ABSTRACT
The nairoviruses include assorted tick-borne bunyaviruses that are emerging as causative agents of infectious diseases among humans and animals. As negative-sense single-stranded RNA (−ssRNA) viruses, nairoviruses encode nucleoprotein (NP) that encapsidates the genomic RNA and further forms ribonucleoprotein (RNP) complex with viral RNA-dependent RNA polymerase (RdRp). We previously revealed that the monomeric NP encoded by Crimean-Congo hemorrhagic fever virus (CCHFV) presents a racket-shaped structure and shows unusual DNA-specific endonuclease activity. To examine the structural and biological variation of nairovirus-encoded NPs, here, we systematically solved the crystal structures of NPs encoded by various nairoviruses, including Hazara virus (HAZV), Kupe virus (KUPV), and Erve virus (ERVEV). Combined with biochemical analysis, our results generate a clearer picture to aid in the understanding of the functional diversity of nairovirus-encoded NPs and the formation of nairovirus RNPs.
IMPORTANCE Nairoviruses comprise several tick-borne bunyaviruses that are emerging as causative agents of infectious diseases among humans and animals; however, little is known of the nairovirus genome assembly and transcription mechanisms. Based on the previous study of CCHFV NP reported by different research groups, we systematically investigate here the structural and functional diversity among three different nairoviruses. This work provides important information on nairovirus nucleoprotein function and the formation of RNPs.
INTRODUCTION
Bunyaviridae is the largest negative-sense, single-stranded (−ssRNA) virus family and contains various pathogens that cause severe infectious diseases in humans, other animals, and plants (1). Over 300 members of the Bunyaviridae family are currently classified into five distinct genera, i.e., Orthobunyavirus, Hantavirus, Nairovirus, Phlebovirus, and Tospovirus (1). Bunyaviruses are characterized by their trisegmented negative-sense genome, in which the large (L) segment encodes RNA-dependent RNA polymerase (RdRp), the medium (M) segment encodes surface glycoproteins, and the small (S) segment encodes nucleocapsid (NP) protein (1). As in other −ssRNA viruses, the genomic RNA of bunyaviruses does not exist as a naked RNA but must be encapsidated by viral NP to form a high-order NP-RNA complex, which further constructs a ribonucleoprotein (RNP) complex with viral RdRp (2, 3).
The Nairovirus genus comprises a number of pathogens that cause infectious diseases in humans and livestock. Nairoviruses are transmitted by ticks, which distinguishes them from other members of the Bunyaviridae family (4). Based on antibody cross-reactivities, nairoviruses are classified into seven serogroups (see Table S1 in the supplemental material) (5). CCHFV, which belongs to the CCHF serogroup, is the most notable pathogen: it causes CCHF in humans, with high mortality, and occurs spontaneously in more than 30 countries in Asia, the Middle East, southeastern Europe, and Africa (6). The high pathogenicity of CCHFV has led to its being considered a severe public health threat and a potential bioterrorism agent worldwide (6). However, as the study of CCHFV is limited by its classification as a biosafety level 4 (BSL-4) level agent, Hazara virus (HAZV), which is closely related to CCHFV and has not been reported to infect humans, is believed to be a good surrogate model for CCHFV infection (7). Viruses in the Nairobi sheep disease group, consisting of Nairobi sheep disease virus (NSDV), Dugbe virus (DUGV), Kupe virus (KUPV), and Finch Creek virus, are frequently isolated from ticks infesting livestock. Erve virus (ERVEV), isolated in 1982 and classified as part of the Thiafora virus group, has been suggested to be one of the causative agents of human thunderclap headaches (8, 9). Understanding the detailed biology of nairoviruses is critical to the development of efficient diagnostics and therapeutics (10).
Although all bunyaviruses require correct genome encapsidation and RNP formation for their proliferation, they have great diversity in the sequence, structure, and function of their NPs (2, 3). For example, the molecular mass of orthobunyavirus-encoded NP is 27 kDa, but the molecular mass of nairovirus-encoded NP is approximately 52 kDa (2). Recent studies have revealed that the NPs of Rift Valley fever virus (RVFV), severe fever with thrombocytopenia syndrome virus (SFTSV), and Toscana virus, which are members of the Phlebovirus genus, present novel protein folding and facilitate oligomerization through N-terminal arms (11-16). Moreover, the NP-RNA complex structures of Bunyamwera virus (BUNV) (17), Leanyer virus (LEAV) (18), La Crosse orthobunyavirus (LACV) (13), and Schmallenberg virus (SBV) (19, 20) revealed that orthobunyavirus-encoded NPs possess N- and C-terminal extensions for interprotomer interaction.
In our previous work, we solved the crystal structure of CCHFV (strain YL04057) NP in a monomeric form, presenting a racket-shaped structure with a head and a stalk domain (PDB accession number 3U3I) (21). Although the head domain of CCHFV NP shares high structural similarities to the N-terminal domain of Lassa fever virus (LASV; belonging to the Arenaviridae family) NP (22, 23), it exhibits an unusual metal-dependent, DNA-specific endonuclease activity (21). Subsequently, Carter et al. reported the structure of NP from CCHFV strain Baghdad-12 (PDB accession number 4AKL) and revealed a significant transposition of the stalk domain through a rotation of 180° and a translation of 40 Å compared to the NP from strain YL04057, indicating the structural flexibility to switch between alternative NP conformations during RNA binding or oligomerization (24). Furthermore, Wang et al. studied the structure of CCHFV (strain IbAr10200) NP pretreated with poly(rU) and revealed another conformational difference in the stalk domain (25) (PDB accession numbers 4AQF and 4AQG). Although a head-to-tail interaction of the stalk domain with the base of the head domain of the adjacent subunit was proposed to stabilize the superhelical organization of the oligomeric form of CCHFV NP (strain IbAr10200), poly(rU) cannot be found in this structure, and the authentic oligomerization state still has not been experimentally observed.
To further explore the structural and functional variability across the Nairovirus genus, in this work, we systematically studied the NPs encoded by representative species from three serogroups. These results provide more information on the distinguishing features and conservation of nairovirus RNP formation.
MATERIALS AND METHODS
Protein production.
Codon-optimized cDNAs for full-length ERVEV NP, KUPV NP, and HAZV NP were synthesized by Genewiz. ERVEV NP [residues 1 to 482, denoted as ERVEV NP(1–482)], KUPV NP (full length), and HAZV NP (full length), were cloned into the pGEX-6p-1 (GE Healthcare) vector with BamHI and XhoI restriction sites using the cloning primers. The sequences of the primers were as follows: for ERVEV NP, forward, 5′-CCGGGATCCATGGAGAATTTAATCG-3′, and reverse, 5′-GGCCTCGAGTTAGATCTTAGGCGGGTC-3′; for KUPV NP, forward, 5′-CGGGATCCATGGAAAATCAGATTAAAGC-3′, and reverse, 5′-GGCCTCGAGTTACACAATCTCCACATTG-3′; and for HAZV NP, forward, 5′-CGGGATCCATGGAGAACAAAATTGT-3′, and reverse, 5′-GGCCTCGAGTTAGATGATGTTAATATTGG-3′. The primary sequences are depicted in Fig. S1 in the supplemental material, and the accuracy of the inserts was verified by sequencing.
