ABSTRACT
Replication of hepatitis C virus (HCV) is dependent on virus-encoded proteins and numerous cellular factors. DDX3 is a well-known host cofactor of HCV replication. In this study, we investigated the role of a DDX3-interacting protein, Y-box binding protein 1 (YB-1), in the HCV life cycle. Both YB-1 and DDX3 interacted with the viral nonstructural protein NS5A. During HCV infection, YB-1 partially colocalized with NS5A and the HCV replication intermediate double-stranded RNA (dsRNA) in HCV-infected Huh-7.5.1 cells. Despite sharing the same interacting partners, YB-1 participated in HCV RNA replication but was dispensable in steady-state HCV RNA replication, different from the action of DDX3. Moreover, knockdown of YB-1 in HCV-infected cells prevented infectious virus production and reduced the ratio of hyperphosphorylated (p58) to hypophosphorylated (p56) forms of NS5A, whereas DDX3 silencing did not affect the ratio of the p58 and p56 phosphoforms of NS5A. Interestingly, silencing of YB-1 severely reduced NS5A protein stability in NS5A-ectopically expressing, replicon-containing, and HCV-infected cells. Furthermore, mutations of serine 102 of YB-1 affected both YB-1–NS5A interaction and NS5A-stabilizing activity of YB-1, indicating that this Akt phosphorylation site of YB-1 plays an important role in stabilizing NS5A. Collectively, our results support a model in which the event of YB-1 phosphorylation-mediated interaction with NS5A results in stabilizing NS5A to sustain HCV RNA replication and infectious HCV production. Overall, our study may reveal a new aspect for the development of novel anti-HCV drugs.
IMPORTANCE Chronic hepatitis C virus (HCV) infection induces liver cirrhosis and hepatocellular carcinoma. The viral nonstructural protein NS5A co-opting various cellular signaling pathways and cofactors to support viral genome replication and virion assembly is a new strategy for anti-HCV drug development. NS5A phosphorylation is believed to modulate switches between different stages of the HCV life cycle. In this study, we identified the cellular protein YB-1 as a novel NS5A-interacting protein. YB-1 is a multifunctional protein participating in oncogenesis and is an oncomarker of hepatocellular carcinoma (HCC). We found that YB-1 protects NS5A from degradation and likely regulates NS5A phosphorylation through its phosphorylation-dependent interaction with NS5A, which might be controlled by HCV-induced signaling pathways. Our observations suggest a model in which HCV modulates NS5A level and the ratio of the p58 and p56 phosphoforms for efficient viral propagation via regulation of cellular signaling inducing YB-1 phosphorylation. Our finding may provide new aspects for developing novel anti-HCV drugs.
INTRODUCTION
Hepatitis C virus (HCV) chronically infects millions of people worldwide (1). Chronic HCV infection induces chronic hepatitis, liver cirrhosis, and hepatocellular carcinoma. HCV infection has become a serious health problem due to the unavailability of an effective vaccine and limited clinical treatment protocols (2).
HCV is a positive-stranded RNA virus that contains a 9.6-kb genome consisting of a single open reading frame flanked by 5′ and 3′ nontranslated regions (NTR). An internal ribosome entry site (IRES) in the 5′NTR directs the translation of a polyprotein, which is processed co- and posttranslationally into 10 or more viral proteins (3, 4). HCV infection is sustained by spatiotemporal interplay between viral proteins and a panel of cellular cofactors to coordinate translation of the viral genome, viral RNA replication, and the production of infectious viral particles. However, there is still limited understanding of the molecular mechanisms underlying the coordinated interactions of these events.
The nonstructural protein 5A (NS5A) is a phosphoprotein highly variable among genotypes of HCV (5). NS5A is recognized as a key modulator of the HCV life cycle, and the factor has emerged as a new target of drug development (2). NS5A, consisting of three domains (6), is a component of the HCV replication complex (7–10) required for infectious virus production (11–13). Domain I of NS5A is essential for HCV RNA replication (14), while most of domain II is not involved (12). Domain III participates in virion assembly (12, 13, 15). NS5A has also been reported to either positively or negatively regulate HCV IRES-mediated translation (16–18). By regulating activity of cellular lipid kinase phosphatidylinositol 4-kinase type III alpha (PI4KIII-α), NS5A has been demonstrated to modulate the formation of a membranous web to support HCV RNA replication (19, 20). A recent study on stilbene 1,2-diamines, small anti-HCV compounds, revealed that NS5A may have a role in the initiation of HCV RNA replication, which is distinct from steady-state HCV RNA replication (21). Moreover, a transient HCV RNA replication occurring early after infection was later recognized and characterized by the colocalization of negative-strand HCV RNA with NS5A but not another replicase component, NS3 (22), underscoring the unique role of NS5A in the early stage of HCV RNA replication. To facilitate HCV propagation, NS5A also regulates multiple cellular signaling pathways, including the phosphoinositol 3-kinase (PI3K)-Akt survival pathway (23). Although NS5A is involved in many steps in the HCV life cycle and host signaling pathways, it does not have known enzymatic activity. NS5A is believed to exhibit its different functions via interactions with specific viral proteins and various host proteins (2). On the other hand, NS5A has been reported to be regulated by ubiquitin-proteasome degradation (24). Administration of zinc mesoporphyrin (ZnMP), a synthetic nonheme metalloporphyrin, induces NS5A ubiquitination and proteasome degradation and, hence, inhibition of HCV RNA replication (24).
Among the cellular factors reported to be involved in the HCV life cycle, DDX3 gets much attention. DDX3 is a multifunctional DEAD box RNA helicase that regulates transcription and translation and may function as either a tumor suppressor or an oncogene in different cell types (25–30). We and others have reported that DDX3 interacts with the HCV core (31–33). Further studies demonstrated that DDX3 is involved in the HCV life cycle and positively regulates HCV RNA replication (34, 35). As DDX3 also accomplishes its multiple functions via intracellular formation of different complexes with a variety of proteins (25, 36), we were interested to know whether DDX3 would act in concert with other cellular factors to modulate HCV replication or viral assembly. The yeast two-hybrid assay was performed to investigate the DDX3 interactome, and Y-box binding protein 1 (YB-1) was identified as one of DDX3-interacting partners (C. H. Chao and Y. H. Wu Lee, unpublished data).
