Background: For some chiral barbiturates, one isomer potentiates and the other inhibits GABA responses by binding to unknown sites.
Results: A photoreactive convulsant barbiturate identifies a transmembrane intersubunit-binding site between the γ and β subunits.
Conclusion: Positive and negative allosteric modulators can bind to a common intersubunit site.
Significance: This study defines a novel mode of regulation of GABAAR responses.
Keywords: allosteric regulation, anesthesia, Cys loop receptor, GABA receptor, homology modeling, photoaffinity labeling
Abstract
In the process of developing safer general anesthetics, isomers of anesthetic ethers and barbiturates have been discovered that act as convulsants and inhibitors of γ-aminobutyric acid type A receptors (GABAARs) rather than potentiators. It is unknown whether these convulsants act as negative allosteric modulators by binding to the intersubunit anesthetic-binding sites in the GABAAR transmembrane domain (Chiara, D. C., Jayakar, S. S., Zhou, X., Zhang, X., Savechenkov, P. Y., Bruzik, K. S., Miller, K. W., and Cohen, J. B. (2013) J. Biol. Chem. 288, 19343–19357) or to known convulsant sites in the ion channel or extracellular domains. Here, we show that S-1-methyl-5-propyl-5-(m-trifluoromethyl-diazirynylphenyl) barbituric acid (S-mTFD-MPPB), a photoreactive analog of the convulsant barbiturate S-MPPB, inhibits α1β3γ2 but potentiates α1β3 GABAAR responses. In the α1β3γ2 GABAAR, S-mTFD-MPPB binds in the transmembrane domain with high affinity to the γ+-β− subunit interface site with negative energetic coupling to GABA binding in the extracellular domain at the β+-α− subunit interfaces. GABA inhibits S-[3H]mTFD-MPPB photolabeling of γ2Ser-280 (γM2–15′) in this site. In contrast, within the same site GABA enhances photolabeling of β3Met-227 in βM1 by an anesthetic barbiturate, R-[3H]methyl-5-allyl-5-(m-trifluoromethyl-diazirynylphenyl)barbituric acid (mTFD-MPAB), which differs from S-mTFD-MPPB in structure only by chirality and two hydrogens (propyl versus allyl). S-mTFD-MPPB and R-mTFD-MPAB are predicted to bind in different orientations at the γ+-β− site, based upon the distance in GABAAR homology models between γ2Ser-280 and β3Met-227. These results provide an explanation for S-mTFD-MPPB inhibition of α1β3γ2 GABAAR function and provide a first demonstration that an intersubunit-binding site in the GABAAR transmembrane domain binds negative and positive allosteric modulators.
Introduction
For over 150 years, drug screens assessing in vivo animal responses have led to the identification of a structurally diverse group of compounds, including simple volatile ethers, alcohols, barbiturates, and steroids, that produce the complex physiological responses desirable for clinical anesthesia (1). In the process of identifying novel anesthetics, comparison of the actions of geometric isomers of certain volatile fluorinated ethers and barbiturate stereoisomers sometimes revealed that one isomer acted as an anesthetic and the other as a convulsant (2–5). Anesthetic barbiturates and other intravenous anesthetics (propofol, etomidate, and steroids), as well as volatile ethers, potentiate inhibitory GABA type A receptors (GABAAR)2 in vitro at concentrations producing anesthesia in vivo (6–8), and the importance of GABAARs for anesthesia is demonstrated by the decreased sensitivity of “knock-in” mice bearing a single amino acid substitution in a GABAAR β subunit to the immobilizing and hypnotic effects of pentobarbital, etomidate, and propofol (9–12).
The convulsant effects of some barbiturates may be mediated by targets other than GABAARs (13). However, the convulsant S-1-methyl-5-phenyl-5-propyl barbituric acid (S-MPPB) inhibits GABAAR responses at the same concentration at which the anesthetic isomer, R-MPPB, potentiates responses (14, 15). Neither anesthetic nor convulsant barbiturates bind directly to the GABA or benzodiazepine-binding sites (16), and the differential effects of R- and S-MPPB on the binding of a GABAAR channel blocker suggest that the convulsant and anesthetic isomers may bind to distinct sites in a GABAAR (14).
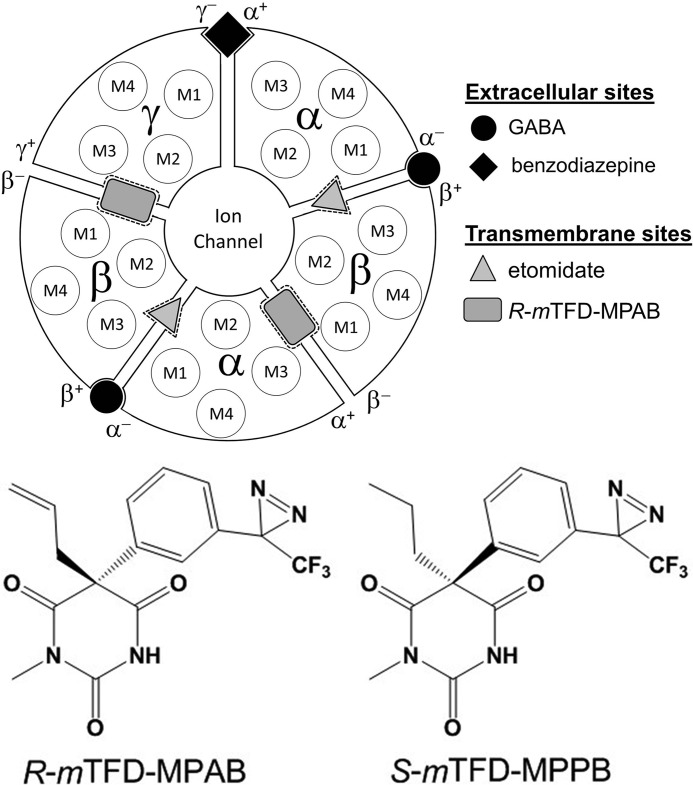
Recently, two classes of general anesthetic-binding sites have been identified in the transmembrane domain (TMD) of α1β3γ2 GABAARs based upon the locations of residues photolabeled by analogs of etomidate, mephobarbital, and propofol in homology models based on published structures of several homologous members of the Cys-loop superfamily of pentameric ligand-gated ion channels (17–19), including one of a human β3 GABAAR (Fig. 1) (20). Photoreactive etomidate analogs identify a high affinity binding site for etomidate at the β+-α− subunit interfaces, based upon the photolabeling of amino acids in the β subunit M3 and α subunit M1 transmembrane helices (21, 22). A mephobarbital analog, R-[3H]mTFD-MPAB, photolabeled amino acids in the βM1, αM3, and γM3 transmembrane helices, identifying a second homologous class of anesthetic-binding sites at the α+-β− and γ+-β− subunit interfaces (23). Although etomidate and R-mTFD-MPAB bind with >50-fold selectivity to the β+- or β−-containing interface sites, respectively, the sites are not strictly etomidate- or barbiturate-specific. Propofol binds with little selectivity to both classes of sites, and a propofol analog photolabels residues in both classes of sites (23, 24). These allosteric anesthetic-binding sites in the TMD show positive energetic coupling to each other and to the GABA-binding sites at the β+-α− subunit interfaces in the extracellular domain (ECD), as etomidate and GABA each enhanced R-[3H]mTFD-MPAB photolabeling.
FIGURE 1.
Locations of binding sites for GABA, benzodiazepines, R-mTFD-MPAB, and etomidate in an (α1)2(β3)2γ GABAAR. GABA-binding sites are in the ECD at the interface between the β and α subunit referred to as the β+-α− subunit interface, and with that nomenclature continued in a counterclockwise direction, the benzodiazepine site is at the α+-γ− subunit interface. Depicted in the TMD are the locations of the four transmembrane helices (M1–M4) in each subunit, the etomidate-binding sites at the β+-α− subunit interfaces that contain the GABA-binding sites in the ECD, and the R-mTFD-MPAB sites at the α+-β− and γ+-β− subunit interfaces.
Similar to R-mTFD-MPAB, S-mTFD-MPAB acts as an anesthetic, but with 10-fold lower potency, and it acts as a low efficacy potentiator of GABA responses (25). In contrast, for mTFD-MPPB, differing from mTFD-MPAB by only two more hydrogen atoms ([1-methyl-5-propyl-5-(m-trifluoromethyl-diazirynylphenyl]barbituric acid, Fig. 1), the R-enantiomer acts as an anesthetic in vivo and potentiated GABA responses for expressed α1β3γ2 GABAAR, whereas the S-enantiomer acts as a convulsant and inhibits GABA responses (26). Thus, the enantiomers of mTFD-MPPB mirror the actions of S- and R-MPPB in vivo and in vitro.
In this report, we prepare S-[3H]mTFD-MPPB and use it as a photoaffinity reagent to determine where a convulsant barbiturate binds in an α1β3γ2 GABAAR. Based upon the direct identification of photolabeled amino acids and the pharmacological specificity of photolabeling, we find that S-[3H]mTFD-MPPB binds in the same γ+-β− interface pocket as R-mTFD-MPAB. However, it binds in a different orientation, and its binding is inhibited allosterically by GABA, indicative of negative energetic coupling between the sites. S-[3H]mTFD-MPPB also binds with lower affinity to the other intersubunit anesthetic sites, with positive energetic coupling to the GABA site. Our results provide a first demonstration that, similar to the benzodiazepine-binding site at the α+-γ− interface in the ECD (27), at least one intersubunit-binding site in the GABAAR's TMD is a target for negative as well as positive allosteric modulators.
Experimental Procedures
Materials
R- and S-mTFD-MPAB ([5-allyl-1-methyl-5-(m-trifluoromethyldiazirynylphenyl]barbituric acid) and R-[3H]mTFD-MPAB (38 Ci/mmol) were prepared previously (25), as was [3H]azietomidate (19 Ci/mmol) (24). Similar to mTFD-MPAB, nonradioactive (±)-mTFD-MPPB (5-propyl-1-methyl-5-(m-trifluoromethyldiazirynylphenyl)barbituric acid) was synthesized by reaction of 5-propyl-1-methyl barbiturate with (4-methoxyphenyl)-[3-(3-trifluoromethyl-3H-diazirin-3-yl)phenyl]iodonium trifluoroacetate, and preparative separation of R- and S-mTFD-MPPB was performed by chiral chromatography on a Chiralpak IC column. R- and S-mTFD-MPPB eluted with retention times of 10.0 and 11.8 min, respectively. Full details of the synthesis and characterization will be presented elsewhere. S-[3H]mTFD-MPPB (50 Ci/mmol, Vitrax, Placentia, CA) was prepared as described (25) by catalytic reduction of S-mTFD-MPAB using tritium gas in the presence of Wilkinson's rhodium catalyst. Bicuculline methochloride was from Abcam. Picrotoxinin, phenobarbital, the FLAG peptide (DYKDDDDK), GABA, soybean asolectin, cyanogen bromide (CNBr), and 3-bromo-3-methyl 2-(2-nitrophenylthio)-3H-indole (BNPS-skatole) were from Sigma. o-Phthalaldehyde (OPA) was from Alfa Aesar. (R)-Etomidate was from Organon Laboratories. Staphylococcus aureus glutamic-C endopeptidase (EndoGlu-C) was from Princeton Separations, and Lysobacter enzymogenes lysine-C endopeptidase (EndoLys-C) was from Roche Applied Science.
Electrophysiology
Whole-cell patch clamp recordings were obtained from induced HEK293-TetR cells expressing either α1β3 or α1β3γ2L GABAA receptors using methods described previously (28, 29). Briefly, cells were seeded on a glass coverslip, and protein expression was induced with tetracycline (2 μg/ml) for 5–26 h before recordings. All experiments were performed at room temperature (20–22 °C). The recording chamber was continuously perfused with the bath solution (in mm) as follows:145 NaCl, 5 KCl, 10 HEPES, 2 CaCl2, 1 MgCl2, and 10 glucose, pH 7.4 (pH adjusted with NaOH). The electrode solution contained (in mm) the following: 140 KCl, 10 HEPES, 1 EGTA, and 2 MgCl2 at pH 7.3 (pH adjusted with KOH). Open pipette resistances ranged from 1.9 to 3 megohms. Cells were voltage-clamped at −50 mV using the patch clamp amplifier (Axopatch 200A, Molecular Devices Corp., Sunnyvale, CA). Whole-cell membrane capacitances and series resistances were compensated electronically by more than 85% with a lag of 10 μs. Series resistances ranged from 0.5 to 2.5 megohms and cell capacitances from 16 to 18.5 picofarads. GABAA receptors were activated using 8-s pulses of GABA delivered via a multichannel superfusion pipette coupled to piezo-electric elements that switched solutions in less than 1 ms. Currents were filtered at 5 kHz and digitized at 10 kHz using pCLAMP version 8.1 (Molecular Devices Corp., Sunnyvale, CA) for off-line analysis with Clampfit 9 (Molecular Devices Corp., Sunnyvale, CA). Statistical analysis was performed in GraphPad Prism version 6 software (GraphPad Software, Inc., San Diego). All data are expressed as mean ± S.D.
