ABSTRACT
Severe acute respiratory syndrome (SARS) emerged in November 2002 as a case of atypical pneumonia in China, and the causative agent of SARS was identified to be a novel coronavirus, severe acute respiratory syndrome coronavirus (SARS-CoV). Bone marrow stromal antigen 2 (BST-2; also known as CD317 or tetherin) was initially identified to be a pre-B-cell growth promoter, but it also inhibits the release of virions of the retrovirus human immunodeficiency virus type 1 (HIV-1) by tethering budding virions to the host cell membrane. Further work has shown that BST-2 restricts the release of many other viruses, including the human coronavirus 229E (hCoV-229E), and the genomes of many of these viruses encode BST-2 antagonists to overcome BST-2 restriction. Given the previous studies on BST-2, we aimed to determine if BST-2 has the ability to restrict SARS-CoV and if the SARS-CoV genome encodes any proteins that modulate BST-2's antiviral function. Through an in vitro screen, we identified four potential BST-2 modulators encoded by the SARS-CoV genome: the papain-like protease (PLPro), nonstructural protein 1 (nsp1), ORF6, and ORF7a. As the function of ORF7a in SARS-CoV replication was previously unknown, we focused our study on ORF7a. We found that BST-2 does restrict SARS-CoV, but the loss of ORF7a leads to a much greater restriction, confirming the role of ORF7a as an inhibitor of BST-2. We further characterized the mechanism of BST-2 inhibition by ORF7a and found that ORF7a localization changes when BST-2 is overexpressed and ORF7a binds directly to BST-2. Finally, we also show that SARS-CoV ORF7a blocks the restriction activity of BST-2 by blocking the glycosylation of BST-2.
IMPORTANCE The severe acute respiratory syndrome coronavirus (SARS-CoV) emerged from zoonotic sources in 2002 and caused over 8,000 infections and 800 deaths in 37 countries around the world. Identifying host factors that regulate SARS-CoV pathogenesis is critical to understanding how this lethal virus causes disease. We have found that BST-2 is capable of restricting SARS-CoV release from cells; however, we also identified a SARS-CoV protein that inhibits BST-2 function. We show that the SARS-CoV protein ORF7a inhibits BST-2 glycosylation, leading to a loss of BST-2's antiviral function.
INTRODUCTION
Severe acute respiratory syndrome coronavirus (SARS-CoV) was identified to be the causative agent of a 2002 to 2004 outbreak of severe respiratory disease that emerged from the Guangdong province of China, resulting in 8,096 cases and 774 deaths across 37 countries (1, 2). SARS-CoV is an enveloped virus with a positive-sense, single-stranded RNA genome of roughly 30,000 nucleotides encoding four structural proteins: the spike (S), envelope (E), membrane (M), and nucleocapsid (N) proteins (3). N protein forms the nucleocapsid, while E and M are minor virion membrane proteins. SARS-CoV entry into the cell is mediated by S-protein binding to angiotensin-converting enzyme 2 (ACE2) on the cell surface (4). In addition to the structural proteins, the SARS-CoV genome encodes several nonstructural and accessory proteins that promote SARS-CoV replication and virulence (5). Some of the nonstructural and accessory proteins function outside of replication as type I interferon antagonists (6–8).
ORF7a is a SARS-CoV genome-encoded accessory protein that is composed of a type I transmembrane protein that localizes primarily to the Golgi apparatus but can be found on the cell surface (9, 10). SARS-CoV ORF7a overlaps ORF7b in the viral genome, where they share a transcriptional regulatory sequence (TRS). ORF7a has a 15-amino-acid (aa) N-terminal signal peptide, an 81-aa luminal domain, a 21-aa transmembrane domain, and a 5-aa cytoplasmic tail (9, 10). To investigate the role of ORF7a in SARS-CoV replication, an ORF7ab deletion virus that replicated to a titer similar to that of wild-type (WT) SARS-CoV in vitro and in vivo was produced (10–12). Characterization of ORF7a in vitro demonstrated the ORF7a-dependent induction of apoptosis in a caspase-dependent pathway (13–15). Analysis of ORF7a evolution during the SARS-CoV outbreak identified several residues in ORF7a that were under positive selection as SARS-CoV evolved during transmission from bat to palm civet to humans (16). These data suggest that ORF7a is vital for SARS-CoV biology and has a yet unidentified role in pathogenesis and disease.
Bone marrow stromal antigen 2 (BST-2; also known as CD317 or tetherin) was initially identified to be a pre-B-cell growth promoter (17, 18). However, BST-2 is also a marker of type I interferon-producing cells (IPC) and is broadly expressed in many cell types when treated with type I interferon (19). BST-2 has an unusual structure, with an N-terminal transmembrane domain, a C-terminal glycosylphosphatidylinositol (GPI) anchor, and two N-linked glycosylation sites in its extracellular domain, and exists as a disulfide-linked homodimer (20, 21). BST-2 traffics through the endoplasmic reticulum (ER) and Golgi apparatus, eventually localizing to the surface and trans-Golgi network (20). Studies have shown evolutionary conservation in three major surface patches of BST-2: near each of the two N-linked glycosylation sites and in the C-terminal region (22).
The antiviral effect of BST-2 was first identified when it was shown that BST-2 inhibits the release of virions of the retrovirus human immunodeficiency virus type 1 (HIV-1) by directly tethering budding virions to the host cell. BST-2 also restricts the release of many other viruses, including alphaviruses, arenaviruses, herpesviruses, paramyxoviruses, and other retroviruses (23–26). BST-2 is thought to restrict virus release by physically tethering the budding enveloped virion to the plasma membrane (27), and a number of mechanism models have been proposed (28, 29). All of the BST-2 restriction models predict that BST-2 functions as a dimer, interfacing through ectodomains that incorporate into both the viral envelope and plasma membrane; however, the models vary in regard to the orientation of the GPI anchor and transmembrane domain. BST-2 has not been shown to interact with any specific viral surface protein but, rather, has been shown to function as an embedded intermembrane physical tether. Therefore, BST-2 is thought to be able to restrict any membrane-budding enveloped virus (28, 29). Previous studies have shown that the ability to form cysteine-linked dimers is necessary for BST-2 function, while conflicting results concerning the importance of the N-linked glycosylation have been reported (29, 30). More recently, it has been suggested that BST-2 is a virus sensor during HIV-1 infection and induces a proinflammatory response through NF-κB (31).
Given the lack of virus specificity in BST-2 restriction, the genomes of numerous viruses encode BST-2 antagonists to allow the release of virions. The first such antagonist was identified to be HIV-1 accessory protein Vpu (27). HIV-1 Vpu binds BST-2 and causes the β-TrCP2-dependent degradation of BST-2 and the efficient release of HIV-1 virions, although it is not clear whether degradation occurs in the lysosome or proteasome (32–34). Other viral antagonists of BST-2 include Chikungunya virus nonstructural protein 1 (nsp1), Ebola virus glycoprotein GP1,2, herpes simplex virus glycoprotein M, HIV-2 envelope glycoprotein, Sendai virus glycoproteins, and simian immunodeficiency virus (SIV) Nef and envelope glycoproteins (23–26, 35–38). HIV-2 and SIV are closely related to HIV-1; however, the envelope glycoproteins from HIV-2 and SIV antagonize BST-2 by sequestration within the trans-Golgi network rather than degradation, suggesting that different mechanisms of BST-2 antagonism exist for different viruses, even within the same virus genus (35, 36). Another example is Ebola virus GP1,2, which antagonizes BST-2 through an unknown mechanism that does not involve surface removal but still leads to BST-2 functional inhibition (39).
Unlike many enveloped viruses, which bud from the cell plasma membrane, coronaviruses bud in the ER-Golgi apparatus intermediate compartment (ERGIC) and are transported to the plasma membrane inside vesicles (40). However, it has recently been shown that BST-2 restricts the release of human coronavirus 229E (hCoV-229E), suggesting that BST-2 can also restrict viruses that bud in the ERGIC and are then released from the cell via vesicle fusion (37).
In this study, we found that BST-2 restricts SARS-CoV virion egress by tethering virions to the plasma membrane. We also identified several SARS-CoV proteins that are putative modulators of BST-2 function. Focusing on ORF7a, we found that ORF7a directly binds BST-2 and, when coexpressed with BST-2, ORF7a localizes to the plasma membrane rather than the ER and Golgi apparatus. Additionally, we demonstrate that the interaction of ORF7a and BST-2 results in inhibition of BST-2 glycosylation, leading to a reduced tethering function in cells and the subsequent loss of BST-2 antiviral function. Together, these data indicate a novel role for SARS-CoV ORF7a as an inhibitor of BST-2, as well as reveal a novel mechanism for altering the function of BST-2.
MATERIALS AND METHODS
Viruses and cells.
icSARS-CoV and SARS-CoV with an ORF7ab deletion (icSARS-ORF7abΔ-CoV) were constructed using the SARS-CoV infectious clone (icSARS-CoV) as previously described (41, 42). All virus stocks were stored at −80°C until they were ready to use. Vero E6 cells were purchased from ATCC (catalog number CRL-1586; Manassas, VA) and were grown in minimal essential medium (MEM; Invitrogen, Carlsbad, CA) with 10% fetal bovine serum (FBS; Atlanta Biologicals, Lawrenceville, GA), 2 mM l-glutamine (Life Technologies, Grand Island, NY), and 1% penicillin-streptomycin (Gemini Bioproducts, West Sacramento, CA). HEK293T cells were grown in Dulbecco's minimal essential medium (DMEM; Invitrogen, Carlsbad, CA) with 10% FBS (Atlanta Biologicals, Lawrenceville, GA), 2 mM l-glutamine (Life Technologies, Grand Island, NY), and 1% penicillin-streptomycin (Gemini Bioproducts, West Sacramento, CA). HEK293T cells expressing ACE2 (HEK293T/ACE2 cells) were a gift from David Wentworth (J. Craig Venter Institute) and were grown in HEK293T medium supplemented with 1 mg/ml G418 (Corning, Manassas, VA).
Plasmids.
We received pCAGGS carrying BST-2–Flag as a gift from Sina Bavari (USAMRIID) (24). We received ORF7a with an Fc fusion tag (ORF7a-Fc) as a gift from Andrew Pekosz (Johns Hopkins University, Baltimore, MD). Plasmids carrying SARS-CoV ORF3a, ORF3b, ORF6, ORF7a, ORF8a, S, E, membrane protein, N, and papain-like protease (PLPro) were produced as described in previous work (6, 7). The nonstructural proteins were cloned into the pCAGGS-green fluorescent protein (GFP) or pCAGGS-hemagglutinin (HA) vector for expression in HEK293T cells as previously described (7). Amplicons were produced using the primers shown in (Table 1). For each construct, an ATG start codon was added as the first codon, but no stop codon was included at the 3′ terminus of each open reading frame (ORF). Rather, an HA or GFP tag was fused in frame to each ORF. The amplicons and vector were digested with EcoRI/XmaI fragments for cloning, and all constructs were verified by sequence analysis.
TABLE 1.