YB-1 has DNA and RNA binding activities and interacts with a number of proteins to participate in almost every aspect of DNA and mRNA metabolism (37). Our recent study demonstrated that YB-1 inhibits mismatch repair through interactions with PCNA (38). Moreover, YB-1 is one of the main packing proteins of mRNPs, positively or negatively regulates cap-dependent translation (37), and activates myc family IRES (39, 40). Several studies have revealed that YB-1 has a role in oncogenesis (37). Many of the YB-1 functions have been demonstrated to be regulated by phosphorylation of the protein at serine 102 (S102) by Akt, p90 ribosomal S6 kinase (RSK) and protein kinase Cα (PKCα) (41). Coincident with its RNA binding activity, YB-1 has been identified as a HCV 3′ NTR-binding protein (42) and interacts with HCV RNA during infection (43). Recent studies suggested that YB-1 interacts and colocalizes with NS3/4A, core, and several HCV cellular cofactors, including DDX3 (43, 44). YB-1 also supports HCV RNA replication while transiently suppressing virion release in an early stage of a plasmid-driven infection system (43, 44).
In this study, we investigated the interaction between YB-1 and the pivotal viral protein NS5A and demonstrated that YB-1 and its cellular interacting partner, DDX3, are novel NS5A-interacting proteins. However, YB-1 regulates HCV RNA replication but not steady-state HCV RNA replication, a function different from that of DDX3. YB-1 silencing also inhibits HCV virus production as well as NS5A phosphorylation. Moreover, NS5A–YB-1 interaction is mediated by the phosphorylation of YB-1 at serine 102. The phosphorylation-mediated interaction between YB-1 and NS5A is essential for sustaining NS5A stability. Findings in the present work suggest a model in which YB-1 maintains the level and phosphorylation state of NS5A in HCV infection to support HCV RNA replication and infectious virus production, which is possibly regulated by HCV-induced cell signaling.
MATERIALS AND METHODS
Cell culture and virus stock preparation.
Huh-7.5.1 cells (kindly provided by Francis V. Chisari, Scripps Research Institute) (45) were cultured in Dulbecco's modified Eagle's medium (DMEM) with 10% fetal bovine serum (FBS). Ava5 cells containing the subgenomic HCV replicon of genotype 1b (kindly provided by Charles M. Rice, Rockefeller University) (46) were cultured in this medium with 1 mg/ml of G418. J6/JFH virus stock was prepared in Huh-7.5.1 cells and titers were determined as previously described (45).
Plasmids and siRNAs.
pFL-J6/JFH (47), J6/JFH(p7-Rluc2A), J6/JFH(p7-Rluc2A)GNN, and J6/JFH(p7-Rluc2A)K33A/R35A (48) plasmids were kindly provided by Charles M. Rice (Rockefeller University). Plasmid pcDNA3/HA-YB-1 or pGFP-YB-1 was generated by inserting the corresponding full-length DNA fragment of YB-1 into BamHI/XhoI-digested pcDNA3-HA (Invitrogen) or BamHI/XbaI-digested pEGFP-C1 (Clontech), respectively. Hemagglutinin (HA)-YB-1(S102A)-, HA-YB-1(S102D)-, green fluorescent protein (GFP)-YB-1(S102A)-, and GFP-YB-1(S102D)-expressing plasmids in which YB-1 serine 102 was replaced by either alanine (S102A mutant) or aspartic acid (S102D mutant) were constructed by site-directed mutagenesis of pcDNA3/HA-YB-1 or pGFP-YB-1 using the QuikChange site-directed mutagenesis system (Stratagene). Small interfering RNA (siRNA)-resistant HA-YB-1-, HA-YB-1(S102A)-, HA-YB-1(S102D)-, GFP-YB-1-, GFP-YB-1(S102A)-, and GFP-YB-1(S102D)-expressing plasmids bearing silent mutations in the target sequence of YB-1-specific siRNA (see below) were generated using QuikChange site-directed mutagenesis system (Stratagene). For construction of plasmid p5′Rluc, a DNA fragment of the EcoRI site to the first 63 nucleotides (nt) of the core coding sequence amplified from pFL-J6/JFH, and the Renilla luciferase (Rluc) coding sequence amplified from pRL-TK (Promega) using a primer containing an XbaI site, were fused by overlapping PCR. The DNA cassette was inserted between the EcoRI and XbaI sites of pFL-J6/JFH to replace the HCV genome. A similar strategy was used to construct plasmid p5′Rluc3′ in which the fused fragment containing 5′ NTR and Rluc coding sequence was further fused with a DNA fragment of the last 13 nt of NS5B to the XbaI site amplified from pFL-J6/JFH by overlapping PCR and inserted into pFL-J6/JFH to replace the HCV genome. Plasmid pcDNA-CMV/FLAG was constructed by inserting a Flag tag into KpnI/EcoRI-digested pcDNA3 (Invitrogen). To generate plasmid pcDNA-CMV/FLAG-NS5A, pcDNA-CMV/FLAG-NS3, or pcDNA-CMV/FLAG-core, a DNA fragment corresponding to NS5A, NS3, or core, respectively, was amplified from pFL-J6/JFH (genotype 2a) and cloned into EcoRI/XbaI-treated pcDNA-CMV/FLAG. Plasmids pFlag-NS5A (49), pFlag-NS3/4A (49), and pFlag-core (genotype 1b, version gi:329763; GenBank accession no. M84754) were kindly provided by Lih-Hwa Hwang (National Yang-Ming University). YB-1-specific siRNA (Silencer Select Validated siRNA s9731) and paired nontargeting negative-control siRNA (Silencer Select Negative Control No. 1) were purchased from Ambion. Pools of siRNAs targeting DDX3 (ON-TARGETplus SMARTpool) and paired control siRNA (ON-TARGETplus Nontargeting siRNA 2) were purchased from Dharmacon.
Antibodies and reagents.
The primary antibodies used for Western blotting or immunofluorescence were anti-YB-1 (rabbit; Abcam), anti-DDX3 (rabbit [26]), anti-NS5A (mouse monoclonal IgG1; Austral Biologicals), anti-NS3 (mouse monoclonal; Abcam), anti-core (mouse monoclonal; Abcam), anti-E2 (mouse monoclonal; Thermo Scientific), anti-double-stranded RNA (dsRNA) (J2 mouse monoclonal IgG2a; English and Scientific Consulting Bt.), anti-GFP (rabbit; Abcam), anti-Flag-horseradish peroxidase (HRP) (Sigma-Aldrich), anti-HA-HRP (Roche), and anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (mouse monoclonal; Sigma-Aldrich) antibodies. The following secondary antibodies were used for immunofluorescence: anti-mouse IgG1 DyLight649 (Jackson ImmunoResearch Laboratories), anti-mouse IgG2a Alexa Fluor 488, and anti-rabbit Alexa Fluor 555 (Molecular Probes, Invitrogen) antibodies. Sodium arsenite, cycloheximide, and MG132 were purchased from Sigma-Aldrich, dimethyl sulfoxide (DMSO) was purchased from Merck, and 4′,6-diamidino-2-phenylindole (DAPI) was purchased from Roche.
