ABSTRACT
An accessory gene between the S and E gene loci is contained in all coronaviruses (CoVs), and its function has been studied in some coronaviruses. This gene locus in human coronavirus OC43 (HCoV-OC43) encodes the ns12.9 accessory protein; however, its function during viral infection remains unknown. Here, we engineered a recombinant mutant virus lacking the ns12.9 protein (HCoV-OC43-Δns12.9) to characterize the contributions of ns12.9 in HCoV-OC43 replication. The ns12.9 accessory protein is a transmembrane protein and forms ion channels in both Xenopus oocytes and yeast through homo-oligomerization, suggesting that ns12.9 is a newly recognized viroporin. HCoV-OC43-Δns12.9 presented at least 10-fold reduction of viral titer in vitro and in vivo. Intriguingly, exogenous ns12.9 and heterologous viroporins with ion channel activity could compensate for the production of HCoV-OC43-Δns12.9, indicating that the ion channel activity of ns12.9 plays a significant role in the production of infectious virions. Systematic dissection of single-cycle replication revealed that ns12.9 protein had no measurable effect on virus entry, subgenomic mRNA (sgmRNA) synthesis, and protein expression. Further characterization revealed that HCoV-OC43-Δns12.9 was less efficient in virion morphogenesis than recombinant wild-type virus (HCoV-OC43-WT). Moreover, reduced viral replication, inflammatory response, and virulence in HCoV-OC43-Δns12.9-infected mice were observed compared to the levels for HCoV-OC43-WT-infected mice. Taken together, our results demonstrated that the ns12.9 accessory protein functions as a viroporin and is involved in virion morphogenesis and the pathogenesis of HCoV-OC43 infection.
IMPORTANCE HCoV-OC43 was isolated in the 1960s and is a major agent of the common cold. The functions of HCoV-OC43 structural proteins have been well studied, but few studies have focused on its accessory proteins. In the present study, we demonstrated that the ns12.9 protein is a newly recognized viroporin, and the ns12.9 gene knockout virus (HCoV-OC43-Δns12.9) presents a growth defect in vitro and in vivo. We identified the important functions of the ns12.9 viroporin in virion morphogenesis during HCoV-OC43 infection. Furthermore, mice infected with HCoV-OC43-Δns12.9 exhibited reduced inflammation and virulence accompanied by a lower titer in the brain than that of wild-type-infected mice, suggesting the ns12.9 viroporin influences virus pathogenesis. Therefore, our findings revealed that the ns12.9 viroporin facilitates virion morphogenesis to enhance viral production, and these results provided a deeper understanding of HCoV-OC43 pathogenesis.
INTRODUCTION
The coronaviruses (CoVs) belong to the Coronaviridae family of the order Nidovirales and are distributed widely among animals, birds, and humans (1). Members of the CoVs are further classified into four genera: Alphacoronavirus, Betacoronavirus, Gammacoronavirus, and Deltacoronavirus (2). Human coronavirus OC43 (HCoV-OC43) was isolated from a patient with upper respiratory tract disease in the 1960s and classified into the Betacoronavirus genus (3). HCoV-OC43 causes mild upper respiratory infection and is identified as a major etiological agent of the common cold (4). Additionally, this virus exhibits neuroinvasive properties that lead to neurological diseases (5–7).
The genome of CoVs is a single-stranded, positive-sense RNA that is 27 to 32 kb in length, and the genome is 5′-capped and 3′-polyadenylated. Approximately two-thirds of the 5′-proximal genome consists of the ORF1a/b replicase gene, whereas the remainder of the genome encodes several accessory proteins and the following four major structural proteins: spike (S), envelope (E), membrane (M), and nucleocapsid (N) proteins (1). The replicase gene encodes two large polyproteins, namely, pp1a and pp1ab, which form a set of nonstructural proteins with autoproteolytic cleavage. These nonstructural proteins are essential for viral transcription, RNA replication, and pathogenesis (8). The S, E, and M proteins are transmembrane proteins embedded in the viral lipid envelope. The S protein interacts with the corresponding host receptors to mediate the virus entry process (9–12). The E and M proteins are critical for viral morphogenesis. Studies have shown that the expression of the E protein with M protein is sufficient to form virus-like particles (VLPs) in vitro (13–15). The major function of the N protein involves binding the viral RNA to form a helical nucleocapsid that is surrounded by the viral envelope (1). The members of lineage A of genus Betacoronavirus, including HCoV-OC43, bovine coronavirus (BCoV), mouse hepatitis virus (MHV), and HCoV-HKU1, possess a hemagglutinin-esterase (HE) gene between the ORF1a/b and S genes. The HE protein is considered the fifth structural protein that incorporates into the viral envelope, and this protein participates in the entry and release process of viral infection (16, 17).
In addition to the structural genes, the genome of CoVs contains accessory genes interspersed among the other genes. The accessory proteins of various CoVs are involved in viral pathogenesis and virulence (18–21), and some of these proteins are dispensable for viral replication (22–24). CoV accessory genes exhibit individual specificity, with numbers varying from one (e.g., HCoV-NL63) to eight (e.g., SARS-CoV). Strikingly, an accessory gene between the S and E gene loci is contained in all CoVs, suggesting that it plays important and conserved roles during CoV infection (1). In our previous study, we found that this accessory protein of SARS-CoV (SARS-3a) and HCoV-229E (229E-ORF4a) forms ion channels (25, 26). These ion channel proteins are identified as viroporins, which is a viral protein family that forms ion channels to permeabilize the membrane and regulate viral infection (27).
Genome nucleotide sequence analysis indicates that the accessory gene between S and E gene loci of HCoV-OC43 encodes a potential protein with a molecular mass of 12.9 kDa, and this protein is named ns12.9 (28). However, the function of the ns12.9 accessory protein during HCoV-OC43 infection is not well known. In the present study, we revealed that the ns12.9 accessory protein is a new member of the viroporins and facilitates virion morphogenesis to enhance viral production. To our knowledge, this is the first study to define the function of an accessory protein in the virion formation stage during CoV infection. Moreover, HCoV-OC43-Δns12.9 is attenuated in virulence, suggesting that HCoV-OC43-Δns12.9 can be developed as a candidate vaccine to protect against HCoV-OC43 infection.
MATERIALS AND METHODS
Cells and viruses.
HEK293T, BHK-21, and RD cells were cultured in Dulbecco's modified Eagle's medium (DMEM; Gibco) supplemented with 10% fetal bovine serum (FBS; Gibco), penicillin (100 U/ml), and streptomycin (100 μg/ml) in a humidified atmosphere of 5% CO2 at 37°C. The recombinant wild-type HCoV-OC43 (HCoV-OC43-WT) and ns12.9 knockout HCoV-OC43 mutant (HCoV-OC43-Δns12.9) were rescued from the infective cDNA clones pBAC-OC43-WT and pBAC-OC43-Δns12.9, respectively. Separate laminar flow hoods and CO2 incubators were used during the experiments to avoid possible cross-contamination. Mutation within the ns12.9 gene of HCoV-OC43-Δns12.9 remained very stable in vitro and in vivo, since no changes were found in the ns12.9 gene during the course of experiments. The recombinant HCoV-OC43-WT and HCoV-OC43-Δns12.9 viral stocks (106 50% tissue culture infective doses [TCID50/ml]) were maintained at −80°C.
Plasmid construction.
Sequences encoding SARS-3a, NL63-ORF3, 229E-ORF4a, OC43-ns12.9, influenza A virus (IAV) M2, hepatitis C virus (HCV) p7, and human enterovirus 71 (EV71) 2B were amplified by PCR and cloned into the pCAGGS vector (a kind gift from Jun-ichi Miyazaki, Osaka University, Japan) with a C-terminal HA tag or Flag tag for expression. ns12.9-HA sequence was cloned into pNWP vector (a kind gift from Jian Fei, Shanghai Institute of Biological Science, China) for cRNA in vitro transcription and cloned into a yeast expression vector, pYES2 (a kind gift from Wei Song, Shanghai Institute of Biological Science, China), for a yeast potassium uptake complementation assay. The infective full-length HCoV-OC43 cDNA clone pBAC-OC43-WT was kindly provided by Pierre J. Talbot (INRS-Institut Armand-Frappier, Québec, Canada). The pBAC-OC43-Δns12.9 cDNA clone was constructed in our laboratory by following a previously described protocol (29). Briefly, a cassette containing a stop codon at the fourth amino acid of the ns12.9 gene followed by the sequence with a selective kanamycin marker flanked by flippase recognition target (FRT) sites was amplified from the pYD-C191 plasmid with a pair of 70-nucleotide (nt) primers as the following primers: OC43-FRT-F (forward), 5′-CTAGCATTTGTTAAAGTTCTTAAGGCCACGCCCTATTAATGGACATTTGAAAGGACGACGACGACAAGTAA-3′; OC43-FRT-R (reverse), 5′-TCTGAGACATTAAAACCGTTAATATAACGGAGATATTTCTTCTCAGGTCTACCACGTCGTGGAATGCCTTC-3′. The underlined 50-nt sequences were homologous to the viral genome sequences immediately upstream or downstream of the mutant nucleotide position (boldface). This cassette subsequently was recombined into the pBAC-OC43-WT cDNA clone by linear recombination in the SW102 bacterial strain. Resulting transformants were selected on LB plates with chloramphenicol and kanamycin to identify the mutant cDNA clone. Finally, arabinose was added to the culture media to induce Flp recombinase expression to remove the kanamycin sequence. Therefore, the resulting pBAC-OC43-Δns12.9 cDNA clone contained a single-nucleotide mutation at position 12 of the ns12.9 gene in addition to an 82-nt segment insertion. This 82-nt sequence consisted of one FRT site (underlined) and an EcoRI restriction site (italic), namely, AAGGACGACGACGACAAGTAAGAAGTTCCTATTCTCTAGAAAGTATAGGAACTTCGAATTCGAAGGCATTCCACGACGTGGT. All plasmids were verified by restriction digestion analysis and direct sequencing.
Antibodies.
The anti-hemagglutinin (HA) mouse monoclonal antibody (MMS-101P) was purchased from Covance. Anti-HA (H6908) and anti-Flag (F7425) rabbit polyclonal antibodies were purchased from Sigma. The Alexa Fluor 488-conjugated goat anti-mouse antibody (A11029) and Cy3-conjugated goat anti-rabbit antibody (111-165-045) were obtained from Molecular Probes. The OC43-N mouse monoclonal antibody (MAB9012) was purchased from Merck Millipore. The horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG antibody and HRP-conjugated goat anti-rabbit IgG antibody were obtained from the Antibody Research Center (Shanghai Institute of Biochemistry and Cellular Biology, Chinese Academy of Sciences, Shanghai, China). The ERGIC53 antibody (H-245), a rabbit polyclonal antibody against the endoplasmic reticulum/Golgi intermediate compartment (ERGIC), was obtained from Santa Cruz Biotechnology. The mouse interleukin-1β (IL-1β; DY401) and IL-6 (DY406) enzyme-linked immunosorbent assay (ELISA) kits were purchased from R&D Systems.
