ABSTRACT
Type I interferons (IFNs) are induced upon viral infection and important mediators of innate immunity. While there is 1 beta interferon (IFN-β) protein, there are 12 different IFN-α subtypes. It has been reported extensively that different viruses induce distinct patterns of IFN subtypes, but it has not been previously shown how the viral multiplicity of infection (MOI) can affect IFN induction. In this study, we discovered the novel finding that human U937 cells infected with 2 different concentrations of Sendai virus (SeV) induce 2 distinct type I IFN subtype profiles. Cells infected at the lower MOI induced more subtypes than cells infected at the higher MOI. We found that this was due to the extent of signaling through the IFN receptor (IFNAR). The cells infected at the lower viral MOI induced the IFNAR2-dependent IFN-α subtypes 4, 6, 7, 10, and 17, which were not induced in cells infected at higher virus concentrations. IFN-β and IFN-α1, -2, and -8 were induced in an IFNAR-independent manner in cells infected at both virus concentrations. IFN-α5, -14, -16, and -21 were induced in an IFNAR-dependent manner in cells infected at lower virus concentrations and in an IFNAR-independent manner in cells infected at higher virus concentrations. These differences in IFN subtype profiles in the 2 virus concentrations also resulted in distinct interferon-stimulated gene induction. These results present the novel finding that different viral MOIs differentially activate JAK/STAT signaling through the IFNAR, which greatly affects the profile of IFN subtypes that are induced.
IMPORTANCE Type I IFNs are pleiotropic cytokines that are instrumental in combating viral diseases. Understanding how the individual subtypes are induced is important in developing strategies to block viral replication. Many studies have reported that different viruses induce distinct type I IFN subtype profiles due to differences in the way viruses are sensed in different cell types. However, we report in our study the novel finding that the amount of virus used to infect a system can also affect which type I IFN subtypes are induced due to the extent of activation of certain signaling pathways. These distinct IFN subtype profiles in cells infected at different MOIs are correlated with differences in interferon-stimulated gene induction, indicating that the same virus can induce distinct antiviral responses depending on the MOI. Because type I IFNs are used as therapeutic agents to treat viral diseases, understanding their antiviral mechanisms can enhance clinical treatments.
INTRODUCTION
Type I interferons (IFNs) are the first line of defense against viral infections. While there is only 1 beta interferon (IFN-β) gene and 1 protein, there are 13 different IFN-α genes and 12 different proteins in humans. IFN-α subtypes 1 and 13 have the same mature protein-coding sequence but have different promoter sequences. All the subtypes have distinct genes controlled by their own promoter regions (1), enabling them to be differentially regulated.
Type I IFNs are induced in response to viral infection in two phases of innate immune signaling. The first occurs when pathogen-associated molecular patterns (PAMPs) activate either cytosolic or membrane-bound pattern recognition receptors (PRRs). These interactions stimulate signaling pathways that eventually converge on the activation of the transcription factors IRF3, IRF7, and/or NF-κB, which lead to the transcription of early type I IFN subtypes. In mice, these early subtypes consist of IFN-β and IFN-α4 (2). However, in humans, it is not known exactly which type I IFN subtypes are induced early. These early subtypes are secreted from the cells and bind to the IFN receptor (IFNAR), which stimulates the phosphorylation of the receptor-associated kinases JAK1 and Tyk2 and subsequently the transcription factors STAT1 and STAT2. Serine phosphorylation of STAT proteins also occurs and is required for optimal transcriptional activity (3). These phosphorylated STATs complex with IRF9 to form the interferon-stimulated gene factor 3 (ISGF3) complex, which translocates into the nucleus to activate the transcription of hundreds of ISGs, IRF7, and additional type I IFNs to amplify the response (4–6). This positive-feedback amplification loop continues until negative regulators of IFN signaling, such as SOCS proteins and IRF2, become activated.
Sendai virus (SeV) has long been used to study type I IFN regulation due to its robust ability to induce large quantities of the type I IFN subtypes (7–9). Reports studying the transcriptional regulation of the IFN-α subtypes in response to SeV infection have indicated that IRF3 and IRF7 play central roles (1). In these studies, it was found that the promoter regions of the subtypes have domains with different binding capabilities for IRF3 and IRF7, and the exact subtypes produced upon infection are dependent on the levels of these transcription factors. However, it is not known how proteins involved in the amplification phase of the IFN pathway, such as STAT1 and -2 and IRF9, contribute to this mechanism. It has been reported that treating human plasmacytoid dendritic cells (pDCs) with Toll-like receptor 9 (TLR9) agonists in concert with IFN-α results in a synergistic induction of IFN-α subtypes that is not seen with either of the two treatments individually (10). This study indicates that signaling downstream of the IFNAR is an important factor to consider when distinguishing the signaling pathways leading to type I IFN subtype induction. However, it is not yet known exactly how the IFNAR-dependent pathways contribute to the profile of IFNs induced during virus infection.
It has been reported extensively that different viruses induce distinct patterns of type I IFN subtype profiles and that this is due mainly to the different combinations of PRRs that are activated when different classes of viruses are present (11, 12). In this study, however, we made the novel observation that the same virus could induce distinct type I IFN subtype profiles in the same cell type when different multiplicities of infection (MOIs) were used. While many reports study type I IFN regulation in plasmacytoid dendritic cells, which are the main IFN-producing cells in circulation, we chose to study the IFN response in the monocytoid cell line U937. Circulating monocytes not only induce type I IFNs upon infection (13, 14), but also home to tissues upon infection, including the brain, so their innate immune response to viruses affects the cytokine milieu and pathology in affected tissues (15, 16). We found that infecting U937 cell cultures at 2 different concentrations of SeV resulted in distinct profiles of type I IFN subtypes, with more subtypes being induced in cultures infected at the lower MOI. This was due to the fact that in cell cultures infected at the lower MOI, there is increased signaling through the IFNAR, which is the second phase of IFN induction. As a result, we found that the subtypes that were uniquely expressed in cultures infected at the lower MOI, IFN-α4, -6, -7, -10, and -17, were induced in an IFNAR-dependent manner. IFN-β and IFN-α1, -2, and -8, on the other hand, were induced early in cultures infected at both virus concentrations in an IFNAR-independent manner. The induction of these subtypes was mostly dependent on NF-κB and PRR signaling. The requirement for IFNAR signaling for the induction of IFN-α5, -14, -16, and -21 was dependent on the virus concentration; in cultures infected at the lower MOI, these subtypes were induced in an IFNAR-dependent manner, while in cultures infected at the higher MOI, the subtypes were induced independently of IFNAR signaling. Importantly, these distinct subtype profiles in cultures infected at 2 different concentrations of SeV resulted in significantly different signatures of ISGs. These results indicate how the MOI used to infect cells greatly affects the extent to which signaling through the IFNAR occurs, and this in turn affects the profile of type I IFN subtypes and ISGs that are induced.
MATERIALS AND METHODS
Cells, viruses, and reagents.
U937 cells (ATCC) were maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS). The cells were passaged every 3 or 4 days when the cell concentration reached about 1 × 106 cells/ml. Only cells between passages 2 and 6 were used for experiments. SeV in allantoic fluid was obtained from Charles River Laboratories at a stock concentration of 4,000 hemagglutination units (HA U)/ml. Encephalomyocarditis virus (EMCV) was grown in Vero cells, and the titer was determined via 50% tissue culture infectious dose (TCID50) assay. The IFNAR2-neutralizing antibody was produced by Precision Antibody (Columbia, MD). The IFN-α-neutralizing antibody was obtained from BEI Resources (Manassas, VA). The IFN-β-neutralizing antibody was obtained from Millipore (Temecula, CA). The inhibitors BX795, chloroquine, and BAY11-7802 were obtained from Invivogen (San Diego, CA). Elutriated primary human monocytes from anonymous healthy donors were obtained from the NIH Clinical Center Department of Transfusion Medicine, with a purity of >90%. The antibodies and dilutions used for the Western blots were as follows: polyclonal rabbit anti-IRF7 (Santa Cruz Biotechnology, Dallas, TX), 1:1,000; monoclonal rabbit anti-pIRF3(S386) (Abcam, Cambridge, MA), 1:2,000; polyclonal rabbit anti-STAT1 (Santa Cruz Biotechnology, Dallas, TX), 1:1,000; monoclonal mouse anti-p(Y701)STAT1 (phosphorylated STAT1 on tyrosine 701; BD Transduction Laboratories, San Jose, CA), 1:5,000; monoclonal rabbit anti-p(S727)STAT1 (Abcam, Cambridge, MA), 1:2,000; monoclonal mouse anti-IRF9(ISGF3γ) (BD Transduction Laboratories, San Jose, CA), 1:1,000; monoclonal mouse anti-actin (Cell Signaling Technology, Danvers, MA), 1:5,000; polyclonal rabbit anti-β-tubulin (Cell Signaling Technology, Danvers, MA), 1:2,000; monoclonal mouse RNA polymerase II (Santa Cruz Biotechnology, Dallas, TX), 1:1,000; monoclonal rabbit p(S172)TBK1 (Cell Signaling Technology, Danvers, MA), 1:1,000; monoclonal mouse anti-p(S32/36)IκBα (Cell Signaling Technology, Danvers, MA), 1:1,000; polyclonal rabbit anti-IκBα (Cell Signaling Technology, Danvers MA), 1:1,000. The antibodies used for immunofluorescence staining were as follows: mouse-anti STAT1-Texas Red (TR) (Santa Cruz Biotechnology, Dallas, TX), 1:50; polyclonal rabbit anti-SeV-fluorescein isothiocyanate (FITC) (Abcam, Cambridge, MA), 1:500.
SeV infection of U937 cells and primary monocytes.
U937 cell and primary monocyte cultures at a concentration of 5 × 105 cells/ml were infected with SeV for the indicated times at the indicated concentrations. A TCID50 assay of U937 cells revealed that the virus concentrations of 1.5 HA U/ml and 150 HA U/ml corresponded to MOIs of 0.05 and 5, respectively. At the indicated time points, the cells were centrifuged at 1,000 rpm for 5 min at 4°C. The supernatants were saved, and the pellets were washed once with phosphate-buffered saline (PBS) (pH 7.4). The pellets were then resuspended in the appropriate buffer for RNA or protein extraction and stored at −80°C until further use. The supernatants were also stored at −80°C.
IFN-α and IFN-β ELISAs.
Supernatants from SeV-infected U937 cells and primary monocytes were collected at the indicated time points. Total IFN-α protein was measured using the VeriKine human IFN-α enzyme-linked immunosorbent assay (ELISA) kit (PBL Assay Science, Piscataway, NJ) following the manufacturer's instructions. Human IFN-β was measured using the Verikine Human IFN-β ELISA kit (PBL Assay Science, Piscataway, NJ) following the manufacturer's instructions.
