Abstract
Borrelia burgdorferi, a causative agent of Lyme borreliosis, is a zoonotic pathogen that survives in nutrient-limited environments within a tick, prior to transmission to its mammalian host. Survival under these prolonged nutrient-limited conditions is thought to be similar to survival during stationary phase, which is characterized by growth cessation and decreased protein production. Multiple ribosome-associated proteins are implicated in stationary-phase survival of Escherichia coli. These proteins include hibernation-promoting factor (HPF), which dimerizes ribosomes and prevents translation of mRNA. Bioinformatic analyses indicate that B. burgdorferi harbors an hpf homolog, the bb0449 gene. BB0449 protein secondary structure modeling also predicted HPF-like structure and function. However, BB0449 protein was not localized in the ribosome-associated protein fraction of in vitro-grown B. burgdorferi. In wild-type B. burgdorferi, bb0449 transcript and BB0449 protein levels are low during various growth phases. These results are inconsistent with patterns of synthesis of HPF-like proteins in other bacterial species. In addition, two independently derived bb0449 mutants successfully completed the mouse-tick infectious cycle, indicating that bb0449 is not required for prolonged survival in the nutrient-limited environment in the unfed tick or any other stage of infection by B. burgdorferi. We suggest either that BB0449 is associated with ribosomes under specific conditions not yet identified or that BB0449 of B. burgdorferi has a function other than ribosome conformation modulation.
INTRODUCTION
Lyme disease, the most common tick-transmitted disease in the United States (1), is caused by infection with the spirochete Borrelia burgdorferi (2, 3). This pathogen's complex infectious cycle includes an Ixodes tick vector and multiple vertebrate hosts. The Ixodes tick vector, like all hard ticks, has three developmental stages: larva, nymph, and adult. Typically, larval ticks become infected with B. burgdorferi by feeding on an infected vertebrate host, often a small mammal. B. burgdorferi survives within the tick midgut (2) as larval ticks molt into nymphs (transstadial passage) and the nymphs wait to feed again, which can include overwintering (4, 5). Following the larval molt, infected nymphs can transmit B. burgdorferi to naive vertebrate hosts, which, in turn, serve as a reservoir of spirochetes for larval ticks. After fed nymphs molt, infected adult ticks can also transmit B. burgdorferi, but adults typically feed on larger animals upon which larvae do not feed. Moreover, little or no transovarial transmission of spirochetes occurs (6, 7). Therefore, adult ticks do not significantly contribute to the maintenance of B. burgdorferi populations (8), and neither do humans, which are incidental hosts of B. burgdorferi. Hence, the larval and nymphal tick life stages are relevant to the zoonotic life cycle of B. burgdorferi.
It has been generally accepted that B. burgdorferi populations are sustained in nature by the long-term infection of vertebrate hosts (9). However, Lindsay et al. (10) suggested that overwintering ticks play an important role in maintaining B. burgdorferi populations. These authors postulated that infected nymphs that survive through the winter can infect vertebrate hosts in the spring, upon which tick larvae can feed in the summer to acquire B. burgdorferi (10). This would result in infected larvae that molt and overwinter as infected nymphs. The midgut of the nymphal tick when overwintering is likely to be a nutrient-limited environment, because the larval tick digests blood meal nutrients prior to molting (11). We are interested in identifying mechanisms that B. burgdorferi uses to survive within the dormant tick vector.
Generally, bacteria employ several mechanisms to combat stressful conditions such as nutrient-limited environments (reviewed in reference 12). One such mechanism found in B. burgdorferi is the stringent response, which relies on the production of an alarmone, guanosine tetraphosphate (ppGpp), to ultimately increase protein recycling (13–18). Another mechanism is the regulation of global protein translation levels by modulating ribosome activity. Three proteins have been implicated in this form of protein translation control in Escherichia coli: ribosome modulation factor (RMF), YfiA (also called pY or RaiA), and hibernation-promoting factor (HPF; previously called YhbH). RMF binds to the 70S ribosome, making a 90S ribosome in which the Shine-Dalgarno (SD)–anti-SD helix region on the 30S ribosomal subunit is covered by RMF and incoming mRNA transcript is blocked (19). HPF subsequently binds to 90S ribosomes and covers the A- and P-sites on the 30S ribosomal subunit, preventing binding of initiator tRNA (19). Ultimately, RMF and HPF dimerize ribosomes, rendering them translationally inactive (12, 19–25). In contrast, YfiA binds to the 30S subunit of 70S ribosomes, overlapping the A- and P-sites, and prevents association of ribosomes with tRNAs (26, 27). YfiA-bound ribosomes are 70S monomers that have been rendered translationally inactive.
The majority of ribosome inactivation/hibernation studies that focused on HPF have studied the biochemistry of ribosome-protein interactions and in vitro ribosome analysis to show that HPF is required for ribosome dimerization (19, 20, 24, 28–31). There have been several other studies that used genetic techniques to address biological phenotypes of hpf-deficient strains. Deletion of hpf in E. coli, and of an hpf homolog in Lactococcus lactis, shows that the gene products are involved in ribosome dimerization, and mutants lacking HPF exhibited subtle effects on bacterial survival in E. coli and recovery of L. lactis following starvation (21, 32). In addition, deletion of the Listeria monocytogenes hpf homolog eliminated ribosome dimerization and impaired tissue colonization in two mouse infection models (33), while deleting the hpf homolog in Erwinia amylovora, a plant pathogen, rendered this pathogen noninfectious to tobacco leaves and immature pear fruits (34). These studies suggest that HPF plays a role in bacterial survival in vitro and perhaps in vivo.
The goal of our study was to determine if B. burgdorferi has RMF, YfiA, or HPF homologs, and if so, whether they regulate ribosome conformation to increase bacterial survival. We also wanted to ascertain if such a mechanism is important in the infectious cycle, specifically, for survival of B. burgdorferi under the nutrient-limited conditions of infected ticks using a simulated overwintering phase.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
All B. burgdorferi strains used were derived from strain B31-S9 (also called B31-A3-68Δbbe02), referred to as wild type (WT) in this study, which is an infectious derivative of the B31 clone A3 that has increased transformation efficiency due to the absence of native restriction-modification systems (35–37) (Table 1). B31-S9 and its derivatives all lack cp9, lp5, and lp56, as determined by PCR analysis. Spirochetes were grown in Barbour-Stoenner-Kelly II (BSKII) medium (38) supplemented with 6% rabbit serum (Pel-Freez Biologicals, Rogers, AR, USA) at 35°C or in solid BSK medium, as described previously (39). All B. burgdorferi-containing plates were incubated at 35°C and in 2.5% CO2. DNA manipulations were performed in E. coli TOP10 cells (Life Technologies, Carlsbad, CA, USA) grown at 37°C in Luria broth.
TABLE 1.
B. burgdorferi strains used in this study
| Strain namea | Full name | Descriptionb | Reference(s) |
|---|---|---|---|
| WT | B31-S9 (also called B31-A3-68Δbbe02) | Transformable, infectious A3 derivative; gene bbe02, encoding a restriction-modification enzyme, replaced with streptomycin resistance cassette | 35, 36 |
| bb0449Δ1-97 | B31-S9bb0449Δ1-97::flaBp-kan | Full open reading frame deletion-insertion of kanamycin resistance cassette at bb0449 locus, deletion of last 3 amino acids of RpoN ORF | This study |
| bb0449Δ4-97 | B31-S9bb0449Δ4-97::flaBp-kan | Deletion of amino acids 4 to 97 and insertion of kanamycin resistance cassette at bb0449 locus, leaving rpoN intact | This study |
| bb0449AR | B31-S9-bb0449+-flgBp-aacC1 | Allelic reconstitution of bb0449 in bb0449Δ4-97, marked with gentamicin resistance cassette | This study |
Strain name denotes an abbreviated name used in this manuscript; full strain names are shown adjacent.
All strains are missing plasmids cp9, lp5, and lp56 as determined by PCR analysis.
Identification of homologous B. burgdorferi proteins.
Amino acid sequences of E. coli RMF (NC_000913.3), YfiA (NC_007779.1), and HPF (NC_000913.3) were compared with the translated B. burgdorferi B31 genome (NC_001318.1) in the NCBI database using BLASTp 2.2.28+. A B. burgdorferi protein sequence identified as similar was aligned with the E. coli protein sequence using the MacVector program (version 12.7.5) (MacVector, Inc., Cary, NC, USA).
BB0449 protein secondary structure modeling.
The Phyre2 open-source modeling program was used to generate secondary structure models of BB0449, which were then compared to HPF cocrystallized with ribosomes from Thermus thermophilus (PDB accession number 3V26) using the open-source Visual Molecular Dynamics (VMD) program (40, 41). Influence of amino acid side chains was estimated by visually overlapping the crystallized HPF and modeled BB0449 proteins in VMD. Secondary structure modeling was also completed with the SWISS-MODEL (42–45), I-Tasser (46–48), RaptorX (49–51), and HHPred (52, 53) modeling programs.
qRT-PCR.
