ABSTRACT
Tetherin is an interferon-inducible restriction factor targeting a broad range of enveloped viruses. Its antiviral activity depends on an unusual topology comprising an N-terminal transmembrane domain (TMD) followed by an extracellular coiled-coil region and a C-terminal glycosylphosphatidylinositol (GPI) anchor. One of the two membrane anchors is inserted into assembling virions, while the other remains in the plasma membrane of the infected cell. Thus, tetherin entraps budding viruses by physically bridging viral and cellular membranes. Although tetherin restricts the release of a large variety of diverse human and animal viruses, only mammalian orthologs have been described to date. Here, we examined the evolutionary origin of this protein and demonstrate that tetherin orthologs are also found in fish, reptiles, and birds. Notably, alligator tetherin efficiently blocks the release of retroviral particles. Thus, tetherin emerged early during vertebrate evolution and acquired its antiviral activity before the mammal/reptile divergence. Although there is only limited sequence homology, all orthologs share the typical topology. Two unrelated proteins of the slime mold Dictyostelium discoideum also adopt a tetherin-like configuration with an N-terminal TMD and a C-terminal GPI anchor. However, these proteins showed no evidence for convergent evolution and failed to inhibit virion release. In summary, our findings demonstrate that tetherin emerged at least 450 million years ago and is more widespread than previously anticipated. The early evolution of antiviral activity together with the high topology conservation but low sequence homology suggests that restriction of virus release is the primary function of tetherin.
IMPORTANCE The continuous arms race with viruses has driven the evolution of a variety of cell-intrinsic immunity factors that inhibit different steps of the viral replication cycle. One of these restriction factors, tetherin, inhibits the release of newly formed progeny virions from infected cells. Although tetherin targets a broad range of enveloped viruses, including retro-, filo-, herpes-, and arenaviruses, the evolutionary origin of this restriction factor and its antiviral activity remained obscure. Here, we examined diverse vertebrate genomes for genes encoding cellular proteins that share with tetherin the highly unusual combination of an N-terminal transmembrane domain and a C-terminal glycosylphosphatidylinositol anchor. We show that tetherin orthologs are found in fish, reptiles, and birds and demonstrate that alligator tetherin efficiently inhibits the release of retroviral particles. Our findings identify tetherin as an evolutionarily ancient restriction factor and provide new important insights into the continuous arms race between viruses and their hosts.
INTRODUCTION
Viruses have most likely existed since the first living cells emerged and infect species from all three domains of life, i.e., archaea, bacteria, and eukaryotes (1). During hundreds of millions of years, cellular organisms have evolved sophisticated and highly diversified antiviral defense strategies to secure their survival. Even prokaryotic bacteria and archaea are able to defend themselves against invading viruses by degrading viral nucleic acids using restriction enzymes (2) and clustered regularly interspaced short palindromic repeats (CRISPR) (3). With the evolution of multicellular eukaryotic organisms more than 600 million years ago (mya), the variety of antiviral defense mechanisms has significantly expanded. The immune system of higher organisms is classically divided into two branches: the innate immune system, which recognizes and counteracts pathogens in a generic and unspecific way, and the adaptive immune system, which is antigen specific and may confer long-lasting immunity against certain pathogens. Whereas the classical adaptive immune system evolved in jawed fish and is unique to vertebrates (4), the innate immune system is substantially older. Effector mechanisms such as the generation of reactive oxygen or nitrogen species, antimicrobial peptides, complement-like proteins, or cytokines can already be found in ancient invertebrates such as arthropods, mollusks, and cnidarians (5–9).
In recent years, several “intrinsic immunity” or “host restriction” factors have been identified (10). These antiviral proteins are not unambiguously defined but usually share several characteristics. For example, their expression is often upregulated by interferons and they represent a first line of defense against viral infections (11). Furthermore, restriction factors often inhibit specific steps of the viral replication cycle by targeting conserved viral components. As a result of the continuous evolutionary arms race between viruses and their hosts, most restriction factors show signatures of positive selection and act in a species-specific manner (12).
Four well-characterized antiretroviral restriction factors are TRIM5α (tripartite motif 5α), APOBEC3G (apolipoprotein B mRNA-editing enzyme, catalytic polypeptide-like 3G), SAMHD1 (sterile alpha motif and histidine/aspartic acid domain-containing protein 1), and tetherin. TRIM5α induces the untimely uncoating of the incoming retroviral capsid (13), APOBEC3G is a deaminase introducing lethal hypermutations into the viral genome (14), SAMHD1 inhibits reverse transcription by depleting intracellular deoxynucleoside triphosphate (dNTP) pools (15, 16), and tetherin restricts the release of newly formed virions from infected cells (17, 18). Whereas the antiviral activities of TRIM5α, APOBEC3G, and SAMHD1 are largely restricted to retroviruses, tetherin inhibits diverse enveloped viruses, including retro-, filo-, rhabdo-, herpes-, arena-, flavi-, corona-, and paramyxoviruses (17, 19–27). This broad activity can be attributed to its ability to target viral membranes instead of a specific viral protein (28). Tetherin has an unusual topology. It comprises two membrane anchors, an α-helical transmembrane domain (TMD) and a C-terminal glycosylphosphatidylinositol (GPI) anchor, which are linked by an extracellular coiled-coil domain (29). Budding virions incorporate one of these two membrane anchors, whereas the other one remains embedded in the membrane of the cell (30). Thus, tetherin inhibits the release of newly formed virions by directly linking viral and cellular membranes (28). Although tetherin has broad activity against different human and animal viruses, only mammalian orthologs have been described. Here, we investigated the deep evolutionary origin of this antiviral protein to further elucidate its role in intrinsic antiviral immunity.
MATERIALS AND METHODS
Expression plasmids.
Tetherin and ponticulin genes were cloned into the cytomegalovirus (CMV) promoter-based pCG expression vector via XbaI and MluI (31). An internal ribosome entry site (IRES) enhanced green fluorescent protein (eGFP) or IRES DsRed cassette was inserted via BamHI so that the gene of interest was expressed together with the fluorophore from a single bicistronic mRNA. The ponticulin A and B genes as well as the coelacanth and alligator orthologs of tetherin were codon optimized to enhance expression in human cells. In some experiments, FLAG-tagged variants of tetherin or ponticulin were used. The PIG-L expression vector (pMEEB-PIG-L-FLAG) was kindly provided by Taroh Kinoshita (32).
Proviral constructs.
Virion release was quantified using a vpu-deficient mutant of human immunodeficiency virus type 1 (HIV-1) group M NL4-3 that has been described previously (33). The lack of vpu renders this virus susceptible to inhibition by human tetherin. The V3 region of 92TH014-12 env was introduced to render this virus R5 tropic (34).
Cell culture and transfections.
Human embryonic kidney 293T (HEK293T) cells (obtained from the American Type Culture Collection [ATCC]) were first described by DuBridge et al. (35). They were maintained in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal calf serum (FCS) and 2 mM glutamine, streptomycin (120 mg/ml), and penicillin (120 mg/ml). The Chinese hamster ovary (CHO) IIIB2A cell line and its PIG-L-deficient derivative (kindly provided by Taroh Kinoshita) were first described by Nakamura and colleagues (32). These cell lines were generated by stably transfecting CHO-K1 cells with expression vectors for DAF and CD59. They were cultured in a mixture of 75% supplemented DMEM and 25% supplemented Ham's F12 medium. HEK293T and CHO cells were transfected using the calcium phosphate method and Lipofectamine LTX, respectively.
Western blotting.
To monitor expression of tetherin and ponticulin, transfected HEK293T cells were lysed in TMPER and M-PER buffer (Thermo Scientific) 2 days posttransfection, respectively. Cell lysates were separated in 4% to 12% Bis-Tris gels (Invitrogen) and transferred to polyvinylidene difluoride (PVDF) membranes. Blots were probed with antibodies against the FLAG tag (F1804; Sigma). For internal controls, blots were incubated with antibodies specific for β-actin (8227; abcam) and GFP (ab290; abcam). Subsequently, membranes were incubated with anti-mouse or anti-rabbit IRDye Odyssey antibodies and proteins were detected using a Li-COR Odyssey scanner.
Flow cytometry.
To determine protein levels at the cell surface, cells were transfected with 5 μg (HEK293T, calcium phosphate, 6 wells) or 1 μg (CHO, Lipofectamine LTX, 12 wells) of vectors coexpressing eGFP and the respective gene. Two days posttransfection, cells were stained extracellularly with an antibody against the FLAG tag (F1804; Sigma) and a secondary allophycocyanin (APC)-conjugated anti-mouse antibody (A-865; Invitrogen). Fluorescence was detected by two-color flow cytometry, and surface expression levels of eGFP-positive cells were calculated.
