of ORF HSPV_002, including the presence of 4 deletions, differing from all other VACV strains (Fig. 9A). These data suggest that HSPV is more closely related to VACV-IOC than to CTGV-like viruses. Consequently, we can assume that the common ancestor of the IOC/CTGV branch was probably an ancient smallpox vaccine sample related to HSPV that escaped to nature, originating feral VACV (CTGV-like viruses).
FIG 9.
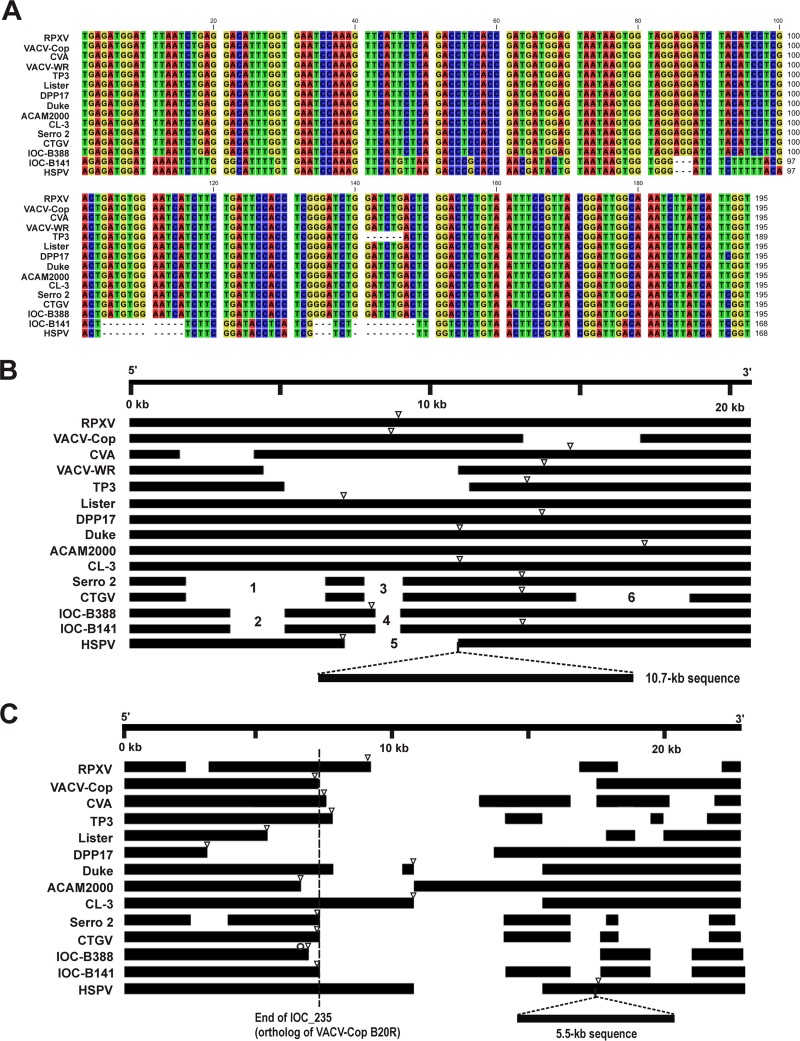
Major differences in the genome terminal regions of B141, B388, other VACV strains, and HSPV. (A) Multialignment of a 168-nt region of orthologs of VACV-Cop C23L (IOC_003). Note that sequences of B141 and HSPV are identical, except for nt 97, and different from all other VACV strains, including B388. (B) Multialignment of the 5′ termini of different VACV genomes. Minor deletions and gaps were omitted in order to highlight major deletion patterns. Deletions marked as 1 to 6 are described in the text. Open triangles indicate the 5′ ITR junction sites in virus genomes. A 10.7-kb deletion, common to all VACV genomes, is represented as an insertion in the HSPV genome to facilitate genome comparison. (C) Scheme of ORF alignment of the 3′ termini based on a blast atlas. Note that, for the sake of clarity, the intergenic regions were removed as well as minor gaps and deletions. Open triangles indicate the 3′ ITR junction sites in virus genomes. The 5.5-kb deletion, common to all VACV, is represented as an insertion in the HSPV genome to facilitate genome comparisons. The dashed line indicates the end of gene IOC_235 (ortholog of VACV-Cop B20R). The open circle indicates the gene IOC_234 (ortholog of VACV-Cop B19R).
This IOC/CTGV/HSPV cluster consistently mapped as the sister group to the Dryvax cluster with high support values using three different reconstruction methods (Fig. 8). Despite that, when fewer Dryvax clones were used for tree reconstructions, HSPV occasionally mapped outside the novel cluster, branching off the base of the Dryvax cluster (data not shown). This tree topology resembled those reported previously by other groups before the IOC and CTGV sequences were available (10–13). This variable relationship between different combinations of Dryvax clones and HSPV probably resulted from inconsistencies previously observed in the Dryvax phylogeny that are related to multiple and past recombination events (11, 39).
Two other features observed in Fig. 8 are in agreement with previous reports: (i) viruses derived from the American Dryvax strain clustered separately from the Eurasian strains, which formed a more dispersed cluster, and (ii) VACV strains WR and IHD-W branched within the Eurasian cluster despite supposedly sharing a common origin with the Dryvax strain, i.e., the New York City Board of Health (NYCBH) strain (1).
