ABSTRACT
The innate immune response is the first line of defense against viruses, and type I interferon (IFN) is a critical component of this response. Similar to other viruses, the gammacoronavirus infectious bronchitis virus (IBV) has evolved under evolutionary pressure to evade and counteract the IFN response to enable its survival. Previously, we reported that IBV induces a delayed activation of the IFN response. In the present work, we describe the resistance of IBV to IFN and the potential role of accessory proteins herein. We show that IBV is fairly resistant to the antiviral state induced by IFN and identify that viral accessory protein 3a is involved in resistance to IFN, as its absence renders IBV less resistant to IFN treatment. In addition to this, we found that independently of its accessory proteins, IBV inhibits IFN-mediated phosphorylation and translocation of STAT1. In summary, we show that IBV uses multiple strategies to counteract the IFN response.
IMPORTANCE In the present study, we show that infectious bronchitis virus (IBV) is resistant to IFN treatment and identify a role for accessory protein 3a in the resistance against the type I IFN response. We also demonstrate that, in a time-dependent manner, IBV effectively interferes with IFN signaling and that its accessory proteins are dispensable for this activity. This study demonstrates that the gammacoronavirus IBV, similar to its mammalian counterparts, has evolved multiple strategies to efficiently counteract the IFN response of its avian host, and it identifies accessory protein 3a as multifaceted antagonist of the avian IFN system.
INTRODUCTION
Infectious bronchitis virus (IBV) is a member of the genus Gammacoronavirus, a group of viruses from the order of Nidovirales characterized by a large positive-stranded RNA genome (1). IBV is the causative agent of infectious bronchitis, which is one of the most important viral diseases in chickens, a highly contagious respiratory disease that can spread to the gastrointestinal or the urogenital tract (2, 3). Despite widespread application of inactivated and live attenuated vaccines, infectious bronchitis remains one of the most reported diseases in poultry farms worldwide. Notwithstanding the widespread nature and economic importance of this virus, interactions between IBV and the host immune response remain poorly understood.
During the immune response to viruses, the type I interferon (IFN) response plays a pivotal role. Recently, we have shown that IBV induces delayed activation of the interferon response (4) in a manner similar to that of several members of the genus Betacoronavirus, including mouse hepatitis virus (MHV), severe acute respiratory syndrome-associated coronavirus (SARS-CoV), and Middle East respiratory syndrome coronavirus (MERS-CoV) (5–8). The observation that coronaviruses delay activation of the IFN response and limit production of IFN suggests that IFN has the ability to hinder their propagation. In apparent contrast, most coronaviruses are relatively resistant to treatment with IFN in vitro (9, 10), one exception being MERS-CoV, which was shown to be highly sensitive to IFN-β in vitro (11, 12). Although previous studies suggest that treatment with IFN could hinder propagation of IBV, based on reduced plaque formation (13) and reduced syncytium formation (14), quantitative data on the resistance of IBV to IFN are lacking.
To date, it is unknown which of the IBV proteins confer resistance to IFN, if any. Various studies have demonstrated that accessory proteins of coronaviruses play an important role in resistance to the IFN-induced antiviral response (10, 12, 15–20). The accessory proteins of coronaviruses are small proteins (50 to 300 amino acids [aa]) that are not essential for virus replication in vitro (21). The number of accessory proteins varies between coronaviruses, and amino acid sequences of accessory proteins from different genera show very limited similarity, suggesting that their function is virus or host specific. IBV has been shown to express at least four accessory proteins, 3a, 3b, 5a, and 5b, which are translated from two polycistronic mRNAs. Recently, we showed that both 3a and 3b limit transcription of Ifnβ and that 3b limits production of IFN protein in vitro (4). Additional roles for IBV accessory proteins have remained elusive.
In the present study, we showed that IBV is relatively resistant to treatment with either IFN-α or IFN-β but that knockout of 3a makes IBV less resistant to treatment with type I IFN. In addition, we showed that IBV inhibits phosphorylation and translocation of the IFN-activated transcription factor STAT1 and inhibits subsequent IFN-mediated activation of an interferon-stimulated gene (ISG) promoter, at least during late stages of the infection. However, using mutant viruses, we demonstrated that the presence of accessory proteins 3a, 3b, 5a, and 5b is not required for either inhibition of STAT1 translocation or activation of an ISG promoter. We discuss two strategies by which IBV counteracts the type I IFN response: one based on counteracting the IFN-mediated antiviral response using accessory protein 3a and another based on blocking of IFN-mediated activation of antiviral genes through inhibition of STAT1 translocation. This study demonstrates that the gammacoronavirus IBV has evolved multiple strategies to counteract activation of and clearance by the type I IFN response.
MATERIALS AND METHODS
Cells.
Chicken embryonic kidneys (CEK) were aseptically removed from 17- to 19-day-old chicken embryos (Charles River, SPAFAS). A cell suspension was obtained by trypsinization of kidneys for 30 min at 37°C and subsequent filtration through a 100-μm mesh. The resulting CEK cells were seeded at 4 × 105 cells/cm2 in a 1:1 mix of medium 199 and F10 medium (Invitrogen) supplemented with 0.5% fetal bovine serum (FBS), 0.1% tryptose phosphate broth, 0.1% sodium bicarbonate, 0.1% HEPES, and 1% penicillin-streptomycin (PenStrep; Gibco, Invitrogen). DF-1 chicken fibroblasts, African green monkey Vero cells, and baby hamster kidney (BHK) cells were cultured in Dulbecco modified Eagle medium (DMEM; Gibco, Invitrogen) supplemented with 10% FBS and 1% PenStrep. All cells were incubated in a humidified incubator at 37°C and 5% CO2.
Viruses.
IBV Beaudette, strain Beau-R, and the generation of the ScAUG3a, ScAUG3b, ScAUG3ab, Δ3ab, and ScAUG5ab viruses were described previously (22–24). In the ScAUG viruses, the start codons of the indicated accessory genes were mutated to stop codons. In the Δ3ab virus, open reading frame (ORF) 3a and all except the final 17 nucleotides of ORF 3b have been deleted (22). The presence of second-site mutations and the absence of protein expression were verified for the applied batch. IBV was amplified on CEK cells and Sindbis virus (SinV) was amplified on BHK cells. All viruses were titrated on the respective cell type on which the experiment was performed using the 50% tissue culture infective dose (TCID50) method as previously described (25).
Immunohistochemistry.
Vero cells were cultured on 8-well Lab-Tek 1.0 borosilicate coverslips (Sigma-Aldrich), whereas CEK cells were cultured in 24-well culture plates. Briefly, cells were fixed with 3.7% paraformaldehyde and permeabilized using 0.1% Triton X-100 in phosphate-buffered saline (PBS). SinV infection was detected using a mouse monoclonal antibody against double-stranded RNA (dsRNA; English & Scientific Consulting), and IBV infection was detected using antibodies against the IBV nucleocapsid (N) protein (Prionics). Tyr701-phosphorylated STAT1 (pSTAT1) was detected using rabbit monoclonal antibody MA5-15071 (Thermo Scientific), and total STAT1 was detected using rabbit polyclonal antibody sc-346 (Santa Cruz Biotechnology). Visualization was performed using Alexa 488- or 568-labeled goat anti-mouse or goat anti-rabbit antibodies (Invitrogen). Antibodies were diluted 1:1,000 in PBS supplemented with 5% FBS, except for anti-pSTAT1, which was diluted 1:500. Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI; 0.5 μg/ml; Sigma). Cells were imaged using a Zeiss Primo Vert microscope and Axiovision software. Image overlays and cross sections were made in ImageJ. To evaluate the effects of IBV on STAT1 translocation to the nucleus, the presence of (phospho-)STAT1 in the nucleus was quantified in wells that were first infected with the appropriate virus strain and then stimulated with IFN. Within these wells, infected cells were identified using the anti-IBV N antibody, and the percentage of nuclei showing translocation of (phospho-)STAT1 in both infected and uninfected cells was calculated based on >500 cells from multiple images.
Interferon sensitivity assay.
CEK, DF-1, or Vero cells at 100% confluence were pretreated for 6 h with different concentrations of recombinant chicken IFN-α or IFN-β produced in HEK293 cells (26) or with recombinant human IFN-α A/D (Sigma-Aldrich) or human IFN-β (CalBioChem). Infections were carried out using different viruses at the desired multiplicity of infection (MOI) for 2 h, after which cells were washed three times with PBS and new medium containing the same concentration of interferon was added. Supernatants were collected for titration at 18 h postinfection (hpi) (CEK cells) or 24 hpi (DF-1 cells). IFN posttreatment was performed in CEK cells that were first infected for 2 h at an MOI of 10, washed three times with PBS, and subsequently incubated with medium containing interferon. Supernatants were collected for titration at 18 hpi.
