Background: Trypanosoma brucei Tim17, the translocase of the mitochondrial inner membrane, is associated with TbTim62.
Results: TbTim62 knockdown reduced stability and assembly of the TbTim17 translocase (TbTIM).
Conclusion: TbTim62 is an essential assembly and stabilization factor for TbTIM and protects TbTim17 from degradation.
Significance: These studies identified a novel component for mitochondrial protein import machinery in T. brucei.
Keywords: mitochondria, protein assembly, protein import, translocation, Trypanosoma brucei, Hsp70, TIM complex, Tim17, Tim62
Abstract
Trypanosoma brucei, the causative agent of human African trypanosomiasis, possesses non-canonical mitochondrial protein import machinery. Previously, we characterized the essential translocase of the mitochondrial inner membrane (TIM) consisting of Tim17 in T. brucei. TbTim17 is associated with TbTim62. Here we show that TbTim62, a novel protein, is localized in the mitochondrial inner membrane, and its import into mitochondria depends on TbTim17. Knockdown (KD) of TbTim62 decreased the steady-state levels of TbTim17 post-transcriptionally. Further analysis showed that import of TbTim17 into mitochondria was not inhibited, but its half-life was reduced >4-fold due to TbTim62 KD. Blue-native gel electrophoresis revealed that TbTim62 is present primarily in ∼150-kDa and also in ∼1100-kDa protein complexes, whereas TbTim17 is present in multiple complexes within the range of ∼300 to ∼1100 kDa. TbTim62 KD reduced the levels of both TbTim62 as well as TbTim17 protein complexes. Interestingly, TbTim17 was accumulated as lower molecular mass complexes in TbTim62 KD mitochondria. Furthermore, depletion of TbTim62 hampered the assembly of the ectopically expressed TbTim17–2X-myc into TbTim17 protein complex. Co-immunoprecipitation analysis revealed that association of TbTim17 with mHSP70 was markedly reduced in TbTim62 KD mitochondria. All together our results demonstrate that TbTim62, a unique mitochondrial protein in T. brucei, is required for the formation of a stable TbTim17 protein complex. TbTim62 KD destabilizes this complex, and unassembled TbTim17 degrades. Therefore, TbTim62 acts as a novel regulatory factor to maintain the levels of TIM in T. brucei mitochondria.
Introduction
Trypanosomatids such as Trypanosoma brucei, Trypanosoma cruzi, and Leishmania species cause deadly human diseases such as African sleeping sickness, American trypanosomiasis or Chagas disease, and leishmaniasis, respectively (1–3). These diseases are spread by insect vectors and affect millions of people worldwide. Trypanosomatids are unicellular parasitic protozoa that belong to the eukaryotic super group excavata (4, 5). These organisms are also classified as the early branching eukaryotes, which possess a single mitochondrion with a large concatenated structure of mitochondrial DNA, known as kinetoplast or kDNA (6–8). Despite its large structure, kDNA only encodes ∼18 mitochondrial proteins (9). Thus, similar to other eukaryotes, a majority of mitochondrial proteins in these parasites (∼1000) are nuclear DNA-encoded and are imported into the mitochondrion after their synthesis in the cytosol (10, 11). Therefore, efficient protein import machinery is essential to maintain mitochondrial functions as well as for the survival of these parasites. However, the structure and function of this important cellular apparatus are poorly understood in these organisms.
Mitochondrial protein import machinery has been extensively characterized in fungi and later in humans and plants (12–15). The translocase of the mitochondrial outer membrane (TOM)2 consists of 7–9 subunits and is the entry gate for most of the mitochondrial proteins (16). Tom40, a β-barrel protein, is the major component of the TOM complex, and it forms the import channel for precursor proteins (17, 18). Once across the mitochondrial outer membrane, mitochondrial inner membrane (MIM) proteins are imported through either one of the two translocases of MIM (TIM), TIM23 or TIM22 (19, 20). Mitochondrial matrix proteins generally have an N-terminal targeting signal (or presequence), and these proteins are translocated via the TIM23 complex in a mitochondrial membrane potential (ΔΨ)-dependent manner (19, 21). Finally these proteins are driven to the mitochondrial matrix by the TIM23-associated and presequence-activated motor complex (PAM), which uses the energy generated by ATP hydrolysis (22). Some MIM proteins with an additional sorting signal are also imported by the TIM23 complex and then laterally sorted to their destination (23). Whereas a certain class of MIM proteins such as mitochondrial metabolite carrier proteins (MCPs), those having multiple internal targeting signals are imported through the TIM22 complex (20, 24). The mitochondrial intermembrane space (IMS)-localized Tims, which are known as small Tims or tiny Tims, chaperone these proteins within the IMS and target them to the TIM22 complex (25, 26). Translocation of proteins through the TIM22 complex also requires ΔΨ but does not depend on ATP hydrolysis in the matrix (24–26).
Both TIM23 and TIM22 are multiprotein complexes in which the protein import channels are formed by the major subunits Tim23 and Tim22, respectively (27, 28). These two proteins and Tim17, an essential structural component of the TIM23 complex, belong to the presequence and amino acid transporter (PRAT) family, which has four transmembrane domains (TMs) in the center of the protein, leaving both the N and C termini in the IMS (29). Tim50, an essential component of the TIM23 complex, acts as the receptor for presequence-containing mitochondrial proteins (30, 31). Homologs for Tim23, Tim17, and Tim50 are conserved in higher eukaryotes (14, 33). On the other hand, besides Tim22, the subunits of the TIM22 complex, such as Tim54 and Tim18, are less conserved in mammals and plants (34, 35).
Many trypanosomatid mitochondrial proteins possess presequences with similar characteristics to those in other eukaryotes and vary in length from 18 to 60 amino acid residues (36, 37). Interestingly, a number of mitochondrial proteins in these parasites possess presequences that can be as short as 8 amino acid residues (38, 39). Trypanosomatids also possess a large repertoire of MCPs (40); those do not have N-terminal mitochondrial targeting signals. Therefore, it can be anticipated that T. brucei possesses an elaborate TIM machinery for import of the matrix and MIM proteins. However, apart from the homologs of Tim17, Tim50, and few small Tims, the counterparts of other Tim proteins are not found in trypanosome genome or its mitochondrial proteome (41–44). We have shown that TbTim17 is essential in T. brucei, and knockdown (KD) of TbTim50 also reduced cell growth significantly (41, 42). Both of these proteins are required for import of presequence-containing proteins into the T. brucei mitochondrion (41, 42, 45). It has been shown that TbTim50 is not involved in the import of MCPs like MCP5 (42). However, the role of TbTim17 in translocation of such proteins has not been elucidated yet. Because trypanosomatids possess a single PRAT-family protein, TbTim17 (46), it is speculated that this protein may possess a broader substrate specificity in comparison to Tim17 found in fungi and higher eukaryotes. We showed that TbTim17 is present in larger molecular mass protein complexes in the MIM. We also have identified several trypanosome proteins by tandem affinity purification of the TbTim17-containing protein complex (45). Among these, two uncharacterized trypanosome proteins, TbTim62 and TbTim54, are found highly significant for T. brucei cell growth and mitochondrial protein import (45). Co-immunoprecipitation analysis confirmed that TbTim62 and TbTim54 interact with TbTim17 in T. brucei (45).
Here, we further characterized the role of TbTim62 in mitochondrial protein import in T. brucei. TbTim62 is a novel trypanosome-specific protein that is localized in the MIM. Blue-native gel electrophoresis (BN-PAGE) revealed that TbTim62 is present in a protein complex of ∼150 kDa. A minor fraction of this protein was also detected in the 1100-kDa protein complex. On the other hand, TbTim17 is found in the multiple complexes within the range of 300–1100 kDa. Two-dimensional BN-SDS-PAGE analysis showed that the ectopically expressed TbTim17-myc is also present in a ∼150-kDa protein complex similar to TbTim62. Interestingly, we found that TbTim62 KD hampered the stability of the TbTim17 protein complexes and reduced the levels of TbTim17 post-transcriptionally. Further analysis revealed that TbTim62 is critical for the association of TbTim17 with mHsp70 and for the assembly of the TbTim17 into the respective protein complexes. From these results we conclude that TbTim62, a novel trypanosome mitochondrial protein, is required for formation of the TbTIM and also to maintain the integrity of this protein complex. Therefore, TbTim62 is essential for mitochondrial protein import in T. brucei.
Experimental Procedures
Strains and Media
The procyclic form of T. brucei 427 cells were grown in SDM-79 medium containing 10% fetal bovine serum. The T. brucei 427 procyclic double-resistant cell line (Tb427 29-13) expressing the tetracycline repressor gene (TetR) and T7RNA polymerase (T7RNAP) (47) was grown in the same medium containing 50 μg/ml hygromycin and 15 μg/ml G418. Cells were harvested at logarithmic phase of growth for different biochemical analyses. Cell numbers were counted in a Neubauer hemocytometer.
T. brucei Transgenic Cell Line and RNA Analysis
TbTim17 and TbTim62 RNAi cell lines were developed as described (45). T. brucei cell line expressing TbTim62 with a tandem affinity purification (TAP) tag at the C terminus was developed previously (45). We also generated the TbTim62 RNAi (Puro) construct by subcloning a portion of the TbTim62 coding region (522–1021 bp) in the p2T7Ti-177 (puromycin) vector (a generous gift from Laurie Read) and used it to transfect the T. brucei cell line containing the pLew100-TbTim17–2X-myc construct, which was developed previously in our laboratory (46). After transfection, cells were selected by puromycin (1 μg/ml). RNA was isolated from the procyclic trypanosomes grown for 4 days with or without doxycycline using TRIzol reagent (Invitrogen) according to the manufacturer's protocol; Northern analysis was performed as described (42, 45). Specific probes were generated using a random primer-labeling protocol (Invitrogen) from the cDNA fragments generated by PCR using the same primer pairs used for preparation of RNAi constructs. Tubulin was used as the loading control.