The phylogenetic inferences of B141 and B388 were based on the alignment of conserved regions of the virus genomes. Nevertheless, recent reports have shown that the structural analysis of the variable genome ends of VACV also can provide relevant information about virus relationships (10, 13). Therefore, we aligned ∼20 kb of the 5′ (Fig. 9B) and 3′ (Fig. 9C) genome termini of different VACV strains, including the five viruses that grouped in the novel VACV cluster. For the sake of clarity, we focused our analyses on large deletions, omitting small gaps. The analysis of HSPV, IOC B141, IOC B388, CTGV, and Serro 2 virus revealed three unique structural patterns in the genome ends consistent with the division of the phylogenetic cluster into three branches: HSPV, IOC clones, and CTGV-like viruses. Nevertheless, the IOC and CTGV-like patterns are more like each other than HSPV (Fig. 9B and C), as expected based on the tree topologies shown in Fig. 8.
Most 5′-end ORFs of the HSPV genome are present in the B141 and B388 genomes (Fig. 9B) with some exceptions, notably the 10.7-kb stretch of HSPV ORFs (HSPV007-HSPV015b), which in fact is absent from all known VACV strains (13, 37). IOC clones and CTGV-like viruses have distinct deletion patterns in the 5′ end not found in the genomes of other VACV strains included in the analysis, especially the Dryvax-derived viruses DPP17, Duke, ACAM2000, and CL-3. The IOC and CTGV-like genomes share the deletions marked 1 to 4, but CTGV-like viruses show a greater loss of genetic information. Deletions 1 and 2 correspond to ORFs encoding ankyrin-like proteins (fragmented genes). Deletion 3 corresponds to the orthologs of VACV-Cop C15L (near-full-length gene in IOC clones), the surface glycoprotein, and VACV-Cop C14L, whereas deletion 4 includes only the last two genes. The HSPV genome has a larger deletion (marked as 5) in the same region. Deletion 6 is unique to the CTGV genome, which has lost several fragmented genes encoding an ankyrin-like protein, as well as most parts of the fragmented ortholog of VACV-Cop C9L. A detailed blast map comparing the 5′-end ORFs of the viruses grouped in the novel cluster is shown in Fig. S1A in the supplemental material.
The structure of the 3′ end of VACV genomes was revealed to be more complex than the 5′ end, as previously noted by others (10, 13) (Fig. 9C). As indicated in Table 1, the genome of B388 has a 4.3-kb deletion in the 3′ end located downstream of IOC_234 (VACV-Cop B19R) (Fig. 9C, open circle). This deletion is not present in the B141 and CTGV-like genomes and accounts for the shorter ITRs of B388 compared with those of B141 (Fig. 9C). Interestingly, Qin et al. (13) suggested that the junction site of the 3′ ITR is a common genetic feature of VACV strains belonging to the same lineage. Accordingly, clone B141, CTGV, and Serro 2 virus have the same 3′ ITR junction site located at the end of genes IOC_235, CTGV_205, and Serro2_201 (VACV-Cop B20R), respectively (Fig. 9C, open triangles and dashed line). In all three viruses this truncated gene corresponds to 19.5% of the length of an intact B20R (DVX_213 in Dryvax clone DPP25 or HSPV_196, for example).
Except for the 4.3-kb deletion in B388, the IOC clones and CTGV-like genomes have distinct patterns in the 3′ end not found in the other VACV strains, including the Dryvax-derived viruses. Some resemblance to the 3′ end structure of the CVA genome is observed. Nevertheless, only IOC B141, CTGV, and Serro 2 virus share the same 3′ ITR junction site as that mentioned above (Fig. 9B, open triangles and dashed line). Similar to all known VACV strains, the IOC clones and CTGV-like viruses also lack the 5.5-kb sequence corresponding to HSPV200 (surface glycoprotein), an ORF present only in the 3′ end of the HSPV genome. A detailed blast map comparing the 3′-end ORFs of the IOC clones CTGV, Serro 2 virus, and HSPV is shown in Fig. S1B in the supplemental material.
DISCUSSION
VACV-IOC was the seed strain of the smallpox vaccine manufactured by the major vaccine producer in Brazil during the smallpox eradication program (1). However, little is known about the biological and immunological features, as well as the phylogenetic relationships of this first-generation vaccine. In this work, we present a comprehensive characterization of two clones of VACV-IOC. We show evidence that both clones could induce a protective immune response against a lethal infection and had low virulence in infected mice. Most notably, this study provides genetic evidence of a novel, independent cluster of VACV phylogeny formed by VACV-IOC, the Brazilian field strains CTGV and Serro 2 virus, and HSPV. Our data strongly support the hypothesis that CTGV-like viruses represent feral VACV that evolved independently of the VACV-IOC used in the 1970s after splitting from a most recent common ancestor, probably an ancient smallpox vaccine sample related to HSPV. Our data revisit the origins of VACV and propose new evolutionary relationships between ancient and extant strains.
The immunization with clone B141 or B388 conferred protection following a lethal challenge, and the profile of protection was comparable to those of the licensed vaccine ACAM2000 and the parental vaccine strain VACV-IOC. It has been reported that ACAM2000 induces immune responses similar to those of its parental strain, Dryvax, which consequently leads to protection (4). We did not investigate the immunogenicity of the parental VACV-IOC in detail; however, our data suggest that B141 and B388 immunogenicity is comparable to that of VACV-IOC, which was successfully used during the Brazilian smallpox vaccination campaign.
An immune response profile similar to that elicited by B141 and B388 generally is observed following infections with VACV and is well established for other vaccine strains (40–43). The humoral immune response induced by smallpox vaccines has been identified as the main arm of the immune response in the protection against orthopoxvirus reinfections (44, 45). Sera of mice immunized with B141 and B388 showed neutralizing titers (1:29) comparable to those obtained for the first-generation vaccine Dryvax (1:22) (43). Nevertheless, MV neutralizing antibody levels of 1:32 or higher are correlated with protection against smallpox in vaccinees (46). Despite the somewhat low levels of MV neutralizing antibodies, both IOC clones induced a robust neutralization response against extracellular virus and protected mice against a lethal challenge, similar to other VACV strains (40, 47).