The recombinant plasmid encoding ERVEV NP(1–482) was transformed into Escherichia coli strain BL21(DE3) and overexpressed as a glutathione S-transferase (GST) fusion protein. The cells were cultured at 37°C in 800 ml LB medium containing 100 μg/ml ampicillin. Once the optical density at 600 nm (OD600) reached ∼0.6, the culture was transferred to 16°C, and protein production was induced by incubating with 0.2 mM isopropyl-β-d-1-thiogalactopyranoside (IPTG) for an additional 16 h. Harvested cells were resuspended in lysis buffer containing 20 mM HEPES (pH 6.8), 500 mM NaCl, 1 μg/ml DNase I, and 1 μg/ml RNase and homogenized with a low-temperature, ultra-high-pressure cell disrupter (JNBIO). The lysate was centrifuged at 25,000 × g for 30 min at 4°C to remove cell debris. The supernatant was then loaded twice onto a GST column preequilibrated with lysis buffer, and the GST tag was removed by digestion with PreScission protease (GE Healthcare) overnight at 4°C. Eluted ERVEV NP(1–482) protein was further purified by Superdex-200 gel filtration chromatography (GE Healthcare). SDS-PAGE analysis revealed over 97% purity of the final purified recombinant protein. The purified protein was then concentrated to 8 mg/ml in a buffer containing 20 mM HEPES (pH 6.8), 200 mM NaCl. The protein production and purification process for KUPV NP and HAZV NP was similar to that for ERVEV NP(1–482) described above.
Crystallization.
The initial crystallization conditions were screened by the hanging-drop vapor diffusion method using commercial crystal screening kits at 16°C, including the Index, Crystal Screen, PEG/Ion, SaltRX, and Crystal Screen cryogenic kits from Hampton Research. Crystals were obtained by mixing 1 μl of the protein solution with an equal volume of a reservoir solution and equilibrating the mixed drop against 300 μl of reservoir solution.
Small crystals of ERVEV NP(1–482) first appeared after 1 day in 200 mM sodium malonate and 20% (wt/vol) polyethylene glycol 3350 (PEG 3350). Further optimization with additive and detergent screens (Hampton Research) was performed, and the final optimized crystals were grown in 200 mM sodium malonate, 20% (wt/vol) PEG 3350, and 7% (vol/vol) 1-butanol. Rodlike crystals grew to a final size of 40 by 40 by 180 μm within 1 day at 4°C. Crystals were harvested and cryoprotected in the well solution containing 4 M sodium formate and cooled in a dry nitrogen stream at 100 K for X-ray data collection.
Meanwhile, another form of crystal appeared after 3 weeks in 1% tryptone, 0.05 M HEPES (pH 7.0), 20% PEG 3350 at 16°C. No further optimization was performed. Crystals were harvested and cryoprotected in the well solution containing an additional 20% (vol/vol) glycerol and cooled in a dry nitrogen stream at 100 K for X-ray data collection. Actually, this crystal proved to be a degraded product of ERVEV NP(1–482) after structure determination and was named NPhead (residues 1 to 180 and 295 to 482).
Tiny crystals of KUPV NP first appeared after 4 h in 4% (vol/vol) Tacsimate (pH 4.0) and 12% (wt/vol) PEG 3350. Further optimization with additive and detergent screens (Hampton Research) was performed, and the final optimized crystals were grown in 200 mM sodium nitrate (pH 8.0), 5 mM cobalt chloride, and 8% (wt/vol) PEG 3350. Cubelike crystals grew to a final size of 80 by 80 by 80 μm within 1 day at 4°C. Crystals were harvested and cryoprotected in the well solution containing an additional 20% (vol/vol) glycerol and cooled in a dry nitrogen stream at 100 K for X-ray data collection.
Small twinned crystal of HAZV NP first appeared after 3 days in 200 mM potassium citrate tribasic monohydrate (pH 8.0) and 20% (wt/vol) PEG 3350. Further optimization with additive and detergent screens (Hampton Research) was performed, and the final optimized crystals were grown in 200 mM potassium citrate tribasic monohydrate (pH 8.0), 5 mM manganese chloride, and 14% (wt/vol) PEG 3350. Crystals grew to a final size of 50 by 40 by 80 μm within 3 days at 20°C. Crystals were harvested and cryoprotected in the well solution containing an additional 20% (vol/vol) glycerol and cooled in a dry nitrogen stream at 100 K for X-ray data collection.
X-ray data collection, processing, and structure determination.
Data were collected under cryogenic conditions at 100 K at beamlines BL17U and BL19U (SSRF, China). Data sets were processed using the HKL2000 package (26). The initial phases were calculated by molecular replacement (MR) in the program CCP4 with CCHFV NP (PDB accession number 3U3I) as the starting model. All the models were manually built into the modified experimental electron density using COOT (27) and further refined in PHENIX (28). The final refinement statistics are summarized in Table 1. Model geometry was verified using the program PROCHECK (29). Structural figures were drawn using the program PyMOL (30). The coordinates and structure factors were deposited in the RCSB.
TABLE 1.
Data collection and refinement statistics
| Parameters | Value(s) for indicated NP |
|||
|---|---|---|---|---|
| ERVEV NP(1–482) | ERVEV NPhead | KUPV NP | HAZV NP | |
| Data collection statistics | ||||
| Cell parameters | ||||
| a (Å) | 49.90 | 46.08 | 73.98 | 67.40 |
| b (Å) | 67.90 | 104.29 | 73.98 | 72.17 |
| c (Å) | 154.08 | 70.54 | 423.98 | 453.67 |
| α, β, γ (°) | 90.0, 93.6, 90.0 | 90.0, 92.4, 90.0 | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 |
| Space group | P21 | P21 | P41212 | P212121 |
| Wavelength used (Å) | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
| Resolution (Å)a | 50.00–2.35 (2.39–2.35) | 50.00–1.80 (1.83–1. 80) | 50.00–2.60 (2.64–2.60) | 50.00–2.90 (2.95–2.90) |
| No. of all reflectionsa | 184,478 (50,568) | 227,184 (11,363) | 421,320 (17,390) | 256,728 (17,338) |
| No. of unique reflectionsa | 42,902 (2,134) | 61,401 (3,071) | 37,285 (1,739) | 42,788 (2,442) |
| Completeness (%) | 99.3 (100.0) | 99.6 (100.0) | 98.5 (93.9) | 84.7 (100) |
| Average I/σ(I) | 9.1 (3.9) | 14.7 (3.6) | 20.9 (4.3) | 11.1 (4.0) |
| Rmergeb (%) | 12.6 (39.9) | 7.7 (35.1) | 9.2 (42.2) | 11.6 (35.2) |
| Refinement statistics | ||||
| No. of reflections used [σ(F) > 0] | 42,875 | 61,367 | 37,162 | 42,731 |
| Rworkc (%) | 17.4 | 16.8 | 21.2 | 20.7 |
| Rfreed (%) | 24.8 | 20.2 | 28.0 | 28.7 |
| RMSD | ||||
| Bond distance (Å) | 0.008 | 0.006 | 0.009 | 0.009 |
| Bond angle (°) | 1.109 | 0.992 | 1.150 | 0.922 |
| Average B value (Å2): | ||||
| For protein atoms | 44.1 | 30.0 | 51.4 | 62.6 |
| For ligand atoms | 0 | 0 | 0 | 0 |
| For solvent atoms | 46.3 | 37.7 | 44.0 | 39.2 |
| No. of: | ||||
| Protein atoms | 7,381 | 5,750 | 7,340 | 14,874 |
| Ligand atoms | 0 | 0 | 0 | 0 |
| Solvent atoms | 270 | 679 | 92 | 104 |
| Ramachandran plot (%) | ||||
| Favored regions | 95.9 | 97.5 | 93.1 | 96.1 |
| Allowed regions | 3.9 | 2.4 | 6.5 | 3.9 |
| Outlier regions | 0.2 | 0.1 | 0.4 | 0 |
Numbers in parentheses are corresponding values for the highest-resolution shell.