Immunofluorescence microscopy.
To analyze the subcellular localization of tetherin, HEK293T or CHO cells were seeded in ibidi 8-well μ-slides and transfected with 0.25 μg of vectors expressing the respective gene using Lipofectamine LTX. At 16 h later, cells were stained with an antibody against the FLAG tag (F1804; Sigma) and a secondary Alexa Fluor 647-conjugated anti-mouse antibody (A21237; Life Technologies). Nuclei were stained with Hoechst 33342 (H1399; Life Technologies). HEK293T cells were additionally stained for the trans-Golgi network using an anti-human TGN46 antibody (AHP500GT; Serotec) and a secondary Alexa Fluor 488-conjugated anti-sheep antibody (A11015; Life Technologies). Cells were analyzed using a confocal laser scanning microscope (LSM 710; Zeiss) with the corresponding software (Zeiss Zen Software [2010]).
Virus release assay.
To determine tetherin-mediated restriction of virion release, HEK293T cells were seeded in 6-well plates and transfected with 5 μg of a proviral construct and increasing amounts of a plasmid coexpressing tetherin or ponticulin. At 40 h posttransfection, cells and supernatants were lysed in Triton X-100 and the relative levels of p24 release were determined using a homemade p24 enzyme-linked immunosorbent assay (ELISA).
Topology prediction software.
TTMHMM Server v. 2.0 (http://www.cbs.dtu.dk/services/TMHMM/), PSIPRED v3.3 (http://bioinf.cs.ucl.ac.uk/psipred/), COILS/PCOILS (http://toolkit.tuebingen.mpg.de/pcoils/), and PredGPI (http://gpcr.biocomp.unibo.it/predgpi/pred.htm) were used to predict the presence and localization of transmembrane domains, β-sheet secondary structures, coiled-coil domains, and GPI anchor addition omega sites, respectively.
Statistical analyses.
Statistical calculations were performed with a two-tailed unpaired Student's t test or a one-sample t test using Graph Pad Prism 5.03. P values of <0.05 were considered statistically significant.
Nucleotide sequence accession numbers.
The GenBank accession numbers for the tetherin ortholog sequence data reported in this article are listed in Table 1.
TABLE 1.
Tetherin orthologs from mammals, reptiles, and fishesa
| Accession no. | Species | Exon length (nt) |
Protein length (aa) | % GPI anchor probability | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Exon 1 | Exon 2 | Exon 3 | Exon 4 | Exon 5 | Exon 6 | Exon 7 | ||||
| NP_004326 | Homo sapiens (human) | 285 | 67 | 61 | 130 | 180 | 99.4 | |||
| NP_001231044 | Cricetulus griseus (Chinese hamster) | 288 | 181 | 40 | 103 | 203 | 99.9 | |||
| XP_007489270 | Monodelphis domestica (gray short-tailed opossum) | 246 | 55 | 61 | 100 | 153 | 100.00 | |||
| XP_012399618 | Sarcophilus harrisiia (Tasmanian devil) | 219 | 76 | 61 | 97 | 150 | 100.00 | |||
| XP_006017475 (isoform X1) | Alligator sinensis (Chinese alligator) | 216 | 97 | 49 | 130 | 163 | 99.9 | |||
| XP_006017476 (isoform X2) | Alligator sinensis (Chinese alligator) | 186 | 88 | 49 | 130 | 150 | 99.9 | |||
| XP_008169758 (isoform X1) | Chrysemis picta bellii (Western painted turtle) | 321 | 97 | 82 | 151 | 216 | 100.0 | |||
| XP_008169759 (isoform X2) | Chrysemis picta bellii (Western painted turtle) | 321 | 85 | 49 | 154 | 202 | 99.9 | |||
| XP_005279003 (isoform X3) | Chrysemis picta bellii (Western painted turtle) | 315 | 85 | 49 | 154 | 200 | 100.0 | |||
| XP_006132368 | Pelodiscus sinensis (Chinese soft-shelled turtle) | 270 | 97 | 53 | 97 | 49 | 174 | 76 | 271 | 95.0 |
| XP_010723297 (isoform X1) | Meleagris gallopavo (turkey) | 330 | 97 | 133 | 81 | 10 | 216 | 81.8 | ||
| XP_010723300 (isoform X2) | Meleagris gallopavo (turkey) | 330 | 97 | 133 | 66 | 10 | 211 | 87.4 | ||
| XP_418228 | Gallus gallus (chicken) | 651 | 97 | 133 | 81 | 10 | 323 | 67.7 | ||
| XP_006001674 | Latimeria chalumnae (West Indian Ocean coelacanth) | 342 | 97 | 154 | 126 | 7 | 241 | 37.3 | ||
| XP_007897024 | Callorhinchus milii (elephant shark) | 258 | 97 | 115 | 115 | 194 | 99.9 | |||
A TMD and a coiled-coil domain were predicted for each of the tetherin orthologs. aa, amino acids; nt, nucleotides.
RESULTS
The tetherin gene arose >450 million years ago.
To date, orthologs of tetherin (also called BST2 or CD317) have been identified in a variety of placental mammals, including primates, rodents, ungulates, and carnivorans (36). With the exception of tetherin from the gray-handed night monkey Aotus lemurinus griseimembra (37), all tetherin orthologs tested are able to inhibit the egress of enveloped viruses from infected cells. While the general topology consisting of an N-terminal TMD, a coiled-coil ectodomain, and a C-terminal GPI anchor is highly conserved, there is often only minimal sequence homology between different tetherin orthologs (36). This is in line with the finding that the overall protein configuration rather than the primary sequence is critical for its antiviral activity (28).
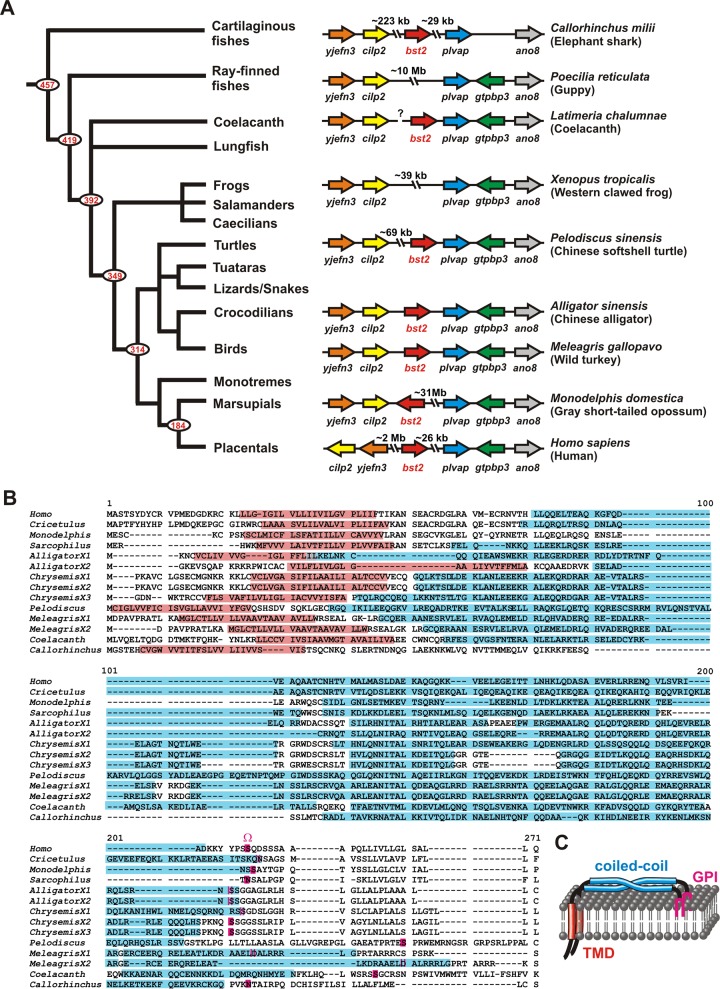
To identify novel tetherin orthologs, we combined sequence analyses with in silico topology predictions and in vitro assays. An initial BLAST search using known tetherin sequences from placental mammals led to the identification of potential tetherin/bst2 orthologs in marsupials (Monodelphis domesticus, Sarcophilus harrisii), crocodilians (Alligator sinensis), turtles (Chrysemis picta, Pelodiscus sinensis), and birds (Meleagris gallopavo) (Fig. 1A and Table 1). Despite very limited sequence homology (Fig. 1B), all genes identified share similar exon-intron structures (Table 1) and are located at a specific locus flanked by genes encoding the plasmalemma vesicle-associated protein (PLVAP) and cartilage intermediate layer protein 2 (CILP2) (Fig. 1A). This strongly suggests that the identified genes represent true orthologs of tetherin. Furthermore, all of these proteins are predicted to have the typical topology of this restriction factor (Fig. 1B and C and Table 1). The TMHMM Server v. 2.0 bioinformatics tool predicted the presence of an N-terminal TMD with a length of 16 to 23 amino acids (aa) (Fig. 1B and Table 1), and, according to the PredGPI analysis software, the probability of the presence of a C-terminal GPI anchor was >95% for most of these species (Fig. 1B and Table 1). Finally, the COILS/PCOILS algorithm predicted the presence of extracellular left-handed coiled-coil domains in all proteins (Fig. 1B and Table 1). Using the MTIDK matrix, probabilities for the presence of coiled coils of above 90% were found for all scanning windows (28, 21, and 14 residues) (see Fig. S1 in the supplemental material). The probabilities determined by the use of weighted and unweighted matrices were similar, indicating that elevated scores were not due to a high incidence of positively charged residues but are indicative of the presence of real coiled-coil structures.
