Abstract
Clostridium difficile is the most common cause of antibiotic-associated diarrhoea worldwide. Over the past 10 years, the incidence and severity of disease have increased in North America and Europe due to the emergence of a hypervirulent clone designated PCR ribotype 027. In this study, we sought to identify phenotypic differences among a collection of 26 presumed PCR ribotype 027 strains from the US and the UK isolated between 1988 and 2008 and also re-evaluated the PCR ribotype. We demonstrated that some of the strains typed as BI by restriction endonuclease analysis, and presumed to be PCR ribotype 027, were in fact other PCR ribotypes such as 176, 198 and 244 due to slight variation in banding pattern compared to the 027 strains. The reassigned 176, 198 and 244 ribotype strains were isolated in the US between 2001 and 2004 and appeared to have evolved recently from the 027 lineage. In addition, the UK strains were more motile and more resistant to most of the antibiotics compared to the US counterparts. We conclude that there should be a heightened awareness of newly identified PCR ribotypes such as 176, 198 and 244, and that they may be as problematic as the notorious 027 strains.
INTRODUCTION
Clostridium difficile is a Gram-positive, anaerobic, spore-forming bacillus that is the causative agent of C. difficile infection (CDI) (Bartlett, 1994). CDI is often caused after broad-spectrum antimicrobial therapy, which disrupts the barrier effect of the endogenous intestinal microflora allowing C. difficile spores to germinate, colonize the gastrointestinal tract and produce toxins, which causes tissue damage (Just et al., 1995, 2001). Antibiotic resistance is likely to be important in infection, as it would provide C. difficile a competitive growth advantage in the gut of patients after antibiotic treatment (Delaney et al., 2007). Colonization is also an important step in C. difficile pathogenesis, which involves various determinants including surface-layer proteins SLPs, adhesins (P47, Cwp66 and Fbp68) (Calabi & Fairweather, 2002; Hennequin et al., 2003; Waligora et al., 2001) and flagella, which have been implicated in adherence of C. difficile to caecal mucus in axenic mice (Tasteyre et al., 2001). Flagella have multiple roles in virulence of other enteric pathogens including motility through the viscous intestinal mucosa, chemotaxis, protein secretion and interaction with the innate immune system (Feuillet et al., 2006; Lee et al., 1986; Milton et al., 1996; Moulton & Montie, 1979; Pruckler et al., 1995).
In the last few years, the appearance of highly virulent and epidemic C. difficile strains has significantly changed the epidemiology of CDI in North America and Europe (Kuijper et al., 2007; Pépin et al., 2005). This is largely due to the emergence of a clonal lineage referred to using different typing methods as BI [restriction endonuclease analysis (REA)], NAP1 (PFGE), 027 (PCR ribotype) (Killgore et al., 2008) and different clusters [multiple-locus variable-number tandem-repeat analysis (MLVA)] (Eckert et al., 2011). The hypervirulent NAP1 types can be subtyped as NAP1, NAP1a or NAP1c variants, whereas there are at least 23 variants of the REA types, BI-1 to BI-23 (Killgore et al., 2008). MLVA can subtype in different clusters (Eckert et al., 2011). All these strains are invariably classified as toxinotype III by toxinotyping (Rupnik et al., 1998).
The first documented PCR ribotype 027 was described as an isolate from a Parisian hospital in 1985 (Popoff et al., 1988). It was occasionally isolated in the 1990s in the US until major outbreaks of 027-related CDI emerged in 2003 (Morris et al., 2002; Redelings et al., 2007; Ricciardi et al., 2007). There has been a threefold increase in CDI in the elderly, largely as a result of the emergence of the 027 strains during 2000–2005 (Jagai & Naumova, 2009). In the UK, major outbreaks emerged in 2006 where the PCR ribotype 027 was identified. The proportion of 027 strains isolated in UK hospitals rose sharply from 25.9 % to 41.3 % between 2005 and 2008 (Brazier et al., 2008). Patients infected with 027 strains have more severe diarrhoea, higher mortality and more recurrences (Goorhuis et al., 2007; Hubert et al., 2007; Loo et al., 2005; Mooney, 2007; Redelings et al., 2007).
A whole genome comparison method using microarrays (comparative phylogenomics) demonstrated that most NAP1/BI/027 isolates formed a single clonal lineage, termed the hypervirulent clade (Griffiths et al., 2010; He et al., 2010; Sebaihia et al., 2006). However, there were exceptions: a single BI-9 (NAP1c) clustered outside the clonal lineage (He et al., 2010; Stabler et al., 2006), identified later as PCR ribotype 001 (He et al., 2010), and BI-14 (NAP1) was an outlier to the hypervirulent clade (Stabler et al., 2006). Interestingly, a previous report showed that not all NAP1 isolates are PCR ribotype 027. For example, strain CA10 was NAP1, but PCR ribotype 019 (Killgore et al., 2008). MLVA has been used to discriminate between isolates with identical PCR ribotypes belonging to types 001, 017 and 027 (van den Berg et al., 2007). Given that many of the PCR-ribotyped C. difficile isolates in the US and UK are 027, it is important to develop more accurate methods to distinguish between these highly virulent strains. Recently, we fully sequenced the genomes of two 027 isolates, an historic strain (CD196, the original strain isolated in Paris in 1985) and a modern strain (R20291, the index 027 strain isolated in the UK in 2006), which revealed some genetic differences between them (Stabler et al., 2009). More recently, Nyč et al. (2011) reported an outbreak of CDI in the Czech Republic in which the strains were PCR ribotype 176 and closely related to 027 strains. In this study, we determined phenotypic differences in 26 presumed 027 strains from the US and the UK, isolated between 1988 and 2008, and re-evaluated their PCR ribotype. We show that the UK and US 027 strains have distinct motility and antibiotic resistance profiles and demonstrate that some strains previously assumed to be 027 are different PCR ribotypes such as 176, 198 and 244.
