Background: Methotrexate targets dihydrofolate reductase (DHFR) and thymidylate synthase (TYMS) to prevent cancer growth, and increases DHFR expression when applied.
Results: TYMS resistant to methotrexate inhibits the methotrexate induced increase in DHFR expression.
Conclusion: Thymidine synthesis regulates post-transcriptional expression of DHFR and TYMS.
Significance: Methotrexate-resistant DHFR and TYMS can be used to regulate cis and trans transgene in primary T cells.
Keywords: chemoresistance, gene therapy, immunology, immunotherapy, post-transcriptional regulation, co-expression, dihydrofolate reductase, drug-inducible, methotrexate, thymidylate synthase
Abstract
Methotrexate (MTX) is an anti-folate that inhibits de novo purine and thymidine nucleotide synthesis. MTX induces death in rapidly replicating cells and is used in the treatment of multiple cancers. MTX inhibits thymidine synthesis by targeting dihydrofolate reductase (DHFR) and thymidylate synthase (TYMS). The use of MTX to treat cancer also causes bone marrow suppression and inhibits the immune system. This has led to the development of an MTX-resistant DHFR, DHFR L22F, F31S (DHFRFS), to rescue healthy cells. 5-Fluorouracil-resistant TYMS T51S, G52S (TYMSSS) is resistant to MTX and improves MTX resistance of DHFRFS in primary T cells. Here we find that a known mechanism of MTX-induced increase in DHFR expression persists with DHFRFS and cis-expressed transgenes. We also find that TYMSSS expression of cis-expressed transgenes is similarly decreased in an MTX-inducible manner. MTX-inducible changes in DHFRFS and TYMSSS expression changes are lost when both genes are expressed together. In fact, expression of the DHFRFS and TYMSSS cis-expressed transgenes becomes correlated. These findings provide the basis for an unrecognized post-transcriptional mechanism that functionally links expression of DHFR and TYMS. These findings were made in genetically modified primary human T cells and have a clear potential for use in clinical applications where gene expression needs to be regulated by drug or maintained at a specific expression level. We demonstrate a potential application of this system in the controlled expression of systemically toxic cytokine IL-12.
Introduction
Antifolate drugs have been in use for seven decades in the treatment of cancer (1). MTX3 is a commonly used agent from this class and inhibits several folate-dependent enzymes including DHFR and TYMS. Inhibition of these proteins adversely affects the de novo synthesis of purine and thymidine nucleotides, which is vital for survival of rapidly replicating cells. MTX has proven valuable for treating rapidly replicating cancers such as leukemia, but the impact of MTX on healthy, replicating tissue leads to dose-limiting toxicities such as bone marrow suppression (2, 3). This disadvantage of MTX has led to intense study into mechanisms of resistance (4) and alternative antifolates for the treatment of cancer (3).
Stemming from the discovery of DHFR mutants that conferred MTX resistance, a mutant that weakly binds MTX, DHFRFS, was developed (5). A strategy was formulated where bone marrow cells would be genetically modified ex vivo with DHFR mutants resistant to MTX (6). The use of DHFRFS was later implemented in genetically modified T cells. T cells modified with DHFRFS are desired following bone marrow transplant for relapsed leukemia to overcome immune suppression of MTX, allowing genetically modified T cells to survive and target cancer (7, 8). An advantage of DHFRFS is that it can be used to select for transgenes useful for therapeutic efficacy such as tumor targeting proteins such as chimeric antigen receptors (8), suicide genes (9), or imaging genes (10). In an effort to broaden the application of DHFRFS, our group added a human TYMS mutant, discovered in a bacterial screen, resistant to 5-fluorouracil (5-FU) (11). Upon the addition of human mutant TYMSSS with DHFRFS in human T cells, we demonstrated improved resistance of T cells to 5-FU and increased resistance to MTX beyond that of DHFRFS alone, in agreement with a series of experiments performed in murine bone marrow cells where linking of DHFRFS to a mutant of TYMS led to improved resistance to 5-FU and the antifolate pemetrexed (12, 13). We suggested that the addition of TYMSSS restores thymidine synthesis in MTX-treated T cells (9).
In working with DHFR and TYMS, it was noted that human DHFR is known to bind and inhibit translation of DHFR mRNA as a post-translational mechanism of regulation. Furthermore, MTX was shown to prevent this binding (14) and lead to an increase in DHFR protein expression. The nucleotide sequence to which DHFR protein binds DHFR mRNA within the coding domain is known (15, 16), and the specific amino acids that DHFR protein utilizes to bind DHFR mRNA have been elucidated (17). However, subsequent studies found that a mutation to mRNA binding amino acids of DHFR still resulted in increased DHFR protein expression in the presence of MTX (18). Conversely, mutations to amino acids unrelated to DHFR binding mRNA (17) prevented MTX-inducible expression of DHFR (18). This suggests that another mechanism of post-transcriptional regulation for DHFR is in effect, and findings detailed in this study lead us to propose that a mechanism independent of mRNA binding is affecting MTX-induced DHFR up-regulation. Notably, we demonstrate that this mechanism can be utilized for regulation of transgene expression when transgenes are located in a 3′ cis arrangement with DHFRFS.