FIG 1.
Identification of previously undescribed tetherin orthologs. (A) Syntenic blocks containing yjefn3 (YjeF N-terminal domain-containing 3), cilp2 (Cartilage Intermediate-Layer Protein 2), bst2/tetherin, plvap (plasmalemma vesicle-associated protein), and ano8 (anoctamin 8) of diverse vertebrate species are shown on the right. Arrows indicate the direction of the ORFs. Gaps represent larger genome regions containing additional ORFs. A phylogenetic tree of vertebrate evolution (72) is shown on the left. Red numbers indicate divergence time estimates (in millions of years ago) for major nodes that are based on Inoue et al. (42). (B) Protein sequence alignment of tetherin orthologs from mammals, reptiles, and fishes. Dashes indicate gaps that were introduced to improve the alignment. Predicted TMDs are highlighted in red and coiled-coil regions in blue. The GPI anchor attachment site (Ω site) is shown in pink. X1 to X3 designate different tetherin isoforms from one species. (C) Cartoon of tetherin illustrating its typical topology comprising an N-terminal TMD (red), an extracellular coiled-coil domain (blue), and a C-terminal GPI lipid raft anchor (pink).
Since some tetherin orthologs may have been missed due to low sequence homology between tetherins from different vertebrate classes, we also manually searched for genes encoding proteins with a predicted tetherin-like configuration. To this end, we took advantage of the high conservation of the plvap and cilp2 genes and screened the respective syntenic blocks in various vertebrate and invertebrate species. In addition to the orthologs identified by the BLAST algorithm, we identified genes in the coelacanth (Latimeria chalumnae) and the elephant shark (Callorhinchus milii) which very likely represent the tetherin orthologs of these species. Like the genes of all other tetherin variants, these piscine genes are adjacent to plvap and encode proteins with a predicted TMD, a coiled-coil ectodomain, and a GPI anchor attachment site (Fig. 1 and Table 1; see also Fig. S1 in the supplemental material). Notably, sequence analyses did not identify any tetherin orthologs in higher invertebrates such as Tunicata, Echinodermata, or Cephalochordata. These findings suggest that a tetherin gene arose early during vertebrate evolution, at least 450 million years ago, before the separation of cartilaginous fish from bony vertebrates (38).
Gene erosion may have resulted in the loss of tetherin in many bird species.
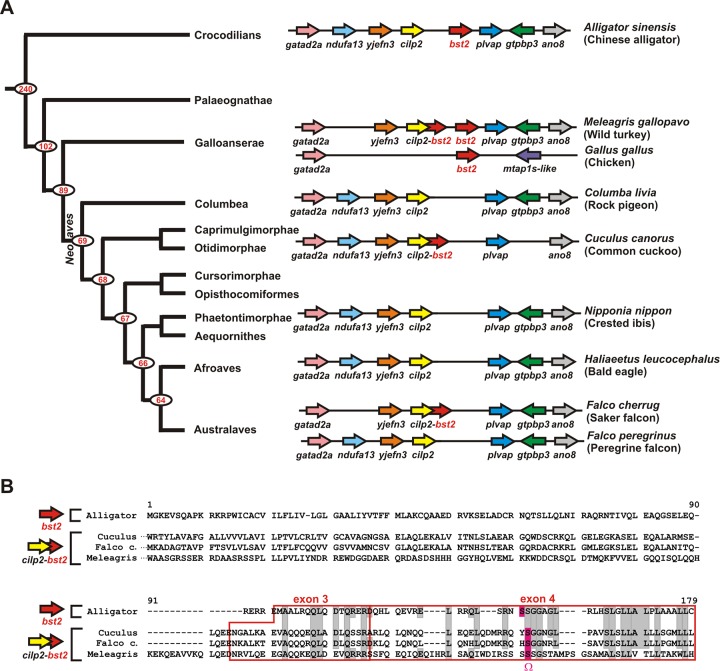
Although tetherin emerged before the divergence of reptiles and birds from mammals and can been found in turkeys (Fig. 1), many bird species lack an obvious tetherin open reading frame (ORF) and seem to contain deletions in the plvap/cilp2 intergenic region (Fig. 2A). In some bird species (e.g., peregrine falcons, rock pigeons, bald eagles, and crested ibises), this genetic erosion seems to have resulted in a complete loss of the tetherin gene. Other species such as the common cuckoo or the saker falcon retained at least exons 3 and 4 (Fig. 2). The latter species are predicted to express fusion proteins consisting of CILP2 and the C-terminal part of tetherin (Fig. 2B). Although these fusion proteins comprise the GPI anchor attachment site encoded by exon 4 of tetherin, the N-terminal CILP2 part is not predicted to form a TMD and it remains to be determined whether these proteins exert any antiviral activity. Interestingly, turkeys also contain such a cilp2-tetherin fusion gene in addition to their regular tetherin ortholog, suggesting that duplication events and/or gene rearrangements may have preceded the gene loss in the cilp2-plvap locus (Fig. 2). Turkeys are morphologically conservative members of the ancient superorder Galloanserae, indicating that the loss of tetherin occurred after the divergence of turkeys from modern birds (i.e., Neoaves). In agreement with this hypothesis, we also identified a putative tetherin ortholog in chickens, another member of the Galloanserae superorder (Fig. 2 and Table 1). In chickens, the cilp2/plvap syntenic block has been disrupted and tetherin is not flanked by the cilp2 and plvap genes (Fig. 2A). The high incidence of deletions and gene rearrangements is in line with the recent observation that birds have experienced a massive reduction in genome size, including the disruption of more than 100 conserved syntenic blocks (39, 40).
FIG 2.
Avian tetherin. (A) The bst2/tetherin gene loci of the indicated bird species are shown. Arrows indicate the direction of the ORFs. Overlapping arrows represent cilp2-bst2 fusion genes. A phylogenetic tree of bird evolution (73) is shown on the left. Red numbers indicate divergence time estimates (in millions of years) for major nodes determined on the basis of data from Green et al. (crocodilian-bird divergence) (74) and Jarvis et al. (intra-avian divergence) (73). (B) Amino acid alignment of alligator X2 tetherin with the CILP2-tetherin fusion proteins of the common cuckoo (XP_009566634), the saker falcon (XP_005444407), and the turkey (XP_010723307). Only the C-terminal ends of the fusion proteins are depicted. Dashes indicate gaps that were introduced to improve the alignment. Sequence comparison revealed that exons 3 and 4 of tetherin (highlighted in red) are fused to the C terminus of cuckoo, saker falcon, and turkey CILP2. Identical amino acids are shown in gray. The GPI anchor attachment site (Ω site) is highlighted in pink.
Piscine, reptilian, and mammalian tetherins are GPI-anchored cell surface proteins.
To functionally characterize distantly related tetherin variants, we selected orthologs of humans and coelacanths as well as both isoforms (X1 and X2) of Chinese alligator tetherin. These species represent the groups of mammals, fish, and Sauropsida. Although alligator isoforms X1 and X2 share exons 3 and 4, they differ substantially in their N-terminal halves, including the short intracellular tail, the TMD, and parts of the ectodomain. Chinese hamster tetherin was also included in the analyses since this ortholog has been suggested to almost exclusively localize to the Golgi apparatus (41), whereas most previously described tetherin variants are mainly found at the cell surface. Western blotting showed that all orthologs are efficiently expressed from CMV promoter-based expression vectors (see Fig. S2A in the supplemental material). The detection of multiple bands ranging from 18 kDa to more than 125 kDa suggested that all tetherins are subject to posttranslational modifications, presumably N-linked glycosylation (see Fig. S2A).
