Background: Autophagy is required in hematopoiesis and protects against leukemogenesis.
Results: When ATG7-dependent canonical autophagy is impaired, ATG7-independent alternative autophagy engages in myeloid cells but not in hematopoietic stem cells.
Conclusion: The integrity of hematopoietic stem cells is jeopardized by a lack of alternative autophagy.
Significance: Learning autophagy organization in hematopoietic system is crucial for understanding hematopoietic stem cell transformation.
Keywords: autophagy, hematopoiesis, hematopoietic stem cells, myeloid cell, stem cells, somatic cells
Abstract
Autophagy is integral to hematopoiesis and protects against leukemogenesis. However, the fundamentals of the required molecular machinery have yet to be fully explored. Using conditional mouse models to create strategic defects in the hematopoietic hierarchy, we have shown that recovery capacities in stem cells and somatic cells differ if autophagy is impaired or flawed. An in vivo Atg7 deletion in hematopoietic stem cells completely ablates the autophagic response, leading to irreversible and ultimately lethal hematopoiesis. However, while no adverse phenotype is manifested in vivo by Atg7-deficient myeloid cells, they maintain active autophagy that is sensitive to brefeldin A, an inhibitor targeting Golgi-derived membranes destined for autophagosome formation in alternative autophagy. Removing Rab9, a key regulatory protein, in alternative autophagy, disables autophagy altogether in Atg7-deficient macrophages. Functional analysis indicates that ATG7-dependent canonical autophagy is physiologically active in both hematopoietic stem cells and in terminally differentiated hematopoietic cells; however, only terminally differentiated cells such as macrophages are rescued by alternative autophagy if canonical autophagy is ineffective. Thus, it appears that hematopoietic stem cells rely solely on ATG7-dependent canonical autophagy, whereas terminally differentiated or somatic cells are capable of alternative autophagy in the event that ATG7-mediated autophagy is dysfunctional. These findings offer new insight into the transformational trajectory of hematopoietic stem cells, which in our view renders the autophagic machinery in stem cells more vulnerable to disruption.
Introduction
Hematopoietic stem cells (HSC),2 armed with the capacity to self-renew and generate somatic/differentiated progenies, are essential for normal hematopoiesis and are largely compelled to transform by imbalances in hematopoietic homeostasis. Autophagy is a conserved catabolic mechanism that protects cells by delivering potentially toxic macromolecular aggregates (e.g. proteins, lipids, and glycans) and damaged or superfluous organelles to lysosomes for degradation (1–6). Various stimuli, such as starvation, endoplasmic reticular stress, DNA damage, and reactive oxygen species, may trigger autophagy. Although studied extensively in somatic cells, our understanding of autophagy in stem cells is very limited. Deletion of the autophagy gene Beclin1 leads to early embryonic lethality (7). Recent research has implicated autophagy in hemostatic control and maintenance of the capacity for self-renewal in stem cells. Autophagy is up-regulated during early differentiation of mouse and human embryonic stem cells (8, 9); is known to regulate maintenance, self-renewal, and differentiation of human mesenchymal stem cells (10, 11); and participates in somatic reprogramming (12, 13) and regulating stem-cell status as well (14). In short, autophagy is required for maintenance of HSCs (15–17). Deletion of essential autophagy genes Atg7 or Fip200 in mouse HSCs leads to defective self-renewal and dysregulated myeloproliferation (15, 17). In addition, recent studies of ours have shown that ATG7-dependent autophagy regulates cell cycles of HSCs and progenitor cells (18), promotes megakaryopoiesis, megakaryocyte differentiation, and thrombopoiesis (19), and regulates hematopoiesis largely via direct targeting Notch (20).
ATG7-dependent autophagy, or canonical autophagy, is characterized by lipidation and processing of microtubule-associated protein light chain 3 (LC3) to form LC3-II, an essential step in autophagosme structuring (2). Previous investigations have documented an ATG5/ATG7-independent alternative autophagic mechanism in mouse embryonic fibroblasts, regulated by proteins such as RAB9, Unc-51-like kinase 1 (ULK1), and Beclin1. Unlike canonical autophagy, autophagosomes are generated in a RAB9-dependent manner by the fusion of isolation membranes with vesicles of trans-Golgi and late endosomal derivation (20, 21). ATG3-independent autophagy, which resembles the ATG7-deletion phenotype, has also been described (21, 22). Although canonical autophagy has been amply and intensively studied, and non-canonical or alternative autophagy similarly has been well documented, the particulars of these mechanisms in differing mammalian systems and the biological significance of their functional heterogeneity remain open to question.
HSCs reside in niche locations and behave differently than differentiated blood cells that are actively exposed to a variety of intra- and extracellular stimuli. Despite a rapidly growing interest in autophagy, the potential divergence in the autophagic profiles of stem cells and somatic/differentiated cells is still fundamentally unknown in mammalian systems. Through the use of conditional mouse models harboring autophagy-essential gene deletions in the hematopoietic hierarchy, we show that two distinct mechanisms of autophagy are operant. HSCs rely solely on canonical autophagy, which is ATG7-dependent and non-recoverable if impaired, whereas disruption of canonical autophagy in myeloid cells triggers an alternative compensatory pathway, thereby maintaining cellular viability and function.
Experimental Procedures
Animals
Atg7f/f mice (kindly from Dr. Komatsu, Japan) (23) were crossed to Vav-Cre mice (Jackson Lab) to obtain Atg7f/f;Vav-Cre and Atgf/+;Vav-Cre mice. Atg7f/f mice was crossed to Lyz-Cre mice (Jackson Lab) to obtain Atg7f/f;Lyz-Cre. Atg7f/f;Lyz-Cre mice was further crossed to GFP-LC3 transgenic mice (Jackson Lab) to obtain Atg7f/f;Lyz-Cre;GFP-LC3 mice. Atg7f/f mice was crossed to Mx1-Cre mice ((Jackson Lab) to obtain Atg7f/f;Mx1-Cre mice. Genotyping was performed on tail genomic DNA. Male and female mice were used equally in all experiments, and littermates were always used as controls. Each group contains at least 6 mice. All experiments with animals are complied with the institutional protocols on animal welfares and approved by the Ethics Committee of Soochow University.
Reagents and Antibodies
CD11b-APC(553312), Ly-6G and Ly-6C-APC, Ter119-FITC, CD71-PE were from BD Biosciences; F4/80-PE(12–4801) was from eBioscience; Ly-6C-FITC was from Biolegend; AnnexinV-FITC PI Apoptosis Kit was from Biouniquer; M-CSF was from Sigma; Anti-Atg7, anti-Beclin1, anti-PI3 Kinase Class III, and anti-GAPDH were from Cell Signaling Technology; anti-LC3 was from Medical & Biological Laboratories.
Flow Cytometry
Flow cytometry experiments were performed with BD fluorescence-activated cell sorting (FACS) Calibur or BD FACS Aria III, followed by magnetic-activated cell sorting (MACS).
Real-time PCR
Total RNA was extracted with TRIzol reagent according to the manufacturer's guidelines (Invitrogen), and reverse transcription was performed with Thermo Scientific Revert Aid First Strand cDNA Synthesis Kit on an ABI 7500 instrument. Samples were run in triplets.
Blood Routine Examination
20 μl of mouse peripheral blood was added into 500 μl CPK-303A (37 °C). Blood routine examination was performed using sysmex KX-21N.
Forced in Vivo or ex Vivo Monocyte-Macrophage Differentiation
For forced in vivo monocyte-macrophage differentiation, mice were treated with thioglycolate. Basically, the thioglycolate-activated macrophages were isolated from Atg7f/f;Lyz-Cre mice or wild-type mice by peritoneal lavage 1–4 days after intraperitoneal injection of 1 ml of 3% sterile thioglycolate medium. The cells from thioglycolate-induced peritonitis were costained with anti-CD11b and anti-F4/80, and F4/80+CD11b+ populations were sorted by FACS. The thioglycolate-induced macrophages were then incubated, respectively, with Baf-A1 (10 nm), BFA (0.1 ng/ml), and cells were cultured in RPMI 1640 with 10% FBS or RPMI 1640 without amino acids and serum for 1.5 h (for starvation). Myeloid cells were finally measured with flow cytometry using specific biomarkers. For forced ex vivo monocyte-macrophage differentiation, monocytes were first isolated from mice by FACS or MACS (Mouse Monocyte Enrichment Kit, Stem Cell), and then cultured in 10% FBS RPMI 1640 with or without M-CSF (25 ng/ml) for 72 h. BFA (0.1 ng/ml) or Baf-A1 (10 nm) was added into culture medium after cultured for 48 h. Differentiation was accessed using flow antibodies indicated.
Confocal Microscopy Analysis of GFP-LC3 Fluorescence Puncta
Macrophages grown on coverslips were incubated with CD11b antibody after removing uncombined antibody using PBS. Cells were fixed in 4% formaldehyde for 30 min at room temperature prior to cell permeabilization with 0.1% Triton X-100 (4 °C, 10 min). The cells were washed in PBS three times for 5 min, incubated on 0.1 μg DAPI for 9 min, and then rinsed with PBS. Fluorescence signals were analyzed using confocal microscope.
Transmission Electron Microscopy of Autophagy in Macrophages
Macrophages were isolated from mice (each treatment sample requires 20 mice of each animal model) and the cell pellets from the macrophage pool were fixed in 3% glutaraldehyde solution in 0.1 m phosphate buffer at 4 °C. After 2 h of fixation in osmium tetroxide (3%), the macrophages were then dehydrated in graded acetone, and embedded in Araldite (Fluka, Buchs, Switzerland). Ultrathin sections were prepared using a diamond knife, collected on copper grids (G 300 Cu), contrasted using both lead citrate and uranyl acetate, and then examined with a transmission electron microscope (JEM-1010, JEOL, Japan).
