Background: Glioblastoma is highly aggressive and incurable by current treatment modalities.
Results: MCSF is regulated by the SYK-PI3K-NFκB pathway in glioma and induces secretion of IGFBP1 from microglia to promote angiogenesis.
Conclusion: Microglial IGFBP1 is a key mediator of MCSF-induced angiogenesis.
Significance: IGFBP1 is a potential target for glioblastoma therapy.
Keywords: angiogenesis, cytokine, endothelial cell, glioblastoma, macrophage, IGFBP1, MCSF, SILAC, microglia
Abstract
Glioblastoma (grade IV glioma/GBM) is the most common primary adult malignant brain tumor with poor prognosis. To characterize molecular determinants of tumor-stroma interaction in GBM, we profiled 48 serum cytokines and identified macrophage colony-stimulating factor (MCSF) as one of the elevated cytokines in sera from GBM patients. Both MCSF transcript and protein were up-regulated in GBM tissue samples through a spleen tyrosine kinase (SYK)-dependent activation of the PI3K-NFκB pathway. Ectopic overexpression and silencing experiments revealed that glioma-secreted MCSF has no role in autocrine functions and M2 polarization of macrophages. In contrast, silencing expression of MCSF in glioma cells prevented tube formation of human umbilical vein endothelial cells elicited by the supernatant from monocytes/microglial cells treated with conditioned medium from glioma cells. Quantitative proteomics based on stable isotope labeling by amino acids in cell culture showed that glioma-derived MCSF induces changes in microglial secretome and identified insulin-like growth factor-binding protein 1 (IGFBP1) as one of the MCSF-regulated proteins secreted by microglia. Silencing IGFBP1 expression in microglial cells or its neutralization by an antibody reduced the ability of supernatants derived from microglial cells treated with glioma cell-conditioned medium to induce angiogenesis. In conclusion, this study shows up-regulation of MCSF in GBM via a SYK-PI3K-NFκB-dependent mechanism and identifies IGFBP1 released by microglial cells as a novel mediator of MCSF-induced angiogenesis, of potential interest for developing targeted therapy to prevent GBM progression.
Introduction
Glioblastoma (grade IV glioma/GBM)4 is the most common, malignant adult primary brain tumor with poor survival (1, 2). Despite advances in treatment strategies, the prognosis is only marginally improved, which shows the need for further understanding of the disease (3). The tumor is surrounded by a microenvironment composed of various stromal elements, which include fibroblasts, leukocytes, endothelial cells, pericytes, and extracellular matrix (4). During cancer progression, the microenvironment also evolves through continuous paracrine communication between tumor and stromal elements, thus suggesting the vital role of tumor-stromal interactions in cancer development (5). In the case of glioma, macrophages/microglial cells are present in abundance, accounting for nearly 30% of tumor mass (6). Moreover, macrophages/microglia have been implicated in glioma pathophysiology (7–9). Macrophages/microglia can be either classically activated (M1 phenotype) or alternatively activated (M2 phenotype). M1 phenotype is considered to be antitumorigenic, whereas M2 is protumorigenic in nature (10). Tumor-associated macrophages belong to the M2 phenotype and promote tumor progression, invasion, and angiogenesis (11, 12). Cytokines are important mediators of tumor-stroma interactions and are deregulated in numerous cancers (13). Alteration in various cytokines and their receptor expression has been reported in GBM (14).
In the current study, we profiled 48 cytokines in the sera of glioma and normal healthy controls and identified 33 cytokines with differential abundance in GBM sera. Cytokines exhibiting an increased level in GBM serum included macrophage colony-stimulating factor (MCSF), which was also up-regulated in GBM tissue via a mechanism dependent on the SYK-PI3K-NFκB pathway. We also established that MCSF is an independent poor prognostic indicator of GBM. Further, we demonstrate that glioma-secreted MCSF induces angiogenesis in vitro and in vivo via macrophage/microglia-secreted factors. These studies were complemented by quantitative proteomics experiments based on stable isotope labeling by amino acids in cell culture (SILAC), to identify in the microglial secretome molecular substrates of angiogenesis elicited by GBM-derived MCSF.
Experimental Procedures
Cell Lines and Reagents
Human glioma cell lines U251, U87, U373, LN299, and A172 were grown in Dulbecco's modified Eagle's medium (DMEM). SVG, an immortalized human fetal glial cell line, was grown in minimal essential medium. CHME-3, an immortalized human microglial cell line (15), was a kind gift from Dr. Anirban Basu (National Brain Research Centre, Manesar, India) and was cultured in DMEM. All media were supplemented with 10% FBS and antibiotics (penicillin, streptomycin, and gentamycin) unless otherwise indicated. Human umbilical vein endothelial cells (HUVEC) were purchased from Life Technologies, Inc., and cultured under company-recommended conditions. For conditioned medium (CM) collection, glioma cells were grown in serum containing growth medium until they reached 80–90% confluence. Then they were washed thoroughly with 1× PBS, and fresh serum-free growth medium was added. The CM was collected after 24 h of incubation, filtered using a 0.2-μm membrane filter, and stored at −20 °C until use. Peripheral blood mononuclear cells were isolated from buffy coat obtained from normal blood donors at Kidwai Memorial Institute of Oncology (Bangalore, India) using the Ficoll gradient method. Later, monocytes were separated from other cells by the plastic adherence method for 2 h and cultured in DMEM under different conditions for 7 days as indicated.
The following reagents were used in this study: recombinant MCSF (Biolegend), MCSF, SYK- and IGFBP1-specific siRNA (Dharmacon), MCSFR inhibitor GW2580 (LC Laboratories), Bay 11-7082 (Sigma-Aldrich), LY294002, U0126 and Bay 61-3606 (Calbiochem), anti-AKT and anti-phospho-AKT (Cell Signaling, 4691 and 4060, respectively), anti-IGFBP1 (R&D Systems, MAB675), anti-MCSFR (Abcam, ab89907), anti-MCSF (Novus Biologicals, NB110-57176), anti-CD68 (Biogenex, MU416-UC), anti-CD86 (Epitomics, 1858-1), anti-CD204(Sigma-Aldrich, HPA000272), MCSF and IGFBP1 ELISA kit (R&D Systems; DY216 and DY871, respectively), and luciferase assay reagent (Promega). The human MCSF cDNA construct was a kind gift from Prof. Richard Stanley (Yeshiva University, New York). The MCSF promoter-dependent luciferase wild type and mutant construct were a kind gift from Prof. Jay Rappaport (Temple University, Philadelphia, PA).
Tumor Samples and Serum Collection
Glioma tumor and blood samples were collected from patients at the National Institute of Mental Health and Neurosciences and the Sri Satya Sai Institute of Higher Medical Sciences (Bangalore, India). As control/normal samples, non-tumorous brain tissue obtained from the non-dominant anterior temporal cortex region during surgery for intractable epilepsy was used. Tissue from tumor as well as normal samples was used for both RNA isolation and immunohistochemistry (IHC) studies. A total of 122 glioma tissue samples (10 grade II/diffuse astrocytoma (DA), 10 grade III/anaplastic astrocytoma (AA), and 102 grade IV/glioblastoma (GBM) and 12 control brain tissues were used in this study. We also used serum samples from 26 normal, 24 DA, 22 AA, and 148 GBM patients. All of the serum samples were collected prior to surgery. Histological specimens were centrally reviewed and confirmed as different grades of glioma by the neuropathologist as per the World Health Organization 2007 classification scheme (16). This study was approved by the ethics committee of the National Institute of Mental Health and Neurosciences and Sri Satya Sai Institute of Higher Medical Sciences. The patient's written informed consent was obtained before collecting samples. Blood samples were collected from normal healthy individuals at the Indian Institute of Science (Bangalore, India) with prior consent and used as normal controls. The patient and normal blood samples were allowed to clot at 4 °C overnight, followed by centrifugation at 4 °C for 5 min at 1000 rpm to separate serum (upper phase) from clot. Serum samples were stored at −80 °C until use.
Serum Cytokine Profiling
Serum cytokine profiling was done using normal (n = 26), DA (n = 24), AA (n = 22), and GBM (n = 148) serum samples by bead array technology. We used commercially available human cytokine kits: 21-plex and 27-plex (Bio-Rad; MF0-005KM11 and M50-0KCAF0Y, respectively) and followed the manufacturer's protocol. The 21-plex included the following cytokines: IFNα2, IL1α, IL2rα, IL3, IL12 (p40), IL16, IL18, CTACK, GROα, HGF, TRAIL, LIF, MCP3, MCSF, MIF, MIG, βNGF, SCF, SCGFβ, SDF1α, and TNFβ. The 27-plex included the following cytokines: IL1β, IL2, IL1rα, IL4, IL5, IL6, IL7, IL8, IL9, IL10, IL12 (p70), IL13, IL15, IL17, eotaxin, basic FGF, GCSF, GMCSF, VEGF, IFNγ, IP10, MCP1, MIP1α, MIP1β, PDGFBB, RANTES, and TNFα. The cytokine levels were log 2-transformed before using for further analysis.
RNA Isolation and qRT-PCR
RNA from cells and tissues were isolated using TRI reagent (Sigma), and cDNA was made using the high capacity cDNA reverse transcription kit (Life Technologies). Gene-specific primers were used to quantify the relative expression by real-time PCR. A gene expression study was performed using ABI PRISM 7900 (Applied Biosystems) sequence detection system and Dynamo kit containing SYBR Green dye (Finnzymes Diagnostics, Espoo, Finland). Expression was analyzed using GAPDH, ACTB, RPL35A, or AGPAT1 as reference genes and followed the ΔΔCt method (17). First, the average Ct value of a gene for a given sample was normalized by subtracting it from the average Ct value of the reference gene, which gave ΔCt. Next, ΔΔCt was calculated by subtracting ΔCt of the test sample from that of control sample for a given gene. Further, the ratio of ΔΔCt was calculated and log 2-transformed to obtain the log 2 ratio.
ELISA
ELISAs for MCSF and IGFBP1 were performed according to the manufacturer's protocol. The cell-free supernatant (100 μl) was used to measure levels of MCSF (pg/ml) in all glioma cell lines. However, in experiments where different pathway inhibitor treatment was given, followed by MCSF level measurement, and in overexpression and silencing conditions, the results were expressed as a percentage of -fold change normalized to the respective control samples. Similarly, IGFBP1 levels were expressed as percentage of fold change normalized to the respective control samples.
Total Protein Isolation and Western Blotting
Total protein extracts were prepared using radioimmune precipitation assay buffer. The extract (100 μg) was resolved on 12% SDS-PAGE and transferred to PVDF membrane (Millipore). The membrane was blocked with 5% skimmed milk powder in 1× PBST buffer for at least 1 h, followed by incubation with primary antibody (1:1000 dilution) at 4 °C overnight. The membrane was washed thoroughly and then incubated with HRP-conjugated secondary antibody (1:10,000 dilution; Sigma) for 2 h at room temperature. The protein was visualized by chemiluminescence (Pierce).