Total RNA was extracted from B. burgdorferi cultures using TRIzol reagent, according to the manufacturer's protocol (Life Technologies, Grand Island, NY, USA), and cDNA was produced as previously described (54). Briefly, RNA was extracted by lysing spirochetes in TRIzol reagent, extracting RNA in chloroform, precipitating it in isopropanol, and resuspending it in RNase-free water. Contaminating DNA was removed by treatment with the Turbo DNA-free kit according to the manufacturer's instructions (Life Technologies), and cDNA was produced using the high-capacity cDNA reverse transcriptase kit (Life Technologies) according to the manufacturer's instructions. Quantitative reverse transcriptase PCRs (qRT-PCRs) were performed as described previously (54) with TaqMan Universal PCR master mix (Life Technologies) and gene-specific primers and probe sets (see Table S1 in the supplemental material) (Integrated DNA Technologies, Coralville, IA, USA, or Sigma-Aldrich, St. Louis, MO, USA). Experiments were performed in biological and technical triplicates, and experimental results were analyzed on a Viia7 real-time PCR system with the Viia7 software package (Life Technologies). The qRT-PCR data were analyzed using a standard curve generated with purified total genomic DNA (Qiagen, Alameda, CA, USA), representing a defined number of genome equivalents, as calculated from the DNA concentration and B. burgdorferi genome size. Transcripts are represented per 1,000 flaB transcripts and were analyzed with a Kruskal-Wallis test with Prism 6 statistical analysis software (GraphPad, La Jolla, CA, USA).
BB0449 polyclonal antiserum production.
The gene sequence of bb0449 was codon optimized for E. coli expression, synthesized, and cloned into the pUC57-Kan vector with added endonuclease restriction sites NdeI and BamHI at the 5′ and 3′ ends of bb0449, respectively (Genewiz, South Plainfield, NJ, USA). Optimized bb0449 was removed from pUC57-Kan by NdeI-BamHI digestion and ligated with similarly digested pET-28a(+) vector, adding an in-frame amino-terminal 6-histidine tag to BB0449. This vector was then transformed into BL21(DE3)pLysS E. coli cells. Recombinant BB0449 protein (rBB0449)-producing E. coli was grown in 1 liter of Overnight Express Instant TB medium (EMD Millipore, Billerica, MA., USA) at 37°C for 16 h. Cultures were harvested by centrifugation and stored at −80°C. Cell pellets were resuspended in 30 ml of lysis buffer (500 mM KCl, 50 mM HEPES [pH 7.5], 2 mM β-mercaptoethanol, 10 mM imidazole, 5 mM MgCl2) and disrupted by sonication, and cell debris was cleared by centrifugation. The soluble protein fraction was combined with 1 ml of preequilibrated nickel-nitrilotriacetic acid (Ni-NTA) agarose (Qiagen) and incubated for 1 h at 4°C with rocking. Subsequently, the Ni-NTA agarose slurry was applied to a chromatography column and washed with 50 bed volumes of lysis buffer. Proteins were eluted from the column in 5-ml fractions in elution buffer (300 mM KCl, 50 mM HEPES [pH 7.5], 2 mM β-mercaptoethanol, 250 mM imidazole, 5 mM MgCl2). Elution fractions were analyzed by SDS-PAGE and Coomassie blue staining. Fractions containing rBB0449 were pooled in an Amicon Ultra centrifugal unit (EMD Millipore) and subjected to buffer exchange and protein concentration into a buffer lacking imidazole (300 mM KCl, 50 mM HEPES [pH 7.5], 2 mM β-mercaptoethanol, 5 mM MgCl2). Production of rBB0449 polyclonal antiserum in rabbits was performed as previously described (55). The Rocky Mountain Laboratories (RML) are accredited by the International Association of Assessment and Accreditation of Laboratory Animal Care. Animal protocols were prepared according to the guidelines of the National Institutes of Health and approved by the RML Animal Care and Use Committee.
SDS-PAGE and immunoblotting.
Total protein lysates were made as previously described (54). Briefly, spirochetes were pelleted, washed twice with HN buffer (50 mM HEPES [pH 7.5], 50 mM NaCl), and resuspended in Laemmli sample buffer (Bio-Rad, Hercules, CA, USA), at a final concentration of 106 or 107 spirochetes per μl. Spirochete cell lysates derived from 107 or 108 spirochetes (as indicated) were separated on 12% or 15% SDS-polyacrylamide gels or 4 to 20% Mini Protean TGX gels (Bio-Rad) in SDS-PAGE running buffer (1.92 M glycine, 26 mM SDS, 250 mM Tris base) and transferred to a nitrocellulose blotting membrane as described previously (56). Nitrocellulose membranes were blocked in 5% nonfat milk (LabScientific Inc., Livingston, NJ, USA) in Tris-buffered saline with 0.1% Tween 20 (TBS-T). Membranes were then incubated with primary antibody or mouse sera at the indicated dilutions: anti-BB0449 polyclonal antibody (1:50), antiflagellin monoclonal antibody H9724 (1:200) (a gift from T. Schwan [57]), or infected mouse sera (1:200). Subsequently, blots were incubated with secondary antibody at the indicated dilutions: anti-rabbit IgG peroxidase-conjugated antibody (1:50,000) (Sigma Life Sciences) or anti-mouse IgG peroxidase-conjugated antibody (1:10,000) (Sigma Life Sciences). Blots were developed with a Super Signal West Pico chemiluminescent substrate kit (Thermo Scientific, Rockford, IL, USA) and X-ray film (LabScientific Inc.).
Crude ribosome preparation.
B. burgdorferi ribosomes were prepared as described with modifications (58). Briefly, WT spirochetes were grown to 2 × 108 cells/ml and incubated for an additional 24 h (referred to as stationary phase + 1 day). A total of 2 × 1011 spirochetes were pelleted by centrifugation (7,000 × g, 10 min, 22°C), resuspended in ice-cold ribosome buffer A (20 mM Tris-HCl [pH 7.5], 300 mM NH4Cl, 10 mM MgCl2, 0.5 mM EDTA, 6 mM β-mercaptoethanol), and lysed with a French press. Cell debris, including membrane blebs and unlysed cells, was removed by ultracentrifugation (30,000 × g, 1 h, 4°C). The supernatant containing cytosolic protein, including ribosomes, was spun through a 32% sucrose cushion (120,000 × g, 21 h, 4°C), resulting in a pellet that should contain ribosomes.
To confirm that the ribosome pellet contained ribosomal proteins, residual DNA and RNA were removed from the B. burgdorferi ribosome protein lysate with Benzonase treatment according to the manufacturer's instructions (Sigma-Aldrich). The ribosome fraction was separated on a 4 to 20% SDS-polyacrylamide gel and stained with Coomassie brilliant blue. Visible bands were excised and sent to the Protein Chemistry Section of the NIAID Research Technologies Branch, NIH, for mass spectrometry identification using the protocol described previously (59, 60).
Inactivation of bb0449.
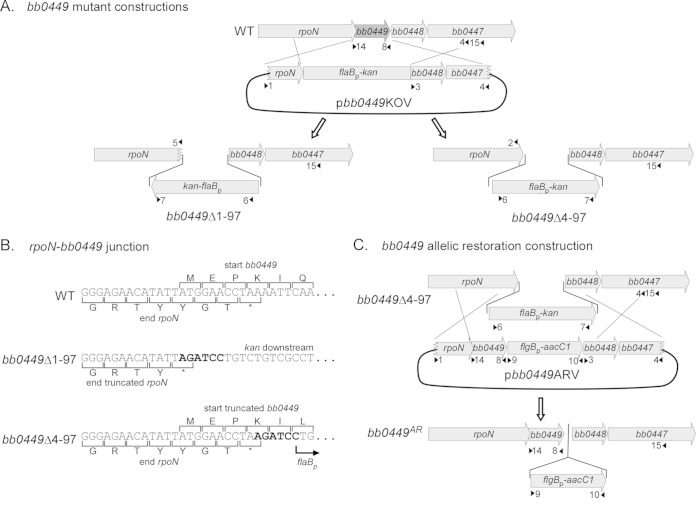
To construct the vector (pbb0449KOVΔ1-97) used to delete the full open reading frame (ORF) of hpf, including the 11 bp overlapping the upstream rpoN gene, flanking upstream and downstream DNA fragments were amplified from genomic DNA with primer pairs 1-2 and 3-4, respectively (Fig. 1; Table 2; see also Table S1 in the supplemental material). Plasmid pbb0449KOVΔ4-97, which did not delete the base pairs encoding the first 3 amino acids (aa) of BB0449, which overlap rpoN, was constructed by amplifying flanking DNA fragments with primer pairs 1-5 and 3-4, respectively (Fig. 1; see also Table S1). PCR products were cloned into pCR-Blunt II-TOPO vector (Fig. 1; see also Table S1). Primers 2, 3, and 5 contained BglII restriction sites. Resulting vectors were digested with the restriction enzymes BglII and NotI. The NotI cleavage site is present in the multicloning site of the vector. Orientations of the upstream and downstream flanking sequences in the pCR-Blunt II-TOPO vector allowed for linearization of the downstream-containing vector and liberation of the upstream fragments using restriction sites contained in the vector sequence. Upstream and downstream flanking sequences were purified from an agarose gel with the MinElute gel extraction kit (Qiagen). Linearized downstream-containing vector was ligated to either of the purified upstream flanking regions. All ligations used T4 DNA ligase (New England BioLabs). The kanamycin resistance cassette (flaBp-kan) (61) was amplified using BamHI restriction site-containing primers 6 and 7, cloned into pCR2.1 TOPO vector, liberated by BamHI restriction digestion, and purified with an agarose gel and a MinElute gel extraction kit (Qiagen) (Fig. 1; see also Table S1). The vectors with adjoining downstream and upstream flanking sequences were digested with BglII to insert the kanamycin resistance cassette between the flanking regions. The structures of the resulting plasmids, called bb0449Δ1-97 knockout vector (pbb0449KOVΔ1-97) and bb0449Δ4-97 knockout vector (pbb0449KOVΔ4-97), were confirmed by restriction digest and sequencing (Table 2). The plasmids have the flaBp-kan insertion in opposite orientations (Fig. 1).