Confocal microscopy.
Cells were washed with phosphate-buffered saline (PBS) at 24 h after infection or transfection, fixed with 4% paraformaldehyde (PFA), and then permeabilized with 0.3% Triton X-100. The cells were blocked with 2% bovine serum albumin (BSA) and immunolabeled with primary antibodies for 2 h at room temperature. Cells were washed with PBS and incubated with Cy3-conjugated goat anti-rabbit antibody or Alexa Fluor 488-conjugated goat anti-mouse antibody for 1 h. Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI) (D9542; Sigma-Aldrich). Colocalization studies were examined using a Leica TCS SP5 confocal microscope (Leica Microsystems).
Coimmunoprecipitation and Western blotting.
Transfected cells were lysed in radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1% NP-40, 0.25% Na-deoxycholate, 1 mM EDTA) with protease inhibitor (Roche, Switzerland). Cell lysates were centrifuged at 15,000 × g for 20 min at 4°C, and the supernatant was incubated with anti-Flag M2 affinity gel (A2220; Sigma) or anti-HA agarose (A2095; Sigma) at 4°C overnight. The gels or agaroses then were washed 5 times with RIPA buffer and lysed in sodium dodecyl sulfate (SDS) loading buffer (50 mM Tris-HCl, pH 6.8, 2% SDS, 10% glycerol, 1% β-mercaptoethanol, and 0.1% bromophenol blue).
The proteins were separated by 15% SDS-PAGE and transferred to nitrocellulose membranes (Bio-Rad). Membranes were blocked with 5% skim milk for 1 h and incubated with primary antibody overnight at 4°C. After washes with TBST (50 mM Tris, 150 mM NaCl, and 0.1% Tween 20, pH 7.5), the membranes were further incubated for 1 h with HRP-conjugated secondary antibody. The protein expression was visualized using SuperSignal West Pico chemiluminescent substrate (Thermo Scientific).
Flow cytometry.
The transfected cells or infected cells were collected and washed with PBS. Cells then were fixed with 4% PFA, immunolabeled with primary antibody, and incubated with an Alexa Fluor 488-conjugated goat anti-mouse antibody. Cells were resuspended in PBS and analyzed by flow cytometry (FACS-LSRII; BD Biosciences). Data analysis was performed using FlowJo software (TreeStar).
Electrophysiological measurements.
The ns12.9-HA cRNA was synthesized using the mMESSAGE mMACHINE high-yield capped RNA transcription SP6 kit (Ambion, USA) from the pNWP-ns12.9-HA plasmid. Healthy oocytes in stages V to VI were injected with 30 ng of cRNA and incubated at 18°C in an ND-96 solution (96 mM NaCl, 2 mM KCl, 1.8 mM CaCl2, 1 mM MgCl2, 2.5 mM pyruvate, and 5 mM HEPES, adjusted to pH 7.4 with NaOH). A two-electrode voltage clamp was used to record the currents mediated by ns12.9 protein from the membranes of Xenopus oocytes as described previously (26). Briefly, the standard voltage clamp protocol consisted of rectangular voltage pulses from −150 to +30 mV in 10-mV increments applied from a holding voltage of −60 mV. During the current recording, the oocytes were bathed in ORi solution (90 mM NaCl, 2 mM KCl, 2 mM CaCl2, and 5 mM HEPES, adjusted to pH 7.4 with NaOH) at room temperature. Current recording and analysis were performed with pClamp 10.0 software (Axon Instruments).
Yeast potassium uptake complementation assay.
Either the empty pYES2 or pYES2-ns12.9-HA vector were transformed into a potassium uptake-deficient yeast strain, W303 R5421 (ura3-52 his3Δ200 leu2Δ1 trp1Δ1 ade2 trk1Δ::HIS3 trk2Δ::HIS3) (a kind gift from Richard F. Gaber, Northwestern University), with the lithium acetate procedure. The pYES2 vector contains a URA3 gene as a selectable marker for positive transformants in ura-negative hosts. The exogenous gene is controlled by the GAL1 promoter, and its expression was induced in the presence of galactose. The complementation of the potassium uptake-deficient yeast was performed as previously described (25). Briefly, yeasts from the same stock were diluted and grown in parallel on YNB ura-negative plates supplemented with 100 mM KCl or 0.2 mM KCl. Plates were kept at 30°C for 3 to 4 days.
Rescue of recombinant HCoV-OC43 viruses.
BHK-21 cells were seeded in 6-well plates and transfected with 5 μg of wild-type or mutant cDNA clone using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. The culture medium was replaced by DMEM supplemented with 2% FBS 24 h after transfection and maintained at 33°C for an additional 72 h. Cell supernatants containing recombinant virus corresponding to the cDNA clone were amplified in RD cells and quantified by the immunoperoxidase assay (IPA).
Titration of rescued virus using IPA.
The titers of infectious HCoV-OC43 virions were determined by IPA as previously reported (30, 31). Briefly, RD cells were seeded in 96-well plates and infected with each virus in a serial dilution of 10−1 to 10−7. Cells were incubated for 4 days, and the cells then were fixed with 4% PFA and permeabilized with 0.3% Triton X-100. The monoclonal mouse antibody specific for the HCoV-OC43 N protein and HRP-conjugated goat anti-mouse IgG antibody were used as the primary and secondary antibodies, respectively. The viral antigens were visualized using TrueBlue peroxidase substrate (KPL), and the viral titers were calculated by the Karber method. For intracellular virions, cells were disrupted by three cycles of freeze-thawing in dry ice and a 37°C water bath, and the viral titers then were quantified as described above.
Viral growth kinetics.
RD or BHK-21 cells were plated into 12-well plates and infected at a multiplicity of infection (MOI) of 0.1. Media (2 ml) were added to each well, and 300 μl of supernatant was collected at 0, 4, 24, 48, 72, 96, 120, and 144 h postinfection (hpi). Viral titers were determined by IPA as described above.
RNA extraction and quantitative RT-PCR.
Viral genomic RNA was extracted from cell supernatants using the TIANamp virus RNA kit (Tiangen) by following the manufacturer's instructions. The ns12.9 gene was checked using the following primers: ns12.9-F (forward), 5′-GGTGGTTGTTGTGATGATTATACTGGATACC-3′; ns12.9-R (reverse), 5′-CCACTACAACTATTGTAACCAATAAACAAATGG-3′. Total RNA from virus-infected cells was extracted using TRIzol reagent (Invitrogen). RNA (1 μg) then was used for reverse transcription using the ReverTra Ace qPCR RT kit (Toyobo). The target gene described above was amplified by PCR from the cDNA. The copies of genomic RNA and subgenomic mRNA (sgmRNA) in infected cells were determined by quantitative real-time PCR (qRT-PCR) using SYBR green real-time PCR master mix (Toyobo). β-Actin was used as an internal control. All of the primers are presented in Table 1.
TABLE 1.
Primers used to test the copies of genomic RNA and sgmRNAs by qRT-PCR
| Primer | Sequence (5′-3′) |
|---|---|
| OC43-RT-F | GTTTGAGGACGCAGAGGAGAAG |
| OC43-RT-R | AAGAAGTCGGCGACAATCCACC |
| Leader-F | CATCCCGCTTCACTGATCTCTTG |
| Rep1a/b-sgmRNA-R | CGCCCACAAGCATAGATTACAGG |
| ns2-sgmRNA-R | AACCCTGAAAATGGGTAAGTGG |
| HE-sgmRNA-R | CGAAACAACATTGGTAGGAGGGT |
| S-sgmRNA-R | TAGGAGGAGGACCGGTGTCTTTATC |
| ns12.9-sgmRNA-R | ACGGAGATATTTCTTCTCAGGTCT |
| E-sgmRNA-R | TTATTTGCCCCACATACCACACAG |
| M-sgmRNA-R | TTTAATAGCTTCATCAGCAGTCCAG |
| N-sgmRNA-R | CCCACTTGAGGATGCCATTACCAG |
| β-actin-F | ACGTTGCTATCCAGGCTGTG |
| β-actin-R | GAGGGCATACCCCTCGTAGA |
Transmission electron microscopy.
RD cells were infected with recombinant WT or Δns12.9 viruses at an MOI of 1 for 24 h. The cells were washed with PBS and fixed with 2% glutaraldehyde overnight at 4°C. The samples then were treated with 2% osmium tetroxide for 1 h, dehydrated gradually through an ethanol series (30, 50, 70, 90, and 100%), and embedded in Epon 812 resin. The infiltrated samples were polymerized at 60°C for 48 h. Ultrathin sections (70 nm) of the cells were produced using an EM UC6 ultramicrotome (Leica) and stained with 2% uranyl acetate and 1% lead citrate. The sections were analyzed with a Tecnai Spirit transmission electron microscope (FEI).
Mice and infection.
Specific-pathogen-free BALB/c mice were obtained from the Jackson Laboratory. Mice were maintained at the Animal Care Facility of the Chinese Academy of Sciences. Groups of 10-day-old mice were intranasally inoculated with 5 μl of the HCoV-OC43-WT or HCoV-OC43-Δns12.9 viral stock (32). The infected mice were monitored daily for weight loss and survival. Four mice of each group were sacrificed at 2, 4, and 6 days postinfection (dpi), and the brain tissues were collected. All protocols complied with the Institutional Animal Care and Use Committee guidelines. The brain tissues were homogenized in 30% (wt/vol) PBS and centrifuged at 12,000 × g for 20 min at 4°C, and the supernatants then were collected to detect the viral titers or cytokine levels.
ELISA.
Mouse proinflammatory cytokines were measured by ELISA according to the manufacturer's instructions.
RESULTS
The ns12.9 accessory protein acts as a viroporin.
