ABSTRACT
Geothermal and hypersaline environments are rich in virus-like particles, among which spindle-shaped morphotypes dominate. Currently, viruses with spindle- or lemon-shaped virions are exclusive to Archaea and belong to two distinct viral families. The larger of the two families, the Fuselloviridae, comprises tail-less, spindle-shaped viruses, which infect hosts from phylogenetically distant archaeal lineages. Sulfolobus spindle-shaped virus 1 (SSV1) is the best known member of the family and was one of the first hyperthermophilic archaeal viruses to be isolated. SSV1 is an attractive model for understanding virus-host interactions in Archaea; however, the constituents and architecture of SSV1 particles remain only partially characterized. Here, we have conducted an extensive biochemical characterization of highly purified SSV1 virions and identified four virus-encoded structural proteins, VP1 to VP4, as well as one DNA-binding protein of cellular origin. The virion proteins VP1, VP3, and VP4 undergo posttranslational modification by glycosylation, seemingly at multiple sites. VP1 is also proteolytically processed. In addition to the viral DNA-binding protein VP2, we show that viral particles contain the Sulfolobus solfataricus chromatin protein Sso7d. Finally, we provide evidence indicating that SSV1 virions contain glycerol dibiphytanyl glycerol tetraether (GDGT) lipids, resolving a long-standing debate on the presence of lipids within SSV1 virions. A comparison of the contents of lipids isolated from the virus and its host cell suggests that GDGTs are acquired by the virus in a selective manner from the host cytoplasmic membrane, likely during progeny egress.
IMPORTANCE Although spindle-shaped viruses represent one of the most prominent viral groups in Archaea, structural data on their virion constituents and architecture still are scarce. The comprehensive biochemical characterization of the hyperthermophilic virus SSV1 presented here brings novel and significant insights into the organization and architecture of spindle-shaped virions. The obtained data permit the comparison between spindle-shaped viruses residing in widely different ecological niches, improving our understanding of the adaptation of viruses with unusual morphotypes to extreme environmental conditions.
INTRODUCTION
Viruses infecting extremophilic archaea have evolved to withstand very high temperatures, low or high pH, or near-saturating salt concentrations (1–5). Remarkably, most of these viruses do not seem to be evolutionarily related to viruses of bacteria or eukaryotes and display a considerable diversity of unique virion morphotypes (3, 4). Indeed, 11 novel viral families have been established by the International Committee for the Taxonomy of Viruses (ICTV) for the classification of archaeal viruses, emphasizing the uniqueness of rod-shaped, spindle-shaped, droplet-shaped, or even bottle-shaped particles that have never been observed among viruses infecting bacteria or eukaryotes (3). Functional studies proved to be highly challenging due to the lack of similarity between the protein sequences and structures of archaeal viruses and those from other viruses and cellular organisms (6–10). Among the morphotypes that are exclusively associated with archaea, spindle-shaped viruses are particularly widespread (11) and have been isolated from highly different environments, including deep-sea hydrothermal vents (12–14), hypersaline environments (15–18), anoxic freshwaters (19), cold Antarctic lakes (20), terrestrial hot springs (21–23), and acidic mines (24).
Recently, we refined the evolutionary relationships among spindle-shaped viruses by assessing the morphological and genomic diversity of all available isolates infecting hosts belonging to phylogenetically distant archaeal lineages, including Thermococcales, Methanococcales, Desulfurococcales, and Sulfolobales (11). The analysis has shown that spindle-shaped viruses can be broadly segregated into two evolutionarily distinct lineages. The first group includes members of the Bicaudaviridae family and several currently unclassified viruses. Viruses of this group have large spindle-shaped virions with one or two long tails and contain circular double-stranded DNA (dsDNA) genomes of ∼70 kb (11, 25). In the case of Acidianus two-tailed virus (ATV), the type species of the Bicaudaviridae family, the two tails develop following release into the environment and completely independently from the host cell (26, 27). Unlike ATV, the unclassified Sulfolobus tengchongensis spindle-shaped viruses 1 and 2 have never been observed to undergo this kind of transformation and contain only one tail (28, 29). Nevertheless, both viruses share a number of genes with ATV, including those encoding unique four-helix bundle major capsid proteins (30, 31).
The second group includes smaller, tail-less spindle-shaped viruses, which have been tentatively classified into seven genera within the family Fuselloviridae (11). Sulfolobus spindle-shaped virus 1 (SSV1) is one of the most extensively studied members of this group and is also among the first archaeal viruses to be isolated (32). SSV1 is a temperate virus, and its circular, positively supercoiled dsDNA genome of 15.4 kb can site specifically integrate into the host genome with the aid of a virus-encoded integrase (33–37). SSV1 has been used as a model to establish the genetic system in hyperthermophilic archaea (38, 39). As a result, the research on SSV1 has focused mainly on the mechanism of viral genome integration into the host chromosome (34, 36, 40) and transcriptional regulation (41, 42). In contrast, only a few studies focused on the organization of SSV1 virions; it has been shown that the SSV1 virion consists of three capsid protein species: two paralogous proteins, VP1 and VP3, and the DNA-binding protein VP2 (32, 43). In addition, the virions were reported to contain a host-derived DNA-binding protein; however, its identity has not been determined (43). Small amounts of viral proteins C792 and D244 also have been reported based on mass spectrometry analysis of viral preparations (7, 44), but the presence of the two proteins in highly purified virions remains to be confirmed. Finally, although SSV1 and fuselloviruses in general are considered to exit the host cell by budding through the cytoplasmic membrane, the actual presence of lipids in SSV1 virions is a matter of debate and has never been rigorously demonstrated (1, 32, 45). Lipids initially detected in SSV1 preparations could be derived from contaminant membrane vesicles which could copurify with the virions (32). Recent attempts to reconstruct the SSV1 virion structure based on cryoelectron microscopy were encumbered by the heterogeneity of the viral particles and provided rather limited insight into the organization of capsid proteins in the virion, whereas the presence of a lipid-containing envelope could not be determined (46).
Although spindle-shaped particles are dominant in hypersaline environments (16, 17), only one such hyperhalophilic archaeal virus, His1, has been isolated to date (15). Recent biochemical and structural studies have shown that His1 virions are composed of one major (VP21) and a few minor capsid protein species (47, 48). Interestingly, a subset of VP21 apparently is modified by lipid moieties, although the lipid bilayer could not be detected by either biochemical or structural approaches (47, 48). Furthermore, treatment of His1 virions with various compounds induced the transformation of spindle-shaped particles into tube-like structures which were devoid of the genomic DNA (47, 49). It has been suggested that such reorganization is biologically relevant and reflects structural changes accompanying virus entry into the host (47). Although SSV1 and His1 infect widely different hosts, thermoacidophilic crenarchaea and hyperhalophilic euryarchaea, respectively, the two viruses display a very similar particle shape, and their major capsid proteins share ∼47% sequence identity, suggesting that they have evolved from a common ancestor (11, 48).
To investigate the evolutionary relationships among spindle-shaped viruses residing in highly different environments, we set out to perform a rigorous biochemical characterization of SSV1 particles. We show that SSV1 virions consist of five structural protein species, among which one, a DNA-binding protein, is encoded by the host. The virus-encoded proteins undergo posttranslational modifications, including proteolytic cleavage and glycosylation. Finally, we put to rest the debate on the presence versus absence of lipids in SSV1 virions by showing that highly purified SSV1 virions contain tetraether lipids selectively recruited from the host cytoplasmic membrane.
MATERIALS AND METHODS
Viruses, strains, and growth conditions.
Sulfolobus shibatae strain B12 (50) and Sulfolobus solfataricus strain P2 (51) were used as hosts for SSV1 (32). All cultures were grown aerobically (120 rpm; Innova 44 Eppendorf) at 78°C. The Sulfolobus growth medium was prepared as described previously (52).
His1 (15) and its host, Haloarcula hispanica strain ATCC 33960, were grown in modified growth medium (MGM) at 37°C as previously described (reference 48 and references therein).
Virus production and purification.
To induce SSV1, cultures of lysogenized S. shibatae B12 at an optical density at 600 nm (OD600) of 0.5 were treated with UV as previously described (32). Twenty-four hours after irradiation, cells and debris were removed by two steps of centrifugation (4,000 rpm, 30 min, 4°C and then 8,000 rpm, 30 min, 4°C; Jouan BR4i rotor AB 50.10A). The cell-free supernatant was mixed with S. solfataricus P2 cells and added to the soft layer of plates prepared as described in Schleper et al. (40), except that Gelzan CM Gelrite (Sigma-Aldrich) was replaced with Phytagel (Sigma-Aldrich). After 72 h at 75°C, the top layers of confluent plates were collected, and 2 ml of medium was added per plate. The suspension was incubated with aeration (120 rpm; Innova 44 Eppendorf) at 78°C for 1 h. Cells and debris were removed by two rounds of centrifugation (8,000 rpm, 30 min, 4°C [Jouan BR4i rotor AB 50.10A], followed by 12,000 rpm, 30 min, 4°C [Avanti J-26XP rotor JLA 16.250]). Virus stocks were stored at 4°C.
