ABSTRACT
The human papillomavirus (HPV) major structural protein L1 composes capsomers that are linked together through interactions mediated by the L1 C terminus to constitute a T=7 icosahedral capsid. H16.U4 is a type-specific monoclonal antibody recognizing a conformation-dependent neutralizing epitope of HPV thought to include the L1 protein C terminus. The structure of human papillomavirus 16 (HPV16) complexed with H16.U4 fragments of antibody (Fab) was solved by cryo-electron microscopy (cryo-EM) image reconstruction. Atomic structures of virus and Fab were fitted into the corresponding cryo-EM densities to identify the antigenic epitope. The antibody footprint mapped predominately to the L1 C-terminal arm with an additional contact point on the side of the capsomer. This footprint describes an epitope that is presented capsid-wide. However, although the H16.U4 epitope suggests the presence of 360 potential binding sites exposed in the capsid valley between each capsomer, H16.U4 Fab bound only to epitopes located around the icosahedral five-fold vertex of the capsid. Thus, the binding characteristics of H16.U4 defined in this study showed a distinctive selectivity for local conformation-dependent interactions with specific L1 invading arms between five-fold related capsomers.
IMPORTANCE Human papillomavirus 16 (HPV16) is the most prevalent oncogenic genotype in HPV-associated anogenital and oral cancers. Here we use cryo-EM reconstruction techniques to solve the structures of the HPV16 capsid complexes using H16.U4 fragment of antibody (Fab). Different from most other antibodies directed against surface loops, H16.U4 monoclonal antibody is unique in targeting the C-terminal arm of the L1 protein. This monoclonal antibody (MAb) is used throughout the HPV research community in HPV serological and vaccine development and to define mechanisms of HPV uptake. The unique binding mode of H16.U4 defined here shows important conformation-dependent interactions within the HPV16 capsid. By targeting an important structural and conformational epitope, H16.U4 may identify subtle conformational changes in different maturation stages of the HPV capsid and provide a key probe to analyze the mechanisms of HPV uptake during the early stages of virus infection. Our analyses precisely define important conformational epitopes on HPV16 capsids that are key targets for successful HPV prophylactic vaccines.
INTRODUCTION
Human papillomavirus (HPV) infections continue to be a significant health burden in patient populations (1, 2). Although commercial vaccines targeting the viral capsid proteins have been applied successfully to protect against high-risk HPV, the efficacy of vaccines is genotype specific, and vaccines provide little therapeutic benefit against existing infections (3). Understanding the antigenic nature of the HPV capsid offers an opportunity to discover structural features that are crucial to capsid integrity and conserved across species. Panels of monoclonal antibodies and mutational analyses have helped to define several antigenic epitopes (4–10); however, determining the conformational epitopes on the capsid surface requires structural analyses, which can be accomplished by cryo-electron microscopy (cryo-EM) technology. Since the HPV life cycle depends on the differentiation of keratinocytes, it is difficult to purify high-titer virus stocks for structural studies. Virus-like particles (VLPs) that are devoid of viral genome (11) have been used successfully for structural studies (8, 12, 13), whereas both pseudovirus (PsV) and quasivirus (QV), which contain expression plasmid DNA (14, 15), have been used for structural studies and infectivity assays (9, 10). For the work presented here, quasivirus has been used throughout.
Papillomaviruses form a nonenveloped T=7 icosahedral capsid that is ∼55 to 60 nm in diameter and contains a circular double-stranded DNA (dsDNA) genome of 8 kb. The capsid is comprised of 360 copies of the L1 major structural protein and an uncertain number of the L2 minor structural protein (15, 16). Five copies of the L1 protein intertwine to form each capsomer, and 72 capsomers interact to constitute a capsid. Twelve capsomers lie on an icosahedral five-fold vertex and are referred to as pentavalent capsomers, whereas the remaining 60 capsomers are each surrounded by six other capsomers and referred to as hexavalent capsomers. The C terminus of each L1 protein, called the C-terminal arm, extends along the capsid floor to interact with the neighboring capsomer before returning to the original donor capsomer (9, 17, 18). Intercapsomer disulfide bonds are formed between cysteine C428 and C175, which stabilize the icosahedral structure and play an important role in virus maturation (18, 19). The core of the capsomer is composed of the common viral structural motif, the antiparallel β-strands BIDG and CHEF (20). Nearly all known conformational epitopes are located on one or more outwardly facing surface-exposed loops that connect the β-strands (21).
We recently reported a cryo-EM study of four different neutralizing monoclonal antibodies (MAbs) that interact with the human papillomavirus 16 (HPV16) capsid (10). Monoclonal antibodies H16.V5, H16.1A, H16.14J, and H263.A2 examined in the previous study all target conformational epitopes located on combinations of the apical surface-exposed loops of L1 proteins. However, a novel neutralizing monoclonal antibody, H16.U4, generated against HPV16 L1 VLP in an earlier study (22) bound capsids differently (21, 23), albeit with a weaker neutralizing ability (13, 21, 23–25) than those of the four previously studied MAbs. A mutational analysis mapped the binding site of H16.U4 to amino acids 427 to 445 of the C-terminal arm of L1 in the canyon between capsomers (21). Additional studies with H16.U4 revealed novel observations of the cellular mechanisms involved in HPV infection (4, 21, 25, 26). For example, H16.U4 blocks cellular attachment and neutralizes VLP and pseudovirus with different efficiencies.
Here we present a cryo-EM structure of HPV16 quasivirus complexed with H16.U4 antibody fragments (Fabs) at 12-Å resolution (see Fig. 1). H16.U4 Fab molecules bound around the pentavalent capsomers at each of the icosahedral five-fold vertices of the capsid. Fitting the atomic structures of the HPV16 L1 capsomer and a Fab variable domain into the cryo-EM complex map identified the conformational epitope. The antibody binding site was comprised predominately of the single invading C-terminal arm connecting each of the hexavalent capsomers related by icosahedral five-fold symmetry. Specifically, the epitope was made up of C-terminal residues 428.CYS, 429.GLN, and 430.LYS. However, the epitope also included an L1 211.THR from the side of the participating hexavalent capsomer. Combined with previous studies, our findings suggest that H16.U4 may neutralize HPV16 by sterically blocking the attachment of an initial cellular receptor that binds around the icosahedral five-fold vertex. In addition, this work illustrates functional differences between hexavalent and pentavalent environments in the capsid due to quasiequivalence.
FIG 1.