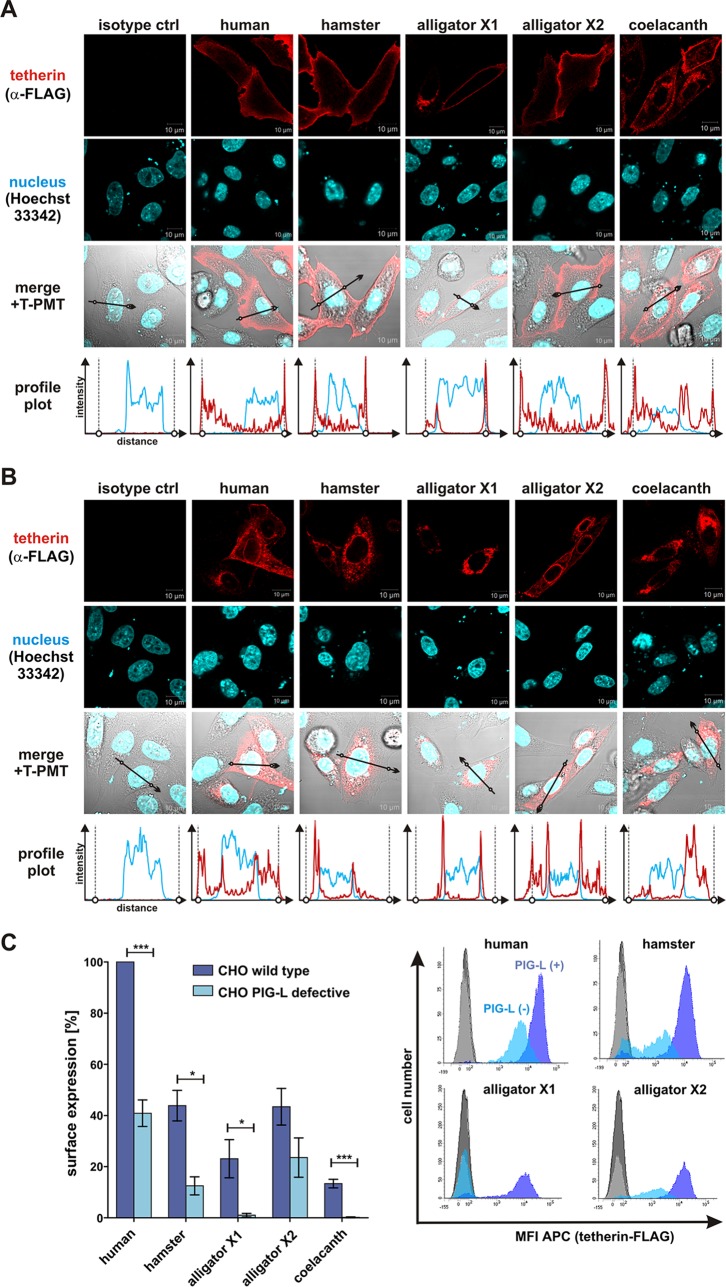
Next, we analyzed the subcellular localization of human, hamster, coelacanth, and alligator tetherin in transfected HEK293T and CHO cells. Flow cytometry revealed that all these proteins are expressed at the cell surface, where virion trapping occurs (17) (see Fig. S2B in the supplemental material). However, surface levels of the coelacanth ortholog and the X1 isoform of alligator tetherin were lower than those of human, hamster, and alligator X2 tetherin (see Fig. S2B). This expression phenotype was confirmed by immunofluorescence microscopy of transfected HEK293T (Fig. S2C) and CHO (Fig. 3A) cells. Whereas the human, hamster, and alligator X2 tetherins were preferentially expressed at the cell surface, substantial amounts of coelacanth and alligator X1 tetherin were also localized to a perinuclear compartment (Fig. 3A; see also Fig. S2C).
FIG 3.
Subcellular localization and GPI anchor dependency of human, hamster, alligator, and coelacanth tetherin. (A and B) Immunofluorescence pictures of CHO wild-type cells (A) or mutant CHO cells lacking a functional pig-l gene, required for GPI anchor synthesis (B). Two days posttransfection with the indicated tetherin expression vectors, cells were permeabilized and incubated with an anti-FLAG antibody. Nuclei were stained using Hoechst 33342. The regions used to generate profile plots are indicated by black arrows. In the profile plots, the localization of the plasma membrane is indicated by circles and vertical dotted lines. T-PMT, transmission-photomultiplier tube. ctrl, control. (C) Flow cytometric analysis of tetherin levels at the surface of transfected CHO cells (wild type [wt] or PIG-L deficient). Means ± standard errors of the means (SEM) of the results of three to five independent experiments are shown on the left (***, P < 0.001; *, P < 0.05). Examples of primary fluorescence-activated cell sorter (FACS) data indicating the mean fluorescence intensity (MFI) of allophycocyanin (APC) are shown on the right.
To verify the presence of a GPI anchor, we took advantage of the availability of a mutant CHO cell line that lacks a functional pig-l gene and thus fails to synthesize this membrane anchor (32). As previously shown (28), the localization of human tetherin was largely restricted to intracellular compartments in GPI anchor-deficient cells (Fig. 3B). A similar redistribution from the cell surface to the cytoplasm was observed for hamster, coelacanth, and both alligator tetherins (Fig. 3B), indicating that all these orthologs represent GPI-anchored proteins. Flow cytometric analyses confirmed that tetherin surface levels were reduced in the absence of a functional GPI anchor (Fig. 3C). Notably, the PIG-L deficiency specifically affected the plasma membrane localization of GPI-anchored proteins, since surface expression of tetherin but not of the CD4 receptor was rescued by exogenous overexpression of PIG-L (see Fig. S3 in the supplemental material).
Tetherin's ability to restrict virus release evolved at least 310 million years ago.
To investigate whether the ability to restrict virus release is an evolutionarily ancient function of tetherin, we analyzed the release of the human immunodeficiency virus 1 (HIV-1) from HEK293T cells cotransfected with increasing amounts of tetherin and a vpu-deficient mutant of HIV-1 NL4-3 that fails to antagonize this restriction factor. Two days posttransfection, p24 levels in the cells and culture supernatants were quantified by ELISA to calculate virus release. Human, hamster, and alligator X2 tetherin efficiently restricted viral particle release in a dose-dependent manner (Fig. 4). In comparison, coelacanth tetherin showed only marginal inhibitory activity and alligator X1 tetherin had no effect. These results demonstrate that not only mammals but also reptiles and possibly fish carry genes that encode tetherin orthologs with antiviral activity. Thus, the ability to restrict the egress of budding virions evolved at least 310 mya, before the divergence of reptiles from mammals (42).
FIG 4.
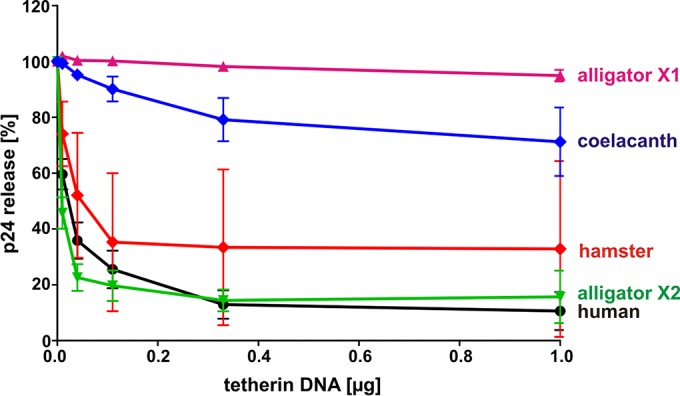
Restriction of HIV-1 release by human, hamster, alligator, and coelacanth tetherin. HEK293T cells were cotransfected with a vpu-deficient proviral construct of HIV-1 NL4-3 and increasing amounts of the indicated tetherin expression vectors. Two days posttransfection, cells and supernatants were harvested and p24 contents were determined by ELISA. Virus release was calculated by dividing the amount of viral capsid in the supernatant by the total amount. The means of the results of three to eight independent experiments ± SEM are shown.
Ponticulins do not restrict virus release, although they share a tetherin-like topology.