Virus inactivation and AV assays. (i) Virus inactivation.
Supernatants from SeV-infected U937 cells that were being assayed for IFN activity were thawed at 37°C, and then β-propiolactone was added at a final concentration of 0.05% (17) to inactivate the virus that remained in the supernatant. The supernatants were then incubated at 4°C overnight and then at 37°C for 2 h prior to pretreatment in the antiviral (AV) assays. We verified that the inactivated virus did not induce type I IFNs in the cells used for the AV assays (data not shown).
(ii) AV assays.
U937 cells were plated in 96-well plates at a concentration of 3 × 104 cells/well (100 μl). The cells were pretreated with 10 μl of virus-inactivated supernatants from infected cells. The cells, other than the controls, were then infected with EMCV for 72 h, and the cytopathic effects (CPE) and cell viability were measured via an MTT [3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide] assay. Briefly, 10 μl of 5-mg/ml MTT in PBS was added to each well and incubated for 4 h at 37°C. The precipitate was then dissolved in 150 μl of acidified isopropanol (100 ml isopropanol plus 660 μl concentrated HCl), and the absorbance was measured at 570 nm using a Hidex Sense plate reader.
Immunofluorescence staining and confocal microscopy.
After 24 h of SeV infection, 1 × 106 cells were removed from suspension and pelleted. The pellets were washed once and then resuspended in 1 ml of serum-free RPMI 1640 medium. The cell suspension was added to a 35-mm glass bottom dish (MatTek Corporation, Ashland, MA) and incubated at room temperature (RT) for 10 min to let the cells adhere. After 10 min, the medium was removed and the plates were washed once with PBS. The cells were fixed in ice-cold methanol for 15 min at −20°C, washed twice with PBS, and then permeabilized in 0.1% Triton X-100 in PBS for 15 min at RT. The plates were then washed twice with PBS and blocked in 5% bovine serum albumin (BSA) in PBS for 30 min at RT. The cells were incubated with primary antibody diluted in 5% BSA (SeV-FITC, 1:500; STAT1-TR, 1:50) for 1 h at RT. After washing 3 times in PBS, secondary-antibody solution diluted in 5% BSA (1:200) was added to the plates and incubated for 30 min at RT. The plates were washed twice with PBS, counterstained with DAPI (4′,6-diamidino-2-phenylindole) at a 1:10,000 dilution for 1 min, and then washed 3 times with PBS. The plates were kept in the dark at 4°C until imaging. The plates were imaged using a Leica SP5 confocal microscope (Leica, Mannheim, Germany).
Treatment with neutralizing antibodies and inhibitors. (i) IFNAR2-neutralizing antibody.
The IFNAR2-neutralizing antibody (clone mA10; Precision Antibody, Columbia, MD) was used at a concentration of 1:100 (antibody/total cell culture volume) for all experiments, as this concentration completely neutralized the biological activities of 1,000 U/ml of IFN-β, all IFN-α subtypes (PBL Assay Science, Piscataway, NJ), and IFN-ω, as well as inhibiting the phosphorylation of STAT1 (Y701) 15 min after treatment with 1,000 U/ml of IFN-α2 (data not shown). For all experiments, cells were incubated with the IFNAR2-neutralizing antibody for 1 h at 37°C prior to infection. If the supernatants were going to be used for subsequent antiviral assays, the antibody was washed off after 1 h. Otherwise, the antibody was left on for the duration of the infection.
(ii) IFN-α-neutralizing antibody.
Polyclonal anti-human leukocyte IFN-α antibody from calf antiserum (BEI Resources, VA) was used at a concentration of 1:100 (antibody/total cell culture volume), as this concentration was 10 times higher than the concentration needed to completely neutralize the biological activities of 1,000 U/ml of all purified IFN-α subtypes (PBL Assay Science, Piscataway, NJ) to ensure an appropriate molar excess of antibody. The antibody was added concomitantly with SeV.
(iii) IFN-β-neutralizing antibody.
Rabbit anti-human IFN-β polyclonal antibody was used at a concentration of 1:10, as this concentration was 10 times higher than the concentration needed to completely neutralize the biological activity of 1,000 U/ml of purified IFN-β (R&D Biosciences, Minneapolis, MN) to ensure an appropriate molar excess of antibody. The antibody was added concomitantly with SeV.
(iv) BX795 treatment.
U937 cells were treated with 1 μM of BX795 (Invivogen, San Diego, CA) for 1 h at 37°C prior to infection. This concentration was chosen based on both the ability to inhibit SeV-induced TBK1 phosphorylation and the limited effects on cell viability.
(v) BAY11-7802.
U937 cells were treated with 7 μM of BAY11-7802 (Invivogen, San Diego, CA) at 37°C for 1 h prior to infection. This concentration was chosen based on both the ability to inhibit tumor necrosis factor alpha (TNF-α)-induced IκB phosphorylation and the limited effects on cell viability.
RNA isolation.
Cells were harvested at the indicated time points and pelleted by spinning at 1,000 rpm at 4°C using a Beckman Coulter Allegra X-15R centrifuge. The supernatants were removed, and the pellets were washed in 1 ml of PBS (pH 7.4). The pellet was then resuspended in 350 μl of RLT buffer plus from the RNeasy plus kit (Qiagen, Germantown, MD). The RNA mixture was then stored at −80°C until RNA isolation. RNA isolation was performed using the RNeasy plus kit according to the manufacturer's instructions. To remove contaminating DNA, two DNase steps were performed. DNA was first removed using the genomic DNA (gDNA) eliminator column (Qiagen, Germantown, MD) prior to RNA binding to the RNeasy column. A second, on-column DNase step was performed using 10 μl of DNase 1 with 70 μl of buffer RDD (Qiagen, Germantown, MD) and 2 μl of Turbo DNase (Thermo Fisher, Waltham, MA) per column. The columns were incubated at room temperature for 30 min. The RNA samples were quantitated using the Nanodrop 1000 (Thermo Scientific, Wilmington, DE).
Reverse transcription.
For each sample, a stock of cDNA was made using 1.5 μg of RNA. The RNA was reverse transcribed using the Superscript III reverse transcriptase kit (Invitrogen, San Diego, CA) according to the manufacturer's protocol. Briefly, the RNA (1.5 μg) was incubated with 1 μl of 10 mM deoxynucleoside triphosphate (dNTP), 50 ng of random primers, and RNase-free water in a total volume of 13.5 μl for 5 min at 65°C; cooled at 4°C for 2 min; and then incubated with 0.5 μl Superscript III reverse transcriptase, 1 μl of 0.1 M dithiothreitol (DTT), 1 μl RNaseOut (Invitrogen, San Diego, CA), and 4 μl of 5× Superscript III buffer according to the following protocol: 50°C for 60 min and 70°C for 15 min for enzyme inactivation. The cDNA stock was then diluted to a concentration of 50 ng of starting RNA/10 μl in water and stored at −20°C.
qRT-PCR analysis (SYBR and TaqMan).
Quantitative reverse transcription (qRT)-PCRs were carried out using the SensiFast SYBR no-Rox kit (Bioline, Taunton, MA) to measure pan-IFN-α in a SYBR green assay and the SensiFast probe No-Rox mastermix (Bioline, Taunton, MA) to measure all other genes in a TaqMan assay. The reactions were run in 96-well plates on a CFX9695 thermocycler (Bio-Rad, Hercules, CA). For the SYBR reactions measuring pan-IFN-α, 10 μl of diluted cDNA was incubated with 10 μl of master mix and 0.075 μl of 100 μM forward and reverse primers according to the following protocol: 95°C for 2 min for polymerase activation, followed by 40 cycles of denaturation at 95°C for 5 s and annealing/extension at 60°C for 15 s. For the TaqMan reactions, 10 μl of diluted cDNA was incubated with 10 μl of master mix, 0.075 μl of 100 μM forward and reverse primers, and 0.05 μl of 100 μM TaqMan probe according to the following protocol: 95°C for 5 min for polymerase activation and then 40 cycles of denaturation at 95°C for 10 s, followed by annealing/extension at 60°C for 50 s. For each reaction, the hypoxanthine-guanine phosphoribosyltransferase (HPRT) housekeeping gene was multiplexed with the genes of interest as the normalizer. Up to 4 genes were multiplexed in a single reaction using the dyes cyanine 5 (CY5), hexachlorofluorescein (HEX), fluorescein amidite (Fam), and Texas Red (TR). The data were analyzed using CFX Manager software, version 2.1 (Bio-Rad). The data are expressed as the percent relative expression of HPRT.
Primers for SYBR green reactions.
The primers chosen to amplify pan-IFN-α were designed based on the most conserved sequences among all IFN-α subtypes. The forward primer is a combination of 6 sequences due to the slight sequence differences in some of the subtypes in the conserved region. The reverse primer is one sequence that is found in every subtype in the conserved region. The sequences are as follows: forward primer mixture, 5′-CCTTCCATGAGATGATCCAGCAGACCT-3′ (IFN-α16), 5′-TCCTCCATGAGATGATGCAGCAGACCT-3′ (IFN-α14), 5′-TCCTCCATGAGGTGATTCAGCAGACCT-3′ (IFN-α6), 5′-TCCTCCATGAGATGATCCAGCAGATCT-3′ (IFN-α2), 5′-TCCTCCATGAGCTGATCCAGCAGATCT-3′ (IFN-α1/13), and 5′-TCCTCCATGAGATGATCCAGCAGACCT-3′ (all other IFN-αs); reverse primer, 5′-GATCTCATGATTTCTGCTCTGACAACC-3′ (boldface and underlining indicate nucleotides that differ among the sequences).
Primers and probes for TaqMan analysis.
For each of the IFN-α subtypes, with the exception of subtypes 1 and 13, a specific primer and TaqMan probe set was designed as previously described (18, 19). One single primer-probe set was designed to amplify subtypes 1 and 13. Specificity was maintained by utilizing a method called the amplification refractory mutation system (ARMS), which was originally used to identify single nucleotide polymorphisms, when designing the primers (19). Briefly, in at least one of the primers, the 3′-most nucleotide was chosen so that a mismatch was formed with all of the subtypes except the subtype of interest. To maintain specificity, the third to last nucleotide in the primer was deliberately mutated to form a mismatch with all of the nontarget subtype sequences. This resulted in a primer sequence that had one mismatch at the third to last nucleotide with the target sequence, which did not significantly alter annealing and extension, and 2 mismatches at the third to last nucleotide and 3′-most nucleotide with the nontarget sequences, which ablated extension. The specificity of each primer-probe set was tested in reactions with all of the IFN-α subtype standards (data not shown). The primer-probe set was deemed specific when the cycle threshold (Ct) for the target gene came up about 15 cycles prior to the nontarget genes.
