Background: Mutations in the voltage-gated sodium channel Nav1.7 cause congenital insensitivity to pain (CIP) in humans.
Results: Missense mutation R907Q in the extracellular disordered loop of Nav1.7 may also cause CIP because of the enhanced MMP-9 proteolysis of the mutant.
Conclusion: Accelerated cleavage of Nav1.7 by MMP-9 explains insensitivity to pain.
Significance: MMP proteolysis of sodium channels is a novel biochemical phenomenon.
Keywords: matrix metalloproteinase (MMP), mutant, pain, protease, proteolysis, sodium channel, voltage-gated sodium channels, MMP-9, Nav1.7, pain signaling, missense mutations
Abstract
Congenital insensitivity to pain (CIP) or congenital analgesia is a rare monogenic hereditary condition. This disorder is characterized by the inability to perceive any form of pain. Nonsense mutations in Nav.1.7, the main pain signaling voltage-gated sodium channel, lead to its truncations and, consequently, to the inactivation of the channel functionality. However, a non-truncating homozygously inherited missense mutation in a Bedouin family with CIP (Nav1.7-R907Q) has also been reported. Based on our currently acquired in-depth knowledge of matrix metalloproteinase (MMP) cleavage preferences, we developed the specialized software that predicts the presence of the MMP cleavage sites in the peptide sequences. According to our in silico predictions, the peptide sequence of the exposed extracellular unstructured region linking the S5–S6 transmembrane segments in the DII domain of the human Nav1.7 sodium channel is highly sensitive to MMP-9 proteolysis. Intriguingly, the CIP R907Q mutation overlaps with the predicted MMP-9 cleavage site sequence. Using MMP-9 proteolysis of the wild-type, CIP, and control peptides followed by mass spectrometry of the digests, we demonstrated that the mutant sequence is severalfold more sensitive to MMP-9 proteolysis relative to the wild type. Because of the substantial level of sequence homology among sodium channels, our data also implicate MMP proteolysis in regulating the cell surface levels of the Nav1.7, Nav1.6, and Nav1.8 channels, but not Nav1.9. It is likely that the aberrantly accelerated MMP-9 proteolysis during neurogenesis is a biochemical rational for the functional inactivation in Nav1.7 and that the enhanced cleavage of the Nav1.7-R907Q mutant is a cause of CIP in the Bedouin family.
Introduction
Congenital insensitivity to pain (CIP)3 or congenital analgesia is a rare hereditary condition. Affected humans are unable to perceive physical pain but retain the ability to feel a touch, heat, or cold stimulus (1). Congenital analgesia is clinically and genetically heterogeneous and caused by mutations in several distinct genes (2, 3). Mutations in SCN9A, encoding the α-subunit of the voltage-gated sodium channel Nav1.7, the predominant voltage-gated sodium channel in peripheral neurons (4), are a cause of autosomal recessive CIP. This disorder is characterized by the inability to perceive any form of pain, in any part of the body, but with intact motor, cognitive, sympathetic, or gastrointestinal and sensory modalities (5–8). Affected individuals cannot normally respond to the environment and are frequently traumatized, manifesting permanent damage and high mortality.
Mammalian 1700–2000-residue-long Nav1.7 and the other eight members of the Nav1.1–Nav1.9 α-subunit family (9) exhibit a common structural motif that includes four homologous domains (DI–DIV) linked by three cytoplasmic loops (L1–L3), with each domain having six transmembrane segments (S1–S6) arranged as a cross (4, 7). In the plasma membrane, the four domains assemble a pore with the sodium channel in the center and with the exposed pore region, encompassing the extracellular sequence linking the S5–S6 transmembrane segments within each of the four homologous domains.
Over the years, numerous genetic, functional, and structural studies have provided convincing insights into the central role of Nav1.7 in pain pathophysiology in both humans and mice (5–8, 10–12). The recent determination of the crystal structure (4BGN) of the bacterial sodium channel has allowed a more precise understanding of the effect of mutations in the structure-function of human Nav1.7 (4). Other voltage-gated sodium channels cannot compensate for a paucity of Nav1.7.