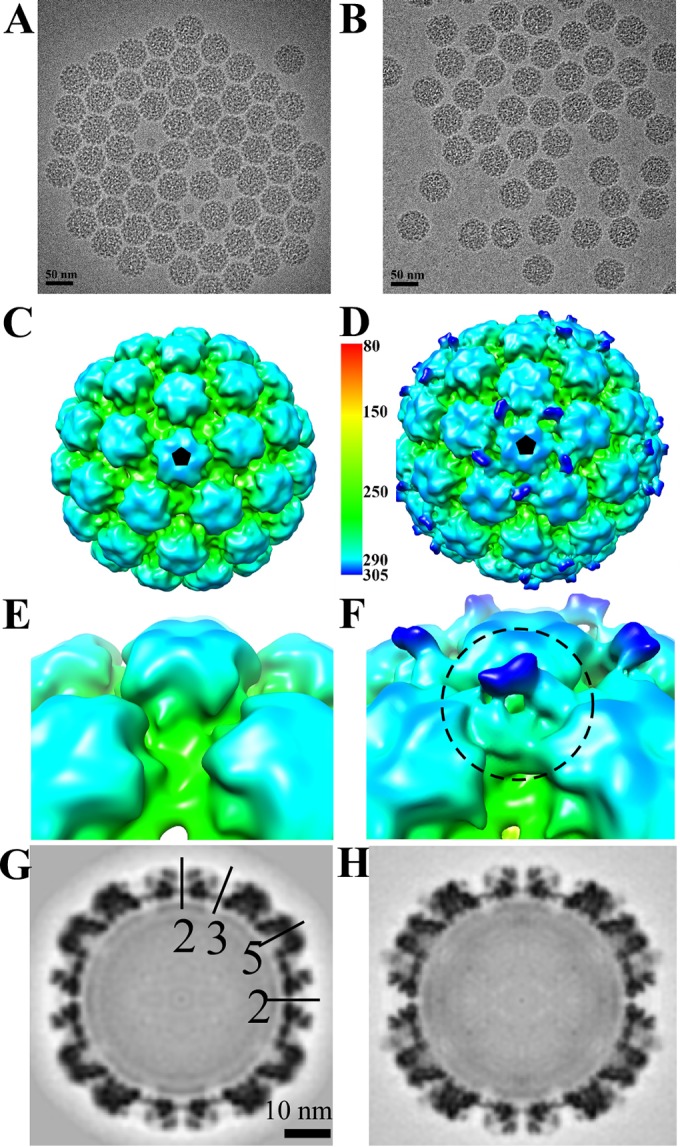
Cryo-EM reconstructions of HPV16 and HPV16-Fab complexes. (A and B) Representative regions of cryo-EM micrographs of the HPV16 particle capsids (A) and HPV16 capsids complexed with H16.U4 Fabs (B). (C and D) The resulting 3D maps of HPV16 (C) and capsid-Fab complex (D) were radially colored according to the distance from the center of the capsid and surface rendered at 1σ. The distance from the center of the capsid (in angstroms) is shown to the right of the color bar. Fab density decorates positions surrounding the five-fold vertices of the capsid (black pentagon). (E and F) The zoomed-in views of the HPV16 alone (E) and the HPV16-U4 complex (F) maps show the pentavalent Fab densities with the constant domain highlighted (dark blue) according to the radially colored map. (G and H) The central sections through the cryo-EM density maps show the quality of the reconstructions. Capsids were cut vertically through the two-, three-, and five-fold icosahedral symmetry axes (black lines), with the central two-fold axis appearing at the 12 o'clock position.
MATERIALS AND METHODS
Preparation of HPV16 quasivirus (QV16).
QV16 particles are comprised of HPV16 L1 and L2 proteins and encapsidate a cottontail rabbit papillomavirus genome (CRPV) containing the simian virus 40 (SV40) origin of replication. QV16 was prepared as described previously (27–29). Briefly, HPV16 sheLL plasmid (kindly provided by John Schiller, NIH) was transfected together with linear CRPV and SV40 origin (SV40ori) DNA into 293TT cells and prepared as described previously (15, 30). QVs were allowed to mature overnight and then pelleted by centrifugation. The centrifuged pellet was resuspended in 1 M NaCl–0.2 M Tris (pH 7.4). After CsCl gradient purification, the lower band was collected, concentrated, and dialyzed against phosphate-buffered saline (PBS), as described previously (9). The concentrated QV16 particles were applied to Formvar-coated copper grids, stained with 2% phosphotungstic acid, and analyzed for integrity and concentration on a JEOL JEM 1400 electron microscope.
Preparation of antibody and Fabs.
H16.U4 antibody was generated in BALB/c mice as described previously (22, 31). Hybridomas were acclimated to animal-component-free medium (BD), and supernatant was purified on protein A or G columns (Pierce). Fab was prepared by digestion with papain in the presence of cysteine (Pierce). The purity of the Fab was assessed by the lack of the fragment crystallizable (Fc) portion, and the integrity of the Fab was determined in an ELISA (enzyme-linked immunosorbent assay). Antibody and Fab protein concentrations were determined by absorbance spectrometry at a wavelength of 280 nm.
Sequencing of H16.U4 antibody heavy and light chains.
The hybridoma cells were pelleted by centrifugation, and RNA was extracted using TRIzol reagent (Life Technologies). cDNAs were synthesized from treated RNA with the RevertAid first strand cDNA synthesis kit (Thermo Scientific). The cDNAs were used as a template for PCR and amplified using Pfu Turbo DNA polymerase (Agilent) or Choice Taq DNA polymerase (Denville). PCR amplification used primers previously described by Wang et al. (32). Immunoglobulin heavy chains were amplified using the isotype-specific constant region 3′ primer and two highly degenerate 5′ primers. The light chains were amplified using the 3′ degenerate kappa chain constant region primer and the 5′ kappa chain framework one region universal degenerate primer. Prior to sequencing, PCR products were purified using the QIAquick PCR purification kit (Qiagen). The same primers used for PCR amplification were also used as sequencing primers to obtain initial sequences. Resolution of the 5′ and 3′ ends of the sequence required sequence-specific primers.
Cryo-electron microscopy.
HPV16 was incubated with 4 Fab molecules per every predicted 360 binding sites for 1 h at room temperature and concentrated to 1.2 mg/ml in PBS buffer. An aliquot of 3 μl of virus-Fab complex was vitrified on Quantifoil holey carbon support grids (Quantifoil, Jena, Germany) that was plunged into liquid ethane using a Cryoplunge 3 (Gatan, Pleasanton, CA). Low-dose conditions were used to record digital images on an Ultrascan 4000 charge-coupled-device (CCD) camera (Gatan, Pleasanton, CA) in a JEOL 2100 LaB6 cryo-EM microscope (JEOL, Peabody, MA) operating at 200 kV and equipped with a Gatan 626 cryo-holder. A sample of HPV quasivirus alone was processed and vitrified in a similar method. The recorded CCD data for HPV16 and HPV16-U4 complexes had calibrated pixel sizes of 1.48 Å/pixel and 2.33 Å/pixel, nominal magnifications of ×80,000 and ×50,000, and defocus ranges of 0.84 to 3.05 μm and 1.60 to 5.43 μm (Table 1), respectively. RELION, AUTO3DEM, and EMAN2 program suites were used for all image processing and three-dimensional (3D) reconstructions (33–35). After the U4 binding site was discovered to map to the epitope surrounding the five-fold vertex, subsequent assays used an excess of two Fab molecules for each binding site of 60 binding sites.
TABLE 1.
Cryo-EM image reconstruction data
| Virus | No. of micrographs | Defocus level range (μm) | No. of particles |
Final resolution (Å) | |
|---|---|---|---|---|---|
| Selected from micrograph | Selected for reconstruction | ||||
| HPV16 | 297 | 0.84–3.05 | 1,895 | 1,718 | 13 |
| HPV16-U4 | 151 | 1.60–5.43 | 5,806 | 2,960 | 12 |
Icosahedral reconstruction.
Virus-Fab complexes were selected from micrographs and used for calculating 3D reconstructions (Table 1). Semiautomatic particle selection was performed using EMAN2's e2boxer.py to obtain the particle coordinates, followed by particle extraction, linearization, normalization, and apodization of the images using RELION (version 1.3-beta) (33, 35). Contrast transfer function parameters were estimated using CTFFIND3 (36). All two-dimensional (2D) and 3D classifications and refinements were performed using RELION. Particle selection used 25 iterations of reference-free 2D class averaging with 20 classes and 57 classes for the HPV16 data set and HPV16-U4 data set, respectively, and 25 iterations of the 3D classification with three classes and seven classes for the two data sets, respectively. The final refinement for the HPV16 and HPV16-U4 complex contained 1,718 and 2,960 particles, respectively. The initial model for the 3D refinement for the two maps was generated from the raw data using script setup_rmc, which is distributed with AUTO3DEM (34, 37). The resolution was estimated where the Fourier shell correlation (FSC) dropped below 0.5 (see Fig. 2).
FIG 2.
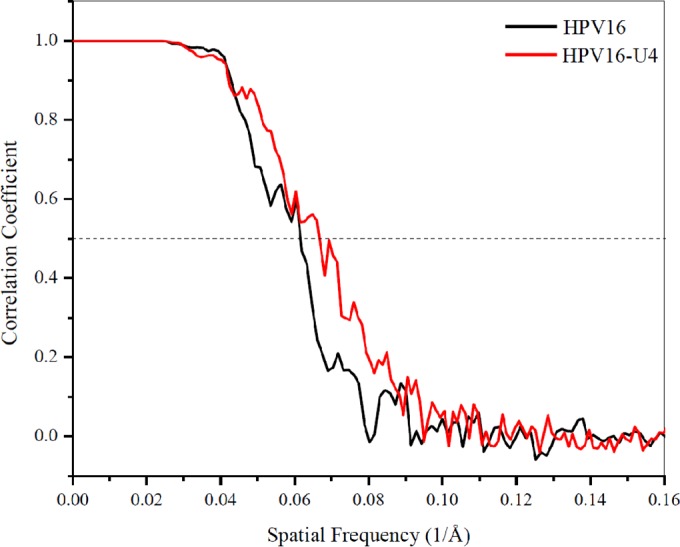
Plot showing the Fourier shell correlation (FSC) versus spatial frequency of the icosahedrally averaged reconstructions for HPV16 and HPV16-Fab complex. The resolution of the reconstructions was assessed where the FSC curve crossed a correlation value of 0.5.