AnnexinV/PI Apoptosis Assays
Cells were costained with FITC-annexin V and propidium iodide (PI) according to the manufacturer's protocol and analyzed with a flow analyzer.
Mouse Primary Monocytes Isolation from Bone Marrow
Bone marrow cells were isolated by removing leg bones from muscles. Cells were detected by flow analysis. In addition, pure mouse primary monocytes were sorted against anti-Ly6C and anti-F4/80.
Western Blotting Analysis
Cells were cultured and lysed in lysis buffer (Cell Signaling) containing protease inhibitor mixture and PhosSTOP phosphatase inhibitor mixture (Roche) on ice for 30 min. Crude lysates were obtained by centrifugation centrifuged at 13,000 × g for 20 min and heated at 95 °C for 5 min at 4 °C. The protein was detected by immunoblotting. Equal amounts of protein were loaded on a 12% SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to polyvinylidene difluoride membrane (Millipore). Anti-Atg7, anti-LC3 polyclonal antibodies were revealed using an appropriate horseradish peroxidase (HRP)-conjugated secondary antibody (Cell Signaling) and detected by an enhanced chemiluminescence kit (Pierce). In conjunction, blots were probed with anti-GAPDH antibody (Cell Signaling) to confirm equal loading of protein.
In Vitro Phagocytosis Assay
Bone marrow CD11b+ cells were combined with reconstituted Escherichia coli that expressed EGFP and incubated at a ratio of 10:1 (E. coli:macrophages) at 37 °C. After 2 and 4 h, macrophage phagocytosis was analyzed by flow cytometry.
Blood Smear
Blood smears are made by placing a drop of blood on one end of a slide, and using a spreader slide to disperse the blood over the slide's length. The slide is left to air dry, after which the blood is fixed to the slide by immersing it briefly in methanol and performed on blood smears stained with Giemsa stain. After staining, the monolayer is viewed under a microscope using magnification up to 400×. Individual cells are examined, and their morphology is characterized and recorded.
Measurement of Reactive Oxygen Species
DCFH-DA dissolved in serum-free medium was added at 10 μm (final concentration). Cells were resuspended in serum-free medium containing DCFH-DA and incubated at 37 °C 15 min, then analyzed by FACS-Calibur Determination of ROS production. Cells were incubated with CM-H 2DCFDA (Molecular Probes) in the dark for 15 min at 37 °C. After washing, cells were analyzed by flow cytometry using a FACS Calibur (BD Biosciences). Data were analyzed by using the CellQuest program (BD Biosciences).
Lentivirus-mediated RNA Interference
Lentiviral vectors to silence Rab9 together with negative control vector (shRNA-NC) were purchased from GenePharma Co, Ltd (Shanghai, China). The sequences to silence Rab9 were shRNA: 5′-GGAGGCAGTTCGAAGAATTCTT-3′. Virus was produced by co-transfection of 293T cells with shRab9 or shRNA-NC and packaging plasmids. For viral infection, viral supernatant were filtered, collected and used to infect macrophages. Monocytes were cultured in M-CSF medium for 48 h, and lentivirus was then added into culture medium. After 48 h, GFP+ macrophages were sorted for further Q-PCR and indicated treatment. The Q-PCR primers used were mouse Rab9, the forward primer was 5′-TACCGGGGTTCTGACTGTTG-3′, and the reverse primer was 3′-GGCTCTTTCACATCTGCGTAATA-5′.
Statistical Analysis
The significance of differences was determined by Student's t test or one-way ANOVA followed by Tukey test. p < 0.05 was considered statistically significant.
Results
Atg7-gene Deletion Confers Differing Phenotypes in HSCs and Terminally Differentiated Hematopoietic Cells
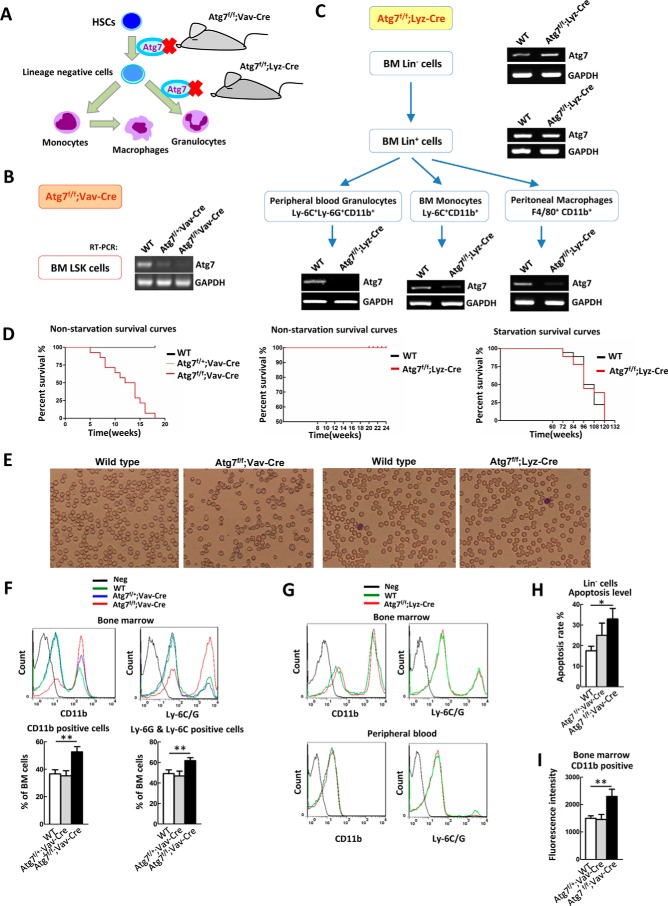
Autophagy in HSCs and in differentiated hematopoietic cells was assessed through conditional mouse models by deleting a critical gene (Atg7) in the hematopoietic hierarchy (Fig. 1A). The Atg7 gene encodes an E1-like enzyme that functions in the two ubiquitin-like conjugation systems essential for autophagosome formation (24). Whole-body homozygous Atg7 knock-out is lethal in mice neonates (23). In our mouse model, Atg7 deletion was selective, limited to the hematopoietic system, and was confirmed by semi-quantitative real-time polymerase chain reaction (RT-PCR). The Vav gene encodes a guanine nucleotide exchange factor in HSCs (25, 26), and its promoter effectively directs exogenous gene expression in HSCs. In Atg7f/f;Vav-Cre mice, Atg7 deletion by Vav promoter-directed Cre expression occurs at stem-cell level. This leads to complete floxing of Atg7 throughout the hematopoietic system (Fig. 1B), given that all descendant hematopoietic cells originate from HSCs (27). In contrast, lysozyme M (Lyz) expression gradually increases during myeloid differentiation, peaking in mature granulocytes and macrophages (28, 29). In Atg7f/f;Lyz-Cre mice, Atg7 deletion is limited to myeloid-cell lineage, including granulocytes, monocytes, and mature macrophages, whereas early hematopoietic cells, including stem cells and progenitors, retain intact Atg7 alleles (Fig. 1C) (30, 31).
FIGURE 1.
Differential deletion of Atg7 in stem cells and terminally differentiated cells of hematopoietic system reveals different phenotypes. A, scheme for deferential deletion of atg7 in mouse hematopoietic hierarchy. B, identification of Atg7f/f;Vav-Cre mice. Atg7 gene is deleted in bone marrow LSK hematopoietic stem and progenitor cells. C, identification of Atg7f/f;Lyz-Cre mice. Atg7 gene is deleted in the myeloid cells including granulocytes, monocytes, and macrophages, but their upstream cells (bone marrow lineage negative and positive cells) maintain atg7 transcript. The designated types of cells from the mouse models were directly sorted from bone marrow cells by FACS, and the presence of Atg7 transcript was determined by RT-PCR. D, Atg7 deletion in hematopoietic stem cells, not myeloid cells, caused mouse death, but atg7 deletion in myeloid cells did not affect mouse death under nutrient-rich or starvation conditions. For starvation, mice were provided with water but without feed. E, blood routine examination of Atg7f/f;Vav-Cre mice and Atg7f/f;Lyz-Cre mice. F, flow cytometric analysis of the percentage of bone marrow myeloid cells (CD11b+) and granulocyte (Ly-6G+ and Ly-6C+) in Atg7f/f;Vav-Cre and wild-type mice. G, flow cytometric analysis of bone marrow and peripheral blood myeloid cells (CD11b+) and granulocyte (Ly-6G+ and Ly-6C+) in Atg7f/f;Lyz-Cre and wild-type mice. H, apoptosis of myeloid cells from the mouse models was assessed using Annexin V/PI staining on flow cytometer. I, flow cytometric analysis of ROS levels in Lin− cells of Atg7f/f;Vav-Cre mice or myeloid cells of Atg7f/f;Lyz-Cre mice. Data are representative or statistical results of three experiments. n ≥ 6. *, p < 0.05; **, p < 0.01.
Phenotypic analysis revealed that the Atg7f/f;Vav-Cre mice developed progressive anemia and splenomegaly, survived for a mean of only 12 weeks, and were sterile (data not shown), consistent with previous reports (16, 17). However, survival rates of wild-type and Atg7f/f:Lyz-Cre mice did not differ under nutrient-rich or starvation conditions (Fig. 1D), suggesting that autophagy in macrophages of Atg7f/f;Lyz-Cre mice remain functional and respond to starvation. As shown by blood smears and cell counts, Atg7f/f;Vav-Cre mice developed worsening anemia, whereas Atg7f/f;Lyz-Cre mice did not differ from wild-type in this regard (Fig. 1E).