Conversely, gain-of-function mutations in Nav1.7 are a causative link to severe neuropathic pain including the inherited erythromelalgia, paroxysmal extreme pain, and small-fiber neuropathy disorders (7). Intriguingly, a non-truncating homozygously inherited missense mutation found in a consanguineous Israeli Bedouin family with CIP (Nav1.7-R907Q in the exposed pore S5–S6 region) has been recently reported (6). This mutation led to both a significant reduction in membrane localization of the R907Q mutant as compared with the wild type and a complete loss of function of Nav1.7, consistent with the absence of pain phenotype. Consistently, the truncated Nav1.7 mutant with the neighboring W908X nonsense mutation (W897X mutation according to the nomenclature of the original publication (5)) was functionally inactive.
Experimental Procedures
MMP-9 Proenzyme and General Reagents
All reagents were purchased from Sigma unless otherwise indicated. The purified MMP-9 proenzyme from human neutrophils was obtained from Enzo Life Sciences (Farmingdale, NY). We also used the purified, recombinant homogenous human MMP-9 proenzyme that was a generous gift of Dr. Rafael Fridman (Wayne State University, Detroit, MI). To generate the mature MMP-9 enzyme, the proenzyme was activated for 3 h at 37 °C using 1 mm p-aminophenylmercuric acetate in 50 mm Tris-HCl, pH 7.5, containing 150 mm NaCl, 5 mm CaCl2, 5 mm MgCl2, and 50 μm ZnCl2. The conversion of the 92-kDa proenzyme into the 84-kDa mature enzyme of MMP-9 was confirmed using gelatin zymography (data not shown) (13).
Cleavage of Synthetic Peptides and Mass Spectrometry Analysis
The Nav1.7 wild-type TLPRWHMNDF (1316.48 Da), CIP R907Q mutant TLPQWHMNDF (1288.43 Da) and control TLPDWHMNDF (1275.39 Da) peptides (the residue 907 is underlined) were synthesized by GenScript (Piscataway, NJ). The peptide sequence was derived from the amino acid residue sequence of the human Nav1.7 voltage-gated sodium channel (GenBankTM accession number Q15858) of either sex. The peptides (1 μg; 40 μm each) were incubated for 2 h at 37 °C with the active MMP-9 enzyme (0.16–0.67 μg, a 1:250–100 enzyme-substrate molar ratio) in 20 μl of 50 mm Tris-HCl, pH 7.5, containing 150 mm NaCl, 5 mm CaCl2, 5 mm MgCl2, and 50 μm ZnCl2.
The spectra of the intact and cleaved peptides were determined by MALDI-TOF MS analysis using an AutoFlex II mass spectrometer (Bruker Daltonics, Billerica, MA). For MS analysis, sample aliquots (2 μl each) were mixed with an equal volume of α-cyano-4-hydroxycinnamic acid (20 mg/ml) in 50% acetonitrile, 0.1% trifluoroacetic acid. The mixtures were spotted directly on a MALDI target plate and then allowed to air-dry and co-crystallize for 15 min. The MALDI mass spectra were analyzed using FlexAnalysis 2.4 software (Bruker Daltonics). The predicted masses of the TLPRW, TLPQW, and HMNDF cleavage products were 671.36, 643.73, and 662.23 Da, respectively.
To quantify MMP-9 proteolysis, we determined the intensities of both the residual intact peptides and common HMNDF digest fragments in the cleavage reactions. To obtain the more accurate quantitative data, the acquisition of the spectral data was narrowed to the 662–665-Da regions that included the common HMNDF cleavage product alone. All the cleavage reactions were repeated at least three times using two independent samples of MMP-9. The results were highly reproducible without any significant day to day variations.
Molecular Modeling
We used the SWISS-MODEL automated structure homology-modeling server (14) for the molecular modeling of the DII domain structure of human Nav1.7. To model the DII domain, we used the Caldalkalibacillus thermarum sodium channel structure (Protein Data Bank (PDB) 4BGN) as a template (15). The Nav1.7 lipid bilayer membrane model has been built using a replacement method (a protein is first packed by lipid-like spheres whose positions are subsequently used to place randomly chosen lipid molecules from the library) using the CHARMM-GUI web interface (16).
