Abstract
Streptococcus pneumoniae is a diverse species causing invasive as well as localized infections that result in massive global morbidity and mortality. Strains vary markedly in pathogenic potential, but the molecular basis is obscured by the diversity and plasticity of the pneumococcal genome. We have previously reported that S. pneumoniae serotype 3 isolates belonging to the same multilocus sequence type (MLST) differed markedly in in vitro and in vivo phenotypes, in accordance with the clinical site of isolation, suggesting stable niche adaptation within a clonal lineage. In the present study, we have extended our analysis to serotype 14 clinical isolates from cases of sepsis or otitis media that belong to the same MLST (ST15). In a murine intranasal challenge model, five ST15 isolates (three from blood and two from ears) colonized the nasopharynx to similar extents. However, blood and ear isolates exhibited significant differences in bacterial loads in other host niches (lungs, ear, and brain) at both 24 and 72 h postchallenge. In spite of these differences, blood and ear isolates were present in the lungs at similar levels at 6 h postchallenge, suggesting that early immune responses may underpin the distinct virulence phenotypes. Transcriptional analysis of lung tissue from mice infected for 6 h with blood isolates versus ear isolates revealed 8 differentially expressed genes. Two of these were exclusively expressed in response to infection with the ear isolate. These results suggest a link between the differential capacities to elicit early innate immune responses and the distinct virulence phenotypes of clonally related S. pneumoniae strains.
INTRODUCTION
Streptococcus pneumoniae (pneumococcus) is one of the foremost bacterial pathogens in terms of global morbidity and mortality. Its disease spectrum includes life-threatening infections such as pneumonia, meningitis, and bacteremia as well as less-serious but highly prevalent infections, including otitis media (OM) and sinusitis. Young children and the elderly are at highest risk, with global estimates of pneumococcal deaths in children under 5 years of age approaching 1 million per year (1). Despite this mortality, S. pneumoniae is a component of the normal microflora of the human nasopharynx. It is estimated that at any given time, approximately 10 to 15% of adults and 25 to 40% of healthy children may be asymptomatically colonized with S. pneumoniae (2). Although only a small fraction of carriers progress to invasive or localized pneumococcal disease, the scale of the denominator results in massive global disease burden.
S. pneumoniae is a diverse, genetically plastic species, and the 93 known capsular serotypes are superimposed on >5,000 sequence types (ST) recognizable by multilocus sequence type (MLST) analysis (3). It has a core genome of roughly 1,500 genes common to all strains, with the remaining 30% of the genome comprising accessory regions (AR) present in some but not all clonal lineages. Individual S. pneumoniae strains differ markedly in their capacity to cause either invasive or localized infections, and both serotype and genetic background influence virulence profile 4, 5). However, the vast heterogeneity has frustrated attempts to identify any association between a given clonal lineage or serotype and the propensity to cause invasive rather than localized infections. Indeed, large molecular epidemiological studies have not yielded consistent findings (6, 7).
We recently identified a link between human tissue tropism, as judged by the source of a clinical isolate (blood versus ear), and virulence profile in a mouse model by comparing multiple ST-matched pairs of S. pneumoniae serotype 3 strains. In the murine intranasal challenge model, only blood isolates caused sepsis, while only otitis media isolates spread to the ear. These findings were indicative of stable niche adaptation within a clonal lineage (8). To further examine the nexus between the clinical source of isolates and their respective virulence-related phenotypes, we have now conducted similar studies in S. pneumoniae serotype 14 strains. Type 14 is a representative serotype conferring high invasive disease potential (9–11). It is also one of the most common etiologic agents of pneumonia in Latin America, the United Kingdom, and Spain and of otitis media in children under 5 years old (12–14). To further understand the basis for the distinct virulence profiles, we have also examined the nature of host innate immune responses elicited by these clinical isolates during the early stages of infection.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
S. pneumoniae strains used in this study are listed in Table 1. Cells were grown in a casein-based, semisynthetic liquid medium (C+Y) or in serum broth (SB), as required (8), or on Columbia agar supplemented with 5% (vol/vol) horse blood (BA) at 37°C in a CO2-enriched atmosphere. Bacterial stocks were prepared from mid-log-phase cultures and stored at −80°C in 20% glycerol.
TABLE 1.
S. pneumoniae serotype 14 isolates used in this study
| Straina | MLST | Source |
|---|---|---|
| ST15/9-47 | ST15 | Ear |
| ST15/51742 | ST15 | Ear |
| ST15/4495 | ST15 | Blood |
| ST15/4534 | ST15 | Blood |
| ST15/4559 | ST15 | Blood |
Strains were isolated between 1988 and 1996 from patients at either the Women's and Children's Hospital, North Adelaide, South Australia, or the Alice Springs Hospital, Northern Territory, Australia. The MLST of each strain was determined as described in Materials and Methods.
MLST.
For MLST analysis, the aroE, gdh, gki, recP, xpt, and ddl genes were PCR amplified and sequenced as described by Spratt (15). The sequence types (ST) of the strains were determined from the MLST database (http://www.pubmlst.org) based on the resulting allelic profiles.
ELISA for CPS quantitation.
Total capsular polysaccharide (CPS) produced by strains was quantified by enzyme-linked immunosorbent assay (ELISA) using a modification of the method described previously (16). Briefly, serial 2-fold dilutions of either purified type 14 CPS standard (American Type Culture Collection, USA) at a starting concentration of 10 μg/ml or total CPS preparations of the strains included in this study were coated on poly-l-lysine-treated Nunc MaxiSorp flat-bottom 96-well plates overnight at 4°C. After blocking with 1% fetal calf serum was performed, the samples were reacted with a 1:10,000 dilution of group 14 typing sera (Statens Seruminstitut, Copenhagen, Denmark) for 2 h. The plates were washed five times in wash buffer (0.05% Tween 20–phosphate-buffered saline [PBS]) and then reacted with a 1:20,000 dilution of goat anti-rabbit IgG alkaline phosphatase conjugate (Sigma-Aldrich) overnight at 4°C. After extensive washing, the plates were developed using alkaline phosphatase substrate (Sigma) in diethanolamine buffer, and the absorbance at 405 nm was read in a Spectramax M2 spectrometer (Molecular Devices, CA, USA).
Animal studies.
Animal experiments were approved by the University of Adelaide Animal Ethics Committee. For each test strain, two groups of five outbred 6-week-old female Swiss mice were anesthetized by intraperitoneal injection of pentobarbital sodium (Nembutal; Rhone-Merieux) at a dose of 66 μg per g of body weight. They were then intranasally challenged with 30 μl of bacterial suspension containing approximately 1 × 108 CFU in SB. The challenge dose was confirmed retrospectively by serial dilution and plating on BA. Groups of five mice were euthanized by CO2 asphyxiation at either 24 or 72 h. Blood and tissue samples (lungs, nasopharynx, brain, and ear) were harvested and pneumococci enumerated in blood or tissue homogenates as described previously (8, 16). For in vivo competition experiments, groups of mice were challenged intranasally with a mixed inoculum comprising approximately 5 × 107 CFU of each strain, as previously described (17). The competitive index (CI) within nasopharyngeal, ear, lung, and brain samples was determined at 24 h and 72 h postchallenge by plating on selective and nonselective media and calculating the ratio of ear isolates to blood isolates relative to the input ratio. For the purposes of CI calculation, when a given strain was not detected in a particular niche, it was assigned a CFU value equivalent to 50% of the detection threshold.
Detroit 562 adherence and invasion assays.