Immunohistochemistry
Paraffin sections (4 μm) from the tumor tissue and control samples were collected on silane-coated slides, and IHC was performed on 66 samples that included 5 normal, 10 DA, 10 AA, and 41 GBM tumors. Antigen retrieval was done by heat treatment in Tris-EDTA buffer (10 mm Tris base, 1 mm EDTA solution, 0.05% Tween 20, pH 9.0) at three different wattages: 850 watts for 5 min, 600 watts for 10 min, and 450 watts for 5 min, respectively. Slides were cooled to room temperature and rinsed in 1× PBS. After the initial processing steps, sections were incubated overnight with primary antibodies: MCSF (1:100 dilution), CD68 (1:40 dilution), CD86 (1:250 dilution), and CD204 (1:200 dilution). This was followed by incubation with secondary antibody (MACH-1 universal HRP-polymer detection kit). 3,3′-Diaminobenzidine (Sigma-Aldrich) was used as the chromogenic substrate. A visual semiquantitative grading scale was applied to assess the intensity of the immunoreactivity as follows: 0 if the staining was absent, 1+ for moderate staining, and 2+ if staining was strong. Only 2+ staining intensity was considered for analysis, in line with our previous studies (18). For each sample, >1000 cells were counted, and the percentage of cells with 2+ staining was taken as the labeling index. The staining pattern for MCSF was cytoplasmic in the tumor cells, whereas CD68, CD86, and CD204 showed positive staining in the tumor-infiltrating microglial/macrophage cells.
Transfection and Luciferase Assay
The LN229 cells were transfected with the construct encoding MCSF or the empty vector using Lipofectamine 2000 (Life Technologies) according to the manufacturer's instructions and grown for 24 h in selection medium containing hygromycin (200 μg/ml). The growth medium with drug was replaced every other day once until distinct colonies appeared (∼3–4 weeks). These resistant cells were pooled and confirmed for MCSF expression by qRT-PCR and ELISA. For silencing experiments, the cells were transfected with a 100 nm concentration of either control non-targeting siRNA (siNT) or gene-specific siRNA, as indicated, using Dharmafect I (Dharmacon) according to the manufacturer's instructions. After 72 h of transfection, cells were harvested and confirmed for MCSF silencing by qRT-PCR and ELISA. In the case of CM collection, 48 h after siRNA transfection, cells were washed and incubated with serum-free growth medium for another 24 h before CM collection.
For the luciferase assay, the cells were transfected with an MCSF promoter-dependent luciferase construct (0.5 μg) along with pCMV-β-Gal (0.5 μg). The MCSF promoter-dependent luciferase construct contains bp −1310 to +48 of the MCSF promoter region cloned into pGL3-Basic vector (Promega). The NFκB mutant has a mutation in four NFκB binding sites at −51, −359, −378, and −438 positions (19). After 6 h of transfection, vehicle control and different pathway inhibitors were added at the indicated concentrations, followed by 24 h of incubation. At the end of the incubation period, cells were harvested, and extracts were made for measuring luciferase activity, which was further normalized to β-galactosidase activity.
HUVEC Tube Formation Assay
In this assay, 96-well plate was precoated with Matrigel (30 μl/well), followed by plating HUVEC cells (passages 2–4) at a concentration of 15,000 cells/well under different conditions, as indicated. After overnight incubation, tube formation was observed, and images were taken from multiple fields of each well using a phase-contrast microscope. Quantification was performed by counting the total number of completely enclosed networks in each well.
Intradermal Angiogenesis Assay
In this assay, male nude mice were injected intradermally at the ventral skin surface with 1 million tumor cells in 100 μl of 1× PBS containing 2% serum. GW2580 inhibitor or vehicle control treatment was given through oral lavage (160 mg/kg body weight daily) following tumor cell inoculation. Five days after tumor cell inoculation, mice were sacrificed, and the tumor-containing skin was dissected and imaged using digital camera. The tumor-directed capillaries were quantified by counting the number of newly formed blood vessels around the tumor-inoculated site.
SILAC Sample Preparation
The CHME-3 cells were grown up to five passages in culture medium containing DMEM depleted of l-arginine and l-lysine (SILAC DMEM, Sigma) instead of DMEM and supplemented with isotope-labeled l-arginine and l-lysine: l-[13C6]arginine (Arg6) and l-[2H4]lysine (Lys4), or l-[13C6-15N4]arginine (Arg10) and l-[13C6-15N2]lysine (Lys8) (all isotopes obtained from Sigma except for Lys4 from Thermo Scientific). Cells were then treated with either U87 CM derived from non-targeting siRNA-transfected cells in the presence of Arg6 and Lys4 (control conditions) or MCSF-specific siRNA transfected cells in the presence of Arg10 and Lys8 (test conditions). After 24 h of incubation with the different CMs (directly added to the culture medium), supernatant from differentially labeled CHME-3 was collected, mixed, and centrifuged at 200 × g for 5 min and then at 20,000 × g for 25 min to remove non-adherent cells and cell debris, respectively. Proteins were precipitated using 10% trichloroacetic acid on ice for 30 min. Precipitated proteins were spun down at 10,000 × g for 20 min and washed three times with diethyl ether to remove any remaining salt from the protein pellets. Precipitated proteins were resuspended in SDS sample buffer (62.5 mm Tris-HCl, pH 6.8, 2% SDS, 10% glycerol, 1% 2-mercaptoethanol, and 0.005% bromphenol blue) for 5 min at 95 °C, followed by 1 h of shaking at room temperature to ensure complete resuspension.
Protein Separation and Identification by LC-MS/MS
Proteins were separated on 12% polyacrylamide gels using the Protean II xi cell system (Bio-Rad). Gels were stained with PageBlue protein staining solution (Fermentas, Life Technologies) and scanned using a computer-assisted densitometer (Epson Perfection V750 PRO). Gel lanes were systematically cut into 16 equal gel pieces and destained with three washes in 50% acetonitrile. After reduction (with 10 mm dithiothreitol at 56 °C for 15 min) and alkylation (55 mm iodoacetamide at room temperature for 30 min), proteins were digested in-gel using trypsin (600 ng/band; Gold, Promega), as described previously (20). Digest products were dehydrated in a vacuum centrifuge and reduced to 2 μl. The generated peptides were analyzed online by nano-flow HPLC-nanoelectrospray ionization using a Q-Exactive mass spectrometer (Thermo Fisher Scientific) coupled to an Ultimate Rapid Separation LC system (Dionex, Thermo Fisher Scientific). Desalting and preconcentration of samples were performed on-line on a Pepmap® precolumn (0.3 × 10 mm; Dionex). A gradient consisting of 0–40% B in A for 90 min, followed by 80% B plus 20% A for 15 min (where A is 0.1% formic acid, 2% acetonitrile in water, and B is 0.1% formic acid in acetonitrile) at 300 nl/min, was used to elute peptides from the capillary reverse-phase column (0.075 × 150 mm, Pepmap®, Dionex). Eluted peptides were electrosprayed online at a voltage of 1.9 kV into the Q-Exactive. A cycle of one full-scan mass spectrum (350–1500 m/z) at a resolution of 70,000 followed by 10 data-dependent MS/MS spectra was repeated continuously throughout the nano-LC separation. All MS/MS spectra were recorded using normalized collision energy at a resolution of 17,500 (automatic gain control target, 1 × 105; 80-ms maximum injection time). Data were acquired using the Xcalibur software (version 2.2). Raw data analysis was performed using the MaxQuant software (version 1.5.0.0) (21). Retention time-dependent mass recalibration was applied with the aid of a first search implemented in the Andromeda software (22). The peak lists were searched against the UniProt human database (release 2014_06, CPS entries), 255 frequently observed contaminants, and reversed sequences of all entries. The following settings were applied: spectra were searched with a mass tolerance of 7 ppm (MS) and 0.5 threshold (MS/MS). Enzyme specificity was set to trypsin. Up to two missed cleavages were allowed, and only peptides of at least 6 amino acids in length were considered. Oxidation on methionine was set as a variable modification. Peptide identifications were accepted based on their false discovery rate (<1%). Accepted peptide sequences were subsequently assembled by MaxQuant into proteins to achieve a false discovery rate of 1% at the protein level. For protein identification, at least two peptides (at least one of which was unique) were required. Relative protein quantifications in samples to be compared were performed based on the median SILAC ratios of at least two peptides, using MaxQuant with standard settings.
BiNGO Network Analysis
Overrepresentation of GO categories among MCSF regulated proteins was analyzed by BiNGO (version 2.44) (23). Of the 67 significantly differentially expressed proteins, 11 proteins corresponding to ribosomal subunits were removed, and the remaining 56 proteins were used as the input set, and the whole annotation set was used as the reference set. Overrepresentation statistics were calculated by hypergeometric analysis and Benjamini and Hochberg false discovery rate correction. The human annotation file was used as a custom annotation file. The full ontology was used as custom ontology file with a level of significance of 0.05.
Statistical Methods
A nonparametric t test was performed to determine the significance of difference between two groups. The comparison among multiple groups was performed by one-way ANOVA (nonparametric test) with Tukey test, which compares all pairs of columns using GraphPad Prism version 5.01. Supervised hierarchical clustering of significantly differentially expressed cytokines was carried out using the MultiExperiment Viewer. The volcano graph for 48 cytokines was drawn using R software (version 3.1.0). The log 2 -fold change expression value of 48 cytokines (x axis) and p value (y axis) were given as input. The prognostic significance was tested by univariate and multivariate Cox proportional hazard analysis using SPSS software (version 19). IHC results were analyzed by using a non-parametric test, the Kruskal-Wallis one-way ANOVA based on ranks, followed by a post hoc test. The results were expressed as mean ± S.D. A p value of <0.05 was considered significant for all analyses.
Results
Cytokine Profiling Identifies Elevated Levels of Serum MCSF in GBM
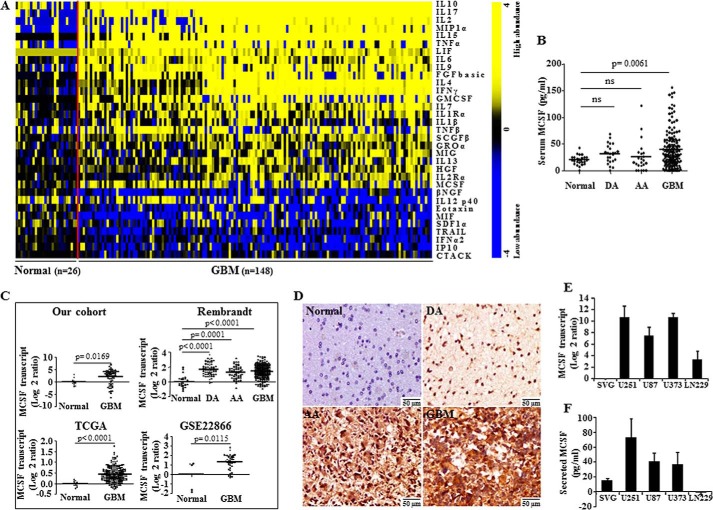
We profiled 48 cytokines in the sera obtained from normal healthy individuals (n = 26), DA (n = 24), AA (n = 22), and GBM (n = 148) patients using a bead array-based platform. A non-parametric t test with false discovery rate correction identified 33 cytokines with different concentrations between GBM and normal sera (Fig. 1A and supplemental Table 1). Further analysis revealed that the transcript levels of 17 cytokines were similarly regulated in GBM tissue compared with normal brain tissue, suggesting that differential expression in GBM is probably the reason for the difference in their concentration in sera (supplemental Table 1).