Isolation and Post-isolation Treatment of Mitochondria
Mitochondria were isolated from the parasite after lysis via nitrogen cavitation in isotonic buffer (42, 45). The isolated mitochondria were stored at a protein concentration of 10 mg/ml in SME buffer (250 mm sucrose, 20 mm MOPS/KOH, pH 7.4, 2 mm EDTA) containing 50% glycerol at −70 °C. Before using, mitochondria were washed twice with nine volumes of SME buffer to remove glycerol. Mitoplasts (i.e. mitochondrial matrix with intact MIM) were isolated after treatment of mitochondria with 3 volumes of water for 15 min on ice to disrupt mitochondrial outer membrane. Afterward the osmolarity was restored with the addition of a ⅓ volume of 2.4 m sucrose. Mitoplasts were recovered by centrifugation at 12,000 × g for 15 min. The supernatant and pellet fractions were analyzed for the release of cytochrome c (Cyt c), an IMS protein, as described (48). For limited proteinase K (PK) digestion, mitochondria in SME buffer (1 mg/ml) were treated with various concentrations of PK (0–200 μg/ml) for 30 min on ice. After incubation, PK was inhibited by phenylmethylsulfonyl fluoride (PMSF, 2 mm), and mitochondria were re-isolated by centrifugation at 10,000 × g at 4 °C for 10 min. For alkali extraction, isolated mitochondria (100 μg) from T. brucei were treated with 100 mm Na2CO3 (100 μl) at pH 11.5 for 30 min on ice. The supernatant and pellet fractions were collected after centrifugation and analyzed by SDS-PAGE and immunoblot.
Determination of Half-life of TbTim17
The wild type T. brucei 427 and TbTim62 RNAi cells grown in the presence of doxycycline were incubated in the presence of cycloheximide (50 μg/ml). Equal numbers of cell were harvested at different time points (0–24 h), and total cellular proteins were analyzed by immunoblot analysis using anti-TbTim17 antibody. Tubulin was used as the loading control.
In Vitro Mitochondrial Protein Import Assay
Isolated mitochondria from T. brucei were used for in vitro assays of protein import as described (42, 45). Mitochondria were pretreated with carbonyl cyanide m-chlorophenyl hydrazone (CCCP) (50 μm) for disruption of mitochondrial ΔΨ as described (42, 45, 49). We have titrated CCCP concentration previously and found 50–100 μm is optimum for dissipation of mitochondrial ΔΨ (49). Other investigators also used similar concentrations of CCCP for ΔΨ degeneration in trypanosomatid mitochondria (50–52). For production of radiolabeled precursor proteins, the coding regions for TbTim17 and TbTim62 were PCR-amplified using sequence-specific primers possessing BamHI and HindIII restriction sites at their 5′ ends, respectively, and the genomic DNA of T. brucei as the template. The PCR products were purified, digested with the respective enzymes, and then subcloned into the pGEM4Z vector between the BamHI and HindIII sites. The cytochrome oxidase subunit 4 (COIV), and a fusion protein of the trypanosome alternative oxidase with dihydrofolate reductase (TAO-DHFR) were also cloned similarly as described (42, 45). Radiolabeled precursor proteins were synthesized in vitro using a coupled transcription/translation rabbit reticulocyte lysate system (TNTR, Promega) according to the manufacturer's protocol using l-[35S]methionine.
Blue-native PAGE (BN-PAGE) and Two-dimensional Gel Electrophoresis
Mitochondrial proteins (100 μg) were solubilized in 50 μl of the ice-cold native buffer (50 mm Bis-Tris, pH 7.2, 50 mm NaCl, 10% w/v glycerol, 1 mm PMSF, 1 μg/ml leupeptin and 1% digitonin). The solubilized mitochondrial proteins were clarified by centrifugation at 100,000 × g for 30 min at 4 °C. The supernatants were mixed with 2.5 μl of native PAGE G250 sample additive (Invitrogen) and electrophoresed on a precast (4–16%) Bis-Tris polyacrylamide gel (Invitrogen) as described (46). Protein complexes were detected by immunoblot analysis. Molecular size marker proteins apoferritin dimer (886 kDa) and apoferritin monomer (443 kDa), β-amylase (200 kDa), alcohol dehydrogenase (150 kDa), and bovine serum albumin (66 kDa) were electrophoresed on the same gel and visualized by Coomassie staining. For two-dimensional BN-SDS-PAGE analysis gel strips representing a single lane on first dimension BN-PAGE gel were excised and sequentially incubated in buffer containing SDS (1%) plus 2-mercaptoethanol (5%) followed by SDS (1%) alone at room temperature for 30 min each to denature proteins in the gel. Next, gel strips were placed on top of the stacking gel and electrophoresed on a second dimension using Tricine SDS-PAGE (12%). After separation, proteins were transferred to a nitrocellulose membrane for immunodecoration.
SDS-PAGE and Immunoblot Analysis
Polyclonal antibodies against the TbTim62 were generated by Antagene Inc. (Sunnyvale, CA) using a peptide consisting of amino acids 43–59 (NGIGPAATKKDGNTD) of the N-terminal region as the antigen. The synthetic peptide was purified and conjugated to keyhole limpet hemocyanin using cysteine. The keyhole limpet hemocyanin-conjugated TbTim62 peptide was then used to generate specific antibodies in rabbits. Antiserum against TbTim62 was affinity-purified using the TbTim62 peptide as the ligand. Proteins from whole cells or isolated mitochondria were separated on a SDS-PAGE and immunoblotted with polyclonal antibodies for TbTim62 and TbTim17 (45), T. brucei voltage-dependent anion channel protein (VDAC) (53), Cyt c (48), cytochrome c1 (Cyt c1) (54), and mitochondrial heat shock protein 70 (mHsp70) (55) or monoclonal antibodies for TAO (56), c-myc epitope (Abcam), and T. brucei β-tubulin (57). Blots were developed with appropriate secondary antibodies and an enhanced chemiluminescence (ECL) kit (Pierce). Intensity of the protein bands was quantitated using Quantity One software (Bio-Rad).
Immunofluorescence Microscopy
T. brucei cells (4 × 106–5 × 106)-expressed TbTim62-TAP was evenly spread over poly-l-lysine (100 μg/ml in H2O)-coated slides as described (42). Once the cells had settled, the slides were washed with cold PBS to remove any unattached cells. The attached cells were fixed with 3.7% paraformaldehyde and permeabilized with 0.1% Triton X-100. After blocking with 5% nonfat milk for 30 min slides were then washed with PBS containing 3% bovine serum albumin. After that, a fluorescein isothiocyanate (FITC)-conjugated anti-mouse IgG was applied as a secondary antibody for visualization under a fluorescent microscope. DNA was stained with 1 μg/ml 4′,6-diamidino-2-phenylindole (DAPI). Cells were imaged using a Nikon TE2000E widefield microscope equipped with a 60 × 1.4 NA Plan Apo VC oil immersion objective. Images were captured using a CoolSNAP HQ2 cooled CCD camera and Nikon Elements Advanced Research software.
Results
TbTim62 Is a Novel Trypanosome-specific Protein
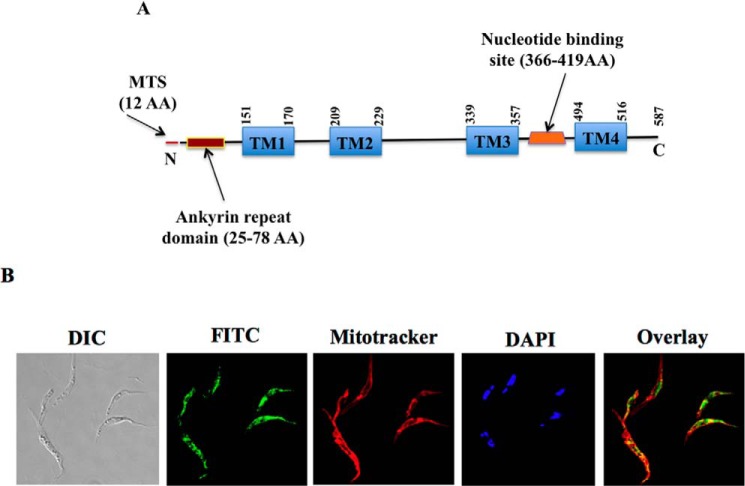
We previously reported that TbTim62 is associated with TbTim17 in T. brucei (45). Analysis of the protein sequence of TbTim62 using the Basic Local Alignment Search Tools (BLAST) (58) revealed that homologs of TbTim62 are only present in trypanosomatids, like T. brucei brucei, Trypanosoma congolense, Trypanosoma cruzi, Leishmania braziliensis, and some species of Phytomonas and Angomonas (data not shown). There are no homologs of Tim62 in fungi and higher eukaryotes. TbTim62 possesses an ankyrin repeat region, which is located near the N terminus of the protein (25–78 amino acids) (Fig. 1A). Ankyrin repeat is responsible for protein-protein interactions and is found in various proteins in different subcellular locations (59). This region of TbTim62 shows partial homology with a protein from Meleagris gallopavo (turkey) (data not shown); however, the function of this protein in turkey is not well characterized. The amino acid region (366–419 amino acids) of TbTim62 also shows weak similarity to the nucleotide binding region of a bacterial nucleotidyltransferase (Fig. 1A). TbTim62 consists of 587 amino acids with an N-terminal mitochondrial-targeting signal of 12 amino acids predicted by the Mitoprot program (60) (Fig. 1A). TMPRED (61) analysis showed that TbTim62 possesses four predicted transmembrane (TM1-TM4) domains in the center of the protein (Fig. 1A), which appear to be similar to the positions of 4 TMs in TbTim17. In addition, TbTim62 (Tb927.8.1740) has been identified in mitochondrial proteomes (10), and Mitocarta analysis also predicted its localization in T. brucei mitochondrion (11). For further verification of the subcellular localization of TbTim62, immunostaining of the T. brucei expressed a C-terminally TAP-tagged TbTim62 was performed using FITC-conjugated anti-IgG antibody. Results showed a reticular staining pattern that was merged with the Mitotracker-stained mitochondria in T. brucei (Fig. 1B). These results thus confirmed that TbTim62 is localized in mitochondria.
FIGURE 1.
TbTim62 is a novel mitochondrial protein in T. brucei. A, schematics of TbTim62 protein. The N-terminal mitochondrial-targeting signal (MTS) is shown in red. Positions of the predicted ankyrin repeat domain and nucleotide binding sites are in brown and orange boxes, respectively. Predicted transmembrane domains (TM1-TM4) are in the blue rectangle. Positions of these TMs are indicated by numbers of the amino acid (AA) residues. B, TbTim62 is localized in mitochondria. T. brucei procyclic form cells expressed TbTim62 with a TAP tag at the C terminus. After induction of expression with doxycycline, cells were stained with sheep IgG antibody followed by FITC-conjugated secondary antibody. Mitotracker and DAPI were used to visualize mitochondria and the nucleus and kinetoplast, respectively. Overlay of the FITC, Mitotracker, and DAPI is shown. DIC, differential interference contrast.
