Abstract
Vesicular stomatitis virus has been shown to bud basolaterally, and the matrix protein, but not glycoprotein, was proposed to mediate this asymmetry. Using polarized T84 monolayers, we demonstrate that no single viral protein is sufficient for polarized budding. Particles are released from the apical and basolateral surfaces and are indistinguishable, indicating that there is no apical assembly defect. We propose that aspects of host cell polarity create a more efficient budding process at the basolateral surface.
TEXT
Many viruses have distinct entry and budding sites in polarized epithelial cells (1–3). Vesicular stomatitis virus (VSV) is the prototypic nonsegmented negative-strand RNA virus and buds preferentially from the basolateral surface of polarized epithelial cells. This budding preference correlates with the basolateral localization of its glycoprotein (G) (4). However, mislocalization of G did not change this phenotype, and thus it was proposed that the matrix protein (M) dictates preferential VSV budding (5). In this study, we demonstrate that neither G nor M alone can account for this bias.
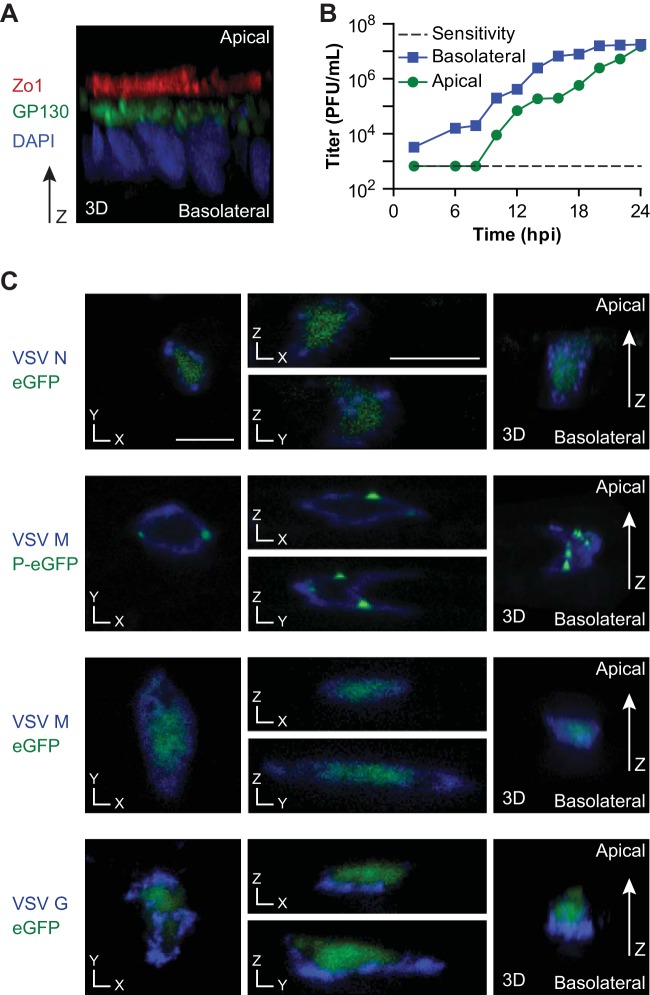
Recombinant VSV (rVSV) was tested for infection and budding preference from the polarized human epithelial T84 cell line. T84 monolayers were infected after polarization on collagen-coated Transwell supports, with polarity demonstrated by a transepithelial resistance (TER) of >1,000 Ω and confirmed by proper localization of the apical tight-junction marker Zo-1 (6), Golgi compartment marker GP130 (7, 8), and the nucleus (7) (Fig. 1A). rVSV infected the basolateral surface 62 ± 14 (n = 4) times more efficiently than the apical surface. Postinfection, rVSV was found to bud preferentially from the basolateral surface (Fig. 1B). Infectious virus appeared initially in the basolateral compartment but was later detected in the apical chamber, with cells maintaining a tight monolayer. Both infection and budding preferences are similar to previous observations with MDCK cells (9). Monolayer integrity was monitored by TER and small-molecule diffusion using medium with or without phenol red in complementary culture compartments. Cells maintained high TER and limited small-molecule diffusion through 12 h postinfection (hpi). Subsequently, TER gradually dropped, reaching 150 Ω at 22 hpi and loss of TER at 24 hpi, when phenol red diffusion was observed (data not shown). Immunocytochemistry analysis with previously characterized antibodies was used to identify the subcellular localization of VSV proteins (10, 11) (Fig. 1C). The nucleocapsid protein (N), phosphoprotein (P), and large polymerase protein (L) have been reported to localize to inclusion bodies throughout the cytoplasm (12). Likewise, we observed N and P in a nonpolarized distribution (Fig. 1C), demonstrating that replication and transcription were not polarized. Similarly, M was found to be cytosolic and membrane bound throughout the apical and basolateral compartments, indicating its nonpolarized distribution. This observation contrasts with previous findings in which M was basolaterally localized in MDCK cells (13). While this localization may not be universally true, it demonstrates that polarized M localization is dispensable for polarized budding. In contrast, G was predominantly basolateral (Fig. 1C), as previously shown (4, 14).
