ABSTRACT
Influenza D virus (FLUDV) is a novel influenza virus that infects cattle and swine. The goal of this study was to investigate the replication and transmission of bovine FLUDV in guinea pigs. Following direct intranasal inoculation of animals, the virus was detected in nasal washes of infected animals during the first 7 days postinfection. High viral titers were obtained from nasal turbinates and lung tissues of directly inoculated animals. Further, bovine FLUDV was able to transmit from the infected guinea pigs to sentinel animals by means of contact and not by aerosol dissemination under the experimental conditions tested in this study. Despite exhibiting no clinical signs, infected guinea pigs developed seroconversion and the viral antigen was detected in lungs of animals by immunohistochemistry. The observation that bovine FLUDV replicated in the respiratory tract of guinea pigs was similar to observations described previously in studies of gnotobiotic calves and pigs experimentally infected with bovine FLUDV but different from those described previously in experimental infections in ferrets and swine with a swine FLUDV, which supported virus replication only in the upper respiratory tract and not in the lower respiratory tract, including lung. Our study established that guinea pigs could be used as an animal model for studying this newly emerging influenza virus.
IMPORTANCE Influenza D virus (FLUDV) is a novel emerging pathogen with bovine as its primary host. The epidemiology and pathogenicity of the virus are not yet known. FLUDV also spreads to swine, and the presence of FLUDV-specific antibodies in humans could indicate that there is a potential for zoonosis. Our results showed that bovine FLUDV replicated in the nasal turbinate and lungs of guinea pigs at high titers and was also able to transmit from an infected animal to sentinel animals by contact. The fact that bovine FLUDV replicated productively in both the upper and lower respiratory tracts of guinea pigs, similarly to virus infection in its native host, demonstrates that guinea pigs would be a suitable model host to study the replication and transmission potential of bovine FLUDV.
INTRODUCTION
Influenza viruses are negative-sense, single-stranded RNA viruses classified in the Orthomyxoviridae family. There are three recognized genera of influenza viruses, designated influenza A virus (IAV or FLUAV), influenza B virus (FLUBV), and influenza C virus (FLUCV). FLUAV and FLUBV have 8 negative-sense, single-stranded RNA segments, whereas FLUCV has only 7 segments. FLUAV proteins include 5 structural proteins, HA (hemagglutinin), NA, M1, M2, and NP (ribonucleoprotein); 3 subunits of the RNA polymerase complex, polymerase basic protein 1 (PB1), polymerase basic protein 2 (PB2), and polymerase acidic protein (PA); and 3 nonstructural proteins, NS1, NS2 (nuclear export protein [NEP]), and PB1-F2 (1). Recent studies have suggested that NS2 and (probably) NS1 of FLUAV are structural proteins that can be detected in virions (2). FLUBV has 6 structural proteins, HA, NA, NB, M2, M1, and NP; 3 subunits of RNA polymerase complex, PA, PB1, and PB2; and 2 non-structural proteins, NS1 and NS2. FLUCV has 4 structural proteins, M2, M1, NP, and the hemagglutinin esterase fusion (HEF) protein that replaces the HA and NA of FLUAV or FLUBV; 3 subunits of RNA polymerase complex, P3, PB1, and PB2; and 2 nonstructural proteins, NS1 and NS2. Depending on the HA and NA proteins, FLUAV has several subtypes and causes severe epidemics and pandemics affecting humans. It also infects various other species of mammals and birds across the world, which can result in an increase in the spread of IAV infection and more-lethal outcomes, especially in poultry, than are seen in humans. FLUBV has no subtypes but possesses two lineages causing localized epidemics and affecting mainly humans and, to some extent, seals (3). The FLUBV genome was also recently detected in domestic pigs, indicating that the virus may infect this agricultural animal (4). Compared to infections by the A and B types, FLUCV infections cause mild disease and were found to have coexisted with FLUAV and FLUBV infections in humans (5, 6).
In 2011, a new influenza virus was isolated in Oklahoma from a 15-week-old swine showing influenza-like symptoms. Electron microscopic studies have shown features similar to those of orthomyxoviruses. Further studies revealed that this virus was negative for neuraminidase and positive for O-acetyl esterase activity, which is a characteristic of FLUCV. Genus-specific real-time reverse transcription-PCR (RT-PCR) failed to detect the virus. However, the new virus showed 50% homology to human FLUCV (7). Deep RNA sequencing (RNA-seq) showed that the HEF protein of the new virus has a conserved enzymatic site but a receptor binding site divergent from that of FLUCV. The virus also exhibited broader cellular tropism than FLUCV. A serological survey showed that the virus is widespread in swine and bovine herds of the United States (8). In addition to a swine isolate, several bovine strains were also isolated from diseased bovines located in different farms of the U.S. Midwest region. Bovines with respiratory disease complex on the west coast were also found to harbor FLUDV (9, 10). The new group of viruses did not cross-react with FLUCV antibodies by hemagglutination inhibition (HI) and agar gel immunodiffusion (AGID) assays and also failed to undergo reassortment with FLUCV in vitro. Phylogenetic analysis also failed to identify reassortment between FLUCV and FLUDV in field isolates. Because these newly discovered viruses shared unique biologic, genetic, and antigenic properties different from those of FLUCV, a proposal was made to classify these viruses in a new genus—influenza D virus, or FLUDV (8).
The bovine and swine FLUDVs shared more than 96% homology in all the segments. The most divergent segments were HEF and P42/M, with 96.7% to 99.0% and 96.9% to 99.2% identity, respectively. The segments with the highest homology were PB1 and NS, with 98.9% to 99.1% and 98.8% to 99.2% homology, respectively (8). Apart from the strains isolated in different regions in the United States, bovine FLUDV was also identified in strains isolated in Shandong Province in China and in France (11, 12). The strains from China shared 94% to 99% homology with the strains in the United States (12). The geographical distribution of bovine FLUDV shows that the virus is prevalent across bovine populations and is circulating in North America, Europe, and Asia at least.
Swine FLUDV was found to infect ferrets and pigs and can also be transmitted to naive animals by contact. Swine FLUDV was able to replicate in nasal turbinates and shed detectable levels of virus in nasal washes. However, swine FLUDV was not detected in trachea and lungs in either ferrets or pigs, indicating its inability to replicate in the lower respiratory tract. The animals seroconverted, but neither ferrets nor pigs developed clinical symptoms and lesions typical of influenza (7). Ferrets have been used as surrogates for human influenza virus infection and pathogenesis. The ability of swine FLUDV to replicate in the upper respiratory tract of ferrets may indicate the zoonotic potential of these viruses.
The objective of this study was to examine viral replication, transmission, and virulence properties of the novel bovine FLUDV in guinea pigs. Our study was based on the following considerations. First, guinea pig has been used extensively as an alternate mammalian animal model for studying the pathogenesis of many influenza viruses, including FLUBV (13–15). Second, this animal model was susceptible to infections by influenza A virus subtypes and demonstrated virus replication in lungs. Third, guinea pigs were able to transmit human influenza viruses from one animal to another (15). Fourth, guinea pigs are easier to handle and house and are less expensive than ferrets. Importantly, guinea pigs share characteristics of airway hyperresponsiveness with and have bronchus-associated lymphoid tissue similar to that of humans (16, 17). Finally, despite being highly similar to a swine FLUDV strain used previously for ferret and pig studies, the bovine FLUDV selected for this study is a representative strain of another antigenic lineage different from swine FLUDV, possessing some distinct variations in the HEF protein, a major mediator of host range and viral tropism (18).
In this study, guinea pigs were divided into three groups: (i) a directly inoculated group; (ii) a contact transmission group; and (iii) an aerosol transmission group. Our results showed that bovine FLUDV efficiently replicated in the lower and upper respiratory tracts of guinea pigs. Detection of FLUDV in lungs of infected animals is novel because our previous studies in ferrets and pigs revealed a lack of virus replication in lungs. We also found that naive guinea pigs acquired infection from the cocaged infected guinea pigs, indicating that bovine FLUDV can be transmitted by direct contact. Taken together, results of our experiments demonstrated that guinea pigs, being good mammalian hosts and widely used in influenza research, could be further explored for mechanistic studies of FLUDV infection and pathogenesis.
MATERIALS AND METHODS
Cells and virus.