TbTim62 Is an Integral MIM Protein
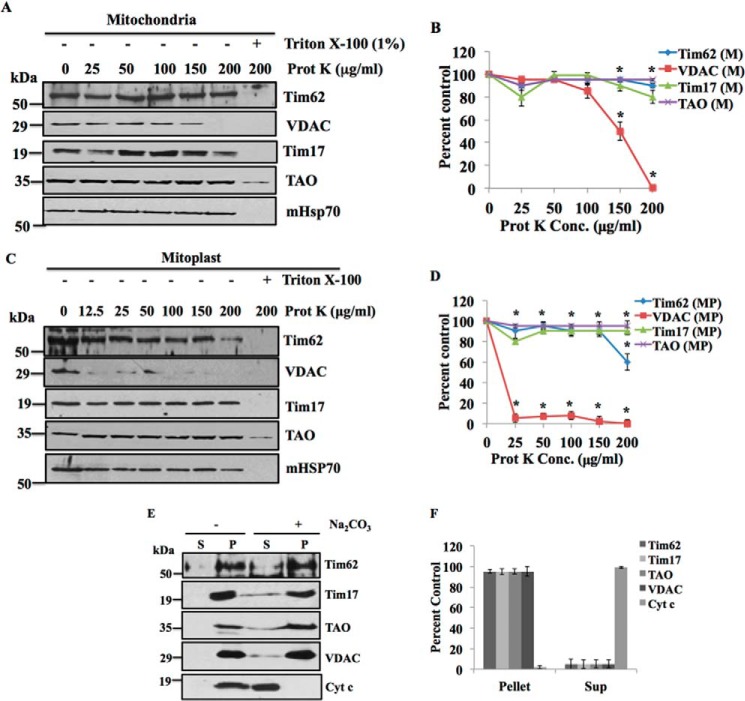
To investigate the submitochondrial location of TbTim62, isolated mitochondria from T. brucei were treated with various concentrations of proteinase K (0–200 μg/ml) (Fig. 2A). TbTim62 was protected from protease treatment similar to MIM proteins like Tim17, TAO, and the matrix-localized mHsp70, whereas VDAC, a mitochondrial outer membrane protein (53), was partially digested as expected by PK at 150–200 μg/ml (Fig. 2, A and B), suggesting that TbTim62 is not an outer membrane protein. Furthermore, TbTim62 was also protected from a similar protease digestion of isolated mitoplast and only partially digested at PK concentration, 200 μg/ml (Fig. 2, C and D), whereas VDAC was completely digested by PK in mitoplast even at the concentration of 12.5 μg/ml (Fig. 2, C and D). TbTim17, TAO, and mHsp70 were also protected in the mitoplast after PK digestion. These results indicated that TbTim62 is not exposed in the IMS and most likely embedded in the inner membrane similar to TbTim17. Alkali extraction of the isolated mitochondria revealed that TbTim62 is present in the alkali-resistant membrane pellet, similar to other mitochondrial membrane proteins, Tim17, TAO, and VDAC (Fig. 2, E and F), whereas Cyt c, a soluble protein of the intermembrane space, is found in the supernatant fraction after alkali treatment, as expected. Therefore, we conclude that TbTim62 is an integral MIM protein.
FIGURE 2.
TbTim62 is localized in the mitochondrial inner membrane. A–D, limited protease digestion of isolated mitochondria (A) and mitoplast (C). Equal amounts of mitochondria and mitoplast isolated from T. brucei procyclic form were treated with various concentrations of proteinase K (Prot K) as indicated. After digestion, mitochondria and mitoplast were re-isolated by centrifugation, and proteins were analyzed by SDS-PAGE and immunoblot using TbTim62, VDAC, TbTim17, TAO, and mHsp70 antibodies as probes. As indicated, some samples were also treated with Triton X-100 (1%) along with proteinase K. Intensity of the respective protein bands for TbTim62, VDAC, Tim17, and TAO in mitochondria (B) and mitoplast (D) after treatment with different concentration of proteinase K were quantitated, normalized with the corresponding mHsp70 intensity, and plotted as the percent of that in no-treatment control. S.E. from three independent experiments are shown. *, p values <0.001. E, sodium carbonate extraction of mitochondrial proteins from T. brucei procyclic form. Mitochondria were treated (+) with Na2CO3 or left untreated (−). Mitochondria were re-isolated by centrifugation. Equal volumes of the supernatant (S) and the pellet (P) fractions were analyzed using TbTim62, TbTim17, TAO, VDAC, and Cyt c antibodies as probes. F, intensity of each protein band in the supernatant and pellet fractions after treatment with Na2CO3 was plotted as percent of that in untreated control pellet. S.E. are calculated from three independent experiments.
Import of TbTim62 into Mitochondria Requires TbTim17
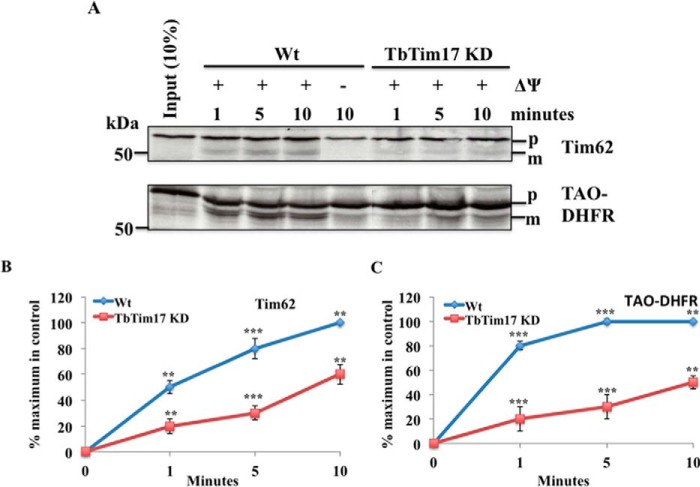
TbTim62 possesses a predicted presequence of 12 amino acids. We wanted to investigate if the import of TbTim62 into mitochondria depends on TbTim17. In vitro import analysis of the radiolabeled TbTim62 into isolated T. brucei wild type mitochondria revealed that TbTim62 is imported into mitochondria (Fig. 3A). Once imported, a matured TbTim62 was formed ina time-dependent manner. The mature protein band was ∼1-kDa smaller than the precursor protein, suggesting that TbTim62 presequence is cleaved after its import into mitochondria (Fig. 3A). Furthermore, pretreatment of mitochondria with CCCP to disrupt mitochondrial membrane potential inhibited formation of the matured band, suggesting that this band is formed due to import and processing of TbTim62. Reduction of TbTim17 protein level significantly hampered the import of TbTim62 (Fig. 3A). Import of TbTim62 was inhibited ∼80% in TbTim17 KD mitochondria in comparison to wild type (Fig. 3, A and B). Import of a presequence-containing fusion protein, TAO-DHFR, was also inhibited at a similar level in TbTim17 KD mitochondria (Fig. 3, A and C) as we reported previously. Therefore, similar to other presequence-containing nucleus-encoded mitochondrial proteins import of TbTim62 depends on TbTim17.
FIGURE 3.
In vitro import of TbTim62 into wild type control and TbTim17 KD T. brucei mitochondria. A, radiolabeled TbTim62 were incubated with isolated mitochondria from the Wt and TbTim17 KD T. brucei procyclic form for different time periods (1–10 min). Import reactions for TAO-DHFR were also run in parallel. Post-import, mitochondria were washed, and proteins were analyzed by SDS-PAGE and autoradiography. Input lanes represent 10% of radiolabeled substrate proteins used for each reaction. The precursor (p) and the mature (m) proteins are shown. For some reactions mitochondrial membrane potential was disrupted by pretreatment of mitochondria with CCCP. Intensity of the imported and matured Tim62 (B) and TAO-DHFR (C) in Wt and TbTim17 KD mitochondria were quantitated using densitometry. The intensity of the matured protein imported into the Wt mitochondria at the longest time point was set as maximum (100%), and the calculated percentage of the maximum import at other time points into mitochondria from the control and TbTim17 KD was plotted versus time. S.E. are calculated from three independent experiments. p values: ***, <0.001; **, <0.05.
TbTim62 KD Decreased the Steady-state Levels of TbTim17 Post-transcriptionally
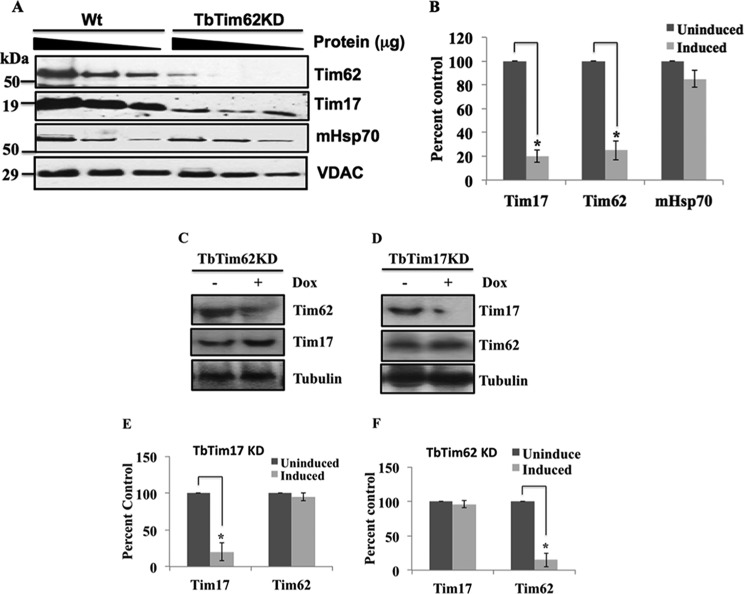
We reported previously that TbTim62 KD reduces cell growth and inhibits import of the N-terminal signal-containing proteins into mitochondria (45). TbTim62 also associates with TbTim17 (45). To investigate if TbTim62 KD has any effect on the levels of TbTim17, immunoblot analysis of mitochondrial proteins isolated from the wild type and TbTim62 RNAi cells at day 4 after induction was performed using anti-TbTim17 and anti-TbTim62 antibodies as probes (Fig. 4A). Results showed that TbTim62 KD reduced the levels of TbTim62 ∼70% after induction of RNAi (Fig. 4, A and B). Interestingly, the level of TbTim17 was also decreased >80% due to knockdown of TbTim62 (Fig. 4, A and B). mHsp70 levels were minimally affected due to TbTim62 KD. VDAC was used as the loading control. Analysis of the transcript levels of TbTim17 and TbTim62 revealed that RNAi specifically reduced the target transcript (Fig. 4, C and D). The levels of TbTim17 transcript remain unaltered in TbTim62 KD cells and vice versa (Fig. 4, C–F). Therefore, these results indicated that TbTim62 KD reduced the level of TbTim17 post-transcriptionally.
FIGURE 4.