Plasmid and siRNA transfection.
Huh-7.5.1 cells were transfected with plasmids by TransIT-LT1 (Mirus) according to the manufacturer's recommendations. Knockdown of YB-1 was done with 100 nM siRNA (unless otherwise indicated), while DDX3 silencing was achieved with 60 nM siRNA using Lipofectamine 2000 (Invitrogen). For both plasmid and siRNA transfections, cells were washed with phosphate-buffered saline (PBS) once at 4 h posttransfection.
In vitro transcription and RNA transfection.
Plasmids encoding the monocistronic reporter RNAs and reporter genomes were linearized by XbaI and purified. In vitro transcription was performed with commercial kits (MEGAscript T7 [Ambion] and RiboMAX T7 [Promega]) according to the manufacturers' protocols. The RNA products were purified with either a MEGAclear kit (Ambion) or TRI reagent (Sigma-Aldrich). Purified monocistronic reporter RNAs were transfected with Lipofectamine 2000 (Invitrogen), whereas reporter genomes were transfected by electroporation as previously described (45).
Western blotting.
Cell lysates or immunoprecipitates were subjected to SDS-PAGE, probed with the desired antibodies, and detected by immunoblotting using enhanced chemiluminescence.
Coimmunoprecipitation.
Cell extracts of Huh-7.5.1 cells cotransfected with either the HA vector or plasmids encoding HA-YB-1 along with the Flag vector, Flag-tagged HCV core, NS3, or NS5A expression plasmids (genotype 2a) were prepared in lysis buffer (0.5% NP-40 and 1× protease inhibitor cocktail in PBS). Cell lysates (800 μg of protein) were immunoprecipitated with 20 μl of anti-Flag M2 magnetic beads (Sigma-Aldrich) in the presence of RNase A at 50 μg/ml. Proteins bound to the beads were then suspended in 60 μl of 2× SDS sample buffer, boiled, and subjected to Western blotting with anti-Flag-HRP and anti-HA-HRP antibodies.
For coimmunoprecipitation with replicon cells, Ava5 cells were lysed in the above-mentioned lysis buffer and incubated (400 μg of protein) with either anti-NS5A antibodies or control anti-Flag-HRP antibodies in the presence of RNase A (50 μg/ml),followed by 20 μl of bovine serum albumin (BSA)-blocked protein G-Sepharose beads (GE Healthcare). Proteins bound to the beads were then suspended in 60 μl of 2× SDS sample buffer, boiled, and subjected to Western blotting with anti-NS5A, anti-YB-1, anti-DDX3, and anti-GAPDH antibodies.
For coimmunoprecipitation with HCV-infected cells, extracts of Huh-7.5.1 cells infected with J6/JFH (genotype 2a) were prepared in the above-mentioned lysis buffer and incubated (2.5 mg of protein) with either anti-YB-1 antibodies or control anti-HA-HRP antibodies in the presence of RNase A (50 μg/ml) followed by 20 μl of BSA-blocked protein G-Sepharose beads (GE Healthcare). Proteins bound to the beads were then suspended in 200 μl of 2× SDS sample buffer, boiled, and subjected to Western blotting with anti-core, anti-E2, anti-NS3, anti-NS5A, anti-YB-1, and anti-GAPDH antibodies.
To examine interactions between NS5A and YB-1 variants, extracts of Huh-7.5.1 cells cotransfected with either the HA vector or plasmids encoding HA-YB-1 variants along with the Flag vector or Flag-tagged HCV NS5A expression plasmid (genotype 2a) were prepared in the lysis buffer described above. Cell lysates (1.2 mg of protein) were immunoprecipitated with 20 μl of anti-Flag M2 magnetic beads (Sigma-Aldrich) in the presence of RNase A at 50 μg/ml. Proteins bound to the beads were then suspended in 90 μl of 2× SDS sample buffer, boiled, and subjected to Western blotting with anti-Flag-HRP and anti-HA-HRP antibodies.
In situ PLA.
Naive Huh-7.5.1 or Huh-7.5.1 cells infected with J6/JFH (multiplicity of infection [MOI] = 0.5) for 48 h were fixed with 4% paraformaldehyde, permeabilized with 0.1% Triton X-100, and blocked with 3% BSA in PBS. After blocking, cells were incubated with anti-YB-1 (rabbit) and either anti-NS5A (mouse) or anti-E2 (mouse) antibodies diluted in PBS with 1% BSA. Recognition of primary antibodies by the two proximity ligation assay (PLA) probes (anti-mouse MINUS and anti-rabbit PLUS) and subsequent ligation and amplification reactions were performed with the Duolink in situ kit (Sigma-Aldrich) according to the manufacturer's instructions. Images were acquired using a Leica DM 6000B fluorescence microscope.
Infection assays.
To examine the role of YB-1 in the HCV life cycle and in HCV replication complex formation, control- or YB-1 siRNA-transfected cells were inoculated with J6/JFH virus at an MOI of 0.5 for 6 h and rinsed twice with fresh medium. Forty-eight hours after inoculation, intracellular protein and RNA samples as well as culture media were collected for Western blotting, quantitative real-time reverse transcription-PCR (RT-PCR), or focus-forming unit analysis (45).
For YB-1 knockdown/rescue experiments, Huh-7.5.1 cells transfected with control or YB-1 siRNA were further transfected 24 h later with either the HA vector or a plasmid encoding siRNA-resistant HA-YB-1, HA-YB-1(S102A), or HA-YB-1(S102D). For examining intracellular viral protein expression and infectious virus production, transfected cells were then inoculated with J6/JFH virus (MOI = 0.5) 24 h after plasmid transfection. Forty-eight hours later, samples were collected and analyzed as described above. For assessing intracellular HCV RNA levels, siRNA- and plasmid-transfected cells were inoculated with J6/JFH virus at 24, 40, or 48 h after plasmid transfection. Forty-eight hours postinoculation, intracellular RNA was extracted for quantitative real-time RT-PCR.