METHODS
The 26 human clinical isolates examined in this study are summarized in Table 1. These include BI-1 to BI-15 (provided by Dale Gerding, Hines VA Hospital, USA) and ten PCR ribotype 027 strains, 027-01 to 027-10 (provided by Derek J. Brown, Glasgow, Scotland). In addition, M120 (provided by Denise Drudy, University College, Dublin) and R20291 (provided by Jon Brazier, Cardiff, Wales) were used as controls. C. difficile was routinely grown on Brazier’s CCEY agar (BioConnections) containing 4 % egg yolk, C. difficile supplement (BioConnections) and 1 % defibrinated horse blood (TCS Biosciences), in brain heart infusion (BHI) broth (Oxoid) supplemented with 2.5 % l-cysteine (Sigma Aldrich) and C. difficile supplement (Fluka) and on blood agar (agar base; Oxoid) supplemented with 7 % defibrinated horse blood (TCS Biosciences). All cultures were grown from glycerol stocks in an anaerobic atmosphere (10 % CO2, 10 % H2, 80 % N2) at 37 °C.
Table 1.
Origin and source of C. difficile isolates (1998–2008)
| Code | Isolate reference number | REA type | Date isolated/received | City, state/province | Ribotype |
|---|---|---|---|---|---|
| BI-1 | 1675 | BI-1 | 26/02/1988 | Minneapolis, MN | 027 |
| BI-2 | 4272 | BI-2 | 14/01/1991 | Tucson, AZ | 027 |
| BI-3 | 4233 | BI-3 | 14/12/1990 | Minneapolis, MN | 027 |
| BI-4 | 5325 | BI-4 | 10/02/1993 | Minneapolis, MN | 027 |
| BI-5 | 5604 | BI-5 | 25/08/1995 | Albany, NY | 027 |
| BI-11 | 6296 | BI-11 | 10/08/2001 | Pittsburgh, PA | 198 |
| BI-10 | 6289 | BI-10 | 10/08/2001 | Pittsburgh, PA | 027 |
| BI-6 | 6336 | BI-6 | 20/05/2003 | Portland, OR | 176 |
| BI-7 | 6335 | BI-7 | 20/05/2003 | Portland, OR | 027 |
| BI-8 | 6367 | BI-8 | 22/01/2004 | Portland, ME | 027 |
| BI-6p | 6413 | BI-6p | 09/09/2004 | Atlanta, GA | 027 |
| BI-6p2 | 6431 | BI-6p2 | 09/09/2004 | New Jersey | 027 |
| BI-12 | 6425 | BI-12 | 09/09/2004 | Camp Hill, PA | 027 |
| BI-13 | 6430 | BI-13 | 09/09/2004 | New Jersey | 027 |
| BI-14 | 6432 | BI-14 | 09/09/2004 | New Jersey | 244 |
| BI-15 | 6436 | BI-15 | 09/09/2004 | New Jersey | 027 |
| R20291 | R20291 | 2006 | Aylesbury, England | 027 | |
| 027-01 | 20070031 | 13/12/2007 | Dundee, Scotland | 027 | |
| 027-02 | 20070036 | 14/12/2007 | Glasgow, Scotland | 027 | |
| 027-03 | 20080090 | 03/04/2008 | Aberdeen, Scotland | 027 | |
| 027-04 | 20080107 | 07/04/2008 | Dumfries, Scotland | 027 | |
| 027-05 | 20080195 | 22/05/2008 | Ayshire, Scotland | 027 | |
| 027-06 | 20080238 | 05/06/2008 | Dumbarton, Scotland | 027 | |
| 027-07 | 20080323 | 26/06/2008 | Edinburgh, Scotland | 027 | |
| 027-08 | 20080533 | 06/08/2008 | Dumbarton, Scotland | 027 | |
| 027-09 | 20080684 | 18/09/2008 | Inverness, Scotland | 027 | |
| 027-10 | 20080783 | 22/10/2008 | Glasgow, Scotland | 027 | |
| M120 | M120 | 2007 | Ireland | 078 |
PCR ribotyping
PCR ribotyping was performed at least in duplicate. Briefly, bacteria were harvested from 48 h anaerobic cultures on blood agar. Cells were resuspended into a 5 % (w/v) solution of Chelex-100 (Bio-Rad) and heated to 100 °C for 12 min. The suspension was separated by centrifugation (13 000 g for 12 min) and the supernatant (10 μl) was added to a 100 μl PCR mixture containing 25 μM each primer (P3, 5′-CTGGGGTGAAGTCGTAACAAGG-3′; and P5, 5′-GCGCCCTTTGTAGCTTGACC-3′), 2.5 units Qiagen HotStar Taq DNA polymerase per reaction, 0.4 mM dNTPs and 3.75 mM MgCl2 per reaction. The reaction mixture was subjected to 30 cycles of 9 °C for 1 min, 92 °C for 1 min, 55 °C for 1 min and 72 °C for 1.5 min. This was followed by 95 °C for 1 min, 55 °C for 45 s and 72 °C for 5 min. The PCR products were concentrated to 40 μl by heating at 75 °C. Electrophoresis was done at 100 mA in 3 % pre-cast Bio-Rad 0.5 % Tris/acetate-EDTA (TAE) agarose gels containing ethidium bromide for 3.5 h at room temperature using pre-chilled TAE buffer. Banding patterns were analysed using GelCompar software (Applied Maths). R20291 was used as a PCR ribotype 027 control.
