ABSTRACT
Human immunodeficiency virus type 1 (HIV-1) replication requires reverse transcription of its RNA genome into a double-stranded cDNA copy, which is then integrated into the host cell chromosome. The essential steps of reverse transcription and integration are catalyzed by the viral enzymes reverse transcriptase (RT) and integrase (IN), respectively. In vitro, HIV-1 RT can bind with IN, and the C-terminal domain (CTD) of IN is necessary and sufficient for this binding. To better define the RT-IN interaction, we performed nuclear magnetic resonance (NMR) spectroscopy experiments to map a binding surface on the IN CTD in the presence of RT prebound to a duplex DNA construct that mimics the primer-binding site in the HIV-1 genome. To determine the biological significance of the RT-IN interaction during viral replication, we used the NMR chemical shift mapping information as a guide to introduce single amino acid substitutions of nine different residues on the putative RT-binding surface in the IN CTD. We found that six viral clones bearing such IN substitutions (R231E, W243E, G247E, A248E, V250E, and I251E) were noninfectious. Further analyses of the replication-defective IN mutants indicated that the block in replication took place specifically during early reverse transcription. The recombinant INs purified from these mutants, though retaining enzymatic activities, had diminished ability to bind RT in a cosedimentation assay. The results indicate that the RT-IN interaction is functionally relevant during the reverse transcription step of the HIV-1 life cycle.
IMPORTANCE To establish a productive infection, human immunodeficiency virus type 1 (HIV-1) needs to reverse transcribe its RNA genome to create a double-stranded DNA copy and then integrate this viral DNA genome into the chromosome of the host cell. These two essential steps are catalyzed by the HIV-1 enzymes reverse transcriptase (RT) and integrase (IN), respectively. We have shown previously that IN physically interacts with RT, but the importance of this interaction during HIV-1 replication has not been fully characterized. In this study, we have established the biological significance of the HIV-1 RT-IN interaction during the viral life cycle by demonstrating that altering the RT-binding surface on IN disrupts both reverse transcription and viral replication. These findings contribute to our understanding of the RT-IN binding mechanism, as well as indicate that the RT-IN interaction can be exploited as a new antiviral drug target.
INTRODUCTION
To establish an infection after entry into a susceptible cell, human immunodeficiency virus type 1 (HIV-1) has to reverse transcribe its RNA genome to double-stranded DNA, followed by integration into the host genome. Reverse transcriptase (RT) and integrase (IN) are the viral enzymes responsible for catalyzing the essential steps of reverse transcription and integration, respectively. Both enzymes are synthesized as part of the Gag-Pol polyprotein, which is later processed by the viral protease to produce active RT and IN during HIV-1 maturation (1, 2). RT is a heterodimeric enzyme consisting of 66- and 51-kDa subunits and catalyzes the RNA- and DNA-dependent reverse transcription of the viral RNA genome into double-stranded cDNA through a complex cascade of events (3, 4). The 32-kDa IN has three domains: an N-terminal zinc-binding domain, a catalytic core domain, and a C-terminal domain (CTD) that binds DNA nonspecifically. IN catalyzes the integration of the viral cDNA into the host genome in two steps: an initial 3′-end processing step that removes two nucleotides at each 3′ end and exposes a highly conserved CA 5′ overhang, followed by a strand transfer step that inserts both processed viral DNA ends into the host cell genome (5, 6). In vitro, IN can also catalyze a reverse reaction, termed disintegration, resolving a DNA mimic of the viral-host DNA intermediate to products corresponding to a 3′ processed viral DNA end and a target duplex DNA (7). IN can multimerize and forms a complex with viral DNA ends, termed the intasome (8–10). Structural studies of the prototype foamy virus (PFV) intasome found the tetramer to be the active IN configuration (9, 11, 12). HIV-1 IN has also been proposed to function as a tetramer (10, 13–17).
Mutations in IN can result in pleiotropic effects throughout the viral life cycle (18, 19), suggesting that it participates in other events during the viral life cycle. Depending on their effects on viral processes, IN mutants are grouped as either class I or class II (20–23). Class I IN mutants are enzymatically inactive in catalyzing the 3′ end processing and strand transfer, and viruses harboring such mutations are specifically blocked at the integration step. Class II IN mutants display partial or full IN activity in vitro but are nonetheless replication defective due to pleiotropic effects at replicative steps other than integration. Several mutagenesis studies have described the effect of IN mutation upon both the early stages of viral replication, such as uncoating (24, 25), reverse transcription (22, 26–34), and nuclear import of preintegration complexes (26, 35–37), and the late steps postintegration, including polyprotein processing, packaging, and maturation (19, 22, 29, 31, 38–41).
Mutational studies support IN involvement during reverse transcription. The IN mutations that disrupt reverse transcription steps are scattered throughout its three domains (19, 29, 30), suggesting that the effect of IN upon reverse transcription may rely on multiple mechanisms that are yet to be fully understood. Some IN pleiotropic effects, especially those associated with late events postintegration, may result from the effect of IN mutations upon proper Gag-Pol polyprotein processing and packaging. The direct involvement of IN on reverse transcription during HIV-1 replication has been demonstrated through transcomplementation experiments using the HIV-1 accessory protein Vpr fused to wild-type (WT) IN, which can be packaged into virions to rescue reverse transcription-defective viruses bearing certain IN mutations (25, 33).
One straightforward explanation for the influence of IN on reverse transcription is that IN binds RT and facilitates viral cDNA synthesis. Under in vitro conditions, HIV-1 IN physically interacts with RT and facilitates the early steps of reverse transcription (27). Deletion analyses of different IN domains and their ability to coimmunoprecipitate with recombinant RT revealed that amino acid residues 220 to 270 within the CTD (IN220–270) are necessary and sufficient to bind RT (28, 34). An RT-binding surface on IN has been previously determined by nuclear magnetic resonance (NMR) spectroscopy using IN220–270 and RT lacking bound primer-template substrate (apo-RT) (42). However, various experimental data suggest that IN can interact with RT in the context of bound nucleic acid substrate: in vitro reverse transcription assays have shown that IN enhances synthesis of early products, stimulating both the initiation and elongation phases of reverse transcription by increasing RT processivity and suppressing the formation of pause products (27, 43). Other studies demonstrate that IN can interact with nucleic acid-bound RT to inhibit DNA-dependent DNA polymerization (44). The precise RT conformation and functional form (either free or nucleic acid bound) that IN initially targets during any particular IN-mediated reverse transcription pathway step remain largely unexamined. To explore the RT-IN interaction under conditions where RT is bound to nucleic acid, we have performed NMR chemical shift perturbation experiments to fully characterize a binding surface on the IN CTD in the presence of RT prebound to a duplex DNA construct that mimics the primer-binding site in the HIV-1 genome.
To determine the functional relevance of the RT-binding surface mapped by the NMR experiment, we analyzed the consequence of selectively substituting different IN residues that form the putative binding surface for both free and DNA-bound RT and characterizing IN functional properties in vitro and in viral replication assays. We found that disruption of the putative binding surface on IN impaired viral replication due to the inability to synthesize viral cDNA, indicating the functional significance of the RT-IN interaction during viral replication.
MATERIALS AND METHODS
Construction of IN mutant viral clones and protein expression plasmids.
Mutations within the IN coding sequence were introduced into a pBluescript cloning vector bearing the AgeI-EcoRI fragment from the infectious HIV-1 NL4-3 molecular clone (nucleotide positions 3486 to 5744) (45) using mutagenic primers and PCR-based site-directed mutagenesis (46). The IN gene containing the desired mutation was then excised from this cloning vector with AgeI and EcoRI and ligated into AgeI/EcoRI-digested NL4-3. Competent TOP10 cells (Invitrogen) were transformed by NL4-3 plasmids encoding the mutant IN sequences, and the resulting colonies were verified for desired constructs by DNA sequencing (Laragen, Inc., Los Angeles, CA). PCR-based site-directed mutagenesis (46) was also used to introduce mutations directly into either a His-tagged pT7-7 expression plasmid containing the full-length HIV-1 IN (pT7-7/H-IN) gene (47) or an expression plasmid encoding HIV-1 IN220–270, which was provided by Robert Craigie, National Institutes of Health (NIH).
Protein expression and purification.
CodonPlus Escherichia coli cells (Agilent) were transformed by expression constructs encoding either full-length IN or amino acids 220 to 270 of IN with an N-terminal His tag for facilitating protein purification. All full-length IN and IN220–270 constructs were purified under nondenaturing conditions as previously described, with few modifications (34). Briefly, transformed cells were grown in LB medium at 32°C until the optical density at 600 nm (OD600) was between 0.8 and 1.0. Protein expression was then induced by adding 0.4 mM isopropyl-1-thio-β-d-galactopyranoside (IPTG), and cells were left to grow for an additional 4 h. Pelleted cells were suspended in a lysis buffer containing 20 mM HEPES-Na, pH 7.5, 5 mM 2-mercaptoethanol, 1 M NaCl, 0.2 mM EDTA, 10% glycerol, 0.5% IGEPAL CA-630 (Sigma-Aldrich), and EDTA-free protease inhibitor tablets (Roche; 1 tablet/10 ml lysis buffer), and further disrupted by sonication. Lysates were then clarified by centrifugation at 100,000 × g for 1 h at 4°C, followed by 16 to 20 h of dialysis at 4°C in 1 L buffer C (20 mM HEPES-Na, pH 7.5, 1 M NaCl, 10% glycerol, 5 mM 2-mercaptoethanol, and 0.1% IGEPAL CA-630). Purified IN or its mutant derivatives were then obtained from clarified lysates using Ni2+-nitrilotriacetic acid (NTA) immobilized metal affinity chromatography (IMAC) and cation exchange chromatography. Purified proteins were quantitated using extinction coefficients that were calculated based upon amino acid sequence, and purity was determined by SDS-PAGE.
For NMR studies, the IN220–270 construct was expressed in CodonPlus E. coli cells at 37°C in minimal M9 medium containing 15N-labeled ammonium chloride. 15N-labeled IN220–270 expression was induced using 0.4 mM IPTG for 4 h after an OD600 of 0.8 had been reached. Purified 15N-labeled IN220–270 was obtained using Ni2+-NTA IMAC, followed by dialysis for 10 to 12 h at 4°C in NMR buffer (50 mM sodium phosphate buffer, pH 6.5, 100 mM NaCl, and 0.5 mM EDTA). Concentration of 15N-labeled IN220–270 for subsequent NMR analysis was performed using a 3,500-molecular-weight-cutoff (MWCO) centrifugal filter unit (Millipore). Recombinant RT was expressed using a plasmid (p6HRT-PROT) with dual inducible expression cassettes for 6×His-tagged p66 and untagged HIV-1 protease to generate the His-tagged p66/p51 RT heterodimer, which was then purified using Ni2+-NTA IMAC and cation exchange chromatography (42, 48). The purity (95%) of the RT heterodimer was confirmed by SDS-PAGE.