To knock down YB-1 or DDX3 after HCV infection, naive Huh-7.5.1 cells were infected with J6/JFH at an MOI of 5 to 7.5 a day after seeding. Six hours after inoculation, cells were washed twice with fresh medium. Two hours later, the infected cells were transfected with control, YB-1, or DDX3 siRNA. Seventy-two hours after transfection, intracellular protein and RNA samples and culture media were collected for Western blotting, quantitative real-time RT-PCR, or focus-forming unit analysis (45), respectively, or the cells were fixed for immunofluorescence.
RNA extraction and quantitative real-time RT-PCR.
Total RNA was extracted using TRI reagent (Invitrogen). cDNAs were synthesized using RevertAid First Strand cDNA synthesis kit (Fermentas). Primers used for reverse transcription were oligo(dT)18 and a previously described primer specific for HCV (50) (5′-CTCCCGGGGCACTCGCAAGC-3′). Real-time PCR was performed using LightCycler FastStart DNA Master SYBR green I (Roche) and LightCycler 2.0 System (Roche). Primers used were as follows: for HCV subgenomic replicon (genotype 1b), 5′-GTCTAGCCATGGCGTTAGTA-3′ (sense) and 5′-CTCCCGGGGCACTCGCAAGC-3′ (antisense) (50); for J6/JFH RNA, 5′-GCCTAGCCATGGCGTTAGTA-3′ (sense) and 5′-CTCCCGGGGCACTCGCAAGC-3′ (antisense), previously described primers (50) with modifications to match the JFH-1 sequence in the 5′ NTR; and for GAPDH, 5′-CAACTACATGGTTTACATGTTC-3′ (sense) and 5′-GCCAGTGGACTCCACGAC-3′ (antisense).
HCV translation assays.
Huh-7.5.1 cells were transfected with control or YB-1 siRNA as described above. siRNA-transfected cells were pretreated with distilled water or sodium arsenite (40 μM) for 1 h and were then transfected with a monocistronic reporter (5′Rluc or 5′Rluc3′) RNA at 96 h post-siRNA transfection. Two hours later, cell lysates were prepared and luciferase activities were detected using the Renilla luciferase assay system (Promega).
HCV replication assay.
Control- or YB-1 siRNA-transfected Huh-7.5.1 cells were transfected with a reporter genome [J6/JFH(p7-Rluc2A), J6/JFH(p7-Rluc2A)K33A/R35A, or J6/JFH(p7-Rluc2A)GNN] by electroporation at 72 h post-siRNA transfection as described above. Cell lysates were collected and assayed for luciferase activities at the desired time points using the Renilla luciferase assay system (Promega).
Immunofluorescence and confocal microscopy.
J6/JFH-infected or naive Huh-7.5.1 cells grown on coverslips were fixed with 4% paraformaldehyde at 4°C for 30 min, permeabilized (0.1% Triton X-100 in PBS), and incubated with 3% BSA in PBS at 4°C for 30 min. Cells were then incubated with primary antibodies diluted in PBS with 1% BSA at 4°C overnight and subsequently secondary antibodies at room temperature for 1 h. After a washing, nuclei were stained with DAPI (Roche). Cells were then mounted in fluorescence mounting medium (Dako). Images were acquired using a Zeiss LSM 700 confocal microscope or a Leica DM 6000B fluorescence microscope. Colocalization coefficients, the ratios of colocalized pixels to all pixels of the proteins examined, were calculated using the Zen 2009 software (Carl Zeiss) and were represented as percentages of colocalization.
Protein half-life assay.
To determine the half-life of the ectopically expressed Flag-tagged NS5A, control- or YB-1 siRNA-transfected Huh-7.5.1 cells were further transfected with the Flag-NS5A-expressing plasmid at 48 h post-siRNA transfection. Forty-eight hours later, siRNA- and plasmid-transfected cells were treated with 100 μg/ml of cycloheximide and collected at the desired time points after cycloheximide treatment.
To assess the half-life of NS5A in Ava5 cells, cycloheximide (100 μg/ml) was added at 72 h after control or YB-1 siRNA transfection, and the protein samples were collected at the desired time points after cycloheximide treatment.
For examining the roles of different YB-1 variants in modulating NS5A half-life, control- or YB-1 siRNA-transfected Huh-7.5.1 cells were further transfected with the Flag-NS5A-expressing plasmid (JFH1, genotype 2a) together with plasmids encoding GFP-YB-1 variants at 48 h post-siRNA transfection. Forty-eight hours later, cells were treated with 100 μg/ml of cycloheximide and collected at the desired time points after cycloheximide treatment.
Statistical analysis.
Statistical analysis was conducted with a two-tailed, unpaired Student's t test.
RESULTS
Interaction of NS5A with YB-1 and DDX3.