Detroit 562 (human nasopharyngeal carcinoma) cells were grown in a 1:1 mix of Dulbecco's modified Eagle medium (Gibco) and Ham's F-12 nutrient mixture (Gibco) supplemented with 5% fetal bovine serum, 2 mM l-glutamine, 50 IU of penicillin, and 50 μg/ml streptomycin. Confluent monolayers in 24-well plates were washed with PBS and infected with pneumococci (approximately 5 × 105 CFU per well) in a 1:1 mixture of the culture medium described above (without antibiotics) and C+Y (pH 7.4). Plates were centrifuged at 500 × g for 5 min and then incubated at 37°C in 5% CO2 for 2.5 h. Monolayers were washed 3 times in PBS, and adherent bacteria were released by treatment with 100 μl trypsin–EDTA followed by 400 μl 0.025% Triton X-100. Lysates were serially diluted and plated on BA to enumerate adherent bacteria. Invasion assays were carried out essentially as described above, except that after the postadherence washing step, cultures were incubated for 1 h in fresh medium supplemented with 200 μg gentamicin and 10 μg benzylpenicillin per ml to kill extracellular bacteria, after which monolayers were again washed, lysed, serially diluted, and plated on BA, as described above.
PCR arrays.
At 6 h postinfection, lung and nasopharyngeal tissue was harvested from infected or sham-infected mice and homogenized. RNA was extracted using a PureLink RNA minikit (Life Technologies), including on-column DNase digestion, according to the manufacturer's instructions. cDNA synthesis was carried out using a RT2 First Strand kit (Qiagen). RNA was analyzed on a LightCycler 480 II system (Roche) by quantitative reverse transcriptase PCR (qRT-PCR) using a RT2 Profiler PCR array mouse innate and adaptive immune responses kit (Qiagen), according to the manufacturer's instructions (18). Data were analyzed using PCR Array data analysis software provided by the manufacturer.
Fluorescence-activated cell sorter (FACS) detection of Ly-6G- and F4/80-positive cells in blood and BAL fluid.
Blood was collected from infected and uninfected mice into heparin tubes by cardiac puncture. Blood leukocytes were extracted by lysing the red blood cells (RBC) with a hypotonic shock. Bronchoalveolar lavage (BAL) was performed using 1 ml of ice-cold PBS.
The blood leukocytes or BAL fluid cells were washed in ice-cold PBS and fixed in 1% paraformaldehyde–PBS over night at 4°C. Before immunofluorescence labeling, the leukocytes were washed with PBS and permeabilized with 0.1 ml of 0.1% Triton X100–PBS for 30 s. The cells were then washed again with PBS and were then incubated with monoclonal rat anti-mouse Ly-6G (BD Biosciences; 551459) or monoclonal rat anti-mouse F4/80 (Santa Cruz Biotechnology; SC-52664) at room temperature for 1 h. Ly-6G (neutrophil marker) or F4/80 (monocyte/macrophage marker) was detected by incubation with Alexa 488-conjugated donkey anti-rat Ig (Invitrogen; A21208) at room temperature for 1 h. Fluorescence data were acquired by the use of a BD FACSCanto system (serial no. V0130) or a BD LSR II system with a high-throughput sampler (serial no. H1169) plus BD FACSDiva software (version 5.0.3) and were analyzed with WEASLE v2.6 software. The fluorescence intensity of Alexa 488 was proportional to the expression level of Ly-6G or F4/80. The data are reported as the product of geometric mean (GM) fluorescence intensity and the total number of Ly-6G- or F4/80-positive cells. The data are presented as means and standard errors (SE), and differences were analyzed using Student's t test.
Examination of lung tissue with HE staining or immunofluorescence labeling.
After the blood and BAL fluid were collected, the lungs of infected and uninfected mice were removed and fixed in 4% formaldehyde overnight at 4°C, embedded in paraffin, sectioned, stained with hematoxylin-eosin (HE), and examined by light microscopy. Alternatively, sections were labeled with rat anti-mouse Ly-6G or F4/80 followed by Alexa-488-conjugated anti-rat IgG and examined by fluorescence microscopy (AX 70; Olympus).
RESULTS
Initial characterization of serotype 14 clinical isolates.
The purpose of this study was to compare pneumococci belonging to the same serotype and clonal lineage derived from distinct host niches. Therefore, we carried out MLST analysis on 18 serotype 14 strains, comprising 9 blood and 9 ear isolates. The two predominant STs were ST15 (n = 5) and ST130 (n = 5). Of the other strains, two belonged to ST124 and ST129, while the others were all assigned to new STs by http://pubmlst.org but were not further investigated. We focused on ST15 because three isolates originated from blood and two from ears (Table 1). All ST15 strains were in the opaque phase, as judged by the opacity phenotype seen during growth on Todd-Hewitt broth supplemented with yeast extract (THY)–catalase plates (16), and the total capsular polysaccharide production levels were similar for all strains, as judged by ELISA (data not shown).