SARS-CoV nonstructural protein cloning primers used in this study
| SARS-CoV gene | Forward primer/EcoRI site | Reverse primer/XmaI site |
|---|---|---|
| nsp1 | 5′-GATCGAATTCACCATGGAGAGCCTTGTTCTTGGTGTCA-3′ | 5′-GATCCCCGGGACCTCCATTGAGCTCACGAGTGAGT-3′ |
| nsp4 | 5′-GATCGAATTCACCATGAAGATTGTTAGTACTTGTTTTA-3′ | 5′-GATCCCCGGGCTGCAGAACAGCAGAAGTGATTGAT-3′ |
| nsp5 | 5′-GATCGAATTCACCATGAGTGGTTTTAGGAAAATGGCAT-3′ | 5′-GATCCCCGGGTTGGAAGGTAACACCAGAGCATTGT-3′ |
| nsp6 | 5′-GATCGAATTCACCATGGGTAAGTTCAAGAAAATTGTTA-3′ | 5′-GATCCCCGGGCTGTACAGTAGCAACCTTGATACAT-3′ |
| nsp7 | 5′-GATCGAATTCACCATGTCTAAAATGTCTGACGTAAAGT-3′ | 5′-GATCCCCGGGCTGAAGAGTAGCACGGTTATCGAGC-3′ |
| nsp8 | 5′-GATCGAATTCACCATGGCTATTGCTTCAGAATTTAGTT-3′ | 5′-GATCCCCGGGCTGTAGTTTAACAGCTGAGTTGGCT-3′ |
| nsp9 | 5′-GATCGAATTCACCATGAATAATGAACTGAGTCCAGTAG-3′ | 5′-GATCCCCGGGCTGAAGACGTACTGTAGCAGCTAAA-3′ |
| nsp10 | 5′-GATCGAATTCACCATGGCTGGAAATGCTACAGAAGTAC-3′ | 5′-GATCCCCGGGCTGCATCAAGGGTTCGCGGAGTTGG-3′ |
| nsp13 | 5′-GATCGAATTCACCATGAGGCTGTAGGTGCTTGTGTATTGT-3′ | 5′-GATCCCCGGGTTGTAATGTAGCCACATTGCGACGTGGTAT-3′ |
| nsp14 | 5′-GATCGAATTCACCATGGCAGAAAATGTAACTGGACTTTTT-3′ | 5′-GATCCCCGGGCTGTAACCTGGTAAATGTATTCCACAGGTT-3′ |
| nsp15 | 5′-GATCGAATTCACCATGAGTTTAGAAAATGTGGCTTATAAT-3′ | 5′-GATCCCCGGGTTGTAGTTTTGGGTAGAAGGTTTCAACATG-3′ |
| nsp16 | 5′-GATCGAATTCACCATGGCAAGTCAAGCGTGGCAACCAG-3′ | 5′-GATCCCCGGGGTTGTTAACAAGAATATCACTTGAA-3′ |
SARS-CoV growth curve.
HEK293T human ACE2 (hACE2) cells were plated in a 24-well plate and grown overnight at 37°C. Cells were transfected, using 2 μl of Lipofectamine LTX (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions, with 700 ng of pCAGGS carrying BST-2–Flag or ORF7a-HA or a control plasmid carrying Mission pLKO.1-puro nonmammalian short hairpin RNA (shRNA) (Sigma-Aldrich, St. Louis, MO). For the glycosylation mutant experiments, pCR3.1-EXN-tetherin-HA and pCR3.1-EXN-tetherin(N65A/N92A)-HA were kindly provided by Paul Bieniasz (29). At 24 h posttransfection, HEK293T/ACE2 cells were infected with icSARS-CoV or icSARS-GFP-CoV at a multiplicity of infection (MOI) of 0.1. Supernatant was taken at 12, 24, and 36 h postinfection to measure the SARS-CoV titer by plaque assay on Vero E6 cells. The supernatant and cell lysate were also analyzed by Western blotting. The growth curve experiments were repeated twice with 6 replicates of each sample.
The products of SARS-CoV RNA replication were assessed by reverse transcription-PCR (RT-PCR). RNA was isolated from cells that had been infected with SARS-CoV for 24 h using the TRIzol reagent (Ambion) according to the manufacturer's instructions. RNA was converted to cDNA using a RevertAid RT-PCR (Thermo Scientific) according to the manufacturer's instructions and treated with RNase H (New England BioLabs) according to the manufacturer's instructions. The levels of SARS-CoV pp1a mRNA (forward primer, GCCGTAGTGTCAGTATCATCACC; reverse primer, AATAGGACCAATCTCTGTAAGAGCC) and N protein mRNA (forward primer, CTCTTGTAGATCTGTTCTCTAAACGAAC; reverse primer, TTACTGTACTAGCAAAGCAATATTGTCG) were quantified using Sybr green PCR master mix (Applied Biosystems) according to the manufacturer's instructions and a 7500 Fast Dx real-time PCR instrument (Applied Biosystems). The levels of SARS-CoV RNA were quantified using the ΔΔCT threshold cycle (CT) method. Means and standard deviations were calculated from 3 independent infections.
Electron microscopy.
Vero E6 cells were plated in a 24-well plate and grown overnight at 37°C. Cells were transfected, using Lipofectamine LTX (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions, with pCAGGS carrying BST-2–Flag or a control plasmid carrying Mission pLKO.1-puro nonmammalian shRNA (Sigma-Aldrich, St. Louis, MO). At 24 h posttransfection, Vero cells were infected with icSARS-CoV or icSARS-GFP-CoV at an MOI of 10. At 24 h postinfection, the cells were fixed and analyzed by electron microscopy. For conventional ultrastructural investigations, infected Vero E6 cells were fixed with 2.5% glutaraldehyde (Electron Microscopy Sciences, Warrington, PA) at 24 h postinfection. After fixation for 72 h, the preserved cells were postfixed in 1.0% osmium tetroxide (Electron Microscopy Sciences), stained en bloc with 2.0% uranyl acetate, dehydrated in a series of graded ethanol, and infiltrated and embedded in Spurr plastic resin (Tousimis Research, Rockville, MD). Embedded blocks were sectioned using a Leica UC7 ultramicrotome, collected thin sections were mounted on 200-mesh copper grids, lead citrate was added as a contrast reagent, and the sections were subsequently viewed at 80 kV with an FEI Tecnai Twin transmission electron microscope.
BST-2–SARS-CoV accessory protein cotransfections.
HEK293T cells were transfected, using Lipofectamine LTX (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions, with 500 ng total DNA. One hundred nanograms of pCAGGS carrying BST-2–Flag, 200 ng or 400 ng of a plasmid carrying GFP- or HA-tagged SARS-CoV proteins, and a control plasmid carrying Mission pLKO.1-puro nonmammalian shRNA (Sigma-Aldrich, St. Louis, MO) were cotransfected into HEK293T cells. After 18 h of expression, the cells were lysed in lysis buffer (20 mM Tris-HCl [pH 7.6], 150 mM NaCl, 1% NP-40, 0.5% SDS, 5 mM EDTA, 1 protease inhibitor tablet). Lysate was combined with 2× Laemmli sample buffer (Bio-Rad, Hercules, CA) before boiling and electrophoresis using Mini-Protean TGX gels (Bio-Rad, Hercules, CA). Protein expression was assessed using rabbit anti-HA antibody (Sigma-Aldrich, St. Louis, MO), rabbit anti-GFP antibody (Sigma-Aldrich, St. Louis, MO), mouse anti-Flag M2 antibody (Sigma-Aldrich, St. Louis, MO), and mouse anti-β-tubulin antibody (Sigma-Aldrich, St. Louis, MO). For the inhibition experiments, cells were transfected as described above, and at 4 h posttransfection, medium was removed and replaced with 20 nM concanamycin A (ConA; Sigma-Aldrich, St. Louis, MO) or 500 nM MG-132 (Sigma-Aldrich, St. Louis, MO). Cell lysate was collected after 18 h of drug treatment. For time course experiments, HEK293T cells were transfected, using Lipofectamine LTX according to the manufacturer's instructions (Invitrogen, Carlsbad, CA), with 500 ng of a plasmid carrying ORF7a or a control plasmid. After 6 h of expression, medium was replaced with fresh HEK293T medium. At 22 h posttransfection, the cells were transfected, using Lipofectamine LTX (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions, with 500 ng of DNA, 100 ng of a plasmid carrying BST-2, and 400 ng of a control plasmid. Cell lysate was collected as described above at 4, 8, 12, and 16 h after the second transfection. Expression was analyzed as described above. Deglycosylation was performed using glycopeptidase F (TaKaRa, Mountain View, CA) according to the manufacturer's instructions for deglycosylating denatured proteins. The ratio of glycosylated to unglycosylated proteins was calculated by measuring the density of the bands with ImageJ software (National Institute of Mental Health, Bethesda, MD). All of the transfection experiments were repeated at least two times.
Anti-Flag immunoprecipitations.
HEK293T cells were transfected, using Lipofectamine LTX (Invitrogen, Carlsbad, CA), with 1,000 ng total DNA. Five hundred nanograms of a plasmid carrying Flag-tagged BST-2 and 500 ng of a plasmid carrying SARS-CoV PLPro-GFP, nsp1-GFP, ORF6-GFP, or ORF7a-HA or a control plasmid carrying Mission pLKO.1-puro nonmammalian shRNA (Sigma-Aldrich, St. Louis, MO) were cotransfected into HEK293T cells. After 18 h of expression, the cells were lysed in lysis buffer (20 mM Tris-HCl [pH 7.6], 150 mM NaCl, 1% NP-40, 0.5% SDS, 5 mM EDTA, 1 protease inhibitor tablet), the extract was centrifuged for 10 min at 4°C, and the supernatant was removed. EZ View Red anti-Flag M2 affinity gel beads (catalog number F2426; Sigma, St. Louis, MO) were added to each extract, and the mixture was rotated overnight at 4°C. The extract was then washed twice with lysis buffer and eluted using 0.1 M glycine (pH 3.5). The eluate was combined with 2× Laemmli sample buffer (Bio-Rad, Hercules, CA) before boiling and electrophoresis using Mini-Protean TGX gels (Bio-Rad, Hercules, CA). Protein levels were assessed using rabbit anti-HA antibody (Sigma-Aldrich, St. Louis, MO) and mouse anti-Flag M2 antibody (Sigma-Aldrich, St. Louis, MO). Coimmunoprecipitation experiments were performed twice.
Confocal microscopy.