FIG 1.
Construction of the bb0449 mutant (bb0449Δ1-97 and bb0449Δ4-97) and allelic restoration (bb0449AR) strains. Gene ORFs are represented by gray arrows, and the bb0449 ORF is in dark gray. Primer locations are shown with small black arrowheads and numbers and correspond to Table S1 in the supplemental material. (A) Construction of the bb0449 mutants. The full or partial bb0449 ORF was replaced with the kanamycin antibiotic resistance cassette, flaBp-kan, from one of the bb0449-knockout vectors (pbb0449KOVΔ1-97 or pbb0449Δ4-97). (B) Closeup of junction between the end of rpoN and the bb0449 deletion-flaBp-kan insertion region. The BglII-BamHI hybrid site creating the junction is shown in bold type. Strain names are indicated to the left of the sequence. (C) Construction of the bb0449 allelic restoration strain (bb0449AR). The bb0449 ORF was restored to bb0449Δ4-97 (linked to a downstream gentamicin antibiotic resistance marker, flgBp-aacC1) by allelic exchange with pbb0449ARV. Figure not drawn to scale.
TABLE 2.
Plasmids used in this study
| Plasmid | Purpose | Strain generated |
|---|---|---|
| pbb0449KOVΔ1-97 | Delete entire bb0449 ORF and insert a kanamycin resistance cassette, truncating rpoN | bb0449Δ1-97 |
| pbb0449KOVΔ4-97 | Delete bb0449 by replacing most of the bb0449 ORF with a kanamycin resistance cassette, leaving rpoN intact | bb0449Δ4-97 |
| pbb0449ARV | Restore bb0449 at its native locus; marked with a gentamicin resistance cassette | bb0449AR |
To inactivate bb0449, WT spirochetes were transformed with either pbb0449KOVΔ1-97 or pbb0449KOVΔ4-97 by electroporation as previously described (62), and allowed to recover overnight in BSKII medium at 35°C. Spirochetes were then plated in BSK plates containing streptomycin (50 μg/ml, to select for retention of streptomycin-marked lp25) and kanamycin (200 μg/ml). The inactivation of bb0449 and the addition and genomic location of the kanamycin resistance cassette were confirmed by PCR (primer pairs 8-14, 6-7, and 6-15, respectively) (Fig. 1; see also Table S1 in the supplemental material). We also confirmed that the plasmid content of transformants was identical to that of the parental WT strain (i.e., missing only cp9, lp5, and lp56) as previously described (63). The bb0449Δ1-97 mutant contains a truncated rpoN gene, which results in a protein missing the last three amino acids. The bb0449Δ4-97 mutant retains a complete rpoN gene and encodes an 18-amino-acid (aa) peptide that starts from the bb0449 ATG but has only 5 amino acids in common with the WT BB0449 protein.
Allelic restoration of bb0449 with the vector pbb0449ARV.
To construct the bb0449 allelic restoration vector (pbb0449ARV), the upstream flanking sequence and the bb0449 open reading frame (ORF) were amplified using primers 1 and 8, which has a BglII restriction site, and cloned into pCR-Blunt II-TOPO vector (Fig. 1; see also Table S1 in the supplemental material). The orientation of the upstream-bb0449 flanking sequences in the pCR-Blunt II-TOPO vector allowed for digestion with BglII and NotI (contained in the vector sequence) to liberate the upstream-bb0449 fragment, which was purified with an agarose gel and the MinElute gel extraction kit (Qiagen). The pCR-Blunt II-TOPO vector containing downstream flanking sequence, which was created when making pbb0449KOVΔ4-97, was linearized by digestion with BglII and NotI and ligated with the upstream-bb0449 fragment. The gentamicin resistance cassette, flgBp-aacC1, was amplified from pBSV2G shuttle vector (64) with primers 9 and 10, which contained BamHI restriction sites; cloned into pCR-Blunt II-TOPO vector; liberated by BamHI digestion; and purified with an agarose gel and the MinElute gel extraction kit (Qiagen) (Fig. 1; see also Table S1). The vector containing the upstream flanking sequence and the bb0449 ORF joined with a BglII restriction site to the downstream flanking sequence was digested with BglII and ligated to a purified gentamicin resistance cassette. The structure of pbb0449ARV was confirmed by restriction digest and sequencing (Table 2).
To reconstitute bb0449, bb0449Δ4-97 spirochetes were transformed with pbb0449ARV as described above. Cultures were plated with streptomycin and gentamicin (40 μg/ml). Reconstitution of bb0449 and the presence and genomic location of the gentamicin resistance cassette were confirmed by PCR (primer pairs 8-14, 9-10, and 9-15, respectively) (Fig. 1). Plasmid content was confirmed to be the same as that of the WT strain (63).
In vitro growth analysis.
Strains were grown to mid-log phase (2 × 107 to 7 × 107 cells/ml) from frozen stocks and diluted to 5 × 105 cells/ml in BSKII medium in triplicate with appropriate antibiotic selection. Spirochete densities were counted in Petroff-Hausser chambers every 24 h for 5 days and again at 8 days.
Experimental mouse-tick infectious cycle.
We determined if bb0449 played a role in the B. burgdorferi infectious cycle with two separate experiments. One experiment used WT and the bb0449Δ1-97 mutant, and the other used WT, the bb0449Δ4-97 mutant, and an allelic restoration strain. We first needle inoculated RML mice, which are outbred derivatives of Swiss Webster mice that are reared at the Rocky Mountain Laboratories breeding facility. Six- to 8-week-old female mice were needle inoculated intraperitoneally (8 × 103 spirochetes) and subcutaneously (2 × 103 spirochetes) for a total of 104 B. burgdorferi spirochetes. Mice were bled 3 weeks postinjection to determine seroconversion, which was assessed by immunoblotting with whole-cell lysates of B. burgdorferi. After ticks fed to repletion, mice were euthanized approximately 5 weeks post-needle inoculation and ear, bladder, and ankle joint tissues were cultured in BSKII medium for isolation of B. burgdorferi.
We also artificially infected larval ticks, as previously described (65), to confirm tick infectivity of B. burgdorferi strains. Briefly, larval ticks (Oklahoma State University) were partially dehydrated for 48 h before immersion and incubation in B. burgdorferi cultures (1 × 108 cells/ml). Immersed larval ticks were washed with phosphate-buffered saline and allowed to recover for 48 to 72 h before feeding on mice (∼150 artificially infected larval ticks/mouse).
To assess spirochete acquisition by ticks, approximately 100 to 150 larval Ixodes scapularis ticks (Oklahoma State University) were fed on seropositive mice 4 weeks post-needle inoculation with B. burgdorferi. Bacterial loads per fed larva were determined by manually homogenizing individual fed larval ticks, plating tick homogenates in BSK plates, and counting CFU. Infection prevalence was calculated by dividing the number of infected ticks by the total number of ticks plated. Remaining larval ticks molted into nymphs and were fed on naive mice to determine transmissibility of B. burgdorferi strains. Unfed and fed nymphal ticks were plated in the same manner as larval ticks to determine transstadial survival and bacterial loads after feeding. Spirochetes were transmissible if the mice upon which infected nymphs fed became infected, as determined by seroconversion and spirochete isolation from ear, bladder, and ankle joint tissues. Bacterial loads in infected mouse tissues were determined using DNA isolation from infected mouse tissues and quantitative PCR (qPCR) analysis as previously described (66).
In addition, a subset of unfed nymphal ticks was held at 4°C for 3 months to simulate overwintering. Nymphal ticks were then fed on naive mice, and transmission was assessed, as described above. Fed nymphal ticks were processed as described above to determine bacterial loads. The remainder of the ticks that had undergone 3 months at 4°C were stored at 23°C for an additional 6 months, fed on mice, and processed as described above.
Statistical significance of B. burgdorferi infectivity and infection prevalence of ticks were determined with Fisher's exact test in a pairwise manner. The reported P values include the lowest and highest of the comparisons. Medians of spirochete tick loads of B. burgdorferi strains were compared using a Kruskal-Wallis test with the GraphPad software Prism 6.
RESULTS
Identification of a B. burgdorferi hpf homolog.
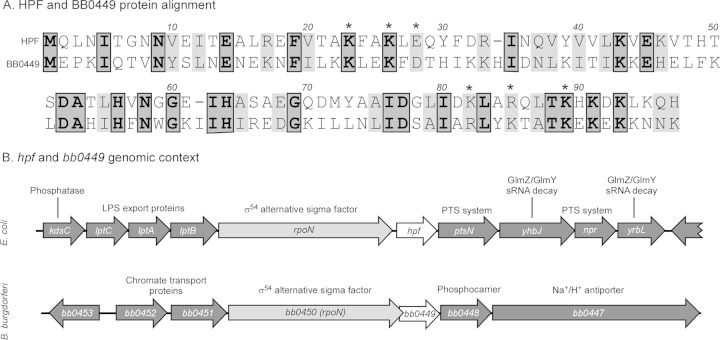
Using the NCBI BLASTP tool, we searched the translated B. burgdorferi B31 genome for homologs of the E. coli proteins RMF, YfiA, and HPF, the three proteins that are directly involved in changing ribosome conformation to control global protein translation. We found an HPF homolog, encoded by the chromosomal gene bb0449, but did not find RMF or YfiA homologs in B. burgdorferi (Fig. 2). Primary amino acid analysis showed that E. coli HPF and B. burgdorferi BB0449 share 26% identity and 46% similarity. BB0449 also shared conserved amino acid residues that were previously inferred to be interaction sites between HPF and ribosomes, based on sequence similarity with known ribosome interaction sites of E. coli YfiA, a protein that shares ribosome binding sites with HPF (24). Of the six HPF-ribosome interaction site amino acid residues, three residues were identical in the B. burgdorferi protein BB0449, while the remaining three residues were similar to those in HPF (e.g., lysine versus arginine) (Fig. 2A). Furthermore, hpf and bb0449 have similar genomic contexts (Fig. 2B). Both genes are directly downstream of rpoN, the gene encoding the alternate sigma factor (σ54), in their respective genomes. The relative positions of rpoN and hpf homologs are conserved in all Borrelia strains (data not shown) and among the Enterobacteriaceae (34). Additionally, the small intergenic spaces in this region of the genome indicate the possibility of coregulation. However, the surrounding regions in E. coli and B. burgdorferi contain genes with different predicted functions, according to NCBI database function assignments (Fig. 2B).