Effect of TbTim62 knockdown on the steady-state levels of TbTim17 protein and transcript. A, mitochondria were isolated from the T. brucei procyclic form Wt and from TbTim62 RNAi cells grown for 4 days in the presence of doxycycline. Mitochondrial proteins (100, 50, and 25 μg) were analyzed by immunoblot analysis using TbTim17, TbTim62, mHsp70, and VDAC antibodies as probes. B, the intensity of the respective protein bands were quantitated using imaging densitometer as described under “Experimental Procedures” normalized with the corresponding VDAC protein bands and plotted as percent in the WT control. S.E. are calculated from three independent experiments. C and D, TbTim17 RNAi and TbTim62 RNAi cells lines were cultured in the presence (+) and absence (−) of doxycycline for 2 days. Total RNAs were isolated and analyzed by Northern blot analysis using cDNAs for TbTim62 and TbTim17 as probes as indicated. Tubulin was used as the control for Northern blot. E and F, the intensity of the respective bands were quantitated using imaging densitometer as described, normalized with the corresponding tubulin band, and plotted as percent in the respective un-induced controls. S.E. are calculated from four independent experiments. *, p values <0.05.
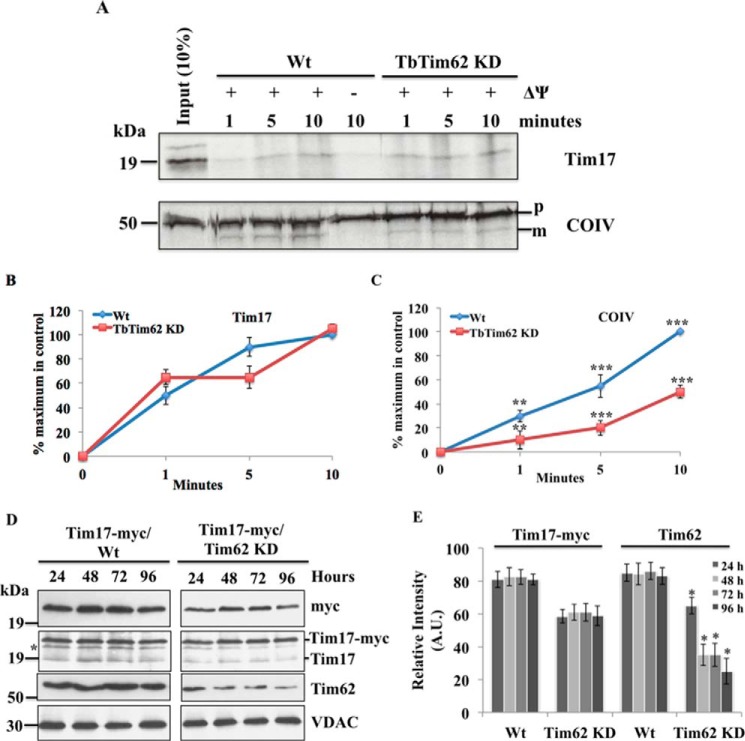
TbTim62 Is Not Required for Import of TbTim17 into T. brucei Mitochondria
As we found that depletion of TbTim62 reduced the steady-state levels of TbTim17, we wanted to investigate if import of TbTim17 into mitochondria is hampered due to a reduction in the levels of TbTim62. To test this assumption, in vitro import analysis was performed using radiolabeled TbTim17 as the substrate. TbTim17 does not have any predicted presequence and thus did not produce any lower molecular mass mature protein after its entry into mitochondria. Therefore, mitochondria samples were treated with PK after import to remove un-imported radiolabeled proteins, and imported TbTim17 that is protected from PK digestion was analyzed by autoradiography. In wild type mitochondria, TbTim17 was imported in a time-dependent manner, and import was mostly inhibited in the absence of mitochondrial membrane potential (Fig. 5A). We did not observe any significant changes in import of TbTim17 into TbTim62 KD mitochondria (Fig. 5, A and B), indicating that TbTim62 is not required for import of TbTim17. We also tested the effect of TbTim62 KD on import of COIV, an N-terminal signal-containing MIM protein. As we reported previously, import of COIV was inhibited ∼40–60% in TbTim62 KD mitochondria in comparison with wild type control (Fig. 5, A and C). Therefore, it suggests that like TbTim17, TbTim62 is also required for import of proteins that have N-terminal mitochondrial-targeting signal, but it is not likely involved in the import of internal signal-containing mitochondrial proteins, like TbTim17.
FIGURE 5.
Import of TbTim17 in the wild type control and TbTim62-depleted mitochondria. A, radiolabeled TbTim17 were incubated with isolated mitochondria from the Wt and TbTim62 KD T. brucei procyclic form for different time periods (1–10 min). Post-import, mitochondria were washed and treated with proteinase K (20 μg/ml) for 30 min at 4 °C, and proteins were analyzed by SDS-PAGE and autoradiography. Import reactions for COIV were also run in parallel. The precursor (p) and the mature (m) proteins are shown. Input lanes represent 10% of radiolabeled substrate proteins used for each reaction. For some reactions mitochondrial membrane potential was disrupted by pretreatment of mitochondria with CCCP. Intensity of the Tim17 (B) and matured COIV (C) imported into Wt and TbTim17 KD mitochondria were quantitated using densitometry. The intensity of the matured protein imported into the Wt mitochondria at the longest time point was set as maximum (100%), and the calculated percentage of the maximum import at other time points into mitochondria from the Wt and TbTim17 KD was plotted versus time. S.E. are calculated from three independent experiments. p values: ***, <0.001; **, <0.05. D, expression and targeting of TbTim17-myc into mitochondria in T. brucei Wt and TbTim62 KD. TbTim17–2X-myc was expressed in the wild type and in T. brucei where TbTim62 levels were reduced simultaneously by RNAi. At different time points (24–96 h) after induction mitochondrial fractions were isolated, and proteins were analyzed by immunoblot using anti-myc, anti-TbTim17, anti-TbTim62, and anti-VDAC antibodies. The ectopically expressed TbTim17–2X-myc and the endogenous TbTim17 were both detected by anti-TbTim17 antibody. E, intensities of the Tim17-myc and Tim62 protein bands were quantitated and normalized with that for the corresponding VDAC band, and values were plotted as relative intensity (arbitrary unit) in Wt and TbTim62 KD mitochondria. The number of experiment was three. Tim62 levels were significantly reduced in Tim17-myc/Tim62 KD relative to TbTim17-myc/Wt mitochondria. *, p values <0.05.
To verify our results that TbTim62 is not involved in the import of TbTim17 into mitochondria, we performed an in vivo mitochondrial protein import assay. For this assay, an ectopic copy of TbTim17 with a 2X-myc epitope at the C terminus was expressed in T. brucei, whereas RNAi reduced the levels of TbTim62 simultaneously upon induction with doxycycline. After induction, cells were harvested at different time points, mitochondrial fractions were isolated, and the levels of TbTim17–2X-myc in the mitochondrial fractions were assessed by immunoblot analysis using anti-myc and anti-TbTim17 antibodies (Fig. 5D). TbTim17–2X-myc was also expressed in the wild type cells to use as the control. We found that anti-myc antibody recognized the ectopically expressed TbTim17–2X-myc (21 kDa), whereas anti-TbTim17 antibody recognized both the endogenous (19 kDa) and the recombinant TbTim17 (21 kDa). In the wild type T. brucei, TbTim17–2X-myc was expressed and accumulated into mitochondria with time (24–96 h) of induction. TbTim17–2X-myc was also accumulated into mitochondria in TbTim62 KD T. brucei, showing that import of TbTim17 into mitochondria was not inhibited due to TbTim62 KD. The levels of TbTim17–2X-myc in mitochondria isolated from TbTim62 KD T. brucei were relatively lower in comparison with those in mitochondria isolated from the wild type cells (Fig. 5, D and E), which could be due to a reduced stability of TbTim17–2X-myc for TbTim62 KD. Immunoblot analysis of these samples with anti-TbTim17 antibody further showed that the newly synthesized TbTim17–2X-myc localized into mitochondria and present at similar levels during 48–96 h after induction of TbTim62 RNAi. As expected, the levels of the endogenous TbTim17 were relatively lower in these mitochondria in comparison to wild type control. Immunoblot analysis of these samples using anti-TbTim62 antibody confirmed that TbTim62 RNAi significantly reduced the levels of the target protein in mitochondria (Fig. 5, D and E). VDAC was used as the loading control. Together these results demonstrate that TbTim62 is not essential for import of TbTim17 into mitochondria but is required for the stability of this protein into the organelle.
TbTim62 KD Reduced the Half-life of TbTim17
To further confirm that TbTim62 KD decreased the stability of TbTim17, we determined the half-life of TbTim17 in the wild type control and TbTim62 KD T. brucei (Fig. 6). For this experiment we inhibited protein synthesis by the addition of cycloheximide (50 μg/ml) and then assessed the steady-state levels of TbTim17 by immunoblot analysis (Fig. 6A). Results showed that in the wild type control cells TbTim17 level was almost unaltered even after inhibition of protein synthesis for 24 h, showing that the half-life of TbTim17 is >24 h, whereas in TbTim62 KD cells the level of TbTim17 was reduced to half within 6 h and was decreased further with longer incubation in the presence of cycloheximide (Fig. 6, A and B), indicating that the half-life of TbTim17 is at least 4-fold less in TbTim62 KD cells in comparison with the wild type control. Therefore, we conclude that a decrease in the levels of TbTim17 in TbTim62 KD cells is due to reduced half-life of this protein.
FIGURE 6.