We also observed that autophagic defects in HSCs of the Atg7f/f;Vav-Cre mouse model culminated in irreversible failure of hematopoiesis, again in agreement with previous findings (16, 17), whereas the ATG7-mediated autophagy defects in Atg7f/f;Lyz-Cre mice did not result in peripheral blood changes relative to wild-type mice (data not shown). To validate these peripheral blood findings, bone marrow samples were analyzed by flow cytometry for specific markers, namely CD11b (monocytes/macrophages) or Ly6G/C (granulocytes). Subsequently, Atg7f/f:Vav-Cre mice displayed marked myeloproliferation (Fig. 1F), but there were no signs of altered differentiation in Atg7f/f;Lyz-Cre mice (Fig. 1G). The genetic and phenotypic data imply that Atg7 deletion in HSCs induces irreversible impairment in hematopoiesis and eventual animal death; however, this outcome does not occur in myeloid cells with this deletion.
We have reported that autophagy is required for terminal differentiation of myeloid cells in vitro (31, 32) and for megakaryocytic differentiation in vivo (19) Another recent study involving exposure to an inducer of monocytic differentiation has demonstrated that induction of autophagy is necessary for ex vivo monocyte-macrophage differentiation (33). Not unexpectedly, loss of autophagy in myeloid cells of the Atg7f/f;Lyz-Cre mouse model would thus have profound consequences in terms of in vivo terminal differentiation. To explain why in vivo Atg7 deletion in myeloid cells did not culminate in dysfunctional terminal differentiation, we examined the potential influence of this deletion on intrinsic myeloid cell function in Atg7f/f;Lyz-Cre mice. To this end, we compared myeloid-cell apoptotic response in Atg7f/f;Vav-Cre and Atg7f/f;Lyz-Cre strains, using wild-type mice as controls. Bone marrow and peripheral blood cells collected from each subset were stained, using anti-CD11b antibody and PI/Annexin V. Although the ratio of apoptotic CD11b-positive cells (monocytes/macrophages) in wild-type and Atg7f/f;Lyz-Cre mice did not differ (data not shown), Atg7 deletion in HSCs of Atg7f/f;Vav-Cre mice clearly boosted apoptosis (Fig. 1H). Previous studies have shown that the absence of autophagy leads to mitochondrial accumulation of reactive oxygen species (ROS) (34, 35). We thus measured ROS level in the CD11b-positive cells of wild-type and Atg7-deficient mice. Again, Atg7f/f;Lyz-Cre and wild-type mice did not differ in ROS clearance (data not shown), suggesting that the capacity for ROS clearance in myeloid cells is not undermined by an ATG7-dependent autophagic defect. In contrast, Atg7 deletion in HSCs from Atg7f/f;Vav-Cre mice ablated ROS clearance, as evident by accumulation of ROS (Fig. 1I).
Given the above outcomes, we then examined myeloid cells in Atg7f/f;Lyz-Cre mice, checking their responsiveness to extracellular stimuli. Phagocytic activity in bone marrow or peritoneal macrophages of Atg7f/f;Lyz-Cre and wild-type mice was investigated by ex vivo addition of GFP-expressing E. coli to macrophage cultures. Uptake of GFP-expressing E. coli by myeloid cells was comparable for both strains (data not shown). This supported our prior observation that survival of Atg7f/f;Lyz-Cre mice harboring an Atg7 deletion in myeloid cells is not compromised under starvation conditions (Fig. 1D).
ATG7-dependent Autophagic Defect in Myeloid Cells Cripples Artificial but Not Physiologic Induction of Monocyte-Macrophage Differentiation
Focusing on canonical autophagy in myeloid cells, the effect of an ATG7-dependent autophagic defect on terminal differentiation of monocytes was examined. Fluorescence-activated cell sorting at monocyte-macrophage stage of differentiation (Ly6C+F4/80+) in bone marrow reached at least 95% of respective myeloid-cell totals for both Atg7f/f:Lyz-Cre and wild-type mice (Fig. 2A), with near-complete floxing of the Atg7 gene in these cells (Fig. 2B). To determine if any lingering ATG7 protein in macrophages of Atg7f/f;Lyz-Cre mice was capable of sustaining canonical autophagy, or if biallelic deletion of the Atg7 gene in macrophages of Atg7f/f;Lyz-Cre mice was sufficient to disrupt this process, comparative analysis of monoallelic Atg7 deletion in HSCs of another conditional Atg7 knock-out mouse model (Atg7f/+;Vav-Cre) was performed. Although >50% of ATG7 protein remained in bone marrow lineage-negative cells of Atg7f/+;Vav-Cre mice (Fig. 2C), monoallelic Atg7 deletion in this mouse model resulted in hematopoietic stem-cell exhaustion, signaled by higher percentages of LSKCD34− cells and LK cells and subsequent reduction in colony-forming ability (Fig. 2D). Hence, HSCs of heterozygous Atg7f/+;Vav-Cre mice proved relatively sensitive to disrupted ATG7-dependent autophagy, even with partial loss of ATG7 protein.
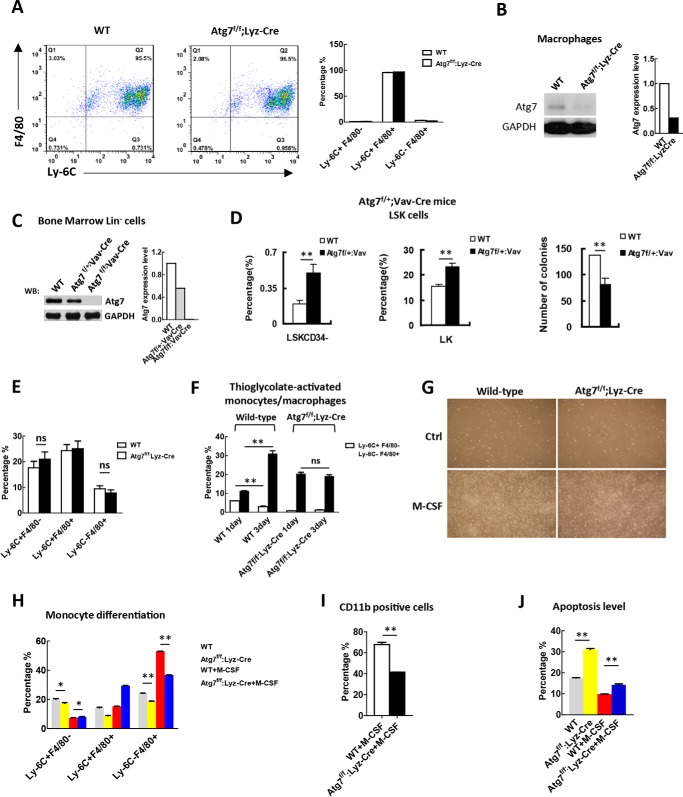
FIGURE 2.
Atg7 deletion in myeloid cells cripples the artificial but not physiological induction of monocyte-macrophage differentiation. A, sorting for myeloid cells at the monocyte-macrophage differentiation stage (Ly6C+F4/80+). Bone marrow cells from Atg7f/f;Lyz-Cre mice and wild-type mice were sorted against the markers with FACS, shown by representative sorting plots (left). The purity of the sorted cells from these two mouse models is shown (right). B, examination of the efficiency for atg7 gene floxing in macrophages of the mouse models as indicated, and the majority of atg7 protein is absent due to the floxing of atg7 gene. To obtain sufficient macrophages for analysis, macrophages were sorted with FACS after M-CSF induction of mononuclear cells isolated with MACS from bone marrow cells. Shown is a representative Western blotting result (left) and quantified data (right). C, examination of the efficiency for atg7 gene floxing in HSPCs of Atg7f/f;Vav-Cre mice and Atg7f/+;Vav-Cre mice by Western analysis (left), and their protein levels were quantified (right). Monoallelic deletion of atg7 only partially reduced atg7 protein level but biallelic deletion caused complete absence of atg7 protein in HSPCs. D, monoallelic deletion of the atg7 gene in Atg7f/+;Vav-Cre mice caused hematopoietic stem cell exhaustion. Percentage of LSKCD34− and LK cells were measured by flow cytometry (left and middle panels). Colony-forming ability of LSK cells from wild-type mice and Atg7f/+;Vav-Cre mice were determined, and the result was quantified (right panel). E, flow cytometric analysis of the percentage of monocytes (Ly-6C+F4/80−) and macrophages (Ly-6C−F4/80+) from bone marrow cells of Atg7f/f;Lyz-Cre mice and wild-type mice. F, examination of artificial induced monocyte-macrophage differentiation by thioglycolate in Atg7f/f;Lyz-Cre mice and wild-type mice. Differentiation was measured by flow cytometry on days 1 and 3 after induction. G, morphological changes associated with or without M-CSF (25 ng/ml)-induced monocyte-macrophage differentiation. H–J, analysis of differentiation and apoptosis of the atg7-deleted monocytes. Primary monocytes from Atg7f/f;Lyz-Cre and wild type mice were sorted against the indicated markers and incubated with or without M-CSF (25 ng/ml) for 72 h, the differentiation and apoptosis levels of monocytes were measured by flow cytometry. Data are representatives or statistical results of three experiments. n ≥ 6. *, p < 0.05; **, p < 0.01.