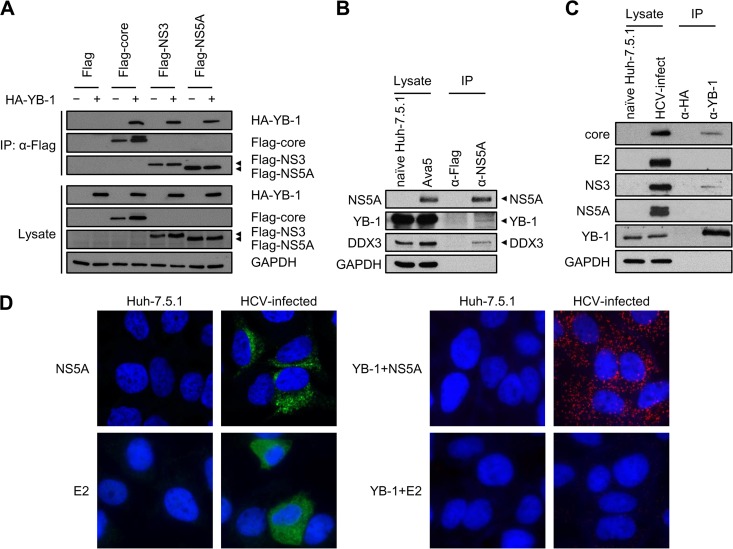
To gain new insights into the mechanism by which YB-1 regulates the HCV life cycle, we attempted to identify novel interactions between HCV proteins and YB-1. Among the HCV proteins, core protein, NS3, and NS5A have notably been shown to have a larger number of cellular interacting partners (51). In view of the fact that both core/YB-1 and NS3/YB-1 interactions have been extensively studied (43, 44), we turned our attention to examining the interaction between NS5A and YB-1. To this end, coimmunoprecipitation was performed with Huh-7.5.1 cell extracts coexpressing HA-YB-1 and Flag-tagged NS5A (JFH1) of genotype 2a in the presence of RNase A. Interaction between NS5A and YB-1 was observed, as were the known NS3–YB-1 and core–YB-1 interactions (Fig. 1A and references 43 and 44. The NS5A–YB-1 interaction was further verified in the context of HCV RNA replication by immunoprecipitating NS5A from HCV subgenomic replicon cells (Ava5, genotype 1b) (46). Specific detection of NS5A by NS5A antibody was validated in naive Huh-7.5.1 and Ava5 cells. Both endogenous YB-1 and its interacting partner DDX3 were coimmunoprecipitated with NS5A (Fig. 1B), indicating that the NS5A/YB-1 interaction is not genotype specific. Next, we examined the NS5A–YB-1 interaction in HCV infection by immunoprecipitating YB-1 from Huh-7.5.1 cells infected with the genotype 2a J6/JFH chimeric virus. Specific detection of the HCV proteins was verified with naive Huh-7.5.1 cells. Although both the known NS3–YB-1 and core–YB-1 interactions were detected, interactions between YB-1 and NS5A or E2 were not observed (Fig. 1C). To further investigate whether NS5A or E2 interacts with YB-1 in HCV infection, the highly sensitive in situ proximity ligation assay (PLA; see Materials and Methods) was performed with HCV-infected Huh-7.5.1 cells (genotype 2a). Specific detection of NS5A and E2 by immunofluorescence with NS5A and E2 antibodies, respectively, was confirmed with naive and HCV-infected Huh-7.5.1 cells (Fig. 1D, left side). The NS5A–YB-1 interaction was observed in HCV-infected cells (red fluorescent dots in Fig. 1D, right side), while interaction between E2 and YB-1 was not detected (Fig. 1D, right side), demonstrating that YB-1 is an authentic NS5A-interacting partner. Collectively, besides the known association with core and NS3 (31–33, 43, 44), both YB-1 and DDX3 interact with the pivotal viral protein NS5A. The results reveal that YB-1 and DDX3 not only interact with each other but also share similar HCV interacting partners.
FIG 1.
YB-1 is a novel NS5A-interacting cellular protein. (A) Coimmunoprecipitation was performed in Huh-7.5.1 cells cotransfected with either an HA vector (−) or expression plasmids for HA-YB-1 (+) along with a Flag vector or Flag-tagged HCV core, NS3, or NS5A of HCV genotype 2a. Immunoprecipitation was performed with lysates treated with RNase A and anti-Flag beads. For detection of HA-YB-1, 10-μg cell lysates (1.25% of total input protein) and one-third of the precipitates (IP) were subjected to Western blotting with an anti-HA antibody. To detect Flag-tagged viral proteins, 40-μg cell lysates (5% of total input protein) and a quarter of the precipitates were subjected to Western blotting with an anti-Flag antibody. GAPDH was included as an internal control. (B) Coimmunoprecipitation was performed in replicon cells (Ava5, genotype 1b). Immunoprecipitation was performed with lysates treated with RNase A and an anti-NS5A antibody. For detection of YB-1, DDX3, and GAPDH, 10 μg of Ava5 cell lysate (2.5% of total input protein) and two-thirds of the precipitates were subjected to Western blotting with anti-YB-1, anti-DDX3, and anti-GAPDH antibodies, respectively. To detect NS5A, 10-μg Ava5 cell lysates (2.5% of total input protein) and a quarter of the precipitates were subjected to Western blotting with an anti-NS5A antibody. (C) Coimmunoprecipitation was performed in J6/JFH-infected Huh-7.5.1 cells (genotype 2a). Immunoprecipitation was performed with lysates treated with RNase A and an anti-YB-1 antibody. For detection of core, E2, NS3, and GAPDH, 10 μg of HCV-infected cell lysate (0.4% of total input protein) and 5% of the precipitates were subjected to Western blotting with anti-core, anti-E2, anti-NS3, and anti-GAPDH antibodies, respectively. To detect NS5A, 10-μg HCV-infected cell lysates (0.4% of total input protein) and 20% of the precipitates were subjected to Western blotting with an anti-NS5A antibody. To detect YB-1, 20-μg HCV-infected cell lysates (0.8% of total input protein) and 15% of the precipitates were subjected to Western blotting with an anti-YB-1 antibody. (D) (Left side) Huh-7.5.1 cells were infected with J6/JFH (genotype 2a) at an MOI of 0.5 for 48 h. Naive or infected Huh-7.5.1 cells were then immunostained for NS5A (green) or E2 (green) as indicated and were observed under a confocal or a fluorescence microscope, respectively. (Right side) Detection of interactions between YB-1 and NS5A or E2 in Huh-7.5.1 cells infected with J6/JFH (genotype 2a) by in situ proximity ligation assay with anti-YB-1 and anti-NS5A or anti-E2 antibodies, as indicated. Images were acquired by a fluorescence microscope. Nuclei were stained with DAPI (blue).
YB-1 silencing inhibits the HCV life cycle.
