ABSTRACT
In 2012, the first cases of infection with the Middle East respiratory syndrome coronavirus (MERS-CoV) were identified. Since then, more than 1,000 cases of MERS-CoV infection have been confirmed; infection is typically associated with considerable morbidity and, in approximately 30% of cases, mortality. Currently, there is no protective vaccine available. Replication-competent recombinant measles virus (MV) expressing foreign antigens constitutes a promising tool to induce protective immunity against corresponding pathogens. Therefore, we generated MVs expressing the spike glycoprotein of MERS-CoV in its full-length (MERS-S) or a truncated, soluble variant of MERS-S (MERS-solS). The genes encoding MERS-S and MERS-solS were cloned into the vaccine strain MVvac2 genome, and the respective viruses were rescued (MVvac2-CoV-S and MVvac2-CoV-solS). These recombinant MVs were amplified and characterized at passages 3 and 10. The replication of MVvac2-CoV-S in Vero cells turned out to be comparable to that of the control virus MVvac2-GFP (encoding green fluorescent protein), while titers of MVvac2-CoV-solS were impaired approximately 3-fold. The genomic stability and expression of the inserted antigens were confirmed via sequencing of viral cDNA and immunoblot analysis. In vivo, immunization of type I interferon receptor-deficient (IFNAR−/−)-CD46Ge mice with 2 × 105 50% tissue culture infective doses of MVvac2-CoV-S(H) or MVvac2-CoV-solS(H) in a prime-boost regimen induced robust levels of both MV- and MERS-CoV-neutralizing antibodies. Additionally, induction of specific T cells was demonstrated by T cell proliferation, antigen-specific T cell cytotoxicity, and gamma interferon secretion after stimulation of splenocytes with MERS-CoV-S presented by murine dendritic cells. MERS-CoV challenge experiments indicated the protective capacity of these immune responses in vaccinated mice.
IMPORTANCE Although MERS-CoV has not yet acquired extensive distribution, being mainly confined to the Arabic and Korean peninsulas, it could adapt to spread more readily among humans and thereby become pandemic. Therefore, the development of a vaccine is mandatory. The integration of antigen-coding genes into recombinant MV resulting in coexpression of MV and foreign antigens can efficiently be achieved. Thus, in combination with the excellent safety profile of the MV vaccine, recombinant MV seems to constitute an ideal vaccine platform. The present study shows that a recombinant MV expressing MERS-S is genetically stable and induces strong humoral and cellular immunity against MERS-CoV in vaccinated mice. Subsequent challenge experiments indicated protection of vaccinated animals, illustrating the potential of MV as a vaccine platform with the potential to target emerging infections, such as MERS-CoV.
INTRODUCTION
In November 2012, a novel coronavirus was identified for the first time in a patient from Saudi Arabia who presented with severe respiratory disease. Later, this virus was termed Middle East respiratory syndrome coronavirus (MERS-CoV) (1). By 26 December 2014, 938 laboratory-confirmed cases of MERS-CoV, mostly from Saudi Arabia and neighboring countries, had been diagnosed and had resulted in 343 casualties (2). A few cases of MERS-CoV were also detected in the United States, the United Kingdom, Netherlands, Austria, France, Greece, Italy, and Germany, indicating the virus's principal potential for spread (2). Fortunately, direct transmission upon contact with human patients seems to be limited, yet is still possible, as determined by analysis of household contact infections among MERS patients' families (3) and as evidenced by a recent cluster of MERS infections in South Korea, with 166 cases between 20 May and 19 June 2015, including 106 third-generation and 11 fourth-generation cases (4, 5). As a natural reservoir, dromedary camels have been identified as the most likely source, based on partially identical genomes detected in viruses isolated from humans or camels (6, 7). Additionally, antibodies against the spike glycoprotein of MERS-CoV with virus-neutralizing capacity were detected in camels (8–10), and infection of humans with MERS-CoV have been reported after contact with infected camels (11, 12). Interestingly, while all other members of the C lineage of the Betacoronavirus genus have been found in different bat species (13, 14), only closely related, most likely precursor viruses of MERS-CoV have been identified in Neoromicia capensis bats (15). Thus, MERS-CoV has a zoonotic origin, but sustained infections, the severity of the disease, and the risk of virus adaption to gain efficient human-to-human transmission mandates the development of effective vaccines to combat local infections and to be prepared for the eventual occurrence of a global pandemic, as previously observed with severe acute respiratory syndrome coronavirus (SARS-CoV) in 2003 (16).
Occurring 10 years before the current MERS-CoV epidemic, SARS-CoV was the first Betacoronavirus of zoonotic origin with potentially fatal outcomes in human patients (1). Experimental vaccines protecting animal models against SARS have been developed (17–19), and the properties of such SARS vaccines may be applicable to vaccines that should protect against MERS-CoV infections. Both neutralizing antibodies and T cell responses are essential for prevention of SARS-CoV infection (17, 18). The spike protein (S), a coronavirus class I fusion protein (20, 21), has been identified as the most immunogenic antigen of SARS-CoV, as it induces a strong humoral as well as cellular immune response (17, 19). Similarly, MERS-S constructs expressed by recombinant modified vaccinia virus Ankara or recombinant adenoviral vectors have already been demonstrated to induce neutralizing antibodies (22, 23). The detected neutralizing capacity of induced antibodies is expected, since the receptor-binding domain (RBD) in the S1 domain of both SARS-CoV and MERS-CoV S proteins mediate host-cell receptor binding as a prerequisite for cell entry (24, 25). Thus, S1 is the main target of neutralizing antibodies (26). Also the RBD of MERS-CoV-S alone has been demonstrated to induce strong neutralizing antibody titers (23, 27–31). In combination with different adjuvants, even induction of T cell responses by the recombinant RBD has been described (31). Thus, a prototypic MERS vaccine should be based on MERS-S expression, since the induction of neutralizing antibodies has been shown to be a direct correlate of protection in cases of SARS-CoV (32).
The measles vaccine is an efficient, live attenuated, replicating virus that induces both humoral and cellular immune responses, has an excellent safety record, and probably provides lifelong protection (33, 34). The vaccine's manufacturing process is extremely well established (35), and millions of doses can be generated quite easily and quickly. Generation of recombinant measles virus (MV) from DNA via reverse genetics is feasible (35) and allows the insertion of additional transcription units (ATU) by duplication of sequences terminated by start and stop sequences (36). Hence, genes expressing foreign antigens up to 6 kb can be cloned into the MV backbone (36) and elicit coexpression of MV proteins and inserted genes. Besides marker genes (37) or immune modulators (38), expression of antigens from foreign pathogens like hepatitis B or C virus (39, 40), HIV (41), West Nile virus (WNV) (42, 43), dengue virus (44), Chikungunya virus (CHIKV) (45), or SARS-CoV (19) by recombinant MVs has already been demonstrated. Thereby, robust immune responses against vector and foreign antigens are induced after vaccination of transgenic, MV-susceptible type I interferon receptor-deficient (IFNAR−/−)-CD46Ge mice (46) or nonhuman primates with recombinant MVs, in general. In particular, protection of vaccinated animals from lethal challenge with WNV (42) or CHIKV (45) was demonstrated and indicated the high efficacy of the system. Interestingly, prevaccinated animals with protective immunity against measles were still amendable to vaccination with the recombinant MV, since significant immune responses against the foreign antigen(s) are consistently induced (41, 45), and the MV-based CHIKV vaccine demonstrated efficacy in phase I trials irrespective of measles virus immunity (47).
Here, we aimed to utilize the efficacy of the MV vaccine platform to generate a live attenuated vaccine against MERS-CoV based on recombinant MVvac2. This recombinant virus reflects the MV vaccine strain Moraten (48), which has been authorized for vaccination against measles. As the antigen, we choose the MERS-CoV S glycoprotein to induce neutralizing antibodies and robust cellular immunity. Two variants of the glycoprotein were analyzed as antigen: the full-length, membrane-anchored MERS-S, and a truncated, soluble form lacking the transmembrane domain (MERS-solS). Both variants include the S1 domain as a target structure. The soluble protein variant should be taken up better by B cells (49–51) and thus should induce humoral immune responses more efficiently (52), potentially boosting virus-neutralizing antibody titers (VNTs). The respective genes were inserted into two different positions of the MV genome to modulate expression of the antigens, and all recombinant MVs were successfully rescued. Cells infected with such viruses expressed the desired antigens. Indeed, immunization of IFNAR−/−-CD46Ge mice induced strong humoral and cellular immune responses directed against MV and MERS-CoV S which were sufficient to protect vaccinated animals from MERS-CoV infection. Thus, MV platform-based vaccines are a powerful option to develop a prepandemic vaccine against MERS-CoV.
MATERIALS AND METHODS
Cells.
Vero cell (African green monkey kidney cells; ATCC CCL-81), 293T cell (ATCC CRL-3216), and EL4 mouse T cell (ATCC TIB-39) lines were purchased from ATCC (Manassas, VA, USA) and cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS; Biochrom, Berlin, Germany) and 2 mM l-glutamine (l-Gln; Biochrom). JAWSII dendritic cells (ATCC CRL-11904) were purchased from ATCC and cultured in minimal essential medium alpha (MEM-α) with ribonucleosides and deoxyribonucleosides (Gibco BRL, Eggenstein, Germany) supplemented with 20% FBS, 2 mM l-Gln, 1 mM sodium pyruvate (Biochrom), and 5 ng/ml murine granulocyte-macrophage colony-stimulating factor (GM-CSF; Peprotech, Hamburg, Germany). DC2.4 and DC3.2 murine dendritic cell lines (53) were cultured in RPMI containing 10% FBS, 2 mM l-Gln, 1% nonessential amino acids (Biochrom), 10 mM HEPES (pH 7,4), and 50 μM 2-mercaptoethanol (Sigma-Aldrich, Steinheim, Germany). All cells were cultured at 37°C in a humidified atmosphere containing 6% CO2 for a maximum of 6 months of culture after thawing of the original stock.
Plasmids.
The codon-optimized gene encoding MERS-CoV-S (GenBank accession number JX869059) flanked with AatII/MluI binding sites in plasmid pMA-RQ-MERS-S was obtained by gene synthesis (Invitrogen Life Technologies, Regensburg, Germany). A truncated form of MERS-S lacking the transmembrane domain was amplified by PCR, flanked with AatII/MluI binding sites, and fully sequenced. Both antigens, as well as the immediate early cytomegalovirus (CMV) promoter (54), were inserted into p(+)BR-MVvac2-GFP(H) or p(+)MVvac2-ATU(P) (48) via AatII/MluI or SfiI/SacII, respectively, to generate p(+)PolII-MVvac2-MERS-S(H), p(+)PolII-MVvac2-MERS-S(P), p(+)PolII-MVvac2-MERS-solS(H), and p(+)PolII-MVvac2-MERS-solS(P), respectively. For construction of lentiviral transfer vectors encoding the MERS-CoV antigens, the open reading frame (ORF) of MERS-S was amplified by PCR with primers encompassing flanking NheI/XhoI restriction sites and template pMA-RQ-MERS-S. Details on primers and PCR procedures are available upon request. PCR products were cloned into pCR2.1-TOPO (Invitrogen Life Technologies) and fully sequenced. Intact antigen ORF was cloned into pCSCW2gluc-IRES-GFP (55) by using NheI/XhoI restriction sites to yield pCSCW2-MERS-S-IRES-GFP.
Production of lentiviral vectors.
Viral vectors were produced using 293T cells and polyethylenimine (PEI; Sigma-Aldrich) transfection (56). A total of 1 × 107 293T cells were seeded per 175-cm2 cell culture flask and cultured overnight. To produce lentivirus vectors pseudotyped with the G protein of vesicular stomatitis virus (VSV-G), these cells were transfected using a standard three-plasmid lentivirus vector system. Cells were transfected with 17.5 μg pCSCW2-MERS-S-IRES-GFP transfer vector, 6.23 μg pMD2.G, and 11.27 μg pCMVΔR8.9 (57), as described previously (58). The medium was exchanged 1 day posttransfection, and HIVMERS-S-IRES-GFP(VSV-G) vector particles were harvested 2 and 3 days after transfection. For harvest of vector particles, the supernatants of three culture flasks were filtered (0.45-μm pore size), pooled, and concentrated by centrifugation (100,000 × g, 3 h, 4°C). Pellets were resuspended in DMEM and stored at −80°C.
Generation of antigen-expressing cell lines.
