Background: Calcium-binding proteins regulate calcium dynamics and downstream signaling events.
Results: Increasing the calcium buffering capacity of the nucleus alters the expression of genes that regulate neuronal architecture.
Conclusion: The nuclear calcium buffering capacity is an important determinant of neuronal morphology.
Significance: Nuclear calcium buffers represent a new target for modulating gene expression and neuronal structure.
Keywords: calcium, calcium binding protein, dendrite, gene expression, signal transduction, synapse
Abstract
Calcium-binding proteins (CaBPs) such as parvalbumin are part of the cellular calcium buffering system that determines intracellular calcium diffusion and influences the spatiotemporal dynamics of calcium signals. In neurons, CaBPs are primarily localized to the cytosol and function, for example, in nerve terminals in short-term synaptic plasticity. However, CaBPs are also expressed in the cell nucleus, suggesting that they modulate nuclear calcium signals, which are key regulators of neuronal gene expression. Here we show that the calcium buffering capacity of the cell nucleus in mouse hippocampal neurons regulates neuronal architecture by modulating the expression levels of VEGFD and the complement factor C1q-c, two nuclear calcium-regulated genes that control dendrite geometry and spine density, respectively. Increasing the levels of nuclear calcium buffers by means of expression of a nuclearly targeted form of parvalbumin fused to mCherry (PV.NLS-mC) led to a reduction in VEGFD expression and, as a result, to a decrease in total dendritic length and complexity. In contrast, mRNA levels of the synapse pruning factor C1q-c were increased in neurons expressing PV.NLS-mC, causing a reduction in the density and size of dendritic spines. Our results establish a close link between nuclear calcium buffering capacity and the transcription of genes that determine neuronal structure. They suggest that the development of cognitive deficits observed in neurological conditions associated with CaBP deregulation may reflect the loss of necessary structural features of dendrites and spines.
Introduction
Calcium-binding proteins (CaBPs),2 often viewed merely as immunohistological markers for certain types of neurons (1, 2), participate in intracellular calcium signaling by modulating basal calcium levels and fine-tuning signal-regulated calcium transients (3). CaBPs can be divided into calcium buffers and calcium sensors, both of which are expressed in various subcellular compartments and allow for the differential regulation of spatially distinct calcium signals and the formation of calcium microdomains with specific functions (3). For example, in nerve terminals, the CaBP Calbindin-D28K is important for paired pulse facilitation, a short-lasting modification of synaptic transmission efficacy (4). A second possible role of calcium buffering systems may be in transcriptional regulation, particularly when CaBPs are expressed in the cell nucleus (5, 6). Nuclear calcium functions as a signal integrator in synapse-to-nucleus communication that relays synaptic activity to the induction or repression of gene transcription (7). The transcription factor cAMP response element-binding protein (CREB) is the prototypical target of the nuclear calcium signaling pathway. However, several other regulators of gene expression participate in the mounting of a nuclear calcium-induced genomic response, including CREB-binding protein, MeCP2, and class IIa histone deacetylases (7). In hippocampal and spinal cord neurons, a large number of nuclear calcium-regulated genes have been identified that are a central part of the genomic programs required for the long-term implementation of several neuroadaptations, including memory formation, acquired neuroprotection, and chronic pain (8–11). At the cellular level, nuclear calcium signaling controls the structural features of neurons. This is mediated by the nuclear calcium-regulated genes encoding VEGFD and the complement protein C1q-c, which regulate the architecture of dendrites and spine density, respectively (9, 11). To determine to what extent this regulatory genomic network specifying neuronal morphology is controlled by the calcium buffering system in the cell nucleus, we expressed a nuclearly targeted form of parvalbumin in mouse hippocampal neurons. The results revealed that changes in the nuclear calcium buffering capacity are linked, through a transcription-dependent process, to robust structural alterations in neurons.
Experimental Procedures
Expression Constructs
The following constructs were used in this study: pAAV-hrGFP, pAAV-mC.NLS, pAAV-PV.NLS-mC, pAAV-shC1q-c, and pAAV-shUNC. pAAV-hrGFP, pAAV-mC.NLS, pAAV-shC1q-c, and pAAV-shUNC have been characterized previously (9, 11–13). For the construction of the nuclearly targeted parvalbumin expression vector, the PV.NLS coding sequence was PCR-amplified from pCMV-PV.NLS-dsRed (14) and then subcloned into pAAV-mC to yield an in-frame PV.NLS-mC fusion. Nuclearly targeted parvalbumin expression vectors have been described previously (14–16).
Virus Production
The method used to construct, package, and purify recombinant adeno-associated viruses (rAAVs) has been described previously (12, 17). The integrity and purity of viral particles were verified by SDS-PAGE.
Hippocampal Cultures and Treatments
Hippocampal neurons from newborn C57BL/6 mice or Sprague-Dawley rats were isolated and cultured as described previously (18, 19). DNA transfection was performed after a culturing period of 8 days in vitro (DIV) using Lipofectamine 2000 (Invitrogen) as described previously (20). Infection of neuronal cultures with rAAVs delivering the different expression constructs was performed on DIV 3–6. Infection rates were determined by analyzing mCherry fluorescence and ranged from 80% to 95% of the neuronal population. Experiments were performed on DIV 10–13.
Immunocytochemistry
Hippocampal neurons were fixed with 4% paraformaldehyde and 4% sucrose in PBS (pH 7.4) at room temperature for 20 min. For the morphometric analyses, fluorescence images were acquired using a confocal laser-scanning microscope (TCS SP2, Leica, Mannheim, Germany) equipped with an inverted fluorescence microscope (DM IRE2, Leica) and Leica confocal scan software. For analysis of dendrites and spines, all images were obtained with sequential acquisition settings and 1024 × 1024 pixel resolution. Each image was a z-series projection of images taken at 1-μm depth intervals for dendrites and 0.5-μm depth intervals for spines.
Calcium Imaging
Hippocampal neurons transfected with either pAAV-mC.NLS or pAAV-PV.NLS-mC were loaded for 30 min at 37 °C with 1 μm Fura-2/AM (Molecular Probes) in CO2-independent culture medium (CICM) consisting of 140 mm NaCl, 2.5 mm KCl, 1.0 mm MgCl2, 2.0 mm CaCl2, 10.0 mm Hepes, 1.0 mm glycine, 35.6 mm d-glucose, and 0.5 mm C3H3NaO3. Neurons were then transferred to fresh warm CICM and allowed to equilibrate for an additional 30–40 min at 37 °C prior to the onset of imaging. Fluorescence was detected using a cooled charge-coupled device camera (iXon, Andor) through a ×40 water immersion objective (LUMPlanFl/IR, Olympus) on an upright microscope (BX51W1, Olympus). Transfected cells (1–3 cells in each field of view containing >15 cells) were identified by the presence of a strongly red fluorescent nucleus (excitation, 570 ± 10 nm; emission, 620 ± 30 nm; Chroma). Fura-2/AM fluorescence (510 ± 40 nm, Chroma) was excited using alternating 340-nm (ET340x, Chroma) and 380-nm (ET380x, Chroma) light provided by a xenon arc lamp with an excitation filter wheel (cellR, Olympus) and imaged at 2 Hz. Data were collected using proprietary software (cellR, Olympus) and analyzed using ImageJ and IgorPro (Wavemetrics, Lake Oswego, OR).