Whether cell-mediated immune responses to VACV are essential for protection against a lethal challenge is still controversial (6, 44, 45). Nevertheless, they are key players in controlling primary infection (e.g., vaccination) (48, 49). In this context, immunization with B141 and B388 efficiently stimulated the production of IFN-γ, TNF-α, or IL-2 by CD4+ and CD8+ T cells in response to VACV. T-cell priming also has been reported following immunization with vaccine strains Dryvax, LC18m6, ACAM2000, and Lister (40, 42, 50, 51). More importantly, a percentage of B141- and B388-primed CD4+ or CD8+ T cells exhibited a polyfunctional phenotype and simultaneously expressed IFN-γ in combination with either TNF-α or IL-2. Interestingly, polyfunctional CD4+ T cells, but not CD8+ T cells, were correlated with smaller lesions in vaccinated individuals following revaccination with VACV-Lister (52). On the other hand, MVA primed highly polyfunctional CD8+ T cells in vaccinees after challenge with Dryvax (53). Based on these findings, we suggest that immunization with B141 and B388 induce a protective T-cell response.
Although equally immunogenic, B388 was, to some extent, more virulent to mice than B141, whereas the parental VACV-IOC showed intermediate virulence compared to that of the clones. In this context, it is interesting that the second-generation vaccine ACAM2000 is less virulent than its parental strain, Dryvax, in intrathalamic infection (4). Therefore, our data suggest that, between the two VACV-IOC clones, B141 is a better alternative candidate for a second-generation vaccine that combines low virulence and immunogenicity.
B141 and B388 had similar spread rates and small-plaque phenotypes in cell culture. Therefore, differences in virulence between B141 and B388 might rely on other characteristics, such as the differential expression of virulence genes. Deletions and truncations of virulence genes usually result in VACV attenuation in animal models (54). Interestingly, clones B141 and B388 have several fragmented virulence genes. Among these are orthologs of the VACV-Cop genes C10L, C9L, C6L, C4L, C2L, M1L, A55R, and F1L and VACV-WR gene B13R.
VACV-Cop C10L encodes a protein recently shown to bind Ku subunits of DNA-PK, leading to the inhibition of cytoplasmic DNA sensing. This protein is intact and predicted to be functional in all extant VACV strains except for MVA (55). The truncated C10 protein encoded by IOC_019/020 (and their ITR paralogs) of B141 and B388 probably is not functional because it lacks the C terminus, which is essential for Ku-binding activity (55).
Remarkably, clones B141 and B388 have two fragmented apoptosis-related genes: the orthologs of VACV-WR B13R and VACV-Cop F1L. B13R (IOC_227/228/229) encodes SPI-2 and also is truncated in other VACV strains (VACV-Cop B13R/B14R). On the other hand, F1L (IOC_063/064) is intact in all VACV strains, except for the IOC clones that retain 30% of the N terminus. F1L encodes a Bcl2 protein that binds to and inhibits caspase 9 activation (56) and Bak/Bax-mediated apoptosis (57). The absence of intact orthologs of B13R and F1L indicates that clones B141 and B388 are devoid of essential mechanisms to inhibit apoptosis. Preliminary results show the presence of cleaved poly(ADP-ribose) polymerase during infection of cells with clones B141 and B388 (L. C. Schnellrath and C. R. Damaso, unpublished results).
Despite these genotypic similarities, B141 and B388 differ by a 4.3-kb deletion in the 3′ end of the B388 genome, which is probably not related to the enhanced virulence of B388. On the other hand, clone B388 has full copies of orthologs of K3L and C3L, whereas these genes are fragmented in B141 (IOC_056/057 and IOC_042/043, respectively). K3L is conserved in all other VACV strains and is an important inhibitor of the type I IFN pathway (58). Nevertheless, the deletion of K3L did not lead to VACV attenuation in the mouse model (59). Therefore, the fragmentation of K3L probably does not account for the lower virulence of B141 compared to that of B388. In contrast, the deletion of C3L caused VACV attenuation in infected mice (60). It encodes a secreted complement control protein (VCP) that binds cellular C3b and C4b through SCR domains and inhibits complement-mediated neutralization of the virus particles (61). The truncated VCP encoded by B141 has one SCR domain, in contrast to four domains present in the full VCP of B388, and this difference probably affects the efficacy of B141 to neutralize the complement during infection. Therefore, C3L will be further investigated as a potential promoter of enhanced virulence in B388, in contrast to B141.
The evolutionary relationships and origins of VACV strains are quite nebulous. In this study, phylogenetic inferences indicated that VACV-IOC branches as a new independent VACV cluster together with CTGV-like viruses and, interestingly, with horsepox virus. The IOC clones and CTGV-like viruses map as subgroups that split from a most recent common ancestor. Most notably, HSPV branches into this novel VACV cluster as an independent subgroup. Sequence alignments and analysis of estimated maximum-likelihood distances indicated that HSPV is more closely related to VACV-IOC than to CTGV-like viruses. Therefore, these data suggest that the common ancestor to VACV-IOC and CTGV-like viruses was an ancient smallpox vaccine strain closely related to horsepox virus. We hypothesize that this ancient vaccine was the sample imported to Rio de Janeiro, Brazil, in 1887 and originated the modern VACV-IOC, but it also escaped to nature and originated feral VACV (CTGV-like viruses). In support of this hypothesis, the transit of infected cows from Rio de Janeiro to other states was reported in the late 1890s as an effort to establish vaccine-producing centers in other states (15, 62). Moreover, reports of poxvirus-related infection in dairy cattle have been described since the beginning of the 20th century, and on several occasions they were related to the transmission of smallpox vaccine from vaccinees (22). In some farms, dairy cows were vaccinated with the smallpox vaccine to prevent further spread of the so-called cow-pox to the entire herd (63). An alternative hypothesis to explain the presence of CTGV-like viruses in the Brazilian wilderness has been recently proposed, suggesting the accidental entry of horses or rodents infected with VACV/HSPV in Brazil and the subsequent establishment of the virus in nature (64). However, our work introduces new evidence based on full-genome sequence and bioinformatics analyses that favor the former hypothesis of an ancient vaccine origin for CTGV-like viruses.