By all accounts, biallelic Atg7 deletion in myeloid cells of Atg7f/f;Lyz-Cre mice should disrupt ATG7-dependent autophagy, having lost the vast majority of ATG7 protein (Fig. 2B). To our surprise, flow cytometry revealed that percentages of monocytes (Ly6C+F4/80−), mature macrophages (Ly6C−F4/80+), and progenitors at monocyte-macrophage stage of differentiation (Ly6C+F4/80+) in bone marrow of Atg7f/f;Lyz-Cre and wild-type mice (Fig. 2E) did not differ. Thus, an ATG7-dependent autophagic defect in Atg7f/f;Lyz-Cre mice did not compromise in vivo physiologic monocyte-macrophage differentiation.
To determine if ATG7-dependent autophagy is actually functional during physiologic monocyte-macrophage differentiation, a forced in vivo differentiation assay was done by treating wild-type and Atg7f/f;Lyz-Cre mice with thioglycolate, an inducer of monocytic differentiation (34). Mice were sacrificed on Days 1 and 3 post-treatment, and thioglycolate-exposed peritoneal cells were collected for flow analysis. In wild-type mice, the percentage of Ly6C−F4/80+ macrophages increased from Day 1 to Day 3, whereas the percentage of Ly6C+F4/80− monocytes decreased, reflecting in vivo physiologic monocyte-macrophage differentiation in response to the induction agent. In Atg7f/f;Lyz-Cre mice, the percentage of Ly6C−F4/80+ macrophages or Ly6C+F4/80− monocytes surprisingly showed no obvious change (Fig. 2F), clearly representing a failure of artificial induction in this setting and underscoring the in vivo physiologic role of ATG7-dependent autophagy in myeloid cells. In turn, this indicated that Atg7 deletion in myeloid cells of Atg7f/f;Lyz-Cre mice had the intended effect. Although percentages of monocytes and macrophages in Atg7f/f;Lyz-Cre and wild-type mice did not differ under physiologic conditions, terminal differentiation patterns were not identical with artificial stimulation. Subsequent to thioglycolate treatment, mice with defective ATG7-dependent autophagy had much lower monocyte counts (Ly6C+F4/80−) and much larger macrophage populations (Ly6C−F4/80+) than wild-type mice on Day 1 of treatment; and because these levels held steady through Day 3 (Fig. 2F), it is likely that autophagy was sustained though an alternate pathway, ensuring homeostasis despite the terminal differentiation failure imposed by Atg7 deletion.
To further test for sustained differentiation of monocytes with ATG7-dependent autophagic defects, we performed an ex vivo monocyte-macrophage differentiation assay. Ly6C+F4/80− monocytes in bone marrow cellular isolates mice were enriched by flow sorting and then selectively treated with macrophage colony-stimulating factor (M-CSF) to induce monocyte-macrophage differentiation. Morphology of cells in Atg7f/f;Lyz-Cre and wild-type mice did not differ. Untreated monocytes from both wild-type and Atg7-deficient mice were rounded and gradually died, whereas monocytes cultured with M-CSF differentiated into clusters of macrophages (Fig. 2G). As evident by the percentage of Ly6C−F4/80+ cells, differentiation in Atg7-deficient myeloid cells, was partially crippled, with or without M-CSF treatment (Fig. 2H). In response, Atg7 deletion in myeloid cells blocked monocyte-macrophage differentiation, resulting in diminished expression of the CD11b (monocyte/macrophage) marker (Fig. 2I) and a higher percentage of apoptotic cells (Fig. 2J). These findings are consistent with those of an earlier study, reporting apoptotic cell death in circulating monocytes that fail to differentiate (34). Likewise, they confirm that ATG7-dependent autophagy is essential for ex vivo induction of monocyte-macrophage differentiation and again illustrate the ATG7-dependent autophagic defect in myeloid cells of Atg7f/f;Lyz-Cre mice.
Collectively, above results provide substantial evidence that an Atg7 deletion in HSCs readily inflicts a functional abnormity. On the other hand, knock-out of Atg7 in myeloid cells does not compromise cell function, although ATG7-dependent autophagy is needed for artificial induction of monocyte-macrophage differentiation in vivo and ex vivo.
Autophagy Is Maintained in Myeloid Cells Not HSCs Despite Impaired or Dysfunctional ATG7-dependent Mechanism
Biallelic Atg7 deletion in HSCs of Atg7f/f;Vav-Cre homologous mice resulted in loss of Atg7 in all descendant cells of the hematopoietic hierarchy (Figs. 1B and 2C), leading to a complete lack of LC3 lipidation, total absence of LC3-II (Fig. 3A), general failure of hematopoiesis, and animal demise (Fig. 1, D and C). However, HSCs of Atg7f/+;Vav-Cre heterozygous mice with monoallelic Atg7 deletion are still capable of active LC3 lipidation (Fig. 3A), despite an overriding functional ablation (Fig. 2D. Thus, even subtotal Atg7 gene deletions impair HSC function. Similarly, macrophages of Atg7f/+;Vav-Cre heterozygous mice with monoallelic Atg7 deletion displayed severely impaired canonical autophagic flux in response to starvation or inhibition by bafilomycin A1 (Baf-A1), which targets the degradation stage of ATG7-dependent autophagy (Fig. 3B). Notably, constitutive conversion of LC3-II from LC3-I was sustained at high levels in Atg7+/− macrophages of the heterozygous mouse model, even without activation of autophagy via starvation (Fig. 3B). Consequently, LC3 conversion is a reasonable index of ATG7-dependent (canonical) autophagy, rather than a bona fide index of ATG7-independent (noncanonical) autophagy. Similarly, crippled canonical autophagic flux was evident in macrophages of Atg7f/f;Lyz-Cre homologous mice with biallelic Atg7 gene deletions; and Atg7 floxing was fairly efficient in these mice, as indicated by Western blot analysis (Fig. 3C).
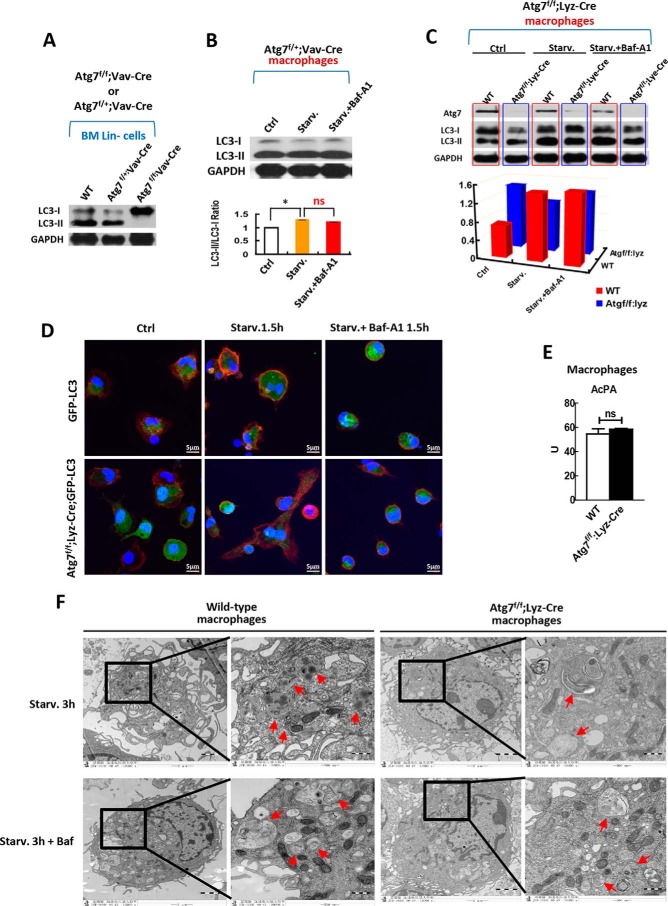
FIGURE 3.
Macrophages but not hematopoietic stem and progenitor cells maintain autophagic response when atg7 is deleted. A, effect of monoallelic or biallelic deletion of atg7 gene in stem and progenitor cells on autophagy response. Western analysis on HSPCs from homogenous Atg7f/f;Vav-Cre mice, Atg7f/+;Vav-Cre heterozygous mice and wild-type mice show that biallelic deletion of agt7 gene in HSPCs caused complete loss of LC3-II conversion from LC3-I, but monoallelic deletion of atg7 gene maintained the LC3 lipidation and processing. B, effect of monoallelic deletion of atg7 gene directed by Vav promoter in macrophages on autophagy response. Western analysis on macrophages from heterozygous Atg7f/+;Vav-Cre mice shows that monoallelic deletion of agt7 gene caused loss of canonical autophagic flux response, albeit constitutive LC3-II conversion from LC3-I maintained. For starvation, serum was deprived for 90 min. For inhibition on atg7-dependent canonical autophagy, bafilomycin A1 of 10 nm was used. The upper panel is a representative Western blotting result, and the lower panel is quantified results from three independent experiments. C, effect of biallelic deletion of atg7 gene directed by Lyz promoter in macrophages on autophagy response. The primary macrophages were lysed after being plated in the culture plate for 2 h under starvation or with/without Baf-A1 (10 nm) treatment and were then immunoblotted with antibodies against Atg7 and LC3 (upper panel). Autophagic flux was quantified (lower panel). Shown is a representative result of three independent experiments. GAPDH served as a loading control in experiments A, B, and C. D, confocal microscopic analysis of GFP-LC3 localization in the macrophages of Atg7f/f;Lyz-Cre;GFP-LC3 mice and GFP-LC3 transgenic mice. The GFP-LC3 transgenic mice served as a control. Representative images are shown for non-starvation (left panel) and for starvation 1.5 h (middle panel) with or without 10 nm of Baf-A1 (right panel). The nucleus was stained with DAPI (blue), CD11b (red) is a marker for macrophages. GFP-LC3 (green) is expressed in all tissue cells including macrophages of the GFP-LC3 transgenic mice. E, macrophages acid phosphatase activity (AcPA) in wild-type and Atg7f/f;Lyz-Cre mice was measured by spectrophotometry. F, transmission electron microscopic analysis of macrophage autophagy in response to starvation treated with or without bafilomycin A1 (Baf, 10 nm), a specific inhibitor on atg7-dependent canonical autophagy. Macrophages were isolated from wild-type mice or Atg7f/f;Lyz-Cre mice and followed by starvation for 3 h. Autophagosomes and autolysosomes are indicated with arrows. Data are representatives or statistical results of three experiments. n≥6. *, p < 0.05; **, p < 0.01.