Virulence phenotype of ST15 blood and ear isolates.
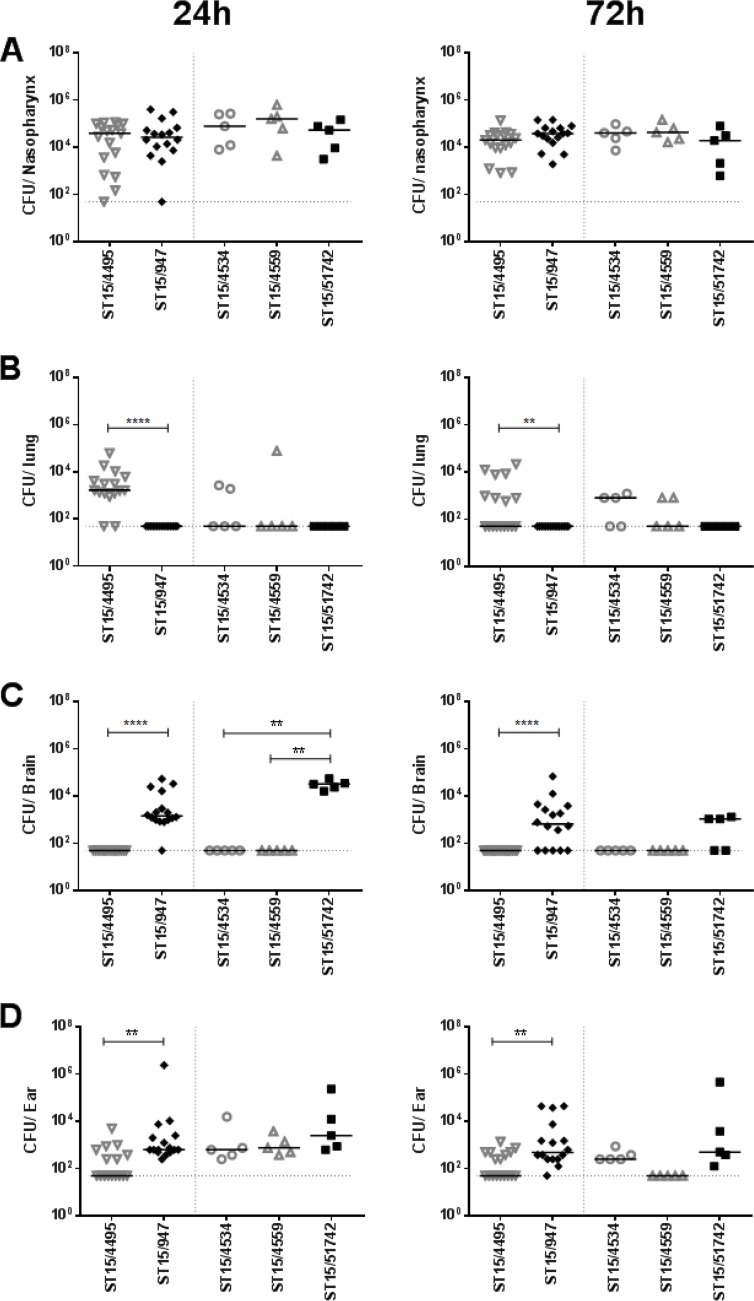
The virulence profiles of the various ST15 strains were then examined in a murine intranasal challenge model. Group sizes were 16 each for one blood (ST15/4495) and one ear isolate (ST15/9-47) (both selected at random) and n = 5 for the remaining strains. Numbers of the respective strains in various host niches (nasopharynx, lungs, blood, brain, and ear) were quantitated at 24 h and 72 h postchallenge (Fig. 1). None of the strains were detected in the blood of any of the mice at either time point (data not presented). The three blood isolates (ST15/4495, ST15/4534, and ST15/4559) and the two ear isolates (ST15/9-47 and ST15/51742) all exhibited similar capacities to colonize the nasopharynx at both 24 h and 72 h postinfection, with the GM bacterial load in this niche in the range of 104 to 105 CFU (Fig. 1A). There were no significant differences in either infection rates or GM bacterial loads. However, significant differences between blood and ear isolates were observed in the other host niches. Neither of the ear isolate strains was detectable in the lungs of any of the infected mice at either time point. In contrast, the ST15/4495 blood isolate strain was present in the lungs of 14/16 mice at 24 h and 8/16 at 72 h postchallenge (P < 0.0001 and P = 0.0032, respectively; Fisher exact test). A similar trend was seen for the other two blood isolates (ST15/4534 and ST15/4559), with 50% of lungs colonized at 72 h (Fig. 1B). The opposite tropism was observed in the brain. None of the three ST15 blood isolates were detected in the brains of any of the mice at either time point. However, the ST15/9-47 ear isolate was detected in the brains of 15/16 mice and 11/16 mice at 24 h and 72 h, respectively (P < 0.0001 in both cases). The other ear isolate (ST15/51742) was present in the brains of 5/5 and 3/5 mice at 24 h and 72 h, respectively (P < 0.01 relative to any of the blood isolates at 24 h) (Fig. 1C). In the infected mice, bacterial loads in the brain were in the range of 103 to 105 CFU. All five ST15 isolates were able to spread to the ear compartment after intranasal challenge, but the proportion of mice whose ears were infected with the ST15/9-47 ear isolate was significantly greater than that of mice challenged with the ST15/4495 blood isolate (16/16 versus 7/16 and 15/16 versus 8/16 at 24 h and 72 h, respectively; P = 0.0008 and P = 0.0155, respectively) (Fig. 1D). The GM bacterial load in the ears for ST15/9-47 was also significantly greater than that for ST15/4495 at both 24 h and 72 h (P = 0.0015 and P = 0.0020, respectively). Similar trends were also seen in the mice challenged with the other ST15 ear isolate and the two other blood isolates. Collectively, these results show that pneumococci of the same serotype and ST exhibit distinct in vivo phenotypes in accordance with the clinical site of isolation.
FIG 1.
Comparative virulence of ST15 blood and ear isolates. Groups of mice were challenged intranasally with the three blood isolates (ST15/4495, ST15/4534, and ST15/4559) and the two ear isolates (ST15/9-47 and ST15/51742), and numbers of pneumococci in the nasopharynx, lungs, brain, and ear at either 24 h or 72 h postchallenge were determined. For ST15/4495 and ST15/9-47, 16 challenged mice were examined at each time point compared with 5 mice at each time point for the remaining strains. Blood isolates are represented by open gray symbols; ear isolates are represented by filled black symbols. The horizontal bars denote the median CFU for each group; the dotted horizontal lines indicate the lower limit of detection, and symbols on the lines denote that no bacteria were detected. Differences in infection rates (number of infected versus number of uninfected mice) were analyzed using the Fisher exact test. **, P < 0.01; ****, P < 0.0001.
In vivo competition between ST15/4495 and ST15/9-47.
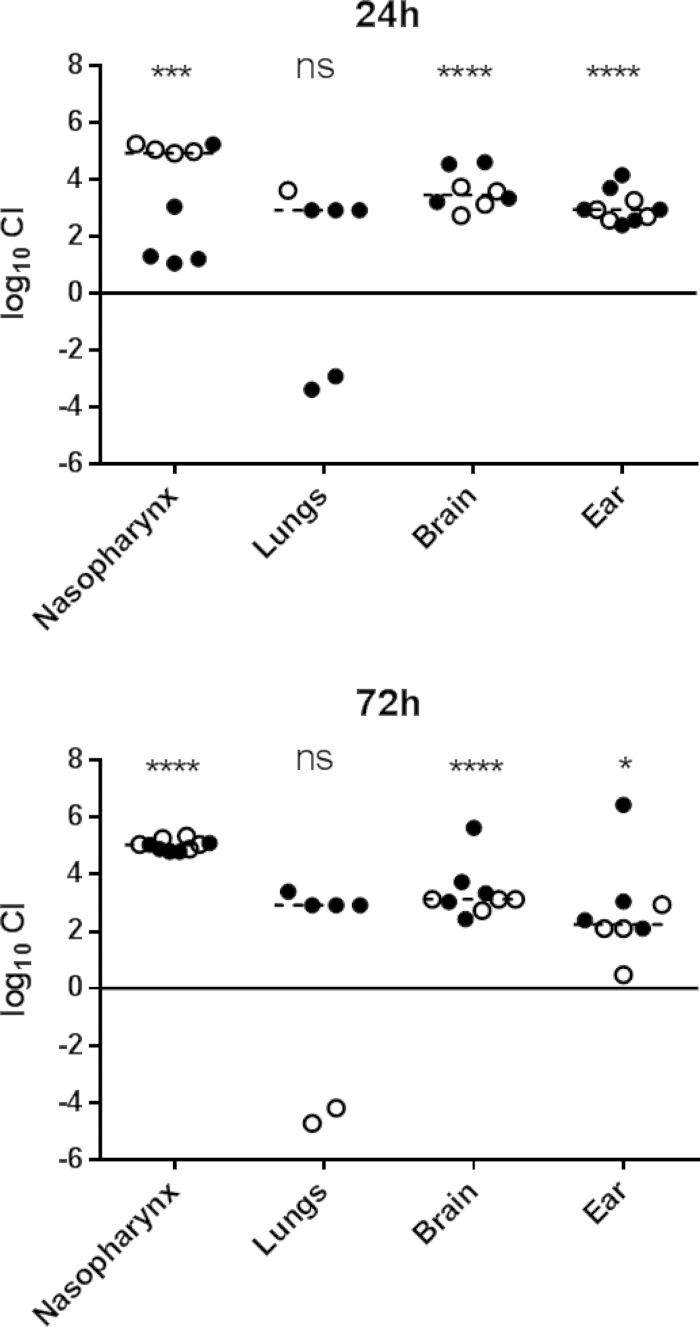
The distinct in vivo tropism observed for the ST15 ear and blood isolates could be attributable to differences in expression of bacterial virulence factors on the bacterial surface or released into the environment and/or to differences in the nature of the host innate immune responses elicited by the respective strains. Thus, the presence of a strain with superior virulence characteristics in a given niche may enhance the virulence of a “passenger” strain. To explore this, we conducted in vivo competition experiments in which two groups of mice were challenged with equal numbers of ST15/4496 and ST15/9-47, and the relative numbers of each strain were determined in various host niches at 24 h and 72 h postchallenge. In order to distinguish the strains, it was necessary to construct erythromycin-resistant derivatives of each strain by transformation with plasmid pAL3 (19). Introduction of the plasmid had no effect on the bacterial growth rate of either strain (data not shown).