Human rectal tumor (HRT-18G) cells and Madin-Darby canine kidney (MDCK) cells maintained in Dulbecco's minimum essential medium supplemented with 10% fetal bovine serum (FBS) (PAA Laboratories Inc., Dartmouth, MA, USA) and penicillin-streptomycin (Life Technologies, Carlsbad, CA, USA) (100 U/ml) were used in this study for cell culture. Influenza D/bovine/Oklahoma/660/2013 virus was originally isolated from the bovine herds of Oklahoma and has been designated a representative strain of a genetic and antigenic lineage different from the swine FLUDV strain that was used for previous ferret and pig studies (8). The virus was grown on human rectal tumor (HRT-18G) cells at a multiplicity of infection (MOI) of 0.1. The cells were allowed to reach only 60% to 70% confluence at the time of infection. The virus was suspended in 2 ml Dulbecco's modified Eagle's medium (DMEM) and incubated at 37°C in 5% CO2 for 1 h. Following infection, fresh DMEM with 0.5 μg/ml tolylsulfonyl phenylalanyl chloromethyl ketone (TPCK)-treated trypsin (Sigma, Saint Louis, MO, USA) was added for further incubation at 37°C in 5% CO2 for 5 days. After 5 days, the infected cell cultures were frozen and thawed. The supernatant was spun at 500 × g for 10 min at 4°C to remove the cellular debris. Determination of virus titers in MDCK cells was done according to the method of Reed and Muench (19). Virus loads in nasal washes and tissue homogenates were titrated using MDCK cells by indirect immunofluorescence assay (IFA). DMEM supplemented with penicillin-streptomycin (Life Technologies, Carlsbad, CA, USA) (200 U/ml) and TPCK-treated trypsin (Sigma, Saint Louis, MO, USA) (0.5 μg/ml) was used as the virus growth and maintenance medium.
Animals.
Specific-pathogen-free (SPF), viral-antibody-free (SPF/VAF), 30-day-old guinea pigs of the Dunkin-Hartley strain (Elm Hill Labs, MA, USA) weighing 300 to 350 g were used for the study. The animals were ear tagged for identification purposes. The duration of the experiment was 3 weeks, which included a 1-week acclimatization period. Animals were provided with food and water ad libitum and kept on a 12-h light/dark cycle. The temperature and relative humidity (RH) of the animal housing ranged from 72 to 75°F and 25% to 33%, respectively. Control animals were housed in a separate room away from the room housing the infected animals and were processed before the inoculated animals. Strict precautionary measures were followed to prevent cross contamination between animals in different cages. Sentinel animals were sampled before inoculated animals in transmission experiments. Gloves were changed between cages during cleaning and handling, and masks and surfaces were disinfected to prevent any possible cross contamination.
Experimental design.
Guinea pigs were divided into three experimental groups for testing growth kinetics of the virus, direct contact transmission, and aerosol transmission. The group for studying virus kinetics consisted of 10 infected animals and 3 mock-infected animals. The animals were infected intranasally with 3 × 105 50% tissue culture infective doses (TCID50)/300 μl of influenza D/bovine/Oklahoma/660/2013 virus (bovine FLUDV). Half (150 μl) of the virus inoculum was delivered in each nostril. The 3 control animals were mock infected with equal volumes of phosphate-buffered saline (PBS). The body weights of all of the animals were recorded before challenge. Guinea pigs were briefly anesthetized using isoflurane prior to infection.
For the contact and aerosol transmission experiments, 6 animals were inoculated intranasally with 3 × 105 TCID50/300 μl of influenza D/bovine/Oklahoma/660/2013 virus (C/660) under conditions of isoflurane anesthesia and the remaining 6 animals were left uninfected. To test contact transmission, 3 infected animals were kept in a cage. At 24 h postinfection (hpi), three sentinel animals were added to the same cage. For studying aerosol transmission, 3 infected animals were housed in a cage with a double-walled wire partition that permitted airflow but prevented direct contact. Three sentinel animals were added at 24 h postinfection. The cages were kept away from the vents to minimize the dilution of aerosols by room ventilation and to provide a conductive environment for virus transmission. The animal experiments were approved by the Institutional Animal Care and Use Committee of South Dakota State University (IACUC no. 14-011A) and were conducted under biosafety level 2 conditions.
Monitoring and sample collection.
Body weight and temperature were monitored daily starting from 2 days before challenge. Prior to challenge, blood was collected from all of the animals from the jugular vein/cranial vena cava under conditions of isoflurane anesthesia. Animals were monitored on a daily basis after the virus challenge for clinical signs, and body temperature and body weight were recorded. Nasal washes were collected from all the infected animals in the directly inoculated group and from three control animals at 1, 3, 5, 7, and 9 days postinfection (dpi). Additionally, two infected animals were euthanized using CO2 at 1, 3, 5, 7, and 9 dpi. Blood, nasal wash, nasal turbinate, trachea, and lung samples were collected. The numbers of infected animals assessed each day from group I were as follows: day 1, n = 10; days 2 and 3, n = 8; days 4 and 5, n = 6; days 6 and 7, n = 4; days 8 and 9, n = 2.
For contact and aerosol transmission experiments, the animals were monitored for weight loss, temperature changes, and other clinical signs on a daily basis. The nasal wash samples were collected from all of the animals at 48-h intervals (2, 4, 6, 8, 10, 12, and 14 dpi) from the time of challenge under conditions of isoflurane anesthesia. These animals were euthanized at 14 dpi, and blood, nasal wash, nasal turbinate, trachea, and lung samples were collected.
Collection of nasal washes.
The nasal washes were collected by instilling 1 ml of PBS using a sterile 25-to-28-gauge cannula into the nostrils and collecting the washes by draining to sterile containers or petri dishes. Animals were anesthetized using isoflurane, and alternate nostrils were used for sample collection on alternate days. Nasal washes collected in the petri dishes were transferred to 1.5-ml tubes and then centrifuged at 500 × g for 6 min at 4°C to remove any debris. The supernatants were stored at −80°C until analysis.
Estimation of virus load in nasal washes and tissues.
Lung, trachea, and nasal turbinates from guinea pigs were collected and stored at −80°C. One gram of tissue was homogenized using DMEM supplemented with penicillin-streptomycin (Life Technologies, Carlsbad, CA, USA) (200 U/ml) and a Stomacher circulator at high speed for 2 min. The homogenized tissue fluid was clarified by spinning at 500 × g for 8 min at 4°C and stored at −80°C. For trachea and nasal turbinate analyses, DMEM with penicillin-streptomycin (Life Technologies, Carlsbad, CA, USA) (200 U/ml) was added and homogenized tissue fluid was collected and stored at −80°C until titration.
For virus isolation, MDCK cells were used for determining the virus titer present in nasal washes and tissue homogenates. A total of 7 × 103 cells were seeded on 96-well tissue culture plates and allowed to grow overnight. When the cells reached 60% to 70% confluence, serial 10-fold dilutions of the sample were inoculated on cell culture plates after the plates were washed with sterile PBS. The inoculated cell culture plates were incubated for 5 days at 37°C. After 5 days, the infected cell culture plates were washed with PBS, fixed in 80% acetone, and allowed to dry. The plates were stained for determinations of virus titers in nasal washes and tissue homogenates by indirect immunofluorescence assay. Virus titration was determined using the Reed and Muench formula to find the 50% endpoints (19).
IFA.
Determinations of virus titers in nasal washes and tissue homogenates were performed by indirect immunofluorescence assay (IFA). The fixed infected cell culture plates were moistened with PBS for 10 min at room temperature before staining. A 100-μl volume of rabbit polyclonal primary antibody (IgG) against bovine FLUDV was added at a 1:200 dilution to each of the wells. The plates were incubated at 37°C for 45 min. Following three washes with PBS, 50 μl of affinity-purified fluorescein-labeled goat anti-rabbit IgG secondary antibody (KPL, Gaithersburg, MD, USA) was added at a 1:1,000 dilution. After the secondary antibody was added, the plates were incubated at 37°C for 45 min.
HI assay.
The preinfection and postinfection sera were treated with receptor-destroying enzyme (RDE; Denka Seiken, Tokyo, Japan) before the HI assay was performed. The RDE treatment was done according to the manufacturer's protocol, and the HI assay was done according to the WHO manual guidelines (20). The HI assay was performed using 1% turkey red blood cells (RBCs) (Lampire Biological Laboratories, Pipersville, PA, USA).
Virus genome sequencing and analysis.
