ABSTRACT
Lytic activation of Kaposi's sarcoma-associated herpesvirus (KSHV) from latency is a critical contributor to pathogenesis and progression of KSHV-mediated disease. Development of targeted treatment strategies and improvement of lytic-phase-directed oncolytic therapies, therefore, hinge on gaining a better understanding of latency-to-lytic-phase transition. A key observation in that regard, also common to other herpesviruses, is the partial permissiveness of latently infected cells to lytic-cycle-inducing agents. Here, we address the molecular basis of why only some KSHV-infected cells respond to lytic stimuli. Since cellular signal transducer and activator of transcription 3 (STAT3) is constitutively active in KSHV-associated cancers, KSHV activates STAT3, and STAT3 has been found to regulate lytic activation of Epstein-Barr virus (EBV)-infected cells, we asked if STAT3 contributes similarly to the life cycle of KSHV. We found that high levels of STAT3 correlate with the refractory state at the single-cell level under conditions of both spontaneous and induced lytic activation; importantly, STAT3 also regulates lytic susceptibility. Further, knockdown of STAT3 suppresses the cellular transcriptional corepressor Krüppel-associated box domain-associated protein 1 (KAP1; also known as TRIM28), and suppression of KAP1 activates lytic genes, including the viral lytic switch RTA, thereby linking STAT3 via KAP1 to regulation of the balance between lytic and latent cells. These findings, taken together with those from EBV-infected and, more recently, herpes simplex virus 1 (HSV-1)-infected cells, cement the contribution of host STAT3 to persistence of herpesviruses and simultaneously reveal an important lead to devise strategies to improve lytic-phase-directed therapies for herpesviruses.
IMPORTANCE Lytic activation of the cancer-causing Kaposi's sarcoma-associated herpesvirus (KSHV) is vital to its life cycle and causation of disease. Like other herpesviruses, however, a substantial fraction of latently infected cells are resistant to lytic-phase-inducing stimuli. Investigating the molecular basis for this refractory state is essential for understanding how the virus persists and how it causes disease and to guide efforts to improve treatment of KSHV-mediated diseases. We found that, like two other herpesviruses, EBV and HSV-1, KSHV exploits the cellular transcription factor STAT3 to regulate the susceptibility of latently infected cells to lytic triggers. These findings highlight a common STAT3-centered strategy used by herpesviruses to maintain persistence in their hosts while also revealing a key molecule to pursue while devising methods to improve herpesvirus lytic-phase-directed therapies.
INTRODUCTION
The oncogenic human gammaherpesvirus human herpesvirus 8 (HHV-8), widely known as Kaposi's sarcoma-associated herpesvirus (KSHV), is the etiologic agent of three human malignancies: Kaposi's sarcoma (KS), primary effusion lymphoma (PEL), and multicentric Castleman's disease (MCD) (1, 2). Like other herpesviruses, KSHV exhibits a dual-phase life cycle that includes latency and lytic infection (3). During latency, the KSHV episome expresses a limited number of viral genes that mediate multiple functions, including maintaining viral genomes, repressing viral lytic gene expression, promoting proliferation of infected cells, and perturbing host immune surveillance (4–6). Periodic switching from latency to the lytic phase results in an orderly expression of a large number of viral genes to produce infectious virions.
Clinicoepidemiologic studies indicate that KSHV lytic activation is a critical contributor to the pathogenesis of KS, PEL, and MCD (7–13). Lytic activation also correlates with disease progression and prognosis (14–16). Indeed treatment with antiviral agents, such as ganciclovir and foscarnet, that target the lytic phase of the KSHV life cycle reduces the risk of development of KS, as well as the progression of KS and MCD (9, 14–16). A few studies have also reported remission of PEL following treatment with the antiviral drug cidofovir (17–19). These studies argue for a better understanding of the molecular mechanisms underlying the latency-to-lytic-phase switch with the ultimate goal of “therapeutically” increasing the number of latently infected cancer cells that switch to the lytic phase and thereby become susceptible to antiviral agents. In fact, a major hurdle to such oncolytic therapy is the presence of large fractions of cells among a population of latently infected cells that are resistant to lytic-cycle-inducing agents (20).
The goals of this study were to detect lytically infected cells using a monoclonal antibody (Ab) and to begin investigating the contribution of cellular factors to susceptibility to lytic-cycle-inducing signals at the single-cell level. The downstream goal is to use our findings to increase the number of lytic cells in response to lytic-cycle-activating agents. Our studies on Epstein-Barr virus (EBV)-infected cells revealed that high levels of signal transducer and activator of transcripton 3 (STAT3), a cellular transcription factor important for oncogenesis and inflammation, correlate with resistance to lytic-phase-inducing signals. Furthermore, STAT3 regulates the susceptibility of latently infected EBV+ cells to lytic-cycle-activating triggers (21, 22). Partly because of the newly discovered contribution of STAT3 to lytic susceptibility in another human gammaherpesvirus (i.e., EBV) and partly because Janus kinase 2 (JAK2)/STAT3 signaling is constitutively active in KSHV+ PEL cells (23), we focused our efforts on STAT3. Using KSHV+ PEL cells that are derived from B cells and provide an excellent model to study the KSHV latency-to-lytic-phase switch, we show here that high levels of STAT3 correlate with and promote the lytic-cycle-refractory state of KSHV-infected cells. We also show that STAT3 regulates the cellular transcriptional corepressor KAP1 (Krüppel-associated box domain-associated protein 1), which represses lytic genes, thus functionally linking STAT3 via KAP1 to lytic susceptibility of KSHV+ latent cells.
MATERIALS AND METHODS
Cell culture and chemical treatment.
The body cavity-based KSHV+ lymphoma cell line BCBL-1 (a kind gift from Shane McAllister, University of Minnesota Medical School) was maintained in RPMI 1640 supplemented with 10% fetal bovine serum (FBS) (Gibco) and 1% penicillin-streptomycin (Gibco). The BCBL-1 cells were subcultured at 3 × 105 cells/ml and treated after 24 h with 20 ng/ml of 12-O-tetradecanoylphorbol-13-acetate (TPA) (catalog no. 79346; Sigma), 0.6 μM valproic acid (VPA) (catalog no. 152064; MP Biomedicals), or 5 μM WP1066 (catalog no. 573097; EMD). Primary human umbilical vein endothelial cells (HUVECs) (Lonza) were cultured in EGM-2 medium (Lonza) in 6-well tissue culture plates coated with 0.1% (wt/vol) gelatin (in phosphate-buffered saline [PBS]) and used at passage 6 for experiments.
Nucleofection of BCBL-1 cells.