Syngeneic target cells based on the C57BL/6-derived DC lines JAWSII, DC2.4, and DC3.2, as well as T cell line EL-4 were transduced with HIVMERS-S-IRES-GFP(VSV-G) vector-containing supernatant to express MERS-S and the green marker protein GFP (JAWSIIgreen-MERS-S, EL-4green-MERS-S, DC2.4green-MERS-S, and DC3.2green-MERS-S), thereby presenting respective peptides via major histocompatibility complex class I (MHC-I). EL-4 cells were alternatively transduced with HIVTurboFP635(VSV-G) vectors (59) to express red fluorescent Katushka protein as a negative control (EL-4red). For this purpose, 1 × 105 target cells were seeded in 24-well plates and transduced with 0.1, 1, or 10 μl of concentrated vector suspension. For analysis of transduction efficiencies, cells were fixed in 1% paraformaldehyde (Merck Millipore, Darmstadt, Germany), and the percentages of GFP-positive or Katushka-positive cells were quantified by flow cytometry using an LSRII flow cytometer (BD, Heidelberg, Germany). Cell populations revealing a 1 to 10% fraction of GFP-positive cells were used for single-cell cloning by limiting dilution. For that purpose, cell dilutions with 50 μl conditioned medium statistically containing 0.3 cells were seeded per well in 96-well plates. Single-cell clones were cultured and analyzed by flow cytometry. GFP-positive clones were selected for further analysis.
Viruses.
The viruses were rescued as described previously (54). In brief, 5 μg of MV genome plasmids with MERS-CoV antigen ORFs were cotransfected with plasmids pCA-MV-N (0.4 μg), pCA-MV-P (0.1 μg), and pCA-MV-L (0.4 μg) encoding MV proteins necessary for genome replication and expression. These plasmids were cotransfected into 293T cells cultured in 6-well plates by using Lipofectamine 2000 (Invitrogen Life Technologies). The transfected 293T cells were overlaid 2 days after transfection onto 50% confluent Vero cells seeded in 10 cm-dishes. Overlay cultures were closely monitored for isolated syncytia, which indicated monoclonal replicative centers. Single syncytia were picked and overlaid onto 50% confluent Vero cells cultured in 6-well plates and harvested as passage 0 (P0) by scraping and a freeze-thaw cycle of cells at the time of maximal infection. Subsequent passages were generated after titration to determine the 50% tissue culture infective dose (TCID50) of infectious virus according to the method of Kaerber and Spaerman (60) and infection of Vero cells at a multiplicity of infeciton (MOI) of 0.03. The viruses were passaged up to P10. MERS vaccine viruses and control viruses MVvac2-GFP(H) and MVvac2-GFP(P) in P3 were used for characterization, and viruses in P4 were used for vaccination. MERS-CoV (isolate EMC/2012) (1) was used for neutralization assays, and challenge virus was propagated in Vero cells and titrated as described above for recombinant MV. All virus stocks were stored in aliquots at −80°C.
Measles virus genome sequence analysis.
The RNA genomes of recombinant MV in P3 or P10 were isolated using the QIAamp RNeasy kit (QIAgen, Hilden, Germany) according to the manufacturer's instructions and resuspended in 50 μl RNase-free water. Viral cDNA was reverse transcribed using the SuperScript II reverse transcription (RT) kit (Invitrogen) with 2 μl viral RNA (vRNA) as the template and random hexamer primers, according to manufacturer's instructions. For specific amplification of antigen ORFs, the respective genomic regions of recombinant MVs were amplified by PCR with primers binding to sequences flanking the regions of interest and cDNA as the template. Detailed descriptions of primers and procedures are available upon request. The PCR products were directly sequenced (Eurofins Genomics, Ebersberg, Germany).
Western blot analysis.
For Western blot analysis, cells were lysed and immunoblotted as previously described (61). A rabbit anti-MERS-CoV serum (1:1,000) was used as the primary antibody for MERS-CoV-S, and a rabbit anti-MV-N polyclonal antibody (1:25,000; Abcam, Cambridge, United Kingdom) was used for MV-N detection. A donkey horseradish peroxidase (HRP)-coupled anti-rabbit IgG (H&L) polyclonal antibody (1:10,000; Rockland, Gilbertsville, PA) served as the secondary antibody for both. Peroxidase activity was visualized with an enhanced chemiluminescence (ECL) detection kit (Thermo Scientific, Bremen, Germany) on Amersham ECL hyperfilm (GE Healthcare, Freiburg, Germany).
Production of recombinant soluble MERS-CoV spike protein.
The S protein lacking the transmembrane domain was genetically tagged with six His residues at its carboxy terminus. The resulting construct was inserted into a Semliki forest virus-derived self-replicating RNA vector (SFV replicon) downstream of the subgenomic promoter. These replicons were transcribed in vitro and purified as previously described (62, 63). The integrity of purified replicon was assessed by on-chip electrophoresis (2100 BioAnalyzer; Agilent, Santa Clara, CA). To produce SFV vector particles, replicon RNA and helper RNA were coelectroporated into BHK21 cells by using a square-wave electroporator (one pulse, 750 V/cm for 16 ms; BTX ECM 830; Harvard Apparatus, Holliston, MA). Particles were harvested after 24 h, frozen in N2 (liquid), and stored at −80°C. For protein production, 2 × 107 BHK21 cells were transduced with SFV particles (MOI, 40) and harvested after 24 h. Cell pellets were lysed (phosphate-buffered saline [PBS], 0.2% Triton X-100, protease inhibitor cocktail [Roche]) for 30 min at 4°C. Afterwards, cells were sonicated, and lysates were cleared by centrifugation (30 min, 21,000 × g, 4°C). The supernatant was filtered (0.2 μm), loaded on a HisTrap high-performance (HP) column (17-5247-01; GE Healthcare), and washed with 10 volumes of binding buffer (20 mM Na2HPO4, 0.5 M NaCl, 10 mM imidazole). S protein was eluted with a gradient of binding buffer containing 0.5 M imidazole followed by buffer exchange to PBS. Protein integrity was checked by Western blotting with a mouse anti-His monoclonal antibody (MAb; 1:50; Dianova, Germany).
Animal experiments.
All animal experiments were carried out in compliance with the regulations of German animal protection laws and as authorized by the RP Darmstadt. Six- to 12-week-old IFNAR−/−-CD46Ge mice expressing human CD46 were inoculated intraperitoneally (i.p.) with 1 × 105 TCID50 of recombinant MV or 200 μl Opti-MEM on days 0 and 28 and bled via the retrobulbar route on days 7, 28, 32, and 49 postinfection (p.i.) under anesthesia. Serum samples were stored at −20°C. Mice were euthanized on day 32 or 49 p.i., and spleens were isolated. For challenge experiments, immunized mice were transduced intranasally (i.n.) on day 63 with 20 μl of an adenovirus vector encoding human DPP4 and mCherry with a final titer of 2.5 × 108 PFU per inoculum (AdV-hDPP4; ViraQuest Inc.) and challenged i.n. with 20 μl of MERS-CoV at a final titer of 7 × 104 TCID50 on day 68. The mice were euthanized 4 days after challenge, and representative left lobe lung samples were prepared for RNA isolation.
Antibody ELISA.
MV bulk antigens (10 μg/ml; Virion Serion, Würzburg, Germany) or recombinant MERS-S protein (20 μg/ml) in 50 μl carbonate buffer (Na2CO3 at 30 mM, NaHCO3 at 70 mM; pH 9.6) was used to coat wells of Nunc Maxisorp 96-well enzyme-linked immunosorbent assay (ELISA) plates (eBioscience) and incubated overnight at 4°C. The plates were washed three times with 150 μl ELISA washing buffer (PBS, 0.1% [wt/vol]Tween 20) and blocked with 50 μl blocking buffer (PBS, 5% bovine serum albumin [BSA], 0.1% Tween 20) for 2 h at room temperature. Mice sera sampled on days −7 or 49 were serially diluted in ELISA dilution buffer (PBS, 1% BSA, 0.1% Tween 20), and 50 μl/well was used for the assay. The plates were incubated at 37°C for 2 h and washed again with ELISA washing buffer. Plates were incubated with 50 μl/well of HRP-conjugated rabbit anti-mouse IgG (Dako; 1:1,000 in ELISA dilution buffer) at room temperature for 1 h. Subsequently, the plates were washed and 100 μl tetramethylbenzidine substrate (eBioscience) was added per well. The reaction was stopped by addition of 50 μl/well H2SO4 (1 N), and the absorbance at 405 nm was measured.
Neutralization assays.
For quantification of VNTs, mouse sera were serially diluted in 2-fold dilutions in DMEM. A total of 50 PFU of MVvac2-GFP(P) or 200 TCID50 of MERS-CoV was mixed with serum dilutions and incubated at 37°C for 1 h. Virus suspensions were added to 1 × 104 Vero cells seeded 4 h prior to assay in 96-well plates and incubated for 4 days at 37°C. VNTs were calculated as the reciprocal of the highest dilution that abolished infection.
ELISpot assays.
Murine gamma interferon (IFN-γ) enzyme-linked immunosorbent spot (ELISpot) assays (eBioscience, Frankfurt, Germany) were performed according to the manufacturer's instructions using multiscreen immunoprecipitation (IP) ELISpot polyvinylidene difluoride 96-well plates (Millipore, Darmstadt, Germany). A total of 5 × 105 splenocytes isolated 4 days after boost immunization were cocultured with 5 × 104 JAWSIIgreen-MERS-S, DC2.4green-MERS-S, or DC3.2green-MERS-S, or untransduced DC cell lines for 36 h in 200 μl RPMI (10% fetal bovine serum [FBS], 2 nM l-Gln, 1% penicillin-streptomycin). Medium alone served as the negative control. Concanavalin A (ConA; Sigma-Aldrich) at 10 μg/ml was used for demonstration of splenocyte reactivity. Recombinant MV bulk antigens (Virion Serion) at 10 μg/ml were used to analyze MV-specific immune responses in vaccinated animals. Cells were removed from the plates, and the plates were incubated with biotin-conjugated anti-IFN-γ antibodies and avidin-HRP according to the manufacturer's instructions. 3-Amino-9-ethyl-carbazole (AEC; Sigma-Aldrich) substrate solution for development of spots was prepared according to the manufacturer's instructions using AEC dissolved in N,N-dimethylformamide (Merck Millipore). Spots were counted using an Eli.Scan ELISpot scanner (AE.L.VIS, Hamburg, Germany) and ELISpot analysis software (AE.L.VIS).
T cell proliferation assay.
Splenocytes isolated 3 weeks after booster immunization were labeled with 0.5 μM carboxyfluorescein succinimidyl ester (CFSE; eBioscience) as previously described (64). In brief, 5 × 105 labeled cells were seeded in RPMI 1640 supplemented with 10% mouse serum, 2 nM l-glutamine, 1 mM HEPES, 1% penicillin-streptomycin, and 100 μM 2-mercaptoethanol in 96-well plates. Aliquots of 200 μl of medium containing ConA (10 μg/ml), MV bulk antigens (10 μg/ml), or 5 × 103 JAWSIIgreen-MERS-S cells were added to each well, and cells were cultured for 6 days. Medium and untransduced JAWSII cells served as controls. Stimulated cells were subsequently stained with CD3-PacBlue (1:50; clone 500A2; Invitrogen Life Technologies) and CD8-allophycocyanin (1:100; clone 53-6.7; eBioscience) antibodies and fixed with 1% paraformaldehyde (PFA) in PBS. Stained cells were analyzed by flow cytometry using an LSR II flow cytometer (BD) and FACSDiva software (BD).
Cytotoxic T lymphocyte (CTL) killing assay.
For restimulation of T cells isolated 4 days after booster immunization, 5 × 106 splenocytes were cocultured with 5 × 104 JAWSIIgreen-MERS-S cells for 6 days in 12-well plates in RPMI 1640 supplemented with 10% FBS, 2 nM l-glutamine, 1 mM HEPES, 1% penicillin-streptomycin, 2-mercaptoethanol (100 μM), and 100 U/ml recombinant interleukin-2 (rIL-2; murine, Peprotech). A total of 2 × 103 EL-4red cells were labeled with 0.5 μM CFSE and mixed with 8 × 103 EL-4green-MERS-S cells per well. Splenocytes were counted and cocultured with EL-4 target cells at the indicated ratios for 4 h. Afterwards, EL-4 cells were labeled with the fixable viability dye eFluor 780 (eBioscience), fixed with 1% PFA, and analyzed by flow cytometry using an LSR II flow cytometer (BD) and FACSDiva software (BD). For determination of the antigen:NC EL-4 ratio, cell counts of living MERS-S-expressing cells were divided by the counts for living negative controls.
Determination of viral RNA copy numbers and infectious virus in mouse tissue.