In Silico Prediction of the Putative Cleavage Sites in Nav Proteins
We used the CleavPredict algorithm to predict the cleavage sites in the full-length Nav proteins. The algorithm is based on the position weight matrices (PWMs) approach. The original cleavage data were derived from the analysis of specific substrates selected from phage display libraries (17, 18).
Results
Recently, using substrate phage display (17, 18), we identified some of the cleavage preferences of the matrix metalloproteinase (MMP) family members (19). Here, we employed a high throughput multiplexed peptide-centric profiling technology involving the cleavage of 18,583 peptides by the 18 individual proteinases from the main subgroups of the MMP family (20). Volumes of the experimental data we acquired, merged with advanced bioinformatics, enabled comparison of the MMP cleavage preferences on a global scale and led to the previously unprecedented level of knowledge in MMP biology. Based on this newly acquired knowledge, we then developed cleavage prediction algorithms and the substrate specificity profiling software (17). This now freely available software (CleavPredict) readily identifies the cleavage site sequences within either individual proteins/peptides or the whole proteome and then ranks the probability of these site cleavages by the individual MMPs. CleavPredict allows us and others to locate in silico, with a nearly 100% accuracy, the MMP cleavage sites in the peptide sequences. The cross-validation tests involving the hundreds of the MMP cleavage sequences identified by others (17, 21–25) validated the accuracy of our cleavage prediction software. Specifically, false prediction rate, accuracy, and specificity were 2–3, 94–96, and 97–98%, respectively, for MMP-9, suggesting that, in addition to the previously known cleavage targets, we can now identify novel, previously unrecognized substrates of this proteinase (17, 20).
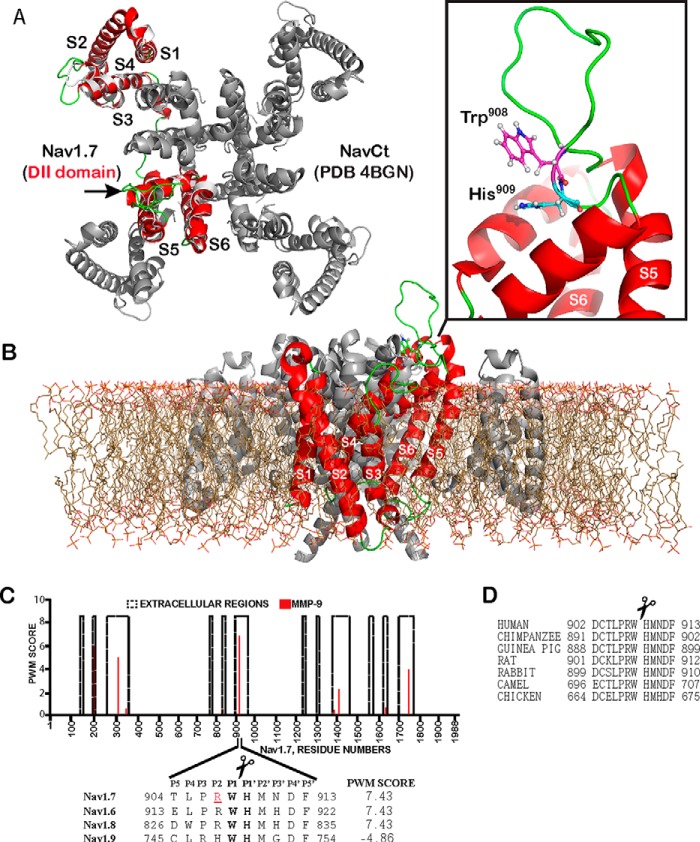
Notably, according to our in silico CleavPredict analysis, the 904–913-peptide sequence of the exposed convex pore region linking the S5–S6 transmembrane segments in the Nav1.7 DII domain (GenBank accession number Q15858) was predicted to be highly sensitive to MMP-9 proteolysis, but not to other MMPs. Intriguingly, a single missense mutation (R907Q) that leads to CIP is currently known in Nav1.7. Even more intriguingly, this CIP-related single amino acid substitution overlaps with the predicted cleavage site of MMP-9 (Fig. 1).
FIGURE 1.