Virus particles were precipitated from the stocks by the addition of ammonium sulfate (Sigma-Aldrich) to 50% (wt/vol) saturation at 4°C as described previously (32). The precipitate (12,000 rpm, 30 min, 4°C; Avanti J-26XP rotor JLA 16.250) was resuspended in SSV1-buffer [20 mM KH2PO4, 1 M NaCl, 2.14 mM MgCl2, 0.43 mM Ca(NO3)2, <0.001% trace elements of Sulfolobales medium, pH 6] (52). In order to remove traces of ammonium sulfate, the virus preparation was dialyzed (Spectra/Por 1; Spectrum Labs) twice against the SSV1 buffer at 4°C.
The virus concentrate was purified in a linear 5 to 20% sucrose gradient (in SSV1 buffer) by rate-zonal centrifugation (24,000 rpm, 20 min, 15°C; Sorvall rotor AH629), and the light-scattering zone was collected. The virus was further concentrated and purified by equilibrium centrifugation (21,000 rpm, 20 h, 15°C; Sorvall rotor AH629) in a CsCl gradient in SSV1 buffer (mean density, 1.30 g/ml). The light-scattering band was collected and diluted 3-fold in SSV1 buffer, followed by concentration by differential centrifugation (21,000 rpm, 20 h, 15°C; Sorvall rotor AH629). The pellet was resuspended in a minimal volume of the SSV1 buffer. The resultant preparation is referred to as the 2× purified sample. The quality of the purification procedure was verified after each step by protein gel analysis (see below), measurement of absorbance at 260 nm, recovery of infectivity (plaque assay), and transmission electron microscopy (TEM; described below).
Production and purification of His1 virions were performed as previously described (48).
Control of SSV1 aggregation.
SSV1 preparations after ammonium sulfate precipitation were dialyzed against the SSV1 buffer containing 0.1, 0.25, 0.5, 1, or 2 M NaCl. SSV1 virions purified in the CsCl density gradient were incubated for 30 min at room temperature in the presence of 1% (vol/vol) ethanol in SSV1 buffer containing 1 M NaCl. The samples were negatively stained and processed for TEM. Virions were ascribed to three different categories: (i) single particles, (ii) rosette-like virion aggregates containing between 2 and 5 particles, and (iii) aggregates with more than 5 particles. The proportion of virions in each of the three categories under the different conditions was determined by TEM. At least 1,000 viral particles from three independent biological replicates were analyzed per condition, and standard deviations were calculated. The infectivity of each sample also was verified by plaque assay as previously described (40).
Protein analyses.
Proteins were analyzed using modified tricine-sodium dodecyl sulfate polyacrylamide gel electrophoresis (tricine-SDS-PAGE) with 4% and 14% (wt/vol) acrylamide concentrations in the stacking and separation gels, respectively (53). After electrophoresis, gels were stained with Coomassie blue (detection limit of >7 ng) or SYPRO Ruby (detection limit of 0.25 to 1 ng) (Life Technologies). Glycosylation of SSV1 proteins was assessed using a Pro-Q Emerald 300 glycoprotein gel stain kit according to the manufacturer's instructions (Life Technologies).
N-terminal sequencing of virion proteins was performed at the Protein Chemistry Core Facility of the Institute of Biotechnology, University of Helsinki, and mass spectrometry (MS) of peptides released by in-gel trypsin digestion was done at Meilahti Clinical and Basic Proteomics Core Facility, University of Helsinki, as described previously (54).
Transmembrane domains and secondary structure elements in viral proteins were predicted using TMHMM (55) and Jpred3 (56), respectively.
The relative quantification of the amount of proteins in each SDS-PAGE gel band was done using ImageJ software (National Institutes of Health). The determined value then was divided by the number of virus particles estimated from the viral DNA absorbance at 260 nm as described below.
Lipid analyses.
S. solfataricus cell pellet and 2× purified SSV1 preparation were freeze-dried, and the biomass was directly acid hydrolyzed by refluxing with 5% HCl in methanol for 3 h by following Pitcher et al. (57) to release glycerol dibiphytanyl glycerol tetraether (GDGTs) lipids. A known amount (10 ng) of a C46 GDGT standard (58) was added to the acid-hydrolyzed fraction, and GDGT lipids were analyzed by high-performance liquid chromatography/atmospheric pressure chemical ionization-mass spectrometry according to Schouten et al. (59). The mass spectrometer was operated in single ion mode (SIM) to monitor GDGTs with 0 to 8 cyclopentane moieties and the C46 GDGT standard. Relative abundances of GDGTs were determined by integrating peak areas of the SIM signal. The signal of the C46 GDGT standard was corrected for the difference in ionization efficiency using a 1:1 mixture of the standard and purified GDGT-0.
To establish the head groups of the GDGTs, S. solfataricus cells were extracted by a modified Bligh-Dyer method and analyzed for intact polar lipids as described by Pitcher et al. (57).
Quantification of viral particles.
The number of infectious particles was determined by plaque assay as described above. Alternatively, SSV1 particles were enumerated by determining the number of genome copies in the preparation. To this end, viral DNA was extracted from the purified virion preparation using the standard phenol-chloroform method, and the number of the genome copies was estimated by measuring the absorbance at 260 nm and considering that the molecular size of the SSV1 genome (15,465 bp; NC_001338) is 9,554,261.87 g/mol.
Electron microscopy.
For conventional negative-stain TEM, samples were prepared as described previously (60). Briefly, 10 μl of sample was adsorbed on grids for 1 min, air dried, and stained with 3% uranyl acetate, pH 4.5 (EuroMedex), for 1 min. Samples were imaged using an FEI Tecnai Biotwin 120 transmission electron microscope operating at 100 kV at the Ultrapole of the Institut Pasteur, Paris, or JEOL JEM-1400 transmission electron microscope operating at 80 kV at the Electron Microscopy Unit of the Institute of Biotechnology, University of Helsinki.
RESULTS
Aggregation of SSV1 particles is modulated by ionic and hydrophobic interactions.
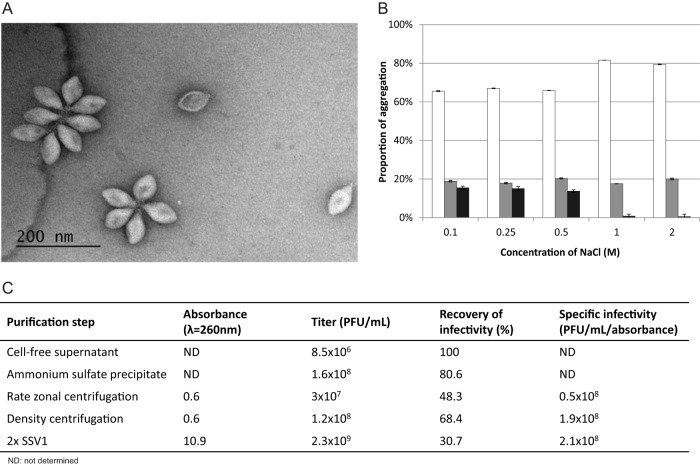
Virions of SSV1 and other fuselloviruses tend to interact with each other by the terminal fibers located at one of the two pointed ends of the viral particles, forming polyvalent aggregates (22, 32, 61). Based on the aggregation state, SSV1 virions can be grouped into one of three categories: (i) individual virions, (ii) rosette-like aggregates containing between 2 and 5 particles, and (iii) aggregates with more than 5 viral particles (Fig. 1A). Since large virion aggregates aggravate virion purification and prevent accurate virion enumeration, we attempted to reduce virion aggregation by varying the ionic strength conditions. Increasing the salt concentration in the SSV1 buffer led to the dissociation of aggregates containing more than 5 particles in a concentration-dependent manner, with a concomitant increase in the proportion of single virions (Fig. 1B), implicating ionic interactions in virion aggregation. However, the portion of rosette-like viral assemblages composed of up to 5 particles remained constant (19 ± 1.5% on average), even at the highest NaCl concentration tested (Fig. 1B). It is noteworthy that SSV1 remained stable and retained infectivity for up to 3 months in a wide range of salt concentrations (0.1 to 2 M NaCl), highlighting the robustness of the viral particles. Considering the highly pronounced hydrophobicity of the protein implicated in the formation of terminal fibers (see below), we tested whether the smaller aggregates could be dissociated by mild treatment with organic solvents. Indeed, in the presence of 1% (vol/vol) ethanol, the proportion of single particles increased to ∼88%, whereas the remaining virion aggregates mainly consisted of 2 particles and no aggregates with 5 particles were observed under TEM. Such treatment reduced the infectivity by ∼50%, while ethanol concentrations above 10% (vol/vol) resulted in complete dissociation of the SSV1 virions (data not shown).