FIG 1.
Polarity of budding of VSV from polarized T84 cells and localization of VSV proteins. (A) T84 polarization was verified by immunocytochemistry analysis, followed by confocal fluorescence microscopy, for the markers indicated. (B) Growth curves of rVSV in polarized T84 monolayers. Cells were infected at a multiplicity of infection of 3, and virus titers were measured at the postinfection times specified. (C) Localization of VSV proteins in polarized T84 cells. Monolayers were infected with rVSV eGFP (24) or rVSV P-eGFP (25), and confocal microscopy for the proteins indicated was performed at 8 hpi. Representative cross sections and three-dimensional reconstructions are shown. The scale bars represent 10 μm.
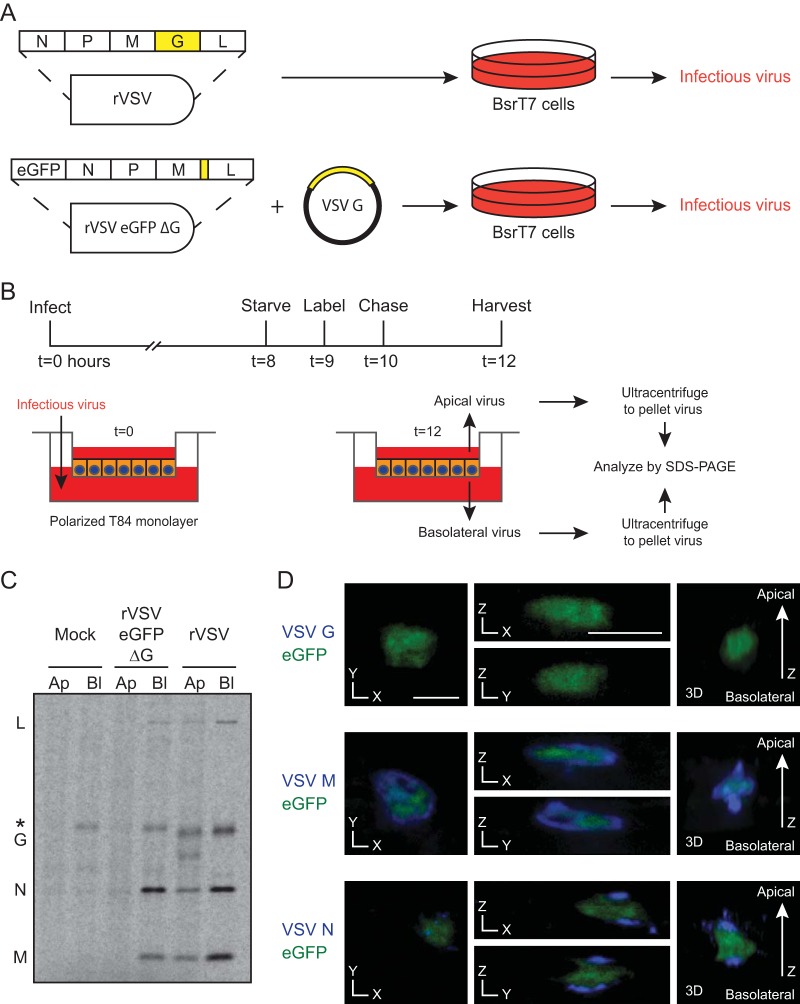
Since G was the only polarized VSV protein, we tested whether its absence might alter VSV budding. We used an rVSV with the G coding sequence deleted and enhanced green fluorescent protein (eGFP) in the first position (rVSV eGFP ΔG). This virus was grown in BSR T7/5 cells with G supplied by transfection (Fig. 2A). To assay budding in T84 monolayers, newly synthesized rVSV eGFP ΔG virions from the apical and basolateral chambers were radiolabeled (Fig. 2B) and analyzed via low-bis SDS-PAGE and autoradiography (Fig. 2C) with the N protein band quantified by ImageQuant TL v7 (GE Healthcare, Piscataway, NJ). This assay revealed that rVSV and rVSV eGFP ΔG preferentially bud basolaterally. To determine if this bias was due to the relative basolateral and apical surface areas, these were measured with ultrathin-section transmission electron microscopy (TEM) of cells (15) and ImageJ (U.S. National Institutes of Health, Bethesda, MD; http://rsb.info.nih.gov/ij/). The basolateral-to-apical area ratio was 3.1 ± 0.2 (n = 31) and cannot account for the differential budding seen with both viruses (Fig. 2C). We confirmed that G deletion does not alter the localization of other viral proteins (Fig. 2D). These data demonstrate that G does not dictate the preferential site of virus production (5).