Antibiotic susceptibility test
A range of antibiotics were used to determine the relative susceptibility to clindamycin, erythromycin, chloramphenicol, tetracycline and fluoroquinolones (moxifloxacin, gatifloxacin and levofloxacin). Strain C. difficile R20291 with known MICs (Drudy et al., 2006; Stabler et al., 2009) was used as a control. The interpretation of MIC results was based on the recommendations given by the guidelines of the Clinical and Laboratory Standards Institute (CLSI, 2008) for all antibiotics used in the present study (Table 2).
Table 2.
Antibiotic breakpoints for anaerobes from Clinical and Laboratory Standards Institute guidelines (CLSI, 2008)
| Antibiotics | Susceptible (μg ml−1) | Resistant (μg ml−1) |
|---|---|---|
| Clindamycin | ⩽2 | ⩾8 |
| Chloramphenicol | ⩽8 | ⩾32 |
| Tetracycline | ⩽8 | ⩾32 |
| Moxifloxacin | ⩽2 | ⩾8 |
| Levofloxacin* | ⩽2 | ⩾8 |
| Gatifloxacin | ⩽2 | ⩾8 |
Levofloxacin breakpoints could be ⩽2 μg ml−1 (susceptible) and ⩾8 μg ml−1 (resistant) and erythromycin breakpoints could be ⩽0.5 μg ml−1 (susceptible) and ⩾32 μg ml−1 (resistant), as no standard has been defined by the CLSI for anaerobes.
Antibiotic susceptibility was determined using the Etest method in all the cases, and the broth dilution method was also used to confirm some of the Etest results. In this case antibiotics were supplied by Sigma-Aldrich.
For the Etest method, a bacterial suspension in BHI to approximately OD600=0.5 (McFarland standard no. 3) was plated on blood agar and allowed to dry for 15–30 min. Etest strips (AB Biodisk) were placed onto agar surfaces. Agar plates were incubated anaerobically at 37 °C for a further 24 h, and MICs were determined following the manufacturer’s instructions. These were performed in triplicate and R20291 was used as a control (Stabler et al., 2009).
In the broth dilution method, bacteria were grown in BHI broth until an OD600 of 0.3. Antibiotics were dissolved according to the manufacturer’s instructions (Sigma) and tested at a range of 1–128 μg ml−1. The MIC was taken as the lowest concentration to inhibit completely visible growth after 24 h growth in an anaerobic chamber at 37 °C. The final MICs were calculated as the mean among the three performed replicates.
Motility assay
Motility assay was performed according to the Stabler et al. (2009) protocol. Briefly, C. difficile cultures were grown anaerobically for 1–2 days on BHI agar. BHI broth plus 0.05 % agar was poured into 30 ml glass vials and placed into an anaerobic chamber. Three single colonies were picked with a loop, inoculated into the top 2–5 mm of BHI agar in the glass vial and left overnight in the anaerobic chamber. The vials were then removed from the chamber and photographed to record the motility. The maximum stalactite length was taken as a measure for motility. The length of those stalactite projections was scored as: <1 cm, non-motile strains; 1–2 cm, motile strain; and >2 cm, highly motile strain. M120 is a non-motile PCR ribotype 078 strain and R20291 is a motile PCR ribotype 027 strain that were used as negative and positive controls, respectively. Experiments were performed in triplicate.
Autoagglutination assay
An autoagglutination assay was performed according to Stabler et al. (2009). C. difficile strains were grown on BHI agar for 1–2 days and then inoculated into pre-reduced 1× PBS to an OD600 of 1.0 (±0.1). Then, 2 ml in triplicate was added to pre-reduced glass tubes and incubated for 24 h at 37 °C, after which 1 ml was removed from the tube surface to measure the OD600. The results were normalized to the starting OD using the equation 100–[(final OD–starting OD)×100] to show the actual autoagglutination percentage. Strain M120 was used as a positive control as this strain shows more than 95 % autoagglutination; R20291 was used as an autoagglutination negative control as it exhibits a low percentage of autoagglutination. Experiments were performed in triplicate.
Statistical analysis
Autoagglutination and motility data were analysed by Tukey’s multiple-comparison test using Prism software 9 version 4.0 (GraphPad Software). P<0.05 was considered statistically significant.
RESULTS
PCR ribotyping
The strains examined in this study are summarized in Table 1. The 16 US isolates were from multiple States between 1988 and 2004 and pre-date the UK strains, which include the UK index strain R20291 isolated in England in 2006 and 10 strains isolated from different regions in Scotland between 2007 and 2008.