FIGURE 1.
MCSF levels in sera, GBM tumor tissue, and glioma cell lines. A, heat map of the 33 differentially abundant cytokines in normal and GBM sera. A dual-color code was used, with yellow and blue indicating high and low abundance, respectively. A total of 24 cytokines were present in higher abundance, and 9 were present at lower abundance in GBM sera compared with normal sera. The red line separates normal from GBM samples. B, scatter plot representation of MCSF levels in normal (n = 26), DA (n = 24), AA (n = 22), and GBM (n = 148) sera estimated by the bead array method. Statistical analysis (one-way ANOVA) was performed, and p values are indicated. The horizontal line represents the mean value. ns, not significant. C, MCSF transcript levels in normal brain tissue (n = 10) and GBM tumor tissue (n = 61) were estimated by qRT-PCR. Microarray data for MCSF gene expression from TCGA consisting of normal (n = 10) and GBM (n = 572) samples; a REMBRANDT data set consisting of normal (n = 28), DA (n = 65), AA (n = 58), and GBM (n = 227) samples; and a GSE22866 data set consisting of normal (n = 6) and GBM (n = 40) samples are shown as a scatter plot. A non-parametric t test was performed, and p values are indicated. The horizontal line in each plot represents the mean value. D, representative images (original magnification, ×160) of MCSF immunostaining in normal brain tissue (n = 5), DA (n = 10), AA (n = 10), and GBM tumor tissue (n = 41). Normal brain is negatively stained, whereas DA, AA, and GBM show variable staining, with maximum staining in GBM. E, MCSF transcript levels in various glioma cell lines estimated by the qRT-PCR method and normalized to SVG (an immortalized glioma cell line). F, MCSF secreted protein levels in the culture supernatant of various glioma cell lines and SVG measured by the ELISA method. Error bars, S.D.
MCSF, one of the significantly elevated cytokines in GBM sera identified in the present study (Fig. 1B), was selected for further investigation given 1) its well suggested role in the development and progression of tumors, including glioma (24, 25); 2) its influence upon survival, proliferation, and differentiation of macrophages/microglial cells, which are present in abundance in the glioma microenvironment (26, 27); and 3) the expression of MCSF receptor (MCSFR) only in microglial cells and not in glioma cells (28), suggesting that glioma cell-secreted MCSF may function through stromal cells and be one key determinant of glioma-stromal cell interactions underlying glioma progression. MCSF transcript levels were up-regulated in GBM, compared with normal brain tissue samples, in our cohort and three more cohorts, namely TCGA, REMBRANDT, and GSE22866 (Fig. 1C). Whereas MCSF transcript levels were significantly higher in all grades of glioma compared with normal brain samples, serum MCSF was only significantly different in GBM when compared with healthy controls (Fig. 1, B and C, REMBRANDT data set). IHC analysis revealed MCSF positivity in the tumor cells of all grades of glioma, with the highest expression in GBM (Fig. 1D and Table 1). Glioma-derived cell lines showed higher but varying levels of MCSF transcript and secreted protein levels (except for LN229), compared with SVG, an immortalized astrocytic cell line (Fig. 1, E and F). These results suggest that MCSF is up-regulated in glioma, particularly in GBM, and that the high levels of MCSF found in GBM serum result probably from its secretion by tumor tissue.
TABLE 1.
Expression of different variables in all grades of glioma by IHC
Values shown are mean ± S.D.
| Variables | Normal (n = 5) | DA (n = 10) | AA (n = 10) | GBM (n = 41) | Kruskal-Wallis p value | Post hoc p value |
|---|---|---|---|---|---|---|
| MCSF | 0.00 ± 0.00 | 12.00 ± 14.76 | 13.50 ± 12.70 | 18.41 ± 13.94 | 0.025 | 0.068a; 0.02b; 0.005c; 0.638d; 0.222e; 0.295f |
| CD68 | 15.00 ± 5.0 | 16.50 ± 6.26 | 21.00 ± 4.59 | 28.54 ± 12.76 | 0.002 | 0.651a; 0.036b; 0.016c; 0.102d; 0.003e; 0.04f |
| CD204 | 0.00 ± 0.00 | 3.50 ± 5.79 | 4.50 ± 5.98 | 25.85 ± 13.96 | <0.001 | 0.188a; 0.114b; 0.001c; 0.687d; <0.001e; <0.001f |
| CD86 | 10.00 ± 0.00 | 2.50 ± 5.4 | 3.5 ± 5.79 | 11.46 ± 10.2 | 0.01 | 0.016a; 0.04b; 0.586c; 0.654d; 0.01e; 0.021f |
a Normal versus DA.
b Normal versus AA.
c Normal versus GBM.
d DA versus AA.
e DA versus GBM.
f AA versus GBM.
MCSF Up-regulation Depends on the SYK-PI3K-NFκB Pathway in GBM
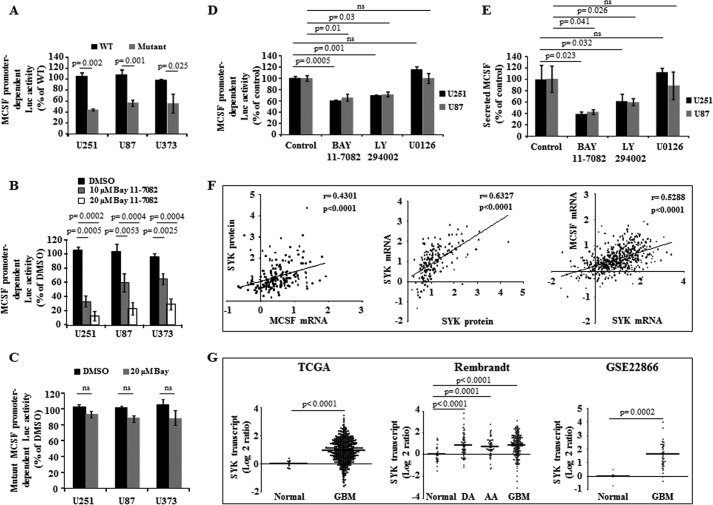
Although MCSF expression is known to be regulated by NFκB pathway in myeloid cell lineage (19), its regulation in GBM remains to be elucidated. We found that the wild type MCSF promoter-dependent luciferase activity decreased significantly upon mutation of NFκB-binding sites in U251, U87, and U373 cells (Fig. 2A; see “Experimental Procedures” for more information). Wild type MCSF promoter-dependent luciferase activity was inhibited by Bay 11-7082, an IKKα inhibitor, in a concentration-dependent manner (Fig. 2B), whereas Bay 11-7082 treatment did not alter mutant MCSF promoter-dependent luciferase activity (Fig. 2C). Further, treatment with a PI3K inhibitor (LY294002), but not a MAPK inhibitor (U0126), inhibited wild type MCSF promoter-dependent luciferase activity and decreased the level of MCSF secreted by U251 and U87 cells, suggesting that MCSF secretion by GBM is regulated by the PI3K-NFκB pathway (Fig. 2, D and E, respectively).
FIGURE 2.
Regulation of MCSF expression in glioma. A, glioma cells were transfected with either a wild type MCSF promoter-dependent luciferase construct or a mutant construct wherein all four NFκB binding sites are mutated. MCSF promoter activity was measured by luciferase assay and normalized to β-galactosidase activity. B, glioma cells were transfected with wild type MCSF promoter-dependent luciferase construct. MCSF promoter activity was measured by luciferase assay and normalized to β-galactosidase activity in presence of vehicle control and two different concentrations of Bay 11-7082. C, glioma cells were transfected with mutant MCSF promoter-dependent luciferase construct. MCSF promoter activity was measured by luciferase assay and normalized to β-Galactosidase activity in the presence of vehicle control and 20 μm Bay 11-7082. D, glioma cells were transfected with wild type MCSF promoter-dependent luciferase construct. MCSF promoter activity was measured by luciferase assay and normalized to β-galactosidase activity in presence of different pathway inhibitors, as indicated. E, MCSF secreted protein levels measured by the ELISA method in the presence of different pathway inhibitors, as indicated, in U251 and U87 cell lines. F, Spearman correlation analysis between MCSF transcript and SYK protein levels and between SYK transcript and SYK protein levels. MCSF and SYK transcript levels were analyzed using the data set derived from TCGA. r and p values are indicated in the graph. The straight line inside the graph indicates the mean. G, scatter plot representation of SYK transcript levels using the TCGA data set consisting of normal (n = 10) and GBM (n = 572) samples; the REMBRANDT data set consisting of normal (n = 28), DA (n = 65), AA (n = 58), and GBM (n = 227) samples; and the GSE22866 data set consisting of normal (n = 6) and GBM (n = 40) samples. A non-parametric t test was performed, and the p values are indicated. The horizontal line in each plot represents the mean value. ns, not significant. Error bars, S.D.