Quantification of viral RNA.
RNA was isolated from tissue culture supernatant on a MagNA Pure 96 instrument using the MagNA Pure 96 DNA and Viral Nucleic Acid Small Volume kit (Roche Diagnostic) and the Viral NA Universal SV 2.0 protocol. Reverse transcription-quantitative PCR (RT-qPCR) was performed on 5 μl of RNA using the SYBR green one-step kit (Bio-Rad) in a Bio-Rad CFX96 PCR apparatus. Primers against the nucleocapsid gene of IBV, based on the sequence with GenBank accession number AY851295, were as previously published (4). The forward primer was GAAGAAAACCAGTCCCAGA, and the reverse primer was TTACCAGCAACCCACAC.
ISG54-luciferase reporter assays.
Vero or DF-1 cells were seeded at 80 to 90% confluence in 96-well plates and transfected using FuGENE HD (Promega) at a 1:3.5 ratio of DNA to FuGENE HD according to manufacturer's specifications. Per well, 100 ng of ISG54-luciferase reporter plasmid (kind gift from David E. Levy [27]) was transfected, together with 2 ng of pRL-SV40 Renilla plasmid (Promega) to correct for differences in transfection efficiency and transcription. At least 24 h later, cells were infected and, at various time points after infection, stimulated with 1,000 U/ml of IFN for an additional 6 h. Firefly and Renilla luciferase activities were quantified using the Dual-Glo luciferase assay (Promega) and a Filtermax F5 luminometer (Molecular Devices). Luciferase activity was calculated relative to the non-IFN-stimulated control showing the maximum activity in noninfected wells and calculating the relative percentage in virus-infected wells.
Western blotting.
Vero cells in 24-well plates at 90% confluence were infected with IBV Beau-R at an MOI of 1. At 18 hpi, cells were stimulated with human IFN-β (10,000 U/ml) for 30 min and subsequently lysed in lysis buffer (20 mM Tris, 100 mM NaCl, 1 mM EDTA, 0.5% Triton X-100, and 1 mM phenylmethylsulfonyl fluoride [PMSF] [pH 8.0]). Samples were boiled for 10 min in Laemmli loading buffer, clarified by centrifugation at 5,000 × g for 5 min, and separated on a 10% SDS-PAGE gel. Proteins were transferred onto a Whatman Protran nitrocellulose membrane (GE Healthcare) by semidry blotting (Trans-Blot SD semidry transfer cell; Bio-Rad). Blotted membranes were blocked overnight in 5% (wt/vol) nonfat dry milk in TBS-Tween (20 mM Tris, 500 mM NaCl, 0.05% Tween 20 [vol/vol] [pH 8.0]) at 4°C. The blotted membranes were incubated with primary antibodies (rabbit anti-STAT1 sc-346, 1:1,000 [Santa Cruz Biotechnology]; rabbit anti-pSTAT1 MA5-15071, 1:500 [Thermo-Scientific]; rabbit anti-β-tubulin Ab6046, 1:2,000 [Abcam]) in 5% nonfat dry milk in TBS-Tween for 1 h at 37°C, followed by incubation with a goat anti-rabbit horseradish peroxidase (HRP)-conjugated antibody (Bio-Rad) at a 1:1,000 dilution in the same buffer for 1 h at 37°C. Chemiluminescence of bound anti-rabbit HRP-conjugated antibody was detected with WesternBright ECL (Advansta) and visualized using Lumni-film (Roche). Quantification of band intensity was performed using ImageJ software.
Statistics.
Statistical analyses were performed in GraphPad Prism 6.0 or IBM SPSS 19. Equality of variance was assessed using Bartlett's test. Significant differences were determined by a one-way analysis of variance (ANOVA) followed by a Bonferroni or Tukey post hoc test or by a two-way ANOVA when indicated.
RESULTS
IBV is relatively resistant to treatment with type I IFN.
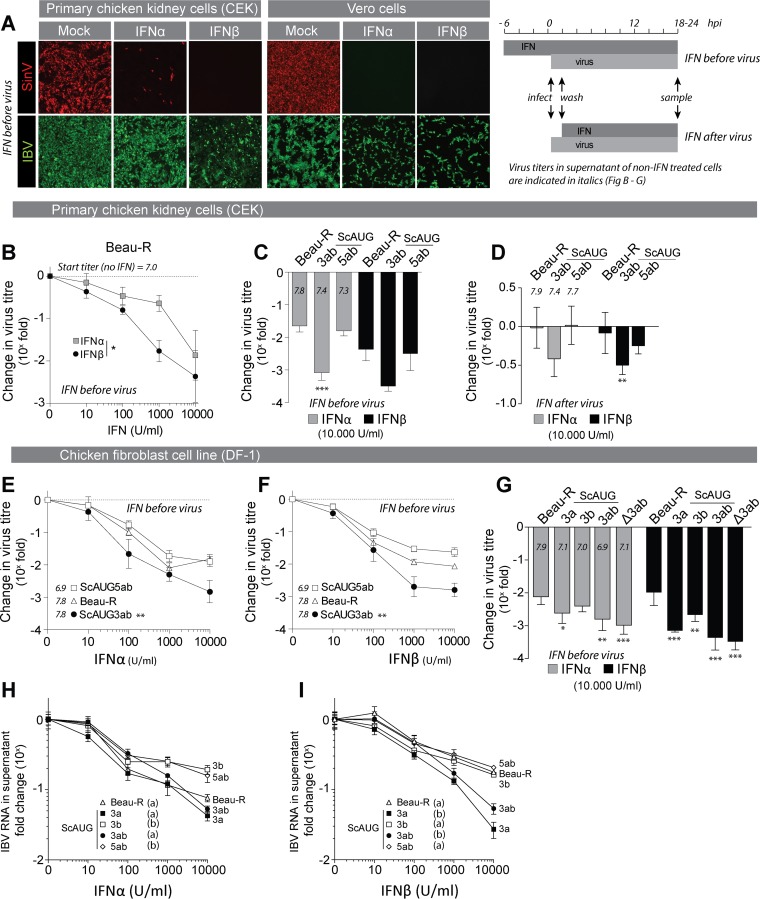
To test resistance of IBV to type I IFN, we treated primary chicken embryo kidney (CEK) cells or Vero cells with recombinant chicken IFN and subsequently infected them with IBV Beau-R or with the IFN-sensitive Sindbis virus as control. Immunofluorescence staining indicated that in both cell types, propagation of IBV was less affected by treatment with IFN-α and IFN-β than propagation of the IFN-sensitive Sindbis virus (Fig. 1A). To investigate the degree of IBV resistance to IFN, we treated CEK cells with increasing concentrations of IFN-α and IFN-β and determined the effect on propagation by titration of Beau-R (Fig. 1B). The titer of Beau-R decreased in a dose-dependent manner, and in CEK cells, the effect of IFN-β on the titer of Beau-R was more pronounced than that of IFN-α. Similar to the case with other coronaviruses, relatively high concentrations of IFN (>1,000 U/ml) were required to hinder propagation of IBV Beau-R, which suggested that IBV, like other coronaviruses, is relatively resistant to IFN and raised the possibility that IBV actively counteracts the type I IFN response.
FIG 1.