FIG 2.
Test of the requirement of G for directional budding. (A) Schematic for growth of rVSV and rVSV eGFP ΔG with G supplied in trans. (B) Time line of infection, radiolabeling, and collection. (C) Concentrated virus from 12 hpi was analyzed by SDS-PAGE. Note that 1/20 of the rVSV, relative to rVSV eGFP ΔG, from basolateral (Bl) budding was loaded for clarity. The basolateral lane of rVSV eGFP ΔG contains a cellular band (*) with mobility similar to that of G that also appears after mock infection. Both rVSV and rVSV eGFP ΔG had significantly more N in the basolateral chamber (Student's t test P values of <0.01 and <0.05, respectively; n = 5). Ap, apical. (D) T84 cells were infected with rVSV eGFP ΔG and analyzed by confocal microscopy for the proteins indicated. Cross sections and three-dimensional renderings are shown. The lack of G was verified by immunocytochemistry analysis, and consequent defective virus production was confirmed by titration (data not shown). The scale bars represent 10 μm.
We tested whether preferential budding was due to an inability of VSV to assemble and bud from the apical surface. Titration of supernatant from the apical surface revealed infectious virions (Fig. 1B). Particles released from both surfaces were indistinguishable by negative-stain TEM (Fig. 3A). To examine budding at the plasma membrane, we limited particle release by using a clone harboring mutations in the M late domains (rVSV M LD−), i.e., PPPY and PSAP mutated to AAPA and AAAA, respectively (16, 17). Ultrathin-section TEM of cells infected with rVSV M LD− revealed bullet-shaped virions on both surfaces (Fig. 3A).
FIG 3.