Samples of immunized and challenged mice (6- by 6-mm tissue slices of approximately 0.035 ± 0.011 g [mean ± standard deviation]) excised from the centers of left lung lobes were homogenized in 1 ml DMEM with ceramic beads (diameter, 1.4 mm) in a FastPrep SP120 instrument three times for 40 s at 6.5 m/s. The homogenate was centrifuged for 3 min at 2,400 rpm in a Mikro 200R centrifuge (Hettich Lab Technology) to remove tissue debris. Live virus titers in supernatant (in TCID50 per milliliter) were determined on Vero cells as described above. Aliquots of 100 μl of the supernatants were used for RNA isolation with the RNeasy minikit (Qiagen) according to the manufacturer's instructions. The RNA amount was measured with the NanoDrop ND-100 spectrophotometer. Total RNA was reverse transcribed and quantified by real-time PCR using the SuperScript III OneStep RT-PCR System (Invitrogen Life Technologies) as described previously (65) with the primer pair upE-Fwd and upE-Rev and the probe upE-Prb on an ABI7900 high-throughput fast real-time PCR system (Life Technologies Instruments).
Additionally, for every sample of the transduced and infected mice, evidence for successful hDPP4 transduction was determined by real-time RT-PCR for mCherry with the OneStep RT-PCR kit on a Rotor Gene Q apparatus (both from Qiagen). Primers and probe (Tib-Molbiol, Berlin, Germany) were as follows: mCherry forward, CATGGTAACGATGAGTTAG; mCherry reverse, GTTGCCTTCCTAATAAGG; mCherry probe, 6-carboxyfluorescein (FAM)–TACCACCTTACTTCCACCAATCGG–BBQ (BlackBerry quencher). Primers and probe were used at final concentrations of 0.4 μM and 0.2 μM, respectively. The quantitative reverse transcription-PCR (qRT-PCR) program was as follows: 50°C for 30 min; 95°C for 15 min; 40 cycles of 95°C for 15 s, 48°C for 30 s, and 72°C for 20 s. All samples for mCherry were evaluated in one run to exclude an impact of different conditions on the results in different runs. Quantification was carried out using a standard curve based on 10-fold serial dilutions of appropriate cloned RNA ranging from 102 to 105 copies. Briefly, PCR fragments were generated using the primers described above. For cloning, the TOPO TA cloning kit with pCR2.1-TOPO plasmid (Invitrogen) and Escherichia coli were used. Inserts were examined for correct orientation and length, amplified with plasmid-specific primers, purified, and transcribed into RNA by using the SP6/T7 transcription kit (Roche).
Histopathological and immunohistochemical examination of lung tissue.
Lungs of vaccinated and mock-vaccinated mice transduced with AdV-hDPP4 were collected on day 4 postchallenge with MERS-CoV. Tissue was fixed in 4% PFA and embedded in paraffin. Sections were cut with a Leica RM2255 microtome (Leica Biosystems) and stained with hematoxylin and eosin (H&E). For detection of MERS-CoV, a rabbit polyclonal antibody against MERS-CoV spike protein S1 (100208-RP; Sino Biological Inc., Beijing, China) diluted 1:50 was used. To monitor adenovirus transduction, a mouse monoclonal antibody against mCherry (ab125096; Abcam) diluted 1:250 was used after antigen retrieval with target retrieval solution (Dako) for 23 min at 97°C. To block unspecific binding, slides were incubated for 10 min with 20% nonimmune pig serum (MERS-CoV) or for 30 min with 20% nonimmune horse serum (mCherry). Primary antibodies were incubated overnight at 4°C. A pig anti-rabbit IgG and a biotinylated horse anti-mouse IgG served as secondary antibodies for MERS-CoV and mCherry, respectively. For detection of antigen-antibody complexes, the ABC method for mCherry and the rabbit PAP method for MERS-CoV were used in combination with diaminobenzidine for staining. Papanicolaou stain was used for counterstaining.
Statistical analysis.
To compare the means of different groups from growth curves, neutralization assays, and ELISpot assays, a nonparametric one-way analysis of variance (one-way ANOVA) was performed. For the proliferation assay, the mean differences between control and vaccinated groups were calculated and analyzed by using an unpaired t test. To all three groups in the CTL killing assays, a linear curve was fitted for antigen versus the log-transformed effector-target ratio (E:T). The P values for differences in slopes were calculated, and MVvac2-MERS-S(H) or MVvac2-MERS-solS(H) was compared with the control, MVvac2-ATU(P). For analysis of challenge data, mean ratios and 95% confidence intervals were calculated based on log-transformed and back-transformed data. The ratio, instead of the difference, was chosen due to the rather log-normal distribution of the data. The widths of the confidence intervals caused high variability of the data, and limited sample sizes were used (n = 10 observations each). For comparisons between groups, the Wilcoxon two-sample test was used. P values were not adjusted for multiple comparisons due to the explorative character of the study.
RESULTS
Generation and expression of MERS-CoV-S by recombinant MVvac2.
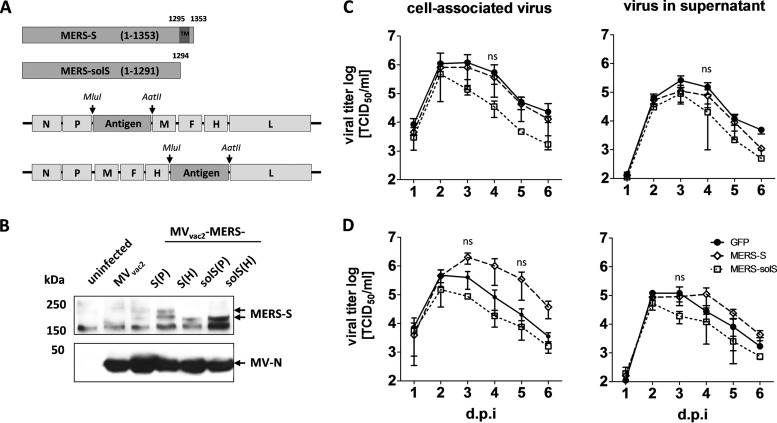
Since the spike protein (S) of SARS-CoV has been shown to potently induce humoral and cellular immune responses, MERS-S was chosen as the appropriate antigen to be expressed by the recombinant MV vaccine platform. In addition to full-length MERS-S, a truncated form lacking the transmembrane and cytoplasmic domains (MERS-solS), was cloned into two different ATUs either behind P (post-P) or H (post-H) cassettes of the vaccine strain MVvac2 genome (Fig. 1A). Virus clones of all recombinant genomes were successfully rescued and amplified up to P10 in Vero cells, with titers of up to 6 × 107 TCID50/ml. The stability of the viral genomes was demonstrated via sequencing of viral genomes after RT-PCR (data not shown). Besides the exclusion of mutations or deletions of the antigen-encoding genes, the verification of antigen expression is essential for vaccine function and, thus, virus characterization. Western blot analysis of Vero cells infected with the different MVvac2-MERS vaccines revealed expression of the antigen (Fig. 1B). Interestingly, the expression of both S and solS was higher when cells were infected with viruses encoding antigens in post-H ATU compared to the post-P constructs. Therefore, growth kinetics were analyzed to check if the insertion or expression of the S antigen variants into or by recombinant MV, respectively, impaired the vaccines' replication (Fig. 1C and D). For that purpose, the vaccine viruses containing the MERS-S or MERS-solS gene in post-H (Fig. 1C) or post-P (Fig. 1D) positions were analyzed in parallel to the corresponding MVvac2-GFP control viruses. MVvac2 encoding full-length, membrane-bound MERS-S grew comparably to the control viruses; only MVvac2-MERS-solS(P) (Fig. 1D) and MVvac2-MERS-solS(H) (Fig. 1C) revealed an approximately 3-fold-reduced maximal virus titer, albeit no statistical significance was observed [1.5 × 105 TCID50/ml for MVvac2-MERS-solS(P) and 4.7 × 105 TCID50/ml for MVvac2-MERS-solS(H) versus 4.7 × 105 for MVvac2-GFP(P) and 1.2 × 106 TCID50/ml for MVvac2-GFP(H)] (Fig. 1C). Thus, cloning and rescue of MVs expressing MERS-CoV antigens, even at the cost of an 4,049 bp additional genome length, were achieved easily and relative quickly. All constructs expressed the inserted antigens without significant impact on viral replication.
FIG 1.
Generation and characterization of MVvac2-MERS-S and MVvac2-MERS-solS. (A) Schematic depiction of full-length MERS-S and a soluble variant lacking the transmembrane and cytoplasmatic region (MERS-solS) (upper schemes) and recombinant MVvac2 genomes used for their expression (lower schemes). Antigen or antigen-encoding genes are depicted in dark gray; MV viral gene cassettes (light gray) are annotated. MluI and AatII restriction sites used for cloning of antigen-encoding genes into post-P or post-H ATU are highlighted (B) Immunoblot analysis of Vero cells infected at an MOI of 0.03 with MVvac2-MERS-S, MVvac2-MERS-solS, or MVvac2-GFP(H) (MVvac2), as depicted above the lanes. Uninfected cells served as mock controls. Blots were probed using rabbit serum reactive against MERS-CoV (upper blot) or mAb reactive against MV-N (lower blot). Arrows indicate specific bands. (C and D) Growth kinetics of recombinant MV on Vero cells infected at an MOI of 0.02 with MVvac2-MERS-S (MERS-S), MVvac2-MERS-solS (MERS-solS), or MVvac2-GFP encoding extra genes in post-H (C) or post-P (D) ATU. Titers of samples prepared at the indicated time points postinfection were determined on Vero cells. Means and standard deviations of three independent experiments are presented. ns, not significant.
Antibodies with neutralizing capacity directed against MV or MERS-CoV are induced by MVvac2-MERS-S and MVvac2-MERS-solS.
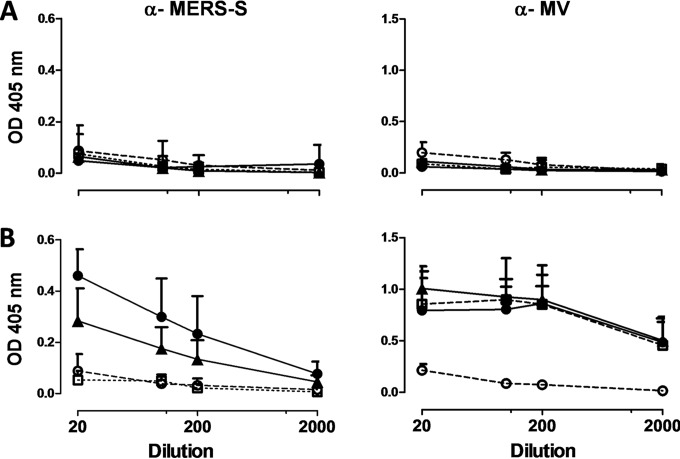
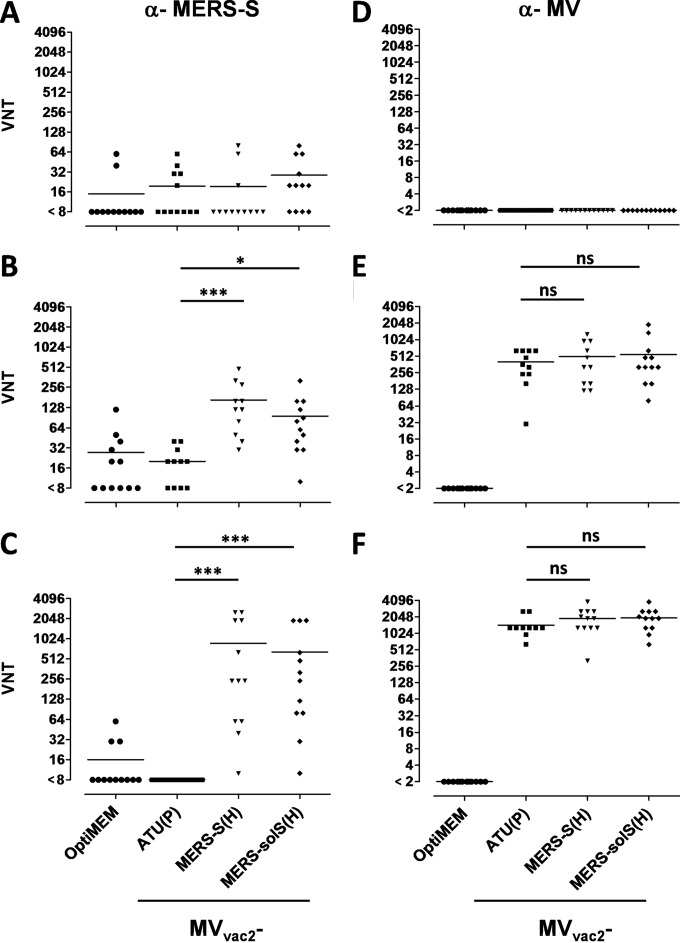
To test the efficacy of the MVvac2-MERS vaccines in vivo, genetically modified IFNAR−/−-CD46Ge mice were chosen, since they are the prime small animal model for analysis of MV-derived vaccines (46). Based on the higher antigen expression of MERS-S and MERS-solS if cloned into the post-H position of the MV genome, the respective viruses were used for vaccination. Thus, 6 mice per group were inoculated via the i.p. route on days 0 and 28, each time with 1 × 105 TCID50 of MVvac2-MERS-S(H), MVvac2-MERS-solS(H), or MVvac2-ATU(P), the latter a recombinant control virus without an insertion of a foreign antigen-encoding gene cassette into an otherwise-empty additional transcription unit. Medium-inoculated mice served as negative controls. At 21 days after boost immunization, sera of immunized mice were compared to prebleed sera by ELISA on antigen-coated plates for antibodies binding to MV bulk antigens or MERS-S (Fig. 2A and B). Indeed, sera of mice vaccinated with MVvac2-MERS-S(H) or MVvac2-MERS-solS(H) clearly encompassed IgG binding to MERS-S (Fig. 2B), whereas no antibodies were found in mice before vaccination (Fig. 2A) or in control mice. Moreover, sera of mice vaccinated with any recombinant MV had IgG in the serum that bound to MV bulk antigens, as expected, indicating successful vaccination with MVs and general mouse reactivity. To determine the neutralizing capacity of the induced antibodies, the potential of serum dilutions to neutralize 200 TCID50 of MERS-CoV or 50 PFU of MVvac2-GFP(H) (Fig. 3A to C) was assayed. All mice immunized with recombinant MV (including the control virus) indeed developed MV VNTs already after the first immunization (Fig. 3B). These titers were boosted approximately 6-fold upon the second immunization (512 to 3,072 VNT) (Fig. 3C). Evidence for induction of neutralizing antibodies against MERS-CoV was only found in mice vaccinated with MVvac2-MERS-S(H) or MVvac2-MERS-solS(H), as expected. The VNT against MERS-CoV reached a titer of 96 to 167 after the first immunization (Fig. 3B) and was boosted about 5- to 7-fold by the second immunization (Fig. 3C). Mice immunized with MVvac2-MERS-S(H) induced slightly higher MERS-CoV VNTs than did MVvac2 expressing the truncated form of the spike protein (167 versus 96 after the first and 874 versus 640 after the second immunization) (Fig. 3B and C). However, this difference was not statistically significant. No VNTs against MV or MERS-CoV were detected in control mice inoculated with medium alone. In summary, both recombinant MVs expressing MERS-S or MERS-solS specifically induced significant amounts of antibodies in immunized mice capable of neutralizing MV as well as MERS-CoV.