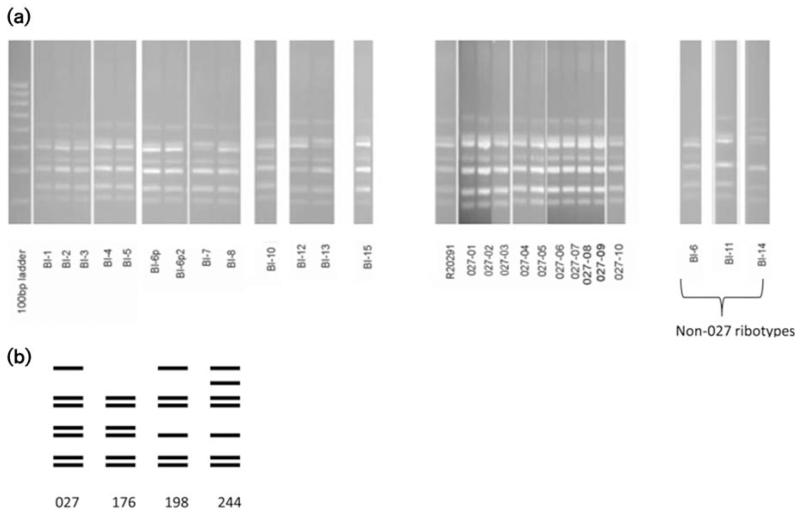
Twenty-three of the 26 isolates were confirmed as PCR ribotype 027. The other three strains were different PCR ribotypes: BI-6 was PCR ribotype 176, BI-11 was PCR ribotype 198 and BI-14 was PCR ribotype 244 (Table 1). The PCR ribotype banding profile of 027 strains consisted of seven distinct bands (Fig. 1a, b). Interestingly, the PCR ribotype 198 (BI-11) and PCR ribotype 176 (BI-6) patterns showed a high level of similarity to the PCR ribotype 027 pattern, differing by just a single band. However, strain BI-14 (PCR ribotype 244) differed by the absence of a band and the addition of a different band (Fig. 1a, b). R20291 was used as a control for the 027 PCR ribotype.
Fig. 1.
PCR ribotype profiles obtained with C. difficile strains. (a) The PCR ribotype banding profile of 027 strains consisted of seven distinct bands (see 027 control). The non-027 strains, PCR ribotype 198 (BI-11) and PCR ribotype 176 (BI-6), differed by just a single band. However, strain BI-14 (PCR ribotype 244) differed by the absence of a band and the addition of a different band. (b) PCR pattern of PCR ribotypes 027, 198, 176 and 244.
Antibiotic resistance
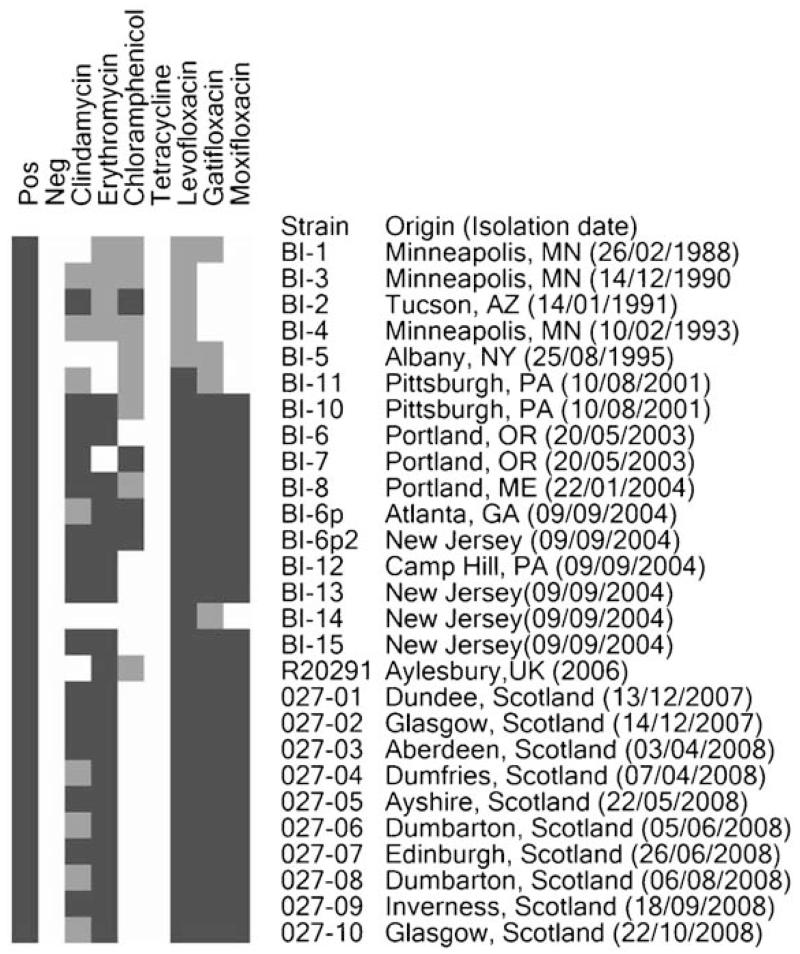
Antibiotic susceptibility was tested for all isolates (Fig. 2). The majority of PCR ribotype 027 strains exhibited either intermediate resistance (8/26 strains) (4.5–6 μg ml−1) or full resistance (15/26 strains) (MIC ≥128 μg ml−1) to clindamycin as well as full resistance to erythromycin (18/26 strains) (MIC ≥128 μg ml−1). The only isolates susceptible to both clindamycin and erythromycin were BI-5 (MIC=2 and 1.75 μg ml−1, respectively) and BI-14 (MIC=2 and 1.5 μg ml−1, respectively). All UK strains and some US strains were highly resistant to fluoroquinolones (20/26 strains) (MIC ≥32 μg ml−1). However, some US strains were fluoroquinolone-susceptible to moxifloxacin (7/26 strains) (MIC=1 μg ml−1), intermediately resistant to levofloxacin (5/26 strains) (MIC=4 μg ml−1) and gatifloxacin-susceptible and/or intermediately resistant (3/26 and 4/26 strains, respectively) (Fig. 2).
Fig. 2.
Antibiotic resistance heat map. All 26 strains were tested for their susceptibility to clindamycin, erythromycin, chloramphenicol, tetracycline and fluoroquinolones (levofloxacin, gatifloxacin, mocifloxacin). Black represents resistant, grey represents intermediately resistant and white represents susceptible.