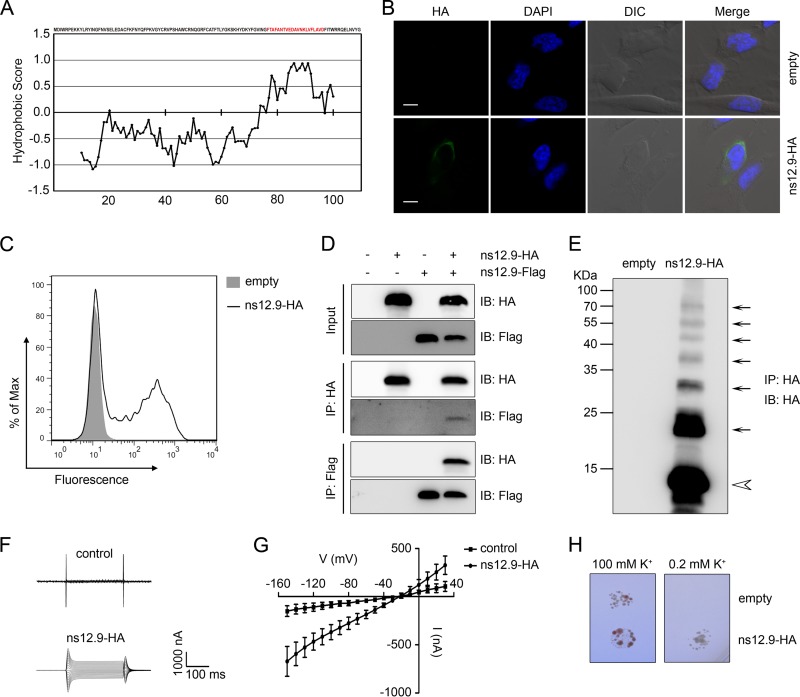
Viroporins are a group of small hydrophobic proteins that tend to oligomerize to form hydrophilic pores or ion channels in cellular membrane (27). Since we previously demonstrated that SARS-3a and 229E-ORF4a are viroporins, we wanted to evaluate whether this accessory protein, ns12.9, in HCoV-OC43 acts as a viroporin. Analysis of the hydrophobicity of ns12.9 protein sequence with the Kyte-Doolittle hydropathy plot (33) revealed that ns12.9 contains a hydrophobic segment with 21 amino acids which could be a potential transmembrane domain (TMD) of integral membrane proteins (Fig. 1A). The plasma membrane localization of ns12.9 then was tested with immunofluorescence staining. As shown in Fig. 1B and C, C-terminally HA-tagged ns12.9 was detected at the cell membrane surfaces of nonpermeabilized cells, suggesting that ns12.9 is a transmembrane protein with an intracellular N terminus and an extracellular C terminus.
FIG 1.
ns12.9 accessory protein of HCoV-OC43 acts as a viroporin. (A) Kyte-Doolittle hydropathy plot of ns12.9 protein. The hydropathy plot was drawn with a window size of 19 using the Kyte-Doolittle method of hydrophilicity calculation. (B) Indirect immunofluorescence analysis of cell membrane expression of ns12.9 in RD cells transfected with pCAGGS-ns12.9-HA or empty vectors. Cells were fixed, nonpermeabilized, and subjected to incubation with anti-HA monoclonal antibody, followed by staining with Alexa Fluor 488-conjugated goat anti-mouse antibody (green). Nuclei were stained with DAPI (blue). Bars represent 15 μm. DIC, differential inference contrast. (C) Flow cytometry analysis of cell membrane expression of ns12.9 in 293T cells transfected with pCAGGS-ns12.9-HA or empty vectors. Cells were nonpermeabilized and stained with anti-HA monoclonal antibody, followed by Alexa Fluor 488-conjugated goat anti-mouse antibody. (D) Reciprocal coimmunoprecipitation assay of ns12.9-HA and ns12.9-Flag in 293T cells. Cells were transfected with pCAGGS-ns12.9-HA or pCAGGS-ns12.9-Flag vectors and lysed at 24 h posttransfection. Cell lysates were subjected to immunoprecipitation (IP) with anti-HA agarose or anti-Flag M2 affinity gel. The immunoprecipitated proteins were determined by Western blotting using polyclonal anti-Flag and anti-HA antibody. IB, immunoblot. (E) Oligomerization of the ns12.9-HA protein. The HA immunoprecipitates from cells transfected with pCAGGS-ns12.9-HA or empty vectors were analyzed by Western blotting with polyclonal anti-HA antibody. The monomer and oligomers are indicated with an arrowhead and arrows, respectively. Representative current traces (F) and I/V curve of the currents (G) in ns12.9-expressing and control oocytes are shown. During the current recording, the oocytes were bathed in ORi solution, and the standard voltage clamp protocol consisted of rectangular voltage pulses from −150 to +30 mV in 10-mV increments applied from a holding voltage of −60 mV. Data represent the means ± standard deviations (SD) (n = 5). (H) Complementation of the potassium uptake-deficient yeast. The yeasts transformed with empty or ns12.9-expressing vector were grown in parallel on plates supplemented with 100 mM or 0.2 mM KCl. Plates were kept at 30°C for 3 to 4 days.
We next sought to investigate whether ns12.9 could form homo-oligomers. Coimmunoprecipitation assays were performed with 293T cells transiently transfected with plasmids encoding HA-tagged and Flag-tagged ns12.9. Notably, coimmunoprecipitation of ns12.9-HA with ns12.9-Flag was detected (Fig. 1D), indicating that ns12.9 is able to oligomerize. To confirm the self-oligomerization of ns12.9, the ns12.9 oligomers were analyzed in anti-HA immunoprecipitates. As shown in Fig. 1E, the monomer and oligomers of ns12.9 were separated by SDS-PAGE, confirming the homo-oligomerization of ns12.9.
Finally, to assess the ion channel activity of ns12.9, membrane currents of ns12.9 cRNA-injected Xenopus oocytes were recorded using a two-electrode voltage clamp (TEVC). A dramatic increase of membrane conductance was detected in the ns12.9-expressing oocytes compared with that of the control oocytes (Fig. 1F and G). In addition, a yeast complementation assay was performed using the potassium uptake-deficient yeast, which grows poorly on low-potassium medium. As shown in Fig. 1H, the growth of ns12.9-expressing transformants could be rescued on the low-potassium (0.2 mM) medium, whereas empty transformants only grew on the high-potassium (100 mM) medium. Therefore, ns12.9 forms ion channels in Xenopus oocytes and yeast.
Collectively, these observations suggest that the ns12.9 accessory protein acts as a viroporin in that it oligomerizes in the cell membrane to form ion channels.
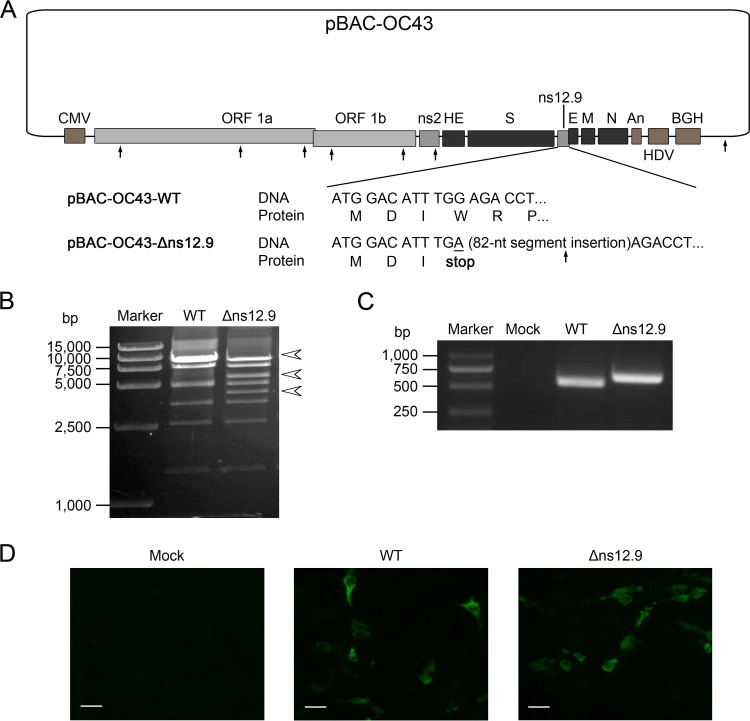
Rescue of the ns12.9-knockout HCoV-OC43 mutant.
To investigate the function of ns12.9 during HCoV-OC43 infection, a recombinant mutant virus defective in the ns12.9 gene was rescued by reverse genetics. For this purpose, we constructed an ns12.9 gene knockout cDNA clone (pBAC-OC43-Δns12.9) in which the fourth codon of the ns12.9 coding sequence was changed to a stop codon. In addition, an 82-nt segment was inserted behind the mutant stop codon to avoid the occurrence of genetic reversion (Fig. 2A). The pBAC-OC43-Δns12.9 mutant cDNA clone was engineered from the infective wild-type clone (pBAC-OC43-WT) by homologous recombination and examined by digestion with the EcoRI restriction enzyme and direct sequencing (Fig. 2B and data not shown). To determine whether the virus was rescued from the cDNA clone in transfected BHK-21 cells, supernatants from transfected cells were harvested and used to infect the RD cells. After three rounds of amplification in RD cells, the existence of viral RNA and N protein in supernatants and RD cells, respectively, was analyzed. As an 82-nt segment was inserted in the ns12.9 gene within pBAC-OC43-Δns12.9, the ns12.9 gene amplified from HCoV-OC43-Δns12.9-infected cells was slightly larger than that amplified from HCoV-OC43-WT-infected cells (Fig. 2C). In addition, similar to the wild-type virus, the OC43-N proteins were detectable in the mutant virus-infected RD cells (Fig. 2D). These results suggest that the HCoV-OC43-Δns12.9 virus was successfully rescued by the mutant cDNA clone.
FIG 2.
Rescue of HCoV-OC43-Δns12.9 from the recombinant mutant pBAC-OC43-Δns12.9 cDNA clone. (A) Schematic representation of the wild-type (pBAC-OC43-WT) and ns12.9 knockout cDNA (pBAC-OC43-Δns12.9) clone. To construct the pBAC-OC43-Δns12.9 clone, the G12A nucleotide (underlined) was changed to a stop codon at Trp4, and an 82-nt segment was inserted behind the mutant stop codon to avoid the possibility of genetic reversion. The EcoRI restriction sites are denoted by arrows. CMV, cytomegalovirus; HDV, hepatitis delta virus ribozyme; BGH, bovine growth hormone termination and polyadenylation sequences. (B) EcoRI restriction digestion analysis of recombinant pBAC-OC43-WT and pBAC-OC43-Δns12.9. The pBAC-OC43-WT plasmid was cleaved into 7 fragments (9802, 9104, 7632, 4916, 3438, 2553, and 1427 bp), whereas the pBAC-OC43-Δns12.9 plasmid was cleaved into 8 fragments with an additional EcoRI restriction site at the 82-nt inserted segment that led to the 9,802-bp fragment being recut into 5,782- and 4,102-bp fragments. The arrowheads indicate different fragments between the pBAC-OC43-WT and pBAC-OC43-Δns12.9 plasmid. (C) Viral RNA was extracted from the supernatants of the infected RD cells and used to amplify the ns12.9 gene. (D) Indirect immunofluorescence analysis of HCoV-OC43 N proteins in RD cells. Cells were labeled with the OC43-N mouse monoclonal antibody, and the Alexa Fluor 488-conjugated goat anti-mouse antibody (green) was used as the secondary antibody. Bars represent 40 μm.