Rmerge = ΣhΣl |Iih − <Ih>|/ΣhΣI <Ih>, where <Ih> is the mean of the observations Iih of reflection h.
Rwork = Σ(||Fp(obs)| − |Fp(calc)||)/Σ|Fp(obs)|.
Rfree is an R factor for a preselected subset (5%) of reflections that was not included in the refinement.
EMSA.
Electrophoretic mobility shift assays (EMSAs) were performed to evaluate potential differences between the nairovirus-encoded NPs and RNA probe, following the conventional method by using a commercial EMSA kit (chemiluminescent EMSA kit; Beyotime Institute of Biotechnology, Haimen, China). Biotin-labeled ssRNA (5′-UCUCAAAGAAAGUUG-3′) was synthesized by TaKaRa Biotechnology (Dalian, China). The reaction mixture was loaded on a 10% native polyacrylamide gel in 0.5% Tris-borate-EDTA, and then the protein-RNA complex was transferred and UV cross-linked for 15 min onto nylon membranes (FFN12; Beyotime Institute of Biotechnology). Bands were visualized using a biotin-labeling probe kit (BeyoECL Plus; Beyotime Institute of Biotechnology).
Endonuclease activity assay.
The endonuclease activity assay was conducted as previously described (21). Purified CCHFV NP, ERVEV NP, HAZV NP, KUPV NP (each at 0.3 μM), and double-stranded DNA (dsDNA) substrate (PCR product, 210 bp in length) were incubated with CoCl2, NiCl2, MgCl2, FeSO4, CaCl2, CuCl2, MnCl2, and ZnCl2 at a concentration of 1 mM. All reactions were stopped by the addition of 10 mM EDTA. For time series digestion of dsDNA, 0.3 μM purified different NPs were reacted with 100 ng/μl DNA substrates in a final volume of 10 μl for 5, 10, 20, 40, and 60 min. The reaction products were loaded onto a 2% (wt/vol) agarose gel and stained with ethidium bromide. For measurement of the specific activity of different NPs, the buffer contained 20 mM HEPES, pH 6.8, 100 mM NaCl, and 5 mM MnCl2. A total of 200 μg/ml calf thymus DNA (purchased from Sigma) was used as a substrate for the reaction at 37°C, and DNase I at various concentrations was used as the control for calibrating a standard curve. CCHFV NP, HAZV NP, KUPV NP, and ERVEV NP were then tested independently at three different concentrations.
Caspase 3 proteolysis assay.
Caspase 3 proteolysis assay of ERVEV NP, HAZV NP, and KUPV NP was conducted as previously described (21). Mixtures of 0.1 μM purified NP and 5 nM caspase 3 in a final volume of 10 μl were reacted at 37°C for 20, 40, and 60 min. The caspase 3 digestion buffer contains 20 mM HEPES, pH 7.0, 200 mM NaCl, 5% (vol/vol) glycerol, 0.2% (wt/vol) 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS), 2 mM EDTA. Reaction products were loaded onto a 15% (wt/vol) sodium dodecyl sulfate polyacrylamide gel and stained with R-250.
Protein structure accession numbers.
The coordinates and structure factors have been deposited with the RCSB under accession numbers 4XZ8 [ERVEV NP(1–482)], 4XZA (ERVEV NPhead), 4XZC (KUPV NP), and 4XZE (HAZV NP).
RESULTS
Structures of the nairovirus-encoded NPs.
The primary sequences of CCHFV, HAZV, KUPV, ERVEV, and DUGV NPs were aligned and show high similarity (see Fig. S1 in the supplemental material). Similar to the expression and purification of CCHFV NP, as previously described (21), the HAZV, KUPV, and ERVEV NPs were expressed as recombinant proteins in Escherichia coli and finally purified in monomeric form with RNase treatment, indicating that all three nairovirus-encoded NPs weakly protected bound nucleic acid.
We next solved the structures of the HAZV, KUPV, and ERVEV NPs (Fig. 1 and Table 1). However, the initial attempt using the whole structures of CCHFV NP monomers (PDB accession number 3U3I) with variable conformations (21, 24) as the search model could not generate interpretable solutions by molecular replacement (MR), indicating different conformations of these NPs. Considering that the head domain and stalk domain of CCHFV NP have relatively flexible orientations, we then used the coordinates of the head domain and stalk domain of CCHFV NP separately as initial search models and successfully solved the structures of these three nairovirus-encoded NPs. Notably, significant variations appeared in the stalk domains, which we actually had to rebuild without the guidance of the CCHFV NP model.
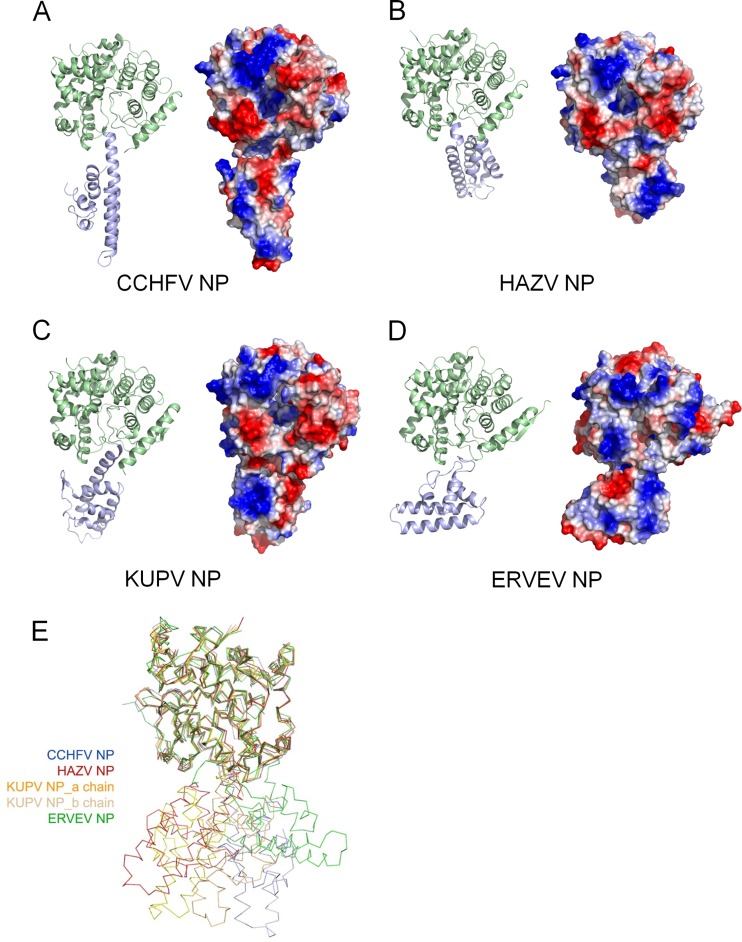
FIG 1.