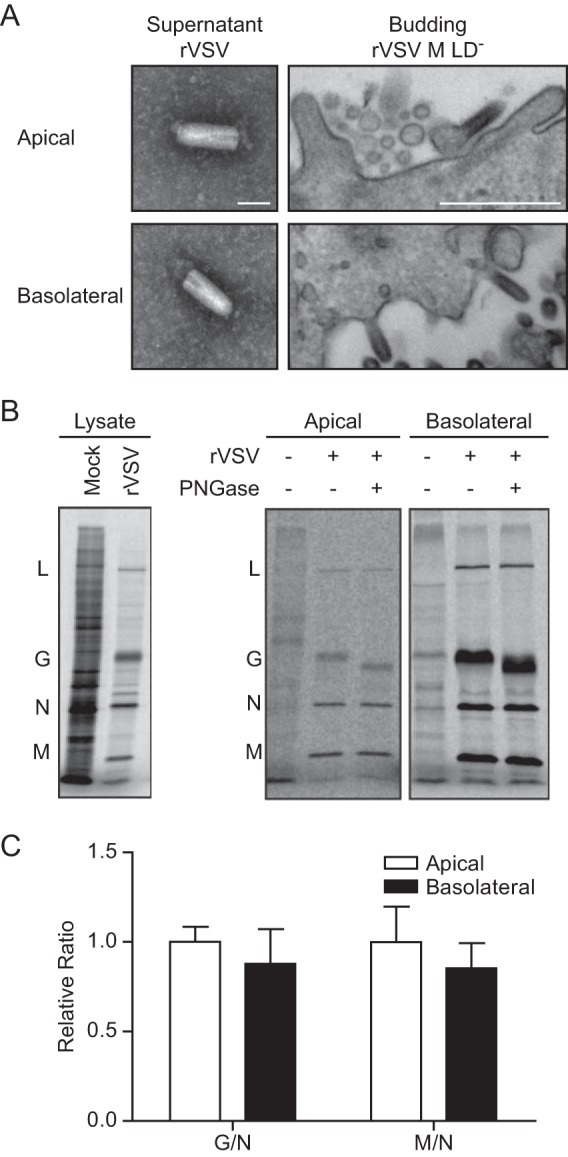
Characterization of virions budded from the apical and basolateral surfaces. (A) Virions from the supernatants of the apical and basolateral compartments of infected T84 cell monolayers at 10 to 12 hpi were imaged by negative-stain TEM. The scale bar represents 100 nm. Monolayers of cells infected with rVSV M LD− were imaged by ultrathin-section TEM, and the apical and basolateral surfaces are shown. The scale bar represents 500 nm. (B) The rVSV from T84 monolayers was analyzed as described in the legend to Fig. 2, except that the pelleted virus was treated with PNGase prior to SDS-PAGE analysis. (C) The G/N and M/N ratios were calculated from non-PNGase-treated virus, and those of apical and basolateral viruses were compared. The differences were not significant by a paired Student t test (P > 0.2; n = 5; the standard error of the mean is indicated).
We next examined the composition of particles from both cell surfaces. Although glycosylation has been shown not to determine the segregation of G (18), it is possible that G glycosylation differs in virions released from the two surfaces. Peptide-N-glycosidase F (PNGase) treatment of particles from the apical and basolateral compartments showed similar glycosylations (Fig. 3B). Using N as a measure of particles, the average amounts of G and M per particle were not found to differ significantly (Fig. 3C) and the relative numbers of basolateral and apical particles per PFU were not significantly different (2.3-fold ± 1.1-fold; P > 0.05 [Student's t test]; n = 5).
The factors that trigger VSV assembly and budding are not known. Virions can be produced without G, demonstrating that G is dispensable for assembly (19). However, sequences in G increase budding efficiency (20, 21), and viral ribonucleoprotein (vRNP) localization to G microdomains has been proposed to initiate assembly (22). In addition, the trigger for condensation of the open vRNP into the nucleocapsid-M protein complex is unknown. We show that budding is polarized in the presence or absence of G and that the localization of other VSV proteins cannot account for this phenotype. Additionally, we demonstrate functional VSV assembly at the apical surface, which does not generate defective particles. We hypothesize that the preferential budding is due to an increase in the efficiency of initiation or completion of assembly at the basolateral surface. A cellular protein(s) required for efficient condensation could be localized basolaterally, or a structural feature, such as tension, of the basolateral region could be more permissive to assembly. The requirements for clathrin-mediated endocytosis differ at the apical and basolateral surfaces in polarized epithelial cells because of tension differences (23). It is possible that generation of the high membrane curvature in VSV particles requires a low-tension environment.