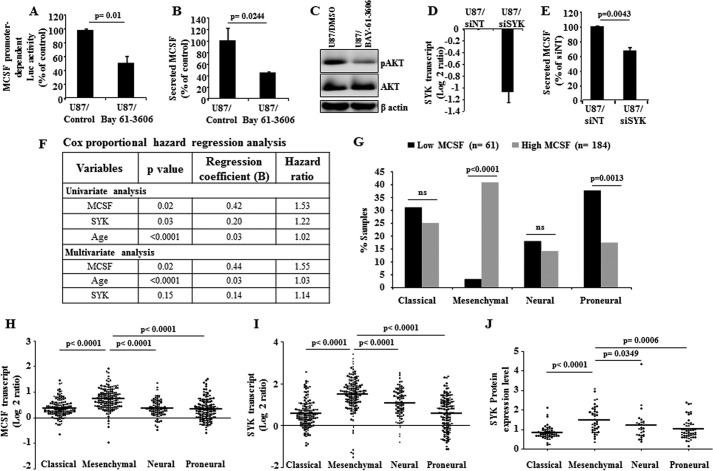
To identify the regulator of MCSF expression upstream from PI3K, we correlated the MCSF transcript levels with the expression level of 171 proteins, measured by reverse phase protein array, in 196 GBMs from the TCGA cohort (29). Among proteins exhibiting significant correlation with MCSF, SYK had the highest significance and positive correlation (p < 0.0001; r = 0.43) (Fig. 2F). Because SYK is shown to activate the PI3K pathway (30–34), we investigated the possibility that SYK could be the activator of PI3K contributing to MCSF up-regulation in glioma. SYK transcript levels were found to be up-regulated in GBM when compared with normal brain samples in TCGA, REMBRANT, and GSE22866 cohorts (Fig. 2G). Because a reverse phase protein array was not done in normal brain samples, SYK protein up-regulation in GBM samples could not be ascertained. However, we found a significant positive correlation between SYK transcript and protein levels (Fig. 2F), suggesting that the SYK protein level may also be up-regulated in GBM. We also found that SYK and MCSF transcript levels had a significant positive correlation (Fig. 2F). Notably, SYK transcript levels were up-regulated in DA and AA (Fig. 2G, REMBRANDT data set), consistent with the up-regulation of MCSF transcript levels in lower grades of glioma (Fig. 1C, REMBRANDT data set). Collectively, up-regulation of SYK in glioma, in particular GBMs, and the positive correlation between SYK and MCSF suggest that SYK may be the upstream regulator of MCSF expression. Further supporting the role of SYK in MCSF up-regulation, treatment of cells with Bay 61-3606, a pharmacological inhibitor of SYK, inhibited MCSF promoter-dependent luciferase activity and decreased secreted MCSF levels as well as AKT phosphorylation (Fig. 3, A–C, respectively). In addition, silencing SYK expression by siRNA also reduced the levels of secreted MCSF (Fig. 3, D and E).
FIGURE 3.
MCSF is regulated by SYK-PI3K-NFκB pathway and acts as poor prognostic indicator in glioma. A, U87 glioma cell line was transfected with wild type MCSF promoter-dependent luciferase construct. MCSF promoter activity was measured by luciferase assay and normalized to β-galactosidase activity in the presence of the SYK inhibitor Bay 61-3606. The luciferase activity was expressed as a percentage of the activity present in untreated control cells. B, MCSF secreted protein levels measured by ELISA in presence of SYK inhibitor Bay 61-3606. MCSF levels were expressed as a percentage of the levels present in untreated control cells. C, Western blot representation of AKT and phospho-AKT (pAKT) proteins in U87 cell line treated with and without SYK inhibitor Bay 61-3606. β-Actin was used as a loading control. D, SYK transcript levels estimated by qRT-PCR method in the U87 glioma cell line transfected with either siNT or siSYK (48 h). The expression values were normalized to the siNT sample. E, MCSF secreted protein levels measured by ELISA in SYK-silenced conditions. MCSF levels were expressed as a percentage of the levels present in cells transfected with siNT. F, Cox proportional hazard regression analysis using MCSF and SYK transcript levels and survival data from the TCGA data set in a cohort of 245 GBM patients. The patient inclusion criteria were as follows: at least 30 days of survival from the time of surgery, Karnosky performance status of ≥70, and any kind of chemotherapy received. G, TCGA patients (n = 245) were divided into gene expression subtypes: classical, mesenchymal, neural, and proneural. Patients of each subtype were further subdivided based on MCSF transcript levels (low MCSF versus high MCSF). The significance of distribution was analyzed by a two-sample z test using R software (version 3.1.0). The patient inclusion criteria were as follows: at least 30 days of survival from the time of surgery, Karnosky performance status of ≥70, and any kind of chemotherapy received. H–J, scatter plot representation of MCSF transcript, SYK transcript, and SYK protein levels in different subtypes of GBM using the TCGA data set, respectively. Statistical analysis (one-way ANOVA) was performed, and the p values are indicated. The horizontal line in each plot represents the mean value. ns, not significant. Error bars, S.D.
Survival correlation using univariate Cox proportional hazard regression analysis revealed that both SYK and MCSF are poor prognostic indicators in GBM (Fig. 3F). However, multivariate survival analysis along with age found that MCSF but not SYK is an independent predictor of survival (Fig. 3F). MCSF transcript, SYK transcript, and SYK protein levels were found significantly higher in the mesenchymal subtype, which is known to be associated with poor survival compared with other subtypes (Fig. 3, H–J, respectively). Corroborating these findings, a subgroup of GBM with high MCSF mRNA was significantly enriched in mesenchymal subtype (Fig. 3G). Collectively, these results confirm that MCSF is an independent poor prognostic indicator in GBM, and MCSF up-regulation in GBM is dependent on the SYK-PI3K-NFκB pathway.
MCSF Secreted by Glioma Cells Induces Angiogenesis through a Macrophage/Microglial Cell-dependent Mechanism
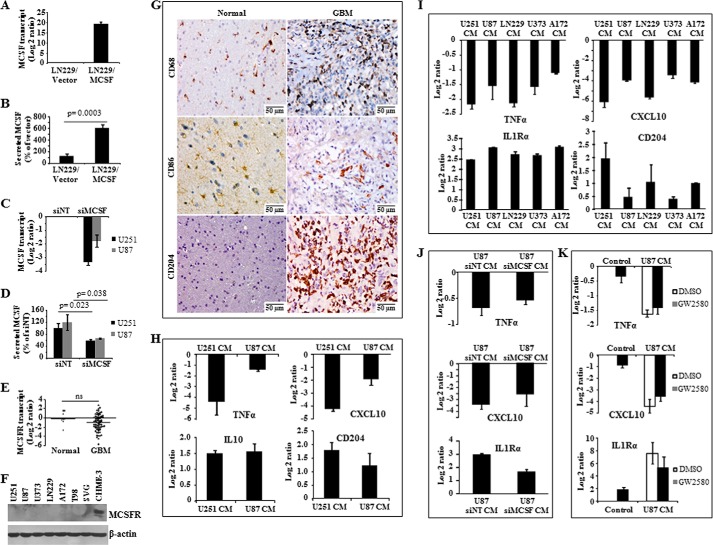
MCSF induces tumor cell proliferation in renal cell carcinoma in an autocrine way and might contribute to M2 polarization of macrophages in glioma (35, 36). To explore the autocrine functions of MCSF in glioma, various properties like proliferation, chemosensitivity, anchorage-independent growth, migration, and invasion were monitored after the addition of recombinant MCSF (rMCSF), ectopic overexpression (Fig. 4, A and B), or silencing of MCSF by siRNA (Fig. 4, C and D). These experiments showed no changes in any of these properties, suggesting the absence of an autocrine function for MCSF in glioma (data not shown). The absence of autocrine function for MCSF in glioma is supported by the unchanged levels of MCSFR transcript in GBM tumor tissue compared with normal brain tissue and also undetectable levels of MCSFR in glioma cell lines compared with the microglial cell line CHME-3 (Fig. 4, E and F, respectively). In line with the possible role of MCSF in M2 polarization of macrophages in glioma (35), we next investigated by IHC the presence of M1 and M2 polarized macrophages in GBM tissue samples. CD68, a macrophage-specific marker, showed expression in the infiltrating macrophages in all grades of glioma with the highest expression in GBM when compared with normal brain samples (Fig. 4G and Table 1). However, IHC staining using M1 macrophage-specific (CD86) and M2 macrophage-specific (CD204) markers showed that the majority of the infiltrated macrophages are of the M2 type in GBM (Fig. 4G and Table 1). To further investigate the role of tumor-secreted MCSF in M2 polarization of macrophages, we monitored the ability of CM derived from glioma cell lines to polarize undifferentiated monocytes derived from human peripheral blood mononuclear cells and microglial cell line CHME-3. Glioma CM (from U251 and U87 cell lines) treatment of monocytes resulted in the down-regulation of M1-specific markers (TNFα and CXCL10) and up-regulation of M2-specific markers (IL10 and CD204) (Fig. 4H). Similarly, treatment of the microglial cell line CHME-3 with glioma CM (from U251, U87, LN229, U373, and A172 cell lines) resulted in the down-regulation of M1-specific markers (TNFα and CXCL10) and up-regulation of M2-specific markers (IL1Rα and CD204) (Fig. 4I). However, when microglial cells were treated with CM derived from glioma cells silenced for MCSF or in the presence of MCSFR inhibitor (GW2580), M2 polarization was not significantly affected (Fig. 4, J and K, respectively). Collectively, these experiments suggest that whereas glioma cell-secreted factors polarize macrophages/microglial cells to the M2 type, tumor-secreted MCSF is not required for M2 polarization. We conclude from these experiments that glioma-secreted MCSF is not required for autocrine functions and M2 polarization of macrophages/microglial cells.
FIGURE 4.
MCSF levels modulation in glioma cell lines and role of MCSF in macrophage polarization. A and B, MCSF transcript and protein levels in LN229 cell line stably expressing vector control or MCSF cDNA estimated by qRT-PCR and ELISA methods, respectively. The expression levels in LN299/MCSF were normalized to respective LN229/vector control samples. C and D, MCSF transcript and protein levels in glioma cell lines transfected with either siNT or siMCSF estimated by qRT-PCR (48 h) and ELISA (72 h) methods, respectively. The expression levels were normalized to the respective siNT samples. E, MCSFR transcript levels in normal brain tissue (n = 8), and GBM tumor tissue (n = 71) estimated by qRT-PCR. A non-parametric t test was performed to find the significance between two groups. The horizontal line represents mean value. ns, non-significant. F, Western blot representation of MCSFR protein levels in various glioma cell lines and microglial cell line CHME-3. β-Actin was used as a loading control. G, representative images (original magnification, ×160) of normal brain and GBM tissue sections stained for macrophage marker (CD68), M1 marker (CD86), and M2 marker (CD204). H, transcript levels of M1 (TNFα and CXCL10) and M2 (IL10 and CD204) phenotype genes measured by qRT-PCR in blood-derived monocytes that were treated with U251 and U87 CM for 7 days. The expression levels were normalized to untreated cells. I, transcript levels of M1 (TNFα and CXCL10) and M2 (IL1Rα and CD204) phenotype genes measured by qRT-PCR in microglial cell line CHME-3, which was treated with various glioma cell line CM for 24 h. The expression levels were normalized to untreated cells. J, transcript levels of M1 (TNFα and CXCL10) and M2 (IL1Rα) phenotype genes measured by qRT-PCR in microglial cell line CHME-3, which was treated with U87 CM derived from either control or MCSF-silenced conditions for 24 h. The expression levels were normalized to untreated cells. K, transcript levels of M1 (TNFα and CXCL10) and M2 (IL1Rα) phenotype genes measured by qRT-PCR in microglial cell line CHME-3, which was pretreated with vehicle or GW2580 for 1 h and later treated with U87 CM for 24 h. The expression levels were normalized to control cells. ns, not significant. Error bars, S.D.