To determine the HEF sequence of viruses isolated from infected animals, we employed HRT-18G cells for cultivation of the infected lung homogenates, which was followed by deep RNA sequencing. Briefly, cells were allowed to reach 60% to 70% confluence and were then infected with bovine FLUDV at an MOI of 0.1 in 1 ml of DMEM. After 1 h of infection, fresh virus infection medium, i.e., DMEM supplemented with penicillin-streptomycin (Life Technologies, Carlsbad, CA, USA) (200 U/ml) and 0.5 μg/ml TPCK-treated trypsin (Sigma, Saint Louis, MO, USA), was added for further incubation at 37°C for 5 days. The virus grown from the infected lung homogenates was sequenced using an Illumina MiSeq system as described previously (7).
The nucleotide sequences of HEF obtained from the inoculum and the lung homogenates of guinea pigs were analyzed against all the homologous sequences in the NCBI database using standard nucleotide BLAST (21). The nucleotide sequences were translated to amino acid sequences using the ExPASy tool (22). Pairwise protein alignments were performed using the multiple-sequence alignment tool from Clustal omega (23).
Gross pathology, histology, and immunohistochemistry (IHC).
Following experimental bovine FLUDV infection of guinea pigs, euthanasia was performed at prescribed time points. A complete necropsy was performed to look for any macroscopic lesion in all the organs. Lung, trachea, and nasal turbinate samples were collected in 10% neutral buffered formalin and embedded in paraffin wax. Sections (5 μm thick) were then cut and stained with hematoxylin and eosin for histopathological examination.
Immunohistochemistry was performed on lung tissue sections stained with a primary rabbit polyclonal antibody developed against purified bovine FLUDV. Sections were deparaffinized, rehydrated, and immersed in 3% H2O2–distilled water for 30 min to block endogenous peroxidase. After washing with Tris-buffered saline (TBS) buffer was performed, the sections were treated with 5% goat serum (Life Technologies, Carlsbad, CA, USA)–TBS buffer for 1 h. The lung sections were then stained in a 1:1,500 dilution of polyclonal rabbit antibody against purified bovine FLUDV for 1 h at room temperature. After rinsing with TBS was performed, the sections were stained with 100 μl of horseradish peroxidase (HRP)-conjugated goat anti-rabbit secondary antibody (Dako EnVision+ System-HRP [DAB], Carpinteria, CA, USA) for 30 min, followed by counterstaining with hematoxylin. Lung sections stained with PBS and isotype antibody instead of specific primary antibody served as negative controls.
RESULTS
Growth kinetics of bovine FLUDV in the upper and lower respiratory tracts of guinea pigs.
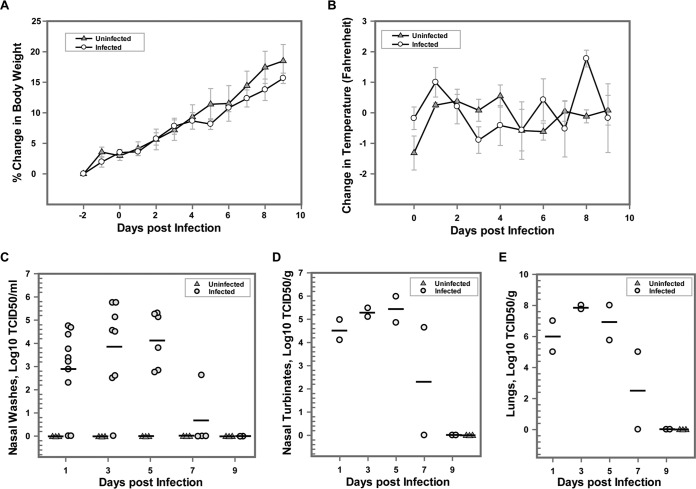
To determine the growth kinetics of bovine FLUDV, 10 animals were intranasally inoculated with 3 × 105 TCID50/300 μl of bovine FLUDV and three animals were mock infected with PBS. The clinical significance of bovine FLUDV in guinea pigs was assessed by regular monitoring of changes in body weight and temperature over a period of 9 days postinfection, while viral replication in the lower and upper respiratory tracts of the guinea pigs was evaluated by determining the amount of virus shed in nasal washes and homogenates of lung and nasal turbinate tissues over a period of 9 days. Overall, there were no obvious clinical signs (body weight and temperature) of influenza virus infection, and the directly infected animals behaved similarly to mock-infected animals. For each animal, the change in body weight postinfection was calculated by comparing the postinfection body weight to the average of the body weights recorded 2 days prior to the infection. The differences were calculated as the percentage of body weight change for each animal plotted for each day. The directly inoculated animals did not show any significant change in body weight compared to the mock-infected animals. An increase in body weight in mock-infected animals compared to directly inoculated animals from 5 dpi to 9 dpi was noted, but this change was not statistically significant (Fig. 1A). Similarly, the difference between the postinfection body temperature and the average preinfection temperature was determined for each animal, and the averages of the differences were plotted for each day (Fig. 1B). Despite the observation of a trend of an increase in the temperature of infected animals on days 0, 1, and 2 postinfection compared to mock-infected animals, the changes in temperature were not significant for the mock-infected animals. In the remaining days of the experiment, infected animals showed little or no change in body temperature. These results indicated that bovine FLUDV infection had no effect on the body weight and temperature changes during the 9-day period in guinea pigs.
FIG 1.
Body weight and temperature changes and virus titers in nasal washes, lungs, and nasal turbinates in guinea pigs after intranasal inoculation of bovine FLUDV in the directly inoculated group. A total of 10 guinea pigs were intranasally inoculated with 3 × 105 TCID50/300 μl of bovine FLUDV (shown as “Infected”) and 3 guinea pigs were mock inoculated with PBS (shown as “Uninfected”). Body weight and temperature were measured daily from 2 days before infection until 14 dpi. (A and B) Percentage changes in body weight (A) and changes in body temperature (B) expressed as means ± standard errors (SE). (C to E) Nasal washes were collected at 1, 3, 5, 7, and 9 dpi. Two inoculated animals were randomly euthanized on each of those days to assess virus load in lungs and nasal turbinates. All mock-inoculated animals were euthanized at 9 dpi. Virus titers in nasal washes are expressed as log10 TCID50 per milliliter (C); virus titers in nasal turbinates (D) and lung (E) are expressed as log10 TCID50 per gram. For panels C, D, and E, each shape represents an individual animal. Horizontal bars show the mean viral titers for each time point.
Next, we determined whether virus replicated in the upper and lower respiratory tracts of infected animals by focusing on quantitative analysis of virus loads in nasal washes, nasal turbinates, and lungs by titrating the samples on MDCK cells. Nasal washes were collected from the directly inoculated and mock-infected animals at 48-h intervals from 1 dpi through 9 dpi. To determine the tissue tropism of the virus, two random animals were selected and euthanized on 1, 3, 5, 7, and 9 dpi and lung and nasal turbinate samples were collected. The results of our experiments showed that FLUDV shedding in the nasal washes started at low levels and then reached a highest titer of 5.75 log10 TCID50/ml on 3 dpi (Fig. 1C). The virus shedding in nasal washes was reduced to undetectable levels on 9 dpi. This clearly showed that bovine FLUDV can replicate in the upper respiratory tract. Virus growth from nasal turbinates ranged from 4.10 to 6.0 log10 TCID50/g on 1, 3, 5, and 7 dpi and further confirmed that bovine FLUDV could successfully replicate in the upper respiratory tract of guinea pigs (Fig. 1D). Robust bovine FLUDV replication was also demonstrated in the lung homogenates of guinea pigs. We noticed that there was a substantial increase in virus titers in lung homogenates of directly inoculated animals starting from 1 dpi through 7 dpi, with a peak of 8 log10 TCID50/g on 5 dpi. By day 9, virus was cleared from the lungs (Fig. 1E). The presence of virus in the lungs as well as in nasal washes and nasal turbinates until 7 dpi indicated that virus was actively replicating in the respiratory tract of the guinea pigs. Previous studies in pigs and ferrets have shown that FLUDV isolated from swine (swine FLUDV) was detected only in the upper respiratory tract and not in the lungs of infected animals.
Seroconversion in directly inoculated guinea pigs.
Pre- and postinfection serum samples were tested for the presence of virus-specific antibodies against bovine FLUDV by the hemagglutination inhibition (HI) assay. All the guinea pigs were seronegative for bovine FLUDV prior to infection as demonstrated by the absence of virus-specific antibodies. The antibody response to bovine FLUDV infection was measured on days 1, 3, 5, 7, and 9 dpi in the directly inoculated group. In the directly inoculated group, virus-specific antibodies were detected by HI at 7 and 9 dpi. One of the 2 infected animals seroconverted on 7 dpi (HI titer, 40), and 2 of the 2 infected animals seroconverted with titers of 20 and 80 on 9 dpi (Table 1). The presence of detectable levels of antibody showed that guinea pigs could be a good model to study the replication and kinetics of bovine FLUDV.
