Background: Glucan phosphatases are essential for glycogen and starch metabolism.
Results: Comparative enzymology of glucan phosphatases defines the mechanism for specific activity versus physiological glucan substrates.
Conclusion: Glucan phosphatases possess a common active site motif but unique specific activities determined by phosphatase and carbohydrate binding domains.
Significance: Defining glucan dephosphorylation is essential for understanding normal plant and animal physiology and human disease.
Keywords: carbohydrate metabolism; carbohydrate-binding protein; glycogen; Lafora disease (Lafora progressive myoclonic epilepsy, MELF); phosphatase; substrate specificity; starch
Abstract
Glucan phosphatases are central to the regulation of starch and glycogen metabolism. Plants contain two known glucan phosphatases, Starch EXcess4 (SEX4) and Like Sex Four2 (LSF2), which dephosphorylate starch. Starch is water-insoluble and reversible phosphorylation solubilizes its outer surface allowing processive degradation. Vertebrates contain a single known glucan phosphatase, laforin, that dephosphorylates glycogen. In the absence of laforin, water-soluble glycogen becomes insoluble, leading to the neurodegenerative disorder Lafora Disease. Because of their essential role in starch and glycogen metabolism glucan phosphatases are of significant interest, yet a comparative analysis of their activities against diverse glucan substrates has not been established. We identify active site residues required for specific glucan dephosphorylation, defining a glucan phosphatase signature motif (CζAGΨGR) in the active site loop. We further explore the basis for phosphate position-specific activity of these enzymes and determine that their diverse phosphate position-specific activity is governed by the phosphatase domain. In addition, we find key differences in glucan phosphatase activity toward soluble and insoluble polyglucan substrates, resulting from the participation of ancillary glucan-binding domains. Together, these data provide fundamental insights into the specific activity of glucan phosphatases against diverse polyglucan substrates.
Introduction
Glucose is stored as large polysaccharides to meet the future metabolic needs of bacteria, fungi, plants, and animals (1, 2). Plants store excess glucose in the form of starch, which is composed of the glucose polymers amylopectin and amylose (2, 3). Amylopectin is the main component of starch, and is formed from α-1,4-glycosidic-linked chains with α-1,6-branches clustered at regular intervals every 20–25 glucose units (2–4). Adjacent amylopectin chains form helices that create crystalline lamellae and cause starch to be water-insoluble, thereby allowing plants to store the maximum amount of energy in the minimum amount of volume (2, 4–6). This form of glucose storage is ideal for the predictable metabolic cycles of photosynthetic plants (2, 7). Conversely, animals, bacteria, and fungi store excess glucose in the form of glycogen, which is also formed from α-1,4-glycosidic linked chains with α-1,6-branches (1, 8). However, the glucose chains within glycogen are shorter and the branches are more evenly spaced, thus prohibiting helix formation (1, 8, 9). These properties allow glycogen to remain water-soluble, which is essential for bursts of metabolic energy in biological systems that do not perform photosynthesis (8). Therefore, each glucose storage structure is adapted to the biological system in which it functions.
Reversible starch phosphorylation by glucan dikinases and phosphatases is critical to control the solubility of starch and permit starch degradation (2, 10, 11). Plant genomes contain two identified glucan kinases, α-glucan water dikinase (GWD)3 and phosphoglucan water dikinase (PWD), and two identified glucan phosphatases, Starch EXcess4 (SEX4) and Like Sex Four2 (LSF2) (12–15). GWD and PWD phosphorylate the hydroxyl group of starch glucose at the C6- and C3-position, respectively, resulting in helical unwinding and local solubilization of the outer starch granule (16–18). This solubilization permits access to hydrolytic enzymes, but the main enzyme that degrades starch, β-amylase, is unable to degrade glucan chains past a phosphate group (19, 20). The glucan phosphatases SEX4 and LSF2 must remove these phosphate groups for cyclical starch degradation to proceed (15, 19, 21). Reversible starch phosphorylation is therefore a dynamic process, with a constant interplay between phosphorylation via glucan dikinases and dephosphorylation via glucan phosphatases (2, 10). In addition, recent work has implicated phosphorylation in starch synthesis, indicating that it may play a role in regulating the superstructure of the starch granule itself (22, 23).
SEX4 contains a chloroplast targeting peptide (cTP), dual-specificity phosphatase (DSP) domain, a carbohydrate binding module (CBM), and a carboxy-terminal (CT) motif (14, 24–26). LSF2 contains a cTP, DSP domain, and a CT motif (15). SEX4 preferentially dephosphorylates the C6-position of starch glucose and LSF2 exclusively dephosphorylates the C3-position (15, 27, 28). Arabidopsis sex4 mutants contain excess leaf starch, decreased plant growth, and an accumulation of short-chain phospho-oligosaccharides (19, 29). Conversely, lsf2 plants display normal plant growth and starch levels, but sex4/lsf2 double mutants contain an exacerbation of the starch excess phenotype compared with sex4 plants (15). These genetic data highlight the critical role glucan phosphatases play in starch metabolism and overall plant viability. Interestingly, investigators have identified an additional Arabidopsis protein that resembles a glucan phosphatase, named Like Sex Four1 (LSF1) (14, 30). Similar to SEX4, LSF1 contains a cTP, DSP, and CBM and lsf1 mutants exhibit a starch excess phenotype (14, 30). However, LSF1 has been reported to lack phosphatase activity, therefore its precise role in starch metabolism is unclear (30).
The human glucan phosphatase laforin dephosphorylates glycogen and is conserved in vertebrates (31–36). Similar to SEX4, laforin utilizes a CBM and DSP domain to achieve glucan phosphatase activity, although the domains are in the opposite orientation (37–39). The importance of laforin in glycogen metabolism is exemplified by its link to the fatal myoclonic epileptic disorder Lafora Disease (LD) (37, 40). Approximately 70% of LD patients harbor mutations in the EPM2A gene encoding laforin and these mutations are distributed throughout both the CBM and DSP domains (36, 37, 41, 42). LD is characterized by the accumulation of cytoplasmic, water-insoluble polyglucan deposits called Lafora bodies (LBs) that result from aberrant glycogen metabolism (43–47). These accumulations of insoluble LBs contain a higher concentration of mono-ester bound phosphate and longer glucose chains, physiochemical properties that are reminiscent of plant amylopectin (44–51). The mechanism for phosphate incorporation into glycogen is currently being disputed, but it is clear that the glucan phosphatase activity of laforin is essential for maintaining normal glycogen metabolism (36, 50, 51).
All three glucan phosphatases are members of the protein tyrosine phosphatase (PTP) superfamily and are classified in the heterogeneous subgroup called dual-specificity phosphatases (DSPs) that share a conserved CX5R catalytic motif (11, 52–56). Most DSPs dephosphorylate pTyr and pSer/Thr residues of proteins, while others dephosphorylate non-proteinaceous substrates such as lipids, nucleic acids, or glucans (53, 54, 56). These differences in substrate specificity are driven, in large part, by the architecture of the DSP (53, 54, 56). Recent structural work has elucidated the primary mechanism underlying the method by which all three glucan phosphatases bind glucan chains and integrate them into the catalytic site (28, 42, 57). The glucan phosphatase DSP domains all contain aromatic/hydrophobic residues that act as platforms to bind glucan chains at the active site (28, 42, 57). In addition, each glucan phosphatase also employs an ancillary glucan-binding domain. The SEX4 structure revealed that its DSP and CBM domains form a continuous binding pocket and simultaneously engage the glucan chain (28). Conversely, the DSP and CBM in laforin are spatially separated and engage glucan chains independently (42). LSF2, lacking a CBM, uses two non-catalytic secondary binding sites associated with its CT domain to interact with starch glucans at a distance of >20 Å from the DSP active site (57). These structural studies demonstrate that the glucan phosphatases are all capable of binding glucan chains via the DSP domain and ancillary glucan-binding domains, but they use distinct variations on this theme to achieve their respective glucan phosphatase functions within their own biological niches.
A comparative analysis of the basis for common glucan phosphatase activity and specific functional differences between family members has not been established. In the present study, we define key determinants of glucan phosphatase activity and position-specific activity. We also determine activity against relevant water-soluble and -insoluble polyglucan substrates. We define the role of secondary glucan-binding domains, and determine how their architectural differences translate into unique enzymatic activities. This information provides comprehensive insights into the biochemical activity of this critical family of enzymes.
