ABSTRACT
Hepatitis C virus (HCV) only infects humans and chimpanzees, while GB virus B (GBV-B), another hepatotropic hepacivirus, infects small New World primates (tamarins and marmosets). In an effort to develop an immunocompetent small primate model for HCV infection to study HCV pathogenesis and vaccine approaches, we investigated the HCV life cycle step(s) that may be restricted in small primate hepatocytes. First, we found that replication-competent, genome-length chimeric HCV RNAs encoding GBV-B structural proteins in place of equivalent HCV sequences designed to allow entry into simian hepatocytes failed to induce viremia in tamarins following intrahepatic inoculation, nor did they lead to progeny virus in permissive, transfected human Huh7.5 hepatoma cells upon serial passage. This likely reflected the disruption of interactions between distantly related structural and nonstructural proteins that are essential for virion production, whereas such cross talk could be restored in similarly designed HCV intergenotypic recombinants via adaptive mutations in NS3 protease or helicase domains. Next, HCV entry into small primate hepatocytes was examined directly using HCV-pseudotyped retroviral particles (HCV-pp). HCV-pp efficiently infected tamarin hepatic cell lines and primary marmoset hepatocyte cultures through the use of the simian CD81 ortholog as a coreceptor, indicating that HCV entry is not restricted in small New World primate hepatocytes. Furthermore, we observed genomic replication and modest virus secretion following infection of primary marmoset hepatocyte cultures with a highly cell culture-adapted HCV strain. Thus, HCV can successfully complete its life cycle in primary simian hepatocytes, suggesting the possibility of adapting some HCV strains to small primate hosts.
IMPORTANCE Hepatitis C virus (HCV) is an important human pathogen that infects over 150 million individuals worldwide and leads to chronic liver disease. The lack of a small animal model for this infection impedes the development of a preventive vaccine and pathogenesis studies. In seeking to establish a small primate model for HCV, we first attempted to generate recombinants between HCV and GB virus B (GBV-B), a hepacivirus that infects small New World primates (tamarins and marmosets). This approach revealed that the genetic distance between these hepaciviruses likely prevented virus morphogenesis. We next showed that HCV pseudoparticles were able to infect tamarin or marmoset hepatocytes efficiently, demonstrating that there was no restriction in HCV entry into these simian cells. Furthermore, we found that a highly cell culture-adapted HCV strain was able to achieve a complete viral cycle in primary marmoset hepatocyte cultures, providing a promising basis for further HCV adaptation to small primate hosts.
INTRODUCTION
Approximately 180 million persons are estimated to be chronically infected by hepatitis C virus (HCV) worldwide, the majority of whom are ignorant of their carrier status until chronic infection progresses toward serious symptomatic liver complications, including fibrosis, cirrhosis, and hepatocellular carcinoma. The recent advent of increasingly efficient and better tolerated, yet costly treatment regimens holds promise for facilitating HCV elimination in a number of patients (1). However, the development of a pangenotypic, cost-effective prophylactic vaccine would arguably help reduce the global HCV burden. A complication for this goal is that HCV is reported to have a very narrow host range, limited to humans and chimpanzees. Efforts to develop murine models mimicking this liver infection are ongoing but have not yet translated into the generation of a small immunocompetent animal model that fully recapitulates human HCV infection (2).
The hepacivirus genus was originally created to uniquely classify HCV within the Flaviviridae family. Interestingly, in the past few years, an increasing number of viruses that are phylogenetically related to HCV have been identified in various mammal species, including rodents (3), bats (4), Old World monkeys (5), and horses (6). Although equine hepacivirus has very recently been detected in the livers of infected horses (7, 8), the liver tropism of most of these recently identified hepaciviruses remains to be assessed. GB virus B (GBV-B) is a hepacivirus that has been conclusively demonstrated to be hepatotropic since 1995 (9). Although its ultimate origin remains unknown, GBV-B does not infect chimpanzees (10), but it does experimentally infect small New World primates that are readily accessible for biomedical research, i.e., tamarins (Saguinus species) and marmosets (Callithrix species), in which it generally causes acute self-resolving hepatitis (11). Interestingly, GBV-B often leads to prolonged viremia for more than 6 months (12), and in some cases, it leads to chronic infections that closely mimic chronic HCV infections (13–15).
Although there is relatively low amino acid conservation between GBV-B and HCV polyproteins (approximately 28%), most enzymatic functions are conserved between these phylogenetically related viruses (16–18), and their shared hepatotropism is of great interest for the development of a GBV-B-based surrogate small primate model to study HCV infection (11, 13, 19). The identification of the viral life cycle step(s) and determinant(s) involved in primate species restriction of HCV and GBV-B would be extremely useful for the successful design of HCV chimeras endowed with minimal GBV-B determinants to allow the completion of the hepaciviral life cycle in small primate hepatocytes. HCV cell entry is the first restricted step in murine hepatocytes, due to the lack of functional murine CD81 and occludin molecules as HCV coreceptors, while their human orthologs have been shown to be essential for HCV infection of human hepatocytes (reviewed in reference 20). Glycoprotein E2 is a key viral determinant of HCV binding to the hepatocyte surface and productive entry into these cells (reviewed in reference 21). GBV-B E2 comprises 264 amino acids and, thus, is about two-thirds the size of HCV E2, and it is less glycosylated (22), features that might translate into different entry mechanisms of these viruses in the respective species-specific hepatocytes.
In the present study, we undertook to determine whether the failure of HCV to infect small New World primates (23) was due to a restriction at the virus cell entry point. As a first approach, we attempted to generate a recombinant virus derived from the cell culture-adapted JFH1 strain of HCV genotype 2a (HCV-2a) (24) and encoding GBV-B envelope glycoproteins (13) in place of equivalent HCV proteins. The engineering of such GBV-B/HCV chimeras led us to explore whether a morphogenesis unit derived from GBV-B and a genomic replication unit derived from HCV may be successfully uncoupled such as to lead to the assembly of infectious viral particles. The replication and particle assembly competences of GBV-B/JFH1 chimeras were studied both in vivo in tamarins and in cell culture. We also compared the in vitro replication properties of GBV-B/JFH1 chimeras to those of intergenotypic HCV genotype 1a and 2a recombinants engineered similarly with phylogenetically closer sequences. As a second approach, virus cell entry was cross studied in hepatic cell lines and primary hepatocyte cultures of human or small primate origin using retroviral particles pseudotyped with HCV or GBV-B glycoproteins.
MATERIALS AND METHODS
Plasmids.
Plasmid pGBV-B/2 contains an infectious, genome-length GBV-B cDNA (GenBank accession number AY243572) (13). Plasmid pGBV-B-3m/SapI has been modified from pGBV-B/2 by site-directed mutagenesis to introduce (i) cDNA substitutions resulting in 3 amino acid changes in the GBV-B polyprotein (Val454Ala [mutation of Val to Ala at position 454], Val1594Thr, and Val2236Ala) and (ii) silent substitutions designed to eliminate 2 existing BspQI restriction sites in nonstructural protein 2 (NS2) and NS5B coding sequences without altering the amino acid polyprotein sequence and (iii) to engineer a BspQI restriction site downstream from the viral cDNA such as to generate RNAs with an exact 3′ extremity by in vitro transcription. Plasmids pH77C and pJFH1, which contain genome-length cDNAs of the H77 strain of HCV genotype 1a (GenBank accession number AF011751) (25) and the JFH1 strain of HCV genotype 2a (GenBank accession number AB047639) (24), respectively, were kindly provided by Robert Purcell (National Institutes of Health, Bethesda, MD, USA) and Takaji Wakita (National Institute of Infectious Diseases, Tokyo, Japan). Plasmid pJad has been modified from pJFH1 to include cDNA substitutions resulting in 3 amino acid changes in the HCV polyprotein (Val2153Ala, Val2440Leu, and Val2941Met) that have been shown to increase viral production in cell culture (26). The mutated sequences were derived from pJFH1-2EI3-adapt, kindly provided by Ralf Bartenschlager (University of Heidelberg, Heidelberg, Germany).
To generate plasmids containing genome-length chimeric GBV-B/HCV or HCV-1a/-2a cDNAs or JFH1 control cDNAs containing point substitutions or deletions, we used a primer-based, overlapping PCR mutagenesis strategy through which HCV-2a JFH1, HCV-1a H77, or GBV-B defined sequences were amplified by PCR and fused to one another prior to being cloned into pJFH1 (24) between two unique restriction sites. The nucleotide sequences of PCR-amplified segments were then verified between the two cloning restriction sites of all recombinant plasmid clones. The following section describes the genetic organization of each plasmid, with the numbering corresponding to the nucleotide (nt) position in the respective viral cDNA. Details of all constructions can be obtained upon request. Plasmid pJ/GAA was derived from pJFH1 by replacing nt 8615 to 8623 [GGC-GCA-GCA], encoding the catalytic site (Gly-Asp-Asp) of the NS5B RNA polymerase, with GGC-GAT-GAC, encoding an inactive amino acid triplet (Gly-Ala-Ala). Plasmid pJ/ΔEp7 was constructed by deleting nt 914 to 2779 of the JFH1 cDNA, corresponding to sequences encoding E1-E2-p7 (envelope proteins 1 and 2 and ion channel protein p7). Plasmid pJ/ΔCp7 was derived from pJ/ΔEp7 and has deletions of nt 398 to 850 and 914 to 2779 of the JFH1 cDNA, corresponding to sequences encoding C-E1-E2-p7, except for the 19 5′-terminal codons of the core (C protein) gene, to preserve intact the HCV internal ribosome entry site [IRES], and the 21 3′-terminal codons of the core gene, the corresponding polypeptide of which serves as a signal peptide for the NS2 downstream protein. Plasmids pJ/E-NS3proGB-Ubi and pJ/C-NS3proGB-Ubi were constructed by replacing JFH1 nt 914 to 3430 and 341 to 3430, encoding E1-E2-p7-NS2 and C-E1-E2-p7-NS2, respectively, with GBV-B nt 914 to 3835 and 446 to 3835, encoding the E1-E2-p13-NS2-NS3pro (serine protease domain of NS3) and C-E1-E2-p13-NS2-NS3pro proteins, respectively, fused to the mouse ubiquitin sequence. GBV-B nt 3266 to 3835 encode the 190 N-terminal amino acids of NS3 that correspond to NS3pro.
Plasmids containing the HCV intergenotypic 1a/2a chimeric cDNAs J/C-NS2H77, J/E-NS3proH77-Ubi, and J/C-NS3proH77-Ubi exhibit replacements of JFH1 nt sequences similar to those in the GBV-B/JFH1 chimeras described above but using sequences derived from HCV H77 cDNA. The NS3proH77 sequence corresponds to nt 3420 to 3986 of pH77C cDNA and encodes the 189 N-terminal amino acids of NS3.
Plasmids pGBV-B-II and pGBV-B-III were derived from phCMV (Genlantis) and contain GBV-B nt 833 to 2284 and 863 to 2284, encoding the 27 and 17 C-terminal amino acids of core, respectively, fused to the full-length sequences of E1 and E2 envelope glycoproteins. All viral sequences were PCR amplified from pGBV-B/2 and included a stop codon at the 3′ end and restriction sites at the 5′ and 3′ extremities. The resulting fragments were cloned into phCMV downstream from the murine encephalomyocarditis virus IRES and AUG initiator codon. The GBV-B sequence cloned into pGBV-B-IIIopt, which is designed similarly to pGBV-B-III, has been codon optimized for enhanced translation in mammalian cells without altering its amino acid sequence (Life Technologies).
