Abstract
To resolve the evolutionary history of rabbit hemorrhagic disease virus (RHDV), we performed a genomic analysis of the viral stocks imported and released as a biocontrol measure in Australia, as well as a global phylogenetic analysis. Importantly, conflicts were identified between the sequences determined here and those previously published that may have affected evolutionary rate estimates. By removing likely erroneous sequences, we show that RHDV emerged only shortly before its initial description in China.
TEXT
The emergence of rabbit hemorrhagic disease virus (RHDV; Caliciviridae) in China in 1984 (1) initiated a wave of severe disease that spread rapidly through rabbit (Oryctolagus cuniculus) populations in Asia, Europe, and the Americas (2–4). As a consequence of its exceptional virulence, high transmissibility, and narrow host range, in 1991, a Czech strain of RHDV, CAPM V351, was imported into Australia, where it was evaluated over the next 4 years at the Australian Animal Health Laboratory as a possible biological control agent against introduced European rabbit populations (5, 6). In 1995, during quarantined field trials on Wardang Island, South Australia, the virus escaped to the mainland (7), starting an epizootic that continues to this day (8, 9). In 1997, this Czech strain was illegally introduced into New Zealand from Australia (10). Despite this well-documented history, little is known about how the virus evolved over the 4 years from its importation to its eventual release, during which time experimental stocks were prepared and passaged in rabbits (9). More broadly, the integrity of such early sequences may have major implications for estimates of the timing of RHDV emergence globally (11), with most previous molecular clock-based studies suggesting that RHDV originated decades before its initial description in China (12–14). Given the extreme virulence of RHDV, such an early emergence is both puzzling and contentious (5, 15). Previous attempts to resolve this paradox have led to conflicting theories about the roles of persistent, long-term infections and cross-immunity from nonpathogenic lagoviruses that may dampen the virulence of pathogenic RHDV (16, 17). The apparently spontaneous emergence of highly pathogenic RHDV has also been utilized in general models of virulence evolution (18).
To help resolve the debate about the origins of RHDV, we sequenced the original stocks used in Australia (8), focusing on single nucleotide polymorphisms (SNPs) to determine the extent of viral heterogeneity and how it was maintained through early passaging. This analysis revealed that the original master stock of imported RHDV contained 15 SNPs (frequency of >1%) at 14 genomic positions (Fig. 1A). Most (53.3%) of these were located within the gene for viral capsid protein VP60 and included four nonsynonymous mutations. The master stock was passaged in rabbits to obtain an intermediate working stock (passage 1), and then a final concentrated stock (passage 2) was used for all subsequent releases of the virus. Although viral diversity was purged during passaging, with most (80%) of the SNPs decreasing in frequency, the SNP frequency increased at three sites—positions 2528 (in the gene for p29), 6217, and 6615 (both in the gene for VP60). Notably, the nonsynonymous SNP C6217A became the dominant variant, increasing in frequency from 41.4% in the master stock to 84.7% in the release stock. Structural mapping onto the VP60 capsid (Fig. 1B) revealed that SNP C6217A (VP60 amino acid Pro-305-Thr) is located on a prominent external loop adjacent to antigenic and receptor-binding sites (19, 20), compatible with viral adaptation during passage. Interestingly, viruses circulating during the initial RHDV epizootic in South Australia contained either Pro or Thr at this position, further suggesting that the release stocks were heterogenous. Two independent sources (dilutions) of the release stock (Elizabeth Macarthur Agricultural Institute batches 1A and 1D) were also sequenced, which revealed remarkable consistency with the same consensus sequence and identical SNPs.
FIG 1.
SNPs in the original stocks of RHDV introduced into Australia. Panel A shows the genome positions (nucleotides) and frequencies (percent) of the SNPs in the original RHDV stocks. Trends in SNP frequency are indicated following passaging from the master stock, through an intermediate working stock, to the release strain with SNPs of increasing and decreasing frequencies plotted above and below the x axis, respectively. The genome positions on a gray background are the SNPs located in the major capsid protein (VP60). Panel B maps the three nonsynonymous SNPs in the capsid gene onto the published crystal structure of the RHDV VP60 protruding (P) domain dimer (Protein Data Bank code 4EGT). A side view is shown with the surface of one subunit (colored gray) in the background, the ribbon structure of the other subunit in the foreground, and the P1 and P2 subdomains colored blue and yellow, respectively. The table provides the codon sequences of all of the SNPs located in the capsid, with the nucleotide (nt) of each SNP in bold and the coding amino acid (aa) in brackets. The prototype sequence corresponds to the majority consensus sequence of the original master stock. Across both panels, shades of green and red indicate trends of increasing and decreasing SNP frequency, respectively.