By codon-optimizing DHFRFS to remove the mRNA binding sequence previously identified (15, 16), we find that MTX continues to induce an increase in DHFRFS expression independent of the mRNA sequence and that the addition of TYMSSS unexpectedly blunts this increase. This finding suggests a novel method of post-transcriptional DHFR expression that is regulated by the enzymatic action of TYMSSS. In performing these experiments in primary human T cells, we also identified that the application of MTX decreases TYMSSS expression in an MTX-dependent manner, likely due to TYMS binding its own mRNA to repress translation (19). Translational repression of TYMS is lost in the presence of tetrahydrofolate (19), and MTX prevents the synthesis of tetrahydrofolate (3). Consistent with this hypothesis, restoration of tetrahydrofolate synthesis in T cells by expressing DHFRFS along with TYMSSS prevented the MTX-induced expression decreases for TYMSSS. Based on these findings, we were led to discover that expression of DHFRFS and TYMSSS is linked post-transcriptionally and results in correlated expression of DHFR and TYMS within human T cells. Here we present methods to increase, decrease, or link the expression of transgenes based on the use of mutant DHFRFS and TYMSSS in a clinically relevant context. This system will permit new forms of post-translational regulation to be utilized for improved control of gene therapy vectors.
Experimental Procedures
Cells
The Jurkat human T cell line was used for controlled expression experiments. Cell line origin and attributes have been previously discussed (9). T cells were derived from healthy donor-derived peripheral blood mononuclear cells (PBMC) from the MDACC Blood Bank, Houston, TX. PBMC were isolated and cryopreserved until use (9).
Genetic Constructs
A mutant DHFRFS containing the original DHFR mRNA codon sequence with point mutations to L22F,F31S was designed to co-express eGFP C terminus to a T2A amino acid sequence. This construct is referred to as FLAG-DHFRFS-2A-eGFP pSBSO (DFSG). The DHFRFS was also codon-optimized to remove the original codon sequence, with specific attention paid to the coding region where DHFR is known to bind its own mRNA (15, 16). This codon-optimized (CoOp) DHFRFS otherwise had the same design as DFSG and is known as FLAG-CoOp DHFRFS-2A-eGFP pSBSO (CoOp DFSG). TYMSSS without codon optimization was co-expressed with a C terminus eGFP or RFP. The constructs are referred to as FLAG-TYMSSS-2A-GFP pSBSO (TSSSG) and FLAG-TYMSSS-2A-RFP pSBSO (TSSSR). A selection-free RFP plasmid NLS-mCherry pSBSO (RFP) was utilized as a control. The construct Myc-ffLuc-NeoR pSBSO (NRF) was electroporated, and G418 (InvivoGen, Ant-gn-1) was used for selection as described below when alternative selection was desired. A construct to select for inducible caspase 9 (iC9) FLAG-DHFRFS-2A-iC9 pSBSO (DFSiC9) was also used. The design and synthesis of these constructs have been described elsewhere (9). The construct FLAG-TYMSSS-2A-IL-12p35-2A-IL-12p40 pSBSO (TSSSIL-12) was synthesized from codon-optimized (GeneArt, Life Technologies) IL-12 p35 and IL-12 p40 transgenes and digested within the 2A regions to ligate IL-12 p35 and IL-12 p40 with a TYMSSS fragment also digested within the 2A region. TSSSG backbone digestion points 5′ to the start site of TYMSSS and 3′ to the IL-12p40 stop site ligated the three components into the TSSSG backbone in a four-part ligation. All constructs described integrate into the cellular genome with the aid of Sleeping Beauty transposase (20).
Culture Conditions
Both Jurkat cells and human PBMC were electroporated, maintained, and selected as described previously (9). Briefly, Jurkat cells were electroporated with transgene, and 48 h later, drug was applied for 2 weeks. Surviving cells were rested in complete media for another 3–5 weeks before testing. Human PBMC were electroporated, and the following day, T cells were propagated with activating and propagating cells at a 1:1 ratio including recombinant human IL-2. 48 h after co-culture was initiated, drug was applied to culture for a 2-week period. Propagation of PBMC with activating and propagating cells and IL-2 selectively expanded T cells and was repeated weekly. The T cells surviving drug treatment from days 2 to 14 were co-cultured without drug for another 3 weeks (9, 20, 21). On day 35 of propagation, T cells were stimulated with anti-CD3, anti-CD28, and IL-2 (9). MTX (Hospira, Lake Forest, IL), pemetrexed (Lilly, Indianapolis, IN), and 5-FU (APP Pharmaceuticals, Schaumburg, IL) were attained from the MDACC pharmacy, whereas raltitrexed (Abcam Biochemicals, Cambridge, MA, catalog number Ab142974) was not.
Flow Cytometry
T cells were prepared for flow cytometry by washing once in FACS buffer, and surface-stained for anti-human CD3-APC (BD Pharmingen, 340661, 1:33 dilution), as described previously (21). Intracellular staining for Myc tag Alexa Fluor 488 (MBL International, M047-A48, 1:33 dilution) and anti-human IL-12 p40/p70-PE (BD Pharmingen, 554575, 1:20 dilution) also proceeded as described previously (21). Transgene expression was analyzed on either a BD LSRFortessa or a BD FACSCalibur (BD Biosciences), with the same flow cytometer used consistently within a given experiment. FlowJo v 10.0.5 (TreeStar Inc., Ashland, OR) was used to analyze flow cytometry data.