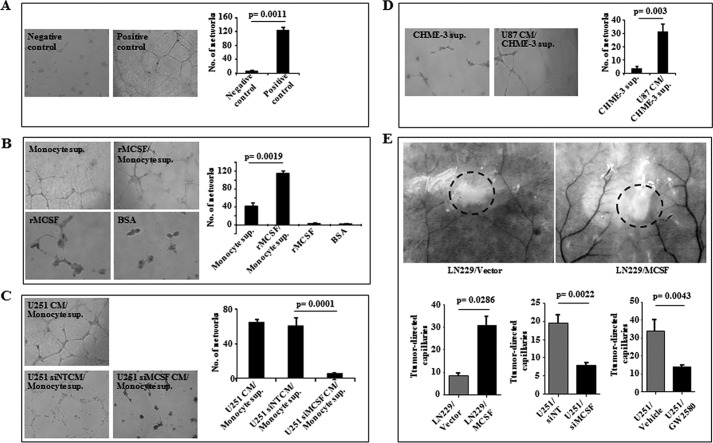
Because rMCSF has been shown to induce VEGFA expression in monocytes, which in turn promotes angiogenesis (37), we next investigated the role of glioma-secreted MCSF in tumor angiogenesis. To address this question, we performed tube formation assay (an in vitro angiogenesis assay) using HUVEC (Fig. 5A, control experiment). The supernatant from rMCSF-treated monocytes was more efficient than the supernatant from untreated monocytes to promote the tube formation in HUVEC (Fig. 5B). The addition of rMCSF or BSA directly to HUVEC failed to induce tube formation (Fig. 5B). Further, the supernatants from monocytes treated with CM derived from either U251 or siNT-transfected U251 cells, but not the supernatant from monocytes treated with CM from siMCSF-transfected U251 cells, induced HUVEC tube formation (Fig. 5C). Similarly, the supernatant from CHME-3 cells treated with U87 CM induced HUVEC tube formation more efficiently, compared with the supernatant from untreated CHME-3 cells (Fig. 5D).
FIGURE 5.
In vitro and in vivo angiogenesis assay. A–D, representative images (original magnification, ×100) of the tube formation assay performed in HUVEC under different conditions as indicated. A, negative control, HUVEC growth medium alone; positive control, HUVEC growth medium with positive angiogenic inducers. B, supernatant from monocytes (Monocyte sup.), supernatant from monocytes treated with recombinant MCSF (rMCSF/Monocyte sup.), rMCSF, and BSA-purified. C, supernatant from monocytes treated with U251 CM (U251 CM/Monocyte sup.), supernatant from monocytes treated with U251 CM derived from non-targeting siRNA-transfected cells (U251 siNT CM/Monocyte sup.), and supernatant from monocytes treated with U251 CM derived from MCSF siRNA-transfected cells (U251 siMCSF CM/Monocyte sup.). D, supernatant from microglia (CHME-3 sup.) and supernatant from microglia treated with U87 CM (U87CM/CHME-3 sup.). The right side of each panel shows the quantification of the corresponding images. A t test was performed, and the p values are indicated. E, representative images of dissected skin of the mice near intradermal glioma tumor showing tumor-directed capillaries and the quantification graphs in different conditions as indicated: cells with vector backbone (LN229/Vector), cells overexpressing MCSF (LN229/MCSF), cells transfected with non-targeting siRNA (U251/siNT), cells transfected with MCSF siRNA (U251/siMCSF), animals treated with vehicle control (U251/Vehicle), and animals treated with MCSFR inhibitor (U251/GW2580) (160 mg/kg body weight/day). A t test was performed, and the p values are indicated. The dashed circles within the images show the tumor location. Error bars, S.D.
To demonstrate the role of MCSF in inducing angiogenesis in vivo, an intradermal angiogenesis assay was performed. LN229/MCSF cells, stably overexpressing MCSF, attracted significantly more blood vessels compared with LN229/vector stable cells (Fig. 5E). Similarly, siMCSF-transfected U251 cells showed a decreased number of tumor-directed capillaries compared with siNT-transfected U251 cells (Fig. 5E). In addition, mice treated with the MCSFR inhibitor GW2580 showed significantly reduced tumor-directed capillaries compared with mice treated with vehicle (Fig. 5E). Collectively, these results suggest that MCSF secreted by glioma cells induces the release by monocytes/microglial cells of extracellular factors, which in turn induce angiogenesis.
IGFBP1 Present in the Microglial Cell Secretome Is a Novel Mediator of MCSF-induced Angiogenesis
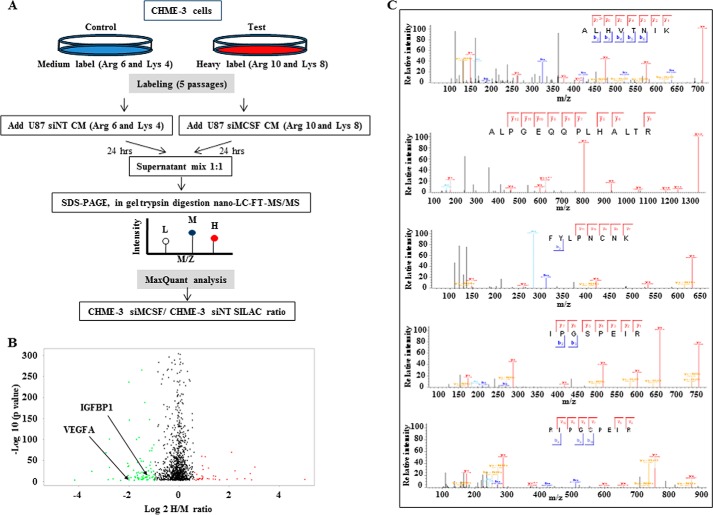
To identify novel factors present in the microglial secretome responsible for MCSF-induced angiogenesis, we used SILAC based on two different sets of heavy amino acids: Arg6 and Lys4 (medium label) for CHME-3 cells treated with U87 siNT CM and Arg10 and Lys8 (heavy label) for CHME-3 cells treated with U87 siMCSF CM (Fig. 6A). The supernatants from differentially labeled CHME-3 cells were mixed in equal proportion and subjected to mass spectrometry. A total of 1196 proteins were identified (supplemental Table 2). Of these, 580 proteins (48.5% of the identified proteins) displayed a SecretomeP score of >0.5 and/or were predicted to have a signal peptide by the Signal P (version 4.1) algorithm (supplemental Table 2). Although most proteins quantified in CHME-3 supernatants did not show significant change between the two experimental conditions, a total of 67 proteins showed significantly different SILAC ratios based on significance B with a p value of 0.01 (Fig. 6B and supplemental Table 3) (21). Thirty-nine of them were down-regulated (negative H/M ratio), whereas 28 proteins were up-regulated (positive H/M ratio) in supernatants derived from CHME-3 cells treated with U87 siMCSF CM compared with supernatants derived from CHME-3 cells treated with U87 siNT CM (supplemental Table 3).
FIGURE 6.
Microglial secretome analysis by SILAC. A, schematic representation of the work flow used to study the variations in the CHME-3 secretome induced by CM of U87 silenced or not for MCSF using SILAC. B, volcano plot representation of SILAC results. The x axis shows the log 2 H/M ratio derived by taking the ratio of mean intensity of heavy label to mean intensity of medium label for each protein. The y axis shows the p value expressed in −log10 scale. A total of 67 proteins were significantly differentially regulated, of which 28 were up-regulated and 39 were down-regulated. Each dot represents one protein in the plot. Red and green dots, up- and down-regulation, respectively. The location of VEGFA and IGFBP1 is indicated. C, annotated MS/MS spectra of five different IGFBP1 peptides.
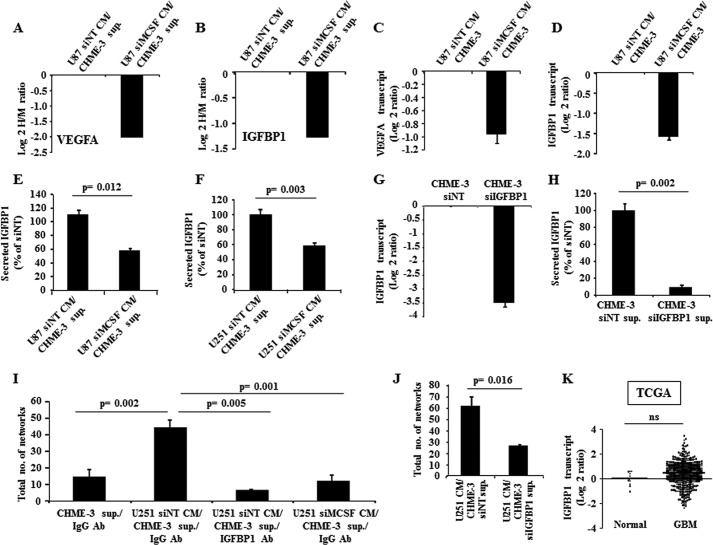
Gene ontology analysis of MCSF-regulated proteins by BINGO showed enrichment in several processes, which emphasizes their important role in regulating angiogenesis (supplemental Table 4). Proteins exhibiting difference in abundance in the supernatant of CHME-3 treated with U87 siMCSF CM and U87 siNT CM included VEGFA, a known angiogenesis inducer (log 2 H/M ratio = −1.998) (Fig. 7A). The reduced VEGFA levels in CHME-3 cells treated with U87 siMCSF CM, compared with CHME-3 cells treated with U87 siNT CM, was further confirmed at the transcript levels (Fig. 7C).
FIGURE 7.
Microglial IGFBP1 is a mediator of MCSF-induced angiogenesis. A and C, VEGFA protein and transcript levels in CHME-3 measured by SILAC and qRT-PCR methods, respectively. The expression levels of VEGFA in CHME-3 upon treatment with CM from MCSF-silenced U87 were normalized to levels present in CHME-3, which was treated with CM from siNT-transfected (control) U87. B and D, IGFBP1 protein and transcript levels in CHME-3 measured by SILAC and qRT-PCR methods, respectively. The expression levels of IGFBP1 in CHME-3 upon treatment with CM from MCSF-silenced U87 were normalized to levels present in CHME-3 that was treated with CM from siNT-transfected (control) U87. E and F, IGFBP1 levels, measured by ELISA, in CHME-3 cell supernatant with different CM treatment as indicated. The IGFBP1 levels were expressed as a percentage of the levels measured in the supernatant of the CHME-3 cells, which was treated with CM from corresponding siNT-transfected glioma cells. A t test was performed, and the p values are indicated. G and H, IGFBP1 transcript and protein levels in the CHME-3 upon transfection with either siNT or siIGFBP1, as measured by qRT-PCR (48 h) and ELISA (72 h) methods, respectively. The expression was normalized to siNT samples. I and J, tube formation assay in various conditions as indicated. I, supernatant from CHME-3 cells in the presence of IgG antibody (CHME-3 sup./IgG Ab); supernatant from CHME-3 cells treated with U251 CM derived from non-targeting siRNA-transfected cells in the presence of IgG antibody (U251 siNT CM/CHME-3 sup./IgG Ab); supernatant from CHME-3 cells treated with U251 CM derived from non-targeting siRNA transfected cells in presence of IGFBP1 antibody (U251 siNT CM/CHME-3 sup./IGFBP1 Ab); and supernatant from CHME-3 cells treated with U251 CM derived from MCSF-silenced cells in the presence of IgG antibody (U251 siMCSF CM/CHME-3 sup./IgG Ab). J, supernatant from CHME-3 cells transfected with siNT and subsequently treated with U251 CM (U251 CM/CHME-3 siNT sup.) and supernatant from CHME-3 cells transfected with siIGFBP1 and subsequently treated with U251 CM (U251 CM/CHME-3 siIGFBP1 sup.). A t test was performed, and the p values are indicated. K, scatter plot representation of IGFBP1 transcript levels using the TCGA data set, which consisted of normal (n = 10) and GBM (n = 572) samples. A non-parametric t test was performed, and the p value is indicated. The horizontal line represents the mean value. ns, non-significant. Error bars, S.D.
