Background: Propeptides regulate the organelle-specific, pH-dependent activation of proprotein convertases.
Results: A histidine residue pair in the propeptide cooperatively defines the activation pH for proprotein convertase 1/3.
Conclusion: Different spatial distributions of histidine residues modulate activation pH of proprotein convertases.
Significance: These results help our understanding of how protease paralogues evolved to regulate activation within specific cellular compartments.
Keywords: histidine, organelle, pH regulation, protease inhibitor, protein folding, serine protease, pH sensor, propeptide-mediated folding, proprotein convertase 1/3, protease activation
Abstract
The propeptides of proprotein convertases (PCs) regulate activation of cognate protease domains by sensing pH of their organellar compartments as they transit the secretory pathway. Earlier experimental work identified a conserved histidine-encoded pH sensor within the propeptide of the canonical PC, furin. To date, whether protonation of this conserved histidine is solely responsible for PC activation has remained unclear because of the observation that various PC paralogues are activated at different organellar pH values. To ascertain additional determinants of PC activation, we analyzed PC1/3, a paralogue of furin that is activated at a pH of ∼5.4. Using biophysical, biochemical, and cell-based methods, we mimicked the protonation status of various histidines within the propeptide of PC1/3 and examined how such alterations can modulate pH-dependent protease activation. Our results indicate that whereas the conserved histidine plays a crucial role in pH sensing and activation of this protease an additional histidine acts as a “gatekeeper” that fine-tunes the sensitivity of the PC1/3 propeptide to facilitate the release inhibition at higher proton concentrations when compared with furin. Coupled with earlier analyses that highlighted the enrichment of the amino acid histidine within propeptides of secreted eukaryotic proteases, our work elucidates how secreted proteases have evolved to exploit the pH of the secretory pathway by altering the spatial juxtaposition of titratable groups to regulate their activity in a spatiotemporal fashion.
Introduction
Eukaryotic cells have evolved an elegant series of membranous compartments to regulate protein synthesis, folding, activation, sorting, and export (1, 2). Beginning with their entry into the endoplasmic reticulum (ER)2 as they exit the ribosome, proteins transit these secretory pathway compartments on their journey to their ultimate destination. Just as each group of compartments has mechanisms for maintaining precise pH and calcium balance (3, 4), each family of proteins has co-evolved means to sense the environmental changes to ensure optimal organismal homeostasis. How eukaryotic proteins have diverged from their prokaryotic ancestors to exploit the unique organellar environment of the secretory pathway for biological function remains a fundamental question in cell biology (5).
Proprotein convertases (PCs) are eukaryotic members of the ubiquitous superfamily of subtilases (6). Comprising nine serine endoproteases (PC1/3, PC2, furin, PC4, PACE4, PC5/PC6, PC7/LPC/PC8, SKI/S1P, and NARC1/PCSK9), PCs are responsible for the conversion of a diverse range of precursor substrates to their active forms and thus play a central role in maintenance of physiologic homeostasis within cells and tissues (5, 7). Furin, the most thoroughly characterized PC, is constitutively expressed in virtually all tissues and catalyzes the maturation of a diverse repertoire of hormones, enzymes, and receptor precursors within the secretory pathway (6). Not surprisingly, misregulation of furin results in both hyper- and hypoactivity and has been associated with cancer invasiveness and metastasis (8–13), susceptibility to viral and parasitic infection (14), and increased severity of cardiovascular disease (15, 16). Additionally, furin-deficient mice die at embryonic day 11 due to cardiac defects resulting from failed chorioallantoic fusion, axial rotation, and ventral closure (17). PC1/3 (also known as PC1 or PC3) along with PC2 is a neuroendocrine convertase responsible for the processing of several critical metabolic regulators, including insulin, glucagon, and proopiomelanocortin (18). Mice with knock-out of pcsk1 or pcsk2, the genes encoding PC1/3 and PC2 respectively, remain viable despite hormonal and/or neuroendocrine deficiencies (19–21). Consistent with this, several studies characterizing mutations in PC1/3 have demonstrated an altered substrate processing that may underlie obesity, type II diabetes mellitus, and endocrine derangements (21–25). Further support for the correlation between polymorphisms in PC1/3 and metabolic disease is offered by reports of both individual patients (25–29) and epidemiologic studies (20).
As unregulated protease activity can have devastating consequences on organismal homeostasis (30), PCs, like all subtilases, are synthesized as zymogens and are activated in a precise spatiotemporal fashion by intramolecular proteolysis (31). A fundamental question has long been how the timing of activation is encoded and recognized by the protease. Our understanding of how this regulation is achieved is based on studies of profurin (32–35). This furin precursor contains an 83-residue N-terminal propeptide that is requisite for folding of its cognate catalytic domain in the ER (36–38) after completion of which the propeptide gets cleaved but subsequently remains associated as an inhibitor until reaching the trans-Golgi network. The necessity of furin to reach the trans-Golgi network before becoming active (39) coupled with the understanding of the unique environments of each compartment of the secretory pathway (40) argues for the presence of an encoded sensor that recognizes and responds to specific environmental signals (32). In fact, studies demonstrate that it is the mildly acidic pH of the trans-Golgi network (pH ∼ 6.5) that triggers the release and degradation of the furin propeptide (PROFUR) and thus release of inhibition of the mature protease domain of furin (MATFUR) (37, 38). Interestingly, although the PC1/3 precursor, pro-PC1/3, transits the secretory pathway in much the same way as the furin precursor, our previous work indicates that PC1/3 requires a lower pH for its activation (pH ∼ 5.5) and thus is likely activated in a later compartment (32).
Organellar pH can alter the protonation status of charged residues, thus altering the structure and stability of the protein both on a local and global scale (41). Given that the pH range of interest in the case of the secretory pathway and PC activation falls within the physiologic range of ∼7.4 in the ER to ∼5.4 in the mature secretory granules, basic residues such as arginine (pKa ∼ 12.5) or acidic residues such as glutamate (pKa ∼ 4.2) are less likely to undergo altered protonation and be candidates for pH sensors; however, the imidazole ring of histidine has a pKa of ∼6.0, making it ideally situated to respond to pH changes in this range. Histidine is used as a pH sensor in a variety of biological molecules, including hemoglobin and class II MHC (42–44), and is enriched within the propeptides of eukaryotic proteases that transit the secretory pathway in contrast to both cytosolic proteases and prokaryotic orthologues (33, 34). Indeed, we identified a histidine (His69) within PROFUR that regulates the compartment-specific activation of MATFUR (45). This histidine is in a loop adjacent to the secondary cleavage site nestled in a solvent-accessible pocket lined by hydrophobic residues. At the near-neutral pH of the ER, the deprotonated histidine acts as a hydrophobic residue, stabilizing the packing within this pocket and keeping the pH-sensitive loop protected against cleavage. Upon entry into the trans-Golgi network, the histidine is exposed to a ∼10-fold higher proton concentration and thus is protonated; as a result, the imidazole side chain becomes polar, disrupting the packing and driving a local conformational change that exposes the secondary cleavage site, allowing for rapid degradation and release of PROFUR from MATFUR (35).
Propeptide domains alone encode sufficient information for regulating the organelle-specific pH-dependent activation of cognate protease domains (32). Thus, when the propeptides of furin and PC1/3 are swapped, the pH-dependent protease activation is transferred in a propeptide-dictated manner both in vitro and in cells. Interestingly, the histidine pH sensor identified in PROFUR is absolutely conserved within the propeptides of all PCs, including PROPC1/3. Despite their structural similarity and the presence of the conserved histidine throughout the family of PCs, it remains unclear how the pH sensitivity of other PCs is encoded.
In this study, we asked whether the pH-sensing histidine is conserved in PROPC1/3 and why does MATPC1/3 not become active at the same pH as furin? We present biochemical and structural data that suggest that the conserved histidine residue plays a critical role in pH-dependent activation similar to what has been established in furin; however, an additional histidine residue modulates this pH sensitivity such that a more acidic environment is required for PC1/3 activation.
Materials and Methods
Cell Culture and Transient Transfection
COS7 cells were maintained in Dulbecco's modified Eagle's high glucose medium (Hyclone) containing 10% (v/v) fetal bovine serum and 1% (v/v) penicillin-streptomycin. Cells were kept at 37 °C in a 5% CO2 environment as described previously (32, 35). For expression of ER-retained constructs, COS7 cells at 60–80% confluence were transfected with pcDNA3.1 expression vectors containing WT PC1/3 truncated at Arg618 with a C-terminal -KDEL sequence (32) or vectors containing the noted PROPC1/3 variants using TransIT LT-1 transfection reagent (Invitrogen) according to the manufacturer's instructions. The constructs also contained an HA tag at the N terminus of the propeptide and a FLAG tag at the N terminus of the protease domain (Fig. 1A).