Statistical Analysis
Statistical analysis and graphical representation were performed as before (9). Briefly, non-Gaussian distributions of a multivariate nature were analyzed by one-way ANOVA Kruskal-Wallis test followed by Dunn's multiple comparison test. Univariate tests (experimental versus control) utilized t tests. Two-way ANOVA was performed when appropriate and followed by Šidák's multiple comparison test. The analysis is denoted in context. Pearson's correlation and linear regression analysis were performed where denoted. Statistical significance was designated as α < 0.05. Means with standard deviations are depicted within each figure
Results and Discussion
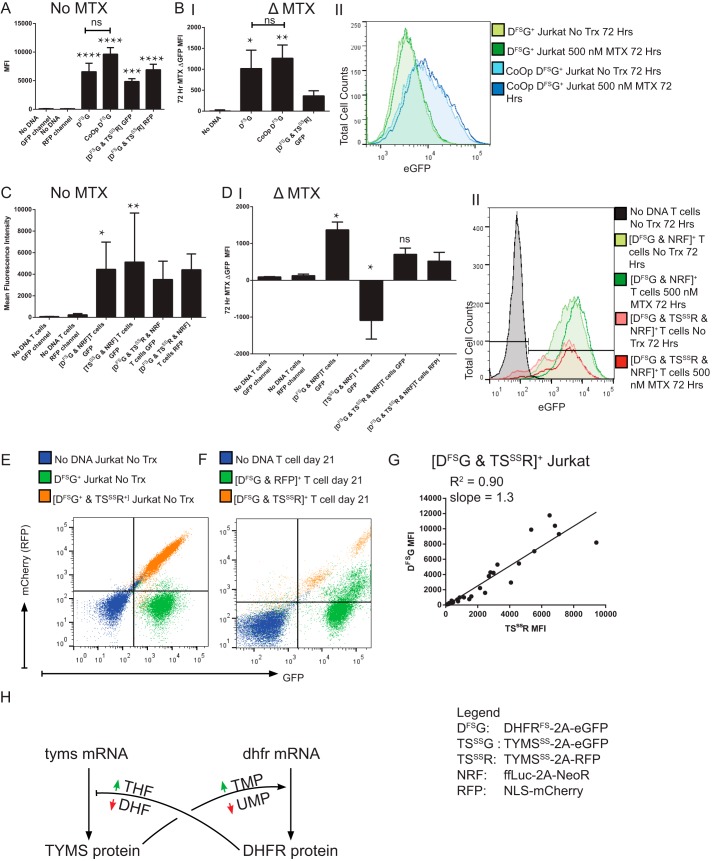
Mayer-Kuckuk et al. (22) demonstrated that DHFR linked to a transgene of interest can be used to induce an increased expression of that transgene in vitro and in vivo when MTX is administered. We attempted to recapitulate an increase in the expression of MTX-resistant DHFRFS after the administration of MTX within human T cells, which Skacel et al. (18) has shown in a hamster cell line. As a control for our studies, DHFRFS transgene was codon-optimized to remove any of the mRNA sequence or structure that contributed to mRNA binding (16). It was hypothesized that this modification would increase DHFRFS expression by removing DHFR protein binding to DHFRFS mRNA and prevent the increase of DHFRFS expression previously noted in the presence of MTX. The expression of DHFRFS and CoOp DHFRFS selected for uniform expression in Jurkat T cell line is shown in Fig. 1A. CoOp DHFRFS did not contribute to a significantly higher expression of DHFRFS as indicated by a cis-expressed eGFP, nor did it prevent MTX-induced increases in transgene expression as noted in Fig. 1B. This was unexpected. However, the loss of MTX-induced increase in DHFRFS expression was noted when TYMSSS was co-expressed with DHFRFS, as seen in Fig. 1, A and B (9). The addition of TYMSSS led to an insignificant reduction in the expression of un-optimized DHFRFS in the absence of MTX. The addition of MTX was unable to induce the same increase in DHFRFS expression seen during the sole expression of either DHFRFS version. Thus, TYMSSS is playing a role in the MTX-inducible increase of DHFRFS, and likely through the restoration of thymidine synthesis.
FIGURE 1.