The phylogenetic relationship of VACV-IOC and CTGV was first hypothesized when CTGV was isolated during an outbreak of pustular disease in cows in 1999 (17). Previous studies have shown that VACV-IOC and CTGV-like viruses share unique genetic features, such as the 18-nt and 15-nt deletions in the orthologs of the A56R gene and K2L gene, respectively (17, 21, 22). Moreover, these viruses share phenotypic features, such as low spreading in cell culture, low virulence in mice (25, 65), and apoptosis induction (S. Reis, L. C. Schnellrath, N. Moussatche, and C. R. Damaso, unpublished data). Nevertheless, we show here that the panel of fragmented virulence genes differs between the IOC clones and CTGV-like viruses, and some sequences present in CTGV-like viruses are missing from the IOC clones. Despite that, both CTGV and Serro 2 virus have smaller genomes than the IOC clones, indicating a greater loss of genetic information during their divergence and coevolution over time. On the other hand, and more importantly, IOC-B141, CTGV, and Serro 2 virus share the same junction site of the 3′ ITR, which has been suggested to be a genetic feature of VACV lineages (13). Together, these phenotypic and genotypic features support the close phylogenetic relatedness of VACV-IOC and CTGV-like viruses.
It is generally accepted that HSPV is closely related to an ancestor of the modern VACV strains (37). Hence, the phylogenetic relatedness of HSPV and VACV-IOC is intriguing and suggests an ancient evolutionary path for the Brazilian smallpox vaccine. Based on this assumption, it would be expected that the novel cluster would branch off the base of the VACV phylogenetic tree. Nevertheless, this topology is observed only when the simplest hierarchical clustering method (unweighted pair group method with arithmetic means [UPGMA]) was used (data not shown), as also noted by others (10, 37). The analysis of more and older strains of HSPV would certainly contribute to this discussion. Nevertheless, the use of equine pustules as smallpox vaccine is an old practice. Jenner and colleagues reported the production of smallpox vaccine using fluid from vesicular eruptions in horses with “horse-pox” in the early 1800s (38, 66). Therefore, this novel cluster raises interesting issues regarding the origins of VACV strains in general and particularly of the Brazilian VACVs.
Historical records indicate that the establishment of an animal-based vaccine in Brazil was accomplished only in 1887, when samples of calf-lymph vaccine were brought to Rio de Janeiro from the Chambon Institute in Paris, France (15, 16). The French Institute used the Beaugency strain (67), which is suggested to be the origin of the VACV-IOC used in the Brazilian smallpox eradication campaign (J. A. Espmark to D. A. Henderson, correspondence on March 31, 1969, file 88-001-10, Sanofi Pasteur Archives [Connaught Campus]). Interestingly, the VACV-IOC cluster always mapped as a sister group to the American Dryvax cluster but not to the cluster of extant European VACV strains. In support of the relatedness of Dryvax and IOC/CTGV strains, HSPV occasionally mapped with the Dryvax cluster when fewer Dryvax genomes were included in the tree reconstructions (data not shown) or in the absence of IOC/CTGV-like virus genomes (10–13). The Dryvax vaccine produced by Wyeth laboratories supposedly was derived from the New York City Board of Health (NYCBH) strain that was brought from England to the United States in 1856 (1).
In an attempt to reconcile our phylogenetic findings, we searched historical records to gain an understanding of the evolutionary connections between VACV-IOC, HSPV, and Dryvax. In 1866, Ernst Chambon and Gustave Lanoix obtained material from spontaneous cases of “cow-pox” in Beaugency, Loire Valley, France, and used it as the seed strain to produce calf-based smallpox vaccine (animal vaccine) at the recently created Institute de Vaccine Animale in Paris, France. A second natural case of “cow-pox” occurred later in 1866 in St. Mandé, suburbs of Paris, and the samples were mixed without changing the “Beaugency” designation. The success of the Beaugency stock led to its distribution to Belgium, Germany, Sweden, Switzerland, French colonies, and the Americas in the next decades (67). In the meantime, spontaneous cases of “horse-pox” were also documented in France from 1860 to the 1880s, and vesicular pustules from infected horses were used as seed material for the production of smallpox vaccine by different French producers (68). The Beaugency strain was introduced into the United States in 1870 by Henry Martin of Boston, who established a private animal vaccine farm. The Beaugency lymph then was widely distributed to several cities, including New York (obtained by Frank Foster, director of the New York Dispensary) (69–71) and Philadelphia, where the Wyeth brothers started an animal vaccine farm in 1885. In the same year, the Wyeth brothers obtained a fresh seed of Beaugency lymph from the Vaccinal Institute in Belgium (71). Interestingly, the NYCBH apparently used different sources of vaccine virus over the years. The annual report of the NYCBH in 1872 indicates the establishment of the animal vaccine in 1871, using in most animals a bovine sample obtained from the Practical Institute for Animal Vaccine in Cuba and Puerto Rico. A few animals were double infected with more scarification points of this Cuban sample and a few points of the Beaugency strain provided by Frank Foster (72). However, in 1874 the NYCBH reported the simultaneous use of humanized (Jennerian) and calf-based vaccines. The former derived from the vaccine stocks of J. P. Loines, who obtained the samples from England in 1856, and the latter were derived from the bovine stocks supposedly produced since 1871 (73). If all these historical records are accurate regarding the strain names and sources, it is possible that the Dryvax vaccine is derived from the Beaugency strain, similar to VACV-IOC, and not (or at least not only) from the NYCBH strain as widely assumed. This hypothesis would account for the relatedness of VACV-IOC and Dryvax and also the mapping of the NYCBH-derived strains WR and IHD-W in a different cluster apart from the Dryvax viruses.