For visual examination of in vivo autophagic response in Atg7-deficient myeloid cells, we created an Atg7f/f;Lyz-Cre;GFP-LC3 mouse model from Atg7f/f;Lyz-Cre conditional mice and GFP-LC3 transgenic mice. Driven by CAG, a ubiquitous promoter, the latter express GFP-LC3 in a host of cell types, including myeloid cells (36). Confocal microscopic results showed punctate dispersion of GFP-LC3 in Atg7+/+ wild-type macrophages of GFP-LC3 transgenic mice, which was not present in Atg7−/− macrophages of Atg7f/f;Lyz-Cre;GFP-LC3 mice under starvation conditions. The puncta observed in wild-type macrophages were due to inhibition of autophagosome-lysosome fusion, with Baf-A1 accumulating in autophagosomes. However, Baf-A1 did not accumulate in GFP-LC3 puncta of Atg7-deficient macrophages (Fig. 3D). These confocal data further confirm that Atg7 deletion in myeloid cells impairs the ATG7-dependent canonical autophagic machinery.
To evaluate whether autophagy is dispensable in macrophages, or if a compensatory autophagic pathway is activated once ATG7-dependent canonical autophagy is impaired in myeloid cells in vivo, we first examined the functional status of lysosomes in Atg7-deficient myeloid cells. Intact lysosomes are essential for cellular degradation of any autophagic cargo. Specifically, the activity of acid phosphatase (a lysosomal marker enzyme) was screened, disclosing that Atg7−/− and wild-type macrophages did not differ significantly. It was therefore apparent that defective ATG7-dependent autophagy had no impact on the lysosomal system in myeloid cells (Fig. 3E). These data provide evidence that a compensatory pathway for autophagy, relying instead on lysosomal degradation, may be activated in Atg7-deficient macrophages.
Finally, transmission electron microscopy (TEM) was engaged to further determine if an autophagic response actually occurs in Atg7-deficient macrophages. We subsequently found that Baf-A1 (a selective inhibitor of canonical autophagy) could suppress formation of autolysosomes induced by starvation in wild-type macrophages. However, Atg7-deficient macrophages did not respond similarly. Baf-A1 failed to suppress starvation-induced autolysosome formation in these cells (Fig. 3F). This TEM data again imply that a compensatory pathway of autophagy may be prompted by dysfunctional ATG7-dependent autophagy in macrophages.
Atg7 Deletion in Macrophages Raises Levels of Key Proteins Regulating Alternative Autophagy
A previous study has demonstrated that in mouse embryonic fibroblast cells, ULK1, Beclin1, VPS34, and RAB9 proteins are involved in an alternative autophagic response that is independent of ATG5/ATG7 (21). Embryonic fibroblasts are differentiated somatic cells that retain limited capacity for further differentiation. To ascertain whether a similar compensatory autophagic mechanism exists in Atg7-deficient myeloid cells, we examined above gene expression levels by real-time PCR, revealing that Vps34, Ulk1, and Beclin1 were transcriptionally up-regulated in Atg7-deficient macrophages of Atg7f/f;Lyz-Cre mice (Fig. 4A, left panel); but this up-regulation was not observed in HSCs and progenitors (LSK and LK cells) of Atg7f/f;Lyz-Cre mice (Fig. 4A, middle and right panels). Given the Lyz promoter specificity, restricting Cre floxing of Atg7 to myeloid lineage (Fig. 1C), Atg7 genes are intact in these upstream cells. Furthermore, in the Atg7f/f;Vav-Cre mouse model, where Atg7 deletion starts at HSC level (Fig. 1A), all Atg7−/− LSK, Atg7−/− LK, and Atg7−/− Lin− cells failed to upregulate above autophagic regulatory genes, and Ulk1 expression was in fact down-regulated (Fig. 4B). However, these genes were up-regulated in splenic cells of Atg7f/f;Vav-Cre mice bearing monoallelic (rather than biallelic) Atg7 deletion (Fig. 4C), indicating differing capacities for compensatory autophagy in somatic cell types.
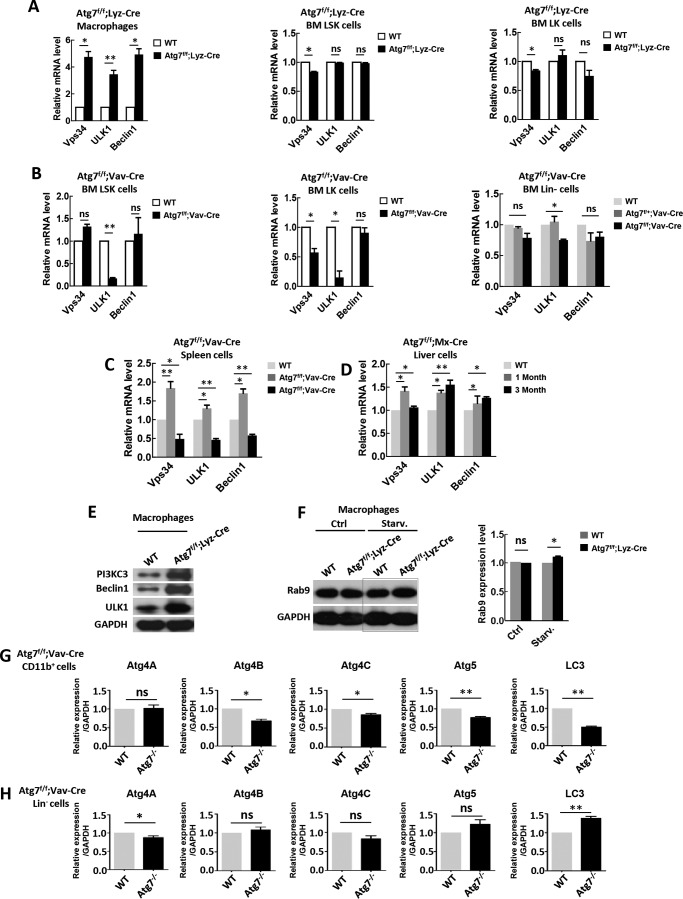
FIGURE 4.
Atg7 deletion leads to an elevated level of key regulatory proteins for alternative autophagy. A–D, RT-PCR determination of transcriptional expression of Vps34, ULK1 and Beclin1 genes in peripheral macrophages (A, left), bone marrow LSK cells (A, middle), and bone marrow LK cells of Atg7f/f;Lyz-Cre mice (A, right); bone marrow LSK cells (B, left), bone marrow LK cells (B, middle), and bone marrow Lin− cells of Atg7f/f;Vav-Cre mice (B, right); spleen cells of Atg7f/f;Vav-Cre mice (C); liver cells of pIpC induced Atg7f/f;Mx-Cre mice (D). Wild-type mice were used as a control. E, Western blotting of PI3KC3, Beclin1, and ULK1 of macrophages from wild-type and Atg7f/f;Lyz-Cre mice. Shown is a representative result. F, Western analysis of Rab9 of the macrophages from Atg7f/f;Lyz-Cre mice and wild-type mice. Representative Western blotting result (left) and quantified data (right) were shown. GAPDH served as a loading control. G and H, quantatative PCR was used to measure the expression of Atg4A, Atg4B, Atg4C, Atg5, and LC3 in myeloid cells (CD11b+) or hematopoietic Lin− negative cells from Atg7f/f;Vav-Cre mice. Relative expression level is normalized to GAPDH. Data are representative or statistical results of three experiments. n ≥ 6. *, p < 0.05; **, p < 0.01.
To further assess autophagic regulatory genes in other Atg7-deficient somatic/differentiated cells, we examined their expression levels in a Mx1 promoter-directed Atg7 conditional knock-out mouse model (Atg7f/f;Mx1-Cre). At 4 weeks of age, these mice were treated with polyinosine-polycytosine (pIpC) three times every other day to induce deletion of Atg7 alleles. In hepatic cells, such treatment did result in up-regulation of Ulk1, Beclin1, and Vps34 genes (Fig. 4D). Consistent with the above transcriptional data, protein levels of PI3KC3, Beclin1, and ULK1 in Atg7-deficient macrophages of Atg7f/f;Lyz-Cre mice were all significantly up-regulated (Fig. 4E). The Rab9 gene, a critical positive regulator of alternative autophagy, appeared to be up-regulated upon starvation in Atg7-deficient macrophages of Atg7f/f;Lyz-Cre mice (Fig. 4F).
A brief look at altered expression of several proteins essential for canonical autophagy was done in the context of Atg7 deletion. In myeloid cells, Atg7 deletion resulted in significant reductions of key proteins (ATG4A, ATG4B, ATG4C, ATG5, and LC3) in canonical autophagy, but in Atg7-deficient lineage-negative cells, expression of these proteins was largely unchanged (Fig. 4, G and H). Atg7-deficient macrophages may augment expression of proteins for alternative autophagy by reducing levels of proteins specific for canonical autophagy. In doing so, the metabolic burden of an Atg7-deficient functional cell is lowered, encouraging its survival under stress.