Generation of viruses.
Virus stocks were generated by transient transfection of 293T cells with NL4-3 constructs encoding the desired IN mutation. Cells were supplemented with 10% fetal bovine serum in RPMI 1640 medium with 10,000 IU/ml penicillin and 10,000 μg/ml streptomycin and transfected at 80 to 90% confluence in T-75 flasks using 8 μg of each viral DNA construct according to a PolyFect-based protocol (Qiagen). Virus-containing medium was collected at 36 to 48 h posttransfection and gravity filtered through 0.45-μm-pore-size cellulose acetate filters. Harvested virus particles were concentrated through a 20% (wt/vol) sucrose cushion by ultracentrifugation at 100,000 × g for 3 h at 4°C, suspended in Cellgro sterile phosphate-buffered saline (PBS; Corning), and stored at −80°C. Viral titers were quantified by levels of HIV-1 p24 antigen, as determined by enzyme-linked immunosorbent assays (ELISA; PerkinElmer).
Western blot analysis of viruses.
SDS-PAGE loading buffer was added to 100 ng of the p24 equivalent of viruses and boiled for 5 min at 95°C. The samples were then electrophoresed on 12% SDS-polyacrylamide gels and transferred onto nitrocellulose membranes (Thermo Fisher). Membranes were blocked with 5% nonfat milk in TBS-T buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, and 0.05% Tween 20) for 1 h and then probed for 1 h with either a 1:500 dilution of anti-HIV human serum (Scripps Laboratory, Inc.), a 1:1,000 dilution of a polyclonal anti-RT antibody, or a 1:500 dilution of a polyclonal anti-IN antibody in TBS-T containing 1% nonfat milk. Blots were subsequently washed three times for 10 min each in TBS-T and then incubated for 1 h with a 1:10,000 dilution of anti-human or anti-rabbit secondary antibodies conjugated with horseradish peroxidase (Santa Cruz Biotechnology, Inc.) in TBS-T with 1% nonfat milk. The signal was detected using a chemiluminescence substrate kit (SuperSignal West Pico; Pierce) according to the manufacturer's instructions. The following reagents were obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH: polyclonal anti-HIV-1 HXB2 IN amino acid residues 23 to 34 from Duane P. Grandgenett (49) and polyclonal anti-HIV-1 RT antibody.
Replication kinetics assays.
Infection assays were carried out as previously described (42). Briefly, 100 ng of the p24 equivalent of viruses was used to infect 2 × 106 CEM cells, and virus-containing medium was carefully collected without disturbing the sedimented cells at 48-h intervals for 10 days postinfection. Harvested viruses were quantified by ELISA against HIV-1 p24 antigen. Following virus collection at each time point, infected cells were split 1:3. Negative controls included mock-infected cells (no virus added) as well as cells infected with heat-inactivated (HI) virus, produced by heating wild-type virions at 95°C for 10 min.
Pulldown assay using RT-conjugated magnetic beads.
RT was conjugated directly to Dynabeads M-270 epoxy using a coupling kit (catalog number 14311D; Life Technologies) at a ratio of 4 μg (34 pmol) of RT to 1 mg of magnetic beads according to the manufacturer's instructions. RT-conjugated beads were stored at a concentration of 1 mg/100 μl in SB buffer (supplied with the kit) at 4°C until use. For the pulldown assay, 100 μl of the RT-conjugated beads (1 mg) was initially washed with 900 μl of binding buffer (20 mM HEPES-Na, pH 7.5, 2 mM MgCl2, 150 mM NaCl, 0.5% Triton X-100, 27.5 mM potassium acetate) and then incubated with 20 pmol of IN in 500 μl of binding buffer at 4°C for 0.5 h. Following incubation, the supernatant fraction containing unbound IN (unbound fraction) was collected. The RT-coupled beads were then washed once with 200 μl of binding buffer, and the supernatant fraction was collected (wash fraction). Any bound IN was then eluted from the RT-conjugated beads by addition of 40 μl of elution buffer (100 mM glycine, pH 2.5) and incubation at room temperature for 5 min. The eluate (bound fraction) was neutralized by the addition of 4 μl of 1 M Tris-HCl, pH 9.5, and resuspended in 30 μl of loading buffer. For ease of analysis, the unbound fractions of some samples were precipitated with 20% trichloroacetic acid (TCA). All samples were heated in loading buffer at 95°C for 3 min before analysis by SDS-PAGE and immunoblotting using polyclonal anti-IN antibody for full-length IN and anti-HIV human serum for IN220–270. In all the samples analyzed, amounts of WT or mutant IN present in the wash fractions were either undetectable or negligible. The percentage of IN binding to RT in these reactions was obtained by the following calculation: [amount bound/(amount bound + amount unbound)] × 100%. The data are expressed as the means ± standard deviations. Differences between WT and mutant means were identified by a two-tailed Student t test, and a P value of 0.01 or less was the criterion for statistical significance.
Viral attachment and entry assays.
Attachment and entry assays were performed as described previously (34). A total of 100 ng of the p24 equivalent of virus was added to 2 × 106 CEM cells, which were then placed at 4°C for 30 min to synchronize virus attachment to the cell surface. Virus-exposed cells were then incubated at 37°C for 4 h to allow infection, pelleted, and washed three times in 1 ml of ice-cold sterile PBS to remove any loosely bound virus from the cell surface. To quantify viral attachment, infected cells were disrupted in a lysis buffer consisting of PBS containing 1% Triton X-100 and 0.1 mM phenylmethylsulfonyl fluoride, and HIV-1 p24 levels in the resulting lysate were subsequently measured by ELISA.
To quantify viral entry, cells were initially treated similarly to the viral attachment assay, except that virus-exposed cells were washed only once in 1 ml of ice-cold sterile PBS following the 37°C incubation step to remove loosely bound virus. Infected cells were then incubated at 37°C for 5 min in 0.5 ml of 0.25% trypsin (Invitrogen) to remove any stably attached virus. Cells were then washed twice more in 1 ml of PBS and then resuspended in 0.5 ml of lysis buffer before viral entry was quantified by measuring the p24 levels using an ELISA.
Real-time qPCR analysis of reverse transcription products.
WT or mutant IN viruses collected from 293T producer cells were treated with 2 U/ml DNase I (New England BioLabs) in the presence of 10 mM MgCl2 for 2 h at room temperature to remove contaminating plasmid DNA. Two million CEM cells were infected with 100 ng of the p24 equivalent of DNase-treated viruses. Infected cells were pelleted by centrifugation at 16 h postinfection, washed twice in 5 ml of PBS, and then lysed in a buffer containing 6.7 mM Tris-HCl, pH 8.0, 0.23 M NaCl, 0.67 mM EDTA, 4.7 M urea, and 1.3% (wt/vol) SDS. Total DNA from the lysed cells was obtained by phenol-chloroform extraction and ethanol precipitation and subsequently used in TaqMan real-time quantitative PCR (qPCR) assays (Applied Biosystems) as previously described (50, 51). The primer pair M667 (5′-GCTAACTAGGGAACCCACTG-3′) and AA55 (5′-CTGCTAGAGATTTTCCACACTGAC-3′) that amplifies the HIV-1 R-U5 region along with the probe ZXF (5′-FAM-TGTGACTCTGGTAACTAGAGATCCCTCAGACCC-TAMRA-3′, where FAM is 6-carboxyfluorescein and TAMRA is 6-carboxytetramethylrhodamine) (34) was used to measure early reverse transcription product levels. The primer/probe pair BGF1 (5′-CAACCTCAAACAGACACCATG-3′) and BGR1 (5′-TCCACGTTCACCTTGCCC-3′) that amplifies the β-globin gene, along with the probe BGX1 (5′-FAM-CTCCTGAGGAGAAGTCTGCCGTTACTGCC-TAMRA-3′), was used as an internal standard for each DNA sample (34). All amplifications were performed in parallel with a set of linearized HIV-1 DNA standards of known concentrations. Viral DNA copy numbers of samples were calculated based on the standard curves and normalized to the internal control.
Endogenous RT activity assay.
Equal amounts of the p24 equivalent of viruses were resuspended in lysis buffer (16.7 mM Tris-HCl, pH 7.8, 80 mM KCl, 0.17 mM EDTA, 6.7 mM dithiothreitol [DTT], 33% glycerol, and 0.3% Triton X-100). All reaction mixtures were incubated at 37°C for 1 h in a final volume of 50 μl containing 0.05 U of poly(rA) · poly(dT) (Amersham Pharmacia) and 2.5 μCi of [3H]dTTP (Amersham Pharmacia). The sample was then spotted on Whatman DE81 cellulose chromatography paper, which was then air dried, washed three times with 10 ml of 2× SSC (0.03 M sodium citrate, 0.3 M NaCl; 1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), and finally washed with 10 ml of 95% ethanol. Levels of radiolabeled reverse transcription products were determined by scintillation counting.
NMR sample preparation, data collection, and analysis.
Synthetic 18-mer (5′-GTCCCTGTTCGGGCGCCA-3′) and 19-mer (5′-ATGGCGCCCGAACAGGGAC-3′) DNA oligonucleotides with sequences corresponding to the 18 3′-terminal nucleotides of the tRNALys3 primer and the viral RNA primer-binding site, respectively, were purchased from Integrated DNA Technologies. Concentrations of these oligodeoxynucleotides were determined using extinction coefficients that were provided by the manufacturer (158,700 and 185,600 M−1 cm−1 for the 18-mer and 19-mer oligonucleotides, respectively). Equimolar amounts of the 19-mer and 18-mer were annealed by heating to 90°C and slow cooling in 10 mM Tris-HCl, pH 8.0, 100 mM NaCl, and 0.02 mM EDTA to form a DNA duplex with a single A nucleotide overhang. Purified RT heterodimer and the 19-mer/18-mer DNA duplex were each dialyzed at 4°C for 12 to 14 h in NMR buffer, concentrated using centrifugal filter units (10,000 and 3,500 MWCOs, respectively; Millipore), and combined in equimolar amounts to give a final RT-DNA complex concentration of 200 μM.