Two competition experiments were performed with 5 mice per group for each time point. In one experiment, mice were challenged with equal numbers of ST15/4496 and ST15/9-47:pAL3; in the other experiment, mice were challenged with equal numbers of ST15/4496:pAL3 and ST15/9-47. Tissue samples were plated on BA or BA plus erythromycin to enable calculation of the ratio of sensitive organisms to resistant organisms and hence of the competitive index (CI). Data pooled from the two experiments are shown in Fig. 2. In spite of the similar capacities of ST15/4495 and ST15/9-47 to colonize the murine nasopharynx when challenged in isolation (Fig. 1A), the ear isolate had a significant competitive advantage over the blood isolate in the mixed-infection model in the nasopharynx at 24 h (P < 0.001) and at 72 h (P < 0.0001), by which time the median CI was approximately 105 (Fig. 2). The ear isolate also predominated in both the ear and brain compartments at both time points (P < 0.0001 for brain at 24 h and 72 h and for ear at 24 h; P < 0.05 for ear at 72 h). Moreover, whereas small numbers of the blood isolate were present along with the dominant ear isolate in the ears of all the infected mice, the blood isolate was never detected in brain tissue of any of the mice at either time point. These findings are understandable, given that access to both the brain and the ear compartments occurs via the nasopharynx, where the ear isolate was present at a 105-fold excess over the blood isolate. A distinct scenario occurred in the lungs. Only 6/10 mice had detectable pneumococci in the lungs at either time point, and in these mice, neither strain had a significant competitive advantage overall (Fig. 2). This was the case in spite of the fact that, when used in isolation for the challenge, only the blood isolate was able to persist in the lungs at either 24 h or 72 h (Fig. 1B). In the competition experiment, the ear isolate outcompeted the blood isolate in 4/6 infected mice, whereas the blood isolate outcompeted the ear isolate in 2/6 mice (Fig. 2). The intranasal challenge inoculum was instilled into the nares under general anesthesia, and a proportion of the dose was aspirated directly into the lungs during the challenge (approximately 106 CFU). When instilled on their own, ST15/9-47 organisms entering the lung during challenge were cleared within 24 h, whereas when ST15/4495 organisms were instilled on their own, the blood isolate was able to resist innate defenses and persist in the lung. However, in the coinfection model, significant numbers of the ear isolate were present in a proportion of the mice at both 24 h and 72 h. These findings indicate that the presence of the ST15/4495 blood isolate, at least during the immediate postchallenge period, is necessary for persistence of the ST15/9-47 ear isolate in the lung compartment.
FIG 2.
In vivo competition between ST15/9-47 (ear isolate) and ST15/4495 (blood isolate). Data shown are pooled from two competition experiments performed with 5 mice per group for each time point (24 h and 72 h postchallenge). In one experiment, mice were challenged with equal numbers (5 × 107 CFU each) of ST15/4495 and ST15/9-47:pAL3 (open symbols); in the other experiment, mice were challenged with equal numbers of ST15/4495:pAL3 and ST15/9-47 (solid symbols). The competitive index (CI) is shown as log10 CI for each tissue of each mouse. The horizontal dotted lines denote the geometric mean CI for each tissue. Statistical differences between the log-transformed geometric mean CI and a hypothetical value of 0 (ratio of 1:1) in each niche were analyzed using the two-tailed Student's t test. ns, not significant; *, P < 0.05; ***, P < 0.001; ****, P < 0.0001.
In vitro adherence and invasion.
To further explore the basis for the enhanced nasopharyngeal colonization fitness of ST15/9-47, the adherence phenotype of all the ST15 isolates was assessed in vitro using Detroit 562 (human nasopharyngeal) cells (Table 2). There were significant differences between several of the strains in total in vitro adherence. Two of the blood isolates (ST15/4495 and ST15/4534) were significantly less adherent than the ST15/9-47 ear isolate (P < 0.01 and P < 0.001, respectively). The other ear isolate (ST15/51742) was even more adherent than ST15/9-47 (P < 0.05). The adherence of the third blood isolate (ST15/4559) was not significantly different from that of ST15/9-47, but that isolate was significantly less adherent than the second ear isolate, ST15/51742 (P < 0.01). The capacity of the ST15/9-47 ear isolate and the ST15/4495 blood isolate to invade Detroit cells was also investigated (Table 2), but no significant difference was observed.
TABLE 2.
Adherence to and invasion of Detroit 562 cells
| Strain | Source | Adherencea (CFU/well ± SEM) | Invasion (CFU/well ± SEM) |
|---|---|---|---|
| ST15/9-47 | Ear | 7.88 × 105 ± 0.78 × 105 | 3.50 × 105 ± 0.65 × 103 |
| ST15/51742 | Ear | 1.19 × 105 ± 0.10 × 106* | Not tested |
| ST15/4495 | Blood | 3.75 × 105 ± 0.56 × 105** | 3.95 × 105 ± 0.28 × 103 |
| ST15/4534 | Blood | 2.85 × 105 ± 0.27 × 105*** | Not tested |
| ST15/4559 | Blood | 7.50 × 105 ± 0.59 × 105 | Not tested |
Significant differences in adherence relative to that of ST15/9-47 are indicated as follows: *, P < 0.05; **, P < 0.01; ***, P < 0.001 (Student's t test, two-tailed).
Transcriptional analysis of host innate responses.
Although only the blood isolates are detectable in the lungs at 24 h, ear isolates are delivered into the lungs during challenge but are cleared by host innate defenses by 24 h. In a pilot experiment, we found similar bacterial loads in the lungs of mice challenged with either ST15/4495 or ST15/9-47 at 6 h postchallenge (107 to 108 CFU in both cases; result not shown). We therefore investigated whether differences in host responses during the immediate postchallenge period might have influenced bacterial clearance from the lungs. Mice were challenged with either the blood isolate (ST15/4495) or the ear isolate (ST15/9-47) or were sham infected (anesthetized and inoculated with 30 μl SB). Total RNA was isolated from lungs and nasopharyngeal tissue 6 h after challenge. Expression of 84 key immune response genes was then quantitated using RT-PCR arrays. Compared to levels in sham-infected samples, a total of 35 genes were induced in the lungs, as judged by a significant increase in the mRNA level following challenge with at least one of the two ST15 strains (Table 3). Overall, the pattern of gene expression indicated a generally proinflammatory response in the lungs at 6 h postchallenge; genes encoding a number of proinflammatory cytokines, including interleukin-1α (IL-1α), IL-1β, IL-23a, IL-6, and tumor necrosis factor alpha (TNF-α), were upregulated. Other genes involved in regulating the immune response were also increased in expression. These included genes encoding the chemokines CCL12, a murine homolog of (human) CCL2, and CXCL10, which is known to be induced by gamma interferon (IFN-γ), a proinflammatory cytokine that also showed increased expression following pneumococcal challenge. Expression of the CCR5 chemokine receptor was also elevated at the mRNA level. Genes encoding elements involved in pathogen recognition and signaling during immune responses, such as Myd88, Toll-like receptor 3 (TLR3), TLR6, TLR7, Nod2, Ddx58, JAK2, and NF-κBIα, were differentially expressed in challenged samples compared to sham-infected samples. Other upregulated genes included those encoding the CD11b integrin (ITGAM) and the associated adhesion molecule ICAM1, inflammasome component NLRP3, and granulocyte-macrophage colony-stimulating factor (GM-CSF) CSF2.
TABLE 3.