Structures of nairovirus-encoded NPs. Structures of CCHFV NP (PDB accession number 3U3I) (A), HAZV NP (B), KUPV NP (C), and ERVEV NP (D) are shown in cartoon diagrams (left) and covered by electrostatic potentials of the surfaces (right). In the cartoon diagrams, the head domains and stalk domains are colored in pale green and blue, respectively. (E) Structural alignment of four nairovirus-encoded NPs. The CCHFV, HAZV, KUPV (a and b chains), and ERVEV NPs are displayed as ribbon diagrams and colored blue, red, gold/brown, and green, respectively. These four structures are shown with superimposed head domains.
The HAZV, KUPV, and ERVEV NPs all share structural topology similar to that of the CCHFV NP, which is characterized by a separate head domain and stalk domain (Fig. 1B to D). Most parts of these three structures were well built and refined, but the different linkages connecting the head domain and stalk domain, i.e., Asn184 to Val196 or Gly185 to Gly194 in HAZV NP, Gly183 to Glu197 or Leu180 to Val194 in KUPV NP, and Leu285 to Val291 in ERVEV NP, could not be built due to the lack of interpretable electron density, indicating their structural flexibility.
The head domains of the HAZV, KUPV, and ERVEV NPs showed very high structural conservation with the head domain of CCHFV NP, with root mean square deviation (RMSD) values of 0.85, 0.79, and 1.09 Å, respectively (Fig. 1E). Notably, slight structural variations can be observed in the head domain. The head domains of the CCHFV and HAZV NPs present the most conserved architecture and consist predominantly of α-helices. In contrast, the residues in the linkage region of α8-α9 and in the C termini of the KUPV and ERVEV NPs constitute a parallel two-β-stranded sheet (Fig. 2A). Interestingly, the head domains of these nairovirus-encoded NPs show low similarity with other viral NPs in the bunyavirus family but high similarity with LASV NP in the arenavirus family (22, 23), with a Z score of around 14 and RMSD value of around 3.5 (see Table S2 in the supplemental material).
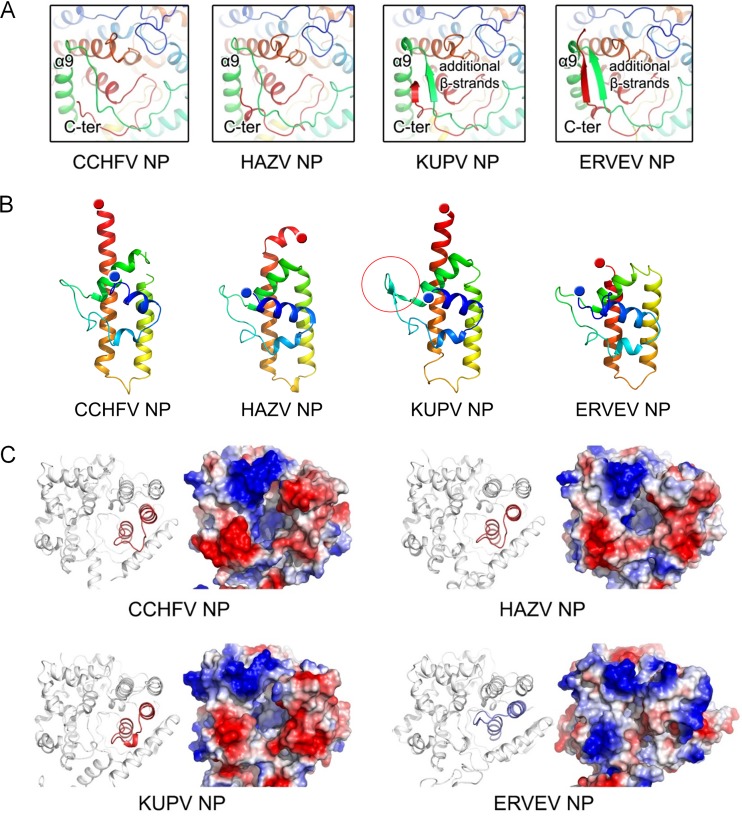
FIG 2.
Structural comparison of nairovirus-encoded NPs. (A) Comparison of the folding of the C terminus and the linkage of α8-α9 in the head domains. All structures are shown as cartoon diagrams in a rainbow color scheme in which the N and C termini are colored blue and red, respectively. (B) Comparison of the stalk domains. A two-β-stranded motif in the KUPV NP stalk domain is highlighted by a red circle. (C) Comparison of the charged pocket of the head domains. The head domain of each NP is shown in a cartoon diagram and a representation of the surface electrostatic potentials.
Comparison of nairovirus-encoded NPs reveals high flexibility of stalk domain.
Although the CCHFV, HAZV, KUPV, and ERVEV NPs all display as head and stalk domains and share the same structural topology, significant conformational differences appear in the orientation between the head domain and the stalk domain (Fig. 1E). In the structure of the CCHFV NP (PDB accession number 3U3I), the head-stalk orientation presents the straightest architecture. However, in the KUPV and ERVEV NPs, the stalk domains exhibit a rotation of approximately 30° to both sides of the CCHFV NP. Remarkably, the stalk domain of HAZV NP is rotated by over 60° compared with the CCHFV NP. Moreover, the KUPV NP stalk domain adopts a different conformation, rotated by approximately 40° within one asymmetric unit (see Fig. S2 in the supplemental material), possibly indicating high structural flexibility during RNA binding or oligomerization.
In addition, the molecular folding of the stalk domains of these nairovirus-encoded NPs presents great variations (Fig. 2B). The stalk domains of all these NPs predominantly consist of four α-helices, with the last α-helix linking to the head domain. In the CCHFV and KUPV NPs, the last α-helix is straight. However, the last 10 residues in the C-terminal end of the stalk domain of the HAZV NP present a bent conformation with an angle of approximately 45°. The C terminus of the last α-helix of ERVEV NP is missing due to the lack of interpretable electron density. Very interestingly, a two-β-strand motif was observed in the stalk domain of KUPV NP, but the same positions in other NPs display loop architectures. It is very likely that these conformational changes are associated with the flexibility of the orientation between the head and the stalk domain.
The structural differences of the head domains reveal distinct DNA-specific endonuclease activities.