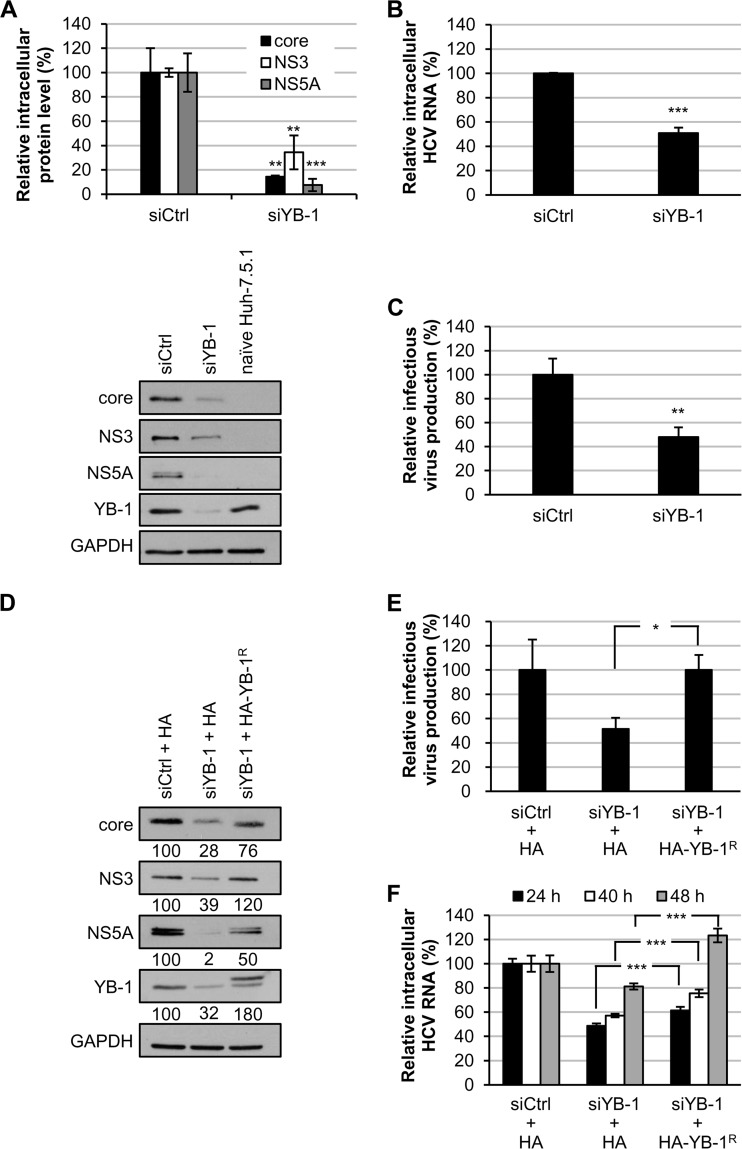
To validate the role of YB-1 in the HCV life cycle, YB-1 siRNA- or control siRNA (100 nM)-transfected Huh-7.5.1 cells were infected with J6/JFH at the low MOI of 0.5. Intracellular HCV protein and RNA levels as well as infectivity in the supernatants were analyzed at 48 h postinfection. YB-1 silencing resulted in downregulation of core, NS3, and NS5A protein expression to 14%, 34% and 8%, respectively, compared with that of control siRNA-transfected cells (Fig. 2A). Intracellular HCV RNA levels and infectious virus titers in the culture supernatant were also reduced by 49% and 52%, respectively, by the knockdown of YB-1 (Fig. 2B and C). YB-1 downregulation was confirmed by immunoblotting, and specific detection of the HCV proteins was validated with naive Huh-7.5.1 cells (Fig. 2A).
FIG 2.
Knockdown of YB-1 inhibits the HCV life cycle. Huh-7.5.1 cells transfected with control siRNA (siCtrl) or YB-1 siRNA (siYB-1) for 48 h were infected with J6/JFH (genotype 2a) at an MOI of 0.5. Cells and culture media were collected at 48 h postinfection. (A) Intracellular protein levels were assessed by Western blotting for core, NS3, NS5A, and YB-1, and GAPDH was used as an internal control. HCV core, NS3, and NS5A levels were quantified by ImageJ and normalized to GAPDH. A representative immunoblot is shown at the bottom. (B) Intracellular HCV RNA levels were analyzed by quantitative RT-PCR and normalized to GAPDH RNA. (C) Infectious virus titers in the culture media were determined by focus-forming unit analysis. Means ± SDs from three independent experiments are shown as amount relative to that of control siRNA-treated cells. Huh-7.5.1 cells transfected with control siRNA (siCtrl) or YB-1 siRNA (siYB-1) for 24 h were transfected with an HA vector or a plasmid encoding siRNA-resistant HA-YB-1 (HA-YB-1R) and infected with J6/JFH (genotype 2a) at an MOI of 0.5 24 h later. Cells and culture media were collected at 48 h postinfection. (D) Intracellular protein expression was detected by Western blotting for core, NS3, NS5A, and YB-1, and GAPDH was used as an internal control. HCV core, NS3, NS5A, and YB-1 levels were quantified by ImageJ and normalized to GAPDH levels. Results are represented as percentages relative to that of cells transfected with control siRNA and the HA vector. (E) Infectious virus titers in the culture media were determined by focus-forming unit analysis in triplicates. Representative results from one of two independent experiments are shown as means ± SDs relative to control siRNA/HA vector-transfected cells. (F) Huh-7.5.1 cells transfected with control siRNA or YB-1 siRNA for 24 h were transfected with the HA vector or a plasmid encoding siRNA-resistant HA-YB-1. At 24, 40, or 48 h, cells were infected with J6/JFH (genotype 2a) at an MOI of 0.5. Cells were collected at 48 h postinfection. Intracellular HCV RNA levels were analyzed by quantitative RT-PCR and normalized to GAPDH RNA in triplicate. Means ± SDs are shown as amount relative to that of control siRNA/HA vector-transfected cells. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
To confirm the specificity of YB-1 siRNA silencing, HCV infection was examined by overexpressing siRNA-resistant YB-1 in YB-1 siRNA-transfected cells (see Materials and Methods). Expression of siRNA-resistant YB-1 restored core, NS3, and NS5A protein levels from 28% to 76%, 39% to 120%, and 2% to 50%, respectively, in YB-1 knockdown cells compared with the control (Fig. 2D). Infectious virus production levels in YB-1 knockdown cells also recovered from 51% to 100% by expressing siRNA-resistant YB-1 (Fig. 2E). Likewise, intracellular HCV RNA levels recovered from 49% to 61%, 57% to 76%, and 81% to 123% with increased intervals (24 to 48 h) between the time points for introducing plasmid encoding siRNA-resistant YB-1 and virus inoculation (Fig. 2F). The results substantiate that YB-1 is essential for the HCV life cycle, as reported by Chatel-Chaix et al. (43, 44).
The effect of YB-1 on HCV IRES-mediated translation does not account for the crucial role of YB-1 in the HCV life cycle.
To examine whether YB-1 participates in the HCV life cycle via regulating HCV IRES-mediated translation, in vivo translation assay was performed with monocistronic reporter RNAs containing the 5′ NTR alone (5′Rluc) or both the 5′ and 3′ NTRs (5′ Rluc3′), as the 3′ NTR promotes HCV translation (52, 53) (Fig. 3A). Interestingly, in contrast to the critical role of YB-1 in HCV propagation, YB-1 silencing moderately enhanced translation of 5′Rluc3′ by 25% (Fig. 3C) but not 5′Rluc (Fig. 3B).