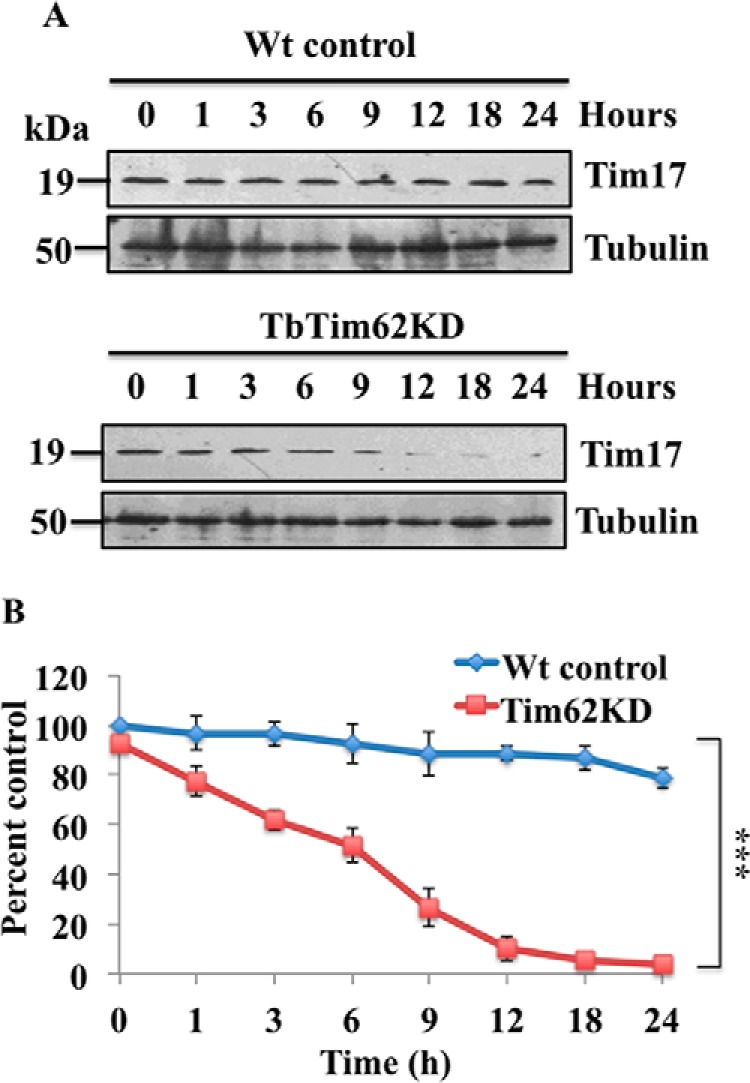
TbTim17 protein decay in wild type control and TbTim62 KD T. brucei. The Wt control and TbTim62 KD cells were grown in the presence of doxycycline for 2 days. Cells were treated with cycloheximide (50 μg/ml) under normal growth conditions. A, at different time points (0–24 h) cells were harvested, and total cellular proteins were analyzed by SDS-PAGE and immunoblot analysis using anti-TbTim17 antibody. Tubulin was used as the loading control. B, intensity of the TbTim17 protein band were quantitated by imaging densitometer, normalized with the intensity of the corresponding tubulin band, and plotted against time. The number of experiments was three. ***, p values <0.001.
Analysis of Mitochondrial Membrane Protein Complexes Consisting of TbTim62 and TbTim17
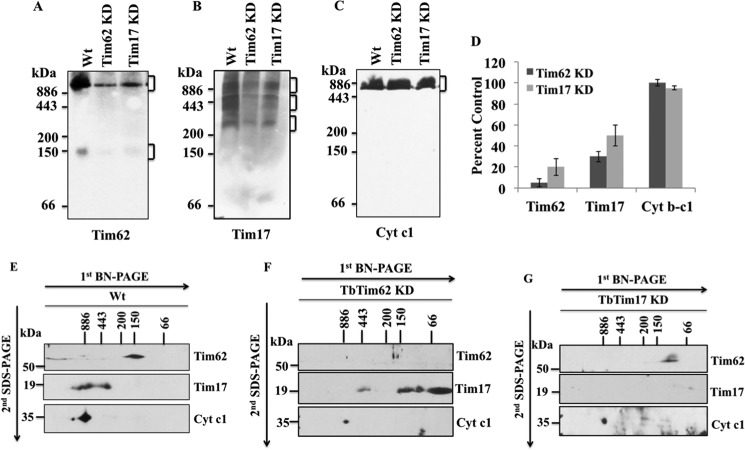
BN-PAGE analysis of the mitochondrial protein complexes followed by immunoblot analysis using anti-TbTim62 antibody revealed that TbTim62 is present in a protein complex of ∼150 kDa (Fig. 7A). A larger complex (∼1100 kDa), which is similar in size to the largest complex recognized by the anti-TbTim17 antibody, was also detected from the mitochondrial samples by this antibody (Fig. 7A). Levels of these complexes were significantly reduced in TbTim62 KD mitochondria (Fig. 7, A and D), showing that these protein complexes indeed contain TbTim62. TbTim17 KD also reduced the levels of these protein complexes in mitochondria in comparison with those in wild type T. brucei. Because TbTim17 is required for the import of TbTim62 into mitochondria, it is expected that depletion of TbTim17 would reduce the levels of the TbTim62 protein complexes. Probing of the duplicate blot with anti-TbTim17 antibody revealed that TbTim17 is present in multiple protein complexes within the range of ∼300 to ∼1100 kDa (Fig. 7B). Levels of these complexes were reduced in TbTim17 KD mitochondria and, more interestingly, also in TbTim62 KD mitochondria (Fig. 7, B and D), indicating that TbTim62 is required for overall stability of the protein complexes consisting of TbTim17. As expected, anti-Cyt c1 antibody recognized the cytochrome b-c1 complex at similar levels in all samples (Fig. 7, C and D).
FIGURE 7.
Analysis of the mitochondrial protein complexes containing TbTim62 and TbTim17. Mitochondria were isolated from T. brucei wild type, TbTim62 KD, and TbTim17 KD cells after 4 days induction of RNAi with doxycycline. Mitochondria (100 μg) samples were solubilized with digitonin (1.0%). The solubilized supernatant were clarified by centrifugation at 100,000 × g and analyzed by BN-PAGE. Protein complexes (as indicated by the bracket at the right side of the blot) were detected by immunoblot analysis using antibodies for TbTim62 (A), TbTim17 (B), and Cyt c1 (C). The Cyt c1 antibody recognized the cytochrome b-c1 reductase complex. Molecular size marker proteins apoferritin dimer (886 kDa), apoferritin monomer (443 kDa), β-amylase (200 kDa), alcohol dehydrogenase (150 kDa), and bovine serum albumin (66 kDa) were run on the gel and visualized by Coomassie Blue staining. D, intensity of the Tim62 (∼150 and ∼1100 kDa), Tim17 (∼300 to ∼1100 kDa), and cytochrome b-c1 (∼700 kDa) protein complexes were quantitated by imaging densitometry. The intensity of these complexes in TbTim62 KD and TbTim17 KD mitochondria were calculated as percent of that in Wt control and plotted. S.E. are calculated from three independent experiments. E–G, gel strips representing individual lanes for Wt (E), TbTim62 KD (F), and TbTim17 KD (H) were excised from the first-dimensional gel and subjected to 12% Tricine SDS-PAGE. Proteins were transferred to a nitrocellulose membrane, and blots were sequentially probed with anti-TbTim17, anti-TbTim62, and anti-Cyt c1 antibodies.
To further resolve the protein complexes detected with the above antibodies, gel strips from the first dimension BN-PAGE for the mitochondrial proteins isolated from the wild type, TbTim62 KD, and TbTim17 KD T. brucei were subjected to Tricine SDS-PAGE, transferred to the nitrocellulose membrane, and sequentially detected with anti-Tim17, anti-Tim62, and anti-Cyt c1 antibodies. Results from the wild type mitochondrial sample further confirmed that TbTim62 is present in the ∼150-kDa protein complex, and TbTim17 is present in a ∼300-kDa and a couple of larger complexes as seen on the one-dimensional PAGE (Fig. 7E). A faint band of TbTim62 was detected above ∼886 kDa, suggesting that a fraction of TbTim62 is also present in a higher molecular mass protein complex, which is similar in size to that of TbTim17. As reported previously, Cyt c1 is found in the cytochrome b-c1 reductase complex (∼700 kDa) (Fig. 7E). In contrast to wild type, two-dimensional-BN-SDS-PAGE analysis of the TbTim62 KD mitochondrial sample showed that TbTim17 is primarily present in much smaller (∼200 to <66 kDa) complexes, and very little was found in the range of 300–1100 kDa (Fig. 7F). These results clearly indicate that TbTim17 complexes are destabilized in TbTim62 KD mitochondria. Interestingly, these smaller size TbTim17 bands were more prominent in two-dimensional than one-dimensional PAGE, which is most likely due to better recognition of the denatured protein than its native form by the anti-TbTim17 antibody. As expected, TbTim17 and TbTim62 proteins were minimally detected in respective KD samples (Fig. 7, F and G). Cyt c1 was detected at similar positions in all three mitochondria samples, although its levels were relatively lower in TbTim17 KD and TbTim62 KD mitochondria than wild type (WT). Together these results showed that TbTim62 KD specifically hampered the stability of the TbTim17 complex.
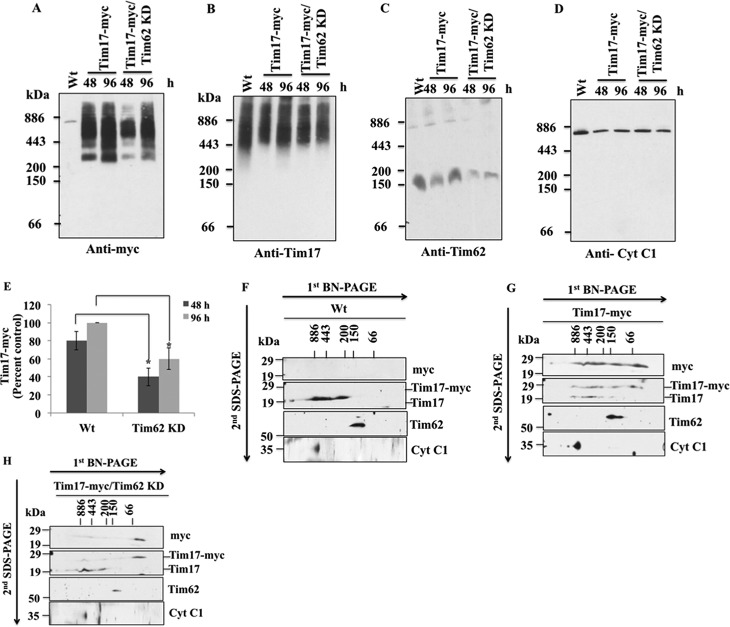
TbTim62 Is Required for the Assembly of TbTim17 Protein Complex
Next we investigated if TbTim62 plays role in the assembly of the newly synthesized TbTim17 into the matured complexes. For this purpose we compared the assembly status of the ectopically expressed TbTim17–2X-myc into TbTim17 complexes in the wild type and TbTim62 KD T. brucei. Mitochondria were isolated from TbTim62 KD and parental T. brucei which expressed the recombinant TbTim17–2X-myc (Fig. 8). Analysis of mitochondrial membrane protein complexes at 48 and 96 h after induction by BN-PAGE followed by immunoblot using anti-myc antibody revealed that TbTim17–2X-myc assembled into the protein complexes similar in size to the endogenous TbTim17 very efficiently in wild type control mitochondria (Fig. 8A). However, in TbTim62 KD T. brucei, TbTim17–2X-myc assembled into the respective mitochondrial protein complexes very poorly at 48 h and also at a significantly lower level at 96 h in comparison to those in the wild type cells (Fig. 8, A and E). Anti-myc antibody did not recognize any complexes from the wild type parasites, which do not express TbTim17–2X-myc. Anti-TbTim17 antibody recognized the TbTim17 protein complexes at comparable levels in mitochondria samples collected at 48 h (Fig. 8B). Therefore, these results demonstrated that TbTim62 is not only needed for the stability of the TbTim17 complexes but also for the assembly of the newly synthesized TbTim17–2X-myc into these complexes. Probing this blot with anti-TbTim62 antibody showed that the levels of the ∼150-kDa protein complex consisting of TbTim62 were greatly reduced due to TbTim62 KD, as expected (Fig. 8C). The levels of the larger complex of TbTim62 (∼1100 kDa) were also reduced in TbTim62 KD T. brucei. Cytochrome b-c1 reductase complex was found at similar levels in all samples (Fig. 8D).