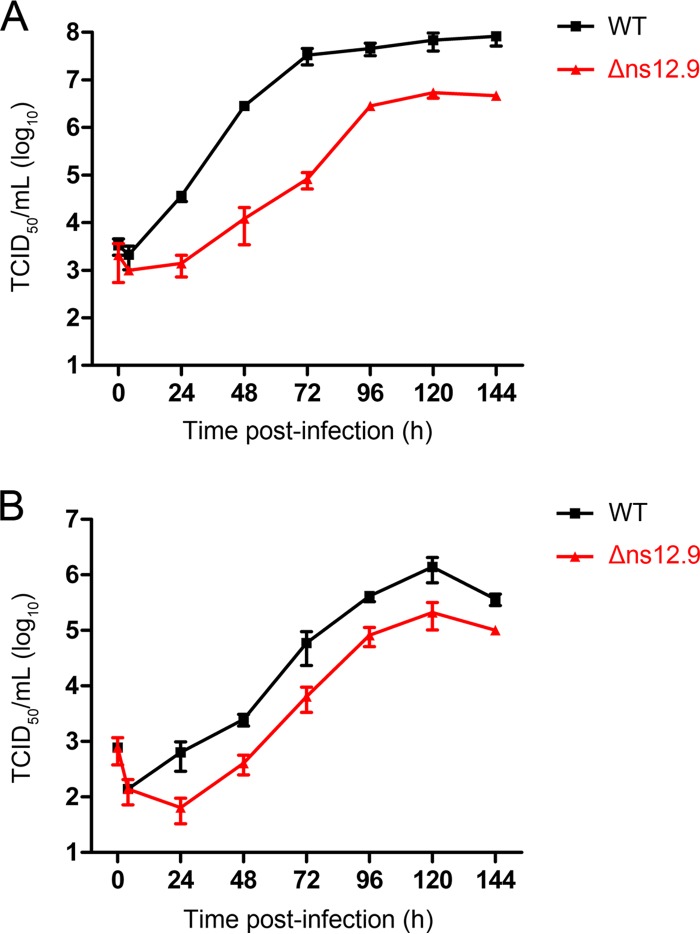
HCoV-OC43-Δns12.9 is defective in growth in vitro.
To evaluate the role of the ns12.9 viroporin contributing to viral production, we analyzed the growth kinetics of the recombinant HCoV-OC43-WT and HCoV-OC43-Δns12.9 mutant viruses in the BHK-21 and RD cells. In BHK-21 cells, the wild-type virus replicated efficiently and reached a maximal titer of approximately 108.0 TCID50/ml at 144 hpi. In contrast, the peak titer of the HCoV-OC43-Δns12.9 virus was approximately 106.6 TCID50/ml at 120 hpi, which was approximately 25-fold reduced compared to that of the wild-type virus (Fig. 3A). In RD cells, although the wild-type and mutant viruses exhibited a similar growth pattern with maximal titers between 96 and 144 hpi, the wild-type viral titer was 10-fold increased compared with that of the mutant virus (Fig. 3B). These data indicate that the ns12.9 viroporin is important for viral propagation in cell cultures.
FIG 3.
ns12.9 accessory protein is important for HCoV-OC43 replication in vitro. Growth kinetic analysis of HCoV-OC43-Δns12.9 in BHK-21 (A) and RD (B) cells is shown. Cells were infected with HCoV-OC43-WT or HCoV-OC43-Δns12.9 at an MOI of 0.1, and supernatants were collected at the indicated times to determine the viral titers using IPA. Data represent the means ± SD and were generated from three independent experiments.
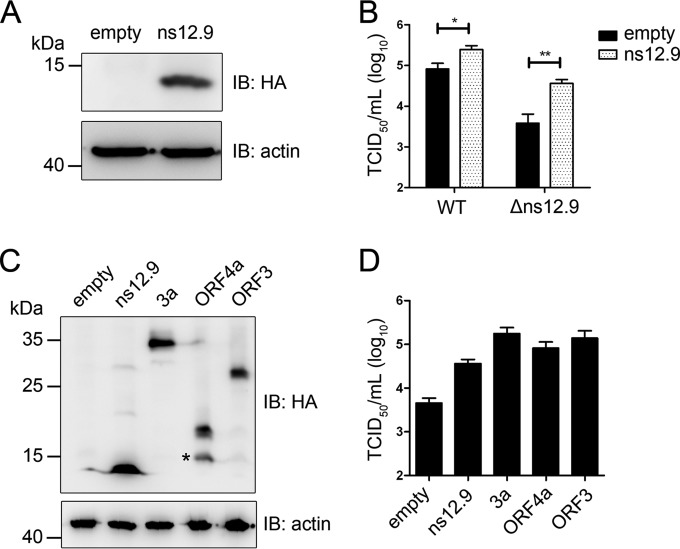
Transient expression of ns12.9 and this accessory protein from other coronaviruses complements HCoV-OC43-Δns12.9.
The reduced viral yield of HCoV-OC43-Δns12.9 in cultured cell lines suggested that the ns12.9 viroporin is important for viral infection. To explore whether the defect is due to the abolishment of ns12.9 viroporin expression, we performed a transient complementation assay. An ns12.9 protein expression vector was constructed, and its expression level was detected by Western blot analysis (Fig. 4A). In this assay, the empty or ns12.9 expression vector was transfected into RD cells, followed by HCoV-OC43-WT or HCoV-OC43-Δns12.9 infection, and the supernatants were collected to determine virus titers. Compared with the empty vector-transfected cells, ns12.9-expressing cells exhibited a significant enhancement of viral production for both HCoV-OC43-Δns12.9 and HCoV-OC43-WT viruses (Fig. 4B). These results confirm that the ns12.9 viroporin plays an important role in the production of infectious virus.
FIG 4.
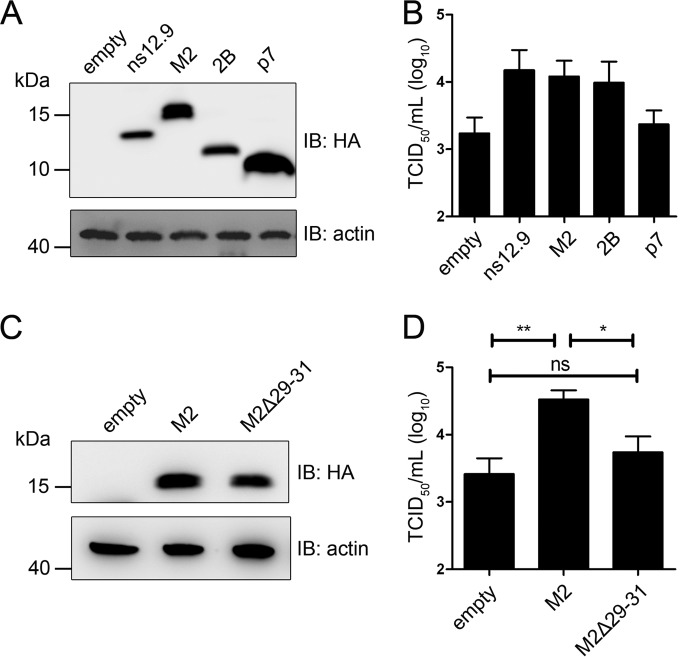
Complementation of HCoV-OC43-Δns12.9 infection in RD cells expressing ns12.9 and this accessory protein from other CoVs. (A) Transient expression of exogenous ns12.9. Cells were transfected with a vector expressing ns12.9-HA or a control vector using Lipofectamine LTX (Life Technologies). The expression levels of ns12.9-HA were determined at 48 h posttransfection by Western blotting. (B) Expression of ns12.9 in RD cells enhances viral production. After 24 h posttransfection, RD cells were infected with HCoV-OC43-WT or HCoV-OC43-Δns12.9 at an MOI of 1. Cell supernatants were collected at 72 hpi, and the viral titers were determined by IPA. (C) Transient expression of ns12.9 and this accessory protein from other CoVs. The expression levels of OC43-ns12.9, SARS-3a, NL63-ORF3, and 229E-ORF4a were determined at 48 h posttransfection by Western blotting. The asterisk indicates a putative truncated variant of full-length 229E-ORF4a. (D) Expression of accessory proteins compensates for the production of HCoV-OC43-Δns12.9. After 24 h posttransfection, RD cells were infected with HCoV-OC43-Δns12.9 at an MOI of 1, and the cell supernatants were collected to determine the viral titers at 72 hpi. Data represent the means ± SD and were generated from three independent experiments. Statistical significance: *, P < 0.05; **, P < 0.01.
Since the accessory gene between the S and E gene loci is contained in all CoVs, we sought to test whether this accessory protein of other HCoVs could compensate for the production of HCoV-OC43-Δns12.9. Vectors expressing SARS-3a, 229E-ORF4a, and NL63-ORF3 were constructed, and their expression levels were detected by Western blot analysis (Fig. 4C). As shown in Fig. 4D, the cells transfected with these vectors led to a significant increase in viral production compared to that of the cells transfected with empty vector, suggesting that these accessory proteins have a conserved function during HCoV infection.
Transient expression of other viroporins complements HCoV-OC43-Δns12.9.
Viroporins are small ion channel proteins encoded by a wide range of viruses and are involved in different aspects of the virus life cycle (27, 34, 35). Therefore, we assessed whether other viroporins could impact HCoV-OC43 infection. Three well-researched viroporins, IAV-M2, HCV-p7, and EV71-2B, were selected and analyzed (Fig. 5A). These viroporins expressing vectors were transfected into RD cells followed by HCoV-OC43-Δns12.9 infection. Similar to the ns12.9-expressing cells, the M2- and 2B-expressing cells showed a significant enhancement of viral production. However, the p7 viroporin failed to compensate for HCoV-OC43-Δns12.9 in replication (Fig. 5B), suggesting a distinct function between ns12.9 and p7 during viral infection.
FIG 5.