Fitting the Fab structures into the corresponding cryo-EM density.
The difference map was calculated by scaling and subtracting the HPV map density from the HPV16-Fab complex map of the same quality, resolution, and size. The Fab variable domain structural model was predicted based on the amino acid sequence using the Rosetta Online Server that Includes Everyone, ROSIE (http://rosie.rosettacommons.org/antibody) (38). The ROSIE model was fitted into the Fab density in the difference map through Chimera (39) following a classic fitting protocol (34–36). The resulting fitted structure was then placed into the complex map, and five Fab molecules were simultaneously refined with five L1 asymmetric units (PDB accession no. or identifier [ID] 3J6R) (18) using Chimera and Situs (39, 40). Contacts between the fitted structures were identified using Chimera with the criteria for van der Waals (VDW) overlap distances set at −0.4 and 0.0 Å. Clashes between atoms were defined by any overlap of 0.6 Å or more. For size measurement, ROSIE model was surface rendered at 12-Å resolution and measured using Chimera.
Accession numbers.
Protein structure Cryo-EM maps for the capsid and capsid-Fab complexes have been deposited in the EM database (www.emdatabank.org/) with accession numbers EMD-6424 (capsid-H16.U4) and EMD-6423 (HPV16). The fitted structures of Fab (ROSIE) and PDB ID 3J6R have been deposited in the PDB under accession no. 3JBA.
RESULTS
The Fab molecules occupy only sites related to icosahedral five-fold symmetry.
HPV16 capsids were incubated with H16.U4 antibody fragments (Fab) in excess of four Fab molecules for every 360 predicted binding sites (see Materials and Methods). The resulting complexes and virus particles alone were vitrified for cryo-EM data collection. The T=7 icosahedral symmetry of the papillomavirus capsid was readily apparent in low-dose cryo-EM images (Fig. 1A and B). Both HPV particles and HPV-Fab complexes displayed spherical shapes with highly uniform diameters of about 60 nm and exhibited internal densities corresponding to the internal density of packaged DNA. The three-dimensional (3D) reconstructions of particles and complex (Fig. 1C and D) had resolutions of 13 and 12 Å, respectively, as estimated where the Fourier shell correlation (FSC) dropped below 0.5 (Fig. 2). Although bound Fab was indistinct in the 2D cryo-EM micrographs, in the 3D HPV16-U4 complex map, densities corresponding to the density of H16.U4 Fab were clearly seen (Fig. 1D) interacting with virus around the pentavalent capsomers at each five-fold vertex of the capsid. Thus, the diameter of the HPV-H16.U4 Fab complex (measuring from bound Fab to Fab) reached 614 Å compared to 590 Å for HPV16 capsids alone. The H16.U4 Fab densities were located in the canyon formed between two hexavalent capsomers and one pentavalent capsomer (Fig. 1E and F), resulting in a total occupancy of 60 Fabs per capsid. The quality of the cryo-EM maps is illustrated in the central sections (Fig. 1G and H) where Fab variable domain density (Fig. 1H) was quantified at about 50% the magnitude of the capsid density, suggesting less than full occupancy of one Fab per asymmetric unit of the virus capsid. Approximately one half of the Fab constant domain could be visualized (Fig. 1F). The magnitude of the Fab constant domain was insufficient to be fully distinguished even at lower contours, suggesting that less than 100% Fab occupancy coupled with the flexibility of the hinge region combined to weaken the constant domain density.
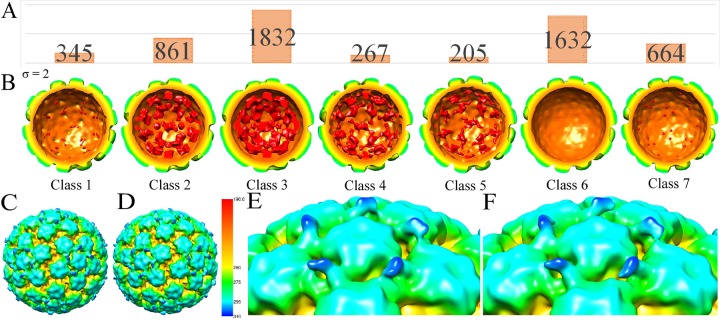
HPV-Fab complexes contained various amounts of packaged DNA.
In general, packaged virus genomes have been shown to affect external virus capsid characteristics; specifically, the amount of DNA within the HPV capsid has been shown to correlate with Fab binding (41–43). There were significant differences in the internal density ranging among the seven classes generated after 3D classification in RELION (33) (Fig. 3). The number of particles assigned to each of the seven classes ranged from 205 to 1,832 particles. The 3D classes ranged in DNA content from entirely empty (class 6) to mostly filled (class 3). Empty capsids (class 6) (Fig. 3D) and those capsids with the most density (classes 2 to 4) (Fig. 3C) were processed separately. The resulting reconstructions showed no differences in the amount of Fab binding; the magnitude of the Fab variable domain was similar when quantified from the central sections (data not shown). At the current resolution, the presence or absence of DNA inside the capsid had no observable effect on the external surface features of the capsid as assessed by binding of Fab (Fig. 3C to F).
FIG 3.
Effect of packaged DNA had little influence on capsid surface features as quantified by Fab binding. (A) HPV16-Fab particles were classified into seven different 3D classes during the reconstruction process. The number of HPV16-Fab particles in each of the seven different 3D classes is shown. (B) The corresponding density maps are colored according to radius and illustrate the amount of DNA density (red). (C and D) The members of classes 2 to 4 with the highest inner density were reconstructed separately to generate a map (C), whereas the particles contained within class 6 with the lowest inner density were used to make a separate map (D). (E and F) Although DNA content is known to affect virus surface features, no discernible differences between full and empty particles (zoomed-in views [E and F)] were seen here. The radial color code used is shown in panel E, which highlights the lower half of the Fab constant domain (blue).
There were differences in the capsomer surface topologies of the virus alone compared to the HPV-Fab complex structure.
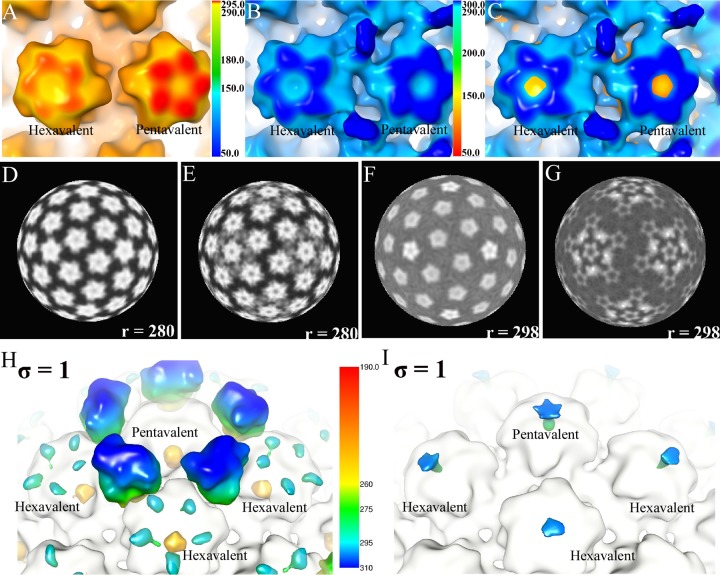
Plotting density against radius verified that relative to the HPV capsid, the diameter of the capsid after Fab binding was not significantly different, suggesting that the Fab did not initiate an expansion of the HPV capsid (Fig. 4). However, a superimposition of the virus and virus-Fab complex maps identified topological differences that mapped to the center of the capsomers (Fig. 5A to C). Differences between the virus and virus-Fab complex structures were illustrated in radially projected maps. At a radius of 280 Å, each capsomer exhibited a slightly fuzzy star-shaped pentamer (Fig. 5D), whereas at the same resolution and at the same radius, the particle-Fab complex capsomer had a sharper density demarcation of the star-shaped capsomer that was especially notable at the tips of the star (BC and EF loops) (Fig. 5E). Compared to the HPV particle map, there appeared to be loss of density from the center of each capsomer in the Fab complex map, which is visualized as a dark center of each capsomer at a radius of 298 Å (Fig. 5F and G). To verify these differences, a difference map was calculated by subtracting capsid density from the HPV-Fab complex map (Fig. 5H). In addition to the obvious Fab densities found around pentavalent capsomers, difference density mapped to the apical surfaces of each capsomer. When the capsid map density was subtracted from the complex map (Fig. 5I), differences in density were found in the center of each capsomer, suggesting a depression might form within the center of each capsomer following H16.U4 binding. One possibility for the distinct differences between capsomer densities of the virus and complex map is a conformational change initiated by the binding of the Fab that could potentially stabilize the capsid and interfere with entry; however, we cannot definitively interpret the cause of the density differences due to the relatively low (12-Å) resolution available at this time.