Accessory protein 3a confers resistance to treatment of IBV with type I IFN. (A) Primary chicken embryo kidney (CEK) cells and Vero cells were prestimulated with IFN (1,000 U/ml) for 6 h and subsequently infected with Sindbis virus (SinV) or IBV (Beau-R) at an MOI of 0.1. At 24 hpi, cells were fixed and stained for dsRNA (red) or IBV-N (green). (B) CEK cells were prestimulated with the indicated concentrations of IFN for 6 h and subsequently infected with Beau-R (MOI of 0.01). At 2 hpi, cells were washed to remove inoculum, and medium with IFN was added. At 18 hpi, supernatant was sampled and titrated (see panel A, graph at right, for sampling time line). Symbols represent the means of triplicate measurements (±SEM) of virus titers from two independent experiments. The asterisk indicates significant differences (P < 0.05) between IFN-α and IFN-β treatment as assessed by a two-way ANOVA. (C) CEK cells were treated with IFN, infected with virus, and sampled at 18 hpi as described for panel B using Beau-R and accessory protein-null viruses (MOI of 0.01). Titers were determined at 18 hpi and are expressed relative to titers of non-IFN-treated wells. The lower the value, the higher the reduction. Symbols indicate the means (±SEM) of triplicate measurements from two independent experiments. Triple asterisks indicate significant difference (P < 0.001) compared to Beau-R as assessed by one-way ANOVA followed by a Bonferroni post hoc test. Titers in non-IFN-treated wells are displayed for each virus. (D) CEK cells were infected with the indicated viruses (MOI of 10), and at 2 hpi, inoculum was removed and cells were incubated with IFN (10,000 U/ml). Virus titers in the supernatant were determined at 18 hpi and are expressed as fold change relative to non-IFN-treated wells infected with the same virus (see Fig. 1A, graph, for sampling time line). (E and F) DF-1 cells were treated with IFN and infected as described for panel B. Symbols indicate the mean relative titers at 24 hpi (±SEM) of triplicate wells from a representative experiment of two biological replicates. Double asterisks indicate significant differences (P < 0.01) between ScAUG3ab virus and the other viruses as assessed by a two-way ANOVA. (G) DF-1 cells were treated with IFN and infected as described for panel B. Bars represent the fold change in virus titer at 24 hpi (±SD) of triplicate wells from two biological replicates. Asterisks indicate significant differences (*, P < 0.05; **, P < 0.01; ***, P < 0.001) from Beau-R, as assessed by a one-way ANOVA followed by a Bonferroni post hoc test. (H and I) DF-1 cells were treated with IFN and infected as described for panel B. At 24 hpi, total RNA was extracted from the cell culture supernatant and virus RNA was quantified by RT-qPCR using primers against the N gene. Values are expressed as fold change relative to non-IFN-treated wells infected with the same virus. The lower the value, the higher the reduction of viral RNA. Symbols represent the means of quadruplicate wells (±SD) from one experiment. Letters indicate significant differences at the highest IFN concentration as assessed by a one-way ANOVA followed by a Tukey post hoc test.
Accessory protein 3a contributes to IFN resistance.
For coronaviruses other than IBV, the accessory proteins have been implicated in counteracting the type I IFN response. To investigate whether the accessory proteins of IBV contribute to resistance to IFN, we stimulated CEK cells with a high concentration of IFN (Fig. 1A, graph; IFN before virus) and infected them with 3a/3b and 5a/5b null viruses (ScAUG3ab and ScAUG5ab). These viruses do not express the indicated accessory proteins owing to a mutation in the AUG start codons. IFN treatment reduced titers of ScAUG3ab more than that of either ScAUG5ab or the parental Beau-R virus (Fig. 1C), suggesting that ScAUG3ab is more sensitive to treatment with IFN. Next, we investigated whether the absence of 3a and 3b would increase sensitivity of IBV to IFN treatment after the infection has been established (Fig. 1A, graph; IFN after virus). We synchronously infected CEK cells using a high MOI of Beau-R, ScAUG3ab, or ScAUG5ab virus. At 2 hpi, cells were incubated with high doses of IFN-α and IFN-β for an additional 16 h, when infectious virus titers were determined by titration of the supernatant (Fig. 1D). The results show that once infection has been established, Beau-R is resistant to IFN treatment and that absence of accessory proteins 3a and 3b leads to a marginal, but significant, increase in sensitivity of IBV to IFN, at least upon IFN-β treatment.
To further investigate IFN sensitivity of ScAUG3ab, we stimulated DF-1 cells with increasing concentrations of IFN-α or IFN-β (Fig. 1E and F). Again, ScAUG3ab was more sensitive to treatment with either IFN-α or IFN-β than ScAUG5ab or the parental Beau-R, indicating that accessory proteins 3a and/or 3b could play an important role in conferring resistance of IBV to treatment with type I IFN in either chicken or mammalian cells. To further investigate whether accessory proteins 3a, 3b, or both are responsible for the observed increase in IFN sensitivity, we stimulated DF-1 cells with 10,000 U/ml of IFN-α or IFN-β and infected them with individual mutants for either accessory protein 3a or 3b (ScAUG3a and ScAUG3b). As a control, we included ScAUG3ab and delta 3a/3b (Δ3ab) viruses. The latter was obtained by deleting the open reading frames of both 3a and 3b (22), and this virus was used to verify that the IFN sensitivity of ScAUG3ab was not due to a second-site mutation in the genome of this virus. Our results show that both ScAUG3a and ScAUG3b were more sensitive to IFN treatment than Beau-R, but the effects on ScAUG3a virus were more pronounced. To further investigate the difference in IFN sensitivity between ScAUG3a and ScAUG3b, we quantified viral RNA in the supernatant of DF-1 cells pretreated with increasing concentration of IFN (Fig. 1H and I). We found that reduction of viral RNA was most prominent in supernatants of cells infected with ScAUG3a and ScAUG3ab, especially after IFN-β treatment. Taken together, our results lead us to conclude that accessory protein 3a is the main contributor to resistance of IBV to type I IFN.
IBV prevents IFN signaling late during infection.
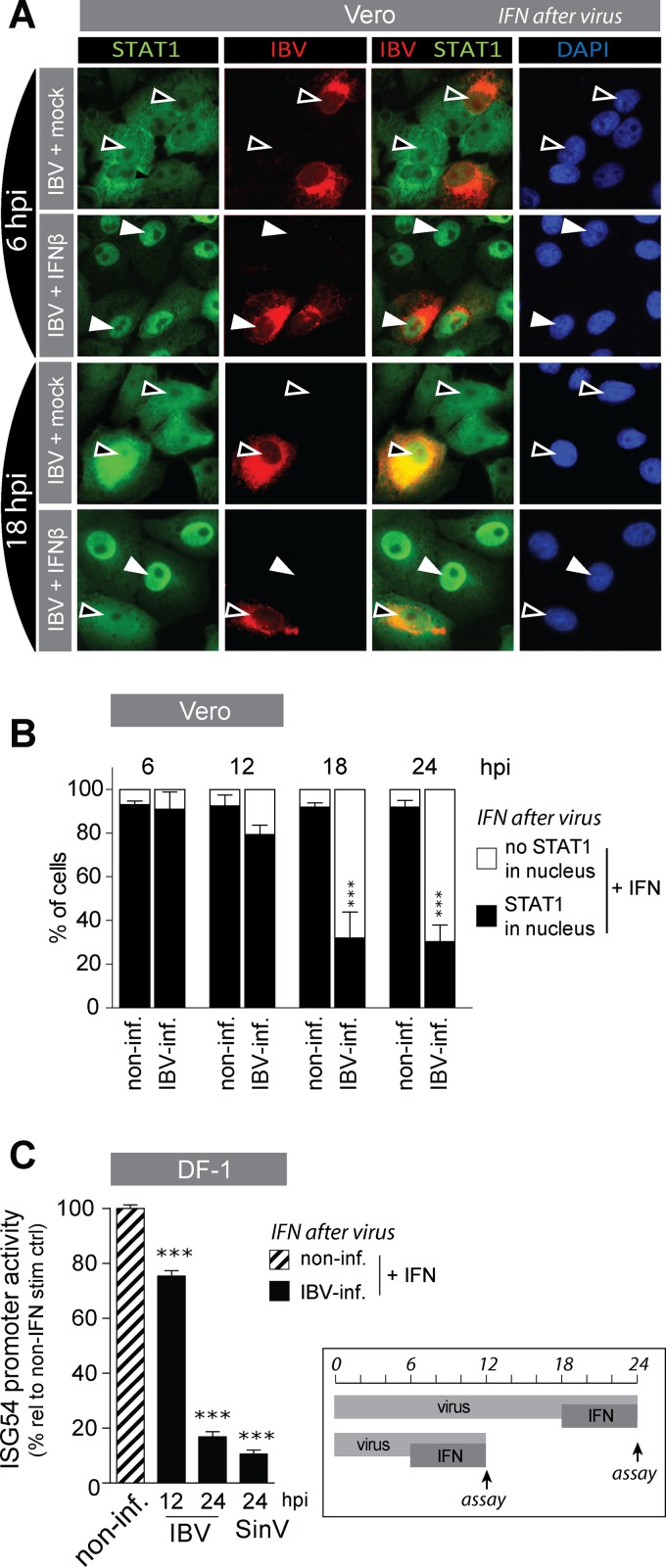
Next, we wanted to investigate how accessory proteins 3a and, to a lesser extent, 3b contribute to IFN resistance. One possibility is that the proteins interfere with signaling of IFN in a manner similar to that of the accessory protein corresponding to ORF 6 of SARS-CoV, which was shown to block IFN signaling through inhibition of nuclear translocation of STAT1 (28). To investigate whether also IBV is able to inhibit nuclear translocation of STAT1, we used Vero cells, as commercially available STAT1 antibodies did not detect chicken STAT1. Vero cells were infected with IBV and translocation of STAT1 was induced at 6 and 18 hpi by stimulation for 30 min with IFN-β. Localization of STAT1 in the nuclei of IBV-infected cells was visualized by immunostaining against STAT1 (Fig. 2A). In mock-treated cells (no stimulation with IFN-β), nuclear translocation of STAT1 was not visible, either in infected or in noninfected cells (black arrowheads), indicating that IBV infection alone does not induce translocation of STAT1. At 6 hpi, IBV did not prevent IFN-β-induced translocation of STAT1 (white arrowheads). At 18 hpi, however, IFN-β-induced translocation of STAT1 was strongly reduced in IBV-infected cells (Fig. 2A, bottom row). This indicated that IBV-mediated inhibition of STAT1 translocation is a time-dependent event.