DHFRFS 3′ cis-transgene expression increases in the presence of MTX independent of mRNA sequence, and the increase is suppressed by restoration of thymidine synthesis. A, Jurkat cells were genetically modified to express FLAG-DHFRFS-2A-eGFP pSBSO (DFSG) with resistance to MTX (n = 4), CoOp DFSG, with known mRNA binding elements DFSG removed (n = 5), and [DFSG & FLAG-TYMSSS-2A-RFP pSBSO (TSSSR)], with enhanced resistance to MTX beyond DFSG alone through the addition of MTX-resistant TYMSSS (n = 7). Genetically modified Jurkat cells were selected for 2 weeks in 1 μm MTX before culturing without MTX for 3–5 weeks. In A, the stable fluorescent protein expression, in the absence of MTX, is depicted by MFI. B, panel I, Jurkat cells were treated for 72 h with 0.5 μm MTX or no treatment. The MFI difference (Δ = eGFP MFI MTX-treated − eGFP MFI-untreated) is depicted. B, panel II, a histogram representative of the data used in panel I demonstrates the MTX-induced change in eGFP MFI for DHFRFS and CoOp DHFRFS in Jurkat cells. No Trx, no treatment. C, in primary T cells, transgenes DHFRFS, TYMSSS, or the combination were selected with ffLuc-NeoR pSBSO (NRF) for 2 weeks in the presence of cytotoxic drug and then propagated without selection for 3 weeks (see “Experimental Procedures”). On day 35, T cells were stimulated with anti-CD3, anti-CD28 antibodies, and 50 IU/ml IL-2 in the presence or absence of MTX for 72 h. The fluorescent protein MFI of untreated cells is shown. D, panel I, depicts the Δ MFI after 72 h of treatment with 0.5 μm MTX in comparison to no treatment. In D, panel II, a representative histogram demonstrates the observed shift in eGFP fluorescence for DHFRFS+ T cells in the presence or absence of MTX (n = 5). E, a trans regulatory pattern of DHFR- and TYMS-linked proteins was observed. A representative flow plot from the 1 μm MTX-selected Jurkat cells left untreated, as in A, demonstrates that in untreated Jurkat cells of either unselected mock-electroporated (No DNA) or DFSG+, there is a globular appearance in the RFP channel. However, co-expression of DHFRFS with TYMSSS in [DFSG & TSSSR]+ Jurkat cells leads to a linear clustering. F, primary T cells were electroporated with DHFRFS and co-transformed with either RFP control or FLAG-TYMSSS-2A-RFP pSBSO (TSSSR) before propagation, as in C. A representative flow plot of primary human T cells from the same donor where [DFSG & RFP], [DFSG & TSSSR], and untransformed T cells is shown on day 21, after 7 days without selection in MTX. A linear clustering of DHFRFS is again noted when co-expressed with TYMSSS that is not noted with RFP alone. G, further studies to identify a trans pattern of linked expression between DHFRFS and TYMSSS were identified in the selection of [DFSG & TSSSR] electroporated Jurkat cells in antifolates MTX (0, 0.01, 0.1, 0.5, 1, 5 μm), pemetrexed (0, 10, 50, 100 μm), and raltitrexed (0, 1, 5, 10 μm). The MFI of DFSG and TSSSR for each expression pattern was plotted after days 2–14 in selection. The values were plotted, and a linear fitting was performed with the R2 from the Pearson's correlation and the slope of the linear regression provided on the graph. These data are assembled from four technical replicates. H, a model of post-transcriptional regulation of DHFR and TYMS mRNA into the respective proteins is depicted with colored arrows indicating increase (green) or decrease (red) in specific small molecules. Black pointed arrows indicate promotion, while black bars indicate inhibition of protein translation. All experiments other than G were independently repeated twice. Kruskal-Wallis test was used to determine significant differences with multivariate analyses. Unmarked statistically significant results refer to comparison between experimental cells and the “No DNA” untreated control cells of equivalent measure. ns = not significant; * = p < 0.05, ** = p < 0.01, *** = p < 0.001, **** = p < 0.0001. TMP, thymidine monophosphate; UMP, uridine monophosphate; DHF, dihydrofolate.
Expression of these transgenes in primary T cells was evaluated to recapitulate the findings of MTX-inducible increases in DHFRFS expression that were prevented by TYMSSS. Expression of DHFRFS, TYMSSS, or [DHFRFS & TYMSSS] was achieved with stability and purity by selecting from days 2–14 of propagation with the respective drugs MTX, 5-FU, and G418 when the selection vector containing neomycin resistance was included (9). The expression of DHFRFS-linked eGFP and TYMSSS-linked RFP can be noted in Fig. 1C for DHFRFS, TYMSSS, or [DHFRFS & TYMSSS]. Again it is noted that DHFRFS expression is increased in the presence of MTX (Fig. 1D), but this increase is blunted and no longer significant when TYMSSS is co-expressed with DHFRFS, as in Jurkat cells. Of note, expression of TYMSSS without DHFRFS was successfully achieved in primary T cells by selection with 5-FU and a trans neomycin resistance gene selected by G418 (9). When TYMSSS was tested for inducible changes in the presence of high doses of MTX (Fig. 1D), it was found that TYMSSS-linked RFP decreased significantly. MTX induced a reduction in the expression of TYMSSS that MTX-resistant DHFRFS restored. Although not definitive, these findings could indicate that TYMSSS is being repressed by a lack of 5,10-methylenetetrahydrofolate (5,10-CH2THF). This is consistent with the findings of Chu et al. (19) that 5,10-CH2THF prevents TYMS-mediated repression of TYMS mRNA translation. Those findings indicate that in a healthy cell where DHFR activity is uninhibited, TYMS is not significantly bound to its mRNA. However, we propose that MTX, which leads to a drop in 5,10-CH2THF, is causing TYMS protein to bind TYMS and TYMSSS mRNA, preventing expression (23). It should be noted that TYMSSS is equivalent to the native sequence with the exception of the point mutations (9).