However, it is important to note that the second half of the 19th century was a period of intense exchange and distribution of European smallpox vaccine lymph of different origins among European countries, the United States, and throughout the world (1, 66, 67, 69). Therefore, our data favor the interesting hypothesis proposed by Evans's group that a heterogeneous ancestor stock of VACV would be the origin of the genetic diversity found in the extant VACV strains (13). Furthermore, our work adds new information to this hypothesis, suggesting that natural cases of so-called horse-pox and cow-pox, as well as the indiscriminate mixture of samples, probably contributed to increased heterogeneity of the subsequent VACV stocks that were randomly sampled during the 19th century.
Supplementary Material
ACKNOWLEDGMENTS
We thank Steven Palmer (University of Windsor, Ontario, Canada) and Christopher Rutty (Sanofi Pasteur Limited, Connaught Campus) for consulting the Connaught Archives and sharing the correspondence from J. A. Espmark to D. A. Henderson. We thank Richard Condit (University of Florida) for helpful discussions and carefully reading the manuscript, Bruno Paredes and Emiliano Medei (Centro Nacional de Biologia Estrutural e Bioimagem–CENABIO-UFRJ) for their support with flow cytometry and the use of the facilities, and Chwang Hong Foo, Gary Cohen, and Geoffrey Smith for antibodies.
This work was supported by grants from CNPq, Faperj, CAPES, and Ministério da Defesa to C.R.D. M.L.G.M. was the recipient of a fellowship from CNPq and is the recipient of a fellowship from Faperj. C.G.O.L. is the recipient of a Faperj fellowship; C.R.D. is the recipient of a research fellowship from CNPq.
The findings and conclusions in this report are those of the author(s) and do not necessarily represent the views of the Centers for Disease Control and Prevention.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.01833-15.
REFERENCES
- 1.Fenner F, Henderson DA, Arita I, Jezek Z, Ladnyi ID. 1988. Smallpox and its eradication. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 2.Rutty C. 2008. Canadian vaccine research, production and international regulation: Connaught Laboratories and smallpox vaccines,1962–1980, p 280–289. In Kroker K, Kealan J, Mazumdar PM (ed), Crafting immunity: working histories of clinical immunology. Ashgate Publishing, Ltd., Burlington, Vermont. [Google Scholar]
- 3.Damon IK, Damaso CR, McFadden G. 2014. Are we there yet? The smallpox research agenda using variola virus. PLoS Pathog 10:e1004108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Monath TP, Caldwell JR, Mundt W, Fusco J, Johnson CS, Buller M, Liu J, Gardner B, Downing G, Blum PS, Kemp T, Nichols R, Weltzin R. 2004. ACAM2000 clonal Vero cell culture vaccinia virus (New York City Board of Health strain)–a second-generation smallpox vaccine for biological defense. Int J Infect Dis 8(Suppl 2):S31–S44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sanchez-Sampedro L, Perdiguero B, Mejias-Perez E, Garcia-Arriaza J, Di Pilato M, Esteban M. 2015. The evolution of poxvirus vaccines. Viruses 7:1726–1803. doi: 10.3390/v7041726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jacobs BL, Langland JO, Kibler KV, Denzler KL, White SD, Holechek SA, Wong S, Huynh T, Baskin CR. 2009. Vaccinia virus vaccines: past, present and future. Antiviral Res 84:1–13. doi: 10.1016/j.antiviral.2009.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moss B. 2011. Smallpox vaccines: targets of protective immunity. Immunol Rev 239:8–26. doi: 10.1111/j.1600-065X.2010.00975.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garcel A, Perino J, Crance JM, Drillien R, Garin D, Favier AL. 2009. Phenotypic and genetic diversity of the traditional Lister smallpox vaccine. Vaccine 27:708–717. doi: 10.1016/j.vaccine.2008.11.063. [DOI] [PubMed] [Google Scholar]
- 9.Osborne JD, Da Silva M, Frace AM, Sammons SA, Olsen-Rasmussen M, Upton C, Buller RM, Chen N, Feng Z, Roper RL, Liu J, Pougatcheva S, Chen W, Wohlhueter RM, Esposito JJ. 2007. Genomic differences of vaccinia virus clones from Dryvax smallpox vaccine: the Dryvax-like ACAM2000 and the mouse neurovirulent clone-3. Vaccine 25:8807–8832. doi: 10.1016/j.vaccine.2007.10.040. [DOI] [PubMed] [Google Scholar]
- 10.Qin L, Liang M, Evans DH. 2013. Genomic analysis of vaccinia virus strain TianTan provides new insights into the evolution and evolutionary relationships between orthopoxviruses. Virology 442:59–66. doi: 10.1016/j.virol.2013.03.025. [DOI] [PubMed] [Google Scholar]