Complementation of HCoV-OC43-Δns12.9 infection in RD cells expressing viroporins. (A) Transient expression of ns12.9 and other viroporins. The expression levels of OC43-ns12.9, IAV-M2, EV71-2B, and HCV-p7 were determined at 48 h posttransfection by Western blotting. (B) The M2 and 2B viroporins compensate for the production of HCoV-OC43-Δns12.9. After 24 h posttransfection, RD cells were infected with HCoV-OC43-Δns12.9 at an MOI of 1. Cell supernatants were collected at 72 hpi, and the viral titers were determined by IPA. (C) Transient expression of M2 and M2Δ29-31 mutant. The expression levels of M2 and M2Δ29-31 mutant were determined at 48 h posttransfection by Western blotting. (D) The M2Δ29-31 mutant does not compensate for the production of HCoV-OC43-Δns12.9. After 24 h posttransfection, RD cells were infected with HCoV-OC43-Δns12.9 at an MOI of 1. Cell supernatants were collected at 72 hpi, and the viral titers were determined by IPA. Data represent the means ± SD and were generated from three independent experiments. Statistical significance: ns, not significant; *, P < 0.05; **, P < 0.01.
To specifically analyze the relevance of ion channel activity in viral production, mutations or deletions within the putative TMD of ns12.9 were introduced to construct an ns12.9 mutant that lacks ion channel activity. However, these mutants failed to change its ion conductivity or to localize at the cell membrane (data not shown). Given that the production of HCoV-OC43-Δns12.9 was compensated for in cells expressing M2 in trans, we attempted to evaluate the contribution of viroporin ion conductivity to viral production by the complementation assay of M2Δ29-31 mutant (Fig. 5C), whose ion channel activity is completely disrupted as a deletion of three amino acids, 29 to 31, in the M2 TMD (36, 37). As shown in Fig. 5D, the transient expression of M2, but not the M2Δ29-31 mutant, complemented HCoV-OC43-Δns12.9 in RD cells, indicating that the ion channel activity of viroporins is closely related to the production of HCoV-OC43.
ns12.9 does not impact viral entry, sgmRNA synthesis, and protein expression.
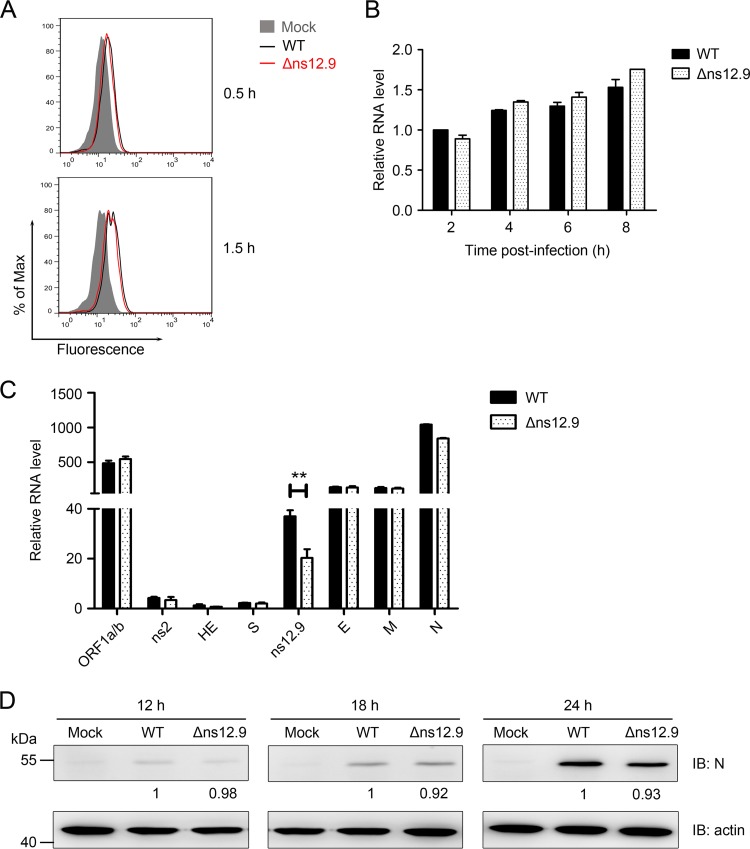
To define the step of the virus life cycle where the ns12.9 viroporin functions, the viral single replication cycle was dissected and analyzed systematically. We first investigated the viral entry process. The efficiency of HCoV-OC43-WT and HCoV-OC43-Δns12.9 infection at 0.5 and 1.5 hpi was analyzed by flow cytometry. As shown in Fig. 6A, similar percentages of N-positive cells were observed in cells infected with the wild-type and mutant viruses, indicating that the ns12.9 protein is not required for the entry process. To further assess genomic replication kinetics, the level of genomic RNA (gRNA) was evaluated by qRT-PCR at early time points in the wild-type- and mutant virus-infected RD cells. Similar levels of gRNA were observed in cells infected with both viruses (Fig. 6B), suggesting that the ns12.9 viroporin does not affect genomic replication of HCoV-OC43.
FIG 6.
ns12.9 accessory protein does not affect the early steps of HCoV-OC43 infection and protein expression. (A) Knockout of ns12.9 gene does not impair the entry process. RD cells were infected with HCoV-OC43-WT or HCoV-OC43-Δns12.9 at an MOI of 1 and fixed at 0.5 and 1.5 hpi. Cells were stained with an OC43-N mouse monoclonal antibody, and the Alexa Fluor 488-conjugated goat anti-mouse antibody was used as the secondary antibody. The stained cells were analyzed by flow cytometry. The results are representative of three independent experiments. (B) Knockout of ns12.9 gene does not impair viral RNA replication. Cells were infected with HCoV-OC43-WT or HCoV-OC43-Δns12.9 at an MOI of 1, and total RNA was extracted at 2, 4, 6, and 8 hpi. Viral RNA levels were analyzed by qRT-PCR with the OC43-RT primers. (C) Knockout of ns12.9 gene does not impair sgmRNA synthesis. Total RNA was extracted from the infected cells at 8 hpi, and HCoV-OC43 sgmRNA levels were analyzed by qRT-PCR using the specific primers presented in Table 1. Data represent the means ± SD and were generated from three independent experiments. Statistical significance: **, P < 0.01. (D) Knockout of ns12.9 gene does not impair OC43-N protein expression. Cell lysates were collected at 12, 18, and 24 hpi, and the expression levels of OC43-N protein were determined by Western blotting. The bands were analyzed and the fold changes of N protein at the indicated times were calculated considering the wild-type infected cells as 1 with actin as an internal control.
As a member of the order Nidovirales, coronavirus generates a series of smaller sgmRNAs that serve as the messenger RNAs for viral protein synthesis. To address whether the knockout of ns12.9 alters sgmRNA synthesis, all HCoV-OC43 sgmRNAs were measured by qRT-PCR. To amplify the sgmRNAs, a leader sequence was selected as a forward primer, and reverse primers were complementary to each gene sequence (Table 1). As shown in Fig. 6C, no significant change was measured in the amount of each sgmRNA between the wild-type and mutant virus, except for ns12.9 sgmRNA. One possible explanation for this observation is that the qPCR amplification efficiency of the ns12.9 gene was suppressed by the extended 82-nt segment in the ns12.9 gene. Because the production of sgmRNA was not affected by ns12.9, deletion of the ns12.9 protein may not influence viral protein synthesis. To confirm that viral protein synthesis was independent of ns12.9, the main structure protein N was selected, and its expression during the time course of infection was detected by Western blot analysis. As expected, comparable levels of N protein expression were observed for cells infected with wild-type and mutant HCoV-OC43 viruses (Fig. 6D).
Taken together, these data demonstrate that the ns12.9 viroporin is dispensable for the early steps and protein expression of HCoV-OC43 infection.
ns12.9 is involved in virion morphogenesis.
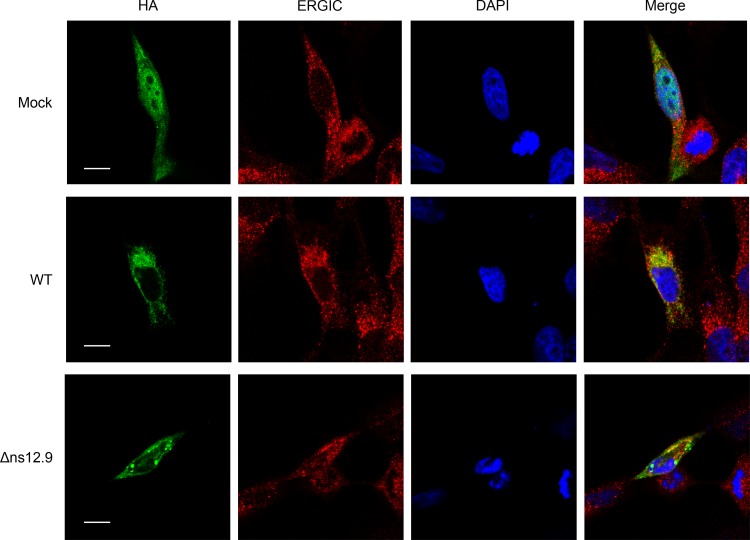
To address whether ns12.9 regulates the late stages of the virus life cycle, we first examined the subcellular localization of ns12.9 during HCoV-OC43 infection. ns12.9 was evenly distributed in the cytoplasm and nucleus of RD cells when ns12.9 was exclusively expressed (Fig. 7, upper). Surprisingly, in wild-type- or mutant virus-infected RD cells, ns12.9 was localized at ERGIC (Fig. 7, middle and lower), the compartment where CoVs assemble and bud intracellularly during viral infection (1). These observations indicate that other viral factors are involved in the localization of ns12.9 during HCoV-OC43 infection.
FIG 7.
Subcellular localization of ns12.9 during HCoV-OC43 infection. RD cells were infected with HCoV-OC43-WT or HCoV-OC43-Δns12.9 at an MOI of 1. At 24 h postinfection, cells were transfected with pCAGGS-ns12.9-HA. Cells were fixed and examined by confocal microscopy analysis at 24 h posttransfection. ns12.9 was stained with anti-HA monoclonal antibody and visualized with Alexa Fluor 488-conjugated goat anti-mouse antibody (green). ERGIC was stained with ERGIC53 antibody (H-245) and visualized with Cy3-conjugated goat anti-rabbit antibody (red). Nuclei were stained with DAPI (blue). Bars represent 20 μm.