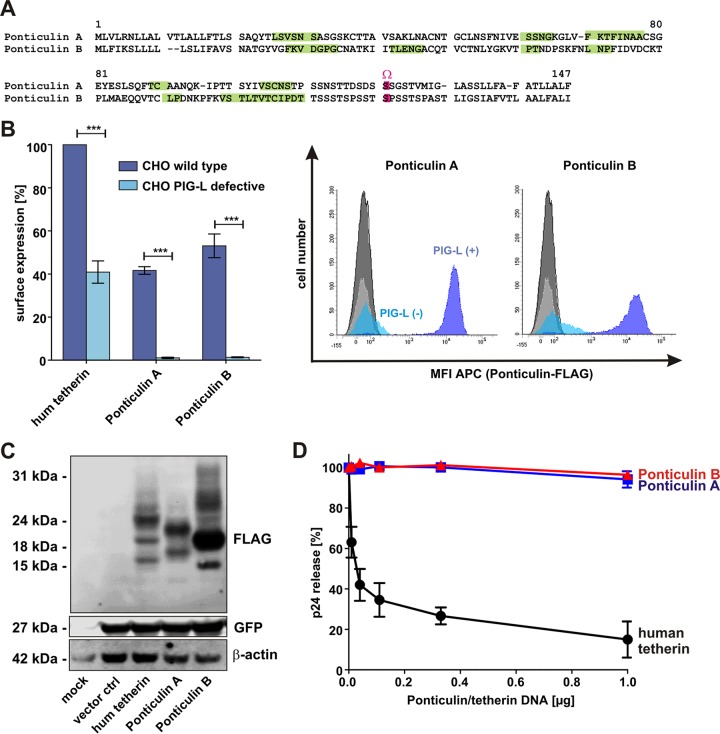
Since the primary amino acid sequence is not critical for the ability of tetherin to restrict virus release (28), we wondered whether completely unrelated proteins with a tetherin-like topology might also show antiviral activity. Only a very small number of proteins containing an N-terminal TMD and a C-terminal GPI anchor have been described. Among them are ponticulin A and B, encoded by genes carried by the slime mold Dictyostelium discoideum (43, 44) (Fig. 5A). In contrast to tetherin, which contains a single α-helical TMD, ponticulins span the membrane multiple times via hydrophobic β-strand structures (43, 45) (Fig. 5A). Flow cytometric analyses of transfected CHO cells confirmed the presence of a GPI anchor, since surface expression was completely abrogated in the absence of a functional pig-l gene (Fig. 5B). Although both proteins were efficiently expressed in HEK293T cells (Fig. 5C), they failed to inhibit the release of retroviral particles (Fig. 5D). Thus, the combination of an N-terminal TMD and a C-terminal GPI anchor is not sufficient to confer antiviral activity to a protein. Notably, the extracellular loop of ponticulin linking the TMD and GPI anchor is only very short and is not predicted to adopt a coiled-coil structure (see Fig. S4 in the supplemental material). Thus, the lack of a flexible ectodomain may account for the absence of antiviral activity in ponticulin A and B.
FIG 5.
Functional characterization of ponticulin A and B. (A) Amino acid alignment of ponticulin A and B from the slime mold Dictyostelium discoideum. Dashes indicate gaps that were introduced to improve the alignment. β-Sheets were predicted using PSIPRED v3.3 and are highlighted in green; the GPI anchor attachment site (Ω site) is shown in pink. (B) Flow cytometric analysis of ponticulin levels at the surface of transfected CHO cells (wt or PIG-L deficient). The means of the results of three to five independent experiments ± SEM are shown on the left (***, P < 0.001). Examples for primary FACS data indicating the mean fluorescence intensity (MFI) of allophycocyanin (APC) are shown on the right. hum, human. (C) Western blot analysis of HEK293T cells transfected with expression vectors for human tetherin or ponticulin A or B. An anti-FLAG antibody was used for detection. β-Actin and eGFP served as loading and transfection controls, respectively. (D) Virus release from HEK293T cells cotransfected with a vpu-deficient proviral construct of HIV-1 NL4-3 and increasing amounts of expression vectors for human tetherin or ponticulin A or B. Two days posttransfection, cells and supernatants were harvested and p24 contents were determined by ELISA. Virus release was calculated by dividing the amount of viral capsid in the supernatant by the total amount. The means of the results of five to seven independent experiments ± SEM are shown.
DISCUSSION
In the present report, we show that orthologs of the restriction factor tetherin are found in various marsupial, bird, reptile, and fish species. The most basal species carrying a tetherin gene was the elephant shark, or Australian ghost shark (Callorhinchus milii), a “living fossil” that is evolutionarily even older than the coelacanth. This indicates that tetherin emerged at least 450 mya, before the divergence of bony vertebrates from the elephant shark (38). The elephant shark is not only one of the oldest but also among the most slowly evolving jawed vertebrate species (38). It has accumulated only a very low number of chromosomal rearrangements and has experienced fewer intron gains or losses than any bony vertebrate (38). We took advantage of this low evolution rate to examine whether tetherin may be the result of a duplication event involving a neighboring gene. Notably, we found some minor sequence homology between tetherin and the plasmalemma vesicle-associated protein (PLVAP) of this species (data not shown), indicating that the two proteins may share a common ancestry. According to the TMHMM Server v. 2.0, COILS/PCOILS, and PredGPI bioinformatics tools, the latter is predicted to have an N-terminal TMD which is followed by a coiled-coil region and a 30% probability of a GPI anchor at its C terminus. However, the overall homology is very low and it remains to be determined whether PLVAP constitutes the true ancestor of tetherin.
The analysis of piscine, reptilian, and mammalian orthologs of tetherin revealed that its ability to restrict virion release evolved at least 310 million years ago. Since tetherin directly targets the budding process rather than a specific viral protein (28), it restricts the egress of a large variety of enveloped viruses. Thus, although only HIV was tested in the present study, these findings are most likely applicable to the restriction of other enveloped viruses as well. The specific viruses that have driven the early evolution of tetherin remain unknown. However, paleovirological analysis of endogenous virus elements (EVEs) has clearly demonstrated that vertebrates have been exposed to diverse virus families for millions of years (46). The most prominent examples of EVEs are endogenous retroviruses that can be found in most vertebrate species (47). Even the coelacanth and elephant shark genomes contain a high diversity of endogenous retroviruses as remnants of ancient infections (48, 49). In addition to retroviral elements, many bird and mammal species also contain EVEs related to hepadna- or filoviruses (46). The extant members of these virus families have a broad host range and are known to be restricted by tetherin (19, 50). Thus, the expression of an active tetherin protein has certainly provided a selection advantage to many vertebrate species.
Notably, tetherin also plays an important role in viral cross-species transmission events, since it is often antagonized in a species-specific manner (51). For example, the sensitivity of influenza A viruses to human tetherin is strain specific (52), suggesting that these viruses may differ in their adaptation to the human host. Influenza A viruses infect a broad range of host species, including mammals, birds, and possibly even reptiles (53, 54). Notably, members of the Galloanserae superorder (e.g., chickens and turkeys) are among the most important reservoirs for human-pathogenic influenza A viruses. Since these bird species encode a tetherin ortholog, it will be interesting to characterize the role of avian and human tetherin in cross-species transmission events involving influenza A viruses.
Despite its long-standing and important role in antiviral immunity, some species seem to have lost their tetherin genes. Most bird species, for example, showed evidence of gene erosion in the cilp2-plvap intergenic region that usually contains the tetherin ORF. These deletions resulted in the complete loss of tetherin or in the emergence of cilp2-tetherin fusion genes. The recent sequencing of 48 bird genomes revealed that birds have experienced a massive gene loss during evolution (39, 40). Thus, tetherin is probably one of more than 1,000 vertebrate genes that have been lost in many bird species (40). Similarly, we did not detect any obvious tetherin orthologs in amphibians or ray-finned fishes, although these vertebrates share an ancestor with tetherin-expressing reptiles, birds, and mammals. This suggests that tetherin may have been lost independently several times during evolution. Ray-finned fish such as the zebra fish, however, show high rates of molecular evolution and evolve(d) much faster than the elephant shark or the coelacanth (38). Thus, we cannot entirely exclude the possibility that orthologs were missed due to the almost complete lack of sequence homology and to incomplete sequencing and/or because they translocated to another gene locus.
Since viruses exert tremendous selection pressure on the evolution of host restriction factors, the respective genes often duplicate and neofunctionalize. Antiretroviral TRIM5 and APOBEC3 proteins, for example, are encoded by large gene clusters of paralogous copies (55, 56). In stark contrast to the TRIM5 and APOBEC families, however, most species seem to carry just a single tetherin gene. The only known exceptions are members of the family of Bovidae which encode two or three paralogs (57, 58). Some species increase their tetherin repertoire by expressing different isoforms from a single gene. Humans, for example, express two alternatively translated isoforms that differ in their antiviral activity and sensitivity toward viral antagonists (59). Similarly, several bird and reptile species may produce different tetherin isoforms via alternative splicing.