Purification of Expressed Human α1β3γ2 GABAARs
α1β3γ2L and α1β3 GABAARs containing a FLAG epitope at the N terminus of the mature α1 subunit (MRK…SYGDYKDDDDKQPS…) were purified from tetracycline-inducible, stably transfected HEK293S cell lines using an anti-FLAG affinity resin as described previously (23, 24, 28, 29). GABAAR was solubilized in 30 mm n-dodecyl β-d-maltopyranoside, and column wash and elution buffers contained 5 mm CHAPS and 0.2 mm asolectin. After elution with 1.5 mm FLAG peptide, aliquots from the eluted fractions were characterized for the number of GABAAR-binding sites, using [3H]muscimol, and for etomidate modulation of [3H]muscimol binding. Starting from membrane fractions containing 4–8 nmol of [3H]muscimol-binding sites, typical purification yields were 0.5–1.5 nmol of purified α1β3γ2 GABAAR (30–60 nm binding sites) and 1.5 nmol of α1β3 GABAAR (60 nm binding sites), each in 15–25 ml of elution buffer. Fractions were flash-frozen in liquid N2 and stored at −80 °C until use.
GABAAR Photoaffinity Labeling
Aliquots of purified FLAG-α1β3γ2 GABAARs in elution buffer were photolabeled at analytical and preparative scales (40–80 μl or 1–2.5 ml of α1β3γ2 GABAAR, per condition, respectively) to either characterize photoincorporation at the subunit level or to identify individual photolabeled amino acids using protein sequencing methods, respectively. Required volumes of stock solutions of radiolabeled, photoreactive anesthetic in methanol were transferred to glass test tubes, and solvent was evaporated under an argon stream before addition of GABAAR. The radioligand was resuspended with occasional vortexing for 30 min on ice. Photolabeling was performed at ∼3 μm S-[3H]mTFD-MPPB (∼11 μCi per analytical and 200 μCi per preparative sample), ∼1 μm R-[3H]mTFD-MPAB (3 μCi per analytical sample), and ∼1.5 μm [3H]azietomidate (2 μCi per analytical sample). Receptors were then equilibrated for 10 min with 300 μm GABA or 30 μm bicuculline, followed by the addition of nonradioactive anesthetics, and solutions were incubated for an additional 30 min on ice. Aliquots were then transferred to 96-well plastic plates (Corning catalog number 2797) for analytical scale or 3.5-cm diameter plastic Petri dishes (Corning catalog number 3001) for preparative scale photolabeling experiments and irradiated on ice for 30 min at a distance of 0.5–1 cm using a 365 nm lamp (Spectroline Model EN-16, Spectronics Corp, Westbury, NJ). Stock solutions of nonradioactive S-mTFD-MPPB (60 mm), R-mTFD-MPAB (60 mm), propofol (1 m), pentobarbital (60 mm), picrotoxinin (60 mm), and etomidate (60 mm) were prepared in methanol. Bicuculline methochloride (6 mm) was prepared in water. Methanol was present in all samples during photolabeling at a concentration of 0.5% (v/v). Photolabeled samples were immediately solubilized in SDS-sample buffer (23) and incubated for 30–60 min at room temperature before SDS-PAGE.
SDS-PAGE and Enzymatic/Chemical Digestion of GABAARs
GABAAR subunits in SDS sample buffer were resolved by SDS-PAGE on 6% Tris-glycine gels, which were constructed as described (23), to accommodate the 150-μl and ∼1.5-ml sample volumes generated in analytical and preparative scale photolabelings, respectively. After electrophoresis, gels were stained with Coomassie Brilliant Blue. In analytical scale experiments, 3H incorporation into subunits was determined by liquid scintillation counting or fluorography, and in preparative scale experiments, subunits were eluted from excised subunit gel bands as described (23). Material eluted from the gel bands was filtered, concentrated, acetone-precipitated (−20 °C), and resuspended in 100–200 μl of digestion buffer (15 mm Tris and 0.1% SDS, pH 8.5). Aliquots (90 or 180 μl) of resuspended subunits were digested at room temperature with EndoLys-C (0.5 units) for 14 days or with EndoGlu-C (2.5 μg) for 2–4 days. Enzymatic digests were fractionated by reversed-phase HPLC (rpHPLC) as described (30), and fractions containing radiolabeled fragments were pooled for N-terminal sequencing or for further chemical fragmentation. Incorporation in γM2 was determined by sequencing a fragment beginning at γ2Asp-260 from an EndoLys-C digest and by sequencing a parallel sample treated after immobilization on the sequencing filter with CNBr as described (31, 32) for cleavage of peptides at the C termini of methionines. Photolabeling in βM2 was determined by sequencing a fragment beginning at β3Ile-242, produced by treating intact subunit immobilized on a sequencing filter with BNPS-skatole as described (22, 24, 33) to cleave at the C terminus of tryptophans. To characterize photolabeling in αM2, we sequenced the fragment beginning at α1Ser-251 at the N terminus of αM2 that can be isolated by rpHPLC fractionation of EndoGlu-C digests of α1 subunit and treatment with OPA at cycle 3 during sequencing (23). Photolabeling in βM1, βM3, αM1, and αM3 was determined by sequencing appropriate rpHPLC fractions from EndoLys-C digests of α1 or β3 subunits (21, 22)
Quantification of Inhibition of Photolabeling
The concentration dependence of inhibition of photolabeling by nonradioactive barbiturates or other drugs was determined in analytical photolabeling experiments. 3H incorporation was determined in the following three stained subunit bands: a 56-kDa band, enriched in the α subunit, and bands of 59 and 61 kDa, enriched in the β subunit but differentially glycosylated (22). The γ2 subunit was distributed more diffusely but centered in the 56-kDa band. For [3H]azietomidate, parameters for the concentration dependence of inhibition were determined for the 56-kDa gel band that reflects photolabeling of α1Met-236 at the β+-α− subunit interface. For R-[3H]mTFD-MPAB, parameters were determined for the 59- and 61-kDa gel bands that reflect photolabeling of β3Met-227 at the β− subunit interfaces (23). For S-[3H]mTFD-MPPB, parameters were determined for the 56-kDa gel band, which in this case reflects photolabeling of γ2Ser-280 (see under “Results”). The concentration dependence of inhibition of subunit photolabeling was fit using nonlinear least squares by SigmaPlot 11.0 (Systat Software) to a single or two-site model using Equations 1 and 2, respectively,
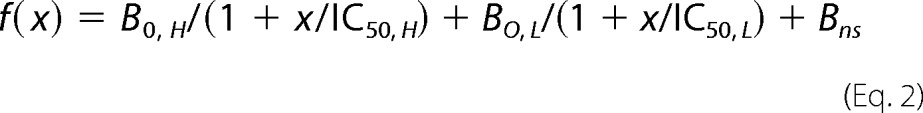 |
where B(x) is the 3H in counts/min (cpm) incorporated into a subunit gel band when the total inhibitor concentration is x, B0 is the specific 3H incorporation in the absence of inhibitor; IC50 is the total concentration of inhibitor that reduces the incorporated 3H by 50%, with H and L denoting the high and low affinity binding sites; nH is the Hill coefficient; and Bns is the nonspecific 3H incorporation in the presence of maximal concentrations of a competitor. Data were fit initially to Equation 1 with variable IC50 values; B0 was equal to the difference between total binding and nonspecific binding, and nH was equal to 1 or variable. When S-mTFD-MPPB inhibition was characterized by n < 1 (see under “Results”) (S-[3H]mTFD-MPPB (+bicuculline) and R-[3H]mTFD-MPAB (+GABA)), data were also fit to Equation 2, with Bo, L equal to specific labeling in the presence of GABA (S-[3H]mTFD-MPPB) or bicuculline (R-[3H]mTFD-MPAB), and Bo, H = B0 − Bo, L. For S-[3H]mTFD-MPPB, R-[3H]mTFD-MPAB, and [3H]azietomidate, Bns was determined in the presence of nonradioactive S-mTFD-MPPB (300 μm), R-mTFD-MPAB (60 μm), or etomidate (300 μm), respectively.
Reversed-phase HPLC and N-terminal Sequence Analysis
Enzymatic digests of GABAAR subunits were fractionated by rpHPLC and subjected to N-terminal sequencing as described (23, 30). Briefly, rpHPLC fractionation was performed using an Agilent 1100 binary HPLC system with a Brownlee Aquapore column. Fractions of 0.5 ml were collected at a flow rate of 200 μl/min, and peptide elution was monitored by the absorbance at 215 nm. Aliquots (10%) of each fraction were counted to determine the 3H distribution. Fractions containing peaks of 3H were pooled and loaded onto Micro TFA glass fiber filters (Applied Biosystems) at 45 °C. Total digests of intact GABAAR subunits and rpHPLC fractions, where indicated, were loaded directly onto Prosorb PVDF filters (Applied Biosystems) according to the manufacturer's directions.
Samples were sequenced using a Procise 492 protein sequencer (Applied Biosystems), with 2/3 of the material from each cycle of Edman degradation used for PTH-derivative quantification and 1/3 collected to measure 3H release by scintillation counting. In some cases, we used o-phthalaldehyde (OPA) treatment during sequencing, as described (34), to chemically isolate a fragment of interest known to contain a proline at a particular cycle of Edman degradation or to test for the presence of a proline. Because OPA reacts with primary amines but not secondary amines (35), OPA treatment at a cycle containing a proline in the peptide of interest allows continued sequencing of that peptide while blocking further sequencing of other peptides not containing a proline at that cycle.
The amount of PTH-derivative released in a given sequencing cycle (in picomoles) was determined by comparing the peak height for the amino acid derivative in the chromatogram to the height of its standard peak. I0, the initial amount of a peptide in a sequencing sample (in picomoles), was determined from the amounts of PTH-derivative in each cycle by nonlinear least squares fit to Equation 3,
where Ix is the background-subtracted mass of the peptide residue in cycle x (in picomoles), and R is the average repetitive yield. For samples containing multiple fragments, only PTH-derivatives unique to the fragment of interest were included in the fit. Amino acid derivatives whose amounts cannot be accurately estimated (His, Trp, Ser, Arg, and Cys) were omitted from the fit. E(x), the efficiency of photolabeling (in cpm/pmol) of the amino acid residue in cycle x was calculated by Equation 4,
where cpmx is the 3H released in cycle x (in cpm).
Molecular Modeling
The locations of the photolabeled residues were visualized in an β3α1β3α1γ2 GABAAR homology model based upon the structure of the homomeric human β3 GABAAR (PDB code 4COF (20)) that was made (Discovery Studio 4.0 (Accelrys, Inc.)) as described for the α1β3 GABAAR (24) with the substitution of the γ2 subunit for the β3 subunit designated E in the PDB model. After construction, the receptor was placed in a membrane force field and minimized (10 cycles) to ease strained interactions. To determine whether the pocket at the γ+-β− interface can accommodate S-mTFD-MPPB, computational docking was performed using the CDocker module. Four randomly oriented S-mTFD-MPPB molecules were placed within the pocket in a binding site sphere of 11 Å radius centered at the level of γ2Ser-280 (γM2–15′), γ2Ser-301 in γM3, and β3Met-227 in βM1. The 100 lowest interaction energy orientations (simulated annealing with full potential minimization) were collected for each molecule from 50 random conformations (high temperature molecular dynamics) and 50 randomized orientations within the spheres (i.e. 2500 initial conditions tested per molecule). 213 of 400 collected solutions predicted stable binding (CDocker interaction energies <0 kcal/mol). The 10 most favored binding solutions had CDocker energies from −35.6 to −38.9 kcal/mol and included orientations with the S-mTFD-MPPB diazirine directed toward γ2Ser-280 and others with the diazirine oriented toward γM3/βM1. This procedure was repeated for the equivalent pockets at the β+-α− and α+-β− interfaces. At the β+-α− interface adjacent to the γ+-β− interface, all 400 collected solutions had interaction energies < −6 kcal/mol. The 10 lowest energies ranged from −33.4 to −37.2 kcal/mol, and for each of these the diazirine was oriented toward βM3/αM1. At the α+-β− interface, all 400 solutions had interaction energies < −25 kcal/mol, with the 10 lowest energy solutions ranging from −38.2 to −40.6 kcal/mol. All 10 orientations were similar, with the diazirine projecting between α1M3 and β3M1.
Results
S-mTFD-MPPB Inhibits α1β3γ2 and Potentiates α1β3 GABAAR Responses
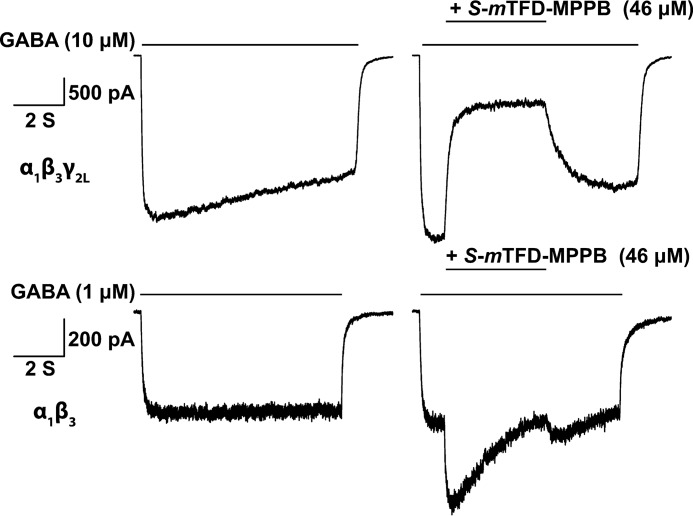
We compared effects of S-mTFD-MPPB on GABA responses in cell lines expressing α1β3γ2 or α1β3 GABAARs. Reponses were measured using approximate EC10 GABA concentrations of 1 μm for α1β3 and 10 μm for α1β3γ2, because at these concentrations sufficiently robust currents are elicited for studying inhibition, while leaving ample room for the observation of current enhancement. As shown in Fig. 2, in α1β3γ2 GABAARs, S-mTFD-MPPB at 46 μm inhibited peak GABA-induced current amplitudes by 72 ± 1.3% (n = 4), whereas in α1β3 GABAARs it enhanced them by 49 ± 18% (n = 6). These results suggest that inhibition by S-mTFD-MPPB requires the presence of the γ2 subunit. The inhibition of α1β3γ2 GABAAR responses by S-mTFD-MPPB is similar to the inhibition of GABA responses in cortical neurons seen for S-MPPB (15) and contrasts with the effects of mTFD-MPAB on α1β3γ2 GABAARs, for which both isomers potentiate responses (25).