ACKNOWLEDGMENTS
We thank Maria Ericsson, Elizabeth Benecchi, and Louise Trakimas for their expertise and preparation of TEM samples at the Harvard Medical School Cell Biology Conventional Electron Microscopy Facility. We thank Wayne Lencer for his expertise on epithelial cells and Meredith O'Hear for the preparation of T84 cells at the Children's Hospital, Boston, Epithelial Cell Biology Core. The anti-N (10G4), anti-M (23H12) and anti-G (IE2) antibodies were kind gifts from Douglas Lyles. We thank the staff of the Nikon Imaging Center at Harvard Medical School, Boston, for their expertise and use of their equipment.
This work was supported by National Institutes of Health grants R01 NS083848 (C.L.C.) and F31 AG041582 (E.D.) and the Natural Sciences and Engineering Research Council of Canada (T.K.S.).
REFERENCES
- 1.Rodriguez Boulan E, Sabatini DD. 1978. Asymmetric budding of viruses in epithelial monlayers [sic]: a model system for study of epithelial polarity. Proc Natl Acad Sci U S A 75:5071–5075. doi: 10.1073/pnas.75.10.5071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Owens RJ, Dubay JW, Hunter E, Compans RW. 1991. Human immunodeficiency virus envelope protein determines the site of virus release in polarized epithelial cells. Proc Natl Acad Sci U S A 88:3987–3991. doi: 10.1073/pnas.88.9.3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schlie K, Maisa A, Freiberg F, Groseth A, Strecker T, Garten W. 2010. Viral protein determinants of Lassa virus entry and release from polarized epithelial cells. J Virol 84:3178–3188. doi: 10.1128/JVI.02240-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pfeiffer S, Fuller SD, Simons K. 1985. Intracellular sorting and basolateral appearance of the G protein of vesicular stomatitis virus in Madin-Darby canine kidney cells. J Cell Biol 101:470–476. doi: 10.1083/jcb.101.2.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zimmer G, Zimmer KP, Trotz I, Herrler G. 2002. Vesicular stomatitis virus glycoprotein does not determine the site of virus release in polarized epithelial cells. J Virol 76:4103–4107. doi: 10.1128/JVI.76.8.4103-4107.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stevenson BR, Siliciano JD, Mooseker MS, Goodenough DA. 1986. Identification of ZO-1: a high molecular weight polypeptide associated with the tight junction (zonula occludens) in a variety of epithelia. J Cell Biol 103:755–766. doi: 10.1083/jcb.103.3.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yeaman C, Grindstaff KK, Nelson WJ. 1999. New perspectives on mechanisms involved in generating epithelial cell polarity. Physiol Rev 79:73–98. [DOI] [PubMed] [Google Scholar]
- 8.Ellis MA, Potter BA, Cresawn KO, Weisz OA. 2006. Polarized biosynthetic traffic in renal epithelial cells: sorting, sorting, everywhere. Am J Physiol Renal Physiol 291:F707–F713. doi: 10.1152/ajprenal.00161.2006. [DOI] [PubMed] [Google Scholar]
- 9.Fuller S, von Bonsdorff CH, Simons K. 1984. Vesicular stomatitis virus infects and matures only through the basolateral surface of the polarized epithelial cell line, MDCK. Cell 38:65–77. doi: 10.1016/0092-8674(84)90527-0. [DOI] [PubMed] [Google Scholar]
- 10.Lefrancois L, Lyles DS. 1982. The interaction of antibody with the major surface glycoprotein of vesicular stomatitis virus. I. Analysis of neutralizing epitopes with monoclonal antibodies. Virology 121:157–167. [PubMed] [Google Scholar]
- 11.Vandepol SB, Lefrancois L, Holland JJ. 1986. Sequences of the major antibody binding epitopes of the Indiana serotype of vesicular stomatitis virus. Virology 148:312–325. doi: 10.1016/0042-6822(86)90328-4. [DOI] [PubMed] [Google Scholar]