Based on findings in Fig. 1, A and B, we propose that DHFRFS expression is also regulated by the synthesis of thymidine. Likewise, based on findings in Fig. 1, C and D, we propose that TYMSSS expression is regulated by the synthesis of tetrahydrofolate (THF). As a derivative of THF is used to make thymidine, a logical conclusion was made that DHFRFS regulates the expression of TYMSSS and TYMSSS regulates the expression of DHFRFS. If this is the case, a correlated expression of DHFRFS and TYMSSS should be noted within individual cells. A correlated expression of DHFRFS and TYMSSS was indeed observed in flow cytometry plots of Jurkat cells in Fig. 1E and primary T cells in Fig. 1F. A control RFP vector co-expressed with DHFRFS, but not modulated by cis expression with TYMSSS, did not appear to have the same co-expression pattern (Fig. 1F). To quantify this observation, Jurkat cells expressing [DHFRFS & TYMSSS] were treated with antifolates MTX, pemetrexed, and raltitrexed at varying concentrations for 2 weeks. DHFRFS-linked eGFP mean fluorescence intensity (MFI) and TYMSSS-linked RFP MFI for each separate well were then plotted and correlated. The linked expression between DHFRFS and TYMSSS was significant and fit a linear regression (Fig. 1G). These findings support a general mechanism for regulation of DHFR and TYMS, which leads to a linear co-expression of DHFRFS and TYMSSS independent of MTX. This model is shown in Fig. 1H.
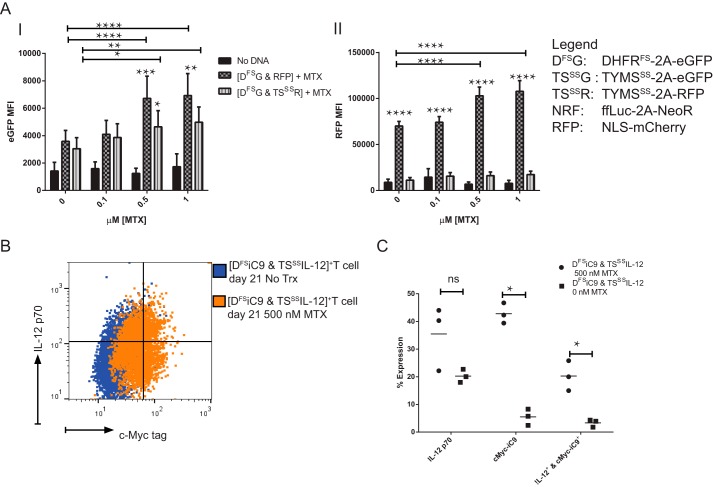
Based on the above model in Fig. 1H, it appears that TYMSSS expression will be stabilized by DHFRFS from strong expression changes in the presence of MTX. This was tested in Fig. 2A with primary T cells expressing DHFRFS along with either RFP or TYMSSS linked to RFP by applying increasing doses of MTX. As expected, DHFRFS-linked eGFP was increased by rising concentrations of MTX, and this increase was blunted by the presence of TYMSSS (Fig. 2A, panel I). This conserves the model in Fig. 1H where restoration of thymidine synthesis prevents the MTX-induced increase in DHFRFS. Further conserving the model, RFP linked to TYMSSS did not significantly increase over any concentration of MTX used (Fig. 2A, panel II). When DHFRFS-linked eGFP increased, so too did the control RFP, and a rise in the expression of RFP alone was not expected. A possible explanation is that this increase was noted above 0.5 μm MTX, and DHFRFS alone is only resistant to 0.5 μm MTX (9). This suggests that higher doses of MTX begin to select for cells with higher gene content of DHFRFS and associated transgenes. This is supported by Kacherovsky et al. (10) where greater DHFRFS expression was selected by the addition of MTX. Notably, DHFRFS co-expressed with TYMSSS is resistant to concentrations of up to 1 μm MTX. This further supports the use of TYMSSS to modulate transgene expression and prevent unwanted selection toward higher gene expression levels of genes expressed in cis or trans with DHFRFS.
FIGURE 2.
Co-expression of DHFRFS with TYMSSS leads to controlled expression of TYMSSS and cis transgenes in the presence of MTX. A, primary T cells from the experiment described in Fig. 1F were propagated to day 35. T cells were stimulated for 72 h with anti-CD3, anti-CD28 antibodies, 50 IU/ml IL-2, and varying concentrations of MTX. The MTX-induced change in eGFP MFI for DHFRFS is shown in panel I, while the influence of MTX on RFP and RFP co-expressed with TYMSSS (TSSSR) is shown in panel II (n = 6, repeated independently twice, analyzed by two-way ANOVA with Šidák's multiple comparison test). B, this regulatory pattern was applied to a clinically relevant problem: the cytokine IL-12 is a strong promoter of anti-tumor activity in T cells, but is highly toxic. A construct expressing IL-12 following TYMSSS, called TSSSIL-12, was used to modulate IL-12 expression in conjunction with the construct DFSiC9. DFSiC9 is capable of selecting T cells with DHFRFS and depleting those T cells with iC9. A representative flow diagram of the same donor depicts intracellular expression of IL-12 and c-Myc-iC9 in [DFSiC9 & TSSSIL-12]-expressing T cells. These cells are shown on day 21 after selection from day 2–14 in 0.1 μm MTX and subsequent treatment with 0.5 μm MTX or no treatment from days 14–21. Cellular secretion of IL-12 was blocked for 6 h before intracellular staining. Gating is based on staining of untransformed, unselected primary T cells stained in the same way. C, three donors were treated as in B, and the change in transgene expression noted after 7 days of treatment with 0.5 μm MTX is shown. Each measure was analyzed by t tests. ns = not significant; * = p < 0.05, ** = p < 0.01, *** = p < 0.001, **** = p < 0.0001.