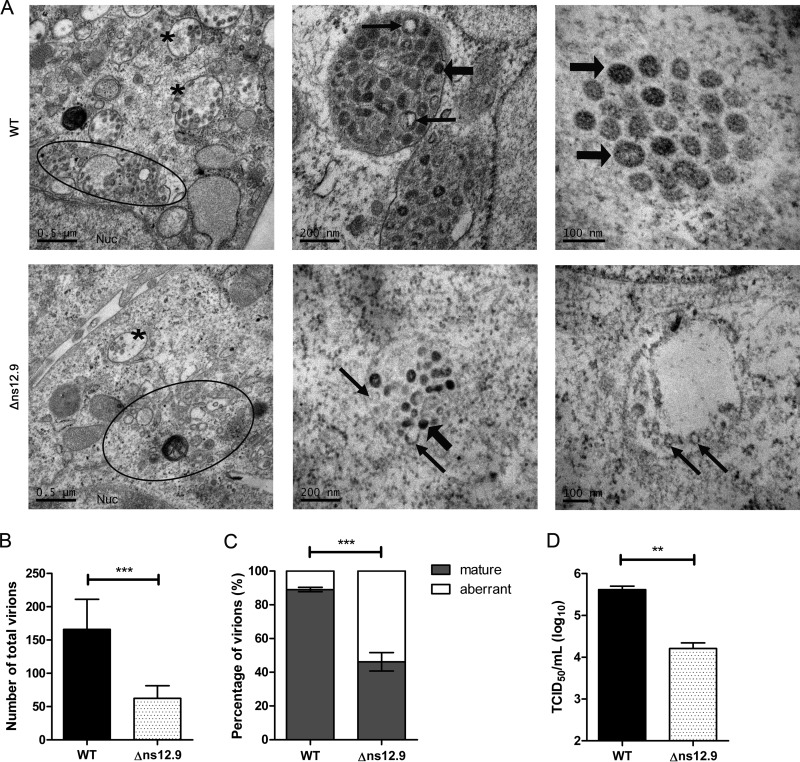
Given that the ns12.9 viroporin is dispensable for the early steps of the virus life cycle and is localized at ERGIC during viral infection, we hypothesized that ns12.9 has an effect on virus assembly. To confirm this hypothesis, the formation of HCoV-OC43 virions in RD cells was examined using transmission electron microscopy. As shown in Fig. 8A, HCoV-OC43 virions were observed at the enlarged and fragmented Golgi cisternae and trafficked in secretory vesicles for egress. The number of virions in cells infected with wild-type virus was higher than that with the mutant virus (Fig. 8B). In addition, in HCoV-OC43-WT-infected cells, mature virions showing a dense core were abundant in vesicles (Fig. 8A, upper), whereas viral particles in HCoV-OC43-Δns12.9-infected cells exhibited an aberrant pattern with a translucent central zone (Fig. 8A, lower). Quantitative analysis of the virions present in the cytoplasm revealed that the mature virions in cells infected with mutant virus were dramatically decreased compared to levels in cells infected with wild-type virus (Fig. 8C). To confirm these observations, the amount of intracellular mature virions was determined. As expected, a lower titer of intracellular infectious virions was measured in cells infected with HCoV-OC43-Δns12.9 than with cells infected with HCoV-OC43-WT (Fig. 8D). This decrease in intracellular viral titers of HCoV-OC43-Δns12.9 was consistent with the decreased viral titers observed in the supernatant (Fig. 3). Therefore, these results demonstrate that ns12.9 viroporin facilitates the formation of viral particles and subsequently increases the production of infectious HCoV-OC43 virions.
FIG 8.
Electron microscopy analysis of HCoV-OC43 virion morphogenesis. (A) Transmission electron micrographs of virions in the infected RD cells. Cells were infected with HCoV-OC43-WT or HCoV-OC43-Δns12.9 at an MOI of 1, fixed at 24 hpi, and processed for transmission electron microscopy. The virions were found in the Golgi complex (circles) and secretory vesicles (asterisks). The thick arrows indicate the mature virions, and the thin arrows indicate the aberrant virions. The quantification of total numbers of virions (B) and the mature or aberrant virions (C) are shown. The virions in HCoV-OC43-WT-infected cells (n = 9) or HCoV-OC43-Δns12.9-infected cells (n = 10) were counted, and data were plotted as the means ± SD. Statistical significance: ***, P < 0.001. (D) Titration of the intracellular infectious virions. RD cells were infected with HCoV-OC43-WT or HCoV-OC43-Δns12.9 at an MOI of 1. Cells were collected at 24 hpi and disrupted by three cycles of freeze-thawing in dry ice and a 37°C water bath. The intracellular infectious viruses were determined by IPA. Data represent the means ± SD and were generated from three independent experiments. Statistical significance: **, P < 0.01.
HCoV-OC43-Δns12.9 is attenuated in vivo.
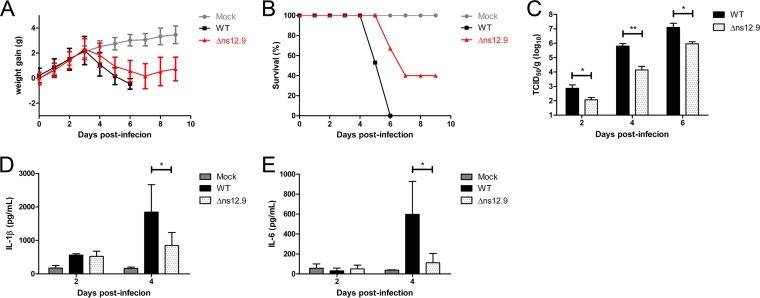
To determine the contribution of ns12.9 viroporin to viral growth in vivo, BALB/c mice were infected intranasally with HCoV-OC43-WT or HCoV-OC43-Δns12.9. The infected mice were monitored daily for weight variations and survival. HCoV-OC43-WT-infected mice gained weight normally during the first 3 days but started to lose weight at 4 dpi. Mice infected with the wild-type virus presented the symptom of less mobility, and all of these mice died at 6 dpi (Fig. 9A and data not shown). However, the HCoV-OC43-Δns12.9-infected mice lost weight more slowly, exhibited mild disease symptoms, and regained weight at 8 dpi with 40% survival (Fig. 9B). The infected mice were sacrificed every 2 dpi to measure the viral titers in brains. As shown in Fig. 9C, the wild-type viral titers were 10- to 100-fold increased compared to the mutant viral titers in brains, indicating that HCoV-OC43-Δns12.9 was attenuated in mice.
FIG 9.
HCoV-OC43-Δns12.9 is attenuated in growth and virulence in vivo. Mice were intranasally inoculated with HCoV-OC43-WT or HCoV-OC43-Δns12.9. Infected mice were monitored daily for weight variations and sacrificed every 2 dpi to measure the viral titers or proinflammatory cytokines in brains. The weight variations (A) and survival curves (B) after infection are presented for each group. (C) The viral titers in the brains of the infected mice at the indicated times were determined by IPA. Statistical significance: *, P < 0.05; **, P < 0.01. The production of IL-1β (D) and IL-6 (E) proinflammatory cytokines at the indicated times was measured by ELISA. Data represent the means ± SD from samples of 4 mice. Statistical significance: *, P < 0.05.
HCoV-OC43 infection and replication in brains causes lethal encephalitis in mice. To assess the immune response in brains after wild-type and mutant viral infection, the production of key proinflammatory cytokines, such as IL-1β and IL-6, was measured by ELISA. Consistent with the viral titer, significantly increased amounts of proinflammatory cytokines were detected in HCoV-OC43-WT-infected mice than in HCoV-OC43-Δns12.9-infected mice (Fig. 9D and E), suggesting that HCoV-OC43-WT infection causes more severe inflammatory response in the brain than HCoV-OC43-Δns12.9 infection, which may contribute to morbidity and mortality.
Collectively, our results indicate that the ns12.9 viroporin is involved in HCoV-OC43 propagation and pathogenesis in vivo.
DISCUSSION
Interspersed among the structural genes, CoV genomes contain accessory genes to encode several accessory proteins that are critical for viral infection (1). An accessory gene located between the S and E structural genes is contained in all CoVs. Thus, this accessory gene may have an important role in CoV infection. Indeed, downregulation of this accessory gene by short interfering RNAs (25, 26, 38), deletion of this gene in a cDNA clone using a reverse genetics system (23, 39), or mutation of this gene via cell culture adaptation (40, 41) results in defective viral production and virulence. Previously, we found that accessory protein 3a in SARS-CoV and ORF4a in HCoV-229E function as viroporins to regulate viral production with an unknown mechanism. In this study, we sought to characterize the functions of this accessory protein, ns12.9, in HCoV-OC43 during viral infection. To this end, we engineered an ns12.9 knockout cDNA clone (pBAC-OC43-Δns12.9) with a genetic approach. We demonstrated that ns12.9 acts as a viroporin in that it has all of the features of viroporins, such as membrane localization, self-oligomerization, and ion channel formation. Furthermore, the absence of ns12.9 led to a lower virus yield in vitro and in vivo, indicating that the ns12.9 viroporin, although not essential, is important for the production of infectious HCoV-OC43 particles.
The complementation of HCoV-OC43-Δns12.9 in RD cells expressing the ns12.9 accessory protein in trans confirmed the importance of ns12.9 in virus replication. Interestingly, this accessory protein from other HCoVs, such as SARS-3a, 229E-ORF4a, and NL63-ORF3, also could rescue the growth of HCoV-OC43-Δns12.9 in trans (Fig. 4D). Although there are large variations among these protein sequences (data not shown), they possess similar properties in forming ion channels and regulating viral production (25, 26). Our results strongly suggest that these accessory proteins exert a conserved viroporin function that is required for virion morphogenesis, which deserves further investigation.
The ion channel activity of viroporins has been reported to associate with viral production, fitness, and pathogenesis (42). In addition to regulating the uncoating stage in the endosome (36), IAV-M2 prevents the excessive acidification of the trans-Golgi network (TGN) to induce the correct maturation of viral hemagglutinin glycoprotein (43). The picornavirus 2B is present primarily in the Golgi compartments and induces calcium efflux, which is important for virus release (44–46). In contrast, although it is needed for late stages of the virus life cycle (47, 48), HCV-p7 localizes predominantly to the ER. In the present study, EV71-2B and IAV-M2 viroporins were efficient at complementing the ns12.9 knockout virus, while HCV-p7 and the M2Δ29-31 mutant, lacking ion conductivity, failed to compensate for it (Fig. 5). Moreover, the ns12.9 viroporin was found to localize at ERGIC in the infected cells (Fig. 7, middle and lower). These results highlight the importance of ion channel activity and ERGIC/Golgi compartment localization of viroporins in HCoV-OC43 infection.