Collectively, above data suggest that somatic/differentiated cells, including myeloid cells, splenic cells, and hepatic cells, but excluding HSCs, are capable of up-regulating the proteins needed to drive alternative autophagy, in the event that ATG7-dependent canonical autophagy is impaired.
Brefeldin A Treatment or Rab9 Knockdown Specifically Inhibits Alternative Autophagic Response in Atg7-deficient Myeloid Cells
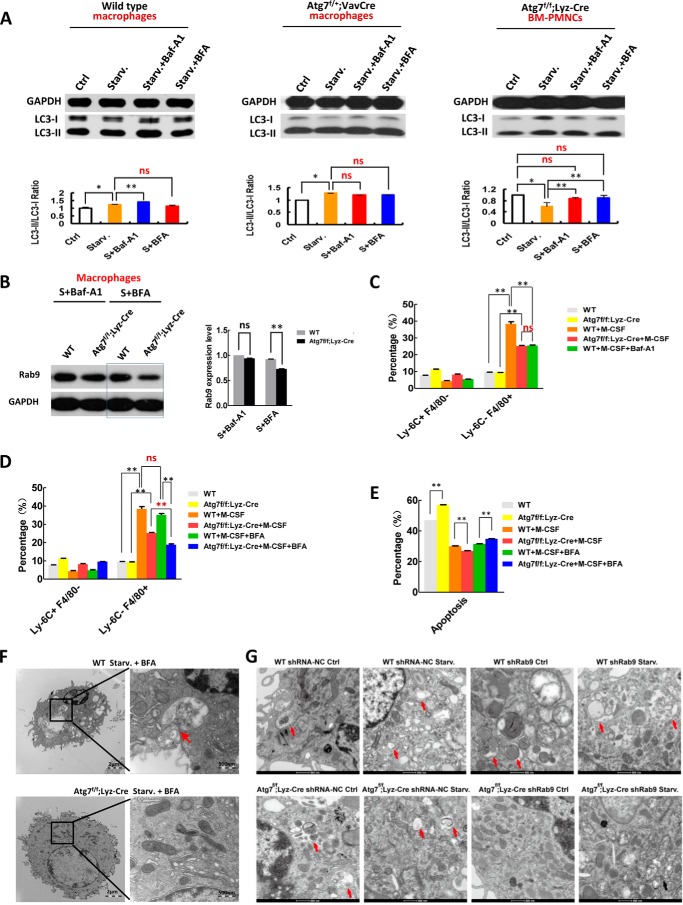
To confirm that a compensatory mechanism of autophagy is operant in Atg7-deficient macrophages, we examined the autophagic response to starvation in mice exposed to selective inhibitors of canonical or alternative autophagic pathways. Baf-A1 alters autophagic flux in wild-type macrophages under starvation, whereas brefeldin A (BFA), an inhibitor targeting an alternative autophagic process (21), has little effect on the dynamics of LC3-II accumulation in macrophages of wild-type mice (Fig. 5A, left panel). Thus, BFA does not inhibit the canonical autophagy pathway in wild-type myeloid cells. However, even in monoallelic Atg7-deficient macrophages of Atg7f/+;Vav-Cre mice, Baf-A1 no longer altered the canonical autophagic flux, nor did BFA (Fig. 5A, middle panel). Examination of peripheral mononuclear cells (PMNCs), as differentiated blood cells in Atg7f/f;Lyz-Cre mice, disclosed severely abnormal autophagic flux; and LC3-II accumulation in these cells was unchanged by BFA treatment (Fig. 5A, right panel). Hence, non-inhibition of canonical autophagy by BFA was confirmed, and unlike wild-type counterparts, BFA, not Baf-A1, noticeably downregulated RAB9 protein, a key regulator of alternative autophagy, in starved Atg7-deficient macrophages of Atg7f/f;Lyz-Cre mice (Fig. 5B). These data further suggest that Atg7-deficient macrophages possess an alternative autophagic mechanism.
FIGURE 5.
Brefeldin A treatment or knockdown of Rab9 inhibits alternative autophagic response in atg7-deleted macrophages. Examination of canonical autophagic flux in response to starvation (1.5 h) with/without bafilomycin A1 (Baf-A1) or brefeldin A (BFA, 0.1 ng/ml, a specific inhibitor on atg7-independent autophagy) in macrophages of wild-type mice (left), Atg7f/+;Vav-Cre heterozygous mice (middle), and in peripheral mononuclear cells (PMNCs) of Atg7f/f;Lyz-Cre homozygous mice (right) by Western analysis. Representative Western blotting results (upper panels) and statistical data of three independent experiments (lower panels) are shown. B, BFA but not Baf-A1 downregulated Rab9 level of macrophages from Atg7f/f;Lyz-Cre mice in response to autophagy stimuli (starvation 1.5 h). Macrophages from wild-type mice served as a control. A representative Western blotting result (left) and quantative data (right) are shown. C, atg7-deleted macrophages of Atg7f/f;Lyz-Cre mice showed an inhibition on M-CSF induced monocyte-macrophage differentiation to the same degree as Baf-A1 inhibition on the M-CSF induced monocyte-macrophage differentiation of wild-type mice. D, BFA but not Baf-A1 inhibits M-CSF induced monocyte-macrophage differentiation in atg7-deleted myeloid cells of Atg7f/f;Lyz-Cre mice. Representative colony-forming results (right) and statistical data (left) are shown. E, BFA inhibition on monocyte differentiation increases apoptosis of atg7-deleted monocytes of Atg7f/f;Lyz-Cre mice. F, transmission electron microscopic analysis of macrophage autophagy in response to starvation treated with or without Brefeldin A (0.1 ng/ml). Macrophages were isolated from wild-type mice or Atg7f/f;Lyz-Cre mice and followed by starvation for 90 min. Autophagosomes and autolysosomes are shown in the representative TEM images. G, knockdown of Rab9 crippled autophagy response in atg7-deleted, but not wild-type macrophages. Red arrows: autophagosmes or autolysosomes; black arrows: damaged mitochondria. Data are representative or statistical results of three experiments. n ≥ 6. *, p < 0.05; **, p < 0.01.
Autophagy is essential for ex vivo monocyte-macrophage differentiation (33). To demonstrate that macrophages use two autophagic processes, relying on the alternative autophagic response if the canonical pathway is impaired, we examined M-CSF-induced monocyte-macrophage differentiation in the presence of selective inhibitors targeting either canonical or alternative autophagy. Monocytes of Atg7f/f;Lyz-Cre and wild-type mice were sorted and cultured with M-CSF to induce differentiation. After 48 h of culture, Baf-A1 was first added, and the cells were harvested at 72 h for microscopic assessment and flow cytometry. M-CSF effectively induced macrophage differentiation ex vivo in wild-type monocytes but was inhibited by Baf-A1, a canonical autophagy inhibitor. Interestingly, M-CSF-induced differentiation of Atg7-deleted monocytes only reached the same level to M-CSF-induced wild-type monocyte differentiation when its canonical autophagy was inhibited with Baf-A1 (Fig. 5C), again indicating Atg7 deletion in myeloid cells of the Atg7f/f;Lyz-Cre mice impairs canonical autophagy. In contrast, BFA did not inhibit M-CSF-induced monocyte-macrophage differentiation in wild-type monocytes, but in Atg7-deficient monocytes, M-CSF-induced monocyte-macrophage differentiation showed significant inhibition (Fig. 5D). Once more, the alternative autophagic response that is predominant in Atg7-deleted monocytes was thus illustrated.
Atg7 deletion in monocytes of Atg7f/f;Lyz-Cre mice increases apoptosis. Consequently, ATG7-dependent autophagy is needed for apoptotic balance in macrophages, as well as for normal progression of cellular differentiation. Forced differentiation with M-CSF inhibited apoptosis in both wild-type and Agt7-deficient monocytes. However, BFA treatment increased apoptosis only in Atg7-deficient monocytes (Fig. 5E), reinforcing our contention that compensatory autophagy is primarily operant in Atg7-deficient monocytes, equating with previously described alternative autophagy (21, 37).
In a further attempt to reconcile compensatory and alternative autophagy (21), transmission electron microscopy was used to examine the autophagic response to BFA in Atg7-deficient macrophages. As anticipated, BFA did not inhibit starvation-induced autolysosome formation in wild-type macrophages, where ATG7-dependent canonical autophagy is functional. However, BFA effectively suppressed the starvation-induced autophagic response in Atg7-deficient macrophages of Atg7f/f;Lyz-Cre mice (Fig. 5F). It is therefore quite apparent that Atg7-deficient macrophages use an alternative mechanism of autophagy if ATG7-dependent canonical autophagy is impaired or dysfunctional.
RAB9 is a critical regulatory protein and a specific requirement in ATG7-independent alternative autophagy, as opposed to ATG7-dependent canonical autophagy (21). Failure of alternative autophagy is then expected with depletion of RAB9. Indeed, knockdown of Rab9 by lentiviral infection of a DNA construct targeting Rab9 crippled the autophagic response, signaled by loss of autolysosomes and accrual of swollen mitochondria in Atg7-deficient macrophages (Fig. 5G). This was not manifested in wild-type macrophages and confirms that alternative autophagy is triggered if canonical autophagy in macrophages is damaged.