FIG 4.
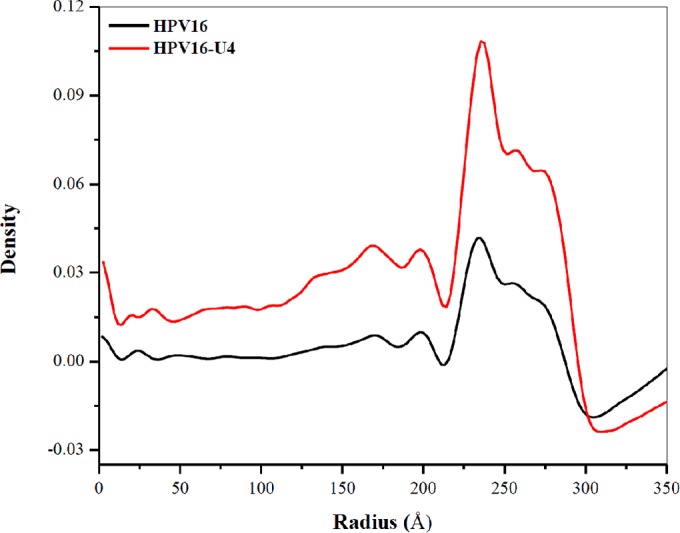
Cryo-EM map density plotted by radii. From the center of each map for HPV16 (black) and HPV16-Fab complex (red), density is plotted to show that although bound Fab extends the radius of the map slightly, there is no expansion of the capsid induced by Fab binding.
FIG 5.
The superimposition and difference maps illustrate significant density disparities between HPV16 particles and HPV16-Fab complexes. (A to C) The HPV map (A) and the HPV16-Fab complex map (B) are superimposed (C) to illustrate the differences between the capsomer densities. Both the hexavalent and pentavalent capsomers have more-filled centers in the HPV map than in the HPV-complex map. (D and E) The density differences between capsomers are also apparent in the radial projections at a radius of 280 Å that show fuzzy star-shaped capsomers in the HPV map alone (D) compared to the HPV-complex map in which the densities are clearer (E). (F and G) When rendered at a radius of 298 Å, the lack of density in the centers of the capsomers in the complex map (G) is seen as a dark center that is significantly different from the presence of density in the HPV16 map (F) (light centers). (H) Difference density calculated from subtracting the HPV16 map from the HPV16-Fab complex map shows clear Fab densities surrounding the pentavalent capsomer. In addition, there are density differences in the apical loop region of each capsomer. (I) The difference density obtained from subtracting the HPV16-Fab complex map from the HPV16 map, or negative difference density, maps to the center of each capsomer, suggesting that binding of Fab causes a conformational change.
A Fab atomic model was fitted to interpret the antibody binding site.
Due to conservation of structure between antibodies, it is reasonable to interpret cryo-EM structures and predict antibody footprints using Fab models (44). The sequence of the variable region of H16.U4 was aligned and annotated so that the complementarity determining regions (CDRs) of both the antibody heavy and light chains could be assigned using the Rosetta online server (ROSIE) (see Materials and Methods). The resulting atomic model generated by ROSIE (38) for the H16.U4 Fab was used for fitting and refinement experiments. This model was also used to simulate Fab density (Fig. 6A) for subsequent measurements of the dimensions of the Fab.
FIG 6.
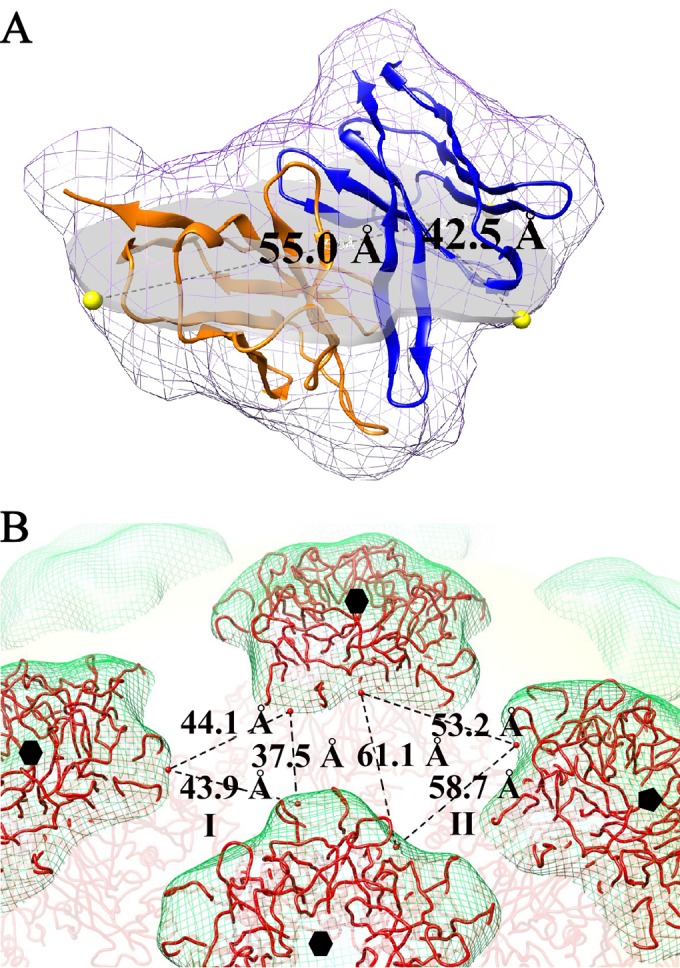
Measurements of steric allowance provided by topology of the HPV16 capsid. (A) The Fab molecule variable domain (heavy and light chains shown in blue and orange, respectively) was surface rendered at 12-Å resolution and measured through the broadest plane, which was selected empirically when positioned parallel to the contact surface with the HPV16 capsid. (B) On the HPV16 capsid surface (green mesh), the two different potential binding areas between capsomers (red wire) (PDB ID 3J6R) (18) that expose the H16.U4 epitope were defined as area I, bordered by three hexavalent capsomers (black hexagons), and area II that is surrounded by two hexavalent capsomers (black hexagons) and one pentavalent capsomer (black pentagon).
The correct orientation of the Fab was quantified from the pseudoatomic structure.
