Abstract
Among the most fascinating virulence attributes of Candida is the ability to transition to a biofilm lifestyle. As a biofilm, Candida cells adhere to a surface, such as a vascular catheter, and become encased in an extracellular matrix. During this mode of growth, Candida resists the normal immune response, often causing devastating disease. Based on scanning electron microscopy images, we hypothesized that host cells and proteins become incorporated into clinical biofilms. As a means to gain an understanding of these host-biofilm interactions, we explored biofilm-associated host components by using microscopy and liquid chromatography-mass spectrometry. Here we characterize the host proteins associated with several in vivo rat Candida albicans biofilms, including those from vascular catheter, denture, and urinary catheter models as well as uninfected devices. A conserved group of 14 host proteins were found to be more abundant during infection at each of the niches. The host proteins were leukocyte and erythrocyte associated and included proteins involved in inflammation, such as C-reactive protein, myeloperoxidase, and alarmin S100-A9. A group of 59 proteins were associated with both infected and uninfected devices, and these included matricellular and inflammatory proteins. In addition, site-specific proteins were identified, such as amylase in association with the denture device. Cellular analysis revealed neutrophils as the predominant leukocytes associating with biofilms. These experiments demonstrate that host cells and proteins are key components of in vivo Candida biofilms, likely with one subset associating with the device and another being recruited by the proliferating biofilm.
INTRODUCTION
Candida causes device-associated infections by adhering to a surface and proliferating as a multicellular community within an extracellular matrix (1, 2). These resilient medical infections occur when microbes colonize foreign material, such as intravascular or urinary catheters, dentures, and other implantable substrates. Approximately half of the 2 million nosocomial infections reported each year in the United States are associated with indwelling device biofilms (3, 4). Unlike planktonic organisms, cells of a biofilm demonstrate exquisite drug resistance, withstanding up to 1,000-fold higher concentrations of antifungals (5–8). Because antifungal therapy is often ineffective in the biofilm setting, treatment of device-associated Candida infections typically requires device removal, which incurs additional morbidity and costs (9).
Infection of medical devices involves a complex process of microbial adherence and proliferation as an adherent mono- or polymicrobial biofilm. One distinct characteristic of biofilm formation is the development of an extracellular matrix (10). Numerous in vitro studies have divulged many of the fungus-derived components of this material, including extracellular proteins and carbohydrates (5, 7, 11–13). However, during the infectious process, Candida biofilms are continuously exposed to various host factors. Although the host is expected to play a role in this process, the roles of specific proteins in biofilm propagation and matrix deposition have not been investigated thoroughly. The composition of host-derived cellular and acellular material in Candida biofilms has been a mystery.
This investigation explores the host contribution to Candida biofilms in clinically relevant animal models representing the most common Candida device infections (14–17). Exposure to host cells and proteins is a dynamic process specific to the infectious niche. For example, Candida vascular catheter biofilms are exposed to the assortment of leukocytes, erythrocytes, platelets, and proteins found in serum. On the other hand, the biofilms associated with denture stomatitis are bathed in saliva. Likewise, urinary catheter biofilms are exposed to urine. Many other factors vary at these locales, including pH, nutrient availability, host defenses, and proximity to epithelial or endothelial cell surfaces. Because of the heterogeneity of these niches, we chose to examine the host contribution to biofilms in three animal models of infection.
In this report, we offer a comprehensive evaluation of the host-derived components of in vivo Candida albicans biofilms. By including three in vivo niches, we were able to identify the incorporation of a recurrent set of proteins, as well as subsets specific to the individual environments. In addition, we analyzed uninfected devices from each of the sites to detect the proteins uniformly interacting with medical devices outside infection. The examination of both the host cells and proteins associating with these biofilms provides insight into novel host-fungus interactions.
MATERIALS AND METHODS
Organism and inoculum.
C. albicans K1 was used for all studies (18). The strain was stored in a 15% (vol/vol) glycerol stock at −80°C and maintained on yeast extract-peptone-dextrose (YPD) medium plus uridine (1% yeast extract, 2% peptone, 2% dextrose, and 80 μg/ml uridine) prior to experiments. Cultures were propagated overnight in YPD supplemented with uridine at 30°C on an orbital shaker at 200 rpm.
Animals.
Specific-pathogen-free Sprague-Dawley rats weighing 350 g (Harlan Sprague-Dawley, Indianapolis, IN) were used for all studies. Animals were maintained in accordance with the Association for Assessment and Accreditation of Laboratory Animal Care criteria, and all studies were approved by the institutional animal care committee.
In vivo venous catheter biofilm model.
A rat jugular vein central venous catheter infection model was used to mimic venous catheter biofilm infections in patients (16, 19). Briefly, following a 24-h conditioning period, the implanted jugular venous catheters were inoculated (106 cells/ml by hemocytometer counts). After 6 h, the inoculum was removed and catheters were instilled with heparinized (100 U/ml) saline for a 48-h growth period. For uninfected controls, an inoculum was not instilled. Catheters from three animals were pooled for each condition.
In vivo rat denture model.
A rat denture biofilm model was used as previously described (15). Briefly, rats were immunosuppressed with cortisone acetate (200 mg/kg of body weight subcutaneously) on the day of inoculation. A stainless steel orthodontic wire (32 gauge) was threaded across the hard palate. A metal spatula was placed on the palate, and cold-cure temporary crown and bridge material was applied over the cheek teeth, spatula, and wire. After solidification, the spatula was removed and the hard palate was inoculated with C. albicans (108 cells/ml by hemocytometer counts). The device was removed after a 48-h growth period. For uninfected controls, an inoculum was not instilled. Gentamicin (80 mg/kg) was administered subcutaneously twice daily throughout the course of the experiment. Rats were housed individually in metabolic cages and were fed a liquid diet. Devices from three animals were pooled for each condition.
In vivo rat urinary model.
A rat urinary catheter biofilm model was used as a model to mirror indwelling urinary catheter infections in patients (14). Briefly, animals received a single dose of cortisone acetate (250 mg/kg) subcutaneously. A silicone catheter threaded onto a guide wire was inserted via the urethra into the bladder and adhered with surgical adhesive and suture. The inoculum (108 cells/ml by hemocytometer counts) was instilled for 2 h. For uninfected controls, an inoculum was not instilled.
Animals received gentamicin (80 mg/kg) subcutaneously twice daily and were given drinking water containing penicillin G sodium (0.9 mg/ml) to prevent bacterial contamination. Catheters were harvested after a 48-h growth period. During the period of catheter placement, animals were maintained in metabolic cages. Catheters from three animals were pooled for each condition.
Ex vivo coverslip biofilm model.