TABLE 1.
Seroconversion of guinea pigs after bovine FLUDV infection
| Group | No. of seroconverted guinea pigs (titer[s])/total no. of guinea pigsa |
||||
|---|---|---|---|---|---|
| Uninfected (9 dpi) | Infected |
Sentinels (14 dpi) | |||
| 7 dpi | 9 dpi | 14 dpi | |||
| Directly inoculated | 0/3 | 1 (40)/2 | 2 (20, 80)/2 | ||
| Contact transmission | 3 (80, 40, 80)/3 | 1 (40)/3 | |||
| Aerosol transmission | 3 (80, 80, 20)/3 | 0/3 | |||
Virus-specific antibody titers determined by HI are given in parentheses.
Transmission of bovine FLUDV in cocaged guinea pigs.
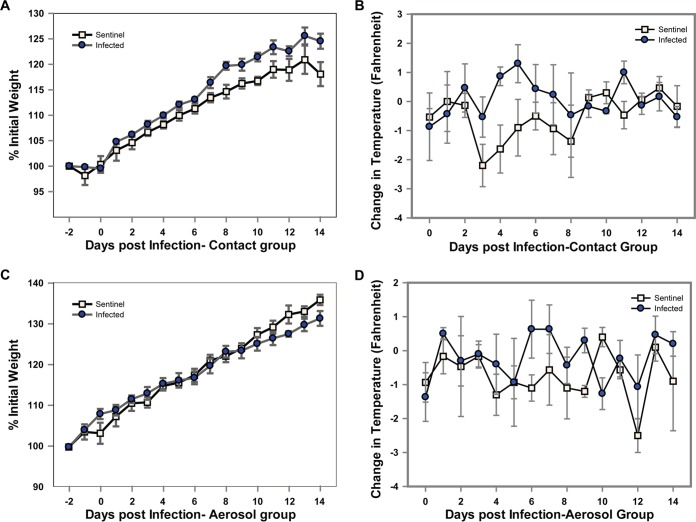
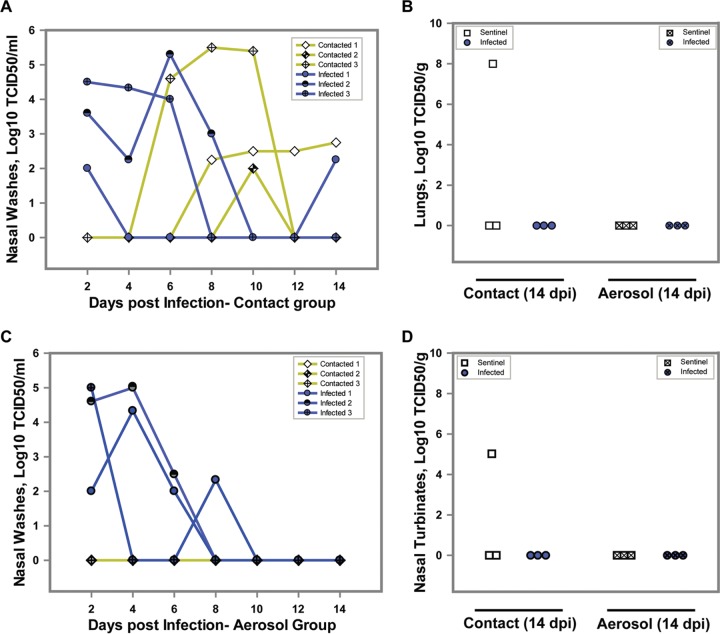
To test whether bovine FLUDV can transmit through contact, 3 directly inoculated guinea pigs at 1 dpi were allowed to cocage with three sentinel animals. The experimental setup for contact transmission facilitated not only direct but also indirect contact (droplets and aerosols). The potential transmission of bovine FLUDV in guinea pigs through contact was investigated by analysis of the changes in the body weight and temperature and also in the virus load in the nasal washes, nasal turbinates, and lungs from the infected and sentinel animals over a period of 14 days. Nasal washes were collected at 48-h intervals from 2 dpi to 14 dpi, and all of the animals were euthanized at 14 dpi. There were no significant differences in body weight and temperature between infected and sentinel animals (Fig. 2A and B). The infected and sentinel animals did not show any observable clinical signs during the 14-day experiment. All 3 infected animals shed the virus in the nasal washes and cleared the virus by 8 dpi (Fig. 3A). Two of three sentinel animals acquired infection from cage mates, and bovine FLUDV was detected in their nasal washes. One sentinel animal shed virus in the nasal washes at 4.6 log10 TCID50/ml starting on 6 dpi and cleared the virus by 10 dpi, while the other sentinel animal started viral shedding on 8 dpi and continued to shed virus at low levels until 14 dpi. Intriguingly, one of the directly inoculated guinea pigs that shed virus once (2 dpi) in the nasal washes and then tested negative until 12 dpi shed virus at 14 dpi with a low titer of 2.25 log10 TCID50/ml. This animal could have acquired infection from the cocaged sentinel guinea pig by contact.
FIG 2.
Changes in body weight and temperature of guinea pigs in the contact transmission group (A and B) and in the aerosol transmission group (C and D). Three guinea pigs were intranasally inoculated with 3 × 105 TCID50/300 μl of bovine FLUDV, and 3 uninfected guinea pigs were added to the cage as sentinels after 24 h. Body weights and temperatures were measured daily from 2 days prior to infection to 14 dpi. (A and B) Changes in percentages of body weights compared to preinfection body weights (A) and changes in temperatures of infected animals compared to temperatures of sentinels (B) in contact transmission group. The data are expressed as means ± SE. (C and D) A similar experiment with the same parameters (body weight and temperature) measured was conducted for the aerosol transmission study. See Materials and Methods for details.
FIG 3.
Virus titer in nasal washes, lungs, and nasal turbinates of guinea pigs exposed to bovine FLUDV by the aerosol or contact route. In both the contact transmission group and aerosol transmission group, 3 guinea pigs were intranasally inoculated with 3 × 105 TCID50/300 μl of bovine FLUDV. After 24 h of infection, 3 uninfected guinea pigs were added to each cage as sentinels. Nasal washes were collected at days 2, 4, 6, 8, 10, 12, and 14 dpi from all animals. All the guinea pigs were euthanized at 14 dpi. (A and C) Virus titers in nasal washes from the contact transmission group (A) and the aerosol transmission group (C) are expressed as log10 TCID50 per milliliter. (B and D) Virus titers in lungs (B) and nasal turbinates (D) of the infected and sentinel animals in the contact and aerosol transmission groups at 14 dpi are expressed as log10 TCID50 per gram. For panels B and D, each shape represents an individual animal.
Further, to confirm the presence of transmission, guinea pig sera from the contact transmission group were tested for the presence of virus-specific HI antibodies. All three directly infected animals in the contact group seroconverted with HI titers of 40 to 80, and one sentinel animal seroconverted with an HI titer of 40 at 14 dpi, thus confirming the occurrence of short-range transmission by contact (Table 1). The absence of serum HI antibody titer in the other contact-infected sentinel animal can be explained by the late commencement of infection (8 dpi), indicating that the animal did not have enough time to develop antibodies by 14 dpi. At 14 dpi, the lungs and nasal turbinates of all 3 infected animals revealed no detectable levels of virus, indicating that the guinea pigs had cleared the virus (Fig. 3B and D). Among the sentinel animals, the animal which acquired late infection without seroconversion showed very high titers of 8 log10 TCID50/g in the lungs and 5.0 log10 TCID50/g in the nasal turbinates at 14 dpi (Fig. 3B and D). Taking the data together, the presence of virus in the nasal washes, lungs, and nasal turbinates and the antibody seroconversion demonstrated that bovine FLUDV can be transmitted from infected guinea pigs to naive guinea pigs by contact.
Aerosol transmission of bovine FLUDV.