FIG 1.
Purification of SSV1. (A) Transmission electron micrograph of negatively stained SSV1 sample. Single particles as well as different aggregates are shown. (B) Depending on the concentration of NaCl in the SSV1 buffer, different stages of aggregation were observed: single particles (white columns), rosette-like structures containing between 2 and 5 particles (gray columns), and aggregates with more than 5 particles (black columns). The number of viruses in each category was determined from negatively stained electron micrographs obtained from three independent experiments. At least 1,000 particles were counted per condition, and error bars represent standard deviations. (C) Analysis of samples taken after each step of the 2× purification procedure. Absorbance at 260 nm, virus titer, recovery of infectivity, and specific infectivity are indicated.
Production of highly purified virions.
In order to ensure high purity of the viral preparation for unambiguous determination of the SSV1 virion constituents, we developed and optimized a multistep purification protocol (see Materials and Methods). S. shibatae lysogens sporadically release SSV1 virions. However, even following UV irradiation, which increased SSV1 production by one order of magnitude (from ca. 105 to 106 PFU/ml), the virus titer was insufficient for robust biochemical virion characterization. To overcome this hurdle, virions were precipitated with ammonium sulfate from the virus stocks obtained by collecting the soft layer of confluent Phytagel plates. The virus preparation subsequently was purified using rate-zonal centrifugation in a linear sucrose gradient to produce 1× purified SSV1. However, the resultant virus preparation contained a substantial amount of impurities, as judged by SDS-PAGE analysis (data not shown), necessitating an additional step of purification. The latter included equilibrium centrifugation in a CsCl gradient and concentration by differential centrifugation, resulting in the production of the 2× purified SSV1 preparation (Fig. 1C). The purification was performed under conditions minimizing the aggregation of virions (1 M NaCl). Virion recovery was monitored throughout the purification procedure, and the final 2× preparation corresponded to ∼31% recovery of infectious particles with a specific infectivity of ∼2 × 108 PFU/ml/unit of absorbance at 260 nm (Fig. 1C). The buoyant density of the purified SSV1 virions in CsCl was estimated to be 1.29 g/ml, which is somewhat higher than that previously reported for SSV1 (1.24 g/ml [32]) but similar to the buoyant density of membrane-containing pleolipoviruses (1.3 g/ml [62]).
Structural proteins of SSV1.
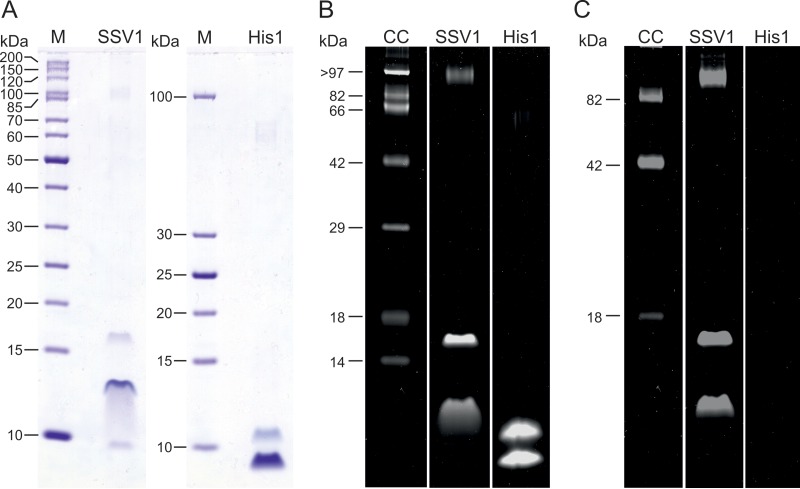
The availability of a highly purified preparation allowed us to assess the biochemical composition of the SSV1 viral particles. Hyperhalophilic spindle-shaped virus His1 was analyzed in parallel as a control and for comparison. 2× Purified SSV1 and His1 virions were examined by tricine-SDS-PAGE. Following Coomassie blue staining, the migration profiles of SSV1 and His1 preparations appeared similar and displayed several major protein bands of low molecular mass (in the range of 7 to 17 kDa) and a minor high-molecular, mass protein band (Fig. 2A); the migration of His1 proteins was similar to that previously reported (48). Unexpectedly, as has also been reported for His1 virus (48), the protein concentration of the purified SSV1 samples could not be determined using the Bradford method (63); it appears that the virions of SSV1 and His1 do not display sufficient reactivity with the Coomassie blue reagent, although the corresponding proteins in the tricine-SDS-PAGE gels could be detected.
FIG 2.
Structural proteins of SSV1. (A) Protein profiles of 2× purified SSV1 virions compared to 2× purified His1 virions in a tricine-SDS-polyacrylamide gel stained with Coomassie blue. Molecular mass markers (M) are shown. The amounts of SSV1 and His1 samples loaded are comparable based on absorbance measurements at 260 nm. (B and C) 2× purified SSV1 and His1 virions analyzed in a tricine-SDS-PAGE gel stained with SYPRO Ruby protein stain (detecting all proteins) (B) and with Pro-Q Emerald 300 glycoprotein detecting reagent (detecting glycosylated proteins) (C). Candy-Cane glycoprotein molecular mass standard (labeled CC) contains a mixture of nonglycosylated and glycosylated proteins. Half of the amount loaded for panel A was added to the gel shown in panels B and C.
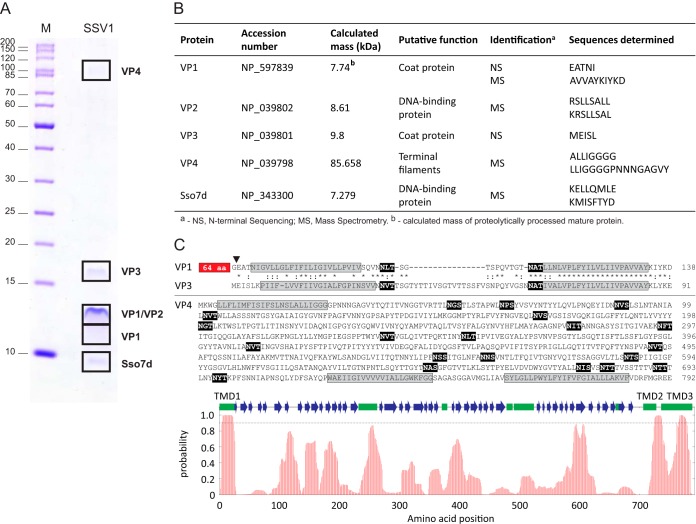
The identity of SSV1 proteins was determined by a combination of N-terminal sequencing and MS techniques. Consistent with previous analysis (43), in the lower-molecular-mass bands we identified the proteins VP1, VP2, and VP3. Proteins VP1 and VP3 are paralogous, highly hydrophobic proteins (each contains two predicted α-helical transmembrane domains [TMDs]) (Fig. 3C). N-terminal sequencing showed that, unlike VP3, VP1 is proteolytically processed, resulting in the removal of 65 N-terminal amino acids (Fig. 3B and C), which also has been shown previously (43). The high-molecular-mass band was identified as a product of open reading frame (ORF) C792 (Fig. 3A). Adhering to the nomenclature used for SSV1 structural proteins (43), we denote the product of ORF C792 as VP4. The presence of VP4 in SSV1 virions was reported previously (44). However, since the SDS-PAGE analysis of the virion preparation was not presented, the possibility of contamination could not be ruled out. Like VP1 and VP3, VP4 is highly hydrophobic; sequence analysis showed that VP4 contains three confidently predicted (probability of higher than 0.9) α-helical TMDs, but high-hydrophobicity regions also are distributed throughout the protein length (Fig. 3C). Notably, the region flanked by TMD1 and TMD2 is predicted to be β-strand rich and is likely to adopt a β-propeller or β-barrel topology.
FIG 3.