Coverslips (13 mm; Thermonax plastic for cell culture) were pretreated with heat-treated serum for 45 min at 30°C in an attempt to incorporate exposure to proteins common to most in vivo infection sites. C. albicans K1 cultures were enumerated by use of a hemocytometer, and cells were resuspended in RPMI-morpholinepropanesulfonic acid (RPMI-MOPS) at 106 cells/ml. For each coverslip, 40 μl of the fungal inoculum was added to the surface. After 1 h of incubation at 30°C, the inoculum was removed and medium (RPMI-MOPS supplemented with 5% EDTA-treated human blood) was added. Biofilms were grown for 24 h at 37°C on an orbital shaker at 200 rpm.
Scanning electron microscopy.
Devices were processed for scanning electron microscopy as previously described (16). Briefly, biofilms were fixed overnight (4% formaldehyde, 1% glutaraldehyde in phosphate-buffered saline [PBS]). Biofilms were then washed with PBS, treated with 1% osmium tetroxide, and washed again. Samples were dehydrated by a series of ethanol washes and critical point drying. Specimens were mounted on aluminum stubs and sputter coated with gold. Samples were imaged in a scanning electron microscope (LEO 1530 or JEOL 6100).
Isolation of biofilm matrix proteins and device-associated proteins.
Biofilms and device-associated proteins were dislodged from infected and uninfected devices by either flushing with phosphate-buffered saline or gentle scraping. Devices and biofilms were gently sonicated at 42 kHz for 20 min (Branson 1510 ultrasonic cleaner sonicator), followed by sonication with a 1-cm by 5-cm probe in an Intrasonic processor (Cole Parmer, Vernon Hills, IL) at an amplitude of 70 for 10 min (13). Soluble proteins were harvested following three centrifugations (4,500 × g for 20 min). Specific proteins were identified by liquid chromatography-mass spectrometry (LC-MS) as previously described (13). Briefly, trypsin-digested matrix was analyzed by nano-LC–tandem MS (MS/MS) by using an Agilent 1100 nanoflow system (Agilent, Palo Alto, CA) connected to a hybrid linear ion trap-Orbitrap mass spectrometer (LTQ-Orbitrap; Thermo Fisher Scientific, San Jose, CA) equipped with a nanoelectrospray ion source (20). Raw MS/MS data were searched against a concatenated Rattus norvegicus amino acid sequence database by using an in-house MASCOT search engine (21). Identified proteins were further annotated and filtered to 1.5% peptide and 0.1% protein false discovery rates with Scaffold Q+, version 4.3.4 (Proteome Software Inc., Portland, OR), using the protein prophet algorithm (22). In order to compare abundances of proteins between samples, Scaffold's unweighted spectrum counts normalization method was applied, which sums mass spectra for each sample (22). These sums were then scaled, and the scaling factor for each sample was applied to each protein group and adjusted its “unweighted” counts to normalized semiquantitative values, which were further used to construct Voronoi tree maps as previously described (23).
Cellular staining.
In vivo biofilms were harvested from the rat venous catheter, rat denture, and rat urinary catheter models. Biofilms were washed and dislodged by flushing of catheters with phosphate-buffered saline and gentle scraping. For urinary catheter biofilms, cells were stained by the thin prep-Papanicolaou method that is conventionally utilized for urinary specimens at our institution. Cells were collected in CytoLyt solution (Cytyc Corporation, a subsidiary of Hologic Corporation, Marlborough, MA). The sample was processed on a ThinPrep 2000 instrument (Cytyc Corporation) using liquid-based methodology per the manufacturer's instructions. Slides were subsequently stained by utilizing the Papanicolaou method. For the vascular and denture model biofilms, samples were collected in phosphate-buffered saline and centrifuged for 5 min at 1,600 rpm in a Thermo Scientific CL2 centrifuge (Thermo Fisher Scientific Inc., Waltham, MA), followed by 10 min at 1,500 rpm on a Cytospin 4 cytocentrifuge (Thermo Fisher Scientific Inc.). Samples were then processed on a Sysmex SP1000-I (Sysmex Corporation, Kobe, Japan) automated slide preparer/stainer per the manufacturer's instructions, stained using the Wright method, and imaged by light microscopy. For calculation of leukocyte-to-Candida ratios, the cells in high-power fields were enumerated.
RESULTS
Imaging of C. albicans in vivo, in vitro, and ex vivo biofilms.
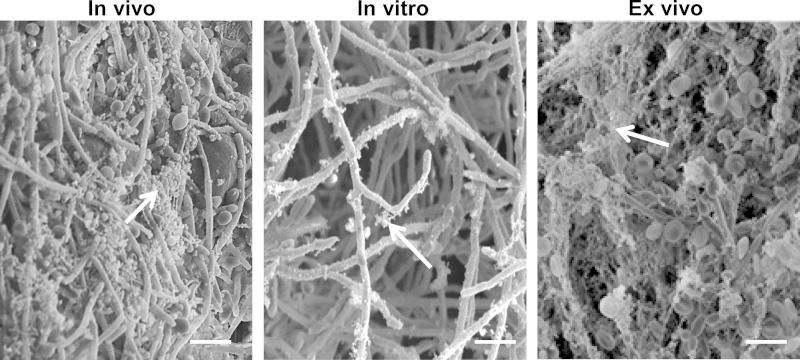
In vivo microbial biofilms are composed of adherent microbes encased in a matrix (1, 24, 25). This extracellular matrix is especially pronounced during in vivo biofilm formation. Examination of a biofilm formed on the surface of a rat vascular catheter revealed a dense, fibrillar coating covering yeast and hyphae (Fig. 1). In contrast, the extracellular material on in vitro biofilms appeared to be less abundant and more granular than fibrillar. Struck by this difference in biofilm structure, we considered the possibility that host proteins may contribute to the biofilm maturation process in vivo. To explore the possibility of host components incorporating into Candida biofilms, we added blood components to our in vitro coverslip model, mimicking a vascular catheter infection. Imaging of the ex vivo model showed a stark contrast to the in vitro biofilm grown on a coverslip (Fig. 1). Numerous host cells were associated with the biofilm. A thick, acellular layer of matrix enveloped the biofilm, similar to that observed for the in vivo biofilm model, suggesting that biofilms incorporate both host proteins and cells during maturation.
FIG 1.
Host factors promote C. albicans biofilm matrix deposition. Candida biofilms were collected from an in vivo rat vascular biofilm infection model or were grown in vitro or ex vivo (in the presence of blood). Images were obtained by scanning electron microscopy to visualize the matrix. This extracellular material, marked by arrows, encased the in vivo biofilms as well as the ex vivo biofilms but was less abundant on in vitro biofilms. Bar, 10 μm.
Mammalian host proteins are integrated into C. albicans biofilms for three in vivo niches.