FIGURE 2.
S-mTFD-MPPB inhibits α1β3γ2 and potentiates α1β3 GABAAR responses. Representative traces of recombinant α1β3 and α1β3γ2L GABAAR-mediated currents expressed by HEK293 cells. Currents were elicited by exposing the cells to an EC10 concentration of GABA (1 or 10 μm, respectively) for 8 s (left panels). A 4-s pulse of S-mTFD-MPPB (46 μm) was co-perfused after 1 s of GABA (right panels). The lengths of the solid lines indicate the duration of GABA or S-mTFD-MPPB application.
mTFD-MPPB Binding to Known General Anesthetic Sites
In initial photolabeling experiments, we tested S- and R-mTFD-MPPB, in the presence of GABA, as inhibitors of α1β3γ2 GABAAR photolabeling by [3H]azietomidate and R-[3H]mTFD-MPAB, photoreactive anesthetics that bind selectively to homologous sites at the GABAAR β+ and β− subunit interfaces (23). Similar to R-mTFD-MPAB, R-mTFD-MPPB was a potent inhibitor of R-[3H]mTFD-MPAB photolabeling (IC50 = 1.8 ± 0.1 μm) and only inhibited [3H]azietomidate photolabeling at high concentrations (IC50 >100 μm). Similar to S-mTFD-MPAB, S-mTFD-MPPB bound weakly to both the R-[3H]mTFD-MPAB (IC50 = 34 ± 9 μm) and [3H]azietomidate (IC50 = 102 ± 11 μm) sites.
S-[3H]mTFD-MPPB Photolabeling of α1β3γ2 GABAAR, Inhibition by GABA but Not by Bicuculline
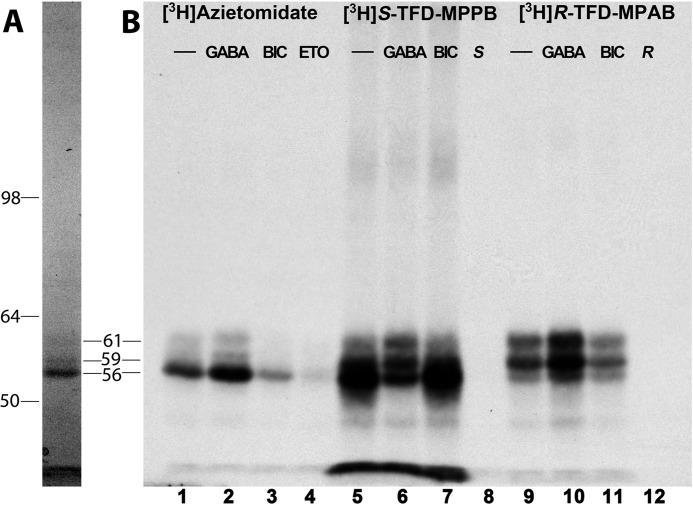
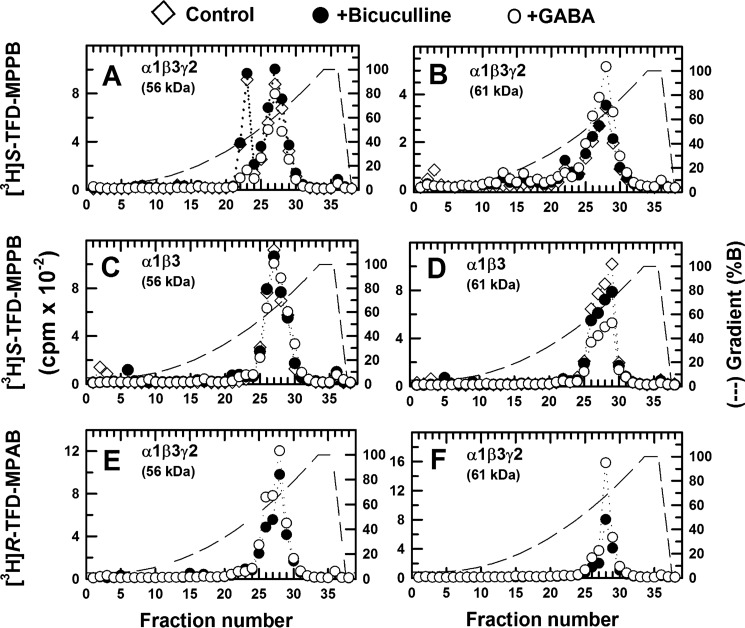
We then examined the effects of GABA and bicuculline, a competitive antagonist, on the covalent incorporation of S-[3H]mTFD-MPPB, [3H]azietomidate, and R-[3H]mTFD-MPAB, as determined by fluorography after SDS-PAGE (Fig. 3). As reported previously (23), [3H]azietomidate photoincorporated primarily into a 56-kDa band, reflecting photolabeling of α1Met-236 in αM1 at the β+-α− subunit interface, and R-[3H]mTFD-MPAB incorporated primarily into 59- and 61-kDa bands, reflecting photolabeling of β3Met-227 in βM1 at the β− subunit interfaces. At both sites, photolabeling was enhanced by GABA but not by bicuculline. In contrast, S-[3H]mTFD-MPPB incorporated most efficiently into a diffusely distributed GABAAR subunit band with mobility of ∼56 kDa. Photolabeling in that band was inhibited by GABA but not by bicuculline, indicating that the GABA-inhibitable photolabeling was not within the GABA-binding site. S-[3H]mTFD-MPPB was photoincorporated at lower levels into the 59- and 61-kDa bands, with that photolabeling enhanced by GABA. All S-[3H]mTFD-MPPB photolabeling appeared inhibitable by nonradioactive S-mTFD-MPPB (60 μm).
FIGURE 3.
Effects of GABA and bicuculline on α1β3γ2 GABAAR photolabeling by S-[3H]mTFD-MPPB, R-[3H]mTFD-MPAB, and [3H]azietomidate. GABAARs (∼5-pmol aliquots) were photolabeled with [3H]azietomidate (2 μm), S-[3H]mTFD-MPPB (4.6 μm), or R-[3H]mTFD-MPAB (1.4 μm) in the absence of other drugs (−) or in the presence of 300 μm GABA, 100 μm bicuculline (BIC), or nonradioactive anesthetics (200 μm etomidate (ETO), 60 μm S-mTFD-MPPB (S), or 60 μm R-mTFD-MPAB (R)). After photolysis, GABAAR subunits were resolved by SDS-PAGE and visualized by Coomassie Blue stain (A, representative lane with the mobilities of the molecular weight markers indicated on the left (Invitrogen SeeBlue Plus2 Pre-Stained Standard). B, 3H incorporation into GABAAR subunits was monitored by fluorography.
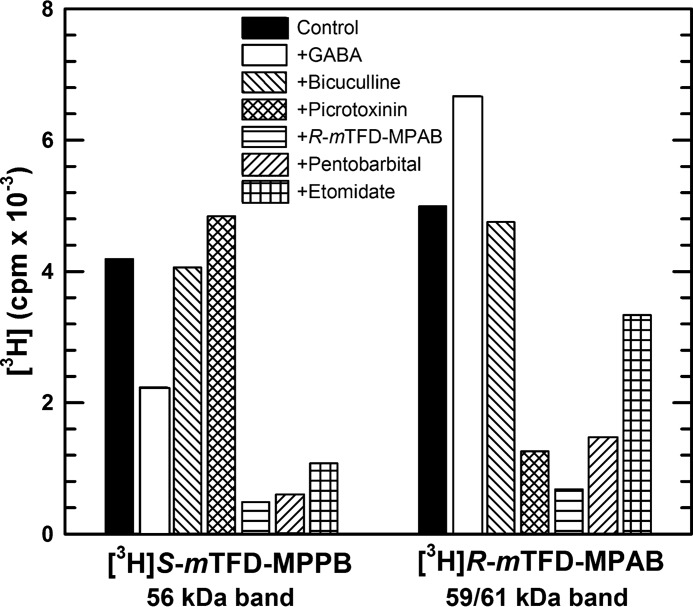
To further characterize the pharmacological specificity of S-[3H]mTFD-MPPB photolabeling, we quantified the effects of anesthetics and convulsants on S-[3H]mTFD-MPPB or R-[3H]mTFD-MPAB photolabeling by liquid scintillation counting of 3H incorporation into the excised 56- or 59/61-kDa gel bands, respectively (Fig. 4). Picrotoxinin, a channel blocker and convulsant, inhibited R-[3H]mTFD-MPAB photolabeling by 75% while causing a small enhancement (∼15%) of S-[3H]mTFD-MPPB photolabeling. At high concentrations, anesthetic barbiturates R-mTFD-MPAB (60 μm) and pentobarbital (2.5 mm), known to inhibit R-[3H]mTFD-MPAB photolabeling (23), each inhibited S-[3H]mTFD-MPPB photolabeling by ∼90%. Etomidate at 300 μm inhibited S-[3H]mTFD-MPPB photolabeling by 75%. That picrotoxinin did not inhibit S-[3H]mTFD-MPPB photolabeling suggested that the GABA-inhibitable labeling is unlikely to be in a site within the ion channel, whereas the inhibition of photolabeling by anesthetic barbiturates and etomidate suggested that the photolabeling may be in the known intersubunit anesthetic-binding sites.
FIGURE 4.
Pharmacological specificity of S-[3H]mTFD-MPPB and R-[3H]mTFD-MPAB photoincorporation into α1β3γ2 GABAAR subunit gel bands. GABAARs were photolabeled with 1.5 μm S-[3H]mTFD-MPPB or 1.2 μm R-[3H]mTFD-MPAB in the absence of other drug (control) or in the presence of GABA (300 μm), bicuculline (100 μm), picrotoxinin (30 μm), R-mTFD-MPAB (100 μm), pentobarbital (2.5 mm), or etomidate (300 μm). After photolabeling, GABAAR subunits were resolved by SDS-PAGE. Subunit bands of 56 kDa and 59/61 kDa were excised from Coomassie Blue-stained gels, and 3H incorporation was determined by liquid scintillation counting. Data are from a single photolabeling experiment.
Initial Localization of S-[3H]mTFD-MPPB-binding Sites within α1β3γ2 GABAAR
Because the γ2 subunit is poorly stained and broadly distributed in the 56/59-kDa region of the SDS-polyacrylamide gel (23), and S-[3H]mTFD-MPPB photolabeling in the ∼56-kDa band appeared more diffusely distributed than the [3H]azietomidate-photolabeled α subunit band, experiments were designed to determine whether S-[3H]mTFD-MPPB was photoincorporated primarily into the α1 or γ2 subunit. Comparison of the distributions of 3H when EndoLys-C subunit digests were fractionated by rpHPLC provided evidence that the GABA-inhibitable photolabeling in the 56-kDa gel band originated from the γ2 subunit rather than the α1 subunit (Fig. 5). For the 56-kDa band from α1β3γ2 GABAAR photolabeled by S-[3H]mTFD-MPPB, there was a GABA-inhibitable hydrophilic peak of 3H eluting at ∼40% organic solvent (Fig. 5A). This peak was not observed in digests of the 59/61-kDa bands (Fig. 5B) or in digests of 56 or 59/61-kDa gel bands from S-[3H]mTFD-MPPB-labeled α1β3 GABAARs (Fig. 5, C and D). S-mTFD-MPPB was photoincorporated into a subunit fragment not labeled by R-[3H]mTFD-MPAB, because there was no hydrophilic peak of 3H in digests of 56 or 59/61-kDa bands derived from α1β3γ2 GABAAR photolabeled by that anesthetic (Fig. 5, E and F). However, in all samples there was a broad peak of 3H in the hydrophobic fractions (60–70% organic) known to contain most of the α1 and β3 subunit transmembrane helices (22, 23).
FIGURE 5.
Reversed-phase HPLC fractionation of EndoLys-C digests of subunits from α1β3γ2 (A and B) and α1β3 (C and D) GABAARs photolabeled by S-[3H]mTFD-MPPB or α1β3γ2 GABAARs photolabeled by R-[3H]mTFD-MPAB (E and F). 3H distribution (○, +300 μm GABA; ●, 30 μm bicuculline; ♢, no drug) upon rpHPLC fractionation of EndoLys-C digests of the 56-kDa (A, C, and E) or 61-kDa (B, D, and F) subunit gel bands from α1β3γ2 or α1β3 GABAARs photolabeled with S-[3H]mTFD-MPPB (3 μm, A–D) or R-[3H]mTFD-MPAB (2 μm, E and F). Photolabeling in a fragment eluting as a hydrophilic peak of 3H (fraction 22 in A), which was seen only in the digest of the 56-kDa gel band from S-[3H]mTFD-MPPB-photolabeled α1β3γ2 GABAAR, was inhibited by GABA but not by bicuculline, a competitive antagonist.