Genes significantly upregulated in infected relative to sham-infected lung tissuea
| Gene product | ST15/4495 infected vs sham infected |
ST15/9-47 infected vs sham infected |
||
|---|---|---|---|---|
| Fold gene expression change | Significance | Fold gene expression change | Significance | |
| APCS | 5.15 | ** | ||
| C5ar1 | 14 | * | 8.47 | ** |
| CCL12 | 36.61 | * | 15.56 | ** |
| CCR5 | 10.68 | * | 5.97 | ** |
| CD14 | 61.12 | ** | ||
| CD40 | 6.21 | * | 2.55 | * |
| CD86 | 3.51 | * | 2.51 | ** |
| CSF2 | 19.34 | * | ||
| CXCL10 | 310.55 | ** | ||
| Ddx58 | 2.36 | * | ||
| ICAM1 | 7.91 | * | 5.13 | * |
| IFN-α2 | 2.50 | * | ||
| IFN-β1 | 96.15 | *** | ||
| IFN-γ | 17.2 | ** | 12.25 | *** |
| IL-10 | 17.09 | * | ||
| IL-1α | 29.81 | ** | 16.64 | *** |
| IL-1β | 88.28 | * | 52.60 | ** |
| IL-1r1 | 2.1 | * | ||
| IL-2 | 4.95 | ** | ||
| IL-23α | 40.9 | ** | ||
| IL-6 | 312.35 | ** | ||
| IRF7 | 4.01 | * | 3.80 | * |
| ITGAM | 3.73 | ** | ||
| JAK2 | 3.29 | ** | 4.41 | * |
| Mx1 | 152.64 | * | ||
| Myd88 | 12.3 | * | 5.22 | * |
| NF-κBIα | 20.34 | * | ||
| NLRP3 | 36.61 | *** | ||
| Nod2 | 9 | ** | ||
| SLC11α1 | 4.26 | * | 5.25 | * |
| TBX21 | 2.4 | ** | ||
| TLR3 | 4.8 | ** | ||
| TLR6 | 3.84 | ** | ||
| TLR7 | 4.80 | ** | ||
| TNF | 119.21 | ** | 50.78 | * |
| IL-1r1 | 2.1 | * | ||
Data for genes upregulated significantly after infection with both ear and blood isolates are highlighted in bold type. Statistical significance is indicated as follows: *, P < 0.05; **, P < 0.01; ***, P < 0.001 (Student's t-test, two-tailed).
Some strain-strain differences were observed, with 8 genes showing significant differential levels of expression in lungs following challenge with ST15/4495 compared to challenge with ST15/9-47 (Table 4). Of these, 6 genes were upregulated by both challenge strains compared to resting samples but to a significantly greater degree following challenge with ST15/4495. These included those encoding the proinflammatory cytokines TNF-α and IL-6, the type I interferon IFN-β, the costimulatory molecule CD40, the transcription factor Tbet (TBX21), and the chemokine CCL12. The remaining 2 genes, encoding APCS and IL-2, were found to be upregulated only following challenge with ST15/9-47.
TABLE 4.
Genes differentially induced by infection with ST15/9-47 versus ST15/4495a
| Gene product | ST15/4495 vs sham infected |
ST15/9-47 vs sham infected |
ST15/9-47 vs ST15/4495 |
|||
|---|---|---|---|---|---|---|
| Fold gene expression change | Significance | Fold gene expression change | Significance | Fold gene expression change | Significance | |
| APCS | −1.10 | ns | 5.15 | ** | 5.67 | ** |
| IL-2 | −1.09 | ns | 4.95 | ** | 5.40 | * |
| CCL12 | 36.61 | * | 15.56 | ** | −2.35 | * |
| CD40 | 6.21 | * | 2.55 | * | −2.43 | * |
| IFN-β1 | 96.15 | *** | 17.84 | ns | −5.39 | *** |
| IL-6 | 1,842.35 | P = 0.06 | 312.35 | ** | −5.90 | * |
| TNF | 119.21 | ** | 50.78 | * | −2.35 | * |
| TBX21 | 2.4 | ** | 1.02 | ns | −2.36 | **** |
Statistical significance is indicated as follows: ns, not significant; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 (Student's t-test, two-tailed).
Since the nasopharynx is the first host environment to which the bacteria are exposed during infection, we examined whether the blood isolate (ST15/4495) and the ear isolate (ST15/9-47) also elicited differential levels of induction of innate immune response genes in this niche at 6 h postinfection. However, we found no significant differences between sham-infected mice and those challenged with either strain in expression of any of the 84 genes analyzed (data not shown).
Cellular and histopathological responses to lung infection.
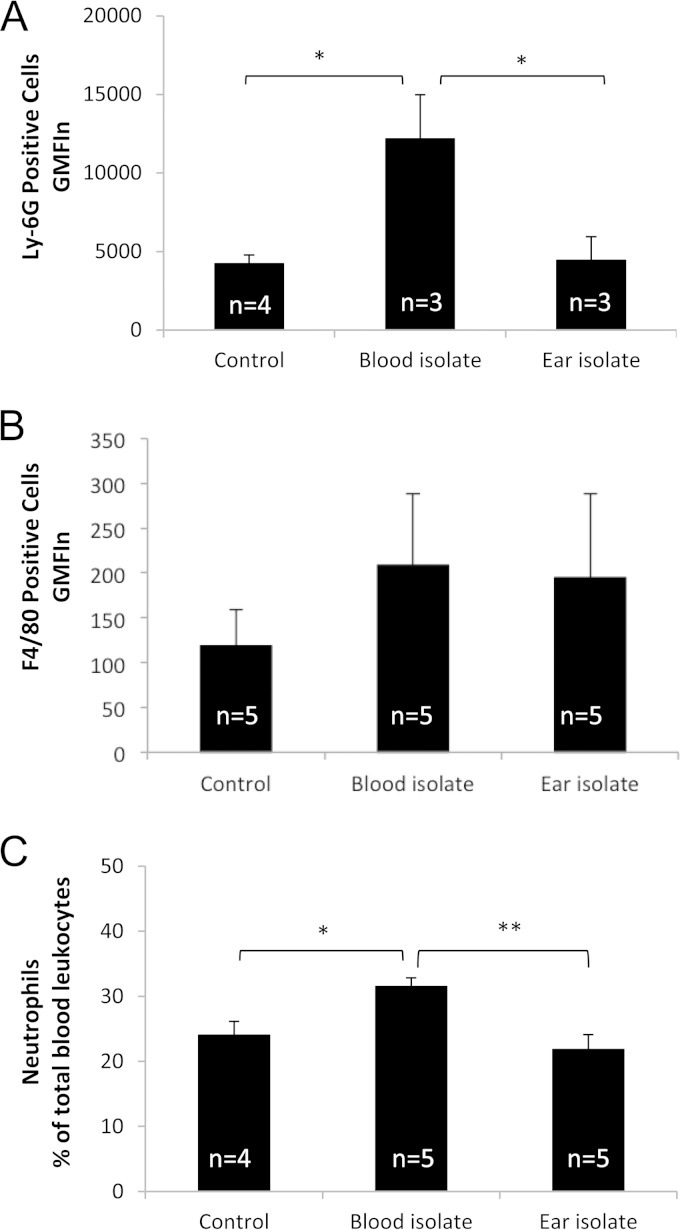
We also examined blood, BAL fluid, and lung tissue for cellular recruitment and histopathological changes. At 6 h postinfection, there were very few cells in the BAL fluid and there were no significant differences between sham-infected mice and those infected with either ST15/9-47 or ST15/4495 in the numbers of BAL fluid neutrophils (Ly-6G-positive cells) or monocyte/macrophages (F4/80-positive cells), as determined by flow cytometry (result not shown). However, a similar FACS analysis of blood leukocytes showed numbers of neutrophils (Fig. 3A), but not monocyte/macrophages (Fig. 3B), in ST15/4495-infected mice that were significantly higher than those seen with either sham-infected or ST15/9-47-infected mice at 6 h (P < 0.05 in both cases). A similar increase in the blood neutrophil count in ST15/4495-infected mice relative to either sham-infected or ST15/9-47-infected mice at 6 h was also evident from differential cell counts of blood films (P < 0.05 and P < 0.01, respectively) (Fig. 3C).