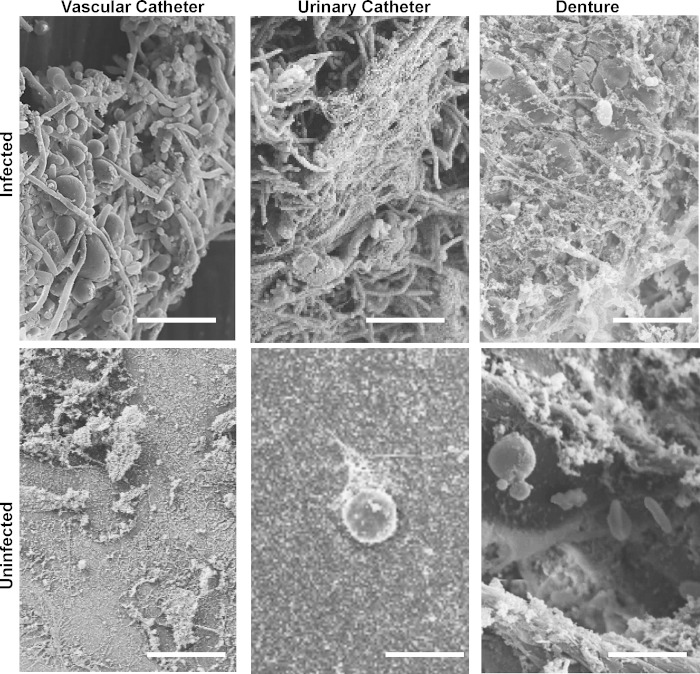
For examination of the extracellular matrix composition of in vivo Candida biofilms, we chose to include three rat models of C. albicans biofilm infection that closely mimic common clinical scenarios (14–16). Mature biofilms formed on the surfaces of these devices, including a vascular catheter, urinary catheter, and denture device, contained abundant extracellular matrix material as visualized by scanning electron microscopy (Fig. 2). Proteins of the extracellular matrix were analyzed by liquid chromatography-mass spectrometry-based proteomics. We used this unbiased approach to capture the involvement of host proteins by searching against a Rattus norvegicus amino acid sequence database. Proteomic analysis of the denture, urinary catheter, and venous catheter models identified 132, 213, and 139 biofilm-associated host proteins, respectively (see Table S1 in the supplemental material). The protein abundance varied considerably by site. For the denture biofilm, the most abundant proteins included Amyl1 (amylase), BPI fold-containing proteins (antimicrobial peptides), and hemoglobin. Keratin, fibrinogen, and hemoglobin were highly represented in the urinary catheter biofilm. For the vascular catheter biofilm, hemoglobin, albumin, and various alpha globulins were the most abundant proteins.
FIG 2.
Imaging of C. albicans-infected and uninfected devices. Devices were collected from rat biofilm infection models (vascular catheter, urinary catheter, and denture) in the presence or absence of C. albicans biofilm infection. Extracellular material and host cells on devices were visualized by scanning electron microscopy. Bar, 10 μm.
We also examined host proteins associating with uninfected devices for each of the niches. Analysis of device-associated proteins revealed 279, 457, and 382 proteins for the denture, urinary catheter, and venous catheter models, respectively (see Table S1 in the supplemental material). The identification of numerous proteins in the absence of infection is consistent with imaging of the devices. By scanning electron microscopy, host cells and debris were adherent to devices without biofilm infection (Fig. 2). Voronoi tree maps were constructed to depict changes in protein abundance in the proteomes of uninfected and Candida-infected medical devices (Fig. 3) (23). These diagrams showed a marked difference between host proteins associating with C. albicans-infected and uninfected devices for each anatomic location.
FIG 3.
Host proteins associate with C. albicans-infected and uninfected devices. Proteins were collected from the extracellular matrix of C. albicans biofilm-infected devices, analyzed by liquid chromatography-mass spectrometry, and searched against a Rattus norvegicus amino acid sequence database. For uninfected samples, proteins associating with the device surface were similarly analyzed for each niche. Abundances were compared using Scaffold's unweighted spectrum normalization, and data are presented as Voronoi tree maps reflecting relative abundances.
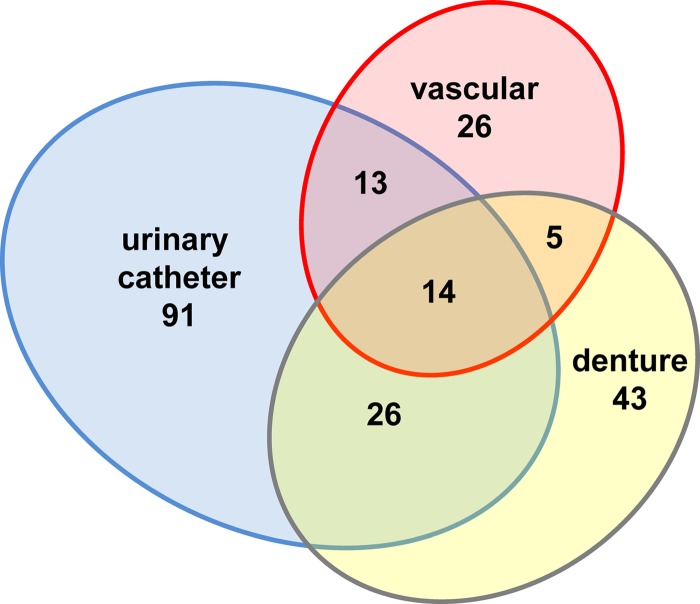
We reasoned that proteins abundant in all three niches may play a role in establishing, maintaining, or mounting an immune response to clinical biofilm infections. We found a subset comprised of 14 host proteins that were more abundant during C. albicans biofilm infection and were conserved among all three niches (Table 1 and Fig. 4). Diverse functional categories were represented in this group. Red blood cell- and heme-related proteins were identified, including hemoglobin, transferrin, and haptoglobin. Also, inflammatory and leukocyte-associated proteins were highly represented, including myeloperoxidase, C-reactive protein, and alarmin S100-A9 (a subunit of calprotectin). In addition, the finding of histones, actin, and myeloperoxidase in this subset suggests the presence of neutrophil extracellular traps (NETs), although these proteins may be deposited by other means as well (26).
TABLE 1.