BI-1 to BI-5 strains, isolated prior to 2003, showed either intermediate resistance (MIC=16 μg ml−1) or were fully resistant (MIC ≥128 μg ml−1) to chloramphenicol, whereas the other strains were susceptible to this antibiotic in most cases (18/26 strains) (MIC=5–12 μg ml−1). Only BI-7, BI-6p and BI-6p2 were resistant (MIC 18–24 μg ml−1) (Fig. 2). All the strains tested were susceptible to tetracycline (MIC <0.7 μg ml−1). R20291 was used as a control for antibiotic resistance.
Motility
There was clear and reproducible evidence of motility among 027 isolates (Fig. 3). US strains (16/26 strains) were motile (stalactite length range: 1.6–1.8 cm) whereas UK strains (10/26 strains) were highly motile (stalactite length range: 2.4–3 cm) (P<0.05) (Fig. 3). The M120 strain is a non-motile negative control (0.7 cm) and R20291 is a highly motile 027 strain positive control (3 cm) (Fig. 3).
Fig. 3.
Relative motility assays for C. difficile strains. Strains were inoculated into 0.05 % BHI agar and incubated for 24 h in an anaerobic chamber. Motility is visualized as stalactite projections. M120 and R20291 are negative and positive controls, respectively. Bar, 1.2 cm.
Autoagglutination
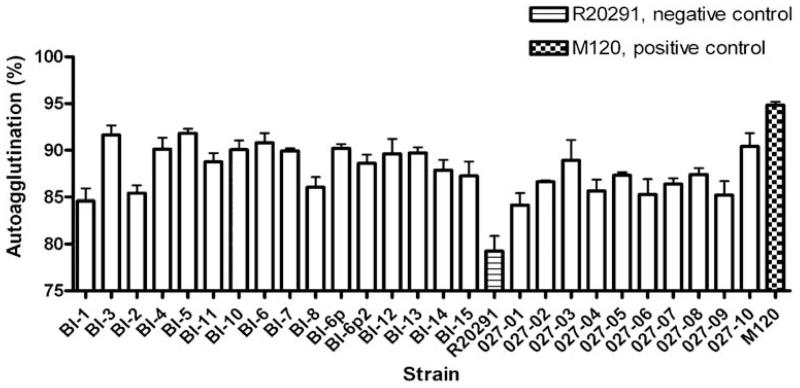
Autoagglutination is often used to demonstrate charge differences on bacterial cells that can be affected by the presence of flagella and how they are modified (Howard et al., 2009). The percentage of autoagglutination was tested for all the isolates. We found a very heterogeneous representation of the autoagglutination phenotype among the strains (Fig. 4).
Fig. 4.
Relative autoagglutination of C. difficile strains. C. difficile strains were grown on BHI plates for 1–2 days, then inoculated into pre-equilibrated PBS to an OD600 of 1.0 (±0.1). These were incubated for 24 h in pre-equilibrated glass tubes, and then the OD600 was measured. The percentage autoagglutination was normalized to the starting OD600. The bars indicate the percentage of cells autoagglutinating. M120 was used as a positive control.
DISCUSSION
Rates and severity of CDI have increased alarmingly in recent years and are in part attributable to the emergence and spread of the 027 clonal lineage (Goorhuis et al., 2007; Hubert et al., 2007; Loo et al., 2005; Mooney, 2007). The 26 presumed PCR ribotype 027 strains investigated in this study were isolated throughout the UK and the US over a 20-year period. The early strains (1988–2004) were from different States in the US and the later strains (2006–2008) were from the UK (Table 1), which presumably emerged from North America. Interestingly, these isolates are generally referred to as BI/NAP1/027, with the assumption that BI types and NAP1 types are PCR ribotype 027. However, we have shown that the REA type ‘BI’ and the PFGE type NAP1 do not always correlate with PCR ribotype 027. PCR ribotype analysis revealed that BI-6, BI-11 and BI-14 were not PCR ribotype 027 as previously presumed. BI-14 was an outlier strain in a previous study (Stabler et al., 2006). In our study, BI-14 had two different bands compared to PCR ribotype 027, being assigned as PCR ribotype 244. However, the similarity of the PCR ribotype banding pattern for BI-11 (PCR ribotype 198) and BI-6 (PCR ribotype 176) was high. Slight band differences between PCR ribotype 176 and presumed PCR ribotype 027 C. difficile isolates have also been observed in Austria (A. Indra, personal communication in 2010) and more recently in the Czech Republic and in Poland (Nyč et al., 2011). Despite being different ribotypes, it is clear that the BI-11 (PCR ribotype 198) and BI-6 (PCR ribotype 176) strains are closely related to 027 strains and have previously been placed in the same clade using comparative genome microarray analyses (Stabler et al., 2006). High similarity in PCR ribotyping pattern between 027 and other ribotypes should be monitored, as strains from these ribotypes may be as problematic as the hypervirulent 027 strains.