Identification of the structural proteins of SSV1. (A) Protein pattern of the 2× purified SSV1 virions in a tricine-SDS-PAGE stained with Coomassie blue. M indicates the molecular mass marker. Locations of protein bands processed for proteomic analyses are depicted with black boxes, and names of proteins identified are indicated on the right. (B) Proteins identified by N-terminal sequencing (NS) and mass spectrometry (MS). The NCBI accession number, theoretical molecular masses, putative functions, and peptide sequences determined during analysis are provided. (C) Sequence analysis of the SSV1 structural proteins VP1, VP3, and VP4. Sequences of the predicted transmembrane domains are highlighted in gray, whereas the theoretical glycosylation consensus motifs (N-X-S/T) are shown on the black background. The position of proteolytic cleavage in VP1 is indicated by a black arrowhead, and the N-terminal 65-amino-acid (aa) residues not shared with VP3 are boxed. Paralogous proteins VP1 and VP3 are aligned; identical and similar amino acid positions are indicated with asterisks and colons, respectively. At the bottom of the panel is the hydrophobicity profile of VP4. The broken line indicates the 0.9 probability threshold for the prediction of the transmembrane domains (TMD1-3). Predicted secondary structure elements are shown with green boxes (α-helixes) and blue arrows (β-strands).
SSV1 virions were found to contain a considerable amount of one host-encoded protein, Sso7d (Fig. 3A and B). Sso7d is a small, basic protein, which belongs to the extensively studied Sul7d family of 7-kDa DNA-binding proteins and represents one of the major chromatin proteins of Sulfolobus solfataricus (64, 65). Notably, SDS-PAGE analysis of fractions collected from the CsCl gradient showed that Sso7 is exclusively detected in the fraction containing SSV1 virions. In addition, a previous study (43) has reported the presence of an unidentified host-encoded DNA-binding protein in SSV1 virions. Consequently, we assign Sso7d as a virion component. Staining of the protein gel with SYPRO Ruby, which is eight times more sensitive than Coomassie brilliant blue stain (66), did not reveal any additional protein bands (Fig. 2B), strongly suggesting that the incorporation of Sso7d into SSV1 particles is specific and biologically relevant rather than accidental.
VP1, VP3, and VP4 are glycosylated.
Molecular masses of SSV1 structural proteins deduced from the gel (Fig. 3A) did not coincide with those calculated from the sequence (Fig. 3B). VP1, VP2, and VP3 migrated in gels as ∼11-, ∼13-, and ∼16-kDa proteins, which is considerably slower than expected based on their calculated molecular masses (i.e., 7.7, 8.6, and 9.8 kDa, respectively) (Fig. 3A and B). Similarly, VP4, with a predicted mass of 85 kDa, migrated as a 100-kDa protein (Fig. 3A). The discrepancy in migration patterns could not be explained by the potential formation of higher-order oligomers. Thus, the possibility of posttranslational modifications was considered. Since virion proteins of several archaeal viruses are known to undergo glycosylation (67–69), we tested whether this modification can be detected in the case of SSV1 virions by staining the proteins with a glycoprotein-specific stain. Indeed, VP1, VP3, and VP4 were found to be glycosylated (Fig. 2C). Unlike many crenarchaeal viruses (6, 8, 70, 71), SSV1 does not encode an identifiable glycosyltransferase; thus, the glycosylation of viral proteins is likely to be performed by cellular enzymes. Protein glycosylation has been studied in several members of Sulfolobales (72), including S. solfataricus (73), which was used in this study for SSV1 production. It has been found that glycosylation in Sulfolobus occurs on the asparagine residue within the consensus motif N-X-S/T (where X is any amino acid except proline). All three SSV1 proteins which we found to be glycosylated (Fig. 2C) contain multiple N-X-S/T motifs: VP1 and VP3 each contain 2 such motifs located in the linker region between the TMDs, whereas VP4 possesses 20 motifs which could undergo glycosylation (Fig. 3C). The extent of glycosylation as well as detailed characterization of the glycans attached to the SSV1 proteins will be the focus of future studies.
SSV1 acquires lipids from the host cytoplasmic membrane.
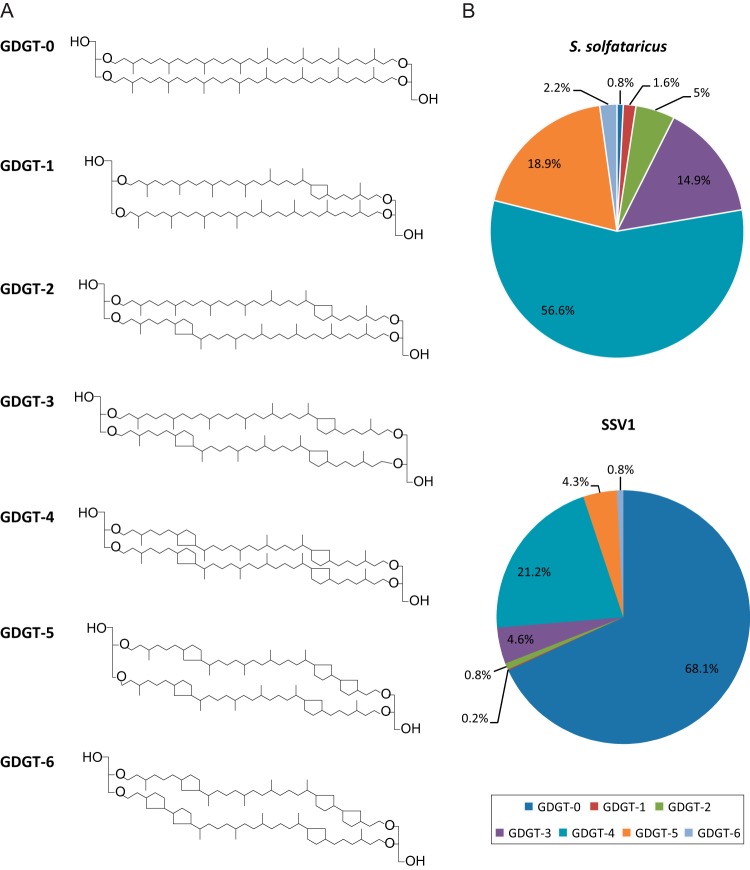
Membranes of organisms from the order Sulfolobales predominantly consist of sn-2,3-dibiphytanyl diglycerol tetraether (also known as glycerol dibiphytanyl glycerol tetraether [GDGT]) lipids, in which the two glycerol moieties are connected by two C40 isoprenoid chains, enabling the formation of monolayer membranes (74, 75). GDGTs differ by the number of cyclopentane moieties within the isoprenoid chains, which can vary from 0 to 8 (i.e., GDGT-0 through GDGT-8).
Preliminary thin-layer chromatography analysis of the material extracted from SSV1 virions and host S. solfataricus cells by chloroform-methanol treatment strongly suggested the presence of phospholipids, although in the case of SSV1 the amount detected using iodine vapor was rather low (data not shown). To determine the exact nature of SSV1 lipids and to compare it to the lipid content of the host, we analyzed GDGTs by liquid chromatography coupled with mass spectrometry on 2× purified SSV1 virions as well as S. solfataricus cells (see Materials and Methods). The analysis revealed that S. solfataricus membrane contains seven GDGT species (Fig. 4A) which are present in different amounts. Under conditions tested, GDGT-4 constituted more than half of the cellular membrane lipids (Fig. 4B). Furthermore, the lipid head groups were found to contain 2 to 3 sugar moieties. This is consistent with the previous analysis which showed that glycolipids of Sulfolobus contain di- and trisaccharides composed of glucose, galactose, or mannose moieties (76, 77). Analysis of the viral particles showed that all 7 GDGT species identified in S. solfataricus membrane also are present in small amounts in SSV1 virions. Interestingly, however, the ratios of different lipids in the viral particles were different from those for the host cytoplasmic membrane. SSV1 virions were strongly enriched in GDGT-0, which represented ∼68% of all viral lipids (Fig. 4B). The proportions of other lipids in the virions roughly followed those in the cellular membrane, i.e., the second most abundant lipid was GDGT-4, followed by GDGT-3 and GDGT-5. Notably, lipid analysis carried out on different SSV1 preparations showed that whereas the proportion of GDGT-0 remained constant in different experiments, the ratios of GDGT-3, GDGT-4, and GDGT-5 were more variable. Unfortunately, the low abundance of lipids in viral particles precluded the detailed analysis of their head groups.
FIG 4.
Analysis of lipids of Sulfolobus solfataricus and SSV1. (A) Structures of lipids analyzed in this study, glycerol dibiphytanyl glycerol tetraethers (GDGTs). The numbers denote how many cyclopentane moieties are present within the isoprenoid chains. (B) Relative distribution of core lipid species identified by HPLC-APCI-MS in S. solfataricus cells and 2× purified SSV1 virions as described in Materials and Methods.
Quantification of the SSV1 virion components.