- 12.Heinrich BS, Cureton DK, Rahmeh AA, Whelan SP. 2010. Protein expression redirects vesicular stomatitis virus RNA synthesis to cytoplasmic inclusions. PLoS Pathog 6:e1000958. doi: 10.1371/journal.ppat.1000958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bergmann JE, Fusco PJ. 1988. The M protein of vesicular stomatitis virus associates specifically with the basolateral membranes of polarized epithelial cells independently of the G protein. J Cell Biol 107:1707–1715. doi: 10.1083/jcb.107.5.1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thomas DC, Brewer CB, Roth MG. 1993. Vesicular stomatitis virus glycoprotein contains a dominant cytoplasmic basolateral sorting signal critically dependent upon a tyrosine. J Biol Chem 268:3313–3320. [PubMed] [Google Scholar]
- 15.Maupin P, Pollard TD. 1983. Improved preservation and staining of HeLa cell actin filaments, clathrin-coated membranes, and other cytoplasmic structures by tannic acid-glutaraldehyde-saponin fixation. J Cell Biol 96:51–62. doi: 10.1083/jcb.96.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jayakar HR, Murti KG, Whitt MA. 2000. Mutations in the PPPY motif of vesicular stomatitis virus matrix protein reduce virus budding by inhibiting a late step in virion release. J Virol 74:9818–9827. doi: 10.1128/JVI.74.21.9818-9827.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Obiang L, Raux H, Ouldali M, Blondel D, Gaudin Y. 2012. Phenotypes of vesicular stomatitis virus mutants with mutations in the PSAP motif of the matrix protein. J Gen Virol 93:857–865. doi: 10.1099/vir.0.039800-0. [DOI] [PubMed] [Google Scholar]
- 18.Green RF, Meiss HK, Rodriguez-Boulan E. 1981. Glycosylation does not determine segregation of viral envelope proteins in the plasma membrane of epithelial cells. J Cell Biol 89:230–239. doi: 10.1083/jcb.89.2.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takada A, Robison C, Goto H, Sanchez A, Murti KG, Whitt MA, Kawaoka Y. 1997. A system for functional analysis of Ebola virus glycoprotein. Proc Natl Acad Sci U S A 94:14764–14769. doi: 10.1073/pnas.94.26.14764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schnell MJ, Buonocore L, Boritz E, Ghosh HP, Chernish R, Rose JK. 1998. Requirement for a non-specific glycoprotein cytoplasmic domain sequence to drive efficient budding of vesicular stomatitis virus. EMBO J 17:1289–1296. doi: 10.1093/emboj/17.5.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robison CS, Whitt MA. 2000. The membrane-proximal stem region of vesicular stomatitis virus G protein confers efficient virus assembly. J Virol 74:2239–2246. doi: 10.1128/JVI.74.5.2239-2246.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Swinteck BD, Lyles DS. 2008. Plasma membrane microdomains containing vesicular stomatitis virus M protein are separate from microdomains containing G protein and nucleocapsids. J Virol 82:5536–5547. doi: 10.1128/JVI.02407-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boulant S, Kural C, Zeeh JC, Ubelmann F, Kirchhausen T. 2011. Actin dynamics counteract membrane tension during clathrin-mediated endocytosis. Nat Cell Biol 13:1124–1131. doi: 10.1038/ncb2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Piccinotti S, Kirchhausen T, Whelan SP. 2013. Uptake of rabies virus into epithelial cells by clathrin-mediated endocytosis depends upon actin. J Virol 87:11637–11647. doi: 10.1128/JVI.01648-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schott DH, Cureton DK, Whelan SP, Hunter CP. 2005. An antiviral role for the RNA interference machinery in Caenorhabditis elegans. Proc Natl Acad Sci U S A 102:18420–18424. doi: 10.1073/pnas.0507123102. [DOI] [PMC free article] [PubMed] [Google Scholar]