Experimental Procedures
Cloning, Expression, and Purification of Recombinant Proteins
Wild type Arabidopsis thaliana SEX4 and LSF2 constructs were designed as previously described (28, 57). Both constructs (Δ89-SEX4, Δ78-LSF2) lack the chloroplast targeting peptide (cTP) along with residues up to the DSP recognition domain, and were subcloned into a pET-28b vector (Novagen) using NdeI and XhoI restriction sites to encode an N-terminal His6 tag, a thrombin cleavage site, and SEX4 or LSF2. Full-length Homo sapiens laforin was subcloned into a pET-21 (Novagen) vector using NdeI and XhoI restriction sites to encode laforin and a C-terminal His6 tag (42). SEX4ΔCBM was designed using the structure of SEX4 and was cloned via a three piece ligation with the DSP domain (residues 89–254) amplified with NdeI and KpnI restriction sites and the CT-motif (residues 338–380) amplified with KpnI and XhoI restriction sites. The fragments were then subcloned into a pET-28b vector to encode an N-terminal His6 tag, a thrombin cleavage site and SEX4ΔCBM. LSF2+CBM was synthesized (GenScript) to contain the LSF2 DSP (LSF2 residues 79–244), the SEX4 CBM (SEX4 residues 250–337), and the LSF2 CT-motif (LSF2 residues 245–282) into a pET-28b vector to encode an N-terminal His6 tag, a thrombin cleavage site, and LSF2+CBM. A point mutation of each wild type construct was created whereby the catalytic cysteine residue was mutated to a serine (C/S), an established technique that generates a catalytically inactive construct for use as a negative control in enzymatic assays (58–60). All point mutants were generated using site-directed mutagenesis (Agilent) and confirmed by DNA sequencing (ACGT, Inc).
All protein constructs were expressed and purified as previously described via the following protocol, except where noted (28, 42, 57). Protein was produced in BL21-Codon Plus Escherichia coli cells (Stratagene). Cells were grown at 37 °C in 2xYT media to an O.D.600 of 0.6–0.8, placed in ice for 20 min, induced with 1 mm isopropyl β-d-thiogalactoside, grown at 16 °C for ∼16 h, and harvested by centrifugation. Cells were lysed in 20 mm Tris-HCl (pH 7.5), 100 mm NaCl, and 2 mm dithiothreitol (DTT), centrifuged, and the proteins were purified using a Profinia IMAC column with Ni2+ beads (Bio-Rad) with a Profinia protein purification system (Bio-Rad). Proteins were eluted in lysis buffer containing 300 mm imidazole. Protein was then dialyzed in 20 mm Tris-HCl (pH 7.5), 100 mm NaCl, and 2 mm DTT overnight in the presence of thrombin. Protein was then reverse purified over the Profinia IMAC column. Protein was purified using a HiLoad 26/60 Superdex 200 size exclusion column. Laforin and laforin-W32G constructs were purified without cleavage of the His6, and protein eluted from the Profinia purification system (Bio-Rad) was immediately purified using a HiLoad 16/60 Superdex 200 size-exclusion column.
Differential Scanning Fluorimetry to Measure Protein Stability
To determine the Tm of proteins 20 μl of 10 μm protein was combined with 20 μl of Differential Scanning Fluorimetry (DSF) buffer (20 μm Tris-HCl pH 7.5, 100 mm NaCl, 2 mm DTT) containing 5x SYPRO Orange Protein Gel Stain (Invitrogen) in a QC PCR 96-well plate (Bio-Rad). Thermal denaturation of proteins was examined with a CFX96 Real-Time PCR instrument (Bio-Rad) from 20 °C to 90 °C at a rate of 1 °C/50 s. The melting temperature (Tm)was calculated by fitting the first derivative of the melting curve with a Gaussian distribution using Prism. Measurements were performed in triplicates for all protein constructs used.
Phosphatase Assays
Generic phosphatase activity assays were performed using para-nitrophenyl phosphate (pNPP) (31, 34, 61). Hydrolysis of pNPP was performed in 50 μl reactions, containing 1x phosphatase buffer (100 mm sodium acetate, 50 mm bis-Tris, 50 mm Tris-HCl, pH 7.0), 2 mm DTT, and 50 mm pNPP. To maintain reactions in the linear phase, SEX4 and LSF2 constructs were performed with 750 ng protein for 20 min and laforin constructs were performed with 200 ng protein for 20 min. Reactions were terminated by the addition of 200 μl of 250 mm NaOH and absorbance was measured at 410 nm. The assay was performed with each protein six times or more to determine specific activity.
Glucan phosphatase activity assays were performed with amylopectin using malachite green (31, 34, 61). Reactions were performed in 20 μl containing 1x phosphatase buffer (100 mm sodium acetate, 50 mm bus-Tris, 50 mm Tris-HCl, pH 7.0) and 2 mm DTT, and 45 μg amylopectin, and 150 ng protein. Amylopectin was solubilized using the acid/base treatment Roach method (62). To maintain enzymatic activity in the linear phase, SEX4 and LSF2 constructs were incubated for 60 min and laforin constructs were incubated for 20 min (11, 42). The reaction was stopped by the addition of 20 μl of 100 mm N-ethylmaleamide and 80 μl of malachite green reagent. Absorbance was measured at 620 nm. The assay was performed with each protein six times or more to determine specific activity. Malachite green assays against purified phospho-oligosaccharides were performed as above with the following modifications. Phospho-oligosaccharides were purified from potato amylopectin as described below. A concentration of phospho-oligosaccharides equivalent to 1 nmol phosphate was used in the reactions. 150 ng protein was used in each reaction. SEX4 constructs were incubated for 8 min, LSF2 constructs were incubated for 7 min, and laforin constructs were incubated for 5 min. Each assay was performed six times to determine specific activity.
Site-specific glucan phosphate activity was determined by release from 33P-labeled starch (15, 27, 28, 57). C6-33P-labeled starch was generated by purifying phosphate-free starch from the Arabidopsis sex1–3 mutant (63), phosphorylating the starch with 33P at the C6-position by incubation with GWD and [β-33P]ATP (Hartmann Analytic). The starch was then washed and phosphorylated at the C3-position by incubation with PWD and unlabeled ATP. C3-33P-labeled starch was generated in the same manner except using GWD and unlabeled ATP for the first step and PWD and [β-33P]ATP for the second resulting in 33P-label at only the C3 position. Recombinant proteins (150 ng) were incubated in dephosphorylation buffer (100 mm sodium acetate, 50 mm bis-Tris, 50 mm Tris-HCl, pH 6.5, 0.5% (v/v) Triton X-100, 1 μg/μl BSA, and 2 mm DTT) with C6- or C3-prelabeled starch (4 mg/ml) in a final volume of 150 μl on a rotating wheel for 5 min at 25 °C. For glucan phosphatase variants, protein concentration and reaction time were increased to 1 μg and 30 min, respectively. The reaction was terminated by the addition of 50 μl of 10% SDS. The reaction tubes were then centrifuged at 13,000 rpm for 5 min to pellet the starch. 33P release into 150 μl of supernatant was determined using a 1900 TR liquid scintillation counter (Packard). The assay was performed with each protein six times to determine specific activity.
Purification of Phospho-oligosaccharides from Potato Amylopectin
Ten grams of amylopectin from potato (Solanum tuberosum) starch (Sigma-Aldrich) were dissolved in 200 ml of 5 mm sodium acetate, pH 4.8, at 60 °C. Partial enzymatic degradation was achieved by addition of isoamylase (25,000 units; from Pseudomonas amyloderamosa, Sigma), pullulanase (3.6 units; from Klebsiella planticola, Megazyme), and β-amylase (1,000 units; from Hordeum vulgare; Megazyme) and incubation at 37 °C for 19 h, followed by addition of α-amylase (3,000 units; from Sus scrofa domestica pancreas, Roche) and additional 3-h incubation at 37 °C. The reaction was stopped by heating to 95 °C for 15 min. Insoluble material was removed by centrifugation for 30 min at 5,000 × g, 4 °C. The supernatant was filtered through a 0.45 μm filter (Sartorius Stedim Biotech), diluted to 1 liter with H2O, the pH was adjusted to 7.0 with NaOH, and the sample was applied at 2 ml per minute to an anion-exchange chromatography column (50 ml Q-Sepharose FF, GE Healthcare) using an ÄKTA Explorer 100 (GE Healthcare) at 4 °C. The column was washed with 2 column volumes of H2O and phospho-oligosaccharides (P-oligos) were eluted into fractions with 3 column volumes of 200 mm NaCl, 10 mm HCl at a flowrate of 5 ml per minute. Positive fractions, as determined by a reducing-end assay (64), were pooled, precipitated in 75% (v/v) ethanol for 30 min on ice and pelleted by centrifugation (30 min, 12,000 × g, 4 °C). The supernatant was removed and the pellet was air-dried and re-dissolved in 2 ml of 2 mm HEPES-KOH, pH 7.0, to a concentration of 45.5 μmol Glc equivalents per ml. P-oligos were analyzed by high-performance anion-exchange chromatography with pulsed amperometric detection (HPAEC-PAD) as described previously (19). The HPAEC-PAD profile of the purified P-oligos resembled that from sex4 mutant plants (19) with an average degree of polymerization of DP7.