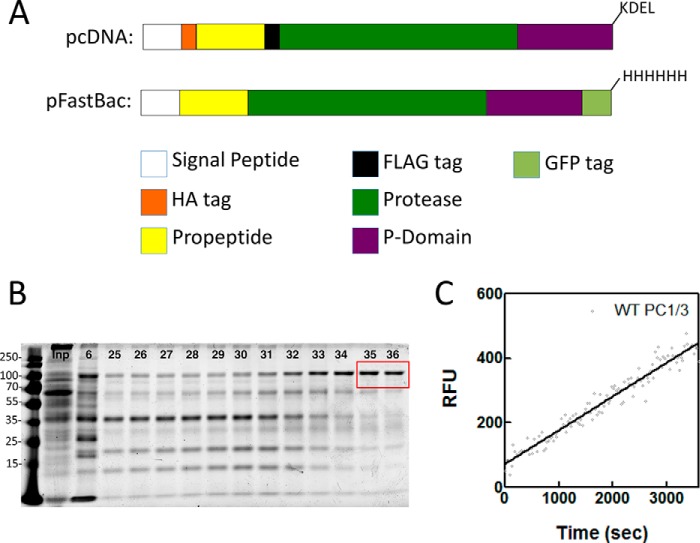
FIGURE 1.
Constructs and characterization of baculovirus-produced PC1/3. A, schematic representation of the constructs used in this study. Top, insert cloned into pcDNA3.1 where the coding sequence for WT mouse PC1/3 has been truncated at Arg618 with the addition of a -KDEL sequence, an HA tag inserted at the N terminus of the propeptide, and a FLAG tag inserted at the N terminus of the protease domain. Bottom, the coding sequence for WT mouse PC1/3 was truncated at Arg618, and a C-terminal GFP tag and His8 tag were added for insertion into the pFastBac vector for baculovirus production. B, representative SDS-polyacrylamide gel showing input loaded (Inp) and fractions collected following elution from a nickel-nitrilotriacetic acid column (6, 25–36). Fractions indicated in the red box were pooled, concentrated, and used for assays. C, representative enzyme progress curve showing baculovirus-produced WT PC1/3 processing of the fluorogenic substrate. The curve suggests that PC1/3 has fully matured and displays normal enzyme characteristics. RFU, relative fluorescence units.
pH-dependent Activation Assays
Twenty-four hours post-transfection with ER-retained PC1/3 constructs, cells were washed two times in PBS and incubated for 15 min in fractionation buffer (270 mm sucrose, 10 mm Tris, pH 7.4) with protease inhibitors (cOmplete EDTA-free, Roche Applied Science). Cells were lysed with three rounds of sonication (10 × 1-s pulses at 30% power) in an ice bath. Unlysed cells and cell debris were pelleted at 800 × g for 15 min at 4 °C, and supernatant was transferred to a clean tube. Samples were incubated with 30% (w/v) PEG 8000 for 16 h at 4 °C with gentle agitation. Precipitated proteins were pelleted by centrifugation at 17000 × g for 30 min at 4 °C. Pellets were resuspended in activity assay buffer (100 mm NaOAc, 0.1% (v/v) Brij-35, 5 mm CaCl2) at a pH of 7.4, 6.4, or 5.4 and incubated at 37 °C for 1 h to allow for activation unless otherwise noted. After 1 h, a 15 μm concentration of the fluorogenic substrate Abz-RVKRGLA-Tyr(3-NO2) was added, and activity was assayed in triplicate on a SpectraMax-M2 spectrofluorometer equipped with a 96-well plate reader (excitation, 320 nm; emission, 425 nm) for 1 h at room temperature. Data were fitted and analyzed using GraphPad Prism. Replicate samples were also processed for Western blotting; the anti-FLAG M2 antibody (Sigma) was used to recognize the N-terminally FLAG-tagged mature protease, and anti-HA.11 (Covance) used to recognize the N-terminally HA-tagged propeptide.
For experiments where limited trypsinization was used to digest the propeptide, PEG 8000-precipitated proteins were resuspended in activity assay buffer, pH 7.4 as above and incubated in the presence or absence of agarose-immobilized tosylphenylalanyl chloromethyl ketone-treated trypsin (∼10 p-toluene-sulfonyl-l-arginine methyl ester (TAME) units per sample; Thermo Scientific) for 1 h 37 °C. Samples were then treated with soybean trypsin inhibitor to a final concentration of 1 μg/ml for 20 min at 37 °C before proceeding with the activity assay as above.
Protein Production and Purification
Codon-optimized sequences encoding either WT-PROPC1/3 (mouse) or leucine and arginine variants were cloned into the pET11b backbone and expressed in BL21(DE3) Escherichia coli as described previously (32, 46). Protein was purified from the soluble fraction by ion exchange after cell lysis via French pressure cell and dialyzed into 6 m guanidinium HCl-containing buffer (50 mm Tris-HCl, 6 m guanidinium HCl, pH 6.5) for long term storage. Before use, proteins were refolded by dialysis against refolding buffer (10 mm Tris, 10 mm cacodylate, 10 mm NaOAc, 150 mm KCl, 5 mm CaCl2) at pH 6.8 unless otherwise noted. Concentration was determined after refolding by absorption at 280 nm.
Enzymatically active mature PC1/3 was expressed via the baculovirus system. The coding sequence of mouse PC1/3 was cloned into the pAcGP67A vector (BD Biosciences) using the primer pair TTTGCGGCGGATCCCGGGAAGAGGCAGTTTGTTAATGAATG and TTCCATGCGGCCGCTCATCATCTCCTGTCATTCTGGACTGTATT into the XmaI and NotI sites, resulting in replacement of the endogenous signal sequence with the gp67 signal sequence and truncation at Arg618. The resulting coding sequence was subcloned into a pFastBac transfer vector (Life Technologies) containing a C-terminal GFP and His8 tag.
The baculovirus genome was generated by transfection into DH10Bac cells according to the manufacturer's protocols (Bac-to-Bac, Life Technologies). P1 virus was generated by infection of adherent Sf9 cells and amplified to the P3 as described in the manual. Virus titer was measured by end point dilution and quantification of infected cells by GFP fluorescence.
Protein was produced by infecting Sf9 suspension cultures in SF900-III medium (Life Technologies) at 4 × 106 cells/ml at a multiplicity of infection of 2. After 24 h, the medium was clarified by two subsequent centrifugation steps at 500 and 10,000 × g, respectively, for 20 min at 4 °C. Protein was precipitated using 20% (w/v) PEG 8000 and resuspended in a 100-fold smaller volume of 50 mm MOPS at pH 6.5, 300 mm NaCl, 5 mm CaCl2, 0.01% (v/v) Brij-35 (Buffer A) mixed with fresh cOmplete EDTA-free protease inhibitor mixture. After clarification by centrifugation at 10,000 × g for 20 min, 10 mm imidazole was added, and the buffer was agitated for 2 h with 1 ml of nickel-nitrilotriacetic acid resin (Thermo Scientific). Afterward the buffer was packed into a column and extensively washed (>20× column volumes) with Buffer A with 25 mm imidazole. PC1/3 was then eluted using a linear imidazole gradient to 500 mm, and fractions containing GFP fluorescence and PC1/3 activity were pooled, flash frozen, and stored at −80 °C (Fig. 1B). The pooled fractions had an estimated yield of 100 μg/liter and a purity of 90%. A representative enzyme progress curve for PC1/3 is depicted in Fig. 1C. Although PC1/3 is known to exhibit complex enzyme kinetics, including a lag phase in activation, we saw minimal evidence of this at the enzyme concentrations used in our studies.
Circular Dichroism Spectroscopy
Circular dichroism spectroscopy was performed on an AVIV model 215 CD spectrometer at 4 °C as described previously (32, 35). Briefly, after centrifugation at 100,000 × g for 30 min, protein concentration was adjusted to ∼0.3 mg/ml, and spectra were obtained using a 1-mm quartz cuvette for far-UV spectra. A minimum of three independent scans were acquired, and data were averaged (31, 47, 48). Structural changes due to substitutions at individual histidine residues in PROPC1/3 were monitored by plotting the changes in ellipticity at 222 nm at different pH values using the WT PROPC1/3 as a control. Equilibrium unfolding can be represented as contributions of two states, the native (N) and the unfolded (U), as described previously (32, 48) and given by Equation 1.
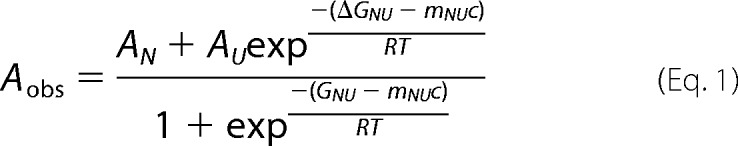 |
Alternately, the Henderson-Hasselbalch equation was used to describe the unfolding as originally described by Tanford and De (49).
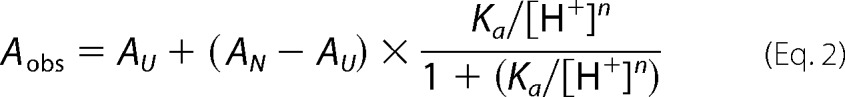 |
where n = 1 or 2 for a one-proton or two-proton model.
Activity Assays and Determination of IC50
Activity assays were performed to determine the inhibitory capabilities of the various propeptides as described previously (32, 35). Briefly, a 15 μm concentration of the fluorogenic substrate (Abz-RVKRGLA-Tyr(3-NO2)) was incubated with serially diluted amounts of either refolded WT or mutant PROPC1/3 in concentrations ranging from 0.15 to 3000 nm in activity assay buffer at pH 6.8, and 0.02 enzyme units (1 enzyme unit of enzyme produces 267 relative fluorescence units/min in the assay buffer) of mature PC1/3 was added to initiate the reaction. Activity was assayed as described above. Data were fitted and analyzed using GraphPad Prism to determine IC50 values as described previously (32, 35).