The structure of Nav1.7 and MMP-9 cleavage site. A, superimposition of the S1–S6 transmembrane segments of the Nav1.7 DII domain (red) and the full-length C. thermarum sodium channel (NavCt, gray; PDB 4BGN). Extracellular regions are in green. An arrow points to the extracellular region connecting the S5–S6 segments. B, side view of the modeled S1–S6 transmembrane segments (red) of the domain DII with the full-length NavCt homologue (gray) immersed in a lipid bilayer. Inset, the loop (green) connecting the S5–S6 segments with the MMP-9 Trp908–His909 cleavage site (sticks). C, the PWM score of the MMP-9 cleavage sites in the Nav1.7 extracellular regions. The high PWM score suggests the presence of a potential cleavage site in the peptide sequence. Inset, the alignment of Nav1.7 with the respective regions of other sodium channels and the PWM score of the predicted MMP-9 cleavage site. The P1 and P1′ residues of the scissile bond are in bold. Arg907 (red and underlined) is mutated in CIP. D, the MMP-9 cleavage site is conserved in Nav1.7 from human, chimpanzee, guinea pig, rat, rabbit, camel, and chicken (GenBank accession numbers Q15858, H2QIX3, H0VMS3, O08562, Q28644, T0NJY4, and E1C4S2, respectively).
The similar potential cleavage sites of MMP-9 were detected in both the 913–922 Nav1.6 (GenBank accession number Q9UQD0) and the 826–835 Nav1.8 (GenBank accession number Q9Y5Y9) sequences, but not in Nav1.9 (GenBank accession number Q9UI33). These sequence regions are a part of the exposed loop in the DII domain structure and, as a result, they appear accessible to secretory soluble proteinases (4).
In contrast with other MMP family members, MMP-9, a unique secretory pro-inflammatory proteinase, has a restricted expression and activation pattern (26). MMP-9 activity is stimulated by a limited set of conditions that are prevalent in inflammation, including peripheral nerve injury, and also in neurogenesis (27, 28). Because our software correctly predicts a vast majority of MMP-9 substrates (17, 20) and because the R907Q missense mutation reduced the presentation of the mutant protein at the plasma membrane, we hypothesized that the mutation enhanced the susceptibility of the membrane Nav1.7 to MMP-9 proteolysis, leading to the fragmented channel incapable of assembling a functional pore at the plasma membrane.
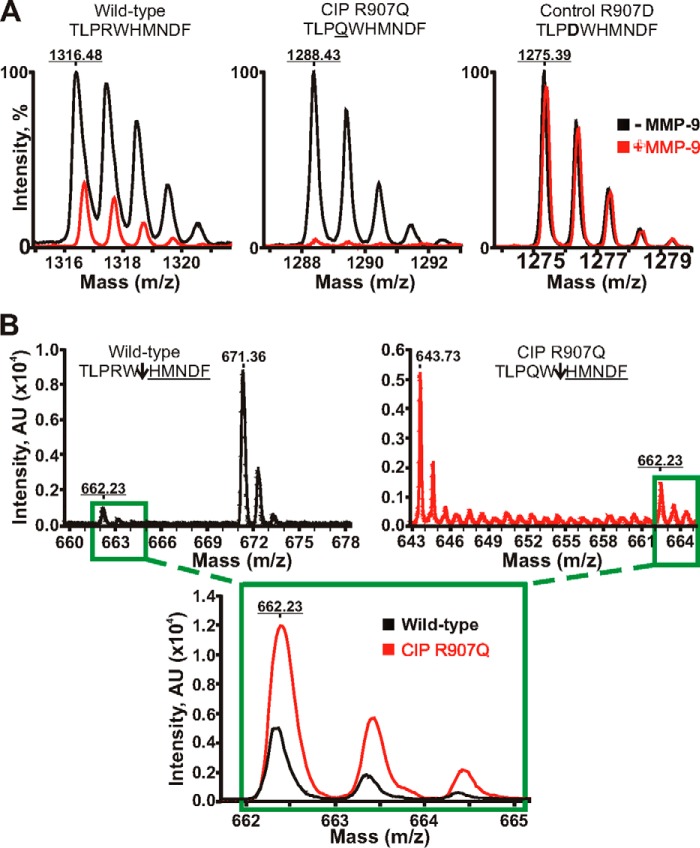
To test our hypothesis, we co-incubated the wild-type and mutant CIP peptides (TLPRWHMNDF and TLPQWHMNDF, respectively; the mutant residue is underlined) with the catalytic amounts of the purified, active MMP-9 mature enzyme. The TLPDWHMNDF peptide with the fully inactivated MMP-9 cleavage site (inactivating mutation is in bold) was used as a control. Cleavage reactions were subjected to the MALDI-TOF MS analysis to identify their MS peptide spectra (Fig. 2). As expected, the control peptide was fully resistant to MMP-9 proteolysis. Our results directly indicated that the CIP mutant sequence was cleaved by MMP-9 about 3-fold more efficiently as compared with the wild-type peptide. These results imply that MMP-9 cleavage of the exposed extracellular unstructured region linking the S5–S6 transmembrane segments in the DII domain encompassing the peptides we studied is enhanced in the CIP mutant Nav1.7 relative to the wild-type channel.