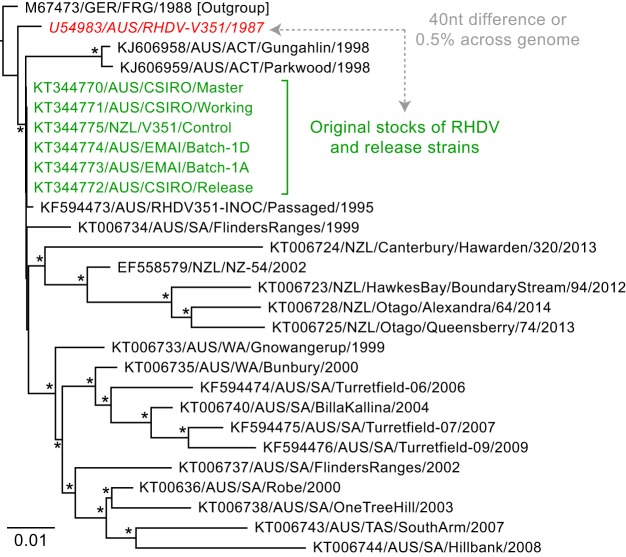
As these sequences represent the most likely founder viruses of the Australian RHDV epizootics, we estimated a genome-wide phylogenetic tree of CAPM V351-derived strains, including the Australian RHDV stocks and another V351 preparation from New Zealand, by using the GTR+Γ model in PhyML (21) (Fig. 2). This confirmed that, with the exception of SNP C6217A in the master stock, the consensus sequences of all of the viral stocks were in agreement yet, importantly, conflicted with the previously published sequence of the virus imported into Australia, known as RHDV V351 (GenBank accession no. U54983). The RHDV V351 genome contains 40 nucleotide differences (0.54%) from those generated here, such that it was either sequenced in error or a minor variant was selectively sequenced from the viral population during cloning. The evidence favors the latter, as an examination of minor viral populations across a small region (positions 3375 to 3732) in the original RHDV V351 publication (22) identified one variable position (nucleotide 3429) that corresponded to one of the differences between RHDV V351 and the stock viruses sequenced here. Regardless, the published RHDV V351 sequence evidently does not reflect the sequence of the founding virus in Australia and should be avoided in comparative analyses.
FIG 2.
Phylogenetic analysis of the original stocks of RHDV from Australia and New Zealand. Shown is a phylogeny of the complete genome sequences of 28 RHDV strains, including six original stocks of RHDV from Australia and New Zealand (green), as well as the previously published sequences of the purported founder virus, RHDV V351 (red). The gray dashed line shows the sequences compared here with the number of mismatches (nucleotides [nt]) and overall genetic difference (percent) indicated. The phylogeny was rooted with an early European RHDV strain (outgroup), and branch lengths are scaled according to the number of nucleotide substitutions per site. Support for individual nodes was estimated from 1,000 bootstrap replicates with asterisks marking nodes with ≥70% bootstrap support.
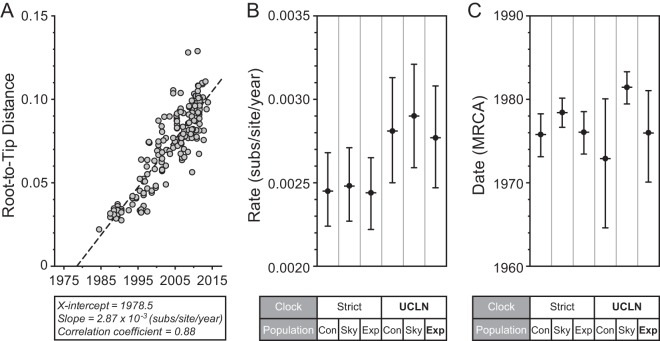
Previous estimates of the evolutionary rate of the VP60 gene of global RHDV range from 7.2 × 10−4 to 1.9 × 10−3 substitutions per site per year (11, 14). Critically, under these rates, RHDV emerged up to 67 years prior to its first description in 1984, which implies that large-scale rabbit deaths went undetected for many years. However, it is likely that these early molecular clock studies incorporated sequences that have been compromised by contamination, poor sequencing, viral heterogeneity, or misdating. Indeed, most previous studies have included the original RHDV V351 sequence (9, 11, 12, 14), even though a single incorrect sequence can have a major impact on evolutionary rate estimates (11). In light of this, we compiled a carefully curated data set of GenBank-derived VP60 sequences (n = 174) to re-estimate the rate and time scale of RHDV evolution (data available upon request). The final alignment included all of the “classic” RHDV isolates (clades A to D) with any sequences removed that contained evidence of recombination, ambiguous collection dates, or large deletions or that were from later passages of vaccine stocks. Importantly, these data exhibited a strong clock-like structure (Fig. 3A). A Bayesian approach (23) was then used to estimate both the evolutionary rate (Fig. 3B) and the time of RHDV origin (Fig. 3C). This incorporated both strict and relaxed (uncorrelated lognormal) molecular clocks, a constant population size, exponential growth, and the Gaussian Markov random field skyride coalescent models (24). Using marginal likelihood estimators (25), the relaxed-clock model with exponential population growth was favored. Accordingly, the mean rate of RHDV evolution was estimated to be 2.77 × 10−3 substitutions per site per year (95% highest probability density [HPD] of 2.47 × 10−3 to 3.08 × 10−3 substitutions per site per year) (Fig. 3B). This rate was significantly higher than previous estimates of global RHDV data (11) but in agreement with more recent and independent studies of RHDV evolution in Australia (8, 9). Importantly, estimates of the evolutionary time scale based on this revised rate place the date of origin of RHDV between 1970 and 1981 (95% HPD intervals) and hence only shortly before its first description in 1984 and without a long “phylogenetic fuse” of undetected transmission.