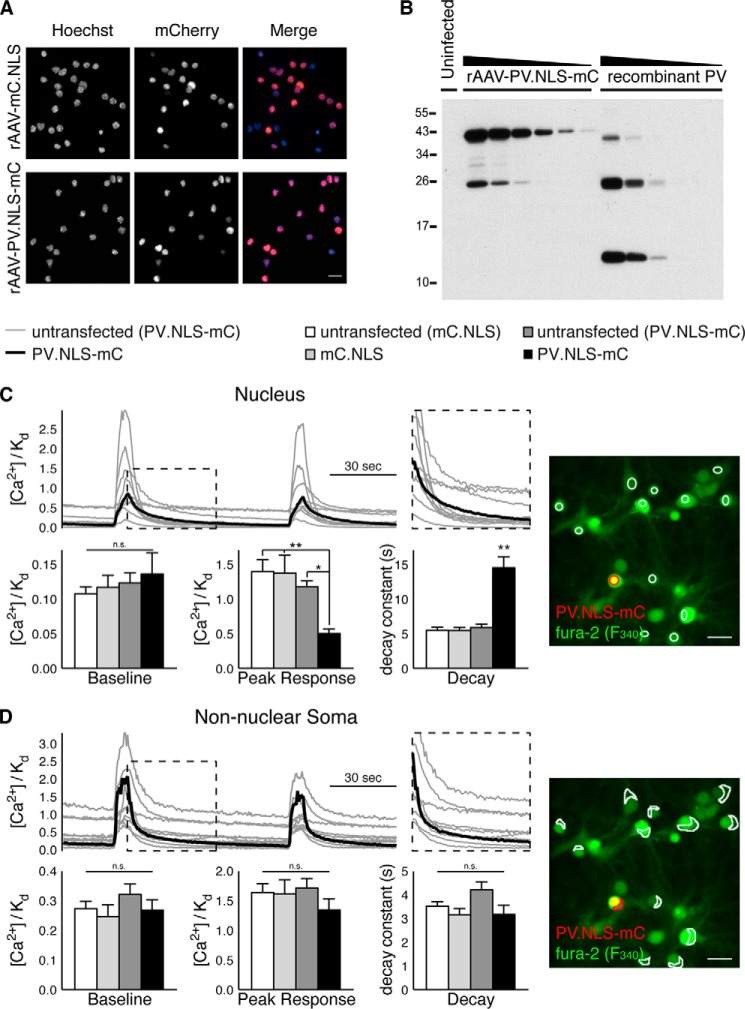
For imaging, cells on coverslips were transferred to an imaging chamber containing room-temperature CICM, and the GABAA receptor antagonist gabazine (100 μm), was applied to induce action potential bursting and associated intracellular calcium rises. The fluorescence ratio r = F340/F380 of Fura-2/AM was converted to [Ca2+]/Kd using the following formula: [Ca2+]/Kd = ((R − Rmin) / (Rmax − R)) × β, where Rmax was measured in CICM containing 10 μm ionomycin, Rmin was measured in calcium-free CICM containing 10 μm ionomycin and 5 mm EGTA, and β was the quotient of F380 obtained in calcium-free CICM containing 10 μm ionomycin and 5 mm EGTA and F380 obtained in CICM containing 10 μm ionomycin (21). Baseline [Ca2+]/Kd levels, gabazine-induced peak [Ca2+]/Kd responses, and the decay rate of [Ca2+]/Kd following the termination of a presumed burst of action potentials (determined by curve fitting to a single exponential) were measured for each neuron having an easily identifiable nucleus and cytoplasm within the visible field. Baseline R and [Ca2+]/Kd values were lower within nuclei than within the cytoplasm (Fig. 1, C and D), and the boundaries of nuclei lying within the plane of focus could be easily discerned in images of R (data not shown). This feature enabled us to draw, with confidence, regions of interest for the analysis of nuclear and non-nuclear somatic calcium responses (Fig. 1, C and 1D). Data points represent means calculated from control and mC.NLS- or PV.NLS-mC-positive cells from a single coverslip. 17–18 coverslips from three independent preparations were analyzed.
FIGURE 1.
Characterization of PV.NLS-mC in primary hippocampal neurons. A, representative images of cultured hippocampal neurons infected with rAAV-mC.NLS or rAAV-PV.NLS-mC. Scale bar = 20 μm. B, representative immunoblot analysis of uninfected hippocampal neurons and of hippocampal neurons infected with rAAV-PV.NLS-mC in comparison with recombinant parvalbumin, which was used for quantification of the nuclear concentration of PV.NLS-mC (see “Experimental Procedures” for details). Loaded amounts were as follows: uninfected, 2.5 μl of total homogenate; rAAV-PV.NLS-mC, 2.5, 1.25, 0.625, 0.3125, 0.156, and 0.078 μl of total homogenate; recombinant PV, 500, 250, 125, 62.5, and 31.25 ng. C, nuclear calcium responses of hippocampal neurons during action potential bursting evoked by treatment with gabazine (100 μm). Top left panel, sample traces showing typical nuclear calcium transients in untransfected cells (gray) and a cell expressing PV.NLS-mC (black). Right panel, corresponding fluorescence overlay image showing nuclear regions of interest for untransfected cells in white and for a PV.NLS-mC-expressing cell in red. Scale bar = 50 μm. Bottom left panel, quantification of calcium responses in hippocampal neuronal nuclei as indicated. Statistical significance was assessed by one-way ANOVA followed by Tukey's post hoc test. Peak response: untransfected (mC.NLS), PV.NLS-mC, p = 0.0011; mC.NLS, PV.NLS-mC, p = 0.0015; untransfected (PV.NLS-mC), PV.NLS-mC, p = 0.0187. Decay: untransfected (mC.NLS), PV.NLS-mC, p < 0.0001; mC.NLS, PV.NLS-mC, p < 0.0001; untransfected (PV.NLS-mC), PV.NLS-mC, p < 0.0001. ns, not significant. D, calcium responses within the non-nuclear somata (cytoplasm) of the same hippocampal neurons as analyzed in C. Top left panel, sample traces showing typical cytoplasmic calcium transients within the non-nuclear somata of untransfected cells (gray) and a cell expressing PV.NLS-mC (black). Right panel, corresponding fluorescence overlay image showing cytoplasmic regions of interest. Scale bar = 50 μm. Bottom left panel, quantification of cytoplasmic calcium responses in the non-nuclear somata of hippocampal neurons as indicated. Statistical significance was assessed by one-way ANOVA followed by Tukey's post hoc test. *, p < 0.05; **, p < 0.01.
Morphometric Analyses
For morphometric analyses, neurons were analyzed 5 days after transfection. Total dendritic length and complexity were calculated using Fiji (22). Briefly, a z-stack acquisition was imported, calibrated, and traced manually using the simple neurite tracer plugin (23). Total dendritic length was then computed. For three-dimensional Sholl analysis (24), the shell interval was set to 5 μm using a plugin available for Fiji. Dendritic spine density, spine width, and spine length were computed manually using Fiji. All analyses were performed blind. For each condition, a minimum of 12 neurons and between 770–2300 spines from at least three independent preparations were analyzed.
Quantitative RT-PCR
Total RNA was extracted from primary hippocampal cultures on DIV 10 using the RNeasy mini kit (Qiagen, Hilden, Germany) with additional on-column DNase I digestion to eliminate genomic DNA contamination according to the instruction of the manufacturer (Qiagen). 1.2 μg of extracted RNA was reverse-transcribed into first-strand cDNA using a high-capacity cDNA reverse transcription kit (Applied Biosystems, Foster City, CA). Quantitative RT-PCR was performed with a sequence detection system model 7300 real-time PCR system using TaqMan Gene Expression Master Mix (Applied Biosystems) and carboxy-fluorescein (FAM) dye-labeled probe sets designed by Applied Biosystems. The expression levels of target genes were normalized to the expression of Gusb.