Host proteins abundant during C. albicans biofilm infection in rat venous catheter, rat urinary catheter, and rat denture models
| Protein | Gene | Accession number |
|---|---|---|
| Actin, cytoplasmic 1 | Actb | P60711 |
| Alpha-actinin-4 | Actn4 | Q9QXQ0 |
| Band 3 anion transport protein | Slc4a1 | F8WFT7 |
| C-reactive protein | Crp | P48199 |
| Haptoglobin | Hp | P06866 |
| Hemoglobin subunit beta-1 | Hbb | P02091 |
| Hemoglobin subunit beta-2 | Hbb2 | P11517 |
| Histone H2A.J | H2afj | A9UMV8 |
| Peroxiredoxin-2 | Prdx2 | P35704 |
| Protein Itih4 | Itih4 | D3ZFC6 |
| Protein LOC100909666 | LOC100909666 | F1LNM4 |
| Protein Mpo | Mpo | D3ZYH8 |
| Protein S100-A9 | S100a9 | P50116 |
| Serotransferrin | Tf | P12346 |
FIG 4.
Host proteins are abundant during C. albicans biofilm infection. Proteins were collected from the extracellular matrix of C. albicans biofilm-infected devices, analyzed by liquid chromatography-mass spectrometry, and searched against a Rattus norvegicus amino acid sequence database. For uninfected samples, proteins associating with the device surface were similarly analyzed for each niche. Abundances were compared using Scaffold's unweighted spectrum normalization. Data are presented as a Venn diagram depicting the number of proteins more abundant during infection for each niche.
We next examined the host proteins present on both uninfected and biofilm-infected devices for each niche. We reasoned that this subgroup of host proteins may be important for initiation of biofilms on medical devices. A total of 58 host proteins recurred for each of the 6 conditions tested (see Table S2 in the supplemental material). Matricellular and coagulation proteins were highly represented and included fibrinogen, plasminogen, fibronectin, and vitronectin. Red blood cell- and heme-associated proteins were found in this group as well (hemoglobin, peroxiredoxin-2, carbonic anhydrase, transferrin, and haptoglobin). Numerous leukocyte-associated and inflammatory proteins were present in this subset, consistent with an inflammatory response to the device alone. Included in this group were myeloperoxidase, neutrophilic granule protein, cathelicidin antimicrobial peptide, C-reactive protein, alarmin S100-A9, alpha-1-antiproteinase, and complement factors (C3 and C9).
The final subset of host proteins analyzed included those less abundant in the infected devices than in the uninfected control devices. This group included 69 proteins with diverse functions (see Table S3 in the supplemental material). One of the striking features of this subgroup was the presence of complement factors, including C3, C4, C8, and C9. This suggests that complement is deposited on uninfected devices but is much less abundant during biofilm infection. One possibility is that factors may be activated and degraded during infection.
Incorporation of host cells into in vivo C. albicans biofilms.
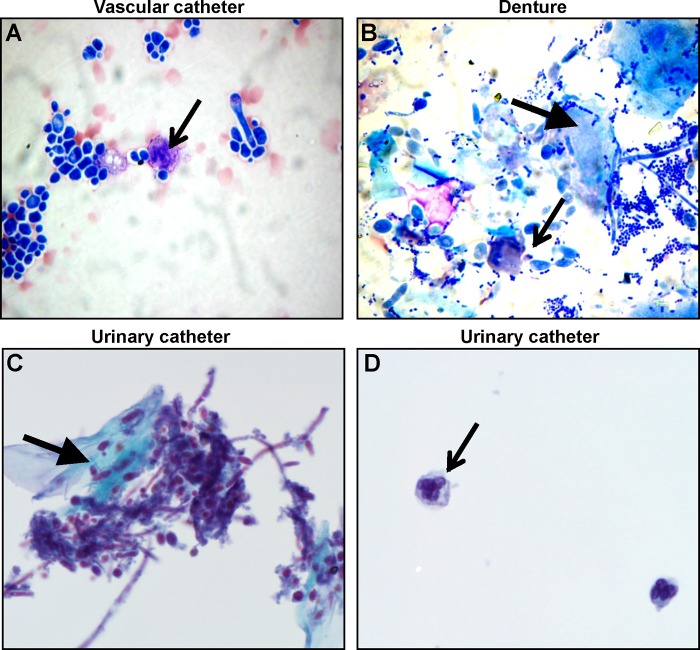
To identify biofilm-associated host cells, in vivo biofilms were dislodged, collected, stained, and imaged. Vascular catheter biofilms were processed by Cytospin centrifugation and stained with Wright stain. The predominate host cell types associated with the vascular catheter biofilm were red blood cells and leukocytes (Fig. 5A). Based on their characteristic polymorphic nuclei, the majority of the incorporated leukocytes were neutrophils. Examination of the denture biofilms by this method revealed the presence of numerous epithelial cells associated with these oral biofilms (Fig. 5B). Although leukocytes were observed as well, the architecture of these cells was not as well preserved during processing. However, the appearance of these cells was most consistent with neutrophils as the predominant leukocytes associating with the denture biofilms. To examine the host cells of the urinary catheter biofilms, the Papanicolaou stain was selected for its ability to preserve and distinguish cells in the urinary environment. Host cells, including urothelial cells and neutrophils, were found to associate with the urinary catheter biofilm (Fig. 5C and D). The thin preparation technique used with the Papanicolaou stain resulted in a more distinct separation of biofilm components, exhibited by separation of neutrophils from fungal components (Fig. 5D). For each of the biofilm niches, the relative abundance of host cells was significantly lower than the fungal burden. The ratios of leukocytes to Candida cells were 1:75, 1:12, and 1:15 for the vascular catheter, denture device, and urinary catheter models, respectively. Given the differences in staining techniques, comparisons among device niches should be interpreted with caution. However, neutrophils were consistently found accompanying the biofilms and were the only immune cell type identified.
FIG 5.
Host cells associate with C. albicans biofilms in vivo. C. albicans biofilms were collected from a rat vascular catheter model (A), a rat denture model (B), and a urinary catheter model (C and D). Following removal from the device, catheter and denture biofilms were loaded on a Cytospin centrifuge, and slides were processed with Wright stain. Urinary catheter biofilms were examined following thin preparation and Papanicolaou staining. Line arrows in panels A, B, and D mark neutrophils. The block arrow in panel B highlights an epithelial cell. The block arrow in panel C highlights a urothelial cell.
DISCUSSION
Numerous host factors have been shown to influence fungal biofilms (27–32). In vitro studies have often included proteins and other factors to mimic the niche site infection (1). Examples include the incorporation of synthetic urine medium to mimic a urinary catheter infection or the addition of saliva to simulate oral biofilm conditions (33–35). However, investigations have not systematically analyzed biofilms to determine the array of mammalian factors involved in the host-biofilm interaction. Here we employed three animal models of Candida biofilm infection to identify biofilm-associated host proteins and cells. Despite the diversity of the surrounding milieus (blood, urine, and saliva), a group of host proteins was found recurrently among the biofilm models. We propose that this group likely encompasses protein subsets that possess both pro- and antibiofilm purposes.