Results
The Glucan Phosphatase Active Site (PTP-loop)
To define the basis for specific glucan phosphatase activity, we analyzed the active site architecture, which critically determines substrate specificity in the PTP superfamily (53, 54, 56). We first examined the sequence and structure of the CX5R active-site PTP-loop of the three known glucan phosphatases SEX4, LSF2, and laforin, along with the homologue LSF1 and the prototypical proteinaceous DSP, Vaccinia H1-related (VHR/DUSP3) (Fig. 1A) (52). In all cases, residues critical for phosphate binding and catalysis are strictly conserved including the Cys and Arg residues as well as Gly at the 3 position. There are clear differences, however, in glucan interacting CX5R motif residues between glucan and non-glucan phosphatases. The glucan phosphatases contain shorter chain Ala and Gly residues at positions 2 and 5, respectively. This conservation is in contrast to Thr/Asp and Glu/Ser in LSF1 and VHR, respectively. Additionally, position 4 is a long chain aliphatic residue in the three glucan phosphatases and an aromatic residue in the two non-glucan phosphatases.
FIGURE 1.
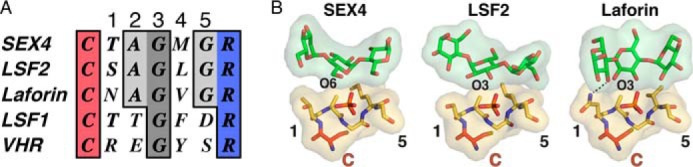
The glucan phosphatase active site. A, multiple sequence alignment of the PTP-loop from At-SEX4-(198–204), At-LSF2-(193–199), Hs-laforin-(266–272), At-LSF1-(390–396), and Hs-VHR-(124–130). The catalytic cysteine and arginine are colored red and blue, respectively. The Gly residue strictly conserved in all PTPs is colored dark gray, and residues conserved in glucan phosphatases are colored light gray. Numbers refer to the amino acid position carboxy-terminal to the catalytic cysteine. B, interactions between the PTP-loop (yellow) and glucan chain (green) in SEX4 (PDB code: 4PYH), LSF2 (PDB code: 4KYR), and laforin (PDB code: 4RKK). The catalytic cysteine (C) is colored red and the phosphate orange. The numbers highlight the residues outlined in the multiple sequence alignment shown in panel A.
Superposition of the glucan-bound structures of SEX4, LSF2, and laforin demonstrates that these positions are critical for glucan substrate engagement (Fig. 1B). The pocket formed by the strictly conserved glycine at position 5 (SEX4-Gly203, LSF2-Gly198, Laforin-Gly271) produces the deep and broad active site architecture that allows engagement of the glucose moiety immediately proximal to the phospho-glucose moiety in the catalytic pocket. The long chain aliphatic at position 4 (SEX4-Met202, LSF2-Leu197, Laforin-Val270) and alanine at position 2 (SEX4-Ala200, LSF2-Ala195, Laforin-Ala268) are both situated to form van der Waals interactions with the face of the glucan chain. Lastly, the short chain hydrophilic residue at position 1 (SEX4-Thr199, LSF2-Ser194, Laforin-Asn267) is positioned to form a hydrogen bond with a hydroxyl of the glucan substrate. Together, these data indicate a consensus for the active-site PTP-loop of glucan phosphatases: Cys, hydrophilic, Ala, Gly, long chain aliphatic, Gly, Arg (CζAGΨGR).
To test the importance of this PTP-loop consensus in determining specific glucan phosphatase activity, we produced SEX4 and LSF2 chimeras with their PTP-loop residues converted to the sequence of LSF1, SEX4 (TAGMG->TTGFD), and LSF2 (SAGLG-> TTGFD). This active site swap is predicted to have significant clashes with a potential glucan substrate due to varied sequences at position 5, conserved as a glycine in glucan phosphatases, and also position 2, conserved as an alanine in glucan phosphatases (Fig. 2A). We first tested the thermal stability of wild type and mutant proteins and found they had equivalent stabilities: 38.94 ± 0.04 °C and 38.60 ± 0.59 °C for SEX4 and 46.61 ± 0.61 °C and 47.34 ± 0.12 for LSF2 °C. Thus, altering the PTP-loop sequence did not affect global protein folding or stability. However, when tested for specific enzymatic activity both chimeric proteins were completely unable to dephosphorylate soluble amylopectin (Fig. 2B) and insoluble starch (Fig. 2C). Position 5 is strictly conserved as a glycine in all known members of the glucan phosphatase family, and very uncommon in other members of the broader DSP family. To further probe the contribution of this position to specific glucan phosphatase activity, we additionally generated the single point mutation SEX4-G203D. Strikingly, this single mutation results in loss of specific glucan phosphatase activity, indicating its importance for specific activity (Fig. 2, B and C). Taken together, these data indicate that glucan phosphatases maintain a signature PTP-loop sequence of CζAGΨGR that is critical for glucan engagement and specific glucan phosphatase activity.
FIGURE 2.
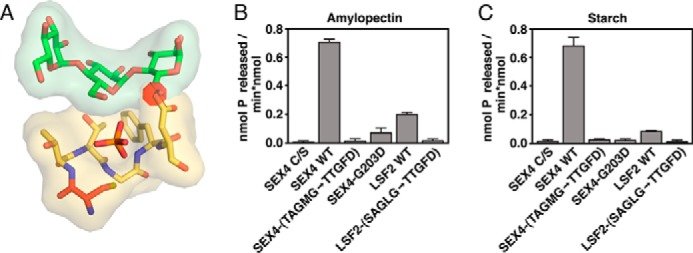
Effects of PTP-loop mutation on glucan phosphatase activity. A, model of the LSF1 PTP-loop (yellow) mutations in SEX4 (PDB code: 4PYH), showing the steric clash (red disk) with the glucan chain caused by the Asp at position 5 for LSF1. B, specific activity of SEX4, LSF2, and PTP-loop variants against soluble amylopectin. Error bars represent the ± S.D. of four replicates. Statistical comparison of wild type and mutant activities demonstrates significant differences, p < 0.001, between all constructs. C, specific activity of SEX4, LSF2, and PTP-loop variants against radiolabeled Arabidopsis starch. Error bars represent the ± S.D. of five replicates. Statistical analysis of wild type and mutant activities demonstrates significant differences, p < 0.001, between all constructs.
Generation of Chimeric Glucan Phosphatases
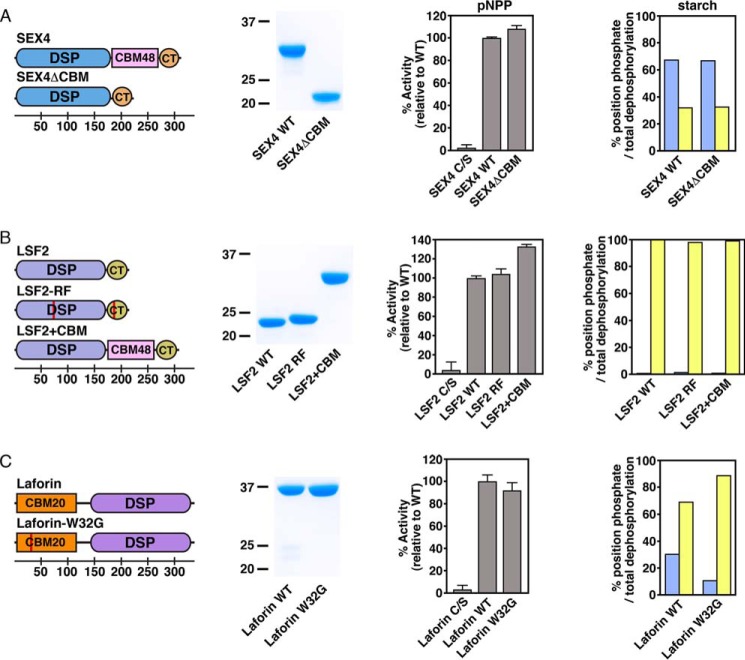
We generated an additional series of chimeric and mutant glucan phosphatase constructs for use in enzymatic analysis (Fig. 3). Three mutant constructs were developed to investigate glucan phosphatase function without the participation of the ancillary glucan-binding domains: SEX4ΔCBM, in which the CBM was removed via fusion of the DSP domain (residues 89–253) to the CT-motif (residues 338–379), LSF2-R157A/F261A (LSF2-RF), which lacks key contacts necessary for CT-motif directed glucan binding (57), and laforin-W32G, which contains an LD patient mutation within the CBM previously shown to be critical for glucan binding (42, 65). Together, these three constructs permitted us to investigate the glucan phosphatase activity and specificity derived from the DSP alone, independent from ancillary glucan-binding domains. In addition, we generated an LSF2+CBM construct in which the SEX4 CBM (residues 250–337) was inserted between the LSF2 DSP domain (residues 79–244) and CT-motif (residues 245–282) to determine how this additional glucan-binding interface affects activity and specificity.