- 11.Qin L, Upton C, Hazes B, Evans DH. 2011. Genomic analysis of the vaccinia virus strain variants found in Dryvax vaccine. J Virol 85:13049–13060. doi: 10.1128/JVI.05779-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Q, Tian M, Feng Y, Zhao K, Xu J, Liu Y, Shao Y. 2013. Genomic sequence and virulence of clonal isolates of vaccinia virus Tiantan, the Chinese smallpox vaccine strain. PLoS One 8:e60557. doi: 10.1371/journal.pone.0060557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qin L, Favis N, Famulski J, Evans DH. 2015. Evolution of and evolutionary relationships between extant vaccinia virus strains. J Virol 89:1809–1824. doi: 10.1128/JVI.02797-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Palmer S, Hochman G, Arbex D. 2010. Smallpox eradication, laboratory visits, and a touch of tourism: travel notes of a Canadian scientist in Brazil. Hist Cienc Saude Manguinhos 17:777–790. doi: 10.1590/S0104-59702010000300012. [DOI] [PubMed] [Google Scholar]
- 15.Fernandes TM. 1999. Vacina antivariólica: ciência, técnica e o poder dos homens, 1808–1920. Editora Fiocruz, Rio de Janeiro, Brazil. [Google Scholar]
- 16.Hochman G. 2009. Priority, invisibility and eradication: the history of smallpox and the Brazilian public health agenda. Med Hist 53:229–252. doi: 10.1017/S002572730000020X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Damaso CR, Esposito JJ, Condit RC, Moussatche N. 2000. An emergent poxvirus from humans and cattle in Rio de Janeiro State: Cantagalo virus may derive from Brazilian smallpox vaccine. Virology 277:439–449. doi: 10.1006/viro.2000.0603. [DOI] [PubMed] [Google Scholar]
- 18.Leite JA, Drumond BP, Trindade GS, Lobato ZI, da Fonseca FG, dos SJ, Madureira MC, Guedes MI, Ferreira JM, Bonjardim CA, Ferreira PC, Kroon EG. 2005. Passatempo virus, a vaccinia virus strain, Brazil. Emerg Infect Dis 11:1935–1938. doi: 10.3201/eid1112.050773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Medaglia ML, Pessoa LC, Sales ER, Freitas TR, Damaso CR. 2009. Spread of Cantagalo virus to northern Brazil. Emerg Infect Dis 15:1142–1143. doi: 10.3201/eid1507.081702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Quixabeira-Santos JC, Medaglia ML, Pescador CA, Damaso CR. 2011. Animal movement and establishment of vaccinia virus Cantagalo strain in Amazon biome, Brazil. Emerg Infect Dis 17:726–729. doi: 10.3201/eid1704.101581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Trindade GS, Guedes MI, Drumond BP, Mota BE, Abrahao JS, Lobato ZI, Gomes JA, Correa-Oliveira R, Nogueira ML, Kroon EG, Da Fonseca FG. 2009. Zoonotic vaccinia virus: clinical and immunological characteristics in a naturally infected patient. Clin Infect Dis 48:e37–40. doi: 10.1086/595856. [DOI] [PubMed] [Google Scholar]
- 22.Moussatche N, Damaso CR, McFadden G. 2008. When good vaccines go wild: feral orthopoxvirus in developing countries and beyond. J Infect Dev Ctries 2:156–173. [DOI] [PubMed] [Google Scholar]
- 23.Afonso PP, Silva PM, Schnellrath LC, Jesus DM, Hu J, Yang Y, Renne R, Attias M, Condit RC, Moussatche N, Damaso CR. 2012. Biological characterization and next-generation genome sequencing of the unclassified Cotia virus SPAn232 (Poxviridae). J Virol 86:5039–5054. doi: 10.1128/JVI.07162-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moussatche N, Keller SJ. 1991. Phosphorylation of vaccinia virus core proteins during transcription in vitro. J Virol 65:2555–2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Santos-Fernandes E, Beltrame CO, Byrd CM, Cardwell KB, Schnellrath LC, Medaglia ML, Hruby DE, Jordan R, Damaso CR. 2013. Increased susceptibility of Cantagalo virus to the antiviral effect of ST-246(R). Antiviral Res 97:301–311. doi: 10.1016/j.antiviral.2012.11.010. [DOI] [PubMed] [Google Scholar]
- 26.National Research Council. 2011. Guide for the care and use of laboratory animals, 8th ed National Academies Press, Washington, DC. [Google Scholar]
- 27.Marques ET Jr, Chikhlikar P, de Arruda LB, Leao IC, Lu Y, Wong J, Chen JS, Byrne B, August JT. 2003. HIV-1 p55Gag encoded in the lysosome-associated membrane protein-1 as a DNA plasmid vaccine chimera is highly expressed, traffics to the major histocompatibility class II compartment, and elicits enhanced immune responses. J Biol Chem 278:37926–37936. doi: 10.1074/jbc.M303336200. [DOI] [PubMed] [Google Scholar]
- 28.Barbosa AV, Medaglia ML, Soares HS, Quixabeira-Santos JC, Gennari SM, Damaso CR. 2014. Presence of neutralizing antibodies to Orthopoxvirus in Capybaras (Hydrochoerus hydrochaeris) in Brazil. J Infect Dev Ctries 8:1646–1649. [DOI] [PubMed] [Google Scholar]
- 29.Arruda LB, Sim D, Chikhlikar PR, Maciel M Jr, Akasaki K, August JT, Marques ET. 2006. Dendritic cell-lysosomal-associated membrane protein (LAMP) and LAMP-1-HIV-1 gag chimeras have distinct cellular trafficking pathways and prime T and B cell responses to a diverse repertoire of epitopes. J Immunol 177:2265–2275. doi: 10.4049/jimmunol.177.4.2265. [DOI] [PubMed] [Google Scholar]