One of the striking groups of proteins observed across all three in vivo niches, in the presence and absence of biofilm infection, was a subset of red blood cell- and heme-related proteins (see Table S2 in the supplemental material). Based on proteomic analysis and scanning electron microscopy images, it appears likely that a layer of red blood cells is deposited on uninfected devices and that these cells may remain intact or become disrupted (Fig. 2). For cells that are disrupted, hemoglobin is recycled or degraded via haptoglobin or peroxiredoxin (36). In addition to the ubiquitous distribution of these proteins, a subset was also more abundant in the infected devices (Table 1). By scanning electron microscopy, it appears that red blood cells incorporate into the biofilm during the maturation process (Fig. 1). Iron scavenging is an important virulence trait for many pathogens, including Candida. C. albicans was recently shown to possess two distinct heme-binding proteins which facilitate iron acquisition from hemoglobin (37). Our studies suggest that hemoglobin is abundant on the surfaces of medical devices. This accessibility to iron may be one reason that device-associated biofilms are so resilient in the presence of host defenses and treatment. The findings also point to the relevance of including blood components in ex vivo studies of Candida biofilms.
Matricellular proteins were among the host protein categories represented in Candida biofilms from all the clinical niches. These proteins, including vitronectin, fibronectin, and fibrinogen, were associated with both infected and uninfected devices across each model system. Matricellular proteins, such as fibronectin, have previously been shown to deposit on medical device surfaces (38–42). The current study confirms the presence of these host proteins on devices from various niches and points to the similarities between the biofilm infection models and common device-associated clinical infections. C. albicans is known to interact with these ubiquitous matricellular proteins through specific interactions, which may enhance tissue invasion and contribute to virulence (43–54). Given the evidence for the likely involvement of these proteins in Candida pathogenicity, pioneering biofilm investigations have emphasized the importance of a host conditioning fluid for optimal biofilm formation and the need to account for the proteins in in vitro biofilm models (29, 35, 55–58). However, little is known about how assimilation of host proteins may affect biofilm structure and function, perhaps even fostering biofilm development.
Several proteins involved in immune response and leukocyte function were identified in the subset of abundant proteins conserved among the biofilm niches (Table 1). The presence of myeloperoxidase and neutrophilic granule protein is consistent with the incorporation of neutrophils or factors released from neutrophils. Indeed, examination of the cellular components of biofilms revealed neutrophils as the major leukocytes present in each of the rat biofilm infections (Fig. 5). However, compared to a previous investigation of nonbiofilm Candida infection, a relatively small number of neutrophils per Candida cell was identified (59). Prior investigations have described leukocytes associating with Candida biofilms (31, 60). In an oral mucosal biofilm model, neutrophils were found to form aggregates near the biofilm surface and even to migrate throughout the biofilm (60). In observing peripheral blood mononuclear cells interacting with in vitro C. albicans biofilms, Chandra et al. demonstrated that not only did the leukocytes not inhibit the biofilm but the cells actively produced factors that augmented biofilm formation (31). The current investigation extends these findings to show that leukocytes, including neutrophils, associate with medical device biofilms at multiple clinical sites of infection.
Little is known about the activation status of the neutrophils. Representation of the components of NETs (myeloperoxidase, histone H2A, neutrophilic granule protein, and actin) in the biofilm matrix samples suggests the release of NETs (Table 1) (26). Since both yeast and hyphal forms of Candida have been shown to induce the release of NETs (NETosis) when incubated with neutrophils (61), it is quite plausible that NETs may be elicited by the biofilm mode of growth as well. However, numerous potential components of NETs were also found in association with uninfected devices, which indicates the possibility of their release in response to the device alone. On microscopic evaluation of the host cells adherent to an uninfected urinary catheter, fibrillar contents were observed in association with cells, consistent with the process of NETosis (Fig. 2).
Clinically, Candida biofilms resist the host immune response. Removal of Candida-infected devices is recommended, even for patients with intact immunity (9). In vitro studies corroborate a difference in immune responses to biofilm and planktonic Candida, but little is known about this mechanism (30, 31, 62, 63). The present study identifies alarmin S100-A9 as a protein abundant among all three device-associated biofilms. Alarmins S100-A8 and S100-A9 have previously been shown to be induced by C. albicans infection in a vaginal model (64). These alarmins appear to promote neutrophil chemotaxis, with a limited impact on the infectious burden (65). Further investigation may be warranted to determine if a similar mechanism of neutrophil response is involved in device-associated Candida infection as well.
We considered the possibility that leukocytes other than neutrophils were recruited to the Candida biofilms. We postulated that these cell types may be present in smaller numbers or may be more difficult to detect by the cellular isolation and staining processes used. Therefore, we searched the host proteomes for the presence of proteins unique to specific leukocyte groups. Our analysis did not detect CD3, the major antigen of T cells, or CD68, an antigenic marker of monocytes (66). Eosinophil peroxidase, a marker of eosinophils, was not represented in the samples. In contrast, neutrophil-associated proteins, including myeloperoxidase and neutrophil granular protein, were present in each of the host proteomic data sets. Note that the current studies did identify lymphocyte cytosolic protein 1 (lcp1) associating with infected urinary and vascular biofilms. This protein has been identified to associate with multiple cell types and has been used as a nonspecific leukocyte marker in other species (67). Taken together, the proteomic analysis was consistent with our cellular microscopy data showing neutrophils associating with Candida biofilms.
There are several similarities among the three models utilized for these experiments. The models all involve biofilm growth on an artificial, implanted device in a rat. In addition to variation in the site of infection, the models differ in terms of immunosuppression. To establish a consistent Candida biofilm, animals with either urinary catheters or denture devices received both corticosteroids and antibiotics. This treatment was not necessary for the vascular catheter infection given the high propensity of Candida to adhere to the catheter surface and the sterility of the blood. It is possible that these factors may have an impact on the host proteome as well. However, uninfected devices received the same corticosteroid and antibiotic treatments, so the influence of these factors should be minimal in comparisons between infected and uninfected devices.
The roles of many of the proteins associating with the Candida biofilm matrix are difficult to predict. Moonlighting proteins may be involved in the host response to biofilm infection. It is also possible that their interaction with the biofilm may be nonspecific, with the biofilm acting as a web to collect host proteins for extracellular deposition. Upon examination of the proteome of an in vitro C. albicans biofilm matrix, Thomas et al. found many similarities between the proteomes of planktonic supernatants and biofilm matrix (11). This suggests that proteins secreted or released from dying cells may become incorporated into biofilms. It is possible that a similar process is in place in vivo to nonspecifically capture host proteins to form the biofilm matrix. Such a process may serve to scavenge proteins for construction of a protective extracellular matrix and to conserve the energy that would otherwise be needed for Candida to assemble and export them. In fact, in this study and a prior investigation, the vast majority of in vivo Candida biofilm matrix proteins were of host origin (13). For example, Candida-derived proteins accounted for only 1.4% of the vascular catheter biofilm extracellular matrix.