Systematic comparison of HCoV-OC43-WT and HCoV-OC43-Δns12.9 replication revealed that the ns12.9 viroporin is not involved in early steps of the virus life cycle. Using electron microscopy, we found that the number of intracellular virions was significantly reduced and that more viral particles exhibited an aberrant pattern when the ns12.9 protein was deleted (Fig. 8). There are several possibilities that can be responsible for ns12.9 viroporin promoting the formation of viral particles. First, ns12.9 may function as a structural protein that enhances the production of infectious virus particles. Numerous viroporins are virion-incorporated proteins (36, 49, 50). However, without an effective antibody, we cannot rule out the possibility that the ns12.9 viroporin is a structural protein. Second, ns12.9 may impair the ionic homeostasis of the ERGIC/Golgi compartments, where CoV morphogenesis takes place. The ionic equilibration between the ERGIC/Golgi compartments and the cytosol could protect the structural proteins and newly formed virions to promote virus assembly and maturation (43, 51). Finally, ns12.9 may be involved in membrane scission and promote the budding of progeny virions. Some viroporins have been shown to enhance membrane permeability, cause membrane depolarization, and facilitate membrane scission to trigger the budding process (52–54).
CoV infection generally exhibits a species-specific property. However, HCoV-OC43 could cross the species barrier to infect mice and cause encephalitis (55, 56). IL-1β is a main proinflammatory cytokine that initiates the pathogenesis of infection (57). Mature IL-1β is cleaved from pro-IL-1β by caspase-1, which is activated by a protein complex termed the inflammasome (58). Inflammasome activation in the central nervous system (CNS) has been studied in various neurological diseases and pathogen infections (59). Here, we identified the robust production of IL-1β in the brain after HCoV-OC43-WT infection (Fig. 9D). This finding may be relevant to the neuropathogenicity of HCoV-OC43. Recently, viroporins were found to activate the inflammasome in macrophages and dendritic cells (60–63). For HCoV-OC43 infection, ongoing studies are investigating whether the ns12.9 viroporin activates the inflammasome in the microglia, the resident macrophages of the CNS. The type I interferons (IFNs) play an essential role in protection against CoV infection (64, 65). MHV 5a, a homologous accessory protein of HCoV-OC43 ns12.9, recently has been reported to be an antagonist of IFN-induced antiviral action (66). Because HCoV-OC43-Δns12.9 presents propagation reduction and virulence attenuation in mice, ns12.9 may be a viral factor to antagonize IFN antiviral responses during HCoV-OC43 infection, which should be investigated further.
In conclusion, we demonstrated that the ns12.9 accessory protein is a viroporin and influences virion morphogenesis during HCoV-OC43 infection. This study on ns12.9 function provides a potential target for developing antiviral drugs and contributes to a deeper understanding of the pathogenesis of HCoV-OC43 or other CoV infections.
ACKNOWLEDGMENTS
We thank Pierre J. Talbot (Laboratory of Neuroimmunovirology, INRS-Institut Armand-Frappier, Laval, Québec, Canada) for providing the pBAC-OC43-WT cDNA clone. We thank Baoqin Xuan (Unit of Herpesvirus and Molecular Virology, Institut Pasteur of Shanghai, Chinese Academy of Sciences, Shanghai, China) for valuable assistance in the construction of plasmid pBAC-OC43-Δns12.9. We thank Tianqing Zhang and Fengling Qin (Core Facility of Cell Biology, Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences, Shanghai, China) for their technical assistance in transmission electron microscopy.
This study was supported by the National Science and Technology Major Project (2013ZX10004-101-005, 2012ZX10002-007-003, and 2013ZX10004-003-003).
REFERENCES
- 1.Masters P. 2006. The molecular biology of coronaviruses. Adv Virus Res 66:193–292. doi: 10.1016/S0065-3527(06)66005-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adams MJ, Carstens EB. 2012. Ratification vote on taxonomic proposals to the International Committee on Taxonomy of Viruses (2012). Arch Virol 157:1411–1422. doi: 10.1007/s00705-012-1299-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McIntosh K, Becker WB, Chanock RM. 1967. Growth in suckling-mouse brain of “IBV-like” viruses from patients with upper respiratory tract disease. Proc Natl Acad Sci U S A 58:2268–2273. doi: 10.1073/pnas.58.6.2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Myint SH. 1994. Human coronaviruses–a brief review. Rev Med Virol 4:35–46. doi: 10.1002/rmv.1980040108. [DOI] [Google Scholar]
- 5.Arbour N, Day R, Newcombe J, Talbot PJ. 2000. Neuroinvasion by human respiratory coronaviruses. J Virol 74:8913–8921. doi: 10.1128/JVI.74.19.8913-8921.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arbour N, Cote G, Lachance C, Tardieu M, Cashman NR, Talbot PJ. 1999. Acute and persistent infection of human neural cell lines by human coronavirus OC43. J Virol 73:3338–3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stewart JN, Mounir S, Talbot PJ. 1992. Human coronavirus gene expression in the brains of multiple sclerosis patients. Virology 191:502–505. doi: 10.1016/0042-6822(92)90220-J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ziebuhr J, Snijder EJ, Gorbalenya AE. 2000. Virus-encoded proteinases and proteolytic processing in the Nidovirales. J Gen Virol 81:853–879. doi: 10.1099/0022-1317-81-4-853. [DOI] [PubMed] [Google Scholar]
- 9.Vlasak R, Luytjes W, Spaan W, Palese P. 1988. Human and bovine coronaviruses recognize sialic acid-containing receptors similar to those of influenza C viruses. Proc Natl Acad Sci U S A 85:4526–4529. doi: 10.1073/pnas.85.12.4526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li W, Moore MJ, Vasilieva N, Sui J, Wong SK, Berne MA, Somasundaran M, Sullivan JL, Luzuriaga K, Greenough TC, Choe H, Farzan M. 2003. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Raj VS, Mou H, Smits SL, Dekkers DH, Muller MA, Dijkman R, Muth D, Demmers JA, Zaki A, Fouchier RA, Thiel V, Drosten C, Rottier PJ, Osterhaus AD, Bosch BJ, Haagmans BL. 2013. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature 495:251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yeager CL, Ashmun RA, Williams RK, Cardellichio CB, Shapiro LH, Look AT, Holmes KV. 1992. Human aminopeptidase N is a receptor for human coronavirus 229E. Nature 357:420–422. doi: 10.1038/357420a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vennema H, Godeke GJ, Rossen JW, Voorhout WF, Horzinek MC, Opstelten DJ, Rottier PJ. 1996. Nucleocapsid-independent assembly of coronavirus-like particles by co-expression of viral envelope protein genes. EMBO J 15:2020–2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang Y, Yang ZY, Kong WP, Nabel GJ. 2004. Generation of synthetic severe acute respiratory syndrome coronavirus pseudoparticles: implications for assembly and vaccine production. J Virol 78:12557–12565. doi: 10.1128/JVI.78.22.12557-12565.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Corse E, Machamer CE. 2000. Infectious bronchitis virus E protein is targeted to the Golgi complex and directs release of virus-like particles. J Virol 74:4319–4326. doi: 10.1128/JVI.74.9.4319-4326.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schultze B, Wahn K, Klenk HD, Herrler G. 1991. Isolated HE-protein from hemagglutinating encephalomyelitis virus and bovine coronavirus has receptor-destroying and receptor-binding activity. Virology 180:221–228. doi: 10.1016/0042-6822(91)90026-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Desforges M, Desjardins J, Zhang C, Talbot PJ. 2013. The acetyl-esterase activity of the hemagglutinin-esterase protein of human coronavirus OC43 strongly enhances the production of infectious virus. J Virol 87:3097–3107. doi: 10.1128/JVI.02699-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ortego J, Sola I, Almazan F, Ceriani JE, Riquelme C, Balasch M, Plana J, Enjuanes L. 2003. Transmissible gastroenteritis coronavirus gene 7 is not essential but influences in vivo virus replication and virulence. Virology 308:13–22. doi: 10.1016/S0042-6822(02)00096-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haijema BJ, Volders H, Rottier PJM. 2004. Live, attenuated coronavirus vaccines through the directed deletion of group-specific genes provide protection against feline infectious peritonitis. J Virol 78:3863–3871. doi: 10.1128/JVI.78.8.3863-3871.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schaecher SR, Touchette E, Schriewer J, Buller RM, Pekosz A. 2007. Severe acute respiratory syndrome coronavirus gene 7 products contribute to virus-induced apoptosis. J Virol 81:11054–11068. doi: 10.1128/JVI.01266-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kopecky-Bromberg SA, Martinez-Sobrido L, Frieman M, Baric RA, Palese P. 2007. Severe acute respiratory syndrome coronavirus open reading frame (ORF) 3b, ORF 6, and nucleocapsid proteins function as interferon antagonists. J Virol 81:548–557. doi: 10.1128/JVI.01782-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Haan CAM, Masters PS, Shen XL, Weiss S, Rottier PJM. 2002. The group-specific murine coronavirus genes are not essential, but their deletion, by reverse genetics, is attenuating in the natural host. Virology 296:177–189. doi: 10.1006/viro.2002.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yount B, Roberts RS, Sims AC, Deming D, Frieman MB, Sparks J, Denison MR, Davis N, Baric RS. 2005. Severe acute respiratory syndrome coronavirus group-specific open reading frames encode nonessential functions for replication in cell cultures and mice. J Virol 79:14909–14922. doi: 10.1128/JVI.79.23.14909-14922.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Youn S, Leibowitz JL, Collisson EW. 2005. In vitro assembled, recombinant infectious bronchitis viruses demonstrate that the 5a open reading frame is not essential for replication. Virology 332:206–215. doi: 10.1016/j.virol.2004.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang RH, Wang K, Lu W, Yu WJ, Xie SQ, Xu K, Schwarz W, Xiong SD, Sun B. 2014. The ORF4a protein of human coronavirus 229E functions as a viroporin that regulates viral production. Biochim Biophys Acta 1838:1088–1095. doi: 10.1016/j.bbamem.2013.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu W, Zheng BJ, Xu K, Schwarz W, Du L, Wong CK, Chen J, Duan S, Deubel V, Sun B. 2006. Severe acute respiratory syndrome-associated coronavirus 3a protein forms an ion channel and modulates virus release. Proc Natl Acad Sci U S A 103:12540–12545. doi: 10.1073/pnas.0605402103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nieva JL, Madan V, Carrasco L. 2012. Viroporins: structure and biological functions. Nat Rev Microbiol 10:563–574. doi: 10.1038/nrmicro2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mounir S, Labonte P, Talbot PJ. 1993. Characterization of the nonstructural and spike proteins of the human respiratory coronavirus OC43: comparison with bovine enteric coronavirus. Adv Exp Med Biol 342:61–67. [DOI] [PubMed] [Google Scholar]