In vitro transcription.
HCV-based plasmid DNAs were linearized by digestion with XbaI and treated with mung bean nuclease (New England BioLabs), while GBV-B-based plasmid DNAs were linearized with BspQI prior to T7 RNA polymerase-mediated in vitro transcription using the T7 RiboMAX express large-scale RNA production system (Promega) according to the manufacturers' instructions. Following RNA synthesis, DNA templates were removed by treatment with DNase (Promega) and RNAs were purified by phenol-chloroform extraction and precipitated with ethanol. The latter steps were omitted for synthetic RNAs used in vivo in tamarins. RNA quality and quantity were monitored by electrophoresis on nondenaturing 1% agarose gels using known quantities of RNA molecular weight markers (RNA Millenium markers-formamide; Ambion) and measurement of the absorbance at 260 nm. Transcribed and purified RNAs were stored at −80°C until use for tamarin intrahepatic inoculations or cell electroporations.
Animals and in vivo infectivity assays.
Purpose-bred red-bellied tamarins (Saguinus labiatus) and marmosets (Callithrix jacchus) were housed and maintained under licenses granted by the United Kingdom Secretary of State for the Home Office in accordance with the Animals (Scientific Procedures) Act 1986 and by the French Ministry of Agriculture, respectively. Experimental protocols were approved by the Animal Welfare and Ethical Review Body (AWERB), the United Kingdom Home Office, the Comité Régional d'Ethique en Matière d'Expérimentation Animale de Strasbourg (CREMEAS), and the Comité Régional d'Ethique pour l'Expérimentation Animale d'Ile de France, Paris 1. Genome-length synthetic RNA transcripts (100 μg) were inoculated into the tamarins' livers, which were exposed by a limited abdominal incision as described previously (27), and tamarin serum-derived GBV-B (107 genome equivalents) was inoculated by the intravenous route. All surgical procedures were performed under anesthesia with recovery. Viremia was followed up in weekly serum samples by extraction and quantification of viral RNA as described below.
Cells.
HEK293T human embryonic kidney cells, simian virus 40 (SV40) T antigen-immortalized small primate TH1 cell lines, and human hepatoma Huh7.5 cells were cultured as previously described (28, 29). Hepatocytes were isolated from livers that were removed from two marmosets (CJ507 and CJ514) at termination, using collagenase perfusion as described previously (30). Primary marmoset hepatocytes (PMH) were frozen in aliquots in 70% University of Wisconsin solution, 20% fetal calf serum, 10% dimethyl sulfoxide (DMSO) and stored in liquid nitrogen until use. PMH were thawed in William's medium E (WME; Life Technologies) containing 5% fetal calf serum and plated in 24-well collagen-coated tissue culture dishes (BioCoat; BD Biosciences) (4 × 105 cells per well). The medium was removed 4 h later and replaced with serum-free medium (SFM) containing hormones and growth factor supplements as previously described (30).
Production of retroviral particles pseudotyped with HCV or GBV-B glycoproteins.
Defective retroviral pseudoparticles (pp) encoding firefly luciferase (FLuc) reporter protein were produced essentially as described previously (31). HEK293T cells (2 × 106 cells) were cotransfected using a calcium phosphate-based transfection kit with three plasmid DNAs, including a plasmid (1 to 4 μg) encoding viral glycoproteins (GBV-B, HCV, vesicular stomatitis virus [VSV], or 4070A amphotropic murine leukemia virus [MLV]), a packaging plasmid (8 μg) encoding Gag and Pol proteins of MLV or lentivirus origin (human immunodeficiency virus [HIV] or simian immunodeficiency virus [SIV]), and an MLV or lentivirus-based transfer vector (8 μg) encoding FLuc. Pseudoparticles released into the culture supernatant of transfected HEK293T cells at 36 h posttransfection were concentrated 100-fold through 25% sucrose cushions by ultracentrifugation at 100,000 × g for 120 min at 4°C, resuspended in phosphate-buffered saline (PBS), and stored at 4°C for ∼24 h until use in infection experiments.
Cell transfection and infection assays.
Huh7.5 cells (4 × 106 cells) were transfected by electroporation with 10 μg of in vitro-transcribed, genome-length RNA as described previously (16) and seeded either in 75-cm2 flasks (2 × 106 cells) for further passage by splitting 1:3 to 1:6 every 3 to 4 days or in 6-well plates (2.5 × 105 cells) for protein expression analysis by immunoblotting at 3 days posttransfection. At 2 to 3 days postplating, PMH cultures were infected with tamarin serum-derived GBV-B at a multiplicity of infection (MOI) of 10 genome equivalents per cell, with cell culture-grown HCV Jad at an MOI of 0.02, 0.04, or 0.14 50% tissue culture infective doses (TCID50) per cell, or with virus treated with 2 successive ultraviolet exposures at 1,000 mJ/cm2. Following incubation for 4 to 6 h at 37°C, the viral inoculum was removed and cells were thoroughly washed (4 to 5 times) with WME. SFM was added, and plates were further incubated at 37°C. Transfected and infected cell extracts were processed for quantification or sequencing analysis of viral RNAs, and culture supernatants were processed for infectivity endpoint dilution titration in Huh7.5 cells or for particle analysis in density gradients as described previously (16) or below.
Huh7.5 and TH1 cells were seeded in 96-well plates at a concentration of 6 × 103 cells per well and infected 24 h later with 3 μl and 15 μl of concentrated pp, either untreated or after heat inactivation for 20 min at 70°C, while PMH cultures were seeded in 24-well plates at a concentration of 4 × 105 cells per well and infected with 100 μl of concentrated pp, either untreated or after heat inactivation for 20 min at 70°C. MLV-pp were used preferentially over VSV-pp in PMH cultures to limit potential luciferase pseudotransduction. For inhibition assays, cells were preincubated with various concentrations of anti-CD81 monoclonal antibody (MAb) or isotype-matched control or pp were preincubated with various concentrations of single-chain antibodies specific for HCV or GBV-B E2 for 1 h at 37°C. Cells were then infected as indicated above in the presence of the respective antibodies. After incubation for 6 h at 37°C, the inoculum was removed and replaced by the appropriate cell culture medium. At 72 h postinfection (p.i.), cells were lysed and relative FLuc activities were calculated as ratios of FLuc activities obtained following pp infection and background FLuc activities measured following infection with matched, heat-inactivated pp stocks. For anti-CD81 antibody inhibition experiments in PMH cultures, the viral inoculum and antibodies were removed 4 h after infection, and cells were washed three times with WME prior to further incubation at 37°C and collection of supernatants at 72 h p.i. for processing by real-time quantitative reverse transcription-PCR (RT-qPCR) or infectivity endpoint dilution titration in Huh7.5 cells.
Quantification and sequence analysis of viral RNAs.
Viral RNA was isolated from transfected cell supernatants, density gradient fractions, or tamarin serum by using the QIAamp viral RNA minikit (Qiagen), and total RNA was isolated from transfected cells by extraction with TRIzol (Invitrogen) in accordance with the manufacturers' instructions. Viral RNA was quantified by RT-qPCR using 50 ng of total intracellular RNA, estimated by absorbance measurement, or aliquots of RNA extracted from fluids. Assays were performed according to a one-step procedure using one-step RT-PCR master mix reagents (Applied Biosystems) with primer/probe sets targeting the HCV 5′ noncoding region (primers, 5′-CGGGAGAGCCATAGTGG-3′ and 5′-AGTACCACAAGGCCTTTCG-3′, and probe, 5′-FAM-CTGCGGAACCGGTGAGTACAC-TAMRA-3′ [FAM is 6-carboxyfluorescein and TAMRA is 6-carboxytetramethylrhodamine]) or the GBV-B NS5A coding sequence (13). Parallel amplification of ribosomal 18S RNA in intracellular RNA extracts using TaqMan ribosomal RNA control reagents (Applied Biosystems) was carried out for purposes of normalization. RNA was reverse transcribed in a 25-μl reaction mixture at 48°C for 30 min, followed by inactivation of reverse transcriptase at 95°C for 10 min. Products were then amplified by PCR for 40 cycles of 15 s at 95°C and 1 min at 60°C. Purified synthetic full-length GBV-B or HCV RNA transcripts whose concentrations (genome equivalents/μl) were determined by absorbance measurements were stored in small aliquots at −80°C and used as standards for quantification. Products were analyzed on a 7500 fast real-time PCR system instrument (Applied Biosystems).
For sequencing purposes, viral RNA was extracted from RNA-transfected or virus-infected cells, converted to cDNA using SuperScript II reverse transcriptase (Invitrogen) and a random-sequence oligodeoxyribonucleotide hexamer [d(N)6] in accordance with the manufacturer's instructions and was amplified by PCR using platinum Taq DNA polymerase (Invitrogen) or One Taq 2× master mix with standard buffer (New England BioLabs) with a series of primer pairs spanning genomic segments of 1,000 to 2,000 bp. The amplified DNAs were purified using the QIAquick PCR purification kit (Qiagen) and subjected to direct sequencing using the BigDye terminator version 1.1 cycle sequencing kit (Applied Biosystems).
Ultracentrifugation of viral particles in density gradients.
Fifteen-milliliter amounts of supernatant fluids were collected at 96 h posttransfection from RNA-transfected cells (2 × 106 cells) seeded in 75-cm2 flasks, clarified by low-speed centrifugation, filtered on 0.45-μm membranes, and concentrated 60-fold using 100-kDa Amicon ultra-15 units (Millipore). Concentrated supernatants (105 μl) were layered on top of preformed 10-to-40% iodixanol gradients (OptiPrep; Sigma-Aldrich) and ultracentrifuged in an SW55Ti rotor (Beckman Coulter) for 16 h at 192,000 × g at 4°C. Ten fractions (490 μl each) were collected from the top of the gradients, and each fraction was analyzed for density by weighing, for viral RNA content following RNA extraction and quantification by RT-qPCR, and for HCV core protein content by using the Ortho HCV enzyme-linked immunosorbent assay (ELISA) antigen test (Ortho Clinical Diagnostics).
Antibodies, immunoblotting, and immunofluorescence.
Immunoblotting and immunofluorescence assays were carried out as described previously (16), using antibodies specific for the following HCV proteins: CH77/CJFH1 (HepC HBcAg 1851; Santa Cruz Biotechnology) at 0.1 μg/ml, E1H77 (A4) diluted 1:3,500 (32), E2H77 (A11) diluted 1:500 (32), and NS3JFH1 (33) diluted 1:300. Polyclonal antibodies specific for GBV-B E2 glycoprotein (anti-E2GB antibody) were raised in rabbits following immunization with a soluble form of GBV-B E2 (comprising amino acids 350 to 564) produced in Drosophila cells as previously described for HCV E2 (34) and diluted 1:2,000 for immunoblots. Monoclonal antibodies specific for GBV-B E2 were selected in mice, and the corresponding recombinant single-chain variable-fragment (scFv) antibodies were produced in Drosophila S2 cells as described previously (35). Monoclonal human antibody 1:7 specific for HCV E2 has been described elsewhere (36). Monoclonal antibodies specific for human CD81 (clone JS-81) and matched, irrelevant IgG1 isotype were purchased from BD Biosciences.
RESULTS
Replication capacities of chimeric HCV JFH1 genomes encoding GBV-B envelope glycoproteins in small primates.