GABA Inhibits S-[3H]mTFD-MPPB Photolabeling of γ2Ser-280 (γM2–15′) at the γ+-β− Subunit Interface
The presence of a novel S-[3H]mTFD-MPPB-photolabeled subunit fragment in digests of the 56-kDa band from α1β3γ2 GABAARs (Fig. 5A) led us to examine the differences in predicted subunit fragmentation patterns for EndoLys-C digests of γ2 compared with the α1 subunit. In the regions of primary structure containing transmembrane helices, the presence of γ2Lys-259 in the M1-M2 loop was notable, because EndoLys-C cleavage there and at either Lys near the C terminus of γM2 would generate a fragment containing only γM2. In contrast, EndoLys-C digestion of α1 or β3 subunits can only produce the fragments beginning before M1 and extending through M2 that had been identified previously in the hydrophobic HPLC fractions (22, 23).
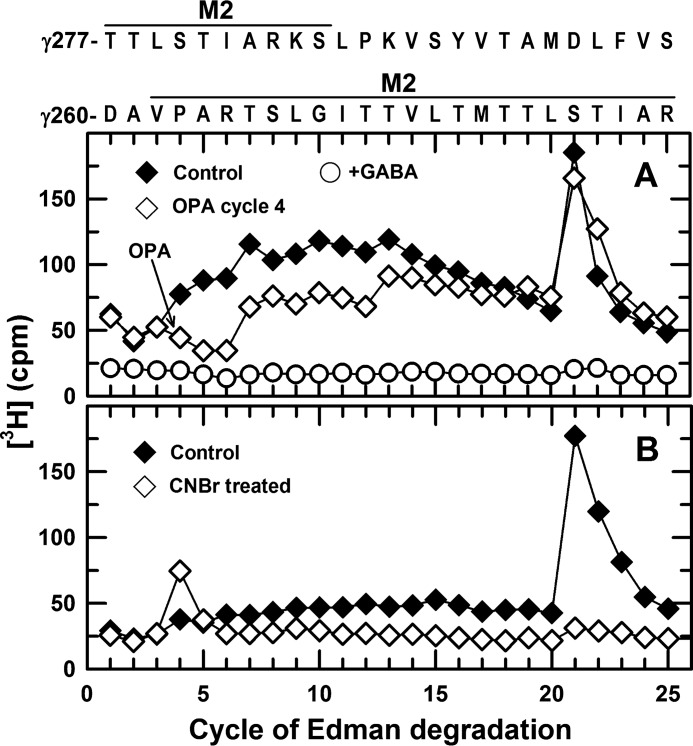
GABA-inhibitable photolabeling of γ2Ser-280 (γM2–15′) was established by N-terminal sequence analyses of material from the hydrophilic rpHPLC peak of 3H from the EndoLys-C digests of the 56-kDa gel band. Because the primary sequences in these fractions originated from the α1 subunit ECD, we used radiochemical sequencing strategies taking advantage of the fact that the γM2 fragment beginning at γ2Asp-260 contains a Pro in cycle 4 and a Met in cycle 17 of Edman degradation. First, two identical samples from GABAAR photolabeled in the absence of GABA were sequenced with sequencing of one sample interrupted at cycle 4 for treatment with OPA to prevent further sequencing of any fragments not containing a Pro at that cycle. For both samples, there was a peak of 3H release in cycle 21 of Edman degradation, consistent with photolabeling of γ2Ser-280 (Fig. 6A). Based on the detected PTH-derivatives, the major fragments present originated from the ECD of the α1 subunit and included peptides beginning at α1Thr-43 and α1Ser-107 at 1–2 pmol. Treatment with OPA reduced sequencing of those fragments by >80%, and the EndoLys-C fragment beginning at γ2Asp-260 was present at a low level (I0 ∼0.2 pmol). No peaks of 3H release were seen when the corresponding fractions were sequenced from GABAAR photolabeled in the presence of GABA (Fig. 6A).
FIGURE 6.
GABA inhibits S-[3H]mTFD-MPPB photolabeling of γ2Ser-280 in γM2 (γM2–15′). 3H release during N-terminal sequencing of aliquots from rpHPLC fractions 22/23 from EndoLys-C digests of the 56-kDa gel band from α1β3γ2 GABAARs photolabeled by S-[3H]mTFD-MPPB (3 μm) in the absence of other drugs (control, ♦, and ♢) or presence (○) of 300 μm GABA. A, when two identical aliquots of the control sample were sequenced with one sample treated with OPA before cycle 4 of Edman degradation, there was a single major peak of 3H release in cycle 21 for both samples. The 3H release in cycle 21 was reduced by >90% for the sample from GABAAR photolabeled in the presence of GABA. B, from a second photolabeling experiment, two identical aliquots from rpHPLC fractions 22/23 were sequenced, with one sample pretreated with CNBr before sequencing to cleave at the C termini of methionines. The single peak of 3H release in cycle 21 of the control sample was shifted to cycle 4 after treatment with CNBr.
Photolabeling of γ2Ser-280 was confirmed by sequencing 2 equivalent samples from another photolabeling experiment, with one sample pretreated with CNBr to cleave at the C termini of methionines. Pretreatment with CNBr shifted the peak of 3H release from cycle 21 to cycle 4 (Fig. 6B), consistent with cleavage at γ2Met-276 in γM2. These radiochemical sequencing strategies established that the GABA-inhibitable photolabeling in the 21st cycle of Edman degradation was in a GABAAR subunit with a defined distribution of Lys, Pro, and Met residues, Lys-Xaa3-Pro-Xaa12-Met-Xaa3, where Xaa is any amino acid. Inspection of the α1, β3, and γ2 subunit sequences revealed only one other fragment consistent with that distribution, a fragment from the γ2 subunit cytoplasmic domain beginning at γ2Asn-336 that also contains a Pro in cycle 2. Because OPA treatment at cycle 2 fully inhibited subsequent release of 3H in cycle 21 (data not shown), the combined radiochemical sequencing strategies established GABA-inhibitable photolabeling of γ2Ser-280 (γM2–15′).
Based on the peak of 3H release in cycle 21 and the mass of the γ2Asp-260 fragment detected after OPA treatment, S-[3H]mTFD-MPPB photolabeled γ2Ser-280 at ∼4000 cpm/pmol, and GABA inhibited that photolabeling by ≥90%. The calculated efficiencies of photolabeling of γ2Ser-280 in different pharmacological conditions and of the amino acids photolabeled in other subunits are tabulated in Table 1. The locations of γ2Ser-280 at the γ+-β− interface and of the other amino acids photolabeled by S-[3H]mTFD-MPPB are depicted in Fig. 7, based upon their locations in a GABAAR homology model described below.
TABLE 1.
Pharmacological specificity of photolabeling of residues in α1β3γ2 and α1β3 GABAARs by S-[3H]mTFD-MPPB, a convulsant, and R-[3H]mTFD-MPAB, an anesthetic (cpm/pmol of PTH-derivative)
The efficiency of photolabeling of a residue (in cpm/pmol) was calculated using Equation 4 (see under “Experimental Procedures”). N, the number of samples sequenced. The data are presented as the mean and individual values when two samples were sequenced or as mean (± S.D.) when three or four samples were sequenced. Other values were determined from the sequencing of single samples, with estimated uncertainties of <25%. The radiochemical specific activities of S-[3H]mTFD-MPPB and R-[3H]mTFD-MPAB are 50 and 38 Ci/mmol, respectively. ND means not determined.
| Subunit–interface | Amino acid |
S-[3H]mTFD-MPPB |
R-[3H]mTFD-MPAB |
||||||
|---|---|---|---|---|---|---|---|---|---|
| α1β3γ2 GABAAR |
α1β3 GABAAR |
α1β3γ2 GABAAR |
|||||||
| Control | GABA | Bicuculline | Control | GABA | Bicuculline | GABA | Bicuculline | ||
| γ+ M2 | Ser-280 | 4000a | 250a | ND | X | X | X | <50 | <50 |
| β+ M3 | Met-286 | 36 ± 10 (n = 4) | 56 ± 17 (n = 4) | 45 ± 8 (n = 3) | 80 | 100 | 70 | 100 | 90 |
| β+ M3 | Phe-289 | 210 ± 10 (n = 4) | 400 ± 40 (n = 4) | 154 ± 27 (n = 3) | 480 | 610 | 440 | 80 | 60 |
| β+ M2 | Thr-262 | ND | 460 | 80 | 220 | 470 | 300 | 170 | 100 |
| β− M1 | Met-227 | 73 (86, 59) | 87 (77, 97) | 67 (59, 74) | 100 | 80 | 60 | 3110 | 2100 |
| β− M1 | Leu-231 | 39 (45, 33) | 57 (57, 56) | 26 (25, 26) | 120 | 150 | 70 | <70 | <50 |
| α+ M3 | Tyr-294 | 100 | ND | 90 | 210 | 220 | 220 | 100 | 70 |
| α− M1 | Met-236 | 70 | ND | 50 | 100 | 90 | ND | <100 | <50 |
a For γ2Ser-280, the cpm/pmol was determined from the data of Fig. 6A. For the control condition, the sample was sequenced with OPA treatment in cycle 4 to allow mass determination. For the +GABA condition, the sample was sequenced without OPA, and the cpm/pmol was calculated from the 3H released in cycle 21 and the mass of the control sample.
FIGURE 7.
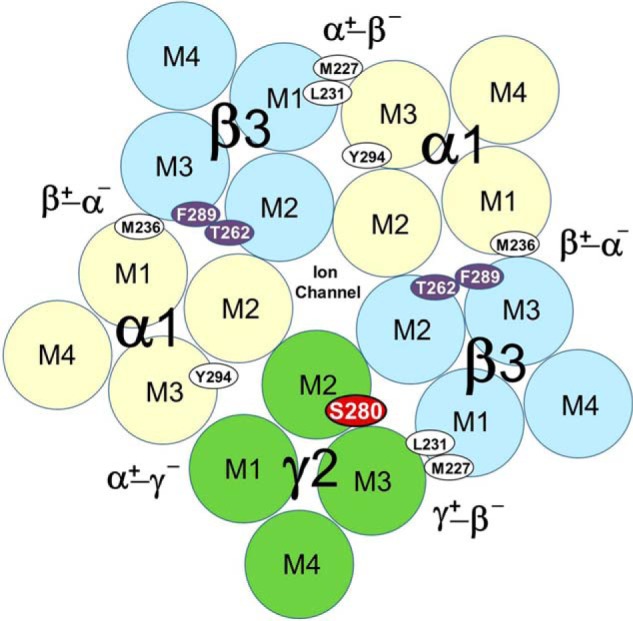
Cross-section of the transmembrane domain of the α1β3γ2 GABAAR showing the locations of the residues photolabeled by S-[3H]mTFD-MPPB. Residues labeled most efficiently in the absence or presence of GABA are highlighted by red or purple backgrounds, respectively. In the absence of GABA, γ2Ser-280 (γM2–15′) is labeled >10-fold more efficiently than any other residue (Table 1). GABA reduces photolabeling of γ2Ser-280 by >90% and enhances photolabeling of β3Thr-262 (βM2–12′) and β3Phe-289 in βM3 by 3–5-fold. The locations of the residues are approximated based upon their locations in an α1β3γ2 GABAAR homology model (see Fig. 12).
Evidence for Additional S-mTFD-MPPB-binding Sites, GABA Enhances S-[3H]mTFD-MPPB Photolabeling of β3Phe-289 (βM3) and β3Thr-262 (βM2–12′) at the β+-α− Interface
We next turned to the identification of S-mTFD-MPPB-binding sites that differ in their GABA sensitivity from the γ+-β− site. In α1β3γ2 GABAARs, GABA enhanced S-[3H]mTFD-MPPB photolabeling in the β3 subunit (Fig. 3; 59/61-kDa band), and rpHPLC fractionation of EndoLys-C digests of α1β3γ2 GABAARs of this gel band enriched in β3 subunits established that all 3H was recovered in the hydrophobic fractions that contain fragments beginning at the N termini of the M1 and M3 helices (Fig. 5B). To identify photolabeled amino acids in βM1 and βM3, we sequenced material from the appropriate rpHPLC fractions isolated from GABAARs photolabeled in the absence or presence of GABA or bicuculline. Representative sequencing data are shown in Fig. 8, A and B, and the calculated efficiencies of amino acid photolabeling in the different pharmacological conditions are tabulated in Table 1.
FIGURE 8.