FIG 3.
Peripheral blood leukocyte counts. (A and B) Leukocytes isolated from peripheral blood collected at 6 h from control (sham-infected) mice or from those infected with the blood isolate (ST15/4495) or the ear isolate (ST15/9-47) were examined by FACS analysis after staining with anti-Ly-6G (A) or anti-F4/80 (B), as described in Materials and Methods. Data are presented as the product of geometric mean fluorescence intensity and the total number of positive cells (GMFIn) for the respective marker (± SE). (C) Alternatively, blood films were subjected to differential cell counts, and neutrophil numbers are expressed as a percentage of total leukocyte levels (mean ± SE). Statistical significance is indicated as follows: *, P < 0.05; **, P < 0.01 (Student's paired, two-tailed t test).
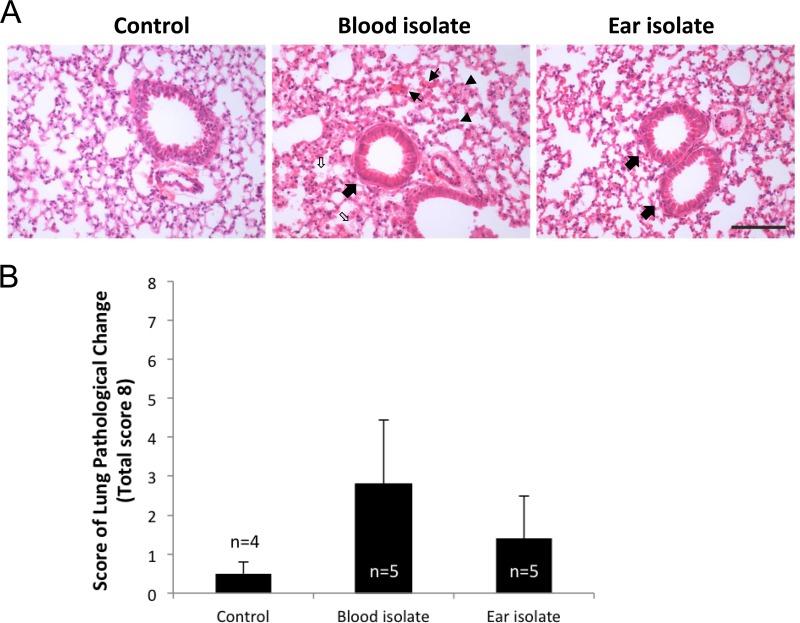
Examination of HE-stained lung sections at 6 h revealed early signs of tissue damage, including congested capillaries, thickened alveolar walls, swollen cuboidal epithelial cells of the bronchioles, and the presence of secretions in the alveolar and bronchiolar spaces, most notably in ST15/4495-infected mice (Fig. 4A). These features were used to develop an 8-point system for scoring histopathological changes (Fig. 4B). The mean aggregate score (± SE) for ST15/4495-infected mice (2.8 ± 1.3) was higher than that for either the ST15/9-47-infected mice (1.4 ± 0.57) or the sham-infected mice (0.5 ± 0.33), but this trend did not reach statistical significance.
FIG 4.
Lung histopathology. HE-stained lung sections from control (sham-infected) mice or from those infected with the blood isolate (ST15/4495) or the ear isolate (ST15/9-47) at 6 h were examined by light microscopy. (A) Representative sections from each group are shown. Bar, 0.1 mm. Slides were also scored (blind) according the following 8-point scheme: congested capillaries (fine arrows) were scored 0 to 2; thickened alveolar wall (arrowheads) were scored 0 to 2; swollen cuboidal epithelial cells of the bronchioles (thick arrows) were scored 0 to 2; and secretions in the alveolar and bronchiole space (open arrows) were scored 0 to 2. (B) Data were examined with Student's unpaired two-tailed t test and are presented as mean ± SE for each group.
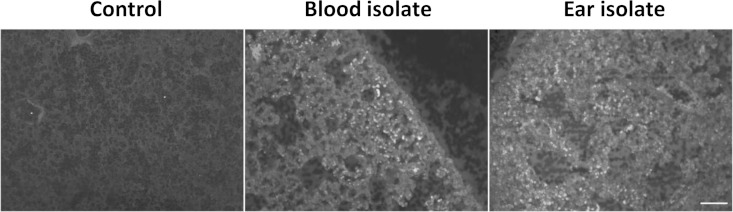
Finally, we examined lung sections for cellular infiltration by immunofluorescence microscopy after staining with anti-Ly-6G (Fig. 5) or anti-F4/80 (not shown). This revealed patchy neutrophil infiltration in 2/5 mice infected with the ST15/4495 blood isolate and 1/5 mice infected with the ST15/9-47 ear isolate, compared with 0/5 for the control mice. No differences between groups in staining for the monocyte/macrophage marker were seen.
FIG 5.
Neutrophil infiltration. Lung tissue from infected or control mice was fixed, sectioned, and labeled with rat anti-mouse Ly-6G or F4/80, followed by Alexa-488-conjugated anti-rat IgG and examined by fluorescence microscopy. Bar, 50 μm.
DISCUSSION
S. pneumoniae is a genetically diverse pathogen, and individual isolates vary markedly in their capacity to progress from asymptomatic nasopharyngeal carriage to causing either localized or systemic disease. The plethora of capsular serotypes and clonal lineages (MLST types) no doubt contribute to their adaptability in different host microenvironments, but they have also complicated previous attempts to unequivocally associate particular serotypes or clonal groups with specific virulence attributes (6, 7). However, we have previously described multiple examples of serotype 3 strains belonging to the same ST that consistently and reproducibly exhibit distinct virulence phenotypes in mice that directly correlate with the original site of isolation from human patients (i.e., ear versus blood) (8). This suggested that, at least in serotype 3, pneumococci exhibit stable niche adaptation within a clonal lineage.
In the present study, we investigated blood and ear isolates belonging to serotype 14, a common cause of OM as well as of invasive disease worldwide (11). This serotype also exhibits high rates of antimicrobial resistance (20). ST15 was one of the dominant STs in our collection and was represented among clinical isolates from cases of sepsis and OM. ST15 blood and ear isolates were able to colonize the murine nasopharynx to roughly similar extents. However, the blood isolate (ST15/4495) was significantly better at colonization and persistence in the lung, while the ear isolate (ST15/9-47) was significantly better able to penetrate and persist in the brain and ears. Similar preferential tropism results for the brain and ear were observed for the other ST15 ear isolate (ST15/51742), while the two other ST15 blood isolates (ST15/4534 and ST15/4559) colonized the lungs in a proportion of infected mice. Significantly, neither of the ear isolates was detected in the lungs of any of the mice at either 24 or 72 h, while none of the three blood isolates were ever detected in the brain. Furthermore, none of the ST15 blood or ear isolates were detected in the blood at any time during the experiment. This is consistent with the long-known fact that mice exhibit a high degree of innate resistance to sepsis after challenge with serotype 14 S. pneumoniae strains. However, these findings also suggest that the very high rate of meningitis that occurs after intranasal challenge with either of the ST15 ear isolates was not a consequence of hematogenous spread. Likely alternative routes for pneumococci from the nasopharynx to the brain include direct invasion by retrograde axonal transport along olfactory neurons into the central nervous system (CNS) (21). Collectively, the findings reported above provide robust evidence for the existence of distinct pathogenic profiles for serotype 14 S. pneumoniae strains belonging to the same ST (in this case, ST15). In a previous study (22), modest differences in bacterial loads in the blood of mice 6 h after intraperitoneal challenge with three serotype 14 blood isolates belonging to ST124 were detected, although the bacteremia was transient and subclinical. However, in the present study, we not only observed highly significant differences in virulence and pathogenic profiles between isolates belonging to the same serotype and clonal lineage but also found a close correlation between pathogenic profiles in mice and the original sites of isolation from human cases of pneumococcal disease. This extends an observation that we have previously reported for serotype 3 strains belonging to ST180, ST232, and ST233 (8). Thus, stable niche adaptation within a clonal lineage appears to be a general phenomenon.