FIG 2.
IBV prevents translocation of STAT1 and IFN signaling at late stages of infection. (A) Vero cells were infected with IBV-Beau-R (MOI of 1 for 6 h and MOI of 0.1 for all other time points) and subsequently stimulated with 1,000 U/ml of IFN-β for 30 min before fixation and staining for IBV-N and STAT1. White arrowheads indicate nuclear accumulation of STAT1, and black arrowheads indicate absence of STAT1 accumulation in the nucleus. (B) Cells were treated as for panel A, and at the indicated time points after IBV infection, the percentage of nuclei showing translocation of STAT1 (black bars) or not (white bars) was determined in noninfected (non-inf.) and in IBV-infected (IBV-inf) cells within IFN-β-treated wells. Each bar indicates the mean percentage of nuclei showing translocation of STAT1 as determined in 50 to 400 cells from multiple images of a representative experiment of two biological replicates. Error bars indicate SD. (C) DF-1 cells were transfected with an ISG54-firefly luciferase construct and 24 h later infected with Beau-R or SinV (MOIs of 5 and 0.5, respectively); at 6 or 18 hpi, cells were stimulated with 1,000 U/ml of IFN-β for an additional 6 h. ISG54 promoter activity was calculated as percentage relative to non-IFN-β-treated wells. Shown is the ISG54 promoter activity in noninfected IFN-β-treated wells (striped bar) and in IBV-infected-IFN-β-treated wells at 12 and 24 hpi (black bars). Firefly luciferase values were normalized to Renilla luciferase to correct for differences in transfection efficiency and protein translation. Bars indicate the means (+SD) of triplicate wells from a representative experiment out of three biological replicates. Triple asterisks indicate significant differences (P < 0.001) with respect to noninfected cells, as assessed by one-way ANOVA followed by a Bonferroni post hoc test.
To substantiate the observed time dependency of IBV-mediated inhibition of STAT1 translocation, we quantified translocation of STAT1 in pictures taken of IBV-infected monolayers, containing both infected and noninfected cells, within IFN-β-treated wells at various time points after IBV infection. In noninfected cells, treatment with IFN-β led to translocation of STAT1 in more than 90% of the cells (Fig. 2B, black bars), regardless of time point (6 to 24 hpi) or the presence of neighboring cells infected with IBV (data not shown). Degrees of translocation of STAT1 in mock-treated cells were comparable between IBV-infected and noninfected cells (<5% [data not shown]), indicating that IBV alone did not induce translocation of STAT1. In contrast, in IBV-infected cells, treatment with IFN-β did not always lead to translocation of STAT1. The inhibition seen in IBV-infected cells was time dependent: at time points between 6 and 12 hpi, translocation of STAT1 was not different from that in noninfected cells, whereas at later time points, between 12 and 18 hpi onwards, STAT1 translocation was strongly inhibited (Fig. 2B, black bars).
To verify whether the observed time-dependent IBV-mediated inhibition of STAT1 translocation would correlate with inhibition of transcription of ISGs, we used an IFN reporter assay, based on the human ISG54 promoter, that contains multiple copies of the STAT1-binding interferon-stimulated response element (ISRE) driving expression of the luciferase gene (27). ISG54-luciferase-transfected DF-1 cells were infected for 12 h or 24 h with IBV and in the last 6 h of infection treated with IFN-β (Fig. 2C, graph at right). Indeed, at early time points after infection (12 hpi), we observed only a marginal inhibition of luciferase production, whereas at later time points (24 hpi), IBV strongly inhibited the IFN-mediated production of luciferase, to the same extent as Sindbis virus, a well-known inhibitor of STAT signaling (Fig. 2C). We interpret inhibition of luciferase activity as the result of a reduction in IFN-mediated ISG54 promoter activity and thus conclude that IBV inhibited the transcription of ISGs by inhibiting translocation of STAT1, but only during later stages of infection.
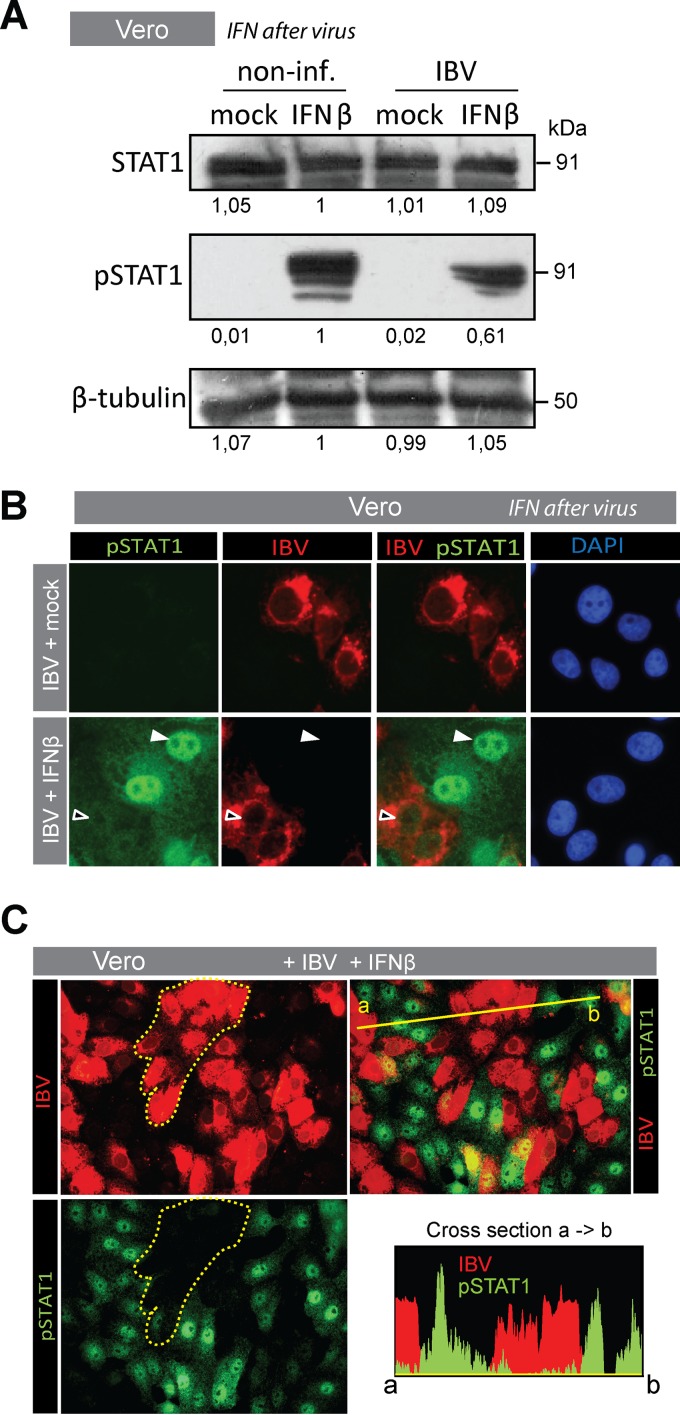
IBV inhibits phosphorylation of STAT1.
A crucial step in IFN-induced translocation of STAT1 is its phosphorylation. Only phosphorylated STAT1 (pSTAT1) can associate with STAT2 and IRF9 to form the transcription factor ISGF3, which binds to ISRE promoter elements. To investigate whether IBV is able to block phosphorylation of STAT1, we first performed a Western blot analysis (Fig. 3A). Levels of total STAT1 were comparable between IBV-infected and noninfected cells, whereas IFN-β-mediated phosphorylation of STAT1 was reduced in infected compared to noninfected cells, confirming that IBV prevents phosphorylation of STAT1 without affecting total STAT1 levels. In the Western blot, we observed a residual signal for pSTAT1 in IFN-β-stimulated IBV-infected cells, which was most likely due to the presence of noninfected cells in the sample.
FIG 3.