Quantification of PV.NLS-mC Expression
Hippocampal neurons infected on DIV 3 with rAAV-PV.NLS-mC were lysed on DIV 10 in boiling SDS sample buffer (160 mm Tris-HCl (pH 6.8), 4% SDS, 30% glycerol, 10 mm dithiothreitol, and 0.02% bromophenol blue). Serially diluted total cell lysates from rAAV-PV.NLS-mC-infected hippocampal neurons and from uninfected control neurons were separated by electrophoresis on 15% SDS-polyacrylamide gels with defined amounts (500, 250, 125, 62.5, and 31.25 ng) of recombinant rat parvalbumin protein (Swant). Proteins were transferred onto a 0.2-μm pore size nitrocellulose membrane (Invitrogen). The lack of staining of the gels with Coomassie Blue confirmed that the transfer of the proteins to the membrane was complete. Membranes were subjected to standard immunoblotting techniques using commercially available antibodies to parvalbumin (Sigma) and HRP-linked secondary antibodies (Dianova), followed by chemiluminescent detection (GE Healthcare Life Sciences). To exclude the possibility of a loss of low molecular weight proteins because of blotting through the 0.2-μm pore membrane, an additional 0.45-μm pore size membrane was placed behind the 0.2-μm pore membrane. However, under the conditions used for the transfer of the proteins to the membrane and their visualization using immunoblotting, the recombinant parvalbumin and the parvalbumin contained in the lysates were present only on the 0.2-μm pore membrane and were not detectable on the second membrane. X-ray films exposed to the chemiluminescence reaction were scanned, and each band was quantified using the gel analyzer tool of ImageJ. Signals were thresholded to fit a linear range by analyzing a series of x-ray films obtained using increasing exposure times. To calculate the amount of PV.NLS-mC present in the neurons, signals derived from the lysates were compared with signals derived from samples containing known amounts of recombinant PV. The amount of PV.NLS-mC present in the lysates was divided by the number of rAAV-PV.NLS-mC-infected neurons corresponding to the amount of lysate. This yielded the protein amount in femtograms per neuron, which equals the protein amount in femtograms per nucleus given that the expression of PV.NLS-mC is restricted to the nucleus. For the conversion from femtograms to moles, we used 42.4 kDa as the molecular mass for PV.NLS-mC. Five independent biological replicates and two independent batches of recombinant PV were used for this analysis, which revealed that, under the conditions used, 3.26 ± 0.4 fmol of PV.NLS-mC are present in a single nucleus from a mouse hippocampal neuron. To express the amount of PV.NLS-mC in the cell nucleus in molarity, we divided 3.26 ± 0.4 fmol by the volume of the nucleus. In a previous study, we quantified three-dimensional image-reconstructed nuclei of cultured rat hippocampal neurons and found that these have an average volume of about 120 fl (25). In a direct comparison, we observed that nuclei of cultured mouse hippocampal neurons were about 50% the size of nuclei of cultured rat hippocampal neurons.3 We therefore used 60 fl to calculate the molarity of nuclear PV.NLS-mC, which amounted to 54 mm. We realize that the aqueous component of the cell nucleus is smaller than the total volume of the nucleus because of the presence of other structures in the nucleus, such as chromatin and components of the transcription and splicing machineries. Therefore, the value obtained for the molarity of nuclear PV.NLS-mC may be an underestimation. However, the error introduced by calculating with a nuclear volume of 60 fl may be counterbalanced by a possible underestimation of the true size of nuclei because fixation procedures used to process cells for the analysis of the size of the nucleus may lead to shrinkage of the nucleus (26).
Antibodies
The following antibodies were used: mouse monoclonal antibodies to parvalbumin and tubulin (Sigma), rabbit polyclonal antibody to cFos (Santa Cruz), mouse monoclonal antibody to phospho-CREB (Upstate-Millipore), rabbit polyclonal antibodies to CREB and ERK1/2 and mouse monoclonal antibody to phospho-ERK1/2 (Cell Signaling Technology), and rabbit polyclonal antibodies to DsRed (Clontech).
Data Analysis
All plotted data represent mean ± S.E. One-way analysis of variance (ANOVA) with Tukey's post hoc test was used for statistical analyses. Results were considered to be statistically significant for significance levels of p < 0.05 (*) or p < 0.01 (**).
Results
Modulation of the Neuronal Nuclear Calcium Buffering Capacity
To investigate the importance of calcium buffers in the cell nucleus for intracellular signal transduction, gene regulation, and neuronal morphology, we increased the nuclear calcium buffering capacity in primary mouse hippocampal neurons by overexpressing a nuclearly targeted version of parvalbumin fused to mCherry (PV.NLS-mC) (14, 15, 27) or, as a control, nuclearly localized mCherry (mC.NLS) using recombinant rAAVs (Fig. 1A). Following infection with rAAV-mC.NLS or rAAV-PV.NLS-mC, expression of mC-NLS or PV.NLS-mC was readily detectable in 80–95% of viable neurons via mCherry fluorescence (Fig. 1A). Using lysates from rAAV-PV.NLS-mC-infected neurons and purified parvalbumin as a standard for the calibration of signals in immunoblot analyses, we first determined the amount of PV.NLS-mC in the nucleus. A detailed description of the quantification of PV.NLS-mC expression can be found in the “Experimental Procedures,” and a typical example of the immunoblots is shown in Fig. 1B. Our analysis revealed a concentration of about 54 mm PV.NLS-mC in the nucleus of primary mouse hippocampal neurons infected with rAAV-PV.NLS-mC. Reports in the literature on precise measurements of intracellular concentrations of CaBPs are rare, and, to the best of our knowledge, no precise information is available on the nuclear calcium buffering capacity. Therefore, it is difficult to make statements about the –fold increase in nuclear calcium buffering capacity achieved using this method. However, a comparison of our results with available data on the expression of parvalbumin (1 mm or more in axons and axon terminals of Purkinje and basket cells (28) and up to 3 mm in outer hair cells (29)) suggests that, using our expression system, the protein-based calcium buffering capacity of the cell nucleus is likely to be increased by at least 1 order of magnitude.
Selective Alteration of Nuclear Calcium Signals by PV.NLS-mC
To determine how overexpressed nuclear parvalbumin influences the amplitude and kinetics of calcium rises triggered by neuronal activity, we performed calcium imaging of primary rat hippocampal neurons transfected with pAAV-PV.NLS-mC or pAAV-mC.NLS. Using the ratiometric chemical calcium indicator Fura-2/AM, we quantified baseline calcium levels, peak calcium responses during bursts of action potentials induced by treatment of the cultures with the GABAA receptor blocker gabazine (30), and the rate at which calcium responses decayed over time (Fig. 1, C and D). No differences in baseline calcium levels were observed. However, consistent with our expectations and with findings published previously (14, 15), we found that peak calcium responses in nuclear but not non-nuclear somatic (cytoplasmic) regions of interest were lower in PV.NLS-mC-transfected cells than in neighboring untransfected cells and that the decay of nuclear but not cytoplasmic calcium transients was slower in PV.NLS-mC-transfected cells than in untransfected control cells. These effects cannot be attributed to the overexpression of an exogenous nuclear protein because no such differences were observed for mC.NLS-transfected cells. Our imaging results therefore confirm that PV.NLS-mC effectively buffers neuronal activity-associated nuclear calcium responses in hippocampal neurons, both decreasing their amplitude and slowing their decay. Moreover, our observation that neither the amplitude nor the kinetics of cytoplasmic calcium responses from the same group of neurons were altered suggest that extranuclear calcium-dependent signaling cascades (i.e. phosphorylation of ERK1/2) should be intact in neurons expressing PV.NLS-mC.