Based on the hypothesis that virus cell entry may be responsible for hepacivirus host restriction, we generated HCV recombinant cDNAs derived from the cell culture-adapted JFH1 strain of HCV (24) and encoding GBV-B envelope glycoproteins in place of the corresponding HCV sequences to mediate entry into small primate hepatocytes. In the majority of engineered or naturally occurring infectious intergenotypic HCV chimeras described to date, the crossover point between the distinct genotype sequences is located at the NS2/NS3 junction, where proteins involved in virus morphogenesis (core C, glycoproteins E1/E2, ion channel protein p7, and nonstructural protein NS2) are uncoupled from nonstructural proteins NS3 to NS5B, which are involved in genome replication (37, 38). We found that GBV-B NS2 protease was unable to ensure cleavage at a heterologous GBV-B NS2/HCV JFH1 NS3 junction, either in the context of truncated chimeric precursors (16) or of full-length chimeric polyproteins (data not shown). This caveat prevented the generation of a functional chimera by simply replacing JFH1 C-NS2 sequences with the corresponding GBV-B sequences. Therefore, we engineered a GBV-B/JFH1 chimera (J/C-NS3proGB-Ubi) in which sequences encoding HCV JFH1 C-E1-E2-p7-NS2 were replaced with the corresponding GBV-B sequences (C-E1-E2-p13-NS2) fused in frame to the GBV-B NS3 protease domain-coding sequence (NS3pro) followed by the ubiquitin gene (Fig. 1A). This design was meant to ensure the release from the recombinant polyprotein of both native JFH1 NS3 via ubiquitin activity and native GBV-B NS2 via its own proteolytic activity (16). A second GBV-B/JFH1 chimeric cDNA (J/E-NS3proGB-Ubi) (Fig. 1A) was engineered similarly but harbored the HCV JFH1 core-coding sequence. We indeed hypothesized that it might be important to preserve the function of the HCV internal ribosome entry site (IRES), which extends into the core coding sequence (39), and/or to maintain putatively essential, HCV-specific interactions between core and RNA or core and nonstructural proteins for particle assembly.
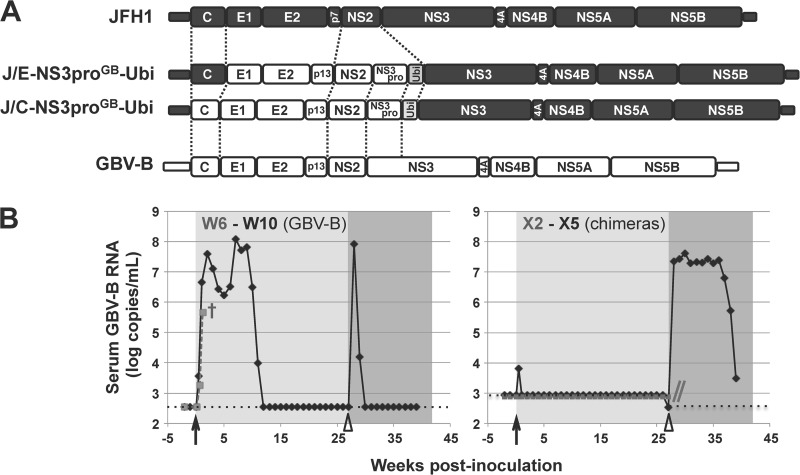
FIG 1.
Infectivity of GBV-B/JFH1 chimeras in tamarins. (A) Schematics of the organization of the engineered chimeric GBV-B/JFH1 cDNAs. HCV JFH1 and GBV-B genomic sequences are represented by black and white boxes, respectively, and the ubiquitin (Ubi) gene by a gray box. Dotted lines delineate the sequences that were exchanged. Recombinants are denoted as follows: “J/” (standing for JFH1) is followed by the range of JFH1 protein sequences replaced (C-NS3pro or E-NS3pro), the virus from which corresponding sequences were derived, in superscript characters (“GB” for GBV-B), and -Ubi. (B) Tamarins W6 (dashed gray line) and W10 (black line) (left) and tamarins X2 (dashed gray line) and X5 (black line) (right) were intrahepatically inoculated (black arrows) with GBV-B-3m/SapI RNA (left) or a mixture of J/C-NS3proGB-Ubi and J/E-NS3proGB-Ubi RNAs (right), respectively. Tamarins W10 and X5 were subsequently challenged (open arrowheads) with an intravenous injection of GBV-B. Serum viral loads were quantified by RT-qPCR assays targeting either GBV-B NS5A sequence (W6, W10, and X5 after GBV-B intravenous injection) or HCV 5′ noncoding region (X2 and X5 after RNA intrahepatic inoculation). Detection limits of both assays are indicated by dotted lines. Tamarin W6 died prematurely 9 days postintrahepatic inoculation (†) due to a cause unrelated to the experiment. The follow-up of tamarin X2 was stopped at 27 weeks postinoculation as indicated by the gray double slash.
While primary small primate hepatocyte cultures are susceptible to GBV-B infection (11), such primary cultures are hard to transfect and do not allow wild-type GBV-B rescue following GBV-B genome-length RNA transfection. Since no known small primate continuous hepatic cell line susceptible to GBV-B for its complete life cycle was available, the infectivity of the chimeric RNAs was first assayed following intrahepatic inoculation into GBV-B-susceptible small primate hosts. The inoculation of a mixture of synthetic, in vitro-transcribed J/C-NS3proGB-Ubi and J/E-NS3proGB-Ubi RNAs into the livers of two tamarins (Saguinus labiatus; animals X2 and X5) did not result in detectable viremia for 39 and 26 weeks postinoculation (p.i.), respectively, with the exception of the transient detection of the chimeric nucleic acid in X5 at 3 days p.i. (Fig. 1B, right). As controls, the intrahepatic inoculation of GBV-B RNA into two tamarins (animals W6 and W10) was shown to lead to high-titer viremia as early as 3 days p.i. and for 11 weeks (Fig. 1B, left). Tamarins W10 and X5 were challenged 27 weeks after the intrahepatic RNA inoculation by intravenous injection of a known infectious dose of GBV-B-positive serum. Tamarin W10 developed a secondary transient viremia for 2 weeks (Fig. 1B, left), demonstrating that the previous GBV-B RNA intrahepatic inoculation conferred partial protection against reinfection, in agreement with the literature (40). In contrast, tamarin X5 developed a typical viremia as observed in naive tamarins, with rapid onset of viral replication within the first 7 days p.i. and detection of viral RNA in serum samples for >12 weeks (Fig. 1B, right). These data were indicative of the failure of previously administered chimeras to grow and demonstrated that this animal was not spuriously refractory to GBV-B infection. The lack of in vivo replication or adaptation of J/C-NS3proGB-Ubi and J/E-NS3proGB-Ubi chimeras in small primates may stem from a defect in infectious particle assembly or release, although we could not exclude at this stage that it might be due to a defect in HCV replication complex activity in simian hepatocytes.
Replication and assembly capacities of GBV-B/JFH1 chimeras in Huh7.5 cells.
The replication and assembly capacities of chimeric GBV-B/JFH1 synthetic RNAs were analyzed following transfection of Huh7.5 cells that are known to support JFH1 replication to high levels (29). Both J/E-NS3proGB-Ubi and J/C-NS3proGB-Ubi RNAs proved equally replication competent, although with approximately 10- to 20-fold-lower efficiency than parental JFH1 RNA and assembly-deficient RNAs lacking structural protein sequences (J/ΔCp7 and J/ΔEp7) (Fig. 2A). These results showed that there was no IRES-driven translation impairment in J/C-NS3proGB-Ubi. The decreased replication activity of these recombinant RNAs was not due to a marked cleavage defect at the Ubi/NS3 junction since mature NS3 but not NS3-containing uncleaved precursors was detected in transfected cell extracts (Fig. 2B). Likewise, mature JFH1 core was readily detected in cells transfected with J/E-NS3proGB-Ubi RNA, demonstrating efficient cleavage at the hybrid junction between JFH1 core and GBV-B E1 (Fig. 2B). The detection of GBV-B E2 glycoprotein expressed from the recombinant polyproteins (Fig. 2B) supported the notion that there was no posttranslational alteration of GBV-B proteins in these contexts.
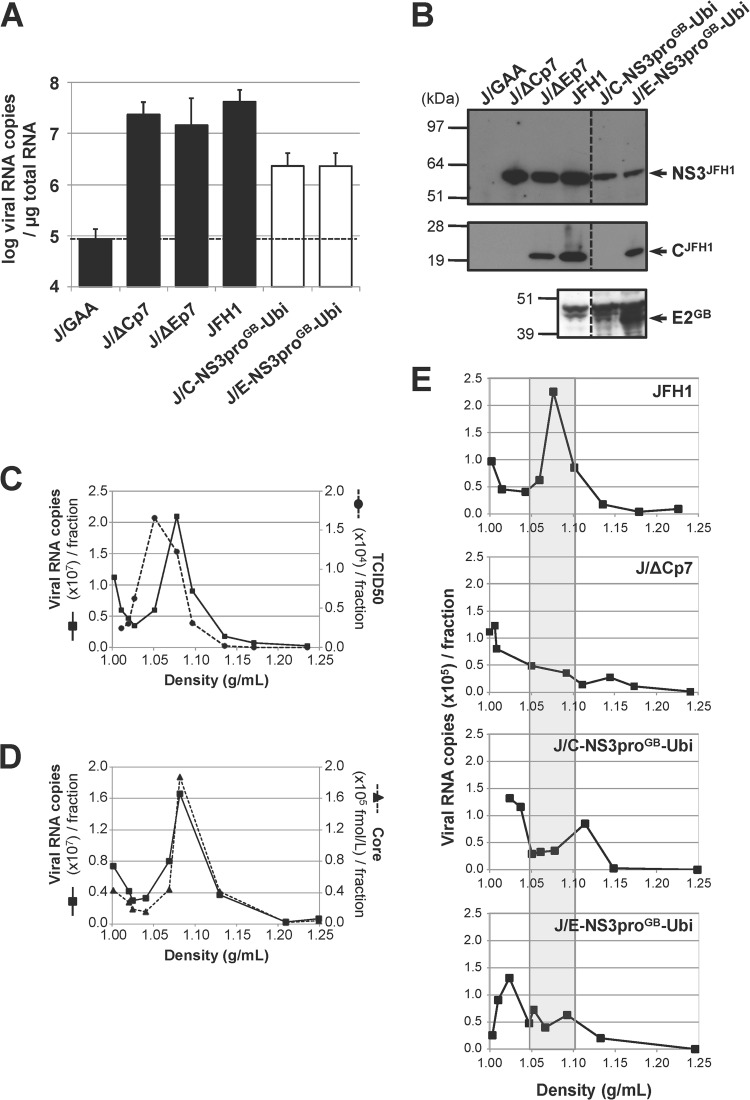
FIG 2.