The atomic model for the H16.U4 Fab variable domain was fitted into the difference map calculated by subtracting the unbound HPV capsid density from the HPV-Fab complex map. The fitted structure was then placed into the corresponding density of the complex map, and five Fab molecules were simultaneously refined with five L1 asymmetric units (one asymmetric unit consists of five L1 proteins from one hexavalent capsomer and one neighboring L1 protein from a pentavalent capsomer) of the capsid (PDB ID 3J6R) (Fig. 7) (18). Due to pseudo-two-fold symmetry, the antibody heavy and light chains could be fitted into the cryo-EM density in either of two orientations related by a 180-degree rotation. Here we defined the two binding modes by the position of the heavy chain relative to the light chain (heavy chain followed by light chain) around the five-fold vertex in a clockwise or counterclockwise direction (Fig. 7B). The Fab variable domain structure was fitted and refined in both possible orientations, providing two possible pseudoatomic structures of the Fab-virus complex. The quality of the two different docking modes was quantified to evaluate the correct binding mode (Table 2). Clockwise and counterclockwise fitted Fab molecules generated a correlation coefficient (cc) of 0.8745 and 0.8772, respectively. The lack of any significant difference may be due to limitations from the resolution of the reconstruction. However, an evaluation of steric collisions in the capsid-Fab interface revealed that the counterclockwise orientation resulted in significantly fewer clashing atoms (calculated to be three clashes with the C-terminal arm) compared to 25 clashes with the neighboring L1 capsomer atoms for the clockwise binding mode (Table 2). Using the pseudoatomic models, residues in the interface between virus particle and Fab were identified on the basis of distance and geometry (Fig. 8C).
FIG 7.
The H16.U4 Fab variable domain fitted into the cryo-EM density. The H16.U4 Fab variable domain with heavy chain (blue) and light chain (orange) is shown fitted with the L1 structural model (red) (PDB ID 3J6R) (18) into the complex map. The unfilled Fab density corresponds to a portion of the constant domain. (A) The zoomed-in image illustrates the interaction between H16.U4 Fab and the C-terminal arm residues C428 to C445 (dark gray). (B) Hexavalent and pentavalent capsomers were indicated to show the directionality of the fitted Fab with the heavy chain following the light chain in a counterclockwise fashion.
TABLE 2.
Statistics for fitting Fab structures into the corresponding cryo-EM densitya
| HPV16-U4 fitting mode | Correlation coefficient | No. of clashes |
|---|---|---|
| Heavy chain counterclockwise | 0.8772 | 3 |
| Heavy chain clockwise | 0.8745 | 25 |
Statistics for fitting Fab structures into the corresponding cryo-EM density are reported for both fitting modes according to the correlation coefficient and number of clashes between atoms within the fitted virus structure.
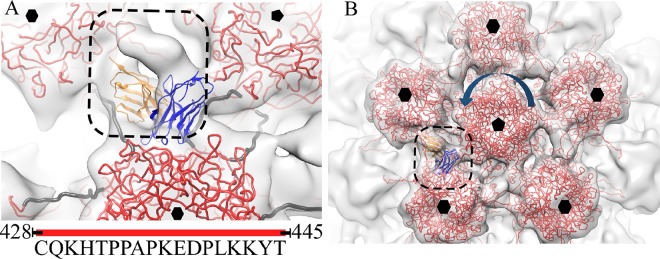
FIG 8.
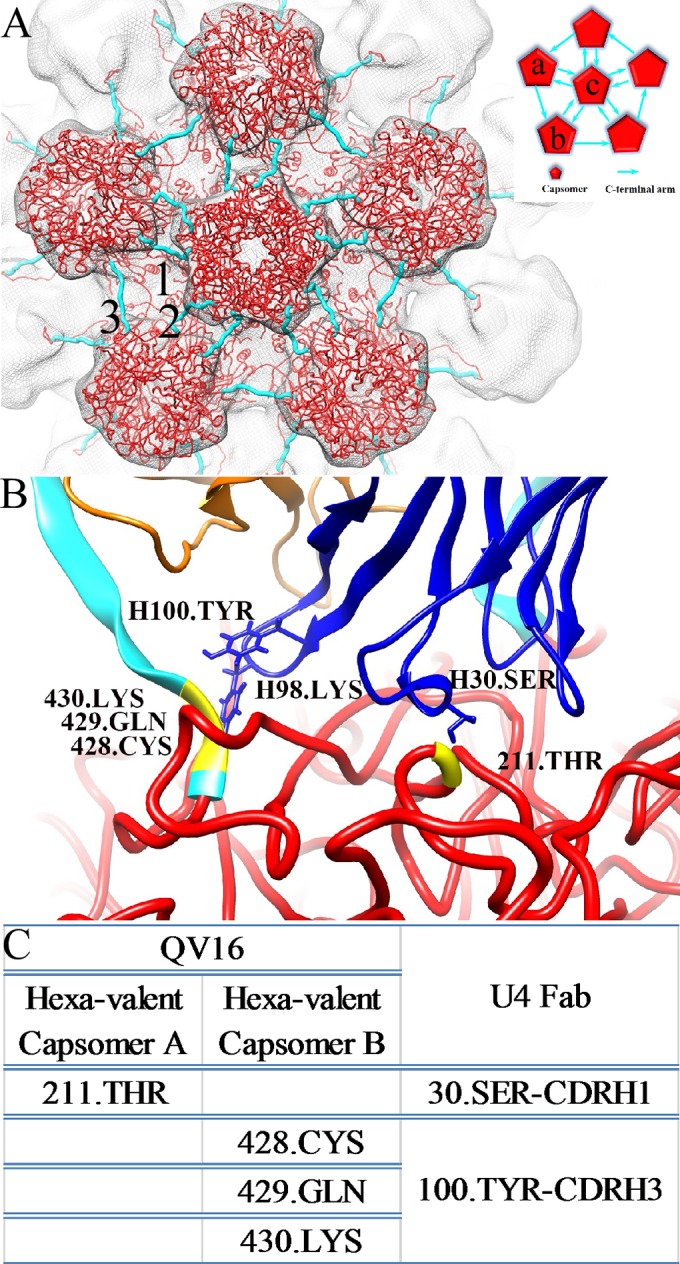
The H16.U4 epitope. The L1 pseudoatomic structure (red) (PDB ID 3J6R) (18) is fitted into a section of the HPV16 capsid to show the five-fold vertex and the orientations of the C-terminal arms (cyan) that connect capsomers. The inset shows the two hexavalent capsomers (labeled a and b) and one pentavalent capsomer (labeled c) that surround the H16.U4 epitope. Between the capsomers (red pentagons), the direction of the arrows (cyan) indicates the C-terminal arm donor and receiver capsomers as described in the text. (B) The fitted Fab variable domain heavy chains (blue) and light chains (orange) are shown to identify the locations of the contacts between Fab and L1 including positions 428, 429, and 430 (yellow) located on the C-terminal arm (cyan) of the capsomer (capsomer a) and position 211 (yellow) on the shoulder of the capsomer (capsomer b). (C) The residues comprising the epitope are listed in the table, along with predicted binding partners.
Two capsomers, termed capsomers a and b, participated in the binding of each H16.U4 Fab (inset in Fig. 8A). The Fab footprint mapped to the L1 C-terminal arm of capsomer a and to the sidewall of capsomer b (Fig. 8B). Three C-terminal arms delineate the space where each H16.U4 bound. Two arms (arms labeled 1 and 2 in Fig. 8A) were located between hexavalent and pentavalent capsomers, whereas the third C-terminal arm (arm labeled 3 in Fig. 8A) joined two hexavalent capsomers (Fig. 8A). Four residues involved in the H16.U4 epitope are predicted to interact with the heavy chain (Fig. 8). Residues 428.CYS, 429.GLN, and 430.LYS are located on the C-terminal arm from donor capsomer a, and residue 211.THR maps to the L1 protein of the recipient capsomer b (Fig. 8C).
Fab molecule dimensions and the distances between capsomers define the H16.U4 binding site around the pentavalent capsomers.
The size of the H16.U4 Fab molecule measured 55.0 Å by 42.5 Å at the broadest portion of the Fab variable domain, with the molecule positioned relative to the binding surface (Fig. 6A) with the CDR loops facing the capsid. Capsomer spacing thus must accommodate this span to allow the Fab CDRs to engage the epitope on the C-terminal arm located between capsomers. The shortest distance between capsomers was found at a radius of 272 Å, at which the largest areas defined by bordering capsomers could be empirically determined (Fig. 6B). Due to icosahedral symmetry, there are two potential areas of binding between capsomers: one area is framed by three hexavalent capsomers (type I); the second space is formed by one pentavalent capsomer and two hexavalent capsomers (type II) (Fig. 6B). Type I sites have an area of 746.3 Å2, which is significantly smaller than the type II area of 1,522.3 Å2 because the distances connecting the capsomers for type I were ∼40% shorter than the type II distances. When comparing the size of H16.U4 (Fig. 6A) Fab, only the type II space is readily accessible for H16.U4 Fab binding.