Notwithstanding the conclusions presented above, the in vivo competition experiments in the present study yielded some unexpected findings. First, although the ear and blood isolates were able to colonize the nasopharynx to similar levels when administered individually, the ST15/9-47 ear isolate massively outcompeted the ST15/4495 blood isolate in this niche in the coinfection model. The reason for this is not clear. Neither strain predominated when the two were cocultured in vitro, and the two strains express the same bacteriocin genes (data not presented). Nevertheless, this finding accounts for the dominance of ST15/9-47 in the brain and ear in the coinfection model, since these host niches are presumably accessed via the nasopharynx, where the ear isolate already enjoyed a 105-fold numeric advantage. On the other hand, the data from the individual challenge experiments might lead one to predict that the ST15/4495 blood isolate would enjoy a competitive advantage in the lungs, but this was not the case, with neither strain predominating. Similar numbers of ear and blood isolates were delivered into the lungs during challenge under general anesthesia, and bacterial loads in lung tissue at 6 h postchallenge were similar. However, in the individual challenge studies, ST15/9-47 was completely cleared from the lungs of all mice by 24 h, while ST15/4495 was able to persist. There are two possible explanations for this. ST15/4495 may secrete a (yet to be identified) virulence factor that can act in trans and support survival of ST15/9-47 in the mixed-infection model. Alternatively, differences in host innate immune responses elicited by the blood and ear isolates could account for the distinct pathogenic profiles. Clearly, responses triggered by ST15/4495 enable this strain to persist in the lung environment. However, this may create a host microenvironment that also enables persistence of coadministered strains such as ST15/9-47 which would otherwise have been cleared.
To establish if the host immune response could play a role in the distinct pathogenic profiles of the ST15 blood or ear isolates, we examined the transcriptional responses of 84 genes representing the major pathways involved in the innate and adaptive immune response to microbial pathogens. Unsurprisingly, infection with either strain induced a variety of proinflammatory genes. However, the strain-specific differences observed provide us with important clues as to how the immune response may vary, yielding very different patterns of infection from two clonally related strains. Six of the eight genes that were differentially expressed in response to infection with ST15/4495 relative to ST15/9-47 are upregulated in response to both strains relative to sham-infected lungs but are induced more strongly in response to ST15/4495. This suggests that while both strains induce a broadly similar response, one of the features distinguishing the two is the strength of the responses induced at similar bacterial loads. Those responses included increased expression of genes encoding CCL12, IL-6, TNF-α, IL-1β, and CD40, which points to an overall stronger macrophage response to the blood isolate than to the ear isolate (23–29). BAL fluid collected at 6 h postinfection contained very few cells, and there were no significant differences between sham-infected mice and those infected with either of the isolates in this regard. However, mice infected with the ST15/4495 blood isolate exhibited significant neutropenia relative to sham-infected or ST15/9-47-infected mice at this early time point. Results of a histopathological examination were also suggestive of increased lung damage in mice infected with ST15/4495.
The findings presented above have parallels with those from a previous study in which we compared host transcriptional responses to highly invasive and noninvasive S. pneumoniae strains belonging to serotype 1 in a similar intranasal challenge model. We showed that a total of 29 genes of the 84 on the array showed significant upregulation; of those, 22 were also upregulated in the present study. The two type 1 strains were present in roughly equal numbers in the lung at 6 h, but the invasive strain triggered a much stronger type I interferon response, which facilitated invasion of the pleural cavity followed by the blood (18).
In the present study, we identified two genes (encoding APCS and IL-2) that were significantly upregulated only in the mice challenged with the ST15/9-47 ear isolate compared to the sham-infected mice; infection with the blood isolate had a negligible effect on their expression. Therefore, it is tempting to speculate that expression of the genes encoding APCS and IL-2 might contribute to the clearance of the ear isolate from the lungs. In particular, serum amyloid P, the protein encoded by the APCS gene, is known to play a role in complement deposition on the pneumococcus, improving phagocyte efficiency and aiding clearance of the pathogen (30).
In this study, we have provided further evidence that S. pneumoniae strains belonging to the same serotype and ST can elicit distinct host innate immune responses and cause different types of disease. Future comparative genomic and transcriptomic analyses of these ST-matched strain pairs with distinct virulence phenotypes should enable identification of specific bacterial determinants of tissue tropism. This knowledge may, in turn, provide opportunities to develop improved vaccine formulations capable of eliciting protection against the full spectrum of pneumococcal disease.
ACKNOWLEDGMENTS
This work was supported by Program Grants 565526 and 1071659 from the National Health and Medical Research Council of Australia (NHMRC). A.W.P. is an Australian Research Council (ARC) DORA Fellow; J.C.P. is a NHMRC Senior Principal Research Fellow; C.T. is an ARC DECRA Fellow.
We thank Layla Mahdi for assistance with animal experiments.