FIGURE 8.
Assembly of TbTim17 into the T. brucei mitochondrial membrane protein complexes. A–D, assembly of the ectopically expressed TbTim17–2X-myc in the mitochondrial protein complexes in T. brucei. Mitochondria were isolated from T. brucei Wt, which expressed TbTim17–2X-myc, and from T. brucei, which expressed this protein along with TbTim62 RNAi at days 2 and 4 after induction with doxycycline. Mitochondrial proteins were solubilized with 1% digitonin and analyzed by BN-PAGE. Proteins were transferred to nitrocellulose membrane and probed with anti-myc (A), anti-TbTim17 (B), anti-TbTim62 (C), and anti-Cyt c1 (D) antibodies. E, intensities of the Tim17-myc complex (∼300 to ∼1100 kDa) in Tim17-myc and Tim17-myc/Tim62 KD cells at 48 and 96 h after induction with doxycycline were quantitated and plotted as the percent of the maximum intensity of the complex found at 96 h in Wt background. S.E. are calculated from three independent experiments. *, p values <0.05. F–H, gel strips representing the individual lanes for Wt (F), TbTim17-myc (G), and TbTim17-myc/Tim62 KD (H) samples collected after 48 h of induction with doxycycline were excised from the first-dimension BN-PAGE gel and subjected to 12% Tricine SDS-PAGE. Proteins were transferred to a nitrocellulose membrane, and blots were sequentially probed with anti-myc, anti-TbTim17, anti-TbTim62, and anti Cyt c1 antibodies.
To further confirm our results, we analyzed these mitochondria samples on two-dimensional-BN-SDS-PAGE followed by immunoblot analysis. TbTim17–2X-myc, as recognized by both anti-myc and anti-TbTim17 antibodies, was found in complexes that are in similar sizes of the complexes formed by the endogenous TbTim17 in WT background (Fig. 8G). A couple of smaller complexes (∼150 kDa and <66 kDa) also formed by TbTim17–2X-myc, which could be the assembly intermediates of TbTim17–2X-myc (Fig. 8G). Specifically the ∼150-kDa TbTim17–2X-myc band is at a similar position with TbTim62, suggesting that TbTim17 most likely associates with TbTim62 first before it assembled to the larger complex. However, we also could not exclude the possibility that these lower molecular mass complexes of TbTim17–2X-myc are the aggregated form of this protein produced due to overexpression. In contrast to the wild type, in TbTim62 KD mitochondria TbTim17–2X-myc was mostly found in the <66-kDa region and was minimally detected in the size similar to the matured complexes (Fig. 8H). Anti-TbTim17 antibody recognized the endogenous TbTim17 complexes similarly in both the TbTim17–2X-myc and TbTim17–2X-myc/TbTim62 KD mitochondria (Fig. 8, G and H). As a control we also analyzed in parallel mitochondria samples from wild type T. brucei cells, which do not express TbTim17–2X-myc (Fig. 8F). Reprobing these blots with anti-TbTim62 antibody showed that the levels of TbTim62 protein complex (∼150 kDa) were reduced in TbTim62 KD mitochondria as expected (Fig. 8H). Cyt c1 was detected at similar positions in all samples (Fig. 8, F–H). Together these results confirmed that TbTim62 KD greatly hampered the assembly of the TbTim17–2X-myc into matured TbTim17 protein complexes in T. brucei.
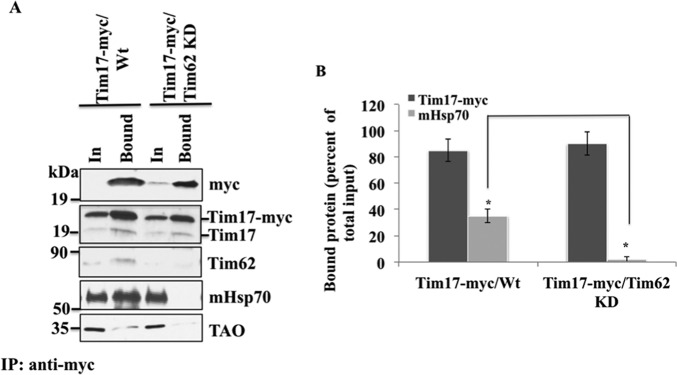
TbTim62 KD Reduced the Association of TbTim17 with mHsp70
Next we investigated if TbTim62 KD has any effect on association of TbTim17 with other proteins. For this purpose we immunoprecipitated TbTim17–2X-myc using anti-myc antibody-coupled agarose beads from the mitochondrial extracts prepared from T. brucei, which expressed this recombinant protein in the presence and absence of TbTim62 RNAi. Immunoblot analysis using anti-myc antibody showed that TbTim17–2X-myc was immunoprecipitated from cells expressing this protein (Fig. 9A). Anti-TbTim17 antibody detected both the TbTim17–2X-myc (21 kDa) and TbTim17 (19 kDa) in the immunoprecipitate from the TbTim17–2X-myc/Wt and TbTim17–2X-myc/TbTim62 KD samples (Fig. 9A), showing that the endogenous protein was associated with the ectopically expressed TbTim17–2X-myc in both types of mitochondria. Reprobing the blots with anti-Hsp70 antibody demonstrated that mitochondrial Hsp70 (30–40% of input) co-precipitated with TbTim17–2X-myc from the TbTim17–2X-myc/Wt mitochondrial extract but not from TbTim17–2X-myc/TbTim62 KD sample (Fig. 9, A and B), indicating that TbTim62 KD hampered the association of mHsp70 with TbTim17–2X-myc. As expected, TbTim62 co-precipitated from the TbTim17–2X-myc/Wt samples, but no protein was detected when RNAi reduced the levels of TbTim62. Together, these results demonstrate that reduction of the levels of TbTim62 reduced the association of mHsp70 with TbTim17 and hampered the stability of the TbTim17 protein complexes.
FIGURE 9.
Co-immunoprecipitation analysis. A, mitochondria isolated from T. brucei expressed TbTim17–2X-myc (TbTim17-myc/WT), as well as from T. brucei, which expressed this protein along with TbTim62 RNAi (TbTim17-myc/TbTim62 KD), were lysed with 1% digitonin. The soluble proteins were immunoprecipitated (IP) with anti-myc-coupled agarose beads. Proteins from the input (In, 10%) and precipitated (Bound, 50%) fractions were analyzed by immunoblotting using anti-myc, anti-TbTim17, anti-TbTim62, anti-mHsp70, and anti-TAO antibodies. Molecular masses for the marker proteins are indicated by the side of the blots. B, amount of bound Tim17-myc and mHsp70 were quantitated as percent of total input from the respective mitochondria samples. S.E. are calculated from three independent experiments. *, p value for precipitated mHsp70 from two types of mitochondrial sample is <0.05.
Discussion
We discovered TbTim62, a novel component of the trypanosomal TIM machinery that has no homologs in other eukaryotes. TbTim62 is an integral MIM protein and possesses a cleavable N-terminal targeting signal. A fraction of TbTim62 is found in an ∼1100-kDa MIM complex, which is similar in size to the largest complex consisting of TbTim17. However, TbTim62 is not present in ∼300–800-kDa protein complexes formed by TbTim17. Depletion of TbTim62 inhibited assembly of TbTim17 into these matured complexes and reduced the association of TbTim17 with mHsp70. The un-assembled TbTim17 was degraded in TbTim62 KD mitochondria, and its steady-state levels were reduced. Therefore, TbTim62 is critical for formation of the stable TbTIM and essential for mitochondrial protein import in T. brucei.
In fungi and animals, Tim17 and Tim23 form modular type complexes (62, 63). BN-PAGE analysis of the fungal mitochondrial protein complexes showed that Tim17 associates with Tim23 and forms a 90-kDa core complex. These two proteins are also found in ∼140- and ∼240 kDa complexes, although at a lower level than the 90-kDa complex. Tim50, a peripheral component of the TIM23 complex, is present in the core as well as in the ∼240-kDa complex along with Tim23 and Tim17. This larger complex also associates with the PAM complex consisting of mHsp70, Pam16, and Pam18 (62–64). Furthermore, the Tim23-17 complex transiently associates with the respiratory complexes during translocation of the MIM proteins (32, 64). Tim23 is the major subunit of this complex, which forms the import channel for the precursor protein.
In trypanosomes, TbTim17 is the only member of the PRAT family protein and most likely the major component of the preprotein import complex (46). We found that TbTim17 also forms modular type complexes, likely by association with different T. brucei proteins. However, sizes of these complexes are different than the fungal Tim23-17 complexes as described above. We showed that TbTim62, TbTim50, TbTim54, mHsp70, and some other proteins are associated with TbTim17 (45). Therefore, these proteins could be the components of the TbTim17 complex. Interestingly, we found that TbTim62, which is an interacting partner of TbTim17, separates as a ∼150-kDa, and TbTim17 is found in the ∼300- to ∼800-kDa protein complexes on BN-PAGE. Therefore, it appears that these two proteins could be present in separate complexes and transiently associate with each other. However, we could not exclude the possibility that the ∼150-kDa TbTim62 protein complex was separated from the larger TbTim17 protein complex during solubilization of the mitochondrial membrane proteins by digitonin or during electrophoresis on a native gel. Overall, we noticed that TbTim17 protein complexes are distinct in sizes and composition than its counterpart in other eukaryotes.
We showed that TbTim62 is essential for the stability of TbTim17- and the TbTim17-containing protein complexes. In the absence of TbTim62, TbTim17 protein complexes destabilized and unassembled or partially assembled TbTim17 accumulated and finally degraded proteolytically, thus the steady-state levels of TbTim17 reduced. As we found that TbTim62 KD did not inhibit the import of TbTim17; therefore, TbTim17 most likely is degraded within mitochondria post-import. TbTim62 possesses an ankyrin repeat motif at the N terminus. This region could be responsible for its interaction with TbTim17. Reduction of the levels of TbTim62 reduced association between these two proteins that may target TbTim17 for degradation by mitochondrial proteases. Therefore, association with TbTim62 is required for the stability of TbTim17 and the TbTim17 protein complexes.