These studies indicate a close relationship between C. albicans biofilms and host components. Defining the host components of biofilms offers new insight into biofilm pathogenesis and host-fungus interactions. However, the functional relevance of the individual proteins largely remains unknown. The current study provides the foundation to explore the impact of host proteins on device-Candida or Candida-Candida adhesion. The study suggests that erythrocytes and neutrophils interact with Candida biofilms, ultimately incorporating into the structure. Further studies are needed to determine the role of these cells during biofilm infection. The identification of host factors in Candida biofilms provides the framework for future host-pathogen interaction studies designed to identify novel drug targets and to augment the immune response against biofilms.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Institutes of Health (grants R01 AI073289-01 and K08 AI108727) and the Burroughs Wellcome Fund (grant 1012299). We acknowledge use of instrumentation supported by the UW MRSEC (grant DMR-1121288) and the UW NSEC (grant DMR-0832760).
Peptide digestion and mass spectrometric analysis were done at the Mass Spectrometry Facility, Biotechnology Center, University of Wisconsin-Madison.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.00931-15.
REFERENCES
- 1.Douglas LJ. 2003. Candida biofilms and their role in infection. Trends Microbiol 11:30–36. doi: 10.1016/S0966-842X(02)00002-1. [DOI] [PubMed] [Google Scholar]
- 2.Kumamoto CA, Vinces MD. 2005. Alternative Candida albicans lifestyles: growth on surfaces. Annu Rev Microbiol 59:113–133. doi: 10.1146/annurev.micro.59.030804.121034. [DOI] [PubMed] [Google Scholar]
- 3.Wenzel RP. 2007. Health care-associated infections: major issues in the early years of the 21st century. Clin Infect Dis 45(Suppl 1):S85–S88. doi: 10.1086/518136. [DOI] [PubMed] [Google Scholar]
- 4.Darouiche RO. 2004. Treatment of infections associated with surgical implants. N Engl J Med 350:1422–1429. doi: 10.1056/NEJMra035415. [DOI] [PubMed] [Google Scholar]
- 5.Al-Fattani MA, Douglas LJ. 2006. Biofilm matrix of Candida albicans and Candida tropicalis: chemical composition and role in drug resistance. J Med Microbiol 55:999–1008. doi: 10.1099/jmm.0.46569-0. [DOI] [PubMed] [Google Scholar]
- 6.Mukherjee PK, Chandra J, Kuhn DM, Ghannoum MA. 2003. Mechanism of fluconazole resistance in Candida albicans biofilms: phase-specific role of efflux pumps and membrane sterols. Infect Immun 71:4333–4340. doi: 10.1128/IAI.71.8.4333-4340.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nett J, Lincoln L, Marchillo K, Massey R, Holoyda K, Hoff B, VanHandel M, Andes D. 2007. Putative role of beta-1,3 glucans in Candida albicans biofilm resistance. Antimicrob Agents Chemother 51:510–520. doi: 10.1128/AAC.01056-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ramage G, Vandewalle K, Wickes BL, Lopez-Ribot JL. 2001. Characteristics of biofilm formation by Candida albicans. Rev Iberoam Micol 18:163–170. [PubMed] [Google Scholar]
- 9.Pappas PG, Kauffman CA, Andes D, Benjamin DK Jr, Calandra TF, Edwards JE Jr, Filler SG, Fisher JF, Kullberg BJ, Ostrosky-Zeichner L, Reboli AC, Rex JH, Walsh TJ, Sobel JD. 2009. Clinical practice guidelines for the management of candidiasis: 2009 update by the Infectious Diseases Society of America. Clin Infect Dis 48:503–535. doi: 10.1086/596757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hawser SP, Baillie GS, Douglas LJ. 1998. Production of extracellular matrix by Candida albicans biofilms. J Med Microbiol 47:253–256. doi: 10.1099/00222615-47-3-253. [DOI] [PubMed] [Google Scholar]
- 11.Thomas DP, Bachmann SP, Lopez-Ribot JL. 2006. Proteomics for the analysis of the Candida albicans biofilm lifestyle. Proteomics 6:5795–5804. doi: 10.1002/pmic.200600332. [DOI] [PubMed] [Google Scholar]
- 12.Vediyappan G, Chaffin WL. 2006. Non-glucan attached proteins of Candida albicans biofilm formed on various surfaces. Mycopathologia 161:3–10. doi: 10.1007/s11046-005-0167-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zarnowski R, Westler WM, Lacmbouh GA, Marita JM, Bothe JR, Bernhardt J, Lounes-Hadj Sahraoui A, Fontaine J, Sanchez H, Hatfield RD, Ntambi JM, Nett JE, Mitchell AP, Andes DR. 2014. Novel entries in a fungal biofilm matrix encyclopedia. mBio 5:e01333-14. doi: 10.1128/mBio.01333-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nett JE, Brooks EG, Cabezas-Olcoz J, Sanchez H, Zarnowski R, Marchillo K, Andes DR. 2014. Rat indwelling urinary catheter model of Candida albicans biofilm infection. Infect Immun 82:4931–4940. doi: 10.1128/IAI.02284-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nett JE, Marchillo K, Spiegel CA, Andes DR. 2010. Development and validation of an in vivo Candida albicans biofilm denture model. Infect Immun 78:3650–3659. doi: 10.1128/IAI.00480-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Andes D, Nett J, Oschel P, Albrecht R, Marchillo K, Pitula A. 2004. Development and characterization of an in vivo central venous catheter Candida albicans biofilm model. Infect Immun 72:6023–6031. doi: 10.1128/IAI.72.10.6023-6031.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kojic EM, Darouiche RO. 2004. Candida infections of medical devices. Clin Microbiol Rev 17:255–267. doi: 10.1128/CMR.17.2.255-267.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andes D, van Ogtrop M. 1999. Characterization and quantitation of the pharmacodynamics of fluconazole in a neutropenic murine disseminated candidiasis infection model. Antimicrob Agents Chemother 43:2116–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nett J, Lincoln L, Marchillo K, Andes D. 2007. Beta-1,3 glucan as a test for central venous catheter biofilm infection. J Infect Dis 195:1705–1712. doi: 10.1086/517522. [DOI] [PubMed] [Google Scholar]