Chemical shift perturbation experiments were performed by adding aliquots of this unlabeled RT-DNA complex to 150 μM 15N-labeled IN220–270 (concentration calculated assuming monomeric IN220–270) in NMR buffer with 93% H2O–7% D2O. To facilitate IN220–270 peak assignments, the same buffer and temperature conditions used in the NMR structure determination of IN220–270 (52) were employed in these NMR experiments. An initial two-dimensional 15N-1H heteronuclear single quantum coherence (HSQC) spectrum (42) of free 15N-labeled IN220–270 was recorded at 25°C on a Bruker 800 MHz Avance spectrometer. Cross-peaks in the NMR spectrum were readily assigned using archived chemical shifts found in the Protein Data Bank (PDB code 1IHV). Successive aliquots of the RT-DNA complex were then added to a solution of 15N-labeled IN220–270 to produce IN220–270:RT-DNA molar ratios of 10:1, 5:1, and 2:1 (calculated assuming monomeric amounts of IN220–270). A 15N-1H HSQC spectrum was acquired for each molar ratio. Spectra were examined, and peak intensities were quantified using the program Sparky (53). Structural representations for IN220–270 were produced using the program MacPyMOL (54).
IN activity assays.
Enzymatic activities of IN mutants were analyzed following previously published protocols (55). Both 3′-end processing and disintegration assays were performed at 37°C for 1 h in 20-μl reaction volumes containing 75 nM purified IN, 5 nM 32P-labeled substrate, 30 mM NaCl, 10 mM MnCl2, 10 mM DTT, and 0.05% IGEPAL. Reaction products were separated on a 15% polyacrylamide–7 M urea gel and quantified using a Typhoon 9410 scanner (GE Healthcare).
RESULTS
Mapping an RT-DNA binding surface on IN by NMR spectroscopy.
An RT-binding surface on IN was previously determined by NMR spectroscopy using recombinant IN220–270 and apo-RT (42). To determine whether the IN-RT interaction changes significantly when RT is bound to nucleic acid, we performed NMR chemical shift perturbation experiments to characterize the binding surface on IN220–270 in the presence of RT that had been prebound to a duplex DNA segment corresponding to the primer-binding site in the HIV-1 genome. X-ray crystal structures of this particular duplex DNA construct bound to RT are available (56, 57), and a dissociation constant of 38 ± 16 nM for RT bound to this DNA duplex has been measured (58). Such a dissociation constant, along with the RT-DNA complex concentrations used in the NMR experiments (14 to 54 μM), suggests that >91% of RT is DNA bound in our NMR experiments.
Successive aliquots of unlabeled RT prebound with duplex DNA (1:1 molar ratio) were added to 150 μM 15N-labeled sample of IN220–270 to produce IN220–270:RT-DNA ratios of 10:1, 5:1, and 2:1. A 15N-1H HSQC spectrum of each sample was acquired. No peaks showed any changes in chemical shift as the titration progressed, and most IN220–270 signals showed relatively gradual reductions in peak intensity during the course of the titration. At a 2:1 IN220–270:RT-DNA molar ratio, most of the peaks in the acquired spectrum became broadened to the noise level, and therefore no further titrations were performed.
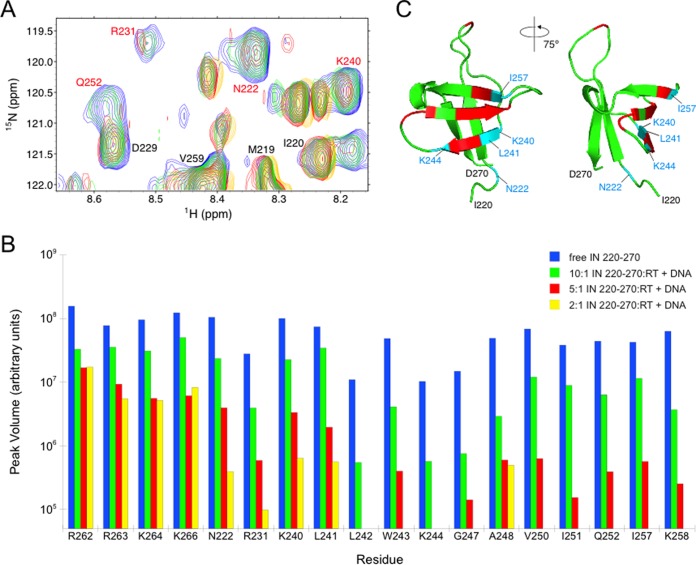
Notably, amide backbone cross-peaks associated with 14 residues showed significant decreases in intensity during the titration compared to intensity levels of other peaks in the spectra (Fig. 1A and B). Nine of the selectively affected residues (R231, L242, W243, G247, A248, V250, I251, Q252, and K258) had been previously mapped to an RT-binding surface on IN using analogous experiments with 15N-labeled IN220–270 and unlabeled apo-RT (42). In the current titration experiment, five additional residues (N222, K240, L241, K244, and I257) also showed significant amide peak decreases (Fig. 1B). These five additional residues clustered with the initial set of nine residues identified in NMR titration using free RT (Fig. 1C). The data suggest that including the DNA substrate in these NMR experiments results in an expanded RT-binding surface on IN, and the selective broadening of the peaks for these particular 14 amino acids suggests that recognition of IN220–270 by the RT-DNA complex is specific (59). Importantly, amide peaks associated with R262, R263, K264, and K266, which are residues that form contacts with free DNA (60–62), were among those peaks showing no significant intensity decreases over the course of the titration (Fig. 1B). The lack of significant amide signal perturbation from these residues suggests that the strong intensity changes in the IN220–270 spectrum observed in the presence of RT and DNA are due to IN220–270 interactions with the RT-DNA complex and not to nonspecific interactions between free DNA and IN220–270. Finally, signal decreases observed during the titration suggest that these 14 residues experience intermediate exchange on the NMR time scale (i.e., chemical exchange occurs at a frequency that is on the order of the difference between free and bound amide group chemical shift) (63, 64).
FIG 1.
Mapping amino acids with significant changes in peak intensity to the IN220–270 structure. (A) Chemical shift perturbation measurements using 15N-labeled IN220–270 and unlabeled RT prebound with DNA. A representative region of the HSQC spectra shows free IN220–270 (blue) and IN220–270 mixed with RT-DNA at molar ratios of 10:1 (green), 5:1(red), and 2:1 (yellow). IN residues labeled in red show significant decreases in amide peak intensity upon addition of RT-DNA. (B) HSQC peak intensities as the titration proceeded for residues from the putative RT-DNA-binding surface on IN220–270, as well as peak intensities from putative DNA-binding residues R262, R263, K264, and K266. Peak intensities for free 15N-labeled IN220–270 and for IN220–270 mixed with unlabeled RT-DNA at molar ratios of 10:1, 5:1, and 2:1 are shown, identified by color as indicated on the figure. Any peak intensities that dropped below the noise threshold of the NMR experiment during the course of the titration are not shown. (C) The apo-RT-binding surface on IN220–270 previously identified is shown in red, and the expanded set of five residues that are newly identified in the current study are shown in blue. Collectively, these 14 residues form the RT-DNA-binding surface on IN220–270.
Characterization of mutant IN viruses to determine functional relevance of RT-IN interaction.
We evaluated the functional relevance of this newly mapped surface on IN by examining the effect of perturbing the putative interacting surface upon viral replication. Since nine IN residues (R231, L242, W243, G247, A248, V250, I251, Q252, and K258) appear to interact with RT with or without DNA, we elected to focus our mutagenesis efforts upon IN residues within this particular set. We have previously investigated the effects of introducing substitutions at two of the putative nine RT-binding residues and found that W243E and V250E substitutions significantly impaired viral replication (42). The exact phase of the viral life cycle affected by these particular amino acid substitutions, as well as substitutions at other IN positions within the putative RT-binding surface, was analyzed in this report. We hypothesize that RT-IN interactions are necessary for reverse transcription during the viral life cycle and that perturbing this interaction will lead to a defect specifically at the reverse transcription step. To determine the functional relevance of the putative RT-binding surface on the IN C-terminal domain, we introduced single amino acid substitutions that alter surface polarity or electrostatic charge using site-directed mutagenesis. If the IN residue is indeed involved in RT binding only and if the binding is critical during viral replication, we expect that the introduced substitution will disrupt RT binding and cause a replication defect specifically at the reverse transcription step without other secondary, off-target effects.
Analysis of replication kinetics.
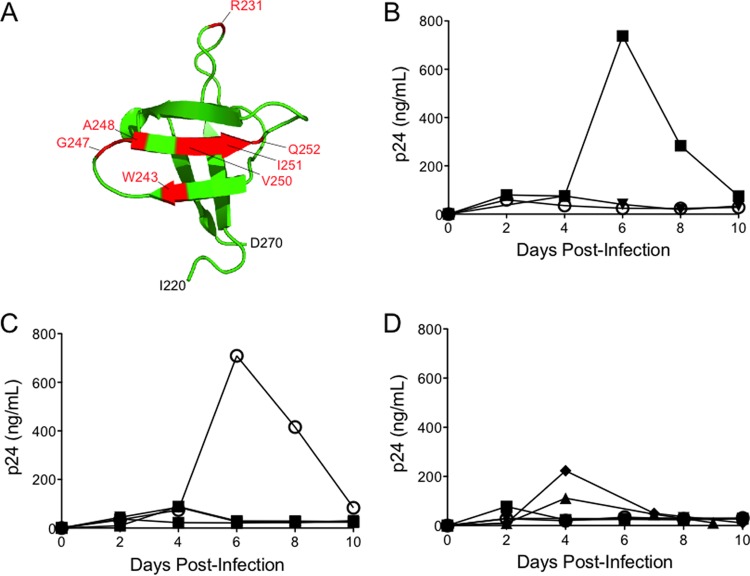
We first investigated the effects of these various substitutions upon viral replication kinetics by infecting CEM cells with equal amounts of p24 equivalent of WT or IN mutant viruses. Virus production was monitored by measuring p24 content in medium every 2 days over a 10-day period (Fig. 2). Negative controls included a mock infection (medium only without virus) and an infection using heat-inactivated wild-type virus. Of the nine IN mutants tested (R231E, W243E, G247A, G247E, A248E, V250A, V250E, I251E, and Q252A), only G247A replicated with kinetics similar to that of the WT virus. In cells infected with either WT or IN mutant G247A, syncytium formation was evident after day 6, which correlated with a peak in the p24 level detected in culture (Fig. 2B and C). IN mutants V250A and Q252A exhibited decreased p24 levels, but levels were above background. Since these two IN mutants still retain a low but detectable level of replication, they were not further characterized. In contrast, mutants R231E, G247E, A248E, and I251E were nonviable (Fig. 2C and D). IN mutants W243E and V250E were also unable to replicate, as previously reported (42).