The primer and probe sequences for the IFN-α subtypes are outlined in Table S2 in the supplemental material. The primer and probe sequences for the remaining genes were as follows: IFN-β, F, 5′-CGAAACTGAAGATCTCCTAGCC-3′, R, 5′-TGCAGTACATTAGCCATCAGTC-3′, and probe, 5′-AATTGTCCAGTCCCAGAGGCACAG-3′; IRF7, F, 5′-TCCCCACGCTATACCATCTAC-3′, R, 5′-GAAGACACACCCTCACGC-3′, and probe, 5′-TTCCAGCTTCACCAGGACCAGG-3′; PKR, F, 5′-TGGAAAGCGAACAAGGAGTAAG-3′, R, 5′-CAGCAAGAATTAGCCCCAAAG-3′, and probe, 5′-TGCGATACATGAGCCCAGAACAGATTT-3′; HPRT, F, 5′-GTATTCATTATAGTCAAGGGCATATCC-3′, R, 5′-AGATGGTCAAGGTCGCAAG-3′, and probe, 5′-TGGTGAAAAGGACCCCACGAAG-3′.
Whole-cell protein lysate preparation.
Cells were harvested at the indicated time points and pelleted by spinning at 1,000 rpm using an Allegra X-15R centrifuge (Beckman Coulter, Pasadena, CA) at 4°C. The supernatants were removed, and the pellets were washed with 1 ml of cold PBS, pH 7.4. The pellets were resuspended in 200 μl of radioimmunoprecipitation assay (RIPA) buffer (0.25% sodium deoxycholate, 0.1% sodium dodecyl chloride, 25 mM Tris [pH 7.4], 150 mM sodium chloride, 1 mM EDTA, and 1% NP-40) with protease and phosphatase inhibitors (Sigma, St. Louis, MO; 1:100 dilution for each) and sonicated for 5 s at an amplitude of 21% using a Vibra cell sonicator (Sonics, Newtown, CT). The samples were then incubated on ice for 30 min and spun at 13,000 rpm using a microcentrifuge (Beckman Coulter, Pasadena, CA) at 4°C for 20 min. The supernatants were collected, and a bicinchoninic acid (BCA) protein assay (Pierce, Waltham, MA) was performed using 10 μl of the protein lysate according to the manufacturer's protocol. Ten-microgram aliquots in RIPA buffer were stored at −80°C and used for Western blot analysis.
Western blot analysis.
For Western blot analysis, 10 μg of protein was mixed with 1 μl of reducing agent (Invitrogen, San Diego, CA) and 4× NuPAGE lithium dodecyl sulfate (LDS) sample loading dye (Invitrogen, San Diego, CA). The samples were heated at 95°C for 5 min and then run on 10% Bis-Tris minigels (Invitrogen, San Diego, CA) at 200 V for 1 h. The protein on the gel was then transferred onto nitrocellulose membranes using the iBlot apparatus (Invitrogen, San Diego, CA). The membranes were then blocked in 5% milk in Tris-buffered saline with 0.1% Tween 20 (TBST) for 1 h at RT. The membranes were then incubated overnight at 4°C with the primary antibody diluted in 5% milk. The following day, the membranes were washed 3 times for 10 min each time with TBST at RT and then incubated with horseradish peroxidase (HRP)-conjugated secondary antibodies (either anti-rabbit or anti-mouse, depending on the species of the primary antibody) at a 1:5,000 dilution in 5% milk for 1 h at RT. The membranes were washed 3 times with TBST for 10 min at RT, and the SuperSignal West Femto Maximum Sensitivity Substrate (ThermoFisher, Waltham, MA) was used to develop the membranes. Chemiluminescence was visualized using a Fujifilm LAS-3000 camera.
Microarray hybridization.
High-quality RNA samples (RNA integrity number score beween 9 and 10) were amplified and labeled using the Illumina TotalPrep RNA amplification kit (Applied Biosystems) with an input of 500 ng of total RNA per sample. Biotinylated antisense RNA (aRNA) was hybridized to an Illumina Human HT-12 Expression BeadChip (NCBI Gene Expression Omnibus [GEO] database accession no. GSE67198) having 47,323 unique probes, using the reagents provided, and imaged using Illumina HiScan.
Microarray expression analysis.
Data extraction was performed using Illumina Genome Studio software without background subtraction or normalization. Signal levels for each array were normalized using quantile normalization after transforming to log base 2. Probes that did not show detectable signal (P < 0.1) on at least two arrays were removed from the data set before normalization. Analysis of variance (ANOVA) was calculated using JMP/Genomics software (SAS Institute, Cary, NC) to report the average fold change and false-discovery rate (Benjamini-Hochberg) adjusted P values. The microarray results were analyzed using Ingenuity Pathway Analysis.
Statistical analysis.
All statistical analyses were conducted using a one-way or two-way ANOVA test with Bonferroni posttest analysis using PRISM6 software (JMP/Genomics software was used for the microarray statistical analyses). The figure legends specify which ANOVA test was used. Statistics were performed for all treatment groups, but only the comparisons relevant to the conclusion for each figure are shown.
Microarray data expression number.
The microarray data were uploaded to the GEO database (accession number GSE67198).
RESULTS
Establishment of SeV-infected U937 cells as a model to study type I IFN subtype regulation.
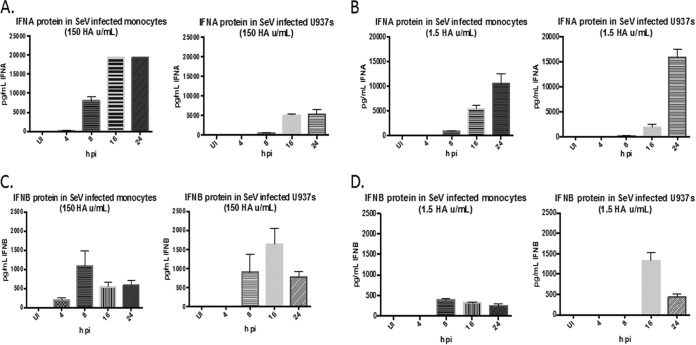
The IFN response to SeV infection was examined in U937 cells and primary monocytes. Both cell types were infected with SeV (150 or 1.5 HA U/ml), and the supernatants were harvested at 4, 8, 16, and 24 h postinfection (p.i.) to assess type I IFN production. Both cell types produced IFN-α and IFN-β proteins at both virus concentrations as measured by ELISA (Fig. 1A to D). The kinetics of IFN-α protein expression were almost identical between the monocytes and U937 cells, although the magnitudes of expression were different (Fig. 1A and B). IFN-β protein was induced at earlier time points in primary monocytes than in U937 cells, but the kinetics of expression were similar between the 2 cell types (Fig. 1C and D). It also should be noted that for both cell types, almost 10-fold more IFN-α than IFN-β was produced. We next measured the production of 34 cytokines by Luminex assay in SeV-infected U937 cells and primary monocytes to compare the cytokine responses to SeV infection in each cell type. The U937 cells and primary monocytes exhibited similar patterns of cytokine production in terms of which cytokines were and were not expressed, with the exception of 2 cytokines (TNF-α and interleukin 6 [IL-6]), which were expressed in monocytes but not U937 cells (see Table S1 in the supplemental material). Because of the immunological similarity between SeV-infected U937 cells and primary monocytes, SeV-infected U937 cells were used as a model system to study type I IFN induction.
FIG 1.
U937 cells and primary monocytes produce IFN-α and IFN-β proteins upon SeV infection. U937 cells and primary monocytes were infected with SeV (150 or 1.5 HA U/ml). Supernatants were harvested at 4, 8, 16, and 24 h p.i. and assayed for IFN-α and IFN-β proteins by ELISA. (A and B) IFN-α protein levels in U937 cells and primary monocytes infected with 150 HA U/ml (A) and 1.5 HA U/ml (B). (C and D) IFN-β protein levels in U937 cells and primary monocytes infected with 150 HA U/ml (C) and 1.5 HA U/ml (D). UI, uninfected. The monocyte data are from 3 different donors. The U937 cell data are from 3 biological replicates. The error bars indicate the standard error of the mean.
Distinct profiles of type I IFN subtypes are induced in cells infected at different virus concentrations.
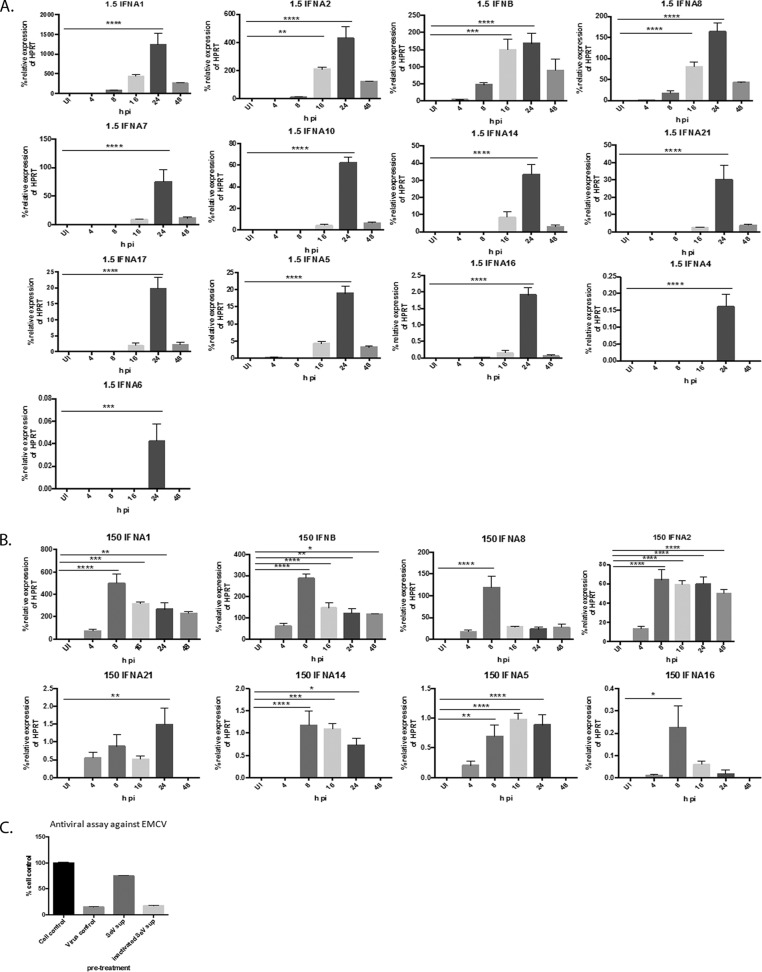
It is well established that different viruses induce distinct profiles of type I IFN subtypes in a cell-type-specific manner. However, it has not yet been determined how infection of cells at varying concentrations of the same virus affects type I IFN subtype induction. Therefore, we infected U937 cell cultures at 2 different concentrations of SeV, 150 HA U/ml and 1.5 HA U/ml, which correspond to MOIs of 5 and 0.05, respectively, for 4, 8, 16, 24, and 48 h and measured type I IFN subtype gene expression. Interestingly, we found that cultures infected with 1.5 HA U/ml induced all 13 IFN-α and IFN-β subtypes as detected by qRT-PCR (Fig. 2A), while cultures infected with 150 HA U/ml of SeV induced only 7 IFN-α subtypes (1, 2, 5, 8, 16, 14, and 21) and IFN-β (Fig. 2B). In other words, IFN-α subtypes 4, 6, 7, 10, and 17 were induced only in the cultures infected at the low concentration of SeV. The kinetics of subtype induction were also different. In the cultures infected at 1.5 HA U/ml, every subtype was significantly upregulated by 24 h p.i., and in cultures infected at the higher SeV concentration, all subtypes except IFN-α21 were significantly upregulated earlier, i.e., by 8 h p.i.; IFN-α21 was significantly upregulated by 24 h p.i. IFN induction upon SeV infection required active virus replication, as treating cells with inactivated virus did not result in any type I IFN in the supernatants as measured by antiviral assay (Fig. 2C). Collectively, these data indicate that cultures infected at different concentrations of SeV resulted in different profiles of type I IFN subtype induction. IFN-α subtypes 4, 6, 7, 10, and 17 were induced in cultures infected at the low but not the high concentration of SeV, suggesting that signaling pathways that are responsible for the induction of these particular subtypes are upregulated in cell cultures infected at the low but not the high concentration of SeV.