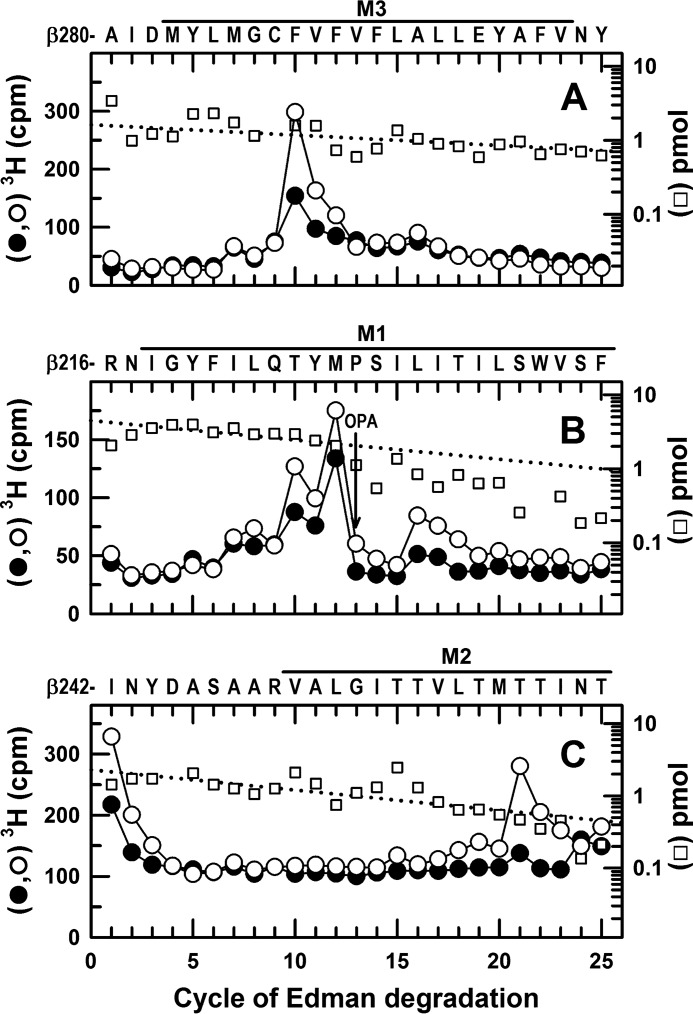
In α1β3γ2 GABAARs, GABA enhances S-[3H]mTFD-MPPB photolabeling at the β+ interface (β3Phe-289 in M3 and β3Thr-262 in M2) and the β− interface (β3Met-227 and β3Leu-231 in M1). 3H cpm (●, ○) and PTH-derivatives (□) released during N-terminal sequencing of fragments before βM3 (A), βM1 (B), and βM2 (C) from GABAARs photolabeled in the presence of bicuculline (●, 30 μm) or GABA (○, 300 μm). From the photolabeling experiment of Fig. 5, A and B, EndoLys-C digests of the 59-kDa gel bands were fractionated by rpHPLC, and materials in fractions 26 and 27 (A) or fractions 28–30 (B) were sequenced. A, primary sequence began at β3Ala-280 (I0 = 1.6 pmol), with a secondary sequence beginning at β3Arg-216 (I0 = ∼1 pmol). The major peak of 3H release at cycle 10, consistent with photolabeling of β3Phe-289, was enhanced by 100% in the presence of GABA (Table 1). B, primary sequence began at β3Arg-216 (I0 = 4.5 pmol), with a secondary sequence beginning at β3Ala-280, present at levels below 1 pmol before OPA treatment in cycle 13 and undetectable after treatment. The peaks of 3H release in cycles 12 and 16 indicated GABA-enhanced photolabeling of β3Met-227 and β3Leu-231 in βM1. The peak of 3H release in cycle 10 corresponds to photolabeling of β3Phe-289 in βM3 of the secondary sequence. C, to identify photolabeling in βM2, aliquots from the 61-kDa gel bands from GABAARs photolabeled in presence of GABA (○) or bicuculline (●) were sequenced after treatment of the sequencing filter with BNPS-skatole to cleave at the C termini of tryptophans. The sequence beginning at β3Ile-242 was present (I0 = 2.3 pmol), along with fragments at 1–4 pmol each beginning at the β3 subunit N terminus, β3Arg-68, β3Val-93, and β3Arg-169, 49 amino acids before βM1, β3Ser-427, and β3Leu-444 in βM4 that are 17 and 4 amino acids in length, respectively. The peak of 3H release in cycle 21 indicated GABA-enhanced photolabeling of β3Thr-262 (βM2–12′). No 3H release was seen when intact β subunit was sequenced, and if cycle 21 of the β3Arg-169 fragment had been photolabeled, it would have been recovered by rpHPLC from EndoLys-C digests as a hydrophilic fragment from the ECD.
Photolabeling of β3Phe-289 in βM3 was identified by the major peak of 3H release in cycle 10 when a fragment was sequenced beginning at β3Ala-280 before βM3 (Fig. 8A). Photolabeling of β3Met-227 and β3Leu-231 in βM1 was identified by the peaks of 3H release in cycles 12 and 16 when a fragment was sequenced beginning at β3Arg-216 (Fig. 8B). Quantification of the efficiencies of photolabeling established that in the control condition (i.e. in the absence of GABA or bicuculline), β3Phe-289 and β3Met-227 were photolabeled at 200 and 70 cpm/pmol, respectively, i.e. at ∼5 and 2% the efficiency of γ2Ser-280 from the same photolabeling experiment. Compared with control, GABA increased photolabeling of β3Phe-289 by 100%, and bicuculline reduced it by 25% (Table 1).
Photolabeling of β3Thr-262 (βM2–12′) was identified by sequencing a fragment beginning at β3Ile-242 in the M2-M3 loop (Fig. 8C). For the sample from GABAAR photolabeled in the presence of GABA, there was a single major peak of 3H release in cycle 21, consistent with photolabeling of β3Thr-262 (βM2–12′) at 460 cpm/pmol, with photolabeling reduced by 80% in the presence of bicuculline (Table 1). β3Thr-262 in βM2 and β3Phe-289 in βM3 are located at the β+-α− subunit interfaces, whereas β3Met-227 and β3Leu-231 in βM1 are located at the α+-β− and γ+-β− interface sites (Fig. 7).
Evidence for Additional S-mTFD-MPPB Sites, S-[3H]mTFD-MPPB Photolabeling in the α Subunit
The presence of similar hydrophobic peaks of 3H in the rpHPLC fractionations of EndoLys-C digests of the 56-kDa gel bands from S-[3H]mTFD-MPPB photolabeled α1β3γ2 and α1β3 GABAARs (Fig. 5, A and C) suggested photolabeling within the fragments eluting there that begin at α1Arg-221 and/or α1Val-280 at the N termini of αM1 and αM3 (21, 22). When we sequenced those fractions from α1β3γ2 GABAARs, the α1Arg-221 and α1Val-280 fragments were present at 4 pmol, and the profiles of 3H release indicated photolabeling of α1Met-236 in αM1 (the amino acid photolabeled by [3H]azietomidate) and α1Tyr-294 (an amino acid in αM3 photolabeled by R-[3H]mTFD-MPAB) at ∼50–100 cpm/pmol, i.e. at a similar efficiency as β3Met-227 in βM1 and ∼2% the efficiency of γ2Ser-280 (+bicuculline) (Table 1). We found no evidence of photolabeling within αM2 in the presence of either GABA or bicuculline. Based upon the mass of the α1Ser-251 fragment sequenced (I0 = 0.5 pmol) and the background levels of 3H release, in the presence of bicuculline, photolabeling of αM2–15′ (α1Ser-270), if it occurred, was at <5% the efficiency of photolabeling of γM2–15′ (γ2Ser-280). Similarly, in the presence of GABA, any photolabeling of αM2–12′ (α1Thr-267) was at <10% the level of labeling of βM2–12′ (β3Thr-262).
S-[3H]mTFD-MPPB Photolabeling in α1β3 GABAAR
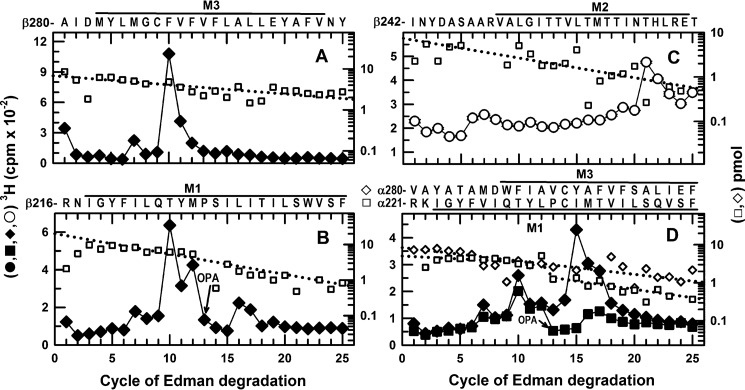
To determine whether the presence of the γ2 subunit altered S-[3H]mTFD-MPPB binding at the β/α intersubunit sites, we used the same procedures to identify the amino acids photolabeled in α1β3 GABAARs, and we established that the same amino acids were photolabeled as follows: β3Phe-289 in βM3 (Fig. 9A); β3Met-227 and β3Leu-231 in βM1 (Fig. 9B); β3Thr-262 (βM2–12′) in βM2 (Fig. 9C); and α1Tyr-294 in αM3 and α1Met-236 in αM1 (Fig. 9D). Furthermore, quantification of the amino acid photolabeling efficiencies (Table 1) established that, just as in the α1β3γ2 GABAAR, the residues photolabeled most efficiently in the α or β subunits were M2 β3Thr-262 and M3 β3Phe-289, and photolabeling of those residues was enhanced in the presence of GABA compared with control or bicuculline. Thus, the γ2 subunit did not alter S-[3H]mTFD-MPPB binding at the β/α intersubunit sites.
FIGURE 9.
In α1β3 GABAARs, S-[3H]mTFD-MPPB photolabels β3Thr-262 (βM2), β3Met-286/β3Phe-289 (βM3), and α1Met-236 (αM1) at the β+-α− interface and β3Met-227/β3Leu-231 (βM1) and α1Tyr-294 (αM3) at the α+-β− interface. 3H cpm (♦,■, ○) and PTH-derivatives (□,♢) released during N-terminal sequencing of fragments beginning before βM3 (A), βM1 (B), βM2 (C), and αM1 and αM3 (D) from α1β3 GABAARs photolabeled with 2.7 μm S-[3H]mTFD-MPPB in the absence (♦, ■) or presence of GABA (○, 300 μm) or bicuculline (data not shown, see Table 1). A, when rpHPLC fractions 26–27 were sequenced from an EndoLys-C digest of the 61-kDa gel band, the fragment beginning at β3Ala-280 was present at 6.8 pmol, and the peaks of 3H release in cycles 7 and 10 indicated photolabeling of β3Met-286 and β3Phe-289. B, when rpHPLC fractions 28 and 29 were sequenced with OPA treatment at cycle 13, corresponding to β3Pro-228 in βM1, the primary sequence began at β3Arg-216 (I0 = 20 pmol) and a secondary sequence began at β3Ala-280, present at 2 pmol before OPA and undetectable after treatment. The peaks of 3H release in cycles 12 and 16 indicated photolabeling of β3Met-227 and β3Leu-231 in βM1. The peaks of release in cycles 7 and 10 resulted from the photolabeling of β3Met-286 and β3Met-289 in the secondary sequence present before OPA treatment in cycle 13. C, to identify photolabeling in βM2, aliquots from the 59-kDa gel bands were sequenced after treatment of the sequencing filter with BNPS-skatole to cleave at the C termini of tryptophans. The sequence beginning at β3Ile-242 was present (I0 = 7pmol), along with fragments beginning at the β3 subunit N terminus, β3Arg-68, β3Val-93, β3Arg-169, and β3Ser-427 at 4–10 pmol each. The peak of 3H release in cycle 21 indicated photolabeling of β3Thr-262 (βM2–12′). D, 2 aliquots of rpHPLC fractions 26–29 from an EndoLys-C digest of the 56-kDa subunit gel band were sequenced with (■) or without (♦) OPA treatment in cycle 13 (at α1Pro-233). For the untreated sample, the fragments beginning at α1Arg-221 (data not shown) and α1Val-280 (♢) were present at 11 and 8 pmol, respectively. For the OPA-treated sample, α1Arg-221 (□) and α1Val-280 (data not shown) were initially present at 5 pmol. After OPA treatment, sequencing of the α1Arg-221 fragment continued, although the α1Val-280 fragment was reduced by >90%. The peak of 3H release in cycle 15, not seen after treatment with OPA, indicated photolabeling of α1Tyr-294 in αM3. After treatment with OPA, the small peak of 3H release in cycle 16 indicated photolabeling of α1Met-236 in αM1. Efficiencies of residue photolabeling in the absence or presence of GABA or bicuculline are included in Table 1.
R-[3H]mTFD-MPAB Photolabeling in α1β3γ2 GABAAR
To allow a more direct comparison of the modes of binding of S-mTFD-MPPB and R-mTFD-MPAB, we also characterized GABAAR photolabeling by R-[3H]mTFD-MPAB in the presence of bicuculline compared with GABA. The efficiencies of photolabeling at the amino acid level are included in Table 1. As noted above, based upon the rpHPLC fractionation of EndoLys-C digests of 56- and 59/61-kDa gel bands (Fig. 5, E and F), there was no evidence of any photolabeling within γM2. Considering just the residues with major photoincorporation in the presence of GABA, S-[3H]mTFD-MPPB photoincorporated into β+ interfaces at 5-fold (β3Phe-289 in βM3) and 3-fold (β3Thr-262 (M2–12′)) the efficiency of [3H]R-mTFD-MPAB, whereas R-[3H]mTFD-MPAB photoincorporated into β− interfaces (β3Met-227 in βM1) with 35-fold higher efficiency than S-[3H]mTFD-MPPB (Table 1). In contrast to S-[3H]mTFD-MPPB, which photolabels γ2Ser-280 in γM2 with high selectivity only in the absence of GABA, R-[3H]mTFD-MPAB photolabeled β3Met-227 in βM1 with high selectivity in the presence of GABA or bicuculline.