FIG 2.
Relationship between E. coli HPF and B. burgdorferi BB0449. (A) E. coli HPF and BB0449 amino acid sequence alignment. Proteins have 26% identity (dark gray) and 46% similarity (light gray). Asterisks indicate amino acids involved in protein-ribosome interaction (23). (B) Genomic contexts of hpf in E. coli genome (strain MG1655, NC_000913.3) and bb0449 in B. burgdorferi genome on the chromosome (strain B31, NC_001318.1). The hpf gene and its homolog, bb0449, are shown in white; the gene for the conserved σ54 alternative sigma factor (rpoN) is in light gray; and other genes are in dark gray. Predicted functions of the gene products are indicated above the gene. LPS, lipopolysaccharide; PTS, phosphotransferase.
Comparison of BB0449 and HPF secondary structure.
The Phyre2 program modeled the BB0449 amino acid sequence as the ribosome hibernation protein YhbH (also called HPF) with a 97.9 confidence level and 29% identity (data not shown). This was reproduced by secondary structure predictions generated by the SWISS-MODEL, I-Tasser, RaptorX, and HHPred programs (data not shown). The predicted model of BB0449 contains 4 β-sheets and 2 α-helices, which is consistent with the structure of HPF cocrystallized with a ribosome from T. thermophilus. Furthermore, comparison of the BB0449 model with crystallized HPF showed almost complete conservation of protein secondary structure, including placement of the six amino acids predicted to interact directly with ribosomes on exposed α-helices (see Fig. S1 in the supplemental material). Secondary structure differences included a shortened β1-sheet and α2-helix in BB0449, but neither of these regions contains amino acids predicted to interact with ribosomes. However, the K82R substitution in BB0449 increases the size of the amino acid R-group, which might interfere with potential ribosomal interaction, although the R85K substitution in BB0449 could compensate by reducing the size of a nearby side chain.
Analysis of in vitro bb0449 transcript and protein levels of WT spirochetes.
In E. coli, the level of hpf transcript increases relative to rRNA in stationary phase, which correlates with a concomitant increase in ribosome dimerization (20). To determine if bb0449 was similarly regulated in B. burgdorferi, we analyzed bb0449 transcript and product levels of cultured WT spirochetes. RNA was extracted, and protein lysates were prepared from WT spirochetes at different growth phases. Transcript and protein levels were determined by qRT-PCR or immunoblotting, respectively. qRT-PCR showed that there was no significant variation in bb0449 transcript levels at different growth phases in cultured spirochetes (P = 0.64) (Fig. 3A). Likewise, immunoblot analysis showed that BB0449 levels do not increase in stationary phase (Fig. 3B). These analyses indicate a lack of differential regulation under these conditions. To visualize BB0449 protein, it was necessary to use cell lysates from 108 spirochetes, which leads to a very strong antiflagellin signal (Fig. 3C). However, we detected only flagellin and no BB0449 when cell lysates from 107 spirochetes were used (data not shown). This likely reflects low levels of BB0449 protein, which is consistent with low transcript levels.
FIG 3.
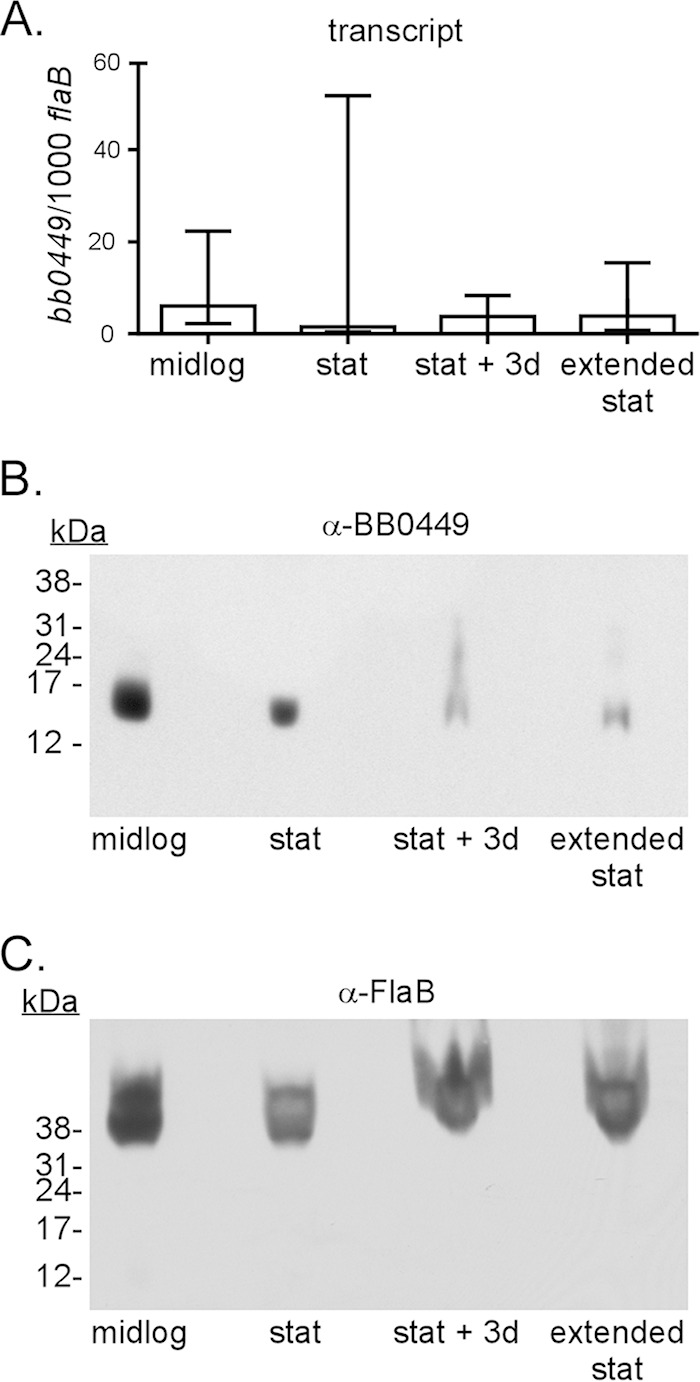
Expression of bb0449 and levels of BB0449 protein in WT B. burgdorferi during growth in culture. (A) bb0449 transcript levels at different growth phases. Graphed are the median number of copies (determined by qRT-PCR) of bb0449 standardized to flaB with the first and third quartile ranges of biological replicates. bb0449 transcript levels do not significantly vary among B. burgdorferi growth phases (P = 0.64). (B) Immunoblot assay showing BB0449 protein in protein lysates prepared at the same growth phases. Lanes were loaded with total cell lysate of 108 B. burgdorferi cells. Growth phases were defined as follows: “midlog,” 2 × 107 to 7 × 107 cells/ml; “stat” (stationary), 2 × 108 cells/ml; “stat + 3d” and “extended stat,” stationary-phase cultures incubated for an additional 3 or 5 to 6 days, respectively. (C) Reprobing of the same immunoblot with monoclonal antiflagellin (FlaB; H9724 [57]).
Cellular localization of BB0449.
Since HPF dimerizes ribosomes to modulate global protein translation levels in E. coli, we used differential centrifugation to selectively enrich for B. burgdorferi ribosomes and probed ribosome preparations with BB0449-specific antiserum to determine if BB0449 was associated with ribosomes. BB0449 protein was visualized predominantly in a fraction containing cytosolic proteins and ribosomes, but not in the ribosome pellet fraction, suggesting that BB0449 is not associated with ribosomes under the conditions tested and is instead a soluble protein (Fig. 4). We confirmed that ribosomal proteins were represented in the ribosome fraction by using mass spectrometry to identify protein bands of the ribosome protein fraction. Fifty-one of 55 ribosomal proteins encoded in B. burgdorferi, as identified in the KEGG database (67), were identified in the tandem mass spectrometry (MS/MS) analysis (see Table S2 in the supplemental material). In addition, BB0449 was not detected by mass spectrometry from the ribosome protein sample. The absence of BB0449 from the ribosome fraction as determined by immunoblotting and mass spectrometry analysis indicated that BB0449 was not associated with ribosomes under the culture conditions tested.
FIG 4.
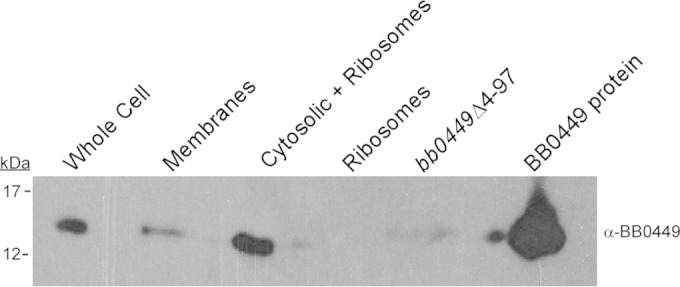
Localization of BB0449 protein by immunoblot analysis of fractions during ribosome preparation from WT B. burgdorferi. Equivalent portions of fractions indicated above lanes were separated by SDS-PAGE, blotted, and hybridized with anti-BB0449 antibody. bb0449Δ4-97, total cell lysate equivalent to 108 cells; BB0449, purified recombinant His-tagged protein; kDa, molecular masses of size standards that migrated at indicated positions.