HEK293T cells were seeded onto a microscope cover glass (Fisher Scientific, Pittsburg, PA) that had been pretreated with fibronectin (Sigma-Aldrich, St. Louis, MO) for 30 min. HEK293T cells were transfected, using Lipofectamine LTX (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions, with 500 ng total DNA. Two hundred fifty nanograms of pCAGGS carrying BST-2–Flag or ORF7a-HA and/or a control plasmid carrying Mission pLKO.1-puro nonmammalian shRNA (Sigma-Aldrich, St. Louis, MO) was transfected. At 24 h posttransfection, cells were fixed with 4% formaldehyde overnight at 4°C and then incubated in cold phosphate-buffered saline (PBS) for 10 min at room temperature. Each sample was permeabilized with permeabilization buffer (PBS, 0.1% Triton X-100, 0.5% bovine serum albumin [BSA]) for 15 min at room temperature and then blocked for 5 min using blocking buffer (PBS, 5% BSA). The cells were washed using wash buffer (PBS, 1% BSA, 0.05% NP-40) and then stained for protein expression. The primary antibodies used were rabbit anti-HA antibody (Sigma-Aldrich, St. Louis, MO) and mouse anti-Flag M2 antibody (Sigma-Aldrich, St. Louis, MO). The cells were incubated with primary antibodies diluted in antibody dilution buffer (PBS, 1% BSA, 0.05% NP-40, 2% normal goat serum) for 45 min at room temperature. The cells were washed three times with wash buffer and then incubated while rocking for 30 min at room temperature with goat anti-rabbit IgG conjugated with aminomethylcoumarin (AMCA) (Vector Laboratories, Burlingame, CA) and/or horse anti-mouse immunoglobulin conjugated with Texas Red (Vector Laboratories, Burlingame, CA). Cells were then washed with wash buffer 3 times and a final time with PBS for 30 min at room temperature. For the ORF7a localization experiments, the ER was stained with concanavalin A and Alexa Fluor 594 conjugate (Invitrogen, Carlsbad, CA) and the Golgi apparatus was stained with Bodipy TR ceramide complexed to BSA (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. The coverslips were then mounted on slides using Vectashield hard-set mounting medium with DAPI (4′,6-diamidino-2-phenylindole; Vector Laboratories, Burlingame, CA). The slides were analyzed by confocal microscopy using a Zeiss LSM 510 microscope. Images were collated and adjusted using ImageJ software (National Institute of Mental Health, Bethesda, MD).
Flow cytometry.
For experiments to determine BST-2 surface expression, HEK293T cells were transfected with BST-2 and ORF7a as described above. After 18 h of expression, the cells were washed with PBS and dissociated with 0.05% trypsin–EDTA (1×), phenol red (Invitrogen, Carlsbad, CA). The cells were washed in HEK293T medium to inactivate the trypsin, resuspended in fluorescence-activated cell sorting (FACS) buffer (PBS with 1% fetal bovine serum), and stained for 20 min with allophycocyanin (APC)-conjugated anti-human CD317 clone RS38E (BioLegend, San Diego, CA) or APC-conjugated mouse IgG1 (Becton Dickinson, Franklin Lakes, NJ) as a control. The cells were then washed, resuspended in FACS buffer, and analyzed on an LSRII flow cytometer (Becton Dickinson, Franklin Lakes, NJ). The data were analyzed using FlowJo software (TreeStar, Ashland, OR).
For experiments to determine mutant BST-2 surface expression, HEK293T cells were plated at 250,000 cells per well in 6-well plates, grown overnight at 37°C with 5% CO2, and then transfected with 1 μg of each DNA (empty vector, empty vector plus WT BST-2, empty vector plus mutant BST-2, empty vector plus ORF7a, WT BST-2 plus ORF7a, mutant BST-2 plus ORF7a). All the following steps were performed at room temperature. At 24 h after transfection, cells were harvested using cell dissociation buffer (Invitrogen). The contents of duplicate transfected wells were pooled, and samples were transferred to a 96-well plate. The cells were pelleted by centrifugation at 2,000 rpm for 2 min, fixed in 4% paraformaldehyde for 5 min, and then washed with 10% FBS in PBS and pelleted as described above. Samples were divided into 2 aliquots and then blocked with 10% FBS in PBS or blocked/permeabilized with 10% saponin in 10% FBS in PBS for 30 min. The cells were pelleted as described above and incubated in primary antibodies for 1 h (HA antibody [1:1,000; catalog number H6908; Sigma] and Flag antibody [1:1,000; catalog number F3165; Sigma]). Cells were washed 2 times with 10% FBS in PBS and pelleted as described above. Secondary antibodies (fluorescein isothiocyanate-conjugated anti-rabbit immunoglobulin [1:1,000; Vector Laboratories] and Alexa Fluor 405-conjugated anti-mouse immunoglobulin [1:1,000; Thermo Fisher]) were added to the cells, and the mixture was incubated for 1 h, followed by washing and pelleting as described above. Cells were resuspended in PBS, and cell surface localization as well as the total cell expression of BST-2 and ORF7a was determined using an LSRII flow cytometer. Compensation controls consisted of cells transfected with no DNA and cells transfected with BST2 or ORF7a alone. Data were analyzed using FlowJo software, and statistical analysis was performed by a t test, with standard errors being based on the results from three individual experiments.
CD of BST-2 and ORF7a-Fc.
The circular dichroism (CD) spectra of 10 μM BST-2 expressed in HEK293T cells, 12 μM BST-2 expressed in Escherichia coli cells, and 8 μM ORF7a-Fc expressed in HEK239T cells were recorded at wavelengths ranging from 200 to 260 nm in 10 mM sodium phosphate buffer (pH 7.5) using a Jasco J-810 instrument. CD melting curves were analyzed at 222 nm by increasing the temperature by 1°C/min starting at 20°C.
SPR analysis.
Twenty-one micrograms of pCAGGS-T7/ORF7a-Fc was transfected into HEK293T cells seeded in 100-mm dishes, using Lipofectamine LTX (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. After 48 h of expression, the supernatant was collected and purified using a HiTrap protein A column (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom) according to the manufacturer's instructions. Purified ORF7a-Fc was subsequently dialyzed into PBS. The codon-optimized sequence of the extracellular domain (residues 47 to 161) of BST-2 with an N-terminal His6 tag and a C-terminal Flag tag was cloned into pET28b. The protein was expressed at 19°C overnight in BL21(DE3)pLysS cells induced at an optical density at 600 nm of 0.6 with 0.4 mM isopropyl-β-d-thiogalactopyranoside. The fusion protein was purified by nickel affinity chromatography (Thermo Scientific, Pittsburg, PA) using a Mono Q 5/50GL anion-exchange column (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom) and finally separated on a Superdex 200 10/300 GL gel filtration column (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom). The extracellular domain (residues 47 to 161) of BST-2 was also cloned into the plGplus vector (R&D Systems, Minneapolis, MN). The resulting BST-2 protein includes a C-terminal His6-Flag tag and, due to the stop codon carried by the protein, was not expressed as an Fc fusion protein. HEK293T cells in suspension culture were transfected with this construct using polyethylenimine (Polysciences, Inc., Warrington, PA). The cell culture supernatant was harvested at 96 h posttransfection and purified using nickel affinity chromatography (Thermo Scientific, Pittsburg, PA), followed by gel filtration on a Superdex 200 10/300 GL column (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom). Direct binding of the extracellular domain of ORF7a-Fc expressed in HEK293T cells to the extracellular domain of BST-2 expressed in E. coli or HEK293T cells was measured by surface plasmon resonance (SPR) analysis using a Biacore T100 instrument (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom). Protein A (1,000 resonance units [RU]) was immobilized by amine coupling on the surface of a CM5 sensor chip. Approximately 170 RU of human IgG (Sigma-Aldrich, St. Louis, MO) as a negative control and also 170 RU of ORF7a-Fc were captured on flow cells 1 and 2, respectively. In single-cycle kinetics experiments, 2-fold dilutions from 80 to 5 μM BST-2 were injected over the surfaces, and the response was recorded after subtraction of the response of the control. HBS-EP was used as a running buffer, and the surfaces were regenerated with 20 mM HCl after each cycle. Steady-state analysis of the data was performed using Biacore T100 evaluation software (version 2.0.3). All of the SPR experiments were repeated at least three times.
Statistical analysis.
Growth curve titers were analyzed by t test using the Holm-Sidak method, with alpha being equal to 5.0%. Prism software (GraphPad Software Inc., La Jolla, CA) was used to perform the analysis.
RESULTS
SARS-CoV proteins antagonize BST-2 expression in vitro.
The genomes of many enveloped viruses, including hCoV-229E, encode proteins that counteract BST-2 (24, 27, 37). We hypothesized that the highly pathogenic SARS-CoV may also inhibit BST-2 function. To investigate this hypothesis, HA- and GFP-tagged SARS-CoV proteins and BST-2 were cotransfected into HEK293T cells and BST-2 expression levels were assessed by Western blotting. Four SARS-CoV proteins, nonstructural protein 1 (nsp1), the papain-like protease (PLPro) domain of nsp3, ORF6, and ORF7a altered BST-2 protein expression or the BST-2 molecular mass. Several proteins encoded by the SARS-CoV genome have been shown to alter other antiviral response pathways during infection (6–8, 43–50). Three of the proteins, PLPro, ORF6, and nsp1, have previously been shown to be interferon antagonists (7, 8, 45). PLPro inhibits interferon regulatory factor 3 (IRF3) and NF-κB activation (45), ORF6 blocks STAT1 nuclear import, and nsp1 blocks beta interferon induction by degrading host mRNAs (6, 7, 45, 48). Because the function of ORF7a is unclear, we decided to further study the interactions between BST-2 and ORF7a.
icSARS-ORF7abΔ-CoV shows defects in replication compared to icSARS-CoV replication when BST-2 is overexpressed.
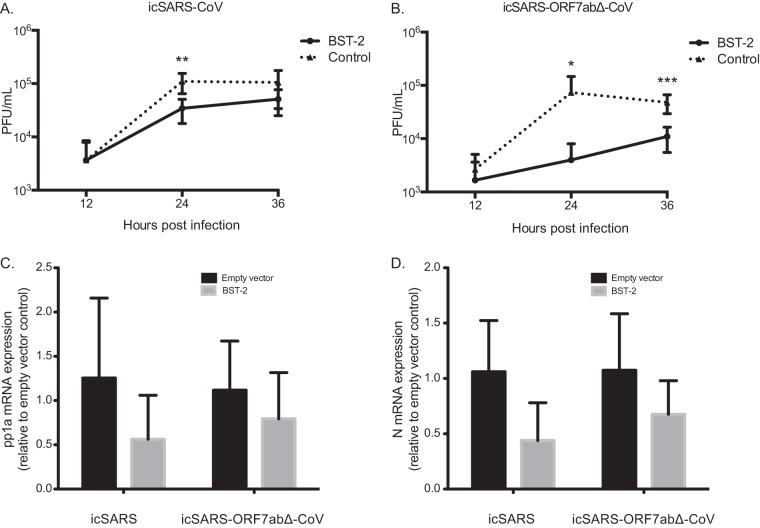
Since ORF7a affects the BST-2 protein, we hypothesized that SARS-CoV with an ORF7ab deletion (icSARS-ORF7abΔ-CoV) would show a greater defect in replication than WT SARS-CoV when BST-2 is overexpressed. Since ORF7a and ORF7b have overlapping open reading frames, a virus with the ORF7ab double deletion was used for the infection experiments. No effect on BST-2 was found in transfection screens when ORF7b was expressed alone in the assays. We transfected HEK293T/ACE2 cells with BST-2 or a control plasmid, and at 24 h posttransfection we infected the cells with either icSARS-CoV or icSARS-ORF7abΔ-CoV at an MOI of 0.1. HEK293T cells do not express endogenous BST-2 (27), so we were able to ensure that any effect was from the transfected BST-2 and not endogenous BST-2 expression. icSARS-CoV replicated to 1.10 × 105 PFU/ml, while in BST-2-expressing cells, icSARS-CoV replicated to significantly lower titers of 3.43 × 104 PFU/ml (Fig. 1A; P < 0.01). icSARS-ORF7abΔ-CoV was also significantly restricted by BST-2 expression at 24 and 36 h. In control transfected cells, icSARS-ORF7abΔ-CoV replicated to 7.37 × 104 PFU/ml and 4.80 × 104 PFU/ml at 24 and 36 h, respectively, while in BST-2-transfected cells, icSARS-ORF7abΔ-CoV replicated to significantly lower titers of 4.00 × 103 PFU/ml (P < 0.05) and 1.10 × 104 PFU/ml (P < 0.001) at 24 and 36 h, respectively (Fig. 1B). While BST-2 restricts SARS-CoV by a small but significant amount, BST-2 restricts icSARS-ORF7abΔ-CoV by a much larger amount, suggesting that ORF7a antagonizes BST-2.