Although tetherin orthologs and isoforms often differ substantially in their primary amino acid sequence, most of them share the typical topology that seems to be sufficient for the ability of tetherin to restrict the release of budding virions (28). In agreement with this, human and alligator X2 tetherin efficiently blocked the egress of HIV-1. Even hamster tetherin restricted virus release, although this ortholog has been reported to primarily localize in Golgi cisternae, where it maintains the structure of the Golgi apparatus (41). While our microscopic analyses in transfected HEK293T cells confirmed colocalization with the Golgi marker TGN46, a considerable amount of hamster tetherin was also detectable at the cell surface. In contrast to the findings by Li et al. (41), this was especially pronounced in CHO cells, where tetherin almost exclusively localized at the plasma membrane. In contrast to human, hamster, and alligator X2 tetherin, the coelacanth ortholog showed only a weak antiviral effect, although it adopts the typical tetherin configuration and was efficiently expressed at the cell surface. Notably, the cell membranes of cold-blooded fish and warm-blooded mammals differ in their lipid composition and fluidity (60). Thus, piscine tetherins may be efficient restriction factors in vivo, and antiviral activities might have been missed in human HEK293T cells due to species-specific adaptations. Surprisingly, isoform X1 of alligator tetherin had no impact on virus release at all, although it shares exons 3 and 4 with the active X2 isoform. Interestingly, the extracellular coiled-coil domain and the TMD are predicted to overlap in isoform X1, whereas they are separated by 10 amino acids in isoform X2. This structural difference might affect the flexibility of the hinge region at the transition of the transmembrane to the ectodomain. Furthermore, isoform X1 contains two sequence stretches (aa 65 to 72 and 96 to 100) in its extracellular region that may interrupt the coiled-coil structure. Whether these structural characteristics and/or other reasons account for the lack of antiviral activity of alligator X1 tetherin remains to be determined.
Only a few cellular proteins containing an N-terminal TMD and a C-terminal GPI anchor have been described. These include an unusual isoform of the prion protein (61–63), Sm23 from Schistosoma mansoni (64), and NcSRS2 from Neospora caninum (65) as well as ponticulin A and B from Dictyostelium discoideum (43–45). Although these proteins are all unrelated to tetherin, they may have acquired the ability to inhibit virus release as a result of convergent evolution. Here, we analyzed the antiviral activity of ponticulin A and B since these proteins not only exhibit a tetherin-like configuration but (like tetherin) also contain N-linked glycosylation sites and conserved cysteine residues that form disulfide bonds (43). Furthermore, tetherin and ponticulins both bind actin via their intracellular domains (44, 66, 67). Although we could confirm that ponticulin A and B are GPI-anchored cell surface proteins, there was no evidence for convergent evolution and ponticulins did not restrict virus release. One possible explanation for the lack of antiviral activity may be the absence of coiled coils in the short extracellular domains of ponticulin A and B.
In summary, our results demonstrate that the host restriction factor tetherin is an evolutionarily ancient protein that is substantially (>450 million years) older and more widespread than, for example, the APOBEC3 and TRIM5 genes, which are unique to (placental) mammals and evolved about 90 to 180 million years ago (55, 56, 68–71). In contrast to most other restriction factors, tetherin does not interact with viral proteins or nucleic acids but targets viral membranes. This antiviral mechanism provides several advantages. First, tetherin has broad activity against diverse enveloped viruses. Second, viruses cannot become resistant by simply acquiring evasion mutations. Third, tetherin tolerates many amino acid substitutions since its antiviral activity does not depend on its primary amino acid sequence but rather on the overall protein configuration. Thus, even if viruses evolve antagonists that remove it from the sites of budding, tetherin may readily mutate the respective interaction sites without losing its antiviral activity. With regard to the selection advantage that tetherin confers to its host, it may come as a surprise that some vertebrate species seem to have lost their tetherin gene. Whether these species encode unknown paralogs of tetherin or whether unrelated proteins have also acquired this simple yet very efficient antiviral activity remains to be investigated.
Supplementary Material
ACKNOWLEDGMENTS
We thank Susanne Engelhart and Martha Mayer for excellent technical assistance. CHO cell lines and a PIG-L expression vector were kindly provided by Taroh Kinoshita. J. Bernd Helms kindly provided an expression vector for hamster tetherin.
This study was funded by the Deutsche Forschungsgemeinschaft (Ki548/11-1), a European Research Council Advanced Grant to F.K., and the FP7 European Union's Research and Innovation funding programme. D.S. was supported by a starting grant of the medical faculty of the University of Ulm (L.SBN.0080). Parts of this work were supported by the Gottfried-Wilhelm Leibniz award to F.K. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.02149-15.
REFERENCES
- 1.Koonin EV, Dolja VV, Krupovic M. 2015. Origins and evolution of viruses of eukaryotes: the ultimate modularity. Virology 479–480:2–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meselson M, Yuan R. 1968. DNA restriction enzyme from E. coli. Nature 217:1110–1114. doi: 10.1038/2171110a0. [DOI] [PubMed] [Google Scholar]
- 3.Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. 2007. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 4.Flajnik MF, Kasahara M. 2010. Origin and evolution of the adaptive immune system: genetic events and selective pressures. Nat Rev Genet 11:47–59. doi: 10.1038/nrg2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hahn UK, Bender RC, Bayne CJ. 2000. Production of reactive oxygen species by hemocytes of Biomphalaria glabrata: carbohydrate-specific stimulation. Dev Comp Immunol 24:531–541. doi: 10.1016/S0145-305X(00)00017-3. [DOI] [PubMed] [Google Scholar]
- 6.Hahn UK, Bender RC, Bayne CJ. 2001. Involvement of nitric oxide in killing of Schistosoma mansoni sporocysts by hemocytes from resistant Biomphalaria glabrata. J Parasitol 87:778–785. doi: 10.1645/0022-3395(2001)087[0778:IONOIK]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 7.Nonaka M. 2014. Evolution of the complement system. Subcell Biochem 80:31–43. doi: 10.1007/978-94-017-8881-6_3. [DOI] [PubMed] [Google Scholar]
- 8.Meister M, Lemaitre B, Hoffmann JA. 1997. Antimicrobial peptide defense in Drosophila. Bioessays 19:1019–1026. doi: 10.1002/bies.950191112. [DOI] [PubMed] [Google Scholar]
- 9.Detournay O, Schnitzler CE, Poole A, Weis VM. 2012. Regulation of cnidarian-dinoflagellate mutualisms: evidence that activation of a host TGFβ innate immune pathway promotes tolerance of the symbiont. Dev Comp Immunol 38:525–537. doi: 10.1016/j.dci.2012.08.008. [DOI] [PubMed] [Google Scholar]
- 10.Bieniasz PD. 2004. Intrinsic immunity: a front-line defense against viral attack. Nat Immunol 5:1109–1115. doi: 10.1038/ni1125. [DOI] [PubMed] [Google Scholar]
- 11.Doyle T, Goujon C, Malim MH. 2015. HIV-1 and interferons: who's interfering with whom? Nat Rev Microbiol 13:403–413. doi: 10.1038/nrmicro3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kirchhoff F. 2010. Immune evasion and counteraction of restriction factors by HIV-1 and other primate lentiviruses. Cell Host Microbe 8:55–67. doi: 10.1016/j.chom.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 13.Stremlau M, Owens CM, Perron MJ, Kiessling M, Autissier P, Sodroski J. 2004. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature 427:848–853. doi: 10.1038/nature02343. [DOI] [PubMed] [Google Scholar]
- 14.Sheehy AM, Gaddis NC, Choi JD, Malim MH. 2002. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418:646–650. doi: 10.1038/nature00939. [DOI] [PubMed] [Google Scholar]