FIG 3.
Estimates of the evolutionary rate of RHDV and the timing of its emergence. A set of 174 complete VP60 capsid sequences from globally circulating RHDV strains was used to estimate a maximum-likelihood phylogeny with PhyML. Panel A shows the root-to-tip genetic distances plotted against the sampling time with linear regression performed to examine the clock-like structure of the data. A Bayesian Markov chain Monte Carlo approach was then used to estimate the evolutionary rate (number of substitutions per site per year), shown in panel B, and the date of emergence (i.e., time to the most recent common ancestor [MRCA]), shown in panel C, for the same VP60 data set and a range of different models. This analysis incorporated both strict and relaxed (uncorrelated lognormal [UCLN]) molecular clocks, as well as constant-size (Con), exponential-growth (Exp), and Gaussian Markov random-field skyride (Sky) coalescent models. In panels B and C, the mean values are shown with black circles with the ranges reporting the 95% highest probability densities and the best-fit model, in this case, the relaxed clock (UCLN) with an exponential-growth population model (bold).
Nucleotide sequence accession numbers.
Newly determined sequences have been submitted to GenBank under accession numbers KT344770 to KT344775.
ACKNOWLEDGMENTS
This work was support by grant DP140103362 from the Australian Research Council and grant R01 AI093804-01A1 from the National Institutes of Health. E.C.H. is supported by an NHMRC Australia Fellowship (AF30). J.-S.E. is supported by an NHMRC Early Career Fellowship (1073466).
REFERENCES
- 1.Liu SJ, Xue HP, Pu BQ, Qian NH. 1984. A new viral disease in rabbits. Anim Husbandry Vet Med 16:253–255. [Google Scholar]
- 2.Cancellotti FM, Renzi M. 1991. Epidemiology and current situation of viral haemorrhagic disease of rabbits and the European brown hare syndrome in Italy. Rev Sci Tech 10:409–422. [DOI] [PubMed] [Google Scholar]
- 3.Gregg DA, House C. 1989. Necrotic hepatitis of rabbits in Mexico: a parvovirus. Vet Rec 125:603–604. [PubMed] [Google Scholar]
- 4.Park NY, Chong CJ, Kim JH, Cho SM, Cha YH, Jung BT, Kim DS, Yoon JB. 1987. An outbreak of viral haemorrhagic pneumonia (tentative name) of rabbits in Korea. J Korean Vet Med Assoc 23:603–610. [Google Scholar]
- 5.Cooke BD. 2014. Australia's war against rabbits: the story of rabbit haemorrhagic disease. CSIRO Publishing, Collingwood, Australia. [Google Scholar]
- 6.Collins BJ, White JR, Lenghaus C, Boyd V, Westbury HA. 1995. A competition ELISA for the detection of antibodies to rabbit haemorrhagic disease virus. Vet Microbiol 43:85–96. doi: 10.1016/0378-1135(94)00082-8. [DOI] [PubMed] [Google Scholar]
- 7.Kovaliski J. 1998. Monitoring the spread of rabbit hemorrhagic disease virus as a new biological agent for control of wild European rabbits in Australia. J Wildl Dis 34:421–428. doi: 10.7589/0090-3558-34.3.421. [DOI] [PubMed] [Google Scholar]
- 8.Eden JS, Kovaliski J, Duckworth JA, Swain G, Mahar JE, Strive T, Holmes EC. 2015. Comparative phylodynamics of rabbit hemorrhagic disease virus in Australia and New Zealand. J Virol 89:9548–9558. doi: 10.1128/JVI.01100-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kovaliski J, Sinclair R, Mutze G, Peacock D, Strive T, Abrantes J, Esteves PJ, Holmes EC. 2014. Molecular epidemiology of rabbit haemorrhagic disease virus in Australia: when one became many. Mol Ecol 23:408–420. doi: 10.1111/mec.12596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O'Hara P. 2006. The illegal introduction of rabbit haemorrhagic disease virus in New Zealand. Rev Sci Tech 25:119–123. [DOI] [PubMed] [Google Scholar]