Significant conformational changes were also observed in the head domain (Fig. 2C). In the crystallographic study of monomeric CCHFV NP, a deep, positively charged pocket that is essential for the endonuclease activity was observed in the head domain (21). Although most parts of this charged pocket presented similar conformations in all four nairovirus-encoded NPs, the loop region of ERVEV NP (residues Q430 to S444), connecting the last two α-helices, displayed great discrepancies (Fig. 2C). In the structures of the CCHFV, HAZV, and KUPV NPs, this loop region extends outward from the pocket to allow an open architecture. In sharp contrast, this loop region folds back toward the pocket and consequently seals it like a lid in the ERVEV NP, and several hydrogen bond interactions are found in the switching of the loop region, such as R51 with Q432 and R434 with A50 and A53. Electrostatic interaction of R379 and R367 with D440 and D438 also appears to contribute to the stabilization of the loop region (see Fig. S3 in the supplemental material). Based on the comparison of electrostatic surface potential, the pockets of the CCHFV, HAZV, and KUPV NP head domains are exposed to solvent, but the pocket of the ERVEV NP head domain is almost completely closed, suggesting a difference in the DNA-specific endonuclease activity of ERVEV NP.
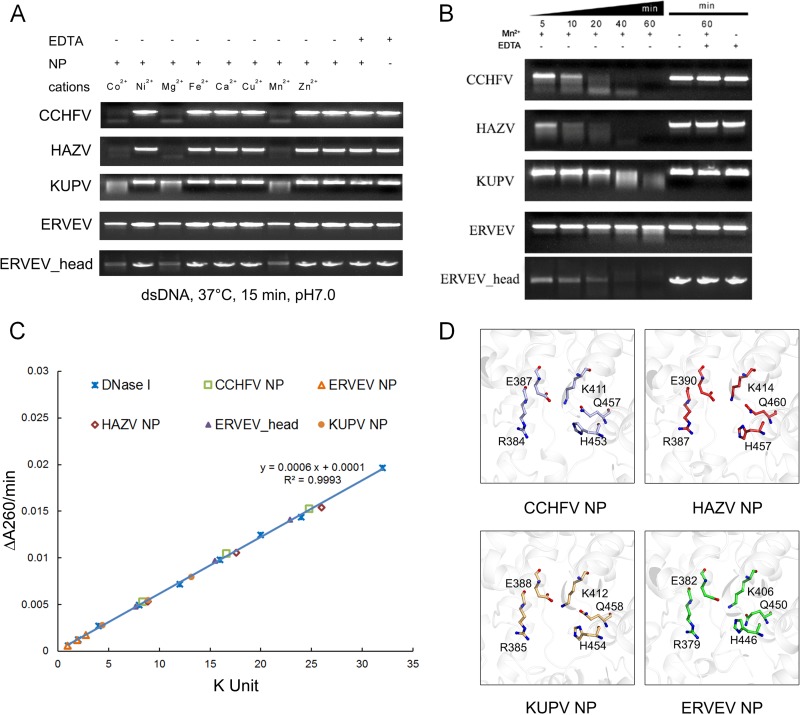
Further experiments were carried out to elucidate the impacts of these structural variations on the endonuclease activities of nairovirus-encoded NPs. We first treated the dsDNA substrate with equal amounts of CCHFV, HAZV, KUPV, and ERVEV NPs (each for 0.3 μM) in the presence of manganese ion, as previously described (21). The HAZV NP and KUPV NP displayed endonuclease activities relatively similar to that of CCHFV NP, whereas ERVEV NP(1–482) presented much lower enzymatic activity (Fig. 3A and B). Because the endonuclease activity of CCHFV NP is metal dependent, we next checked the impacts of different cations on these nairovirus-encoded NPs and verified that the variations in their endonuclease activities were not due to divalent cations in the reaction buffer (Fig. 3A). We quantified the endonuclease activities of the different nairovirus-encoded NPs and found that the CCHFV, HAZV, and KUPV NPs showed activities of 118.8 kU/mg, 125.3 kU/mg, and 63.1 kU/mg, respectively, while ERVEV NP(1–482) exhibited an extremely attenuated, if not absent, activity of 13.3 kU/mg (Fig. 3C).
FIG 3.
Endonuclease activities of nairovirus-encoded NPs. (A) Effects of different divalent cations on NP nuclease activities. (B) Time series of in vitro dsDNA degradation assay. (C) Measurement of the specific activities of different NPs. (D) Domains of the CCHFV, HAZV, KUPV, and ERVEV NPs are aligned and shown in the same orientation for comparison. The residues with the most essential impacts on endonuclease activities of nairovirus-encoded NPs are represented as colored sticks, while the polypeptides of the four NPs are displayed as white cartoons.
Interestingly, we noted that the enzymatic activity of the ERVEV NP increased slowly if placed on ice for a long time during the measurement of enzymatic activity. Considering the possibility that the ERVEV NP(1–482) could be degraded to some state that regained the enzymatic activity, time gradient degradation assays were conducted to confirm this speculation (see Fig. S4 in the supplemental material). Further enzymatic assays quantified the specific activity of the ERVEV NP(1–482) degradation product to 109.8 kU/mg (with storage at 16°C for 14 days) (Fig. 3B). With the additional use of peptide mass fingerprinting analysis data, the site of the degradation was located and was shown to comprise residues 1 to 180 and 295 to 482. The degraded protein is part of the head domain and was named ERVEV NPhead.
The structures of ERVEV NP(1–482) (head domain alone) and ERVEV NPhead show very high similarity, with an RMSD value of 0.39 Å. As both of these structures display the closed form of the loop region and generate almost the same completely closed substrate pockets and electrostatic surface potential, the discrepancy in their enzymatic activities should not result from the locale of the loop region; however, we note that the ERVEV NPhead displays another channel to the active site through the bottom of the head domain (see Fig. S5 in the supplemental material), and therefore, we speculate that the degradation of the stalk domain creates this alternative channel for the entry of the DNA substrate.
RNA binding affinities of recombinant nairovirus-encoded NPs in vitro.
Negative-strand RNA viruses encode NP to encapsidate the viral genome and, further, form a ribonucleoprotein complex with genomic RNA. Analysis of the electrostatic surface potential of HAZV, KUPV, and ERVEV NPs reveals a conserved deep positively charged pocket in the head domain, like that in CCHFV NP (see Fig. S1 in the supplemental material). This pocket also displays high structural similarity with the RNA binding pocket of LASV NP (23), suggesting its potential role in RNA binding. However, according to the reports of previous work on CCHFV NP by three independent research groups, the recombinant CCHFV NP displays weak binding affinity with various ssRNA probes in vitro. To investigate the diversity of RNA binding affinities of nairoviral NPs, a series of assays was carried out to check this function. First, during the protein purification process, all of these NPs were monitored by UV absorption, which gave an A280/A260 ratio of around 1.8, indicating that no cellular nucleic acid binds with these recombinant proteins. We next employed an electrophoretic mobility shift assay (EMSA) by using a short ssRNA probe derived from the nairovirus genomic S segment (probe sequence, 5′-UCUCAAAGAAAGUUG-3′, modified with biotin at the 5′ end) to test the RNA binding abilities of CCHFV, HAZV, ERVEV, and KUPV NPs. The results showed that the CCHFV, HAZV, ERVEV, and KUPV NPs all bound to the RNA probe and that KUPV NP showed a higher binding affinity with the probe than the other three viral NPs. Notably, the RNA binding affinity of all of these nairovirus-encoded NPs is modest, since free RNA can still be visualized even when the NP/RNA ratio is 2:1 (see Fig. S6). Taken together, considering all of these biochemical and structural results reported by us and other research groups, we believe that the recombinant nairovirus-encoded NPs expressed from E. coli display weak binding with RNA in vitro.