FIGURE 2.
Mass spectrometry analysis of MMP-9 proteolysis of the Nav1.7 peptides. A, MALDI monoisotopic MS spectra of the peptides. The wild-type TLPRWHMNDF (1316.48 Da), mutant TLPQWHMNDF (the CIP R907Q mutation is underlined; 1288.43 Da), and control TLPDWHMNDF (the inactivating R907D mutation is in bold; 1275.39 Da) peptides were incubated with MMP-9 at a 1:100 enzyme-substrate molar ratio. The graphs show the superimposed monoisotopic spectra of the respective intact peptides incubated with or without MMP-9 (red and black, respectively). The average mass of the intact peptides is underlined. Although the wild-type peptide and, especially, the CIP mutant were efficiently cleaved by MMP-9, the control peptide was fully resistant. B, quantification of the common HMNDF cleavage product. The wild-type peptide (black) and the R907D CIP mutant (red) were incubated with MMP-9 at a 1:250 enzyme-substrate molar ratio. Upper panels, the spectra of the TLPRW (671.36 Da), TLPQW (643.73 Da), and HMNDF (662.23 Da) cleavage products. An arrow points to the scissile bond. The molecular mass of the common HMNDF fragment is underlined. Inset, the superimposed spectra of the common HMNDF fragment generated by MMP-9 proteolysis of the wild-type and mutant CIP R907Q peptides. AU, arbitrary units.
Furthermore, as a result of both these experimental findings and the considerable level of sequence homology among sodium channels, we now confidently expect that the Nav1.6 and Nav1.8 voltage-gated sodium channels, but not Nav1.9, are also similarly cleaved by MMP-9 (Fig. 2). In agreement, in our earlier immunostaining studies, we have already observed the high levels of Nav1.6 in teased nerve fibers isolated from the MMP-9-null mice relative to those obtained from the wild-type animals (29).
Discussion
In human genome, the SCN9A gene codes for the α-subunit of the voltage-gated Nav1.7 sodium channel, the main pain signaling channel that is expressed in peripheral pain-sensing neurons (10). A number of gain-of-function mutations in SCN9A result in severe episodic neuropathic pain and the three distinct disorders known in humans such as paroxysmal extreme pain disorder, inherited/primary erythromelalgia, and small-fiber neuropathy (reviewed in Ref. 7). In turn, multiple recently identified bi-allelic loss-of-function truncating mutations in SCN9A lead to either the dysfunctional Nav1.7 protein or, probably, to no protein being produced, and, consequently, to a disorder named autosomal recessive CIP (1, 3, 11, 12, 30). CIP patients are incapable of perceiving any form of pain, in any part of the body, but other modalities are normal. It is not entirely surprising that some of affected individuals make a living as street performers (5).
A non-truncating Nav1.7-R896Q mutation, however, has been found in a consanguineous Israeli Bedouin family (6). So far, this is the only missense mutation in Nav1.7 that causes CIP. This missense mutation was mapped to the exposed extracellular unstructured region (the pore region) linking the S5–S6 transmembrane segments in the DII domain of the human Nav1.7. The Nav1.7 channels exist in multiple, if not all, vertebrate species, and the vertebrate homologues are not fully sequence homologous to the human Nav1.7 (9, 31). Nevertheless, these pain-sensing channels are likely to be fully functional in vertebrates regardless of their multiple sequence dissimilarities relative to the human protein. Naturally, it is intriguing that the single missense Nav1.7-R896Q mutation resulted in the severe loss of function phenotype consistent with CIP in humans. In addition, the Nav1.7-R896Q mutation caused a significant reduction in membrane localization of the mutant protein as compared with the wild type (6).