- 29.Qian Z, Xuan B, Gualberto N, Yu D. 2011. The human cytomegalovirus protein pUL38 suppresses endoplasmic reticulum stress-mediated cell death independently of its ability to induce mTORC1 activation. J Virol 85:9103–9113. doi: 10.1128/JVI.00572-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lambert F, Jacomy H, Marceau G, Talbot PJ. 2008. Titration of human coronaviruses, HcoV-229E and HCoV-OC43, by an indirect immunoperoxidase assay. Methods Mol Biol 454:93–102. doi: 10.1007/978-1-59745-181-9_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.St-Jean JR, Desforges M, Almazan F, Jacomy H, Enjuanes L, Talbot PJ. 2006. Recovery of a neurovirulent human coronavirus OC43 from an infectious cDNA clone. J Virol 80:3670–3674. doi: 10.1128/JVI.80.7.3670-3674.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.St-Jean JR, Jacomy H, Desforges M, Vabret A, Freymuth F, Talbot PJ. 2004. Human respiratory coronavirus OC43: genetic stability and neuroinvasion. J Virol 78:8824–8834. doi: 10.1128/JVI.78.16.8824-8834.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kyte J, Doolittle RF. 1982. A simple method for displaying the hydropathic character of a protein. J Mol Biol 157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 34.Wang K, Xie SQ, Sun B. 2011. Viral proteins function as ion channels. Biochim Biophys Acta 1808:510–515. doi: 10.1016/j.bbamem.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gonzalez ME, Carrasco L. 2003. Viroporins. FEBS Lett 552:28–34. doi: 10.1016/S0014-5793(03)00780-4. [DOI] [PubMed] [Google Scholar]
- 36.Pinto LH, Holsinger LJ, Lamb RA. 1992. Influenza virus M2 protein has ion channel activity. Cell 69:517–528. doi: 10.1016/0092-8674(92)90452-I. [DOI] [PubMed] [Google Scholar]
- 37.Watanabe T, Watanabe S, Ito H, Kida H, Kawaoka Y. 2001. Influenza a virus can undergo multiple cycles of replication without M2 ion channel activity. J Virol 75:5656–5662. doi: 10.1128/JVI.75.12.5656-5662.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Åkerström S, Mirazimi A, Tan Y-J. 2007. Inhibition of SARS-CoV replication cycle by small interference RNAs silencing specific SARS proteins, 7a/7b, 3a/3b, and S. Antiviral Res 73:219–227. doi: 10.1016/j.antiviral.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Donaldson EF, Yount B, Sims AC, Burkett S, Pickles RJ, Baric RS. 2008. Systematic assembly of a full-length infectious clone of human coronavirus NL63. J Virol 82:11948–11957. doi: 10.1128/JVI.01804-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Woods RD. 2001. Efficacy of a transmissible gastroenteritis coronavirus with an altered ORF-3 gene. Can J Vet Res 65:28–32. [PMC free article] [PubMed] [Google Scholar]
- 41.Song DS, Yang JS, Oh JS, Han JH, Park BK. 2003. Differentiation of a Vero cell adapted porcine epidemic diarrhea virus from Korean field strains by restriction fragment length polymorphism analysis of ORF 3. Vaccine 21:1833–1842. doi: 10.1016/S0264-410X(03)00027-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nieto-Torres JL, Verdia-Baguena C, Castano-Rodriguez C, Aguilella VM, Enjuanes L. 2015. Relevance of viroporin ion channel activity on viral replication and pathogenesis. Viruses 7:3552–3573. doi: 10.3390/v7072786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ciampor F, Bayley PM, Nermut MV, Hirst EMA, Sugrue RJ, Hay AJ. 1992. Evidence that the amantadine-induced, M2-mediated conversion of influenza A virus hemagglutinin to the low PH conformation occurs in an acidic transgolgi compartment. Virology 188:14–24. doi: 10.1016/0042-6822(92)90730-D. [DOI] [PubMed] [Google Scholar]
- 44.Campanella M, de Jong AS, Lanke KWH, Melchers WJG, Willems PHGM, Pinton P, Rizzuto R, van Kuppeveld FJM. 2004. The coxsackievirus 2B protein suppresses apoptotic host cell responses by manipulating intracellular Ca2+ homeostasis. J Biol Chem 279:18440–18450. doi: 10.1074/jbc.M309494200. [DOI] [PubMed] [Google Scholar]
- 45.de Jong AS, de Mattia F, Van Dommelen MM, Lanke K, Melchers WJG, Willems PHGM, van Kuppeveld FJM. 2008. Functional analysis of picornavirus 2B proteins: effects on calcium homeostasis and intracellular protein trafficking. J Virol 82:3782–3790. doi: 10.1128/JVI.02076-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van Kuppeveld FJ, Hoenderop JG, Smeets RL, Willems PH, Dijkman HB, Galama JM, Melchers WJ. 1997. Coxsackievirus protein 2B modifies endoplasmic reticulum membrane and plasma membrane permeability and facilitates virus release. EMBO J 16:3519–3532. doi: 10.1093/emboj/16.12.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Steinmann E, Pietschmann T. 2010. Hepatitis C virus P7–a viroporin crucial for virus assembly and an emerging target for antiviral therapy. Viruses 2:2078–2095. doi: 10.3390/v2092078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Steinmann E, Penin F, Kallis S, Patel AH, Bartenschlager R, Pietschmann T. 2007. Hepatitis C virus p7 protein is crucial for assembly and release of infectious virions. PLoS Pathog 3:e103. doi: 10.1371/journal.ppat.0030103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wilson L, McKinlay C, Gage P, Ewart G. 2004. SARS coronavirus E protein forms cation-selective ion channels. Virology 330:322–331. doi: 10.1016/j.virol.2004.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ito N, Mossel EC, Narayanan K, Popov VL, Huang C, Inoue T, Peters CJ, Makino S. 2005. Severe acute respiratory syndrome coronavirus 3a protein is a viral structural protein. J Virol 79:3182–3186. doi: 10.1128/JVI.79.5.3182-3186.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Michelangeli F, Liprandi F, Chemello ME, Ciarlet M, Ruiz MC. 1995. Selective depletion of stored calcium by thapsigargin blocks rotavirus maturation but not the cytopathic effect. J Virol 69:3838–3847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hsu K, Han J, Shinlapawittayatorn K, Deschenes I, Marban E. 2010. Membrane potential depolarization as a triggering mechanism for Vpu-mediated HIV-1 release. Biophys J 99:1718–1725. doi: 10.1016/j.bpj.2010.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huang DTN, Chi NW, Chen SC, Lee TY, Hsu K. 2011. Background K(2P) channels KCNK3/9/15 limit the budding of cell membrane-derived vesicles. Cell Biochem Biophys 61:585–594. doi: 10.1007/s12013-011-9241-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.DeDiego ML, Alvarez E, Almazan F, Rejas MT, Lamirande E, Roberts A, Shieh WJ, Zaki SR, Subbarao K, Enjuanes L. 2007. A severe acute respiratory syndrome coronavirus that lacks the E gene is attenuated in vitro and in vivo. J Virol 81:1701–1713. doi: 10.1128/JVI.01467-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jacomy H, Talbot PJ. 2003. Vacuolating encephalitis in mice infected by human coronavirus OC43. Virology 315:20–33. doi: 10.1016/S0042-6822(03)00323-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Butler N, Pewe L, Trandem K, Perlman S. 2006. Murine encephalitis caused by HCoV-OC43, a human coronavirus with broad species specificity, is partly immune-mediated. Virology 347:410–421. doi: 10.1016/j.virol.2005.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tisoncik JR, Korth MJ, Simmons CP, Farrar J, Martin TR, Katze MG. 2012. Into the eye of the cytokine storm. Microbiol Mol Biol Rev 76:16–32. doi: 10.1128/MMBR.05015-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Martinon F, Burns K, Tschopp J. 2002. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol Cell 10:417–426. doi: 10.1016/S1097-2765(02)00599-3. [DOI] [PubMed] [Google Scholar]
- 59.Chakraborty S, Kaushik DK, Gupta M, Basu A. 2010. Inflammasome signaling at the heart of central nervous system pathology. J Neurosci Res 88:1615–1631. [DOI] [PubMed] [Google Scholar]
- 60.Ichinohe T, Pang IK, Iwasaki A. 2010. Influenza virus activates inflammasomes via its intracellular M2 ion channel. Nat Immunol 11:404–410. doi: 10.1038/ni.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nieto-Torres JL, DeDiego ML, Verdiá-Báguena C, Jimenez-Guardeño JM, Regla-Nava JA, Fernandez-Delgado R, Castaño-Rodriguez C, Alcaraz A, Torres J, Aguilella VM, Enjuanes L. 2014. Severe acute respiratory syndrome coronavirus envelope protein ion channel activity promotes virus fitness and pathogenesis. PLoS Pathog 10:e1004077. doi: 10.1371/journal.ppat.1004077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ito M, Yanagi Y, Ichinohe T. 2012. Encephalomyocarditis virus viroporin 2B activates NLRP3 inflammasome. PLoS Pathog 8:e1002857. doi: 10.1371/journal.ppat.1002857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Triantafilou K, Kar S, Vakakis E, Kotecha S, Triantafilou M. 2013. Human respiratory syncytial virus viroporin SH: a viral recognition pathway used by the host to signal inflammasome activation. Thorax 68:66–75. doi: 10.1136/thoraxjnl-2012-202182. [DOI] [PubMed] [Google Scholar]
- 64.Cervantes-Barragan L, Zust R, Weber F, Spiegel M, Lang KS, Akira S, Thiel V, Ludewig B. 2007. Control of coronavirus infection through plasmacytoid dendritic-cell-derived type I interferon. Blood 109:1131–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ireland DDC, Stohlman SA, Hinton DR, Atkinson R, Bergmann CC. 2008. Type I interferons are essential in controlling neurotropic coronavirus infection irrespective of functional CD8 T cells. J Virol 82:300–310. doi: 10.1128/JVI.01794-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Koetzner CA, Kuo LL, Goebel SJ, Dean AB, Parker MM, Masters PS. 2010. Accessory protein 5a is a major antagonist of the antiviral action of interferon against murine coronavirus. J Virol 84:8262–8274. doi: 10.1128/JVI.00385-10. [DOI] [PMC free article] [PubMed] [Google Scholar]