FIG 1.
icSARS-CoV and icSARS-ORF7abΔ-CoV infection of cells with and without BST-2 expression. (A and B) HEK293T/ACE2 cells were transfected with pCAGGS carrying BST-2–Flag or a control plasmid. At 24 h posttransfection, HEK293T/ACE2 cells were infected with icSARS-CoV (A) or icSARS-ORF7abΔ-CoV (B) at an MOI of 0.1. Supernatant and cell lysate were taken at 12, 24, and 36 h postinfection. The virus titer in supernatants taken at 12, 24, and 36 h was determined. (C and D) RNA extracted from icSARS-CoV- and icSARS-ORF7abΔ-CoV-infected HEK293T/ACE2 cells was analyzed by real-time PCR for genomic RNA (C) or leader-containing N mRNA (D) levels as a signature of replicating virus. *, significant at a P value of <0.05; **, significant at a P value of <0.01; ***, significant at a P value of <0.001. The data shown are representative of those from two independent experiments.
We confirmed that BST-2 does not affect another step in the SARS-CoV replication cycle by assessing the accumulation of SARS-CoV RNA products of replication in the presence of BST-2 at 24 h postinfection. There was no significant effect of BST-2 expression on SARS-CoV pp1a mRNA (Fig. 1C) or SARS-CoV N mRNA (Fig. 1D), regardless of ORF7a expression. These data suggest that BST-2 does not affect SARS-CoV RNA accumulation, even in the absence of ORF7a expression.
The defect in icSARS-ORF7abΔ-CoV replication is due to direct tethering of SARS-CoV virions to the plasma membrane.
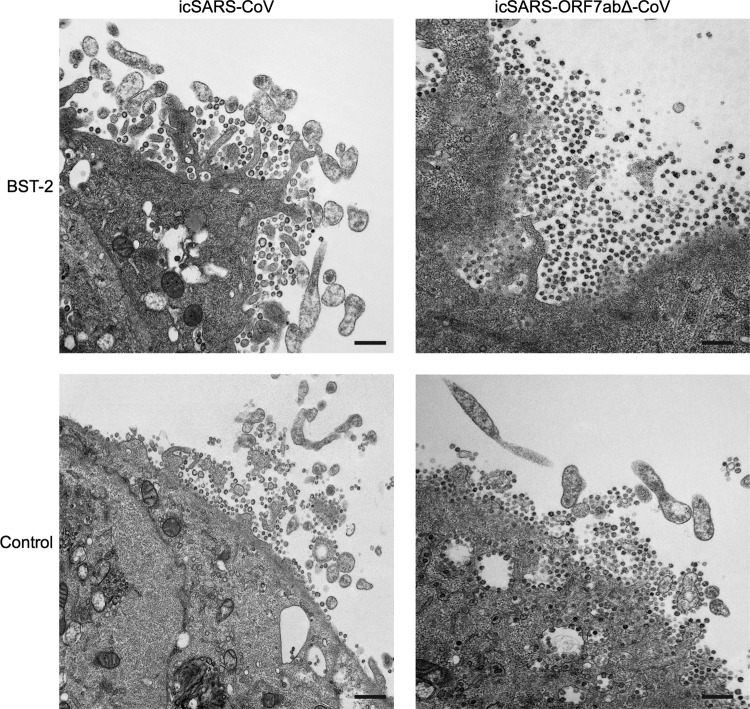
Since BST-2 has been shown to restrict virus replication by directly tethering HIV-1 virions to the plasma membrane (27, 51), we sought to determine if overexpression of BST-2 was preventing the release of SARS-CoV and if icSARS-ORF7abΔ-CoV was more susceptible to BST-2 restriction. To determine if BST-2 was affecting release, we transfected either BST-2 or a control plasmid into Vero E6 cells and subsequently infected the cells with either icSARS-CoV or icSARS-ORF7abΔ-CoV at an MOI of 10. At 24 h postinfection, the cells were fixed and imaged using electron microscopy. When the Vero E6 cells were transfected with the control plasmid, the minimal accumulation of both icSARS-CoV and icSARS-ORF7abΔ-CoV virions was found at the plasma membrane (Fig. 2). Transfection of BST-2 led to the accumulation of a small amount of icSARS-CoV on the plasma membrane (Fig. 2, top left). BST-2 transfection showed a much greater effect on icSARS-ORF7abΔ-CoV, with a large amount of virions accumulating at the plasma membrane (Fig. 2, top right). These results confirm that, as in many other viruses, BST-2 restricts SARS-CoV by preventing virus release. The increased effect of BST-2 on icSARS-ORF7abΔ-CoV further suggests that ORF7a acts as an inhibitor of BST-2-mediated restriction of SARS-CoV.
FIG 2.
BST-2 tethers SARS-CoV to the plasma membrane. BST-2 or control plasmid was transfected into Vero E6 cells and infected with either icSARS-CoV or icSARS-ORF7abΔ-CoV at an MOI of 10. At 24 h postinfection, cells were fixed and imaged by electron microscopy. Transfection of BST-2 results in a large increase in the amount of icSARS-ORF7abΔ-CoV virions retained at the surface compared to that for control transfected cells. Bars, 500 nm.
ORF7a expression leads to lower-molecular-mass BST-2 within the cells but not reduced BST-2 surface expression.
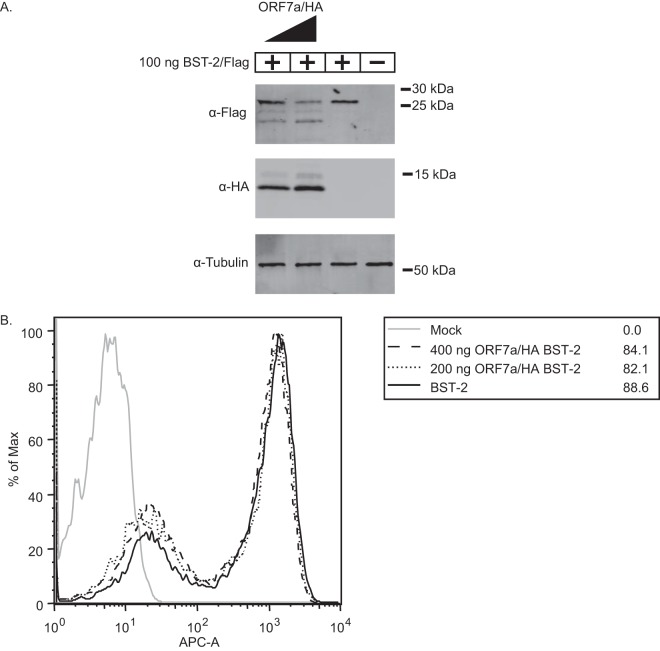
Since ORF7a appears to be a BST-2 antagonist, we aimed to determine if SARS-CoV ORF7a causes BST-2 surface removal and subsequent degradation, as seen in HIV-1 Vpu protein antagonism (33, 51). SARS-CoV ORF7a was cotransfected with increasing amounts of BST-2 to assay the effect of ORF7a on BST-2 expression. Increasing the amount of ORF7a cotransfected with BST-2 led to decreased levels of BST-2 expression and lower-molecular-mass products, suggesting that the BST-2 protein is affected by ORF7a expression (Fig. 3A). Next, we sought to determine if, like HIV-1 Vpu, the expression of ORF7a leads to a reduction in BST-2 surface expression (51). To assay the effect of ORF7a on BST-2 surface expression, we transfected BST-2 either alone or in combination with an ORF7a expression plasmid to compare BST-2 surface expression by flow cytometry. Untransfected cells exhibited little to no expression of surface BST-2. Cells transfected with BST-2 alone were 88.2% positive for surface BST-2 expression, with the majority of cells being in a population expressing larger amounts and a smaller percentage being in a population expressing smaller amounts (Fig. 3B). Interestingly, increasing amounts of ORF7a had no effect on the surface expression of BST-2 (Fig. 3B). These data demonstrate that ORF7a coexpression leads to lower-molecular-mass BST-2 within cells but does not lead to the removal of BST-2 from the surface, suggesting that ORF7a may antagonize BST-2 through a mechanism other than removal from the surface.
FIG 3.
ORF7a expression leads to lower-molecular-mass BST-2 within the cells but not reduced BST-2 surface expression. BST-2 was transfected into HEK293T cells with increasing amounts of ORF7a. At 18 h posttransfection, the cells were either lysed and analyzed by Western blotting or stained with an anti-BST-2 antibody conjugated to APC (APC-A) and analyzed using flow cytometry. Untransfected cells did not express BST-2. Cells transfected with BST-2 showed high levels of BST-2 expression, and cotransfection of ORF7a and BST-2 led to decreased levels of the high-molecular-mass BST-2 band and increasing levels of a lower-molecular-mass BST-2 band in a dose-dependent manner (A) but did not lead to reduced surface expression of BST-2 (B). The data shown are representative of those from two independent experiments.
Lysosomal and proteasomal inhibitors do not affect BST-2 antagonism by ORF7a.
While we did not observe the ORF7a-dependent removal of surface BST-2, we did observe the appearance of lower-molecular-mass bands of BST-2, suggesting degradation of intracellular BST-2. Many other viruses, such as HIV-1, antagonize BST-2 by degradation through either the lysosome or the proteasome, and thus, we assessed whether lysosomal or proteasomal inhibitors could block BST-2 antagonism by ORF7a (32–34). First, to demonstrate that concanamycin A (ConA) and MG-132 inhibit proteasome and lysosomal degradation, respectively, at the concentrations used in HEK293T cells, we treated cells and assayed for ubiquitin and LC3B levels by Western blotting (Fig. 4A and B). As expected, MG-132 treatment increased total ubiquitin levels in the cell (Fig. 4A) and ConA treatment blocked lysosomal degradation, as shown by an increase in the lower-molecular-mass LC3B product. To test for the effect of proteosomal or lysosomal effects on BST-2 antagonism, we transfected HEK293T cells with plasmids carrying BST-2 and ORF7a or a control plasmid and at 4 h posttransfection replaced the medium with medium containing either 20 nM concanamycin A (to inhibit lysosomal degradation) or 500 nM MG-132 (to inhibit proteasome function). At 18 h posttransfection, the cells were lysed and analyzed by Western blotting to determine if BST-2 was degraded. After treatment, lower-molecular-mass bands were still observed. Treatment with neither concanamycin A nor MG-132 blocked the ability of ORF7a to antagonize BST-2 (Fig. 4C). The 2 lanes on the far right in Fig. 4C contain a background band with a molecular mass similar to that of HA-tagged ORF7a that did not affect the experiment. These data demonstrate that the appearance of lower-molecular-mass bands of BST-2 is not due to lysosomal or proteasomal degradation and suggest that ORF7a antagonizes BST-2 through an alternative mechanism.
FIG 4.