Infectivity of chimeric GBV-B/JFH1 RNAs in Huh7.5 cells. (A) Replication activities of chimeric GBV-B/JFH1 RNAs. Huh7.5 cells were transfected with the parental JFH1 RNA, control RNAs (black bars), including the replication-deficient RNA carrying inactivating mutations in the polymerase active-site codons (J/GAA) and the assembly-deficient RNAs devoid of structural protein sequences (J/ΔCp7 and J/ΔEp7), or the indicated chimeric GBV-B/JFH1 RNAs (white bars). Viral RNA extracted from transfected cells at 96 h posttransfection was quantified by RT-qPCR relative to total cellular RNA normalized with respect to 18S RNA quantification. Mean results ± standard deviations from 3 independent transfections are shown. The dashed line indicates the threshold under which RNAs are nonreplicative. (B) Maturation of chimeric polyproteins. Protein extracts prepared from RNA-transfected cells at 96 h posttransfection were analyzed by SDS-PAGE followed by immunoblotting with anti-NS3JFH1, anti-CH77/JFH1, and anti-E2GB antibodies. (C to E) Viral particle production in transfected-cell supernatants. (C, D) The supernatant from JFH1 RNA-transfected Huh7.5 cells collected at 96 h posttransfection was concentrated over a 100-kDa exclusion membrane, loaded on a 10-to-40% iodixanol gradient, and ultracentrifuged to equilibrium. The RNA content (black lines, panels C and D), the infectivity (dashed line, panel C), and the core antigen content (dashed line, panel D) were determined in each of the 10 fractions collected, using RT-qPCR, TCID50 titration in Huh7.5 cells, and ELISA, respectively, and plotted as a function of density, measured by weighing each fraction. (E) Viral RNAs in the supernatants from cell cultures transfected with the indicated RNAs were collected at 96 h posttransfection, concentrated, and quantified by RT-qPCR. Adjusted volumes of concentrated supernatants corresponding to equal quantities of viral RNA were loaded on iodixanol gradients. Following ultracentrifugation, the RNA content of each fraction was determined by RT-qPCR. The shaded area delineates the density range at which RNA associated with JFH1 particles is expected to peak. The profiles shown are representative of 5 independent experiments.
To determine whether GBV-B/JFH1 RNAs led to viral production, the infectivity of supernatants from RNA-transfected cells was first examined following infection of naive Huh7.5 cells. No viral antigen expression (core or NS3) was detected in infected cells at 3 days p.i. (data not shown). Since it was unknown whether GBV-B glycoproteins could drive entry into human hepatocytes, the presence of viral particles in transfected cell supernatants was next examined on the basis of viral RNA density profiles in 10-to-40% iodixanol gradients. Following equilibrium ultracentrifugation, the viral RNA content of each of 10 fractions was determined by RT-qPCR. The peak of parental JFH1 RNA sedimentation occurred at a density of 1.05 to 1.10 g/ml, fully overlapping the peak for core protein and most of the infectivity peak (Fig. 2C and D). Of note, infectivity was also associated with slightly lower densities (1.03 to 1.05 g/ml) (Fig. 2C), in agreement with the highest infectivity of viral particles associated with cellular lipoproteins (41). The analysis of a 1:100 dilution of JFH1 RNA-transfected cell supernatant demonstrated a clear RNA peak in the same density fraction as from undiluted supernatants, unambiguously signaling the detection of viral particles (Fig. 2E, top). This observation confirmed that the density approach could successfully be applied to supernatants anticipated to contain reduced viral titers. However, supernatants from cells transfected with either J/C-NS3proGB-Ubi or J/E-NS3proGB-Ubi RNA showed no predominant association of RNA with a specific density, similar to the results for the assembly-deficient deletion mutant J/ΔCp7 (Fig. 2E). These findings suggested an absence or low levels of secreted viral particles.
Comparative analysis of the potential for cell culture adaptation of GBV-B/JFH1 and H77/JFH1 chimeras.
Engineered HCV intergenotypic recombinants bearing heterologous C-NS2 sequences often require compensatory substitution(s) in their genomes to yield fully infectious virus (37). To explore whether this would also be the case for GBV-B/JFH1 chimeras, we undertook to attempt adaptation of GBV-B/JFH1 chimeras by serial passage of transfected cells. Two similar HCV intergenotypic chimeras were also created (J/C-NS3proH77-Ubi and J/E-NS3proH77-Ubi), in which the heterologous C-E1-E2-p7-NS2-NS3pro or E1-E2-p7-NS2-NS3pro sequences were derived from the H77 strain of HCV genotype 1a (25) (Fig. 3A). A third chimera (J/C-NS2H77) contained the H77 C-NS2 sequence and mirrored previously described, cell culture-adaptable HCV-1a/JFH1 intergenotypic recombinants (37, 42) (Fig. 3A). Altogether, these intergenotypic chimeras were aimed at determining whether the ubiquitin gene insertion was not detrimental for the recombinants' infectivity and at facilitating the identification of putative adaptive mutations (e.g., in the ubiquitin gene) that could be applied to boost the replication of both GBV-B/JFH1 and H77/JFH1 chimeras.
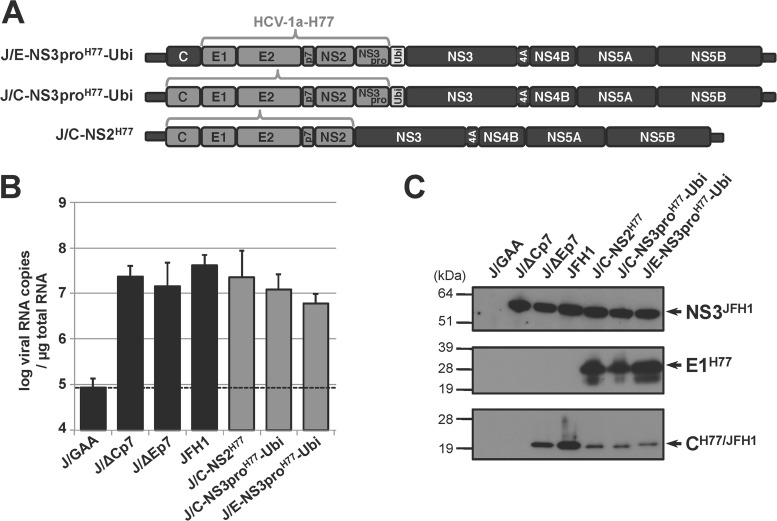
FIG 3.
Replication of H77/JFH1 recombinant RNAs and polyprotein processing in human hepatoma Huh7.5 cells. (A) Schematic organization of engineered intergenotypic H77/JFH1 cDNAs. HCV-2a strain JFH1 and HCV-1a strain H77 genomic sequences are represented by black and dark gray boxes, respectively, while the sequence encoding ubiquitin (Ubi) is represented by a light gray box. Constructs were denominated as described in the legend to Fig. 1, with “H77” in superscript characters indicating the HCV-1a H77 origin of the grafted sequences. (B) Replication activities of intergenotypic H77/JFH1 recombinant RNAs. Viral RNAs extracted at 96 h posttransfection following transfection of Huh7.5 cells with the indicated control (black bars; see the legend to Fig. 2) or recombinant (gray bars) RNAs were quantified by RT-qPCR relative to total cellular RNA normalized with respect to 18S RNA quantification. Mean results ± standard deviations of 3 independent transfections are shown. (C) Maturation of chimeric polyproteins. Protein extracts prepared from RNA-transfected cells at 96 h posttransfection were analyzed by SDS-PAGE followed by immunoblotting with anti-NS3JFH1, anti-E1H77, anti-CH77/JFH1 antibodies.
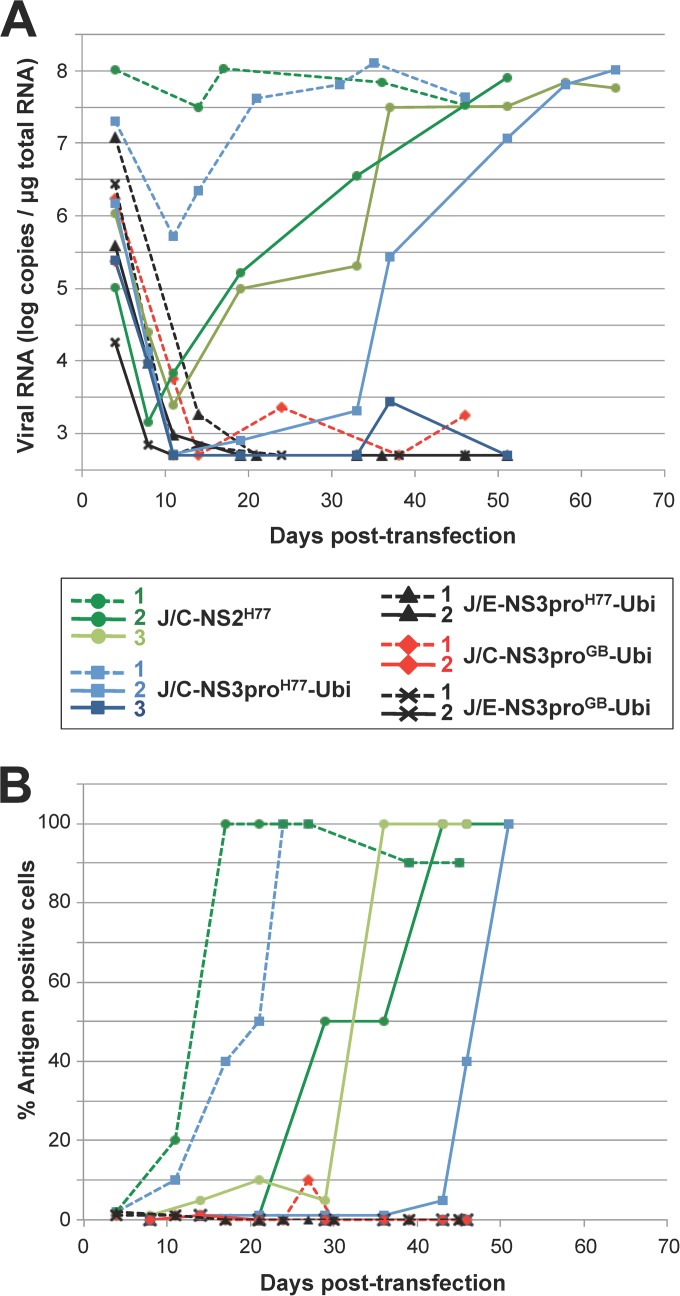
The three intergenotypic H77/JFH1 RNAs replicated efficiently, and the resulting polyproteins were accurately proteolytically processed (Fig. 3B and C). No RNA peak at the expected particle-associated density was observed for these intergenotypic recombinant RNAs following fractionation of the supernatants collected at 4 days posttransfection (data not shown), as is described above for the GBV-B/JFH1 chimeras. Virus adaptation was monitored by quantifying intracellular viral RNA extracted from transfected cells by RT-qPCR, and virus spreading in the cultured cells based on the percentages of HCV NS3-positive cells at various times posttransfection. Intracellular viral RNA quantities decreased for all chimeras during the first 3 or 4 passages (Fig. 4A). Chimeric RNAs in which the core sequence was from HCV JFH1 (J/E-NS3proH77-Ubi and J/E-NS3proGB-Ubi) were undetectable from ∼20 days posttransfection in two independent experiments, suggesting that these chimeras failed to adapt to viral particle production. In contrast, J/C-NS3proH77-Ubi and J/C-NS2H77 intergenotypic RNAs reaccumulated in transfected cultures over time (Fig. 4A). Concomitantly, in three independent transfection experiments, virus spread progressively within cell monolayers, which were fully infected within 17 to 51 days posttransfection depending both on the recombinant RNA and Huh7.5 cell permissiveness (Fig. 4B and data not shown). Interestingly, GBV-B/JFH1 chimeric RNA J/C-NS3proGB-Ubi was sporadically detected in significant quantities at various times posttransfection, including at 24 to 27 days posttransfection, when it was accompanied by the detection of antigen-positive cell foci in the culture, suggesting that this chimera might have a potential for adaptation. However, the RNA levels were too low to retrieve sequence information and adaptation was not successfully achieved at the conclusion of two independent experiments (Fig. 4).