- 11.Hicks AL, Duffy S. 2012. One misdated sequence of rabbit hemorrhagic disease virus prevents accurate estimation of its nucleotide substitution rate. BMC Evol Biol 12:74. doi: 10.1186/1471-2148-12-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kinnear M, Linde CC. 2010. Capsid gene divergence in rabbit hemorrhagic disease virus. J Gen Virol 91:174–181. doi: 10.1099/vir.0.014076-0. [DOI] [PubMed] [Google Scholar]
- 13.Moss SR, Turner SL, Trout RC, White PJ, Hudson PJ, Desai A, Armesto M, Forrester NL, Gould EA. 2002. Molecular epidemiology of rabbit haemorrhagic disease virus. J Gen Virol 83:2461–2467. doi: 10.1099/0022-1317-83-10-2461. [DOI] [PubMed] [Google Scholar]
- 14.Kerr PJ, Kitchen A, Holmes EC. 2009. Origin and phylodynamics of rabbit hemorrhagic disease virus. J Virol 83:12129–12138. doi: 10.1128/JVI.01523-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Forrester NL, Trout RC, Turner SL, Kelly D, Boag B, Moss S, Gould EA. 2006. Unravelling the paradox of rabbit haemorrhagic disease virus emergence, using phylogenetic analysis; possible implications for rabbit conservation strategies. Biol Conservation 131:296–306. doi: 10.1016/j.biocon.2006.05.005. [DOI] [Google Scholar]
- 16.Gould EA. 2012. First case of rabbit haemorrhagic disease in Canada: contaminated flying insect, vs. long-term infection hypothesis. Mol Ecol 21:1042–1047. doi: 10.1111/j.1365-294X.2012.05462.x. [DOI] [PubMed] [Google Scholar]
- 17.Peacock D, Mutze G, Sinclair R, Kovaliski J, Cooke B. 2012. Rabbit haemorrhagic disease: applying Occam's razor to competing hypotheses. Mol Ecol 21:1038–1041. doi: 10.1111/j.1365-294X.2011.05466.x. [DOI] [PubMed] [Google Scholar]
- 18.Boots M, Hudson PJ, Sasaki A. 2004. Large shifts in pathogen virulence relate to host population structure. Science 303:842–844. doi: 10.1126/science.1088542. [DOI] [PubMed] [Google Scholar]
- 19.Wang X, Xu F, Liu J, Gao B, Liu Y, Zhai Y, Ma J, Zhang K, Baker TS, Schulten K, Zheng D, Pang H, Sun F. 2013. Atomic model of rabbit hemorrhagic disease virus by cryo-electron microscopy and crystallography. PLoS Pathog 9:e1003132. doi: 10.1371/journal.ppat.1003132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thouvenin E, Laurent S, Madelaine MF, Rasschaert D, Vautherot JF, Hewat EA. 1997. Bivalent binding of a neutralising antibody to a calicivirus involves the torsional flexibility of the antibody hinge. J Mol Biol 270:238–246. doi: 10.1006/jmbi.1997.1095. [DOI] [PubMed] [Google Scholar]
- 21.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 22.Gould AR, Kattenbelt JA, Lenghaus C, Morrissy C, Chamberlain T, Collins BJ, Westbury HA. 1997. The complete nucleotide sequence of rabbit haemorrhagic disease virus (Czech strain V351): use of the polymerase chain reaction to detect replication in Australian vertebrates and analysis of viral population sequence variation. Virus Res 47:7–17. doi: 10.1016/S0168-1702(96)01399-8. [DOI] [PubMed] [Google Scholar]
- 23.Drummond AJ, Rambaut A. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol 7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Minin VN, Bloomquist EW, Suchard MA. 2008. Smooth skyride through a rough skyline: Bayesian coalescent-based inference of population dynamics. Mol Biol Evol 25:1459–1471. doi: 10.1093/molbev/msn090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baele G, Lemey P, Bedford T, Rambaut A, Suchard MA, Alekseyenko AV. 2012. Improving the accuracy of demographic and molecular clock model comparison while accommodating phylogenetic uncertainty. Mol Biol Evol 29:2157–2167. doi: 10.1093/molbev/mss084. [DOI] [PMC free article] [PubMed] [Google Scholar]