Caspase 3 cleavage site is not conserved.
A previous study revealed that the host performs caspase 3 cleavage at the remote end of the CCHFV stalk domain in the specific sequence 266DEVD269, prohibiting the proliferation of CCHFV and functioning as a host defense mechanism against lytic CCHFV infection (31).
It is well known that caspase 3 exhibits substrate specificity by recognizing a tetrapeptide motif, D-X-X-D (32), where the C-terminal aspartic acid residue (P1 position) is absolutely required while variations at the other three positions can be tolerated (33). In caspase 3, the P1 Asp is bound in a deep basic S1 pocket formed by the conserved residues R64-R207-Q161 of caspase 3, and a peptide with Glu at P1 instead of Asp was hydrolyzed at a 20,000-fold-lower kcat/Km (34). Furthermore, by screening with a combinational peptide library, caspase 3 was revealed to prefer the peptide sequences DE(M/L)D-(S/G) and DE(V/I)D (35). In particular, caspase 3 exhibits 170% faster hydrolysis on a sequence of DLVD than on the canonical DEVD peptide (36).
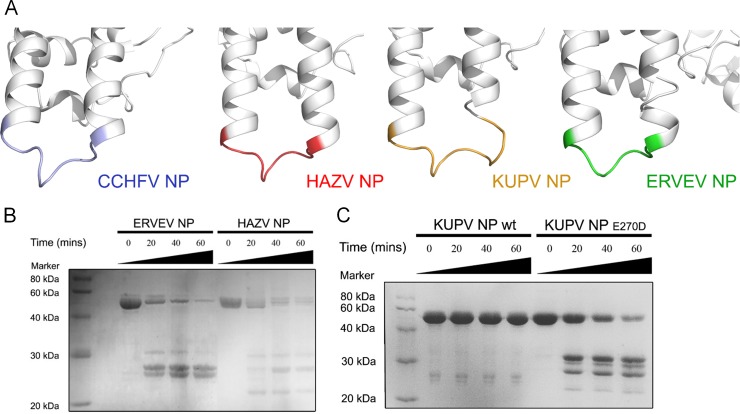
By comparing the primary protein sequences and the three-dimensional structures of nairovirus-encoded NPs, we found that the CCHFV, HAZV, and ERVEV NPs have a conserved caspase 3 cleavage site at the remote end of the stalk domains. In particular, the most essential P1 positions are strictly conserved Asp residues in these three nairoviruses (Fig. 4A). In contrast, although KUPV NP displays a similar loop between α-helix 11 and α-helix 12 at the remote end of the stalk domain, the P1 position of the potential caspase 3 cleavage site of KUPV NP is a Glu residue instead of an Asp residue, suggesting that KUPV NP may not be a target for caspase 3 cleavage.
FIG 4.
Variations on caspase 3 cleavage. (A) Comparison of the caspase 3 cleavage sites of nairovirus-encoded NPs. The stalk domains of the CCHFV, HAZV, KUPV, and ERVEV NPs are aligned and shown in white cartoon diagrams with the same orientation. The residues covering the caspase 3 cleavage sites in each NP are highlighted in different colors. (B) Caspase 3 proteolysis of ERVEV NP and HAZV NP in vitro. (C) Caspase 3 proteolysis of KUPV NP wild type (wt) and E270D mutant in vitro.
To verify this speculation, we treated the CCHFV, KUPV, HAZV, and ERVEV NPs with caspase 3 and examined the products using SDS-PAGE (Fig. 4B). The results showed that the HAZV and ERVEV NPs can be well hydrolyzed, and caspase 3 displayed higher activity with ERVEV NP as the substrate than with HAZV NP. Meanwhile, KUPV NP exhibited no reaction with caspase 3, which is consistent with the sequence alignment result. We also produced the Dugbe virus (DUGV) NP from the Nairobi sheep disease group and found that DUGV NP cannot be hydrolyzed by caspase 3 either. As the P1 position of DUGV NP is a Glu residue, as in KUPV NP, this result is not surprising. Moreover, we also generated a single mutation of Glu270 to Asp, and the SDS-PAGE result shows that KUPV NPE270D can be hydrolyzed by caspase 3 (Fig. 4C). All these results indicated that the host innate immune defense mechanism found for CCHFV most likely cannot be employed against all of the viruses in the Nairovirus genus.
DISCUSSION
We systematically solved the structures of NPs encoded by three nairoviruses: HAZV, KUPV, and ERVEV. All of these NPs share similar structural topology with the CCHFV NP, which is characterized by an overall racket-shaped structure consisting of head and stalk domains. Moreover, structural alignment revealed that the nairovirus-encoded NPs show low similarity to NPs in other genera (such as Phlebovirus and Orthobunyavirus) of the Bunyaviridae family but are more like the NP of LASV in the Arenaviridae family (see Table S2 in the supplemental material). Previous work that focused on virus entry processes shows some similarities between nairovirus and arenavirus (37, 38), such as the dependence for cellular protease during the glycoprotein maturation process, and further phylogenetic analyses based on the L sequence (39) and the NP sequence (24) both reported a close relationship between nairovirus and arenavirus. Taken together, these results may suggest the possibility of reevaluation of the classification of the Bunyaviridae and Arenaviridae families.
Previous work has shown that CCHFV NPs reported by three research groups display close correspondence of all folds but significant transition of the stalk domain through different rotations and translations (21, 24, 25); considering the tiny differences in their primary sequences, the different conformations are proposed to be a result of the high flexibility of the stalk domain and being trapped in different orientations by crystal packing. Significant conformational differences also appeared in the HAZV, KUPV, and ERVEV NPs reported in this work. Especially for KUPV NP, we captured different conformations in which the stalk domain rotated by approximately 40° within one asymmetric unit, indicating high structural flexibility of the two domains. Moreover, the first linkage region (the residues around L181 to S194 for CCHFV NP) connecting the head domain and stalk domain could not be built due to the lack of interpretable electron density, indicating their structural flexibility. This linkage region corresponds to helix α6 in LASV NP, which adopted an open state when bound to the RNA probe during the so-called “gating mechanism.” Another remaining linkage region of these two domain is helix α12 (around residues A271 to R298 for CCHFV NP), but this single helix is insufficient to establish a rigid link between the head and stalk domains. Taken together, we propose that the linkage region between these two domains is highly flexible across nairovirus-encoded NPs, allowing interchangeable conformational changes for some key function of NPs, such as oligomerization of RNP or the binding with viral RNA.
Significant conformational differences were also observed in the head domain. Unlike the CCHFV, HAZV, and KUPV NPs, the positively charged pocket was observed to be closed in the head domain of the ERVEV NP, suggesting a different endonuclease activity of ERVEV NP. The results of endonuclease activity assays of ERVEV NP also confirm this speculation. Interestingly, degraded ERVEV NP regains endonuclease activity, possibly by creating an alternative DNA substrate entry channel. However, further work on the level of cell infection experiments is required to determine the authentic state and function of the ERVEV NP in the virus replication cycle.