Strain characterization.
To determine whether bb0449 played a role in the infectious cycle of B. burgdorferi, we constructed two bb0449 mutants (bb0449Δ1-97 and bb0449Δ4-97) and an allelic restoration strain in bb0449Δ4-97 (bb0449AR) (Fig. 1). One mutant (bb0449Δ1-97) lacks the entire BB0449 coding sequence and removes the last 3 amino acids of the preceding RpoN open reading frame (ORF), and the other deletion (in bb0449Δ4-97) begins after the end of the rpoN gene, yielding a truncated version of the BB0449 protein that is 18 aa in length, of which only the first 5 amino acids are the same as in the native BB0449 product (Fig. 1A and B). Loss of BB0449 protein production in the two mutants and restoration of protein production in the allelic restoration strain were confirmed by SDS-PAGE and immunoblotting (Fig. 5 and data not shown). Analysis of growth in culture showed that there was no significant difference in the growth rates or the final spirochete densities of bb0449Δ1-97, bb0449Δ4-97, or bb0449AR compared to WT (data not shown).
FIG 5.
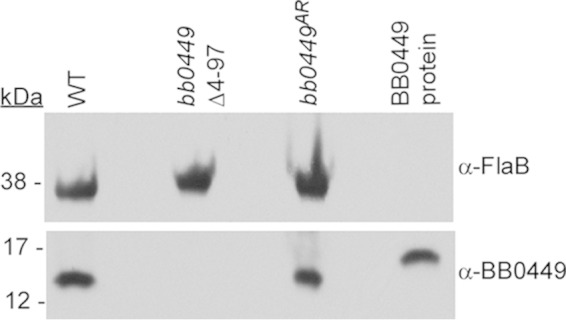
Immunoblot assay demonstrating absence of BB0449 protein in the bb0449Δ4-97 mutant. Lysates of indicated strains and purified BB0449 were separated by SDS-PAGE, blotted, and probed with antibody to flagellin (α-FlaB, to standardize protein loading) and to BB0449. BB0449 protein, recombinant BB0449 protein with a 6×His tag, which increased the protein size. Protein lysates equivalent to 108 spirochetes were loaded in each lane. kDa, molecular masses of size standards that migrated at indicated positions.
Infectious cycle analysis of WT, mutant, and allelic restoration strains.
To ascertain if bb0449 is required for mouse infection, we performed two separate infectious cycle experiments with combinations of WT, bb0449 mutants, and bb0449AR. We needle inoculated mice with approximately 104 cells of each strain. Three weeks postinoculation, we assessed seroreactivity of mice by immunoblotting, and larval ticks were fed on a subset of seropositive mice. We euthanized the mice 5 weeks post-needle inoculation, after tick feeding, and collected ear, bladder, and joint tissues for spirochete isolation. Infected mice were defined as those that seroconverted and for which spirochetes were isolated from mouse tissues. There was no significant difference in the number of mice infected by any of the B. burgdorferi strains in analyses of either separate or combined experiments (P = 0.33 to 1.00) (Table 3). We also used quantitative PCR to assess loads of spirochetes in tissues of infected animals and found no significant differences in tissue burden for any strain (data not shown). Therefore, bb0449 is not required for mouse infection by needle inoculation.
TABLE 3.
Infectivity and transmissibility of B. burgdorferi strains
| Infection route of mice | B. burgdorferi strain | No. of infected mice/total no. of micea |
|---|---|---|
| Needle inoculationb | WTc | 9/10 |
| Δbb0449 cumulativec | 8/10 | |
| bb0449Δ1-97 | 5/5 | |
| bb0449Δ4-97 | 3/5 | |
| bb0449AR | 4/5 | |
| Tick bited | WT | 4/4 |
| Δbb0449 cumulative | 3/4 | |
| bb0449Δ1-97 | 1/2 | |
| bb0449Δ4-97 | 2/2 | |
| bb0449AR | 1/1 | |
| Tick bite (4°C, 3 mo)e | WT | 2/2 |
| bb0449Δ4-97f | 4/4 | |
| bb0449ARg | 2/2 |
Infected mice are defined as mice that seroconverted and from which spirochetes were reisolated.
Mice were needle inoculated subcutaneously and intraperitoneally with 1 × 104 B. burgdorferi cells.
WT and Δbb0449 cumulative data are pooled from two infection experiments that used either WT and bb0449Δ1-97 or WT, bb0449Δ4-97, and bb0449AR (allelic restoration) strains.
Ten to 20 infected nymphs were fed on naive mice approximately 12 weeks after the larval feeding.
4°C for 3 months simulates “overwintering” prior to feeding.
The ticks placed on two of these mice had been incubated for an additional 6 months at 23°C prior to feeding.
The ticks that fed on one of these mice had been incubated for an additional 6 months at 23°C prior to feeding.
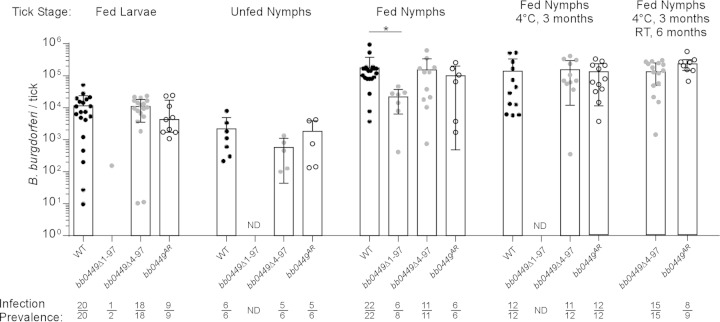
Acquisition of spirochetes and colonization of larval ticks were determined by individually plating a subset of replete larval ticks that fed on serologically positive mice. There was no significant difference in either the proportion of infected ticks or the number of CFU in tick homogenates from ticks infected with WT, bb0449Δ4-97, or bb0449AR (P = 0.67) (Fig. 6). These data suggest comparable acquisition of spirochetes by larval ticks among B. burgdorferi strains. In contrast to the large numbers of fed larval ticks obtained in the first larval feeding, we obtained only low numbers of fed larvae during the WT and bb0449Δ1-97 larval tick feeding, so we plated two larval ticks to confirm the presence of infected larvae. Only one of the ticks tested that fed on a bb0449Δ1-97-infected mouse was infected (Fig. 6). We concluded that some proportion of larval ticks acquired bb0449Δ1-97 spirochetes. We also confirmed that both of the bb0449 mutants and the allelic restoration strain were capable of infecting larval ticks by artificially infecting larval ticks, feeding those larval ticks on naive mice, and then individually plating them to determine if the ticks were infected. Consistent with naturally infected larvae, we found no statistically significant difference in the spirochete loads of artificially infected larval ticks (P = 0.87) (data not shown).
FIG 6.
Spirochete loads per tick at various tick developmental stages. Ticks infected with WT, bb0449Δ1-97, bb0449Δ4-97, or bb0449AR were plated either before or 5 to 10 days after feeding on mice, to determine spirochete CFU. Data represent two independent infection experiments. WT data are pooled from the two experiments. “Fed Larvae,” larval ticks infected by feeding on infected mice to determine acquisition; “Unfed Nymphs,” infected nymphs plated before feeding to determine transstadial survival; “Fed Nymphs,” infected nymphs fed to repletion on naive mice; “Fed Nymphs 4°C, 3 months,” infected nymphs held at 4°C for 3 months before feeding on naive mice; “Fed Nymphs 4°C, 3 months RT, 6 months,” infected nymphs held for 3 months at 4°C and a further 6 months at 23°C before feeding. Bacterial loads were compared within a specific tick developmental stage with a Kruskal-Wallis test. There was no statistical difference in bacterial loads in fed larvae, unfed nymphs, fed nymphs held at 4°C for 3 months, or fed nymphs held at 4°C for 3 months and 23°C for 6 months (P = 0.67, 0.38, 0.56, and 0.15, respectively). WT-infected fed nymphs did have higher bacterial loads than bb0449Δ1-97-infected fed nymphs (P = 0.02). Points indicate values from individual ticks. Bars represent median tick bacterial loads, and error bars indicate the first and third interquartile ranges. Infection prevalence is shown as number of infected ticks divided by number of ticks plated. ND, not determined.
To determine the importance of bb0449 in transstadial passage, we individually plated unfed nymphs from cohorts of larvae that fed upon mice infected by needle inoculation with WT, bb0449Δ4-97, or bb0449AR. There was no significant difference in the prevalence of infection in ticks (P = 0.44 to 1.00) or in unfed nymph spirochete loads (P = 0.13) (Fig. 6), indicating that bb0449 is not required for successful transstadial passage.