FIG 2.
Induction of antibodies that specifically bind MERS-S (α-MERS-S) or MV (α-MV) antigens. Sera of mice vaccinated on days 0 and 28 with indicated viruses were sampled on days −7 (prebleed, A) and 49 (B) and analyzed for antibodies that bound MERS-S or MV bulk antigens by ELISA. Medium-inoculated mice served as mock controls. Antibodies binding to recombinant MERS-S or MV bulk antigens were detected at an optical density of 405 nm in the ELISA. Means and standard deviations of each group are depicted (n = 6). Filled triangles, MVvac2-MERS-S(H); filled circles, MVvac2-MERS-solS(H); open circles, mock controls; open squares, MVvac2-ATU(P).
FIG 3.
Analysis of neutralizing antibodies. VNTs for animals vaccinated on days 0 and 28 with the indicated viruses and sampled on day −7 (A and D), 28 (B and E), or 49 (C and F) for complete neutralization of 200 TCID50 of MERS-CoV or 50 PFU of MV. Medium-inoculated mice served as mock controls. VNTs were calculated as reciprocals of the highest dilution abolishing infectivity. Dots represent single animals (n = 10); horizontal lines represents mean per group. The y axis starts at the detection limit; all mice with VNTs at the detection limit had no detectable VNT. ns, not significant; *, P < 0.05; ***, P < 0.0001.
Splenocytes of animals vaccinated with MVvac2-MERS-S or MVvac2-MERS-solS secrete IFN-γ upon MERS-S-specific stimulation.
To analyze the ability of MV-based vaccine viruses to induce MERS-CoV-specific cellular immune responses, splenocytes of animals vaccinated with MVvac2-MERS-S(H) or MVvac2-MERS-solS(H), or control animals inoculated with medium or MVvac2-ATU(P), were analyzed for antigen-specific IFN-γ secretion by ELISpot assay. For this purpose, mice were immunized following the described prime-boost scheme, and splenocytes were isolated 4 days after the second immunization. To restimulate the antigen-specific T cells in vitro, syngeneic murine DC cell lines (JAWSII, DC2.4, and DC3.2) had been genetically modified by lentiviral vector transduction to stably express MERS-S protein and thereby presented the respective T cell MHC epitopes. Single-cell clones were derived by flow cytometric sorting of single GFP-positive cells. Antigen expression by transduced DCs was verified by Western blot analysis (data not shown).
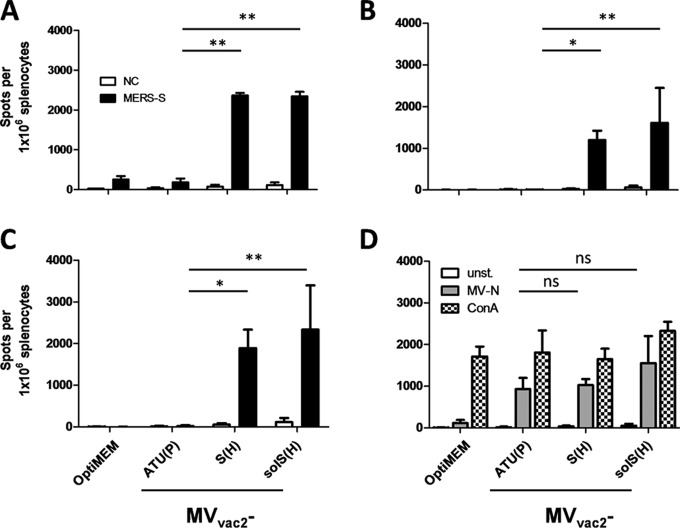
ELISpot assays using splenocytes of vaccinated animals in coculture with JAWSII-MERS-S revealed about 2,400 IFN-γ-secreting cells per 1 × 106 splenocytes after immunization with MVvac2-MERS-S or MVvac2-MERS-solS (Fig. 4A). In contrast, control mice revealed a background response of about 200 IFN-γ-producing cells per 1 × 106 splenocytes. As expected, restimulation of T cells by JAWSII presenting no exogenous antigen revealed only reactivity in the background range (Fig. 4A). To rule out clonal or cell line-associated artifacts, antigen-specific IFN-γ secretion by splenocytes of MVvac2-MERS-S- or MVvac2-MERS-solS-vaccinated mice was confirmed by stimulation with transgenic DC2.4 (Fig. 4B) or DC3.2 (Fig. 4C) cell clones expressing MERS-S. These cell lines stimulated 1,200 to 2,300 IFN-γ-secreting cells per 1 × 106 splenocytes in animals receiving the recombinant MERS vaccines, whereas no background stimulation of respective controls was observed. The differences between MV control and MVvac2-MERS-S- or MVvac2-MERS-solS-vaccinated mice were significant for all cell lines. Additionally, cellular immune responses targeting MV antigens were detected upon stimulation with MV bulk antigens in vaccinated mice that had received any recombinant virus, as expected. However, MV bulk antigens stimulated only about 930 to 1,500 IFN-γ-secreting cells per 1 × 106 splenocytes of MV-vaccinated animals. Finally, splenocytes of all mice revealed a similar basic reactivity to unspecific T cell stimulation, as confirmed by similar numbers of IFN-γ-secreting cells upon ConA treatment (Fig. 4D). Remarkably, both stimulation by ConA or MV bulk antigens resulted in lower numbers of IFN-γ+ cells than stimulation by DCs expressing MERS-S, indicating an extremely robust induction of cellular immunity against this antigen. Thus, the generated MV-based vaccine platform expressing MERS-S or MERS-solS not only induces humoral but also strong MERS S-specific cellular immune responses.
FIG 4.
Secretion of IFN-γ after antigen-specific restimulation of splenocytes. (A to C) IFN-γ ELISpot analysis results with splenocytes of mice vaccinated on days 0 and 28 with indicated viruses, isolated 4 days after boost immunization and after coculture with JAWSII (A), DC2.4 (B), or DC3.2 (C) dendritic cell lines transgenic for MERS-S or untransduced controls (NC). (D) To analyze cellular responses directed against MV, splenocytes were stimulated with 10 μg/ml MV bulk antigens (MV-N) or left unstimulated (unst.). The reactivity of splenocytes was confirmed by ConA treatment (10 μg/ml). Presented are means and standard deviation per group (n = 6). ns, not significant; *, P < 0.05; **, P < 0.01.
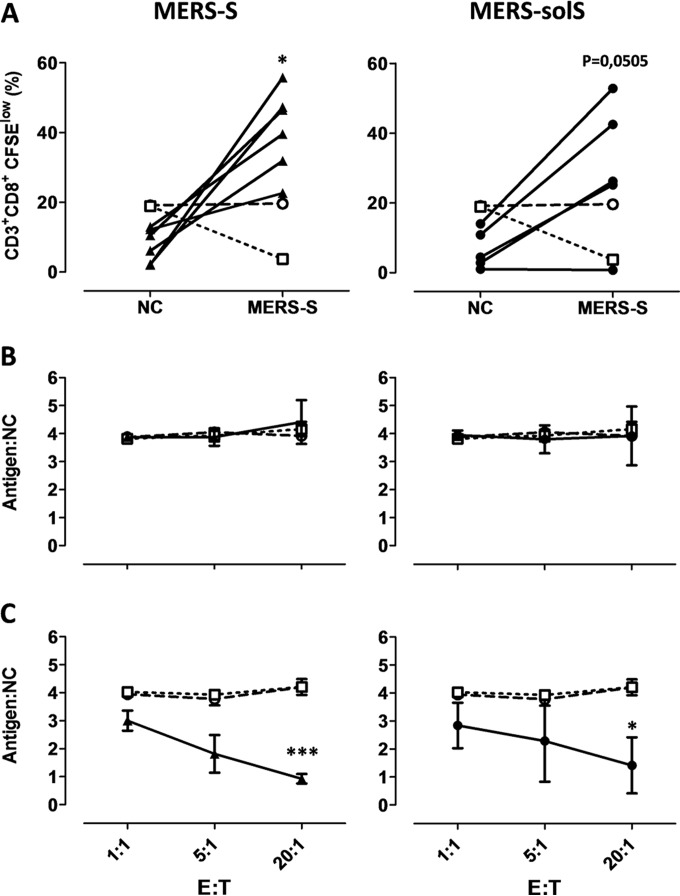
MVvac2-MERS-S(H) or MVvac2-MERS-solS(H) induce antigen-specific CD8+ CTLs.
While ELISpot analyses revealed antigen-specific IFN-γ secretion by vaccinated mice's T cells, we next aimed at detecting antigen-specific CD8+ CTLs, which would be important for clearance of virus-infected cells. For that purpose, proliferation of CD8+ T cells upon stimulation with MERS-S was analyzed 3 weeks after the boost via a flow cytometric assay. Mice were immunized as described above, and splenocytes were isolated 21 days after the boost. JAWSII cells expressing MERS-S were used for restimulation of MERS-S-specific T cells. The splenocytes were labeled with CFSE and subsequently cocultured with JAWSII-MERS-S cells or, as a control, with parental JAWSII cells for 6 days and finally stained for CD3 and CD8 before being analyzed by fluorescence-activated cell sampling for proliferation, detectable by the dilution of the CFSE stain due to cell division.
T cells of mice vaccinated with MVvac2-MERS-S or MVvac2-MERS-solS revealed an increase in the population of CD3+ CD8+ CFSElow cells after restimulation with JAWSII-MERS-S cells compared to restimulation with parental JAWSII without MERS antigens (Fig. 5A). In contrast, T cells of control mice did not reveal this pattern, but the CFSElow population remained rather constant, as expected. This specific increase in CD3+ CD8+ CFSElow cells, which was significant for MVvac2-MERS-S-vaccinated and nearly significant (P = 0.0505) for MVvac2-MERS-solS-vaccinated mice, indicated that CD3+ CD8+ CTLs specific for MERS-S proliferated upon respective stimulation. Thus, MERS-specific cytotoxic memory T cells are induced in mice after vaccination with MVvac2-MERS-S(H) or MVvac2-MERS-solS(H).
FIG 5.