In addition, several research groups have reported various functions of LASV NP. The C-terminal domain of LASV NP is well known as a 3′-to-5′ dsRNA-specific exonuclease (22, 40, 41). However, the function of the N-terminal domain is more complicated. Qi et al. reported a cap-binding ability of the LASV N-terminal domain based on structural information (22), while Hastie et al. reported the LASV NP N-terminal domain structure with a bound ssRNA that revealed a gating mechanism of helix α6 (23). Interestingly, Qi et al., in their supplementary information (Fig. S3 of reference 22), also reported that LASV NP can digest dsDNAs both in trimeric and hexameric forms. Since the head domains of nairovirus-encoded NPs show high similarity with the LASV NP N-terminal domain, it is very interesting to investigate the subtle regulation mechanisms that coordinate these distinct functions during different replication stages of the virus life cycle and different locations in the host cell.
Previous results have shown that a DEVD motif is located at the apex of CCHFV NP stalk domain, and CCHFV NP can be catalyzed into two fragments by host cell caspase 3 but still kept as an intact unit (24). On the one hand, this finding could be interpreted as an immune defense of the host cell against CCHFV infection; on the other hand, it is possible that CCHFV might have evolved this strategy to act as a suicide decoy substrate for caspase 3, hence delaying or inhibiting host cell apoptosis. By comparing the three-dimensional structures of nairovirus-encoded NPs, we found that the CCHFV, HAZV, and ERVEV NPs have a conserved potential caspase 3 cleavage site at the remote end of the stalk domain, which is one of the most accessible sites of the whole protein. However, the KUPV NP and DUGV NP cannot be recognized and hydrolyzed by caspase 3. These results indicated that the host innate immune interaction found in CCHFV infection that acts through the stalk domain of the NP may not be employed against other nairovirus serogroups.
Combining all of the structural and biochemical results, our new data provide deeper insight into the biological role of NPs in the transcription and replication processes of nairoviruses. We hope this work will provide valuable information for understanding the formation of nairovirus RNPs and be helpful for the development of intervention strategies to lessen the pathogenic burden of nairovirus infections.
Supplementary Material
ACKNOWLEDGMENTS
We gratefully acknowledge the staffs of beamlines BL17U and BL19U, SSRF, China, for their assistance with diffraction data collection.
This work was supported by the National Natural Science Foundation of China (grants 31300606, 31200641, and 31200586), Tianjin Municipal Natural Science Foundation (grants 14JCQNJC10100, 10ZCKFSY08700, 10ZCKFSY08800, and 13ZXCXSY13500), the Ministry of Science and Technology of China 973 Project (grant no. 2013CB911103), and the Doctoral Fund of the Ministry of Education of China.
The authors declare no competing financial interest.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.01680-15.
REFERENCES
- 1.Guu TS, Zheng W, Tao YJ. 2012. Bunyavirus: structure and replication. Adv Exp Med Biol 726:245–266. doi: 10.1007/978-1-4614-0980-9_11. [DOI] [PubMed] [Google Scholar]
- 2.Zhou H, Sun Y, Guo Y, Lou Z. 2013. Structural perspective on the formation of ribonucleoprotein complex in negative-sense single-stranded RNA viruses. Trends Microbiol 21:475–484. doi: 10.1016/j.tim.2013.07.006. [DOI] [PubMed] [Google Scholar]
- 3.Sun Y, Guo Y, Lou Z. 2012. A versatile building block: the structures and functions of negative-sense single-stranded RNA virus nucleocapsid proteins. Protein Cell 3:893–902. doi: 10.1007/s13238-012-2087-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marriott AC, Nuttall PA. 1996. Molecular biology of nairoviruses, p 91–104. In Elliott EM. (ed), The Bunyaviridae. Plenum Press, London, United Kingdom. [Google Scholar]
- 5.Lasecka L, Baron MD. 2014. The molecular biology of nairoviruses, an emerging group of tick-borne arboviruses. Arch Virol 159:1249–1265. doi: 10.1007/s00705-013-1940-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Flick R, Whitehouse CA. 2005. Crimean-Congo hemorrhagic fever virus. Curr Mol Med 5:753–760. doi: 10.2174/156652405774962335. [DOI] [PubMed] [Google Scholar]
- 7.Dowall SD, Findlay-Wilson S, Rayner E, Pearson G, Pickersgill J, Rule A, Merredew N, Smith H, Chamberlain J, Hewson R. 2012. Hazara virus infection is lethal for adult type I interferon receptor-knockout mice and may act as a surrogate for infection with the human-pathogenic Crimean-Congo hemorrhagic fever virus. J Gen Virol 93:560–564. doi: 10.1099/vir.0.038455-0. [DOI] [PubMed] [Google Scholar]
- 8.Dilcher M, Koch A, Hasib L, Dobler G, Hufert FT, Weidmann M. 2012. Genetic characterization of Erve virus, a European Nairovirus distantly related to Crimean-Congo hemorrhagic fever virus. Virus Genes 45:426–432. doi: 10.1007/s11262-012-0796-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Treib J, Dobler G, Haass A, von Blohn W, Strittmatter M, Pindur G, Froesner G, Schimrigk K. 1998. Thunderclap headache caused by Erve virus? Neurology 50:509–511. doi: 10.1212/WNL.50.2.509. [DOI] [PubMed] [Google Scholar]
- 10.Lou Z, Sun Y, Rao Z. 2014. Current progress in antiviral strategies. Trends Pharmacol Sci 35:86–102. doi: 10.1016/j.tips.2013.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Raymond DD, Piper ME, Gerrard SR, Skiniotis G, Smith JL. 2012. Phleboviruses encapsidate their genomes by sequestering RNA bases. Proc Natl Acad Sci U S A 109:19208–19213. doi: 10.1073/pnas.1213553109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Raymond DD, Piper ME, Gerrard SR, Smith JL. 2010. Structure of the Rift Valley fever virus nucleocapsid protein reveals another architecture for RNA encapsidation. Proc Natl Acad Sci U S A 107:11769–11774. doi: 10.1073/pnas.1001760107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reguera J, Malet H, Weber F, Cusack S. 2013. Structural basis for encapsidation of genomic RNA by La Crosse Orthobunyavirus nucleoprotein. Proc Natl Acad Sci U S A 110:7246–7251. doi: 10.1073/pnas.1302298110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiao L, Ouyang S, Liang M, Niu F, Shaw N, Wu W, Ding W, Jin C, Peng Y, Zhu Y, Zhang F, Wang T, Li C, Zuo X, Luan CH, Li D, Liu ZJ. 2013. Structure of severe fever with thrombocytopenia syndrome virus nucleocapsid protein in complex with suramin reveals therapeutic potential. J Virol 87:6829–6839. doi: 10.1128/JVI.00672-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Olal D, Dick A, Woods VL Jr, Liu T, Li S, Devignot S, Weber F, Saphire EO, Daumke O. 