FIG 2.
Replication kinetics of WT and IN mutant HIV-1 viral clones. CEM cells were infected with 100 ng of the p24 equivalent of WT or IN mutant viruses, and the culture medium was tested for p24 production at the indicated time points postinfection. Data representative of three replicates are shown. (A) The positions of the six IN substitution mutants on the RT-binding surface of IN220–270 are as labeled. (B) Replication kinetics of the WT (■), heat-inactivated WT (HI; ○), and mock control (no virus added; ▼). (C) Replication kinetics of IN mutants R231E (■), W243E (▼), G247A (○), and G247E (●). (D) Replication kinetics of IN mutants A248E (▼), V250A (▲), V250E (○), I251E (■), and Q252A (◆).
Analysis of polyprotein packaging and processing.
Several IN mutants have been previously reported to be replication defective due to impaired polyprotein processing (22, 65). We examined the effects of the six replication-incompetent IN mutants on viral protein packaging and precursor processing by Western blot analysis. The G247A IN mutant, which replicates similarly to the WT (Fig. 2C), was included as another positive control. One hundred nanograms of the p24 equivalent of WT or IN mutant virus was lysed, separated on a 12% denaturing polyacrylamide gel, and transferred to nitrocellulose membranes. Blots were probed with either human anti-HIV serum to detect all packaged viral proteins (Fig. 3A) or with antibodies specific for RT or IN (Fig. 3B and C). All IN mutants exhibited proper Gag-Pol packaging and processing at levels comparable to those of the WT control (Fig. 3A and B). The virion incorporation of IN in mutants W243E, V250E, and I251E was further confirmed by probing with an anti-IN antibody (Fig. 3C). Mutants W243E, V250E, and I251E contained less incorporated IN than that found in the WT and less incorporated IN than the amount of incorporated RT. One possible explanation is that the processed INs of mutants W243E, V250E, and I251E were subjected to enhanced degradation. However, we found through direct examination using immunoblotting that purified mutated INs W243E, V250E, and I251E reacted poorly with the anti-HIV human serum and the polyclonal anti-IN antibody (data not shown). The low sensitivity of these particular detection reagents against the mutated INs during immunoblotting represents at least a contributing factor to the apparent poor incorporation of processed IN with these IN mutants.
FIG 3.
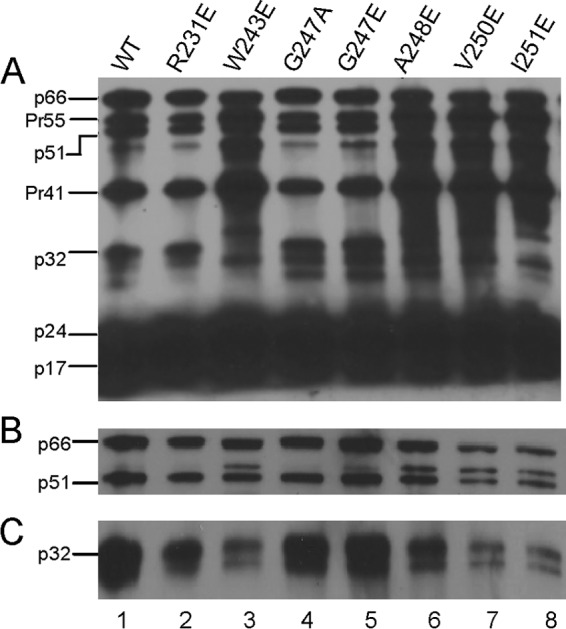
Western blot analysis of HIV-1 proteins packaged in WT and IN mutant virus particles. Each lane contains 100 ng of the p24 equivalent of the viral lysate. Blots were probed with human anti-HIV-1 serum (A), anti-RT antibody (B), or anti-IN antibody (C). Labels on the left correspond to positions of viral proteins with their molecular masses in kilodaltons. p66 and p51 correspond to the subunits of the RT heterodimer, Pr55 and Pr41 represent Gag precursor proteins, p32 corresponds to IN, p24 corresponds to capsid, and p17 corresponds to matrix.
Replication-deficient IN mutants show decreased binding to RT.
To determine if the replication defect of the IN mutants was caused by interference with RT-IN complex formation, we expressed and purified the recombinant WT IN and various single-amino-acid-substituted IN derivatives and measured their abilities to bind immobilized RT by carrying out a pulldown assay using RT-conjugated magnetic beads. Based on their WT levels of viral protein packaging and processing, we selected IN mutants R231E, W243E, G247E, and A248E for further analyses. A reaction mixture containing 20 pmol of WT IN and magnetic beads without any conjugated RT served as a negative control to assess any nonspecific interaction, and IN binding to the beads was not detected under these conditions (data not shown). When the reaction mixture contained magnetic beads conjugated with RT, incubation with WT IN resulted in 29% ± 11% of the IN being cosedimented (Fig. 4A). When the reaction mixture was incubated with IN bearing the G247A substitution, which has no effect on the replication kinetics (Fig. 2C), the amount of IN pulled down by the beads was 107% of the WT level (Fig. 4A). In the presence of an identical amount of RT-conjugated magnetic beads, the levels of mutated INs R231E, W243E, G247E, and A248E cosedimented with the beads were 39% ± 19%, 34% ± 14%, 36% ± 10%, and 43% ± 18%, respectively, of the WT IN level (Fig. 4A). The levels of cosedimentation of these four IN mutants were all significantly less than the WT IN level (P ≤ 0.01).
FIG 4.
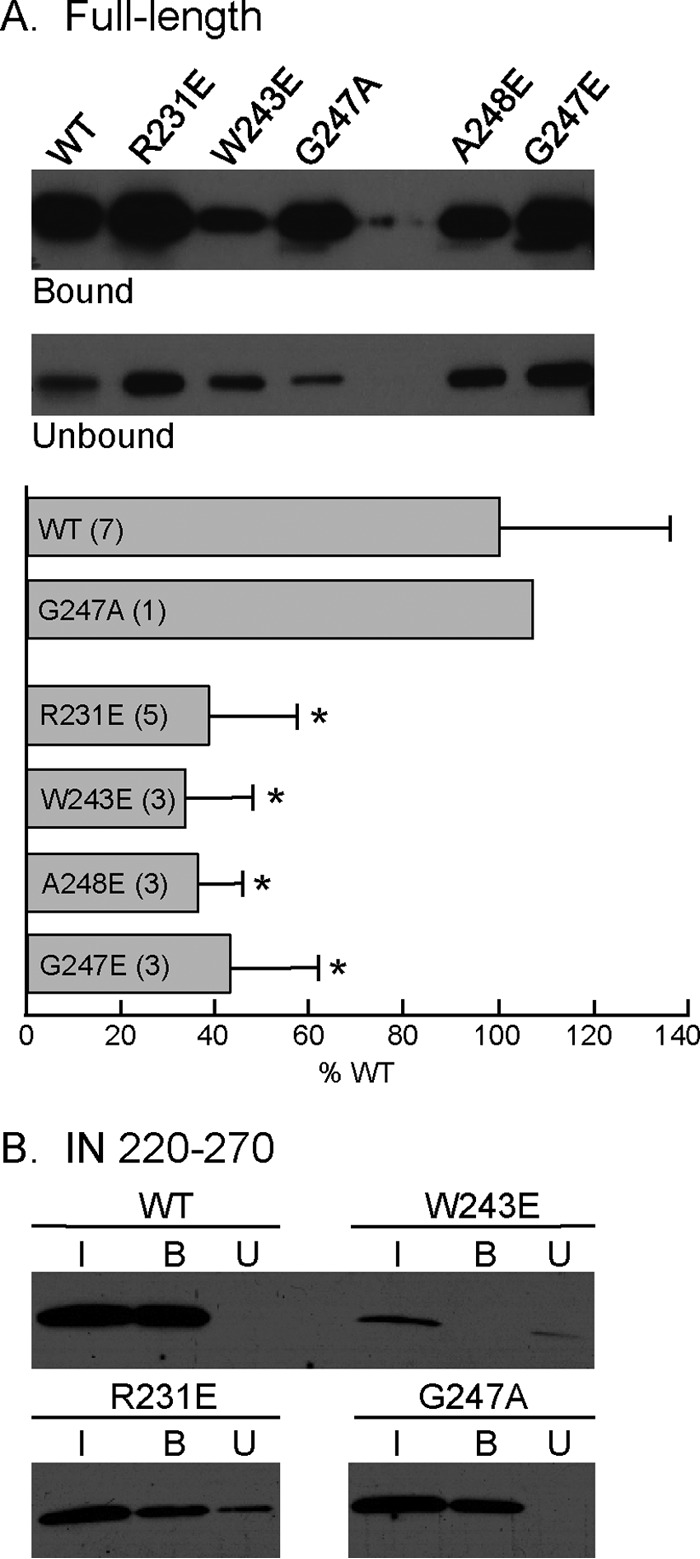
Binding of HIV-1 RT-conjugated magnetic beads with the WT or the indicated substitutions within the context of full-length IN or IN220–270. (A) Full-length IN. Bound and unbound fractions were obtained as described in Materials and Methods. The final volume of the unbound fraction was 500 μl, and a 40-μl aliquot was analyzed by SDS-PAGE and immunoblotting. The level of WT IN binding to RT was normalized to 100%, and the values of mutant INs are expressed as percentages of the binding of the WT IN. Data are expressed as the means ± standard deviations, and the numbers in parentheses represent the number of experiments. Asterisks denote significant differences from WT (P ≤ 0.01). (B) IN220–270. The amounts of IN in the input, bound, and unbound fractions are denoted I, B, and U, respectively. The unbound fraction was TCA precipitated, and the pellet was resuspended in 30 μl of loading buffer before analysis by SDS-PAGE and immunoblotting.
We have shown previously that the C-terminal domain of IN (IN220–270) is necessary and sufficient in binding RT, and a K258A substitution in the context of IN220–270 shows an 8-fold decrease in binding affinity to RT compared to that of WT IN (42). To further confirm the decreased ability of the mutated INs to bind RT, we repeated the pulldown assay using purified IN220–270 containing the R231E or W243E substitution (Fig. 4B). IN220–270 bearing the G247A substitution was used as a positive control. Compared to the WT IN, the cosedimented amounts of IN220–270 containing the R231E, W243E, and G247A substitutions were 65%, 34%, and 98%, respectively, of the WT IN220–270 (Fig. 4B).