Results
Sequence and Structural Analysis of PROFUR and PROPC1/3 Orthologues
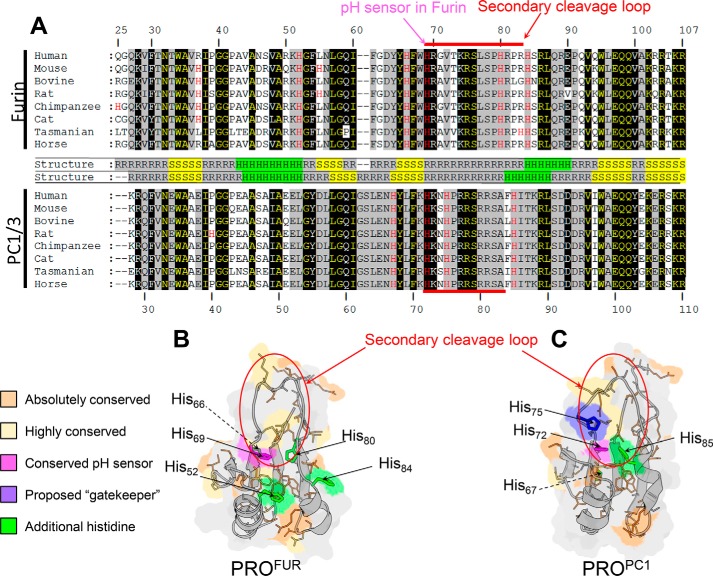
Multiple sequence alignment of PROFUR and PROPC1/3 provides several insights that serve as a starting point to generate a hypothesis about the mechanism of pH sensing in PC1/3 and how differences in sequence can encode different sensitivities to pH between the two homologues. The alignment of eight orthologues of PROFUR and PROPC1/3 indicates a high degree of sequence similarity, including stretches of completely conserved residues distributed throughout the entirety of both propeptides (Fig. 2A). A structural comparison establishes that both propeptides adopt almost identical folds and comprise two α helices packed against four β sheets with a flexible loop region containing the secondary cleavage site between helices 1 and 2 (Fig. 2, B and C). As we have previously established that the pH sensor in furin is a histidine residue located at position 69 in the sequence (45), we paid particular attention to the distribution of histidines within PROPC1/3 compared with PROFUR. Although PROFUR has histidines distributed throughout, PROPC1/3 has only four, all of which are clustered within 10 residues of the secondary cleavage site. Of these, His72 in PROPC1/3 aligns with His69, the pH sensor identified in PROFUR, and is positioned similarly at the N terminus of the cleavage loop, and thus was of primary interest. Notably, however, His72 appears more buried within the core of PROPC1/3 than His69 in the PROFUR (Fig. 2B), and His75 in PROPC1/3 that does not have a corresponding histidine in PROFUR is also located within the cleavage loop of PROPC1/3. Based on the structure (Protein Data Bank code 1KN6 (50)), His75 in PROPC1/3 is surface-exposed and appears to overlie His72, leading us to speculate that His75 may modulate the solvent accessibility of His72. Two other histidines, His67 and His85, are also located within close proximity of the cleavage site loop. His67, which corresponds to His66 in furin, is N-terminal to the cleavage loop and is situated at the interface that makes contact with the protease domain. Although His66 was proposed as a potential pH sensor (50, 51), subsequent biochemical and cell-based experiments demonstrated that this residue does not play a role in the pH-dependent activation of furin (45), making the corresponding His67 in PROPC1/3 a less likely player in the pH sensing mechanism of this protease. The remaining histidine residue (His85) does not contribute significantly to the packing of the core of the propeptide and is solvent-exposed within helix 2 located at the C-terminal side of the cleavage loop. Because local loop movement is critical for facilitating accessibility of the secondary cleavage site for proteolysis, His85 appears to be an unlikely candidate for a pH sensor, a supposition that is consistent with our preliminary IC50 values and circular dichroism data that showed the protonation at these positions does not have a significant impact on the binding, structure, or stability of the isolated PROPC1/3 (data not shown). Hence, here we focus on His72 and His75 as putative pH sensors within PROPC1/3.
FIGURE 2.
Comparative analysis of PROFUR and PROPC1/3. A, multiple sequence alignment of eight orthologues of PROFUR (top) and PROPC1/3. Names of the species are denoted on the figure, and their corresponding accession numbers for furin are as follows: human, P09958; mouse, P23188; bovine, Q28193; rat, P23377; chimpanzee, H2QA34; cat, M3W594; Tasmanian devil, G3W614; and horse, F7DHR9. Similarly, the species and their corresponding accession numbers for PC1/3 are as follows: human: P29120; mouse: P63239; bovine, Q9GLR1; rat, P28840; chimpanzee, H2QR92; cat, M3W5Q0; Tasmanian devil, G3VJH4; and horse, F6TCF0. Residues are numbered with respect to the human homologue of each peptide. Residues that are absolutely conserved in orthologues of either PROFUR or PROPC1/3 are shaded in black, whereas those residues conserved in orthologues of both PROFUR and PROPC1/3 are in yellow text with black shading. Highly conserved residues are shaded in gray. Histidine residues are highlighted in red text with the conserved pH sensor indicated by the magenta arrow. Residues comprising the secondary cleavage loop are indicated by the red bar. In between sequence alignments, secondary structures are indicated: R indicates random coil, S indicates β sheet, and H indicates α helix. B, homology model for PROFUR (32). C, solution structure (Protein Data Bank code 1KN6) of PROPC1/3. The absolutely conserved residues in B and C are colored orange, and highly conserved residues are colored yellow. The conserved pH-sensing histidines in PROFUR and PROPC1/3 are colored magenta, and additional histidines are colored green with the proposed gatekeeper histidine residue in PROPC1/3 is colored purple. The secondary cleavage loop is circled in red.
Analysis of the Role of Histidine Residues in the pH-dependent Activation of the Precursor Protein Pro-PC1/3
To examine the role that the two histidine residues (His72 and His75) may play in the pH-dependent activation of the precursor pro-PC1/3 in vivo, we first measured the ability of propeptide variants of PC1/3 to undergo activation using ER-localized constructs as described previously (Fig. 1A and Ref. 32). Here pro-PC1/3 (∼78 kDa) undergoes efficient autoprocessing to produce MATPC1/3 (∼67 kDa) that remains non-covalently associated with PROPC1/3 (∼10 kDa) to form a stable inhibition complex (32). Because the KDEL motif restricts proteins to the neutral environs of the ER, PROPC1/3 in the inhibition complex does not undergo the second autoprocessing step that is necessary for degradation and release of the propeptide. Hence, at pH values of 7.4 and 6.4, the ER extracts of MATPC1/3 failed to degrade the PROPC1/3 from the inhibition complex (Fig. 3A, inset) and displayed negligible catalytic activity against the fluorogenic peptide substrate (Fig. 3A). However, preincubation of the ER extracts of MATPC1/3 at pH 5.4 caused a robust increase in the catalytic activity (Fig. 3A) that coincided with degradation of PROPC1/3 as seen by the Western blot analysis (Fig. 3A, inset). It should be noted that this second autoprocessing event is time-dependent as degradation of the propeptide did not take place immediately upon exposure to low pH and at longer times of incubation continued to be degraded (Fig. 3E).
FIGURE 3.
pH-dependent activation of the ER-retained precursor variants of PC1/3. Activity from ER extracts of cells transfected with KDEL-tagged pro-PC1/3 after preincubation at the indicated pH is shown. The activity profiles for WT precursor activated using various pH values are shown in black bars, variants at position 72 are shown in blue, at variants at position 75 are shown in red, and variants at both positions 72 and 75 are shown in green. Histidines at these positions were mutated to leucine (A) or arginine (B). Cells transfected with empty vector (Cont) were treated identically with the results shown by gray bars. Gray dashed lines indicate the threshold for baseline activity, below which an enzyme is considered inactive. Results are the mean ± S.D. (error bars) of three independent experiments performed in triplicate. Insets show Western blots detecting the product of the primary cleavage, which results in a 9-kDa fragment of PROPC1/3. The asterisk and plus indicate the uncleaved (∼9-kDa) and cleaved (∼4-kDa) forms of WT PROPC1/3 along with the leucine and arginine variants of His72 and His75. C, enzymatic activity using ER extracts from cells transfected with WT PROPC1/3 along with H72L-PROPC1/3, H75L-PROPC1/3, and H72L/H75L-PROPC1/3 after preincubation at pH 7.4 followed by incubation in the presence or absence of trypsin. D, Western blot showing the uncleaved precursor (pro-PC1/3; 78 kDa) and MATPC1/3 (67 kDa) for the WT and histidine variants. E, time course of processing of the WT PROPC1/3 after incubation at pH 5.4 for the indicated number of minutes. The asterisk and plus indicate the uncleaved (∼9-kDa) and cleaved (∼4-kDa) forms of PROPC1/3. RFU, relative fluorescence units.
We then compared the WT pro-PC1/3 with variants in which His72 or His75 individually or both His72 and His75 together were substituted with either a leucine or arginine to mimic the constitutively deprotonated or protonated states of histidine, respectively, in accordance with our earlier work on the pH sensor in PROFUR (32, 35). Substituting either His72 or His75 with leucine allowed for pro-PC1/3 variants to undergo processing at the primary cleavage site to produce MATPC1/3 (Fig. 3D) but abrogated degradation of the histidine to leucine variants of PROPC1/3 at pH of 5.4, thereby preventing MATPC1/3 from cleaving the fluorogenic substrate (Fig. 3, A and inset). Furthermore, no additional decrease in activity was observed when both histidines were mutated to leucine.