Nuclear Calcium Buffering Capacity and Neuronal Morphology
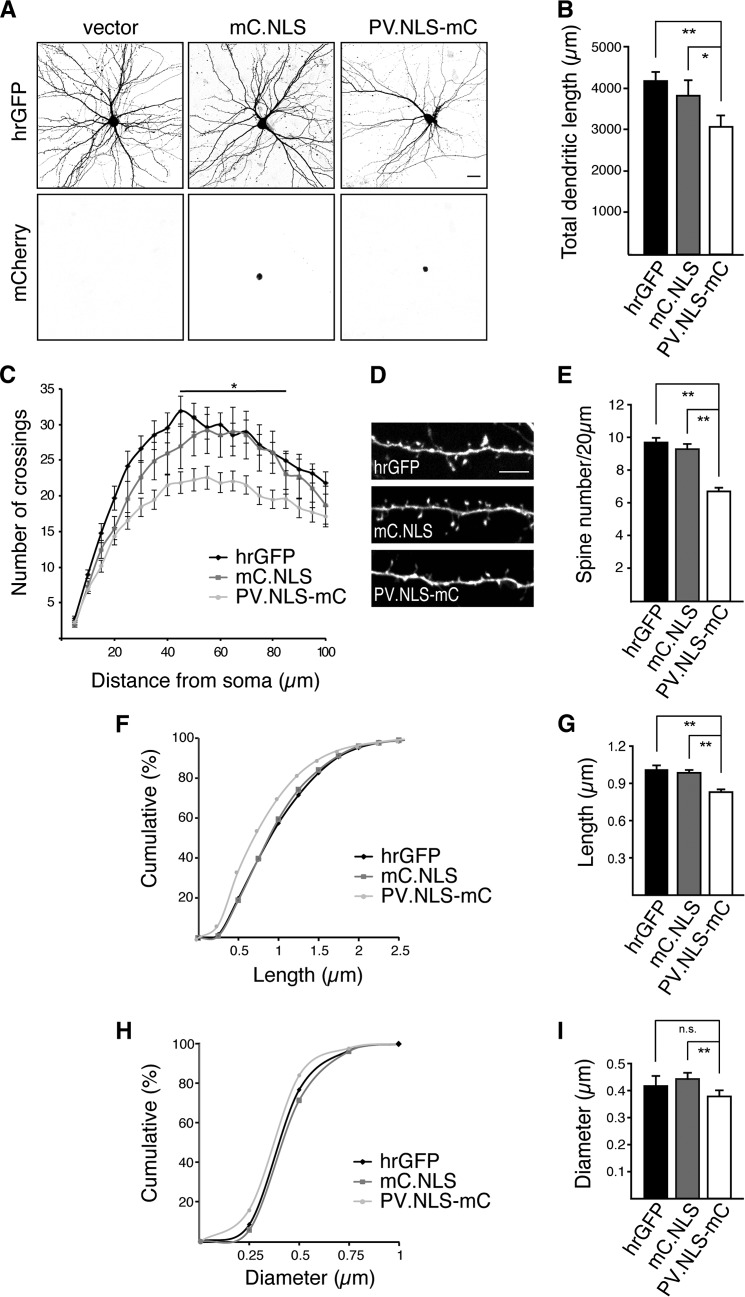
We next investigated the morphology of hippocampal neurons expressing PV.NLS-mC. The structure of neurons was visualized by expressing a humanized Renilla reniformis GFP (hrGFP) alongside PV.NLS-mC or, as a control, mC.NLS (Fig. 2A). A morphometric assessment of transfected neurons using Sholl analysis (9) revealed that, compared with controls, neurons expressing PV.NLS-mC showed a significant decrease in both the total length and complexity of their dendritic trees (Fig. 2, B and C). This alteration in the geometry of dendrites of PV.NLS-mC-expressing hippocampal neurons was accompanied by a significant decrease in the density of dendritic spines (Fig. 2, D and E). Moreover, the remaining spines of PV.NLS-mC-expressing neurons were considerably shorter and thinner than those of control neurons (Fig. 2, F–I). These results indicate that nuclear calcium buffering capacity is functionally linked to the regulation of important structural features of hippocampal pyramidal neurons.
FIGURE 2.
Nuclear expression of the calcium buffer parvalbumin affects neuronal morphology. A, representative micrographs of cultured hippocampal neurons transfected with an expression vector for hrGFP or cotransfected with expression vectors for hrGFP and mC.NLS or PV.NLS-mC as indicated. GFP fluorescence (top row) reveals neuronal architecture. mCherry fluorescence (bottom row) shows nuclear localization of mC.NLS and PV.NLS-mC. Scale bar = 20 μm. B, quantification of the total dendritic length of hippocampal neurons transfected as indicated. Statistically significant differences were determined by one-way ANOVA followed by Tukey's post hoc test. hrGFP, PV.NLS-mC, p = 0.009557; mC.NLS, PV.NLS-mC, p = 0.04021. C, Sholl analysis of hippocampal neurons transfected as indicated. D, representative micrographs of dendritic spines of hippocampal neurons transfected as in A. Scale bar = 5 μm. E, quantification of dendritic spine density of neurons transfected as in A. Statistically significant differences were determined by one-way ANOVA followed by Tukey's post hoc test. hrGFP, PV.NLS-mC, p = 0.0000103; mC.NLS, PV.NLS-mC, p = 0.00001033. F and H, cumulative frequency plots of spine length and width from neurons transfected as indicated. G and I, average spine lengths and widths from neurons transfected with the indicated constructs. Statistically significant differences were determined by one-way ANOVA followed by Tukey's post hoc test: Spine length: hrGFP, PV.NLS-mC, p = 0.0006633; mC.NLS, PV.NLS-mC, p = 0.003244. Spine width: hrGFP, PV.NLS-mC, p = 0.1066; mC.NLS, PV.NLS-mC, p = 0.002185. More than 2300 spines and 12 neurons from a minimum of three independent preparations were examined for each construct. ns, not significant.*, p < 0.05; **, p < 0.01.
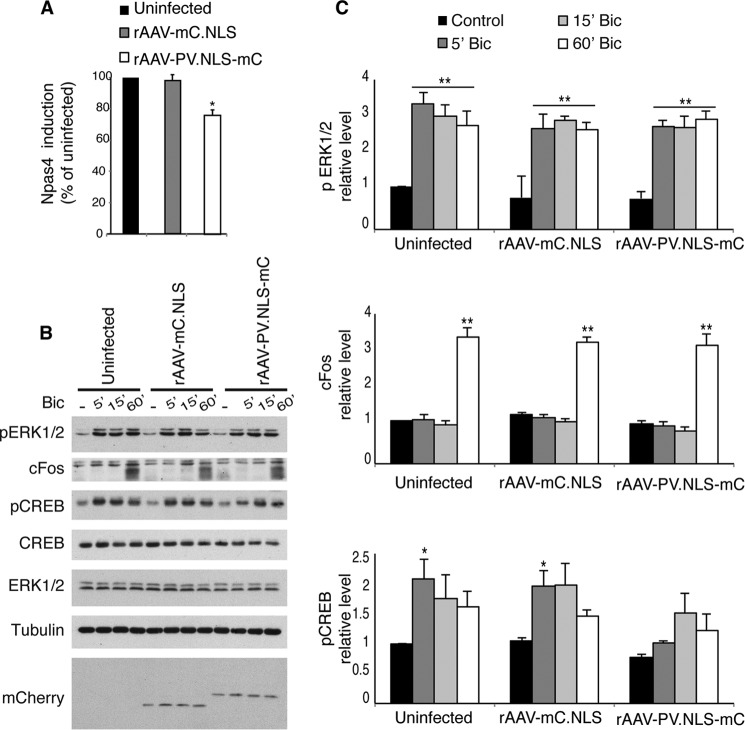
As an underlying mechanism, we considered the possibility that PV.NLS-mC causes changes in the expression of neuronal morphology-regulating genes by interfering with transcription-relevant calcium signaling pathways. Indeed, in PV.NLS-mC-expressing hippocampal neurons, induction of the calcium-regulated gene Npas4 (induced by action potential bursting following treatment of the cultures with the GABAA receptor blocker bicuculline (8)) was impaired (Fig. 3A). We also investigated the effect of nuclear calcium buffers on calcium-dependent signaling to the transcription factor CREB. We found that hippocampal neurons expressing PV.NLS-mC fail to undergo an increase in CREB phosphorylation on its activator site, serine 133, at an early time point (5 min) after bicuculline treatment (Fig. 3, B and C). At later time points (i.e. 15 and 60 min), however, we did observe a bicuculline-induced increase in CREB phosphorylation on serine 133 (Fig. 3, B and C). This is expected because bicuculline treatment leads to periodically occurring bursts of action potentials, each of which generates a robust calcium transient that invades the cell nucleus (31, 32), leading to saturation of the nuclear calcium buffering capacity. Consistent with saturation of nuclear calcium buffering, we observed that the increase in c-Fos levels detected 60 min after bicuculline treatment was unaffected by the expression of PV.NLS-mC (Fig. 3, B and C). We also analyzed the regulation (i.e. phosphorylation) of ERK1/2, which is activated in synaptically stimulated neurons via a Ras-dependent process that is initiated by submembraneous calcium signals and operates independently of nuclear calcium (31). As expected, the activation of ERK1/2 following bicuculline treatment was not impaired by the expression of PV.NLS-mC (Fig. 3, B and C).
FIGURE 3.
PV.NLS impairs the initial phase of CREB activation. A, quantitative RT-PCR analysis of Npas4 mRNA levels in uninfected hippocampal neurons and in hippocampal neurons infected with rAAVs driving expression of the indicated proteins (n = 5) and treated with bicuculline (2 h, 50 μm). Statistically significant differences were determined by one-way ANOVA followed by Tukey's post hoc test. Uninfected, PV.NLS-mC, p = 0.0175; mC.NLS, PV.NLS-mC, p = 0.0265. B, immunoblot analysis of uninfected hippocampal neurons and of hippocampal neurons infected with rAAV-mC.NLS or rAAV-PV.NLS-mC and treated with bicuculline (Bic, 50 μm) for the indicated times. A tubulin immunoblot is shown as a control for protein loading and mCherry for detection of mC.NLS and PV.NLS-mC expression. C, quantification of the experiment shown in B. All values are normalized to uninfected controls (n = 3). Statistically significant differences were determined by one-way ANOVA followed by Tukey's post hoc test. pCREB: uninfected, 5′ Bic, control, p = 0.041; mC.NLS, 5′ Bic, control, p = 0.0372; PV.NLS-mC, 5′ Bic, control, p = 0.684. *, p < 0.05; **, p < 0.01.