REFERENCES
- 1.O'Brien KL, Wolfson LJ, Watt JP, Henkle E, Deloria-Knoll M, McCall N, Lee E, Mulholland K, Levine OS, Cherian T, Hib and Pneumococcal Global Burden of Disease Study Team . 2009. Burden of disease caused by Streptococcus pneumoniae in children younger than 5 years: global estimates. Lancet 374:893–902. doi: 10.1016/S0140-6736(09)61204-6. [DOI] [PubMed] [Google Scholar]
- 2.Crook DW, Brueggemann AB, Sleeman KL, Peto TEA. 2004. Pneumococcal carriage, p 136–147. In Tuomanen EI, Mitchell TJ, Morrison DA (ed), The pneumococcus. ASM Press, Washington, DC. [Google Scholar]
- 3.Enright MC, Spratt BG. 1998. A multilocus sequence typing scheme for Streptococcus pneumoniae: identification of clones associated with serious invasive disease. Microbiology 144:3049–3060. doi: 10.1099/00221287-144-11-3049. [DOI] [PubMed] [Google Scholar]
- 4.Kelly T, Dillard JP, Yother J. 1994. Effect of genetic switching of capsular type on virulence of Streptococcus pneumoniae. Infect Immun 62:1813–1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McAllister LJ, Ogunniyi AD, Stroeher UH, Leach AJ, Paton JC. 2011. Contribution of serotype and genetic background to virulence of serotype 3 and serogroup 11 pneumococcal isolates. Infect Immun 79:4839–4849. doi: 10.1128/IAI.05663-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sjöström K, Spindler C, Ortqvist A, Kalin M, Sandgren A, Kühlmann-Berenzon S, Henriques-Normark B. 2006. Clonal and capsular types decide whether pneumococci will act as a primary or opportunistic pathogen. Clin Infect Dis 42:451–459. doi: 10.1086/499242. [DOI] [PubMed] [Google Scholar]
- 7.Sandgren A, Sjostrom K, Olsson-Liljequist B, Christensson B, Samuelsson A, Kronvall G, Henriques Normark B. 2004. Effect of clonal and serotype-specific properties on the invasive capacity of Streptococcus pneumoniae. J Infect Dis 189:785–796. doi: 10.1086/381686. [DOI] [PubMed] [Google Scholar]
- 8.Trappetti C, van der Maten E, Amin Z, Potter AJ, Chen AY, van Mourik PM, Lawrence AJ, Paton AW, Paton JC. 2013. Site of isolation determines biofilm formation and virulence phenotypes of Streptococcus pneumoniae serotype 3 clinical isolates. Infect Immun 81:505–513. doi: 10.1128/IAI.01033-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sleeman KL, Griffiths D, Shackley F, Diggle L, Gupta S, Maiden MC, Moxon ER, Crook DW, Peto TE. 2006. Capsular serotype-specific attack rates and duration of carriage of Streptococcus pneumoniae in a population of children. J Infect Dis 194:682–688. doi: 10.1086/505710. [DOI] [PubMed] [Google Scholar]
- 10.Kronenberg A, Zucs P, Droz S, Mühlemann K. 2006. Distribution and invasiveness of Streptococcus pneumoniae serotypes in Switzerland, a country with low antibiotic selection pressure, from 2001 to 2004. J Clin Microbiol 44:2032–2038. doi: 10.1128/JCM.00275-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brueggemann AB, Griffiths DT, Meats E, Peto T, Crook DW, Spratt BG. 2003. Clonal relationships between invasive and carriage Streptococcus pneumoniae and serotype- and clone-specific differences in invasive disease potential. J Infect Dis 187:1424–1432. doi: 10.1086/374624. [DOI] [PubMed] [Google Scholar]
- 12.Burgos J, Falcó V, Borrego A, Sordé R, Larrosa MN, Martinez X, Planes AM, Sánchez A, Palomar M, Rello J, Pahissa A. 2013. Impact of the emergence of non-vaccine pneumococcal serotypes on the clinical presentation and outcome of adults with invasive pneumococcal pneumonia. Clin Microbiol Infect 19:385–391. doi: 10.1111/j.1469-0691.2012.03895.x. [DOI] [PubMed] [Google Scholar]
- 13.Bewick T, Sheppard C, Greenwood S, Slack M, Trotter C, George R, Lim WS. 2012. Serotype prevalence in adults hospitalised with pneumococcal non-invasive community-acquired pneumonia. Thorax 67:540–545. doi: 10.1136/thoraxjnl-2011-201092. [DOI] [PubMed] [Google Scholar]
- 14.Gentile A, Bardach A, Ciapponi A, Garcia-Marti S, Aruj P, Glujovsky D, Calcagno JI, Mazzoni A, Colindres RE. 2012. Epidemiology of community-acquired pneumonia in children of Latin America and the Caribbean: a systematic review and meta-analysis. Int J Infect Dis 16:e5–e15. doi: 10.1016/j.ijid.2011.09.013. [DOI] [PubMed] [Google Scholar]
- 15.Spratt BG. 1999. Multilocus sequence typing: molecular typing of bacterial pathogens in an era of rapid DNA sequencing and the internet. Curr Opin Microbiol 2:312–316. doi: 10.1016/S1369-5274(99)80054-X. [DOI] [PubMed] [Google Scholar]
- 16.Trappetti C, Ogunniyi AD, Oggioni MR, Paton JC. 2011. Extracellular matrix formation enhances the ability of Streptococcus pneumoniae to cause invasive disease. PLoS One 6:e19844. doi: 10.1371/journal.pone.0019844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harvey RM, Stroeher UH, Ogunniyi AD, Smith-Vaughan HC, Leach AJ, Paton JC. 2011. A variable region within the genome of Streptococcus pneumoniae contributes to strain-strain variation in virulence. PLoS One 6:e19650. doi: 10.1371/journal.pone.0019650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hughes CE, Harvey RM, Plumptre CD, Paton JC. 2014. Development of primary invasive pneumococcal disease caused by serotype 1 pneumococci is driven by early increased type I interferon response in the lung. Infect Immun 82:3919–3926. doi: 10.1128/IAI.02067-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Macrina FL, Tobian JA, Jones KR, Evans RP, Clewell DB. 1982. A cloning vector able to replicate in Escherichia coli and Streptococcus sanguis. Gene 19:345–353. doi: 10.1016/0378-1119(82)90025-7. [DOI] [PubMed] [Google Scholar]
- 20.Hall-Stoodley L, Nistico L, Sambanthamoorthy K, Dice B, Nguyen D, Mershon WJ, Johnson C, Hu FZ, Stoodley P, Ehrlich GD, Post JC. 2008. Characterization of biofilm matrix, degradation by DNase treatment and evidence of capsule downregulation in Streptococcus pneumoniae clinical isolates. BMC Microbiol 8:173. doi: 10.1186/1471-2180-8-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Ginkel FW, McGhee JR, Watt JM, Campos-Torres A, Parish LA, Briles DE. 2003. Pneumococcal carriage results in ganglioside-mediated olfactory tissue infection. Proc Natl Acad Sci U S A 100:14363–14367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Silva NA, McCluskey J, Jefferies JM, Hinds J, Smith A, Clarke SC, Mitchell TJ, Paterson GK. 2006. Genomic diversity between strains of the same serotype and multilocus sequence type among pneumococcal clinical isolates. Infect Immun 74:3513–3518. doi: 10.1128/IAI.00079-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mosser DM. 2003. The many faces of macrophage activation. J Leukoc Biol 73:209–212. doi: 10.1189/jlb.0602325. [DOI] [PubMed] [Google Scholar]
- 24.van der Poll T, Keogh CV, Guirao X, Buurman WA, Kopf M, Lowry SF. 1997. Interleukin-6 gene-deficient mice show impaired defense against pneumococcal pneumonia. J Infect Dis 176:439–444. [DOI] [PubMed] [Google Scholar]
- 25.Mackay F, Loetscher H, Stueber D, Gehr G, Lesslauer W. 1993. Tumor necrosis factor alpha (TNF-alpha)-induced cell adhesion to human endothelial cells is under dominant control of one TNF receptor type, TNF-R55. J Exp Med 177:1277–1286. doi: 10.1084/jem.177.5.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Collins T, Read MA, Neish AS, Whitley MZ, Thanos D, Maniatis T. 1995. Transcriptional regulation of endothelial cell adhesion molecules: NF-kappa B and cytokine-inducible enhancers. FASEB J 9:899–909. [PubMed] [Google Scholar]
- 27.Lee PY, Li Y, Kumagai Y, Xu Y, Weinstein JS, Kellner ES, Nacionales DC, Butfiloski EJ, van Rooijen N, Akira S, Sobel ES, Satoh M, Reeves WH. 2009. Type I interferon modulates monocyte recruitment and maturation in chronic inflammation. Am J Pathol 175:2023–2033. doi: 10.2353/ajpath.2009.090328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alderson MR, Armitage RJ, Tough TW, Strockbine L, Fanslow WC, Spriggs MK. 1993. CD40 expression by human monocytes: regulation by cytokines and activation of monocytes by the ligand for CD40. J Exp Med 178:669–674. doi: 10.1084/jem.178.2.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Portillo JA, Feliciano LM, Okenka G, Heinzel F, Subauste MC, Subauste CS. 2012. CD40 and tumour necrosis factor-alpha co-operate to up-regulate inducuble nitric oxide synthase expression in macrophages. Immunology 135:140–150. doi: 10.1111/j.1365-2567.2011.03519.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yuste J, Botto M, Bottoms SE, Brown JS. 2007. Serum amyloid P aids complement-mediated immunity to Streptococcus pneumoniae. PLoS Pathog 3:1208–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]