To gain a better understanding of SSV1 virion organization, we performed a relative quantitation of lipids and proteins. Due to various reasons, not all virions released from the cell are infectious (i.e., plaque forming); the ratio between noninfectious and infectious particles can vary greatly between different viruses (78). Thus, we have established the correspondence between the number of infectious SSV1 particles determined by the plaque assay and the number of genome copies estimated from the purified viral DNA absorbance at 260 nm. The particle-to-PFU ratio was estimated to be around 5, which is consistent with the values (5 to 10 particles/PFU) determined previously by quantitative TEM (40). Since there are more particles than PFU (i.e., the ratio is more than 1), for subsequent calculations, we used the number of particles estimated from the number of genome copies rather than PFU. Quantitation of virion components (see Materials and Methods) showed that each virion contains ∼6 fg of lipids and ∼14 fg of proteins, i.e., the two components are present in a 1:2.4 ratio. Given that the estimated total number of SSV1 particles might be slightly skewed due to potential artifacts (e.g., the presence of empty virions, etc.), these values should be considered with caution.
DISCUSSION
Large-scale production and high-level purification procedures are a prerequisite for comprehensive biochemical and structural characterization of any virus. Here, we have optimized a purification protocol for the hyperthermophilic spindle-shaped virus SSV1, which allowed its biochemical characterization. Environmental distribution of spindle-shaped viruses is particularly broad (11). Interestingly, although the natural habitats of SSV1 are characterized by very low pH and high temperatures, we found that SSV1 virions also are stable in high-salinity conditions; prolonged incubation in the presence of 2 M NaCl had no effect on virion stability or infectivity. This indicates that the design of spindle-shaped virions is inherently robust, which might explain the success of this virus group in colonizing very diverse ecological niches where their archaeal hosts are found (11, 48).
Our analyses have shown that the SSV1 virion consists of four virus-encoded (VP1-4) and one host-derived (Sso7d) protein (Fig. 3). Paralogous proteins VP1 and VP3 have homologs in all spindle-shaped viruses characterized thus far (11), including hyperhalophilic virus His1 (48), and represent a signature protein in this group of viruses. Protein VP4 has been suggested previously to be involved in the formation of terminal fibers based on correlation between the fiber morphology and the presence of vp4-like genes in different members of the Fuselloviridae (22). Here, we have demonstrated that VP4 is indeed a part of the virion, confirming a previous report by Menon et al. (44). Electron microscopy analysis indicates that terminal fibers are implicated in virion aggregation (Fig. 1A). Two groups of virion aggregates can be defined: (i) aggregates composed of up to five virions and (ii) those containing more than five viral particles. The latter assemblages seem to be dependent on ionic interactions, whereas the former ones are not (Fig. 1B). Instead, the smaller rosette-like aggregates apparently are held together by hydrophobic interactions, presumably involving VP4, and can be dispersed by mild treatment with organic solvents. The high hydrophobicity of VP4 (Fig. 3C) is in line with this conclusion.
VP2 has been shown previously to be tightly bound to dsDNA, suggesting a role in organizing the SSV1 genome (43). Unexpectedly, VP2 is conserved in only four (SSV1, SSV6, ASV1, and SMF1) out of 10 fuselloviruses for which complete genome sequences are available. Moreover, in-frame deletion of the VP2-encoding gene had no observable effect on virion assembly or infectivity, indicating that the gene is dispensable under laboratory growth conditions (79). Interestingly, unlike other SSV1 VPs, VP2 is not specific to fuselloviruses but also is encoded by unrelated proviruses of the euryarchaeon Archaeoglobus veneficus SNP6 (80), Sulfolobus turreted icosahedral virus 2 (81), and bacterial viruses of the recently established family Sphaerolipoviridae (82). Such patchy phyletic distribution of vp2-like genes in fuselloviruses, its conservation in other archaeal and bacterial viruses, and the dispensability of VP2 for SSV1 infectivity indicate that the vp2 gene has been acquired relatively late in the history of fuselloviruses from a different group of viruses.
Identification of the host-encoded DNA-binding protein Sso7d in SSV1 virions suggests that Sso7d plays an important role in the organization and condensation of the viral genome prior to packaging. Sso7d, a member of the Sul7d family, is one of the major chromatin proteins responsible for chromosome organization in Sulfolobus (65). This small basic protein is known to bind dsDNA nonspecifically and induces negative supercoiling (83) as well as compaction of relaxed or positively supercoiled DNA in vitro (64). SSV1 DNA is highly positively supercoiled in SSV1 virions (33). This positive supercoiling might result from the activity of Sulfolobus reverse gyrase, an enzyme that introduces positive supercoiling in vitro in topologically closed DNA (33). Alternatively, positive supercoiling might be induced by stoichiometric binding of a DNA-binding protein, followed by the relaxation of compensatory negative superturns by cellular DNA topoisomerases. In the latter hypothesis, this DNA binding protein cannot be SSo7d, since this protein induces negative supercoiling in vitro (83). Thus, SSV1 DNA might be positively supercoiled first by reverse gyrase and later is condensed during the packaging process by interaction between Sso7d and positively supercoiled viral DNA. However, the effects of VP2 on DNA supercoiling as well as condensation of SSV1 DNA by VP2 and Sso7d remain to be investigated.
Notably, previous analysis also has suggested that viral particles contain protein D244 (44), which, however, is not essential for virion assembly and infectivity (79). The X-ray structure of the D244 orthologue from the Sulfolobus spindle-shaped virus Ragged Hills revealed that the protein is a member of the PD-(D/E)XK nuclease superfamily (84), arguing against the possibility that D244 plays a structural role in virion formation. We could not detect D244 in our virus preparation, although its presence in amounts that were below our detection limit cannot be ruled out.
Protein glycosylation is one of the most common posttranslational modifications in archaeal viruses, particularly in viruses infecting hyperthermophilic hosts (67–69), and it could play an important role in virion stability and/or interaction with the host cell. Accordingly, many hyperthermophilic archaeal viruses encode their own glycosyltransferases (6, 8, 70, 71), with some viruses containing as many as five different glycosyltransferase genes per genome (8). However, this is not the case for SSV1 or any other known fusellovirus. Nevertheless, three of the SSV1 virion proteins, VP1, VP3, and VP4, are glycosylated (Fig. 2C). To the best of our knowledge, the glycosylation of virion proteins has never been observed for any spindle-shaped virus. Somewhat paradoxically, hyperhalophilic spindle-shaped virus His1 encodes a putative glycosyltransferase but does not seem to glycosylate its virion proteins, at least not under laboratory conditions (48) (Fig. 2C). In the absence of dedicated virus-encoded glycosyltransferases, the glycosylation of SSV1 VPs is likely to be performed by the host enzymes. Consistent with this, multiple consensus glycosylation motifs (N-X-S/T) (72, 73) are present in all three SSV1 glycoproteins; VP4 contains a particularly high number of such motifs (Fig. 3C). A recent study of protein glycosylation in S. solfataricus has shown that some cell surface proteins can be heavily glycosylated (73). For example, protein SSO1273 contains 20 N-X-S/T motifs, and all of them were found to be modified with a glycan, which has a mass of 1,298.4 Da (73). The slower migration of VP4 in tricine-SDS-PAGE gel (as 100 kDa instead of the calculated 85 kDa) would be consistent with glycosylation on most of the theoretical glycosylation sites. The detailed characterization of the glycan structures and the extent of the SSV1 VP glycosylation, as well as the biological significance of this modification, will be an exciting area of future research.
The presence of lipids in SSV1 virions has been a matter of debate for many years (1, 32, 45). Indeed, even recent low-resolution (∼32-Å) reconstructions of SSV1 virion structure provided no insight concerning this issue (46). Here, we resolve this long-lasting dispute by providing evidence for the presence of lipids in highly purified SSV1 virions. Furthermore, for the first time, we determined the molecular composition of the lipid content and show that SSV1 virions contain seven different species of GDGT lipids (Fig. 4). This result is consistent with the early electron microscopic observations of SSV1 budding through the cell membrane (32). Notably, the ratio of different lipid species in the virions was different from that found in the cytoplasmic membrane of the host cells, suggesting a selective incorporation of lipids into the virion. Similarly, lipid composition of Sulfolobus turreted icosahedral virus, which has an internal membrane, and lipid content of membrane vesicles produced by S. solfataricus were found to differ considerably from that of the cellular membrane (68, 85). Interestingly, the hyperhalophilic spindle-shaped virus His1 does not appear to contain free lipids; instead, its major capsid protein, homologous to VP1/VP3 of SSV1, was concluded to be covalently modified by a lipid moiety (48), suggesting that distinct spindle-shaped viruses use different mechanisms for lipid acquisition from the host.