PV.NLS Influences the Transcription of Genes Important for Neuronal Morphology
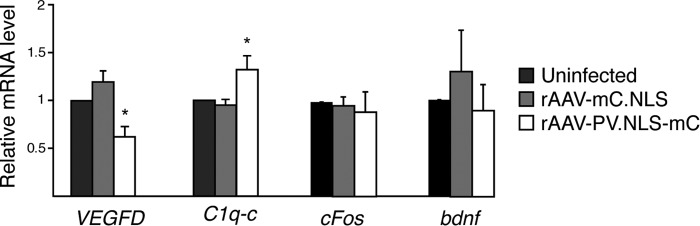
Given that the observed changes in neuronal structure take place under conditions of basal synaptic activity, we next investigated the expression of morphology-relevant genes that are controlled, under basal conditions, by a nuclear calcium signaling pathway. We focused on two nuclear calcium signaling targets, VEGFD and the complement factor C1q-c, that can function as regulators of neuronal structure (9, 11). In the mouse hippocampus, VEGFD is critical for dendritic tree maintenance and is required for the ability of mice to form long-term memories (9). The complement factor C1q-c, an initiator of the complement cascade, functions as a synapse pruning factor in spinal cord and hippocampal neurons (11, 33–35). To investigate whether, under basal conditions, the nuclear calcium buffering capacity affects the expression of VEGFD or C1q-c, primary hippocampal neurons were infected with rAAV-PV.NLS-mC or rAAV-mC.NLS (Figs. 1A and 4). Quantitative RT-PCR analysis revealed that, indeed, PV.NLS-mC expression caused alterations in the mRNA levels of both VEGFD and C1q-c. Compared with rAAV-mC.NLS-infected controls, rAAV-PV.NLS-mC-infected neurons had significantly lower levels of VEGFD mRNA (Fig. 4). In contrast, and consistent with previous observations that C1q-c expression is suppressed by nuclear calcium signals, the mRNA levels of C1q-c in hippocampal neurons expressing PV.NLS-mC were increased compared with controls (Fig. 4). The basal expression levels of two other synaptic activity-regulated genes that were analyzed in parallel, c-fos and bdnf, were found to be unaltered by the expression of PV.NLS-mC (Fig. 4).
FIGURE 4.
Increasing the calcium buffering capacity of the nucleus alters the expression of genes that regulate neuronal architecture. Quantitative RT-PCR analysis of VEGFD, C1q-c, cFOS, and bdnf mRNA levels in uninfected hippocampal neurons and in hippocampal neurons infected with rAAVs driving the expression of the indicated proteins (n = 5–9). Statistically significant differences were determined by one-way ANOVA followed by Tukey's post hoc test. VEGFD: uninfected, PV.NLS-mC, p = 0.03578; mC.NLS, PV.NLS-mC, p = 0.0009283. C1q-c: uninfected, PV.NLS-mC. p = 0.04019; mC.NLS, PV.NLS-mC. p = 0.05287. *, p < 0.05.
PV.NLS Regulates Neuronal Morphology via the Modulation of VEGFD and C1q-c
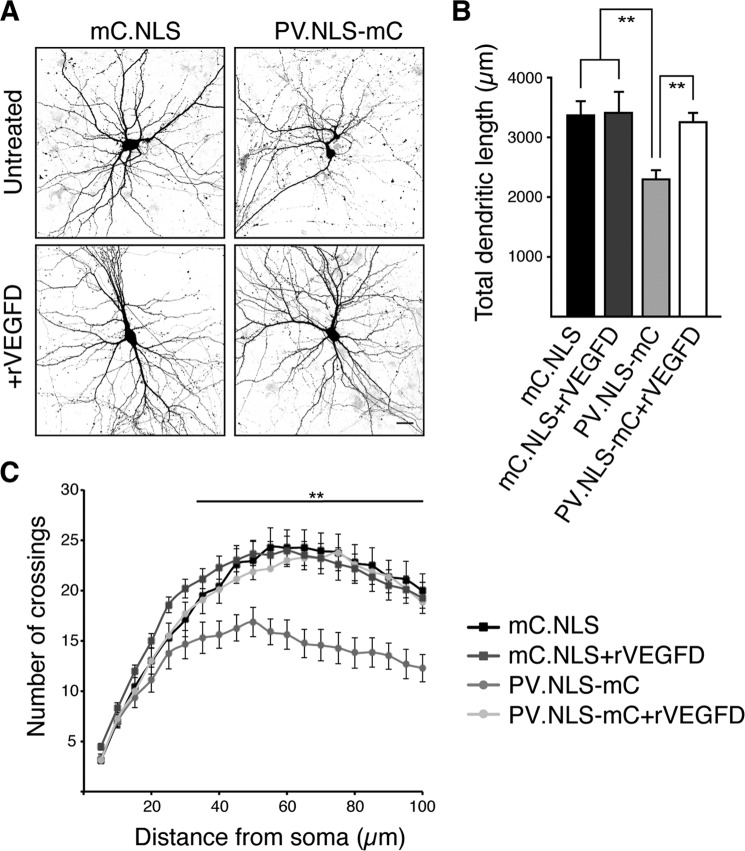
To investigate a possible causal link between the altered expression levels of VEGFD and C1q-c and the observed structural changes of neurons, we carried out rescue experiments. We first determined whether the observed decrease in expression levels of VEGFD in PV.NLS-mC-expressing neurons is responsible for the observed changes in dendrite geometry by exposing hippocampal neurons expressing PV.NLS-mC to recombinant VEGFD (rVEGFD) (Fig. 5A). A morphometric assessment of these neurons revealed that rVEGFD treatment had no detectable effect on the morphology of control neurons expressing mC.NLS, which is consistent with a previous study (9). However, rVEGFD rescued the reduction in dendritic length and complexity caused by the expression of PV.NLS-mC (Fig. 5, B and C). These results indicate that the nuclear calcium buffering capacity can modulate dendritic length and complexity through the regulation of VEGFD expression levels.
FIGURE 5.
Nuclear expression of the calcium buffer parvalbumin regulates dendritic morphology via VEGFD. A, representative micrographs of cultured hippocampal neurons cotransfected with expression vectors for hrGFP and mC.NLS or PV.NLS-mC with or without treatment for 3 days with rVEGFD (100 ng/ml). Scale bar = 20 μm. B, quantification of the total dendritic length of hippocampal neurons transfected as in A and, where indicated, treated with rVEGFD. C, Sholl analysis of hippocampal neurons transfected and treated as indicated. Statistically significant differences were determined by one-way ANOVA followed by Tukey's post hoc test. mC.NLS, PV.NLS-mC, p = 0.001785; mC.NLS + rVEGFD, PV.NLS-mC, p = 0.00472; PV.NLS-mC, PV.NLS-mC + rVEGFD, p = 0.002428. More than 12 neurons from a minimum of three independent preparations were examined for each construct. **, p < 0.01.