FIG 4.
Cell culture adaptation of GBV-B/JFH1 and HCV intergenotypic chimeras. Cells transfected with the indicated GBV-B/JFH1 or H77/JFH1 chimeric RNAs were serially passaged. Graphs combine data from two independent passage experiments, represented by dashed and plain lines, respectively, each with the results for 1 or 2 independently transcribed chimeric RNAs shown in the colors indicated in the key. (A) Genome replication was monitored by RT-qPCR quantification of intracellular viral RNA at various times posttransfection. (B) Virus spreading within the cell monolayers was evaluated by quantifying the percentage of HCV NS3-positive cells by indirect immunofluorescence.
The open reading frames of four adapted J/C-NS2H77 viruses and three adapted J/C-NS3proH77-Ubi viruses were sequenced. Remarkably, at least one nonsynonymous substitution was found in the JFH1 NS3 coding sequence in all infectious recombinants (Table 1). Interestingly, the four infectious J/C-NS2H77 recombinants sequenced harbored a substitution in the NS3 helicase (Q1251L or R1412W), whereas the three viable J/C-NS3proH77-Ubi chimeras harbored distinct substitutions in the NS3 protease or in the ubiquitin sequence, in addition to one of the helicase mutations in some cases (Table 1). When introduced singly into the J/C-NS3proH77-Ubi cDNA backbone, the P1032L and P1159Q substitutions in the NS3 protease, as well as the Q1251L mutation in the NS3 helicase, were each subsequently found to be sufficient to yield parentlike infectious virus titers at 3 to 4 days posttransfection, whereas the substitution in the ubiquitin sequence (I44T) did not rescue the chimera's infectivity (Table 2). Additional substitutions in E1 or E2 and NS5A or NS5B coding sequences that likely provide additional fitness to the viruses were selected in some infectious recombinants (Table 1). Together, these data demonstrated that the replacement of JFH1 C-NS2 sequences with H77 sequences fused to H77 NS3pro and ubiquitin gave rise to chimeric viruses that could be efficiently adapted in cell culture. In contrast, we could not successfully adapt similarly designed GBV-B/JFH1 chimeras in Huh7.5 cells. This failure could be explained by a lack of assembly coordination between GBV-B and HCV proteins or the inability of GBV-B surface proteins to mediate human hepatocyte infection.
TABLE 1.
Mutations identified in infectious J/C-NS2H77 and J/C-NS3proH77-Ubi pseudorevertants rescued following passage of transfected cells
| Protein | Amino acid substitution(s) ina: |
||||||
|---|---|---|---|---|---|---|---|
| J/C-NS2H77 virus (day posttransfection): |
J/C-NS3proH77-Ubi virus (day posttransfection): |
||||||
| 1 (30) | 2 (65) | 3 (50) | 4 (35) | 1 (40) | 2 (65) | 4 (35) | |
| E1H77 | I348T | M345T | M345T | M345T | |||
| E2H77 | E591G | E591D | Y718S | ||||
| Ubi | I44T | ||||||
| NS3JFH1 | Q1251L | R1412W | Q1251L | Q1251L | P1032L, Q1251L | P1159Q | S1328T, R1412W |
| NS5AJFH1 | N2156K | S2358R | L2276P | ||||
| NS5BJFH1 | S2552G | S2457G | |||||
Amino acid numbering corresponds to amino acid position within H77 polyprotein, ubiquitin (Ubi), or JFH1 polyprotein, as indicated in the protein name. Amino acid substitutions found in several viruses (viruses 1 to 4) rescued independently at the indicated numbers of days posttransfection are represented in bold characters. Substitutions present in 100% of bulk virus populations are highlighted by gray shading.
TABLE 2.
Effects of point mutations on J/C-NS3proH77-Ubi viral production
| Backbone | Amino acid substitution ina: |
Mean infectious titer ± SD (log TCID50/ml)b | ||
|---|---|---|---|---|
| Ubi | NS3JFH1 protease | NS3JFH1 helicase | ||
| JFH1 | — | — | — | 3.29 ± 0.40 |
| J/C-NS3proH77-Ubi | — | — | — | < |
| I44T | — | — | < | |
| — | — | Q1251L | 2.94 ± 0.49 | |
| — | P1032L | — | 3.47 ± 0.63 | |
| — | P1032L | Q1251L | 2.79 ± 0.62 | |
| — | P1159Q | — | 3.02 ± 0.48 | |
A substitution introduced by site-directed mutagenesis into the J/C-NS3proH77-Ubi cDNA within the sequence encoding the indicated protein is denoted by the native residue followed by its numbering within JFH1 polyprotein and the mutant residue (except for the substitution in the ubiquitin gene, which is numbered according to the amino acid position within the Ubi gene). —, none.
Infectious titers (mean results ± standard deviations from 3 transfection experiments, each with 2 or 3 independently transcribed RNAs) were determined 3 days posttransfection. <, below infectivity threshold (1.60 log TCID50/ml).
Virus cell entry is not responsible for hepacivirus host restriction.
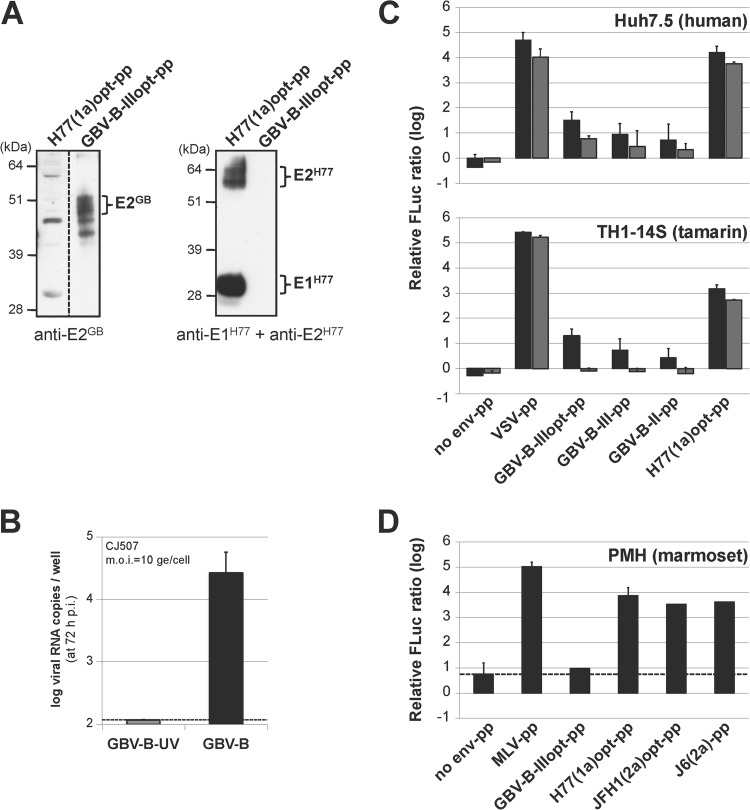
To focus on the virus entry step only, cross-entry into human and small primate hepatocytes mediated by HCV and GBV-B glycoproteins was analyzed by using defective retroviral particles pseudotyped with GBV-B or HCV glycoproteins. Eukaryotic-codon-optimized GBV-B sequences encoding the 17 C-terminal amino acids of core protein fused to E1 and E2 (GBV-B-IIIopt-pp) were used such as to increase glycoprotein expression levels and incorporation at the surface of the pseudoparticles (data not shown). HCV pseudoparticles were assembled using codon-optimized E1-E2 glycoprotein sequences from the H77 strain of HCV genotype 1a [H77(1a)opt-pp] downstream from the corresponding core signal peptide, as described previously (43). The incorporation of GBV-B and HCV glycoproteins at the surface of pseudoparticles was verified by immunoblotting with polyclonal GBV-B E2-specific antibodies or monoclonal antibodies directed against HCV E1 and E2 (32), respectively (Fig. 5A).
FIG 5.
HCV and GBV-B pseudoparticle cross-entry into human and small primate cells. (A) HCV and GBV-B glycoprotein incorporation at the surface of pseudoparticles. Aliquots of the indicated pseudoparticle (pp) stocks were analyzed by SDS-PAGE followed by immunoblotting with anti-E2GB polyclonal antibodies (left) and a mixture of anti-E1H77 and anti-E2H77 monoclonal antibodies (right). (B) Primary marmoset hepatocyte (PMH) cultures are susceptible to GBV-B infection. PMH cultures derived from the liver of marmoset CJ507 were infected with tamarin-derived GBV-B or with UV-inactivated GBV-B at an MOI of 10 genome equivalents (ge) per cell. Virus production was evaluated by RT-qPCR quantification of GBV-B RNA extracted from culture supernatants at 72 h p.i. (mean results ± standard deviations from 5 experiments). (C, D) Analysis of HCV-pp and GBV-B-pp entry into human and small primate cells. Human (Huh7.5) and tamarin (TH1-14S) hepatic cell lines were infected with high doses (15 μl, black bars) and 5-fold-lower doses (3 μl, gray bars) of the indicated untreated or heat-inactivated pp stocks with MLV cores (C), and PMH cultures were infected with 100 μl (black bars) of untreated or heat-inactivated pp stocks with lentivirus cores (D). Mean log10 values ± standard deviations of the ratios of firefly luciferase (FLuc) activities obtained 72 h after infection with the untreated and heat-inactivated pp in 3 independent experiments are shown. The dashed line represents the threshold below which entry is not considered to be specific with respect to the FLuc ratio obtained for pseudoparticles devoid of envelope glycoprotein (no env-pp).
The human hepatoma cell line Huh7.5 and a small primate hepatic cell line derived from SV40 T-antigen immortalization of tamarin hepatocytes (TH1-14S) (28) were infected with pseudoparticles (pp) assembled on the murine leukemia virus (MLV) core. We established cultures of primary hepatocytes from marmoset livers (PMH) that were first shown to be fully susceptible to GBV-B following infection with a viral inoculum derived from the serum of a viremic tamarin (Fig. 5B). PMH cultures were transduced with pseudoparticles assembled on a lentivirus core due to the quiescent nature of these cells. Pseudoparticle transduction activity was evaluated in infected cells at 72 h p.i. by quantifying relative pp-encoded firefly luciferase reporter activities (FLuc ratios, normalized with respect to background activities obtained with heat-inactivated pp). Pseudotypes harboring glycoprotein G from vesicular stomatitis virus (VSV) or Env from amphotropic MLV that bind to ubiquitous cellular receptors expressed at the surface of a large variety of cells generated highly significant relative FLuc activities (Fig. 5C and D), confirming productive entry via these glycoproteins in all cells tested. GBV-B-IIIopt-pp generated significant and dose-dependent, although relatively low, FLuc ratios in both Huh7.5 and TH1-14S cells, suggesting that GBV-B glycoproteins mediated significant entry into human as well as simian hepatocytes (Fig. 5C). The very low entry or lack of entry of GBV-B-IIIopt-pp into PMH (Fig. 5D) may be explained by higher nonspecific FLuc ratios (Fig. 5D, no env-pp) and by the fact that the glycoprotein sequences used in these pseudotypes were derived from a tamarin GBV-B isolate and may be less efficient at interacting with cognate receptors at the surface of marmoset hepatocytes. The use of GBV-B-pp expressing wild-type E1-E2 downstream from core sequences of two different lengths, containing the putative E1 signal peptide, corroborated GBV-B glycoprotein-mediated entry into human hepatic cells, although the transduction levels with these pseudoparticles were lower than those of GBV-B-IIIopt-pp (Fig. 5C). Remarkably, HCV H77(1a)opt-pp were conversely able to transduce not only Huh7.5 cells, as previously described in the literature (31), but also all tamarin hepatic cell lines tested (Fig. 5C and data not shown), as well as PMH cultures (Fig. 5D). PMH cultures were also efficiently transduced with HCV-pp harboring glycoproteins from HCV genotype 2a (Fig. 5D). Altogether, these results strongly suggest that there is no blockage of HCV entry into small primate hepatocytes.