BCBL-1 cells were subcultured at 5 × 105 cells/ml 24 h prior to transfection. Cells (2 × 106) were transfected with either 20 μg of plasmid (pEGFPN1 or pEGFPN1-STAT3; from Nancy Reich, Stony Brook University [24, 25]) or 130 pmol of small interfering RNA (siRNA) (30 pmol fluorescein isothiocyanate-positive [FITC+] scrambled siRNA plus 100 pmol scrambled siRNA or 100 pmol siRNA targeting human STAT3; Santa Cruz) in Cell Line Nucleofection Solution V (catalog no. VCA-1003; Lonza) using an Amaxa Nucleofector II (program T-001). For certain experiments (see Fig. 3A, B, D, and E and Fig. 5), 150 pmol of siRNA (scrambled siRNA and siRNAs targeting human STAT3 or KAP1; all from Santa Cruz) was used.
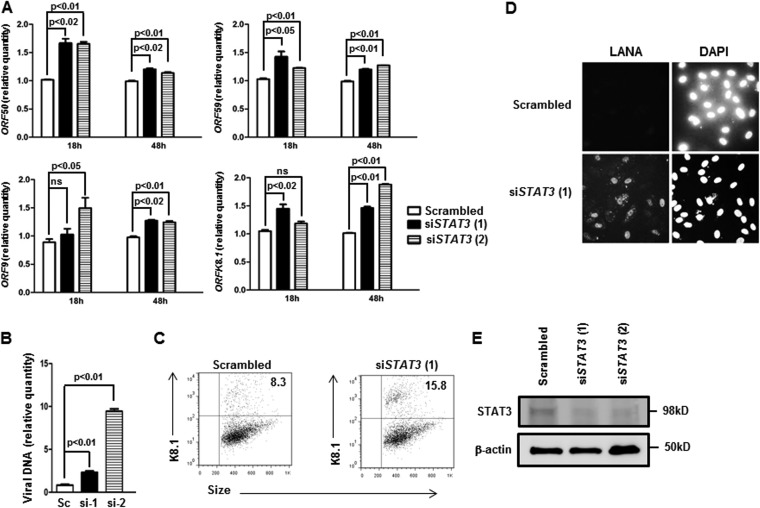
FIG 3.
Knockdown of endogenous STAT3 results in increased spontaneous KSHV lytic activation. (A) BCBL-1 cells were transfected with scrambled siRNA or two different siRNAs to STAT3 [siSTAT3 (1) and siSTAT3 (2)] and harvested 18 h and 48 h later for determination of relative amounts of transcripts from KSHV lytic genes ORF50, ORF59, ORF9, and ORFK8.1 by qRT-PCR after normalization to 18S rRNA using the ΔΔCT method. Error bars indicate SEM of 3 technical replicates from each of 2 transfection experiments. (B) BCBL-1 cells were transfected with scrambled siRNA (Sc) or siRNAs to STAT3 (si-1 and si-2) and harvested 48 h later for determination of relative amounts of cell-associated KSHV DNA by qPCR. Error bars indicate SEM of 3 technical replicates from each of 2 transfection experiments. (C) BCBL-1 cells were transfected with either STAT3 siRNA and FITC+ scrambled siRNA (to mark transfected cells) at a 3:1 ratio [siSTAT3 (1)] or with FITC− and FITC+ scrambled siRNA at a 3:1 ratio (Scrambled). After 36 h, the FITC+ (i.e., transfected) population was examined by flow cytometry for K8.1+ cells. Numbers indicate percentages of transfected cells that were spontaneously lytic. A representative of the results of two experiments is shown. (D) BCBL-1 cells were treated as for panel B, and cell supernatants were used to inoculate primary HUVECs. After 48 h, HUVECs were fixed, permeabilized, stained with anti-LANA antibody and DAPI, and visualized at ×40 magnification. (E) BCBL-1 cells were transfected with scrambled siRNA or siRNA to STAT3, harvested at 48 h posttransfection, and subjected to Western blot analysis using anti-STAT3 and anti-β-actin antibodies.
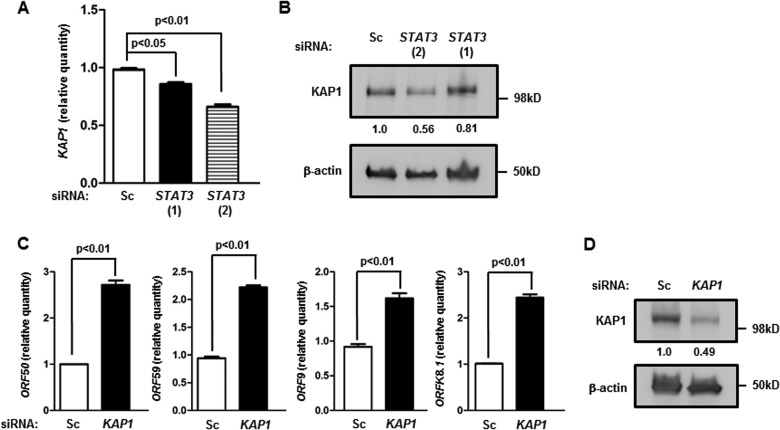
FIG 5.
Knockdown of STAT3 suppresses KAP1, and suppression of KAP1 causes an increase in the levels of lytic transcripts. BCBL-1 cells were transfected with scrambled siRNA (Sc) or two different siRNAs to STAT3 [STAT3 (1) and STAT3 (2)] (A and B) or scrambled siRNA (Sc) or siRNA to KAP1 (C and D) and harvested 24 h (C and D) or 48 h (A and B) later for determination of the relative amounts of transcripts from cellular KAP1 (A) and the KSHV lytic genes ORF50, ORF59, ORF9, and ORFK8.1 (C) by qRT-PCR after normalization to 18S rRNA using the ΔΔCT method or subjected to Western blotting using anti-KAP1 and anti-β-actin antibodies (B and D). Error bars indicate SEM of 3 technical replicates from each of 2 transfection experiments. Numbers in panels B and D indicate relative amounts of KAP1 protein determined by densitometry after normalization to β-actin.
Infection of HUVECs with KSHV.
HUVECs were plated on gelatin-coated 6-well plates with glass coverslips at 1 × 105 cells/well. The next day, monolayers were inoculated for 2 h at 37°C with 1.8 ml of cell supernatants derived from BCBL-1 cells left untreated or treated for 48 h with 0.6 μM VPA or 5 μM WP1066. This was followed by spinoculation at 2,000 rpm for 15 min at 25°C, after which the inoculum was removed and cells were incubated at 37°C and 5% CO2 in fresh EGM-2 medium for 48 h.
Immunoblotting and immunofluorescence.
Total cell extracts were electrophoresed in 10% SDS-polyacrylamide gels, transferred to nitrocellulose membranes, and reacted with antibodies. Mouse monoclonal Abs were used to detect K8.1 (1:50; a kind gift from Bala Chandran, Rosalind Franklin University) and β-actin (AC-15; 1:10,000 dilution; Sigma), a rabbit polyclonal Ab was used to detect STAT3 (sc-482; 1:500 dilution; Santa Cruz), and a rabbit polyclonal Ab was used to detect KAP1 (A300-274A; 1:3,000 dilution; Bethyl Laboratories). Signals were detected using enhanced chemiluminescence.