- 30.Li H, Durbin R. 2010. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tcherepanov V, Ehlers A, Upton C. 2006. Genome Annotation Transfer Utility (GATU): rapid annotation of viral genomes using a closely related reference genome. BMC Genomics 7:150. doi: 10.1186/1471-2164-7-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Drummond AJ, Suchard MA, Xie D, Rambaut A. 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol 29:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith GL, Law M. 2004. The exit of vaccinia virus from infected cells. Virus Res 106:189–197. doi: 10.1016/j.virusres.2004.08.015. [DOI] [PubMed] [Google Scholar]
- 37.Tulman ER, Delhon G, Afonso CL, Lu Z, Zsak L, Sandybaev NT, Kerembekova UZ, Zaitsev VL, Kutish GF, Rock DL. 2006. Genome of horsepox virus. J Virol 80:9244–9258. doi: 10.1128/JVI.00945-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baxby D. 1996. Should smallpox virus be destroyed? The relevance of the origins of vaccinia virus. Soc Hist Med 9:117–119. [DOI] [PubMed] [Google Scholar]
- 39.Smithson C, Kampman S, Hetman BM, Upton C. 2014. Incongruencies in vaccinia virus phylogenetic trees. Computation 2:182–198. doi: 10.3390/computation2040182. [DOI] [Google Scholar]
- 40.Berhanu A, King DS, Mosier S, Jordan R, Jones KF, Hruby DE, Grosenbach DW. 2010. Impact of ST-246 on ACAM2000 smallpox vaccine reactogenicity, immunogenicity, and protective efficacy in immunodeficient mice. Vaccine 29:289–303. doi: 10.1016/j.vaccine.2010.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Melamed S, Paran N, Katz L, Ben-Nathan D, Israely T, Schneider P, Levin R, Lustig S. 2007. Tail scarification with vaccinia virus Lister as a model for evaluation of smallpox vaccine potency in mice. Vaccine 25:7743–7753. doi: 10.1016/j.vaccine.2007.09.023. [DOI] [PubMed] [Google Scholar]
- 42.Meseda CA, Mayer AE, Kumar A, Garcia AD, Campbell J, Listrani P, Manischewitz J, King LR, Golding H, Merchlinsky M, Weir JP. 2009. Comparative evaluation of the immune responses and protection engendered by LC16m8 and Dryvax smallpox vaccines in a mouse model. Clin Vaccine Immunol 16:1261–1271. doi: 10.1128/CVI.00040-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meseda CA, Garcia AD, Kumar A, Mayer AE, Manischewitz J, King LR, Golding H, Merchlinsky M, Weir JP. 2005. Enhanced immunogenicity and protective effect conferred by vaccination with combinations of modified vaccinia virus Ankara and licensed smallpox vaccine Dryvax in a mouse model. Virology 339:164–175. doi: 10.1016/j.virol.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 44.Panchanathan V, Chaudhri G, Karupiah G. 2010. Antiviral protection following immunization correlates with humoral but not cell-mediated immunity. Immunol Cell Biol 88:461–467. doi: 10.1038/icb.2009.110. [DOI] [PubMed] [Google Scholar]
- 45.Xu R, Johnson AJ, Liggitt D, Bevan MJ. 2004. Cellular and humoral immunity against vaccinia virus infection of mice. J Immunol 172:6265–6271. doi: 10.4049/jimmunol.172.10.6265. [DOI] [PubMed] [Google Scholar]
- 46.Mack TM, Noble J Jr, Thomas DB. 1972. A prospective study of serum antibody and protection against smallpox. Am J Trop Med Hyg 21:214–218. [DOI] [PubMed] [Google Scholar]
- 47.Jentarra GM, Heck MC, Youn JW, Kibler K, Langland JO, Baskin CR, Ananieva O, Chang Y, Jacobs BL. 2008. Vaccinia viruses with mutations in the E3L gene as potential replication-competent, attenuated vaccines: scarification vaccination. Vaccine 26:2860–2872. doi: 10.1016/j.vaccine.2008.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Belyakov IM, Earl P, Dzutsev A, Kuznetsov VA, Lemon M, Wyatt LS, Snyder JT, Ahlers JD, Franchini G, Moss B, Berzofsky JA. 2003. Shared modes of protection against poxvirus infection by attenuated and conventional smallpox vaccine viruses. Proc Natl Acad Sci U S A 100:9458–9463. doi: 10.1073/pnas.1233578100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gordon SN, Cecchinato V, Andresen V, Heraud JM, Hryniewicz A, Parks RW, Venzon D, Chung HK, Karpova T, McNally J, Silvera P, Reimann KA, Matsui H, Kanehara T, Shinmura Y, Yokote H, Franchini G. 2011. Smallpox vaccine safety is dependent on T cells and not B cells. J Infect Dis 203:1043–1053. doi: 10.1093/infdis/jiq162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Grosenbach DW, Jordan R, King DS, Berhanu A, Warren TK, Kirkwood-Watts DL, Tyavanagimatt S, Tan Y, Wilson RL, Jones KF, Hruby DE. 2008. Immune responses to the smallpox vaccine given in combination with ST-246, a small-molecule inhibitor of poxvirus dissemination. Vaccine 26:933–946. doi: 10.1016/j.vaccine.2007.11.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ferrier-Rembert A, Drillien R, Meignier B, Garin D, Crance JM. 2007. Safety, immunogenicity and protective efficacy in mice of a new cell-cultured Lister smallpox vaccine candidate. Vaccine 25:8290–8297. doi: 10.1016/j.vaccine.2007.09.050. [DOI] [PubMed] [Google Scholar]