Discussion
ATG7 mediates ubiquitin-like protein systems where ATG8 and ATG12 are conjugated with phosphatidylethanolamine and ATG5, respectively, and ATG16 interacts with ATG5.37 ATG7 is thus considered critical for autophagic machinery (23, 24). However, other research has indicated that autophagy in mouse embryonic fibroblasts functions independently of ATG5/ATG7 (21). Our data here suggest that biallelic Atg7 deletion leads to irreversible loss of autophagy in HSCs that rely solely on ATG7-dependent canonical autophagy, whereas the same Atg7 deletion in myeloid cells triggers an alternative autophagic pathway.
Theoretically, Atg7−/− myeloid cells of Atg7f/f;Vav-Cre homologous mice would be optimal for studying ATG7 dependence in differentiated hematopoietic cells. The floxing of Atg7 in HSCs is truly complete, so all descendant cells, including myeloid lines, should lack Atg7 alleles. Unfortunately, multilineage differentiation in such HSCs is simply not feasible, so Atg7−/− myeloid cells cannot be generated for analysis, Western blotting in particular.
Examining the monoallelic Atg7+/− macrophages of Atg7f/+;Vav-Cre mice shows that constitutive conversion of LC3-II from LC3-I remains at high levels in the heterozygous mouse model, even without starvation-induced autophagy (Fig. 3B). This suggests that LC3 conversion is not a bona fide indicator of functional autophagy in instances of Atg7 mutation or deletion. In our hands, partial loss (50%) of ATG7 protein through monoallelic Atg7 deletion produced dysfunctional autophagic flux, given that LC3-II did not accumulate in Atg7+/− macrophages of Atg7f/+;Vav-Cre mice upon exposure to Baf-A1 (Fig. 3B). This suggests that ATG7-dependent autophagy in Atg7−/− macrophages of Atg7f/f;Lyz-Cre mice should be disabled if a majority of ATG7 protein is absent.
Although autophagosome accumulation corresponds with LC3 lipidation, autophagic degradation is not guaranteed, unless LC3-II further accrues upon inhibiting autophagic degradation. Therefore, autophagy may still be defective, even if LC3 lipidation is maintained (39, 40). In this study, autophagic flux analysis suggested that residual ATG7 protein and LC3 lipidation/processing in Atg7+/− macrophages of Atg7f/+;Vav-Cre mice are insufficient to maintain a functional ATG7-dependent autophagic response. In other words, a major reduction in the AGT7 protein within myeloid cells of Atg7f/f;Lyz-Cre homologous mice should suffice to impair ATG7-dependent autophagy in these differentiated cells.
Because ATG7-dependent autophagy impairment in myeloid cells scarcely displaced functional defect, nor lost autophagy response, one might think that functional somatic/differentiated cells may compensate in some way, resuming obligatory ATG7-dependent canonical autophagy. Surprisingly, M-CSF-induced differentiation in Atg7-deficient monocytes of Atg7f/f;Lyz-Cre mice only reached the same level as wild-type monocytic counterparts when canonical autophagy was inhibited by Baf-A1 (Fig. 5C), thus supporting our contention that Atg7 deletion in myeloid cells impairs canonical autophagy. Unlike Baf-A1, which inhibits endoplasmic reticulum-derived autophagosomes essential for canonical autophagy (38), BFA inhibits Golgi-derived membranes, leading to inhibition of alternative autophagy (21). BFA inhibited differentiation in M-CSF-stimulated Atg7-deficient monocytes of Atg7f/f;Lyz-Cre mice, but comparably treated monocytes of wild-type mice were unaffected (Fig. 5D), reinforcing the concept that an alternative autophagic pathway is operant in Atg7-deficient myeloid cells of Atg7f/f;Lyz-Cre mice to compensate for functional loss of canonical autophagy.
Our transmission electron microscopy data indicate that Baf-A1, a selective inhibitor on ATG7-dependent canonical autophagy, did not prevent autolysosome formation in starved macrophages of Atg7f/f;Lyz-Cre mice (Fig. 3F, right panel) but did so in starved wild-type macrophages (Fig. 3F, left panel), where ATG7-dependent canonical autophagy functions. Moreover, the selective inhibitor of alternative autophagy, BAF (21), inhibited autolysosome formation in starved Atg7-deficient macrophages of Atg7f/f;Lyz-Cre mice (Fig. 5F, lower left panel). The specificity of BFA was supported by its inability to inhibit autolysosome formation in starved macrophages of wild-type mice, retaining ATG7-dependent canonical autophagy (Fig. 5F, upper left panel).
Knockdown of Rab9, removing a key regulatory protein required in alternative autophagy, disrupted alternative autophagy in Atg7-deficient macrophages but not in wild-type macrophages (Fig. 5G), attesting to our impression that alternative autophagy in macrophages is active only if canonical autophagy becomes dysfunctional. Nevertheless, it should be noted that starvation-induced autophagy in Atg7-deficient macrophages of Atg7f/f;Lyz-Cre mice is not as robust as that seen in wild-type macrophages. Hence, canonical autophagy is likely the mainstay, whereas alternative autophagy is complementary.
Collectively, the experiments conducted here indicate that the Atg7f/f;Lyz-Cre mouse model is valid for assessing biomolecular dynamics in response to loss of ATG7-dependent canonical autophagy in macrophages. A recent study with the same Atg7f/f;Lyz-Cre mouse model has shown that when infected with Mycobacterium tuberculosis or M. tuberculosis var. bovis, macrophages lacking Atg7 may accumulate p62, thereby inducing expression of Nrf2 target genes that encode scavenger receptors, MARCO and MSR1, for an increase in phagocytosis (41). Our observations and those of others indicate that macrophages lacking Atg7 actually mount a varied response to differing infectious pathogens. While indispensable in HSCs, macrophages are capable of instigating a compensatory autophagic mechanism when ATG7-dependent autophagy is impaired.
Our results further demonstrate that in myeloid cells, splenic cells, and hepatic cells, albeit not stem cells, Atg7 deletion results in up-regulation of PI3K, Beclin1, Ulk1, and Rab9 (Fig. 4), and BFA down-regulates Rab9 (Fig. 5B), all of which are required for alternative autophagy (21, 37). Such divergence within the hematopoietic system may be applicable to all types of cellular hierarchies in mammalian tissues.
Hence, unlike stem cells that rely solely on AGT7-dependent canonical autophagy, somatic/differentiated/functional cells have at least two autophagic options. Stem cells are largely dominant figures with low rates of metabolism, generating the least intracellular stress and residing where extracellular stress is minimized (42). Thus, only one autophagic pathway may suffice in this context. In the absence of autophagy, mitochondrial ROS accumulate (34, 43), ultimately leading to malignant transformation from cumulative DNA damage. Somatic/differentiated cells are exposed to various extracellular and environmental stresses through their respective functions and thus require more innate protection. Such divergence of autophagy within the hematopoietic hierarchy enables functional blood cells to deal with stress more efficiently and effectively. Our findings indicate that autophagy is more vulnerable in stem and progenitor cells, and if damaged, is only recoverable in somatic/differentiated cells. These revelations expand our understanding of malignant transformation, which is more apt to involve cells of lesser differentiation.
Author Contributions
Y. C. and J. W. designed the experiment, Y. C. did most of the experiments; S. Z., N. Y., X. L., W. L., Z. W., F. X., L. S., Z. W., Y. F., H. Z., Y. Z., W. Z., S. H., and X. Z. conducted experiments. J. W. wrote the paper, and all authors read and approved the paper.
This work was supported by grants from the National Natural Science Foundation of China (No.31071258, No.81272336, No.31271526, and No.31201073), The Ministry of Science and Technology of China (No. 2011CB512101), The Department of Science and Technology of Jiangsu Province of China (No. BK20130333), and a project funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions. The authors declare no conflicts of interest.
- HSC
- hematopoietic stem cells
- MACS
- magnetic-activated cell sorting
- LC
- light chain
- ROS
- reactive oxygen species
- pIpC
- polyinosine-polycytosine
- BFA
- brefeldin A
- HSPC
- hematopoietic stem and progenitor cells.