Immunofluorescence for KSHV latency-associated nuclear antigen 1 (LANA-1) was carried out in HUVECs treated with cell supernatants from treated or untreated BCBL-1 cells. At 48 h posttreatment, HUVECs were washed in PBS, fixed in 4% paraformaldehyde for 10 min at room temperature, washed, and permeabilized in 0.1% Triton X-100. Coverslips were blocked in 1% human AB serum in 5% BSA-PBS for 30 min and then stained using rabbit anti-LANA Ab (1:500; a kind gift from Craig McCormick, Dalhousie University). After primary-antibody binding, cell monolayers were washed and incubated for 45 min with Alexa Fluor 488 anti-rabbit Ab (1:2,000; Molecular Probes) and mounted using Vectashield with DAPI (4′,6-diamidino-2-phenylindole) (Molecular Probes).
Flow cytometry.
To stain for K8.1 alone, BCBL-1 cells were washed with 1× PBS and stained with mouse anti-K8.1 Ab diluted at 1:10 in fluorescence-activated cell sorter (FACS) buffer (1× PBS with 0.5% FBS), followed by staining with allophycocyanin (APC)-conjugated anti-mouse IgG (sc-3818; Santa Cruz) diluted at 1:200 in FACS buffer. For double staining of K8.1 and STAT3, BCBL-1 cells were washed with 1× PBS and stained with mouse anti-K8.1 Ab, followed by staining with FITC-conjugated anti-mouse IgG (F0257; Sigma) diluted at 1:200. Cells were then fixed with BD Cytofix/Cytoperm solution and stained with rabbit anti-STAT3 Ab diluted at 1:50 in 1× BD Perm/Wash solution, followed by staining with Alexa Fluor 647-conjugated anti-rabbit IgG (A21245; Life Technologies). Events were acquired using FACSCalibur (BD), and data were analyzed using FlowJo software (Tree Star). Gating was performed by comparison with similarly treated cells that were stained with isotype control antibodies or secondary antibodies alone.
Quantitative reverse transcription-PCR (qRT-PCR).
Total RNA was isolated from BCBL-1 cells using an RNeasy kit (Qiagen), followed by DNase digestion (Promega). RNA was quantitated with a NanoDrop (Thermo Scientific). RNA (1 μg) was converted to cDNA using qScript cDNA SuperMix (Quanta BioSciences). Relative transcript levels of selected cellular and viral genes were determined with gene-specific primers using Fast SYBR green master mix on a StepOne Plus thermocycler (Applied Biosystems). The sequences of the primers used are as follows: forward primer, GTAACCCGTTGAACCCCATT, and reverse primer, CCATCCAATCGGTAGTAGCG for 18S rRNA; forward primer, TTGATTTTGGAGGGATCTCG, and reverse primer, GAGTCAACGGATTTGGTCGT for GAPDH; forward primer, GGCGTCGTGATTAGTGATGAT, and reverse primer, GGGCTACAATGTGATGGCCT for HPRT1; forward primer, GAGTATGCCTGCCGTGTG, and reverse primer, AATCCAAATGCGGCATCT for the β2 microglobulin gene; forward primer, GCCTCTGTGTGAGACCTGTG, and reverse primer, AGTACGTTCACCATCCCGAG for KAP1; forward primer, CCCTGAGCCAGTTTGTCATT, and reverse primer, ATGGGTTAAAGGGGATGATG for ORF50; forward primer, TGGTCGGCGGTTCAGTCATCAA, and reverse primer, GCGGCCGCTAAGAAAATCGA for ORFK8.1; forward primer, TTAGAAGTGGAAGGTGTGCC, and reverse primer, TCCTGGAGTCCGGTATAGAATC for ORF59; and forward primer, TAGGCGCTTCGTGCTGG, and reverse primer, CCGGATTGCTGCACTCGTA, for ORF9.
Relative expression levels were calculated using the ΔΔCT method after normalization to 18S rRNA, HPRT1, or GAPDH. Individual samples were assayed in triplicate.
KSHV viral load assay.
One million BCBL-1 cells transfected with siRNAs or plasmids and treated with VPA (or untreated) were resuspended in 100 μl of 0.2× PBS, heated to 95°C for 1 h, and then treated with 10 mg/ml of proteinase K (catalog no. 19131; Qiagen) at 56°C overnight. The enzyme was inactivated by heating at 95°C for 1 h. Aliquots of the lysate (10 μl from siRNA-transfected cells and 0.01 μl from plasmid-transfected cells) were used to amplify the KSHV ORFK9 gene by real-time PCR using the following primers: ORFK9-F, 5′-GTCTCTGCGCCATTCAAAAC-3′, and ORFK9-R, 5′-CCGGACACGACAACTAAGAA-3′. Relative KSHV load was quantified based on a standard curve PCR generated using VPA-treated BCBL-1 cells; the standard quantitative-PCR (qPCR) curve gave linear detection over 5 log units of target dilutions.
Statistical analysis.
P values were calculated by comparing the means of two groups of interest using an unpaired Student t test.
RESULTS
High levels of STAT3 mark KSHV+ cells that are refractory to lytic activation.
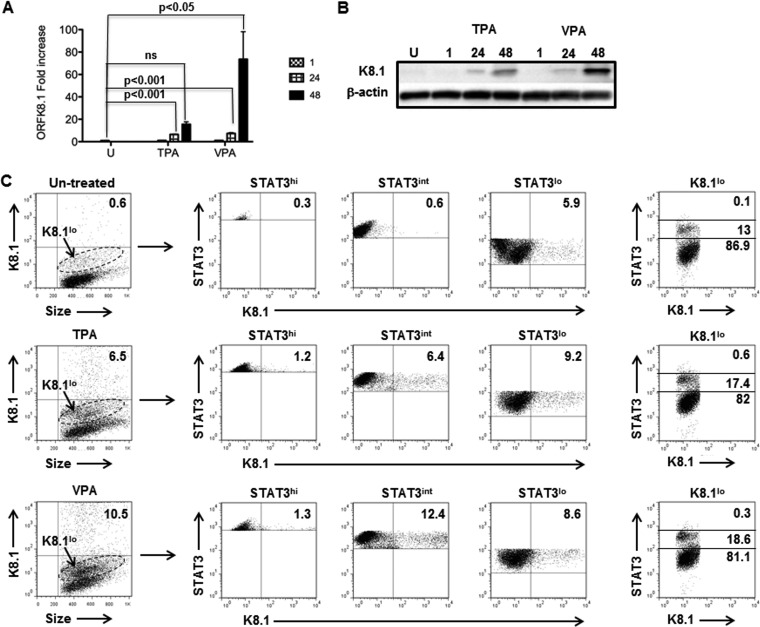
To determine the relationship between cellular STAT3 and KSHV lytic activation, we used a well-validated monoclonal antibody to the KSHV glycoprotein K8.1 (a late lytic protein) (26) in flow cytometric analysis of single cells. To establish the optimal time for detection of K8.1+ lytic cells, we treated BCBL-1 cells with the lytic-cycle-inducing agent TPA or VPA. We found that ORFK8.1 message and protein levels were increased at 24 h and even more so at 48 h posttreatment with both agents compared to untreated cells (Fig. 1A and B). Further, VPA treatment showed a more pronounced increase in steady-state ORFK8.1 RNA and protein levels than TPA treatment after 48 h (Fig. 1A and B). We therefore expected to be able to detect K8.1+ lytic cells as early as 24 h. However, to ensure maximal detection of lytic cells by flow cytometry, we treated cells with TPA or VPA for 48 h. Figure 1C shows that after TPA and VPA treatment, 6.5% and 10.5% of the cells, respectively, expressed high levels of K8.1 compared to 0.6% of the cells in the untreated population. Notably, higher levels of K8.1 protein in 48-hour VPA-treated cells (Fig. 1B) than in TPA-treated cells (Fig. 1C) corresponded to an increase in the number of K8.1+ lytic cells.