IBV prevents translocation and phosphorylation of STAT1 Vero cells were infected for 18 h with IBV Beau-R (MOI of 0.1) and subsequently stimulated with 1,000 U/ml of IFN-β for 30 min. (A) Western blot analysis of noninfected and IBV-infected monolayers that were either mock treated or treated with IFN-β. Staining was performed using antibodies against STAT1 and Tyr701-phosphorylated STAT1. Staining against β-tubulin was included as a loading control. Numbers below the blots indicate the intensities of the bands, expressed as fold ratio relative to the IFN-β-stimulated, noninfected sample. (B) Vero cells treated as described above were fixed and stained for IBV-N and pSTAT1. White arrowheads indicate translocation of pSTAT1, and black arrowheads indicate absence of pSTAT1 from the nucleus. (C) To verify the overall decrease of pSTAT1, an area containing IBV-infected cells within an IFN-β-stimulated monolayer is delineated by a dotted line in the top left image and is overlaid on the bottom left image to illustrate the absence of pSTAT1 in IBV-infected cells. Cross section: fluorescence intensity plot of pSTAT1 and IBV-N along the yellow line indicated in the top right image.
To better quantify the reduction in STAT1 phosphorylation observed in the Western blot analysis, we visualized IFN-β-induced phosphorylation of STAT1 in IBV-infected cells (18 hpi), using a pSTAT1-specific antibody. pSTAT1 could not be detected in mock-treated cells, even when infected with IBV (Fig. 3B, top row, left). Cells treated with IFN-β, however (Fig. 3B, bottom row), showed nuclear translocation of pSTAT1, but mostly in noninfected cells. In IBV-infected cells, in contrast, translocation of pSTAT1 was severely reduced. In addition to reduced levels of nuclear pSTAT1 (i.e., reduced translocation), we also observed reduced levels of cytoplasmic pSTAT1 in IFN-β-stimulated cells infected with IBV (Fig. 3C, delineated area). A cross section of IBV-infected areas versus noninfected areas confirmed the general lack of pSTAT1 signal in IBV-infected cells (Fig. 3C). Taken together, our data suggest that IBV prevents IFN-induced phosphorylation of STAT1.
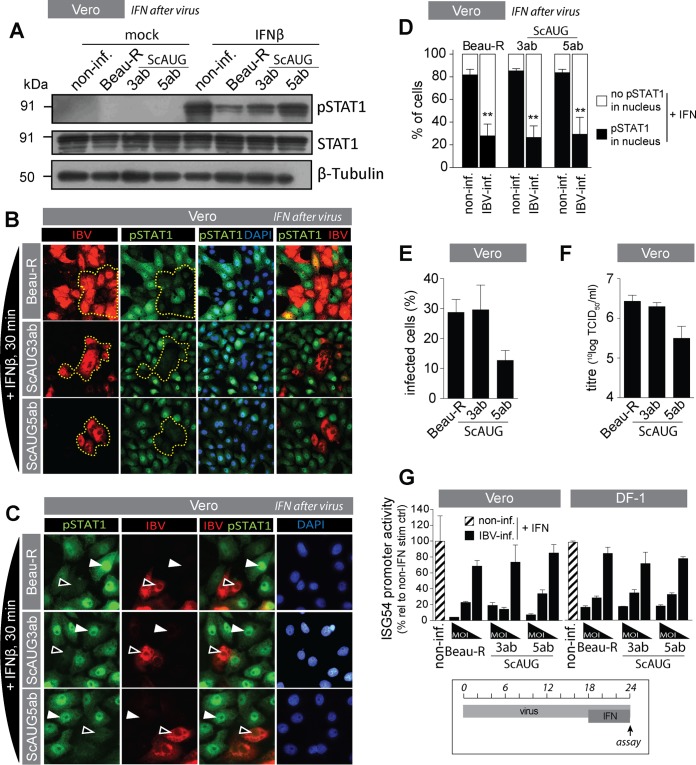
IBV accessory proteins are not required for inhibition of phosphorylation and translocation of STAT1.
The betacoronavirus SARS-CoV mediates inhibition of STAT1 translocation by its accessory protein ORF 6 (28, 29). To test whether the IBV accessory proteins are also involved in inhibition of phosphorylation and translocation of STAT1, we used ScAUG3ab and ScAUG5ab viruses. First, we investigated whether the accessory proteins of IBV are involved in inhibition of STAT1 phosphorylation. Western blot analysis indicated that wild-type Beau-R had a more pronounced inhibitory effect on STAT1 phosphorylation than ScAUG5ab, whereas the inhibitory effect on pSTAT1 of ScAUG3ab was intermediate (Fig. 4A). To confirm the increased phosphorylation of STAT1 in ScAUG3ab- and ScAUG5ab-infected cells, we performed immunostaining for pSTAT1. We found that, contrary to the Western blot analysis, both phosphorylation (Fig. 4B) and translocation (Fig. 4C) of pSTAT1 appeared to be inhibited to the same extents by ScAUG3ab, ScAUG5ab, and Beau-R. To better compare inhibition of pSTAT1 translocation between ScAUG3ab, ScAUG5ab, and Beau-R, we performed image analysis of infected and noninfected cells within infected monolayers after stimulation with IFN. Our results show that nuclear translocation of pSTAT1 was inhibited to the same extents by all three viruses (Fig. 4D, black bars, IBV-inf.). Degrees of nuclear translocation of pSTAT1 in noninfected cells within infected monolayers (non-inf.) were comparable between the three viruses. To explain the apparent discrepancy between the levels of STAT1 phosphorylation observed in the Western blot (Fig. 4A) and in the STAT1 immunostaining (Fig. 4B), we investigated the efficiency of replication of Beau-R, ScAUG3ab, and ScAUG5ab in Vero cells. To do so, we quantified the percentage of infected cells in microscopic images (Fig. 4E) in parallel to quantification of virus titer in supernatants of infected cells (Fig. 4F). These experiments indicated that replication of ScAUG5ab was less efficient than that of Beau-R and ScAUG3ab, which is in agreement with a previous report showing that replication of ScAUG5ab is reduced in Vero cells but not in CEK cells (30). Reduced replication of ScAUG5ab in Vero cells provides an explanation for its reduced inhibitory effect on IFN-mediated phosphorylation of STAT1 in the Western blot analysis. In short, we conclude that phosphorylation and nuclear translocation of pSTAT1 are inhibited to the same extents by ScAUG3ab, ScAUG5ab, and the parental Beau-R virus. Next, we investigated to which extent ScAUG3ab and ScAUG5ab would inhibit IFN-mediated activation of the ISG54 promoter and found no differences between ScAUG3ab, ScAUG5ab, and Beau-R, in both Vero and DF-1 cells (Fig. 4G). Taken together, our data indicate that the inhibition of phosphorylation and translocation of STAT1 as well as activation of the ISG54 promoter, observed after infection with IBV, are independent of the accessory proteins 3a, 3b, 5a, and 5b.
FIG 4.
IBV accessory proteins are not required for inhibition of STAT1 translocation and ISG promoter activation. (A) Western blot analysis of IBV-infected (MOI of 1; 18 hpi) and noninfected Vero cells that were either mock treated or treated with IFN-β for 30 min. Staining was performed using an antibody against Tyr701-phosphorylated STAT1, and an antibody against β-tubulin was used as a loading control. (B) Vero cells were infected with the indicated viruses (MOI of 0.1) and, at 18 hpi, stimulated with 1,000 U/ml of IFN-β for 30 min and then stained for IBV-N and pSTAT1. The area delineated by the yellow dotted line indicates the overall decrease in pSTAT1 staining in IBV-infected cells. (C) Vero cells were infected with Beau-R, ScAUG3ab, or ScAUG5ab virus (MOI of 0.1) and, at 18 hpi, stimulated with 1,000 U/ml of IFN-β for 30 min and then stained for IBV-N and pSTAT1. White arrowheads indicate translocation of pSTAT1, and black arrowheads indicate absence of accumulation of pSTAT1 in the nucleus. (D) In parallel, the percentage of nuclei showing translocation of STAT1 (black bars) or not (white bars) was determined in noninfected and in IBV-infected cells within IFN-β-treated wells. Each bar indicates the mean (+SD) percentage of nuclei showing translocation based on 100 to 300 cells from multiple images of a representative experiment from two biological replicates. Double asterisks indicate significant differences (P < 0.01) with respect to noninfected cells, as assessed by one-way ANOVA followed by a Bonferroni post hoc test. (E) Quantification of the percentage of IBV-infected cells in microscopic images of cells infected with the indicated viruses at an MOI of 0.1 and stained using IBV-N-specific antibody at 18 hpi. For each virus, at least 500 cells divided over 10 microscopic fields were analyzed. (F) Virus titers in supernatants from Vero cells infected for 18 h with the indicated viruses at an MOI of 0.01. (G) Vero and DF-1 cells were transfected with an ISG54-firefly luciferase construct and 24 h later infected with Beau-R, ScAUG3ab, or ScAUG5ab at MOIs 5, 0.5, and 0.05. At 18 hpi, cells were stimulated with 1,000 U/ml of IFN-β for an additional 6 h. After a total of 24 h, firefly and Renilla luciferase activities were quantified. ISG54 promoter activity was calculated as a percentage relative to non-IFN-β-treated wells. Shown is the ISG54 promoter activity in noninfected, IFN-β-treated wells (striped bar) and in IBV-infected, IFN-β-treated wells (black bars). Firefly luciferase values were normalized to Renilla luciferase to correct for differences in transfection efficiency and protein translation. Bars indicate the means (+SD) of triplicate wells of a representative example of 3 biological replicates.