Similar to the observation made earlier for virion incorporated of IN in different mutants, the recovery (bound plus unbound) variation among WT and mutated INs, especially W243E, was likely attributable to their different reactivities to antibodies during immunoblotting (data not shown).
Replication-defective and poor-RT-binding IN mutants are impaired in early viral cDNA synthesis.
Our analyses thus far have indicated that several amino acid substitutions on the purported RT-binding surface of IN results in replication-incompetent viruses and disruption of the RT-IN interaction. To confirm the biological relevance of the interaction, we examined the ability of the four replication-defective and poor-RT-binding IN mutants to synthesize viral cDNA. Experiments using mock-infected cells and cells infected with heat-inactivated WT virus served as negative controls. Viruses were treated with DNase I prior to infection assays to eliminate contamination from any residual NL4-3 plasmid DNA carried over from transfection. Total DNA was extracted from target cells at 16 h postinfection and analyzed for the early reverse transcription product, minus-strand strong stop DNA (−sssDNA), using qPCR as previously described (34, 51). DNA input from each sample was normalized to the level of the human β-globin gene, which was amplified under identical conditions. All four replication-defective IN mutants were significantly impaired for −sssDNA synthesis, exhibiting activity ranging from 0 to 25% of the WT level (Fig. 5A).
FIG 5.
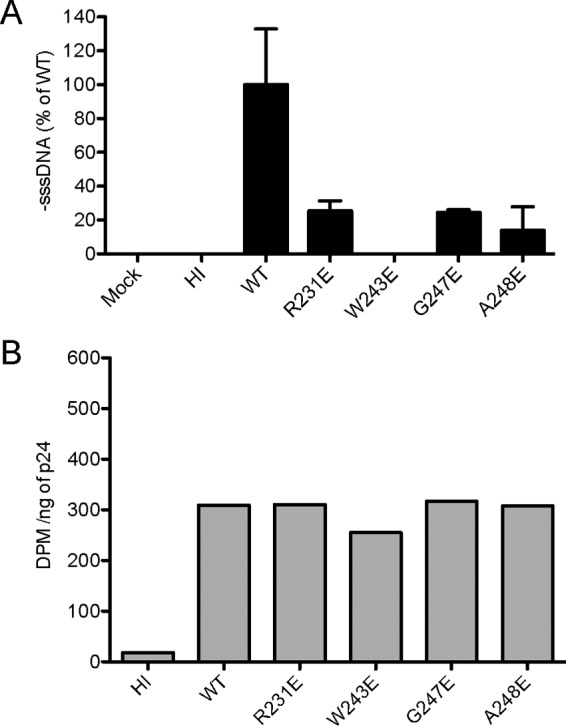
qPCR analysis of viral cDNA synthesis in cells infected with IN mutant viral clones. Values in the bar graphs represent the means of two replicates. (A) Amounts of early reverse-transcribed viral cDNA present in infected cells were measured 16 h after infection. DNA input of each sample was normalized by the amount of human β-globin gene amplified under identical conditions. (B) Endogenous RT activity assay of cell lysates prepared from cells infected by WT or IN mutants. Radioactivity incorporated into reverse transcription products was determined by scintillation counting and expressed in disintegrations per minute (DPM). Viruses are described in the legend to Fig. 2.
To account for the possibility of a defective RT resulting in the lack of reverse transcription products, we examined endogenous reverse transcriptase activity in each IN mutant virus. Equal amounts of detergent-lysed IN mutant virions were supplied with an exogenous template and radiolabeled TTP as substrates, and the amount of newly transcribed radioactive transcripts was measured. All IN mutants packaged RT at levels comparable to the level of the wild-type virus (Fig. 3B). Based on the amount of [3H]TTP incorporated into the reverse transcription product, we observed no significant difference in RT-catalyzed polymerization activity in any of the mutant IN viruses relative to that of the WT virus, ruling out the possibility that the observed lack of reverse transcription products was due to a defective RT (Fig. 5B). These results indicate that IN substitutions at the RT-binding surface adversely impact reverse transcription, leading to defects in viral replication without affecting endogenous RT activity.
Replication-defective and poor-RT-binding IN mutants are competent in viral attachment and entry.
To verify that the defect of the replication-incompetent IN mutants occurs specifically during the step of reverse transcription, we determined the ability of the four replication-defective and poor-RT-binding mutant viruses to attach and enter cells. CEM cells were incubated with equal amounts of the p24 equivalent of WT virus or each of the four IN mutant viruses. As a positive control, we included the replication-incompetent IN C130S mutant, which is normal in cell attachment and entry and has a specific defect at the step of reverse transcription (28, 34). Negative controls included both mock-infected cells and cells infected with heat-inactivated WT virus. Following a 4-h incubation with virus, cells were either treated with trypsin to facilitate subsequent detection of intracellular p24 levels in cell lysates or washed and lysed without trypsin treatment to quantify total amounts of both cell-associated and intracellular p24. All IN mutants showed amounts of cell-associated and intracellular p24 comparable to those of the WT virus (Fig. 6). These results showed that all four IN mutants deficient in replication and reverse transcription have no defects in virus attachment and entry.
FIG 6.
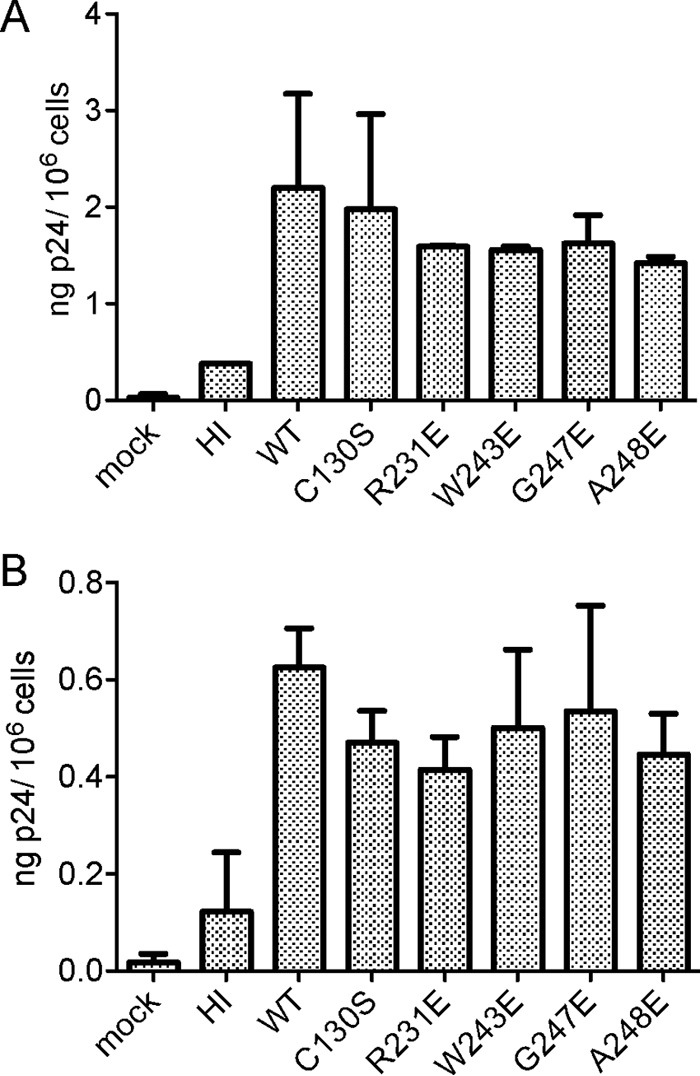
Attachment and entry cell assay analysis of replication-defective IN mutants. CEM cells were infected with 100 ng of the p24 equivalent of WT or IN mutant viruses and analyzed for viral attachment (A) and entry (B) as described in Materials and Methods. Cells exposed to HI virus and a mock control were used as negative controls. Viruses are described in the legend to Fig. 2.
Recombinant mutant INs that bind RT poorly are catalytically active.
To investigate whether the IN substitutions affected IN enzymatic activities, we assessed in vitro the ability of the various recombinant INs to catalyze the 3′ end processing (Fig. 7A) and disintegration reactions (Fig. 7B). All replication-defective mutant INs maintained in vitro 3′-end processing and disintegration activities similar to those of the WT enzyme (Fig. 7). Since the four substitutions in the IN CTD did not adversely affect catalysis, we infer that these substitutions do not grossly perturb protein folding and IN dimerization required for proper 3′ end processing and disintegration.
FIG 7.
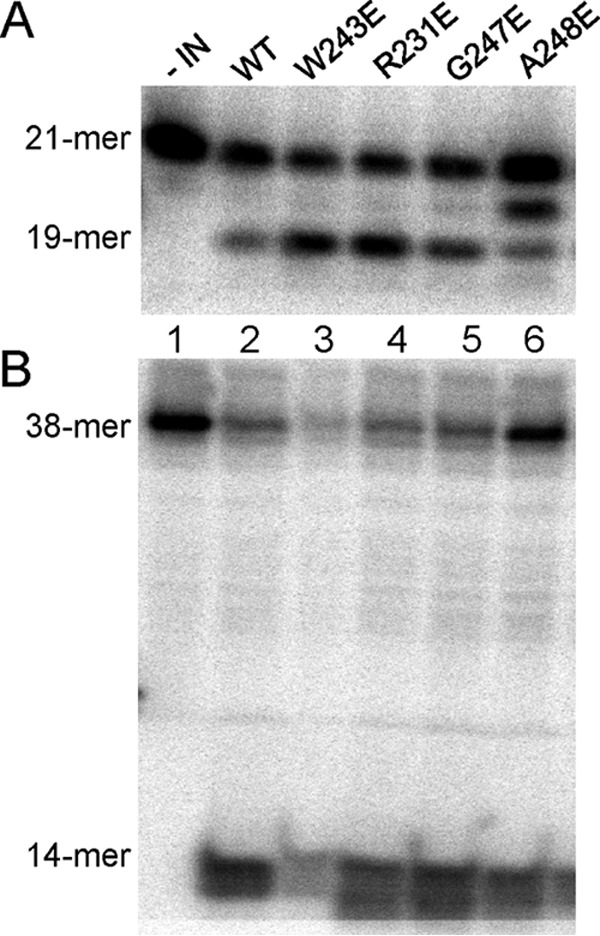
In vitro IN activity assays for WT and IN mutants. (A) 3′ End processing. 5′-end 32P-labeled 21-mer substrate was incubated with 40 pmol of WT (lane 2) or mutated INs (lanes 3 to 6) to assess dinucleotide cleavage activity, which produces a 19-mer product. (B) Disintegration activity catalyzed by WT or mutated INs using a 5′-end 32P-labeled disintegration substrate. The assay measures the ability of IN to resolve the substrate mimicking the integration intermediate into a 14-nucleotide labeled hairpin product (viral DNA) and an unlabeled 24-nucleotide circular molecule (target DNA). In both panels, lane 1 represents a negative control that contained the labeled substrate without IN.