FIG 1.
Cells expressing high levels of STAT3 protein are refractory to spontaneous and induced KSHV lytic activation. BCBL-1 cells were treated with TPA or VPA or left untreated (U). Cells were harvested at 1, 24, and 48 h (A and B) and at 48 h (C) posttreatment. (A) RNA was isolated and subjected to qRT-PCR to determine the relative levels of lytic ORFK8.1 transcripts. Fold changes were calculated by the ΔΔCT method, normalized to three housekeeping genes, GAPDH, HPRTI, and B2M (β2 microglobulin). The data are presented as means and standard errors of the mean (SEM) and are representative of the results of two experiments with 3 technical replicates. (B) Cell lysates were immunoblotted using antibodies to K8.1 and β-actin. (C) Cells were immunostained for K8.1 and STAT3 and subjected to flow cytometry. The numbers within the dot plots in the left column indicate the percent K8.1hi cells under untreated or TPA- or VPA-treated conditions after comparison with similarly treated cells stained with isotype control antibody; the dashed oval gates indicate K8.1lo cells. The numbers within the dot plots in the three middle columns indicate the percent K8.1hi cells in subpopulations expressing different levels of STAT3, i.e., STAT3hi (high), STAT3int (intermediate), and STAT3lo (low). Numbers in the dot plots on the right indicate the percent K8.1lo cells (from the oval gates in the left-hand dot plots) expressing low, intermediate, and high levels of STAT3.
To correlate the levels of STAT3 with lytic activation, we divided the cells into STAT3hi (high), STAT3int (intermediate), and STAT3lo (low) based on the relative levels of expression of endogenous STAT3 and then examined them for K8.1 expression (Fig. 1C). We found that STAT3hi cells were predominantly refractory, with only 0.3% to 1.3% of the cells expressing high levels of K8.1 whether under spontaneous lytic conditions (i.e., untreated) or treated with lytic-cycle-inducing agents. In contrast, lytic cells, spontaneous or induced by chemicals, arose primarily from STAT3int and STAT3lo cells. Of note, the majority (81.1% to 86.9%) of cells expressing low levels of K8.1, and likely to be early lytic cells, also expressed low levels of STAT3. Thus, high levels of STAT3 correlate with the refractory state in KSHV-infected cells.
Chemical inhibition of STAT3 results in KSHV lytic activation.
STAT3 is a convergence point of multiple signaling pathways, especially cytokine and growth factor signaling pathways (27). Receptor engagement typically results in activation of STAT3 via members of the JAK family. Activated STAT3 then functions as a transcription activator of many genes, including itself (28). Changes in STAT3 protein levels would be expected to occur under a variety of physiological and pathological conditions, leading us to investigate the effect of a STAT3 inhibitor on KSHV lytic activation. We found that exposing BCBL-1 cells to WP1066, an agent that inhibits JAK2-mediated activation and nuclear localization of STAT3 (29), resulted in substantial increases in steady-state levels of messages from the KSHV lytic genes ORF50 (an immediate-early lytic gene), ORF9 and ORF59 (early lytic genes), and ORFK8.1 (a late lytic gene) (Fig. 2A). Compared to the kinetics in VPA-treated cells, the increase in ORF50 and ORF59 RNA levels showed a slight lag following treatment with WP1066 alone; this was not the case for ORFK8.1 message. Also, by 18 h, exposure to VPA and WP1066 resulted in substantial and synergistic increases in steady-state levels of lytic messages beyond that caused by VPA alone. Thus, chemical inhibition of STAT3 resulted in an increase in the levels of messages from KSHV lytic genes belonging to all 3 kinetic classes.
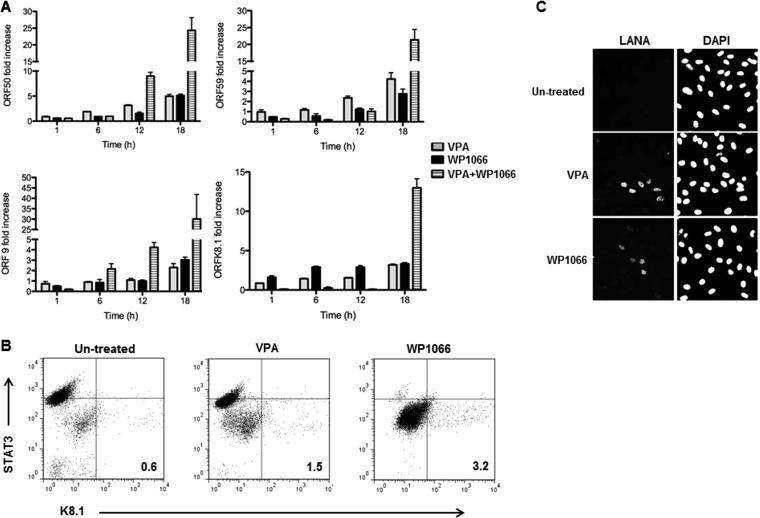
FIG 2.
Chemical inhibition of STAT3 results in KSHV lytic activation, increase in the number of lytic cells, and increase in production of infectious virions. BCBL-1 cells were treated with VPA, WP1066, or VPA plus WP1066 or left untreated. Cells were harvested at 1, 6, 12, and 18 h posttreatment (A), at 24 h posttreatment (B), and at 48 h posttreatment (C). (A) RNA was analyzed at different times by qRT-PCR for relative transcript levels of 4 lytic genes, ORF50, ORF59, ORF9, and ORFK8.1, compared to untreated cells. KSHV-specific transcript levels were normalized to GAPDH, HRPTI, and B2M (β2 microglobulin), and fold changes were determined by the ΔΔCT method. Data are presented as means and SEM and are representative of the results of two separate experiments with 3 technical replicates. (B) Cells were immunostained for K8.1 and STAT3 and subjected to flow cytometry. Numbers indicate the percent K8.1+ (lytic) cells; these percentages were determined after comparison with similarly treated cells stained with isotype control antibody. (C) Cell supernatants were used to inoculate primary HUVECs. After 48 h, HUVECs were fixed, permeabilized, stained with anti-LANA antibody and DAPI, and visualized at ×40 magnification.