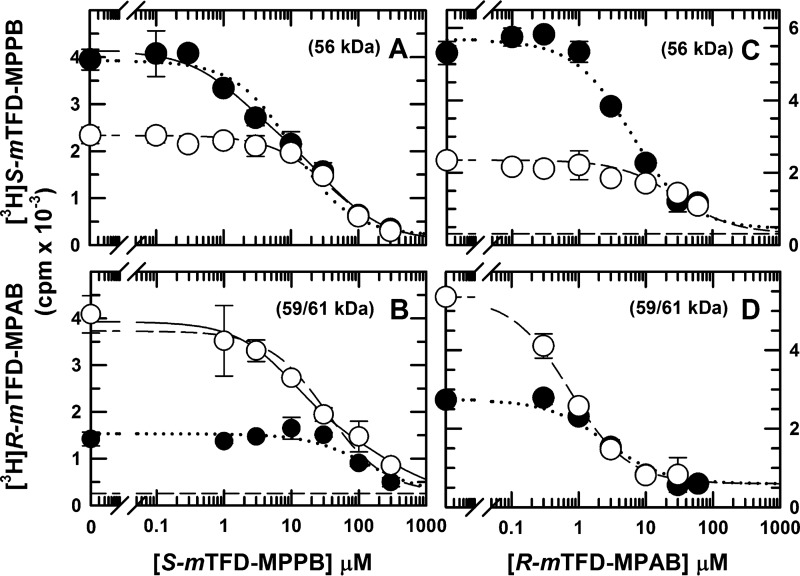
S-mTFD-MPPB Inhibition of S-[3H]mTFD-MPPB and R-[3H]mTFD-MPAB Photolabeling
To characterize S-mTFD-MPPB binding affinity at the γ+-β− interface, we compared the concentration dependence of nonradioactive S-mTFD-MPPB inhibition of S-[3H]mTFD-MPPB photoincorporation in the 56-kDa gel band from α1β3γ2 GABAARs photolabeled in the presence of bicuculline or GABA (Fig. 10A). In the presence of bicuculline, S-[3H]mTFD-MPPB photoincorporation in γ2Ser-280 accounts for ∼50% of the 3H incorporated in the ∼56-kDa gel band containing the α1 and γ2 subunits (Fig. 5A). In contrast, in the presence of GABA, photolabeling of γ2Ser-280 is inhibited by ≥90%, and 3H in the 56-kDa gel band results primarily from photolabeling in αM3/αM1. In the presence of GABA, S-mTFD-MPPB inhibition of S-[3H]mTFD-MPPB photolabeling was well fit by a one-site model with IC50 = 39 ± 5 μm. In contrast, in the presence of bicuculline, the concentration dependence of inhibition was fit by a Hill coefficient, nH = 0.74 ± 0.10. Inhibition was consistent with a two-site model as follows: a high affinity component (IC50, H = 1.7 ± 0.5 μm) that reduced the level of photolabeling to that observed in the presence of GABA, and a low affinity component with IC50, L = 38 ± 8 μm, similar to the affinity seen in the presence of GABA (Table 2). These results indicate that S-mTFD-MPPB binds in the presence of bicuculline to the γ+-β− site with ∼10-fold higher affinity than it binds to other intersubunit sites in the presence of bicuculline or GABA.
FIGURE 10.
S-mTFD-MPPB and R-mTFD-MPAB inhibition of S-[3H]mTFD-MPPB or R-[3H]mTFD-MPAB photolabeling of α1β3γ2 GABAARs. GABAARs were photolabeled by S-[3H]mTFD-MPPB (A and C) or R-[3H]mTFD-MPAB (B and D) in the presence of bicuculline (●, 30 μm) or GABA (○, 300 μm) and nonradioactive S-mTFD-MPPB (A and B) or R-mTFD-MPAB (C and D). 3H incorporation into the 56-kDa (A and C) or 59/61-kDa (B and D) subunit bands was determined by liquid scintillation counting. Data are average (± S.D.) of two separate experiments. Data were fit to single-site (nH = 1, dot or dash traces) or two-site (solid traces) binding models as described under “Experimental Procedures.” Parameter fits are tabulated in Table 2.
TABLE 2.
S-mTFD-MPPB and R-mTFD-MPAB inhibition of α1β3γ2 GABAAR photolabeling by S-[3H]mTFD-MPPB, R-[3H]mTFD-MPAB, and R-[3H]azietomidate
IC50 values, the total anesthetic concentrations resulting in 50% inhibition of GABAAR photolabeling, were determined as described under “Experimental Procedures.” Parameters were determined from fits of data from two independent experiments, each carried out in parallel in the presence of GABA or bicuculline. ND means not determined.
| Drug |
S-[3H]mTFD-MPPB IC50 |
R-[3H]mTFD-MPAB IC50 |
R-[3H]Azietomidate IC50 |
|||
|---|---|---|---|---|---|---|
| +GABA | +Bicuculline | +GABA | +Bicuculline | +GABA | +Bicuculline | |
| μm | μm | μm | ||||
| S-mTFD-MPPB | 39 ± 5a | 1.7 ± 0.5/38 ± 8b | 10 ± 2/310 ± 130b | 130 ± 60a | 102 ± 11 | >100 |
| R-mTFD-MPAB | 29 ± 6c | 7.5 ± 1.6 | 0.7 ± 0.1 | 2.3 ± 0.3 | 76 ± 14d | >100 |
| R-mTFD-MPPB | ND | ND | 1.8 ± 0.1 | 7.9 ± 1.4 | >100 | >100 |
a +GABA and +bicuculline data from Fig. 10, A and B, were fit to a one-site model (Equation 1, nH = 1).
b +Bicuculline (Fig. 10A) and +GABA (Fig. 10B) data were fit to a two-site model (Equation 2).
c Value of IC50 when data fit to Equation 1, nH = 1. When fit to Equation 1 with variable nH, IC50 = 38 ± 6 μm, nH = 0.6 ± 0.1.
d Data were from Ref. 23.
To further clarify the state-dependence of S-mTFD-MPPB binding at β− interface sites, we also determined its inhibition of R-[3H]mTFD-MPAB photolabeling (Fig. 10B). In the presence of bicuculline, S-mTFD-MPPB inhibited R-[3H]mTFD-MPAB photolabeling with low affinity (IC50 = 130 ± 60 μm), not with the IC50 of 1.7 μm characteristic of its binding to the γ+-β− site based upon inhibition of S-[3H]mTFD-MPPB photolabeling. In contrast, R-mTFD-MPAB inhibition of R-[3H]mTFD-MPAB photolabeling was consistent with a single site model (nH = 1) in the presence of bicuculline (IC50 = 2.3 ± 0.3 μm) or GABA (IC50 = 0.7 ± 0.04 μm) (Fig. 10D and Table 2). The difference in IC50 values for S-mTFD-MPPB inhibition of S-[3H]mTFD-MPPB and R-[3H]mTFD-MPAB photolabeling was unexpected. However, β3Met-227, the amino acid that dominates β subunit photolabeling by R-[3H]mTFD-MPAB in the presence of GABA or bicuculline (Table 1), is present at both the α+-β− and γ+-β− interfaces (Fig. 7). Thus, the difference in IC50 values seen for S-mTFD-MPPB inhibition suggests that in the presence of bicuculline R-[3H]mTFD-MPAB photolabels primarily β3Met-227 at the α+-β− site and that the increase of photolabeling of β3Met-227 in the presence of GABA reflects photolabeling at the γ+-β− site as a result of increased binding affinity at that site. Similar to R-mTFD-MPAB and in contrast to S-mTFD-MPPB, the R-enantiomer of mTFD-MPPB was a potent inhibitor of R-[3H]mTFD-MPAB photolabeling in the presence of bicuculline (IC50 = 7.9 ± 1.4 μm) and 4-fold more potent in the presence of GABA (IC50 = 1.8 ± 0.1 μm) (Table 2).
The concentration dependence of S-mTFD-MPPB inhibition of R-[3H]mTFD-MPAB photolabeling (Fig. 10B) in the presence of GABA was fit by a Hill coefficient, nH = 0.6 ± 0.04, consistent with site heterogeneity. This inhibition was consistent with a two-site model as follows: 1) a high affinity component (IC50, H = 10 ± 2 μm) that reduced photolabeling to the level observed in the presence of bicuculline, and 2) a low affinity component with IC50, L = 310 ± 130 μm, similar to the affinity seen in the presence of bicuculline. In the presence of bicuculline, R-mTFD-MPAB inhibition of S-[3H]mTFD-MPPB photolabeling (Fig. 10C) was consistent with a single-site model (nH = 1) with an IC50 (7.5 ± 1.6 μm) close to that seen for inhibition of R-[3H]mTFD-MPAB photolabeling (Table 2). However, in the presence of GABA, inhibition was characterized by nH = 0.6 ± 0.1 (IC50 = 38 ± 6 μm), a consequence of S-[3H]mTFD-MPPB photolabeling of amino acids in αM3 (α+ interface) and αM1 (α− interface).
S-[3H]mTFD-MPPB Photolabeling of γ2Ser-280 (γM2–15′) Is Inhibited Allosterically by Etomidate but Not by Picrotoxinin
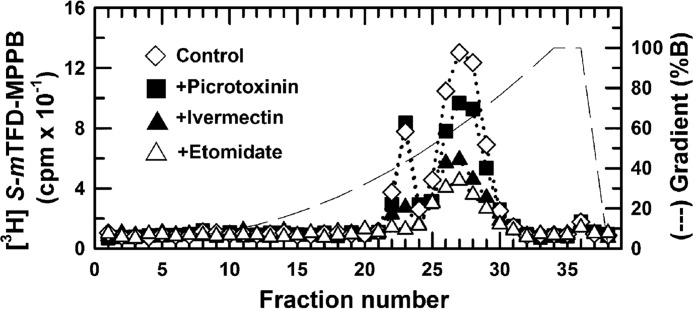
To further characterize the pharmacological specificity of S-[3H]mTFD-MPPB binding at the γ+-β− interface, we used rpHPLC fractionation of EndoLys-C digests of the 56-kDa gel bands to monitor the effects of bicuculline, GABA, picrotoxinin, etomidate, and ivermectin on the photolabeling of γ2Ser-280 (Figs. 5A and 11; Table 3). Compared with control, neither bicuculline (30 μm, Fig. 5A) nor picrotoxinin (30 μm) inhibited photolabeling in the hydrophilic fractions containing γM2. With one preparation of purified GABAAR, they increased photolabeling by ∼200 and 80%, respectively, when GABA (300 μm) inhibited photolabeling by 80%. In two other preparations, they increased photolabeling by 5–40%, whereas GABA inhibited by 80%. Etomidate (300 μm), which binds with high affinity and selectivity at the β+-α− interface sites, inhibited photolabeling by >90%. This inhibition must reflect strong negative allosteric coupling, because at the same concentration etomidate enhances R-[3H]mTFD-MPAB binding at this β− interface site (23). Ivermectin (30 μm), which binds at subunit interfaces (36), inhibited photolabeling by ∼75%.
FIGURE 11.
S-[3H]mTFD-MPPB photolabeling of γ2Ser-280 is inhibited allosterically by etomidate but not by picrotoxinin. EndoLys-C digests of the 56-kDa subunit gel bands from α1β3γ2 GABAARs photolabeled by S-[3H]mTFD-MPPB in the absence of other drugs (♢), in the presence of 30 μm picrotoxinin (■), 30 μm ivermectin (▴), or 300 μm etomidate (▵) were fractionated by rpHPLC. Photolabeling of γ2Met-280, as assayed by the amount of 3H cpm in rpHPLC fractions 21–23, is quantified in Table 3.
TABLE 3.
Pharmacological specificity of S-[3H]mTFD-MPPB photolabeling of γ2Ser-280 in α1β3γ2 GABAAR (% control)
Incorporation in γ2-Ser-280 was quantified in experiments including those in Figs. 5A and 11 by measuring the 3H cpm recovery in fractions 21–23 from rpHPLC fractionations of EndoLys-C digests of 56-kDa gel bands from GABAARs photolabeled with S-[3H]mTFD-MPPB in the absence of other drugs (control) or in the presence of drugs. n is the number of photolabeling experiments.
| +Bicuculline | +GABA | +Etomidate | +S-mTFD-MPPB | +Ivermectin | +Picrotoxinin |
|---|---|---|---|---|---|
| 190 ± 110 (n = 3) | 24 ± 5 (n = 3) | 5 (n = 1) | 5 (n = 1) | 30 (n = 1) | 140 ± 50 (n = 2) |
Discussion
In this study, we demonstrate that the S-isomer of mTFD-MPPB, an α1β3γ2 GABAAR inhibitor, stabilizes the receptor in a closed channel state by binding with high affinity to a TMD site in the γ+-β− interface previously identified as a binding site for the anesthetic barbiturate R-mTFD-MPAB (23). S-mTFD-MPPB binding at this site shows negatively energetic coupling to GABA binding in the ECD. In contrast, R-mTFD-MPAB binding is positively coupled to GABA. Our results provide the first demonstration that subtle changes in structure determine whether a drug acts as a positive or negative GABAAR allosteric modulator when binding at a TMD intersubunit site, as occurs at the ECD α+-γ− interface benzodiazepine-binding site (27, 37).