In order to test the transmissibility of bovine FLUDV via aerosol or droplets, three guinea pigs were inoculated intranasally with 3 × 105 TCID50/300 μl and three noncontact sentinel animals were placed in the same cages after 24 h. The infected and sentinel animals were separated with double-walled wire mesh, thus facilitating only airborne transmission and preventing all types of direct contact. The duration of the experiment and the collection of nasal washes were similar to the conditions used for the contact transmission experiment. Similarly to the contact transmission group, the body weight and temperature changes between the infected and sentinel animals were not significant (Fig. 2C and D). In the aerosol transmission group, 3/3 directly infected animals showed detectable virus replication and shed the virus in their nasal washes until 6 dpi, with a peak of 5.0 log10 TCID50/ml on 2 and 4 dpi (Fig. 3C). None of the three sentinel animals shed any detectable level of virus in nasal washes or had virus present in lungs and nasal turbinates during the study. This suggested that bovine FLUDV cannot be transmitted by the aerosol route. At 14 dpi, none of three infected animals had detectable virus in the lungs or nasal turbinates, indicating that the virus had cleared by day 14 (Fig. 3B and D). In the HI assay, all three infected animals in the aerosol transmission group had seroconverted with HI titers of 20 to 80 at 14 dpi (Table 1). In marked contrast, neither virus replication nor virus-specific HI antibodies were found in three sentinel animals. Our data suggested that bovine FLUDV was incapable of transmitting infection in guinea pigs by the aerosol route under the experimental conditions used in this study. A future study is needed to draw a conclusion regarding whether it can be transmitted by the aerosol route in guinea pigs.
Pathology of bovine FLUDV in the respiratory tract of guinea pigs.
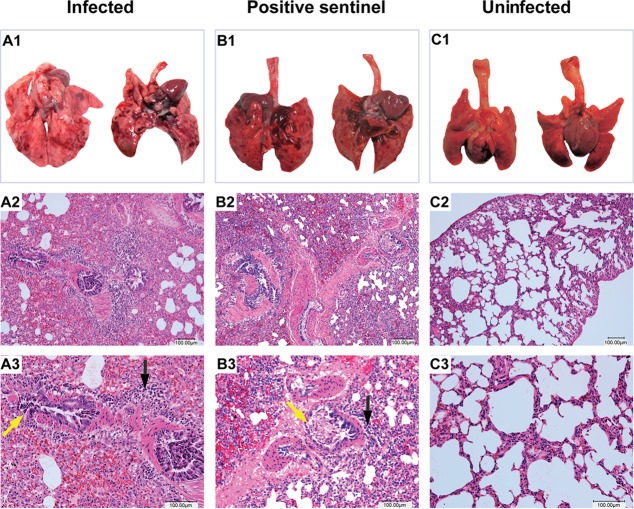
Neither experimentally infected guinea pigs nor contact-infected guinea pigs showed any clinical signs of disease after bovine FLUDV infection. However, mild to moderate macroscopic changes were observed in the lungs of the directly inoculated animals and contact-infected animals during necropsy. No other lesions were observed in any other organs. On necropsy, primary gross lung lesions showed areas of congestion and hemorrhage on 1 dpi that progressed to areas of pulmonary consolidation at 3, 5, and 7 dpi. Lungs from an uninfected animal did not show any macroscopic lesions; however, lungs from the sentinel animal infected by contact showed similar areas of pulmonary consolidation comparable to the macroscopic lung lesions of directly inoculated animals on 5 and 7 dpi (Fig. 4A1, B1, and C1). Histopathological examination of lungs showed minimal to mild inflammation in the trachea with infiltration of lymphocytes and plasma cells, mild hyperplasia of tracheobronchial lymph nodes (not shown), mild to moderate atelectasis in lungs, vascular congestion, bronchitis with desquamation of bronchial epithelium with exudate, and peribronchial and perivascular cuffing by lymphocytes and plasma cells in all of the directly inoculated animals at 3, 5, and 7 dpi. In the directly inoculated animals, alveolar septa were thickened by neutrophils and RBCs, and bronchopneumonia with accumulation of luminal exudate was also observed (Fig.4A1 to A3). The sentinel animal that acquired the infection from a cocaged infected animal by contact developed similar inflammatory changes with thickened alveolar septa, bronchitis, peribronchial and perivascular lymphocyte and plasma cell infiltration, and mild neutrophilic bronchopneumonia at 14 dpi (Fig. 4B1 to B3). One of the three uninfected control animals showed the presence of foreign material possibly derived from the bedding material and demonstrated mild sloughing of pulmonary epithelium and suppurative inflammation within the examined lung.
FIG 4.
Pathological changes of the lungs in infected, positive-testing sentinel guinea pigs and uninfected guinea pigs after bovine FLUDV infection. Macroscopic lung lesions as indicated as follows: A1, a directly inoculated animal at 7 dpi; B1, a positive sentinel infected by contact at 14 dpi; C1, an uninfected animal at 9 dpi. Panels A1 and B1 showed multifocal areas of pulmonary consolidation in directly inoculated and contact-infected sentinel animals. Microscopic lung lesions are indicated as follows: detailed histology results are shown at magnifications of ×100 for the second row (A2, B2, and C2) and ×200 for the third row (A3, B3, and C3). Hematoxylin and eosin (H&E) staining of lung sections from a directly inoculated animal at 7 dpi (A2 and A3) and from a positive-testing sentinel infected by contact at 14 dpi (B2 and B3) showed multifocal areas of alveolar inflammation with infiltration of lymphocytes, plasma cells (black arrows), and RBCs in the lung parenchyma. Bronchiolar inflammation with desquamation of the epithelial cells (yellow arrows) and peribronchial infiltration of lymphocytes were also seen. (C2 and C3) Uninfected lung sections showed clear alveolar spaces without any inflammatory cell infiltration. Bars, 100 μm.
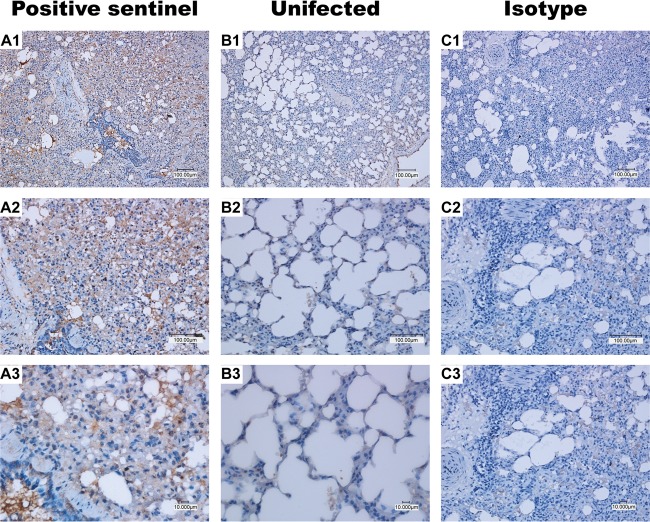
Immunohistochemistry of the lung sections demonstrated the presence of bovine FLUDV-specific antigen in the lungs of directly inoculated and infected sentinel animals (Fig. 5 and 6). The presence of the viral antigen in lungs correlated with the severity of lung macroscopic pathology. Bovine FLUDV antigens were present in larger amounts within alveolar septum and within bronchi and the bronchiolar epithelium in the lungs of directly inoculated animals at 3, 5, and 7 dpi compared to 1 dpi (Fig. 5). At 14 dpi, the amount of viral antigen in the lungs of a sentinel animal which acquired infection by contact was comparable to the amount of viral antigen present in those of the directly inoculated animals (Fig. 6A1 to A3), which further confirmed that bovine FLUDV transmission had occurred by contact. No viral antigen was detected from the lungs of the uninfected control animals (Fig. 6B1 to B3). Further, an isotype antibody from a directly inoculated bovine FLUDV-positive lung showed no staining, confirming the specificity of the primary antibody used for IHC staining (Fig. 6C1 to C3).
FIG 5.
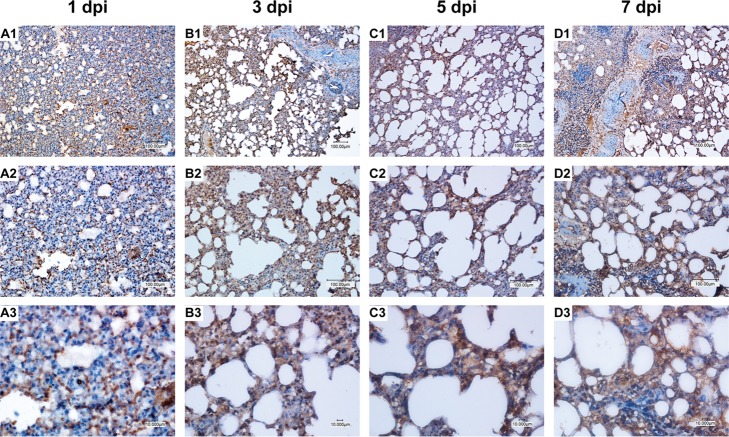
Bovine FLUDV antigen in the lungs of directly inoculated animals. Brown staining indicates cells positive for bovine FLUDV antigen in the infected lung tissue. Magnifications are ×100 for first row (panels A1, B1, C1, and D1), ×200 for the second row (panels A2, B2, C2, and D2), and ×400 for the third row (panels A3, B3, C3, and D3). Viral antigens were present in the alveolar and bronchial epithelial cells. Viral antigens present in the infected lung tissue at 1 dpi (A1, A2, and A3), 3 dpi (B1, B2, and B3), 5 dpi (C1, C2, and C3), and 7 dpi (D1, D2, and D3) are shown. Bars, 100 μm and 10 μm.