Inhibition of STAT3 results in lytic activation.
We next asked if the increase in lytic gene expression actually resulted in an increase in the number of lytically infected cells. Figure 2B shows that there were 3.2% K8.1+ lytic cells after 24 h of treatment with WP1066 compared to 1.5% in VPA-treated cells. As expected, treatment of cells with WP1066 resulted in lower levels of STAT3 in the majority of cells (Fig. 2B, far-right dot plot compared to the 2 dot plots on the left). Also, nearly all lytic cells arose from cells expressing low levels of STAT3, whereas cells with higher levels of STAT3 remained strictly refractory. To further investigate if lytic activation secondary to STAT3 impairment results in the production and release of infectious particles, we harvested cell culture supernatants 48 h after treatment of cells with VPA or WP1066. Exposure of HUVECs to culture supernatants from VPA- and WP1066-treated cells resulted in greater numbers of LANA-1-positive HUVECs than exposure to supernatants from untreated cells (Fig. 2C). These data indicate that functionally inhibiting STAT3 enhances productive lytic activation in KSHV-infected B cells.
STAT3 regulates KSHV lytic activation.
Since chemical inhibitors are often associated with off-target effects, we used previously validated siRNAs (22, 30) to specifically inhibit STAT3. Figure 3A shows that, compared to scrambled-siRNA-treated cells, introduction of 2 different siRNAs targeting STAT3 into latently infected cells resulted in significant increases in mRNA levels from ORF50, ORF59, ORF9, and ORFK8.1. Furthermore, these increases in lytic gene expression corresponded to increases in the cell-associated KSHV load (Fig. 3B), the percentage of K8.1+ lytically infected cells (Fig. 3C) (15.8% in siSTAT3 compared to 8.3% in scrambled siRNA), and infectious virions in the cell supernatant (Fig. 3D); treatment of siRNA-transfected cells with VPA did not cause an additional increase in lytic cells (data not shown), likely due to the combined stresses of lowering STAT3 levels and exposure to an HDAC inhibitor. Figure 3E shows that, as expected, siRNAs to STAT3 suppressed the level of STAT3 compared to scrambled-siRNA treatment. Thus, suppressing the levels of STAT3 is sufficient to induce the KSHV lytic cycle.
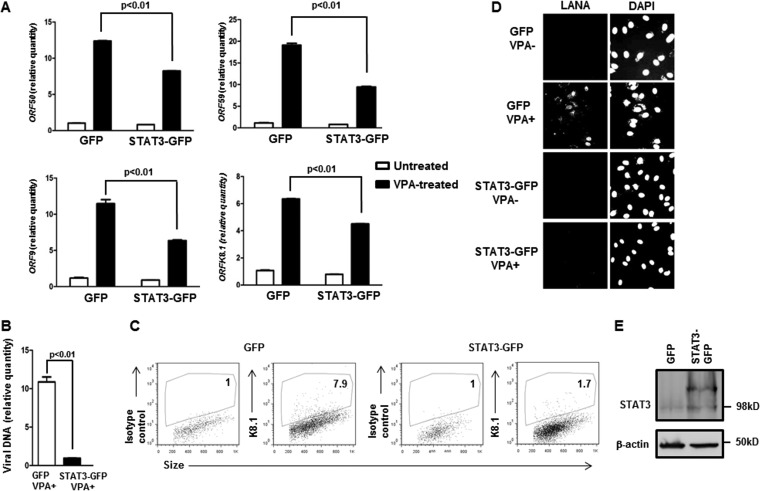
To determine if increasing the levels of STAT3 had an opposite effect on lytic activation, i.e., restrained the KSHV lytic cycle, we introduced a green fluorescent protein (GFP)-tagged STAT3 plasmid into latently infected cells and then induced the lytic cycle with VPA. Figure 4A shows that, compared to empty-vector (GFP)-transfected cells, cells transfected with STAT3 overexpression plasmid demonstrated statistically significant reductions in message levels from ORF50, ORF59, ORF9, and ORFK8.1. Overexpression of STAT3 also suppressed the KSHV load in VPA-treated cells (Fig. 4B) and the amount of infectious virions in the cell-free supernatant (Fig. 4D). To determine if STAT3 overexpression caused fewer cells to support the KSHV lytic cycle, we gated on GFP+ (i.e., transfected) cells and then enumerated K8.1-expressing cells. As shown in Fig. 4C, there was a nearly 90% reduction (6.9% for GFP versus 0.7% for STAT3-GFP) in the percentage of lytic cells among those that had been successfully transfected. Also as expected, introduction of a STAT3 plasmid resulted in an increase in the levels of STAT3 (Fig. 4E). Taken together, our data demonstrate that the levels of cellular STAT3 regulate susceptibility of latently infected cells to KSHV lytic-cycle activation.
FIG 4.
Overexpression of STAT3 restrains susceptibility to KSHV lytic activation. (A to D) BCBL-1 cells were transfected with the pEGFPN1 plasmid (GFP) or the pEGFPN1-STAT3 plasmid (STAT3-GFP), exposed to VPA after 12 h, and harvested after another 24 h for determination of relative amounts of transcripts from KSHV lytic genes ORF50, ORF59, ORF9, and K8.1 by qRT-PCR after normalization to 18S rRNA using the ΔΔCT method (A), for determination of relative amounts of cell-associated KSHV DNA by qPCR (B), for determination of K8.1+ cells by flow cytometry (C), or assayed for infectious virions in the cell supernatant by inoculation of HUVECs and staining with anti-LANA antibody and DAPI 48 h later (D). (A and B) Error bars indicate SEM of 3 technical replicates from each of 2 transfection experiments. (C) Numbers indicate the percentages of GFP+ (i.e., transfected) cells that were lytic. Lytic (i.e., K8.1+) gates were placed based on a 1% cutoff in similarly treated cells that were stained with isotype control antibody. A representative of the results of two experiments is shown. (E) BCBL-1 cells were transfected with GFP plasmid or STAT3-GFP plasmid (as in panels A to D) and harvested at 24 h posttransfection for Western blot analysis using anti-STAT3 and anti-β-actin antibodies.
Cellular KAP1 links STAT3 to KSHV lytic activation.