We next assessed the ability of spirochetes to be transmitted to naive mice and to replicate within a nymphal tick. We fed infected nymphs on naive mice and individually plated fed nymphs to determine spirochete loads. There was no significant difference in the prevalence of spirochete infection in fed nymphs (P = 0.06 to 1.00). However, the median spirochete load in WT-infected fed nymphs was approximately five times higher than that in bb0449Δ1-97-infected fed nymphs (P = 0.02), which were infected with spirochetes that produced truncated RpoN, but there was no statistically significant difference in bacterial loads among any of the other strains (Fig. 6). Furthermore, all of the B. burgdorferi strains, including bb0449Δ1-97, infected naive mice by nymphal tick bite, regardless of spirochete burdens in ticks. This shows that bb0449 is not required for transmission (Table 3). Bacterial loads in infected mouse tissues (ear, heart, and ankle joint) were not different among B. burgdorferi strains, indicating that bb0449 is not required for spirochete dissemination (data not shown).
To evaluate the effects on transmissibility of extended periods within a tick vector, we placed cohorts of unfed nymphs infected with WT, bb0449Δ4-97, or bb0449AR at 4°C for 3 months, to simulate overwintering, before feeding them on naive mice. There was no statistically significant difference in the bacterial loads of fed infected nymphs held at 4°C (P = 0.56) (Fig. 6). In addition, all three B. burgdorferi strains from ticks kept at 4°C were able to be transmitted to naive mice by tick bite (Table 3). These data indicate that survival through the infectious cycle and experimental overwintering process does not require bb0449. After an additional 6 months of incubation at 23°C, an additional two cohorts of ticks infected with bb0449Δ4-97 and one cohort infected with bb0449AR were fed on mice. Again, the spirochete loads in the ticks were similar (Fig. 6), and all three mice became infected (Table 3). These data underscore the ability of spirochetes lacking bb0449 to survive an extended period in unfed ticks and to recover after entering the nutrient-rich mammalian environment.
DISCUSSION
The infectious cycle of B. burgdorferi includes extended periods of time within the tick midgut during which spirochetes must survive, such as within overwintering nymphal ticks. Therefore, B. burgdorferi likely has a mechanism to cope with the nutrient-limited conditions found in an unfed tick midgut. In our study, we found that B. burgdorferi harbors a protein, BB0449, that is homologous to E. coli HPF, which facilitates the formation of translationally inactive ribosomes in E. coli during stationary phase (Fig. 2). Despite protein homology, we determined that BB0449 is not found in ribosome protein fractions of B. burgdorferi grown to stationary phase, suggesting that BB0449 may not function as an HPF-like protein or that BB0449 associates with ribosomes under specific culture conditions not yet identified (Fig. 4). In addition, we constructed two independent bb0449 mutants that differed both in the direction of the inserted selectable marker and by either leaving an intact rpoN ORF or deleting the last three codons of the rpoN gene. Although the bb0449Δ1-97 mutant might have been expected to have a stronger phenotype, based on reduced RpoN function, both mutants completed the experimental tick-mouse infectious cycle, indicating that bb0449 is not required at any point during experimental infection (Table 3; Fig. 6). In fact, bb0449Δ4-97-infected nymphs were able to transmit spirochetes to naive mice after only 72 h of feeding (unpublished data). We have also made strains that overproduce the bb0449 product in culture, but we have found no defect in growth that correlates with the amount of protein produced (L. Fazzino and K. Tilly, unpublished data), which might be expected if BB0449 dimerized ribosomes and blocked global translation.
Although our results are inconsistent with BB0449 having a role in ribosome dimerization, we did not empirically show that BB0449 does not influence ribosome dimerization in all circumstances, specifically within the infectious cycle at stages where we were unable to measure levels of the protein or transcript. It is possible that BB0449 functions as a ribosome dimerization protein in B. burgdorferi in a specific set of conditions not recapitulated during growth in culture. The conservation of the secondary structure of BB0449 supports this hypothesis (see Fig. S1 in the supplemental material). The only indication of changes in secondary structure that could influence the ability of BB0449 to bind to ribosomes was the substitution of lysine for arginine at a key residue introducing a larger R-group. However, determining if this change affects the ability of BB0449 to bind ribosomes is imprecise without BB0449 cocrystallized with B. burgdorferi ribosomes. Overall, our secondary structure analyses of BB0449 did not detect large differences between HPF and BB0449 secondary structure, suggesting that BB0449 should be able to interact with ribosomes in B. burgdorferi. Biochemical studies, such as those performed previously in other bacteria (19, 21, 24, 28, 31, 32, 68), may help exclude the possibility of, or elucidate, a molecular role for BB0449 in B. burgdorferi ribosome modulation. Despite these caveats, the ability of B. burgdorferi totally lacking the BB0449 product to complete the mouse-tick infectious cycle, including spending approximately 9 months in unfed ticks, argues against a major impact on ribosome modulation by BB0449, if it exists.
Two additional pieces of data support the alternative hypothesis that BB0449 does not function as an HPF-like protein: (i) the lack of regulation seen in bb0449 transcript expression and protein production patterns (Fig. 3) and (ii) the absence of homologs of additional E. coli ribosome modulation proteins, such as RMF and YfiA. The low levels of bb0449 transcript and protein in B. burgdorferi do not change in various growth phases (Fig. 3), which is incongruent with the observed increase in hpf transcript in relation to ribosome copy number in stationary-phase E. coli (21). In addition to these data, we have been unable to detect bb0449 transcript in RNA derived from infected ticks, either when unfed or at several time points during and after feeding (Fazzino and Tilly, unpublished). Moreover, BB0449 protein levels were no higher in spirochetes that had been cultured at 25°C than in those grown at 35°C (Fazzino and Tilly, unpublished). This low-level gene expression and protein production in B. burgdorferi may reflect a fundamentally different protein function. Ultimately, the molecular role of BB0449 is nonessential, as the protein is not required to complete the experimental infectious cycle.
Moreover, the amount of BB0449 detected in cultured spirochetes is likely to be inadequate to bind sufficient numbers of ribosomes to reduce global protein translation. In contrast to bb0449 transcript levels, E. coli grown to stationary phase has approximately 1 copy of hpf transcript per ribosome (21). In addition, HPF-mediated ribosome dimerization in E. coli is a transient and easily reversible process. In fact, Ueta et al. observed the disassociation of RMF, HPF, and YfiA from stationary-phase translationally inactive ribosomes within 1 min of transferring E. coli to fresh medium (21). This reversible attribute, again, signifies that relatively high levels of protein, which are not present in B. burgdorferi when grown in culture (or likely in ticks, where transcript was below the level of detection in the stages that we examined), would be needed to impact global protein translation levels.
The family of HPF-like proteins has either a short (97-amino-acid) or a long (200-amino-acid) form (24). B. burgdorferi BB0449 is homologous to the short HPF protein. Short HPF proteins are generally found in bacteria that also harbor RMF and YfiA (to which B. burgdorferi does not appear have homologs) and are not able to dimerize ribosomes in vitro without the RMF protein (21, 24). In comparison, long HPF-like proteins are thought to functionally replace both RMF and short HPF because of an extended C terminus and have been shown to dimerize ribosomes in vitro (22, 68). The lack of association between BB0449 and ribosomes corresponds with the observation that short HPF proteins are unable to dimerize ribosomes without additional proteins (Fig. 4). Although a previous study by Ueta et al. (24) reported that B. burgdorferi harbored a long HPF-like protein found by searching the KEGG database, current searches of the KEGG database do not yield any long HPF homologs in the B. burgdorferi genomes and likely indicate that the previously described long HPF was an incorrect annotation and has since been removed.
It is also possible that BB0449 has a function other than ribosome modulation. Other studies suggest that HPF-like proteins may modulate RpoN, also called alternative sigma factor 54 (34, 69). In fact, the B. burgdorferi bb0449 ORF is directly downstream of rpoN, making the involvement of BB0449 in modulation of RpoN an attractive hypothesis. Furthermore, the relative positions of rpoN and hpf homologs are conserved in the genus Borrelia (data not shown) and in Enterobacteriaceae (34). However, in B. burgdorferi, RpoN helps regulate multiple virulence genes encoding outer surface proteins (e.g., OspC and DbpA) by directing transcription of the gene for a second alternative sigma factor, RpoS (reviewed in reference 70) (71–73). A lack of functional RpoN, RpoS, or OspC in B. burgdorferi results in strains that are noninfectious or severely attenuated in mammals (reviewed in reference 70) (71–73). The ability of the bb0449 mutants to complete the entire experimental infectious cycle indicates that BB0449 is not a crucial component controlling RpoN/RpoS activity, although it does not exclude the possibility of involvement.
In conclusion, we have shown that bb0449 is not required to complete the experimental mouse-tick infectious cycle. In addition, despite homology to E. coli HPF at the primary and secondary structure levels, BB0449 protein did not appear to bind to ribosomes in cultured spirochetes. Although it remains a possibility that BB0449 is associated with ribosomes only under conditions that we have not examined in this study, our findings that BB0449 is not required for survival in the nutrient-deprived unfed tick environment, not associated with ribosomes under the conditions examined, and not made in sufficient quantities to bind ribosomes stoichiometrically indicate that BB0449 may serve a role other than ribosome dimerization in B. burgdorferi.
Supplementary Material
ACKNOWLEDGMENTS
We thank the following people: Amitava Roy of the Bioinformatics and Computational Biosciences Branch (NIH) for assistance with secondary structure analysis of BB0449; Craig Martens with bioinformatics analysis of putative bb0449 promoter locations; Glenn Nardone, Ming Zhao, and L. Renee Olano of the NIH Research Technologies Branch, Mass Spectrometry and Mass Spectrometry-Bioinformatics sections for performing the MS/MS and data analysis; Leonard Evans for kindly allowing us access to his ultracentrifuge; Kevin Lawrence for assistance with the French press protocol; and Tom Schwan for the gift of the monoclonal antibody against flagellin. We thank Phil Stewart and Jay Carroll for helpful discussions, and we thank Nick Noriea, Iman Chouikha, Irene Kasumba, Bharti Bhatia, and Phil Stewart for critical reading of the manuscript. We acknowledge the expert assistance of Anita Mora in figure preparation.