FIGURE 3.
Ancillary glucan binding domains in glucan phosphatase position-specific activity of A) SEX4, B) LSF2, and C) Laforin. Domain organization of glucan phosphatase constructs used in this study, including SEX4ΔCBM, LSF2-RF, LSF2+CBM, and laforin-W32G (first column). Red lines represent point mutations. Bottom scale represents residue number. Coomassie-stained SDS-PAGE gel showing purified wild-type and variants (second column). Activity of glucan phosphatases and variants against para-nitrophenyl phosphate (pNPP) represented as percent activity relative to the wild type construct (third column). Error bars represent the ± S.D. of five replicates. Relative specific activity of wild type glucan phosphatases and variants at the C6 (blue) and C3 (yellow) positions of radiolabeled Arabidopsis starch represented as the percentage of total position dephosphorylation relative to total dephosphorylation (fourth column). Statistical analysis of wild type and mutant activities demonstrated no statistically significant differences, except for laforin wild type and laforin-W32G (p < 0.005).
All of the glucan phosphatase variants were expressed as soluble protein and purified to near homogeneity (Fig. 3). We tested each variant's ability to dephosphorylate para-nitrophenyl phosphate (pNPP), a generic PTP substrate. We found that each variant possesses activity comparable to the corresponding wild type constructs with no statistically significant decreases in activity (Fig. 3). Therefore, the variant proteins are functional phosphatases suitable for elucidating the relative contribution of the DSP and ancillary glucan-binding domains to specific activity and specificity.
Substrate Specificity of Glucan Phosphatases
In a physiological context, glucan phosphatases dephosphorylate carbohydrate substrates in a position-specific manner. Starch is phosphorylated at both the C6- and C3-positions (2, 11) while glycogen is phosphorylated at the C6-, C3-, and C2-positions (50, 51, 66). Therefore, we tested the physical basis for position-specific activity of glucan phosphatases. Previous studies have established that SEX4 preferentially dephosphorylates the C6-position of starch. We found that the protein lacking the CBM domain maintained position-specific activity (Fig. 3A). Similarly, LSF2 exclusively dephosphorylates the C3-position of starch glucans (15), and removal of the ancillary glucan binding domains did not affect the position-specific activity (Fig. 3B). Moreover, the addition of a glucan binding domain also did not affect specificity, since LSF2+CBM showed no change (Fig. 3B). The substrate specificity of laforin has not previously been determined. We found that laforin preferentially dephosphorylates the C3-position, the inverse of what is seen with SEX4 (Fig. 3C). These data are consistent with the C3-specific orientation of the glucan chain observed in the recently reported laforin glucan-bound structure (42). The only significant change in preference was observed for laforin, where the laforin-W32G mutant exhibited slightly greater C3-specificity than wild type (Fig. 3C). Cumulatively, these data indicate that glucan phosphatase site-specific activity is determined by the DSP domain.
Comparative Glucan Phosphatase Activity Against Soluble Amylopectin and Phospho-oligosaccharides
Data involving these chimeric proteins led to the question of the role of ancillary glucan-binding domains, found in all known glucan phosphatases (28, 42, 57, 65). Laforin and SEX4 each contain a CBM and LSF2 contains two non-catalytic glucan binding sites associated with its CT-motif. These domains bind glucans independently from glucan-interacting platforms within the DSP domain itself (28, 42, 57). We hypothesized that these domains may be critical to glucan dephosphorylation against water-soluble versus -insoluble substrates among the different glucan phosphatases.
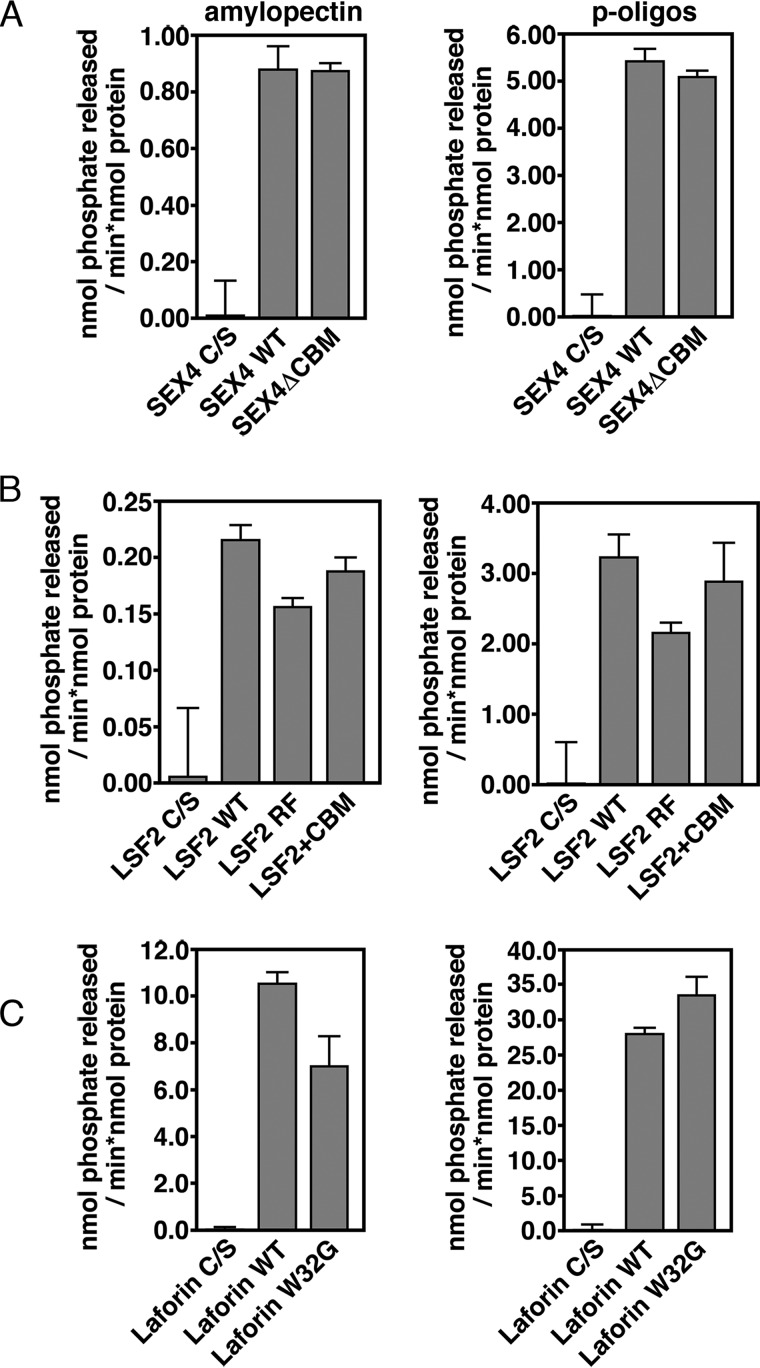
We first compared glucan phosphatase activity against solubilized potato amylopectin, which has a relatively high level of glucose-bound phosphate estimated at 1 phosphate molecule per 317 glucose units (67). The amylopectin was solubilized and phosphate release by glucan phosphatases measured. SEX4 (0.88 ± 0.08 nmol Pi released × min−1 × nmol protein−1), LSF2 (0.22 ± 0.01 nmol Pi released × min−1 × nmol protein−1), and laforin (10.6 ± 0.4 nmol Pi released × min−1 × nmol protein−1), all displayed robust specific activity (Fig. 4, A–C), consistent with previous studies (61).
FIGURE 4.
Glucan phosphatase activity against non-crystalline, water-soluble polyglucan substrates. Activity of A) SEX4, B) LSF2, and C) Laforin wild-type and variants against solubilized amylopectin (left) and soluble phospho-oligosaccharides purified from potato (right). Error bars represent the ± S.D. of five replicates. Statistical analysis of wild type and mutant activities demonstrates significant differences (p < 0.005), between all constructs except between SEX4 wild type and SEX4ΔCBM with amylopectin and LSF2 wild type and LSF2+CBM for soluble phospho-oligosaccharides.