To confirm whether MATPC1/3 obtained using the His72 or His75 PROPC1/3 variants are catalytically active, the inhibition complexes were treated with trypsin (Fig. 3C) to cleave the arginine- and lysine-rich propeptides. The results establish that the inhibition complexes, which failed to undergo autoactivation at pH 7.4, can be activated by incubation with trypsin at this pH, indicating that the protease itself is functional and that the His72 or His75 PROPC1/3 variants are incapable of undergoing pH-dependent activation.
In contrast with the leucine substitutions, when His72 or His75 were replaced with arginine, the activity of the resulting MATPC1/3 was more nuanced (Fig. 3B). Although the double mutant H72R/H75R appeared to be constitutively active at all pH values, the maximal activity of the H72R variant was only marginally greater than the control at all pH values, suggesting that protonation at this position, although necessary (as evidenced by the inability of the H72L variant to be activated), must be appropriately timed. The Western blot depicting the processing of the precursor (Fig. 3D) demonstrates that the H72R pro-PC1/3 variant did not undergo appreciable processing and suggests that inappropriate protonation of this residue may impair folding and/or processing similar to results seen in furin (45). An alternate explanation is that this mutation in the propeptide impairs autocatalytic activity, resulting in reduced propeptide excision in the ER similar to the previously reported S307L mutation, which also displayed diminished activity in trans against a peptide substrate (25). Interestingly, the H75R variant showed activity at pH 7.4 that was higher than that of the wild type and appeared to be completely active at pH 6.4, making the resulting MATPC1/3 more furin-like. Not surprisingly, the Western blot analysis established that the H75R variant of pro-PC1/3 was efficiently autoprocessed and that H75R-PROPC1/3 was completely destroyed at pH of 7.4 on a time scale that we were not able to capture in our analysis (Fig. 3, B and inset). Thus, our mutational analysis suggests that both histidines at position 72 and 75 within PROPC1/3 may be playing roles in the pH-dependent activation of pro-PC1/3 as it transits the secretory pathway. A final caveat of these studies is that retaining the proteins in the ER/cis-Golgi prevents terminal glycosylation, which has been shown to play a role in processing and secretion of mature PC1/3. The extent of glycosylation of these constructs has not been determined; however, one report suggests that it is the addition of the precursor glycan (which occurs in the ER), not the terminal trimming, that is indispensable for PC1/3 function (52).
Effect of pH on Structural Changes in PROPC1/3
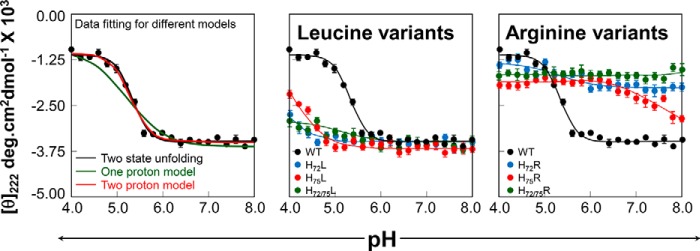
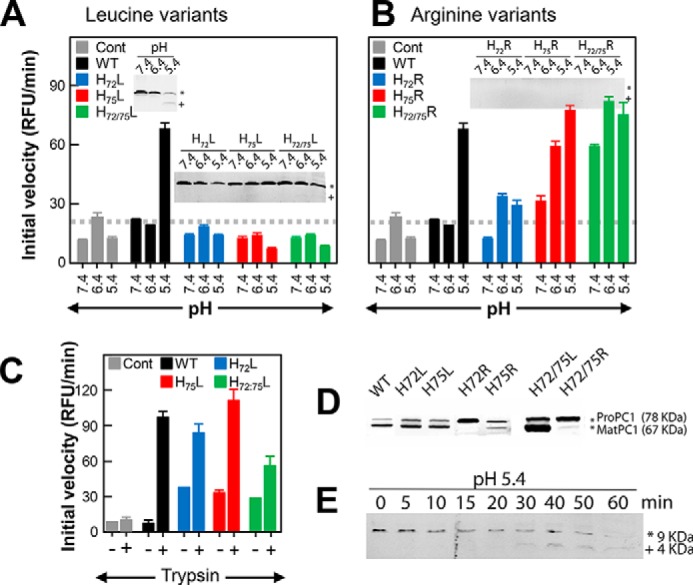
Becausethe protonation status of the pH sensor alters the structure of PROFUR (32, 35), we next analyzed how the protonation of His72 and His75 affects the secondary structure of isolated PROPC1/3 by circular dichroism spectroscopy using leucine and arginine variants as mimics for the constitutively protonated and deprotonated states, respectively. The CD spectra reveal the presence of significant secondary structure within isolated PROPC1/3 that was not significantly perturbed when the histidines were substituted by leucines (Fig. 4). This is consistent with our hypothesis that at pH 6.8 the histidine residues are in their deprotonated forms and thus demonstrates that leucine can act as a faithful representation of the constitutively deprotonated state.
FIGURE 4.
Effect of His substitution on the secondary structure of isolated PROPC1/3. Circular dichroism spectra of histidine to leucine (left) and histidine to arginine (right) variants of isolated PROPC1/3 at pH 6.8 and plotted as molar ellipticity (θ) degrees (deg)·cm2 dmol−1. Spectra for the wild type are shown as a black line (pH 6.8) or dashes (pH 4.0), mutations at position 72 are shown in blue, mutations at position 75 are shown in red, and double mutants are shown in green.
When His72 and/or His75 was replaced with arginine, an amino acid that mimics a constitutively protonated histidine, the secondary structure of the variant PROPC1/3 decreased significantly as seen from the changes in ellipticity at 222 and 208 nm (Fig. 4). It is important to note two key points. First, the effects of the H72R substitution alone were greater than the H75R substitution alone, and second, the effects of the substitutions are cumulative wherein there was a greater loss of secondary structure in the H72R/H75R double mutant than in either of the single mutants. For comparison, the spectrum of WT-PROPC1/3 at pH 4.0 is shown (Fig. 4, black dashed line). We can see that the behavior of H72R/H75R-PROPC1/3 at pH 6.8 recapitulates the behavior of the WT at pH 4.0 where we would expect both histidines to be protonated and thus the propeptide to be maximally unfolded. With respect to the shifts in the troughs of the spectra around 208 nm, an indicator of α helicity, we can see that the spectra of H75R is slightly left-shifted toward 194 nm with respect to the wild type spectra, indicating a higher percentage of random coil. In contrast, the trough in H72R is right-shifted but significantly decreased in intensity, suggesting that the although the propeptide has more α helical content it is losing overall structure; this perhaps reflects a transition from a more structured to a less structured form similar to what has been observed in subtilisin (31). The cumulative effect of the arginine substitutions can be seen in the spectra of the double mutant where the spectral trough is roughly equivalent to that of WT-PROPC1/3 at pH 4.0.
The loss in structure is consistent with our earlier observations of the in vitro catalytic activities. Notably, because the double mutant (H72R/H75R) is the only variant that exhibited constitutive activity at pH 7.4 (Fig. 3, A and B), these results strongly suggest that protonation of either His72 or His75 causes destabilization of the isolated PROPC1/3; however, there is likely a requisite role for both residues in the pH-dependent activation of PC1/3.
A limitation of the above biophysical descriptions is the fact that they are done at static pH, whereas the PCs experience a broad range of pH as they transit the secretory pathway. Therefore, to further investigate the effect of these substitutions on the structural changes in the propeptide in a physiologically relevant way, we monitored unfolding of the isolated PROPC1/3 via changes in secondary structure between pH 8.0 and pH 4.0 (Fig. 5). The WT-PROPC1/3 underwent a cooperative sigmoidal transition from a well structured state at pH 8.0 to a less structured state at pH 4.0 (Fig. 5, A and B, black points). Data were fit to various models, including two-state unfolding (black line) and the Henderson-Hasselbalch equation accounting for either a single protonation event (green line) or two protonation events (red line). In each case, the midpoint of the transition between the well structured state and the less structured state occurred at ∼pH 5.3, coincident with the pH of the later, more acidic compartment in which MATPC1/3 is activated in the secretory pathway.
FIGURE 5.
Effect of pH on the secondary structure of isolated IMCPC1/3. A, pH-induced unfolding of the isolated WT-PROPC1/3 (black dots). Unfolding was monitored by changes in ellipticity at 222 nm as a function of pH and plotted as molar ellipticity (θ) degrees (deg)·cm2 dmol−1. Data were fit to equations describing two-state unfolding (black line) or the Henderson-Hasselbalch equation for one proton (green line) or two protons (red line). B, pH-induced unfolding of histidine to leucine (left) and histidine to arginine (right) variants compared with the WT PROPC1/3 (black) with mutations at position 72 shown in blue, mutations at position 75 shown in red, and double mutants shown in green. Error bars represent S.D.