After import, TbTim17 is required to be assembled with other proteins to form the translocase complex. TbTim62 clearly plays a role in this process. We showed that the ectopically expressed TbTim17–2X-myc was assembled into the endogenous TbTim17 complex at a significantly reduced rate in TbTim62 RNAi cells in comparison to the wild type parasite. Furthermore, TbTim17–2X-myc was mostly found in a >66 kDa complex in TbTim62 KD mitochondria, showing that its assembly is inhibited. In addition, TbTim17–2X-myc was also present in an ∼150-kDa complex that is similar in size to the complex formed by TbTim62, suggesting that association of TbTim62 could be prerequisite for TbTim17 to be assembled into the matured complexes. Therefore, all together these results showed that TbTim62 is critical for the assembly of TbTim17 with other components, which is required for formation of stable TbTim17 complexes.
In fungi and higher eukaryotes, mHsp70 associates with the preprotein translocase, Tim23-17 (12, 13). We also found that TbTim17 stably associates with mHsp70 in wild type T. brucei mitochondria. TbTim62 was also co-precipitated with TbTim17–2X-myc and TbTim17, showing its association with the TbTim17 complex as reported previously (46). In contrast to the wild type mitochondria, the association of TbTim17 with mHSP70 was greatly reduced when the levels of TbTim62 were decreased by RNAi, showing that TbTim62 is required for association of TbTim17 with mHsp70. However, the reduction of TbTim62 did not inhibit the homotypic association of TbTim17, as we observed the endogenous TbTim17 co-precipitated with TbTim17–2X-myc from the mitochondrial extract obtained from both the wild type as well as TbTim62 KD T. brucei. From these results it can be anticipated that TbTim62 is responsible for recruiting mHsp70 to the TbTim17 complex, which facilitates its assembly into the matured complexes consisting of TbTim17. Therefore, TbTim62 is needed not only for the stability of the existing TbTim17 complex but also for the formation of a stable TbTim17 complex. Tandem affinity purification of TbTim62 protein complex revealed that TbTim62 indeed associates with several chaperone proteins, such as Hsp70, Hsp60, Hsp94, and a Clp-protease like chaperone (data not shown). However, further investigation is required to clarify the role of TbTim62 in the assembly of the TbTim17 protein complexes. Together, our results showed that TbTim62 is a unique and important partner of the TIM complex in T. brucei. In the absence of TbTim62, the translocase complex is not formed or dissociates rapidly, and TbTim17 is targeted for degradation. Therefore, TbTim62 levels could be an important parameter for regulation of mitochondrial activities in T. brucei.
Author Contributions
U. K. S., V. H., and M. C. planned and executed the experiments. A major part of the work was done by U. K. S. V. H. performed and analyzed the experiments shown in Figs. 3 and 4, A–C. M. C. wrote the manuscript. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgments
We thank George Cross for the pLew100 vectors and the T. brucei 427 29-13 cell line, Paul Englund for anti-mHSP70, Steve Hajduk for anti-Cyt c1, Andre Schneider for anti-Cyt c, and Keith Gull for p2T7Ti-177 RNAi vector and β-tubulin antibodies. We thank Joseph Smith and Ebony Weems for critically reviewing the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants 2SC1GM081146, T32HL007727, U54RR026140, U54MDO7593, and G12RR003032. The authors declare that they have no conflicts of interest with the contents of this article.
- TOM
- translocase of the mitochondrial outer membrane
- MIM
- mitochondrial inner membrane
- TIM
- translocase of MIM
- TbTIM
- T. brucei TIM
- PAM
- presequence-activated motor complex
- MCP
- metabolite carrier protein
- IMS
- mitochondrial intermembrane space
- PRAT
- presequence and amino acid transporter
- TM
- transmembrane
- KD
- knockdown
- BN-PAGE
- Blue-native polyacrylamide gel electrophoresis
- TAP
- tandem affinity purification
- Cyt c
- cytochrome c
- CCCP
- carbonyl cyanide m-chlorophenyl hydrazone
- COIV
- cytochrome oxidase subunit 4
- TAO
- trypanosome alternative oxidase
- DHFR
- dihydrofolate reductase
- Bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine
- VDAC
- voltage-dependent anion channel protein
- Hsp70
- heat shock protein 70
- mHsp70
- mitochondrial Hsp70.
References
- 1. Cecílio P., Pérez-Cabezas B., Santarém N., Maciel J., Rodrigues V., Cordeiro da Silva A. (2014) Deception and manipulation: the arms of Leishmania, a successful parasite. Front. Immunol. 5, 480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rodrigues J. C., Godinho J. L., de Souza W. (2014) Biology of human pathogenic trypanosomatids: epidemiology, lifecycle, and ultrastructure. Subcell. Biochem. 74, 1–42 [DOI] [PubMed] [Google Scholar]
- 3. Beschin A., Van Den Abbeele J., De Baetselier P., Pays E. (2014) African trypanosome control in the insect vector and mammalian host. Trends Parasitol. 30, 538–547 [DOI] [PubMed] [Google Scholar]
- 4. Brennand A., Rico E., Michels P. A. (2012) Autophagy in trypanosomatids. Cells 1, 346–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rezende A. M., Assis L. A., Nunes E. C., da Costa Lima T. D., Marchini F. K., Freire E. R., Reis C. R., de Melo Neto O. P. (2014) The translation initiation complex eIF3 in trpanosmatids and other pathogenic excavates: identification of conserved and divergent features based on orthologue analysis. BMC Genomics 15, 1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Povelones M. L. (2014) Beyond replication: division and segregation of mitochondrial DNA in kinetoplastids. Mol. Biochem. Parasitol. 196, 53–60 [DOI] [PubMed] [Google Scholar]
- 7. Jensen R. E., Englund P. T. (2012) Network news: the replication of kinetoplastid DNA. Annu. Rev. Microbiol. 66, 473–491 [DOI] [PubMed] [Google Scholar]
- 8. Motta M. C. (2008) Kinetoplast as a potential chemotherapeutic target of trypanosomatids. Curr. Pharm. Des. 14, 847–854 [DOI] [PubMed] [Google Scholar]
- 9. Aphasizhev R., Aphasizheva I. (2011) Mitochondrial RNA processing in trypanosomes. Res. Microbiol. 162, 655–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Acestor N., Panigrahi A. K., Ogata Y., Anupama A., Stuart K. D. (2009) Protein composition of Trypanosoma brucei mitochondrial membrane. Proteomics. 9, 5497–5508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang X., Cui J., Nilsson D., Gunasekera K., Chanfon A., Song X., Wang H., Xu Y., Ochsenreiter T. (2010) The Trypanosoma brucei MitoCarta and its regulation and splicing pattern during development. Nucleic Acids Res. 38, 7378–7387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Neupert W., Herrmann J. M. (2007) Translocation of proteins into mitochondria. Annu. Rev. Biochem. 76, 723–749 [DOI] [PubMed] [Google Scholar]
- 13. Schmidt O., Pfanner N., Meisinger C. (2010) Mitochondrial protein import: from proteomics to functional mechanisms. Nat. Rev. Mol. Cell Biol. 11, 655–667 [DOI] [PubMed] [Google Scholar]
- 14. Bauer M. F., Gempel K., Reichert A. S., Rappold G. A., Lichtner P., Gerbitz K. D., Neupert W., Brunner M., Hofmann S. (1999) Genetic and structural characterization of the human mitochondrial inner membrane translocase. J. Mol. Biol. 289, 69–82 [DOI] [PubMed] [Google Scholar]
- 15. Lister R., Hulett J. M.., Lithgow T., Whelan J. (2005) Protein import into mitochondria: origins and functions today. Mol. Membr. Biol. 22, 87–100 [DOI] [PubMed] [Google Scholar]
- 16. Rapaport D. (2005) How does the TOM complex mediates insertion of precursor proteins into mitochondrial outer membrane. J. Cell Biol. 171, 419–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Melin J., Schulz C., Wrobel L., Bernhard O., Chacinska A., Jahn O., Schmidt B., Rehling P. (2014) Presequence recognition by the Tom40 channel contributes to precursor translocation into the matrix. Mol. Cell. Biol. 34, 3473–3485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hill K., Model K., Ryan M. T., Dietmeier K., Martin F., Wagner R., Pfanner N. (1998) Tom40 forms the hydrophilic channel of the mitochondrial import pore for preproteins. Nature 395, 516–521 [DOI] [PubMed] [Google Scholar]
- 19. van der Laan M., Hutu D. P., Rehling P. (2010) On the mechanism of preprotein import by the mitochondrial preprotein translocase. Biochim. Biophys. Acta 1803, 732–739 [DOI] [PubMed] [Google Scholar]
- 20. Kerscher O., Holder J., Srinivasan M., Leung R. S., Jensen R. E. (1997) The Tim54p-Tim22p complex mediates insertion of proteins into the mitochondrial inner membrane. J. Cell Biol. 139, 1663–1675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mokranjac D., Neupert W. (2008) Energetics of protein translocation into mitochondria. Biochim. Biophys. Acta 1777, 758–762 [DOI] [PubMed] [Google Scholar]
- 22. Neupert W., Brunner M. (2002) The protein import motor of mitochondria. Nat. Rev. Mol. Cell Biol. 3, 555–565 [DOI] [PubMed] [Google Scholar]
- 23. Herrmann J. M., Neupert W. (2003) Protein insertion into the inner membrane of mitochondria. IUBMB Life 55, 219–225 [DOI] [PubMed] [Google Scholar]
- 24. Ferramosca A., Zara V. (2013) Biogenesis of mitochondrial carrier proteins: molecular mechanisms of import into mitochondria. Biochim. Biophys. Acta 1833, 494–502 [DOI] [PubMed] [Google Scholar]
- 25. Koehler C. M., Jarosch E., Tokatlidis K., Schmid K., Schweyen R. J., Schatz G. (1998) Import of mitochondrial carriers mediated by essential proteins of the intermembrane space. Science 279, 369–373 [DOI] [PubMed] [Google Scholar]