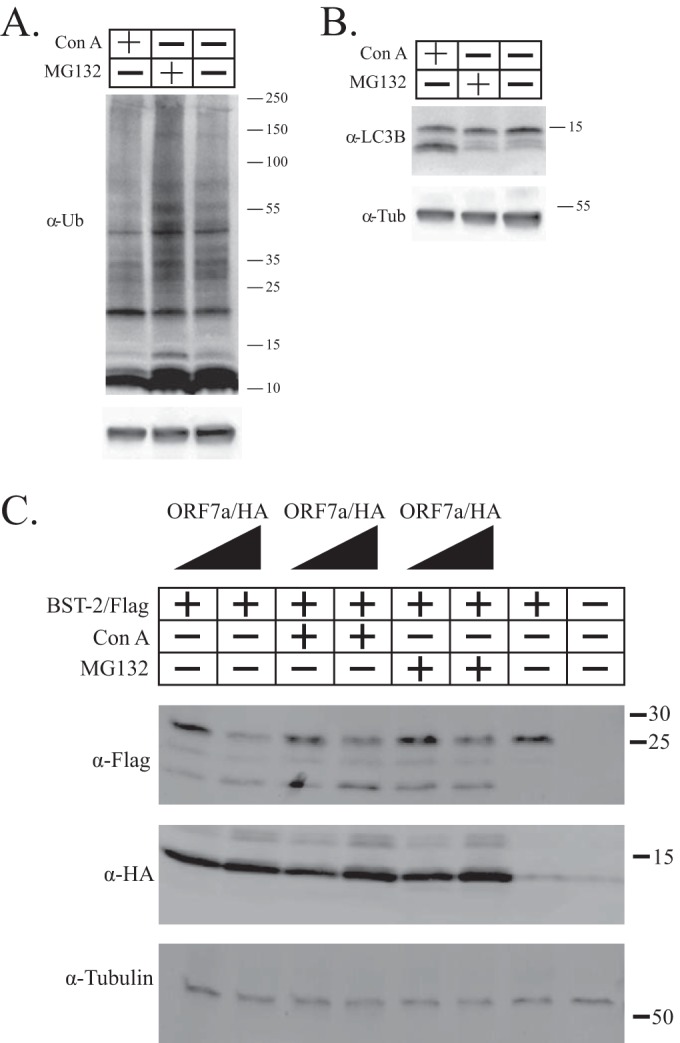
Effects of proteasomal and lysosomal inhibitors on ORF7a antagonism of BST-2. (A and B) HEK293T cells were treated with either ConA or MG-132 to demonstrate that the concentrations used are inhibitory to proteasomal or lysosomal degradation, respectively. The lysate was analyzed by Western blotting with antibodies against ubiquitin (α-Ub) (A) and LC3B (B) to demonstrate the efficacies of the compounds. (C) Increasing amounts of ORF7a were transfected into HEK293T cells. The cells were subsequently treated with the lysosome inhibitor concanamycin A or the proteasome inhibitor MG-132. Neither inhibitor prevented BST-2 antagonism by ORF7a. The data shown are representative of those from three independent experiments. The numbers next to the gels in panels A to C are molecular masses (in kilodaltons).
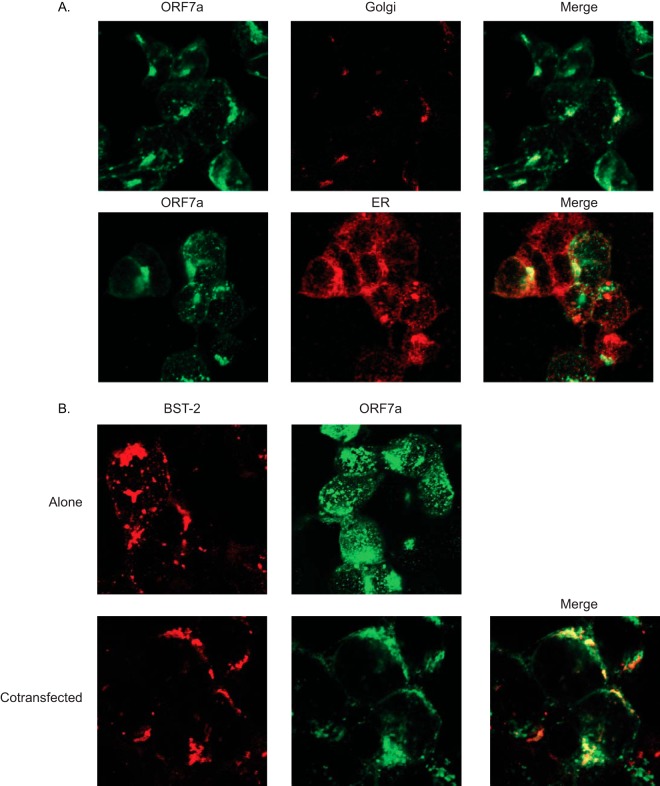
BST-2 colocalizes with and alters the localization of SARS-CoV ORF7.
Since icSARS-ORF7abΔ-CoV is more susceptible to BST-2 restriction and ORF7a appears to cause the appearance of a low-molecular-mass BST-2 band, we hypothesized that BST-2 may alter the ORF7a localization within the cell. ORF7a was transfected into HEK293T cells, and the cells were stained for ORF7a as well as the ER and Golgi apparatus (9, 10). ORF7a normally localizes to the Golgi apparatus and was also detectable in the ER, as would be expected for a protein that passes through the ER to the Golgi apparatus (Fig. 5A). To determine if BST-2 and ORF7a colocalize, we performed confocal microscopy. When transfected alone, ORF7a localized primarily to the Golgi apparatus, whereas BST-2 localized to the plasma membrane (Fig. 5B). When BST-2 and ORF7a were cotransfected, ORF7a appeared to localize to the plasma membrane, coincident with BST-2 (Fig. 5B). These data suggest that BST-2 and ORF7a may be interacting in cells.
FIG 5.
ORF7a colocalizes with BST-2. HEK293T cells were transfected with BST-2, ORF7a, or both. (A) BST-2 was stained with mouse anti-Flag primary antibody and goat anti-mouse Texas Red-conjugated secondary antibody. ORF7a was stained with rabbit anti-HA primary antibody and horse anti-rabbit AMCA secondary antibody. Localization was analyzed by confocal microscopy (merged images are yellow). (B) When BST-2 and ORF7a are cotransfected, the two proteins display an overlapping signal, as seen in yellow in the merged image.
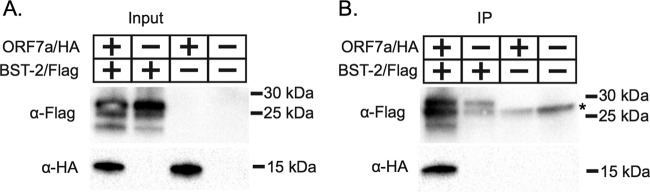
SARS-CoV ORF7a coimmunoprecipitates with BST-2.
Having shown that ORF7a both alters protein mobility and localizes to the plasma membrane when coexpressed with BST-2, we sought to determine if there is a molecular interaction between the two proteins. We cotransfected BST-2 and ORF7a into HEK293T cells, and at 18 h posttransfection the cells were lysed. We immunoprecipitated proteins from the transfected cells and performed immunoblotting for both BST-2 and ORF7a. We found that BST-2 and ORF7a were present in both the input and the coimmunoprecipitate (Fig. 6), suggesting an interaction between BST-2 and ORF7a either directly or within a larger multicomponent complex.
FIG 6.
ORF7a coimmunoprecipitates with BST-2. (A) HEK293T cells were sham transfected or transfected with ORF7a-HA and BST-2–Flag separately or together. After expression for 18 h, cells were lysed and analyzed by Western blotting for expression. (B) BST-2 was immunoprecipitated (IP) with anti-Flag beads. Bound protein was eluted and analyzed by Western blotting. BST-2 was detected with a mouse anti-Flag M2 antibody. ORF7a was detected with rabbit anti-HA antibodies. ORF7a was detected in the eluate from the coimmunoprecipitation, suggesting an interaction between ORF7a and BST-2. The data shown are representative of those from two independent experiments. *, a nonspecific band.
The direct interaction between ORF7a and BST-2 is regulated by BST-2 glycosylation.
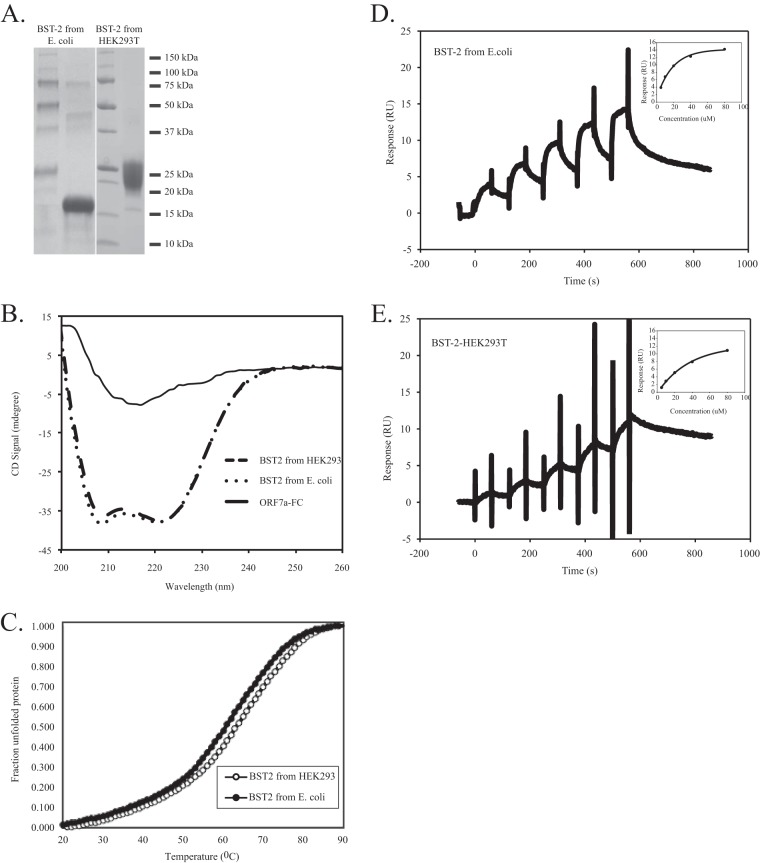
To assess whether the extracellular domain of ORF7a interacts directly with the extracellular domain of BST-2, we performed surface plasmon resonance (SPR) analysis of ORF7a–BST-2 binding. SPR analysis allows direct quantitation of protein-protein interactions by measuring the affinity between two proteins. One protein is immobilized on a sensor chip, and the other is flowed over the sensor chip in increasing concentrations. Binding of proteins causes changes in refraction, which is detected and recorded as the number of resonance units (RU). Affinity can then be calculated from changes in the numbers of RU (52). ORF7a with an Fc fusion tag (ORF7a-Fc) was expressed and purified from HEK293T cells, and BST-2 was expressed and purified from both E. coli and HEK293T cells (Fig. 7A). The CD spectra of BST-2 expressed in both HEK239T and E. coli cells revealed the expected pattern for a protein primarily containing α-helical folds (Fig. 7B). ORF7a-Fc, in contrast, showed a spectrum typical of proteins formed dominantly by β sheets (Fig. 7B). Melting temperatures were deduced from the melting curves (Fig. 7C), and tetrameric BST-2 expressed in E. coli cells had a slightly lower melting temperature of 61.95°C than dimeric BST-2 expressed in HEK293T cells, which had a melting temperature of 65.3°C (Fig. 7C). These data suggest that both BST-2 and ORF7a-Fc are folded correctly, and therefore, they were used for the SPR analysis.