Induction of MERS-S-specific CTLs. (A) Proliferation assay using splenocytes of mice vaccinated on days 0 and 28 with MVvac2-MERS-S(H) or MVvac2-MERS-solS(H) and isolated 21 days after boost immunization, after coculture with JAWSII dendritic cell lines transgenic for MERS-S (right, filled triangles), or untransduced controls (left, filled circles). Depicted are the percentages of CD8+ T cells with low CFSE staiing, indicating proliferation in the samples. Results for splenocytes of vaccinated mice are displayed individually and the trend between paired unstimulated and restimulated samples is outlined. Splenocytes of control vaccinated mice [open circles, mock; open squares, MVvac2-ATU(P)] were pooled. (B and C) Killing assay using splenocytes of mice vaccinated on days 0 and 28 and isolated 4 days after boost immunization. Splenocytes were cocultured with untransduced JAWSII (B) or with antigen-presenting JAWSII-MERS-S (C) or for 6 days. Activated CTLs were then cocultured with EL-4-MERS-S target cells (antigen) and EL-4red control cells (NC) at the indicated effector:target (E:T) ratios for 4 h. Ratios of living target versus nontarget cells (antigen:NC) were determined by flow cytometry. Filled triangles, MVvac2-MERS-S(H); filled circles, MVvac2-MERS-solS(H); open circles, mock; open squares, MVvac2-ATU(P). Results shown are means and standard deviation of each group (n = 6). ns, not significant; *, P < 0.05; ***, P < 0.0001.
Induced T cells revealed antigen-specific cytotoxicity.
To demonstrate the effector ability of induced CTLs, a killing assay was performed to directly analyze antigen-specific cytotoxicity (Fig. 5B). Splenocytes of immunized mice isolated 4 days post-booster vaccination were cocultured with JAWSII-MERS-S or the nontransduced control JAWSII cells for 6 days to restimulate antigen-specific T cells. When these restimulated T cells were coincubated with a defined mixture of EL-4green-MERS-S target and EL-4red control cells (ratio, 4:1), only T cells from MVvac2-MERS-S(H)-vaccinated or MVvac2-MERS-solS(H)-vaccinated mice significantly shifted the ratio of live MERS-S-expressing target cells to control cells in a dose-dependent manner (Fig. 5B). This antigen-dependent killing was also dependent on restimulation with JAWSII-CoV-S cells, since naive T cells did not shift significantly the ratios of target to nontarget cells.
These results indicated that CTLs isolated from MVvac2-MERS-S(H)- or MVvac2-MERS-solS(H)-vaccinated mice are capable of lysing cells expressing MERS-S. Neither splenocytes of control mice restimulated with JAWSII-MERS-S nor splenocytes of MERS-S-vaccinated mice restimulated with control JAWSII cells showed such an antigen-specific killing activity. These results demonstrated that the MV-based vaccine platform induces fully functional antigen-specific CD8+ CTLs in vaccinated mice when applied as a MERS-CoV vaccine.
Vaccination of mice with MVvac2-MERS-S(H) or MVvac2-MERS-solS(H) rescues animals from challenge with MERS-CoV.
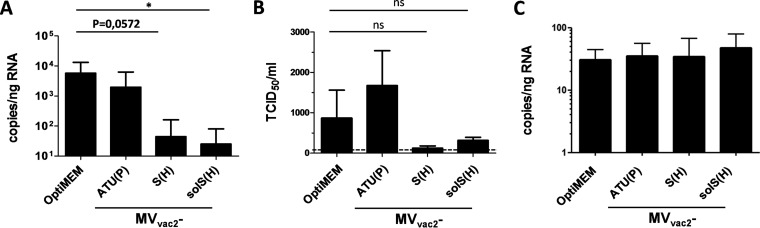
The induction of strong humoral and cellular immune responses directed against MERS-CoV in mice vaccinated with MVvac2-MERS-S(H) or MVvac2-MERS-solS(H) indicated that those animals are possibly protected against a challenge with MERS-CoV. To investigate the efficacy of the candidate vaccines, two independent experiments were performed in which groups of five mice were either vaccinated with MVvac2-MERS-S(H), MVvac2-MERS-solS(H), or control MV [MVvac2-ATU(P)] or left untreated. All mice immunized with MVvac2-MERS-S(H) or MVvac2-MERS-solS(H) showed VNTs directed against MERS-CoV, with titers up to 1,280 for MERS-S and up to 960 for MERS-solS. No MERS-CoV-neutralizing antibodies were detected in control mice (data not shown). Since the murine DPP4 does not serve as a functional MERS-CoV entry receptor (66) and mice are therefore not susceptible to MERS-CoV infection, the vaccinated mice were i.n. transduced with a recombinant adenoviral vector to express human DPP4 (AdV-hDPP4) in murine airways. At 5 days after airway transduction with AdV-hDPP4, mice were infected i.n. with 7 × 104 TCID50 MERS-CoV. Four days later, animals were euthanized, lungs were isolated, the tissue was homogenized, and homogenates were used for purification of total RNA and virus titration. In the lungs of mock-infected control mice, MERS-CoV RNA was detected by qRT-PCR (9,649 ± 3,045 MERS-CoV genome copies/ng RNA) (Fig. 6A). Mice vaccinated with control MVvac2-ATU(P) showed slightly lower copy numbers of viral RNA (5,923 ± 3,045 MERS-CoV genome copies/ng RNA) (Fig. 6A). Vaccination with MVvac2-MERS-S(H) or MVvac2-MERS-solS(H) resulted in near-complete reduction of viral loads to 74 ± 60 genome copies/ng RNA or 51 ± 32 genome copies/ng RNA, respectively (Fig. 6A). Next, titers of infectious virus were determined in the lung tissue. While the titers were generally low, they corresponded to the qRT-PCR data. In mock-treated control mice, titers of up to 5,000 TCID50/ml were determined (mean, 868 ± 692 TCID50/ml) and in lungs of mice vaccinated with the vaccine backbone without MERS antigen [MVvac2-ATU(P)], 1,673 ± 866 TCID50/ml were detected. A considerable albeit statistically not significant reduction of infectious virus titers was found in mice vaccinated with MVvac2-MERS-S(H) or MVvac2-MERS-solS(H) compared to mock control mice (Fig. 6B). These results revealed that, indeed, vaccination with the recombinant measles viruses was able to protect mice against a challenge with MERS-CoV.
FIG 6.
Viral loads after MERS-CoV challenge in vivo. (A and B) Viral loads, determined as genome copies per nanogram of RNA (A) or infectious virus titers (B) in the lungs of prevaccinated mice after transduction with DPP4-encoding AdV 21 days after boost and challenge with MERS-CoV 25 days after boost. Two independent experiments (n = 4 to 5 per group). Error bars, standard errors of the means (SEM). Dotted line, limit of detection (LOD of qPCR, <1.7 copies/ng RNA). ns, not significant; *, P < 0.05. (C) AdV transduction control mCherry mRNA results (in copies per nanogram of RNA). Error bars, SEM.
MERS-CoV infection of transduced mice was not always successful, which was indicated by a completely negative PCR result for viral genomes in about 40% of all animals. In approximately 30% of MERS-CoV-negative animals, PCR for the mCherry gene was negative, indicating that transduction was not successful and explaining why these mice were not susceptible. Why the remaining transduced mice were not infected is currently unclear. However, even when the dropout animals were included in statistical analysis, the difference between mean viral loads of the medium control group and MVvac2-MERS-solS(H)-treated mice (ratio, 278.2; 95% confidence interval, 1.52 to 50,904) stayed significant (P = 0.0329). Protection of the MVvac2-MERS-S(H)-vaccinated group was close to significance (P = 0.057) compared to mock-treated animals (ratio, 149.2; 95% CI, 0.82 to 27,301).
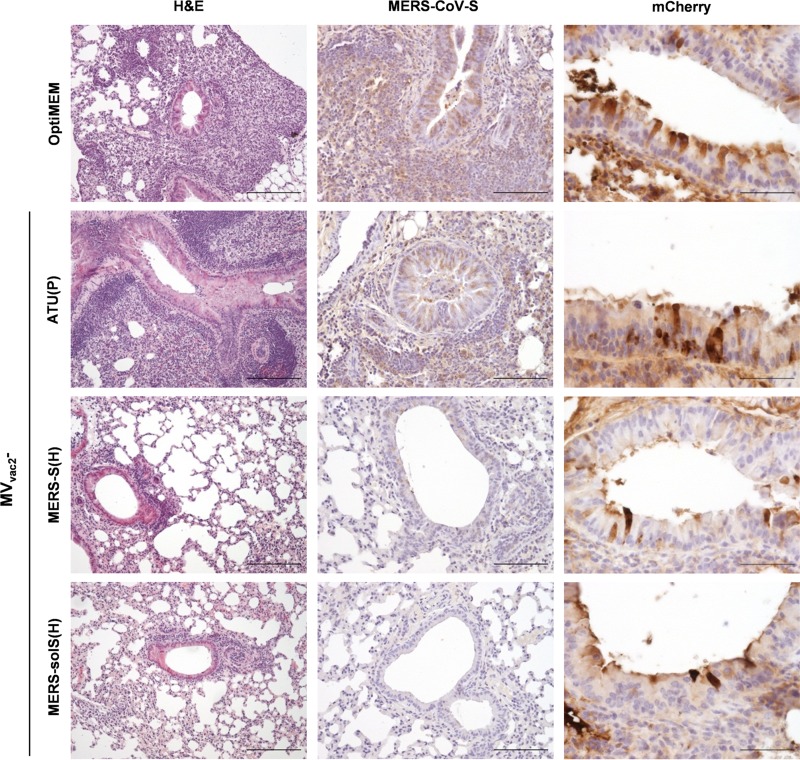
Histological analyses were performed to analyze if the reduced viral load in mice vaccinated with MVvac2-MERS-solS(H) or MVvac2-MERS-S(H) was matched by decreased pathological changes in mouse lungs (Fig. 7). For this purpose, lungs were examined with H&E staining to visualize inflammation. Additionally, MERS-S and mCherry expression was determined by immunohistochemistry using antigen-specific antibodies. Consistent with the qRT-PCR results, all mice that were positive in the qRT-PCR for the mCherry gene expressed mCherry in epithelia of the lungs, demonstrating successful transduction (Fig. 7, right column). The histopathological examination of H&E-stained lung tissues clearly showed differences between the vaccinated mice and controls (Fig. 7, left column). In the mock (Opti-MEM) as well as vector control [MVvac2-ATU(P)] groups, large areas of inflamed tissue were observed to be densely packed with lymphocytes, macrophages and, to a lesser extent, neutrophils and eosinophils. Moreover, hyperplasia of the bronchus-associated lymphoid tissue was present to various degrees. These inflamed areas colocalized with expression of MERS-CoV spike protein (Fig. 7, middle column). Mice that were vaccinated with recombinant MV expressing MERS-S showed fewer signs of inflammation and consistently lower MERS-S expression after challenge with MERS. These differences were most obvious in lungs of MVvac2-MERS-solS(H)-vaccinated animals, where only small foci of inflammation could be observed. These results revealed that vaccination with recombinant MV expressing MERS S reduced pathological changes in the lungs of MERS-CoV-infected mice.
FIG 7.
Histopathological changes and immunohistochemical analysis of lungs after challenge. Analysis of lung tissue of representative prevaccinated mice (as indicated) after transduction with hDPP4-encoding AdV and challenge with MERS-CoV. Pictures arranged in a row were from samples of the same individual mouse. Paraffin-fixed tissue was stained with H&E (first column; scale bar, 200 μm), with Ab against MERS-CoV-S antigen (middle column; scale bar, 100 μm), and as a control of AdV transduction against mCherry (right column; scale bar, 50 μm).