STAT3 is generally considered a transcription activator, yet it represses lytic genes. When we examined a publically available STAT3 chromatin immunoprecipitation-DNA sequencing (ChIP-seq) data set (22, 31), we identified cellular KAP1, a transcriptional corepressor, as a potential STAT3 transcriptional target. Importantly, KAP1 has been previously found to regulate lytic susceptibility in cytomegalovirus (CMV)- and KSHV-infected cells (32–35); however, how KAP1 itself is regulated in virus-infected cells is not entirely clear. Introduction of the 2 siRNAs targeting STAT3 in BCBL-1 cells resulted in suppression of KAP1 transcript and protein levels (Fig. 5A and B), indicating that STAT3 regulates KAP1 in BCBL-1 cells. Furthermore, transfection of siRNA to KAP1 resulted in increases in the levels of transcripts from lytic genes belonging to all 3 kinetic classes (Fig. 5C). As expected, siRNA to KAP1 caused suppression of the KAP1 protein level (Fig. 5D). These results collectively implicate cellular KAP1 in STAT3-mediated regulation of KSHV lytic susceptibility.
DISCUSSION
The lytic phase of KSHV is critical to its pathogenesis; in this study, we provide evidence for STAT3 as a mechanistic link between cells that are susceptible versus those that are refractory to lytic stimuli. We make this connection between a host transcription factor and a key event in the life cycle of the virus at the single-cell level. Not only do high levels of STAT3 correlate with the refractory state regardless of whether cells respond to spontaneous lytic triggers in culture or to exogenously provided chemical triggers, but suppression of STAT3 makes KSHV-infected cells more permissive to signals that trigger lytic activation; this culminates in production of infectious virus. In contrast, experimental overexpression of STAT3 causes maintenance of the refractory/latency state. Importantly, we found that STAT3 functions via KAP1, a cellular transcriptional corepressor, to regulate lytic susceptibility. These findings implicate STAT3, an important cellular prosurvival and proproliferative protein constitutively active in KSHV+ tumors, in a regulatory role that limits the exit of KSHV from latency.
Although KSHV lytic activation is linked to pathogenesis and often to progression of KS, hemophagocytic lymphohistiocytosis, MCD, and PEL (7–13), a host of disparate diseases arising from a multitude of cell types, KSHV+ B cell lines, such as BCBL-1 cells that were originally derived from clinical specimens of PEL, provide the most robust tissue culture models for understanding molecular events underlying viral latency and lytic activation. Another important reason for selecting BCBL-1 cells is that they are not coinfected with EBV, thereby avoiding the confounding presence of another gammaherpesvirus while examining this mechanistic question. Indeed while most PELs are coinfected with EBV (36), the existence of EBV-negative PELs indicates that KSHV infection is sufficient for disease. Nonetheless, future studies need to address the contributions of STAT3 and KAP1 to regulation of the KSHV lytic cycle in other cell lines and, more importantly, in KSHV-infected tissues.
STAT3 is an important molecular node for many cellular signaling pathways and can potentially regulate the expression of thousands of genes (22, 31). This study and our earlier studies demonstrate that, in addition to phosphorylation and dimerization of STAT3, modulation of the protein level of STAT3 also regulates its function (22, 30, 37–39). Our data also indicate that STAT3 functions at least partly by repressing the immediate-early gene product RTA, the key latency-to-lytic-phase viral switch. While it is generally thought to be a transactivator, STAT3 can suppress gene expression in some instances either directly (40) or indirectly. Indeed, we found that STAT3 exploits a transcriptional corepressor to repress lytic genes. Whether the effects on transcript levels of early and late lytic genes are secondary to repression of RTA or due to direct repression of these genes by KAP1 remains to be determined.
KSHV LANA and cellular Nrf2 have been found to repress ORF50 by recruiting KAP1 during the early stage of KSHV primary infection and in latently infected KSHV+ cell lines, respectively (33, 35). Also, phosphorylation of KAP1 at S824 has been linked to derepression of lytic genes in CMV- and KSHV-infected cells (32, 34). Our results now reveal modulation of KAP1 levels by STAT3 as a mechanism to regulate KSHV lytic susceptibility. However, whether KAP1 is recruited to ORF50 (and potentially the other 3 lytic genes) by LANA, cellular Nrf2, or some other means is presently unclear.
Dysregulation of STAT3 is linked to most human cancers, including KSHV+ PEL and KS (23, 41, 42). Notably, infection of endothelial cells with KSHV results in phosphorylation of STAT3 at S727 and phosphorylation of KAP1 at S473; the latter relieves STAT3 from KAP1-mediated repression (37). Furthermore, KSHV-encoded viral interleukin 6 (IL-6) is able to activate STAT3 via binding to gp130, the common IL-6 receptor (43); though it is a lytic gene product, viral IL-6 could contribute to STAT3 activation and therefore to increased STAT3 levels in a paracrine manner in latent cells. Our findings now indicate that, apart from contributing to the inflammatory milieu and subsequent growth transformation (37, 44), this increased STAT3 also contributes to viral persistence. This exploitation of cellular STAT3 by both of the human oncogenic herpesviruses EBV and KSHV to ensure maintenance of a persistent pool of latently infected cells despite the presence of lytic activation triggers appears to extend beyond the gammaherpesvirus subfamily. A recent report showed that STAT3 restricts herpes simplex virus 1 (HSV-1) lytic activation (45). Thus, STAT3-mediated regulation of viral lytic activation may be a common theme among human herpesviruses. This knowledge could guide novel strategies to prevent lytic activation of HSV-1 and therefore prevent reactivation-associated alphaherpesvirus disease. On the other hand, increasing the number of latently infected cells undergoing lytic activation could greatly enhance the efficacy of oncolytic therapies directed against KSHV+ and EBV+ cancers.
ACKNOWLEDGMENTS
This study was supported by a collaborative grant from the SUNY REACH (Research Excellence in Academic Health) to C.A.K. and S.B.-M., NIH grant AI113134 to S.B.-M., and funds from The Research Foundation for The State University of New York to S.B.-M.
We thank Bala Chandran at Rosalind Franklin University of Medicine and Science for providing us with the monoclonal antibody to KSHV K8.1 and Nancy Reich for plasmids pEGFPN1 and pEGFPN1-STAT3.
C.A.K., X.L., and S.B.-M. designed the study; C.A.K., A.B.-G., and X.L. carried out the experiments; C.A.K., X.L., and S.B.-M. analyzed data and interpreted the findings; and X.L. and S.B.-M. wrote the manuscript.
We declare no conflict of interest.