FIG 3.
YB-1 silencing moderately enhances and slightly inhibits HCV IRES-mediated translation under normal and stress conditions, respectively. (A) Diagram of the reporter RNAs. 5′Rluc consists of the HCV 5′ NTR (genotype 2a) and Renilla luciferase. 5′Rluc3′ contains the same structure at the 5′ end but with the HCV 3′ NTR (genotype 2a) following Renilla luciferase. Huh-7.5.1 cells transfected with control or YB-1 siRNA were pretreated with distilled water or 40 μM sodium arsenite (SA)for 1 h before transfection with 5′Rluc (B) or 5′Rluc3′ (C) RNA. Luciferase activities were analyzed at 2 h posttransfection. Results are presented as means ± SDs from three independent experiments relative to cells treated with the control siRNA. *, P < 0.05; ***, P < 0.001.
HCV has been reported to induce translational stress at various periods of infection (54) and uses different translation initiation factors under stress than normal conditions (55, 56). To further explore the role of YB-1 in HCV translation, an in vivo translation assay was performed under stress conditions induced by sodium arsenite. Relative to the translation of 5′Rluc, translation of 5′Rluc3′ was more resistant to stress (78% versus 55% retention, Fig. 3B and C), implying a previously undefined role of the HCV 3′ NTR in the stress resistance of HCV IRES translation. Knockdown of YB-1 had no effects on HCV IRES-mediated translation of 5′Rluc (Fig. 3B), and there was only a 10% decrease of translation of 5′Rluc3′ under stress conditions (Fig. 3C). Taken together, our results suggest that YB-1 has either a moderate or a minor effect that inhibits or promotes HCV IRES-mediated translation under normal or stress conditions, respectively, which are insufficient to account for the crucial role of YB-1 in the HCV life cycle.
YB-1 is involved in HCV RNA replication.
The effects of YB-1 on HCV RNA replication were next investigated by luciferase assay with the infectious HCV genome J6/JFH(p7-Rluc2A) (Fig. 4A) (48). The incorporated Renilla luciferase reporter allows sensitive detection of primary translation of HCV genome and establishment of HCV RNA replication in the HCV life cycle. These two stages of the viral life cycle can be distinguished by comparing the replication kinetics of J6/JFH(p7-Rluc2A) with that of the replication-defective genome harboring mutations in NS5B [J6/JFH(p7-Rluc2A)GNN] (Fig. 4A) (48). To exclude the effects of YB-1 on HCV infectivity, we further included an assembly-defective variant, J6/JFH(p7-Rluc2A)K33A/R35A, with mutations in p7 (Fig. 4A), which leads to defective virion morphogenesis without impairing HCV RNA replication (48).
FIG 4.
Downregulation of YB-1 reduces HCV RNA replication. (A) Diagram of the reporter genome (genotype 2a). The p7 mutation, K33A/R35A, and the GNN mutation are shown. Huh-7.5.1 cells transfected with control siRNA (siCtrl) or YB-1 siRNA (siYB-1) were electroporated with the wild-type reporter genome [J6/JFH(p7-Rluc2A)] (B), the reporter genome with the p7 mutation [J6/JFH(p7-Rluc2A)K33A/R35A)](C), or the replication-deficient reporter genome [J6/JFH(p7-Rluc2A)GNN] (D). Cell lysates were harvested at the indicated time points posttransfection, and luciferase activities were analyzed. For panels B and C, graphs of luciferase activity along with the posttransfection time points are shown on the right, while the luciferase activity at 2 to 12 h is focused on in left graphs. Means ± SDs for triplicate wells are shown. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
YB-1 transient knockdown cells or control cells were transfected with each of the reporter genome variants, and luciferase activities were determined at various time points. Knockdown of YB-1 led to a slight increase in luciferase activity in cells transfected with each of three genomes at 2 and/or 4 h posttransfection (Fig. 4B and C [left side] and D), implying that YB-1 might repress primary translation of the HCV genome, consistent with the results from the in vivo translation assay (Fig. 3C). Notably, luciferase activities were elevated at 10 to 12 h posttransfection in cells transfected with J6/JFH(p7-Rluc2A) (Fig. 4B) or J6/JFH(p7-Rluc2A)K33A/R35A (Fig. 4C), but not in cells transfected with J6/JFH(p7-Rluc2A)GNN (Fig. 4D), indicating that the initiation of HCV RNA replication had occurred at 10 to 12 h posttransfection. YB-1 silencing inhibited luciferase activities in cells transfected with J6/JFH(p7-Rluc2A) and J6/JFH(p7-Rluc2A)K33A/R35A to up to 48% and 52%, respectively (Fig. 4B and C) at 10 to 30 h posttransfection, suggesting that YB-1 is required for HCV RNA replication.
YB-1 colocalizes with HCV replication complexes.