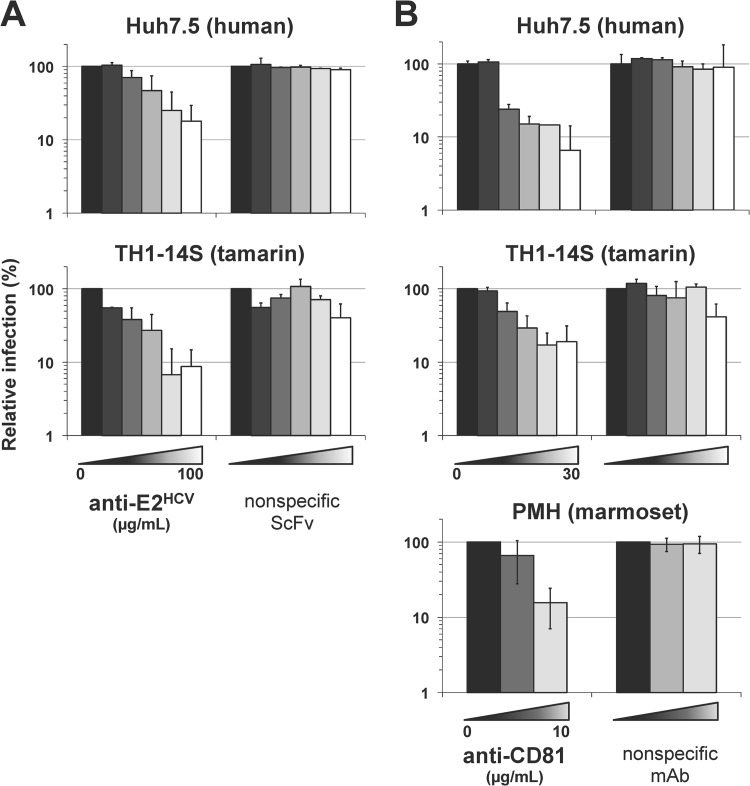
HCV-pp entry into small primate cells is mediated by HCV E2 and the simian ortholog of CD81.
We demonstrated a dose-dependent inhibition of H77(1a)opt-pp infection in TH1-14S cells in the presence of a single-chain variable fragment derived from an HCV E2-specific monoclonal antibody (36), while an irrelevant single-chain monoclonal antibody raised against a soluble form of GBV-B E2 had no significant effect (Fig. 6A). These data confirmed the specific involvement of HCV E2 glycoprotein in HCV-pp entry into small primate hepatocytes. Using a monoclonal antibody recognizing tamarin tetraspanin CD81, as well as the human ortholog, which is an essential coreceptor for HCV in human hepatocytes (31), we next observed dose-dependent inhibition of H77(1a)opt-pp infection in TH1-14S cells and in PHM cultures, as in Huh7.5 cells, whereas a nonspecific isotypic monoclonal antibody had no effect (Fig. 6B). This indicated that the interaction of HCV glycoproteins with simian CD81 is likely to be equally as important for HCV entry into small primate hepatocytes as it is for entry into human hepatocytes. In addition, it is noteworthy that H77(1a)opt-pp, in contrast to VSV-pp, did not show any sign of entry into fibroblastic cells isolated from tamarins (data not shown). Yet, we found that tamarin fibroblastic cells do express CD81 at their surface, as determined by flow cytometry analysis (data not shown), indicating that simian CD81 is not sufficient to mediate HCV entry into small primate cells and that additional coreceptors are likely required, as in human cells. This demonstrates the specificity of HCV entry into hepatic cells of small primates, like its entry into those of human origin (31).
FIG 6.
E2- and CD81-mediated entry of HCV-pp into small primate hepatic cells. (A) H77(1a)opt-pp were preincubated with increasing concentrations (0.2, 1, 5, 25, and 100 μg/ml, from dark gray to white bars) of anti-HCV E2 or anti-GBV-B E2 (nonspecific scFv) single-chain antibodies or in the absence of antibody (black bars) prior to infection of Huh7.5 (top) or TH1-14S (bottom) cells. (B) Huh7.5 and TH1-14S cell lines and PMH cultures were preincubated with increasing concentrations (0.1, 1, 3, 10, and 30 μg/ml for cell lines or 1 and 10 μg/ml for PMH, from dark gray to white bars) of anti-CD81 monoclonal antibody (MAb) or nonspecific isotype-matched MAb or in the absence of MAb (black bars) prior to infection with H77(1a)opt-pp. Relative infection ratios were determined at 72 h p.i. by scoring FLuc activities in the presence of antibodies relative to FLuc activities in the absence of antibodies set at 100% and are represented with a logarithmic scale.
Productive infection of PMH cultures with HCV Jad.
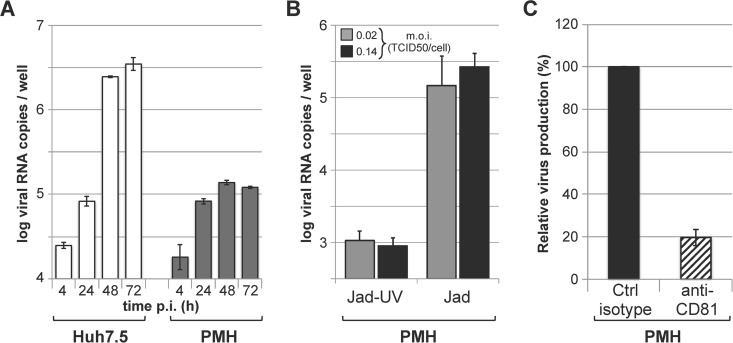
The efficient entry of JFH1(2a)opt-pp into small primate hepatocytes prompted us to investigate whether PMH cultures could support the entire infectious life cycle of JFH1. For these experiments, we used a highly adapted variant of JFH1 (Jad) that contained cell-culture adaptive mutations in NS5A and NS5B, as previously described (44). PMH cultures were infected with Jad at various multiplicities of infection and washed extensively at 4 h p.i. to remove viral inoculum, and newly secreted virus in the culture supernatants was quantified at various times p.i. The quantification of viral production by RT-qPCR analysis of PMH supernatants revealed accumulation of Jad RNA during the first 48 h p.i., suggestive of Jad particle production (Fig. 7A). Comparative kinetic studies in Huh7.5 cells following Jad infection under similar conditions showed that higher RNA levels were reached at 48 h and, more markedly, at 72 h p.i. in Huh7.5 cells than in PMH cultures (Fig. 7A). We hypothesize that a robust interferon response in PMH may limit the extent of viral replication over time, unlike the case for Huh7.5 cells, which are deficient in interferon response pathways. Using two multiplicities of infection, we found that the levels of Jad RNA recovered in the supernatants at 3 days p.i. were 2.0 to 2.5 log units above the levels of RNA quantified from supernatants of cells infected with UV-treated Jad (Fig. 7B). The infectious Jad production in PMH culture supernatants was titrated in Huh7.5 cells and found to be 2.96 ± 0.39 log TCID50/ml (mean ± standard deviation from 2 independent experiments each performed in duplicate or triplicate), while no infectivity was associated with cells infected with UV-treated Jad. Comparatively, the infectious Jad production in Huh7.5 cells infected under similar conditions reached 4.58 ± 0.02 log TCID50/ml, confirming that these human cells are more permissive to HCV. In addition, anti-CD81 antibodies were shown to decrease viral yields in PMH culture supernatants down to ∼20% compared to the levels found with isotype-matched control antibodies (Fig. 7C), supporting the conclusion that Jad is able to productively enter and replicate in cells of PMH cultures by making use of CD81 as a (co)receptor. Altogether, these results are strongly indicative of a productive, although low-yield HCV infection in these PMH cultures, suggesting that all steps of the HCV Jad life cycle, including virus entry, genome replication, and particle assembly, may take place in primary small primate hepatocytes.
FIG 7.
Infection of PMH cultures with HCV Jad. (A) PMH cultures (gray bars) or Huh7.5 cells (white bars) were infected with Jad at an MOI of 0.04 TCID50/cell, and viral production was determined at the indicated times p.i. by RT-qPCR titration of the culture supernatants. (B) PMH cultures were infected with untreated or UV-inactivated Jad at an MOI of 0.02 (gray bars) or 0.14 (black bars) TCID50 per cell. Viral production was determined at 3 days p.i. by RT-qPCR titration of the culture supernatants. Mean results ± standard deviations from two experiments each performed in duplicates or triplicates are represented. (C) PMH cultures were incubated with Jad at a MOI of 0.14 TCID50 per cell in the presence of anti-CD81 (hatched bar) or control isotype-matched (black bar) antibodies for 4 h and further incubated in the absence of antibodies for 72 h. Viral production was quantified at 72 h p.i. by RT-qPCR titration of the culture supernatants and expressed with respect to viral titers obtained in the presence of control antibodies set at 100%.
DISCUSSION
This work explored the possibility of developing GBV-B/HCV chimeras comprising a morphogenesis unit derived from GBV-B and a genomic replication unit derived from HCV to further the development of a small primate in vivo model for HCV. We found that chimeric J/C-NS3proGB-Ubi RNA, encoding appropriately processed GBV-B C, E1, E2, p13, and NS2, did not lead to viremia in GBV-B-susceptible tamarins (Fig. 1). This defect could not be attributed to a lack of function of HCV replicase complex in small primate hepatocytes, since we further documented RNA replication following infection of primary marmoset hepatocyte cultures with HCV Jad (Fig. 7). Furthermore, J/C-NS3proGB-Ubi RNA replicated efficiently but was unable to lead to viral particle production in Huh7.5 cells at early times posttransfection (Fig. 2) or following passage of transfected cells (Fig. 4). We showed that this defect could not be explained by the lack of infection of human hepatocyte cultures by GBV-B glycoprotein-containing particles since (i) retroviral particles pseudotyped with GBV-B glycoproteins were able to transduce Huh7.5 cells (Fig. 5) and (ii) GBV-B isolated from the serum of a viremic tamarin was able to efficiently infect Huh7.5 cells and subsequently proceed to efficient genome replication in these cells (data not shown). Altogether, these results strongly suggest that the defect of J/C-NS3proGB-Ubi likely resides in the inability of this chimeric RNA to lead to infectious particle assembly and/or release. This contrasts with the properties of a similar HCV intergenotypic chimera (J/C-NS3proH77-Ubi), which reproducibly led to efficient virus production through the acquisition of compensatory mutations within its genome following serial passage of transfected cells (Fig. 4 and Tables 1 and 2). In HCV, essential cross talk between structural proteins, p7, NS2, and nonstructural proteins of the replication complex has been shown to be essential for virus morphogenesis and release (reviewed in reference 45). Our data suggest that similar protein interactions are likely to be equally important to yield GBV-B particles and that the greater genetic distance between GBV-B and HCV than between HCV subtypes 1a and 2a renders restoration of these interactions more unlikely. In line with these findings, we also failed to obtain infectious reverse HCV-1a/GBV-B chimeras derived from GBV-B and encoding HCV-H77 C-E1-E2-p7 or C-E1-E2-p7-NS2 proteins upon intrahepatic inoculation into small primates (data not shown). Surprisingly, however, Li et al. recently described HCV-1b/GBV-B chimeras encoding E1-E2-p7 or C-E1-E2-p7 of an HCV-1b isolate within the replicative backbone of GBV-B that are infectious in marmosets (46). Whether these discrepancies reflect morphogenesis requirements specific to the HCV subtype used in the construct necessitates further investigation.