DISCUSSION
The conformational epitope of the H16.U4 antibody is significantly different from those of other surface loop targeting antibodies, such as H16.V5 and H16.1A (23). On the basis of previous immunological studies, the binding site of the H16.U4 antibody was predicted to be located in the L1 C-terminal arm between positions 427 and 445 and was thought to be accessible at the cleft in the canyon between neighboring capsomers (21). Subsequent structural studies have refined the C-terminal arm arrangement in HPV capsids (Fig. 8A) (9, 17, 18). Of the two types of available binding spaces (defined as type I and II in Results) for H16.U4 (Fig. 6), only the larger type II site could accommodate H16.U4 binding. To reach the binding sites on the C-terminal arm, the entire variable domain has to dock into the canyon (Fig. 7A) with a binding angle of 71° measured between the pseudo-two-fold symmetry axis of the Fab variable domain and the pseudoicosahedral five-fold symmetry axis of the hexavalent capsomer to which Fab is bound (capsomer b in Fig. 8A). One explanation for the constrained binding of the H16.U4 Fab to the five-fold vertices of the capsid is steric allowances and size restrictions of the available epitope surface area between capsomers. It is conceivable that the conformation of the five-fold epitope is slightly different due to the quasiequivalence of the T=7 icosahedral capsid.
Although the type II space is large enough to allow binding of the H16.U4 Fab, it appears to be a snug fit. Any conformational changes to the capsid that decrease the type II regional space would likely abrogate H16.U4 binding. Thus, the accessibility of H16.U4 Fab binding will be sensitive to conformational variations and potential changes that are the result of the maturation of the capsid, which may therefore explain U4 binding properties. The monoclonal antibody of H16.U4 was generated against HPV16 L1 only virus-like particles (VLPs) and showed weaker affinity (10 to 20%) to pseudovirus (PsV) particles (22, 26). Furthermore, MAb H16.U4 has the ability to neutralize PsV in vitro, but not authentic HPV16 (generated in xenografts implanted in severe combined immunodeficiency mice) (23). We have noted, however, that high concentrations of H16.U4 antibody are needed to neutralize successfully HPV16 PsV (19). Collectively, these results suggest that there may be conformational differences between VLP, pseudotype, and authentic HPV16 capsids that map to the U4 binding site. Differences have not been identified between HPV capsid types (VLP, pseudovirus, quasivirus, and authentic virus) when using H16.V5, H16.1A, and other antibodies that target the surface loops mapping to the capsomers (7–10, 22). This disparity suggests that the capsomer structure itself is conserved, and it is the packing of capsomers mediated by the structure of the C-terminal arm that confers differences in the intercapsomeric canyon. Specifically, that the packing of capsomers among capsid types may differ is supported by our recent characterization of indistinguishable capsomers between three different capsid diameters of quasivirus in a high-resolution data set (data not shown). The epitope of H16.U4 determined here implies that the L1 VLP has the largest type II regional space among the three particles, whereas authentic HPV16 virions have the smallest. Pseudo- and quasi-HPV16 likely have an intermediate type II steric space allowing for H16.U4 binding; a difference perhaps conferred by the packaged DNA. Nevertheless, the steric requirements of H16.U4 binding and differences between capsid types may allow H16.U4 to be used as a unique tracker molecule to differentiate capsids with minor structural differences.
The H16.U4 Fab constant domain density was weak compared to that of the variable domain (Fig. 1F), which is likely attributable to a combination of factors, including the incomplete occupation of Fab, the inherent flexibility of the H16.U4 Fab molecule, and the low resolution of the reconstruction. The Fab density was quantified at about half the magnitude of that of the capsid, indicating that only about half the potential 60 binding sites are occupied by Fab. This low occupancy is likely due to low affinity (25), since the Fab was incubated with capsids in great excess. Fab has two regions of flexibility that allow binding to sites with variable distances (45, 46); the joint between the variable domain-constant domain junction, also called the “elbow,” and a flexible stretch of polypeptide chain which connects two Fabs and the Fc portion, called the “hinge.” This combination of factors resulted in the lack of full Fab density in the complex map after the reconstruction process.
The correct orientation along the pseudodyad axis for the fitted Fab was identified according to steric interference between fitted molecules (Table 2). Thus, the best binding mode was with the heavy chain arranged counterclockwise relative to the light chain around the five-fold symmetry axis of the capsid (Fig. 7). There are two modes of interaction for the intercapsomeric C-terminal arms located at each icosahedral five-fold vertex (Fig. 8A). The pentavalent capsomer donates a C-terminal arm to each of five neighbors and receives five arms from every recipient. Thus, there are five pairs of closely associated arms around the pentavalent capsomer (Fig. 8A, inset). The five hexavalent capsomers that surround the pentavalent capsomer each provide one C-terminal arm to a hexavalent neighbor, forming a counterclockwise circle of solitary single arms around the five-fold symmetry axis (Fig. 8A, inset). Only these single arms have the conformation and are sufficiently separated in topological space to allow the H16.U4 epitope to be engaged.
The epitope analysis of the C-terminal arm corroborated our previous mutational and immunological studies in which the H16.U4 binding site was located near L1 residue 430 and between residues 427 and 445 (21). Included within the epitope is residue 428.CYS that participates in the formation of an intercapsomeric disulfide bond (19, 47, 48), although there is no indication that the disulfide bond was affected by Fab binding in this study. The interaction of one Fab with two capsomers may have allowed the H16.U4 Fab to bind and pull neighboring hexavalent capsomers together, which may have changed the capsid structure. A depression at the center on each capsomer is present in the HPV-Fab complex map (Fig. 4), suggesting that the H16.U4 binding may alter the surface loop arrangements and induce a central conformational change. However, such a conformational change cannot be confirmed at the current resolution and requires further study for verification.
The epitope of H16.U4 Fab on the intercapsomeric C-terminal arm may overlap with receptor binding sites. It has been reported that preincubation with H16.U4 MAbs prevented subsequent HPV16 binding to the cell surface (25). Virus initially binds heparan sulfate proteoglycans (HSPG) (49–53), and the HSPG binding sites include residues on the C-terminal arm (442.LYS and 443.LYS) and BC loop (54.LYS, 57.ASN, and 59.LYS) near the intercapsomeric canyon (54). Our results indicated that H16.U4 may neutralize HPV16 by sterically blocking HSPG receptor binding, which also presupposes that HSPG may bind preferentially around the five-fold vertex. Collectively, these results suggest that there may be functional differences between hexavalent and pentavalent environments. We believe that higher-resolution maps of HPV16 and virus-Fab complexes based on innovative cryo-EM technology will reveal more features of the capsids, and we are currently working on this in our lab.
The unique binding mode of H16.U4 defined here shows important conformation-dependent interactions with the HPV16 capsid. Based on the established differences in affinity and neutralization, H16.U4 has the ability to discriminate between the capsid types of VLP, pseudo- or quasicapsids, and authentic virus. Our study also suggested that compared with the likely conserved structure of the capsomer, the structure of the C-terminal arm varies among capsids. We have established here that the C-terminal arm structure does vary within the same capsid as shown by H16.U4 preferential binding. By targeting an important structural and conformational epitope, H16.U4 can label subtle conformational changes in different maturation stages of the HPV capsid and provide a key probe to analyze the mechanisms of HPV uptake during the early stages of virus infection.
ACKNOWLEDGMENTS
We thank John Schiller for kindly providing HPV16 sheLL and pYSEAP plasmids. We thank the Microscopy Imaging Core Facility at The Pennsylvania State University College of Medicine.
This project is funded, in part, under a grant with the Pennsylvania Department of Health using Tobacco CURE Funds. This work was also supported by Penn State COM institutional research grant 124171-IRG-13-042-01-IRG (S.H.), NIH SIG 1S10RR031780-01A1 (S.H.), and the Jake Gittlen Memorial Golf Tournament (N.D.C.).