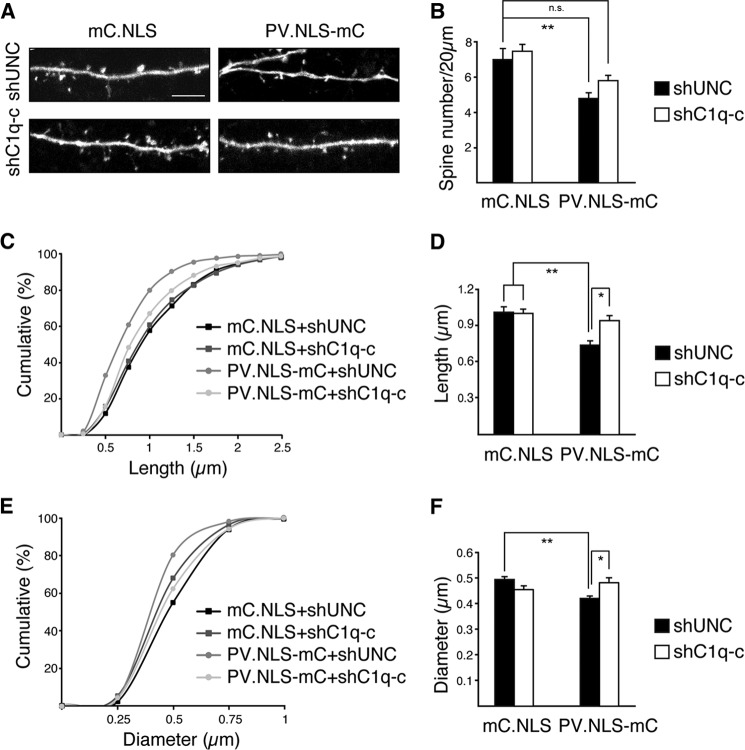
We finally investigated whether the observed increased expression of C1q-c is responsible for the observed spine phenotype of PV.NLS-mC-expressing neurons via loss-of-function experiments using RNAi. Mouse hippocampal neurons were transfected with expression vectors for mC.NLS or PV.NLS-mC alongside previously characterized expression vectors for a C1q-c-specific shRNA (shC1q-c) or a control shRNA (shUNC, Fig. 6A) (11). The RNAi constructs also contained an expression cassette for enhanced GFP to visualize the neuronal architecture. We found that the difference in spine density in mC.NLS- versus PV.NLS-mC-expressing neurons observed in the control group expressing shUNC was lost in neurons transfected with expression vectors for shC1q-c (Fig. 6B). Within the groups of PV.NLS-mC-expressing neurons, the spine density of shC1q-c-expressing neurons was higher than that of the neurons expressing shUNC, although this difference did not reach statistical significance (p = 0.16, Fig. 6B). Therefore, knockdown of C1q-c can partially restore the decrease in spine density induced by increasing nuclear calcium buffering. We obtained full rescue of the PV.NLS-mC-induced spine morphology phenotype in the C1q-c loss-of-function experiments. Both the length and width of spines of hippocampal neurons expressing PV.NLS-mC were restored to control levels through transfection with the shC1q-c expression vector (Fig. 6, C–F). These results indicate that the observed alterations in the density and morphology of dendritic spines caused by increases in nuclear calcium buffering are mediated, at least in part, by a concomitant up-regulation of C1q-c.
FIGURE 6.
PV.NLS expression affects dendritic spine density and size via C1q-c expression. A, representative micrographs of enhanced GFP-labeled dendritic spines on hippocampal neurons cotransfected with pAAV-shC1q-c or pAAV-shUNC and pAAV-mC.NLS or pAAV-PV.NLS-mC. Scale bar = 5 μm. B, quantification of the dendritic spine density of neurons transfected as in A. Statistically significant differences were determined by one-way ANOVA followed by Tukey's post hoc test. mC.NLS + shUNC, PV.NLS-mC + shUNC, p = 0.009762; mC.NLS + shUNC, PV.NLS-mC + shC1q-c, p = 0.4084; mC.NLS + shC1q-c, PV.NLS-mC + shUNC, p = 0.0006115; PV.NLS-mC + shUNC, PV.NLS-mC + shC1q-c, p = 0.15762. ns, not significant. C and E, cumulative frequency plots of spine length and width in neurons transfected with the indicated constructs. D and F, average spine lengths and widths from neurons transfected as indicated. Statistically significant differences were determined by one-way ANOVA followed by Tukey's post hoc test. Spine length: mC.NLS + shUNC, PV.NLS-mC + shUNC, p = 0.0006208; mC.NLS + shC1q-c, PV.NLS-mC + shUNC, p = 0.001089; PV.NLS-mC + shUNC, PV.NLS-mC + shC1q-c, p = 0.01785. Spine width: mC.NLS + shUNC, PV.NLS-mC + shUNC, p = 0.005149; mC.NLS + shC1q-c, PV.NLS-mC + shUNC, p = 0.5191; PV.NLS-mC + shUNC, PV.NLS-mC + shC1q-c, p = 0.02638. More than 770 spines and 12 neurons from a minimum of three independent preparations were examined for each construct. *, p < 0.05; **, p < 0.01.
Discussion
In this study, we established a functional link between the calcium buffering capacity of the cell nucleus, the expression of neuronal morphology-relevant genes, and structural alterations of dendrites and spines.
Nuclear Calcium Buffering in Transcriptional Regulation
Unlike calcium sensors such as calmodulin or caldendrin (36–38), CaBPs that act as calcium buffers primarily affect the spatiotemporal characteristics of calcium transients. They have been less at the center of attention because, despite their generally accepted role in shaping cellular calcium signals, only limited information is available regarding their relevance for physiological processes. The discovery that calcium buffering in the cell nucleus is an important determinant of neuronal architecture may at first seem surprising. However, it does fit into the general concept according to which nuclear calcium signaling serves as a key regulator of various brain functions (7). Given that nuclear calcium signaling not only regulates genes that specify neuronal structure, one might expect that the calcium buffering capacity of the nucleus also modulates other cellular processes or network functions. These may include, in particular, neuronal survival, memory formation, and the development of chronic pain, for all of which nuclear calcium-regulated gene programs have been identified (8–12, 39).
Important mediators of nuclear calcium-regulated genomic responses include the nuclear calcium/calmodulin-dependent enzymes calcium/calmodulin-dependent kinase II (CaMKII) and calcium/calmodulin-dependent kinase IV (CaMKIV), which control the activity of several transcriptional regulators, including CREB, CREB-binding protein, and MeCP2 (7, 40, 41). Our experiments uncovered that the levels of calcium buffering in the nucleus, which have a direct influence on the amplitude and kinetics of neuronal activity-induced nuclear calcium rises, can also influence the kinetics of CREB activation following synaptic activity so that high levels of nuclear calcium buffering eliminate the fast component (Fig. 3, B and C). In a previous study, we could also show that nuclear calcium buffering affects the nucleocytoplasmic shuttling of class IIa histone deacetylases (13), key players in the epigenetic control of gene expression (42, 43). Nuclear calcium buffering is also likely to alter the activity of the downstream regulatory element (DRE)-binding protein, DREAM (also known as calsenilin or KChIP3 (44)), a multifunctional protein with several EF-hand calcium binding domains that can function as a calcium-regulated transcriptional repressor (5, 45). Therefore, the consequences of changes in the nuclear calcium buffering capacity are diverse and widespread and, collectively, are expected to have an impact on the rate of transcription of numerous genes. In this study, we describe the regulation of two morphology-relevant genes, VEGFD and C1q-c, by nuclear calcium buffering. An unbiased transcriptome analysis is likely to uncover many more genes.
Nuclear Calcium Buffering and Disease
The link between nuclear calcium buffering and neuronal structure may help us understand the etiology of neurological dysfunctions associated with alterations in calcium buffering. For example, in Alzheimer disease and schizophrenia, but also in aging, both structural remodeling of neuronal circuitry (46–48) and abnormal expression or deregulation of neuronal calcium buffers have been reported (49–51). It is unknown, however, whether the observed alterations in the calcium buffer content of affected brain regions result from changes in the expression levels of CaBPs or are, instead, caused by the loss of interneurons, which typically express high levels of certain CaBPs (3). If indeed the level of calcium buffering is altered, then it would be important to determine the precise cellular and spatial distribution of affected calcium buffers and to analyze the expression of nuclear calcium signaling-dependent morphology-relevant target genes, including VEGFD and C1q-c.
Modulation of Nuclear Calcium Buffering as a Therapeutic Strategy
One possible application of our experimental approach may be to use modulators of nuclear calcium buffering to induce structural and functional changes in neurons and networks. Increases in the nuclear buffering capacity of neurons can be generated by the overexpression of nuclearly targeted calcium buffers, which, as reported here, is accompanied by a reduction in the length and complexity of dendritic trees and in the number of dendritic spines. Conceptually, this could be useful for the treatment of autism spectrum disorders in which failures in obligatory spine pruning during development result in an abnormally high number of dendritic spines that inversely correlates with cognitive functions (48). On the other hand, it may be equally desirable to generate tools that reduce the nuclear calcium buffering capacity because these would be expected to increase the number of large spines and the complexity of dendritic trees. In particular, these types of changes in neuronal geometry may prove to be beneficial in various neurodegenerative conditions associated with spine loss and dendrite atrophy.