All cell culture-adapted HCV-1a/HCV-2a recombinants described in our study contain a substitution leading to an amino acid change in JFH1 NS3. The Q1251L and R1412W substitutions in the NS3 helicase, which were observed in the genomes of most J/C-NS2H77 and J/C-NS3proH77-Ubi viruses rescued (Table 1), were also reported in JFH1 recombinants encoding HCV-1a C-NS2 proteins that have been described in the literature (37, 42), highlighting preferential ways of rescuing functional H77 insertions. The Q1251L substitution was found to be sufficient to compensate for the J/C-NS3proH77-Ubi chimera's defect (Table 2), likely by restoring important cross talk between NS2 and NS3-4A, which have been shown by imaging and genetic methods to be essential for the recruitment of core protein from cytosolic lipid droplets into maturing particles (47). Our data also demonstrate that the insertion of H77 NS3pro and ubiquitin sequences had no negative impact on the recombinants' infectivity. This provides an effective way to create infectious HCV JFH1-based recombinants encoding C-NS2 proteins from hepaciviruses with phylogenetically divergent NS2 protease, e.g., from newly discovered mammalian hepaciviruses (3). Interestingly, while no substitution was selected in the NS3 protease-coding sequences of the four J/C-NS2H77 viruses, each of the three infectious J/C-NS3proH77-Ubi viruses harbored a substitution targeting either the NS3 protease domain of JFH1 (P1032L and P1159Q) or the inserted ubiquitin sequence. The substitutions in JFH1 NS3 protease but not the one in ubiquitin were found to be individually compensatory for the J/C-NS3proH77-Ubi chimera (Table 2). We speculate that these substitutions may modulate the cleavage kinetics at the Ubi/NS3 junction, in line with the fact that cleavage at the NS2/NS3 junction has recently been reported to be a rate-limiting step in JFH1 replication (48).
The development of retroviral particles pseudotyped with GBV-B glycoproteins allowed us to provide the first evidence that GBV-B glycoproteins could mediate significant virus entry into human hepatoma cell lines (Fig. 5). It would be of interest to introduce E2 mutations selected in vivo during prolonged or persistent GBV-B infections in marmosets (14, 15, 49) to determine whether they increase GBV-B-pp cell entry. Efficient GBV-B-pp would represent an invaluable tool to seek GBV-B receptors at the surface of human and simian hepatocytes and determine whether differences in E2 glycoproteins of GBV-B and HCV translate into different mechanisms of hepacivirus cell entry. The fact that HCV genotype 1a or 2a glycoproteins can mediate productive, CD81-dependent entry into small primate hepatic cell lines or primary hepatocyte cultures in the absence of all human coreceptors (Fig. 5 and 6) strongly suggests that there is no blockage at HCV cell entry into simian hepatocytes. Recently, it has also been reported that HCV-pp can productively transduce primary cultures of macaque hepatocytes (50). Importantly, we showed that a highly replicating JFH1 variant (Jad) was capable of not only entry but also genome replication and low-level virus production in primary marmoset hepatocyte cultures (Fig. 7). These ex vivo data provide an encouraging indication of possible further adaptation of Jad to small primates, which would provide an invaluable and accessible in vivo model system to study HCV vaccine candidates.
ACKNOWLEDGMENTS
This work was supported in part by a grant from the ANRS-FRENSH (France REcherche, Nord & Sud, Sida-HIV, Hépatites), an autonomous agency at Inserm, France, and by the Institut Pasteur, Paris, and the Centre National de la Recherche Scientifique (CNRS), France. C.M., C.B., and L.W. were the recipients of doctoral fellowships from the ANRS-FRENSH, Université Paris Diderot, and CNRS, respectively, and A.S. was the recipient of a postdoctoral fellowship from the ANRS-FRENSH.
We gratefully acknowledge the expertise of the veterinary and husbandry staff and J. Mitchell (NIBSC, United Kingdom) for early GBV-B molecular analyses, Fanélie Wanert, Jean-Marie Helies, Pierrick Regnard, and Nicolas Herrenschmidt (Primate Center, Université Strasbourg, France) for housing of marmosets and help in preparing primary marmoset hepatocytes, David Ghibaudo (I.P. Paris, France) for helpful suggestions in the initial stages of this work, Brigitte Blumen and Hô Tinh Tâm Châu (I.P. Paris, France) for excellent technical assistance, Matthieu Fritz and Stephanie Aicher (I.P. Paris, France) for sharing their experimental expertise, and the following colleagues for sharing valuable reagents: Takaji Wakita (NIID, Tokyo, Japan) for pJFH1, pSGR-JFH1, and anti-JFH1 NS3 antibodies, Ralf Bartenschlager (Universität Heidelberg, Germany) for cell culture-adapted pJFH1-2EI3-adapt cDNA, Robert Purcell (NIH, Bethesda, MD, USA) for pH77C, Mats Persson (Karolinska Institutet, Stockholm, Sweden) for anti-H77 E2 scFv antibody, Charles Rice (Rockefeller University, New York, NY, USA) for Huh7.5 cells, and Jean Dubuisson (IBL, Lille, France) for anti-H77 E1 and E2 antibodies.
REFERENCES
- 1.Feeney ER, Chung RT. 2014. Antiviral treatment of hepatitis C. BMJ 348:g3308. doi: 10.1136/bmj.g3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.von Schaewen M, Ploss A. 2014. Murine models of hepatitis C: what can we look forward to? Antiviral Res 104:15–22. doi: 10.1016/j.antiviral.2014.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Drexler JF, Corman VM, Muller MA, Lukashev AN, Gmyl A, Coutard B, Adam A, Ritz D, Leijten LM, van Riel D, Kallies R, Klose SM, Gloza-Rausch F, Binger T, Annan A, Adu-Sarkodie Y, Oppong S, Bourgarel M, Rupp D, Hoffmann B, Schlegel M, Kummerer BM, Kruger DH, Schmidt-Chanasit J, Setien AA, Cottontail VM, Hemachudha T, Wacharapluesadee S, Osterrieder K, Bartenschlager R, Matthee S, Beer M, Kuiken T, Reusken C, Leroy EM, Ulrich RG, Drosten C. 2013. Evidence for novel hepaciviruses in rodents. PLoS Pathog 9:e1003438. doi: 10.1371/journal.ppat.1003438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Quan PL, Firth C, Conte JM, Williams SH, Zambrana-Torrelio CM, Anthony SJ, Ellison JA, Gilbert AT, Kuzmin IV, Niezgoda M, Osinubi MO, Recuenco S, Markotter W, Breiman RF, Kalemba L, Malekani J, Lindblade KA, Rostal MK, Ojeda-Flores R, Suzan G, Davis LB, Blau DM, Ogunkoya AB, Alvarez Castillo DA, Moran D, Ngam S, Akaibe D, Agwanda B, Briese T, Epstein JH, Daszak P, Rupprecht CE, Holmes EC, Lipkin WI. 2013. Bats are a major natural reservoir for hepaciviruses and pegiviruses. Proc Natl Acad Sci U S A 110:8194–8199. doi: 10.1073/pnas.1303037110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lauck M, Sibley SD, Lara J, Purdy MA, Khudyakov Y, Hyeroba D, Tumukunde A, Weny G, Switzer WM, Chapman CA, Hughes AL, Friedrich TC, O'Connor DH, Goldberg TL. 2013. A novel hepacivirus with an unusually long and intrinsically disordered NS5A protein in a wild Old World primate. J Virol 87:8971–8981. doi: 10.1128/JVI.00888-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lyons S, Kapoor A, Sharp C, Schneider BS, Wolfe ND, Culshaw G, Corcoran B, McGorum BC, Simmonds P. 2012. Nonprimate hepaciviruses in domestic horses, United kingdom. Emerg Infect Dis 18:1976–1982. doi: 10.3201/eid1812.120498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pfaender S, Cavalleri JM, Walter S, Doerrbecker J, Campana B, Brown RJ, Burbelo PD, Postel A, Hahn K, Anggakusuma, Riebesehl N, Baumgartner W, Becher P, Heim MH, Pietschmann T, Feige K, Steinmann E. 2015. Clinical course of infection and viral tissue tropism of hepatitis C virus-like nonprimate hepaciviruses in horses. Hepatology 61:447–459. doi: 10.1002/hep.27440. [DOI] [PubMed] [Google Scholar]
- 8.Scheel TK, Kapoor A, Nishiuchi E, Brock KV, Yu Y, Andrus L, Gu M, Renshaw RW, Dubovi EJ, McDonough SP, Van de Walle GR, Lipkin WI, Divers TJ, Tennant BC, Rice CM. 2015. Characterization of nonprimate hepacivirus and construction of a functional molecular clone. Proc Natl Acad Sci U S A 112:2192–2197. doi: 10.1073/pnas.1500265112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Simons JN, Pilot-Matias TJ, Leary TP, Dawson GJ, Desai SM, Schlauder GG, Muerhoff AS, Erker JC, Buijk SL, Chalmers ML, Van Sant CL, Mushahwar IK. 1995. Identification of two flavivirus-like genomes in the GB hepatitis agent. Proc Natl Acad Sci U S A 92:3401–3405. doi: 10.1073/pnas.92.8.3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bukh J, Apgar CL, Govindarajan S, Purcell RH. 2001. Host range studies of GB virus-B hepatitis agent, the closest relative of hepatitis C virus, in New World monkeys and chimpanzees. J Med Virol 65:694–697. doi: 10.1002/jmv.2092. [DOI] [PubMed] [Google Scholar]
- 11.Lanford RE, Chavez D, Notvall L, Brasky KM. 2003. Comparison of tamarins and marmosets as hosts for GBV-B infections and the effect of immunosuppression on duration of viremia. Virology 311:72–80. doi: 10.1016/S0042-6822(03)00193-4. [DOI] [PubMed] [Google Scholar]
- 12.Weatherford T, Chavez D, Brasky KM, Lanford RE. 2009. The marmoset model of GB virus B infections: adaptation to host phenotypic variation. J Virol 83:5806–5814. doi: 10.1128/JVI.00033-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martin A, Bodola F, Sangar DV, Goettge K, Popov V, Rijnbrand R, Lanford RE, Lemon SM. 2003. Chronic hepatitis associated with GB virus B persistence in a tamarin after intrahepatic inoculation of synthetic viral RNA. Proc Natl Acad Sci U S A 100:9962–9967. doi: 10.1073/pnas.1731505100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iwasaki Y, Mori K, Ishii K, Maki N, Iijima S, Yoshida T, Okabayashi S, Katakai Y, Lee YJ, Saito A, Fukai H, Kimura N, Ageyama N, Yoshizaki S, Suzuki T, Yasutomi Y, Miyamura T, Kannagi M, Akari H. 2011. Long-term persistent GBV-B infection and development of a chronic and progressive hepatitis C-like disease in marmosets. Front Microbiol 2:240. doi: 10.3389/fmicb.2011.00240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takikawa S, Engle RE, Faulk KN, Emerson SU, Purcell RH, Bukh J. 2010. Molecular evolution of GB virus B hepatitis virus during acute resolving and persistent infections in experimentally infected tamarins. J Gen Virol 91:727–733. doi: 10.1099/vir.0.015750-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boukadida C, Marnata C, Montserret R, Cohen L, Blumen B, Gouttenoire J, Moradpour D, Penin F, Martin A. 2014. NS2 proteins of GB virus B and hepatitis C virus share common protease activities and membrane topologies. J Virol 88:7426–7444. doi: 10.1128/JVI.00656-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ranjith-Kumar CT, Santos JL, Gutshall LL, Johnston VK, Lin-Goerke J, Kim MJ, Porter DJ, Maley D, Greenwood C, Earnshaw DL, Baker A, Gu B, Silverman C, Sarisky RT, Kao C. 2003. Enzymatic activities of the GB virus-B RNA-dependent RNA polymerase. Virology 312:270–280. doi: 10.1016/S0042-6822(03)00247-2. [DOI] [PubMed] [Google Scholar]