The sequence analysis of Nav1.7 using the highly accurate cleavage prediction software we developed (17) pointed to the presence of the highly efficient MMP-9 cleavage site in the Nav1.7 pore region. Furthermore, the sequence of this cleavage site overlapped the R896Q mutation, suggesting that the mutation affected the rate of MMP-9 proteolysis of Nav1.7. To mimic the proteolysis of the unstructured Nav1.7 pore region by MMP-9 in vivo, we synthesized the peptides whose sequence was derived from the pore region, and then we digested the peptide samples in vitro using the purified, active MMP-9 enzyme. The digests were next analyzed by mass spectrometry to determine the efficiency of proteolysis, the peptide mass and, consequently, the scissile bond. According to our data, the R896Q mutant sequence was severalfold more sensitive to MMP-9 proteolysis relative to the wild type. These results allow us to suggest that the excessive MMP-9 proteolysis is a biochemical rationale for the functional inactivation in the Nav1.7 missense mutant and CIP in the Bedouin family (6, 27). Consistent with our data, the truncated Nav1.7 mutant with the neighboring W908X nonsense mutation (W897X mutation according to the nomenclature of the original publication (5)) is also functionally inactive. The presence of this inactive W908X mutant implies that cleavage at the Tyr908–His909 site by MMP-9 may also cause the inactivation of Nav1.7. Based on the sequence homology in the Nav family, our data also suggest that MMP-9 proteolysis likely targets the Nav1.6 and Nav1.8 channels (but not Nav1.9), rather than Nav.1.7 alone.
Overall, our focused results highlight the primary role of the Nav1.7 channel in pain signaling. Our data also suggest that, similarly to the rate of the protein synthesis and presentation of the Nav1.7 protein on the cell membrane, MMP proteolysis is an important element in the regulation of both Nav1.7 function and pain control mechanisms. Likewise, the accelerated MMP-9 proteolysis during neuroinflammation may contribute to the functional alterations in Nav1.7 and aberrant pain processing (28, 29).
Author Contributions
A. G. R., S. K., K. M., and P. C. designed, performed, and analyzed the experiments shown in Figures 1 and 2. S. H. and J. D. provided technical assistance and contributed to the preparation of the manuscript. V. I. S. and A. Y. S. designed the study and wrote the paper. All authors reviewed the results and approved the final version of the manuscript.
The work was supported, in whole or in part, by National Institutes of Health Grants R01 DE022757 (to V. I. S. and A. Y. S.), RO1 CA157328 (to A. Y. S.) and R01 GM098835 (to P. C.). The work was also supported by Department of Veterans Affairs Merit Review Award 5I01BX000638 (to V. I. S.). The authors declare that they have no conflicts of interest with the contents of this article.
- CIP
- congenital insensitivity to pain
- MMP
- matrix metalloproteinase
- PWM
- position weight matrix.