FIG 6.
Bovine FLUDV antigen in the lung tissue of the sentinel animal infected through contact. Panels A1, A2, and A3 represent lungs from the sentinel animal infected by contact. Brown staining shows cells positive for bovine FLUDV antigen. (B1 to B3) Uninfected animals did not show any brown stained infected cells for bovine FLUDV. (C1 to C3) Isotype antibody staining did not show any brown staining, confirming the specificity of the primary antibody used for immunohistochemistry. Magnifications are ×100 for the first row (panels A1, B1, and C1), ×200 for the second row (panels A2, B2, and C2), and ×400 for the third row (panels A3, B3, and C3). Bars, 100 μm and 10 μm.
Full-length-genome analysis of viruses isolated from the lungs of directly inoculated and cocaged sentinel guinea pigs.
To determine whether bovine FLUDV had evolved some interesting mutations that promoted viral adaption to guinea pigs, we determined the full-length genome of viruses isolated from the lungs of directly inoculated and cocaged sentinel animals. The sequence analysis was conducted to examine and determine the similarities and differences of the full genome of the parent inoculum and the full genomes of the viruses propagated from lung homogenates of directly inoculated or sentinel animals. For deep RNA sequencing, viral RNA was extracted from the parent inoculum and also from the viruses recovered from lung homogenates of directly inoculated and cocaged sentinel guinea pigs. HRT-18G cells were used for the virus propagation.
Full-genome-sequence analysis showed that both of the recovered virus isolates from guinea pig lungs exhibited mutations in HEF, P42, and NP proteins compared to the parent inoculum. HEF protein sequence analysis of the parent inoculum and of virus from the lung of sentinel animal which acquired infection by contact revealed 1 mutation, A746G, with an amino acid change of N249S. Compared to the inoculum, HEF protein sequence analysis of the bovine FLUDV that originated from the directly inoculated guinea pig revealed one mutation, C755T, with an amino acid change of A252V. While protein PB1 mutated only in a contact-infected sentinel-derived virus with D232N, PB2 mutated only in a directly inoculated animal with E44D and N668K mutations (Table 2). In contrast to proteins HEF, PB1, and PB2 lacking uniform mutations, the other proteins exhibited mutations common to the two virus samples (derived from directly infected and sentinel animals). These mutations consisted of E41R and L316F in P42 and E247D in NP. No mutations were observed in NS protein (Table 2). Interestingly, we found that most of the observed amino acid changes had occurred at frequencies of approximately 93% to 99% in our RNA-seq reads. There were two changes (HEF A252V and PB1 D232N) occurring at around 50% frequency, indicating strong polymorphism at these two positions.
TABLE 2.
Viral genome and protein sequence changes in directly inoculated and sentinel contact-infected animal lung isolates compared to parent inoculum of FLUDVa
| Segment | Sequence category | Sequence change(s) |
|
|---|---|---|---|
| Directly inoculated animals | Sentinel infected by contact | ||
| HEF | nt | C755T | A746G |
| aa | A252V (45.2) | N249S (99.1) | |
| P42 | nt | G121A; *A122G; C946T | G121A; *A122G; C946T |
| aa | E41R (98.6); L316F (98.5) | E41R (99.5); L316F (98.5) | |
| NP | nt | G741C; *T760C; *T768C; *A792G | G741C; *T760C; *T768C; *A792G |
| aa | E247D (99.8) | E247D (99.7) | |
| P3 | nt | *A297G; *A804G | *A297G; *A804G |
| aa | |||
| PB1 | nt | *T18G; *T21C | *T18G; *T21C; G694A |
| aa | D232N (52.4) | ||
| PB2 | nt | G132C; C2004G | |
| aa | E44D (94.1); N668K (92.9) | ||
Asterisks (*) indicate silent mutations. nt, nucleotide; aa, amino acid. Note that no mutations were observed in the NS segment. Percent frequency of each amino acid mutation is given in parentheses. Unique sequence changes in sentinel infected animals are highlighted in boldface characters. Also note that, because the sequences for the PB1 and PB2 segments were not resolved in high quality in RNA-seq, the PB1 and PB2 sequence changes were calculated based on the sequence of D/660, which was used to produce the parent inoculum used for animal infection experiments.
DISCUSSION
Bovine and swine FLUD viruses are prevalent in cattle and swine populations, as demonstrated by the serological studies conducted earlier (7, 12, 18). Since FLUD is a novel virus, different from the existing influenza virus genera, a laboratory animal model would help us to understand the viral and host factors responsible for the virulence, pathogenesis, and transmissibility of this virus. In this study, we evaluated the virulence of the unadapted bovine FLUDV in guinea pigs by testing its ability to replicate and transmit by means of aerosol spread and contact. Generally, influenza viruses need to be adapted to the host species before they can replicate to high titers and induce disease in a model host (24). Adaptation of the virus in vivo results in mutations that can facilitate better receptor binding and promote viral fitness, thereby increasing replication efficiency and the virulence of the virus. These adapted viruses could be different from the parent strains. Previous studies in guinea pig and ferret models have shown exceptions and demonstrated high titers of influenza A virus in the upper respiratory tract and high transmissibility to cocaged animals without any adaptation (13, 15, 25).
Selection of an infectious dose of 3 × 105 TCID50/300 μl for guinea pig infection experiments was based on an earlier study of swine FLUDV in pigs and ferrets (7). After intranasal inoculation of bovine FLUDV, guinea pigs showed no clinical signs and behaved normally. However, detection of virus replication in the nasal washes from 1 to 7 dpi indicated that guinea pigs were susceptible to bovine FLUDV infection. Intranasal inoculation of 106 TCID50/ml swine FLUDV in pigs and ferrets also resulted in no clinical symptoms or shed virus in nasal washes from 3 dpi (7). Following intranasal inoculation of bovine FLUDV, all the infected guinea pigs seroconverted with HI antibody titers ranging from 20 to 80 within a period of 14 days. Taken together, these findings are similar to those of previous reported studies of influenza A viruses in guinea pigs, indicating that guinea pigs can act as a natural susceptible-animal model for studying the antigenicity and pathogenicity of bovine FLUDV.
Earlier studies of influenza A viruses in guinea pig models also showed that virus replication largely occurred in the upper respiratory tract with high viral titers and showed no or low detectable levels of virus in lungs (13, 15, 26). Surprisingly, bovine FLUDV replicated at high titers of 7.75 and 8 log10 TCID50/g in the lungs of directly inoculated guinea pigs at 3 and 5 dpi, respectively, and hence showed that guinea pigs can support FLUDV replication in the lower respiratory tract in a very efficient manner. The sentinel animal that acquired the infection from the infected cage mate by contact transmission also showed 8 log10 TCID50/g virus in the lungs. Similarly, a pathogenicity study of bovine FLUDV using an isolate designated D/Zoetis in gnotobiotic calves also showed 7 log10 TCID50/ml virus in the lungs with no observable clinical signs (27). Another study designed to evaluate the pathogenicity and virulence of bovine FLUDV in 8-week-old pigs demonstrated viral replication in both the upper and lower respiratory tracts, including lung, and the presence of seroconversion in all inoculated pigs and also showed transmission of the infection by contact (27). Our findings in guinea pigs are in a good agreement with those findings obtained in pigs and gnotobiotic calves with bovine FLUDV, further strengthening its validity as an animal model to study this newly emerging influenza virus.
Intriguingly, bovine FLUDV, as shown here and reported elsewhere, replicated in both the lower and upper respiratory tracts of animals (guinea pigs or pigs or calves), while swine FLUDV replicated only in the upper respiratory tract and not in the lower respiratory tract and lung of ferrets and pigs. These contrasting results indicate that bovine and swine viruses appear to differ in lung tropism in different animals. This observation is also consistent with our clinical data showing that bovine FLUDV is often isolated from lungs of diseased calves, whereas swine virus is isolated only from the upper respiratory tract of diseased pigs (data not shown). We reasoned that the difference in lung tropism between swine D/OK and two bovine viruses (D/660 and D/Zoetis) might be a result of differences in the receptor recognition of the HEF protein. Further experimental verification is required to test this prediction.