Our group previously described a construct of DHFRFS cis expressing an inducible suicide gene, iC9 (9). This construct, called DFSiC9, selects for T cells expressing DFSiC9 in the presence of MTX and ablates DFSiC9+ T cells in the presence of drug that activates iC9 to induce apoptosis (24). Based on the above findings, the DHFRFS in DFSiC9 could be used to modulate and potentially ablate the expression of a transgene of interest that is otherwise too toxic to express without regulation. IL-12 is such a transgene. IL-12 is a cytokine capable of inducing a strong immune response against tumor from tumor-specific T cells (25). However, systemic IL-12 is highly toxic and hence of low efficacy (26). Groups seeking to apply IL-12 to a clinical setting have sought inducible systems to overcome the toxicity related to this cytokine where T cells express IL-12 after activation (27). Presented here is an alternative approach where IL-12 is expressed cis to TYMSSS to decrease and stabilize the expression level of IL-12. In Fig. 2B, a flow plot demonstrates the expression of IL-12 cis-expressed with TYMSSS and iC9 cis-expressed with DHFRFS expression. The primary T cells were either left untreated or treated with high doses of MTX for 7 days. This expression pattern appears to indicate that IL-12 can be stably expressed even in toxic doses of MTX. A further analysis of similarly manipulated donors (Fig. 2C) demonstrates the potential of TYMSSS when co-expressed with DHFRFS to stabilize the expression of the potentially toxic transgenes of interest (here IL-12).
As for the endogenous regulatory mechanism proposed in Fig. 1H, there is evidence to support a natural regulatory pathway leading to linked expression of TYMS and DHFR. Regulation of TYMS expression by TYMS binding its own mRNA (19) and regulation of DHFR expression by DHFR binding its own mRNA (14) are both translationally regulated by the presence of folates. Our findings indicate that a DHFR mRNA-independent translational or post-translational mechanism is also involved in directing the co-expression of DHFR and TYMS. In this study, DHFRFS and TYMSSS expression was tracked by fluorescent proteins linked by a 2A ribosomal “slip” site. These linkages express two independent proteins due to slippage of the completed N terminus peptide at the 2A sequence. In this study, the N terminus peptide was DHFRFS and TYMSSS (28). The above findings of MTX-inducible changes suggest that if the fluorescent protein is regulated by either DHFRFS or TYMSSS, then this regulation is occurring before or during translation rather than after the proteins separate post-translationally. Thus, our findings add complexity to the translational regulation of DHFR and TYMS to meet an unknown biological need for combined expression of DHFR and TYMS. Findings by Anderson et al. (29) demonstrated that DHFR and TYMS co-localize as a multi-enzyme complex within the nucleus during replication of DNA. Stabilization and construction of this multi-enzyme complex within the cell may explain the need for tight translational control of DHFR and TYMS.
In conclusion, we have demonstrated an endogenous mechanism of post-transcriptional regulation for DHFR and TYMS that can be co-opted with either DHFRFS or TYMSSS to regulate expression of cis- or trans-expressed transgenes. The purpose of this study is to describe a novel system for transgene regulation in gene therapy. This system is of significant clinical interest as the genes described are human transgenes of low immunogenicity that utilize MTX to regulate transgene expression. MTX is readily available for use in both hospital and clinical settings. As these studies demonstrate that this phenomenon occurs in primary human T cells, this system can readily be used in the field of transgenic T cell immunotherapy to improve the safety and efficacy of T cell therapies used to treat cancer (30). What remains to be determined is how an improved understanding of the regulatory mechanism surrounding DHFR and TYMS expression may lead to a better understanding of developmental disorders and improved chemotherapeutics.
Author Contributions
D. R. designed, acquired, analyzed, and interpreted experiments and wrote the manuscript. A. M. designed experiments, acquired data, and was involved in critical revisions of the manuscript. A. A. acquired data and was involved in critical revisions of the manuscript. L. J. N. C. designed experiments and approved the final version of the manuscript for publication. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgments
We extend gratitude to the dedicated work of Helen Huls, Tiejuan Mi, and Drs. Kumar, Belousova, and Singh for laboratory support. We thank Drs. Dat Tran, Dean Lee, and George McNamara for assistance in manuscript preparation, and Dr. David Spencer for assistance in developing and utilizing iC9. We thank the MDACC South Campus Flow Cytometry Facility, supported by the NCI Cancer Center Support Grant P30CA16672 through the National Institutes of Health and Cancer Center Core Grant (CA16672), for use of their facilities.
This work was supported in part by National Institutes of Health Grants RO1 (CA124782, CA120956, and CA141303) and P01 (CA148600); Burroughs Wellcome Fund; Cancer Prevention and Research Institute of Texas; Gillson Longenbaugh Foundation; Harry T. Mangurian, Jr., Fund for Leukemia Immunotherapy; Institute of Personalized Cancer Therapy; Leukemia and Lymphoma Society; Lymphoma Research Foundation; MDACC's Sister Institution Network Fund; Miller Foundation; National Foundation for Cancer Research; Pediatric Cancer Research Foundation; and William Lawrence and Blanche Hughes Children's Foundation. Some of the technology described was advanced through research conducted at the MDACC by Laurence J. N. Cooper, M.D., Ph.D. In January 2015, some technology generated by Dr. Cooper was licensed for commercial application to ZIOPHARM Oncology, Inc., and Intrexon Corporation in exchange for equity interests in each of these companies. Hence MDACC, Dr. Cooper, Dr. Rushworth, and Mr. Alpert have a financial interest in ZIOPHARM Oncology, Inc., and Intrexon Corporation. On May 7, 2015, Dr. Cooper was appointed as the Chief Executive Officer at ZIOPHARM. Dr. Cooper is now a Visiting Scientist at MDACC where he will continue to supervise the development of this technology.