References
- 1. Klionsky D. J., Emr S. D. (2000) Autophagy as a regulated pathway of cellular degradation. Science 290, 1717–1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Levine B., Klionsky D. J. (2004) Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev. Cell 6, 463–477 [DOI] [PubMed] [Google Scholar]
- 3. Singh R., Kaushik. S., Wang Y., Xiang Y., Novak I., Komatsu M., Tanaka K., Cuervo A. M., Czaja M. J. (2009) Autophagy regulates lipid metabolism. Nature 458, 1131–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Singh R., Xiang Y., Wang Y., Baikati K., Cuervo A. M., Luu Y. K., Tang Y., Pessin J. E., Schwartz G. J., Czaja M. J. (2009) Autophagy regulates adipose mass and differentiation in mice. J. Clin. Invest. 119, 3329–3339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Heaton N. S., Randall G. (2010) Dengue virus-induced autophagy regulates lipid metabolism. Cell Host and Microbe 8, 422–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Seino J., Wang L., Harada Y., Huang C., Ishii K., Mizushima N., Suzuki T. (2013) Basal autophagy is required for the efficient catabolism of sialyl oligosaccharides. J. Biol. Chem. 288, 26898–26907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yue Z., Jin S., Yang C., Levine A. J., Heintz N. (2003) Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci U.S.A. 100, 15077–15082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tra T., Gong L., Kao L. P., Li X. L., Grandela C., Devenish R. J., Wolvetang E., Prescott M. (2011) Autophagy in human embryonic stem cells. PloS One 2011; 6, e27485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Vessoni A. T., Muotri A. R., Okamoto O. K. (2012) Autophagy in stem cell maintenance and differentiation. Stem Cells Dev 21, 513–520 [DOI] [PubMed] [Google Scholar]
- 10. Oliver L., Hue E., Priault M., Vallette F. M. (2012) Basal autophagy decreased during the differentiation of human adult mesenchymal stem cells. Stem Cells Dev 21, 2779–2788 [DOI] [PubMed] [Google Scholar]
- 11. Lee Y., Jung J., Cho K. J., Lee S. K., Park J. W., Oh I. H., Kim G. J. (2013) Increased SCF/c-kit by hypoxia promotes autophagy of human placental chorionic plate-derived mesenchymal stem cells via regulating the phosphorylation of mTOR. J. Cell. Biochem. 114, 79–88 [DOI] [PubMed] [Google Scholar]
- 12. Menendez J. A., Vellon L., Oliveras-Ferraros C., Cufí S., Vazquez-Martin A. (2010) mTOR-regulated senescence and autophagy during reprogramming of somatic cells to pluripotency: a roadmap from energy metabolism to stem cell renewal and aging. Cell Cycle 10, 3658–3677 [DOI] [PubMed] [Google Scholar]
- 13. Wang S., Xia P., Ye B., Huang G., Liu J., Fan Z. (2013) Transient Activation of Autophagy via Sox2-Mediated Suppression of mTOR is an important early step in reprogramming to pluripotency. Cell Stem Cell 13, 617–625 [DOI] [PubMed] [Google Scholar]
- 14. Pan H., Cai N., Li M., Liu G. H., Izpisua Belmonte J. C. (2013) Autophagic control of cell 'stemness'. EMBO Mol Med 5, 327–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Liu F., Lee J. Y., Wei H., Tanabe O., Engel J. D., Morrison S. J. (2010) FIP200 is required for the cell-autonomous maintenance of fetal hematopoietic stem cells. Blood 116, 4806–4814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mortensen M., Ferguson D. J., Edelmann M., Kessler B., Morten K. J., Komatsu M., Simon A. K. (2010) Loss of autophagy in erythroid cells leads to defective removal of mitochondria and severe anemia in vivo. Proc Natl Acad Sci U.S.A. 107, 832–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mortensen M., Soilleux E. J., Djordjevic G., Tripp R., Lutteropp M., Sadighi-Akha E., Stranks A. J., Glanville J., Knight S., Jacobsen S. E., Kranc K. R., Simon A. K. (2011) The autophagy protein Atg7 is essential for hematopoietic stem cell maintenance. J. Exp. Med. 208, 455–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cao Y., Zhang A., Cai J., Yuan N., Lin W., Liu S., Xu F., Song L., Li X., Fang Y., Wang Z., Zhang H., Zhao W., Hu S., Zhang S., Wang J. (2015) Autophagy regulates hematopoietic stem and progenitor cell cycle in a nutrient-dependent manner. Exp. Hematol. 43, 229–242 [DOI] [PubMed] [Google Scholar]
- 19. Cao Y., Cai J., Zhang S., Yuan N., Li X., Fang Y., Song L., Shang M., Liu S., Zhao W., Hu S., Wang J. (2015) Loss of autophagy leads to failure in megakaryopoiesis, megakaryocyte differentiation and thrombopoiesis. Exp. Hematol. 43, 488–494 [DOI] [PubMed] [Google Scholar]
- 20. Cao Y., Cai J., Zhang S., Yuan N., Fang Y., Wang Z., Li X., Cao D., Xu F., Lin W., Song L., Wang Z., Wang J., Xu X., Zhang Y., Zhao W., Hu S., Zhang X., Wang J. (2015) Autophagy sustains hematopoiesis through targeting Notch. Stem Cell Dev. 10.1089/scd.2015.0176 [DOI] [PubMed] [Google Scholar]
- 21. Nishida Y., Arakawa S., Fujitani K., Yamaguchi H., Mizuta T., Kanaseki T., Komatsu M., Otsu K., Tsujimoto Y., Shimizu S. (2009) Discovery of Atg5/Atg7-independent alternative macrophagy. Nature 461, 654–658 [DOI] [PubMed] [Google Scholar]
- 22. Chang T. K., Shravage B. V., Hayes S. D., Powers C. M., Simin R. T., Wade Harper J., Baechrecke E. H. (2013) Uba1 functions in Atg7 and Atg3-independent autophagy. Nat Cell Biol. 15, 1067–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Komatsu M., Waguri S., Ueno T., Iwata J., Murata S., Tanida I., Ezaki J, Mizushima N., Ohsumi Y., Uchiyama Y., Kominami E., Tanaka K., Chiba T. (2005) Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J. Cell Biol. 169, 425–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Komatsu M., Wang Q. J., Holstein G. R., Friedrich V. L. Jr., Iwata J. I., Kominami E., Chait B. T., Tanaka K., Yue Z. (2007) Essential role for autophagy protein Atg7 in the maintenance of axonal homeostasis and the prevention of axonal degeneration, Proc. Natl. Acad. Sci. U.S.A. 104, 14489–14494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cancelas J. A., Lee A. W., Prabhakar R., Stringer K. F., Zheng Y., Williams D. A. (2005) Rac GPTases differentially integrate signals regulating hematopoietic stem cell localization. Nat. Med. 11, 886–891 [DOI] [PubMed] [Google Scholar]
- 26. Sanchez-Aguilera A., Lee Y. J., Lo Celso C., Ferraro F., Brumme K., Mondal S., Kim C., Dorrance A., Luo H. R., Scadden D. T., Williams D. A. (2011) Guanine nucleotide exchange factor Vav1 regulates perivascular homing and bone marrow retention of hematopoietic stem and progenitor cells. Proc. Natl. Acad. Sci. U.S.A. 108, 9607–9612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Weissman I. L., Shizuru J. A. (2009) The origins of the identification and isolation of hematopoietic stem cells, and their capability to induce donor-specific transplantation tolerance and treat autoimmune diseases. Blood 112, 3543–3553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cross M., Mangelsdorf I., Wedel A., Renkawitz R. (1988) Mouse lysozyme M gene: isolation, characterization, and expression studies. Proc. Natl. Acad. Sci. U.S.A. 85, 6232–6236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Goren I., Allmann N., Yogev N., Schürmann C., Linke A., Holdener M., Waisman A., Pfeilschifter J., Frank S. A. (2009) Transgenic Mouse Model of Inducible Macrophage Depletion: Effects of Diphtheria Toxin-Driven Lysozyme M-Specific Cell Lineage Ablation on Wound Inflammatory, Angiogenic, and Contractive Processes. Am. J. Pathol. 175, 132–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Clausen B. E., Burkhardt C., Reith W., Renkawitz R., Förster I. (1999) Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res. 8, 265–277 [DOI] [PubMed] [Google Scholar]
- 31. Colosetti P., Puissant A., Robert G., Luciano F., Jacquel A., Gounon P., Cassuto J. P., Auberger P. (2009) Autophagy is an important event for megakaryocytic differentiation of the chronic myelogenous leukemia K562 cell line. Autophagy 5, 1092–1098 [DOI] [PubMed] [Google Scholar]
- 32. Wang J., Lian H., Zhao Y., Kauss M. A., Spindel S. (2008) Vitamin D3 induces autophagy of human myeloid leukemia cells. J. Biol. Chem. 283, 25596–25605 [DOI] [PubMed] [Google Scholar]
- 33. Zhang Y., Morgan M. J., Chen K., Choksi S., Liu Z. G. (2012) Induction of autophagy is essential for monocyte-macrophage differentiation. Blood 119, 2895–2905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tal M. C., Sasai M., Lee H. K., Yordy B., Shadel G. S., Iwasaki A. (2009) Absence of autophagy results in reactive oxygen species-dependent amplification of RLR signaling. Proc. Natl. Acad. Sci. U.S.A. 106, 2770–2775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhang Y., Qi H., Taylor R., Xu W., Liu L. F., Jin S. (2007) The role of autophagy in mitochondria maintenance: characterization of mitochondrial functions in autophagy-deficient S. cerevisiae strains. Autophagy 3, 337–346 [DOI] [PubMed] [Google Scholar]
- 36. Mizushima N., Yamamoto A., Matsui M., Yoshimori T., Ohsumi Y. (2004) In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol. Biol. Cell 15, 1101–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Shimizu S., Arakawa S., Nishida Y. (2010) Autophagy takes an alternative pathway. Autophagy 6, 290–291 [DOI] [PubMed] [Google Scholar]
- 38. Mizushima N., Yoshimori T. (2007) How to interpret LC3 immunoblotting. Autophagy 3, 542–545 [DOI] [PubMed] [Google Scholar]
- 39. Klionsky D. J. (2012) Guideline for the use and interpretation of assays for monitoring autophagy. Autophagy 8, 445–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Xie Z., Klionsky D. J. (2007) Autophagosome formation: core machinery and adaptations. Nat. Cell Biol. 9, 1102–1109 [DOI] [PubMed] [Google Scholar]
- 41. Bonilla D., Bhattacharya A., Sha Y., Xu Y., Xiang Q., Kan A., Jagannath C., Komatsu M., Eissa N. T. (2013) Autophagy Regulates Phagocytosis by Modulating the Expression of Scavenger Receptors. Immunity 39, 537–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Moore K., Lemischka I. R. (2006) Stem cells and their niches. Science 311, 880–1885 [DOI] [PubMed] [Google Scholar]
- 43. Warr M. R., Binnewies M., Flach J., Reynaud D., Garg T., Malhotra R., Debnath J., Passegue E. (2013) FOXO3A directs a protective autophagy program in hematopoietic stem cells. Nature 409, 323–327 [DOI] [PMC free article] [PubMed] [Google Scholar]