FIG 2.
Infection with 1.5 and 150 HA U/ml of SeV results in distinct profiles of IFN-α subtype induction. U937 cells were infected with either 1.5 or 150 HA U/ml of SeV for 4, 8, 16, 24, and 48 p.i. RNA was harvested at each time point, and IFN-α and IFN-β subtype RNA was measured via qRT-PCR. (A) IFN-β and IFN-α subtypes in cells infected with 1.5 HA U/ml. (B) IFN-β and IFN-α subtypes in cells infected with 150 HA U/ml. The data are from 3 biological replicates performed in duplicate. Statistics were performed using one-way ANOVA with Bonferroni posttest analysis. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. qRT-PCR data are shown as the percent relative expression of HPRT. (C) U937 cells were infected with either live or inactivated SeV (150 HA U/ml) for 24 h. The supernatants were harvested and then used to pretreat fresh U937 cells prior to EMCV infection. After 72 h, cell viability was measured via MTT assay, and the ability of the supernatants to restrict EMCV-induced CPE was analyzed. The error bars indicate the standard error of the mean.
JAK/STAT signaling through the IFNAR occurs to a greater extent in cell cultures infected at the lower virus concentration.
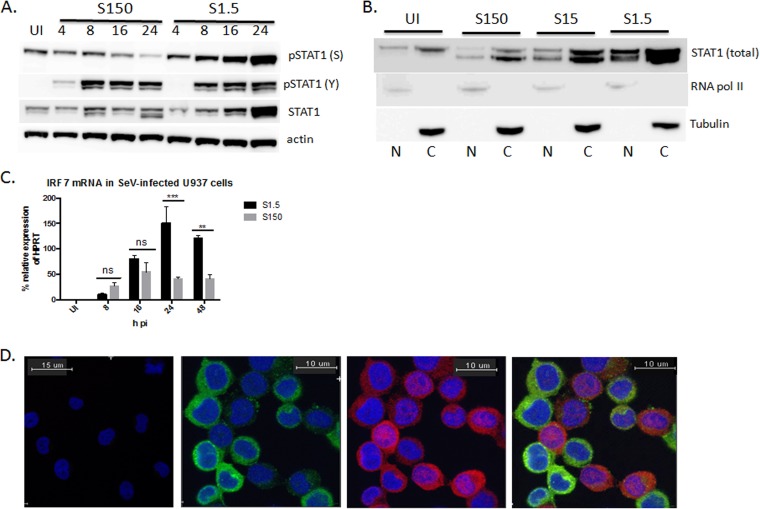
We next sought to determine which signaling pathways might be responsible for the difference in the IFN subtype expression profile in cultures infected at both SeV concentrations. Type I IFN subtypes are induced during two phases of the innate immune response. The first phase occurs following virus sensing and PRR activation. The IFN subtypes induced during this phase are secreted and then bind to the IFNAR in an autocrine and paracrine manner and activate the second phase of IFN induction, also known as amplification. We sought to determine whether this second phase of signaling was responsible for the difference in subtype profiles under the two virus conditions, because it can be assumed that the way this cell type senses the virus would be the same at both concentrations. Furthermore, it has been previously reported that the SeV C protein inhibits STAT1 signaling (20, 21). To determine if this was the case, we first compared the extent of JAK/STAT signaling in cultures infected at both virus concentrations. We compared protein expression and activation of STAT1 in cultures infected with either 1.5 or 150 HA U/ml of SeV for 4, 8, 16, and 24 h p.i. We found that cultures infected at the lower concentration of virus displayed higher overall protein expression of STAT1 by 16 and 24 h p.i. (Fig. 3A) and increased nuclear accumulation of total STAT1, which includes both unphosphorylated and phosphorylated forms, by 24 h p.i. (Fig. 3B). Interestingly, while the levels of p(Y701)STAT1 were the same between cultures infected at the two virus concentrations at 8, 16, and 24 h p.i., increased levels of p(S727)STAT1 were found in cultures infected with 1.5 HA U/ml, especially by 16 and 24 h p.i., compared to cultures infected with 150 HA U/ml (Fig. 3A). The levels of p(S727)STAT1 increased over time in cultures infected at the lower MOI. However, in contrast, levels decreased over time in cultures infected at the higher MOI. IRF7 mRNA levels were also significantly higher in cultures infected at the lower versus higher MOI of virus at 24 and 48 h p.i. (Fig. 3C). This suggests that some sort of JAK/STAT-inhibitory mechanism may exist in cultures infected at the higher SeV concentration. To determine if this was the case, we infected U937 cell cultures with 1.5 HA U/ml of SeV and costained for both SeV and STAT1 to determine, using confocal microscopy, if STAT1 nuclear localization was occurring in the infected or uninfected cells. We found that the vast majority of STAT1 nuclear localization (red) occurred in cells that had the least or no virus expression (green); STAT1 nuclear localization was almost absent in cells positive for SeV expression (Fig. 3D). These results suggest that SeV may inhibit STAT signaling in the infected cells. Because cell cultures infected at the low MOI have a higher percentage of uninfected cells, it is not surprising that JAK/STAT signaling is occurring to a greater extent in cultures infected at the lower concentration of virus. The extent of JAK/STAT signaling through the IFNAR in the cell cultures could therefore potentially be responsible for the differing type I IFN induction patterns.
FIG 3.
JAK/STAT signaling is increased in cells infected with 1.5 HA U/ml of SeV. (A) U937 cells were infected with either 1.5 (S1.5) or 150 (S150) HA U/ml of SeV and harvested at 4, 8, 16, and 24 h p.i. Whole-cell protein lysates were prepared, and Western blot analysis was performed to assess STAT1, p(Y701)STAT1, p(S727)STAT1, and actin expression. UI, uninfected. The Western blot is representative of 3 biological replicates. (B) U937 cells were infected with 150 (S150), 15 (S15), or 1.5 (S1.5) HA U/ml of SeV for 24 h, and nuclear and cytoplasmic protein preparations were made. Western blot analysis for STAT1, RNA Pol II, and tubulin was performed. RNA Pol II and tubulin were the nuclear and cytoplasmic loading controls, respectively. The Western blot is representative of 2 biological replicates. N, nuclear; C, cytoplasmic. (C) qRT-PCR analysis of IRF7 mRNA in cells infected with either 1.5 or 150 HA U/ml for 4, 8, 16, 24, and 48 h p.i. A two-way ANOVA with a Bonferroni posttest analysis was used for statistical analysis. **, P < 0.01; ***, P < 0.001; ns, not significant. qRT-PCR data are shown as the percent relative expression of HPRT. (D) U937 cells were either not infected or infected with SeV (1.5 HA U/ml) for 16 h, fixed on glass dishes, and then stained for SeV (green), STAT1 (red), and DAPI (blue). (Left) Uninfected. (Middle and right) Infected with SeV alone (green) (left middle), STAT1 alone (red) (right middle), and merge of SeV and STAT1 (green and red) (right). The images are representative of 2 biological replicates. For each experiment, 4 field views were imaged with similar results. The error bars indicate the standard error of the mean.
Type I IFN mRNA and protein induction depend more on IFNAR signaling in cell cultures infected at the lower virus concentration.