References
- 1. de Lera Ruiz M., Kraus R. L. (2015) Voltage-gated sodium channels: structure, function, pharmacology and clinical indications. J. Med. Chem. 10.1021/jm501981g [DOI] [PubMed] [Google Scholar]
- 2. Bennett D. L., Woods C. G. (2014) Painful and painless channelopathies. Lancet Neurol. 13, 587–599 [DOI] [PubMed] [Google Scholar]
- 3. Brouwer B. A., Merkies I. S., Gerrits M. M., Waxman S. G., Hoeijmakers J. G., Faber C. G. (2014) Painful neuropathies: the emerging role of sodium channelopathies. J. Peripher. Nerv. Syst. 19, 53–65 [DOI] [PubMed] [Google Scholar]
- 4. Payandeh J., Scheuer T., Zheng N., Catterall W. A. (2011) The crystal structure of a voltage-gated sodium channel. Nature 475, 353–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cox J. J., Reimann F., Nicholas A. K., Thornton G., Roberts E., Springell K., Karbani G., Jafri H., Mannan J., Raashid Y., Al-Gazali L., Hamamy H., Valente E. M., Gorman S., Williams R., McHale D. P., Wood J. N., Gribble F. M., Woods C. G. (2006) An SCN9A channelopathy causes congenital inability to experience pain. Nature 444, 894–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cox J. J., Sheynin J., Shorer Z., Reimann F., Nicholas A. K., Zubovic L., Baralle M., Wraige E., Manor E., Levy J., Woods C. G., Parvari R. (2010) Congenital insensitivity to pain: novel SCN9A missense and in-frame deletion mutations. Hum. Mutat. 31, E1670–1686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dib-Hajj S. D., Yang Y., Black J. A., Waxman S. G. (2013) The NaV1.7 sodium channel: from molecule to man. Nat. Rev. Neurosci. 14, 49–62 [DOI] [PubMed] [Google Scholar]
- 8. Goldberg Y. P., MacFarlane J., MacDonald M. L., Thompson J., Dube M. P., Mattice M., Fraser R., Young C., Hossain S., Pape T., Payne B., Radomski C., Donaldson G., Ives E., Cox J., Younghusband H. B., Green R., Duff A., Boltshauser E., Grinspan G. A., Dimon J. H., Sibley B. G., Andria G., Toscano E., Kerdraon J., Bowsher D., Pimstone S. N., Samuels M. E., Sherrington R., Hayden M. R. (2007) Loss-of-function mutations in the Nav1.7 gene underlie congenital indifference to pain in multiple human populations. Clin. Genet. 71, 311–319 [DOI] [PubMed] [Google Scholar]
- 9. Catterall W. A., Goldin A. L., Waxman S. G. (2005) International Union of Pharmacology. XLVII. Nomenclature and structure-function relationships of voltage-gated sodium channels. Pharmacol. Rev. 57, 397–409 [DOI] [PubMed] [Google Scholar]
- 10. Toledo-Aral J. J., Moss B. L., He Z. J., Koszowski A. G., Whisenand T., Levinson S. R., Wolf J. J., Silos-Santiago I., Halegoua S., Mandel G. (1997) Identification of PN1, a predominant voltage-dependent sodium channel expressed principally in peripheral neurons. Proc. Natl. Acad. Sci. U.S.A. 94, 1527–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lee J. H., Park C. K., Chen G., Han Q., Xie R. G., Liu T., Ji R. R., Lee S. Y. (2014) A monoclonal antibody that targets a NaV1.7 channel voltage sensor for pain and itch relief. Cell 157, 1393–1404 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 12. Gingras J., Smith S., Matson D. J., Johnson D., Nye K., Couture L., Feric E., Yin R., Moyer B. D., Peterson M. L., Rottman J. B., Beiler R. J., Malmberg A. B., McDonough S. I. (2014) Global Nav1.7 knockout mice recapitulate the phenotype of human congenital indifference to pain. PLoS One 9, e105895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Vandooren J., Geurts N., Martens E., Van den Steen P. E., Opdenakker G. (2013) Zymography methods for visualizing hydrolytic enzymes. Nat. Methods 10, 211–220 [DOI] [PubMed] [Google Scholar]
- 14. Biasini M., Bienert S., Waterhouse A., Arnold K., Studer G., Schmidt T., Kiefer F., Cassarino T. G., Bertoni M., Bordoli L., Schwede T. (2014) SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 42, W252–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tsai C. J., Tani K., Irie K., Hiroaki Y., Shimomura T., McMillan D. G., Cook G. M., Schertler G. F., Fujiyoshi Y., Li X. D. (2013) Two alternative conformations of a voltage-gated sodium channel. J. Mol. Biol. 425, 4074–4088 [DOI] [PubMed] [Google Scholar]