DISCUSSION
In this study, we investigated the in vitro sensitivity of the gammacoronavirus IBV to treatment with IFN and the potential role of IBV accessory proteins in conferring resistance to the host's type I IFN response. We found IBV to be relatively resistant to either pre- or posttreatment with IFN and showed that simultaneous knockout of the accessory proteins 3a and 3b decreased resistance of IBV to IFN treatment. In addition, we present evidence that accessory protein 3a is primarily responsible for the observed IFN resistance by IBV. Finally, we found that IBV interferes with IFN signaling by inhibition of phosphorylation and nuclear translocation of STAT1 in a time-dependent manner and that both 3a and 3b are dispensable for this activity. In summary, this study demonstrates that the gammacoronavirus IBV has evolved multiple strategies to antagonize the innate immune response.
The coronaviruses MHV, SARS-CoV, MERS-CoV, and IBV have all been shown to induce modest and delayed transcription of Ifnβ (8, 31). Alpha- and Betacoronaviruses (not Gamma- or Deltacoronaviruses) encode the nsp1 protein, which decreases transcription of Ifnβ and inhibits synthesis of host proteins, thereby further reducing production of IFN (4, 32–35). The observation that coronaviruses employ multiple strategies to limit production of IFN seems to suggest that IFN could be detrimental to the propagation of coronaviruses. However, treatment of both MHV and feline coronavirus (FCoV) with IFN (1,000 U) reduces their propagation by approximately 1 log only, indicating that these viruses are relatively resistant to IFN (9, 36). In comparison, SARS-CoV is at least 10 times more sensitive (37–39) and MERS-CoV even 1,000 times more sensitive to IFN treatment than MHV (11, 29). We found that propagation of IBV was reduced by 0.5 to 2.5 logs upon pretreatment with IFN (1,000 U) and less than 0.5 log upon IFN posttreatment, suggesting that IBV is relatively resistant to IFN especially, when the infection has already been established. Our results indicate that both the ScAUG3a and ScAUG3ab viruses are less resistant to IFN treatment than the parental virus, whereas IFN resistance of ScAUG3b was comparable to that of the parental virus. These results indicate that of the four accessory proteins of IBV, 3a is the protein that primarily contributes to the resistance of IBV to IFN. Interestingly, it was previously shown that during infection of primary chicken trachea organ culture (TOC), the titers of both ScAUG3a and ScAUG3ab viruses declined more rapidly than that of the parental virus or ScAUG3b (22). In view of our findings, the decrease in titer of ScAUG3a and ScAUG3ab in TOC could be the result of increased sensitivity of both viruses to IFN produced by cells of the TOC.
Compared to MHV and FCoV, SARS-CoV is relatively sensitive to IFN treatment. However, MERS-CoV is 50 to 100 times more sensitive than SARS-CoV (11, 29). The difference in sensitivity between the latter two viruses has been ascribed to the ability of SARS-CoV to inhibit nuclear translocation of pSTAT1 (29). Considering the relative resistance of IBV to treatment with IFN, we investigated whether IBV, similar to SARS-CoV, would inhibit nuclear translocation of pSTAT1. We observed that at time points earlier than 18 hpi, IBV did not inhibit nuclear translocation of pSTAT1 or activation of a STAT1-responsive promoter (ISG54). In contrast, from 18 hpi onwards, IBV inhibited both IFN-mediated pSTAT1 translocation and activation of the ISG54 promoter. Of interest, SARS-CoV has been shown to inhibit STAT1 translocation as early as 8 hpi, whereas MERS-CoV did not inhibit STAT1 translocation (29). In another study, MHV did not inhibit IFN-mediated translocation of STAT1-GFP at 9 hpi but inhibited IFN-mediated ISG expression at 11 hpi and rescued Sendai virus (SeV) from the antiviral effects of IFN-β when MHV was present prior to SeV infection and for a total period of 16 h (5). Our data indicate a time-dependent inhibition of IFN signaling by IBV, a phenomenon that has not been reported for other coronaviruses, although it cannot be excluded that for the betacoronaviruses MHV and possibly MERS-CoV, inhibition of pSTAT1 translocation could be a relatively late event, similar to what we observed for the gammacoronavirus IBV.
For SARS-CoV, it has been shown that accessory protein ORF 6 is responsible for blocking nuclear translocation of STAT1 by tethering nuclear import factors at the endoplasmic reticulum (ER)/Golgi membrane, inhibiting expression of STAT1-activated genes (19, 28, 40). In the present study, we showed that IBV inhibits phosphorylation of STAT1 and that in contrast to the case with SARS-CoV, the presence of accessory proteins of IBV was not required for inhibition of STAT1-mediated signaling. Our data suggest that IBV and SARS-CoV may exploit different strategies to inhibit translocation of STAT1.
Taking together the ability of IBV to significantly delay transcription of Ifnβ, up until 12 to 18 hpi, and delay subsequent translation of IFN until 36 hpi (4) and the inhibition of pSTAT1 translocation at times points after 18 hpi, we suggest there could be a correlation between the timing of Ifnβ transcription by the host cell and inhibition of IFN signaling induced by IBV. Although there is no proof of causality, we hypothesize that changes in the host cell trigger the relocation of or conformational changes in IBV proteins, which, in turn, activates their anti-IFN activity. Further research is needed to test this hypothesis.
In general, coronavirus accessory proteins have been shown to antagonize the IFN response at various steps. For example, proteins 4a and 4b of MERS and 3b of SARS inhibit activation of Ifnβ (12, 16, 19), whereas protein 7 of transmissible gastroenteritis virus (TGEV) and 3b of IBV inhibit transcription and translation of Ifnβ (4, 17, 41). Notwithstanding these and other steps to counteract and/or avoid activation of the IFN response (reviewed in reference 42), accessory proteins not only inhibit activation of the IFN response but also antagonize the antiviral effect of IFN. The ORF 6 protein of SARS-CoV inhibits IFN signaling by blocking translocation of STAT1 (28), ns2 of MHV inhibits the IFN-activated OAS-RNase L antiviral pathway (20), and 5a of MHV and 7a of FCoV also confer resistance to IFN treatment but via presently unknown mechanisms (10, 18). Using IBV accessory protein null viruses, we showed that knockout of protein 3a renders IBV more sensitive to IFN treatment. In a previous study, we found that 3a decreases transcription of Ifnβ and modulates production of IFN protein (4). The mechanism by which accessory protein 3a confers resistance to IFN treatment remains unclear, although in the present study, we were able to show that 3a does not interfere with STAT1-mediated signaling.
To explain the role of 3a in counteracting the type I IFN response, we hypothesize that 3a might interact with host proteins involved in both the induction of Ifnβ as well as the IFN-induced antiviral response. Host proteins that meet these criteria are, for example, the dsRNA-activated antiviral proteins protein kinase R (PKR) and 2′-5′-oligoadenylate synthetase (OAS). For MHV, it was demonstrated that accessory protein ns2 antagonizes the OAS RNase L pathway (20), a potent antiviral response activated by double-stranded RNA. Accessory protein 3a of IBV, however, does not contain the canonical HxT/S catalytic 2H-phosphoesterase motifs that are essential for the IFN-antagonistic activity of ns2 (43). Interestingly, accessory protein 3a has been shown to partially colocalize with dsRNA in IBV-infected chicken cells (44), which could indicate that 3a may prevent the dsRNA-mediated activation of the OAS RNase L pathway.
Coronaviruses induce extensive remodelling of intracellular membranes (45–47), a process that is essential for coronavirus replication (48–50). It has been suggested that these membrane structures shield dsRNA from the host cell (45, 51, 52), to avoid activation of the IFN response and to simultaneously shield nascent viral RNA from the activity of antiviral proteins (45, 52). The shielding of dsRNA by membrane structures could explain both the delayed transcription of Ifnβ during MHV and IBV infections (8, 31) and the inability of these two coronaviruses to inhibit Ifnβ transcription induced by poly I·C or other RNA viruses (3, 32, 33). An alternative explanation for their involvement in limiting IFN production and in resistance to IFN would be that 3a of IBV could stabilize IBV-induced membrane structures. The absence of 3a would then lead to destabilization of the membrane structures, allowing replicating IBV to be detected by antiviral proteins and pattern recognition receptors. Additional research is required to identify how exactly the IBV accessory protein 3a counteracts the type I IFN response.