This research was supported by the Division of Intramural Research of the NIAID, NIH.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.00925-15.
REFERENCES
- 1.Bacon RM, Kugeler KJ, Mead PS. 2008. Surveillance for Lyme disease—United States, 1992–2006. MMWR Surveill Summ 57:1–9. [PubMed] [Google Scholar]
- 2.Burgdorfer W, Barbour AG, Hayes SF, Benach JL, Grunwaldt E, Davis JP. 1982. Lyme disease—a tick-borne spirochetosis? Science 216:1317–1319. doi: 10.1126/science.7043737. [DOI] [PubMed] [Google Scholar]
- 3.Steere AC, Grodzicki RL, Kornblatt AN, Craft JE, Barbour AG, Burgdorfer W, Schmid GP, Johnson E, Malawista SE. 1983. The spirochetal etiology of Lyme disease. N Engl J Med 308:733–740. doi: 10.1056/NEJM198303313081301. [DOI] [PubMed] [Google Scholar]
- 4.Lindsay LR, Barker IK, Surgeoner GA, McEwen SA, Gillespie TJ, Addison EM. 1998. Survival and development of the different life stages of Ixodes scapularis (Acari: Ixodidae) held within four habitats on Long Point, Ontario, Canada. J Med Entomol 35:189–199. doi: 10.1093/jmedent/35.3.189. [DOI] [PubMed] [Google Scholar]
- 5.Porco TC. 1999. A mathematical model of the ecology of Lyme disease. IMA J Math Appl Med Biol 16:261–296. doi: 10.1093/imammb/16.3.261. [DOI] [PubMed] [Google Scholar]
- 6.Nefedova VV, Korenberg EI, Gorelova NB, Kovalevskii YV. 2004. Studies on the transovarial transmission of Borrelia burgdorferi sensu lato in the taiga tick Ixodes persulcatus. Folia Parasitol (Praha) 51:67–71. doi: 10.14411/fp.2004.010. [DOI] [PubMed] [Google Scholar]
- 7.Rollend L, Fish D, Childs JE. 2013. Transovarial transmission of Borrelia spirochetes by Ixodes scapularis: a summary of the literature and recent observations. Ticks Tick Borne Dis 4:46–51. doi: 10.1016/j.ttbdis.2012.06.008. [DOI] [PubMed] [Google Scholar]
- 8.Lane RS, Piesman J, Burgdorfer W. 1991. Lyme borreliosis: relation of its causative agent to its vectors and hosts in North America and Europe. Annu Rev Entomol 36:587–609. doi: 10.1146/annurev.en.36.010191.003103. [DOI] [PubMed] [Google Scholar]
- 9.Donahue JG, Piesman J, Spielman A. 1987. Reservoir competence of white-footed mice for Lyme disease spirochetes. Am J Trop Med Hyg 36:92–96. [DOI] [PubMed] [Google Scholar]
- 10.Lindsay LR, Barker IK, Surgeoner GA, McEwen SA, Campbell GD. 1997. Duration of Borrelia burgdorferi infectivity in white-footed mice for the tick vector Ixodes scapularis under laboratory and field conditions in Ontario. J Wildl Dis 33:766–775. doi: 10.7589/0090-3558-33.4.766. [DOI] [PubMed] [Google Scholar]
- 11.Sonenshine DE, Anderson JM. 2014. Mouthparts and digestive system: anatomy and molecular biology of feeding and digestion, p 122–162. In Sonenshine DE, Roe RM (ed), Biology of ticks, 2nd ed, vol 1 Oxford University Press, New York, NY. [Google Scholar]
- 12.Starosta AL, Lassak J, Jung K, Wilson DN. 2014. The bacterial translation stress response. FEMS Microbiol Rev 38:1172–1201. doi: 10.1111/1574-6976.12083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bugrysheva J, Dobrikova EY, Sartakova ML, Caimano MJ, Daniels T, Radolf JD, Godfrey HP, Cabello F. 2003. Characterization of the stringent response and rel(Bbu) expression of Borrelia burgdorferi. J Bacteriol 185:957–965. doi: 10.1128/JB.185.3.957-965.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bugrysheva J, Dobrikova EY, Godfrey HP, Sartakova ML, Cabello FC. 2002. Modulation of Borrelia burgdorferi stringent response and gene expression during extracellular growth with tick cells. Infect Immun 70:3061–3067. doi: 10.1128/IAI.70.6.3061-3067.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bugrysheva JV, Bryksin AV, Godfrey HP, Cabello FC. 2005. Borrelia burgdorferi rel is responsible for generation of guanosine-3′-diphosphate-5′-triphosphate and growth control. Infect Immun 73:4972–4981. doi: 10.1128/IAI.73.8.4972-4981.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bugrysheva JV, Godfrey HP, Schwartz I, Cabello FC. 2011. Patterns and regulation of ribosomal RNA transcription in Borrelia burgdorferi. BMC Microbiol 11:17. doi: 10.1186/1471-2180-11-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Concepcion MB, Nelson DR. 2003. Expression of spoT in Borrelia burgdorferi during serum starvation. J Bacteriol 185:444–452. doi: 10.1128/JB.185.2.444-452.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Drecktrah D, Lybecker M, Popitsch N, Rescheneder P, Hall LS, Samuels DS. 2015. The Borrelia burgdorferi RelA/SpoT homolog and stringent response regulate survival in the tick vector and global gene expression during starvation. PLoS Pathog 11:e1005160. doi: 10.1371/journal.ppat.1005160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Polikanov YS, Blaha GM, Steitz TA. 2012. How hibernation factors RMF, HPF, and YfiA turn off protein synthesis. Science 336:915–918. doi: 10.1126/science.1218538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maki Y, Yoshida H, Wada A. 2000. Two proteins, YfiA and YhbH, associated with resting ribosomes in stationary phase Escherichia coli. Genes Cells 5:965–974. doi: 10.1046/j.1365-2443.2000.00389.x. [DOI] [PubMed] [Google Scholar]
- 21.Ueta M, Yoshida H, Wada C, Baba T, Mori H, Wada A. 2005. Ribosome binding proteins YhbH and YfiA have opposite functions during 100S formation in the stationary phase of Escherichia coli. Genes Cells 10:1103–1112. doi: 10.1111/j.1365-2443.2005.00903.x. [DOI] [PubMed] [Google Scholar]
- 22.Yoshida H, Wada A. 2014. The 100S ribosome: ribosomal hibernation induced by stress. Wiley Interdiscip Rev RNA 5:723–732. doi: 10.1002/wrna.1242. [DOI] [PubMed] [Google Scholar]
- 23.Yoshida H, Yamamoto H, Uchiumi T, Wada A. 2004. RMF inactivates ribosomes by covering the peptidyl transferase centre and entrance of peptide exit tunnel. Genes Cells 9:271–278. doi: 10.1111/j.1356-9597.2004.00723.x. [DOI] [PubMed] [Google Scholar]
- 24.Ueta M, Ohniwa RL, Yoshida H, Maki Y, Wada C, Wada A. 2008. Role of HPF (hibernation promoting factor) in translational activity in Escherichia coli. J Biochem 143:425–433. doi: 10.1093/jb/mvm243. [DOI] [PubMed] [Google Scholar]
- 25.Ortiz JO, Brandt F, Matias VR, Sennels L, Rappsilber J, Scheres SH, Eibauer M, Hartl FU, Baumeister W. 2010. Structure of hibernating ribosomes studied by cryoelectron tomography in vitro and in situ. J Cell Biol 190:613–621. doi: 10.1083/jcb.201005007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Agafonov DE, Kolb VA, Spirin AS. 2001. Ribosome-associated protein that inhibits translation at the aminoacyl-tRNA binding stage. EMBO Rep 2:399–402. doi: 10.1093/embo-reports/kve091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vila-Sanjurjo A, Schuwirth BS, Hau CW, Cate JH. 2004. Structural basis for the control of translation initiation during stress. Nat Struct Mol Biol 11:1054–1059. doi: 10.1038/nsmb850. [DOI] [PubMed] [Google Scholar]
- 28.Ueta M, Wada C, Daifuku T, Sako Y, Bessho Y, Kitamura A, Ohniwa RL, Morikawa K, Yoshida H, Kato T, Miyata T, Namba K, Wada A. 2013. Conservation of two distinct types of 100S ribosome in bacteria. Genes Cells 18:554–574. doi: 10.1111/gtc.12057. [DOI] [PubMed] [Google Scholar]
- 29.De Bari H, Berry EA. 2013. Structure of Vibrio cholerae ribosome hibernation promoting factor. Acta Crystallogr Sect F Struct Biol Cryst Commun 69:228–236. doi: 10.1107/S1744309113000961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yoshida H, Ueta M, Maki Y, Sakai A, Wada A. 2009. Activities of Escherichia coli ribosomes in IF3 and RMF change to prepare 100S ribosome formation on entering the stationary growth phase. Genes Cells 14:271–280. doi: 10.1111/j.1365-2443.2008.01272.x. [DOI] [PubMed] [Google Scholar]
- 31.Sato A, Watanabe T, Maki Y, Ueta M, Yoshida H, Ito Y, Wada A, Mishima M. 2009. Solution structure of the E. coli ribosome hibernation promoting factor HPF: implications for the relationship between structure and function. Biochem Biophys Res Commun 389:580–585. doi: 10.1016/j.bbrc.2009.09.022. [DOI] [PubMed] [Google Scholar]
- 32.Puri P, Eckhardt TH, Franken LE, Fusetti F, Stuart MC, Boekema EJ, Kuipers OP, Kok J, Poolman B. 2014. Lactococcus lactis YfiA is necessary and sufficient for ribosome dimerization. Mol Microbiol 91:394–407. doi: 10.1111/mmi.12468. [DOI] [PubMed] [Google Scholar]