The Pennsylvania Department of Health specifically disclaims responsibility for any analyses, interpretations, or conclusions.
J.G., S.M.B., and S.A.B. performed research. R.E.A. performed microscopy and collected cryo-EM data. J.G. processed and analyzed cryo-EM data. H.L. provided computer software support. J.G., N.D.C., and S.H. wrote the paper.
REFERENCES
- 1.Gilmer LS. 2015. Human papillomavirus vaccine update. Prim Care 42:17–32. doi: 10.1016/j.pop.2014.09.011. [DOI] [PubMed] [Google Scholar]
- 2.Dickson EL, Vogel RI, Luo X, Downs LS. 2015. Recent trends in type-specific HPV infection rates in the United States. Epidemiol Infect 143:1042–1047. doi: 10.1017/S0950268814001538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McKee SJ, Bergot A-S, Leggatt GR. 2015. Recent progress in vaccination against human papillomavirus-mediated cervical cancer: HPV vaccination. Rev Med Virol 25:54–71. doi: 10.1002/rmv.1824. [DOI] [PubMed] [Google Scholar]
- 4.Christensen ND, Cladel NM, Reed CA, Budgeon LR, Embers ME, Skulsky DM, McClements WL, Ludmerer SW, Jansen KU. 2001. Hybrid papillomavirus L1 molecules assemble into virus-like particles that reconstitute conformational epitopes and induce neutralizing antibodies to distinct HPV types. Virology 291:324–334. doi: 10.1006/viro.2001.1220. [DOI] [PubMed] [Google Scholar]
- 5.Wang Z, Christensen N, Schiller JT, Dillner J. 1997. A monoclonal antibody against intact human papillomavirus type 16 capsids blocks the serological reactivity of most human sera. J Gen Virol 78:2209–2215. doi: 10.1099/0022-1317-78-9-2209. [DOI] [PubMed] [Google Scholar]
- 6.Varsani A, Williamson A-L, de Villiers D, Becker I, Christensen ND, Rybicki EP. 2003. Chimeric human papillomavirus type 16 (HPV-16) L1 particles presenting the common neutralizing epitope for the L2 minor capsid protein of HPV-6 and HPV-16. J Virol 77:8386–8393. doi: 10.1128/JVI.77.15.8386-8393.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rizk RZ, Christensen ND, Michael KM, Muller M, Sehr P, Waterboer T, Pawlita M. 2008. Reactivity pattern of 92 monoclonal antibodies with 15 human papillomavirus types. J Gen Virol 89:117–129. doi: 10.1099/vir.0.83145-0. [DOI] [PubMed] [Google Scholar]
- 8.Zhao Q, Modis Y, High K, Towne V, Meng Y, Wang Y, Alexandroff J, Brown M, Carragher B, Potter CS, Abraham D, Wohlpart D, Kosinski M, Washabaugh MW, Sitrin RD. 2012. Disassembly and reassembly of human papillomavirus virus-like particles produces more virion-like antibody reactivity. Virol J 9:52. doi: 10.1186/1743-422X-9-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee H, Brendle SA, Bywaters SM, Guan J, Ashley RE, Yoder JD, Makhov AM, Conway JF, Christensen ND, Hafenstein S. 2015. A cryo-electron microscopy study identifies the complete H16.V5 epitope and reveals global conformational changes initiated by binding of the neutralizing antibody fragment. J Virol 89:1428–1438. doi: 10.1128/JVI.02898-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guan J, Bywaters SM, Brendle SA, Lee H, Ashley RE, Makhov AM, Conway JF, Christensen ND, Hafenstein S. 2015. Structural comparison of four different antibodies interacting with human papillomavirus 16 and mechanisms of neutralization. Virology 483:253–263. doi: 10.1016/j.virol.2015.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kirnbauer R, Booy F, Cheng N, Lowy DR, Schiller JT. 1992. Papillomavirus L1 major capsid protein self-assembles into virus-like particles that are highly immunogenic. Proc Natl Acad Sci U S A 89:12180–12184. doi: 10.1073/pnas.89.24.12180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao Q, Potter CS, Carragher B, Lander G, Sworen J, Towne V, Abraham D, Duncan P, Washabaugh MW, Sitrin RD. 2014. Characterization of virus-like particles in GARDASIL® by cryo transmission electron microscopy. Hum Vaccin Immunother 10:734–739. doi: 10.4161/hv.27316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deschuyteneer M, Elouahabi A, Plainchamp D, Plisnier M, Soete D, Corazza Y, Lockman L, Giannini S, Deschamps M. 2010. Molecular and structural characterization of the L1 virus-like particles that are used as vaccine antigens in CervarixTM, the AS04-adjuvanted HPV-16 and -18 cervical cancer vaccine. Hum Vaccin 6:407–419. doi: 10.4161/hv.6.5.11023. [DOI] [PubMed] [Google Scholar]
- 14.Christensen ND. 2005. Cottontail rabbit papillomavirus (CRPV) model system to test antiviral and immunotherapeutic strategies. Antivir Chem Chemother 16:355–362. doi: 10.1177/095632020501600602. [DOI] [PubMed] [Google Scholar]
- 15.Buck CB, Pastrana DV, Lowy DR, Schiller JT. 2005. Generation of HPV pseudovirions using transfection and their use in neutralization assays. Methods Mol Med 119:445–462. [DOI] [PubMed] [Google Scholar]
- 16.Baker TS, Newcomb WW, Olson NH, Cowsert LM, Olson C, Brown JC. 1991. Structures of bovine and human papillomaviruses. Analysis by cryoelectron microscopy and three-dimensional image reconstruction. Biophys J 60:1445–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wolf M, Garcea RL, Grigorieff N, Harrison SC. 2010. Subunit interactions in bovine papillomavirus. Proc Natl Acad Sci U S A 107:6298–6303. doi: 10.1073/pnas.0914604107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cardone G, Moyer AL, Cheng N, Thompson CD, Dvoretzky I, Lowy DR, Schiller JT, Steven AC, Buck CB, Trus BL. 2014. Maturation of the human papillomavirus 16 capsid. mBio 5:e01104–14. doi: 10.1128/mBio.01104-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Buck CB, Thompson CD, Pang Y-YS, Lowy DR, Schiller JT. 2005. Maturation of papillomavirus capsids. J Virol 79:2839–2846. doi: 10.1128/JVI.79.5.2839-2846.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rossmann MG, Johnson JE. 1989. Icosahedral RNA virus structure. Annu Rev Biochem 58:533–569. doi: 10.1146/annurev.bi.58.070189.002533. [DOI] [PubMed] [Google Scholar]
- 21.Carter JJ, Wipf GC, Benki SF, Christensen ND, Galloway DA. 2003. Identification of a human papillomavirus type 16-specific epitope on the C-terminal arm of the major capsid protein L1. J Virol 77:11625–11632. doi: 10.1128/JVI.77.21.11625-11632.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Christensen ND, Dillner J, Eklund C, Carter JJ, Wipf GC, Reed CA, Cladel NM, Galloway DA. 1996. Surface conformational and linear epitopes on HPV-16 and HPV-18 L1 virus-like particles as defined by monoclonal antibodies. Virology 223:174–184. doi: 10.1006/viro.1996.0466. [DOI] [PubMed] [Google Scholar]