- 18.Scarselli E, Urbani A, Sbardellati A, Tomei L, De Francesco R, Traboni C. 1997. GB virus B and hepatitis C virus NS3 serine proteases share substrate specificity. J Virol 71:4985–4989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rijnbrand R, Yang Y, Beales L, Bodola F, Goettge K, Cohen L, Lanford RE, Lemon SM, Martin A. 2005. A chimeric GB virus B with 5′ nontranslated RNA sequence from hepatitis C virus causes hepatitis in tamarins. Hepatology 41:986–994. doi: 10.1002/hep.20656. [DOI] [PubMed] [Google Scholar]
- 20.Sandmann L, Ploss A. 2013. Barriers of hepatitis C virus interspecies transmission. Virology 435:70–80. doi: 10.1016/j.virol.2012.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zeisel MB, Felmlee DJ, Baumert TF. 2013. Hepatitis C virus entry. Curr Top Microbiol Immunol 369:87–112. doi: 10.1007/978-3-642-27340-7_4. [DOI] [PubMed] [Google Scholar]
- 22.Ghibaudo D, Cohen L, Penin F, Martin A. 2004. Characterization of GB virus B polyprotein processing reveals the existence of a novel 13 kDa protein with partial homology to hepatitis C virus p7 protein. J Biol Chem 279:24965–24975. doi: 10.1074/jbc.M401148200. [DOI] [PubMed] [Google Scholar]
- 23.Garson JA, Whitby K, Watkins P, Morgan AJ. 1997. Lack of susceptibility of the cottontop tamarin to hepatitis C infection. J Med Virol 52:286–288. doi:. [DOI] [PubMed] [Google Scholar]
- 24.Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z, Murthy K, Habermann A, Krausslich HG, Mizokami M, Bartenschlager R, Liang TJ. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med 11:791–796. doi: 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yanagi M, Purcell RH, Emerson SU, Bukh J. 1997. Transcripts from a single full-length cDNA clone of hepatitis C virus are infectious when directly transfected into the liver of a chimpanzee. Proc Natl Acad Sci U S A 94:8738–8743. doi: 10.1073/pnas.94.16.8738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaul A, Woerz I, Meuleman P, Leroux-Roels G, Bartenschlager R. 2007. Cell culture adaptation of hepatitis C virus and in vivo viability of an adapted variant. J Virol 81:13168–13179. doi: 10.1128/JVI.01362-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Warter L, Cohen L, Benureau Y, Chavez D, Yang Y, Bodola F, Lemon SM, Traboni C, Lanford RE, Martin A. 2009. A cooperative interaction between nontranslated RNA sequences and NS5A protein promotes in vivo fitness of a chimeric hepatitis C/GB virus B. PLoS One 4:e4419. doi: 10.1371/journal.pone.0004419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen Z, Benureau Y, Rijnbrand R, Yi J, Wang T, Warter L, Lanford RE, Weinman SA, Lemon SM, Martin A, Li K. 2007. GB virus B disrupts RIG-I signaling by NS3/4A-mediated cleavage of the adaptor protein MAVS. J Virol 81:964–976. doi: 10.1128/JVI.02076-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blight KJ, McKeating JA, Marcotrigiano J, Rice CM. 2003. Efficient replication of hepatitis C virus genotype 1a RNAs in cell culture. J Virol 77:3181–3190. doi: 10.1128/JVI.77.5.3181-3190.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lanford RE, Estlack L. 1999. A cultivation method for highly differentiated primary chimpanzee hepatocytes permissive for hepatitis C virus replication. Methods in molecular medicine: hepatitis C. 19:501–515. doi: 10.1385/0-89603-521-2:501. [DOI] [PubMed] [Google Scholar]
- 31.Lavillette D, Tarr AW, Voisset C, Donot P, Bartosch B, Bain C, Patel AH, Dubuisson J, Ball JK, Cosset FL. 2005. Characterization of host-range and cell entry properties of the major genotypes and subtypes of hepatitis C virus. Hepatology 41:265–274. doi: 10.1002/hep.20542. [DOI] [PubMed] [Google Scholar]
- 32.Dubuisson J, Hsu HH, Cheung RC, Greenberg HB, Russell DG, Rice CM. 1994. Formation and intracellular localization of hepatitis C virus envelope glycoprotein complexes expressed by recombinant vaccinia and Sindbis viruses. J Virol 68:6147–6160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kato T, Date T, Miyamoto M, Furusaka A, Tokushige K, Mizokami M, Wakita T. 2003. Efficient replication of the genotype 2a hepatitis C virus subgenomic replicon. Gastroenterology 125:1808–1817. doi: 10.1053/j.gastro.2003.09.023. [DOI] [PubMed] [Google Scholar]
- 34.Krey T, d'Alayer J, Kikuti CM, Saulnier A, Damier-Piolle L, Petitpas I, Johansson DX, Tawar RG, Baron B, Robert B, England P, Persson MA, Martin A, Rey FA. 2010. The disulfide bonds in glycoprotein E2 of hepatitis C virus reveal the tertiary organization of the molecule. PLoS Pathog 6:e1000762. doi: 10.1371/journal.ppat.1000762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gilmartin AA, Lamp B, Rumenapf T, Persson MA, Rey FA, Krey T. 2012. High-level secretion of recombinant monomeric murine and human single-chain Fv antibodies from Drosophila S2 cells. Protein Eng Des Sel 25:59–66. doi: 10.1093/protein/gzr058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Johansson DX, Voisset C, Tarr AW, Aung M, Ball JK, Dubuisson J, Persson MA. 2007. Human combinatorial libraries yield rare antibodies that broadly neutralize hepatitis C virus. Proc Natl Acad Sci U S A 104:16269–16274. doi: 10.1073/pnas.0705522104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scheel TK, Gottwein JM, Carlsen TH, Li YP, Jensen TB, Spengler U, Weis N, Bukh J. 2011. Efficient culture adaptation of hepatitis C virus recombinants with genotype-specific core-NS2 by using previously identified mutations. J Virol 85:2891–2906. doi: 10.1128/JVI.01605-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Noppornpanth S, Lien TX, Poovorawan Y, Smits SL, Osterhaus AD, Haagmans BL. 2006. Identification of a naturally occurring recombinant genotype 2/6 hepatitis C virus. J Virol 80:7569–7577. doi: 10.1128/JVI.00312-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rijnbrand R, Bredenbeek PJ, Haasnoot PC, Kieft JS, Spaan WJ, Lemon SM. 2001. The influence of downstream protein-coding sequence on internal ribosome entry on hepatitis C virus and other flavivirus RNAs. RNA 7:585–597. doi: 10.1017/S1355838201000589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bukh J, Engle RE, Govindarajan S, Purcell RH. 2008. Immunity against the GBV-B hepatitis virus in tamarins can prevent productive infection following rechallenge and is long-lived. J Med Virol 80:87–94. doi: 10.1002/jmv.21013. [DOI] [PubMed] [Google Scholar]
- 41.Gastaminza P, Kapadia SB, Chisari FV. 2006. Differential biophysical properties of infectious intracellular and secreted hepatitis C virus particles. J Virol 80:11074–11081. doi: 10.1128/JVI.01150-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yi M, Ma Y, Yates J, Lemon SM. 2007. Compensatory mutations in E1, p7, NS2, and NS3 enhance yields of cell culture-infectious intergenotypic chimeric hepatitis C virus. J Virol 81:629–638. doi: 10.1128/JVI.01890-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maurin G, Fresquet J, Granio O, Wychowski C, Cosset FL, Lavillette D. 2011. Identification of interactions in the E1E2 heterodimer of hepatitis C virus important for cell entry. J Biol Chem 286:23865–23876. doi: 10.1074/jbc.M110.213942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kaul A, Stauffer S, Berger C, Pertel T, Schmitt J, Kallis S, Zayas M, Lohmann V, Luban J, Bartenschlager R. 2009. Essential role of cyclophilin A for hepatitis C virus replication and virus production and possible link to polyprotein cleavage kinetics. PLoS Pathog 5:e1000546. doi: 10.1371/journal.ppat.1000546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lindenbach BD, Rice CM. 2013. The ins and outs of hepatitis C virus entry and assembly. Nat Rev Microbiol 11:688–700. doi: 10.1038/nrmicro3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li T, Zhu S, Shuai L, Xu Y, Yin S, Bian Y, Wang Y, Zuo B, Wang W, Zhao S, Zhang L, Zhang J, Gao GF, Allain JP, Li C. 2014. Infection of common marmosets with hepatitis C virus/GB virus-B chimeras. Hepatology 59:789–802. doi: 10.1002/hep.26750. [DOI] [PubMed] [Google Scholar]
- 47.Counihan NA, Rawlinson SM, Lindenbach BD. 2011. Trafficking of hepatitis C virus core protein during virus particle assembly. PLoS Pathog 7:e1002302. doi: 10.1371/journal.ppat.1002302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Madan V, Paul D, Lohmann V, Bartenschlager R. 2014. Inhibition of HCV replication by cyclophilin antagonists is linked to replication fitness and occurs by inhibition of membranous web formation. Gastroenterology 146:1361–1372. doi: 10.1053/j.gastro.2014.01.055. [DOI] [PubMed] [Google Scholar]
- 49.McGarvey MJ, Iqbal M, Nastos T, Karayiannis P. 2008. Restricted quasispecies variation following infection with the GB virus B. Virus Res 135:181–186. doi: 10.1016/j.virusres.2008.03.013. [DOI] [PubMed] [Google Scholar]
- 50.Sourisseau M, Goldman O, He W, Gori JL, Kiem HP, Gouon-Evans V, Evans MJ. 2013. Hepatic cells derived from induced pluripotent stem cells of pigtail macaques support hepatitis C virus infection. Gastroenterology 145:966–969. doi: 10.1053/j.gastro.2013.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]