- 52.Puissant-Lubrano B, Bossi P, Gay F, Crance JM, Bonduelle O, Garin D, Bricaire F, Autran B, Combadiere B. 2010. Control of vaccinia virus skin lesions by long-term-maintained IFN-gamma+ TNF-alpha+ effector/memory CD4+ lymphocytes in humans. J Clin Investig 120:1636–1644. doi: 10.1172/JCI38506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Precopio ML, Betts MR, Parrino J, Price DA, Gostick E, Ambrozak DR, Asher TE, Douek DC, Harari A, Pantaleo G, Bailer R, Graham BS, Roederer M, Koup RA. 2007. Immunization with vaccinia virus induces polyfunctional and phenotypically distinctive CD8(+) T cell responses. J Exp Med 204:1405–1416. doi: 10.1084/jem.20062363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Perdiguero B, Esteban M. 2009. The interferon system and vaccinia virus evasion mechanisms. J Interferon Cytokine Res 29:581–598. doi: 10.1089/jir.2009.0073. [DOI] [PubMed] [Google Scholar]
- 55.Peters NE, Ferguson BJ, Mazzon M, Fahy AS, Krysztofinska E, Arribas-Bosacoma R, Pearl LH, Ren H, Smith GL. 2013. A mechanism for the inhibition of DNA-PK-mediated DNA sensing by a virus. PLoS Pathog 9:e1003649. doi: 10.1371/journal.ppat.1003649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhai D, Yu E, Jin C, Welsh K, Shiau CW, Chen L, Salvesen GS, Liddington R, Reed JC. 2010. Vaccinia virus protein F1L is a caspase-9 inhibitor. J Biol Chem 285:5569–5580. doi: 10.1074/jbc.M109.078113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Taylor JM, Quilty D, Banadyga L, Barry M. 2006. The vaccinia virus protein F1L interacts with Bim and inhibits activation of the pro-apoptotic protein Bax. J Biol Chem 281:39728–39739. doi: 10.1074/jbc.M607465200. [DOI] [PubMed] [Google Scholar]
- 58.Beattie E, Tartaglia J, Paoletti E. 1991. Vaccinia virus-encoded eIF-2 alpha homolog abrogates the antiviral effect of interferon. Virology 183:419–422. doi: 10.1016/0042-6822(91)90158-8. [DOI] [PubMed] [Google Scholar]
- 59.Rice AD, Turner PC, Embury JE, Moldawer LL, Baker HV, Moyer RW. 2011. Roles of vaccinia virus genes E3L and K3L and host genes PKR and RNase L during intratracheal infection of C57BL/6 mice. J Virol 85:550–567. doi: 10.1128/JVI.00254-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Isaacs SN, Kotwal GJ, Moss B. 1992. Vaccinia virus complement-control protein prevents antibody-dependent complement-enhanced neutralization of infectivity and contributes to virulence. Proc Natl Acad Sci U S A 89:628–632. doi: 10.1073/pnas.89.2.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Girgis NM, Dehaven BC, Fan X, Viner KM, Shamim M, Isaacs SN. 2008. Cell surface expression of the vaccinia virus complement control protein is mediated by interaction with the viral A56 protein and protects infected cells from complement attack. J Virol 82:4205–4214. doi: 10.1128/JVI.02426-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Teixeira LA, Almeida M. 2003. The beginnings of the smallpox vaccine in São Paulo: a little-known story. Hist Cienc Saude Manguinhos 10:23. [PubMed] [Google Scholar]
- 63.Vellini LL. 1953. A varíola bovina (cowpox) no Estado de São Paulo e sua transmissão ao homem. Hospital 44:247–252. [PubMed] [Google Scholar]
- 64.Shchelkunov SN. 2013. An increasing danger of zoonotic orthopoxvirus infections. PLoS Pathog 9:e1003756. doi: 10.1371/journal.ppat.1003756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ferreira JM, Drumond BP, Guedes MI, Pascoal-Xavier MA, Almeida-Leite CM, Arantes RM, Mota BE, Abrahao JS, Alves PA, Oliveira FM, Ferreira PC, Bonjardim CA, Lobato ZI, Kroon EG. 2008. Virulence in murine model shows the existence of two distinct populations of Brazilian vaccinia virus strains. PLoS One 3:e3043. doi: 10.1371/journal.pone.0003043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Edwardes EJ. 1902. A concise history of small-pox and vaccination in Europe. H. K. Lewis, London, United Kingdom. [Google Scholar]
- 67.Copeman SM. 1899. Vaccination; its natural history and pathology. Macmillan and Co., London, United Kingdom. [Google Scholar]
- 68.Crookshank EM. 1889. History and pathology of vaccination. P. Blackiston, Philadelphia, PA. [Google Scholar]
- 69.Lindsley CA. 1882. Vaccination. The case. Lockwood & Brainard Co., Hartford, CT. [Google Scholar]
- 70.Foster FP. 1872. A report on animal vaccination at the New-York dispensary, in the year 1871: presented to the board of trustees by Frank P. Foster. John W. Amerman, New York, NY. [Google Scholar]
- 71.Willrich M. 2011. Pox: an American history. The Penguin Press, New York, NY. [Google Scholar]
- 72.NYCBH. 1872. Second annual report of the Board of Health of the Health Department of the City of New York, vol 1871/1872 New York Printing Co., New York, NY. [Google Scholar]
- 73.NYCBH. 1876. Fifth and sixth annual report of the Board of Health of the Health Department of the City of New York, vol s1874/1875 New York Printing Co., New York, NY. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.