FIG 7.
Binding of ORF7a to BST-2. (A) Purified BST-2 expressed in E. coli and HEK293T cells was stained with Coomassie blue. The BST-2 expressed in E. coli has a lower molecular mass than the BST-2 expressed in HEK293T cells due to a lack of glycosylation. (B and C) CD spectra (B) and melting curves (C) of BST-2 and ORF7a-Fc. The CD spectra of 10 μM BST-2 expressed in HEK293T cells, 12 μM BST-2 expressed in E. coli cells, and 8 μM ORF7a-Fc expressed in HEK239T cells are shown. (D and E) Representative sensograms obtained in SPR experiments analyzing the direct interaction of ORF7a-Fc with unglycosylated BST-2 expressed in E. coli (D) and glycosylated BST-2 expressed in HEK293T cells (E) are shown. For SPR experiments, ORF7a-Fc was captured via protein A immobilized on a CM5 chip. Single-cycle kinetics were performed by injection of 5 μM, 10 μM, 20 μM, 40 μM, and 80 μM BST-2. The data shown are representative of those from three independent experiments.
By SPR analysis, we observed that unglycosylated BST-2 expressed in E. coli was able to bind to ORF7a-Fc with an affinity (equilibrium dissociation constant [KD]) of 10 μM (Fig. 7D). Binding of glycosylated BST-2 expressed in HEK293T cells, though, exhibited markedly weaker responses in identical SPR experiments, in which responses did not reach equilibrium and, therefore, did not allow us to quantify an accurate KD for this interaction (Fig. 7E). We did attempt to fit the data to estimate the KD for this interaction, and the binding of ORF7a to glycosylated BST-2 was at least 4 times weaker than that to unglycosylated BST-2. These data indicate that ORF7a binds directly to unglycosylated BST-2 with an affinity at micromolar concentrations and that the presence of N-linked glycosylation at positions 65 and 92 of BST-2 significantly weakens this interaction.
ORF7a expression interferes with BST-2 glycosylation.
Given that ORF7a-dependent BST-2 antagonism is unaffected by lysosomal or proteasomal inhibitors and that ORF7a binds unglycosylated BST-2 with a substantially higher affinity than glycosylated BST-2, we hypothesized that ORF7a may bind to BST-2 before it is glycosylated in the ER and interfere with the glycosylation of BST-2. To determine if ORF7a interferes with glycosylation, we transfected HEK293T cells with increasing amounts of ORF7a. Cotransfection of increasing amounts of ORF7a led to lower-molecular-mass bands of BST-2 in a dose-dependent manner (Fig. 8). To confirm that the lower-molecular-mass bands were unglycosylated, we treated the lysate from cells expressing BST-2 with glycopeptidase F. Previous studies have shown that treatment with glycopeptidase F removes all the glycosylation from BST-2 (30). The BST-2 lysate treated with glycopeptidase F showed a shift to a lower molecular mass, and its size was identical to that of the lower-molecular-mass band present when BST-2 was cotransfected with ORF7a (Fig. 8A). To further confirm that cotransfection of ORF7a leads to decreased levels of unglycosylated BST-2, we measured the density of each band and calculated the ratio of glycosylated to unglycosylated BST-2. As the levels of ORF7a increase, the levels of glycosylated BST-2 decrease (Fig. 8B). These data suggest that ORF7a interferes with the glycosylation of BST-2.
FIG 8.
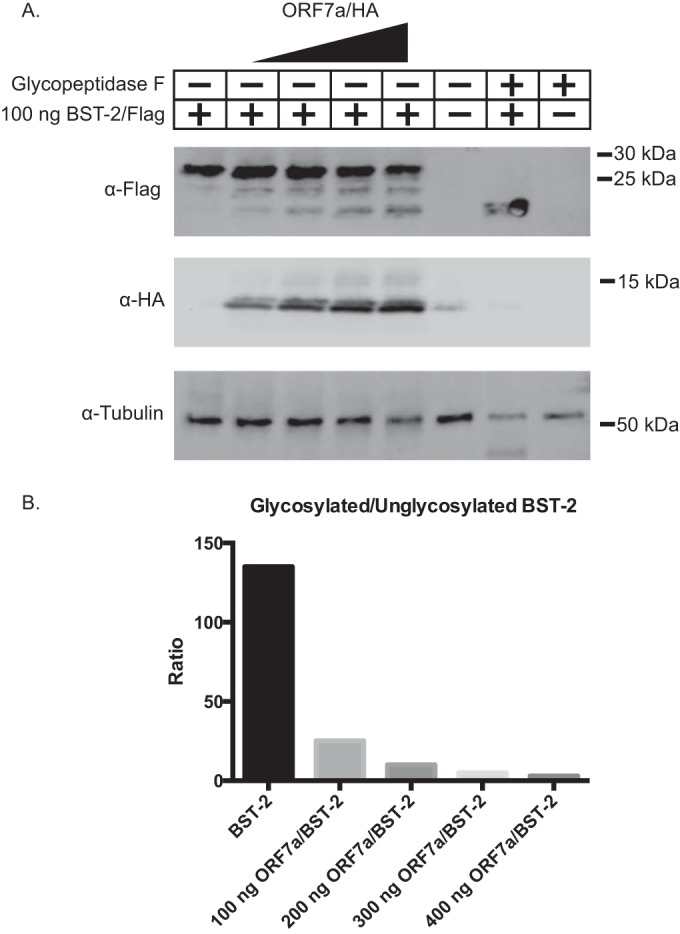
ORF7a interferes with the glycosylation of BST-2. BST-2 was transfected into HEK293T cells with increasing amounts of ORF7a. At 18 h posttransfection, cells were lysed and analyzed by Western blotting. (A) Increasing levels of ORF7a lead to increased levels of a lower-molecular-mass band of BST-2, which we hypothesized to be unglycosylated BST-2. To confirm that the lower-molecular-mass band was unglycosylated BST-2, we treated the lysate from BST-2-transfected cells with glycopeptidase F, which deglycosylates proteins. When treated with glycopeptidase F, the molecular mass of BST-2 decreases to that of the lower-molecular-mass band of BST-2 when it is cotransfected with ORF7, suggesting that ORF7a leads to increased levels of unglycosylated BST-2. (B) The density of each band was measured, and the ratio of glycosylated to unglycosylated BST-2 was calculated and graphed. The data shown are representative of those from three independent experiments.
Unglycosylated BST-2 no longer restricts icSARS-ORF7abΔ-CoV release.
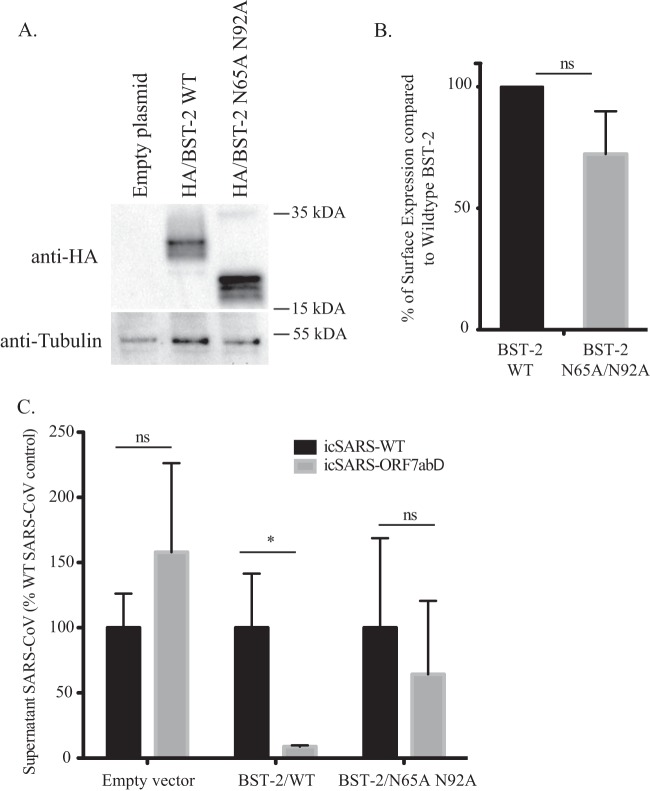
To confirm that the glycosylation of BST-2 is necessary for restriction of icSARS-ORF7abΔ-CoV, we transfected HEK293T/ACE2 cells with a mutant of BST-2 (called BST-2 N65A/N92A) that does not undergo N-linked glycosylation (29). We confirmed the expression of the N65A/N92A mutant BST-2 by Western blotting, where we observed expression only of the expected 19-kDa unglycosylated form of BST-2 (Fig. 9A). We then confirmed that the N65A/N92A mutant was still able to localize to the cell surface by quantifying the amount of WT and mutant BST-2 on nonpermeabilized cells by flow cytometry. Surface labeling showed that mutant BST-2 expression on the plasma membrane was not significantly different from WT BST-2 surface expression (Fig. 9B). We transfected cells with plasmids carrying WT and N65A/N92A mutant BST-2 and then infected those cells with icSARS-CoV or icSARS-ORF7abΔ-CoV. In these experiments, the plasmid carrying BST-2 has an HA tag inserted at amino acid 463, so we first confirmed that HA–BST-2 is still able to significantly (10-fold, P = 0.0046) restrict icSARS-ORF7abΔ-CoV compared to its level of restriction of icSARS-CoV Urbani (Fig. 9C). However, there was no significant difference in the levels of icSARS-CoV Urbani and icSARS-ORF7abΔ-CoV in the supernatants of cells transfected with N65A/N92A mutant BST-2 (Fig. 9C; P = 0.274), suggesting that N-linked glycosylation is required for the BST-2-mediated restriction of icSARS-ORF7abΔ-CoV.
FIG 9.
Unglycosylated BST-2 fails to inhibit SARS-CoV egress. HEK293T ACE2 cells were transfected with either a control plasmid or a plasmid carrying wild-type HA-tagged BST-2 or a mutant HA-tagged BST-2 containing the N65A and N92A mutations. (A) The expression levels of wild-type BST-2 and N65A/N92A BST-2 were analyzed by Western blotting with anti-HA antibody and anti-tubulin antibody as a loading control. The BST-2 N65A/N92A mutant runs noticeably more slowly due to its loss of glycosylation. (B) HEK293T ACE2 cells were transfected with each plasmid, and the levels of the BST-2 protein on the surface of the cells were analyzed by flow cytometry with an anti-HA antibody. The percentage of surface expression of WT BST-2-transfected cells displaying surface localization of the B65A/N92A mutant BST-2 is graphed. (C) HEK293T ACE2 cells were transfected with each plasmid and infected with either icSARS-CoV or icSARS-ORF7abΔ-CoV. Cell supernatants were analyzed by plaque assay, and the amount of icSARS-CoV released is graphed as the percentage of the wild-type icSARS-CoV released. Notice the loss of inhibition of icSARS-ORF7abΔ-CoV release in the mutant BST-2-transfected cells compared to the inhibition of icSARS-ORF7abΔ-CoV release in wild-type BST-2-transfected cells. *, P < 0.005; ns, not significant.