REFERENCES
- 1.Boshoff C, Weiss R. 2002. AIDS-related malignancies. Nat Rev Cancer 2:373–382. doi: 10.1038/nrc797. [DOI] [PubMed] [Google Scholar]
- 2.Cesarman E, Chang Y, Moore PS, Said JW, Knowles DM. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N Engl J Med 332:1186–1191. doi: 10.1056/NEJM199505043321802. [DOI] [PubMed] [Google Scholar]
- 3.Fields BN, Knipe DM, Howley PM. 1996. Fundamental virology, 3rd ed. Lippincott-Raven, Philadelphia, PA. [Google Scholar]
- 4.Okada S, Goto H, Yotsumoto M. 2014. Current status of treatment for primary effusion lymphoma. Intractable Rare Dis Res 3:65–74. doi: 10.5582/irdr.2014.01010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lan K, Kuppers DA, Verma SC, Robertson ES. 2004. Kaposi's sarcoma-associated herpesvirus-encoded latency-associated nuclear antigen inhibits lytic replication by targeting Rta: a potential mechanism for virus-mediated control of latency. J Virol 78:6585–6594. doi: 10.1128/JVI.78.12.6585-6594.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li X, Liang D, Lin X, Robertson ES, Lan K. 2011. Kaposi's sarcoma-associated herpesvirus-encoded latency-associated nuclear antigen reduces interleukin-8 expression in endothelial cells and impairs neutrophil chemotaxis by degrading nuclear p65. J Virol 85:8606–8615. doi: 10.1128/JVI.00733-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boneschi V, Brambilla L, Berti E, Ferrucci S, Corbellino M, Parravicini C, Fossati S. 2001. Human herpesvirus 8 DNA in the skin and blood of patients with Mediterranean Kaposi's sarcoma: clinical correlations. Dermatology 203:19–23. doi: 10.1159/000051697. [DOI] [PubMed] [Google Scholar]
- 8.Campbell TB, Borok M, Gwanzura L, MaWhinney S, White IE, Ndemera B, Gudza I, Fitzpatrick L, Schooley RT. 2000. Relationship of human herpesvirus 8 peripheral blood virus load and Kaposi's sarcoma clinical stage. AIDS 14:2109–2116. doi: 10.1097/00002030-200009290-00006. [DOI] [PubMed] [Google Scholar]
- 9.Casper C, Nichols WG, Huang ML, Corey L, Wald A. 2004. Remission of HHV-8 and HIV-associated multicentric Castleman disease with ganciclovir treatment. Blood 103:1632–1634. doi: 10.1182/blood-2003-05-1721. [DOI] [PubMed] [Google Scholar]
- 10.Chen J, Ueda K, Sakakibara S, Okuno T, Parravicini C, Corbellino M, Yamanishi K. 2001. Activation of latent Kaposi's sarcoma-associated herpesvirus by demethylation of the promoter of the lytic transactivator. Proc Natl Acad Sci U S A 98:4119–4124. doi: 10.1073/pnas.051004198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fardet L, Blum L, Kerob D, Agbalika F, Galicier L, Dupuy A, Lafaurie M, Meignin V, Morel P, Lebbe C. 2003. Human herpesvirus 8-associated hemophagocytic lymphohistiocytosis in human immunodeficiency virus-infected patients. Clin Infect Dis 37:285–291. doi: 10.1086/375224. [DOI] [PubMed] [Google Scholar]
- 12.Grandadam M, Dupin N, Calvez V, Gorin I, Blum L, Kernbaum S, Sicard D, Buisson Y, Agut H, Escande JP, Huraux JM. 1997. Exacerbations of clinical symptoms in human immunodeficiency virus type 1-infected patients with multicentric Castleman's disease are associated with a high increase in Kaposi's sarcoma herpesvirus DNA load in peripheral blood mononuclear cells. J Infect Dis 175:1198–1201. doi: 10.1086/593567. [DOI] [PubMed] [Google Scholar]
- 13.Oksenhendler E, Carcelain G, Aoki Y, Boulanger E, Maillard A, Clauvel JP, Agbalika F. 2000. High levels of human herpesvirus 8 viral load, human interleukin-6, interleukin-10, and C reactive protein correlate with exacerbation of multicentric Castleman disease in HIV-infected patients. Blood 96:2069–2073. [PubMed] [Google Scholar]
- 14.Jones JL, Hanson DL, Chu SY, Ward JW, Jaffe HW. 1995. AIDS-associated Kaposi's sarcoma. Science 267:1078–1079. doi: 10.1126/science.7855583. [DOI] [PubMed] [Google Scholar]
- 15.Martin DF, Kuppermann BD, Wolitz RA, Palestine AG, Li H, Robinson CA. 1999. Oral ganciclovir for patients with cytomegalovirus retinitis treated with a ganciclovir implant. Roche Ganciclovir Study Group. N Engl J Med 340:1063–1070. [DOI] [PubMed] [Google Scholar]
- 16.Robles R, Lugo D, Gee L, Jacobson MA. 1999. Effect of antiviral drugs used to treat cytomegalovirus end-organ disease on subsequent course of previously diagnosed Kaposi's sarcoma in patients with AIDS. J Acquir Immune Defic Syndr Hum Retrovirol 20:34–38. doi: 10.1097/00042560-199901010-00005. [DOI] [PubMed] [Google Scholar]
- 17.Halfdanarson TR, Markovic SN, Kalokhe U, Luppi M. 2006. A non-chemotherapy treatment of a primary effusion lymphoma: durable remission after intracavitary cidofovir in HIV negative PEL refractory to chemotherapy. Ann Oncol 17:1849–1850. doi: 10.1093/annonc/mdl139. [DOI] [PubMed] [Google Scholar]
- 18.Hocqueloux L, Agbalika F, Oksenhendler E, Molina JM. 2001. Long-term remission of an AIDS-related primary effusion lymphoma with antiviral therapy. AIDS 15:280–282. doi: 10.1097/00002030-200101260-00023. [DOI] [PubMed] [Google Scholar]
- 19.Luppi M, Trovato R, Barozzi P, Vallisa D, Rossi G, Re A, Ravazzini L, Potenza L, Riva G, Morselli M, Longo G, Cavanna L, Roncaglia R, Torelli G. 2005. Treatment of herpesvirus associated primary effusion lymphoma with intracavity cidofovir. Leukemia 19:473–476. doi: 10.1038/sj.leu.2403646. [DOI] [PubMed] [Google Scholar]
- 20.Li Q, He M, Zhou F, Ye F, Gao SJ. 2014. Activation of Kaposi's sarcoma-associated herpesvirus (KSHV) by inhibitors of class III histone deacetylases: identification of sirtuin 1 as a regulator of the KSHV life cycle. J Virol 88:6355–6367. doi: 10.1128/JVI.00219-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Daigle D, Megyola C, El-Guindy A, Gradoville L, Tuck D, Miller G, Bhaduri-McIntosh S. 2010. Upregulation of STAT3 marks Burkitt lymphoma cells refractory to Epstein-Barr virus lytic cycle induction by HDAC inhibitors. J Virol 84:993–1004. doi: 10.1128/JVI.01745-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hill ER, Koganti S, Zhi J, Megyola C, Freeman AF, Palendira U, Tangye SG, Farrell PJ, Bhaduri-McIntosh S. 2013. Signal transducer and activator of transcription 3 limits Epstein-Barr virus lytic activation in B lymphocytes. J Virol 87:11438–11446. doi: 10.1128/JVI.01762-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aoki Y, Feldman GM, Tosato G. 2003. Inhibition of STAT3 signaling induces apoptosis and decreases survivin expression in primary effusion lymphoma. Blood 101:1535–1542. doi: 10.1182/blood-2002-07-2130. [DOI] [PubMed] [Google Scholar]