- 26. Curran S. P., Leuenberger D., Schmidt E., Koehler C. M. (2002) The role of the Tim8p-Tim13p complex in a conserved import pathway for mitochondrial polytopic inner membrane proteins. J. Cell Biol. 158, 1017–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Truscott K. N., Kovermann P., Geissler A., Merlin A., Meijer M., Driessen A. J., Rassow J., Pfanner N., Wagner R. (2001) A presequence- and voltage-sensitive channel of the mitochondrial preprotein translocase formed by Tim23. Nat. Struct. Biol. 8, 1074–1082 [DOI] [PubMed] [Google Scholar]
- 28. Rehling P., Model K., Brandner K., Kovermann P., Sickmann A., Meyer H. E., Kühlbrandt W., Wagner R., Truscott K. N., Pfanner N. (2003) Protein insertion into the mitochondrial inner membrane by a twin-pore translocase. Science 299, 1747–1751 [DOI] [PubMed] [Google Scholar]
- 29. Rassow J., Dekker P. J., van Wilpe S., Meijer M., Soll J. (1999) The preprotein translocase of the mitochondrial inner membrane: function and evolution. J. Mol. Biol. 286, 105–120 [DOI] [PubMed] [Google Scholar]
- 30. Gevorkyan-Airapetov L., Zohary K., Popov-Celeketic D., Mapa K., Hell K., Neupert W., Azem A., Mokranjac D. (2009) Interaction of Tim23 with Tim50 is essential for protein translocation by the mitochondrial TIM23 complex. J. Biol. Chem. 284, 4865–4872 [DOI] [PubMed] [Google Scholar]
- 31. Geissler A., Chacinska A., Truscott K. N., Wiedemann N., Brandner K., Sickmann A., Meyer H. E., Meisinger C., Pfanner N., Rehling P. (2002) The mitochondrial preprotein translocase: an essential role of Tim50 in directing preproteins to the import channel. Cell 111, 507–518 [DOI] [PubMed] [Google Scholar]
- 32. Saddar S., Dienhart M. K., Stuart R. A. (2008) The F1F0-ATP synthase complex influences the assembly state of the cytochrome BC1-cytochrome oxidase supercomplex and its association with the TIM23 machinery. J. Biol. Chem. 283, 6677–6686 [DOI] [PubMed] [Google Scholar]
- 33. Murcha M. W., Wang Y., Narsai R., Whelan J. (2014) The plant mitochondrial protein import apparatus: the differences make it interesting. Biochim. Biophys. Acta 1840, 1233–1245 [DOI] [PubMed] [Google Scholar]
- 34. Hewitt V. L., Gabriel K., Traven A. (2014) The ins and outs of the intermembrane space: diverse mechanisms and evolutionary rewiring of mitochondrial protein import routes. Biochim. Biophys. Acta 1840, 1246–1253 [DOI] [PubMed] [Google Scholar]
- 35. Baker M. J., Webb C. T., Stroud D. A., Palmer C. S., Frazier A. E., Guiard B., Chacinska A., Gulbis J. M., Ryan M. T. (2009) Structural and functional requirements for activity of the Tim9-Tim10 complex in mitochondrial protein import. Mol. Biol. Cell 20, 769–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Häusler T., Stierhof Y.-D., Blattner J., Clayton C. (1997) Conservation of mitochondrial targeting sequence function in mitochondrial and hydrogenosomal proteins from the early-branching eukaryotes Crithidia, Trypanosoma, and Trichomonas. Eur. J. Cell Biol. 73, 240–251 [PubMed] [Google Scholar]
- 37. Hamilton V., Singha U. K., Smith J. T., Weems E., Chaudhuri M. (2014) Trypanosome alternative oxidase possesses both an N-terminal and internal mitochondrial targeting signal. Eukaryot. Cell 13, 539–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Saxowsky T. T., Choudhary G., Klingbeil M. M., Englund P. T. (2003) Trypanosoma brucei has two distinct mitochondrial DNA polymerase beta enzymes. J. Biol. Chem. 278, 49095–49101 [DOI] [PubMed] [Google Scholar]
- 39. Hines J. C., Ray D. S. (1998) The Crithidia fasciulata KAP1 gene encodes a highly basic protein associated with kinetoplast DNA. Mol. Biochem. Parasitol. 94, 41–52 [DOI] [PubMed] [Google Scholar]
- 40. Colasante C., Peña Diaz P., Clayton C., Voncken F. (2009) Mitochondrial carrier family inventory of Trypanosoma brucei brucei: Identification, expression and subcellular localisation. Mol. Biochem. Parasitol. 167, 104–117 [DOI] [PubMed] [Google Scholar]
- 41. Singha U. K., Peprah E., Williams S., Walker R., Saha L., Chaudhuri M. (2008) Characterization of the mitochondrial inner membrane translocator Tim17 from Trypanosoma brucei. Mol. Biochem. Parasitol. 159, 30–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Duncan M. R., Fullerton M., Chaudhuri M. (2013) Tim50 in Trypanosoma brucei possesses a dual specificity phosphatase activity and is critical for mitochondrial protein import. J. Biol. Chem. 288, 3184–3197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gentle I. E., Perry A. J., Alcock F. H., Likić V. A., Dolezal P., Ng E. T., Purcell A. W., McConnville M., Naderer T., Chanez A. L., Charrière F., Aschinger C., Schneider A., Tokatlidis K., Lithgow T. (2007) Conserved motifs reveal details of ancestry and structure in the small TIM chaperones of the mitochondrial inter membrane space. Mol. Biol. Evol. 24, 1149–1160 [DOI] [PubMed] [Google Scholar]
- 44. Eckers E., Cyrklaff M., Simpson L., Deponte M. (2012) Mitochondrial protein import pathways are functionally conserved among eukaryotes despite compositional diversity of the import machineries. Biol. Chem. 393, 513–524 [DOI] [PubMed] [Google Scholar]
- 45. Singha U. K., Hamilton V., Duncan M. R., Weems E., Tripathi M. K., Chaudhuri M. (2012) The Translocase of Mitochondrial Inner Membrane in Trypanosoma brucei. J. Biol. Chem. 287, 14480–14493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Weems E., Singha U. K., Hamilton V., Smith J. T., Waegemann K., Mokranjac D., Chaudhuri M. (2015) Functional complementation analyses reveal that the single PRAT-family protein of Trypanosoma brucei is a divergent homolog of Tim17 in Saccharomyces cerevisiae. Eukaryot. Cell. 14, 286–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wirtz E., Leal S., Ochatt C., Cross G. A. (1999) A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol. Biochem. Parasitol. 99, 89–101 [DOI] [PubMed] [Google Scholar]
- 48. Esseiva A. C., Chanez A. L., Bochud-Allemann N., Martinou J. C., Hemphill A., Schneider A. (2004) Temporal dissection of Bax-induced events leading to fission of the single mitochondrion in Trypanosoma brucei. EMBO Rep. 5, 268–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Williams S., Saha L., Singha U. K., Chaudhuri M. (2008) Trypanosoma brucei: Differential requirement of membrane potential for import of proteins into mitochondria in two developmental stages. Exp. Parasitol. 118, 420–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. de Macêdo J. P., Schumann Burkard G., Niemann M., Barrett M. P., Vial H., Mäser P., Roditi I., Schneider A., Bütikofer P. (2015) An atypical mitochondrial carrier that mediates drug action in Trypanosoma brucei. PLoS Pathog. 11, e1004875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hasne M. P., Barrett M. P. (2000) Transport of methionine in Trypanosoma brucei brucei. Mol. Biochem. Parasitol. 111, 299–307 [DOI] [PubMed] [Google Scholar]
- 52. Volpato H., Desoti V. C., Valdez R. H., Ueda-Nakamura T., Silva Sde O., Sarragiotto M. H., Nakamura C. V. (2015) Mitochondrial dysfunction induced by N-butyl-1-(4-dimethylamino)phenyl-1,2,3,4-tetrahydro-β-carboline-3-carboximide is required for cell death of Trypanosoma cruzi. PLoS ONE 10, e0130652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Singha U. K., Sharma S., Chaudhuri M. (2009) Down regulation of mitochondrial porin inhibits cell growth and alters respiratory phenotype in Trypanosoma brucei. Eukaryot. Cell 8, 1418–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Priest J. W., Hajduk S. L. (1994) Developmental regulation of Trypanosoma brucei cytochrome c reductase during bloodstream to procyclic differentiation. Mol. Biochem. Parasitol. 65, 291–304 [DOI] [PubMed] [Google Scholar]
- 55. Effron P. N., Torri A. F., Engman D. M., Donelson J. E., Englund P. T. (1993) A mitochondrial heat shock protein from Crithidia fasciculata. Mol. Biochem. Parasitol. 59, 191–200 [DOI] [PubMed] [Google Scholar]
- 56. Chaudhuri M., Ajayi W., Hill G. C. (1998) Biochemical and molecular properties of the Trypanosoma brucei alternative oxidase. Mol. Biochem. Parasitol. 95, 53–68 [DOI] [PubMed] [Google Scholar]
- 57. Woods A., Sherwin T., Sasse R., MacRae T. H., Baines A. J., Gull K. (1989) Definition of individual components within the cytoskeleton of Trypanosoma brucei by a library of monoclonal antibodies. J. Cell Sci. 93, 491–500 [DOI] [PubMed] [Google Scholar]
- 58. Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990) Basic local alignment search tool. J. Mol. Biol. 215, 403–410 [DOI] [PubMed] [Google Scholar]
- 59. Ni X., Li X., Tao S., Xu M., Ma H., Wang X. (2013) Blockaed of ankyrin repeat-rich membrane spanning protein modulates extracellular signal-regulated kinase expression and inhibits allergic inflammation in ovalbumin-sensitizde mice. Biomed. Rep. 1, 674–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Claros M. G., Vincens P. (1996) Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur. J. Biochem. 241, 779–786 [DOI] [PubMed] [Google Scholar]
- 61. Bagos P. G., Liakopoulos T. D., Hamodrakas S. J. (2006) Algorithms for incorporating prior topological information in HMMS: application to transmembrane proteins. BMC Bioinformatics 7, 189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Milisav I., Moro F., Neupert W., Brunner M. (2001) Modular structure of the TIM23 preprotein translocase of mitochondria. J. Biol. Chem. 276, 25856–25861 [DOI] [PubMed] [Google Scholar]
- 63. Tamura Y., Harada Y., Yamano K., Watanabe K., Ishikawa D., Ohshima C., Nishikawa S., Yamamoto H., Endo T. (2006) Identification of Tam41 maintaining integrity of the TIM23 protein translocase complex in mitochondria. J. Cell Biol. 174, 631–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Wiedemann N., van der Laan M., Hutu D. P., Rehling P., Pfanner N. (2007) Sorting switch of mitochondrial presequence translocase involves coupling of motor module to respiratory chain. J. Cell Biol. 179, 1115–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]