DISCUSSION
Our studies further expand the role of BST-2 in the restriction of enveloped viruses. We screened selected genes from the SARS-CoV genome and identified four potential BST-2 modulators, of which one was SARS-CoV ORF7a. While ORF7a has been shown to induce apoptosis, a definitive role for ORF7a during infection has not been established (13–15). Through overexpression, infection, and transfection studies, we demonstrate that BST-2 blocks the release of SARS-CoV virions, that ORF7a overcomes this inhibition, and that viruses in which ORF7ab is deleted display increased sensitivity to BST-2. Importantly, the inhibition of BST-2 is not by protein degradation but is by inhibition of its activity through the inhibition of glycosylation at two key sites on the protein that are required for its antiviral function. We demonstrate that a BST-2 mutant protein in which the two glycosylation sites are removed still traffics to the plasma membrane but is unable to inhibit SARS-CoV release. Our data also demonstrate that, unlike HIV-1 Vpu, which removes the BST-2 protein from the surface and induces degradation (32, 33, 51), SARS-CoV ORF7a does not remove BST-2 from the plasma membrane.
We have confirmed the interaction of ORF7a and BST-2 using multiple assays, including immunoprecipitation, colocalization, and surface plasmon resonance assays, which showed that ORF7a directly binds to unglycosylated BST-2 but not to glycosylated BST-2. Previous studies have suggested that the glycosylation of BST-2 is required for BST-2 antiviral activity (29) and the amino acid residues surrounding the N-linked glycosylation sites are evolutionarily conserved in BST-2, suggesting that these amino acids may be important for BST-2 function (22). We further demonstrated that N-linked glycosylation is required for the restriction of SARS-CoV lacking ORF7a, suggesting that the blocking of glycosylation by ORF7a is directly responsible for the antagonism of BST-2. BST-2 N-linked glycosylation has been proposed to effect the HIV-1 restriction activity of BST-2 (27, 29, 33, 51, 53); however, we have demonstrated for the first time that a virus encodes a BST-2 antagonist that inhibits BST-2 glycosylation, providing a potential mechanism for other putative viral BST-2 antagonists.
Taken together, the data suggest that ORF7a may function by binding to and preventing N-linked glycosylation of BST-2, preventing the tethering of SARS-CoV virions to the cytoplasmic membrane after they are released from the cell. We hypothesize that while BST-2 is trafficking through the ER and Golgi apparatus to the surface, ORF7a and BST-2 interact in the Golgi apparatus, where the extracellular domain of ORF7a binds the unglycosylated extracellular domain of BST-2 and either directly prevents glycosylation of BST-2 or binds to the evolutionarily conserved sites and as a side effect blocks N-linked glycosylation. SARS-CoV virions form in the ERGIC during virion maturation, and it has yet to be determined whether ORF7a or BST-2 is present in those compartments. BST-2 is potentially binding newly released SARS-CoV virions at the plasma membrane; however, most models of BST-2 function predict that BST-2 is inserted into the membrane as the virion forms (20), so we would predict that BST-2 first interacts with SARS-CoV virions in the ERGIC.
While the genomes of a variety of enveloped viruses encode BST-2 antagonists, those antagonists function by different mechanisms. Both HIV-1 Vpu and Kaposi's sarcoma-associated herpesvirus K5 ubiquitinate BST-2, leading to surface removal and subsequent lysosomal degradation (27, 51, 54). HIV-2 Env also removes BST-2 from the surface, but rather than being degraded, BST-2 is relocated to the trans-Golgi network and cannot function as a cytoplasmic membrane tether (36). SIV Env removes BST-2 from the surface through BST-2 internalization by endocytosis (38, 55). Ebola virus GP1,2 does not remove BST-2 from the surface but antagonizes BST-2 through an as yet unknown mechanism (39). The diverse mechanisms of known BST-2 antagonists demonstrate that viruses have independently evolved many different ways of antagonizing BST-2, an important restriction factor for any enveloped virus. It is possible that the genomes of all enveloped viruses encode BST-2 antagonists that act by a variety of mechanisms, but in most viruses these remain undiscovered.
In this study, we have identified BST-2 to be a potential inhibitor of SARS-CoV release. Our studies suggest that SARS-CoV ORF7a antagonizes the function of BST-2 by interfering with its N-linked glycosylation while binding it in the Golgi apparatus and then trafficking with it from the Golgi apparatus to the plasma membrane. From this we predict that therapeutics designed to inhibit the interaction between BST-2 and ORF7a may inhibit virus growth in vitro and in vivo.
ACKNOWLEDGMENTS
This work was supported by the Division of Intramural Research of the National Institute of Allergy and Infectious Diseases (NIAID) with grant RO1AI1095569 (to M.B.F.) and by NIAID grant R01AI087452 (to E.J.S.).
REFERENCES
- 1.World Health Organization. 2003. WHO summary of probable SARS cases with onset of illness from 1 November 2002 to 31 July 2003. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 2.Drosten C, Preiser W, Günther S, Schmitz H, Doerr HW. 2003. Severe acute respiratory syndrome: identification of the etiological agent. Trends Mol Med 9:325–327. doi: 10.1016/S1471-4914(03)00133-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marra MA, Jones SJM, Astell CR, Holt RA, Brooks-Wilson A, Butterfield YSN, Khattra J, Asano JK, Barber SA, Chan SY, Cloutier A, Coughlin SM, Freeman D, Girn N, Griffith OL, Leach SR, Mayo M, McDonald H, Montgomery SB, Pandoh PK, Petrescu AS, Robertson AG, Schein JE, Siddiqui A, Smailus DE, Stott JM, Yang GS, Plummer F, Andonov A, Artsob H, Bastien N, Bernard K, Booth TF, Bowness D, Czub M, Drebot M, Fernando L, Flick R, Garbutt M, Gray M, Grolla A, Jones S, Feldmann H, Meyers A, Kabani A, Li Y, Normand S, Stroher U, Tipples GA, Tyler S, et al. 2003. The genome sequence of the SARS-associated coronavirus. Science 300:1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- 4.Li W, Moore MJ, Vasilieva N, Sui J, Wong SK, Berne MA, Somasundaran M, Sullivan JL, Luzuriaga K, Greenough TC, Choe H, Farzan M. 2003. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rota PA, Oberste MS, Monroe SS, Nix WA, Campagnoli R, Icenogle JP, Peñaranda S, Bankamp B, Maher K, Chen M-H, Tong S, Tamin A, Lowe L, Frace M, DeRisi JL, Chen Q, Wang D, Erdman DD, Peret TCT, Burns C, Ksiazek TG, Rollin PE, Sanchez A, Liffick S, Holloway B, Limor J, McCaustland K, Olsen-Rasmussen M, Fouchier R, Günther S, Osterhaus ADME, Drosten C, Pallansch MA, Anderson LJ, Bellini WJ. 2003. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- 6.Frieman M, Yount B, Heise M, Kopecky-Bromberg SA, Palese P, Baric RS. 2007. Severe acute respiratory syndrome coronavirus ORF6 antagonizes STAT1 function by sequestering nuclear import factors on the rough endoplasmic reticulum/Golgi membrane. J Virol 81:9812–9824. doi: 10.1128/JVI.01012-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kopecky-Bromberg SA, Martínez-Sobrido L, Frieman M, Baric RA, Palese P. 2007. Severe acute respiratory syndrome coronavirus open reading frame (ORF) 3b, ORF 6, and nucleocapsid proteins function as interferon antagonists. J Virol 81:548–557. doi: 10.1128/JVI.01782-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wathelet MG, Orr M, Frieman MB, Baric RS. 2007. Severe acute respiratory syndrome coronavirus evades antiviral signaling: role of nsp1 and rational design of an attenuated strain. J Virol 81:11620–11633. doi: 10.1128/JVI.00702-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nelson CA, Pekosz A, Lee CA, Diamond MS, Fremont DH. 2005. Structure and intracellular targeting of the SARS-coronavirus Orf7a accessory protein. Structure 13:75–85. doi: 10.1016/j.str.2004.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schaecher SR, Touchette E, Schriewer J, Buller M, Pekosz A. 2007. Severe acute respiratory syndrome coronavirus gene 7 products contribute to virus-induced apoptosis. J Virol 81:11054–11068. doi: 10.1128/JVI.01266-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frieman MB, Yount B, Sims AC, Deming DJ, Morrison TE, Sparks J, Denison M, Heise M, Baric RS. 2006. SARS coronavirus accessory ORFs encode luxury functions. Adv Exp Med Biol 581:149–152. doi: 10.1007/978-0-387-33012-9_26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yount B, Roberts RS, Sims AC, Deming D, Frieman MB, Sparks J, Denison MR, Davis N, Baric RS. 2005. Severe acute respiratory syndrome coronavirus group-specific open reading frames encode nonessential functions for replication in cell cultures and mice. J Virol 79:14909–14922. doi: 10.1128/JVI.79.23.14909-14922.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schaecher SR, Touchette E, Schriewer J, Buller RM, Pekosz A. 2007. Severe acute respiratory syndrome coronavirus gene 7 products contribute to virus-induced apoptosis. J Virol 81:11054–11068. doi: 10.1128/JVI.01266-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tan Y-J, Fielding BC, Goh P-Y, Shen S, Tan THP, Lim SG, Hong W. 2004. Overexpression of 7a, a protein specifically encoded by the severe acute respiratory syndrome coronavirus, induces apoptosis via a caspase-dependent pathway. J Virol 78:14043–14047. doi: 10.1128/JVI.78.24.14043-14047.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tan Y-X, Tan THP, Lee MJR, Tham P-Y, Gunalan V, Druce J, Birch C, Catton M, Fu NY, Yu VC, Tan Y-J. 2007. Induction of apoptosis by the severe acute respiratory syndrome coronavirus 7a protein is dependent on its interaction with the Bcl-XL protein. J Virol 81:6346–6355. doi: 10.1128/JVI.00090-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tang X, Li G, Vasilakis N, Zhang Y, Shi Z, Zhong Y, Wang L-F, Zhang S. 2009. Differential stepwise evolution of SARS coronavirus functional proteins in different host species. BMC Evol Biol 9:52. doi: 10.1186/1471-2148-9-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goto T, Kennel SJ, Abe M, Takishita M, Kosaka M, Solomon A, Saito S. 1994. A novel membrane antigen selectively expressed on terminally differentiated human B cells. Blood 84:1922–1930. [PubMed] [Google Scholar]
- 18.Ishikawa J, Kaisho T, Tomizawa H, Lee BO, Kobune Y, Inazawa J, Oritani K, Itoh M, Ochi T, Ishihara K, Hirano T. 1995. Molecular cloning and chromosomal mapping of a bone marrow stromal cell surface gene, BST2, that may be involved in pre-B-cell growth. Genomics 26:527–534. doi: 10.1016/0888-7543(95)80171-H. [DOI] [PubMed] [Google Scholar]