Generally, an abnormal dendritic architecture can negatively influence neuronal electrical properties and compromise neuronal signal processing and network computations (52), ultimately resulting in debilitating cognitive deficits (9). Therefore, neurons may have to adopt the right level of structural complexity and connectivity to fulfill their functions. Disease- or aging-associated aberrations away from an “optimal” neuronal architecture, either too little or too much complexity and connectivity, may lower the level of network performance. Modulators of nuclear calcium buffering offer a means to achieve a rebalancing of structural features to regain or improve brain function. The development of such therapeutic tools would be facilitated greatly by the identification of CaBPs that are expressed endogenously in the neuronal cell nucleus.
Taken together, our results indicate that the calcium buffering capacity of neuronal nuclei is an important determinant of neuronal architecture. The development of tools for increasing or decreasing nuclear calcium buffer expression may represent a valid and novel strategy for the treatment of disease- or aging-related conditions that are expected to benefit from changes in the morphology of dendrites and spines.
Author Contributions
H. B. conceived the project. H. B., D. M., A. M. H., K. S., and U. W. designed the experiments. D. M., A. M. H., K. S., and U. W. performed all experiments. H. B., D. M., A. M. H., K. S., and U. W. analyzed and interpreted the data. H. B., D. M., A. M. H., and K. S. wrote the paper.
Acknowledgments
We thank Iris Bünzli-Ehret for help with the preparation of hippocampal cultures, Dr. Yan Yu for observations regarding the size of mouse and rat hippocampal nuclei, and Drs. Barbara Ehrlich and Wayne Grant for the parvalbumin.NLS plasmid.
This work was supported by an ERC Advanced Grant (to H. B.), by Sonderforschungsbereich 636 of the Deutsche Forschungsgemeinschaft, by the Excellence Cluster CellNetworks at Heidelberg University, and by German Ministry of Education and Research Grant 01GQ1003A. The authors declare that they have no conflicts of interest with the contents of this article.
Y. Yu and H. Bading, unpublished observation.
- CaBP
- calcium-binding protein
- CREB
- cAMP response element-binding protein
- rAAV
- recombinant adeno-associated virus
- DIV
- day(s) in vitro
- CICM
- CO2-independent culture medium
- ANOVA
- analysis of variance
- hrGFP
- humanized Renilla reniformis GFP
- rVEGFD
- recombinant VEGFD
- PV
- parvalbumin.
References
- 1. Baimbridge K. G., Celio M. R., Rogers J. H. (1992) Calcium-binding proteins in the nervous system. Trends Neurosci. 15, 303–308 [DOI] [PubMed] [Google Scholar]
- 2. Andressen C., Blümcke I., Celio M. R. (1993) Calcium-binding proteins: selective markers of nerve cells. Cell Tissue Res. 271, 181–208 [DOI] [PubMed] [Google Scholar]
- 3. Schwaller B. (2009) The continuing disappearance of “pure” Ca2+ buffers. Cell. Mol. Life Sci. 66, 275–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Blatow M., Caputi A., Burnashev N., Monyer H., Rozov A. (2003) Ca2+ buffer saturation underlies paired pulse facilitation in calbindin-D28k-containing terminals. Neuron 38, 79–88 [DOI] [PubMed] [Google Scholar]
- 5. Carrión A. M., Link W. A., Ledo F., Mellström B., Naranjo J. R. (1999) DREAM is a Ca2+-regulated transcriptional repressor. Nature 398, 80–84 [DOI] [PubMed] [Google Scholar]
- 6. Mueller A., Schäfer B. W., Ferrari S., Weibel M., Makek M., Höchli M., Heizmann C. W. (2005) The calcium-binding protein S100A2 interacts with p53 and modulates its transcriptional activity. J. Biol. Chem. 280, 29186–29193 [DOI] [PubMed] [Google Scholar]
- 7. Bading H. (2013) Nuclear calcium signalling in the regulation of brain function. Nat. Rev. Neurosci. 14, 593–608 [DOI] [PubMed] [Google Scholar]
- 8. Zhang S. J., Zou M., Lu L., Lau D., Ditzel D. A., Delucinge-Vivier C., Aso Y., Descombes P., Bading H. (2009) Nuclear calcium signaling controls expression of a large gene pool: identification of a gene program for acquired neuroprotection induced by synaptic activity. PLoS Genet. 5, e1000604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mauceri D., Freitag H. E., Oliveira A. M., Bengtson C. P., Bading H. (2011) Nuclear calcium-VEGFD signaling controls maintenance of dendrite arborization necessary for memory formation. Neuron 71, 117–130 [DOI] [PubMed] [Google Scholar]
- 10. Oliveira A. M., Hemstedt T. J., Bading H. (2012) Rescue of aging-associated decline in Dnmt3a2 expression restores cognitive abilities. Nat. Neurosci. 15, 1111–1113 [DOI] [PubMed] [Google Scholar]
- 11. Simonetti M., Hagenston A. M., Vardeh D., Freitag H. E., Mauceri D., Lu J., Satagopam V. P., Schneider R., Costigan M., Bading H., Kuner R. (2013) Nuclear calcium signaling in spinal neurons drives a genomic program required for persistent inflammatory pain. Neuron 77, 43–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhang S. J., Steijaert M. N., Lau D., Schütz G., Delucinge-Vivier C., Descombes P., Bading H. (2007) Decoding NMDA receptor signaling: identification of genomic programs specifying neuronal survival and death. Neuron 53, 549–562 [DOI] [PubMed] [Google Scholar]
- 13. Schlumm F., Mauceri D., Freitag H. E., Bading H. (2013) Nuclear calcium signaling regulates nuclear export of a subset of class IIa histone deacetylases following synaptic activity. J. Biol. Chem. 288, 8074–8084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rodrigues M. A., Gomes D. A., Leite M. F., Grant W., Zhang L., Lam W., Cheng Y. C., Bennett A. M., Nathanson M. H. (2007) Nucleoplasmic calcium is required for cell proliferation. J. Biol. Chem. 282, 17061–17068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pusl T., Wu J. J., Zimmerman T. L., Zhang L., Ehrlich B. E., Berchtold M. W., Hoek J. B., Karpen S. J., Nathanson M. H., Bennett A. M. (2002) Epidermal growth factor-mediated activation of the ETS domain transcription factor Elk-1 requires nuclear calcium. J. Biol. Chem. 277, 27517–27527 [DOI] [PubMed] [Google Scholar]
- 16. Estrada M., Uhlen P., Ehrlich B. E. (2006) Ca2+ oscillations induced by testosterone enhance neurite outgrowth. J. Cell Sci. 119, 733–743 [DOI] [PubMed] [Google Scholar]
- 17. Klugmann M., Symes C. W., Leichtlein C. B., Klaussner B. K., Dunning J., Fong D., Young D., During M. J. (2005) AAV-mediated hippocampal expression of short and long Homer 1 proteins differentially affect cognition and seizure activity in adult rats. Mol. Cell. Neurosci. 28, 347–360 [DOI] [PubMed] [Google Scholar]
- 18. Bading H., Greenberg M. E. (1991) Stimulation of protein tyrosine phosphorylation by NMDA receptor activation. Science 253, 912–914 [DOI] [PubMed] [Google Scholar]
- 19. Zhang S. J., Buchthal B., Lau D., Hayer S., Dick O., Schwaninger M., Veltkamp R., Zou M., Weiss U., Bading H. (2011) A signaling cascade of nuclear calcium-CREB-ATF3 activated by synaptic NMDA receptors defines a gene repression module that protects against extrasynaptic NMDA receptor-induced neuronal cell death and ischemic brain damage. J. Neurosci. 31, 4978–4990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wiegert J. S., Bengtson C. P., Bading H. (2007) Diffusion and not active transport underlies and limits ERK1/2 synapse-to-nucleus signaling in hippocampal neurons. J. Biol. Chem. 282, 29621–29633 [DOI] [PubMed] [Google Scholar]