DISCUSSION
In this study, we have mapped the putative RT-binding surface on HIV-1 IN by NMR and determined the biological relevance of the IN-RT interaction during viral replication. Using the structural information as a guide, we have prepared and characterized nine mutant viruses, with each containing a substitution of an IN residue located within the RT-binding surface. Several IN mutants are completely replication incompetent with the defect occurring specifically during synthesis of early reverse transcription products. Purified recombinant HIV-1 INs containing such substitutions, though catalytically active, are deficient in binding RT. The specific impairment of early reverse transcription in viruses harboring IN substitutions at the putative RT-binding surface, together with the disruption of RT-binding by IN derivatives containing such substitutions, supports our hypothesis that the RT-IN interaction is biologically significant during reverse transcription.
The mechanistic basis by which IN is specifically required for reverse transcription during HIV-1 replication is not known. Reverse transcription catalyzed by recombinant HIV-1 RT is inefficient in vitro (66, 67). This is partly due to poor processivity and a propensity for RT to dissociate from the primer-template at pause sites (68). The early events of reverse transcription include two distinct modes: initiation, which corresponds to the incorporation of the first five nucleotides, has a primer-specific and distributive mode of polymerization, while elongation has a nonspecific and processive mode of DNA synthesis (69). Using recombinant proteins, we previously found that IN stimulates both the initiation and elongation modes of reverse transcription by increasing RT processivity and suppressing formation of pause products (27). Since all four replication-defective IN mutants examined have a specific defect at the step of early reverse transcription and since recombinant INs containing the same substitutions are impaired in RT binding, we speculate that the IN substitutions disrupt RT-IN complex formation required in the early steps of reverse transcription, rendering the virus replication incompetent.
The RT-binding surface on IN has been determined previously using IN220–270 and apo-RT alone (42), and the effect of the presence of a DNA substrate on the RT-IN complex was not known. Binding kinetics between IN and RT are consistent with a two-state reaction process, suggesting that recognition of IN involves a conformational change in RT (42). Indeed, X-ray crystal structures of apo-RT and DNA-bound RT (56, 57) and molecular dynamics simulations (70) indicate that conformational flexibility and mobility are prominent features of both p66 and p51 subunits upon substrate recognition and binding. In particular, the p66 subunit adopts an open conformation when bound to nucleic acid substrate, with the thumb subdomain rotating away from the fingers subdomain to expose a large cleft, while the p66 of apo-RT assumes a closed conformation, with the thumb domain filling the cleft (56, 71–73). RT-catalyzed DNA-dependent DNA polymerization studies suggest that IN has greater affinity for RT bound to a DNA template-primer pair than for RT alone (44), implying that IN interacts with RT in the open conformation in this particular setting.
To examine the RT-IN interaction under a more physiological condition, we have performed the same NMR approach to identify the RT-binding surface on IN220–270 in the presence of RT that was prebound with equimolar amounts of a 19-mer/18-mer duplex DNA that mimics the primer-template substrate. In addition to the nine amino acids identified earlier with apo-RT (42), the introduction of the DNA substrate resulted in an expanded binding surface that included five more IN residues. With the exception of R231, the residues comprising this expanded binding surface mapped to one side of the IN220–270 structure (Fig. 1C). While not located precisely at the RT-binding surface, R231 is positioned within a flexible loop connecting the β1 and β2 strands near the RT-binding surface (52) and can conceivably extend to form RT contacts with or without conformational adjustment.
We attribute the expanded binding surface to IN220–270 interactions with the RT-DNA complex rather than to any free DNA species present in our NMR titration experiment. In support of this interpretation, the previously measured dissociation constant of ∼38 nM for this RT-DNA template-primer complex (58) suggests that free DNA levels in our system should be relatively low (<9% of the total DNA used in these NMR experiments). Furthermore, we expect that any free DNA in this system would bind nonspecifically to the positively charged residues R262, R263, and K264 that are located on the face opposite to the putative RT-binding surface of IN220–270. Previous work with the HIV-2 IN CTD indicates that R262, R263, and K264 are involved in nonspecific DNA binding (61, 62), and homology modeling of HIV-1 IN onto the PFV intasome structure, along with cross-linking assays with DNA and the K266E mutant IN, suggests that HIV-1 IN K266 is involved in forming DNA contacts (60). We observed no strong peak intensity decreases for R262, R263, K264, or K266 during the course of our NMR titration experiments. Instead, peak intensities for residues from the putative RT-binding surface drop off much more quickly than those residues expected to form DNA contacts (Fig. 1B). This result is consistent with a view that the set of residues showing rapid peak changes form interactions with the RT-DNA complex and not free DNA, as any free DNA present in the system should produce peak changes on the opposite side of the molecule (i.e., at R262, R263, K264, and K266). In all, our data support a model that includes a specific RT-binding surface on IN that expands when it interacts with DNA-bound RT. Moreover, identifying the same protein surface on IN220–270 in the presence of the RT-DNA complex or apo-RT alone further validates the biological relevance of this IN surface in binding RT.
IN mutations can be pleiotropic, affecting various steps in the virus life cycle (18, 19). Besides integration and reverse transcription, other steps include nuclear import of preintegration complexes (26, 35–37), polyprotein processing, assembly, and maturation (22, 31, 38–40, 65, 74). Since IN is synthesized and packaged into the immature virion as part of the Gag-Pol polyprotein, many alterations in virion morphology and defects at the late steps of replication may be caused by the effect of IN mutations on the Gag-Pol precursor protein (22, 38, 40, 65, 74). However, certain IN mutations, such as His12Ala and Phe185Ala substitutions and deletion of the C-terminal 22 residues, specifically impair reverse transcription with no apparent effects on the Gag-Pol polyprotein and other steps in the life cycle (29, 30, 33). Also, many class II mutations involving residues in the IN CTD share a reverse transcription defect, with no apparent alteration to polyprotein processing and packaging by the IN substitutions (22). Mutagenesis experiments involving some of the RT-binding IN residues identified in this study have been reported previously. In support of our finding that the 14 residues identified by the NMR represent the RT-binding surface, replacing the conserved Leu residue at position 241 with Ala (L241A) resulted in a replication-defective virus at the cDNA synthesis step (22). Substituting Glu for the important DNA-interacting residue Lys244, which is among the five additional residues constituting the expanded RT-binding surface on IN when RT is prebound with DNA, led to a replication-defective virus with WT levels of late reverse-transcribed products and defective IN enzymatic activity (75); however, a double IN mutant, K240A/K244E, exhibited a significant decrease in this reverse transcription product (26). Also, the identity of the amino acid substitution plays an important role in the phenotype of the resulting mutant. In this study, we showed that replication of the mutant virus G247A is indistinguishable from that of the WT, whereas the G247E mutant is replication defective and has impaired RT binding. Similarly, we found that the viral clone R231E is replication incompetent and defective in binding RT, whereas an earlier study reported that an R231A mutant is replication competent (22). Additionally, virus harboring Gly247 with a Trp (G247W) substitution is replication incompetent, with a Gag processing defect caused by activation of an alternative splice site (65). We did not observe any effects on Gag-Pol processing when the same amino acid was replaced with either Ala or Glu (G247A or G247E) (Fig. 3).
Of note, the host protein Gemin2/SIP1, a member of the survival motor neuron complex that mediates the assembly of spliceosomal small nuclear ribonucleoproteins (76), has been reported to interact with HIV-1 IN to facilitate reverse transcription (77). Knockdown of Gemin2 with small interfering RNA reduces HIV-1 infectivity and lowers early reverse transcription products but has no effect on HIV-1 expression from integrated proviruses (77). Further analyses showed that disrupting the IN-Gemin2 interaction abrogates reverse transcription, and Gemin2 functions to promote reverse transcription by stabilizing IN multimerization and enhancing IN-RT assembly on viral RNA (43). Therefore, in addition to direct binding, IN may impact reverse transcription in the context of an IN-Gemin2 complex. Residues critical for Gemin2 interaction are located within the central core and C-terminal domains of IN (43, 77). In addition to Gemin2, several other host proteins have been shown to interact with HIV-1 IN (for a review, see reference 78). Some of these host proteins, such as Ku70 and dynein, can bind to the IN CTD, and their interactions with IN appear to play a role during reverse transcription (79, 80). We do not know at present if these IN-binding host proteins can affect positively the RT-IN interaction or compete with RT for IN binding. Notably, we have observed that several IN mutants exhibit a greater defect on replication than RT binding (compare Fig. 2 and 4). It is possible that the cosedimentation assay does not fully recapitulate the RT-IN interaction in vivo or that the IN substitutions tested in this study may additionally affect the binding of IN with other host proteins, which consequently contribute to the overall defect in viral replication.
Our results showing that substitutions at the RT-binding surface on IN significantly impair viral replication in tissue culture, specifically at the reverse transcription step, indicate that the RT-IN interaction is functionally significant during viral replication. Considering the emergence of viral resistance observed against many clinically relevant anti-HIV drugs that target the enzymatic active sites of either RT or IN, the critical interaction between RT and IN may function as an allosteric target for designing and developing new antiviral drugs.
ACKNOWLEDGMENTS
The ELISA for p24 was carried out by the Center for AIDS Research (CFAR) Virology Core Facility of the University of California, Los Angeles (UCLA), which is supported by UCLA CFAR grant 5P30 AI028697 and the UCLA AIDS Institute and by the UCLA Council of Bioscience Resources. This work was partly supported by NIH grant GM101416 and an International Collaborative grant (number 2012C24015) from the Science Technology Department of Zhejiang Province, China, to S.A.C. S.S.T. was partly supported by Research Training in Pharmacological Sciences (NIH T32GM008652) and a UCLA AIDS Institute/CFAR Fellowship Award. J.T.M. and S.F.J.L.G. are supported by the Intramural Research Program of the National Cancer Institute, NIH, Department of Health and Human Services.