The biochemical characterization of SSV1 virions presented here provides a foundation for future investigations of different aspects of biology and structure of hyperthermophilic spindle-shaped viruses. Of special interest is the role of lipids during the entry and exit stages of fusellovirus infection; the comparison of these strategies with those employed by eukaryotic membrane-containing viruses might provide particularly important insights into the evolution of mechanisms mediating membrane fusion and virus budding.
ACKNOWLEDGMENTS
This work was supported by the Agence Nationale de la Recherche (ANR) program BLANC, project EXAVIR (D.P.), and by Academy Professor (Academy of Finland) funding grants 256518, 283072, and 255342 (D.H.B.). E.R.J.Q. was supported by a fellowship from the Ministère de l'Enseignement Supérieur et de la Recherche of France and the Université Pierre et Marie Curie, Paris, France, as well as the European Molecular Biology Organization (ASTF 62-2014) and a CIMO Fellowship, Finland (TM-13-9054). We thank Academy of Finland (grants 271413 and 272853) and the University of Helsinki for support of the EU ESFRI Instruct Centre for Virus Production (ICVIR) used in this study.
We thank Sari Korhonen, Päivi Hannuksela, and Helin Veskiväli for excellent technical assistance.
REFERENCES
- 1.Atanasova NS, Senčilo A, Pietilä MK, Roine E, Oksanen HM, Bamford DH. 2015. Comparison of lipid-containing bacterial and archaeal viruses. Adv Virus Res 92:1–61. [DOI] [PubMed] [Google Scholar]
- 2.Pietilä MK, Demina TA, Atanasova NS, Oksanen HM, Bamford DH. 2014. Archaeal viruses and bacteriophages: comparisons and contrasts. Trends Microbiol 22:334–344. doi: 10.1016/j.tim.2014.02.007. [DOI] [PubMed] [Google Scholar]
- 3.Prangishvili D. 2013. The wonderful world of archaeal viruses. Annu Rev Microbiol 67:565–585. doi: 10.1146/annurev-micro-092412-155633. [DOI] [PubMed] [Google Scholar]
- 4.Prangishvili D. 2015. Archaeal viruses: living fossils of the ancient virosphere? Ann N Y Acad Sci 1341:35–40. doi: 10.1111/nyas.12710. [DOI] [PubMed] [Google Scholar]
- 5.Prangishvili D, Koonin EV, Krupovic M. 2013. Genomics and biology of Rudiviruses, a model for the study of virus-host interactions in Archaea. Biochem Soc Trans 41:443–450. doi: 10.1042/BST20120313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krupovic M, White MF, Forterre P, Prangishvili D. 2012. Postcards from the edge: structural genomics of archaeal viruses. Adv Virus Res 82:33–62. doi: 10.1016/B978-0-12-394621-8.00012-1. [DOI] [PubMed] [Google Scholar]
- 7.Lawrence CM, Menon S, Eilers BJ, Bothner B, Khayat R, Douglas T, Young MJ. 2009. Structural and functional studies of archaeal viruses. J Biol Chem 284:12599–12603. doi: 10.1074/jbc.R800078200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Prangishvili D, Garrett RA, Koonin EV. 2006. Evolutionary genomics of archaeal viruses: unique viral genomes in the third domain of life. Virus Res 117:52–67. doi: 10.1016/j.virusres.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 9.Quemin ER, Prangishvili D, Krupovic M. 2014. Hard out there: understanding archaeal virus biology. Future Virol 9:703–706. doi: 10.2217/fvl.14.52. [DOI] [Google Scholar]
- 10.DiMaio F, Yu X, Rensen E, Krupovic M, Prangishvili D, Egelman EH. 2015. Virology A virus that infects a hyperthermophile encapsidates A-form DNA. Science 348:914–917. doi: 10.1126/science.aaa4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krupovic M, Quemin ER, Bamford DH, Forterre P, Prangishvili D. 2014. Unification of the globally distributed spindle-shaped viruses of the Archaea. J Virol 88:2354–2358. doi: 10.1128/JVI.02941-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Geslin C, Le Romancer M, Erauso G, Gaillard M, Perrot G, Prieur D. 2003. PAV1, the first virus-like particle isolated from a hyperthermophilic euryarchaeote, “Pyrococcus abyssi.” J Bacteriol 185:3888–3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gorlas A, Koonin EV, Bienvenu N, Prieur D, Geslin C. 2012. TPV1, the first virus isolated from the hyperthermophilic genus Thermococcus. Environ Microbiol 14:503–516. doi: 10.1111/j.1462-2920.2011.02662.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Geslin C, Le Romancer M, Gaillard M, Erauso G, Prieur D. 2003. Observation of virus-like particles in high temperature enrichment cultures from deep-sea hydrothermal vents. Res Microbiol 154:303–307. doi: 10.1016/S0923-2508(03)00075-5. [DOI] [PubMed] [Google Scholar]
- 15.Bath C, Dyall-Smith ML. 1998. His1, an archaeal virus of the Fuselloviridae family that infects Haloarcula hispanica. J Virol 72:9392–9395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oren A, Bratbak G, Heldal M. 1997. Occurrence of virus-like particles in the Dead Sea. Extremophiles 1:143–149. doi: 10.1007/s007920050027. [DOI] [PubMed] [Google Scholar]
- 17.Porter K, Russ BE, Dyall-Smith ML. 2007. Virus-host interactions in salt lakes. Curr Opin Microbiol 10:418–424. doi: 10.1016/j.mib.2007.05.017. [DOI] [PubMed] [Google Scholar]
- 18.Sime-Ngando T, Lucas S, Robin A, Tucker KP, Colombet J, Bettarel Y, Desmond E, Gribaldo S, Forterre P, Breitbart M, Prangishvili D. 2011. Diversity of virus-host systems in hypersaline Lake Retba, Senegal. Environ Microbiol 13:1956–1972. doi: 10.1111/j.1462-2920.2010.02323.x. [DOI] [PubMed] [Google Scholar]
- 19.Borrel G, Colombet J, Robin A, Lehours AC, Prangishvili D, Sime-Ngando T. 2012. Unexpected and novel putative viruses in the sediments of a deep-dark permanently anoxic freshwater habitat. ISME J 6:2119–2127. doi: 10.1038/ismej.2012.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.López-Bueno A, Tamames J, Velazquez D, Moya A, Quesada A, Alcami A. 2009. High diversity of the viral community from an Antarctic lake. Science 326:858–861. doi: 10.1126/science.1179287. [DOI] [PubMed] [Google Scholar]
- 21.Bize A, Peng X, Prokofeva M, Maclellan K, Lucas S, Forterre P, Garrett RA, Bonch-Osmolovskaya EA, Prangishvili D. 2008. Viruses in acidic geothermal environments of the Kamchatka Peninsula. Res Microbiol 159:358–366. doi: 10.1016/j.resmic.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 22.Redder P, Peng X, Brugger K, Shah SA, Roesch F, Greve B, She Q, Schleper C, Forterre P, Garrett RA, Prangishvili D. 2009. Four newly isolated fuselloviruses from extreme geothermal environments reveal unusual morphologies and a possible interviral recombination mechanism. Environ Microbiol 11:2849–2862. doi: 10.1111/j.1462-2920.2009.02009.x. [DOI] [PubMed] [Google Scholar]
- 23.Rice G, Stedman K, Snyder J, Wiedenheft B, Willits D, Brumfield S, McDermott T, Young MJ. 2001. Viruses from extreme thermal environments. Proc Natl Acad Sci U S A 98:13341–13345. doi: 10.1073/pnas.231170198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baker BJ, Comolli LR, Dick GJ, Hauser LJ, Hyatt D, Dill BD, Land ML, Verberkmoes NC, Hettich RL, Banfield JF. 2010. Enigmatic, ultrasmall, uncultivated Archaea. Proc Natl Acad Sci U S A 107:8806–8811. doi: 10.1073/pnas.0914470107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hochstein R, Bollschweiler D, Engelhardt H, Lawrence CM, Young M. 2015. Large tailed spindle viruses of archaea: a new way of doing viral business. J Virol 89:9146–9149. doi: 10.1128/JVI.00612-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Häring M, Vestergaard G, Rachel R, Chen L, Garrett RA, Prangishvili D. 2005. Virology: independent virus development outside a host. Nature 436:1101–1102. doi: 10.1038/4361101a. [DOI] [PubMed] [Google Scholar]