- 15.Laguette N, Sobhian B, Casartelli N, Ringeard M, Chable-Bessia C, Ségéral E, Yatim A, Emiliani S, Schwartz O, Benkirane M. 2011. SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature 474:654–657. doi: 10.1038/nature10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hrecka K, Hao C, Gierszewska M, Swanson SK, Kesik-Brodacka M, Srivastava S, Florens L, Washburn MP, Skowronski J. 2011. Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature 474:658–661. doi: 10.1038/nature10195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Neil SJD, Zang T, Bieniasz PD. 2008. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature 451:425–430. doi: 10.1038/nature06553. [DOI] [PubMed] [Google Scholar]
- 18.Van Damme N, Goff D, Katsura C, Jorgenson RL, Mitchell R, Johnson MC, Stephens EB, Guatelli J. 2008. The interferon-induced protein BST-2 restricts HIV-1 release and is downregulated from the cell surface by the viral Vpu protein. Cell Host Microbe 3:245–252. doi: 10.1016/j.chom.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jouvenet N, Neil SJD, Zhadina M, Zang T, Kratovac Z, Lee Y, McNatt M, Hatziioannou T, Bieniasz PD. 2009. Broad-spectrum inhibition of retroviral and filoviral particle release by tetherin. J Virol 83:1837–1844. doi: 10.1128/JVI.02211-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sakuma T, Noda T, Urata S, Kawaoka Y, Yasuda J. 2009. Inhibition of Lassa and Marburg virus production by tetherin. J Virol 83:2382–2385. doi: 10.1128/JVI.01607-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Radoshitzky SR, Dong L, Chi X, Clester JC, Retterer C, Spurgers K, Kuhn JH, Sandwick S, Ruthel G, Kota K, Boltz D, Warren T, Kranzusch PJ, Whelan SPJ, Bavari S. 2010. Infectious Lassa virus, but not filoviruses, is restricted by BST-2/tetherin. J Virol 84:10569–10580. doi: 10.1128/JVI.00103-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weidner JM, Jiang D, Pan X-B, Chang J, Block TM, Guo J-T. 2010. Interferon-induced cell membrane proteins, IFITM3 and tetherin, inhibit vesicular stomatitis virus infection via distinct mechanisms. J Virol 84:12646–12657. doi: 10.1128/JVI.01328-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sarojini S, Theofanis T, Reiss CS. 2011. Interferon-induced tetherin restricts vesicular stomatitis virus release in neurons. DNA Cell Biol 30:965–974. doi: 10.1089/dna.2011.1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kong W-S, Irie T, Yoshida A, Kawabata R, Kadoi T, Sakaguchi T. 2012. Inhibition of virus-like particle release of Sendai virus and Nipah virus, but not that of mumps virus, by tetherin/CD317/BST-2. Hiroshima J Med Sci 61:59–67. [PubMed] [Google Scholar]
- 25.Blondeau C, Pelchen-Matthews A, Mlcochova P, Marsh M, Milne RSB, Towers GJ. 2013. Tetherin restricts herpes simplex virus 1 and is antagonized by glycoprotein M. J Virol 87:13124–13133. doi: 10.1128/JVI.02250-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang S-M, Huang K-J, Wang C-T. 2014. BST2/CD317 counteracts human coronavirus 229E productive infection by tethering virions at the cell surface. Virology 449:287–296. doi: 10.1016/j.virol.2013.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pan X-B, Qu X-W, Jiang D, Zhao X-L, Han J-C, Wei L. 2013. BST2/Tetherin inhibits hepatitis C virus production in human hepatoma cells. Antiviral Res 98:54–60. doi: 10.1016/j.antiviral.2013.01.009. [DOI] [PubMed] [Google Scholar]
- 28.Perez-Caballero D, Zang T, Ebrahimi A, McNatt MW, Gregory DA, Johnson MC, Bieniasz PD. 2009. Tetherin inhibits HIV-1 release by directly tethering virions to cells. Cell 139:499–511. doi: 10.1016/j.cell.2009.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kupzig S, Korolchuk V, Rollason R, Sugden A, Wilde A, Banting G. 2003. Bst-2/HM1.24 is a raft-associated apical membrane protein with an unusual topology. Traffic 4:694–709. doi: 10.1034/j.1600-0854.2003.00129.x. [DOI] [PubMed] [Google Scholar]
- 30.Venkatesh S, Bieniasz PD. 2013. Mechanism of HIV-1 virion entrapment by tetherin. PLoS Pathog 9:e1003483. doi: 10.1371/journal.ppat.1003483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tanaka M, Herr W. 1990. Differential transcriptional activation by Oct-1 and Oct-2: interdependent activation domains induce Oct-2 phosphorylation. Cell 60:375–386. doi: 10.1016/0092-8674(90)90589-7. [DOI] [PubMed] [Google Scholar]
- 32.Nakamura N, Inoue N, Watanabe R, Takahashi M, Takeda J, Stevens VL, Kinoshita T. 1997. Expression cloning of PIG-L, a candidate N-acetylglucosaminyl-phosphatidylinositol deacetylase. J Biol Chem 272:15834–15840. doi: 10.1074/jbc.272.25.15834. [DOI] [PubMed] [Google Scholar]
- 33.Rücker E, Grivel J-C, Münch J, Kirchhoff F, Margolis L. 2004. Vpr and Vpu are important for efficient human immunodeficiency virus type 1 replication and CD4+ T-cell depletion in human lymphoid tissue ex vivo. J Virol 78:12689–12693. doi: 10.1128/JVI.78.22.12689-12693.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Münch J, Ständker L, Pöhlmann S, Baribaud F, Papkalla A, Rosorius O, Stauber R, Sass G, Heveker N, Adermann K, Escher S, Klüver E, Doms RW, Forssmann W-G, Kirchhoff F. 2002. Hemofiltrate CC chemokine 1[9-74] causes effective internalization of CCR5 and is a potent inhibitor of R5-tropic human immunodeficiency virus type 1 strains in primary T cells and macrophages. Antimicrob Agents Chemother 46:982–990. doi: 10.1128/AAC.46.4.982-990.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.DuBridge RB, Tang P, Hsia HC, Leong PM, Miller JH, Calos MP. 1987. Analysis of mutation in human cells by using an Epstein-Barr virus shuttle system. Mol Cell Biol 7:379–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sauter D. 2014. Counteraction of the multifunctional restriction factor tetherin. Front Microbiol 5:163. doi: 10.3389/fmicb.2014.00163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wong SK, Connole M, Sullivan JS, Choe H, Carville A, Farzan M. 2009. A New World primate deficient in tetherin-mediated restriction of human immunodeficiency virus type 1. J Virol 83:8771–8780. doi: 10.1128/JVI.00112-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Venkatesh B, Lee AP, Ravi V, Maurya AK, Lian MM, Swann JB, Ohta Y, Flajnik MF, Sutoh Y, Kasahara M, Hoon S, Gangu V, Roy SW, Irimia M, Korzh V, Kondrychyn I, Lim ZW, Tay B-H, Tohari S, Kong KW, Ho S, Lorente-Galdos B, Quilez J, Marques-Bonet T, Raney BJ, Ingham PW, Tay A, Hillier LW, Minx P, Boehm T, Wilson RK, Brenner S, Warren WC. 2014. Elephant shark genome provides unique insights into gnathostome evolution. Nature 505:174–179. doi: 10.1038/nature12826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lovell PV, Wirthlin M, Wilhelm L, Minx P, Lazar NH, Carbone L, Warren WC, Mello CV. 2014. Conserved syntenic clusters of protein coding genes are missing in birds. Genome Biol 15:565. doi: 10.1186/s13059-014-0565-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang G, Li C, Li Q, Li B, Larkin DM, Lee C, Storz JF, Antunes A, Greenwold MJ, Meredith RW, Ödeen A, Cui J, Zhou Q, Xu L, Pan H, Wang Z, Jin L, Zhang P, Hu H, Yang W, Hu J, Xiao J, Yang Z, Liu Y, Xie Q, Yu H, Lian J, Wen P, Zhang F, Li H, Zeng Y, Xiong Z, Liu S, Zhou L, Huang Z, An N, Wang J, Zheng Q, Xiong Y, Wang G, Wang B, Wang J, Fan Y, da Fonseca RR, Alfaro-Núñez A, Schubert M, Orlando L, Mourier T, Howard JT, Ganapathy G, et al. 2014. Comparative genomics reveals insights into avian genome evolution and adaptation. Science 346:1311–1320. doi: 10.1126/science.1251385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li X, Kaloyanova D, van Eijk M, Eerland R, van der Goot G, Oorschot V, Klumperman J, Lottspeich F, Starkuviene V, Wieland FT, Helms JB. 2007. Involvement of a Golgi-resident GPI-anchored protein in maintenance of the Golgi structure. Mol Biol Cell 18:1261–1271. doi: 10.1091/mbc.E06-03-0236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Inoue JG, Miya M, Lam K, Tay B-H, Danks JA, Bell J, Walker TI, Venkatesh B. 2010. Evolutionary origin and phylogeny of the modern holocephalans (Chondrichthyes: Chimaeriformes): a mitogenomic perspective. Mol Biol Evol 27:2576–2586. doi: 10.1093/molbev/msq147. [DOI] [PubMed] [Google Scholar]