The distribution of the receptors in the mammalian host tissue is an important factor determining the host susceptibility and tissue tropism of influenza virus. Earlier studies in guinea pigs conducted to determine the distribution of α2,6 and α2,3 sialylated glycoproteins, which are the main receptors of influenza A viruses, have been reported. While both α2,6 and α2,3 sialylated glycoproteins were present in nasal epithelia and trachea, only α2,3 sialylated glycoproteins were present in the lung (28, 29). Similarly to human influenza C virus, swine and bovine FLUDV utilizes 9-O-acetyl-N-acetylneuraminic acid (Neu5,9Ac2) as the cellular receptor to trigger an infection and encodes sialate-O-acetylesterase to cleave the 9-O-acetyl group to release the virions (7). However, it is not clear whether FLUDV can bind to Neu5,9Ac2 with both linkages with similar levels of efficiency. It can be argued that bovine viruses such as D/660 and D/Zoetis bind well to the Neu5,9Ac2 receptor regardless of the specific linkage, whereas swine D/OK preferably binds only to Neu5,9Ac2 with α2,6 linkage and not to that with the α2,3 linkage present in the lungs of various animals. This hypothesis should be tested in future investigations.
The transmissibility characteristics of influenza viruses were previously studied using various animal models such as mice, ferrets, guinea pigs, pigs, and nonhuman primates (24). The presence of bovine FLUDV in the bovine populations of the United States, France, and China could be a potential threat to public health. To determine the transmissibility of bovine FLUDV, we tested two different models: contact transmission and aerosol/respiratory droplet transmission. A high level (2/3 animals) of bovine FLUDV transmission from infected guinea pigs to sentinel animals occurred by contact as demonstrated by the seroconversion (1/3 animals) and high titer of virus (1/3 animals) in the lungs in sentinels, while the aerosol route failed to cause infection. The airborne transmission experiments largely depended on environmental factors such as temperature, humidity, airflow of the experiment room, and infectious dose used. Among these factors, the rate of change in the velocity and direction of air currents in the facility are important variables, especially in aerosol transmission experiments. Previous experiments using influenza type A and B viruses in guinea pigs conducted in environmental test chambers demonstrated efficient transmission lasting for a longer period at 5°C and 20% relative humidity (RH) than at 20°C and 20% RH and also showed that high temperatures and high humidity significantly reduced the transmission efficiency (30). The use of large droplets of >5 to 10 μm in diameter facilitates only short-range transmission, while small droplets of <5 μm are responsible for long-range transmission (31). The temperature and humidity levels of our facility ranged from 22 to 24°C and from 21% to 30%, respectively, during the study. Further, we kept the cage away from the airflow of the room and did not use any controlled-air chambers for aerosol transmission study. The absence of infection in the sentinels in the aerosol transmission group can be attributed to the factors detailed above. A similar aerosol transmission study done in guinea pigs using a pandemic H1N1 2009 influenza A virus demonstrated virus shedding between days 2 and 4 dpi and also reported no transmission upon reversal of the cage positions relative to airflow, showing that the efficiency of infection increased when the orientation of the cages with infected animals to the cages with uninfected animals was in the direction of the airflow (15). The absence of persistent infection and productive replication in the sentinel animals in the aerosol group can thus be explained under the given experimental conditions. The low aerosol transmission ability of FLUDV in general puzzled us when we initially studied FLUDV in pigs. RT-PCR screening of approximately 3,000 nasal swabs of pigs with influenza-like symptoms resulted in only 4 positive-testing samples (data not shown). The low-aerosol-transmission issue was observed also in our ferret study, where FLUDV was not detected in the aerosol group despite the fact that 1 of 3 sentinel ferrets exhibited seroconversion (7). It should be noted that the inefficient transmission observed in pigs, ferrets, and guinea pigs is in a marked contrast with that observed in cattle in that approximately 15% of nasal swabs from cattle that manifested influenza-like symptoms tested positive for FLUDV (8). Thus, our current hypothesis is that FLUDV is unique among influenza viruses in that it transmits by the aerosol route efficiently only in its primary host, cattle, and not in other tested animal species, including guinea pigs. This working model will be tested in our future experiment.
Transmission experiments using influenza types A and B in various animal models also showed higher efficiency in transmission by the contact route than by the aerosol route (14). Swine FLUDV was also reported to transmit by contact in pigs and ferrets (7). In our study, two sentinel animals acquired infection by contact as demonstrated by the serum hemagglutination inhibition (HI) titer of one of the sentinel animals (HI titer, 80) and the high titer of virus present in the lung and nasal washes in the other. Interestingly, the sentinel guinea pig infected by contact acquired an infection strong enough to spread to the cocaged seropositive directly inoculated animal, causing it to start a secondary round of virus shedding in nasal washes at 14 dpi (nasal wash titer at 14 dpi, 2.25 log10 TCID50/ml); however, there was no detectable amount of virus in the lungs at 14 dpi. A serum HI titer of 40 or above is considered to be the gold standard for protection by reducing the pathogenesis of influenza viruses in humans. Our data showed that a serum HI titer of 80 in that cocaged seropositive animal definitely alleviated the pathogenesis caused by virus reactivation by reducing the amount of virus shedding in nasal washes (2.25 log10 TCID50/ml) and also prevented virus replication in the lungs at 14 dpi. Earlier studies in guinea pigs and ferrets using a potent monoclonal IgG antibody administered intramuscularly against A/California/04/2009 (H1N1) virus hemagglutinin failed to protect the animals from airborne transmission. However, the same monoclonal IgG antibody administered intranasally and also administered intramuscularly as recombinant IgA significantly reduced the virus shedding in nasal washes in a dose-dependent manner and prevented virus replication in lungs (26). This explains the low level of virus shedding in the seropositive animal and also underlines the role of mucosal immune responses versus systemic immune responses in the pathogenesis and prevention of the influenza virus infection.
Since we first described this new influenza virus in cattle, designated influenza D virus (FLUDV), in 2013, there have been published reports on the detection of this virus in diseased calves in China and France. Also, a report from a recent study included the observation that FLUDV was frequently detected in calves with acute respiratory disease and was not identified in clinically healthy animals. In addition, recognized viral etiological pathogens commonly associated with bovine respiratory disease (BRD), namely, bovine viral diarrhea virus, bovine coronavirus, bovine herpesvirus 1, and bovine respiratory syncytial virus, were not detected. These findings suggest an important role of FLUDV in the BRD complex and challenge our understanding of BRD. Despite the progress, it is not yet clear whether FLUDV can spread from bovines to humans. The zoonotic potential of bovine FLUDV has not yet been reported and is an area worth study. A serum survey conducted in humans earlier with respect to swine FLUDV showed seroprevalence in only 1.3% of the human samples tested (7). Considering the pathogenicity of bovine FLUDV and host tropism in ferrets, guinea pigs, pigs, and calves, the zoonotic potential of these viruses cannot be dismissed. Further serological studies have to be done to study the prevalence of these viruses in personnel and occupational workers closely associated with animals and animal facilities such as farmers, veterinarians, animal technicians, laboratory staff, and animal keepers.
Overall, guinea pigs can act as a good feasible model for studying the pathogenesis of influenza D viruses for the following reasons: (i) the virus was able to replicate in the nasal turbinates, and virus was shed through nasal washes; (ii) unlike other influenza types, bovine FLUDV replicated to a high titer in lungs and therefore can productively replicate in the lower respiratory tract; (iii) bovine FLUDV was transmitted from the infected guinea pigs to naive animals by contact; (iv) no adaptation is required for in vivo pathogenicity experiments; and (v) all of the directly inoculated and contact-infected guinea pigs seroconverted. The data we present here show that guinea pigs are naturally susceptible to FLUDV infection and therefore represent an excellent animal model to study host-virus interactions that would help in devising future strategies for in-depth study of the pathogenicity, antigenicity, and immunogenicity of this novel influenza virus.
ACKNOWLEDGMENTS
We thank Michele Mucciante, Diane Baker, and Amanda Sondag (Animal Resource Wing, South Dakota State University [SDSU]) for all the assistance in conducting the animal experiments. We also thank Runxia Liu and Megan Quast of the laboratory of F.L. (SDSU), Amanda Brock (SDSU), and Andrew Demers (University of Nebraska—Lincoln) for all the technical help and advice.