- 19.Blasius AL, Giurisato E, Cella M, Schreiber RD, Shaw AS, Colonna M. 2006. Bone marrow stromal cell antigen 2 is a specific marker of type I IFN-producing cells in the naive mouse, but a promiscuous cell surface antigen following IFN stimulation. J Immunol 177:3260–3265. doi: 10.4049/jimmunol.177.5.3260. [DOI] [PubMed] [Google Scholar]
- 20.Kupzig S, Korolchuk V, Rollason R, Sugden A, Wilde A, Banting G. 2003. Bst-2/HM1.24 is a raft-associated apical membrane protein with an unusual topology. Traffic 4:694–709. doi: 10.1034/j.1600-0854.2003.00129.x. [DOI] [PubMed] [Google Scholar]
- 21.Ohtomo T, Sugamata Y, Ozaki Y, Ono K, Yoshimura Y, Kawai S, Koishihara Y, Ozaki S, Kosaka M, Hirano T, Tsuchiya M. 1999. Molecular cloning and characterization of a surface antigen preferentially overexpressed on multiple myeloma cells. Biochem Biophys Res Commun 258:583–591. doi: 10.1006/bbrc.1999.0683. [DOI] [PubMed] [Google Scholar]
- 22.Swiecki M, Scheaffer SM, Allaire M, Fremont DH, Colonna M, Brett TJ. 2011. Structural and biophysical analysis of BST-2/tetherin ectodomains reveals an evolutionary conserved design to inhibit virus release. J Biol Chem 286:2987–2997. doi: 10.1074/jbc.M110.190538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones PH, Maric M, Madison MN, Maury W, Roller RJ, Okeoma CM. 2013. BST-2/tetherin-mediated restriction of chikungunya (CHIKV) VLP budding is counteracted by CHIKV non-structural protein 1 (nsP1). Virology 438:37–49. doi: 10.1016/j.virol.2013.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Radoshitzky SR, Dong L, Chi X, Clester JC, Retterer C, Spurgers K, Kuhn JH, Sandwick S, Ruthel G, Kota K, Boltz D, Warren T, Kranzusch PJ, Whelan SPJ, Bavari S. 2010. Infectious Lassa virus, but not filoviruses, is restricted by BST-2/tetherin. J Virol 84:10569–10580. doi: 10.1128/JVI.00103-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blondeau C, Pelchen-Matthews A, Mlcochova P, Marsh M, Milne RSB, Towers GJ. 2013. Tetherin restricts herpes simplex virus type 1 and is antagonised by glycoprotein M. J Virol 87:13124–13133. doi: 10.1128/JVI.02250-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bampi C, Rasga L, Roux L. 2013. Antagonism to human BST-2/tetherin by Sendai virus glycoproteins. J Gen Virol 94:1211–1219. doi: 10.1099/vir.0.051771-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neil SJD, Zang T, Bieniasz PD. 2008. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature 451:425–430. doi: 10.1038/nature06553. [DOI] [PubMed] [Google Scholar]
- 28.Fitzpatrick K, Skasko M, Deerinck TJ, Crum J, Ellisman MH, Guatelli J. 2010. Direct restriction of virus release and incorporation of the interferon-induced protein BST-2 into HIV-1 particles. PLoS Pathog 6:e1000701. doi: 10.1371/journal.ppat.1000701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perez-Caballero D, Zang T, Ebrahimi A, McNatt MW, Gregory DA, Johnson MC, Bieniasz PD. 2009. Tetherin inhibits HIV-1 release by directly tethering virions to cells. Cell 139:499–511. doi: 10.1016/j.cell.2009.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Andrew AJ, Miyagi E, Kao S, Strebel K. 2009. The formation of cysteine-linked dimers of BST-2/tetherin is important for inhibition of HIV-1 virus release but not for sensitivity to Vpu. Retrovirology 6:80. doi: 10.1186/1742-4690-6-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Galão RP, Le Tortorec A, Pickering S, Kueck T, Neil SJD. 2012. Innate sensing of HIV-1 assembly by tetherin induces NFκB-dependent proinflammatory responses. Cell Host Microbe 12:633–644. doi: 10.1016/j.chom.2012.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Douglas JL, Viswanathan K, McCarroll MN, Gustin JK, Früh K, Moses AV. 2009. Vpu directs the degradation of the human immunodeficiency virus restriction factor BST-2/tetherin via a βTrCP-dependent mechanism. J Virol 83:7931–7947. doi: 10.1128/JVI.00242-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mangeat B, Gers-Huber G, Lehmann M, Zufferey M, Luban J, Piguet V. 2009. HIV-1 Vpu neutralizes the antiviral factor tetherin/BST-2 by binding it and directing its beta-TrCP2-dependent degradation. PLoS Pathog 5:e1000574. doi: 10.1371/journal.ppat.1000574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mitchell RS, Katsura C, Skasko MA, Fitzpatrick K, Lau D, Ruiz A, Stephens EB, Margottin-Goguet F, Benarous R, Guatelli JC. 2009. Vpu antagonizes BST-2-mediated restriction of HIV-1 release via β-TrCP and endo-lysosomal trafficking. PLoS Pathog 5:e1000450. doi: 10.1371/journal.ppat.1000450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gupta RK, Mlcochova P, Pelchen-Matthews A, Petit SJ, Mattiuzzo G, Pillay D, Takeuchi Y, Marsh M, Towers GJ. 2009. Simian immunodeficiency virus envelope glycoprotein counteracts tetherin/BST-2/CD317 by intracellular sequestration. Proc Natl Acad Sci U S A 106:20889–20894. doi: 10.1073/pnas.0907075106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Le Tortorec A, Neil SJ. 2009. Antagonism to and intracellular sequestration of human tetherin by the human immunodeficiency virus type 2 envelope glycoprotein. J Virol 83:11966–11978. doi: 10.1128/JVI.01515-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang S-M, Huang K-J, Wang C-T. 2014. BST2/CD317 counteracts human coronavirus 229E productive infection by tethering virions at the cell surface. Virology 449:287–296. doi: 10.1016/j.virol.2013.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang F, Wilson SJ, Landford WC, Virgen B, Gregory D, Johnson MC, Munch J, Kirchhoff F, Bieniasz PD, Hatziioannou T. 2009. Nef proteins from simian immunodeficiency viruses are tetherin antagonists. Cell Host Microbe 6:54–67. doi: 10.1016/j.chom.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kaletsky RL, Francica JR, Agrawal-Gamse C, Bates P. 2009. Tetherin-mediated restriction of filovirus budding is antagonized by the Ebola glycoprotein. Proc Natl Acad Sci U S A 106:2886–2891. doi: 10.1073/pnas.0811014106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stertz S, Reichelt M, Spiegel M, Kuri T, Martínez-Sobrido L, García-Sastre A, Weber F, Kochs G. 2007. The intracellular sites of early replication and budding of SARS-coronavirus. Virology 361:304–315. doi: 10.1016/j.virol.2006.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yount B, Curtis KM, Fritz EA, Hensley LE, Jahrling PB, Prentice E, Denison MR, Geisbert TW, Baric RS. 2003. Reverse genetics with a full-length infectious cDNA of severe acute respiratory syndrome coronavirus. Proc Natl Acad Sci U S A 100:12995–13000. doi: 10.1073/pnas.1735582100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sims AC, Baric RS, Yount B, Burkett SE, Collins PL, Pickles RJ. 2005. Severe acute respiratory syndrome coronavirus infection of human ciliated airway epithelia: role of ciliated cells in viral spread in the conducting airways of the lungs. J Virol 79:15511–15524. doi: 10.1128/JVI.79.24.15511-15524.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pewe L, Zhou H, Netland J, Tangudu C, Olivares H, Shi L, Look D, Gallagher T, Perlman S. 2005. A severe acute respiratory syndrome-associated coronavirus-specific protein enhances virulence of an attenuated murine coronavirus. J Virol 79:11335–11342. doi: 10.1128/JVI.79.17.11335-11342.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tangudu C, Olivares H, Netland J, Perlman S, Gallagher T. 2007. Severe acute respiratory syndrome coronavirus protein 6 accelerates murine coronavirus infections. J Virol 81:1220–1229. doi: 10.1128/JVI.01515-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Frieman M, Ratia K, Johnston RE, Mesecar AD, Baric RS. 2009. Severe acute respiratory syndrome coronavirus papain-like protease ubiquitin-like domain and catalytic domain regulate antagonism of IRF3 and NF-kappaB signaling. J Virol 83:6689–6705. doi: 10.1128/JVI.02220-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barretto N, Jukneliene D, Ratia K, Chen Z, Mesecar AD, Baker SC. 2005. The papain-like protease of severe acute respiratory syndrome coronavirus has deubiquitinating activity. J Virol 79:15189–15198. doi: 10.1128/JVI.79.24.15189-15198.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Clementz MA, Chen Z, Banach BS, Wang Y, Sun L, Ratia K, Baez-Santos YM, Wang J, Takayama J, Ghosh AK, Li K, Mesecar AD, Baker SC. 2010. Deubiquitinating and interferon antagonism activities of coronavirus papain-like proteases. J Virol 84:4619–4629. doi: 10.1128/JVI.02406-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kamitani W, Narayanan K, Huang C, Lokugamage K, Ikegami T, Ito N, Kubo H, Makino S. 2006. Severe acute respiratory syndrome coronavirus nsp1 protein suppresses host gene expression by promoting host mRNA degradation. Proc Natl Acad Sci U S A 103:12885–12890. doi: 10.1073/pnas.0603144103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Devaraj SG, Wang N, Chen Z, Chen Z, Tseng M, Barretto N, Lin R, Peters CJ, Tseng CT, Baker SC, Li K. 2007. Regulation of IRF-3-dependent innate immunity by the papain-like protease domain of the severe acute respiratory syndrome coronavirus. J Biol Chem 282:32208–32221. doi: 10.1074/jbc.M704870200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Basu D, Walkiewicz MP, Frieman M, Baric RS, Auble DT, Engel DA. 2009. Novel influenza virus NS1 antagonists block replication and restore innate immune function. J Virol 83:1881–1891. doi: 10.1128/JVI.01805-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Van Damme N, Goff D, Katsura C, Jorgenson RL, Mitchell R, Johnson MC, Stephens EB, Guatelli J. 2008. The interferon-induced protein BST-2 restricts HIV-1 release and is downregulated from the cell surface by the viral Vpu protein. Cell Host Microbe 3:245–252. doi: 10.1016/j.chom.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hoa XD, Kirk AG, Tabrizian M. 2007. Towards integrated and sensitive surface plasmon resonance biosensors: a review of recent progress. Biosens Bioelectron 23:151–160. doi: 10.1016/j.bios.2007.07.001. [DOI] [PubMed] [Google Scholar]
- 53.McNatt MW, Zang T, Hatziioannou T, Bartlett M, Ben Fofana I, Johnson WE, Neil SJD, Bieniasz PD. 2009. Species-specific activity of HIV-1 Vpu and positive selection of tetherin transmembrane domain variants. PLoS Pathog 5:e1000300. doi: 10.1371/journal.ppat.1000300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mansouri M, Viswanathan K, Douglas JL, Hines J, Gustin J, Moses AV, Früh K. 2009. Molecular mechanism of BST2/tetherin downregulation by K5/MIR2 of Kaposi's sarcoma-associated herpesvirus. J Virol 83:9672–9681. doi: 10.1128/JVI.00597-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jia HP, Look DC, Shi L, Hickey M, Pewe L, Netland J, Farzan M, Wohlford-Lenane C, Perlman S, McCray PB Jr. 2005. ACE2 receptor expression and severe acute respiratory syndrome coronavirus infection depend on differentiation of human airway epithelia. J Virol 79:14614–14621. doi: 10.1128/JVI.79.23.14614-14621.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]