- 27.Prangishvili D, Vestergaard G, Haring M, Aramayo R, Basta T, Rachel R, Garrett RA. 2006. Structural and genomic properties of the hyperthermophilic archaeal virus ATV with an extracellular stage of the reproductive cycle. J Mol Biol 359:1203–1216. doi: 10.1016/j.jmb.2006.04.027. [DOI] [PubMed] [Google Scholar]
- 28.Erdmann S, Chen B, Huang X, Deng L, Liu C, Shah SA, Le Moine Bauer S, Sobrino CL, Wang H, Wei Y, She Q, Garrett RA, Huang L, Lin L. 2014. A novel single-tailed fusiform Sulfolobus virus STSV2 infecting model Sulfolobus species. Extremophiles 18:51–60. doi: 10.1007/s00792-013-0591-z. [DOI] [PubMed] [Google Scholar]
- 29.Xiang X, Chen L, Huang X, Luo Y, She Q, Huang L. 2005. Sulfolobus tengchongensis spindle-shaped virus STSV1: virus-host interactions and genomic features. J Virol 79:8677–8686. doi: 10.1128/JVI.79.14.8677-8686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goulet A, Vestergaard G, Felisberto-Rodrigues C, Campanacci V, Garrett RA, Cambillau C, Ortiz-Lombardia M. 2010. Getting the best out of long-wavelength X-rays: de novo chlorine/sulfur SAD phasing of a structural protein from ATV. Acta Crystallogr D Biol Crystallogr 66:304–308. doi: 10.1107/S0907444909051798. [DOI] [PubMed] [Google Scholar]
- 31.Krupovic M, Bamford DH. 2011. Double-stranded DNA viruses: 20 families and only five different architectural principles for virion assembly. Curr Opin Virol 1:118–124. doi: 10.1016/j.coviro.2011.06.001. [DOI] [PubMed] [Google Scholar]
- 32.Martin A, Yeats S, Janekovic D, Reiter WD, Aicher W, Zillig W. 1984. SAV 1, a temperate u.v.-inducible DNA virus-like particle from the archaebacterium Sulfolobus acidocaldarius isolate B12. EMBO J 3:2165–2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nadal M, Mirambeau G, Forterre P, Reiter WD, Duguet M. 1986. Positively supercoiled DNA in a virus-like particle of an archaebacterium. Nature 321:256–258. doi: 10.1038/321256a0. [DOI] [Google Scholar]
- 34.Clore AJ, Stedman KM. 2007. The SSV1 viral integrase is not essential. Virology 361:103–111. doi: 10.1016/j.virol.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 35.Muskhelishvili G. 1994. The archaeal SSV integrase promotes intermolecular excisive recombination in vitro. Syst Appl Microbiol 16:605–608. [Google Scholar]
- 36.Serre MC, Letzelter C, Garel JR, Duguet M. 2002. Cleavage properties of an archaeal site-specific recombinase, the SSV1 integrase. J Biol Chem 277:16758–16767. doi: 10.1074/jbc.M200707200. [DOI] [PubMed] [Google Scholar]
- 37.Palm P, Schleper C, Grampp B, Yeats S, McWilliam P, Reiter WD, Zillig W. 1991. Complete nucleotide sequence of the virus SSV1 of the archaebacterium Sulfolobus shibatae. Virology 185:242–250. doi: 10.1016/0042-6822(91)90771-3. [DOI] [PubMed] [Google Scholar]
- 38.Jonuscheit M, Martusewitsch E, Stedman KM, Schleper C. 2003. A reporter gene system for the hyperthermophilic archaeon Sulfolobus solfataricus based on a selectable and integrative shuttle vector. Mol Microbiol 48:1241–1252. doi: 10.1046/j.1365-2958.2003.03509.x. [DOI] [PubMed] [Google Scholar]
- 39.Stedman KM, Schleper C, Rumpf E, Zillig W. 1999. Genetic requirements for the function of the archaeal virus SSV1 in Sulfolobus solfataricus: construction and testing of viral shuttle vectors. Genetics 152:1397–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schleper C, Kubo K, Zillig W. 1992. The particle SSV1 from the extremely thermophilic archaeon Sulfolobus is a virus: demonstration of infectivity and of transfection with viral DNA. Proc Natl Acad Sci U S A 89:7645–7649. doi: 10.1073/pnas.89.16.7645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fröls S, Gordon PM, Panlilio MA, Schleper C, Sensen CW. 2007. Elucidating the transcription cycle of the UV-inducible hyperthermophilic archaeal virus SSV1 by DNA microarrays. Virology 365:48–59. doi: 10.1016/j.virol.2007.03.033. [DOI] [PubMed] [Google Scholar]
- 42.Fusco S, She Q, Bartolucci S, Contursi P. 2013. T(lys), a newly identified Sulfolobus spindle-shaped virus 1 transcript expressed in the lysogenic state, encodes a DNA-binding protein interacting at the promoters of the early genes. J Virol 87:5926–5936. doi: 10.1128/JVI.00458-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reiter WD, Palm P, Henschen A, Lottspeich F, Zillig W, Grampp B. 1987. Identification and characterization of the genes encoding three structural proteins of the Sulfolobus virus-like particle SSV1. Mol Gen Genet 206:144–153. doi: 10.1007/BF00326550. [DOI] [Google Scholar]
- 44.Menon SK, Maaty WS, Corn GJ, Kwok SC, Eilers BJ, Kraft P, Gillitzer E, Young MJ, Bothner B, Lawrence CM. 2008. Cysteine usage in Sulfolobus spindle-shaped virus 1 and extension to hyperthermophilic viruses in general. Virology 376:270–278. doi: 10.1016/j.virol.2008.03.026. [DOI] [PubMed] [Google Scholar]
- 45.Reiter WD, Zillig W, Palm P. 1988. Archaebacterial viruses. Adv Virus Res 34:143–188. doi: 10.1016/S0065-3527(08)60517-5. [DOI] [PubMed] [Google Scholar]
- 46.Stedman KM, DeYoung M, Saha M, Sherman MB, Morais MC. 2015. Structural insights into the architecture of the hyperthermophilic fusellovirus SSV1. Virology 474:105–109. doi: 10.1016/j.virol.2014.10.014. [DOI] [PubMed] [Google Scholar]
- 47.Hong C, Pietilä MK, Fu CJ, Schmid MF, Bamford DH, Chiu W. 2015. Lemon-shaped halo archaeal virus His1 with uniform tail but variable capsid structure. Proc Natl Acad Sci U S A 112:2449–2454. doi: 10.1073/pnas.1425008112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pietilä MK, Atanasova NS, Oksanen HM, Bamford DH. 2013. Modified coat protein forms the flexible spindle-shaped virion of haloarchaeal virus His1. Environ Microbiol 15:1674–1686. doi: 10.1111/1462-2920.12030. [DOI] [PubMed] [Google Scholar]
- 49.Hanhijärvi KJ, Ziedaite G, Pietilä MK, Haeggstrom E, Bamford DH. 2013. DNA ejection from an archaeal virus–a single-molecule approach. Biophys J 104:2264–2272. doi: 10.1016/j.bpj.2013.03.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yeats S, McWilliam P, Zillig W. 1982. A plasmid in the archaebacterium Sulfolobus acidocaldarius. EMBO J 1:1035–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.She Q, Singh RK, Confalonieri F, Zivanovic Y, Allard G, Awayez MJ, Chan-Weiher CC, Clausen IG, Curtis BA, De Moors A, Erauso G, Fletcher C, Gordon PM, Heikamp-de Jong I, Jeffries AC, Kozera CJ, Medina N, Peng X, Thi-Ngoc HP, Redder P, Schenk ME, Theriault C, Tolstrup N, Charlebois RL, Doolittle WF, Duguet M, Gaasterland T, Garrett RA, Ragan MA, Sensen CW, Van der Oost J. 2001. The complete genome of the crenarchaeon Sulfolobus solfataricus P2. Proc Natl Acad Sci U S A 98:7835–7840. doi: 10.1073/pnas.141222098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zillig W, Kletzin A, Schleper C, Holz I, Janekovic D, Hain J, Lanzendorfer M, Kristjansson JK. 1994. Screening for Sulfolobales, their plasmids and their viruses in Icelandic solfataras. Syst Appl Microbiol 16:609–628. [Google Scholar]
- 53.Schägger H, von Jagow G. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem 166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 54.Pietilä MK, Roine E, Paulin L, Kalkkinen N, Bamford DH. 2009. An ssDNA virus infecting archaea: a new lineage of viruses with a membrane envelope. Mol Microbiol 72:307–319. doi: 10.1111/j.1365-2958.2009.06642.x. [DOI] [PubMed] [Google Scholar]