REFERENCES
- 1.Hill M, Tachedjian G, Mak J. 2005. The packaging and maturation of the HIV-1 Pol proteins. Curr HIV Res 3:73–85. doi: 10.2174/1570162052772942. [DOI] [PubMed] [Google Scholar]
- 2.Telesnitsky A, Goff SP. 1997. Reverse transcriptase and the generation of retroviral DNA, p 121–160. In Coffin JM, Hughes SH, Varmus HE (ed), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor Laboratory, NY. [PubMed] [Google Scholar]
- 3.Hu WS, Hughes SH. 2012. HIV-1 reverse transcription. Cold Spring Harb Perspect Med 2:a006882. doi: 10.1101/cshperspect.a006882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jacobo-Molina A, Arnold E. 1991. HIV reverse transcriptase structure-function relationships. Biochemistry 30:6351–6356. doi: 10.1021/bi00240a001. [DOI] [PubMed] [Google Scholar]
- 5.Engelman A, Mizuuchi K, Craigie R. 1991. HIV-1 DNA integration: mechanism of viral DNA cleavage and DNA strand transfer. Cell 67:1211–1221. doi: 10.1016/0092-8674(91)90297-C. [DOI] [PubMed] [Google Scholar]
- 6.Li X, Krishnan L, Cherepanov P, Engelman A. 2011. Structural biology of retroviral DNA integration. Virology 411:194–205. doi: 10.1016/j.virol.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chow SA, Vincent KA, Ellison V, Brown PO. 1992. Reversal of integration and DNA splicing mediated by integrase of human immunodeficiency virus. Science 255:723–726. doi: 10.1126/science.1738845. [DOI] [PubMed] [Google Scholar]
- 8.Hare S, Di Nunzio F, Labeja A, Wang J, Engelman A, Cherepanov P. 2009. Structural basis for functional tetramerization of lentiviral integrase. PLoS Pathog 5:e1000515. doi: 10.1371/journal.ppat.1000515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hare S, Gupta SS, Valkov E, Engelman A, Cherepanov P. 2010. Retroviral intasome assembly and inhibition of DNA strand transfer. Nature 464:232–236. doi: 10.1038/nature08784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li M, Mizuuchi M, Burke TR Jr, Craigie R. 2006. Retroviral DNA integration: reaction pathway and critical intermediates. EMBO J 25:1295–1304. doi: 10.1038/sj.emboj.7601005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hare S, Maertens GN, Cherepanov P. 2012. 3′-Processing and strand transfer catalysed by retroviral integrase in crystallo. EMBO J 31:3020–3028. doi: 10.1038/emboj.2012.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maertens GN, Hare S, Cherepanov P. 2010. The mechanism of retroviral integration from X-ray structures of its key intermediates. Nature 468:326–329. doi: 10.1038/nature09517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bera S, Pandey KK, Vora AC, Grandgenett DP. 2009. Molecular Interactions between HIV-1 integrase and the two viral DNA ends within the synaptic complex that mediates concerted integration. J Mol Biol 389:183–198. doi: 10.1016/j.jmb.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Faure A, Calmels C, Desjobert C, Castroviejo M, Caumont-Sarcos A, Tarrago-Litvak L, Litvak S, Parissi V. 2005. HIV-1 integrase crosslinked oligomers are active in vitro. Nucleic Acids Res 33:977–986. doi: 10.1093/nar/gki241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kotova S, Li M, Dimitriadis EK, Craigie R. 2010. Nucleoprotein intermediates in HIV-1 DNA integration visualized by atomic force microscopy. J Mol Biol 399:491–500. doi: 10.1016/j.jmb.2010.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krishnan L, Engelman A. 2012. Retroviral integrase proteins and HIV-1 DNA integration. J Biol Chem 287:40858–40866. doi: 10.1074/jbc.R112.397760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang JY, Ling H, Yang W, Craigie R. 2001. Structure of a two-domain fragment of HIV-1 integrase: implications for domain organization in the intact protein. EMBO J 20:7333–7343. doi: 10.1093/emboj/20.24.7333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cannon PM, Wilson W, Byles E, Kingsman SM, Kingsman AJ. 1994. Human immunodeficiency virus type 1 integrase: effect on viral replication of mutations at highly conserved residues. J Virol 68:4768–4775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Engelman A, Englund G, Orenstein JM, Martin MA, Craigie R. 1995. Multiple effects of mutations in human immunodeficiency virus type 1 integrase on viral replication. J Virol 69:2729–2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Engelman A. 1999. In vivo analysis of retroviral integrase structure and function. Adv Virus Res 52:411–426. doi: 10.1016/S0065-3527(08)60309-7. [DOI] [PubMed] [Google Scholar]
- 21.Li X, Koh Y, Engelman A. 2012. Correlation of recombinant integrase activity and functional preintegration complex formation during acute infection by replication-defective integrase mutant human immunodeficiency virus. J Virol 86:3861–3879. doi: 10.1128/JVI.06386-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu R, Ghory HZ, Engelman A. 2005. Genetic analyses of conserved residues in the carboxyl-terminal domain of human immunodeficiency virus type 1 integrase. J Virol 79:10356–10368. doi: 10.1128/JVI.79.16.10356-10368.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu R, Limon A, Ghory HZ, Engelman A. 2005. Genetic analyses of DNA-binding mutants in the catalytic core domain of human immunodeficiency virus type 1 integrase. J Virol 79:2493–2505. doi: 10.1128/JVI.79.4.2493-2505.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Briones MS, Chow SA. 2010. A new functional role of HIV-1 integrase during uncoating of the viral core. Immunol Res 48:14–26. doi: 10.1007/s12026-010-8164-z. [DOI] [PubMed] [Google Scholar]
- 25.Briones MS, Dobard CW, Chow SA. 2010. Role of human immunodeficiency virus type 1 integrase in uncoating of the viral core. J Virol 84:5181–5190. doi: 10.1128/JVI.02382-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ao Z, Fowke KR, Cohen EA, Yao X. 2005. Contribution of the C-terminal tri-lysine regions of human immunodeficiency virus type 1 integrase for efficient reverse transcription and viral DNA nuclear import. Retrovirology 2:62. doi: 10.1186/1742-4690-2-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dobard CW, Briones MS, Chow SA. 2007. Molecular mechanisms by which human immunodeficiency virus type 1 integrase stimulates the early steps of reverse transcription. J Virol 81:10037–10046. doi: 10.1128/JVI.00519-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hehl EA, Joshi P, Kalpana GV, Prasad VR. 2004. Interaction between human immunodeficiency virus type 1 reverse transcriptase and integrase proteins. J Virol 78:5056–5067. doi: 10.1128/JVI.78.10.5056-5067.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leavitt AD, Robles G, Alesandro N, Varmus HE. 1996. Human immunodeficiency virus type 1 integrase mutants retain in vitro integrase activity yet fail to integrate viral DNA efficiently during infection. J Virol 70:721–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Masuda T, Planelles V, Krogstad P, Chen ISY. 1995. Genetic analysis of human immunodeficiency virus type 1 integrase and the U3 att site: unusual phenotype of mutants in the zinc finger-like domain. J Virol 69:6687–6696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shin C-G, Taddeo B, Haseltine WA, Farnet CM. 1994. Genetic analysis of the human immunodeficiency virus type 1 integrase protein. J Virol 68:1633–1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsurutani N, Kubo M, Maeda Y, Ohashi T, Yamamoto N, Kannagi M, Masuda T. 2000. Identification of critical amino acid residues in human immunodeficiency virus type 1 IN required for efficient proviral DNA formation at steps prior to integration in dividing and nondividing cells. J Virol 74:4795–4806. doi: 10.1128/JVI.74.10.4795-4806.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu X, Liu H, Xiao H, Conway JA, Hehl E, Kalpana GV, Prasad V, Kappes JC. 1999. Human immunodeficiency virus type 1 integrase protein promotes reverse transcription through specific interactions with the nucleoprotein reverse transcription complex. J Virol 73:2126–2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhu K, Dobard C, Chow SA. 2004. Requirement for integrase during reverse transcription of human immunodeficiency virus type 1 and the effect of cysteine mutations of integrase on its interactions with reverse transcriptase. J Virol 78:5045–5055. doi: 10.1128/JVI.78.10.5045-5055.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ikeda T, Nishitsuji H, Zhou X, Nara N, Ohashi T, Kannagi M, Masuda T. 2004. Evaluation of the functional involvement of human immunodeficiency virus type 1 integrase in nuclear import of viral cDNA during acute infection. J Virol 78:11563–11573. doi: 10.1128/JVI.78.21.11563-11573.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jayappa KD, Ao Z, Yang M, Wang J, Yao X. 2011. Identification of critical motifs within HIV-1 integrase required for importin α3 interaction and viral cDNA nuclear import. J Mol Biol 410:847–862. doi: 10.1016/j.jmb.2011.04.011. [DOI] [PubMed] [Google Scholar]
- 37.Woodward CL, Prakobwanakit S, Mosessian S, Chow SA. 2009. Integrase interacts with nucleoporin NUP153 to mediate the nuclear import of human immunodeficiency virus type 1. J Virol 83:6522–6533. doi: 10.1128/JVI.02061-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bukovsky A, Gottlinger H. 1996. Lack of integrase can markedly affect human immunodeficiency virus type 1 particle production in the presence of an active viral protease. J Virol 70:6820–6825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lu R, Limon A, Devroe E, Silver PA, Cherepanov P, Engelman A. 2004. Class II integrase mutants with changes in putative nuclear localization signals are primarily blocked at a postnuclear entry step of human immunodeficiency virus type 1 replication. J Virol 78:12735–12746. doi: 10.1128/JVI.78.23.12735-12746.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Quillent C, Borman AM, Paulous S, Dauguet C, Clavel F. 1996. Extensive regions of pol are required for efficient human immunodeficiency virus polyprotein processing and particle maturation. Virology 219:29–36. doi: 10.1006/viro.1996.0219. [DOI] [PubMed] [Google Scholar]