2014. Structural insights into RNA encapsidation and helical assembly of the Toscana virus nucleoprotein. Nucleic Acids Res 42:6025–6037. doi: 10.1093/nar/gku229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou H, Sun Y, Wang Y, Liu M, Liu C, Wang W, Liu X, Li L, Deng F, Wang H, Guo Y, Lou Z. 2013. The nucleoprotein of severe fever with thrombocytopenia syndrome virus processes a stable hexameric ring to facilitate RNA encapsidation. Protein Cell 4:445–455. doi: 10.1007/s13238-013-3901-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li B, Wang Q, Pan X, Fernandez de Castro I, Sun Y, Guo Y, Tao X, Risco C, Sui SF, Lou Z. 2013. Bunyamwera virus possesses a distinct nucleocapsid protein to facilitate genome encapsidation. Proc Natl Acad Sci U S A 110:9048–9053. doi: 10.1073/pnas.1222552110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Niu F, Shaw N, Wang YE, Jiao L, Ding W, Li X, Zhu P, Upur H, Ouyang S, Cheng G, Liu ZJ. 2013. Structure of the Leanyer orthobunyavirus nucleoprotein-RNA complex reveals unique architecture for RNA encapsidation. Proc Natl Acad Sci U S A 110:9054–9059. doi: 10.1073/pnas.1300035110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dong H, Li P, Bottcher B, Elliott RM, Dong C. 2013. Crystal structure of Schmallenberg orthobunyavirus nucleoprotein-RNA complex reveals a novel RNA sequestration mechanism. RNA 19:1129–1136. doi: 10.1261/rna.039057.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dong H, Li P, Elliott RM, Dong C. 2013. Structure of Schmallenberg orthobunyavirus nucleoprotein suggests a novel mechanism of genome encapsidation. J Virol 87:5593–5601. doi: 10.1128/JVI.00223-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guo Y, Wang W, Ji W, Deng M, Sun Y, Zhou H, Yang C, Deng F, Wang H, Hu Z, Lou Z, Rao Z. 2012. Crimean-Congo hemorrhagic fever virus nucleoprotein reveals endonuclease activity in bunyaviruses. Proc Natl Acad Sci U S A 109:5046–5051. doi: 10.1073/pnas.1200808109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qi X, Lan S, Wang W, Schelde LM, Dong H, Wallat GD, Ly H, Liang Y, Dong C. 2010. Cap binding and immune evasion revealed by Lassa nucleoprotein structure. Nature 468:779–783. doi: 10.1038/nature09605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hastie KM, Liu T, Li S, King LB, Ngo N, Zandonatti MA, Woods VL Jr, de la Torre JC, Saphire EO. 2011. Crystal structure of the Lassa virus nucleoprotein-RNA complex reveals a gating mechanism for RNA binding. Proc Natl Acad Sci U S A 108:19365–19370. doi: 10.1073/pnas.1108515108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carter SD, Surtees R, Walter CT, Ariza A, Bergeron E, Nichol ST, Hiscox JA, Edwards TA, Barr JN. 2012. Structure, function, and evolution of the Crimean-Congo hemorrhagic fever virus nucleocapsid protein. J Virol 86:10914–10923. doi: 10.1128/JVI.01555-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Y, Dutta S, Karlberg H, Devignot S, Weber F, Hao Q, Tan YJ, Mirazimi A, Kotaka M. 2012. Structure of Crimean-Congo hemorrhagic fever virus nucleoprotein: superhelical homooligomers and the role of caspase 3 cleavage. J Virol 86:12294–12303. doi: 10.1128/JVI.01627-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Otwinowski Z, Minor W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 27.Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 28.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Laskowski R, MacArthur M, Moss D, Thornton J. 1993. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr 26:283–291. doi: 10.1107/S0021889892009944. [DOI] [Google Scholar]
- 30.DeLano W. 2002. The PyMOL molecular graphics system. Delano Scientific LLC, San Carlos, CA: http://www.pymol.org. [Google Scholar]
- 31.Karlberg H, Tan YJ, Mirazimi A. 2011. Induction of caspase activation and cleavage of the viral nucleocapsid protein in different cell types during Crimean-Congo hemorrhagic fever virus infection. J Biol Chem 286:3227–3234. doi: 10.1074/jbc.M110.149369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Agniswamy J, Fang B, Weber IT. 2007. Plasticity of S2-S4 specificity pockets of executioner caspase 7 revealed by structural and kinetic analysis. FEBS J 274:4752–4765. doi: 10.1111/j.1742-4658.2007.05994.x. [DOI] [PubMed] [Google Scholar]
- 33.Fang B, Boross PI, Tozser J, Weber IT. 2006. Structural and kinetic analysis of caspase 3 reveals role for s5 binding site in substrate recognition. J Mol Biol 360:654–666. doi: 10.1016/j.jmb.2006.05.041. [DOI] [PubMed] [Google Scholar]
- 34.Stennicke HR, Renatus M, Meldal M, Salvesen GS. 2000. Internally quenched fluorescent peptide substrates disclose the subsite preferences of human caspases 1, 3, 6, 7 and 8. Biochem J 350(Pt 2):563–568. doi: 10.1042/0264-6021:3500563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thornberry NA, Rano TA, Peterson EP, Rasper DM, Timkey T, Garcia-Calvo M, Houtzager VM, Nordstrom PA, Roy S, Vaillancourt JP, Chapman KT, Nicholson DW. 1997. A combinatorial approach defines specificities of members of the caspase family and granzyme B. Functional relationships established for key mediators of apoptosis. J Biol Chem 272:17907–17911. [DOI] [PubMed] [Google Scholar]
- 36.Lien S, Pastor R, Sutherlin D, Lowman HB. 2004. A substrate-phage approach for investigating caspase specificity. Protein J 23:413–425. doi: 10.1023/B:JOPC.0000039555.92058.51. [DOI] [PubMed] [Google Scholar]
- 37.Lenz O, ter Meulen J, Klenk HD, Seidah NG, Garten W. 2001. The Lassa virus glycoprotein precursor GP-C is proteolytically processed by subtilase SKI-1/S1P. Proc Natl Acad Sci U S A 98:12701–12705. doi: 10.1073/pnas.221447598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vincent MJ, Sanchez AJ, Erickson BR, Basak A, Chretien M, Seidah NG, Nichol ST. 2003. Crimean-Congo hemorrhagic fever virus glycoprotein proteolytic processing by subtilase SKI-1. J Virol 77:8640–8649. doi: 10.1128/JVI.77.16.8640-8649.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vieth S, Torda AE, Asper M, Schmitz H, Gunther S. 2004. Sequence analysis of L RNA of Lassa virus. Virology 318:153–168. doi: 10.1016/j.virol.2003.09.009. [DOI] [PubMed] [Google Scholar]
- 40.Hastie KM, King LB, Zandonatti MA, Saphire EO. 2012. Structural basis for the dsRNA specificity of the Lassa virus NP exonuclease. PLoS One 7:e44211. doi: 10.1371/journal.pone.0044211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hastie KM, Kimberlin CR, Zandonatti MA, MacRae IJ, Saphire EO. 2011. Structure of the Lassa virus nucleoprotein reveals a dsRNA-specific 3′ to 5′ exonuclease activity essential for immune suppression. Proc Natl Acad Sci U S A 108:2396–2401. doi: 10.1073/pnas.1016404108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.