DISCUSSION
In this study, we have demonstrated the capacity of recombinant MV encoding different forms of the MERS-CoV S glycoprotein to induce both strong humoral and cellular immune responses that revealed a protective capacity in a challenge model of mice vaccinated with these stable live-attenuated vaccines. So far, different strategies to develop vaccines against MERS-CoV have been proposed, including recombinant full-length S protein (67) or the receptor-binding domain (RBD) of MERS-S (27, 28, 30, 31, 68), as well as platform-based approaches using modified vaccinia virus Ankara (MVA) (22) or adenoviral vectors (AdV) (23) encoding MERS-S. Similar to our MV-based vaccine, these experimental vaccines induced humoral immune responses with virus-neutralizing capacities. Among vectored vaccines, immunization with MVA- or AdV-expressing MERS-S resulted in VNTs of approximately 1,800 or 1,024, respectively, when used to immunize BALB/c mice. Vaccination with MVvac2-MERS-S or MVvac2-MERS-solS induced somewhat lower VNTs of about 840, which is an extremely robust titer if it is taken into account that mice were immunized with 103-fold fewer virus particles than with MVA and 106-fold lower particles than with replication-deficient AdV. Moreover, transgenic IFNAR−/−-CD46Ge mice have been used in our study with defects in type I IFN receptor signaling. Knockout of the type I IFN receptor results in reduced adaptive immune responses (68–70), since type I IFNs are an important link between the innate and adaptive immunity via, among others factors, activation of DCs (71), leading to a disadvantage for the mouse adaptive immune system. Nevertheless, these mice have to be used routinely to analyze efficacy of MV-based vaccines in a small animal model (46), since wild-type mice are not susceptible to MV infection for mainly two reasons. First, murine homologues of MV receptors cannot be used for cell entry (69), with the exception of nectin-4 (70). Second, MV replication is strongly impaired by type I IFN responses (71, 72), and mice with an intact IFNAR feedback loop failed to be susceptible to MV infection (46). Therefore, the IFNAR−/−-CD46Ge mouse strain transgenic for human MV vaccine receptor CD46 and with a knockout of the IFNAR is used to analyze MV-based vaccines. Additionally, the mouse strain backgrounds (BALB/c versus C57BL/6) differ in T helper cell responses (BALB/c, predominantly Th2; C57BL/6, predominantly Th1 responses [73]), which reflects the different balance of cellular versus humoral immunity (74, 75). Thus, the mouse model which had to be used in this study certainly is disadvantageous with respect to VNTs. To directly compare efficacy of the different vector systems, all vectors should ideally be used side by side in the same animal model. This may be a focus of future studies. The VNTs of about 1,000 induced by three immunizations with recombinant RBD are hardly comparable to our results, since other protocols for determination of VNTs were used in the other studies (27, 31). Interestingly, the expression of the soluble version of S by MV did not enhance VNTs. This is consistent with humoral immunity induced by DNA vaccines targeting SARS-CoV. Plasmids encoding soluble SARS-S lacking the transmembrane domain provoked lower VNTs than membrane-bound variants (32). An altered, less physiological conformation of the S protein has been proposed to result from deletion of the transmembrane domain, which should be responsible for worse immune recognition and lower antibody titers binding to the native, correctly folded S proteins in virus particles. In contrast, the soluble S1 domain of MERS-S expressed by AdV actually induced slightly higher VNTs than did full-length S (23). However, soluble constructs consisting of the MERS S1 and S2 domains have not been compared to the soluble S1 domain yet. Interestingly, recombinant MV expressing soluble MERS-S revealed slightly impaired replication in comparison to control MV, in contrast to MV expressing full-length MERS-S. This impaired viral replication might be based on cytotoxicity of MERS-solS, probably as a result of an altered folding or the solubility of the S protein. Cytotoxic effects of the S protein have already been observed for the S2 domain of SARS-S (76–80), but not for other coronaviruses, like mouse hepatitis coronavirus (81). However, both MV-based vaccines encoding either the soluble or the full-length variant of MERS-S did induce strong VNTs and cellular immune responses.
The protective capacity of humoral immune responses against CoV infection is controversial. Neutralizing antibodies have been identified as correlates of protection against SARS-CoV challenge, since passive serum transfer was sufficient to rescue animals from challenge (32, 82) and T cell depletion did not impair protection (32). In contrast, immunization with the nucleocapsid protein resulted in protection against the coronavirus infectious bronchitis virus (IBV) without induction of neutralizing antibodies (83, 84), indicating the capacity of cellular immune responses for IBV protection. Anyway, the antigenic potential of S for induction of CD4+ or CD8+ T cell immunity has already been demonstrated for SARS-CoV (32, 85) by using recombinant protein or DNA vaccines. Also for MERS-CoV, application of RBD protein together with adjuvants has been shown to induce cellular immunity (27, 31). We demonstrated in this study that induction of cellular immunity by a vectored vaccine works independently from adjuvants or the application strategy. The MV-based vaccine induced very strong MERS-S-specific CD8+ T cell responses, revealed by ELISpot, killing, and proliferation assays. The broad repertoire of reactivity, in the case of antigen-specific proliferation also 21 days after the booster immunization, indicated induction of both a functional effector and memory T cell repertoire by MVvac2-MERS-S and MVvac2-MERS-solS. Thereby, the extraordinary high number of IFN-γ-secreting T cells in vaccinated mice both stresses the potential of the vaccine platform and underlines the immunogenicity of MERS-S.
On top, the present study tested whether the induced immune responses protected mice against a challenge infection with MERS-CoV. Indeed, vaccination with MVvac2-MERS-S or MVvac2-MERS-solS significantly reduced viral loads in the lungs of vaccinated mice after challenge with MERS-CoV. As expected, this reduction of viral load correlated with reduced pathological alterations in the lung, indicating that MV-derived MERS vaccines were able to confer protection against MERS-CoV infection. At least 4 mice out of each group did not reveal any MERS-CoV infection nor any pathological lung alterations indicating failure of infection in these individuals. In 30% of those mice, transduction with the recombinant adenovirus expressing human DPP4 did not appear to be successful. However, the majority of mice with no signs of MERS-CoV infection at all showed expression of mCherry, indicating that transduction was successful. Currently, the reason for the failure to infect these animals is unclear.
The direct correlates of protection in the vaccinated mice remain to be determined in future studies. Most recently, mice transgenic for human DPP4 have been developed that allow analysis of MERS-CoV infection on a more robust and physiologic basis (86). These could only be used for analysis of MV-based vaccines after intercrossing them with IFNAR−/−-CD46Ge or similar mouse strains to obtain mice simultaneously susceptible to MV and MERS-CoV, which may also be a focus of future work.
Efficacy of MVvac2-based MERS vaccines has been demonstrated in MV-naive mice. Theoretically, preexisting antivector immunity against the MV backbone may be considered a potential limitation both for the specific MERS vaccines tested in this study and for use of recombinant MV as a vaccine platform, in general, for MV-immunized patients (87). However, it has been clearly demonstrated both in mice (41, 45) and nonhuman primates (41) with humoral immune responses regarded to be protective against measles that vaccination with recombinant MVs encoding antigens of HIV-1 (41) or Chikungunya virus (45) still induced surprisingly robust antigen-specific immune responses. Most interestingly, when the efficacy of recombinant MV-CHIKV vaccine in a phase I trial in human volunteers was analyzed, the vaccine was recently shown to be effective in inducing anti-CHIKV immune responses irrespective of preexisting antimeasles immunity (47). These data question the “sterilizing” character of measles immunity and clearly indicate the potential of recombinant MV as a promising vaccine platform for vaccination against MERS-CoV or other infectious agents, in general. Indeed, the efficacy of MV-based recombinant vaccines has been demonstrated preclinically with quite a range of different pathogen antigens, e.g., HBV (39), dengue virus (44), WNV (42), and CHIKV (45). Additionally, the efficacy of MV to induce immune responses against coronaviruses has been shown for the S protein and nucleocapsid protein of SARS-CoV (19). All these recombinant vaccines have in common that they are based on a very-well-known platform: MV vaccines have been shown to exhibit an extremely beneficial safety profile in light of the millions of applied doses over the last 40 years. Only heavily immune-suppressed patients are excluded from measles vaccination campaigns, but the protection holds over decades and is thought to be most likely for life (33, 34).
Most interestingly, a quite similar recombinant vaccine based on a rhabdovirus, a member of another family within the Mononegavirales order, is currently being tested in the clinic as an experimental vaccine against Ebola virus (EBOV) infection. Recombinant VSV encoding the Ebola Zaire strain glycoprotein. replacing VSV-G (VSV-ZEBOV) was shown to be effective in animal models (88, 89) and is now being tested in phase I trials for safety in human patients (90), in preparation to being moved to the field to combat current EBOV epidemics. Thereby, the potential interest in such platform-based vaccines to combat emerging or reemerging infections is impressively highlighted.
Taken together, MV vaccine strain Moraten-derived recombinant MVvac2 vaccines are effective vaccines against MERS-CoV, inducing both humoral and cellular immune responses protective for vaccinated animals. Thereby, the capacity of the recombinant MV-based vaccine platform for fast generation of effective vaccines has been demonstrated also with a more general view to future emerging or reemerging infections, but also with a view on MERS-CoV. MV-MERS-S provides an opportunity for further development of this experimental vaccine to be prepared especially to reduce the risk of pandemic spread of this disease.
ACKNOWLEDGMENTS
We thank Vivian Koch, Daniela Müller, Michael Schmidt, and Gotthard Ludwig for excellent technical assistance, Sonja Witzel, Manuel Gimmel, Tina Hempel, Aileen Laubenheimer, and Tobias Blödel for excellent technical support in production of recombinant MERS-S protein, and Roland Plesker for assistance in animal experiments. We thank Lucie Sauerhering and Erik Dietzel for help with animal studies under biosafety level 4 conditions. We are indebted to Ron Fouchier for providing MERS-CoV strain EMC/2012, to Kenneth Rock for DC2.4 and DC3.2 cells, and to Christian Drosten and Doreen Muth for PCR protocols. We further thank Bakhos Tannous for providing pCSCW2gluc-IRES-GFP and Kenneth Lundström for the SFV vector system.
TWINCORE is a joint venture between the Hannover Medical School and the Helmholtz Centre for Infection Research, in Hannover, Germany.
This work was supported by the German Center for Infection Research (DZIF; TTU 01.802, TTU 01.904) and the Deutsche Forschungsgemeinschaft (SFB1021, SFB593).
REFERENCES
- 1.Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RM. 2012. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med 367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 2.World Health Organization. 2014. Middle East respiratory syndrome coronavirus (MERS-CoV)—Saudi Arabia. World Health Organization, Geneva, Switzerland: http://www.who.int/csr/don/17-december-2014-mers/en/. [Google Scholar]
- 3.Drosten C, Meyer B, Müller MA, Corman VM, Al-Masri M, Hossain R, Madani H, Sieberg A, Bosch BJ, Lattwein E, Alhakeem RF, Assiri AM, Hajomar W, Albarrak AM, Al-Tawfiq JA, Zumla AI, Memish ZA. 2014. Transmission of MERS-coronavirus in household contacts. N Engl J Med 371:828–835. doi: 10.1056/NEJMoa1405858. [DOI] [PubMed] [Google Scholar]