Identical treatment of PROPC1/3 variants yielded some interesting insights. At pH values above ∼6.5, the leucine variants of PROPC1/3 appeared similarly well folded as the WT PROPC1/3; however, as pH dropped, the behavior of the mutants diverged. The H72L-PROPC1/3 variant was largely stable, retaining its secondary structure at pH 5.0 and only undergoing insignificant unfolding (∼20% when compared with WT-PROPC1/3) as the pH approached 4.0 potentially due to the influence of secondary residues. Under identical conditions, the H75L variant also appeared unaffected by pH until pH 5.0 after which it lost about 60% of its secondary structure when compared with WT PROPC1/3 at pH 4.0. Thus, our results indicate that the overall pH-dependent stability of the H72L variant is greater than that of H75L as well as WT PROPC1/3 and suggest that the His72 residue contributes more toward the pH sensitivity. When the two mutations were combined, we observed that the variant PROPC1/3 was largely unaffected by the changing proton concentrations, reflecting the replacement of both histidines with nonprotonatable mimics. This suggests that although the effects of protonation and/or salt bridge formation on the structure of PROPC1/3 cannot be ruled out the histidine protonation status is the primary driver of these structural changes.
In contrast, we noted a striking effect of making the alternate substitutions, replacing histidines with positively charged arginine. Both the single H72R and double H72R/H75R exhibited significantly less secondary structure than WT-PROPC1/3 even at pH 8.0, and although the H72R variant did undergo some pH-dependent unfolding at pH ∼6.0, again suggesting a minor role for protonation of secondary residues in the lower pH range, these changes were small. The H75R exhibited the most striking of pH-dependent changes: at pH 8.0, the H75R mutant had significant secondary structure and appeared to undergo a sigmoidal transition to a less folded state with a midpoint of transition at pH ∼7.0, again making this variant the most furin-like of those examined. Taken as a whole, these data again suggest a role for both His72 and His75; in replacing both histidines with leucine, the pH-dependent loss of secondary structure could be abrogated, whereas the single substitutions merely attenuated this loss. Similarly, although replacing both residues with arginine rendered PROPC1/3 likewise impervious to changes in pH, it existed in a baseline less well folded state. The single arginine variants retained a degree of structural responsiveness to pH, undergoing structural transitions at distinct pH values, suggesting underlying differences in their pKa values.
Effect of His Substitution on the Interactions of PROPC1/3 with MATPC1/3
Although studies of the isolated PROPC1/3 are useful to understand the role of changing proton concentration on the conformational dynamics of this critical domain, ultimately PROPC1/3 exists as a complex with its cognate protease domain and exerts its regulatory effects via inhibition. Therefore, we next determined the IC50 values of PROPC1/3 and its histidine variants for MATPC1/3 (Fig. 6). The WT PROPC1/3 inhibited MATPC1/3 with an IC50 of 2.3 nm, which is consistent with previously published values (Fig. 6, A and B). Substituting either histidine singly or in combination with leucine did not significantly perturb the IC50 values of PROPC1/3 (Fig. 6B) consistent with our observation that there were not significant changes in secondary structure of these at pH 5.5 (Fig. 3A). Likewise, consistent with our earlier observations, both the single H75R and the double H72R/H75R variants were poor inhibitors with IC50 values ∼10–15-fold higher than that of WT PROPC1/3. However, inhibition studies with H72R-PROPC1/3 variant yielded some surprising results: unlike the other arginine variants, the IC50 of H72R-PROPC1/3 was similar to that of the WT PROPC1/3 (Fig. 6B) consistent with our prior observation that in vitro activity of this variant is only ∼50% of the wild type. As we are yet unable to directly examine how pH affects the PROPC1/3-MATPC1/3 inhibition complex directly because of the high concentrations of mature PC1/3 required to create stoichiometric complexes and difficulty in obtaining a truly catalytically inactive enzyme for obtaining stable complexes, we are left to speculate that perhaps there is interplay of additional residues at the interface of the protease and PROPC1/3 that allow the propeptide to remain tightly associated despite having less structure and lower stability at acidic pH.
FIGURE 6.
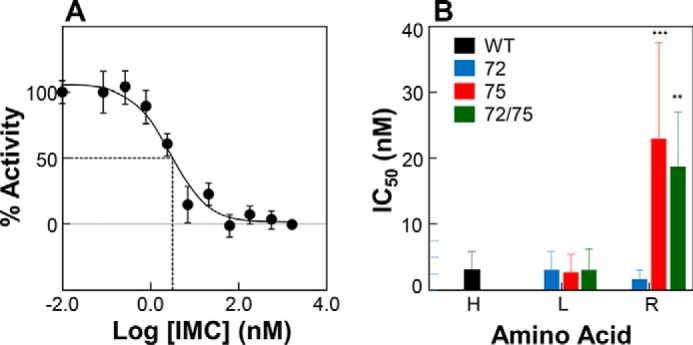
IC50 values of WT PROPC1/3 and its variants for interactions with MATPC1/3. A, representative plot of activity of WT MATPC1/3 as a function of concentration of WT-PROPC1/3. Values are mean ± S.D. (error bars) of one experiment performed in triplicate. B, IC50 values determined for WT PROPC1/3 and the histidine variants at pH 6.8 and plotted as the mean ± S.D. (error bars) of three independent experiments performed in triplicate. Those values that are statistically different from the WT are indicated by asterisks (***, p < 0.001; **, p < 0.01).
Discussion
Organellar pH is a critical regulator of a wide range of intracellular events, including zymogen activation, signaling cascades, ion channel activity, and receptor-ligand interactions. Each of these events must be tightly regulated both temporally and spatially to maintain organismal homeostasis, and many eukaryotic proteins have evolved to exploit environmental pH as a cue to regulate their activation via alterations in the protonation status of titratable amino acid side chains. Not surprisingly then, protonation with increasing organellar acidification is the major biochemical cue that regulates the final activation step of PCs. We are only just beginning to understand how the differing pH sensitivity of individual PCs is encoded and how the addition of a single proton can drive the biochemical and biophysical changes required to initiate protease activation. We have earlier demonstrated that protonation of His69 can disrupt a hydrophobic pocket within PROFUR to drive local unfolding of the secondary cleavage site loop to facilitate PROFUR proteolysis that results in furin activation (35, 45). Although His69 is conserved within all PCs, individual PCs nonetheless undergo activation at different pH values with actual proton concentrations that vary over 10-fold, suggesting that additional complexities remain to be determined.
Our results demonstrate that although histidine residues are mostly conserved within orthologues of PC1/3 and furin individually they show significant differences between the two families with only the primary pH sensor residue (His69 in PROFUR and His72 in PROPC1/3) absolutely conserved throughout the alignment (Fig. 2). The remaining histidines, His67, His75, and His85, in PROPC1/3 are located within proximity of the cleavage site loop. His67 (which corresponds to His66 in furin) is situated at the interface with the protease domain and has been demonstrated to have no influence on pH sensing in furin (45). Furthermore, preliminary studies using leucine and arginine substitutions at positions 67 and 85 in PC1/3 suggest that these variants do not affect the secondary structure of the isolated propeptide domain or their IC50 values (data not shown). It is noteworthy that (i) the counterpart to His69, which is solvent-accessible in furin, appears more buried within PC1/3; (ii) the imidazole side chain of His75 is solvent-exposed and does not have an analogous counterpart in furin; and (iii) His75 precedes a proline residue in PC1/3. Experimental studies have established that when a histidine side chain precedes a proline residue the proline isomerization rates increase up to 10-fold when the imidazole side chain is protonated relative to the deprotonated state (53, 54). In this study, we therefore analyzed the role of the conserved histidine in PC1/3 (His72) as well as that of a second histidine (His75), which we propose acts as a “gatekeeper” that regulates solvent accessibility and thus protonation of the primary pH sensing histidine.
To decipher the role of the two histidine residues in regulating the pH-dependent activation of PC1/3, we used constructs with a C-terminal KDEL sequence that retains the noncovalently bound inhibition complex within the ER; in this case, pro-PC1/3 has undergone the pH-independent primary processing subsequent to folding, but the propeptide remains associated, acting as an inhibitor of protease activity. Our results indicated that mutation of either His72 or His75 to a nonprotonatable mimic was sufficient to block the second processing event and thus activation of PC1/3 at all pH values assayed, indicating that the protonation of both histidine residues is necessary and that neither alone are sufficient to drive pH-dependent activation of PC1/3. It is noteworthy that when replaced with an arginine, a surrogate that mimics a constitutively protonated state, the H72R-PROPC1/3 did not promote activation, whereas the H75R-PROPC1/3 underwent marginal activation at pH 7.4. Although activation at pH 6.4 and 5.4 increased for both H72R-PROPC1/3 and H75R-PROPC1/3, only the double variant (H72R/H75R-PROPC1/3) mediated robust activation at all pH values. On this basis, we propose that both His72 and His75 appear to work cooperatively to mediate the pH-dependent activation of PC1/3 (Fig. 3).