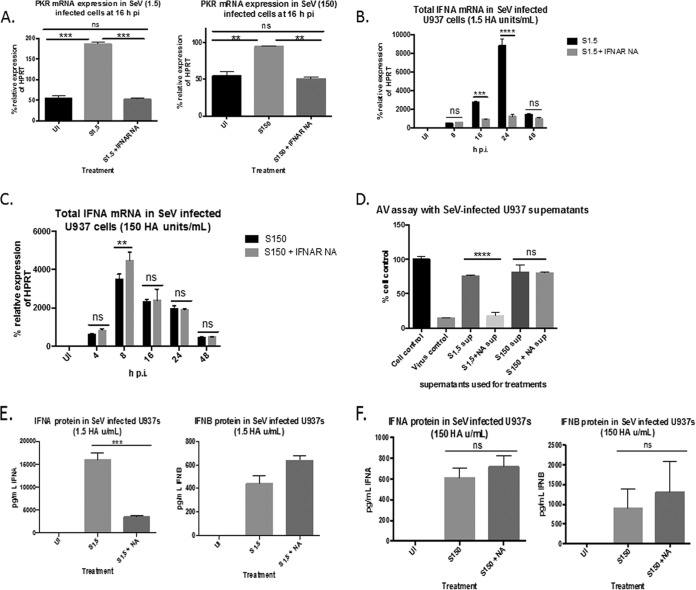
To determine if this increased JAK/STAT signaling in cultures infected at the lower concentration of virus affected overall IFN expression, we infected U937 cell cultures at both virus concentrations in the presence or absence of a neutralizing antibody to IFNAR2. Total IFN-α mRNA was measured by qRT-PCR at 4, 8, 16, 24, and 48 h to determine how blocking IFNAR signaling would affect overall IFN-α levels. At both virus concentrations, the mRNA expression of the ISG protein kinase R (PKR) was significantly downregulated to uninfected levels in the presence of the IFNAR2 antibody (Fig. 4A), indicating that the antibody was effectively inhibiting downstream signaling. We found that in the cultures infected with 1.5 HA U/ml, the initial induction levels of IFN-α at 8 h p.i. were comparable between cultures treated or not with the IFNAR2-neutralizing antibody. However, by 24 h p.i., IFN-α levels in cultures infected without the IFNAR-neutralizing antibodies increased 10-fold, while levels in cultures infected in the presence of IFNAR2-neutralizing antibody remained stable (Fig. 4B). In the cultures infected with 150 HA U/ml, however, there was no reduction of IFN-α expression levels at any of the time points in cells infected in the presence or absence of the neutralizing antibody. At 8 h p.i., there was actually an increase in expression in the presence of the antibody (Fig. 4C). This indicates that the IFNs being produced in cultures infected with 150 HA U/ml of SeV were most likely due to earlier IFNAR-independent mechanisms. We observed the same effect when looking at biologically active type I IFN protein in the supernatants as measured by the ability of the supernatant to protect fresh U937 cells against EMCV-mediated CPE. The amounts of biologically active type I IFN in the supernatants from cultures infected with 1.5 HA U/ml in the presence of IFNAR2-neutralizing antibody were significantly smaller than the amounts in cultures infected in the absence of the IFNAR2-neutralizing antibody (Fig. 4D). However, there was no difference in the amounts of biologically active type I IFN in the supernatants from cultures infected with 150 HA U/ml in the presence or absence of antibody (Fig. 4D). This decrease in biologically active type I IFN in cultures infected with 1.5 HA U/ml was due to a decrease in IFN-α protein rather than IFN-β as measured by ELISA (Fig. 4E). As expected, there was no decrease in IFN-α or IFN-β in cultures infected with 150 HA U/ml as measured by ELISA (Fig. 4F). These results indicate that the induction of IFN-α RNA and protein is more dependent on IFNAR2 signaling in cultures infected at the lower concentration of virus.
FIG 4.
Total IFN-α mRNA and protein induction is more reliant on IFNAR2 signaling in cells infected with 1.5 HA U/ml of SeV than in cells infected with 150 HA U/ml. (A) qRT-PCR analysis of PKR gene expression in cells infected with either 1.5 or 150 HA U/ml of SeV in the presence or absence of IFNAR2-neutralizing antibody. (B and C) Total IFN-α mRNA in cells infected with either 1.5 (B) or 150 (C) HA U/ml of SeV in the presence or absence of IFNAR-neutralizing antibody (NA). (D) Supernatants (sup) from cells infected with either 1.5 or 150 HA U/ml of SeV in the presence or absence of IFNAR NA (NA) for 24 h were used to pretreat fresh U937 cells in an antiviral assay using EMCV as the challenge virus. Cell viability was measured 72 h post-EMCV infection. Cell control, no pretreatment or infection; virus control, EMCV infected, no pretreatment. (E and F) ELISA measuring IFN-α protein (left) and IFN-β protein (right) in supernatants from cells infected with 1.5 (E) or 150 (F) HA U/ml of SeV in the presence or absence of IFNAR2 NA. Statistics were performed using a two-way ANOVA (B and C) and one-way ANOVA (A and D to F). Bonferroni posttest analyses were performed. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, not significant. The data are representative of 3 biological replicates performed in duplicate. qRT-PCR data are shown as the percent relative expression of HPRT. The error bars indicate the standard error of the mean.
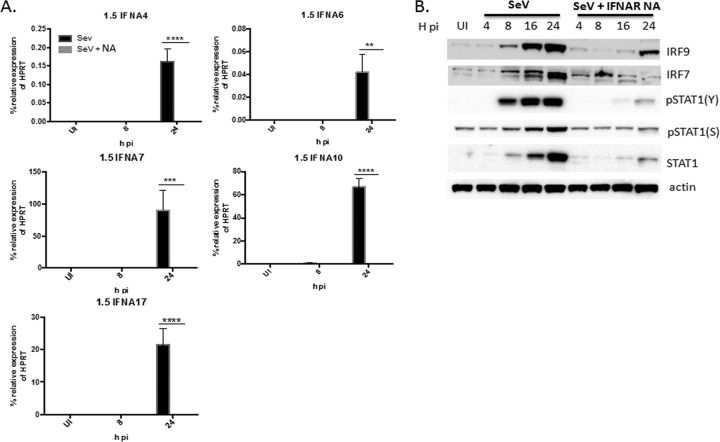
IFN-α subtypes 4, 6, 7, 10, and 17 are induced in an IFNAR-dependent manner in cell cultures infected at the lower virus concentration.
Because we found that overall IFN-α induction is more reliant on IFNAR signaling in cultures infected with 1.5 versus 150 HA U/ml of SeV, we sought to determine if the subtypes that were induced in cultures infected with 1.5 but not 150 HA U/ml, which included IFN-α subtypes 4, 6, 7, 10, and 17, were induced in an IFNAR-dependent manner. We infected U937 cell cultures with 1.5 HA U/ml in the presence or absence of IFNAR2-neutralizing antibody and measured the mRNA expression of IFN-α subtypes 4, 6, 7, 10, and 17 via qRT-PCR at 24 h p.i. We found that while each of these subtypes was significantly upregulated at 24 h p.i. in the absence of the IFNAR2-neutralizing antibody, expression was completely abolished in the presence of the IFNAR2 antibody (Fig. 5A). This inhibition of subtypes in the presence of the IFNAR2-neutralizing antibody was associated with a decrease in IRF7, IRF9, STAT1, p(S727)STAT1, and p(Y701)STAT1 protein expression (Fig. 5B), further suggesting that JAK/STAT signaling downstream of the IFNAR may be necessary for induction of these subtypes.
FIG 5.
IFN-α subtypes 4, 6, 7, 10, and 17 are induced in an IFNAR-dependent manner. (A) U937 cells were infected with 1.5 HA U/ml of SeV in the presence or absence of IFNAR2-neutralizing antibody (NA) for 8 and 24 h. qRT-PCR analysis was performed measuring IFN-α4, -6, -7, -10, and -17 mRNA. Statistics were performed using one-way ANOVA and Bonferroni posttest analysis. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. The data shown are from 3 biological replicates performed in duplicate. (B) Western blot analysis measuring IRF9, IRF7, p(Y701)STAT1, p(S727)STAT1, STAT1, and actin protein expression in cells infected with 1.5 HA U/ml in the presence or absence of IFNAR2-neutralizing antibody for 4, 8, 16, and 24 h p.i. UI, uninfected. The Western blot is representative of 3 biological replicates. qRT-PCR data are shown as the percent relative expression of HPRT. The error bars indicate the standard error of the mean.
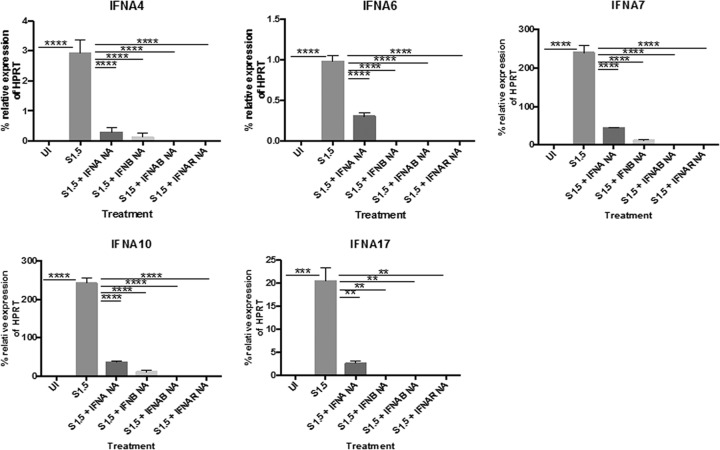
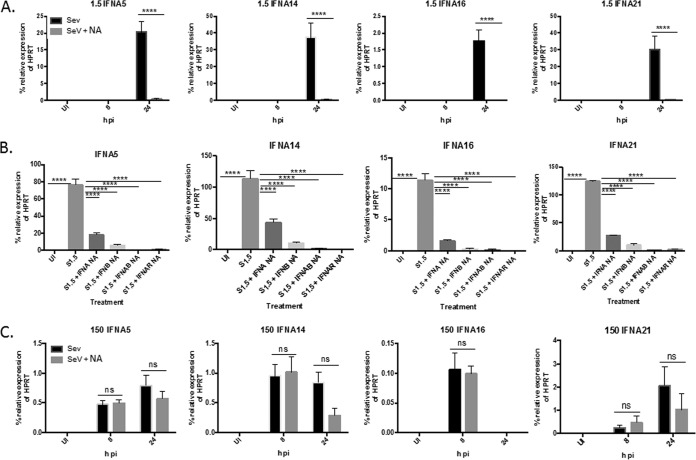
In addition to neutralizing IFNAR2, we further confirmed these findings by infecting U937 cell cultures with SeV (1.5 HA U/ml) in the presence or absence of neutralizing antibodies to IFN-β and/or total IFN-α. U937 cells were infected with 1.5 HA U/ml of SeV in the presence or absence of at least a 100-fold molar excess of neutralizing antibodies to total IFN-α, IFN-β, or both for 24 h, and then mRNA expression of IFN-α subtypes 4, 6, 7, 10, and 17 was measured. For all subtypes, neutralizing either IFN-α or IFN-β resulted in a significant downregulation of RNA. However, neutralizing both IFN-α and IFN-β completely blocked expression, which recapitulated the effects of blocking the IFNAR (Fig. 6). These results indicate that IFN-α subtypes 4, 6, 7, 10, and 17 are induced in an IFNAR-dependent manner. Both IFN-α and IFN-β are required to signal through the IFNAR to induce expression of these subtypes.
FIG 6.
Neutralizing IFN-α and IFN-β abolishes mRNA induction of IFN-α4, -6, -7, -10, and -17. U937 cells were infected with 1.5 HA U/ml of SeV for 24 h in the presence or absence of neutralizing antibodies to total IFN-α, IFN-β, IFN-α plus IFN-β (IFNAB) or IFNAR2 (IFNAR NA). mRNA expression of IFN-α4, -6, -7, -10, and -17 was measured via qRT-PCR. One-way ANOVA with Bonferroni posttest analysis was performed. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. The data shown are from 2 biological replicates performed in duplicate. qRT-PCR data are shown as the percent relative expression of HPRT. The error bars indicate the standard error of the mean.