In order to identify novel mediators of MCSF-induced angiogenesis in CHME-3 supernatant, we chose IGFBP1 (identified with five peptides by MS/MS analysis; Fig. 6C) for further investigation for the following reasons: 1) it was not differentially expressed between GBM and control brain tissues, 2) its level was reduced in microglial supernatant treated with U87 CM derived from the MCSF-silenced condition (log 2 H/M ratio = −1.266), and 3) it is known to be a secreted protein (Fig. 7, B and K, and supplemental Table 3). The reduced IGFBP1 expression in CHME-3 cells treated with U87 siMCSF CM was first verified at the level of transcripts (Fig. 7D) and then confirmed by ELISA in an independent set of experiments (Fig. 7, E and F). Next, we carried out experiments to explore the role of CHME-3-secreted IGFBP1 in angiogenesis. Adding an IGFBP1 antibody substantially inhibited the ability of the supernatant from CHME-3 cells treated with U251 siNT CM to induce tube formation compared with the addition of IgG control antibody (Fig. 7I, compare third bar with second bar). As expected, the supernatant derived from CHME-3 cells treated with U251 siMCSF CM failed to show any increase in tube formation (Fig. 7I, compare fourth bar with second bar). To further confirm the role of IGFBP1 in angiogenesis, we carried out experiments using supernatant from IGFBP1-silenced CHME-3 cells. The supernatant derived from CHME-3 cells transfected with siNT and subsequently treated with U251 CM was more efficient to promote tube formation, compared with the supernatant from CHME-3 cells transfected with siIGFBP1 and subsequently treated with U251 CM (Fig. 7, G, H, and J). Collectively, these results identify IGFBP1 secreted by microglial cells in response to MCSF as an important mediator of angiogenesis.
Discussion
The study of the stromal components and the host immune response in addition to tumor cells has led to a better understanding of cancer development (38). Cytokines play an important role in tumor-stroma interaction, thus facilitating tumor progression and aggressiveness (39). In the present study, we identified 33 cytokines exhibiting differences in abundance in GBM sera, 17 of which are likely to be secreted by tumor tissue. The high proportion of profiled cytokines exhibiting differences in abundance in sera from patients with GBM found in the present study is consistent with previous investigations aimed at establishing distinctive cytokine signatures of various pathological situations, including breast cancer, which showed a deregulation of more than half of the cytokines tested (40–42).
MCSF, one of the cytokines present at high levels in glioma sera, in particular GBM, was further studied in detail in line with its important role in survival, proliferation, and differentiation of macrophages/microglial cells, which are present in abundance in the glioma microenvironment (26, 27). Although several reports showed elevated levels of MCSF in various cancers, including glioma, the regulation and the function of MCSF in GBM remain poorly characterized (25, 43, 44). In this study, we confirm that MCSF is up-regulated in glioma, particularly in GBM, both tumor tissue and serum. It is interesting to note that whereas elevated levels of MCSF transcript and protein are seen in all grades of glioma (DA, AA, and GBM), serum MCSF was found elevated only in GBM. This might be due to the fact that there is a selective blood-brain barrier disruption reported in GBM compared with low grade glioma (45).
In an effort to identify the mechanism underlying MCSF up-regulation in tumor cells, we found that it is critically dependent on the SYK-PI3K-NFκB cascade. SYK plays a major role in survival signaling through multiple classes of immune recognition receptors in hematopoietic cells (46). However, SYK has been shown to act as both tumor promoter and tumor suppressor in different cancers (47). Whereas SYK activation generally involves recruitment through its Src homology 2 domains to a phosphorylated immune receptor tyrosine-based activation motif, it can also be activated by other mechanisms, such as elevated levels and its association with integrins (47). However, the impact of SYK in glioma has not been reported so far. Here, we demonstrate that both SYK transcript and protein levels are up-regulated in glioma. To identify the possible upstream activators of SYK, analysis of expression of various integrins, which are known to be activated in GBM (48), from TCGA data revealed 11 integrin α, 7 integrin β, and 3 integrin-interacting proteins, including integrin-linked kinase, to be up-regulated in GBM compared with normal brain samples (supplemental Table 5). The PI3K-AKT pathway plays a major role downstream of activated SYK (47). Further, several reports suggest that PI3K and NFκB pathways are activated by SYK (30–34). Our study demonstrates that SYK activates the PI3K-AKT pathway, which further leads to NFκB-dependent up-regulation of MCSF in GBM. Collectively, we conclude that the SYK-PI3K-NFκB pathway, probably activated through integrins, is essential for MCSF up-regulation and may play a tumor promoter role in GBM.
We also found that both SYK and MCSF levels are higher in the mesenchymal subtype of GBM compared with other subtypes. A very high level of MCSF seen in the mesenchymal subtype is particularly interesting because mesenchymal GBMs are significantly enriched in high risk GBMs with poor survival (49). Correspondingly, we also found that MCSF mRNA level was a poor prognostic indicator in GBM and that a subgroup with high MCSF mRNA is enriched with mesenchymal GBM patients significantly. The high levels of MCSF seen in the mesenchymal subtype, which result from SYK up-regulation, may favor angiogenesis (see below) and explain the poor prognostics of mesenchymal GBMs.
In breast cancer and renal cell carcinoma, MCSF and its receptor MCSFR are co-expressed in tumor cells, allowing an autocrine action and thereby increasing malignancy and proliferation (36, 50). Likewise, data indicate expression of both MCSF and its receptor MCSFR in glioma, suggesting autocrine and paracrine effects (43). However, we found that glioma-secreted MCSF has no function in promoting tumor cell properties directly in an autocrine fashion. Consistently, we found that the MCSFR is not expressed in glioma tumor and cell lines. However, the CHME-3 microglial cell line expresses high levels of MCSFR, suggesting that MCSF may function by activating certain pathways in microglial cells. A role for MCSF-MCSFR signaling in M2 polarization of macrophages has also been proposed, based on a report indicating that MCSFR inhibition resulted in reduced tumor growth with decreased M2 markers in macrophages (28). Although this report proves the importance of MCSFR in M2 polarization of macrophages, a direct role for MCSF in M2 polarization remained to be established. This is particularly important because MCSFR is also activated by IL34 (51). Our results demonstrate that although glioma cell CM contained factors that could induce M2 polarization of monocytes/microglial cells, MCSF present in the glioma CM is not required for M2 polarization.
Recombinant human MCSF induces angiogenesis through macrophages by promoting VEGFA expression (37). Our results demonstrate that MCSF present in the glioma cell CM acts on monocytes or microglial cells and promotes secretion of factors, which induce angiogenesis in vitro. We also provide evidence that angiogenesis elicited by monocyte/microglia exposed to glioma cell CM is dependent on activation of MCSF-MCSFR signaling in macrophages. Further, a search of novel mediators of angiogenesis in the microglial secretome using an unbiased proteomic strategy identified 67 proteins significantly regulated by MCSF.
Gene ontology analysis of the MCSF-regulated proteins in the microglial secretome underscored their important role in angiogenesis. Whereas the cellular component category revealed the importance of extracellular matrix, extracellular region, extracellular space, and proteins therein, biological process ontology recognized the proliferation of endothelial cells, angiogenesis, and wound healing in addition to the process of coagulation. Temporal and spatial regulation of extracellular matrix remodeling events plays a key role in angiogenesis (52). Tissue factor-initiated coagulation through protease-activated receptor family of G-protein-coupled receptors has been found to promote tumor angiogenesis (53, 54). The molecular function gene ontology identified the importance of cell surface receptors, calcium ion, and phospholipid binding. Intracellular calcium and downstream signal transduction have been shown to regulate angiogenesis (55, 56). Phospholipids have been shown to regulate tumor angiogenesis (57).
Consistent with previous findings (37), we demonstrate that the VEGFA level is increased in the microglial cell secretome through a MCSF-dependent mechanism. Likewise, glioma-derived MCSF increased the IGFBP1 level in the secretome of microglial cells. Furthermore, neutralizing IGFBP1 by a specific antibody in the microglial secretome after treatment with glioma cell CM strongly reduced tube formation in HUVEC, indicating that MCSF-induced increase in IGFBP1 secretion by microglial cells was essential for angiogenesis. Similarly, silencing IGFBP1 in microglial cells before treatment with glioma cell CM significantly reduced the ability of microglial cell secretome to induce tube formation in HUVEC. Notably, there was no significant difference in IGFBP1 transcript levels between GBM and normal brain (Fig. 7K), suggesting that IGFBP1 secreted by microglial cells in response to glioma-secreted MCSF is probably the major source for tumor angiogenesis.
There are several reports in the literature connecting IGFBP1 to angiogenesis. IGF1 (insulin-like growth factor 1), which is regulated by its binding to IGFBP1, regulates migration and angiogenesis of human endothelial cells (58). IGFBP1, which can also function in an insulin-like growth factor-independent manner, induces angiogenesis and binds to the prosurvival factor BAK, thereby inhibiting growth-inhibitory functions of p53 (59). IGFBP1 also induces endothelial nitric-oxide synthase activity through PI3K signaling, resulting in increased nitric oxide (NO) production (60–63). IGFBP1 secreted from human chondrocytes upon activation by lysophosphatidic acid through the lysophosphatidic acid receptor has been found to induce angiogenesis (57).
Collectively, these findings and our data suggest that IGFBP1 released by microglial cells is an important effector contributing to angiogenesis elicited by glioma-derived MCSF. Therefore, they point to the possible role of factors other than VEGF that may promote angiogenesis and cancer stem cell-derived endothelial cells through transdifferentiation (64). This is of potential clinical interest because bevacizumab, a VEGF-targeting antibody approved by the United States Food and Drug Administration, failed to show any significant improvement in overall and progression-free survival in the recently conducted trials (65, 66). Thus, IGFBP1 could be a potential alternate candidate for developing a targeted therapy for GBM.