The biophysical analysis of the isolated propeptides further supported the idea that both histidine residues were important in the mechanism of activation for this protease. Here, the pH-dependent structural transitions seen in the various mutant propeptides are particularly insightful. The H72L-PROPC1/3, H75L-PROPC1/3, and H72L/H75L-PROPC1/3 variants behaved similarly to WT PROPC1/3 between pH 6.0 and 8.0. However, at pH less than 6.0, the WT PROPC1/3 began to unfold with a calculated pKa of ∼5.2 when the data were fit to a two-proton model (Fig. 5A). Under these conditions, all histidine to leucine PROPC1/3 variants were significantly more stable than the WT PROPC1/3, and although the H75L-PROPC1/3 lost about 50% of its ellipticity at 222 nm when compared with WT PROPC1/3, H72L-PROPC1/3 and H72L/H75L-PROPC1/3 variants underwent only marginal unfolding (∼20%) under identical conditions (Fig. 5B). Although these results indicate that a single substitution of histidine with nonprotonatable leucine residues at position 72 or 75 blunts the ability of PROPC1/3 variant to respond to more acidic pH, the subtle differences in the behavior of the individual variants allows one to begin teasing apart the differing roles of these residues.
First, given that the single H72L variant was sufficient to attenuate the pH-dependent structural changes within the propeptide (Fig. 5B) and block pH-dependent activation (Fig. 3A), we propose that the protonation status of His72 is a major driver of the requisite unfolding of the cleavage loop of PROPC1/3. Second, the observation that the H75L underwent partial unfolding, albeit to a lesser degree than the WT PROPC1/3, indicates that although protonation of His72 is necessary it is not sufficient, and thus the combinatorial effect of protonation of both residues appears to be necessary for activation of PC1/3. As a final note, the small change in secondary structural content in the double H72R/H75R-PROPC1/3 variant was likely due to the influence of other titratable groups, which at low pH values may enhance the local changes required for propeptide processing and release, and thus further studies are needed to more fully investigate this possibility.
By introducing an arginine in place of the histidines of interest, we were able to assess the effect of a positive charge at these positions within the propeptides. Again, the differences between the variants shed light on the differing role that each of these histidines play and lends further support to the notion that there is a synergistic effect between His72 and His75 that allows for the precise spatiotemporal regulation of the loosening of secondary structure required for PROPC1/3 processing. Even at pH 8, H72R-PROPC1/3 displayed markedly less secondary structure than the WT-PROPC1/3, reflecting its role as a major driver of destabilization. There was an additional loss of α helicity as pH dropped below ∼6 either due to the aggregate effect of protonation of His75 or the influence of other side chains. The behavior of H75R-PROPC1/3 is again of particular interest: although somewhat destabilized relative to WT-PROPC1/3, it was nonetheless well structured at pH 8 when compared with H72R-PROPC1/3 and H72R/H75R-PROPC1/3. As pH was lowered, H75R-PROPC1/3 immediately began to lose secondary structure but plateaued at pH 6. This again highlights our assertion that the positive charge at either of these positions singly is not sufficient to account for the activation behavior of PC1/3 but rather that cooperative action between early protonation of His75 followed by later protonation of His72 is required.
A final piece of insight is offered by the ability of the propeptides to inhibit the protease domain (Fig. 6). As would be expected, IC50 values (at pH 6.8) for the leucine variant propeptides roughly approximated those of the wild type, whereas those for the H75R and H72R/H75R variant were ∼15-fold higher, indicating that they are notably weaker inhibitors. At first, it may seem surprising that the H72R variant has an IC50 that is comparable with that of the wild type; however, we propose that this reflects a critical role of the histidine at position 75: in addition to modulating the solvent accessibility of the hydrophobic pocket it overlays, protonation of His75 may be involved in a destabilization of the propeptide-protease complex. As we are yet unable to produce the mature protease in sufficient quantities, we are unable to more directly determine kon/koff and more thoroughly characterize the interaction between the protease and its cognate propeptide.
Taking the above as a whole, we can then propose a mechanism by which PC1/3 is activated (Fig. 7). We believe that the two histidine residues within the cleavage loop are in proximity of one another and require sequential protonation to allow for activation. His75, which sits at the top of the hydrophobic pocket, is likely protonated first and begins the local unfolding of the cleavage loop. It is tempting to speculate that protonation of His75 facilitates local unfolding by enhancing the kinetics of cis-trans isomerization of Pro76 as demonstrated by systematic studies in a pentapeptide series, Ac-Ala-Xaa-Pro-Ala-Lys-NH2 (53). These studies, which examined the influence of each of the 20 amino acids at position Xaa on the energetics of proline isomerization using NMR spectroscopy, demonstrated that the rates of proline isomerization are enhanced severalfold only when the side chains of tyrosine and histidine residues are protonated. The occurrence of histidine residues preceding a proline is higher than the overall frequency of the individual amino acids would predict (55). Thus we propose that the protonation of His75 enhances local unfolding of the loop, which allows His72 to become more solvent-accessible and simultaneously disrupt the interface between the propeptide and protease. Once solvent-accessible, His72 can then be protonated, thus delivering the second and final blow that drives the processing and dissociation of the propeptide, thus releasing inhibition from the protease. In this model, His75 acts as a gatekeeper residue, blocking access to the hydrophobic pocket in its deprotonated state, a restriction that can be removed via protonation. The model for two protonation events is also supported by CD data on the WT-PROPC1/3. Notably, when we fit the CD data from pH-dependent unfolding experiments to a modified form of the Henderson-Hasselbalch equation (49), we found that a model of two titratable groups better fit the conformational changes in PROPC1/3 in response to changing pH than one that only allowed for a single protonation (Fig. 5A). We also speculate that the close proximity of His72 and His75 may also be critical to understanding the mechanism of activation and reason that PC1/3 is only activated in acidic environs. When both residues are unprotonated, hydrophobic packing of the core of the propeptide is the major stabilizing force similar to the furin propeptide. One must ask then is it simply a greater degree of stability in the structure of PROPC1/3 that requires the stronger disrupting force of two positive charges to drive its unfolding, or does the protonation of one residue influence the protonation of the second? Concomitant work in our laboratory has been focused on defining pKa values of the histidines within PROFUR and PROPC1/3. Although the pKa values of His67, His75, and His85 are all ∼6.0, the pKa of His72 is acid-shifted with a value of ∼5.5 (56), which similar to the pH of activation of PC1/3. Therefore, we believe that the spatial arrangement and interaction of His72 and His75 can explain why PC1/3 is activated by a 10-fold higher proton concentration than its paralogue furin.
FIGURE 7.
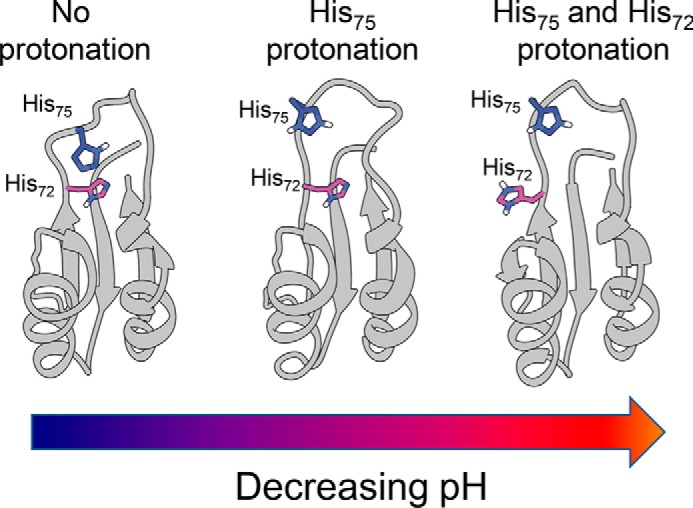
Model of histidine protonation in PROPC1/3. Two-dimensional schematic representations of the NMR solution structures of the PC1/3 propeptide (Protein Data Bank code 1KN6) displaying histidines side chains of interest are shown. Left, both histidines are deprotonated, maintaining packing of core and protecting cleavage site. Center, His75 is protonated, causing partial unfolding of the cleavage loop and exposing His72. Right, both histidines are protonated, causing cleavage loop to unfold completely and exposing cleavage site to allow processing.
This work offers significant insight into the question of how each member of the PC family encodes unique organelle-specific information about activation and processing. We believe that although the primary pH sensor (His69 in furin and His72 in PC1/3) has been evolutionarily conserved throughout PCs, concomitant with histidine enrichment has been a tuning of the pH sensitivity of these proteases. As seen in the multiple sequence alignment, only the position of the primary pH sensor has been absolutely conserved; thus domain-specific conservation as opposed to position-specific conservation seems to be crucial to encoding pH sensitivity in the propeptides of proteases in this family. Looking more broadly at secreted proteases across eukaryotes and prokaryotes, we had reported previously that there is an enrichment of histidine residues in the propeptides of secreted eukaryotic proteases that is not seen in their prokaryotic paralogues. Again, we see that although there is a bias for more histidines within this particular domain there is no appreciable bias for specific positions (33). These observations highlight the importance of spatial juxtapositioning of titratable groups in regions of proteins to tune their pH sensitivity, an evolutionary adaptation that was likely concomitant with the emergence of compartmentalization and specialization allowed by the secretory pathway within eukaryotic cells.