- 16. Jo S., Kim T., Iyer V. G., Im W. (2008) CHARMM-GUI: a web-based graphical user interface for CHARMM. J. Comput. Chem. 29, 1859–1865 [DOI] [PubMed] [Google Scholar]
- 17. Kumar S., Ratnikov B. I., Kazanov M. D., Smith J. W., Cieplak P. (2015) CleavPredict: a platform for reasoning about matrix metalloproteinases proteolytic events. PLoS One 10, e0127877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ratnikov B. I., Cieplak P., Gramatikoff K., Pierce J., Eroshkin A., Igarashi Y., Kazanov M., Sun Q., Godzik A., Osterman A., Stec B., Strongin A., Smith J. W. (2014) Basis for substrate recognition and distinction by matrix metalloproteinases. Proc. Natl. Acad. Sci. U.S.A. 111, E4148–4155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Egeblad M., Werb Z. (2002) New functions for the matrix metalloproteinases in cancer progression. Nat. Rev. Cancer 2, 161–174 [DOI] [PubMed] [Google Scholar]
- 20. Kukreja M., Shiryaev S. A., Cieplak P., Muranaka N., Routenberg D. A., Chernov A. V., Kumar S., Remacle A. G., Smith J. W., Kozlov I. A., Strongin A. Y. (2015) The high-throughput multiplexed peptide-centric profiling illustrates both the substrate cleavage redundancy and specificity in the MMP family. Chem. Biol. 10.1016/j.chembiol.2015.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Prudova A., auf dem Keller U., Butler G. S., Overall C. M. (2010) Multiplex N-terminome analysis of MMP-2 and MMP-9 substrate degradomes by iTRAQ-TAILS quantitative proteomics. Mol. Cell. Proteomics 9, 894–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Schilling O., Barré O., Huesgen P. F., Overall C. M. (2010) Proteome-wide analysis of protein carboxy termini: C terminomics. Nat. Methods 7, 508–511 [DOI] [PubMed] [Google Scholar]
- 23. Schilling O., Overall C. M. (2007) Proteomic discovery of protease substrates. Curr. Opin. Chem. Biol. 11, 36–45 [DOI] [PubMed] [Google Scholar]
- 24. Schilling O., Overall C. M. (2008) Proteome-derived, database-searchable peptide libraries for identifying protease cleavage sites. Nat. Biotechnol. 26, 685–694 [DOI] [PubMed] [Google Scholar]
- 25. Turk B. E., Huang L. L., Piro E. T., Cantley L. C. (2001) Determination of protease cleavage site motifs using mixture-based oriented peptide libraries. Nat. Biotechnol. 19, 661–667 [DOI] [PubMed] [Google Scholar]
- 26. Vandooren J., Van den Steen P. E., Opdenakker G. (2013) Biochemistry and molecular biology of gelatinase B or matrix metalloproteinase-9 (MMP-9): the next decade. Crit. Rev. Biochem. Mol. Biol. 48, 222–272 [DOI] [PubMed] [Google Scholar]
- 27. Ethell I. M., Ethell D. W. (2007) Matrix metalloproteinases in brain development and remodeling: synaptic functions and targets. J. Neurosci. Res. 85, 2813–2823 [DOI] [PubMed] [Google Scholar]
- 28. Liu H., Shiryaev S. A., Chernov A. V., Kim Y., Shubayev I., Remacle A. G., Baranovskaya S., Golubkov V. S., Strongin A. Y., Shubayev V. I. (2012) Immunodominant fragments of myelin basic protein initiate T cell-dependent pain. J. Neuroinflammation 9, 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kim Y., Remacle A. G., Chernov A. V., Liu H., Shubayev I., Lai C., Dolkas J., Shiryaev S. A., Golubkov V. S., Mizisin A. P., Strongin A. Y., Shubayev V. I. (2012) The MMP-9/TIMP-1 axis controls the status of differentiation and function of myelin-forming Schwann cells in nerve regeneration. PLoS One 7, e33664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Waxman S. G., Merkies I. S., Gerrits M. M., Dib-Hajj S. D., Lauria G., Cox J. J., Wood J. N., Woods C. G., Drenth J. P., Faber C. G. (2014) Sodium channel genes in pain-related disorders: phenotype-genotype associations and recommendations for clinical use. Lancet Neurol. 13, 1152–1160 [DOI] [PubMed] [Google Scholar]
- 31. Smith E. S., Omerbašić D., Lechner S. G., Anirudhan G., Lapatsina L., Lewin G. R. (2011) The molecular basis of acid insensitivity in the African naked mole-rat. Science 334, 1557–1560 [DOI] [PubMed] [Google Scholar]