Author Contributions
A. S. H., B. A. C., A. A., V. S., and K. Somasundaram conceived and coordinated the study. M. B. N., P. M., and K. S. wrote the paper. M. B. N., S. U., and P. M. designed, performed, and analyzed the experiments shown in Fig. 6. M. B. N. and V. P. designed, performed, and analyzed the experiments shown in Figs. 1–3. S. D. S. and K. Sravani designed, performed, and analyzed the experiments shown in Figs. 1D and 4G. All authors reviewed the results and approved the final version of the manuscript.
Supplementary Material
Acknowledgments
The results published here are in whole or in part based upon data generated by the Cancer Genome Atlas (TCGA) pilot project established by NCI and NHGRI, National Institutes of Health. The use of data sets from Institute NC and GSE22866 is acknowledged. The Central Animal Facility, Indian Institute of Science, is acknowledged for animal experiments. LC-MS/MS analyses were performed using the facilities of the Functional Proteomics Platform of Montpellier Languedoc-Roussillon.
The authors declare that they have no conflicts of interest with the contents of this article.

This article contains supplemental Tables 1–5.
- GBM
- glioblastoma
- MCSF
- macrophage colony-stimulating factor
- SYK
- spleen tyrosine kinase
- SILAC
- stable isotope labeling by amino acids in cell culture
- VEGFA
- vascular endothelial growth factor A
- IGFBP1
- insulin-like growth factor-binding protein 1
- HUVEC
- human umbilical vein endothelial cell(s)
- CM
- conditioned medium
- IHC
- immunohistochemistry
- DA
- diffuse astrocytoma
- AA
- anaplastic astrocytoma
- qRT-PCR
- quantitative RT-PCR
- siNT
- non-targeting siRNA
- ANOVA
- analysis of variance
- MCSFR
- MCSF receptor
- rMCSF
- recombinant MCSF
- H/M
- ratio of mean intensity of heavy label to mean intensity of medium label
- Arg6
- l-[13C6]arginine
- Lys4
- l-[2H4]lysine
- Arg10
- l-[13C6-15N4]arginine
- Lys8
- l-[13C6-15N2]lysine.
References
- 1. Holland E. C. (2000) Glioblastoma multiforme: the terminator. Proc. Natl. Acad. Sci. U.S.A. 97, 6242–6244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wen P. Y., Kesari S. (2008) Malignant gliomas in adults. N. Engl. J. Med. 359, 492–507 [DOI] [PubMed] [Google Scholar]
- 3. Stupp R., Hegi M. E., Mason W. P., van den Bent M. J., Taphoorn M. J., Janzer R. C., Ludwin S. K., Allgeier A., Fisher B., Belanger K., Hau P., Brandes A. A., Gijtenbeek J., Marosi C., Vecht C. J., Mokhtari K., Wesseling P., Villa S., Eisenhauer E., Gorlia T., Weller M., Lacombe D., Cairncross J. G., Mirimanoff R. O., European Organisation for Research and Treatment of Cancer Brain Tumour and Radiation Oncology Groups, and National Cancer Institute of Canada Clinical Trials Group (2009) Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 10, 459–466 [DOI] [PubMed] [Google Scholar]
- 4. Pietras K., Ostman A. (2010) Hallmarks of cancer: interactions with the tumor stroma. Exp. Cell Res. 316, 1324–1331 [DOI] [PubMed] [Google Scholar]
- 5. Ungefroren H., Sebens S., Seidl D., Lehnert H., Hass R. (2011) Interaction of tumor cells with the microenvironment. Cell Commun. Signal. 9, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Charles N. A., Holland E. C., Gilbertson R., Glass R., Kettenmann H. (2011) The brain tumor microenvironment. Glia 59, 1169–1180 [DOI] [PubMed] [Google Scholar]
- 7. Badie B., Schartner J. (2001) Role of microglia in glioma biology. Microsc. Res. Tech. 54, 106–113 [DOI] [PubMed] [Google Scholar]
- 8. Gutmann D. H. (2015) Microglia in the tumor microenvironment: taking their TOLL on glioma biology. Neuro. Oncol. 17, 171–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yang I., Han S. J., Kaur G., Crane C., Parsa A. T. (2010) The role of microglia in central nervous system immunity and glioma immunology. J. Clin. Neurosci. 17, 6–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bingle L., Brown N. J., Lewis C. E. (2002) The role of tumour-associated macrophages in tumour progression: implications for new anticancer therapies. J. Pathol. 196, 254–265 [DOI] [PubMed] [Google Scholar]
- 11. Pollard J. W. (2004) Tumour-educated macrophages promote tumour progression and metastasis. Nat. Rev. Cancer 4, 71–78 [DOI] [PubMed] [Google Scholar]
- 12. Sica A., Schioppa T., Mantovani A., Allavena P. (2006) Tumour-associated macrophages are a distinct M2 polarised population promoting tumour progression: potential targets of anti-cancer therapy. Eur. J. Cancer 42, 717–727 [DOI] [PubMed] [Google Scholar]
- 13. Dranoff G. (2004) Cytokines in cancer pathogenesis and cancer therapy. Nat. Rev. Cancer 4, 11–22 [DOI] [PubMed] [Google Scholar]
- 14. Hao C., Parney I. F., Roa W. H., Turner J., Petruk K. C., Ramsay D. A. (2002) Cytokine and cytokine receptor mRNA expression in human glioblastomas: evidence of Th1, Th2 and Th3 cytokine dysregulation. Acta Neuropathol. 103, 171–178 [DOI] [PubMed] [Google Scholar]
- 15. Janabi N., Peudenier S., Héron B., Ng K. H., Tardieu M. (1995) Establishment of human microglial cell lines after transfection of primary cultures of embryonic microglial cells with the SV40 large T antigen. Neurosci. Lett. 195, 105–108 [DOI] [PubMed] [Google Scholar]
- 16. Louis D. N., Ohgaki H., Wiestler O. D., Cavenee W. K., Burger P. C., Jouvet A., Scheithauer B. W., Kleihues P. (2007) The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 114, 97–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Reddy S. P., Britto R., Vinnakota K., Aparna H., Sreepathi H. K., Thota B., Kumari A., Shilpa B. M., Vrinda M., Umesh S., Samuel C., Shetty M., Tandon A., Pandey P., Hegde S., Hegde A. S., Balasubramaniam A., Chandramouli B. A., Santosh V., Kondaiah P., Somasundaram K., Rao M. R. (2008) Novel glioblastoma markers with diagnostic and prognostic value identified through transcriptome analysis. Clin. Cancer Res. 14, 2978–2987 [DOI] [PubMed] [Google Scholar]
- 18. Santosh V., Arivazhagan A., Sreekanthreddy P., Srinivasan H., Thota B., Srividya M. R., Vrinda M., Sridevi S., Shailaja B. C., Samuel C., Prasanna K. V., Thennarasu K., Balasubramaniam A., Chandramouli B. A., Hegde A. S., Somasundaram K., Kondaiah P., Rao M. R. (2010) Grade-specific expression of insulin-like growth factor-binding proteins-2, -3, and -5 in astrocytomas: IGFBP-3 emerges as a strong predictor of survival in patients with newly diagnosed glioblastoma. Cancer Epidemiol. Biomarkers Prev. 19, 1399–1408 [DOI] [PubMed] [Google Scholar]
- 19. Kogan M., Haine V., Ke Y., Wigdahl B., Fischer-Smith T., Rappaport J. (2012) Macrophage colony stimulating factor regulation by nuclear factor κB: a relevant pathway in human immunodeficiency virus type 1 infected macrophages. DNA Cell Biol. 31, 280–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Thouvenot E., Urbach S., Dantec C., Poncet J., Séveno M., Demettre E., Jouin P., Touchon J., Bockaert J., Marin P. (2008) Enhanced detection of CNS cell secretome in plasma protein-depleted cerebrospinal fluid. J. Proteome Res. 7, 4409–4421 [DOI] [PubMed] [Google Scholar]
- 21. Cox J., Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 22. Cox J., Neuhauser N., Michalski A., Scheltema R. A., Olsen J. V., Mann M. (2011) Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10, 1794–1805 [DOI] [PubMed] [Google Scholar]
- 23. Maere S., Heymans K., Kuiper M. (2005) BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21, 3448–3449 [DOI] [PubMed] [Google Scholar]
- 24. Bender A. M., Collier L. S., Rodriguez F. J., Tieu C., Larson J. D., Halder C., Mahlum E., Kollmeyer T. M., Akagi K., Sarkar G., Largaespada D. A., Jenkins R. B. (2010) Sleeping beauty-mediated somatic mutagenesis implicates CSF1 in the formation of high-grade astrocytomas. Cancer Res. 70, 3557–3565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nowicki A., Szenajch J., Ostrowska G., Wojtowicz A., Wojtowicz K., Kruszewski A. A., Maruszynski M., Aukerman S. L., Wiktor-Jedrzejczak W. (1996) Impaired tumor growth in colony-stimulating factor 1 (CSF-1)-deficient, macrophage-deficient op/op mouse: evidence for a role of CSF-1-dependent macrophages in formation of tumor stroma. Int. J. Cancer 65, 112–119 [DOI] [PubMed] [Google Scholar]
- 26. Brugger W., Kreutz M., Andreesen R. (1991) Macrophage colony-stimulating factor is required for human monocyte survival and acts as a cofactor for their terminal differentiation to macrophages in vitro. J. Leukoc. Biol. 49, 483–488 [DOI] [PubMed] [Google Scholar]
- 27. Sawada M., Suzumura A., Yamamoto H., Marunouchi T. (1990) Activation and proliferation of the isolated microglia by colony stimulating factor-1 and possible involvement of protein kinase C. Brain Res. 509, 119–124 [DOI] [PubMed] [Google Scholar]
- 28. Pyonteck S. M., Akkari L., Schuhmacher A. J., Bowman R. L., Sevenich L., Quail D. F., Olson O. C., Quick M. L., Huse J. T., Teijeiro V., Setty M., Leslie C. S., Oei Y., Pedraza A., Zhang J., Brennan C. W., Sutton J. C., Holland E. C., Daniel D., Joyce J. A. (2013) CSF-1R inhibition alters macrophage polarization and blocks glioma progression. Nat. Med. 19, 1264–1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Akbani R., Ng P. K., Werner H. M., Shahmoradgoli M., Zhang F., Ju Z., Liu W., Yang J. Y., Yoshihara K., Li J., Ling S., Seviour E. G., Ram P. T., Minna J. D., Diao L., Tong P., Heymach J. V., Hill S. M., Dondelinger F., Städler N., Byers L. A., Meric-Bernstam F., Weinstein J. N., Broom B. M., Verhaak R. G., Liang H., Mukherjee S., Lu Y., Mills G. B. (2014) A pan-cancer proteomic perspective on The Cancer Genome Atlas. Nat. Commun. 5, 3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chen L., Monti S., Juszczynski P., Ouyang J., Chapuy B., Neuberg D., Doench J. G., Bogusz A. M., Habermann T. M., Dogan A., Witzig T. E., Kutok J. L., Rodig S. J., Golub T., Shipp M. A. (2013) SYK inhibition modulates distinct PI3K/AKT- dependent survival pathways and cholesterol biosynthesis in diffuse large B cell lymphomas. Cancer Cell 23, 826–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hatton O., Lambert S. L., Krams S. M., Martinez O. M. (2012) Src kinase and Syk activation initiate PI3K signaling by a chimeric latent membrane protein 1 in Epstein-Barr virus (EBV)+ B cell lymphomas. PLoS One 7, e42610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jiang K., Zhong B., Gilvary D. L., Corliss B. C., Vivier E., Hong-Geller E., Wei S., Djeu J. Y. (2002) Syk regulation of phosphoinositide 3-kinase-dependent NK cell function. J. Immunol. 168, 3155–3164 [DOI] [PubMed] [Google Scholar]