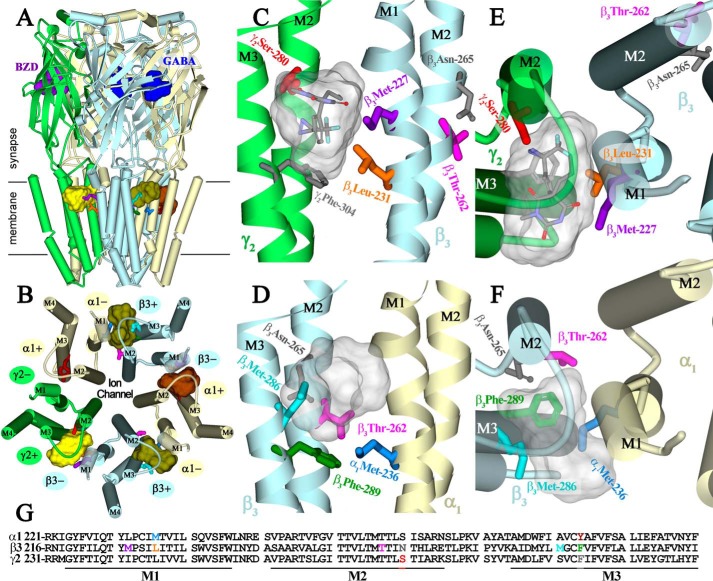
S-[3H]mTFD-MPPB binding at the γ+-β− interface was identified by the efficient and GABA-inhibitable photolabeling of γ2Ser-280 in γM2. No other GABA-inhibitable photolabeling was observed either in α1β3γ2 or α1β3 GABAARs (Table 1). This observation, together with the inhibition detected in α1β3γ2 but not α1β3 GABAARs (Fig. 2), ties this site to the inhibition seen in vitro and convulsant activity seen in vivo (26). Interestingly, γ2Ser-280 is homologous to β3Asn-265 (both M2–15′ residues), a determinant of etomidate and propofol potency as GABAAR potentiators in vitro and as anesthetics in vivo (9, 38). In a GABAAR homology model based upon the structure of a homomeric β3 GABAAR (Fig. 12), γ2Ser-280 is located at the γ+-β− interface, lining a pocket in which R-[3H]mTFD-MPAB binds and photolabels β3Met-227 in βM1 and, at lower efficiency, γ2Ser-301 in γM3 (Fig. 12, C and E) (23).
FIGURE 12.
S-mTFD-MPPB-binding sites at subunit interfaces within the transmembrane domain of an α1β3γ2 GABAAR. A–F, views are shown of an α1β3γ2 GABAAR homology model built using the β3 GABAAR crystal structure (PDB 4COF), with subunits color-coded (α1, light yellow; β3, light blue; and γ2, green), and amino acids of interest shown in stick representation, color-coded to match their colors in the primary structure alignment of the M1-M3 region of the three subunits (G). Views from the side (A) and of the TMD from the ECD-TMD interface (B) include the locations of GABA- (blue) and benzodiazepine (BZD, purple)-binding sites in the ECD and the pharmacologically distinct binding sites in the TMD for S-mTFD-MPPB at the γ+-β− (yellow), β+-α− (olive), and α+-β− (brown) interfaces. Illustrated in C, E and D, F are details of the pockets at γ+-β− and β+-α− interfaces, respectively, viewed from the lipid (C and D) and from the base of the ECD (E and F). The residues photolabeled by S-[3H]mTFD-MPPB (γ2Ser-280 (γM2–15′), β3Met-227 and β3Leu-231, β3Thr-262, β3Met-286, β3Phe-289, and α1Met-236) are shown in stick representation, as well as two residues (gray) that are not photolabeled as follows: γ2Phe-304, at the γ+-α− interface in a position equivalent to β3Phe-289 at the β+-α− interface, and β3Asn-265 (βM2–15′) at the β+-α− interface, an in vitro and in vivo sensitivity determinant for the GABAAR potentiating and anesthetic effects of etomidate and propofol (9, 38). Also included in C–F are the Connolly surfaces enclosing the 10 lowest energy docking solutions for S-mTFD-MPPB at each interface pocket. The volume within the Connolly surfaces at the γ+-α− and β+-α− interfaces are 590 and 420 Å3, respectively, and 315 Å3 for a single molecule. C and E, S-mTFD-MPPB is shown in stick representation at the γ+-β− interface in a low energy solution (CDocker interaction energy −38.3 kcal/mol) with the diazirine ∼3 Å from γ2Ser-280 (M2–15′), the GABAAR amino acid photolabeled most efficiently (+bicuculline). R-[3H]mTFD-MPAB photolabels β3Met-227 but not γ2Ser-280. At the β+-α− interface, [3H]azietomidate photolabels β3Met-286 and α1Met-236 (21, 22).
In addition to the γ+-β− site, S-mTFD-MPPB also bound with ∼10-fold lower affinity to the intersubunit TMD site in the β+-α− interface, photolabeling residues that overlap with those photolabeled by [3H]azietomidate (Fig. 12, D and F). Similar to etomidate, S-mTFD-MPPB binding at the β+-α− site is positively coupled to GABA binding. However, inhibition of [3H]azietomidate photolabeling establishes that even in the presence of GABA, S-mTFD-MPPB binds weakly to those sites.
State Dependence of S-mTFD-MPPB and R-mTFD-MPAB Binding
The differences in the IC50 values of S-mTFD-MPPB in the presence of GABA or bicuculline provide evidence that agonist/antagonist binding at the orthosteric site in the purified α1β3γ2 GABAAR shifts the receptor conformational equilibrium, presumably between desensitized and closed states in our photolabeling assays. Our results provide a simple explanation for why S-mTFD-MPPB inhibits α1β3γ2 and potentiates α1β3 GABAARs. S-mTFD-MPPB binds at the γ+-β− site with ≥10-fold higher affinity in the bicuculline-stabilized state than the GABA-stabilized state or to the β+-α− site in its preferred GABA-stabilized state. Hence, S-mTFD-MPPB binding at the γ+-β− site will result in negative allosteric modulation of GABA responses in the α1β3γ2 GABAAR. In an α1β3 GABAAR that has no γ+-β−-binding site, S-[3H]mTFD-MPPB photolabeling of residues at the β+-α− and α+-β− intersubunit sites is enhanced in the presence of GABA. This enhanced photolabeling is consistent with positive energetic coupling between S-mTFD-MPPB and GABA binding, with S-mTFD-MPPB acting as a positive allosteric modulator of GABA responses.
Our results also indicate that the positive energetic coupling between R-mTFD-MPAB and GABA binding is mediated primarily by strong state dependence of binding at the γ+-β− site. In the presence of bicuculline, S-mTFD-MPPB binds with high affinity at the γ+-β− site (IC50, H = 1.7 μm), but it inhibits R-[3H]mTFD-MPAB photolabeling with an IC50 of 130 μm. This discrepancy indicates that in the presence of bicuculline, R-[3H]mTFD-MPAB photolabels primarily β3Met-227 at the α+-β− site, and the 50% increase of β3Met-227 photolabeling in the presence of GABA compared with bicuculline results primarily from enhanced binding affinity at the γ+-β− site. In the presence of GABA, R-mTFD-MPAB binds to both β− sites with similar affinity (IC50 = 0.7 μm). Further studies will be required to quantify the asymmetry of R-mTFD-MPAB state dependence between the γ+-β− and α+-β− sites, similar to the asymmetry seen for agonist binding at the nonequivalent transmitter-binding sites in the muscle-type nicotinic acetylcholine receptor (39–41).
S-mTFD-MPPB and R-mTFD-MPAB Bind in Different Orientations at the γ+-β− Interface
S-mTFD-MPPB and R-mTFD-MPAB, which differ in structure only by chirality and the presence of either a 5-propyl or 5-allyl substituent (Fig. 1), bind with high affinity (IC50 values of <3 μm) at the γ+-β− interface, in the presence of bicuculline or GABA, respectively. Therefore, the selective photolabeling of γ2Ser-280 by S-[3H]mTFD-MPPB compared with that of β3Met-227 by R-[3H]mTFD-MPAB provides direct experimental evidence that the two drugs must bind in different orientations within this interface pocket. γ2Ser-280 and β3Met-227 are on opposite surfaces of the pocket with a distance of 11 Å between α-carbons in the GABAAR homology model (Fig. 12, C and E). S-[3H]mTFD-MPPB and R-[3H]mTFD-MPAB, with extended lengths of ∼10 Å, must bind in opposite but overlapping orientations with their diazirines oriented toward γM2 and βM1, respectively. In contrast, at the β+-α− interface both S-mTFD-MPPB and R-mTFD-MPAB bind with low affinity (enhanced by GABA) and photolabel the same amino acids.
Based upon computational docking, S-mTFD-MPPB is predicted to bind stably and with similar energies in the pockets at each of the subunit interfaces. The predicted locations of bound S-mTFD-MPPB are shown in Fig. 12 as Connolly surface representations of the 10 lowest energy solutions, with S-mTFD-MPPB shown in stick representation in Fig. 12, C and E, at the γ+-β−-binding site in an orientation with the reactive diazirine in proximity to γ2Ser-280. As seen previously in computational docking studies of TDBzl-etomidate or R-mTFD-MPAB (22, 23), S-mTFD-MPPB is also predicted to bind with similar energies at each of the intersubunit interfaces in homology models based upon other homomeric pentameric ligand-gated ion channels. Thus, docking studies cannot yet provide any explanation for the preferential binding of S-mTFD-MPPB at the γ+-β− interface or of the observed state dependence.
Pharmacological Specificity of Binding at the γ+-β− Site
Etomidate, at a concentration where it binds selectively at the β+-α− interface, allosterically inhibited photolabeling by ∼90%. This allosteric inhibition is predicted because GABA and etomidate stabilize the same receptor state (42, 43). Because R-mTFD-MPAB also binds to the γ+-β−-intersubunit pocket with highest affinity in the presence of GABA, it may also inhibit S-mTFD-MPPB binding allosterically. However, in view of the proximity of the residues in the γ+-β−-intersubunit pocket photolabeled by S-mTFD-MPPB and R-mTFD-MPAB, competitive inhibition is the simplest interpretation. Picrotoxinin, which binds at the cytoplasmic end of the ion channel (17, 44), did not inhibit S-[3H]mTFD-MPPB photolabeling. This establishes that in the purified α1β3γ2 GABAAR, picrotoxinin binds preferentially to the same closed channel state as bicuculline, a result consistent with its allosteric inhibition of [3H]muscimol binding to rat brain membrane fractions (45) and with recent mutational analyses (46). The strong negative coupling between S-mTFD-MPPB binding at the γ+-β− site and GABA binding in the ECD or anesthetic binding at the β+-α− site necessitates care in the use of S-[3H]mTFD-MPPB to identify other drugs that bind preferentially in the closed channel state to sites in the TMD. However, the differential binding properties of S-[3H]mTFD-MPPB, R-[3H]mTFD-MPAB, and [3H]azietomidate now allow the development of assays to determine whether drugs such as the volatile convulsant and GABAAR inhibitor fluorothyl (bis[2,2,2-trifluoroethy] ether) or its anesthetic isomer and GABAAR potentiator “iso-fluorothyl” (1,1,1,3,3,3-hexafluoro-2-methoxypropane) (4, 47, 48) bind selectively to intersubunit sites in the presence of bicuculline or GABA, respectively.
Conclusions
Our novel finding is that in a α1β3γ2 GABAAR the binding pocket in the TMD at the γ+-β− interface is the binding site for S-mTFD-MPPB, a negative allosteric modulator in vitro and a convulsant in vivo, although R-mTFD-MPAB, an anesthetic, binds with high affinity to the same intersubunit pocket but with a different orientation and with positive coupling to GABA binding. Intersubunit-binding sites in the TMD for positive and negative allosteric modulators have been identified in nicotinic acetylcholine receptors and serotonin 5-HT3 receptors containing cation-selective channels (49, 50). Also, general anesthetics of diverse chemical structure that act as GABAAR-positive allosteric modulators bind with varying selectivities to each of the intersubunit sites in the GABAAR TMD. Further studies are required to determine whether the γ+-β− binding pocket has unique structural features that result in negative as well as positive allosteric modulation or whether other drugs can inhibit GABA responses by binding to the homologous sites at the other subunit interfaces.
Author Contributions
J. B. C. and K. W. M. conceived and coordinated the study. S. S. J. and J. B. C. designed and analyzed the experiments of Figs. 3–8 and 10–11 that were performed by S. S. J. X. Z. expressed and purified GABAARs. P. Y. S. and K. S. B. synthesized chemical reagents used in the study. D. C. C. conducted the homology modeling and computational docking studies. R. D. and K. W. M. designed, performed, and analyzed electrophysiology experiments shown in Fig. 2. J. B. C. and S. S. J. wrote the paper with input from all authors. All authors approved the final version of the manuscript.
Acknowledgment
We thank Dr. Ayman Hamouda for useful comments on the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant GM-58448 from USPHS. The authors declare that they have no conflicts of interest with the contents of the article.
- GABAAR
- GABA type A receptors
- MPPB
- 1-methyl-5-phenyl-5-propyl barbituric acid
- mTFD-MPPB
- 1-methyl-5-propyl-5-(m-trifluoromethyl-diazirynylphenyl)barbituric acid
- mTFD-MPAB
- 1-methyl-5-allyl-5-(m-trifluoromethyl-diazirynylphenyl)barbituric acid
- ECD
- extracellular domain
- TMD
- transmembrane domain
- EndoGlu-C
- S. aureus endoproteinase Glu-C
- EndoLys-C
- Lysobacter enzymogenes endoproteinase Lys-C
- BNPS-skatole
- 3-bromo-3-methyl-2-(2-nitrophenylthio)-3H-indole
- rpHPLC
- reversed-phase high-performance liquid chromatography
- OPA
- o-phthalaldehyde
- PTH-derivative
- phenylthiohydantoin-derivative
- PDB
- Protein Data Bank.