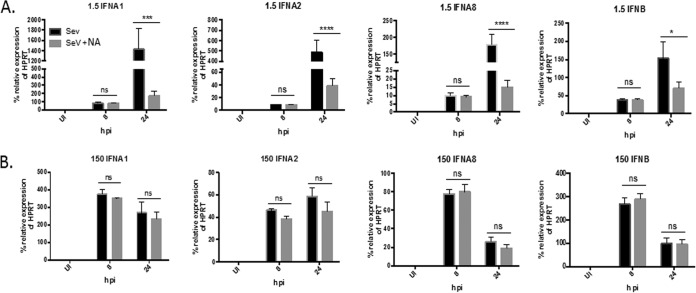
IFN-β and IFN-α subtypes 1, 2, and 8 are induced early in an IFNAR-independent manner in cell cultures infected at both virus concentrations.
In order for the induction of IFN-α subtypes 4, 6, 7, 10, and 17 to take place downstream of the IFNAR, the subtypes must first be induced during the early virus-sensing phase so that stimulation of the IFNAR can occur in an autocrine and paracrine manner. We observed that in cultures infected with 1.5 HA U/ml, IFN-β and IFN-α subtypes 1, 2, and 8 were the only subtypes that were induced early at 8 h p.i. (Fig. 2A). Expression of the rest of the subtypes occurred by 16 to 24 h p.i. Therefore, we hypothesized that IFN-β and IFN-α subtypes 1, 2, and 8 were induced early during PRR sensing in an IFNAR-independent manner. To test this hypothesis, U937 cell cultures were infected with 1.5 HA U/ml of SeV in the presence or absence of an IFNAR2-blocking antibody. mRNA expression of IFN-β and IFN-α subtypes 1, 2, and 8 was measured via qRT-PCR at 8 and 24 h p.i. These two time points were chosen based on our previous time course in cultures infected with 1.5 HA U/ml; 8 h p.i. is when these subtypes are first induced, and 24 h p.i. is when peak levels are induced. Interestingly, we found that for each of the subtypes, there was no significant difference in mRNA expression between cultures infected in the presence or absence of IFNAR2-neutralizing antibody at 8 h p.i. (Fig. 7A). At 24 h p.i., however, the presence of the neutralizing antibody significantly inhibited, but did not abolish, the mRNA expression of all of these subtypes, suggesting that amplification of the subtypes may be occurring at the later time point. These results suggest that in cultures infected with 1.5 HA U/ml of SeV, initial induction of IFN-β and IFN-α subtypes 1, 2, and 8 occurs in an IFNAR-independent manner, while amplification at later time points is IFNAR dependent. As expected, in cultures infected with 150 HA U/ml of SeV where induction of type I IFNs occurs in an IFNAR-independent manner (Fig. 4C), there is no significant difference in expression of these subtypes between cells infected in the presence or absence of IFNAR2-neutralizing antibody (Fig. 7B).
FIG 7.
IFN-β and IFN-α subtypes 1, 2 and 8 are initially induced early in an IFNAR-independent manner in both virus infections. U937 cells were infected with either 1.5 (A) or 150 (B) HA U/ml of SeV for 8 and 24 h in the presence or absence of IFNAR NA. mRNA expression of IFN-α1, -2, and -8 and IFN-β was measured via qRT-PCR. A one-way ANOVA with Bonferroni posttest analysis was performed. *, P < 0.05; ***, P < 0.001; ****, P < 0.0001; ns, not significant. The data shown are from 3 biological replicates performed in duplicate. qRT-PCR data are shown as the percent relative expression of HPRT. The error bars indicate the standard error of the mean.
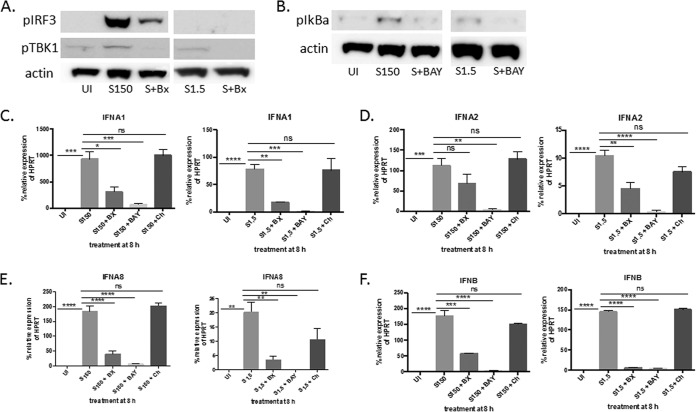
We established that human IFN-β and IFN-α subtypes 1, 2, and 8 are induced early in infection in an IFNAR-independent manner. We next determined if early PRR activation during virus sensing might be responsible for the early induction of these subtypes. Previous reports have indicated that SeV can be sensed by the RIG-I/MDA5 cytosolic receptors or TLR7 (22–24). We therefore sought to determine if inhibiting these pathways inhibited the induction of the subtypes in cells infected at both virus concentrations. Since the subtypes are induced at both high and low virus concentrations, we hypothesized that the way the cell senses the virus should be the same under both conditions. U937 cell cultures were infected in the presence or absence of the following inhibitors: BX795, which inhibits TBK1/IRF3 phosphorylation downstream of RIG-I/MDA5; chloroquine, which inhibits TLR7 signaling; and BAY11-7082, which inhibits IκB phosphorylation and the subsequent activation of NF-κB, which is a transcription factor that has been reported to be necessary for PRR-activated type I IFN induction (25). Cultures that were infected with 1.5 or 150 HA U/ml SeV for 3 h in the presence of BX795 showed successful inhibition of TBK1 phosphorylation (Fig. 8A). The cultures infected with 150 HA U/ml and treated with BX795 also showed inhibition of IRF3 phosphorylation (Fig. 8A). pIRF3 was not detected in cultures infected with 1.5 HA U/ml, most likely because the levels required for detection by Western blotting had not been reached by 3 h p.i. By 8 h p.i., pIRF3 was clearly detected (data not shown). Cell cultures infected in the presence of BAY11-7802 for 3 h showed successful inhibition of IκB phosphorylation (Fig. 8A and B). U937 cells were then infected with 1.5 or 150 HA U/ml of SeV in the presence or absence of these inhibitors for 8 h. mRNA expression of IFN-β and IFN-α1, -2, and -8 was measured via qRT-PCR. The inhibition of NF-κB resulted in the significant downregulation of every subtype at both virus concentrations (Fig. 8C to F). These subtypes were also significantly downregulated in the presence of the TBK1 inhibitor at both virus concentrations (Fig. 8C to F), with the exception of IFN-α2, where the downregulation of mRNA was not significant in the cultures infected with 150 HA U/ml of SeV (Fig. 8D). These results were confirmed when we used a more specific inhibitor of TBK1, MRT67307 (data not shown). Inhibiting TLR7 signaling with chloroquine did not significantly alter the expression of any of these subtypes at either concentration. Increasing the amount of chloroquine did not change the results (data not shown). These results suggest that the TBK1/IRF3 pathway and NF-κB may be responsible for the induction of the IFNAR-independent subtypes.
FIG 8.
IFN-β and IFN-α subtypes 1, 2, and 8 are induced in an NF-κB- and TBK1-dependent manner. (A) U937 cells were infected with 150 or 1.5 HA U/ml of SeV (S) for 3 h in the presence or absence of BX795 (BX), and pTBK1 and pIRF3 protein expression was measured via Western blotting. Actin was measured as a loading control. (B) Same as panel A, except cells were infected in the presence or absence of BAY11-7802 (BAY), and pIκB protein expression was measured. (C) U937 cells were infected with either 150 or 1.5 HA U/ml of SeV for 8 h in the presence or absence of the following inhibitors: BX795 (BX), BAY11-7802 (BAY), or chloroquine (Ch). (C to F) mRNA expression of IFN-α1 (C), IFN-α2 (D), IFN-α8 (E), and IFN-β (F) was measured via qRT-PCR. A one-way ANOVA with Bonferroni posttest analysis was performed. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, not significant. The data shown are from 3 biological replicates performed in duplicate. qRT-PCR data are shown as the percent relative expression of HPRT. The error bars indicate the standard error of the mean.
IFN-α subtypes 5, 14, 16, and 21 are induced in either an IFNAR-dependent or -independent manner, depending on the MOI.
Our previous data suggest that IFN-β and IFN-α subtypes 1, 2, and 8 are induced early by an IFNAR-independent mechanism and IFN-α subtypes 4, 6, 7, 10, and 17 are induced later by an IFNAR-dependent mechanism. We next determined how the remaining subtypes, IFN-α5, -14, -16, and -21, were induced. These subtypes were induced in cultures infected at both the high and low virus concentrations. To test this, we infected cultures with either 1.5 or 150 HA U/ml of SeV in the presence or absence of the IFNAR2-neutralizing antibody and measured induction at 8 and 24 h p.i. Surprisingly, we found that the induction of the subtypes displayed both IFNAR-dependent and -independent characteristics, depending on the MOI. In U937 cell cultures infected with 1.5 HA U/ml of SeV, the expression of the subtypes occurs at 24 h p.i. and is almost completely dependent on IFNAR signaling, as pretreatment with the IFNAR2-neutralizing antibody resulted in significant inhibition of mRNA expression at 24 h p.i. (Fig. 9A). These results were recapitulated when the cultures were infected in the presence or absence of neutralizing antibodies to both IFN-β and total IFN-α (Fig. 9B). However, in cultures infected with 150 HA U/ml, the subtypes were induced early at 8 h p.i., and there was no significant difference in expression levels in the presence or absence of the IFNAR2-neutralizing antibody at any of the time points tested (Fig. 9C). These data indicate that different pathways can induce IFN-α subtypes 5, 14, 16, and 21, depending on the amount of infecting virus. With low concentrations of infecting virus, the subtypes are induced later in an IFNAR-dependent manner. However, at high concentrations of infecting virus, the subtypes are induced early and by an IFNAR-independent mechanism.
FIG 9.