- MTX
- methotrexate
- DHFR
- dihydrofolate reductase
- TYMS
- thymidylate synthase
- 5-FU
- 5-fluorouracill
- PBMC
- peripheral blood mononuclear cell
- CoOp
- codon-optimized
- iC9
- inducible caspase 9
- ANOVA
- analysis of variance
- THF
- tetrahydrofolate
- 5,10-CH2THF
- 5,10-methylenetetrahydrofolate
- MFI
- mean fluorescence intensity.
References
- 1. Farber S., Diamond L. K. (1948) Temporary remissions in acute leukemia in children produced by folic acid antagonist, 4-aminopteroyl-glutamic acid. N. Engl. J. Med. 238, 787–793 [DOI] [PubMed] [Google Scholar]
- 2. McGuire J. J. (2003) Anticancer antifolates: current status and future directions. Curr. Pharm. Des. 9, 2593–2613 [DOI] [PubMed] [Google Scholar]
- 3. Walling J. (2006) From methotrexate to pemetrexed and beyond. A review of the pharmacodynamic and clinical properties of antifolates. Invest. New Drugs 24, 37–77 [DOI] [PubMed] [Google Scholar]
- 4. Abali E. E, Skacel N. E., Celikkaya H., Hsieh Y. C. (2008) Regulation of human dihydrofolate reductase activity and expression. Vitam. Horm. 79, 267–292 [DOI] [PubMed] [Google Scholar]
- 5. Ercikan-Abali E. A., Mineishi S., Tong Y., Nakahara S., Waltham M. C., Banerjee D., Chen W., Sadelain M., Bertino J. R. (1996) Active site-directed double mutants of dihydrofolate reductase. Cancer Res. 56, 4142–4145 [PubMed] [Google Scholar]
- 6. Sorrentino B. P. (2002) Gene therapy to protect haematopoietic cells from cytotoxic cancer drugs. Nat. Rev. Cancer 2, 431–441 [DOI] [PubMed] [Google Scholar]
- 7. Jonnalagadda M., Brown C. E., Chang W. C., Ostberg J. R., Forman S. J., Jensen M. C. (2013) Engineering human T cells for resistance to methotrexate and mycophenolate mofetil as an in vivo cell selection strategy. PLoS One 8, e65519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jonnalagadda M., Brown C. E., Chang W. C., Ostberg J. R., Forman S. J., Jensen M. C. (2013) Efficient selection of genetically modified human T cells using methotrexate-resistant human dihydrofolate reductase. Gene Therapy 20, 853–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rushworth D., Alpert A., Santana-Carrero R., Olivares S., Spencer D., Cooper L. (2015) Anti-thymidylate resistance enables transgene selection and cell survival for T cells in the presence of 5-fluorouracil and anti-folates. Gene Therapy 10.1038/gt.2015.88 [DOI] [PubMed] [Google Scholar]
- 10. Kacherovsky N., Liu G. W., Jensen M. C., Pun S. H. (2015) Multiplexed gene transfer to a human T-cell line by combining Sleeping Beauty transposon system with methotrexate selection. Biotechnol. Bioeng. 112, 1429–1436 [DOI] [PubMed] [Google Scholar]
- 11. Landis D. M., Heindel C. C., Loeb L. A. (2001) Creation and characterization of 5-fluorodeoxyuridine-resistant Arg50 loop mutants of human thymidylate synthase. Cancer Res. 61, 666–672 [PubMed] [Google Scholar]
- 12. Capiaux G. M., Budak-Alpdogan T., Alpdogan O., Bornmann W., Takebe N., Banerjee D., Maley F., Bertino J. R. (2004) Protection of hematopoietic stem cells from pemetrexed toxicity by retroviral gene transfer with a mutant dihydrofolate reductase-mutant thymidylate synthase fusion gene. Cancer Gene Ther. 11, 767–773 [DOI] [PubMed] [Google Scholar]
- 13. Capiaux G. M., Budak-Alpdogan T., Takebe N., Mayer-Kuckuk P., Banerjee D., Maley F., Bertino J. R. (2003) Retroviral transduction of a mutant dihydrofolate reductase-thymidylate synthase fusion gene into murine marrow cells confers resistance to both methotrexate and 5-fluorouracil. Hum. Gene Ther. 14, 435–446 [DOI] [PubMed] [Google Scholar]
- 14. Chu E., Takimoto C. H., Voeller D., Grem J. L., Allegra C. J. (1993) Specific binding of human dihydrofolate reductase protein to dihydrofolate reductase messenger RNA in vitro. Biochemistry 32, 4756–4760 [DOI] [PubMed] [Google Scholar]
- 15. Ercikan-Abali E. A., Banerjee D., Waltham M. C., Skacel N., Scotto K. W., Bertino J. R. (1997) Dihydrofolate reductase protein inhibits its own translation by binding to dihydrofolate reductase mRNA sequences within the coding region. Biochemistry 36, 12317–12322 [DOI] [PubMed] [Google Scholar]
- 16. Tai N., Schmitz J. C., Chen T. M., Chu E. (2004) Characterization of a cis-acting regulatory element in the protein-coding region of human dihydrofolate reductase mRNA. Biochem. J. 378, 999–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tai N., Ding Y., Schmitz J. C., Chu E. (2002) Identification of critical amino acid residues on human dihydrofolate reductase protein that mediate RNA recognition. Nucleic Acids Res. 30, 4481–4488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Skacel N., Menon L. G., Mishra P. J., Peters R., Banerjee D., Bertino J. R., Abali E. E. (2005) Identification of amino acids required for the functional up-regulation of human dihydrofolate reductase protein in response to antifolate treatment. J. Biol. Chem. 280, 22721–22731 [DOI] [PubMed] [Google Scholar]