- 4.Petersen E, Hui DS, Perlman S, Zumla A. 2015. Middle East respiratory syndrome: advancing the public health and research agenda on MERS. Lessons from the South Korea outbreak. Int J Infect Dis 36:54–55. doi: 10.1016/j.ijid.2015.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cowling BJ, Park M, Fang VJ, Wu P, Leung GM, Wu JT. 2015. Preliminary epidemiological assessment of MERS-CoV outbreak in South Korea, May to June. Euro Surveill 20:pii=2113 http://dx.doi.org/10.28907/1560-7917.ES2015.25.21163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Raj VS, Farag EA, Reusken CB, Lamers MM, Pas SD, Voermans J, Smits SL, Osterhaus AD, Al-Mawlawi N, Al-Romaihi HE, Al Hajri MM, El-Sayed AM, Mohran KA, Ghobashy H, Al Hajri F, Al-Thani M, Al-Marri SA, El-Maghraby MM, Koopmans MP, Haagmans BL. 2014. Isolation of MERS coronavirus from a dromedary camel, Qatar, 2014. Emerg Infect Dis 20:1339–1342. doi: 10.3201/eid2008.140663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferguson NM, Van Kerkhove MD. 2014. Identification of MERS-CoV in dromedary camels. Lancet Infect Dis 14:93–94. doi: 10.1016/S1473-3099(13)70691-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Müller MA, Corman VM, Jores J, Meyer B, Younan M, Liljander A, Bosch B, Lattwein E, Hilali M, Musa BE, Bornstein S, Drosten C. 2014. MERS coronavirus neutralizing antibodies in camels, Eastern Africa, 1983-1997. Emerg Infect Dis 20:2093–2095. doi: 10.3201/eid2012.141026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meyer B, Müller MA, Corman VM, Reusken CB, Ritz D, Godeke G, Lattwein E, Kallies S, Siemens A, van Beek J, Drexler JF, Muth D, Bosch B, Wernery U, Koopmans MP, Wernery R, Drosten C. 2014. Antibodies against MERS coronavirus in dromedary camels, United Arab Emirates, 2003 and 2013. Emerg Infect Dis 20:552–559. doi: 10.3201/eid2004.131746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Corman VM, Jores J, Meyer B, Younan M, Liljander A, Said MY, Gluecks I, Lattwein E, Bosch B, Drexler JF, Bornstein S, Drosten C, Müller MA. 2014. Antibodies against MERS coronavirus in dromedary camels, Kenya, 1992-2013. Emerg Infect Dis 20:1319–1322. doi: 10.3201/eid2008.140596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Memish ZA, Cotten M, Meyer B, Watson SJ, Alsahafi AJ, Al Rabeeah Abdullah A, Corman VM, Sieberg A, Makhdoom HQ, Assiri A, Al Masri M, Aldabbagh S, Bosch B, Beer M, Müller MA, Kellam P, Drosten C. 2014. Human infection with MERS coronavirus after exposure to infected camels, Saudi Arabia, 2013. Emerg Infect Dis 20:1012–1015. doi: 10.3201/eid2006.140402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Azhar EI, El-Kafrawy SA, Farraj SA, Hassan AM, Al-Saeed MS, Hashem AM, Madani TA. 2014. Evidence for camel-to-human transmission of MERS coronavirus. N Engl J Med 370:2499–2505. doi: 10.1056/NEJMoa1401505. [DOI] [PubMed] [Google Scholar]
- 13.Lau SK, Li KS, Tsang AK, Lam CS, Ahmed S, Chen H, Chan K, Woo Patrick C, Yuen KY. 2013. Genetic characterization of betacoronavirus lineage C viruses in bats reveals marked sequence divergence in the spike protein of pipistrellus bat coronavirus HKU5 in Japanese pipistrelle: implications for the origin of the novel Middle East respiratory syndrome coronavirus. J Virol 87:8638–8650. doi: 10.1128/JVI.01055-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Annan A, Baldwin HJ, Corman VM, Klose SM, Owusu M, Nkrumah EE, Badu EK, Anti P, Agbenyega O, Meyer B, Oppong S, Sarkodie YA, Kalko Elisabeth KV, Lina Peter HC, Godlevska EV, Reusken C, Seebens A, Gloza-Rausch F, Vallo P, Tschapka M, Drosten C, Drexler JF. 2013. Human betacoronavirus 2c EMC/2012-related viruses in bats, Ghana and Europe. Emerg Infect Dis 19:456–459. doi: 10.3201/eid1903.121503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Corman VM, Ithete NL, Richards LR, Schoeman MC, Preiser W, Drosten C, Drexler JF. 2014. Rooting the phylogenetic tree of Middle East respiratory syndrome coronavirus by characterization of a conspecific virus from an African bat. J Virol 88:11297–11303. doi: 10.1128/JVI.01498-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peiris JSM, Guan Y, Yuen KY. 2004. Severe acute respiratory syndrome. Nat Med 10:S88–S97. doi: 10.1038/nm1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bai B, Lu X, Meng J, Hu Q, Mao P, Lu B, Chen Z, Yuan Z, Wang H. 2008. Vaccination of mice with recombinant baculovirus expressing spike or nucleocapsid protein of SARS-like coronavirus generates humoral and cellular immune responses. Mol Immunol 45:868–875. doi: 10.1016/j.molimm.2007.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Du L, Zhao G, Chan Chris CS, Sun S, Chen M, Liu Z, Guo H, He Y, Zhou Y, Zheng B, Jiang S. 2009. Recombinant receptor-binding domain of SARS-CoV spike protein expressed in mammalian, insect and E. coli cells elicits potent neutralizing antibody and protective immunity. Virology 393:144–150. doi: 10.1016/j.virol.2009.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liniger M, Zuniga A, Tamin A, Azzouz-Morin TN, Knuchel M, Marty RR, Wiegand M, Weibel S, Kelvin D, Rota PA, Naim HY. 2008. Induction of neutralising antibodies and cellular immune responses against SARS coronavirus by recombinant measles viruses. Vaccine 26:2164–2174. doi: 10.1016/j.vaccine.2008.01.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dimitrov DS. 2004. Virus entry: molecular mechanisms and biomedical applications. Nat Rev Microbiol 2:109–122. doi: 10.1038/nrmicro817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hofmann H, Pöhlmann S. 2004. Cellular entry of the SARS coronavirus. Trends Microbiol 12:466–472. doi: 10.1016/j.tim.2004.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Song F, Fux R, Provacia LB, Volz A, Eickmann M, Becker S, Osterhaus Albert DME, Haagmans BL, Sutter G. 2013. Middle East respiratory syndrome coronavirus spike protein delivered by modified vaccinia virus Ankara efficiently induces virus-neutralizing antibodies. J Virol 87:11950–11954. doi: 10.1128/JVI.01672-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim E, Okada K, Kenniston T, Raj VS, AlHajri MM, Farag Elmoubasher ABA, AlHajri F, Osterhaus Albert DME, Haagmans BL, Gambotto A. 2014. Immunogenicity of an adenoviral-based Middle East respiratory syndrome coronavirus vaccine in BALB/c mice. Vaccine 32:5975–5982. doi: 10.1016/j.vaccine.2014.08.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gierer S, Bertram S, Kaup F, Wrensch F, Heurich A, Krämer-Kühl A, Welsch K, Winkler M, Meyer B, Drosten C, Dittmer U, Hahn T von, Simmons G, Hofmann H, Pöhlmann S. 2013. The spike protein of the emerging betacoronavirus EMC uses a novel coronavirus receptor for entry, can be activated by TMPRSS2, and is targeted by neutralizing antibodies. J Virol 87:5502–5511. doi: 10.1128/JVI.00128-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raj VS, Mou H, Smits SL, Dekkers DH, Müller MA, Dijkman R, Muth D, Demmers JA, Zaki A, Fouchier RA, Thiel V, Drosten C, Rottier PJ, Osterhaus AD, Bosch BJ, Haagmans BL. 2013. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus EMC. Nature 495:251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He Y, Lu H, Siddiqui P, Zhou Y, Jiang S. 2005. Receptor-binding domain of severe acute respiratory syndrome coronavirus spike protein contains multiple conformation-dependent epitopes that induce highly potent neutralizing antibodies. J Immunol 174:4908–4915. doi: 10.4049/jimmunol.174.8.4908. [DOI] [PubMed] [Google Scholar]
- 27.Ma C, Li Y, Wang L, Zhao G, Tao X, Tseng CK, Zhou Y, Du L, Jiang S. 2014. Intranasal vaccination with recombinant receptor-binding domain of MERS-CoV spike protein induces much stronger local mucosal immune responses than subcutaneous immunization: implication for designing novel mucosal MERS vaccines. Vaccine 32:2100–2108. doi: 10.1016/j.vaccine.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Du L, Kou Z, Ma C, Tao X, Wang L, Zhao G, Chen Y, Yu F, Tseng CK, Zhou Y, Jiang S. 2013. A truncated receptor-binding domain of MERS-CoV spike protein potently inhibits MERS-CoV infection and induces strong neutralizing antibody responses: implication for developing therapeutics and vaccines. PLoS One 8:e81587. doi: 10.1371/journal.pone.0081587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Du L, Zhao G, Kou Z, Ma C, Sun S, Poon VK, Lu L, Wang L, Debnath AK, Zheng B, Zhou Y, Jiang S. 2013. Identification of a receptor-binding domain in the S protein of the novel human coronavirus Middle East respiratory syndrome coronavirus as an essential target for vaccine development. J Virol 87:9939–9942. doi: 10.1128/JVI.01048-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mou H, Raj VS, van Kuppeveld FJ, Rottier PJ, Haagmans BL, Bosch BJ. 2013. The receptor binding domain of the new Middle East respiratory syndrome coronavirus maps to a 231-residue region in the spike protein that efficiently elicits neutralizing antibodies. J Virol 87:9379–9383. doi: 10.1128/JVI.01277-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lan J, Deng Y, Chen H, Lu G, Wang W, Guo X, Lu Z, Gao GF, Tan W. 2014. Tailoring subunit vaccine immunity with adjuvant combinations and delivery routes using the Middle East respiratory coronavirus (MERS-CoV) receptor-binding domain as an antigen. PLoS One 9:e112602. doi: 10.1371/journal.pone.0112602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang Z, Kong W, Huang Y, Roberts A, Murphy BR, Subbarao K, Nabel GJ. 2004. A DNA vaccine induces SARS coronavirus neutralization and protective immunity in mice. Nature 428:561–564. doi: 10.1038/nature02463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hilleman MR. 2001. Current overview of the pathogenesis and prophylaxis of measles with focus on practical implications. Vaccine 20:651–665. doi: 10.1016/S0264-410X(01)00384-X. [DOI] [PubMed] [Google Scholar]
- 34.Griffin DE. 2002. Measles virus. In Encyclopedia of molecular medicine. John Wiley & Sons, Inc, Hoboken, NJ. doi: 10.1002/0471203076.emm0739. [DOI] [Google Scholar]
- 35.Radecke F, Spielhofer P, Schneider H, Kaelin K, Huber M, Dötsch C, Billeter MA. 1995. Rescue of measles viruses from cloned DNA. EMBO J 14:5773–5784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Billeter MA, Naim HY, Udem SA. 2009. Reverse genetics of measles virus and resulting multivalent recombinant vaccines: applications of recombinant measles viruses. Curr Top Microbiol Immunol 329:129–162. doi: 10.1007/978-3-540-70523-9_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Duprex WP, Rima BK. 2002. Using green fluorescent protein to monitor measles virus cell-to-cell spread by time-lapse confocal microscopy. Methods Mol Biol 183:297–307. doi: 10.1385/1-59259-280-5:297. [DOI] [PubMed] [Google Scholar]
- 38.Singh M, Billeter MA. 1999. A recombinant measles virus expressing biologically active human interleukin-12. J Gen Virol 80:101–106. doi: 10.1099/0022-1317-80-1-101. [DOI] [PubMed] [Google Scholar]
- 39.Singh M, Cattaneo R, Billeter MA. 1999. A recombinant measles virus expressing hepatitis B virus surface antigen induces humoral immune responses in genetically modified mice. J Virol 73:4823–4828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reyes-del Valle J, de la Fuente Cynthia Turner MA, Springfeld C, Apte-Sengupta S, Frenzke ME, Forest A, Whidby J, Marcotrigiano J, Rice CM, Cattaneo R. 2012. Broadly neutralizing immune responses against hepatitis C virus induced by vectored measles viruses and a recombinant envelope protein booster. J Virol 86:11558–11566. doi: 10.1128/JVI.01776-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lorin C, Mollet L, Delebecque F, Combredet C, Hurtrel B, Charneau P, Brahic M, Tangy F. 2004. A single injection of recombinant measles virus vaccines expressing human immunodeficiency virus (HIV) type 1 clade B envelope glycoproteins induces neutralizing antibodies and cellular immune responses to HIV. J Virol 78:146–157. doi: 10.1128/JVI.78.1.146-157.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Desprès P, Combredet C, Frenkiel M, Lorin C, Brahic M, Tangy F. 2005. Live measles vaccine expressing the secreted form of the West Nile virus envelope glycoprotein protects against West Nile virus encephalitis. J Infect Dis 191:207–214. doi: 10.1086/426824. [DOI] [PubMed] [Google Scholar]
- 43.Brandler S, Marianneau P, Loth P, Lacôte S, Combredet C, Frenkiel M, Desprès P, Contamin H, Tangy F. 2012. Measles vaccine expressing the secreted form of West Nile virus envelope glycoprotein induces protective immunity in squirrel monkeys, a new model of West Nile virus infection. J Infect Dis 206:212–219. doi: 10.1093/infdis/jis328. [DOI] [PubMed] [Google Scholar]
- 44.Brandler S, Ruffie C, Najburg V, Frenkiel M, Bedouelle H, Desprès P, Tangy F. 2010. Pediatric measles vaccine expressing a dengue tetravalent antigen elicits neutralizing antibodies against all four dengue viruses. Vaccine 28:6730–6739. doi: 10.1016/j.vaccine.2010.07.073. [DOI] [PubMed] [Google Scholar]
- 45.Brandler S, Ruffié C, Combredet C, Brault J, Najburg V, Prevost M, Habel A, Tauber E, Desprès P, Tangy F. 2013. A recombinant measles vaccine expressing Chikungunya virus-like particles is strongly immunogenic and protects mice from lethal challenge with Chikungunya virus. Vaccine 31:3718–3725. doi: 10.1016/j.vaccine.2013.05.086. [DOI] [PubMed] [Google Scholar]