- 20.Martin SE, Shabanowitz J, Hunt DF, Marto JA. 2000. Subfemtomole MS and MS/MS peptide sequence analysis using nano-HPLC micro-ESI Fourier transform ion cyclotron resonance mass spectrometry. Anal Chem 72:4266–4274. doi: 10.1021/ac000497v. [DOI] [PubMed] [Google Scholar]
- 21.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. 1999. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20:3551–3567. doi:. [DOI] [PubMed] [Google Scholar]
- 22.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. 2002. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem 74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 23.Bernhardt J, Funke S, Hecker M, Siebourg J. 2009. Visualizing gene expression data via Voronoi treemaps, p 233–241. Proceedings of the Sixth International Symposium on Voronoi Diagrams, ISVD 2009, Copenhagen, Denmark, 23 to 26 June, 2009. [Google Scholar]
- 24.Donlan RM, Costerton JW. 2002. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin Microbiol Rev 15:167–193. doi: 10.1128/CMR.15.2.167-193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Costerton JW, Stewart PS, Greenberg EP. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 26.Brinkmann V, Reichard U, Goosmann C, Fauler B, Uhlemann Y, Weiss DS, Weinrauch Y, Zychlinsky A. 2004. Neutrophil extracellular traps kill bacteria. Science 303:1532–1535. doi: 10.1126/science.1092385. [DOI] [PubMed] [Google Scholar]
- 27.Ramage G, Martinez JP, Lopez-Ribot JL. 2006. Candida biofilms on implanted biomaterials: a clinically significant problem. FEMS Yeast Res 6:979–986. doi: 10.1111/j.1567-1364.2006.00117.x. [DOI] [PubMed] [Google Scholar]
- 28.Nett J, Andes D. 2006. Candida albicans biofilm development, modeling a host-pathogen interaction. Curr Opin Microbiol 9:340–345. doi: 10.1016/j.mib.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 29.Frade JP, Arthington-Skaggs BA. 2011. Effect of serum and surface characteristics on Candida albicans biofilm formation. Mycoses 54:e154–e162. doi: 10.1111/j.1439-0507.2010.01862.x. [DOI] [PubMed] [Google Scholar]
- 30.Katragkou A, Simitsopoulou M, Chatzimoschou A, Georgiadou E, Walsh TJ, Roilides E. 2011. Effects of interferon-gamma and granulocyte colony-stimulating factor on antifungal activity of human polymorphonuclear neutrophils against Candida albicans grown as biofilms or planktonic cells. Cytokine 55:330–334. doi: 10.1016/j.cyto.2011.05.007. [DOI] [PubMed] [Google Scholar]
- 31.Chandra J, McCormick TS, Imamura Y, Mukherjee PK, Ghannoum MA. 2007. Interaction of Candida albicans with adherent human peripheral blood mononuclear cells increases C. albicans biofilm formation and results in differential expression of pro- and anti-inflammatory cytokines. Infect Immun 75:2612–2620. doi: 10.1128/IAI.01841-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harriott MM, Lilly EA, Rodriguez TE, Fidel PL Jr, Noverr MC. 2010. Candida albicans forms biofilms on the vaginal mucosa. Microbiology 156:3635–3644. doi: 10.1099/mic.0.039354-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Uppuluri P, Dinakaran H, Thomas DP, Chaturvedi AK, Lopez-Ribot JL. 2009. Characteristics of Candida albicans biofilms grown in a synthetic urine medium. J Clin Microbiol 47:4078–4083. doi: 10.1128/JCM.01377-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nikawa H, Nishimura H, Hamada T, Kumagai H, Samaranayake LP. 1997. Effects of dietary sugars and, saliva and serum on Candida biofilm formation on acrylic surfaces. Mycopathologia 139:87–91. doi: 10.1023/A:1006851418963. [DOI] [PubMed] [Google Scholar]
- 35.Chandra J, Mukherjee PK, Leidich SD, Faddoul FF, Hoyer LL, Douglas LJ, Ghannoum MA. 2001. Antifungal resistance of candidal biofilms formed on denture acrylic in vitro. J Dent Res 80:903–908. doi: 10.1177/00220345010800031101. [DOI] [PubMed] [Google Scholar]
- 36.Low FM, Hampton MB, Winterbourn CC. 2008. Peroxiredoxin 2 and peroxide metabolism in the erythrocyte. Antioxid Redox Signal 10:1621–1630. doi: 10.1089/ars.2008.2081. [DOI] [PubMed] [Google Scholar]
- 37.Kuznets G, Vigonsky E, Weissman Z, Lalli D, Gildor T, Kauffman SJ, Turano P, Becker J, Lewinson O, Kornitzer D. 2014. A relay network of extracellular heme-binding proteins drives C. albicans iron acquisition from hemoglobin. PLoS Pathog 10:e1004407. doi: 10.1371/journal.ppat.1004407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Francois P, Schrenzel J, Stoerman-Chopard C, Favre H, Herrmann M, Foster TJ, Lew DP, Vaudaux P. 2000. Identification of plasma proteins adsorbed on hemodialysis tubing that promote Staphylococcus aureus adhesion. J Lab Clin Med 135:32–42. doi: 10.1016/S0022-2143(00)70018-7. [DOI] [PubMed] [Google Scholar]
- 39.Proctor RA. 2000. Toward an understanding of biomaterial infections: a complex interplay between the host and bacteria. J Lab Clin Med 135:14–15. doi: 10.1016/S0022-2143(00)70015-1. [DOI] [PubMed] [Google Scholar]
- 40.Jenney CR, Anderson JM. 2000. Adsorbed serum proteins responsible for surface dependent human macrophage behavior. J Biomed Mater Res 49:435–447. [DOI] [PubMed] [Google Scholar]
- 41.Brash JL, Ten Hove P. 1993. Protein adsorption studies on ‘standard’ polymeric materials. J Biomater Sci Polym Ed 4:591–599. [DOI] [PubMed] [Google Scholar]
- 42.Yanagisawa N, Li DQ, Ljungh A. 2004. Protein adsorption on ex vivo catheters and polymers exposed to peritoneal dialysis effluent. Perit Dial Int 24:264–273. [PubMed] [Google Scholar]
- 43.Jakab E, Paulsson M, Ascencio F, Ljungh A. 1993. Expression of vitronectin and fibronectin binding by Candida albicans yeast cells. APMIS 101:187–193. doi: 10.1111/j.1699-0463.1993.tb00100.x. [DOI] [PubMed] [Google Scholar]
- 44.Bouali A, Robert R, Tronchin G, Senet JM. 1987. Characterization of binding of human fibrinogen to the surface of germ-tubes and mycelium of Candida albicans. J Gen Microbiol 133:545–551. [DOI] [PubMed] [Google Scholar]