The issue of “mistuning” via mutation is also interesting to consider. The most obvious case to consider would be the loss of the conserved pH sensor, yielding either an inactivatable or constitutively active protease depending on the type of mutation. More interesting, however, would be the mutation of one of the surrounding residues and its effect on the local environs and protonatability of not only the primary pH sensor but also the gatekeeper or tuning residues. Evidence for the plausibility of this notion is lent by several single nucleotide polymorphisms in PROFUR and PROPC1/3. Preliminary characterization of one of these polymorphisms, ΔR80Q (57), indicates that changes in the propeptide can affect the biosynthesis, secretion, and catalytic activity similar to other previously described SNPs in the catalytic domain, and thus it is hypothesized that it may contribute to cancer (including head and neck carcinoma and lung adenocarcinoma (58)) or metabolic disease. Further characterization of these polymorphisms would be insightful not only in understanding the basis on which they are able to cause disease but also in the broader sense of understanding what role propeptides have in dictating proteolytic activity and processing. Similarly, environmental homeostasis is required for optimal proteolytic function; perturbation of cellular pH through disease (59) may cause activation of a PC prematurely or prevent activation all together. The role of the pH gradient in the secretory pathway in orchestrating proteolytic processing of substrates cannot be understated. Thus as we continue to understand the interplay among the proprotein convertases, proton concentration, and pH sensors, we hope to gain better insight into how things go wrong in the disease state and how we can better approach solutions to restore homeostasis.
Author Contributions
D. M. W. made substantial contributions to conception, design, and acquisition of data. J. E. helped with the analysis. U. S. helped draft and revise the article and helped with the analysis of data. All authors reviewed the results and approved the final version of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Ruth L. Kirschstein National Research Service Award 5F30DK096752 (to D. M. W.). This work was also supported by National Science Foundation Grant MCB0746589 (to U. S.) and American Heart Association Predoctoral Training Grant 12PRE11470005 (to J. E.). The authors declare that they have no conflicts of interest with the contents of this article.
- ER
- endoplasmic reticulum
- PC
- proprotein convertase
- PROFUR
- furin propeptide
- MATFUR
- mature protease domain of furin
- PROPC1/3
- PC1/3 propeptide
- Abz
- aminobenzoic acid.
References
- 1. Soskine M., Tawfik D. S. (2010) Mutational effects and the evolution of new protein functions. Nat. Rev. Genet. 11, 572–582 [DOI] [PubMed] [Google Scholar]
- 2. Embley T. M., Martin W. (2006) Eukaryotic evolution, changes and challenges. Nature 440, 623–630 [DOI] [PubMed] [Google Scholar]
- 3. Levin L. R., Buck J. (2015) Physiological roles of acid-base sensors. Annu. Rev. Physiol. 77, 347–362 [DOI] [PubMed] [Google Scholar]
- 4. Srivastava J., Barber D. L., Jacobson M. P. (2007) Intracellular pH sensors: design principles and functional significance. Physiology 22, 30–39 [DOI] [PubMed] [Google Scholar]
- 5. Shinde U., Thomas G. (2011) Insights from bacterial subtilases into the mechanisms of intramolecular chaperone-mediated activation of furin. Methods Mol. Biol. 768, 59–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Thomas G. (2002) Furin at the cutting edge: from protein traffic to embryogenesis and disease. Nat. Rev. Mol. Cell Biol. 3, 753–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Seidah N. G., Sadr M. S., Chrétien M., Mbikay M. (2013) The multifaceted proprotein convertases: their unique, redundant, complementary, and opposite functions. J. Biol. Chem. 288, 21473–21481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rashid S., Tavori H., Brown P. E., Linton M. F., He J., Giunzioni I., Fazio S. (2014) Proprotein convertase subtilisin kexin type 9 promotes intestinal overproduction of triglyceride-rich apolipoprotein B lipoproteins through both low-density lipoprotein receptor-dependent and -independent mechanisms. Circulation 130, 431–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fu J., Bassi D. E., Zhang J., Li T., Nicolas E., Klein-Szanto A. J. (2012) Transgenic overexpression of the proprotein convertase furin enhances skin tumor growth. Neoplasia 14, 271–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Arsenault D., Lucien F., Dubois C. M. (2012) Hypoxia enhances cancer cell invasion through relocalization of the proprotein convertase furin from the trans-Golgi network to the cell surface. J. Cell Physiol. 227, 789–800 [DOI] [PubMed] [Google Scholar]
- 11. Bassi D. E., Zhang J., Cenna J., Litwin S., Cukierman E., Klein-Szanto A. J. (2010) Proprotein convertase inhibition results in decreased skin cell proliferation, tumorigenesis, and metastasis. Neoplasia 12, 516–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Scamuffa N., Sfaxi F., Ma J., Lalou C., Seidah N., Calvo F., Khatib A. M. (2014) Prodomain of the proprotein convertase subtilisin/kexin Furin (ppFurin) protects from tumor progression and metastasis. Carcinogenesis 35, 528–536 [DOI] [PubMed] [Google Scholar]
- 13. López de Cicco R., Bassi D. E., Zucker S., Seidah N. G., Klein-Szanto A. J. (2005) Human carcinoma cell growth and invasiveness is impaired by the propeptide of the ubiquitous proprotein convertase furin. Cancer Res. 65, 4162–4171 [DOI] [PubMed] [Google Scholar]
- 14. Pasquato A., Ramos da Palma J., Galan C., Seidah N. G., Kunz S. (2013) Viral envelope glycoprotein processing by proprotein convertases. Antiviral Res. 99, 49–60 [DOI] [PubMed] [Google Scholar]
- 15. Seidah N. G., Awan Z., Chrétien M., Mbikay M. (2014) PCSK9: a key modulator of cardiovascular health. Circ. Res. 114, 1022–1036 [DOI] [PubMed] [Google Scholar]
- 16. Susan-Resiga D., Essalmani R., Hamelin J., Asselin M. C., Benjannet S., Chamberland A., Day R., Szumska D., Constam D., Bhattacharya S., Prat A., Seidah N. G. (2011) Furin is the major processing enzyme of the cardiac-specific growth factor bone morphogenetic protein 10. J. Biol. Chem. 286, 22785–22794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Roebroek A. J., Umans L., Pauli I. G., Robertson E. J., van Leuven F., Van de Ven W. J., Constam D. B. (1998) Failure of ventral closure and axial rotation in embryos lacking the proprotein convertase Furin. Development 125, 4863–4876 [DOI] [PubMed] [Google Scholar]
- 18. Zhu X., Zhou A., Dey A., Norrbom C., Carroll R., Zhang C., Laurent V., Lindberg I., Ugleholdt R., Holst J. J., Steiner D. F. (2002) Disruption of PC1/3 expression in mice causes dwarfism and multiple neuroendocrine peptide processing defects. Proc. Natl. Acad. Sci. U.S.A. 99, 10293–10298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Scamuffa N., Calvo F., Chrétien M., Seidah N. G., Khatib A. M. (2006) Proprotein convertases: lessons from knockouts. FASEB J. 20, 1954–1963 [DOI] [PubMed] [Google Scholar]
- 20. Stijnen P., Tuand K., Varga T. V., Franks P. W., Aertgeerts B., Creemers J. W. (2014) The association of common variants in PCSK1 with obesity: a HuGE review and meta-analysis. Am. J. Epidemiol. 180, 1051–1065 [DOI] [PubMed] [Google Scholar]
- 21. Benzinou M., Creemers J. W., Choquet H., Lobbens S., Dina C., Durand E., Guerardel A., Boutin P., Jouret B., Heude B., Balkau B., Tichet J., Marre M., Potoczna N., Horber F., Le Stunff C., Czernichow S., Sandbaek A., Lauritzen T., Borch-Johnsen K., Andersen G., Kiess W., Körner A., Kovacs P., Jacobson P., Carlsson L. M., Walley A. J., Jørgensen T., Hansen T., Pedersen O., Meyre D., Froguel P. (2008) Common nonsynonymous variants in PCSK1 confer risk of obesity. Nat. Genet. 40, 943–945 [DOI] [PubMed] [Google Scholar]
- 22. Blanco E. H., Peinado J. R., Martín M. G., Lindberg I. (2014) Biochemical and cell biological properties of the human prohormone convertase 1/3 Ser357Gly mutation: a PC1/3 hypermorph. Endocrinology 155, 3434–3447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Creemers J. W., Choquet H., Stijnen P., Vatin V., Pigeyre M., Beckers S., Meulemans S., Than M. E., Yengo L., Tauber M., Balkau B., Elliott P., Jarvelin M. R., Van Hul W., Van Gaal L., Horber F., Pattou F., Froguel P., Meyre D. (2012) Heterozygous mutations causing partial prohormone convertase 1 deficiency contribute to human obesity. Diabetes 61, 383–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Prabhu Y., Blanco E. H., Liu M., Peinado J. R., Wheeler M. C., Gekakis N., Arvan P., Lindberg I. (2014) Defective transport of the obesity mutant PC1/3 N222D contributes to loss of function. Endocrinology 155, 2391–2401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Farooqi I. S., Volders K., Stanhope R., Heuschkel R., White A., Lank E., Keogh J., O'Rahilly S., Creemers J. W. (2007) Hyperphagia and early-onset obesity due to a novel homozygous missense mutation in prohormone convertase 1/3. J. Clin. Endocrinol. Metab. 92, 3369–3373 [DOI] [PubMed] [Google Scholar]