- 19. Chu E., Koeller D. M., Casey J. L., Drake J. C., Chabner B. A., Elwood P. C., Zinn S., Allegra C. J. (1991) Autoregulation of human thymidylate synthase messenger RNA translation by thymidylate synthase. Proc. Natl. Acad. Sci. U.S.A. 88, 8977–8981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Singh H., Figliola M. J., Dawson M. J., Olivares S., Zhang L., Yang G., Maiti S., Manuri P., Senyukov V., Jena B., Kebriaei P., Champlin R. E., Huls H., Cooper L. J. (2013) Manufacture of clinical-grade CD19-specific T cells stably expressing chimeric antigen receptor using Sleeping Beauty system and artificial antigen presenting cells. PLoS One 8, e64138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rushworth D., Jena B., Olivares S., Maiti S., Briggs N., Somanchi S., Dai J., Lee D., Cooper L. J. (2014) Universal artificial antigen presenting cells to selectively propagate T cells expressing chimeric antigen receptor independent of specificity. J. Immunother. 37, 204–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mayer-Kuckuk P., Banerjee D., Malhotra S., Doubrovin M., Iwamoto M., Akhurst T., Balatoni J., Bornmann W., Finn R., Larson S., Fong Y., Gelovani Tjuvajev J., Blasberg R., Bertino J. R. (2002) Cells exposed to antifolates show increased cellular levels of proteins fused to dihydrofolate reductase: a method to modulate gene expression. Proc. Natl. Acad. Sci. U.S.A. 99, 3400–3405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lin X., Parsels L. A., Voeller D. M., Allegra C. J., Maley G. F., Maley F., Chu E. (2000) Characterization of a cis-acting regulatory element in the protein coding region of thymidylate synthase mRNA. Nucleic Acids Res. 28, 1381–1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Straathof K. C., Pulè M. A., Yotnda P., Dotti G., Vanin E. F., Brenner M. K., Heslop H. E., Spencer D. M., Rooney C. M. (2005) An inducible caspase 9 safety switch for T-cell therapy. Blood 105, 4247–4254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kerkar S. P., Muranski P., Kaiser A., Boni A., Sanchez-Perez L., Yu Z., Palmer D. C., Reger R. N., Borman Z. A., Zhang L., Morgan R. A., Gattinoni L., Rosenberg S. A., Trinchieri G., Restifo N. P. (2010) Tumor-specific CD8+ T cells expressing interleukin-12 eradicate established cancers in lymphodepleted hosts. Cancer Res. 70, 6725–6734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Leonard J. P., Sherman M. L., Fisher G. L., Buchanan L. J., Larsen G., Atkins M. B., Sosman J. A., Dutcher J. P., Vogelzang N. J., Ryan J. L. (1997) Effects of single-dose interleukin-12 exposure on interleukin-12-associated toxicity and interferon-γ production. Blood 90, 2541–2548 [PubMed] [Google Scholar]
- 27. Zhang L., Morgan R. A., Beane J. D., Zheng Z., Dudley M. E., Kassim S. H., Nahvi A. V., Ngo L. T., Sherry R. M., Phan G. Q., Hughes M. S., Kammula U. S., Feldman S. A., Toomey M. A., Kerkar S. P., Restifo N. P., Yang J. C., Rosenberg S. A. (2015) Tumor-infiltrating lymphocytes genetically engineered with an inducible gene encoding interleukin-12 for the immunotherapy of metastatic melanoma. Clin. Cancer Res. 21, 2278–2288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. de Felipe P., Luke G. A., Hughes L. E., Gani D., Halpin C., Ryan M. D. (2006) E unum pluribus: multiple proteins from a self-processing polyprotein. Trends Biotechnol. 24, 68–75 [DOI] [PubMed] [Google Scholar]
- 29. Anderson D. D., Woeller C. F., Chiang E. P., Shane B., Stover P. J. (2012) Serine hydroxymethyltransferase anchors de novo thymidylate synthesis pathway to nuclear lamina for DNA synthesis. J. Biol. Chem. 287, 7051–7062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Corrigan-Curay J., Kiem H. P., Baltimore D., O'Reilly M., Brentjens R. J., Cooper L., Forman S., Gottschalk S., Greenberg P., Junghans R., Heslop H., Jensen M., Mackall C., June C., Press O., Powell D., Ribas A., Rosenberg S., Sadelain M., Till B., Patterson A. P., Jambou R. C., Rosenthal E., Gargiulo L., Montgomery M., Kohn D. B. (2014) T-cell immunotherapy: looking forward. Mol. Ther. 22, 1564–1574 [DOI] [PMC free article] [PubMed] [Google Scholar]