We then tested the ability for the ancillary glucan-binding variants to dephosphorylate water-soluble amylopectin (Fig. 4). Surprisingly, we found that removal of the ancillary glucan-binding interfaces had little effect on their specific activity. Strikingly, SEX4ΔCBM had effectively the same activity against water-soluble amylopectin as wild type SEX4, with no statistically significant decrease (Fig. 4A). LSF2-RF, LSF2+CBM, and laforin-W32G showed moderate but significant decreases in activity compared with their respective wild type counterparts, ranging from 12–33% (Fig. 4, B and C).
We also compared the ability of the glucan phosphatases to dephosphorylate short-chain, soluble phospho-oligosaccharides. The relative activity of the wild type glucan phosphatases against soluble phospho-oligosaccharides was similar to that seen against amylopectin, with SEX4 (5.5 ± 0.2 nmol Pi released × min−1 × nmol protein−1), LSF2 (3.2 ± 0.3 nmol Pi released × min−1 × nmol protein−1), and laforin (28.1 ± 0.8 nmol Pi released × min−1 × nmol protein−1) all showing robust specific activity (Fig. 4, A–C). Similarly, the activity of the glucan phosphatase variants with altered glucan-binding domains showed activity levels equivalent to that observed for wild-type proteins (Fig. 4, A–C). These results indicate that the DSP domain is necessary and sufficient for specific activity against water-soluble polyglucan substrates, while the ancillary glucan-binding domains are dispensable.
Glucan Phosphatase Activity Against Insoluble Starch
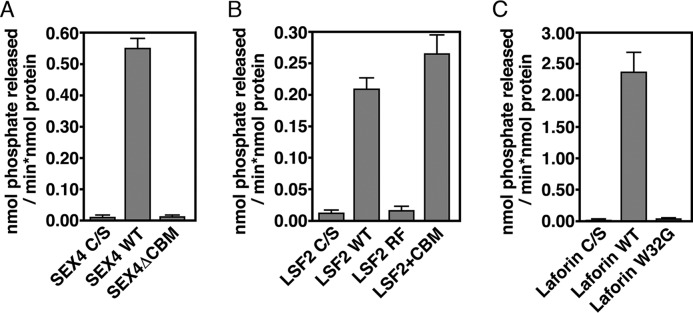
We next performed a comparative enzymatic analysis against the complex water-insoluble glucan substrate starch. Starch differs significantly from the soluble polyglucan substrates due to the glucan chains forming helices and semi-crystalline lamellae, which directly contribute to its insolubility (2, 3). Phosphatase activity was determined using 33P-radiolabeled starch. All three wild type glucan phosphatases showed robust specific activity versus starch: SEX4 (0.55 ± 0.03 nmol Pi released × min−1 × nmol protein−1), LSF2 (0.21 ± 0.02 nmol Pi released × min−1 × nmol protein−1), and laforin (2.38 ± 0.31 nmol Pi released × min−1 × nmol protein−1) (Fig. 5). As seen with soluble glucan substrates laforin again had the highest absolute activity indicating that the overall trend in total activity between all three glucan phosphatases is consistent between soluble and insoluble polyglucan substrates.
FIGURE 5.
Glucan phosphatase activity against the crystalline, water-insoluble polyglucan substrate starch. Activity of A) SEX4, B) LSF2, and C) Laforin wild-type and variants against radiolabeled Arabidopsis starch. Error bars represent the ± S.D. of six replicates. Statistical analysis demonstrates a significant difference between wild type and mutant for all variants (p < 0.005).
Next, we tested the glucan phosphatase variants to determine the contribution of ancillary glucan-binding domains on specific activity of an insoluble glucan (Fig. 5). In striking contrast to soluble glucan substrates, we saw a dramatic effect. SEX4ΔCBM activity was reduced 97%, comparable to the level of background. LSF2-RF activity against starch was also drastically reduced by 91%. Interestingly, LSF2+CBM had a statistically significant increase in activity of 26% against starch compared with wild type, indicating increased interaction facilitated by the glucan-binding domain. Laforin-W32G activity was also dramatically reduced by 98% compared with wild type. In contrast to soluble polyglucan substrates, these data demonstrate the necessary role for ancillary glucan-binding domain for the specific activity of glucan phosphatases against insoluble polyglucan substrates.
Discussion
Glucan phosphatases are critical for regulating the metabolism of complex carbohydrates in plants and animals, influencing the utilization and solubility of starch and glycogen, respectively (11, 36). The current study defines the comparative enzymology of glucan phosphatases in terms of specific activity against both water-soluble and -insoluble polyglucan substrates (Fig. 6). We also identify both key common elements of glucan phosphatase enzymology and intriguing differences between family members.
FIGURE 6.
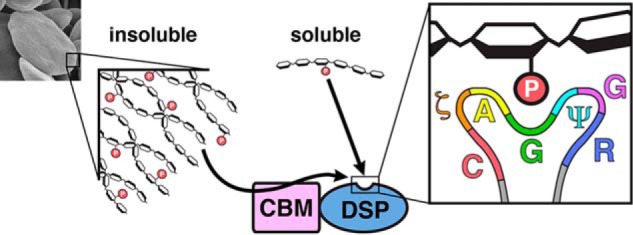
Summary of glucan phosphatase activity against polyglucan substrates. Soluble phospho-glucan substrates (phospho-oligosaccharrides and solubilized amylopectin) are dephosphorylated directly by the DSP-domain. Complex, insoluble polyglucan substrates (starch, image at left) must be engaged by the CBM before dephosphorylation can occur by the DSP-domain. The inset at right depicts the glucan phosphatase PTP-loop, which contains a CζAGΨGR motif necessary for engaging glucans at the catalytic site.
Our results demonstrate that the PTP-loop of glucan phosphatases forms a conserved structural motif required for engagement of glucan substrates. We identify a glucan phosphatase PTP-loop signature sequence of CζAGΨGR that is uniquely adapted for engaging glucan chains and directing them toward the active site (Fig. 6). This signature motif not only helps to define bona fide glucan phosphatases but also explains the lack of LSF1 activity. Although LSF1 contains both phosphatase and carbohydrate binding domains it does not possess the identified CζAGΨGR motif and thus lacks glucan phosphatase activity (11). Additionally, previous studies have identified the lack of a histidine residue N-terminal to the catalytic cysteine (11, 30). While LSF1 binds starch granules efficiently, these data together strongly indicate that its target substrate is not the starch glucans themselves as is the case with both SEX4 and LSF2.
We find that the DSP domain is responsible for engagement and dephosphorylation of glucan substrates in an orientation-specific fashion. The three glucan phosphatases show significantly different total activity and position-specific preference. SEX4 shows a C6 preference, laforin shows a C3 preference, and LSF2 is C3 specific. Previously, the substrate specificity of laforin was entirely unknown. Phosphate monoesters have been reported in glycogen at the C6-, C3-, and C2-positions (50, 51, 66). We have determined that laforin preferentially dephosphorylates the C3-position, consistent with both structural studies (42) and the observation that C6-phosphates represent a minority (20%) of glycogen-bound phosphate (66). Further, the W32G Lafora disease mutant showed both reduced activity as well as an alteration in C3/C6 specificity, with increased C3-specificity. These data suggest that phosphorylation of the different positions may have differential effects in animals, as has previously been demonstrated in plants (15). Thus, glycogen phosphorylated on different hydroxyl positions could be analogous to phosphoinositols where phosphorylation of different hydroxyls results in diverse downstream effects. Future studies will be necessary to understand this additional dimension of glycogen metabolism and the molecular pathology of LD.
The present study also demonstrates that the DSP domain of glucan phosphatases is sufficient for specific activity against water-soluble polyglucans (Fig. 6). These glucans are freely soluble and lack the higher-ordered crystalline features of starch. Thus, the rapid diffusion of the polyglucan and glucan phosphatase in solution likely enables the glucan-interacting motifs present at the active site to readily engage and remove phosphate groups. While the ancillary binding domains are not required for activity against soluble glucan substrates, we establish that the ancillary binding domains are absolutely critical for activity against insoluble starch (Fig. 6). This difference highlights the distinct environment of glucans at the outer starch granule, which has physiochemical properties unfavorable to enzymatic modification as a crucial feature of its cellular function. In this water-insoluble microenvironment, the ancillary glucan-binding domains are necessary for glucan binding and dephosphorylation.