References
- 1. Patel P. M., Patel H. H., Roth D. (2011) Goodman and Gilman's The Pharmacological Basis of Experimental Therapeutics (Brunton L., Chabner B., Knollman B., eds) pp. 527–564, McGraw Hill, New York [Google Scholar]
- 2. Krantz J. C. Jr., Truitt E. B. Jr., Ling A. S., Speers L. (1957) Anesthesia. LV. The pharmacologic response to hexafluorodiethyl ether. J. Pharmacol. Exp. Ther. 121, 362–368 [PubMed] [Google Scholar]
- 3. Downes H., Perry R. S., Ostlund R. E., Karler R. (1970) A study of the excitatory effects of barbiturates. J. Pharmacol. Exp. Ther. 175, 692–699 [PubMed] [Google Scholar]
- 4. Koblin D. D., Eger E. I. 2nd, Johnson B. H., Collins P., Terrell R. C., Speers L. (1981) Are convulsant gases also anesthetics? Anesth. Analg. 60, 464–470 [PubMed] [Google Scholar]
- 5. Büch H. P., Schneider-Affeld F., Rummel W., Knabe J. (1973) Stereochemical dependence of pharmacological activity in a series of optically active N-methylated barbiturates. Naunyn Schmiedeberg's Arch. Pharmacol. 277, 191–198 [DOI] [PubMed] [Google Scholar]
- 6. Macdonald R. L., Olsen R. W. (1994) GABAA receptor channels. Annu. Rev. Neurosci. 17, 569–602 [DOI] [PubMed] [Google Scholar]
- 7. Hemmings H. C. Jr., Akabas M. H., Goldstein P. A., Trudell J. R., Orser B. A., Harrison N. L. (2005) Emerging molecular mechanisms of general anesthetic action. Trends Pharmacol. Sci. 26, 503–510 [DOI] [PubMed] [Google Scholar]
- 8. Franks N. P. (2008) General anaesthesia: from molecular targets to neuronal pathways of sleep and arousal. Nat. Rev. Neurosci. 9, 370–386 [DOI] [PubMed] [Google Scholar]
- 9. Jurd R., Arras M., Lambert S., Drexler B., Siegwart R., Crestani F., Zaugg M., Vogt K. E., Ledermann B., Antkowiak B., Rudolph U. (2003) General anesthetic actions in vivo strongly attenuated by a point mutation in the GABA(A) receptor β3 subunit. FASEB J. 17, 250–252 [DOI] [PubMed] [Google Scholar]
- 10. Reynolds D. S., Rosahl T. W., Cirone J., O'Meara G. F., Haythornthwaite A., Newman R. J., Myers J., Sur C., Howell O., Rutter A. R., Atack J., Macaulay A. J., Hadingham K. L., Hutson P. H., Belelli D., et al. (2003) Sedation and anesthesia mediated by distinct GABAA receptor isoforms. J. Neurosci. 23, 8608–8617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zeller A., Arras M., Jurd R., Rudolph U. (2007) Identification of a molecular target mediating the general anesthetic actions of pentobarbital. Mol. Pharmacol. 71, 852–859 [DOI] [PubMed] [Google Scholar]
- 12. Drexler B., Antkowiak B., Engin E., Rudolph U. (2011) Identification and characterization of anesthetic targets by mouse molecular genetics approaches. Can. J. Anesth. 58, 178–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Holland K. D., Canney D. J., Rothman S. M., Ferrendelli J. A., Covey D. F. (1990) Physiological modulation of the GABA receptor by convulsant and anticonvulsant barbiturates in cultured rat hippocampal neurons. Brain Res. 516, 147–150 [DOI] [PubMed] [Google Scholar]
- 14. Ticku M. K., Rastogi S. K., Thyagarajan R. (1985) Separate site(s) of action of optical isomers of 1-methyl-5-phenyl-5-propylbarbituric acid with opposite pharmacological activities at the GABA receptor complex. Eur. J. Pharmacol. 112, 1–9 [DOI] [PubMed] [Google Scholar]
- 15. Kamiya Y., Andoh T., Furuya R., Hattori S., Watanabe I., Sasaki T., Ito H., Okumura F. (1999) Comparison of the effects of convulsant and depressant barbiturate stereoisomers on AMPA-type glutamate receptors. Anesthesiology 90, 1704–1713 [DOI] [PubMed] [Google Scholar]
- 16. Leeb-Lundberg F., Olsen R. W. (1982) Interactions of barbiturates of various pharmacological categories with benzodiazepine receptors. Mol. Pharmacol. 21, 320–328 [PubMed] [Google Scholar]
- 17. Hibbs R. E., Gouaux E. (2011) Principles of activation and permeation in an anion-selective Cys-loop receptor. Nature 474, 54–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Corringer P. J., Poitevin F., Prevost M. S., Sauguet L., Delarue M., Changeux J. P. (2012) Structure and pharmacology of pentameric receptor channels: from bacteria to brain. Structure 20, 941–956 [DOI] [PubMed] [Google Scholar]
- 19. Althoff T., Hibbs R. E., Banerjee S., Gouaux E. (2014) X-ray structures of GluCl in apo states reveal a gating mechanism of Cys-loop receptors. Nature 512, 333–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Miller P. S., Aricescu A. R. (2014) Crystal structure of a human GABAA receptor. Nature 512, 270–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li G.-D., Chiara D. C., Sawyer G. W., Husain S. S., Olsen R. W., Cohen J. B. (2006) Identification of a GABAA receptor anesthetic-binding site at subunit interfaces by photolabeling with an etomidate analog. J. Neurosci. 26, 11599–11605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chiara D. C., Dostalova Z., Jayakar S. S., Zhou X., Miller K. W., Cohen J. B. (2012) Mapping general anesthetic-binding site(s) in human α1β3 γ-aminobutyric acid type A receptors with [3H]TDBzl-etomidate, a photoreactive etomidate analog. Biochemistry 51, 836–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chiara D. C., Jayakar S. S., Zhou X., Zhang X., Savechenkov P. Y., Bruzik K. S., Miller K. W., Cohen J. B. (2013) Specificity of intersubunit general anesthetic-binding sites in the transmembrane domain of the human α1β3γ2 γ-aminobutyric acid type A (GABAA) receptor. J. Biol. Chem. 288, 19343–19357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jayakar S. S., Zhou X., Chiara D. C., Dostalova Z., Savechenkov P. Y., Bruzik K. S., Dailey W. P., Miller K. W., Eckenhoff R. G., Cohen J. B. (2014) Multiple propofol-binding sites in a γ-aminobutyric acid type A receptor (GABAAR) identified using a photoreactive propofol analog. J. Biol. Chem. 289, 27456–27468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Savechenkov P. Y., Zhang X., Chiara D. C., Stewart D. S., Ge R., Zhou X., Raines D. E., Cohen J. B., Forman S. A., Miller K. W., Bruzik K. S. (2012) Allyl m-trifluoromethyldiazirine mephobarbital: an unusually potent enantioselective and photoreactive barbiturate general anesthetic. J. Med. Chem. 55, 6554–6565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Desai R., Savechenkov P., Ge R., Bruzik K., Raines D., Miller K. (2013) R- and S-mTFD-MPPB, an anesthetic and a convulsant, have opposing effect on γ-aminobutyric acid (GABA) type A receptors. FASEB J. 27, lb556 [Google Scholar]
- 27. Sigel E. (2005) The benzodiazepine recognition site on GABAA receptors. Med. Chem. Rev. 2, 251–256 [DOI] [PubMed] [Google Scholar]
- 28. Dostalova Z., Liu A., Zhou X., Farmer S. L., Krenzel E. S., Arevalo E., Desai R., Feinberg-Zadek P. L., Davies P. A., Yamodo I. H., Forman S. A., Miller K. W. (2010) High-level expression and purification of Cys-loop ligand-gated ion channels in a tetracycline-inducible stable mammalian cell line: GABA(A) and serotonin receptors. Protein Sci. 19, 1728–1738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dostalova Z., Zhou X., Liu A., Zhang X., Zhang Y., Desai R., Forman S. A., Miller K. W. (2014) Human α1β3γ2L γ-aminobutyric acid type A receptors: high-level production and purification in a functional state. Protein Sci. 23, 157–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ziebell M. R., Nirthanan S., Husain S. S., Miller K. W., Cohen J. B. (2004) Identification of binding sites in the nicotinic acetylcholine receptor for [3H]azietomidate, a photoactivatable general anesthetic. J. Biol. Chem. 279, 17640–17649 [DOI] [PubMed] [Google Scholar]
- 31. Scott M. G., Crimmins D. L., McCourt D. W., Tarrand J. J., Eyerman M. C., Nahm M. H. (1988) A simple in situ cyanogen bromide cleavage method to obtain internal amino acid sequence of proteins electroblotted to polyvinyldifluoride membranes. Biochem. Biophys. Res. Commun. 155, 1353–1359 [DOI] [PubMed] [Google Scholar]
- 32. Hamouda A. K., Kimm T., Cohen J. B. (2013) Physostigmine and galanthamine bind in the presence of agonist at the canonical and noncanonical subunit interfaces of a nicotinic acetylcholine receptor. J. Neurosci. 33, 485–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Crimmins D. L., McCourt D. W., Thoma R. S., Scott M. G., Macke K., Schwartz B. D. (1990) In situ chemical cleavage of proteins immobilized to glass-fiber and polyvinylidenedifluoride membranes: cleavage at tryptophan residues with 2-(2′-nitrophenylsulfenyl)-3-methyl-3′-bromoindolenine to obtain internal amino acid sequence. Anal. Biochem. 187, 27–38 [DOI] [PubMed] [Google Scholar]
- 34. Middleton R. E., Cohen J. B. (1991) Mapping of the acetylcholine binding site of the nicotinic acetylcholine receptor: [3H]nicotine as an agonist photoaffinity label. Biochemistry 30, 6987–6997 [DOI] [PubMed] [Google Scholar]
- 35. Brauer A. W., Oman C. L., Margolies M. N. (1984) Use of o-phthalaldehyde to reduce background during automated Edman degradation. Anal. Biochem. 137, 134–142 [DOI] [PubMed] [Google Scholar]
- 36. Lynagh T., Lynch J. W. (2012) Ivermectin binding sites in human and invertebrate Cys-loop receptors. Trends Pharmacol. Sci. 33, 432–441 [DOI] [PubMed] [Google Scholar]
- 37. Richter L., de Graaf C., Sieghart W., Varagic Z., Mörzinger M., de Esch I. J., Ecker G. F., Ernst M. (2012) Diazepam-bound GABAA receptor models identify new benzodiazepine binding-site ligands. Nat. Chem. Biol. 8, 455–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Belelli D., Lambert J. J., Peters J. A., Wafford K., Whiting P. J. (1997) The interaction of the general anesthetic etomidate with the γ-aminobutyric acid type A receptor is influenced by a single amino acid. Proc. Natl. Acad. Sci. U.S.A. 94, 11031–11036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Prince R. J., Sine S. M. (1999) Acetylcholine and epibatidine binding to muscle acetylcholine receptors distinguish between concerted and uncoupled models. J. Biol. Chem. 274, 19623–19629 [DOI] [PubMed] [Google Scholar]
- 40. Andreeva I. E., Nirthanan S., Cohen J. B., Pedersen S. E. (2006) Site specificity of agonist-induced opening and desensitization of the Torpedo californica nicotinic acetylcholine receptor. Biochemistry 45, 195–204 [DOI] [PubMed] [Google Scholar]
- 41. Nayak T. K., Auerbach A. (2013) Asymmetric transmitter binding sites of fetal muscle acetylcholine receptors shape their synaptic response. Proc. Natl. Acad. Sci. U.S.A. 110, 13654–13659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rüsch D., Zhong H., Forman S. A. (2004) Gating allosterism at a single class of etomidate sites on α(1)β(2)γ(2L) GABA(A) receptors accounts for both direct activation and agonist modulation. J. Biol. Chem. 279, 20982–20992 [DOI] [PubMed] [Google Scholar]
- 43. Guitchounts G., Stewart D. S., Forman S. A. (2012) Two etomidate sites in the α1β2γ2 γ-aminobutyric acid type A receptors contribute equally and noncooperatively to modulation of channel gating. Anesthesiology 116, 1235–1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Erkkila B. E., Sedelnikova A. V., Weiss D. S. (2008) Stoichiometric pore mutations of the GABA(A)R reveal a pattern of hydrogen bonding with picrotoxin. Biophys. J. 94, 4299–4306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Quast U., Brenner O. (1983) Modulation of [3H]muscimol binding in rat cerebellar and cerebral cortical membranes by picrotoxin, pentobarbitone, and etomidate. J. Neurochem. 41, 418–425 [DOI] [PubMed] [Google Scholar]
- 46. Gielen M., Thomas P., Smart T. G. (2015) The desensitization gate of inhibitory Cys-loop receptors. Nat. Commun. 6, 6829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ling A. S., Truitt E. B. Jr., Krantz J. C. Jr. (1959) Further pharmacologic studies on hexafluorodiethyl ether (HFE). J. Pharmacol. Exp. Ther. 126, 366–373 [PubMed] [Google Scholar]
- 48. Krasowski M. D. (2000) Differential modulatory actions of the volatile convulsant flurothyl and its anesthetic isomer at inhibitory ligand-gated ion channels. Neuropharmacology 39, 1168–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hamouda A. K., Stewart D. S., Chiara D. C., Savechenkov P. Y., Bruzik K. S., Cohen J. B. (2014) Identifying barbiturate binding sites in a nicotinic acetylcholine receptor with [3H]allyl m-trifluoromethyldiazirine mephobarbital, a photoreactive barbiturate. Mol. Pharmacol. 85, 735–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Trattnig S. M., Harpsøe K., Thygesen S. B., Rahr L. M., Ahring P. K., Balle T., Jensen A. A. (2012) Discovery of a novel allosteric modulator of 5-HT3 receptors. J. Biol. Chem. 287, 25241–25254 [DOI] [PMC free article] [PubMed] [Google Scholar]