- 46.Mrkic B, Pavlovic J, Rülicke T, Volpe P, Buchholz CJ, Hourcade D, Atkinson JP, Aguzzi A, Cattaneo R. 1998. Measles virus spread and pathogenesis in genetically modified mice. J Virol 72:7420–7427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ramsauer K, Schwameis M, Firbas C, Müllner M, Putnak RJ, Thomas SJ, Desprès P, Tauber E, Jilma B, Tangy F. 2015. Immunogenicity, safety, and tolerability of a recombinant measles-virus-based Chikungunya vaccine: a randomised, double-blind, placebo-controlled, active-comparator, first-in-man trial. Lancet Infect Dis 15:519–527. doi: 10.1016/S1473-3099(15)70043-5. [DOI] [PubMed] [Google Scholar]
- 48.del Valle JR, Devaux P, Hodge G, Wegner NJ, McChesney MB, Cattaneo R. 2007. A vectored measles virus induces hepatitis B surface antigen antibodies while protecting macaques against measles virus challenge. J Virol 81:10597–10605. doi: 10.1128/JVI.00923-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bajenoff M, Germain RN. 2009. B-cell follicle development remodels the conduit system and allows soluble antigen delivery to follicular dendritic cells. Blood 114:4989–4997. doi: 10.1182/blood-2009-06-229567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pape KA, Catron DM, Itano AA, Jenkins MK. 2007. The humoral immune response is initiated in lymph nodes by B cells that acquire soluble antigen directly in the follicles. Immunity 26:491–502. doi: 10.1016/j.immuni.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 51.De Becker G, Moulin V, Tielemans F, De Mattia F, Urbain J, Leo O, Moser M. 1998. Regulation of T helper cell differentiation in vivo by soluble and membrane proteins provided by antigen-presenting cells. Eur J Immunol 28:3161–3171. doi:. [DOI] [PubMed] [Google Scholar]
- 52.Enriquez-Rincon F, Klaus GG. 1984. Differing effects of monoclonal anti-hapten antibodies on humoral responses to soluble or particulate antigens. Immunology 52:129–136. [PMC free article] [PubMed] [Google Scholar]
- 53.Shen Z, Reznikoff G, Dranoff G, Rock KL. 1997. Cloned dendritic cells can present exogenous antigens on both MHC class I and class II molecules. J Immunol 158:2723–2730. [PubMed] [Google Scholar]
- 54.Martin A, Staeheli P, Schneider U. 2006. RNA polymerase II-controlled expression of antigenomic RNA enhances the rescue efficacies of two different members of the Mononegavirales independently of the site of viral genome replication. J Virol 80:5708–5715. doi: 10.1128/JVI.02389-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hewett JW, Tannous B, Niland BP, Nery FC, Zeng J, Li Y, Breakefield XO. 2007. Mutant torsin A interferes with protein processing through the secretory pathway in DYT1 dystonia cells. Proc Natl Acad Sci U S A 104:7271–7276. doi: 10.1073/pnas.0701185104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Boussif O, Lezoualc'h F, Zanta MA, Mergny MD, Scherman D, Demeneix B, Behr JP. 1995. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proc Natl Acad Sci U S A 92:7297–7301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zufferey R, Nagy D, Mandel RJ, Naldini L, Trono D. 1997. Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo. Nat Biotechnol 15:871–875. doi: 10.1038/nbt0997-871. [DOI] [PubMed] [Google Scholar]
- 58.Münch RC, Mühlebach MD, Schaser T, Kneissl S, Jost C, Plückthun A, Cichutek K, Buchholz CJ. 2011. DARPins: an efficient targeting domain for lentiviral vectors. Mol Ther 19:686–693. doi: 10.1038/mt.2010.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bach P, Abel T, Hoffmann C, Gal Z, Braun G, Voelker I, Ball CR, Johnston Ian CD, Lauer UM, Herold-Mende C, Mühlebach MD, Glimm H, Buchholz CJ. 2013. Specific elimination of CD133+ tumor cells with targeted oncolytic measles virus. Cancer Res 73:865–874. doi: 10.1158/0008-5472.CAN-12-2221. [DOI] [PubMed] [Google Scholar]
- 60.Kärber G. 1931. Beitrag zur kollektiven Behandlung pharmakologischer Reihenversuche. Arch Exp Pathol Pharmakol 162:480–483. doi: 10.1007/BF01863914. [DOI] [Google Scholar]
- 61.Funke S, Maisner A, Mühlebach MD, Koehl U, Grez M, Cattaneo R, Cichutek K, Buchholz CJ. 2008. Targeted cell entry of lentiviral vectors. Mol Ther 16:1427–1436. doi: 10.1038/mt.2008.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kuhn AN, Diken M, Kreiter S, Selmi A, Kowalska J, Jemielity J, Darzynkiewicz E, Huber C, Tureci O, Sahin U. 2010. Phosphorothioate cap analogs increase stability and translational efficiency of RNA vaccines in immature dendritic cells and induce superior immune responses in vivo. Gene Ther 17:961–971. doi: 10.1038/gt.2010.52. [DOI] [PubMed] [Google Scholar]
- 63.Kreiter S, Konrad T, Sester M, Huber C, Tureci O, Sahin U. 2007. Simultaneous ex vivo quantification of antigen-specific CD4+ and CD8+ T cell responses using in vitro transcribed RNA. Cancer Immunol Immunother 56:1577–1587. doi: 10.1007/s00262-007-0302-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lyons AB, Parish CR. 1994. Determination of lymphocyte division by flow cytometry. J Immunol Methods 171:131–137. doi: 10.1016/0022-1759(94)90236-4. [DOI] [PubMed] [Google Scholar]
- 65.Corman VM, Eckerle I, Bleicker T, Zaki A, Landt O, Eschbach-Bludau M, van Boheemen S, Gopal R, Ballhause M, Bestebroer TM, Muth D, Muller MA, Drexler JF, Zambon M, Osterhaus AD, Fouchier RM, Drosten C. 2012. Detection of a novel human coronavirus by real-time reverse-transcription polymerase chain reaction. Euro Surveill 17:pii-20285 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20285. [DOI] [PubMed] [Google Scholar]
- 66.Coleman CM, Matthews KL, Goicochea L, Frieman MB. 2014. Wild-type and innate immune-deficient mice are not susceptible to the Middle East respiratory syndrome coronavirus. J Gen Virol 95:408–412. doi: 10.1099/vir.0.060640-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Coleman CM, Liu YV, Mu H, Taylor JK, Massare M, Flyer DC, Glenn GM, Smith GE, Frieman MB. 2014. Purified coronavirus spike protein nanoparticles induce coronavirus neutralizing antibodies in mice. Vaccine 32:3169–3174. doi: 10.1016/j.vaccine.2014.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ma C, Wang L, Tao X, Zhang N, Yang Y, Tseng CK, Li F, Zhou Y, Jiang S, Du L. 2014. Searching for an ideal vaccine candidate among different MERS coronavirus receptor-binding fragments: the importance of immunofocusing in subunit vaccine design. Vaccine 32:6170–6176. doi: 10.1016/j.vaccine.2014.08.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dhiman N, Jacobson RM, Poland GA. 2004. Measles virus receptors: SLAM and CD46. Rev Med Virol 14:217–229. doi: 10.1002/rmv.430. [DOI] [PubMed] [Google Scholar]
- 70.Mühlebach MD, Mateo M, Sinn PL, Prüfer S, Uhlig KM, Leonard VH, Navaratnarajah CK, Frenzke M, Wong XX, Sawatsky B, Ramachandran S, McCray PB, Cichutek K, von Messling V, Lopez M, Cattaneo R. 2011. Adherens junction protein nectin-4 (PVRL4) is the epithelial receptor for measles virus. Nature 480:530–533. doi: 10.1038/nature10639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.De Maeyer E, Enders JF. 1961. An interferon appearing in cell cultures infected with measles virus. Exp Biol Med 107:573–578. doi: 10.3181/00379727-107-26692. [DOI] [PubMed] [Google Scholar]
- 72.Leopardi R, Hyypiä T, Vainionpää R. 1992. Effect of interferon-alpha on measles virus replication in human peripheral blood mononuclear cells. APMIS 100:125–131. doi: 10.1111/j.1699-0463.1992.tb00850.x. [DOI] [PubMed] [Google Scholar]
- 73.Mills CD, Kincaid K, Alt JM, Heilman MJ, Hill AM. 2000. M-1/M-2 macrophages and the Th1/Th2 paradigm. J Immunol 164:6166–6173. doi: 10.4049/jimmunol.164.12.6166. [DOI] [PubMed] [Google Scholar]
- 74.Scott P. 1993. Selective differentiation of CD4+ T helper cell subsets. Curr Opin Immunol 5:391–397. doi: 10.1016/0952-7915(93)90058-Z. [DOI] [PubMed] [Google Scholar]
- 75.Brenner GJ, Cohen N, Moynihan JA. 1994. Similar immune response to nonlethal infection with herpes simplex virus-1 in sensitive (BALB/c) and resistant (C57BL/6) strains of mice. Cell Immunol 157:510–524. doi: 10.1006/cimm.1994.1246. [DOI] [PubMed] [Google Scholar]
- 76.Chow Ken YC, Yeung YS, Hon CC, Zeng F, Law KM, Leung Frederick CC. 2005. Adenovirus-mediated expression of the C-terminal domain of SARS-CoV spike protein is sufficient to induce apoptosis in Vero E6 cells. FEBS Lett 579:6699–6704. doi: 10.1016/j.febslet.2005.10.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yeung Y, Yip C, Hon C, Chow Ken YC, Ma Iris CM, Zeng F, Leung Frederick CC. 2008. Transcriptional profiling of Vero E6 cells over-expressing SARS-CoV S2 subunit: insights on viral regulation of apoptosis and proliferation. Virology 371:32–43. doi: 10.1016/j.virol.2007.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liu R, Wu L, Huang B, Huang J, Zhang Y, Ke M, Wang J, Tan W, Zhang R, Chen H, Zeng Y, Huang W. 2005. Adenoviral expression of a truncated S1 subunit of SARS-CoV spike protein results in specific humoral immune responses against SARS-CoV in rats. Virus Res 112:24–31. doi: 10.1016/j.virusres.2005.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Weingartl H, Czub M, Czub S, Neufeld J, Marszal P, Gren J, Smith G, Jones S, Proulx R, Deschambault Y, Grudeski E, Andonov A, He R, Li Y, Copps J, Grolla A, Dick D, Berry J, Ganske S, Manning L, Cao J. 2004. Immunization with modified vaccinia virus Ankara-based recombinant vaccine against severe acute respiratory syndrome is associated with enhanced hepatitis in ferrets. J Virol 78:12672–12676. doi: 10.1128/JVI.78.22.12672-12676.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Czub M, Weingartl H, Czub S, He R, Cao J. 2005. Evaluation of modified vaccinia virus Ankara based recombinant SARS vaccine in ferrets. Vaccine 23:2273–2279. doi: 10.1016/j.vaccine.2005.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.An S, Chen CJ, Yu X, Leibowitz JL, Makino S. 1999. Induction of apoptosis in murine coronavirus-infected cultured cells and demonstration of E protein as an apoptosis inducer. J Virol 73:7853–7859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Subbarao K, McAuliffe J, Vogel L, Fahle G, Fischer S, Tatti K, Packard M, Shieh W, Zaki S, Murphy B. 2004. Prior infection and passive transfer of neutralizing antibody prevent replication of severe acute respiratory syndrome coronavirus in the respiratory tract of mice. J Virol 78:3572–3577. doi: 10.1128/JVI.78.7.3572-3577.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Seo SH, Wang L, Smith R, Collisson EW. 1997. The carboxyl-terminal 120-residue polypeptide of infectious bronchitis virus nucleocapsid induces cytotoxic T lymphocytes and protects chickens from acute infection. J Virol 71:7889–7894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yu L, Liu W, Schnitzlein WM, Tripathy DN, Kwang J. 2001. Study of protection by recombinant fowl poxvirus expressing C-terminal nucleocapsid protein of infectious bronchitis virus against challenge. Avian Dis 45:340–348. doi: 10.2307/1592973. [DOI] [PubMed] [Google Scholar]
- 85.Du L, Zhao G, Lin Y, Sui H, Chan C, Ma S, He Y, Jiang S, Wu C, Yuen K, Jin D, Zhou Y, Zheng B. 2008. Intranasal vaccination of recombinant adeno-associated virus encoding receptor-binding domain of severe acute respiratory syndrome coronavirus (SARS-CoV) spike protein induces strong mucosal immune responses and provides long-term protection against SARS-CoV infection. J Immunol 180:948–956. doi: 10.4049/jimmunol.180.2.948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Agrawal AS, Garron T, Tao X, Peng B, Wakamiya M, Chan T, Couch RB, Tseng CK. 2015. Generation of transgenic mouse model of Middle East respiratory syndrome-coronavirus infection and disease. J Virol 89:3659–3670. doi: 10.1128/JVI.03427-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tangy F, Naim HY. 2005. Live attenuated measles vaccine as a potential multivalent pediatric vaccination vector. Viral Immunol 18:317–326. doi: 10.1089/vim.2005.18.317. [DOI] [PubMed] [Google Scholar]
- 88.Garbutt M, Liebscher R, Wahl-Jensen V, Jones S, Moller P, Wagner R, Volchkov V, Klenk H, Feldmann H, Stroher U. 2004. Properties of replication-competent vesicular stomatitis virus vectors expressing glycoproteins of filoviruses and arenaviruses. J Virol 78:5458–5465. doi: 10.1128/JVI.78.10.5458-5465.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jones SM, Feldmann H, Stroher U, Geisbert JB, Fernando L, Grolla A, Klenk H, Sullivan NJ, Volchkov VE, Fritz EA, Daddario KM, Hensley LE, Jahrling PB, Geisbert TW. 2005. Live attenuated recombinant vaccine protects nonhuman primates against Ebola and Marburg viruses. Nat Med 11:786–790. doi: 10.1038/nm1258. [DOI] [PubMed] [Google Scholar]
- 90.World Health Organization. 2014. WHO welcomes approval of a second Ebola vaccine trial in Switzerland. World Health Organization, Geneva, Switzerland: http://www.who.int/mediacentre/news/statements/2014/second-ebola-vaccine/en/. [Google Scholar]