Taken together, the results of the present study indicate that infectious bronchitis virus is relatively resistant to treatment with IFN, at least in vitro, and suggest that IBV resists the antiviral activity of IFN via at least two mechanisms. First, IBV inhibits IFN-mediated activation of antiviral genes through inhibition of STAT1 phosphorylation and subsequent nuclear translocation in a time-dependent manner. This inhibition occurs at relatively late time points after infection, correlating with upregulation of Ifnβ transcription (3). Second, IBV counteracts the IFN response primarily through the action of the 3a protein. This study demonstrated that the gammacoronavirus IBV, similar to its mammalian counterparts, has evolved multiple strategies to efficiently counteract the IFN response of its avian host, and it identifies accessory protein 3a as an antagonist of the avian IFN system.
ACKNOWLEDGMENTS
This work was financially supported by MSD Animal Health, Bioprocess Technology & Support, Boxmeer, The Netherlands. Helena Maier and Paul Britton were supported by The Pirbright Institute and the Biotechnology and Biological Sciences Research Council (BBSRC).
We thank E. J. Bakker from the Mathematical and Statistical Methods Group of Wageningen University and E. van den Born from MSD Animal health for assistance with statistical analysis and quantification of IBV RNA, respectively.
REFERENCES
- 1.Gorbalenya AE, Enjuanes L, Ziebuhr J, Snijder EJ. 2006. Nidovirales: evolving the largest RNA virus genome. Virus Res 117:17–37. doi: 10.1016/j.virusres.2006.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cavanagh D. 2007. Coronavirus avian infectious bronchitis virus. Vet Res 38:281–297. doi: 10.1051/vetres:2006055. [DOI] [PubMed] [Google Scholar]
- 3.Britton P, Cavanagh D. 2007. Avian coronavirus diseases and infectious bronchitis vaccine development, p 161–181. In Thiel V. (ed), Coronaviruses: molecular and cellular biology. Caister Academic Press, Norfolk, United Kingdom. [Google Scholar]
- 4.Kint J, Fernandez-Gutierrez M, Maier HJ, Britton P, Langereis MA, Koumans J, Wiegertjes GF, Forlenza M. 2015. Activation of the chicken type I interferon response by infectious bronchitis coronavirus. J Virol 89:1156–1167. doi: 10.1128/JVI.02671-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rose KM, Elliott R, Martinez-Sobrido L, Garcia-Sastre A, Weiss SR. 2010. Murine coronavirus delays expression of a subset of interferon-stimulated genes. J Virol 84:5656–5669. doi: 10.1128/JVI.00211-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou H, Zhao J, Perlman S. 2010. Autocrine interferon priming in macrophages but not dendritic cells results in enhanced cytokine and chemokine production after coronavirus infection. mBio 1:e00219-10. doi: 10.1128/mBio.00219-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Devaraj SG, Wang N, Chen Z, Tseng M, Barretto N, Lin R, Peters CJ, Tseng CT, Baker SC, Li K. 2007. Regulation of IRF-3-dependent innate immunity by the papain-like protease domain of the severe acute respiratory syndrome coronavirus. J Biol Chem 282:32208–32221. doi: 10.1074/jbc.M704870200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Menachery VD, Eisfeld AJ, Schafer A, Josset L, Sims AC, Proll S, Fan S, Li C, Neumann G, Tilton SC, Chang J, Gralinski LE, Long C, Green R, Williams CM, Weiss J, Matzke MM, Webb-Robertson BJ, Schepmoes AA, Shukla AK, Metz TO, Smith RD, Waters KM, Katze MG, Kawaoka Y, Baric RS. 2014. Pathogenic influenza viruses and coronaviruses utilize similar and contrasting approaches to control interferon-stimulated gene responses. mBio 5:e01174-14. doi: 10.1128/mBio.01174-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roth-Cross JK, Martinez-Sobrido L, Scott EP, Garcia-Sastre A, Weiss SR. 2007. Inhibition of the alpha/beta interferon response by mouse hepatitis virus at multiple levels. J Virol 81:7189–7199. doi: 10.1128/JVI.00013-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dedeurwaerder A, Olyslaegers DA, Desmarets LM, Roukaerts ID, Theuns S, Nauwynck HJ. 4 November 2013. The ORF7-encoded accessory protein 7a of feline infectious peritonitis virus as a counteragent against interferon-alpha induced antiviral response. J Gen Virol doi: 10.1099/vir.0.058743-0. [DOI] [PubMed] [Google Scholar]
- 11.Hart BJ, Dyall J, Postnikova E, Zhou H, Kindrachuk J, Johnson RF, Olinger GG, Frieman MB, Holbrook MR, Jahrling PB, Hensley L. 9 December 2013. Interferon-beta and mycophenolic acid are potent inhibitors of Middle East respiratory syndrome coronavirus in cell-based assays. J Gen Virol doi: 10.1099/vir.0.061911-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matthews KL, Coleman CM, van der Meer Y, Snijder EJ, Frieman MB. 17 January 2014. The ORF4b-encoded accessory proteins of MERS-coronavirus and two related bat coronaviruses localize to the nucleus and inhibit innate immune signaling. J Gen Virol doi: 10.1099/vir.0.062059-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pei J, Sekellick MJ, Marcus PI, Choi IS, Collisson EW. 2001. Chicken interferon type I inhibits infectious bronchitis virus replication and associated respiratory illness. J Interferon Cytokine Res 21:1071–1077. doi: 10.1089/107999001317205204. [DOI] [PubMed] [Google Scholar]
- 14.Otsuki K, Maeda J, Yamamoto H, Tsubokura M. 1979. Studies on avian infectious bronchitis virus (IBV). III. Interferon induction by and sensitivity to interferon of IBV. Arch Virol 60:249–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang Y, Zhang L, Geng H, Deng Y, Huang B, Guo Y, Zhao Z, Tan W. 2013. The structural and accessory proteins M, ORF 4a, ORF 4b, and ORF 5 of Middle East respiratory syndrome coronavirus (MERS-CoV) are potent interferon antagonists. Protein Cell 4:951–961. doi: 10.1007/s13238-013-3096-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Niemeyer D, Zillinger T, Muth D, Zielecki F, Horvath G, Suliman T, Barchet W, Weber F, Drosten C, Muller MA. 2013. Middle East respiratory syndrome coronavirus accessory protein 4a is a type I interferon antagonist. J Virol 87:12489–12495. doi: 10.1128/JVI.01845-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cruz J, Sola I, Becares M, Alberca B, Plana J, Enjuanes L, Zuniga S. 2011. Coronavirus gene 7 counteracts host defenses and modulates virus virulence. PLoS Pathog 7:e1002090. doi: 10.1371/journal.ppat.1002090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koetzner CA, Kuo L, Goebel SJ, Dean AB, Parker MM, Masters PS. 2010. Accessory protein 5a is a major antagonist of the antiviral action of interferon against murine coronavirus. J Virol 84:8262–8274. doi: 10.1128/JVI.00385-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kopecky-Bromberg SA, Martínez-Sobrido L, Frieman M, Baric RA, Palese P. 2007. Severe acute respiratory syndrome coronavirus open reading frame (ORF) 3b, ORF 6, and nucleocapsid proteins function as interferon antagonists. J Virol 81:548–557. doi: 10.1128/JVI.01782-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao L, Jha BK, Wu A, Elliott R, Ziebuhr J, Gorbalenya AE, Silverman RH, Weiss SR. 2012. Antagonism of the interferon-induced OAS-RNase L pathway by murine coronavirus ns2 protein is required for virus replication and liver pathology. Cell Host Microbe 11:607–616. doi: 10.1016/j.chom.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Narayanan K, Huang C, Makino S. 2008. Coronavirus accessory proteins, p 235–244. In Perlman S, Gallagher T, Snijder E (ed), Nidoviruses. ASM Press, Washington, DC. doi: 10.1128/9781555815790.ch15. [DOI] [Google Scholar]