We establish that laforin has a significantly higher level of activity than the other glucan phosphatases, which may be related to the function of laforin in preventing the accumulation of neuro-toxic, insoluble Lafora bodies. The higher specific activity of laforin may arise from cooperativity, as recent evidence demonstrates that laforin forms a functional dimer (42, 68). Additionally, activity data from the LD patient mutation W32G significantly advances our understanding of the mechanism of laforin mutations. Strikingly, there is an increasing loss of activity for the W32G mutation comparing amylopectin (33% loss), to glycogen (57% loss) (42), to starch (98% loss). Thus, the CBM domain has an enhanced contribution to activity as the carbohydrate increases in complexity and/or insolubility. These differences have significant implications for laforin's role in human disease, since it has been demonstrated that LBs are insoluble starch-like inclusions. Moreover, laforin preferentially binds insoluble starch-like Lafora bodies compared with glycogen (65, 69, 70). These data suggest that laforin requires its ancillary glucan-binding domain to avidly engage and dephosphorylate nascent LBs, which are aberrant starch-like polyglucan substrates. Multiple LD patient point mutations, including W32G, exist in the laforin CBM, suggesting a common mechanism for this class of mutations.
The lower activity of SEX4 and LSF2, relative to laforin, is likely to be advantageous in their biological context. The current model of reversible starch phosphorylation describes a complex interplay involving glucan phosphorylation, amylases, and glucan dephosphorylation, which are essential steps for efficient solubilization and degradation (2, 11). Changes in the dynamics between phosphorylation and dephosphorylation in vivo impact both starch synthesis and catabolism, including the inability to degrade starch and alterations in granule morphology (13, 15, 19, 22, 30, 63, 71–73). Thus, the lower glucan phosphatase activity of SEX4 and LSF2 may provide an essential kinetic mechanism controlling the balance between phosphorylation and dephosphorylation to permit degradation before dephosphorylation drives the outer starch granule back into an insoluble state.
Plants store insoluble starch and animals store soluble glycogen. While plants store insoluble starch, sex4-3 plants accumulate soluble phospho-oligosaccharides whose role in normal plant metabolism is unclear. Humans accumulate aberrant polyglucosan bodies in both LD and during the aging process, as corpora amylacea is observed in multiple tissues and accumulates with age (74). Thus, there are many avenues to explore the role of glucan (de)phosphorylation in both plant and animal systems. The differences observed between position-specific glucan phosphatase activity against soluble and insoluble substrates opens up new avenues for understanding glucan phosphatase function and provides a critical variable to consider when measuring and comparing glucan phosphatase activity.
Author Contributions
D. A. M., M. S. G., and C. W. V. K. designed experiments, interpreted data, and wrote the manuscript. D. A. M. generated constructs, performed mutagenesis, purified protein, and performed experiments. M. R., K. D. A., and B. D. T. purified protein and performed experiments. M. R. interpreted data. D. S. and O. K. purified starch from gwd-deficient plants and interpreted data. All authors edited and approved the final version of the manuscript.
Acknowledgments
We thank Drs. Samuel Zeeman, Martin Chow, and Yvonne Fondufe-Mittendorf and members of the Gentry laboratory for fruitful discussion and technical assistance. The contents are solely the responsibility of the authors and do not necessarily represent the official views of the NSF, NIH, or other funding agencies.
This work was supported by National Institutes of Health Grants R01NS070899 (to M. S. G), P20GM103486 (to M. S. G. and C. W. V. K.); KSEF grants KSEF-2268RDE-014 and KSEF-2971-RDE-017 (to M. S. G.); Mitzutani Foundation for Glycoscience Award (to M. S. G.); NSF Grants IIA-1355438 (to M. S. G.) and MCB-1252345 (to M. S. G.); the ETH-Zürich (to O. K.), and Swiss-South African Joint Research Program Grant IZ LS X3122916 (to O. K.); Swiss National Science Foundation SNSF-Grant 31003A_147074 (to D. S.). M. S. G. and C. W. V. K. are founders of OptiMol Enzymes LLC.
- GWD
- α-glucan water dikinase
- SEX4
- Starch EXcess4
- LSF2
- Like Sex Four2
- PWD
- phosphoglucan water dikinase
- DSP
- dual-specificity phosphatase
- CBM
- carbohydrate binding module
- CT
- carboxy-terminal motif
- LSF1
- Like Sex Four1
- LD
- Lafora Disease
- LB
- Lafora Body
- PTP
- protein tyrosine phosphatase
- pNPP
- para-nitrophenyl phosphate
- VHR
- vaccinia H1-related phosphatase
- LSF2-RF
- LSF2-R157A/F261A.
References
- 1. Roach P. J., Depaoli-Roach A. A., Hurley T. D., Tagliabracci V. S. (2012) Glycogen and its metabolism: some new developments and old themes. Biochem. J. 441, 763–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Streb S., Zeeman S. C. (2012) Starch metabolism in Arabidopsis. Arabidopsis Book 10, e0160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tester R. F., Karkalas J., Qi X. (2004) Starch - Composition, fine structure and architecture. J. Cereal Sci. 39, 151–165 [Google Scholar]
- 4. Buléon A., Colonna P., Planchot V., Ball S. (1998) Starch granules: structure and biosynthesis. Int. J. Biol. Macromol. 23, 85–112 [DOI] [PubMed] [Google Scholar]
- 5. Gallant D. J., Bouchet B., Baldwin P. M. (1997) Microscopy of starch: Evidence of a new level of granule organization. Carbohydr. Polym. 32, 177–191 [Google Scholar]
- 6. Blennow A., Engelsen S. B. (2010) Helix-breaking news: fighting crystalline starch energy deposits in the cell. Trends Plant Sci. 15, 236–240 [DOI] [PubMed] [Google Scholar]
- 7. Zeeman S. C., Smith S. M., Smith A. M. (2007) The diurnal metabolism of leaf starch. Biochem. J. 401, 13–28 [DOI] [PubMed] [Google Scholar]
- 8. Meléndez R., Mélendez-Hevia E., Cascante M. (1997) How did glycogen structure evolve to satisfy the requirement for rapid mobilization of glucose? A problem of physical constraints in structure building. J. Mol. Evol. 45, 446–455 [DOI] [PubMed] [Google Scholar]
- 9. Meléndez-Hevia E., Waddell T. G., Shelton E. D. (1993) Optimization of molecular design in the evolution of metabolism: the glycogen molecule. Biochem. J. 295, 477–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kötting O., Kossmann J., Zeeman S. C., Lloyd J. R. (2010) Regulation of starch metabolism: the age of enlightenment? Curr. Opin. Plant Biol. 13, 321–329 [DOI] [PubMed] [Google Scholar]
- 11. Silver D. M., Kotting O., Moorhead G. B. (2014) Phosphoglucan phosphatase function sheds light on starch degradation Trends Plant Sci. 7, 471–478 [DOI] [PubMed] [Google Scholar]
- 12. Ritte G., Lloyd J. R., Eckermann N., Rottmann A., Kossmann J., Steup M. (2002) The starch-related R1 protein is an alpha -glucan, water dikinase. Proc. Natl. Acad. Sci. U.S.A. 99, 7166–7171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kötting O., Pusch K., Tiessen A., Geigenberger P., Steup M., Ritte G. (2005) Identification of a novel enzyme required for starch metabolism in Arabidopsis leaves. The phosphoglucan, water dikinase. Plant Physiol. 137, 242–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fordham-Skelton A. P., Chilley P., Lumbreras V., Reignoux S., Fenton T. R., Dahm C. C., Pages M., Gatehouse J. A. (2002) A novel higher plant protein tyrosine phosphatase interacts with SNF1-related protein kinases via a KIS (kinase interaction sequence) domain. The Plant J. 29, 705–715 [DOI] [PubMed] [Google Scholar]