IFN-α subtypes 5, 14, 16, and 21 are induced in an IFNAR-dependent manner in cells infected with 1.5 HA U/ml and an IFNAR-independent manner in cells infected with 150 HA U/ml. (A and C) U937 cells were infected with either 1.5 (A) or 150 (C) HA U/ml of SeV for 8 and 24 h in the presence or absence of IFNAR NA. mRNA expression of IFN-α5, -14, -16, and -21 was measured via qRT-PCR. The data shown are from 3 biological replicates performed in duplicate technical replicates. (B) U937 cells were infected with 1.5 HA U/ml of SeV for 24 h in the presence or absence of neutralizing antibodies to total IFN-α, IFN-β, IFN-α plus IFN-β (IFNAB), or IFNAR2 (IFNAR NA). mRNA expression of IFN-α5, -14, -16, and -21 was measured via qRT-PCR. The data are from 2 biological replicates performed in duplicate. A one-way ANOVA with a Bonferroni posttest analysis was performed. ****, P < 0.0001. qRT-PCR data are shown as the percent relative expression of HPRT. The error bars indicate the standard error of the mean.
Distinct profiles of ISGS are induced in cell cultures infected at different virus concentrations.
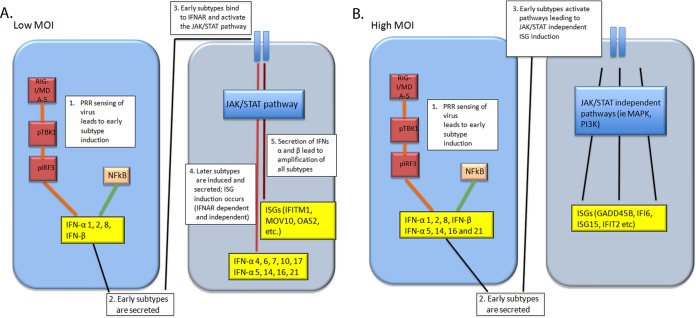
Our results thus far indicated that different concentrations of the same virus not only affect which type I IFN subtypes are induced, but also the extent of JAK/STAT signaling that takes place downstream of the IFNAR. We next determined if these differences affected the signature of antiviral ISGs that are induced. We infected U937 cell cultures with either 1.5 or 150 HA U/ml of SeV for 24 h and then analyzed the profile of ISGs using a gene expression microarray under the 2 virus conditions, as well as in uninfected cells. We found that 1,008 ISGs were differentially regulated between the 2 virus concentrations according to the Interferome database (www.interferome.org). Both qualitative and quantitative differences were found in the types and levels of ISGs induced in response to the 2 virus conditions. Some ISGs, including GADD45B, TXNIP, CCR7, TSC22D3, HIST1H2AC, IFI6, and TRAF1 genes, were induced only in the cultures infected with 150 HA U/ml of SeV, while other genes, including IFITM1, IFI27, NEXN, IFII44L, TRIM22, NMI, and MOV10 genes, were induced only in cultures infected with 1.5 HA U/ml of SeV. There was also a subset of genes (encoding IFIT2, IFIT1, OASL, ISG15, IFIH1, OAS2, IFI44, PMAIP1, and DDX58) that were significantly induced to similar levels under both virus conditions compared to uninfected cells (see Table S3 in the supplemental material). These results indicate that the differential induction of type I IFNs by different concentrations of the same virus can significantly affect which ISGs are induced and, consequently, the antiviral response. Figure 10 summarizes these results.
FIG 10.
Schematic illustrating type I IFN and ISG induction in cell cultures infected with a high or low MOI of SeV.
DISCUSSION
The induction and regulation of type I IFN subtypes has been extensively studied due to their importance in mounting an effective antiviral response. It is known that different viruses lead to the induction of different type I IFN subtype profiles. It is thought that this is due to different viral PRR sensors being activated, depending on the type of infecting virus. These sensors then activate different signaling pathways and transcription factors that have different capabilities to bind to the promoter regions of distinct type I IFN subtypes. This idea has been supported by the fact that stimulating human leukocytes with agonists for TLR3, -7, -8, and -9 resulted in different IFN-α subtype patterns (11, 12). Furthermore, different type I IFN subtype profiles can also be induced in a cell-type-specific manner (26), as different cell types express varying levels of both PRRs and transcription factors that can affect subtype promoter regulation. Our present study, however, presents the novel finding that the same virus can induce different type I IFN profiles in the same cell type, depending on the MOI used to infect the cells. This is primarily due to differences in the extent of signaling through the IFNAR, which is a process that has not been previously attributed to the differential induction of individual subtypes. Not only does the MOI affect which subtypes are induced, it also affects the mechanism by which certain subtypes are induced. Finally, these distinct subtype profiles induced by the same virus in the same cell type also result in differential ISG induction. Taking the data together, by looking at the MOI-specific differences in type I IFN subtype induction profiles, we were able to uncover an additional mechanism by which individual type I IFN subtypes are regulated. We also discovered differential expression of ISGs, which could be important in the pleiotropic effects of IFNs.
We found that in cell cultures infected with 1.5 HA U/ml of SeV, induction of type I IFNs mirrored the classical model of IFN induction, where early subtypes are induced and then bind to the IFNAR to induce later subtypes (2–6). Cultures infected with 1.5 HA U/ml of SeV exhibited higher dependency on IFNAR signaling for the induction of IFN-α mRNAs than cultures infected with 150 HA U/ml. This dependency could be due to a number of factors. First, in cultures infected with 1.5 HA U/ml, there is a much higher frequency of uninfected bystander cells that are capable of being primed by the IFN that is secreted from the virus-infected cells than in cultures infected with 150 HA U/ml. Because these cells are uninfected, proteins that are directly or indirectly activated by the virus may not be activated in the cells, resulting in a different immunological microenvironment. Our confocal microscopy results appear to support this conclusion. It is possible that virus infection in a cell triggers proteins that inhibit JAK/STAT signaling, which would explain why infections at a low MOI induce more overall IFN subtypes that are IFNAR dependent, since not as many virus-infected cells that contain these negative regulators would be present; the majority of IFN-producing cells would be primed rather than infected. Our microarray analysis revealed that SOCS1, SOCS2, PIAS4, and PIAS1, which all inhibit STAT protein function, are all upregulated to a greater extent in cultures infected with 150 HA U/ml than in cultures infected with 1.5 HA U/ml (microarray data not shown). Second, it is also possible that viral proteins that inhibit JAK/STAT signaling downstream of the IFNAR inhibit induction of IFNAR-dependent, but not -independent, subtypes. The C protein of SeV has been reported to interfere with JAK/STAT signaling by degrading STAT1 and -2 (20, 21). This would have an effect on signaling only in cells that are directly infected. We found, using confocal microscopy, that STAT1 nuclear localization occurred only in cells that were uninfected (Fig. 3D). Because there would be far more virus-infected cells in the 150- versus the 1.5-HA U/ml group, a disruption in IFNAR signaling would be more apparent in the cultures infected at the higher versus the lower concentration. We found in our studies that serine phosphorylation of STAT1 at residue 727 was inhibited in cultures infected with 150 HA U/ml, while p(S727)STAT1 levels continually increased in cultures infected with 1.5 HA U/ml over time. It is possible that serine phosphorylation of STAT1 could be required for the induction of IFNAR-dependent subtypes and that virus infection inhibits this process, thus leading to inhibition of IFNAR-dependent subtypes in cells infected with more virus.
The varying dependence of IFNAR signaling in the cultures infected at the 2 virus concentrations enabled us to discover that the induction of IFN-α subtypes 4, 6, 7, 10, and 17 was completely dependent on IFNAR signaling at 24 h p.i. IFN-β and IFN-α subtypes 1, 2, and 8, on the other hand, were induced early, by 8 h p.i., in an IFNAR-independent manner at both virus concentrations. Amplification of these subtypes at the lower virus concentration, however, was IFNAR dependent. It is likely that in SeV infection, IFN-β and IFN-α subtypes 1, 2, and 8 are the first subtypes that are induced in SeV-infected cells and then prime surrounding uninfected cells through the IFNAR. However, this result cannot be confirmed because specific antibodies that neutralize the individual subtypes do not exist. The induction of IFN-α subtypes 5, 14, 16, and 21, however, exhibited both IFNAR-dependent and -independent properties according to how much virus was used to infect the cells. This suggests that the virus may be directly or indirectly activating certain factors that are needed for the transcriptional induction of the subtypes. When a very low concentration of virus infects a cell, these factors may not be present in sufficient amounts to induce transcription, so induction is purely dependent on the amplification response through the IFNAR. The amount of virus used in an infection is therefore an extremely important factor to take into consideration when studying the regulation of IFN subtypes.
The induction of type I IFN subtypes and the signaling pathways that regulate them are highly dependent on the regulatory and transcription factor binding sequences in their promoter regions. Type I IFN genes are distinct, intronless genes that are clustered on chromosome 9 in humans. There are 13 different IFN-α genes but only 12 different proteins; IFN-α1 and -13 share the same protein-coding sequences, but their regulatory regions are different. An alignment of the promoter regions of IFN-β and the IFN-α subtypes revealed that subtypes 4, 7, 10, and 17, which are all dependent on IFNAR2 signaling, share between 89 and 94% sequence similarity. IFN-α6, which is also an IFNAR-dependent subtype, shares about 60% sequence similarity with the other members of this group. Interestingly, the type I IFN subtypes that we found to be IFNAR independent, which are IFN-β and IFN-α1, -2, and -8, share only 39 to 60% sequence similarity, much less than the IFNAR-dependent subtypes. This may not be surprising, since it has been reported extensively in the literature that the activation of different PRRs induces distinct patterns of type I IFN subtypes, as stimulating human and nonhuman primate leukocytes with agonists for TLR3, -7, -8, and -9 resulted in different IFN-α subtype patterns (11, 12). Therefore, subtypes that are induced early following PRR stimulation may have distinct sets of transcription factor binding sites for proteins that are activated in the different PRR signaling pathways. The main signaling pathway responsible for the amplification of the IFN response downstream of the IFNAR, however, is the JAK/STAT pathway, which leads to the activation of IRF7 and the ISGF3 complex. This transcription complex has been reported to further induce IFN-α expression by binding to the PRDI region in the IFN-α promoter region (6), as well as enhancing IRF-7 gene expression (27), both of which amplify the IFN response. Therefore, it is not surprising that the subtypes that are IFNAR dependent have a much higher degree of sequence similarity in their promoter regions than the IFNAR-independent subtypes; there is less variability in the transcription factors and complexes that activate their transcription.