CDI is frequently linked to treatment with antibiotics which provide an advantage for C. difficile survival (Delaney et al., 2007). In this study, we showed that UK strains were resistant to most of the antibiotics tested. However US strains showed a variable range of antibiotic resistance depending on the antibiotic. It is a reasonable hypothesis that these changes in antibiotic resistance profiles could be linked to the changes in antibiotic prescription policy over time and in different countries. The most noteworthy is the rise in resistance to fluoroquinolones such as levofloxacin, gatifloxacin and moxifloxacin. With few exceptions, antibiotic resistance in C. difficile is transposon-mediated: erythromycin resistance (ermB) is carried on Tn5398 (Hussain et al., 2005; Sebaihia et al., 2006) or CTnCD11 (He et al., 2010), chloramphenicol resistance (catD) is carried on Tn4453 (Lyras et al., 2004) and tetracycline resistance is carried on Tn5397 (Hussain et al., 2005). Previous studies identified a novel transposon in R20291, CTn-027, encoding a chloramphenicol resistance gene (CDR20291_3461) in some PCR ribotype 027 strains (Stabler et al., 2009). However, all UK strains with the exception of R20291 (used as a control) and some US strains were chloramphenicol-susceptible with some exceptions, indicating that the transmission of this transposon-mediated antibiotic resistance might not be uniform throughout the 027 lineage. On the other hand, tetracycline resistance (tetM), usually carried on Tn5397, was absent in all 26 strains tested. Generally, resistance to macrolides (e.g. erythromycin) and lincosamides (e.g. clindamycin) is mediated via the presence of the ermB gene and it is more common in 027 isolates (Solomon et al., 2011). Although 14 strains were erythromycin- and clindamycin-resistant, TnCD11 carrying ermB was only present in five of the BI isolates (BI-6, BI-7, BI-10, BI-13 and BI-15) analysed by whole genome analysis (He et al., 2010). This suggests an alternative mechanism for ermB resistance in the 027 lineage. The development of antibiotic resistance in modern 027 strains emphasizes the importance of antibiotic susceptibility testing for the emergence of antibiotic resistance.
The flagella-associated genes in strain 630 (PCR ribotype 012) are found in two loci, F1 and F3, separated by an interflagella locus, F2 (Stabler et al., 2009). The level of sequence identity in 027 strains compared to the 630 strain for the F1 and F3 region was high, but there were significant differences in the F2 region (Stabler et al., 2009). This may provide a genetic basis for the motility differences among 027 isolates used in this study, but to date available genetic information on the strains in this study cannot explain the difference in motility between the 027 strains. Modifications in motility and antibiotic resistance between the 027 strains may reflect a genetic change with respect to the flagella and glycosylation loci as well as the loss and gain of transposons and the accumulation of mutations over time. Such changes could be beneficial for invasion, adhesion, access to nutrients and general survival and transmissibility of C. difficile.
Understanding the evolution of clones such as the PCR ribotype 027 will be important in predicting the early emergence (or disappearance) of highly virulent C. difficile strains and represents a current public health imperative. In this study, phenotypic differences between the 26 UK and US strains suggest that the PCR ribotype 027 is genetically variable. It appears that the 176 and 198 ribotypes have evolved recently from the 027 lineage. Thus there should be a heightened awareness that the recently identified PCR ribotypes 176 and 198 may be as problematic as the 027 strains.
ACKNOWLEDGEMENTS
We acknowledge the Wellcome Trust for funding this research. M. D. C. and E. V. acknowledge the Health Protection Agency Collaborating Centre at UCLH and the Fundación Alfonso Martinez Escudero, respectively, for funding. We thank Derek Brown, Dale Gerding, Jon Brazier and Denise Drudy for provision of C. difficile strains.
Abbreviations
- CDI
Clostridium difficile infection
- MLVA
multiple-locus variable-number tandem-repeat analysis
- REA
restriction endonuclease analysis
REFERENCES
- Bartlett JG. Clostridium difficile: history of its role as an enteric pathogen and the current state of knowledge about the organism. Clin Infect Dis. 1994;18(Suppl. 4):S265–S272. doi: 10.1093/clinids/18.supplement_4.s265. [DOI] [PubMed] [Google Scholar]
- Brazier JS, Raybould R, Patel B, Duckworth G, Pearson A, Charlett A, Duerden BI, HPA Regional Microbiology Network Distribution and antimicrobial susceptibility patterns of Clostridium difficile PCR ribotypes in English hospitals, 2007-08. Euro Surveill. 2008;13 doi: 10.2807/ese.13.41.19000-en. pii:19000. [DOI] [PubMed] [Google Scholar]
- Calabi E, Fairweather N. Patterns of sequence conservation in the S-layer proteins and related sequences in Clostridium difficile. J Bacteriol. 2002;184:3886–3897. doi: 10.1128/JB.184.14.3886-3897.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CLSI . Performance Standards for Antimicrobial Susceptibility Testing; 18th Informational Supplement. Clinical and Laboratory Standards Institute; Wayne, PA: 2008. M100-S18. [Google Scholar]
- Delaney JA, Dial S, Barkun A, Suissa S. Antimicrobial drugs and community-acquired Clostridium difficile-associated disease, UK. Emerg Infect Dis. 2007;13:761–763. doi: 10.3201/eid1305.061124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drudy D, Quinn T, O’Mahony R, Kyne L, O’Gaora P, Fanning S. High-level resistance to moxifloxacin and gatifloxacin associated with a novel mutation in gyrB in toxin-A-negative, toxin-B-positive Clostridium difficile. J Antimicrob Chemother. 2006;58:1264–1267. doi: 10.1093/jac/dkl398. [DOI] [PubMed] [Google Scholar]
- Eckert C, Vromman FO, Halkovich A, Barbut F. Multilocus variable-number tandem repeat analysis: a helpful tool for subtyping French Clostridium difficile PCR ribotype 027 isolates. J Med Microbiol. 2011;60:1088–1094. doi: 10.1099/jmm.0.029009-0. [DOI] [PubMed] [Google Scholar]
- Feuillet V, Medjane S, Mondor I, Demaria O, Pagni PP, Galán JE, Flavell RA, Alexopoulou L. Involvement of Toll-like receptor 5 in the recognition of flagellated bacteria. Proc Natl Acad Sci U S A. 2006;103:12487–12492. doi: 10.1073/pnas.0605200103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goorhuis AVKT, Van der Kooi T, Vaessen N, Dekker FW, Van den Berg R, Harmanus C, van den Hof S, Notermans DW, Kuijper EJ. Spread and epidemiology of Clostridium difficile polymerase chain reaction ribotype 027/toxinotype III in The Netherlands. Clin Infect Dis. 2007;45:695–703. doi: 10.1086/520984. [DOI] [PubMed] [Google Scholar]
- Griffiths D, Fawley W, Kachrimanidou M, Bowden R, Crook DW, Fung R, Golubchik T, Harding RM, Jeffery KJ, et al. Multilocus sequence typing of Clostridium difficile. J Clin Microbiol. 2010;48:770–778. doi: 10.1128/JCM.01796-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He M, Sebaihia M, Lawley TD, Stabler RA, Dawson LF, Martin MJ, Holt KE, Seth-Smith HM, Quail MA, et al. Evolutionary dynamics of Clostridium difficile over short and long time scales. Proc Natl Acad Sci U S A. 2010;107:7527–7532. doi: 10.1073/pnas.0914322107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Healthcare Commission Healthcare Commission Investigation into outbreaks of Clostridium difficile at Stoke Mandeville Hospital, Buckinghamshire Hospitals NHS Trust. 2006 http://www.cqc.org.uk/_db/_documents/Stoke_Mandeville.pdf.