- 15. Santelia D., Kötting O., Seung D., Schubert M., Thalmann M., Bischof S., Meekins D. A., Lutz A., Patron N., Gentry M. S., Allain F. H., Zeeman S. C. (2011) The phosphoglucan phosphatase like sex four2 dephosphorylates starch at the c3-position in Arabidopsis. Plant Cell 23, 4096–4111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ritte G., Heydenreich M., Mahlow S., Haebel S., Kötting O., Steup M. (2006) Phosphorylation of C6- and C3-positions of glucosyl residues in starch is catalysed by distinct dikinases. FEBS Lett. 580, 4872–4876 [DOI] [PubMed] [Google Scholar]
- 17. Edner C., Li J., Albrecht T., Mahlow S., Hejazi M., Hussain H., Kaplan F., Guy C., Smith S. M., Steup M., Ritte G. (2007) Glucan, water dikinase activity stimulates breakdown of starch granules by plastidial beta-amylases. Plant Physiol. 145, 17–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hejazi M., Fettke J., Haebel S., Edner C., Paris O., Frohberg C., Steup M., Ritte G. (2008) Glucan, water dikinase phosphorylates crystalline maltodextrins and thereby initiates solubilization. Plant J. 55, 323–334 [DOI] [PubMed] [Google Scholar]
- 19. Kötting O., Santelia D., Edner C., Eicke S., Marthaler T., Gentry M. S., Comparot-Moss S., Chen J., Smith A. M., Steup M., Ritte G., Zeeman S. C. (2009) STARCH-EXCESS4 is a laforin-like Phosphoglucan phosphatase required for starch degradation in Arabidopsis thaliana. Plant Cell 21, 334–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Takeda Y., Hizukuri S. (1981) Studies on starch phosphate. Part 5. Reexamination of the action of sweet-potato beta-amylase on phophorylated (1->4)-a-D-glucan. Carbohydr. Res. 39, 151–165 [Google Scholar]
- 21. Gentry M. S., Dixon J. E., Worby C. A. (2009) Lafora disease: insights into neurodegeneration from plant metabolism. Trends Biochem. Sci. 34, 628–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hejazi M., Mahlow S., Fettke J. (2014) The glucan phosphorylation mediated by alpha-glucan, water dikinase (GWD) is also essential in the light phase for a functional transitory starch turn-over. Plant Signal Behav. 9, e28892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Skeffington A. W., Graf A., Duxbury Z., Gruissem W., Smith A. M. (2014) Glucan, Water Dikinase Exerts Little Control over Starch Degradation in Arabidopsis Leaves at Night. Plant Physiol. 165, 866–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kerk D., Conley T. R., Rodriguez F. A., Tran H. T., Nimick M., Muench D. G., Moorhead G. B. (2006) A chloroplast-localized dual-specificity protein phosphatase in Arabidopsis contains a phylogenetically dispersed and ancient carbohydrate-binding domain, which binds the polysaccharide starch. Plant J. 46, 400–413 [DOI] [PubMed] [Google Scholar]
- 25. Niittylä T., Comparot-Moss S., Lue W.-L., Messerli G., Trevisan M., Seymour M. D. J., Gatehouse J. A., Villadsen D., Smith S. M., Chen J., Zeeman S. C., Smith A. M. (2006) Similar protein phosphatases control starch metabolism in plants and glycogen metabolism in mammals. J. Biol. Chem. 281, 11815–11818 [DOI] [PubMed] [Google Scholar]
- 26. Vander Kooi C. W., Taylor A. O., Pace R. M., Meekins D. A., Guo H. F., Kim Y., Gentry M. S. (2010) Structural basis for the glucan phosphatase activity of Starch Excess4. Proc. Natl. Acad. Sci. U.S.A. 107, 15379–15384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hejazi M., Fettke J., Kötting O., Zeeman S. C., Steup M. (2010) The Laforin-like dual-specificity phosphatase SEX4 from Arabidopsis hydrolyzes both C6- and C3-phosphate esters introduced by starch-related dikinases and thereby affects phase transition of alpha-glucans. Plant Physiol. 152, 711–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Meekins D. A., Raththagala M., Husodo S., White C. J., Guo H. F., Kötting O., Vander Kooi C. W., Gentry M. S. (2014) Phosphoglucan-bound structure of starch phosphatase Starch Excess4 reveals the mechanism for C6 specificity. Proc. Natl. Acad. Sci. U.S.A. 111, 7272–7277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zeeman S. C., Northrop F., Smith A. M., Rees T. (1998) A starch-accumulating mutant of Arabidopsis thaliana deficient in a chloroplastic starch-hydrolysing enzyme. Plant J. 15, 357–365 [DOI] [PubMed] [Google Scholar]
- 30. Comparot-Moss S., Kötting O., Stettler M., Edner C., Graf A., Weise S. E., Streb S., Lue W. L., MacLean D., Mahlow S., Ritte G., Steup M., Chen J., Zeeman S. C., Smith A. M. (2010) A putative phosphatase, LSF1, is required for normal starch turnover in Arabidopsis leaves. Plant Physiol. 152, 685–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Worby C. A., Gentry M. S., Dixon J. E. (2006) Laforin, a dual specificity phosphatase that dephosphorylates complex carbohydrates. J. Biol. Chem. 281, 30412–30418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tagliabracci V. S., Turnbull J., Wang W., Girard J. M., Zhao X., Skurat A. V., Delgado-Escueta A. V., Minassian B. A., Depaoli-Roach A. A., Roach P. J. (2007) Laforin is a glycogen phosphatase, deficiency of which leads to elevated phosphorylation of glycogen in vivo. Proc. Natl. Acad. Sci. U.S.A. 104, 19262–19266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tagliabracci V. S., Girard J. M., Segvich D., Meyer C., Turnbull J., Zhao X., Minassian B. A., Depaoli-Roach A. A., Roach P. J. (2008) Abnormal metabolism of glycogen phosphate as a cause for Lafora disease. J. Biol. Chem. 283, 33816–33825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gentry M. S., Dowen R. H. 3rd, Worby C. A., Mattoo S., Ecker J. R., Dixon J. E. (2007) The phosphatase laforin crosses evolutionary boundaries and links carbohydrate metabolism to neuronal disease. J. Cell Biol. 178, 477–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gentry M. S., Pace R. M. (2009) Conservation of the glucan phosphatase laforin is linked to rates of molecular evolution and the glucan metabolism of the organism. BMC Evol. Biol. 9, 138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gentry M. S., Romá-Mateo C., Sanz P. (2013) Laforin, a protein with many faces: glucan phosphatase, adapter protein, et alii. FEBS J. 280, 525–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Minassian B. A., Lee J. R., Herbrick J. A., Huizenga J., Soder S., Mungall A. J., Dunham I., Gardner R., Fong C. Y., Carpenter S., Jardim L., Satishchandra P., Andermann E., Snead O. C. 3rd, Lopes-Cendes I., Tsui L. C., Delgado-Escueta A. V., Rouleau G. A., Scherer S. W. (1998) Mutations in a gene encoding a novel protein tyrosine phosphatase cause progressive myoclonus epilepsy. Nat. Genet. 20, 171–174 [DOI] [PubMed] [Google Scholar]
- 38. Serratosa J. M., Gómez-Garre P., Gallardo M. E., Anta B., de Bernabé D. B., Lindhout D., Augustijn P. B., Tassinari C. A., Malafosse R. M., Topcu M., Grid D., Dravet C., Berkovic S. F., de Córdoba S. R. (1999) A novel protein tyrosine phosphatase gene is mutated in progressive myoclonus epilepsy of the Lafora type (EPM2). Hum. Mol. Genet. 8, 345–352 [DOI] [PubMed] [Google Scholar]
- 39. Wang J., Stuckey J. A., Wishart M. J., Dixon J. E. (2002) A unique carbohydrate binding domain targets the lafora disease phosphatase to glycogen. J. Biol. Chem. 277, 2377–2380 [DOI] [PubMed] [Google Scholar]
- 40. Minassian B. A. (2001) Lafora's disease: towards a clinical, pathologic, and molecular synthesis. Pediatr. Neurol. 25, 21–29 [DOI] [PubMed] [Google Scholar]
- 41. Minassian B. A., Ianzano L., Meloche M., Andermann E., Rouleau G. A., Delgado-Escueta A. V., Scherer S. W. (2000) Mutation spectrum and predicted function of laforin in Lafora's progressive myoclonus epilepsy. Neurology 55, 341–346 [DOI] [PubMed] [Google Scholar]
- 42. Raththagala M., Brewer M. K., Parker M. W., Sherwood A. R., Wong B. K., Hsu S., Bridges T. M., Paasch B. C., Hellman L. M., Husodo S., Meekins D. A., Taylor A. O., Turner B. D., Auger K. D., Dukhande V. V., Chakravarthy S., Sanz P., Woods V. L. Jr., Li S., Vander Kooi C. W., Gentry M. S. (2015) Structural mechanism of laforin function in glycogen dephosphorylation and lafora disease. Mol. Cell 57, 261–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lafora G. R. (1911) Uber des Vorkommen amyloider KJrperchen im innern der Ganglianzellen Virchows. Arch. f. Path. Anat., 205–295 [Google Scholar]