- 24.Cimica V, Chen HC, Iyer JK, Reich NC. 2011. Dynamics of the STAT3 transcription factor: nuclear import dependent on Ran and importin-beta1. PLoS One 6:e20188. doi: 10.1371/journal.pone.0020188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu S, Bekker-Jensen S, Mailand N, Lukas C, Bartek J, Lukas J. 2006. Claspin operates downstream of TopBP1 to direct ATR signaling towards Chk1 activation. Mol Cell Biol 26:6056–6064. doi: 10.1128/MCB.00492-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu L, Puri V, Chandran B. 1999. Characterization of human herpesvirus-8 K8.1A/B glycoproteins by monoclonal antibodies. Virology 262:237–249. doi: 10.1006/viro.1999.9900. [DOI] [PubMed] [Google Scholar]
- 27.Yu H, Kortylewski M, Pardoll D. 2007. Crosstalk between cancer and immune cells: role of STAT3 in the tumour microenvironment. Nat Rev Immunol 7:41–51. doi: 10.1038/nri1995. [DOI] [PubMed] [Google Scholar]
- 28.Yang J, Chatterjee-Kishore M, Staugaitis SM, Nguyen H, Schlessinger K, Levy DE, Stark GR. 2005. Novel roles of unphosphorylated STAT3 in oncogenesis and transcriptional regulation. Cancer Res 65:939–947. [PubMed] [Google Scholar]
- 29.Iwamaru A, Szymanski S, Iwado E, Aoki H, Yokoyama T, Fokt I, Hess K, Conrad C, Madden T, Sawaya R, Kondo S, Priebe W, Kondo Y. 2007. A novel inhibitor of the STAT3 pathway induces apoptosis in malignant glioma cells both in vitro and in vivo. Oncogene 26:2435–2444. doi: 10.1038/sj.onc.1210031. [DOI] [PubMed] [Google Scholar]
- 30.Koganti S, Hui-Yuen J, McAllister S, Gardner B, Grasser F, Palendira U, Tangye SG, Freeman AF, Bhaduri-McIntosh S. 2014. STAT3 interrupts ATR-Chk1 signaling to allow oncovirus-mediated cell proliferation. Proc Natl Acad Sci U S A 111:4946–4951. doi: 10.1073/pnas.1400683111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.ENCODE Project Consortium. 2011. A user's guide to the encyclopedia of DNA elements (ENCODE). PLoS Biol 9:e1001046. doi: 10.1371/journal.pbio.1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chang PC, Fitzgerald LD, Van Geelen A, Izumiya Y, Ellison TJ, Wang DH, Ann DK, Luciw PA, Kung HJ. 2009. Kruppel-associated box domain-associated protein-1 as a latency regulator for Kaposi's sarcoma-associated herpesvirus and its modulation by the viral protein kinase. Cancer Res 69:5681–5689. doi: 10.1158/0008-5472.CAN-08-4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gjyshi O, Roy A, Dutta S, Veettil MV, Dutta D, Chandran B. 2015. Activated Nrf2 Interacts with Kaposi's sarcoma-associated herpesvirus latency protein LANA-1 and host protein KAP1 to mediate global lytic gene repression. J Virol 89:7874–7892. doi: 10.1128/JVI.00895-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rauwel B, Jang SM, Cassano M, Kapopoulou A, Barde I, Trono D. 7 April 2015. Release of human cytomegalovirus from latency by a KAP1/TRIM28 phosphorylation switch. eLife. doi: 10.7554/eLife.06068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun R, Liang D, Gao Y, Lan K. 2014. Kaposi's sarcoma-associated herpesvirus-encoded LANA interacts with host KAP1 to facilitate establishment of viral latency. J Virol 88:7331–7344. doi: 10.1128/JVI.00596-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nador RG, Cesarman E, Chadburn A, Dawson DB, Ansari MQ, Sald J, Knowles DM. 1996. Primary effusion lymphoma: a distinct clinicopathologic entity associated with the Kaposi's sarcoma-associated herpes virus. Blood 88:645–656. [PubMed] [Google Scholar]
- 37.King CA. 2013. Kaposi's sarcoma-associated herpesvirus kaposin B induces unique monophosphorylation of STAT3 at serine 727 and MK2-mediated inactivation of the STAT3 transcriptional repressor TRIM28. J Virol 87:8779–8791. doi: 10.1128/JVI.02976-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koganti S, Clark C, Zhi J, Li X, Chen EI, Chakrabortty S, Hill ER, Bhaduri-McIntosh S. 2015. Cellular STAT3 functions via PCBP2 to restrain Epstein-Barr virus lytic activation in B lymphocytes. J Virol 89:5002–5011. doi: 10.1128/JVI.00121-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koganti S, de la Paz A, Freeman AF, Bhaduri-McIntosh S. 2014. B lymphocytes from patients with hypomorphic mutation in STAT3 resist Epstein-Barr virus-driven cell proliferation. J Virol 88:516–524. doi: 10.1128/JVI.02601-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang H, Hu H, Greeley N, Jin J, Matthews AJ, Ohashi E, Caetano MS, Li HS, Wu X, Mandal PK, McMurray JS, Moghaddam SJ, Sun SC, Watowich SS. 2014. STAT3 restrains RANK- and TLR4-mediated signalling by suppressing expression of the E2 ubiquitin-conjugating enzyme Ubc13. Nat Commun 5:5798. doi: 10.1038/ncomms6798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Darnell JE., Jr 2002. Transcription factors as targets for cancer therapy. Nat Rev Cancer 2:740–749. doi: 10.1038/nrc906. [DOI] [PubMed] [Google Scholar]
- 42.Yu H, Jove R. 2004. The STATs of cancer—new molecular targets come of age. Nat Rev Cancer 4:97–105. doi: 10.1038/nrc1275. [DOI] [PubMed] [Google Scholar]
- 43.Molden J, Chang Y, You Y, Moore PS, Goldsmith MA. 1997. A Kaposi's sarcoma-associated herpesvirus-encoded cytokine homolog (vIL-6) activates signaling through the shared gp130 receptor subunit. J Biol Chem 272:19625–19631. doi: 10.1074/jbc.272.31.19625. [DOI] [PubMed] [Google Scholar]
- 44.Punjabi AS, Carroll PA, Chen L, Lagunoff M. 2007. Persistent activation of STAT3 by latent Kaposi's sarcoma-associated herpesvirus infection of endothelial cells. J Virol 81:2449–2458. doi: 10.1128/JVI.01769-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Du T, Zhou G, Roizman B. 2013. Modulation of reactivation of latent herpes simplex virus 1 in ganglionic organ cultures by p300/CBP and STAT3. Proc Natl Acad Sci U S A 110:E2621–E2628. doi: 10.1073/pnas.1309906110. [DOI] [PMC free article] [PubMed] [Google Scholar]