- 23.White WI, Wilson SD, Palmer-Hill FJ, Woods RM, Ghim SJ, Hewitt LA, Goldman DM, Burke SJ, Jenson AB, Koenig S, Suzich JA. 1999. Characterization of a major neutralizing epitope on human papillomavirus type 16 L1. J Virol 73:4882–4889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roden RB, Armstrong A, Haderer P, Christensen ND, Hubbert NL, Lowy DR, Schiller JT, Kirnbauer R. 1997. Characterization of a human papillomavirus type 16 variant-dependent neutralizing epitope. J Virol 71:6247–6252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Day PM, Thompson CD, Buck CB, Pang Y-YS, Lowy DR, Schiller JT. 2007. Neutralization of human papillomavirus with monoclonal antibodies reveals different mechanisms of inhibition. J Virol 81:8784–8792. doi: 10.1128/JVI.00552-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Culp TD, Spatz CM, Reed CA, Christensen ND. 2007. Binding and neutralization efficiencies of monoclonal antibodies, Fab fragments, and scFv specific for L1 epitopes on the capsid of infectious HPV particles. Virology 361:435–446. doi: 10.1016/j.virol.2006.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brendle SA, Culp TD, Broutian TR, Christensen ND. 2010. Binding and neutralization characteristics of a panel of monoclonal antibodies to human papillomavirus 58. J Gen Virol 91:1834–1839. doi: 10.1099/vir.0.017228-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mejia AF, Culp TD, Cladel NM, Balogh KK, Budgeon LR, Buck CB, Christensen ND. 2006. Preclinical model to test human papillomavirus virus (HPV) capsid vaccines in vivo using infectious HPV/cottontail rabbit papillomavirus chimeric papillomavirus particles. J Virol 80:12393–12397. doi: 10.1128/JVI.01583-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pyeon D, Lambert PF, Ahlquist P. 2005. Production of infectious human papillomavirus independently of viral replication and epithelial cell differentiation. Proc Natl Acad Sci U S A 102:9311–9316. doi: 10.1073/pnas.0504020102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pastrana DV, Buck CB, Pang Y-YS, Thompson CD, Castle PE, FitzGerald PC, Krüger Kjaer S, Lowy DR, Schiller JT. 2004. Reactivity of human sera in a sensitive, high-throughput pseudovirus-based papillomavirus neutralization assay for HPV16 and HPV18. Virology 321:205–216. doi: 10.1016/j.virol.2003.12.027. [DOI] [PubMed] [Google Scholar]
- 31.Christensen ND, Kreider JW, Kan NC, DiAngelo SL. 1991. The open reading frame L2 of cottontail rabbit papillomavirus contains antibody-inducing neutralizing epitopes. Virology 181:572–579. doi: 10.1016/0042-6822(91)90890-N. [DOI] [PubMed] [Google Scholar]
- 32.Wang Z, Raifu M, Howard M, Smith L, Hansen D, Goldsby R, Ratner D. 2000. Universal PCR amplification of mouse immunoglobulin gene variable regions: the design of degenerate primers and an assessment of the effect of DNA polymerase 3′ to 5′ exonuclease activity. J Immunol Methods 233:167–177. doi: 10.1016/S0022-1759(99)00184-2. [DOI] [PubMed] [Google Scholar]
- 33.Scheres SHW. 2012. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol 180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yan X, Sinkovits RS, Baker TS. 2007. AUTO3DEM—an automated and high throughput program for image reconstruction of icosahedral particles. J Struct Biol 157:73–82. doi: 10.1016/j.jsb.2006.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, Ludtke SJ. 2007. EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol 157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 36.Mindell JA, Grigorieff N. 2003. Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol 142:334–347. doi: 10.1016/S1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- 37.Yan X, Dryden KA, Tang J, Baker TS. 2007. Ab initio random model method facilitates 3D reconstruction of icosahedral particles. J Struct Biol 157:211–225. doi: 10.1016/j.jsb.2006.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lyskov S, Chou F-C, Conchúir SÓ, Der BS, Drew K, Kuroda D, Xu J, Weitzner BD, Renfrew PD, Sripakdeevong P, Borgo B, Havranek JJ, Kuhlman B, Kortemme T, Bonneau R, Gray JJ, Das R. 2013. Serverification of molecular modeling applications: the Rosetta Online Server that Includes Everyone (ROSIE). PLoS One 8:e63906. doi: 10.1371/journal.pone.0063906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera: a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 40.Wriggers W, Milligan RA, McCammon JA. 1999. Situs: a package for docking crystal structures into low-resolution maps from electron microscopy. J Struct Biol 125:185–195. doi: 10.1006/jsbi.1998.4080. [DOI] [PubMed] [Google Scholar]
- 41.Hafenstein S, Fane BA. 2002. phi X174 genome-capsid interactions influence the biophysical properties of the virion: evidence for a scaffolding-like function for the genome during the final stages of morphogenesis. J Virol 76:5350–5356. doi: 10.1128/JVI.76.11.5350-5356.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bernal RA, Hafenstein S, Esmeralda R, Fane BA, Rossmann MG. 2004. The phiX174 protein J mediates DNA packaging and viral attachment to host cells. J Mol Biol 337:1109–1122. doi: 10.1016/j.jmb.2004.02.033. [DOI] [PubMed] [Google Scholar]
- 43.Hafenstein SL, Chen M, Fane BA. 2004. Genetic and functional analyses of the øX174 DNA binding protein: the effects of substitutions for amino acid residues that spatially organize the two DNA binding domains. Virology 318:204–213. doi: 10.1016/j.virol.2003.09.018. [DOI] [PubMed] [Google Scholar]
- 44.Hafenstein S, Bowman VD, Sun T, Nelson CDS, Palermo LM, Chipman PR, Battisti AJ, Parrish CR, Rossmann MG. 2009. Structural comparison of different antibodies interacting with parvovirus capsids. J Virol 83:5556–5566. doi: 10.1128/JVI.02532-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sotriffer CA, Rode BM, Varga JM, Liedl KR. 2000. Elbow flexibility and ligand-induced domain rearrangements in antibody Fab NC6.8: large effects of a small hapten. Biophys J 79:614–628. doi: 10.1016/S0006-3495(00)76320-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Janeway CA Jr, Travers P, Walport M, Shlomchik MJ. 2001. The structure of a typical antibody molecule, p 3:2–3:11. In Immunobiology: the immune system in health and disease, 2nd ed Garland Science, New York, NY. [Google Scholar]
- 47.Li M, Beard P, Estes PA, Lyon MK, Garcea RL. 1998. Intercapsomeric disulfide bonds in papillomavirus assembly and disassembly. J Virol 72:2160–2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Conway MJ, Cruz L, Alam S, Christensen ND, Meyers C. 2011. Differentiation-dependent interpentameric disulfide bond stabilizes native human papillomavirus type 16. PLoS One 6:e22427. doi: 10.1371/journal.pone.0022427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Giroglou T, Florin L, Schafer F, Streeck RE, Sapp M. 2001. Human papillomavirus infection requires cell surface heparan sulfate. J Virol 75:1565–1570. doi: 10.1128/JVI.75.3.1565-1570.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Joyce JG, Tung JS, Przysiecki CT, Cook JC, Lehman ED, Sands JA, Jansen KU, Keller PM. 1999. The L1 major capsid protein of human papillomavirus type 11 recombinant virus-like particles interacts with heparin and cell-surface glycosaminoglycans on human keratinocytes. J Biol Chem 274:5810–5822. doi: 10.1074/jbc.274.9.5810. [DOI] [PubMed] [Google Scholar]
- 51.Johnson KM, Kines RC, Roberts JN, Lowy DR, Schiller JT, Day PM. 2009. Role of heparan sulfate in attachment to and infection of the murine female genital tract by human papillomavirus. J Virol 83:2067–2074. doi: 10.1128/JVI.02190-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shafti-Keramat S, Handisurya A, Kriehuber E, Meneguzzi G, Slupetzky K, Kirnbauer R. 2003. Different heparan sulfate proteoglycans serve as cellular receptors for human papillomaviruses. J Virol 77:13125–13135. doi: 10.1128/JVI.77.24.13125-13135.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Selinka H-C, Florin L, Patel HD, Freitag K, Schmidtke M, Makarov VA, Sapp M. 2007. Inhibition of transfer to secondary receptors by heparan sulfate-binding drug or antibody induces noninfectious uptake of human papillomavirus. J Virol 81:10970–10980. doi: 10.1128/JVI.00998-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Richards KF, Bienkowska-Haba M, Dasgupta J, Chen XS, Sapp M. 2013. Multiple heparan sulfate binding site engagements are required for the infectious entry of human papillomavirus type 16. J Virol 87:11426–11437. doi: 10.1128/JVI.01721-13. [DOI] [PMC free article] [PubMed] [Google Scholar]