- 26. Benjannet S., Rondeau N., Paquet L., Boudreault A., Lazure C., Chrétien M., Seidah N. G. (1993) Comparative biosynthesis, covalent post-translational modifications and efficiency of prosegment cleavage of the prohormone convertases PC1 and PC2: glycosylation, sulphation and identification of the intracellular site of prosegment cleavage of PC1 and PC2. Biochem. J. 294, 735–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jackson R. S., Creemers J. W., Farooqi I. S., Raffin-Sanson M. L., Varro A., Dockray G. J., Holst J. J., Brubaker P. L., Corvol P., Polonsky K. S., Ostrega D., Becker K. L., Bertagna X., Hutton J. C., White A., Dattani M. T., Hussain K., Middleton S. J., Nicole T. M., Milla P. J., Lindley K. J., O'Rahilly S. (2003) Small-intestinal dysfunction accompanies the complex endocrinopathy of human proprotein convertase 1 deficiency. J. Clin. Investig. 112, 1550–1560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jackson R. S., Creemers J. W., Ohagi S., Raffin-Sanson M. L., Sanders L., Montague C. T., Hutton J. C., O'Rahilly S. (1997) Obesity and impaired prohormone processing associated with mutations in the human prohormone convertase 1 gene. Nat. Genet. 16, 303–306 [DOI] [PubMed] [Google Scholar]
- 29. Martín M. G., Lindberg I., Solorzano-Vargas R. S., Wang J., Avitzur Y., Bandsma R., Sokollik C., Lawrence S., Pickett L. A., Chen Z., Egritas O., Dalgic B., Albornoz V., de Ridder L., Hulst J., Gok F., Aydoğan A., Al-Hussaini A., Gok D. E., Yourshaw M., Wu S. V., Cortina G., Stanford S., Georgia S. (2013) Congenital proprotein convertase 1/3 deficiency causes malabsorptive diarrhea and other endocrinopathies in a pediatric cohort. Gastroenterology 145, 138–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. López-Otín C., Bond J. S. (2008) Proteases: multifunctional enzymes in life and disease. J. Biol. Chem. 283, 30433–30437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Subbian E., Yabuta Y., Shinde U. P. (2005) Folding pathway mediated by an intramolecular chaperone: intrinsically unstructured propeptide modulates stochastic activation of subtilisin. J. Mol. Biol. 347, 367–383 [DOI] [PubMed] [Google Scholar]
- 32. Dillon S. L., Williamson D. M., Elferich J., Radler D., Joshi R., Thomas G., Shinde U. (2012) Propeptides are sufficient to regulate organelle-specific pH-dependent activation of furin and proprotein convertase 1/3. J. Mol. Biol. 423, 47–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Elferich J., Dillon S., Shinde U. (2012) Proceeding of the 6th International Conference on Bioinformatics and Biomedical Engineering, Shanghai, China, May 17–20, 2012, Abstract Number 71241, IEEE, New York [Google Scholar]
- 34. Elferich J., Williamson D. M., Krishnamoorthy B., Shinde U. (2013) Propeptides of eukaryotic proteases encode histidines to exploit organelle pH for regulation. FASEB J. 27, 2939–2945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Williamson D. M., Elferich J., Ramakrishnan P., Thomas G., Shinde U. (2013) The mechanism by which a propeptide-encoded pH sensor regulates spatiotemporal activation of furin. J. Biol. Chem. 288, 19154–19165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Creemers J. W., Vey M., Schäfer W., Ayoubi T. A., Roebroek A. J., Klenk H. D., Garten W., Van de Ven W. J. (1995) Endoproteolytic cleavage of its propeptide is a prerequisite for efficient transport of furin out of the endoplasmic reticulum. J. Biol. Chem. 270, 2695–2702 [DOI] [PubMed] [Google Scholar]
- 37. Anderson E. D., Molloy S. S., Jean F., Fei H., Shimamura S., Thomas G. (2002) The ordered and compartment-specific autoproteolytic removal of the furin intramolecular chaperone is required for enzyme activation. J. Biol. Chem. 277, 12879–12890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Anderson E. D., VanSlyke J. K., Thulin C. D., Jean F., Thomas G. (1997) Activation of the furin endoprotease is a multiple-step process: requirements for acidification and internal propeptide cleavage. EMBO J. 16, 1508–1518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Vey M., Schäfer W., Berghöfer S., Klenk H. D., Garten W. (1994) Maturation of the trans-Golgi network protease furin: compartmentalization of propeptide removal, substrate cleavage, and COOH-terminal truncation. J. Cell Biol. 127, 1829–1842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Paroutis P., Touret N., Grinstein S. (2004) The pH of the secretory pathway: measurement, determinants, and regulation. Physiology 19, 207–215 [DOI] [PubMed] [Google Scholar]
- 41. Schönichen A., Webb B. A., Jacobson M. P., Barber D. L. (2013) Considering protonation as a posttranslational modification regulating protein structure and function. Annu. Rev. Biophys. 42, 289–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zachos C., Blanz J., Saftig P., Schwake M. (2012) A critical histidine residue within LIMP-2 mediates pH sensitive binding to its ligand β-glucocerebrosidase. Traffic 13, 1113–1123 [DOI] [PubMed] [Google Scholar]
- 43. Baird F. E., Pinilla-Tenas J. J., Ogilvie W. L., Ganapathy V., Hundal H. S., Taylor P. M. (2006) Evidence for allosteric regulation of pH-sensitive System A (SNAT2) and System N (SNAT5) amino acid transporter activity involving a conserved histidine residue. Biochem. J. 397, 369–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rötzschke O., Lau J. M., Hofstätter M., Falk K., Strominger J. L. (2002) A pH-sensitive histidine residue as control element for ligand release from HLA-DR molecules. Proc. Natl. Acad. Sci. U.S.A. 99, 16946–16950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Feliciangeli S. F., Thomas L., Scott G. K., Subbian E., Hung C. H., Molloy S. S., Jean F., Shinde U., Thomas G. (2006) Identification of a pH sensor in the furin propeptide that regulates enzyme activation. J. Biol. Chem. 281, 16108–16116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yabuta Y., Subbian E., Oiry C., Shinde U. (2003) Folding pathway mediated by an intramolecular chaperone. A functional peptide chaperone designed using sequence databases. J. Biol. Chem. 278, 15246–15251 [DOI] [PubMed] [Google Scholar]
- 47. Subbian E., Yabuta Y., Shinde U. (2004) Positive selection dictates the choice between kinetic and thermodynamic protein folding and stability in subtilases. Biochemistry 43, 14348–14360 [DOI] [PubMed] [Google Scholar]
- 48. Subbian E., Williamson D.M., Shinde U. (2015) Protein folding mediated by an intramolecular chaperone: the energy landscape for unimolecular pro-subtilisin E maturation. Adv. Biosci. Biotechnol. 6, 73–88 [Google Scholar]
- 49. Tanford C., De P. K. (1961) The unfolding of β-lactoglobulin at pH 3 by urea, formamide, and other organic substances. J. Biol. Chem. 236, 1711–1715 [PubMed] [Google Scholar]
- 50. Tangrea M. A., Bryan P. N., Sari N., Orban J. (2002) Solution structure of the pro-hormone convertase 1 pro-domain from Mus musculus. J. Mol. Biol. 320, 801–812 [DOI] [PubMed] [Google Scholar]
- 51. Tangrea M. A., Alexander P., Bryan P. N., Eisenstein E., Toedt J., Orban J. (2001) Stability and global fold of the mouse prohormone convertase 1 pro-domain. Biochemistry 40, 5488–5495 [DOI] [PubMed] [Google Scholar]
- 52. Zandberg W. F., Benjannet S., Hamelin J., Pinto B. M., Seidah N. G. (2011) N-Glycosylation controls trafficking, zymogen activation and substrate processing of proprotein convertases PC1/3 and subtilisin kexin isozyme-1. Glycobiology 21, 1290–1300 [DOI] [PubMed] [Google Scholar]
- 53. Reimer U., Scherer G., Drewello M., Kruber S., Schutkowski M., Fischer G. (1998) Side-chain effects on peptidyl-prolyl cis/trans isomerisation. J. Mol. Biol. 279, 449–460 [DOI] [PubMed] [Google Scholar]
- 54. Texter F. L., Spencer D. B., Rosenstein R., Matthews C. R. (1992) Intramolecular catalysis of a proline isomerization reaction in the folding of dihydrofolate reductase. Biochemistry 31, 5687–5691 [DOI] [PubMed] [Google Scholar]
- 55. Reimer U., el Mokdad N., Schutkowski M., Fischer G. (1997) Intramolecular assistance of cis/trans isomerization of the histidine-proline moiety. Biochemistry 36, 13802–13808 [DOI] [PubMed] [Google Scholar]
- 56. Elferich J., Williamson D. M., David L. L., Shinde U. (2015) Determination of histidine pKa values in the propeptides of furin and PC1/3 using histidine hydrogen-deuterium exchange mass spectrometry. Anal. Chem. 87, 7909–7917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Pickett L. A., Yourshaw M., Albornoz V., Chen Z., Solorzano-Vargas R. S., Nelson S. F., Martín M. G., Lindberg I. (2013) Functional consequences of a novel variant of PCSK1. PLoS One 8, e55065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Forbes S. A., Beare D., Gunasekaran P., Leung K., Bindal N., Boutselakis H., Ding M., Bamford S., Cole C., Ward S., Kok C. Y., Jia M., De T., Teague J. W., Stratton M. R., McDermott U., Campbell P. J. (2015) COSMIC: exploring the world's knowledge of somatic mutations in human cancer. Nucleic Acids Res. 43, D805–D811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Weisz O. A. (2003) Organelle acidification and disease. Traffic 4, 57–64 [DOI] [PubMed] [Google Scholar]