- 55.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 56.Cole C, Barber JD, Barton GJ. 2008. The Jpred 3 secondary structure prediction server. Nucleic Acids Res 36:W197–W201. doi: 10.1093/nar/gkn238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pitcher A, Hopmans EC, Mosier AC, Park SJ, Rhee SK, Francis CA, Schouten S, Damsté JS. 2011. Core and intact polar glycerol dibiphytanyl glycerol tetraether lipids of ammonia-oxidizing archaea enriched from marine and estuarine sediments. Appl Environ Microbiol 77:3468–3477. doi: 10.1128/AEM.02758-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huguet C, Hopmans EC, Febo-Ayala W, Thompson DH, Sinninghe Damsté JS, Schouten S. 2006. An improved method to determine the absolute abundance of glycerol dibiphytanyl glycerol tetraether lipids. Org Geochem 37:1036–1041. doi: 10.1016/j.orggeochem.2006.05.008. [DOI] [Google Scholar]
- 59.Schouten S, Huguet C, Hopmans EC, Kienhuis MV, Damsté JS. 2007. Analytical methodology for TEX86 paleothermometry by high-performance liquid chromatography/atmospheric pressure chemical ionization-mass spectrometry. Anal Chem 79:2940–2944. doi: 10.1021/ac062339v. [DOI] [PubMed] [Google Scholar]
- 60.Quemin ER, Lucas S, Daum B, Quax TE, Kuhlbrandt W, Forterre P, Albers SV, Prangishvili D, Krupovic M. 2013. First insights into the entry process of hyperthermophilic archaeal viruses. J Virol 87:13379–13385. doi: 10.1128/JVI.02742-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wiedenheft B, Stedman K, Roberto F, Willits D, Gleske AK, Zoeller L, Snyder J, Douglas T, Young M. 2004. Comparative genomic analysis of hyperthermophilic archaeal Fuselloviridae viruses. J Virol 78:1954–1961. doi: 10.1128/JVI.78.4.1954-1961.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pietilä MK, Atanasova NS, Manole V, Liljeroos L, Butcher SJ, Oksanen HM, Bamford DH. 2012. Virion architecture unifies globally distributed pleolipoviruses infecting halophilic archaea. J Virol 86:5067–5079. doi: 10.1128/JVI.06915-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 64.Napoli A, Zivanovic Y, Bocs C, Buhler C, Rossi M, Forterre P, Ciaramella M. 2002. DNA bending, compaction and negative supercoiling by the architectural protein Sso7d of Sulfolobus solfataricus. Nucleic Acids Res 30:2656–2662. doi: 10.1093/nar/gkf377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang Z, Guo L, Huang L. 2012. Archaeal chromatin proteins. Sci China Life Sci 55:377–385. doi: 10.1007/s11427-012-4322-y. [DOI] [PubMed] [Google Scholar]
- 66.Berggren K, Chernokalskaya E, Steinberg TH, Kemper C, Lopez MF, Diwu Z, Haugland RP, Patton WF. 2000. Background-free, high sensitivity staining of proteins in one- and two-dimensional sodium dodecyl sulfate-polyacrylamide gels using a luminescent ruthenium complex. Electrophoresis 21:2509–2521. doi:. [DOI] [PubMed] [Google Scholar]
- 67.Kandiba L, Aitio O, Helin J, Guan Z, Permi P, Bamford DH, Eichler J, Roine E. 2012. Diversity in prokaryotic glycosylation: an archaeal-derived N-linked glycan contains legionaminic acid. Mol Microbiol 84:578–593. doi: 10.1111/j.1365-2958.2012.08045.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Maaty WS, Ortmann AC, Dlakic M, Schulstad K, Hilmer JK, Liepold L, Weidenheft B, Khayat R, Douglas T, Young MJ, Bothner B. 2006. Characterization of the archaeal thermophile Sulfolobus turreted icosahedral virus validates an evolutionary link among double-stranded DNA viruses from all domains of life. J Virol 80:7625–7635. doi: 10.1128/JVI.00522-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mochizuki T, Yoshida T, Tanaka R, Forterre P, Sako Y, Prangishvili D. 2010. Diversity of viruses of the hyperthermophilic archaeal genus Aeropyrum, and isolation of the Aeropyrum pernix bacilliform virus 1, APBV1, the first representative of the family Clavaviridae. Virology 402:347–354. doi: 10.1016/j.virol.2010.03.046. [DOI] [PubMed] [Google Scholar]
- 70.Mochizuki T, Krupovic M, Pehau-Arnaudet G, Sako Y, Forterre P, Prangishvili D. 2012. Archaeal virus with exceptional virion architecture and the largest single-stranded DNA genome. Proc Natl Acad Sci U S A 109:13386–13391. doi: 10.1073/pnas.1203668109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Larson ET, Reiter D, Young M, Lawrence CM. 2006. Structure of A197 from Sulfolobus turreted icosahedral virus: a crenarchaeal viral glycosyltransferase exhibiting the GT-A fold. J Virol 80:7636–7644. doi: 10.1128/JVI.00567-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jarrell KF, Ding Y, Meyer BH, Albers SV, Kaminski L, Eichler J. 2014. N-linked glycosylation in Archaea: a structural, functional, and genetic analysis. Microbiol Mol Biol Rev 78:304–341. doi: 10.1128/MMBR.00052-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Palmieri G, Balestrieri M, Peter-Katalinic J, Pohlentz G, Rossi M, Fiume I, Pocsfalvi G. 2013. Surface-exposed glycoproteins of hyperthermophilic Sulfolobus solfataricus P2 show a common N-glycosylation profile. J Proteome Res 12:2779–2790. doi: 10.1021/pr400123z. [DOI] [PubMed] [Google Scholar]
- 74.Villanueva L, Damste JS, Schouten S. 2014. A re-evaluation of the archaeal membrane lipid biosynthetic pathway. Nat Rev Microbiol 12:438–448. doi: 10.1038/nrmicro3260. [DOI] [PubMed] [Google Scholar]
- 75.Schouten S, Hopmans EC, Sinninghe Damsté JS. 2013. The organic geochemistry of glycerol dialkyl glycerol tetraether lipids: a review. Org Geochem 54:19–61. doi: 10.1016/j.orggeochem.2012.09.006. [DOI] [Google Scholar]
- 76.Langworthy TA. 1977. Comparative lipid composition of heterotrophically and autotrophically grown Sulfolobus acidocaldarius. J Bacteriol 130:1326–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Langworthy TA, Mayberry WR, Smith PF. 1974. Long-chain glycerol diether and polyol dialkyl glycerol triether lipids of Sulfolobus acidocaldarius. J Bacteriol 119:106–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Flint SJ, Enquist LW, Racaniello VR, Skalka AM. 2009. Principles of virology, 3rd ed ASM Press, Washington, DC. [Google Scholar]
- 79.Iverson E, Stedman K. 2012. A genetic study of SSV1, the prototypical fusellovirus. Front Microbiol 3:200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Makarova KS, Wolf YI, Forterre P, Prangishvili D, Krupovic M, Koonin EV. 2014. Dark matter in archaeal genomes: a rich source of novel mobile elements, defense systems and secretory complexes. Extremophiles 18:877–893. doi: 10.1007/s00792-014-0672-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Happonen LJ, Redder P, Peng X, Reigstad LJ, Prangishvili D, Butcher SJ. 2010. Familial relationships in hyperthermo- and acidophilic archaeal viruses. J Virol 84:4747–4754. doi: 10.1128/JVI.02156-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pawlowski A, Rissanen I, Bamford JK, Krupovic M, Jalasvuori M. 2014. Gammasphaerolipovirus, a newly proposed bacteriophage genus, unifies viruses of halophilic archaea and thermophilic bacteria within the novel family Sphaerolipoviridae. Arch Virol 159:1541–1554. doi: 10.1007/s00705-013-1970-6. [DOI] [PubMed] [Google Scholar]
- 83.López-García P, Knapp S, Ladenstein R, Forterre P. 1998. In vitro DNA binding of the archaeal protein Sso7d induces negative supercoiling at temperatures typical for thermophilic growth. Nucleic Acids Res 26:2322–2328. doi: 10.1093/nar/26.10.2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Menon SK, Eilers BJ, Young MJ, Lawrence CM. 2010. The crystal structure of D212 from sulfolobus spindle-shaped virus ragged hills reveals a new member of the PD-(D/E)XK nuclease superfamily. J Virol 84:5890–5897. doi: 10.1128/JVI.01663-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ellen AF, Albers SV, Huibers W, Pitcher A, Hobel CF, Schwarz H, Folea M, Schouten S, Boekema EJ, Poolman B, Driessen AJ. 2009. Proteomic analysis of secreted membrane vesicles of archaeal Sulfolobus species reveals the presence of endosome sorting complex components. Extremophiles 13:67–79. doi: 10.1007/s00792-008-0199-x. [DOI] [PubMed] [Google Scholar]