In our study, we found that the presence of different IFN subtype profiles in cultures infected with either 1.5 or 150 HA U/ml of SeV induced different ISG signatures. About 1,000 ISGs were differentially induced according to the Interferome database, and of these, some were uniquely expressed in cultures infected with 1.5 HA U/ml, some were uniquely expressed in cultures with 150 HA U/ml, some were expressed in both infections at the same levels, and some were expressed in both infections at differing levels. This indicates that although the extents of signaling through the IFNAR were extremely different between cultures infected with 1.5 versus 150 HA U/ml of SeV, both infections resulted in the induction of hundreds of ISGs. This is most likely due to the fact that canonical JAK/STAT signaling is not the only pathway that induces ISGs and an antiviral response. Alternative JAK/STAT-independent pathways induced by type I IFNs that lead to ISG induction have been studied extensively in the literature and include the MAPK and PKR pathways (28, 29). Our data support these previous findings. Ingenuity Pathway Analysis was performed on our microarray data and revealed that in cells infected with 150 HA U/ml, two of the most highly regulated pathways included phosphatidylinositol 3-kinase (PI3K)/AKT signaling and the role of PKR in IFN signaling. In cell cultures infected with 1.5 HA U/ml, however, the two most highly regulated pathways included IRF/PRR signaling and JAK/STAT IFN signaling (data not shown). It is likely that different IFNs present in a system can stimulate distinct JAK/STAT-dependent and -independent pathways, and this would in turn affect the ISGs that are induced. For example, in mouse J2E cells, IFN-α subtypes 1, 2, 4, and 5 all induced tyrosine phosphorylation of STAT1, while only subtypes 1 and 4 induced phosphorylation of STAT5a/5b. None of the IFN-α subtypes induced phosphorylation of JNK, while all IFN-α subtypes, but not IFN-β, induced phosphorylation of p38 MAPK (30). Therefore, it is possible that both canonical and noncanonical pathways can lead to the induction of ISGs, and the presence of different type I IFN subtypes could potentially affect which of these pathways are activated. These results further demonstrate that the amount of virus in a system can have major effects on the type of antiviral response that is initiated.
Supplementary Material
ACKNOWLEDGMENTS
We thank the Cytokine Biology Section for their assistance with this study. We also thank the Microarray Core Facility and Biological Imaging Core Facility in the Research Technology Branch at NIAID for their assistance with experiments.
This work was supported by the Intramural Research Training Program (IRTA) and the Division of Intramural Research (DIR) at the National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.01727-15.
REFERENCES
- 1.Génin P, Lin R, Hiscott J, Civas A. 2009. Differential regulation of human interferon A gene expression by interferon regulatory factors 3 and 7. Mol Cell Biol 29:3435–3450. doi: 10.1128/MCB.01805-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marie I, Durbin JE, Levy DE. 1998. Differential viral induction of distinct interferon-alpha genes by positive feedback through interferon regulatory factor-7. EMBO J 17:6660–6669. doi: 10.1093/emboj/17.22.6660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Su L, David M. 2000. Distinct mechanisms of STAT phosphorylation via the interferon-alpha/beta receptor. Selective inhibition of STAT3 and STAT5 by piceatannol. J Biol Chem 275:12661–12666. [DOI] [PubMed] [Google Scholar]
- 4.Honda K, Yanai H, Negishi H, Asagiri M, Sato M, Mizutani T, Shimada N, Ohba Y, Takaoka A, Yoshida N, Taniguchi T. 2005. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature 434:772–777. doi: 10.1038/nature03464. [DOI] [PubMed] [Google Scholar]
- 5.Sato M, Hata N, Asagiri M, Nakaya T, Taniguchi T, Tanaka N. 1998. Positive feedback regulation of type I IFN genes by the IFN-inducible transcription factor IRF-7. FEBS Lett 441:106–110. doi: 10.1016/S0014-5793(98)01514-2. [DOI] [PubMed] [Google Scholar]
- 6.Yoneyama M, Suhara W, Fukuhara Y, Sato M, Ozato K, Fujita T. 1996. Autocrine amplification of type I interferon gene expression mediated by interferon stimulated gene factor 3 (ISGF3). J Biochem 120:160–169. doi: 10.1093/oxfordjournals.jbchem.a021379. [DOI] [PubMed] [Google Scholar]
- 7.Zoon KC, Buckler CE, Bridgen PJ, Gurari-Rotman D. 1978. Production of human lymphoblastoid interferon by Namalva cells. J Clin Microbiol 7:44–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cantell K, Hirvonen S, Kauppinen HL, Myllylä G. 1981. Production of interferon in human leukocytes from normal donors with the use of Sendai virus. Methods Enzymol 78:29–38. doi: 10.1016/0076-6879(81)78094-7. [DOI] [PubMed] [Google Scholar]
- 9.Civas A, Génin P, Morin P, Lin R, Hiscott J. 2006. Promoter organization of the interferon-A genes differentially affects virus-induced expression and responsiveness to TBK1 and IKKepsilon. J Biol Chem 281:4856–4866. doi: 10.1074/jbc.M506812200. [DOI] [PubMed] [Google Scholar]
- 10.Liao AP, Salajegheh M, Morehouse C, Nazareno R, Jubin RG, Jallal B, Yao Y, Greenberg SA. 2010. Human plasmacytoid dendritic cell accumulation amplifies their type 1 interferon production. Clin Immunol 136:130–138. doi: 10.1016/j.clim.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hillyer P, Mane VP, Schramm LM, Puig M, Verthelyi D, Chen A, Zhao Z, Navarro MB, Kirschman KD, Bykadi S, Jubin RG, Rabin RL. 2012. Expression profiles of human interferon-alpha and interferon-lambda subtypes are ligand- and cell-dependent. Immunol Cell Biol 90:774–783. doi: 10.1038/icb.2011.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Puig M, Tosh KW, Schramm LM, Grajkowska LT, Kirschman KD, Tami C, Beren J, Rabin RL, Verthelyi D. 2012. TLR9 and TLR7 agonists mediate distinct type I IFN responses in humans and nonhuman primates in vitro and in vivo. J Leukoc Biol 91:147–158. doi: 10.1189/jlb.0711371. [DOI] [PubMed] [Google Scholar]
- 13.Palmer P, Tovey MG, Raschilas F, Brassart L, Meritet J-F, Porcher R, Lebon P. 2007. Type I interferon subtypes produced by human peripheral mononuclear cells from one normal donor stimulated by viral and non-viral inducing factors. Eur Cytokine Netw 18:108–114. [DOI] [PubMed] [Google Scholar]
- 14.Barbalat R, Lau L, Locksley RM, Barton GM. 2009. Toll-like receptor 2 on inflammatory monocytes induces type I interferon in response to viral but not bacterial ligands. Nat Immunol 10:1200–1207. doi: 10.1038/ni.1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clay CC, Rodrigues DS, Ho YS, Fallert BA, Janatpour K, Reinhart TA, Esser U. 2007. Neuroinvasion of fluorescein-positive monocytes in acute simian immunodeficiency virus infection. J Virol 81:12040–12048. doi: 10.1128/JVI.00133-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fischer-Smith T, Rappaport J. 2005. Evolving paradigms in the pathogenesis of HIV-1-associated dementia. Expert Rev Mol Med 7:1–26. [DOI] [PubMed] [Google Scholar]
- 17.Barrett AD, Hunt N, Dimmock NJ. 1984. A rapid method for the inactivation of virus infectivity prior to assay for interferons. J Virol Methods 8:349–351. doi: 10.1016/0166-0934(84)90072-7. [DOI] [PubMed] [Google Scholar]
- 18.Szubin R, Chang WLW, Greasby T, Beckett L, Baumgarth N. 2008. Rigid interferon-alpha subtype responses of human plasmacytoid dendritic cells. J Interferon Cytokine Res 28:749–763. doi: 10.1089/jir.2008.0037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Newton CR, Graham A, Heptinstall LE, Powell SJ, Summers C, Kalsheker N, Smith JC, Markham AF. 1989. Analysis of any point mutation in DNA. The amplification refractory mutation system (ARMS). Nucleic Acids Res 17:2503–2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gotoh B, Takeuchi K, Komatsu T, Yokoo J. 2003. The STAT2 activation process is a crucial target of Sendai virus C protein for the blockade of alpha interferon signaling. J Virol 77:3360–3370. doi: 10.1128/JVI.77.6.3360-3370.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Komatsu T, Takeuchi K, Yokoo J, Tanaka Y, Gotoh B. 2000. Sendai virus blocks alpha interferon signaling to signal transducers and activators of transcription. J Virol 74:2477–2480. doi: 10.1128/JVI.74.5.2477-2480.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lund JM, Alexopoulou L, Sato A, Karow M, Adams NC, Gale NW, Iwasaki A, Flavell RA. 2004. Recognition of single-stranded RNA viruses by Toll-like receptor 7. Proc Natl Acad Sci U S A 101:5598–5603. doi: 10.1073/pnas.0400937101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gitlin L, Benoit L, Song C, Cella M, Gilfillan S, Holtzman MJ, Colonna M. 2010. Melanoma differentiation-associated gene 5 (MDA5) is involved in the innate immune response to paramyxoviridae infection in vivo. PLoS Pathog 6:e1000734. doi: 10.1371/journal.ppat.1000734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baum A, García-Sastre A. 2011. Differential recognition of viral RNA by RIG-I. Virulence 2:166–169. doi: 10.4161/viru.2.2.15481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kawai T, Akira S. 2010. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol 11:373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- 26.Li L, Sherry B. 2010. IFN-alpha expression and antiviral effects are subtype and cell type specific in the cardiac response to viral infection. Virology 396:59–68. doi: 10.1016/j.virol.2009.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Génin P, Morin P, Civas A. 2003. Impairment of interferon-induced IRF-7 gene expression due to inhibition of ISGF3 formation by trichostatin A. J Virol 77:7113–7119. doi: 10.1128/JVI.77.12.7113-7119.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Uddin S, Majchrzak-Kita B, Woodson J, Arunkumar P, Alsayed Y, Pine R, Young PR, Fish EN, Platanias LC. 1999. Activation of the p38 mitogen-activated protein kinase by type I interferons. J Biol Chem 274:30127–30131. doi: 10.1074/jbc.274.42.30127. [DOI] [PubMed] [Google Scholar]
- 29.Kaur S, Sassano A, Joseph AM, Majchrzak-Kita B, Eklund EA, Verma A, Brachmann SM, Fish EN, Platanias LC. 2008. Dual regulatory roles of phosphatidylinositol 3-kinase in IFN signaling. J Immunol 181:7316–7323. doi: 10.4049/jimmunol.181.10.7316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cull VS, Tilbrook PA, Bartlett EJ, Brekalo NL, James CM. 2003. Type I interferon differential therapy for erythroleukemia: specificity of STAT activation. Blood 101:2727–2735. doi: 10.1182/blood-2002-05-1521. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.