- 22.Hodgson T, Britton P, Cavanagh D. 2006. Neither the RNA nor the proteins of open reading frames 3a and 3b of the coronavirus infectious bronchitis virus are essential for replication. J Virol 80:296–305. doi: 10.1128/JVI.80.1.296-305.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Casais R, Davies M, Cavanagh D, Britton P. 2005. Gene 5 of the avian coronavirus infectious bronchitis virus is not essential for replication. J Virol 79:8065–8078. doi: 10.1128/JVI.79.13.8065-8078.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Casais R, Thiel V, Siddell SG, Cavanagh D, Britton P. 2001. Reverse genetics system for the avian coronavirus infectious bronchitis virus. J Virol 75:12359–12369. doi: 10.1128/JVI.75.24.12359-12369.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kint J, Maier HJ, Jagt E. 2015. Quantification of infectious bronchitis coronavirus by titration in vitro and in ovo. Methods Mol Biol 1282:89–98. doi: 10.1007/978-1-4939-2438-7_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schultz U, Rinderle C, Sekellick MJ, Marcus PI, Staeheli P. 1995. Recombinant chicken interferon from Escherichia coli and transfected COS cells is biologically active. Eur J Biochem 229:73–76. doi: 10.1111/j.1432-1033.1995.0073l.x. [DOI] [PubMed] [Google Scholar]
- 27.Paulson M, Press C, Smith E, Tanese N, Levy DE. 2002. IFN-stimulated transcription through a TBP-free acetyltransferase complex escapes viral shutoff. Nat Cell Biol 4:140–147. doi: 10.1038/ncb747. [DOI] [PubMed] [Google Scholar]
- 28.Frieman M, Yount B, Heise M, Kopecky-Bromberg SA, Palese P, Baric RS. 2007. Severe acute respiratory syndrome coronavirus ORF6 antagonizes STAT1 function by sequestering nuclear import factors on the rough endoplasmic reticulum/Golgi membrane. J Virol 81:9812–9824. doi: 10.1128/JVI.01012-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Wilde AH, Raj VS, Oudshoorn D, Bestebroer TM, van Nieuwkoop S, Limpens RW, Posthuma CC, van der Meer Y, Barcena M, Haagmans BL, Snijder EJ, van den Hoogen BG. 2013. MERS-coronavirus replication induces severe in vitro cytopathology and is strongly inhibited by cyclosporin A or interferon-alpha treatment. J Gen Virol 94:1749–1760. doi: 10.1099/vir.0.052910-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dent S. 2014. Proteomic analysis of IBV Infection to study the effect of the accessory proteins 5a and 5b. Ph.D. thesis. University of Reading, Reading, United Kingdom. [Google Scholar]
- 31.Kindler E, Jonsdottir HR, Muth D, Hamming OJ, Hartmann R, Rodriguez R, Geffers R, Fouchier RA, Drosten C, Muller MA, Dijkman R, Thiel V. 2013. Efficient replication of the novel human betacoronavirus EMC on primary human epithelium highlights its zoonotic potential. mBio 4:e00611-12. doi: 10.1128/mBio.00611-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kamitani W, Huang C, Narayanan K, Lokugamage KG, Makino S. 2009. A two-pronged strategy to suppress host protein synthesis by SARS coronavirus Nsp1 protein. Nat Struct Mol Biol 16:1134–1140. doi: 10.1038/nsmb.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang C, Lokugamage KG, Rozovics JM, Narayanan K, Semler BL, Makino S. 2011. Alphacoronavirus transmissible gastroenteritis virus nsp1 protein suppresses protein translation in mammalian cells and in cell-free HeLa cell extracts but not in rabbit reticulocyte lysate. J Virol 85:638–643. doi: 10.1128/JVI.01806-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Züst R, Cervantes-Barragan L, Kuri T, Blakqori G, Weber F, Ludewig B, Thiel V. 2007. Coronavirus non-structural protein 1 is a major pathogenicity factor: implications for the rational design of coronavirus vaccines. PLoS Pathog 3:e109. doi: 10.1371/journal.ppat.0030109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vervelde L, Matthijs MG, van Haarlem DA, de Wit JJ, Jansen CA. 2013. Rapid NK-cell activation in chicken after infection with infectious bronchitis virus M41. Vet Immunol Immunopathol 151:337–341. doi: 10.1016/j.vetimm.2012.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou H, Perlman S. 2006. Preferential infection of mature dendritic cells by mouse hepatitis virus strain JHM. J Virol 80:2506–2514. doi: 10.1128/JVI.80.5.2506-2514.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cinatl J, Morgenstern B, Bauer G, Chandra P, Rabenau H, Doerr HW. 2003. Treatment of SARS with human interferons. Lancet 362:293–294. doi: 10.1016/S0140-6736(03)13973-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zheng B, He ML, Wong KL, Lum CT, Poon LL, Peng Y, Guan Y, Lin MC, Kung HF. 2004. Potent inhibition of SARS-associated coronavirus (SCOV) infection and replication by type I interferons (IFN-alpha/beta) but not by type II interferon (IFN-gamma). J Interferon Cytokine Res 24:388–390. doi: 10.1089/1079990041535610. [DOI] [PubMed] [Google Scholar]
- 39.Yoshikawa T, Hill TE, Yoshikawa N, Popov VL, Galindo CL, Garner HR, Peters CJ, Tseng CT. 2010. Dynamic innate immune responses of human bronchial epithelial cells to severe acute respiratory syndrome-associated coronavirus infection. PLoS One 5:e8729. doi: 10.1371/journal.pone.0008729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wathelet MG, Orr M, Frieman MB, Baric RS. 2007. Severe acute respiratory syndrome coronavirus evades antiviral signaling: role of nsp1 and rational design of an attenuated strain. J Virol 81:11620–11633. doi: 10.1128/JVI.00702-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cruz JL, Becares M, Sola I, Oliveros JC, Enjuanes L, Zuniga S. 2013. Alphacoronavirus protein 7 modulates host innate immune response. J Virol 87:9754–9767. doi: 10.1128/JVI.01032-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kindler E, Thiel V. 2014. To sense or not to sense viral RNA: essentials of coronavirus innate immune evasion. Curr Opin Microbiol 20:69–75. doi: 10.1016/j.mib.2014.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang R, Jha BK, Ogden KM, Dong B, Zhao L, Elliott R, Patton JT, Silverman RH, Weiss SR. 2013. Homologous 2′,5′-phosphodiesterases from disparate RNA viruses antagonize antiviral innate immunity. Proc Natl Acad Sci U S A doi: 10.1073/pnas.1306917110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Davies MT. 2009. Subcellular location and protein interactions of the infectious bronchitis virus gene 3 and 5 accessory proteins. Ph.D. thesis. University of Warwick, Coventry, United Kingdom. [Google Scholar]
- 45.Knoops K, Kikkert M, Worm SH, Zevenhoven-Dobbe JC, van der Meer Y, Koster AJ, Mommaas AM, Snijder EJ. 2008. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol 6:e226. doi: 10.1371/journal.pbio.0060226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ulasli M, Verheije MH, de Haan CA, Reggiori F. 2010. Qualitative and quantitative ultrastructural analysis of the membrane rearrangements induced by coronavirus. Cell Microbiol 12:844–861. doi: 10.1111/j.1462-5822.2010.01437.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maier HJ, Hawes PC, Cottam EM, Mantell J, Verkade P, Monaghan P, Wileman T, Britton P. 2013. Infectious bronchitis virus generates spherules from zippered endoplasmic reticulum membranes. mBio 4:e00801-13. doi: 10.1128/mBio.00801-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lundin A, Dijkman R, Bergström T, Kann N, Adamiak B, Hannoun C, Kindler E, Jónsdóttir HR, Muth D, Kint J, Forlenza M, Müller MA, Drosten C, Thiel V, Trybala E. 2014. Targeting membrane-bound viral RNA synthesis reveals potent inhibition of diverse coronaviruses including the Middle East respiratory syndrome virus. PLoS Pathog 10:e1004166. doi: 10.1371/journal.ppat.1004166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van Hemert MJ, van den Worm SH, Knoops K, Mommaas AM, Gorbalenya AE, Snijder EJ. 2008. SARS-coronavirus replication/transcription complexes are membrane-protected and need a host factor for activity in vitro. PLoS Pathog 4:e1000054. doi: 10.1371/journal.ppat.1000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Neuman BW, Angelini MM, Buchmeier MJ. 2014. Does form meet function in the coronavirus replicative organelle? Trends Microbiol doi: 10.1016/j.tim.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Versteeg GA, Bredenbeek PJ, van den Worm SH, Spaan WJ. 2007. Group 2 coronaviruses prevent immediate early interferon induction by protection of viral RNA from host cell recognition. Virology 361:18–26. doi: 10.1016/j.virol.2007.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou H, Perlman S. 2007. Mouse hepatitis virus does not induce beta interferon synthesis and does not inhibit its induction by double-stranded RNA. J Virol 81:568–574. doi: 10.1128/JVI.01512-06. [DOI] [PMC free article] [PubMed] [Google Scholar]