- Hennequin C, Janoir C, Barc MC, Collignon A, Karjalainen T. Identification and characterization of a fibronectin-binding protein from Clostridium difficile. Microbiology. 2003;149:2779–2787. doi: 10.1099/mic.0.26145-0. [DOI] [PubMed] [Google Scholar]
- Howard SL, Jagannathan A, Soo EC, Hui JP, Aubry AJ, Ahmed I, Karlyshev A, Kelly JF, Jones MA, et al. Campylobacter jejuni glycosylation island important in cell charge, legionaminic acid biosynthesis, and colonization of chickens. Infect Immun. 2009;77:2544–2556. doi: 10.1128/IAI.01425-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubert BLV, Loo VG, Bourgault AM, Poirier L, Dascal A, Fortin E, Dionne M, Lorange M. A portrait of the geographic dissemination of the Clostridium difficile North American pulsed-field type 1 strain and the epidemiology of C. difficile-associated disease in Québec. Clin Infect Dis. 2007;44:238–244. doi: 10.1086/510391. [DOI] [PubMed] [Google Scholar]
- Hussain HA, Roberts AP, Mullany P. Generation of an erythromycin-sensitive derivative of Clostridium difficile strain 630 (630ΔΔerm) and demonstration that the conjugative transposon Tn916ΔE enters the genome of this strain at multiple sites. J Med Microbiol. 2005;54:137–141. doi: 10.1099/jmm.0.45790-0. [DOI] [PubMed] [Google Scholar]
- Jagai J, Naumova E. Clostridium difficile-associated disease in the elderly, United States. Emerg Infect Dis. 2009;15:343–344. doi: 10.3201/eid1502.080785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Just I, Wilm M, Selzer J, Rex G, von Eichel-Streiber C, Mann M, Aktories K. The enterotoxin from Clostridium difficile (ToxA) monoglucosylates the Rho proteins. J Biol Chem. 1995;270:13932–13936. doi: 10.1074/jbc.270.23.13932. [DOI] [PubMed] [Google Scholar]
- Just I, Hofmann F, Genth H, Gerhard R. Bacterial protein toxins inhibiting low-molecular-mass GTP-binding proteins. Int J Med Microbiol. 2001;291:243–250. doi: 10.1078/1438-4221-00127. [DOI] [PubMed] [Google Scholar]
- Killgore G, Thompson A, Johnson S, Brazier J, Kuijper E, Pepin J, Frost EH, Savelkoul P, Nicholson B, et al. Comparison of seven techniques for typing international epidemic strains of Clostridium difficile: restriction endonuclease analysis, pulsed-field gel electrophoresis, PCR-ribotyping, multilocus sequence typing, multilocus variable-number tandem-repeat analysis, amplified fragment length polymorphism, and surface layer protein A gene sequence typing. J Clin Microbiol. 2008;46:431–437. doi: 10.1128/JCM.01484-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuijper EJ, Coignard B, Brazier JS, Suetens C, Drudy D, Wiuff C, Pituch H, Reichert P, Schneider F, et al. Update of Clostridium difficile-associated disease due to PCR ribotype 027 in Europe. Euro Surveill. 2007;12:E1–E2. doi: 10.2807/esm.12.06.00714-en. [DOI] [PubMed] [Google Scholar]
- Lee A, O’Rourke JL, Barrington PJ, Trust TJ. Mucus colonization as a determinant of pathogenicity in intestinal infection by Campylobacter jejuni: a mouse cecal model. Infect Immun. 1986;51:536–546. doi: 10.1128/iai.51.2.536-546.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo VGPL, Poirier L, Miller MA, Oughton M, Libman MD, Michaud S, Bourgault AM, Nguyen T, Frenette C, et al. A predominantly clonal multi-institutional outbreak of Clostridium difficile-associated diarrhea with high morbidity and mortality. N Engl J Med. 2005;353:2442–2449. doi: 10.1056/NEJMoa051639. [DOI] [PubMed] [Google Scholar]
- Lyras D, Adams V, Lucet I, Rood JI. The large resolvase TnpX is the only transposon-encoded protein required for transposition of the Tn4451/3 family of integrative mobilizable elements. Mol Microbiol. 2004;51:1787–1800. doi: 10.1111/j.1365-2958.2003.03950.x. [DOI] [PubMed] [Google Scholar]
- Milton DL, O’Toole R, Horstedt P, Wolf-Watz H. Flagellin A is essential for the virulence of Vibrio anguillarum. J Bacteriol. 1996;178:1310–1319. doi: 10.1128/jb.178.5.1310-1319.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooney H. Annual incidence of MRSA falls in England, but C. difficile continues to rise. BMJ. 2007;335:958. doi: 10.1136/bmj.39388.597569.DB. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris AM, Jobe BA, Stoney M, Sheppard BC, Deveney CW, Deveney KE. Clostridium difficile colitis: an increasingly aggressive iatrogenic disease? Arch Surg. 2002;137:1096–1100. doi: 10.1001/archsurg.137.10.1096. [DOI] [PubMed] [Google Scholar]