- 33. Lowell C. A. (2011) Src-family and Syk kinases in activating and inhibitory pathways in innate immune cells: signaling cross talk. Cold Spring Harb. Perspect. Biol. 10.1101/cshperspect.a002352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Takada Y., Aggarwal B. B. (2004) TNF activates Syk protein tyrosine kinase leading to TNF-induced MAPK activation, NF-κB activation, and apoptosis. J. Immunol. 173, 1066–1077 [DOI] [PubMed] [Google Scholar]
- 35. Komohara Y., Ohnishi K., Kuratsu J., Takeya M. (2008) Possible involvement of the M2 anti-inflammatory macrophage phenotype in growth of human gliomas. J. Pathol. 216, 15–24 [DOI] [PubMed] [Google Scholar]
- 36. Menke J., Kriegsmann J., Schimanski C. C., Schwartz M. M., Schwarting A., Kelley V. R. (2012) Autocrine CSF-1 and CSF-1 receptor coexpression promotes renal cell carcinoma growth. Cancer Res. 72, 187–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Eubank T. D., Galloway M., Montague C. M., Waldman W. J., Marsh C. B. (2003) M-CSF induces vascular endothelial growth factor production and angiogenic activity from human monocytes. J. Immunol. 171, 2637–2643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Li H., Fan X., Houghton J. (2007) Tumor microenvironment: the role of the tumor stroma in cancer. J. Cell Biochem. 101, 805–815 [DOI] [PubMed] [Google Scholar]
- 39. Sung S. Y., Hsieh C. L., Wu D., Chung L. W., Johnstone P. A. (2007) Tumor microenvironment promotes cancer progression, metastasis, and therapeutic resistance. Curr. Probl. Cancer 31, 36–100 [DOI] [PubMed] [Google Scholar]
- 40. Bozza F. A., Salluh J. I., Japiassu A. M., Soares M., Assis E. F., Gomes R. N., Bozza M. T., Castro-Faria-Neto H. C., Bozza P. T. (2007) Cytokine profiles as markers of disease severity in sepsis: a multiplex analysis. Crit. Care 11, R49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lyon D. E., McCain N. L., Walter J., Schubert C. (2008) Cytokine comparisons between women with breast cancer and women with a negative breast biopsy. Nurs. Res. 57, 51–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Steuerwald N. M., Foureau D. M., Norton H. J., Zhou J., Parsons J. C., Chalasani N., Fontana R. J., Watkins P. B., Lee W. M., Reddy K. R., Stolz A., Talwalkar J., Davern T., Saha D., Bell L. N., Barnhart H., Gu J., Serrano J., Bonkovsky H. L. (2013) Profiles of serum cytokines in acute drug-induced liver injury and their prognostic significance. PLoS One 8, e81974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Alterman R. L., Stanley E. R. (1994) Colony stimulating factor-1 expression in human glioma. Mol. Chem. Neuropathol. 21, 177–188 [DOI] [PubMed] [Google Scholar]
- 44. Gadducci A., Ferdeghini M., Castellani C., Annicchiarico C., Prontera C., Facchini V., Bianchi R., Genazzani A. R. (1998) Serum macrophage colony-stimulating factor (M-CSF) levels in patients with epithelial ovarian cancer. Gynecol. Oncol. 70, 111–114 [DOI] [PubMed] [Google Scholar]
- 45. Schneider S. W., Ludwig T., Tatenhorst L., Braune S., Oberleithner H., Senner V., Paulus W. (2004) Glioblastoma cells release factors that disrupt blood-brain barrier features. Acta Neuropathol. 107, 272–276 [DOI] [PubMed] [Google Scholar]
- 46. Monroe J. G. (2006) ITAM-mediated tonic signalling through pre-BCR and BCR complexes. Nat. Rev. Immunol. 6, 283–294 [DOI] [PubMed] [Google Scholar]
- 47. Krisenko M. O., Geahlen R. L. (2015) Calling in SYK: SYK's dual role as a tumor promoter and tumor suppressor in cancer. Biochim. Biophys. Acta 1853, 254–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mattern R. H., Read S. B., Pierschbacher M. D., Sze C. I., Eliceiri B. P., Kruse C. A. (2005) Glioma cell integrin expression and their interactions with integrin antagonists: research article. Cancer Ther. 3A, 325–340 [PMC free article] [PubMed] [Google Scholar]
- 49. Arimappamagan A., Somasundaram K., Thennarasu K., Peddagangannagari S., Srinivasan H., Shailaja B. C., Samuel C., Patric I. R., Shukla S., Thota B., Prasanna K. V., Pandey P., Balasubramaniam A., Santosh V., Chandramouli B. A., Hegde A. S., Kondaiah P., Sathyanarayana Rao M. R. (2013) A fourteen gene GBM prognostic signature identifies association of immune response pathway and mesenchymal subtype with high risk group. PLoS One 8, e62042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Elmlinger M. W., Deininger M. H., Schuett B. S., Meyermann R., Duffner F., Grote E. H., Ranke M. B. (2001) In vivo expression of insulin-like growth factor-binding protein-2 in human gliomas increases with the tumor grade. Endocrinology 142, 1652–1658 [DOI] [PubMed] [Google Scholar]
- 51. Chihara T., Suzu S., Hassan R., Chutiwitoonchai N., Hiyoshi M., Motoyoshi K., Kimura F., Okada S. (2010) IL-34 and M-CSF share the receptor Fms but are not identical in biological activity and signal activation. Cell Death Differ. 17, 1917–1927 [DOI] [PubMed] [Google Scholar]
- 52. Eming S. A., Hubbell J. A. (2011) Extracellular matrix in angiogenesis: dynamic structures with translational potential. Exp. Dermatol. 20, 605–613 [DOI] [PubMed] [Google Scholar]
- 53. Belting M., Ahamed J., Ruf W. (2005) Signaling of the tissue factor coagulation pathway in angiogenesis and cancer. Arterioscler. Thromb. Vasc. Biol. 25, 1545–1550 [DOI] [PubMed] [Google Scholar]
- 54. Nash G. F., Walsh D. C., Kakkar A. K. (2001) The role of the coagulation system in tumour angiogenesis. Lancet Oncol. 2, 608–613 [DOI] [PubMed] [Google Scholar]
- 55. Hamdollah Zadeh M. A., Glass C. A., Magnussen A., Hancox J. C., Bates D. O. (2008) VEGF-mediated elevated intracellular calcium and angiogenesis in human microvascular endothelial cells in vitro are inhibited by dominant negative TRPC6. Microcirculation 15, 605–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kohn E. C., Alessandro R., Spoonster J., Wersto R. P., Liotta L. A. (1995) Angiogenesis: role of calcium-mediated signal transduction. Proc. Natl. Acad. Sci. U.S.A. 92, 1307–1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Chuang Y. W., Chang W. M., Chen K. H., Hong C. Z., Chang P. J., Hsu H. C. (2014) Lysophosphatidic acid enhanced the angiogenic capability of human chondrocytes by regulating Gi/NF-κB-dependent angiogenic factor expression. PLoS One 9, e95180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lee O. H., Bae S. K., Bae M. H., Lee Y. M., Moon E. J., Cha H. J., Kwon Y. G., Kim K. W. (2000) Identification of angiogenic properties of insulin-like growth factor II in in vitro angiogenesis models. Br. J. Cancer 82, 385–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Leu J. I., George D. L. (2007) Hepatic IGFBP1 is a prosurvival factor that binds to BAK, protects the liver from apoptosis, and antagonizes the proapoptotic actions of p53 at mitochondria. Genes Dev. 21, 3095–3109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Cooke J. P., Losordo D. W. (2002) Nitric oxide and angiogenesis. Circulation 105, 2133–2135 [DOI] [PubMed] [Google Scholar]
- 61. Fiedler L. R., Wojciak-Stothard B. (2009) The DDAH/ADMA pathway in the control of endothelial cell migration and angiogenesis. Biochem. Soc. Trans. 37, 1243–1247 [DOI] [PubMed] [Google Scholar]
- 62. Leiper J., Nandi M., Torondel B., Murray-Rust J., Malaki M., O'Hara B., Rossiter S., Anthony S., Madhani M., Selwood D., Smith C., Wojciak-Stothard B., Rudiger A., Stidwill R., McDonald N. Q., Vallance P. (2007) Disruption of methylarginine metabolism impairs vascular homeostasis. Nat. Med. 13, 198–203 [DOI] [PubMed] [Google Scholar]
- 63. Rajwani A., Ezzat V., Smith J., Yuldasheva N. Y., Duncan E. R., Gage M., Cubbon R. M., Kahn M. B., Imrie H., Abbas A., Viswambharan H., Aziz A., Sukumar P., Vidal-Puig A., Sethi J. K., Xuan S., Shah A. M., Grant P. J., Porter K. E., Kearney M. T., Wheatcroft S. B. (2012) Increasing circulating IGFBP1 levels improves insulin sensitivity, promotes nitric oxide production, lowers blood pressure, and protects against atherosclerosis. Diabetes 61, 915–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Olar A., Aldape K. D. (2014) Using the molecular classification of glioblastoma to inform personalized treatment. J. Pathol. 232, 165–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Chinot O. L., Wick W., Mason W., Henriksson R., Saran F., Nishikawa R., Carpentier A. F., Hoang-Xuan K., Kavan P., Cernea D., Brandes A. A., Hilton M., Abrey L., Cloughesy T. (2014) Bevacizumab plus radiotherapy-temozolomide for newly diagnosed glioblastoma. N. Engl. J. Med. 370, 709–722 [DOI] [PubMed] [Google Scholar]
- 66. Gilbert M. R., Dignam J. J., Armstrong T. S., Wefel J. S., Blumenthal D. T., Vogelbaum M. A., Colman H., Chakravarti A., Pugh S., Won M., Jeraj R., Brown P. D., Jaeckle K. A., Schiff D., Stieber V. W., Brachman D. G., Werner-Wasik M., Tremont-Lukats I. W., Sulman E. P., Aldape K. D., Curran W. J. Jr., Mehta M. P. (2014) A randomized trial of bevacizumab for newly diagnosed glioblastoma. N. Engl. J. Med. 370, 699–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.