- 44. Yokoi S., Austin J., Witmer F. (1967) Isolation and characterization of Lafora bodies in two cases of myoclonus epilepsy. J. Neuropathol. Exp. Neurol. 26, 125–127 [PubMed] [Google Scholar]
- 45. Yokoi S., Austin J., Witmer F., Sakai M. (1968) Studies in myoclonus epilepsy (Lafora body form). I. Isolation and preliminary characterization of Lafora bodies in two cases. Arch. Neurol. 19, 15–33 [DOI] [PubMed] [Google Scholar]
- 46. Sakai M., Austin J., Witmer F., Trueb L. (1970) Studies in myoclonus epilepsy (Lafora body form). II. Polyglucosans in the systemic deposits of myoclonus epilepsy and in corpora amylacea. Neurology 20, 160–176 [DOI] [PubMed] [Google Scholar]
- 47. Ganesh S., Delgado-Escueta A. V., Suzuki T., Francheschetti S., Riggio C., Avanzini G., Rabinowicz A., Bohlega S., Bailey J., Alonso M. E., Rasmussen A., Thomson A. E., Ochoa A., Prado A. J., Medina M. T., Yamakawa K. (2002) Genotype-phenotype correlations for EPM2A mutations in Lafora's progressive myoclonus epilepsy: exon 1 mutations associate with an early-onset cognitive deficit subphenotype. Hum. Mol. Genet. 11, 1263–1271 [DOI] [PubMed] [Google Scholar]
- 48. Collins G. H., Cowden R. R., Nevis A. H. (1968) Myoclonus epilepsy with Lafora bodies. An ultrastruc- tural and cytochemical study. Arch. Pathol. 86, 239–254 [PubMed] [Google Scholar]
- 49. Turnbull J., Wang P., Girard J. M., Ruggieri A., Wang T. J., Draginov A. G., Kameka A. P., Pencea N., Zhao X., Ackerley C. A., Minassian B. A. (2010) Glycogen hyperphosphorylation underlies lafora body formation. Ann. Neurol. 68, 925–933 [DOI] [PubMed] [Google Scholar]
- 50. Tagliabracci V. S., Heiss C., Karthik C., Contreras C. J., Glushka J., Ishihara M., Azadi P., Hurley T. D., DePaoli-Roach A. A., Roach P. J. (2011) Phosphate incorporation during glycogen synthesis and Lafora disease. Cell Metab. 13, 274–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Nitschke F., Wang P., Schmieder P., Girard J. M., Awrey D. E., Wang T., Israelian J., Zhao X., Turnbull J., Heydenreich M., Kleinpeter E., Steup M., Minassian B. A. (2013) Hyperphosphorylation of glucosyl C6 carbons and altered structure of glycogen in the neurodegenerative epilepsy Lafora disease. Cell Metab. 17, 756–767 [DOI] [PubMed] [Google Scholar]
- 52. Yuvaniyama J., Denu J. M., Dixon J. E., Saper M. A. (1996) Crystal structure of the dual specificity protein phosphatase VHR. Science 272, 1328–1331 [DOI] [PubMed] [Google Scholar]
- 53. Alonso A., Rojas A., Godzik A., Mustelin T. (2003) The Dual-Specific Protein Tyrosine Phosphatse Family. Berlin, Springer [Google Scholar]
- 54. Tonks N. K. (2006) Protein tyrosine phosphatases: from genes, to function, to disease. Nat. Rev. Mol. Cell Biol. 7, 833–846 [DOI] [PubMed] [Google Scholar]
- 55. Moorhead G. B., De Wever V., Templeton G., Kerk D. (2009) Evolution of protein phosphatases in plants and animals. Biochem. J. 417, 401–409 [DOI] [PubMed] [Google Scholar]
- 56. Tonks N. K. (2013) Protein tyrosine phosphatases–from housekeeping enzymes to master regulators of signal transduction. FEBS J. 280, 346–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Meekins D. A., Guo H. F., Husodo S., Paasch B. C., Bridges T. M., Santelia D., Kötting O., Vander Kooi C. W., Gentry M. S. (2013) Structure of the Arabidopsis glucan phosphatase like sex four2 reveals a unique mechanism for starch dephosphorylation. Plant Cell 25, 2302–2314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zhou G., Denu J. M., Wu L., Dixon J. E. (1994) The catalytic role of Cys124 in the dual specificity phosphatase VHR. J. Biol. Chem. 269, 28084–28090 [PubMed] [Google Scholar]
- 59. Jia Z., Barford D., Flint A. J., Tonks N. K. (1995) Structural basis for phosphotyrosine peptide recognition by protein tyrosine phosphatase 1B. Science 268, 1754–1758 [DOI] [PubMed] [Google Scholar]
- 60. Begley M. J., Taylor G. S., Brock M. A., Ghosh P., Woods V. L., Dixon J. E. (2006) Molecular basis for substrate recognition by MTMR2, a myotubularin family phosphoinositide phosphatase. Proc. Natl. Acad. Sci. U.S.A. 103, 927–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Sherwood A. R., Paasch B. C., Worby C. A., Gentry M. S. (2013) A malachite green-based assay to assess glucan phosphatase activity. Anal. Biochem. 435, 54–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wang W., Roach P. J. (2004) Glycogen and related polysaccharides inhibit the laforin dual-specificity protein phosphatase. Biochem. Biophys. Res. Commun. 325, 726–730 [DOI] [PubMed] [Google Scholar]
- 63. Yu T. S., Kofler H., Häusler R. E., Hille D., Flügge U. I., Zeeman S. C., Smith A. M., Kossmann J., Lloyd J., Ritte G., Steup M., Lue W. L., Chen J., Weber A. (2001) The Arabidopsis sex1 mutant is defective in the R1 protein, a general regulator of starch degradation in plants, and not in the chloroplast hexose transporter. Plant Cell 13, 1907–1918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Anthon G. E., Barrett D. M. (2002) Determination of reducing sugars with 3-methyl-2-benzothiazolinonehydrazone. Anal. Biochem. 305, 287–289 [DOI] [PubMed] [Google Scholar]
- 65. Ganesh S., Tsurutani N., Suzuki T., Hoshii Y., Ishihara T., Delgado-Escueta A. V., Yamakawa K. (2004) The carbohydrate-binding domain of Lafora disease protein targets Lafora polyglucosan bodies. Biochem. Biophys. Res. Commun. 313, 1101–1109 [DOI] [PubMed] [Google Scholar]
- 66. DePaoli-Roach A. A., Contreras C. J., Segvich D. M., Heiss C., Ishihara M., Azadi P., Roach P. J. (2015) Glycogen phosphomonoester distribution in mouse models of the progressive myoclonic epilepsy, Lafora disease. J. Biol. Chem., 290, 841–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Muhrbeck P E. A. (1991) Influence of the naturally-occuring phosphate esters on the crystallinity of potato starch. J. Sci. Food Agric. 55, 13–18 [Google Scholar]
- 68. Liu Y., Wang Y., Wu C., Liu Y., Zheng P. (2006) Dimerization of Laforin is required for its optimal phosphatase activity, regulation of GSK3β phosphorylation, and Wnt signaling. J. Biol. Chem. 281, 34768–34774 [DOI] [PubMed] [Google Scholar]
- 69. Chan E. M., Ackerley C. A., Lohi H., Ianzano L., Cortez M. A., Shannon P., Scherer S. W., Minassian B. A. (2004) Laforin preferentially binds the neurotoxic starch-like polyglucosans, which form in its absence in progressive myoclonus epilepsy. Hum. Mol. Genet. 13, 1117–1129 [DOI] [PubMed] [Google Scholar]
- 70. Ianzano L., Young E. J., Zhao X. C., Chan E. M., Rodriguez M. T., Torrado M. V., Scherer S. W., Minassian B. A. (2004) Loss of function of the cytoplasmic isoform of the protein laforin (EPM2A) causes Lafora progressive myoclonus epilepsy. Hum. Mutat. 23, 170–176 [DOI] [PubMed] [Google Scholar]
- 71. Zeeman S. C., Umemoto T., Lue W. L., Au-Yeung P., Martin C., Smith A. M., Chen J. (1998) A mutant of Arabidopsis lacking a chloroplastic isoamylase accumulates both starch and phytoglycogen. Plant Cell 10, 1699–1712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Carciofi M., Shaif S. S., Jensen S. L., Blennow A., Svensson J. T., Vincze E., Hebelstrup K. H. (2011) Hyperphosphorylation of cereal starch. J. Cereal Sci. 54, 339–346 [Google Scholar]
- 73. Mahlow S., Hejazi M., Kuhnert F., Garz A., Brust H., Baumann O., Fettke J. (2014) Phosphorylation of transitory starch by alpha-glucan, water dikinase during starch turnover affects the surface properties and morphology of starch granules. New Phytol. 203, 495–507 [DOI] [PubMed] [Google Scholar]
- 74. Cavanagh J. B. (1999) Corpora-amylacea and the family of polyglucosan diseases. Brain Res. Brain Res. Rev. 29, 265–295 [DOI] [PubMed] [Google Scholar]