- 33.Kline BC, McKay SL, Tang WW, Portnoy DA. 2015. The Listeria monocytogenes hibernation-promoting factor is required for the formation of 100S ribosomes, optimal fitness, and pathogenesis. J Bacteriol 197:581–591. doi: 10.1128/JB.02223-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ancona V, Li W, Zhao Y. 2014. Alternative sigma factor RpoN and its modulation protein YhbH are indispensable for Erwinia amylovora virulence. Mol Plant Pathol 15:58–66. doi: 10.1111/mpp.12065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rego RO, Bestor A, Rosa PA. 2011. Defining the plasmid-encoded restriction-modification systems of the Lyme disease spirochete Borrelia burgdorferi. J Bacteriol 193:1161–1171. doi: 10.1128/JB.01176-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kawabata H, Norris SJ, Watanabe H. 2004. BBE02 disruption mutants of Borrelia burgdorferi B31 have a highly transformable, infectious phenotype. Infect Immun 72:7147–7154. doi: 10.1128/IAI.72.12.7147-7154.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Elias AF, Schmutzhard J, Stewart PE, Schwan TG, Rosa P. 2002. Population dynamics of a heterogeneous Borrelia burgdorferi B31 strain in an experimental mouse-tick infectious cycle. Wien Klin Wochenschr 114:557–561. [PubMed] [Google Scholar]
- 38.Barbour AG. 1984. Isolation and cultivation of Lyme disease spirochetes. Yale J Biol Med 57:521–525. [PMC free article] [PubMed] [Google Scholar]
- 39.Rosa P, Samuels DS, Hogan D, Stevenson B, Casjens S, Tilly K. 1996. Directed insertion of a selectable marker into a circular plasmid of Borrelia burgdorferi. J Bacteriol 178:5946–5953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. 2015. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Humphrey W, Dalke A, Schulten K. 1996. VMD: visual molecular dynamics. J Mol Graphics 14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 42.Arnold K, Bordoli L, Kopp J, Schwede T. 2006. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 43.Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, Schmidt T, Kiefer F, Cassarino TG, Bertoni M, Bordoli L, Schwede T. 2014. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res 42:W252–W258. doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guex N, Peitsch MC, Schwede T. 2009. Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: a historical perspective. Electrophoresis 30(Suppl 1):S162–S173. doi: 10.1002/elps.200900140. [DOI] [PubMed] [Google Scholar]
- 45.Kiefer F, Arnold K, Kunzli M, Bordoli L, Schwede T. 2009. The SWISS-MODEL repository and associated resources. Nucleic Acids Res 37:D387–D392. doi: 10.1093/nar/gkn750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roy A, Kucukural A, Zhang Y. 2010. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc 5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y. 2015. The I-TASSER suite: protein structure and function prediction. Nat Methods 12:7–8. doi: 10.1038/nmeth.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang Y. 2008. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kallberg M, Margaryan G, Wang S, Ma J, Xu J. 2014. RaptorX server: a resource for template-based protein structure modeling. Methods Mol Biol 1137:17–27. doi: 10.1007/978-1-4939-0366-5_2. [DOI] [PubMed] [Google Scholar]
- 50.Kallberg M, Wang H, Wang S, Peng J, Wang Z, Lu H, Xu J. 2012. Template-based protein structure modeling using the RaptorX web server. Nat Protoc 7:1511–1122. doi: 10.1038/nprot.2012.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Peng J, Xu J. 2011. RaptorX: exploiting structure information for protein alignment by statistical inference. Proteins 79(Suppl 10):S161–S171. doi: 10.1002/prot.23175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hildebrand A, Remmert M, Biegert A, Soding J. 2009. Fast and accurate automatic structure prediction with HHpred. Proteins 77(Suppl 9):S128–S132. doi: 10.1002/prot.22499. [DOI] [PubMed] [Google Scholar]
- 53.Soding J, Biegert A, Lupas AN. 2005. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res 33:W244–W248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dulebohn DP, Hayes BM, Rosa PA. 2014. Global repression of host-associated genes of the Lyme disease spirochete through post-transcriptional modulation of the alternative sigma factor RpoS. PLoS One 9:e93141. doi: 10.1371/journal.pone.0093141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bestor A, Rego RO, Tilly K, Rosa PA. 2012. Competitive advantage of Borrelia burgdorferi with outer surface protein BBA03 during tick-mediated infection of the mammalian host. Infect Immun 80:3501–3511. doi: 10.1128/IAI.00521-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Towbin H, Staehelin T, Gordon J. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A 76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Barbour AG, Hayes SF, Heiland RA, Schrumpf ME, Tessier SL. 1986. A Borrelia-specific monoclonal antibody binds to a flagellar epitope. Infect Immun 52:549–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sundermeier T, Ge Z, Richards J, Dulebohn D, Karzai AW. 2008. Studying tmRNA-mediated surveillance and nonstop mRNA decay. Methods Enzymol 447:329–358. doi: 10.1016/S0076-6879(08)02217-9. [DOI] [PubMed] [Google Scholar]
- 59.Nowalk AJ, Nolder C, Clifton DR, Carroll JA. 2006. Comparative proteome analysis of subcellular fractions from Borrelia burgdorferi by NEPHGE and IPG. Proteomics 6:2121–2134. doi: 10.1002/pmic.200500187. [DOI] [PubMed] [Google Scholar]
- 60.Stewart PE, Carroll JA, Dorward DW, Stone HH, Sarkar A, Picardeau M, Rosa PA. 2012. Characterization of the Bat proteins in the oxidative stress response of Leptospira biflexa. BMC Microbiol 12:290–303. doi: 10.1186/1471-2180-12-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bono JL, Elias AF, Kupko JJ III, Stevenson B, Tilly K, Rosa P. 2000. Efficient targeted mutagenesis in Borrelia burgdorferi. J Bacteriol 182:2445–2452. doi: 10.1128/JB.182.9.2445-2452.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Samuels DS. 1995. Electrotransformation of the spirochete Borrelia burgdorferi. Methods Mol Biol 47:253–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bunikis I, Kutschan-Bunikis S, Bonde M, Bergstrom S. 2011. Multiplex PCR as a tool for validating plasmid content of Borrelia burgdorferi. J Microbiol Methods 86:243–247. doi: 10.1016/j.mimet.2011.05.004. [DOI] [PubMed] [Google Scholar]
- 64.Elias AF, Bono JL, Kupko JJ, Stewart PE, Krum JG, Rosa PA. 2003. New antibiotic resistance cassettes suitable for genetic studies in Borrelia burgdorferi. J Mol Microbiol Biotechnol 6:29–40. doi: 10.1159/000073406. [DOI] [PubMed] [Google Scholar]
- 65.Policastro PF, Schwan TG. 2003. Experimental infection of Ixodes scapularis larvae (Acari: Ixodidae) by immersion in low passage cultures of Borrelia burgdorferi. J Med Entomol 40:364–370. doi: 10.1603/0022-2585-40.3.364. [DOI] [PubMed] [Google Scholar]
- 66.Morrison TB, Ma Y, Weis JH, Weis JJ. 1999. Rapid and sensitive quantification of Borrelia burgdorferi-infected mouse tissues by continuous fluorescent monitoring of PCR. J Clin Microbiol 37:987–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kanehisa M, Goto S. 2000. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ueta M, Wada C, Wada A. 2010. Formation of 100S ribosomes in Staphylococcus aureus by the hibernation promoting factor homolog SaHPF. Genes Cells 15:43–58. doi: 10.1111/j.1365-2443.2009.01364.x. [DOI] [PubMed] [Google Scholar]
- 69.Merrick MJ, Coppard JR. 1989. Mutations in genes downstream of the rpoN gene (encoding sigma 54) of Klebsiella pneumoniae affect expression from sigma 54-dependent promoters. Mol Microbiol 3:1765–1775. doi: 10.1111/j.1365-2958.1989.tb00162.x. [DOI] [PubMed] [Google Scholar]
- 70.Samuels DS. 2011. Gene regulation in Borrelia burgdorferi. Annu Rev Microbiol 65:479–499. doi: 10.1146/annurev.micro.112408.134040. [DOI] [PubMed] [Google Scholar]
- 71.Tilly K, Krum JG, Bestor A, Jewett MW, Grimm D, Bueschel D, Byram R, Dorward D, Stewart P, Rosa P. 2006. Borrelia burgdorferi OspC protein required exclusively in a crucial early stage of mammalian infection. Infect Immun 74:3554–3564. doi: 10.1128/IAI.01950-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yang XF, Lybecker MC, Pal U, Alani SM, Blevins J, Revel AT, Samuels DS, Norgard MV. 2005. Analysis of the ospC regulatory element controlled by the RpoN-RpoS regulatory pathway in Borrelia burgdorferi. J Bacteriol 187:4822–4829. doi: 10.1128/JB.187.14.4822-4829.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hübner A, Wang X, Nolen DM, Popova TG, Cabello FC, Norgard M. 2001. Expression of Borrelia burgdorferi OspC and DbpA is controlled by a RpoN-RpoS regulatory pathway. Proc Natl Acad Sci U S A 98:12724–12729. doi: 10.1073/pnas.231442498. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.