- 41.Taddeo B, Haseltine WA, Farnet CM. 1994. Integrase mutants of human immunodeficiency virus type 1 with a specific defect in integration. J Virol 68:8401–8405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wilkinson TA, Januszyk K, Phillips ML, Tekeste SS, Zhang M, Miller JT, Le Grice SF, Clubb RT, Chow SA. 2009. Identifying and characterizing a functional HIV-1 reverse transcriptase-binding site on integrase. J Biol Chem 284:7931–7939. doi: 10.1074/jbc.M806241200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nishitsuji H, Hayashi T, Takahashi T, Miyano M, Kannagi M, Masuda T. 2009. Augmentation of reverse transcription by integrase through an interaction with host factor, SIP1/Gemin2 is critical for HIV-1 infection. PLoS One 4:e7825. doi: 10.1371/journal.pone.0007825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tasara T, Maga G, Hottiger MO, Hubscher U. 2001. HIV-1 reverse transcriptase and integrase enzymes physically interact and inhibit each other. FEBS Lett 507:39–44. doi: 10.1016/S0014-5793(01)02945-3. [DOI] [PubMed] [Google Scholar]
- 45.Adachi A, Gendelman HE, Koenig S, Folks T, Willey R, Rabson A, Martin MA. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol 59:284–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fisher CL, Pei GK. 1997. Modification of a PCR-based site-directed mutagenesis method. Biotechniques 23:570–574. [DOI] [PubMed] [Google Scholar]
- 47.Shibagaki Y, Chow SA. 1997. Central core domain of retroviral integrase is responsible for target site selection. J Biol Chem 272:8361–8369. doi: 10.1074/jbc.272.13.8361. [DOI] [PubMed] [Google Scholar]
- 48.Le Grice SF, Gruninger-Leitch F. 1990. Rapid purification of homodimer and heterodimer HIV-1 reverse transcriptase by metal chelate affinity chromatography. Eur J Biochem 187:307–314. doi: 10.1111/j.1432-1033.1990.tb15306.x. [DOI] [PubMed] [Google Scholar]
- 49.Grandgenett DP, Goodarzi G. 1994. Folding of the multidomain human immunodeficiency virus type-I integrase. Protein Sci 3:888–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Khan M, Garcia-Barrio M, Powell MD. 2001. Restoration of wild-type infectivity to human immunodeficiency virus type 1 strains lacking nef by intravirion reverse transcription. J Virol 75:12081–12087. doi: 10.1128/JVI.75.24.12081-12087.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Korin YD, Zack JA. 1998. Progression to the G1b phase of the cell cycle is required for completion of human immunodeficiency virus type 1 reverse transcription in T cells. J Virol 72:3161–3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lodi PJ, Ernst JA, Kuszewski J, Hickman AB, Engelman A, Craigie R, Clore GM, Gronenborn AM. 1995. Solution structure of the DNA binding domain of HIV-1 integrase. Biochemistry 34:9826–9833. doi: 10.1021/bi00031a002. [DOI] [PubMed] [Google Scholar]
- 53.Goddard TD, Kneller DG. 2008. Sparky 3. University of California, San Francisco, CA. [Google Scholar]
- 54.Schrodinger LLC. 2012. The PyMOL molecular graphics system, version 1.5.0.5. Schrodinger, LLC, New York, NY. [Google Scholar]
- 55.Chow SA. 1997. In vitro assays for activities of retroviral integrase. Methods 12:306–317. doi: 10.1006/meth.1997.0484. [DOI] [PubMed] [Google Scholar]
- 56.Ding J, Das K, Hsiou Y, Sarafianos SG, Clark AD Jr, Jacobo-Molina A, Tantillo C, Hughes SH, Arnold E. 1998. Structure and functional implications of the polymerase active site region in a complex of HIV-1 RT with a double-stranded DNA template-primer and an antibody Fab fragment at 2.8 Å resolution. J Mol Biol 284:1095–1111. doi: 10.1006/jmbi.1998.2208. [DOI] [PubMed] [Google Scholar]
- 57.Jacobo-Molina A, Ding J, Nanni RG, Clark AD Jr, Lu X, Tantillo C, Williams RL, Kamer G, Ferris AL, Clark P, et al. 1993. Crystal structure of human immunodeficiency virus type 1 reverse transcriptase complexed with double-stranded DNA at 3.0 Å resolution shows bent DNA. Proc Natl Acad Sci U S A 90:6320–6324. doi: 10.1073/pnas.90.13.6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Patel PH, Jacobo-Molina A, Ding J, Tantillo C, Clark AD Jr, Raag R, Nanni RG, Hughes SH, Arnold E. 1995. Insights into DNA polymerization mechanisms from structure and function analysis of HIV-1 reverse transcriptase. Biochemistry 34:5351–5363. doi: 10.1021/bi00016a006. [DOI] [PubMed] [Google Scholar]
- 59.Zuiderweg ER. 2002. Mapping protein-protein interactions in solution by NMR spectroscopy. Biochemistry 41:1–7. doi: 10.1021/bi011870b. [DOI] [PubMed] [Google Scholar]
- 60.Krishnan L, Li X, Naraharisetty HL, Hare S, Cherepanov P, Engelman A. 2010. Structure-based modeling of the functional HIV-1 intasome and its inhibition. Proc Natl Acad Sci U S A 107:15910–15915. doi: 10.1073/pnas.1002346107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lutzke RA, Plasterk RH. 1998. Structure-based mutational analysis of the C-terminal DNA binding domain of human immunodeficiency virus type 1 integrase: critical residues for protein oligomerization and DNA binding. J Virol 72:4841–4848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lutzke RA, Vink C, Plasterk RH. 1994. Characterization of the minimal DNA-binding domain of the HIV integrase protein. Nucleic Acids Res 22:4125–4131. doi: 10.1093/nar/22.20.4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lian L-Y, Roberts GCK. 1993. Effects of chemical exchange on NMR spectra, p 153–182. In Roberts GCK. (ed), NMR of macromolecules: a practical approach. Oxford University Press, Inc., New York, NY. [Google Scholar]
- 64.Palmer AG III, Kroenke CD, Loria JP. 2001. Nuclear magnetic resonance methods for quantifying microsecond-to-millisecond motions in biological macromolecules. Methods Enzymol 339:204–238. doi: 10.1016/S0076-6879(01)39315-1. [DOI] [PubMed] [Google Scholar]
- 65.Mandal D, Feng Z, Stoltzfus CM. 2008. Gag-processing defect of human immunodeficiency virus type 1 integrase E246 and G247 mutants is caused by activation of an overlapping 5′ splice site. J Virol 82:1600–1604. doi: 10.1128/JVI.02295-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gotte M, Li X, Wainberg MA. 1999. HIV-1 reverse transcription: a brief overview focused on structure-function relationships among molecules involved in initiation of the reaction. Arch Biochem Biophys 365:199–210. doi: 10.1006/abbi.1999.1209. [DOI] [PubMed] [Google Scholar]
- 67.Kati WM, Johnson KA, Jerva LF, Anderson KS. 1992. Mechanism and fidelity of HIV reverse transcriptase. J Biol Chem 267:25988–25997. [PubMed] [Google Scholar]
- 68.Klarmann GJ, Schauber CA, Preston BD. 1993. Template-directed pausing of DNA synthesis by HIV-1 reverse transcriptase during polymerization of HIV-1 sequences in vitro. J Biol Chem 268:9793–9802. [PubMed] [Google Scholar]
- 69.Lanchy JM, Ehresmann C, Le Grice SF, Ehresmann B, Marquet R. 1996. Binding and kinetic properties of HIV-1 reverse transcriptase markedly differ during initiation and elongation of reverse transcription. EMBO J 15:7178–7187. [PMC free article] [PubMed] [Google Scholar]
- 70.Temiz NA, Bahar I. 2002. Inhibitor binding alters the directions of domain motions in HIV-1 reverse transcriptase. Proteins 49:61–70. doi: 10.1002/prot.10183. [DOI] [PubMed] [Google Scholar]
- 71.Hsiou Y, Ding J, Das K, Clark AD Jr, Hughes SH, Arnold E. 1996. Structure of unliganded HIV-1 reverse transcriptase at 2.7 Å resolution: implications of conformational changes for polymerization and inhibition mechanisms. Structure 4:853–860. doi: 10.1016/S0969-2126(96)00091-3. [DOI] [PubMed] [Google Scholar]
- 72.Rodgers DW, Gamblin SJ, Harris BA, Ray S, Culp JS, Hellmig B, Woolf DJ, Debouck C, Harrison SC. 1995. The structure of unliganded reverse transcriptase from the human immunodeficiency virus type 1. Proc Natl Acad Sci U S A 92:1222–1226. doi: 10.1073/pnas.92.4.1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sarafianos SG, Das K, Tantillo C, Clark AD Jr, Ding J, Whitcomb JM, Boyer PL, Hughes SH, Arnold E. 2001. Crystal structure of HIV-1 reverse transcriptase in complex with a polypurine tract RNA:DNA. EMBO J 20:1449–1461. doi: 10.1093/emboj/20.6.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mohammed KD, Topper MB, Muesing MA. 2011. Sequential deletion of the integrase (Gag-Pol) carboxyl terminus reveals distinct phenotypic classes of defective HIV-1. J Virol 85:4654–4666. doi: 10.1128/JVI.02374-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Williams KL, Zhang Y, Shkriabai N, Karki RG, Nicklaus MC, Kotrikadze N, Hess S, Le Grice SF, Craigie R, Pathak VK, Kvaratskhelia M. 2005. Mass spectrometric analysis of the HIV-1 integrase-pyridoxal 5′-phosphate complex reveals a new binding site for a nucleotide inhibitor. J Biol Chem 280:7949–7955. doi: 10.1074/jbc.M413579200. [DOI] [PubMed] [Google Scholar]
- 76.Battle DJ, Kasim M, Yong J, Lotti F, Lau CK, Mouaikel J, Zhang Z, Han K, Wan L, Dreyfuss G. 2006. The SMN complex: an assembly machine for RNPs. Cold Spring Harbor Symp Quant Biol 71:313–320. doi: 10.1101/sqb.2006.71.001. [DOI] [PubMed] [Google Scholar]
- 77.Hamamoto S, Nishitsuji H, Amagasa T, Kannagi M, Masuda T. 2006. Identification of a novel human immunodeficiency virus type 1 integrase interactor, Gemin2, that facilitates efficient viral cDNA synthesis in vivo. J Virol 80:5670–5677. doi: 10.1128/JVI.02471-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Engelman A. 2003. The roles of cellular factors in retroviral integration. Curr Top Microbiol Immunol 281:209–238. [DOI] [PubMed] [Google Scholar]
- 79.Jayappa KD, Ao Z, Wang X, Mouland AJ, Shekhar S, Yang X, Yao X. 2015. Human immunodeficiency virus type 1 employs the cellular dynein light chain 1 protein for reverse transcription through interaction with its integrase protein. J Virol 89:3497–3511. doi: 10.1128/JVI.03347-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zheng Y, Ao Z, Wang B, Jayappa KD, Yao X. 2011. Host protein Ku70 binds and protects HIV-1 integrase from proteasomal degradation and is required for HIV replication. J Biol Chem 286:17722–17735. doi: 10.1074/jbc.M110.184739. [DOI] [PMC free article] [PubMed] [Google Scholar]