- 43.Hitt AL, Lu TH, Luna EJ. 1994. Ponticulin is an atypical membrane protein. J Cell Biol 126:1421–1431. doi: 10.1083/jcb.126.6.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hitt AL, Hartwig JH, Luna EJ. 1994. Ponticulin is the major high affinity link between the plasma membrane and the cortical actin network in Dictyostelium. J Cell Biol 126:1433–1444. doi: 10.1083/jcb.126.6.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hitt AL, Iijima-Shimizu M, DuBay MJ, Antonette LL, Urushihara H, Wilkerson CG. 2003. Identification of a second member of the ponticulin gene family and its differential expression pattern. Biochim Biophys Acta 1628:79–87. doi: 10.1016/S0167-4781(03)00115-5. [DOI] [PubMed] [Google Scholar]
- 46.Katzourakis A, Gifford RJ. 2010. Endogenous viral elements in animal genomes. PLoS Genet 6:e1001191. doi: 10.1371/journal.pgen.1001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Herniou E, Martin J, Miller K, Cook J, Wilkinson M, Tristem M. 1998. Retroviral diversity and distribution in vertebrates. J Virol 72:5955–5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Han G-Z, Worobey M. 2012. An endogenous foamy-like viral element in the coelacanth genome. PLoS Pathog 8:e1002790. doi: 10.1371/journal.ppat.1002790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Han G-Z. 2015. Extensive retroviral diversity in shark. Retrovirology 12:34. doi: 10.1186/s12977-015-0158-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yan R, Zhao X, Cai D, Liu Y, Block TM, Guo J-T, Guo H. 2015. The interferon-inducible protein tetherin inhibits hepatitis B virus virion secretion. J Virol 89:9200–9212. doi: 10.1128/JVI.00933-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sauter D, Specht A, Kirchhoff F. 2010. Tetherin: holding on and letting go. Cell 141:392–398. doi: 10.1016/j.cell.2010.04.022. [DOI] [PubMed] [Google Scholar]
- 52.Gnirß K, Zmora P, Blazejewska P, Winkler M, Lins A, Nehlmeier I, Gärtner S, Moldenhauer A-S, Hofmann-Winkler H, Wolff T, Schindler M, Pöhlmann S. 24 June 2015. Tetherin sensitivity of influenza A viruses is strain specific: role of hemagglutinin and neuraminidase. J Virol doi: 10.1128/JVI.00615-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Temple BL, Finger JW, Jones CA, Gabbard JD, Jelesijevic T, Uhl EW, Hogan RJ, Glenn TC, Tompkins SM. 2015. In ovo and in vitro susceptibility of American alligators (Alligator mississippiensis) to avian influenza virus infection. J Wildl Dis 51:187–198. doi: 10.7589/2013-12-321. [DOI] [PubMed] [Google Scholar]
- 54.Davis LM, Spackman E. 2008. Do crocodilians get the flu? Looking for influenza A in captive crocodilians. J Exp Zool A Ecol Genet Physiol 309:571–580. doi: 10.1002/jez.454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Münk C, Willemsen A, Bravo IG. 2012. An ancient history of gene duplications, fusions and losses in the evolution of APOBEC3 mutators in mammals. BMC Evol Biol 12:71. doi: 10.1186/1471-2148-12-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Johnson WE, Sawyer SL. 2009. Molecular evolution of the antiretroviral TRIM5 gene. Immunogenetics 61:163–176. doi: 10.1007/s00251-009-0358-y. [DOI] [PubMed] [Google Scholar]
- 57.Arnaud F, Black SG, Murphy L, Griffiths DJ, Neil SJ, Spencer TE, Palmarini M. 2010. Interplay between ovine bone marrow stromal cell antigen 2/tetherin and endogenous retroviruses. J Virol 84:4415–4425. doi: 10.1128/JVI.00029-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Takeda E, Nakagawa S, Nakaya Y, Tanaka A, Miyazawa T, Yasuda J. 2012. Identification and functional analysis of three isoforms of bovine BST-2. PLoS One 7:e41483. doi: 10.1371/journal.pone.0041483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cocka LJ, Bates P. 2012. Identification of alternatively translated tetherin isoforms with differing antiviral and signaling activities. PLoS Pathog 8:e1002931. doi: 10.1371/journal.ppat.1002931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cossins AR, Prosser CL. 1978. Evolutionary adaptation of membranes to temperature. Proc Natl Acad Sci U S A 75:2040–2043. doi: 10.1073/pnas.75.4.2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stewart RS, Drisaldi B, Harris DA. 2001. A transmembrane form of the prion protein contains an uncleaved signal peptide and is retained in the endoplasmic reticulum. Mol Biol Cell 12:881–889. doi: 10.1091/mbc.12.4.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hegde RS, Mastrianni JA, Scott MR, DeFea KA, Tremblay P, Torchia M, DeArmond SJ, Prusiner SB, Lingappa VR. 1998. A transmembrane form of the prion protein in neurodegenerative disease. Science 279:827–834. doi: 10.1126/science.279.5352.827. [DOI] [PubMed] [Google Scholar]
- 63.Hegde RS, Tremblay P, Groth D, DeArmond SJ, Prusiner SB, Lingappa VR. 1999. Transmissible and genetic prion diseases share a common pathway of neurodegeneration. Nature 402:822–826. doi: 10.1038/45574. [DOI] [PubMed] [Google Scholar]
- 64.Köster B, Strand M. 1994. Schistosoma mansoni: Sm23 is a transmembrane protein that also contains a glycosylphosphatidylinositol anchor. Arch Biochem Biophys 310:108–117. doi: 10.1006/abbi.1994.1146. [DOI] [PubMed] [Google Scholar]
- 65.Nishikawa Y, Tragoolpua K, Makala L, Xuan X, Nagasawa H. 2002. Neospora caninum NcSRS2 is a transmembrane protein that contains a glycosylphosphatidylinositol anchor in insect cells. Vet Parasitol 109:191–201. doi: 10.1016/S0304-4017(02)00256-X. [DOI] [PubMed] [Google Scholar]
- 66.Chia CP, Hitt AL, Luna EJ. 1991. Direct binding of F-actin to ponticulin, an integral plasma membrane glycoprotein. Cell Motil Cytoskeleton 18:164–179. doi: 10.1002/cm.970180303. [DOI] [PubMed] [Google Scholar]
- 67.Rollason R, Korolchuk V, Hamilton C, Jepson M, Banting G. 2009. A CD317/tetherin-RICH2 complex plays a critical role in the organization of the subapical actin cytoskeleton in polarized epithelial cells. J Cell Biol 184:721–736. doi: 10.1083/jcb.200804154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Conticello SG, Thomas CJF, Petersen-Mahrt SK, Neuberger MS. 2005. Evolution of the AID/APOBEC family of polynucleotide (deoxy)cytidine deaminases. Mol Biol Evol 22:367–377. [DOI] [PubMed] [Google Scholar]
- 69.Tareen SU, Sawyer SL, Malik HS, Emerman M. 2009. An expanded clade of rodent Trim5 genes. Virology 385:473–483. doi: 10.1016/j.virol.2008.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sawyer SL, Emerman M, Malik HS. 2007. Discordant evolution of the adjacent antiretroviral genes TRIM22 and TRIM5 in mammals. PLoS Pathog 3:e197. doi: 10.1371/journal.ppat.0030197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Conticello SG. 2008. The AID/APOBEC family of nucleic acid mutators. Genome Biol 9:229. doi: 10.1186/gb-2008-9-6-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Meyer A, Zardoya R. 2003. Recent advances in the (molecular) phylogeny of vertebrates. Annu Rev Ecol Evol Syst 34:311–338. doi: 10.1146/annurev.ecolsys.34.011802.132351. [DOI] [Google Scholar]
- 73.Jarvis ED, Mirarab S, Aberer AJ, Li B, Houde P, Li C, Ho SY, Faircloth BC, Nabholz B, Howard JT, Suh A, Weber CC, da Fonseca RR, Li J, Zhang F, Li H, Zhou L, Narula N, Liu L, Ganapathy G, Boussau B, Bayzid MS, Zavidovych V, Subramanian S, Gabaldón T, Capella-Gutiérrez S, Huerta-Cepas J, Rekepalli B, Munch K, Schierup M, Lindow B, Warren WC, Ray D, Green RE, Bruford MW, Zhan X, Dixon A, Li S, Li N, Huang Y, Derryberry EP, Bertelsen MF, Sheldon FH, Brumfield RT, Mello CV, Lovell PV, Wirthlin M, Schneider MP, Prosdocimi F, et al. 2014. Whole-genome analyses resolve early branches in the tree of life of modern birds. Science 346:1320–1331. doi: 10.1126/science.1253451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Green RE, Braun EL, Armstrong J, Earl D, Nguyen N, Hickey G, Vandewege MW, St John JA, Capella-Gutiérrez S, Castoe TA, Kern C, Fujita MK, Opazo JC, Jurka J, Kojima KK, Caballero J, Hubley RM, Smit AF, Platt RN, Lavoie CA, Ramakodi MP, Finger JW, Suh A, Isberg SR, Miles L, Chong AY, Jaratlerdsiri W, Gongora J, Moran C, Iriarte A, McCormack J, Burgess SC, Edwards SV, Lyons E, Williams C, Breen M, Howard JT, Gresham CR, Peterson DG, Schmitz J, Pollock DD, Haussler D, Triplett EW, Zhang G, Irie N, Jarvis ED, Brochu CA, Schmidt CJ, McCarthy FM, Faircloth BC, et al. 2014. Three crocodilian genomes reveal ancestral patterns of evolution among archosaurs. Science 346:1254449. doi: 10.1126/science.1254449. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.