The study was funded in part by SDSU AES 3AH-477 (F.L.), SD00H547-15 (R.S.K.), BCAPP 3SJ-163 (F.L.), and NIH/NIAID AI107379 (F.L.).
REFERENCES
- 1.Zhang N, Zheng B, Lu L, Zhou Y, Jiang S, Du L. 18 December 2014, posting date Advancements in the development of subunit influenza vaccines. Microbes Infect doi: 10.1016/j.micinf.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hutchinson EC, Charles PD, Hester SS, Thomas B, Trudgian D, Martínez-Alonso M, Fodor E. 2014. Conserved and host-specific features of influenza virion architecture. Nat Commun 5:4816–4816. doi: 10.1038/ncomms5816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ng AK, Lam MK, Zhang H, Liu J, Au SW, Chan PK, Wang J, Shaw PC. 2012. Structural basis for RNA binding and homo-oligomer formation by influenza B virus nucleoprotein. J Virol 86:6758–6767. doi: 10.1128/JVI.00073-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ran Z, Shen H, Lang Y, Kolb EA, Turan N, Zhu L, Ma J, Bawa B, Liu Q, Liu H, Quast M, Sexton G, Krammer F, Hause BM, Christopher-Hennings J, Nelson EA, Richt J, Li F, Ma W. 2015. Domestic pigs are susceptible to infection with influenza B viruses. J Virol 89:4818–4826. doi: 10.1128/JVI.00059-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Antón A, Marcos MA, Codoner FM, de Molina P, Martinez A, Cardenosa N, Godoy P, Torner N, Martinez MJ, Ramon S, Tudo G, Isanta R, Gonzalo V, de Anta MT, Pumarola T. 2011. Influenza C virus surveillance during the first influenza A (H1N1) 2009 pandemic wave in Catalonia, Spain. Diagn Microbiol Infect Dis 69:419–427. doi: 10.1016/j.diagmicrobio.2010.11.006. [DOI] [PubMed] [Google Scholar]
- 6.Matsuzaki Y, Abiko C, Mizuta K, Sugawara K, Takashita E, Muraki Y, Suzuki H, Mikawa M, Shimada S, Sato K, Kuzuya M, Takao S, Wakatsuki K, Itagaki T, Hongo S, Nishimura H. 2007. A nationwide epidemic of influenza C virus infection in Japan in 2004. J Clin Microbiol 45:783–788. doi: 10.1128/JCM.01555-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hause BM, Ducatez M, Collin EA, Ran Z, Liu R, Sheng Z, Armien A, Kaplan B, Chakravarty S, Hoppe AD, Webby RJ, Simonson RR, Li F. 2013. Isolation of a novel swine influenza virus from Oklahoma in 2011 which is distantly related to human influenza C viruses. PLoS Pathog 9:e1003176. doi: 10.1371/journal.ppat.1003176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hause BM, Collin EA, Liu R, Huang B, Sheng Z, Lu W, Wang D, Nelson EA, Li F. 2014. Characterization of a novel influenza virus in cattle and swine: proposal for a new genus in the Orthomyxoviridae family. mBio 5:e00031-14. doi: 10.1128/mBio.00031-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collin EA, Sheng Z, Lang Y, Ma W, Hause BM, Li F. 2015. Cocirculation of two distinct genetic and antigenic lineages of proposed influenza D virus in cattle. J Virol 89:1036–1042. doi: 10.1128/JVI.02718-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ng TF, Kondov NO, Deng X, Van Eenennaam A, Neibergs HL, Delwart E. 2015. A metagenomics and case-control study to identify viruses associated with bovine respiratory disease. J Virol 89:5340–5349. doi: 10.1128/JVI.00064-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ducatez MF, Pelletier C, Meyer G. 2015. Influenza D virus in cattle, France, 2011–2014. Emerg Infect Dis 21:368–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang WM, Wang SC, Peng C, Yu JM, Zhuang QY, Hou GY, Liu S, Li JP, Chen JM. 2014. Identification of a potential novel type of influenza virus in Bovine in China. Virus Genes 49:493–496. doi: 10.1007/s11262-014-1107-3. [DOI] [PubMed] [Google Scholar]
- 13.Gabbard JD, Dlugolenski D, Van Riel D, Marshall N, Galloway SE, Howerth EW, Campbell PJ, Jones C, Johnson S, Byrd-Leotis L, Steinhauer DA, Kuiken T, Tompkins SM, Tripp R, Lowen AC, Steel J. 2014. Novel H7N9 influenza virus shows low infectious dose, high growth rate, and efficient contact transmission in the guinea pig model. J Virol 88:1502–1512. doi: 10.1128/JVI.02959-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lowen AC, Bouvier NM, Steel J. 2014. Transmission in the Guinea pig model. Current topics in microbiology and immunology 385:157–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lowen AC, Mubareka S, Tumpey TM, Garcia-Sastre A, Palese P. 2006. The guinea pig as a transmission model for human influenza viruses. Proc Natl Acad Sci U S A 103:9988–9992. doi: 10.1073/pnas.0604157103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang L, Chitano P, Murphy TM. 2005. Length oscillation induces force potentiation in infant guinea pig airway smooth muscle. Am J physiology Lung cellular and molecular physiology 289:L909–L915. doi: 10.1152/ajplung.00128.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Azoulay-Dupuis E, Lambre CR, Soler P, Moreau J, Thibon M. 1984. Lung alterations in guinea-pigs infected with influenza virus. J comparative pathology 94:273–283. doi: 10.1016/0021-9975(84)90046-X. [DOI] [PubMed] [Google Scholar]
- 18.Collin EA, Sheng Z, Lang Y, Ma W, Hause BM, Li F. 29 October 2014. Co-circulation of two distinct genetic and antigenic lineages of proposed influenza D virus in cattle. J Virol doi: 10.1128/JVI.02718-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reed L, Muench H. 1938. A simple method of estimating fifty percent endpoints. Am J Hyg 27:493–497. [Google Scholar]
- 20.World Health Organization. 2011. Global Influenza Surveillance Network. Manual for the laboratory diagnosis and virological surveillance of influenza. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 21.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 22.Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A. 2003. ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res 31:3784–3788. doi: 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, Thompson JD, Higgins DG. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Margine I, Krammer F. 2014. Animal models for influenza viruses: implications for universal vaccine development. Pathogens 3:845–874. doi: 10.3390/pathogens3040845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Belser JA, Katz JM, Tumpey TM. 2011. The ferret as a model organism to study influenza A virus infection. Dis Model Mech 4:575–579. doi: 10.1242/dmm.007823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seibert CW, Rahmat S, Krause JC, Eggink D, Albrecht RA, Goff PH, Krammer F, Duty JA, Bouvier NM, Garcia-Sastre A, Palese P. 2013. Recombinant IgA is sufficient to prevent influenza virus transmission in guinea pigs. J Virol 87:7793–7804. doi: 10.1128/JVI.00979-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rapp-Gabrielson VJ. 2013. Abstr 94th Annu Meet Conf Res Workers Anim Dis (CRWAD), Chicago, IL, USA, p 165. [Google Scholar]
- 28.Gao Y, Zhang Y, Shinya K, Deng G, Jiang Y, Li Z, Guan Y, Tian G, Li Y, Shi J, Liu L, Zeng X, Bu Z, Xia X, Kawaoka Y, Chen H. 2009. Identification of amino acids in HA and PB2 critical for the transmission of H5N1 avian influenza viruses in a mammalian host. PLoS Pathog 5:e1000709. doi: 10.1371/journal.ppat.1000709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun Y, Bi Y, Pu J, Hu Y, Wang J, Gao H, Liu L, Xu Q, Tan Y, Liu M, Guo X, Yang H, Liu J. 2010. Guinea pig model for evaluating the potential public health risk of swine and avian influenza viruses. PLoS One 5:e15537. doi: 10.1371/journal.pone.0015537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pica N, Chou YY, Bouvier NM, Palese P. 2012. Transmission of influenza B viruses in the guinea pig. J Virol 86:4279–4287. doi: 10.1128/JVI.06645-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mubareka S, Lowen AC, Steel J, Coates AL, Garcia-Sastre A, Palese P. 2009. Transmission of influenza virus via aerosols and fomites in the guinea pig model. J Infect Dis 199:858–865. doi: 10.1086/597073. [DOI] [PMC free article] [PubMed] [Google Scholar]