- 45.Saville SP, Thomas DP, Lopez Ribot JL. 2006. A role for Efg1p in Candida albicans interactions with extracellular matrices. FEMS Microbiol Lett 256:151–158. doi: 10.1111/j.1574-6968.2006.00109.x. [DOI] [PubMed] [Google Scholar]
- 46.Sturtevant J, Calderone R. 1997. Candida albicans adhesins: biochemical aspects and virulence. Rev Iberoam Micol 14:90–97. [PubMed] [Google Scholar]
- 47.Lopez CM, Wallich R, Riesbeck K, Skerka C, Zipfel PF. 2014. Candida albicans uses the surface protein Gpm1 to attach to human endothelial cells and to keratinocytes via the adhesive protein vitronectin. PLoS One 9:e90796. doi: 10.1371/journal.pone.0090796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pendrak ML, Krutzsch HC, Roberts DD. 2000. Structural requirements for hemoglobin to induce fibronectin receptor expression in Candida albicans. Biochemistry 39:16110–16118. doi: 10.1021/bi0012585. [DOI] [PubMed] [Google Scholar]
- 49.Lopez-Ribot JL, Monteagudo C, Sepulveda P, Casanova M, Martinez JP, Chaffin WL. 1996. Expression of the fibrinogen binding mannoprotein and the laminin receptor of Candida albicans in vitro and in infected tissues. FEMS Microbiol Lett 142:117–122. [DOI] [PubMed] [Google Scholar]
- 50.Santoni G, Spreghini E, Lucciarini R, Amantini C, Piccoli M. 2001. Involvement of alpha(v)beta3 integrin-like receptor and glycosaminoglycans in Candida albicans germ tube adhesion to vitronectin and to a human endothelial cell line. Microb Pathog 31:159–172. doi: 10.1006/mpat.2001.0459. [DOI] [PubMed] [Google Scholar]
- 51.Donohue DS, Ielasi FS, Goossens KV, Willaert RG. 2011. The N-terminal part of Als1 protein from Candida albicans specifically binds fucose-containing glycans. Mol Microbiol 80:1667–1679. doi: 10.1111/j.1365-2958.2011.07676.x. [DOI] [PubMed] [Google Scholar]
- 52.Yan S, Rodrigues RG, Cahn-Hidalgo D, Walsh TJ, Roberts DD. 1998. Hemoglobin induces binding of several extracellular matrix proteins to Candida albicans. Identification of a common receptor for fibronectin, fibrinogen, and laminin. J Biol Chem 273:5638–5644. [DOI] [PubMed] [Google Scholar]
- 53.Pendrak ML, Rodrigues RG, Roberts DD. 2007. Induction of a high affinity fibronectin receptor in Candida albicans by caspofungin: requirements for beta (1,6) glucans and the developmental regulator Hbr1p. Med Mycol 45:157–168. doi: 10.1080/13693780601164314. [DOI] [PubMed] [Google Scholar]
- 54.Gaur NK, Smith RL, Klotz SA. 2002. Candida albicans and Saccharomyces cerevisiae expressing ALA1/ALS5 adhere to accessible threonine, serine, or alanine patches. Cell Commun Adhes 9:45–57. doi: 10.1080/15419060212187. [DOI] [PubMed] [Google Scholar]
- 55.Nikawa H, Nishimura H, Makihira S, Hamada T, Sadamori S, Samaranayake LP. 2000. Effect of serum concentration on Candida biofilm formation on acrylic surfaces. Mycoses 43:139–143. doi: 10.1046/j.1439-0507.2000.00564.x. [DOI] [PubMed] [Google Scholar]
- 56.Nikawa H, Nishimura H, Hamada T, Yamashiro H, Samaranayake LP. 1999. Effects of modified pellicles on Candida biofilm formation on acrylic surfaces. Mycoses 42:37–40. doi: 10.1046/j.1439-0507.1999.00270.x. [DOI] [PubMed] [Google Scholar]
- 57.Byers JD, Ratner BD. 2004. Bioinspired implant materials befuddle bacteria. ASM News 70:232–237. [Google Scholar]
- 58.Nemet B, Zagar Z. 2000. Fibronectin concentrations in catheter sepsis. Clin Microbiol Infect 6:121–124. doi: 10.1046/j.1469-0691.2000.00029.x. [DOI] [PubMed] [Google Scholar]
- 59.Lionakis MS, Lim JK, Lee CC, Murphy PM. 2011. Organ-specific innate immune responses in a mouse model of invasive candidiasis. J Innate Immun 3:180–199. doi: 10.1159/000321157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dongari-Bagtzoglou A, Kashleva H, Dwivedi P, Diaz P, Vasilakos J. 2009. Characterization of mucosal Candida albicans biofilms. PLoS One 4:e7967. doi: 10.1371/journal.pone.0007967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Urban CF, Reichard U, Brinkmann V, Zychlinsky A. 2006. Neutrophil extracellular traps capture and kill Candida albicans yeast and hyphal forms. Cell Microbiol 8:668–676. doi: 10.1111/j.1462-5822.2005.00659.x. [DOI] [PubMed] [Google Scholar]
- 62.Xie Z, Thompson A, Sobue T, Kashleva H, Xu H, Vasilakos J, Dongari-Bagtzoglou A. 2012. Candida albicans biofilms do not trigger reactive oxygen species and evade neutrophil killing. J Infect Dis 206:1936–1945. doi: 10.1093/infdis/jis607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Katragkou A, Chatzimoschou A, Simitsopoulou M, Georgiadou E, Roilides E. 2011. Additive antifungal activity of anidulafungin and human neutrophils against Candida parapsilosis biofilms. J Antimicrob Chemother 66:588–591. doi: 10.1093/jac/dkq466. [DOI] [PubMed] [Google Scholar]
- 64.Yano J, Lilly E, Barousse M, Fidel PL Jr. 2010. Epithelial cell-derived S100 calcium-binding proteins as key mediators in the hallmark acute neutrophil response during Candida vaginitis. Infect Immun 78:5126–5137. doi: 10.1128/IAI.00388-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yano J, Noverr MC, Fidel PL Jr. 2012. Cytokines in the host response to Candida vaginitis: identifying a role for non-classical immune mediators, S100 alarmins. Cytokine 58:118–128. doi: 10.1016/j.cyto.2011.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rehg JE, Bush D, Ward JM. 2012. The utility of immunohistochemistry for the identification of hematopoietic and lymphoid cells in normal tissues and interpretation of proliferative and inflammatory lesions of mice and rats. Toxicol Pathol 40:345–374. doi: 10.1177/0192623311430695. [DOI] [PubMed] [Google Scholar]
- 67.Meijer AH, van der Sar AM, Cunha C, Lamers GE, Laplante MA, Kikuta H, Bitter W, Becker TS, Spaink HP. 2008. Identification and real-time imaging of a myc-expressing neutrophil population involved in inflammation and mycobacterial granuloma formation in zebrafish. Dev Comp Immunol 32:36–49. doi: 10.1016/j.dci.2007.04.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.