- 21. Grynkiewicz G., Poenie M., Tsien R. Y. (1985) A new generation of Ca2+ indicators with greatly improved fluorescence properties. J. Biol. Chem. 260, 3440–3450 [PubMed] [Google Scholar]
- 22. Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J. Y., White D. J., Hartenstein V., Eliceiri K., Tomancak P., Cardona A. (2012) Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Longair M. H., Baker D. A., Armstrong J. D. (2011) Simple Neurite Tracer: open source software for reconstruction, visualization and analysis of neuronal processes. Bioinformatics 27, 2453–2454 [DOI] [PubMed] [Google Scholar]
- 24. Sholl D. A. (1953) Dendritic organization in the neurons of the visual and motor cortices of the cat. J. Anatomy 87, 387–406 [PMC free article] [PubMed] [Google Scholar]
- 25. Wittmann M., Queisser G., Eder A., Wiegert J. S., Bengtson C. P., Hellwig A., Wittum G., Bading H. (2009) Synaptic activity induces dramatic changes in the geometry of the cell nucleus: interplay between nuclear structure, histone H3 phosphorylation, and nuclear calcium signaling. J. Neurosci. 29, 14687–14700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kasischke K., Büchner M., Ludolph A. C., Riepe M. W. (2001) Nuclear shrinkage in live mouse hippocampal slices. Acta Neuropathol. 101, 483–490 [DOI] [PubMed] [Google Scholar]
- 27. Guatimosim S., Amaya M. J., Guerra M. T., Aguiar C. J., Goes A. M., Gómez-Viquez N. L., Rodrigues M. A., Gomes D. A., Martins-Cruz J., Lederer W. J., Leite M. F. (2008) Nuclear Ca2+ regulates cardiomyocyte function. Cell Calcium 44, 230–242 [DOI] [PubMed] [Google Scholar]
- 28. Kosaka T., Kosaka K., Nakayama T., Hunziker W., Heizmann C. W. (1993) Axons and axon terminals of cerebellar Purkinje cells and basket cells have higher levels of parvalbumin immunoreactivity than somata and dendrites: quantitative analysis by immunogold labeling. Exp. Brain Res. 93, 483–491 [DOI] [PubMed] [Google Scholar]
- 29. Hackney C. M., Mahendrasingam S., Penn A., Fettiplace R. (2005) The concentrations of calcium buffering proteins in mammalian cochlear hair cells. J. Neurosci. 25, 7867–7875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pegoraro S., Broccard F. D., Ruaro M. E., Bianchini D., Avossa D., Pastore G., Bisson G., Altafini C., Torre V. (2010) Sequential steps underlying neuronal plasticity induced by a transient exposure to gabazine. J. Cell Physiol. 222, 713–728 [DOI] [PubMed] [Google Scholar]
- 31. Hardingham G. E., Arnold F. J., Bading H. (2001) A calcium microdomain near NMDA receptors: on switch for ERK-dependent synapse-to-nucleus communication. Nat. Neurosci. 4, 565–566 [DOI] [PubMed] [Google Scholar]
- 32. Hardingham G. E., Fukunaga Y., Bading H. (2002) Extrasynaptic NMDARs oppose synaptic NMDARs by triggering CREB shut-off and cell death pathways. Nat. Neurosci. 5, 405–414 [DOI] [PubMed] [Google Scholar]
- 33. Chu Y., Jin X., Parada I., Pesic A., Stevens B., Barres B., Prince D. A. (2010) Enhanced synaptic connectivity and epilepsy in C1q knockout mice. Proc. Natl. Acad. Sci. U.S.A. 107, 7975–7980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Stevens B., Allen N. J., Vazquez L. E., Howell G. R., Christopherson K. S., Nouri N., Micheva K. D., Mehalow A. K., Huberman A. D., Stafford B., Sher A., Litke A. M., Lambris J. D., Smith S. J., John S. W., Barres B. A. (2007) The classical complement cascade mediates CNS synapse elimination. Cell 131, 1164–1178 [DOI] [PubMed] [Google Scholar]
- 35. Stephan A. H., Barres B. A., Stevens B. (2012) The complement system: an unexpected role in synaptic pruning during development and disease. Annu. Rev. Neurosci. 35, 369–389 [DOI] [PubMed] [Google Scholar]
- 36. Haeseleer F., Imanishi Y., Sokal I., Filipek S., Palczewski K. (2002) Calcium-binding proteins: intracellular sensors from the calmodulin superfamily. Biochem. Biophys. Res. Commun. 290, 615–623 [DOI] [PubMed] [Google Scholar]
- 37. Burgoyne R. D. (2007) Neuronal calcium sensor proteins: generating diversity in neuronal Ca2+ signalling. Nat. Rev. Neurosci. 8, 182–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. McCue H. V., Haynes L. P., Burgoyne R. D. (2010) The diversity of calcium sensor proteins in the regulation of neuronal function. Cold Spring Harb. Perspect. Biol. 2, a004085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Limbäck-Stokin K., Korzus E., Nagaoka-Yasuda R., Mayford M. (2004) Nuclear calcium/calmodulin regulates memory consolidation. J. Neurosci. 24, 10858–10867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chawla S., Hardingham G. E., Quinn D. R., Bading H. (1998) CBP: a signal-regulated transcriptional coactivator controlled by nuclear calcium and CaM kinase IV. Science 281, 1505–1509 [DOI] [PubMed] [Google Scholar]
- 41. Buchthal B., Lau D., Weiss U., Weislogel J. M., Bading H. (2012) Nuclear calcium signaling controls methyl-CpG-binding protein 2 (MeCP2) phosphorylation on serine 421 following synaptic activity. J. Biol. Chem. 287, 30967–30974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bertos N. R., Wang A. H., Yang X. J. (2001) Class II histone deacetylases: structure, function, and regulation. Biochem. Cell Biol. 79, 243–252 [PubMed] [Google Scholar]
- 43. de Ruijter A. J., van Gennip A. H., Caron H. N., Kemp S., van Kuilenburg A. B. (2003) Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem. J. 370, 737–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Osawa M., Tong K. I., Lilliehook C., Wasco W., Buxbaum J. D., Cheng H. Y., Penninger J. M., Ikura M., Ames J. B. (2001) Calcium-regulated DNA binding and oligomerization of the neuronal calcium-sensing protein, calsenilin/DREAM/KChIP3. J. Biol. Chem. 276, 41005–41013 [DOI] [PubMed] [Google Scholar]
- 45. Mellström B., Naranjo J. R. (2001) Ca2+-dependent transcriptional repression and derepression: DREAM, a direct effector. Semin. Cell Dev. Biol. 12, 59–63 [DOI] [PubMed] [Google Scholar]
- 46. Dickstein D. L., Kabaso D., Rocher A. B., Luebke J. I., Wearne S. L., Hof P. R. (2007) Changes in the structural complexity of the aged brain. Aging Cell 6, 275–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Dickstein D. L., Weaver C. M., Luebke J. I., Hof P. R. (2013) Dendritic spine changes associated with normal aging. Neuroscience 251, 21–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Penzes P., Cahill M. E., Jones K. A., VanLeeuwen J. E., Woolfrey K. M. (2011) Dendritic spine pathology in neuropsychiatric disorders. Nat. Neurosci. 14, 285–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Heizmann C. W., Braun K. (1992) Changes in Ca2+-binding proteins in human neurodegenerative disorders. Trends Neurosci. 15, 259–264 [DOI] [PubMed] [Google Scholar]
- 50. Eyles D. W., McGrath J. J., Reynolds G. P. (2002) Neuronal calcium-binding proteins and schizophrenia. Schizophr. Res. 57, 27–34 [DOI] [PubMed] [Google Scholar]
- 51. Riascos D., de Leon D., Baker-Nigh A., Nicholas A., Yukhananov R., Bu J., Wu C. K., Geula C. (2011) Age-related loss of calcium buffering and selective neuronal vulnerability in Alzheimer's disease. Acta Neuropathol. 122, 565–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Šišková Z., Justus D., Kaneko H., Friedrichs D., Henneberg N., Beutel T., Pitsch J., Schoch S., Becker A., von der Kammer H., Remy S. (2014) Dendritic structural degeneration is functionally linked to cellular hyperexcitability in a mouse model of Alzheimer's disease. Neuron 84, 1023–1033 [DOI] [PubMed] [Google Scholar]