- Moulton RC, Montie TC. Chemotaxis by Pseudomonas aeruginosa. J Bacteriol. 1979;137:274–280. doi: 10.1128/jb.137.1.274-280.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyč O, Pituch H, Matějková J, Obuch-Woszczatynski P, Kuijper EJ. Clostridium difficile PCR ribotype 176 in the Czech Republic and Poland. Lancet. 2011;377:1407. doi: 10.1016/S0140-6736(11)60575-8. [DOI] [PubMed] [Google Scholar]
- Pépin J, Valiquette L, Cossette B. Mortality attributable to nosocomial Clostridium difficile-associated disease during an epidemic caused by a hypervirulent strain in Quebec. CMAJ. 2005;173:1037–1042. doi: 10.1503/cmaj.050978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popoff MR, Rubin EJ, Gill DM, Boquet P. Actin-specific ADP-ribosyltransferase produced by a Clostridium difficile strain. Infect Immun. 1988;56:2299–2306. doi: 10.1128/iai.56.9.2299-2306.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruckler JM, Benson RF, Moyenuddin M, Martin WT, Fields BS. Association of flagellum expression and intracellular growth of Legionella pneumophila. Infect Immun. 1995;63:4928–4932. doi: 10.1128/iai.63.12.4928-4932.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redelings MD, Sorvillo F, Mascola L. Increase in Clostridium difficile-related mortality rates, United States, 1999-2004. Emerg Infect Dis. 2007;13:1417–1419. doi: 10.3201/eid1309.061116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricciardi R, Rothenberger DA, Madoff RD, Baxter NN. Increasing prevalence and severity of Clostridium difficile colitis in hospitalized patients in the United States. Arch Surg. 2007;142:624–631. doi: 10.1001/archsurg.142.7.624. discussion 631. [DOI] [PubMed] [Google Scholar]
- Rupnik M, Avesani V, Janc M, von Eichel-Streiber C, Delmée M. A novel toxinotyping scheme and correlation of toxinotypes with serogroups of Clostridium difficile isolates. J Clin Microbiol. 1998;36:2240–2247. doi: 10.1128/jcm.36.8.2240-2247.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebaihia M, Wren BW, Mullany P, Fairweather NF, Minton N, Stabler R, Thomson NR, Roberts AP, Cerdeño-Tárraga AM, et al. The multidrug-resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome. Nat Genet. 2006;38:779–786. doi: 10.1038/ng1830. [DOI] [PubMed] [Google Scholar]
- Solomon K, Fanning S, McDermott S, Murray S, Scott L, Martin A, Skally M, Burns K, Kuijper E, et al. PCR ribotype prevalence and molecular basis of macrolide-lincosamide-streptogramin B (MLSB) and fluoroquinolone resistance in Irish clinical Clostridium difficile isolates. J Antimicrob Chemother. 2011;66:1976–1982. doi: 10.1093/jac/dkr275. [DOI] [PubMed] [Google Scholar]
- Stabler RA, Gerding DN, Songer JG, Drudy D, Brazier JS, Trinh HT, Witney AA, Hinds J, Wren BW. Comparative phylogenomics of Clostridium difficile reveals clade specificity and microevolution of hypervirulent strains. J Bacteriol. 2006;188:7297–7305. doi: 10.1128/JB.00664-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stabler RA, He M, Dawson L, Martin M, Valiente E, Corton C, Lawley TD, Sebaihia M, Quail MA, et al. Comparative genome and phenotypic analysis of Clostridium difficile 027 strains provides insight into the evolution of a hypervirulent bacterium. Genome Biol. 2009;10:R102. doi: 10.1186/gb-2009-10-9-r102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasteyre A, Barc MC, Collignon A, Boureau H, Karjalainen T. Role of FliC and FliD flagellar proteins of Clostridium difficile in adherence and gut colonization. Infect Immun. 2001;69:7937–7940. doi: 10.1128/IAI.69.12.7937-7940.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg RJ, Schaap I, Templeton KE, Klaassen CH, Kuijper EJ. Typing and subtyping of Clostridium difficile isolates by using multiple-locus variable-number tandem-repeat analysis. J Clin Microbiol. 2007;45:1024–1028. doi: 10.1128/JCM.02023-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waligora AJ, Hennequin C, Mullany P, Bourlioux P, Collignon A, Karjalainen T. Characterization of a cell surface protein of Clostridium difficile with adhesive properties. Infect Immun. 2001;69:2144–2153. doi: 10.1128/IAI.69.4.2144-2153.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]