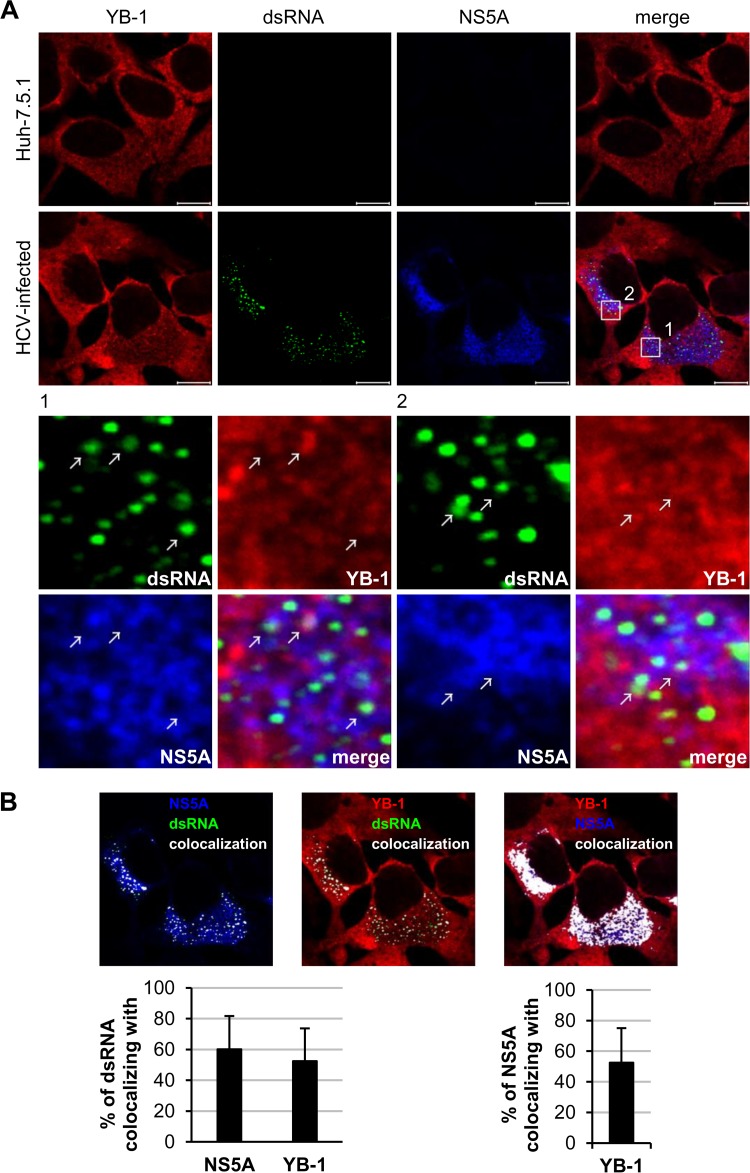
To examine whether YB-1 colocalizes with the HCV replication markers, dsRNA and NS5A, immunofluorescence analysis was performed on J6/JFH-infected Huh-7.5.1 cells. dsRNA is the intermediate of HCV RNA replication representing the site of replication complex (57), while NS5A is a component of the HCV replication complex (7–10). Triple staining with antibodies against YB-1, dsRNA, and NS5A revealed that YB-1 did colocalize with NS5A in the HCV replication complex (Fig. 5A). Quantitative analysis showed that about 52% of the dsRNA colocalized with YB-1, while approximately 60% of dsRNA colocalized with NS5A (Fig. 5B). Consistent with results of coimmunoprecipitation and in situ proximity ligation assays (Fig. 1), 53% of NS5A colocalized with YB-1 in both the replication complex and the cytoplasm (Fig. 5B), confirming that YB-1 is an NS5A-interacting protein.
FIG 5.
YB-1 colocalizes with HCV replication complexes in HCV-infected cells. (A) Huh-7.5.1 cells were infected with J6/JFH (genotype 2a) at an MOI of 0.5. Forty-eight hours postinfection, cells were immunostained for dsRNA (green), NS5A (blue), and YB-1 (red) and were observed under a confocal microscope. The lower portion shows the enlarged images of the boxed regions. The colocalization sites of YB-1, dsRNA, and NS5A are indicated by white arrows. (B) Colocalization of NS5A and dsRNA (left), YB-1 and dsRNA (middle), and YB-1 and NS5A (right) as analyzed by Zen 2009 software (Zeiss). Colocalization sites are indicated in white. Percentages of dsRNA that colocalized with NS5A or YB-1 or of NS5A that colocalized with YB-1 are shown as means ± SDs (graphs). n = 44; scale bars = 10 μm.
To further confirm the role of YB-1 in HCV RNA replication, control- or YB-1 siRNA-transfected Huh-7.5.1 cells were infected with J6/JFH (genotype 2a) for 48 h, and the cells were subjected to immunofluorescence detection with dsRNA, NS5A, and YB-1 antibodies. YB-1 was found to be downregulated to 13% that of control cells by siRNA-mediated silencing (Fig. 6A). Consistent with the positive regulatory role of YB-1 in HCV RNA replication (Fig. 4), YB-1 knockdown significantly reduced dsRNA and NS5A levels, to 51% and 42%, respectively, compared with that of the control cells (Fig. 6A).
FIG 6.
YB-1 silencing reduces HCV replication complexes. (A) Huh-7.5.1 cells transfected with control siRNA (siCtrl) or YB-1 siRNA (siYB-1) for 48 h were infected with J6/JFH (genotype 2a) at an MOI of 0.5. Cells were fixed at 48 h postinfection and subjected to immunostain for YB-1 (red), dsRNA (green), and NS5A (blue) and were observed under a confocal microscope. Intensities of YB-1, dsRNA, and NS5A were analyzed by Zen 2009 software (Zeiss) and are shown as means ± SDs relative to that of control siRNA-treated cells (graphs). (B) Colocalizations of NS5A and dsRNA (left), YB-1 and dsRNA (middle), and YB-1 and NS5A (right) from panel A were analyzed by Zen 2009 software (Zeiss) and are displayed in white. Percentages of dsRNA that colocalized with NS5A or YB-1 or of NS5A that colocalized with YB-1 are shown as means ± SDs relative to control siRNA-treated cells (graphs). n = 45. ***, P < 0.001.
The inhibitory effect of YB-1 silencing on HCV replication complex formation was then affirmed by quantitative analysis of dsRNA and NS5A colocalization, which was reduced to 65% of the control by YB-1 silencing (Fig. 6B), further substantiating that YB-1 silencing disturbs the formation of HCV replication complexes. Colocalization of dsRNA and NS5A with YB-1 was also analyzed. The percentages of dsRNA and NS5A that colocalized with YB-1 were decreased to 27% and 12% of the control, respectively, by knockdown of YB-1 (Fig. 6B), as a consequence of a more dramatic decline of YB-1 levels than that of dsRNA and NS5A (Fig. 6A). Taken together, these results reveal that YB-1 colocalizes with HCV replication complexes in HCV-infected cells, which further substantiates the role of YB-1 in HCV RNA replication.
YB-1 is dispensable for steady-state HCV RNA replication but is involved in infectious virus production.
In addition to steady-state HCV RNA replication, the initial phase of HCV RNA replication has previously been identified (21, 22). Moreover, besides its role in the steady-state HCV RNA replication, unique roles of NS5A in the early stage of HCV RNA replication, including membranous web formation and transient RNA replication after infection, have been recognized (19–22). Given that YB-1 is an NS5A-interacting protein, we examined whether YB-1 functions in the initiation of HCV RNA replication or in steady-state HCV RNA replication by investigating the effects of silencing YB-1 in HCV-infected cells (Fig. 7A). The HCV replication complex is protected by a membrane (8, 58, 59) and is stable, with limited interchange of internal and periphery proteins (60, 61). To verify the elimination of preincorporated YB-1 in the replication complex in HCV-infected cells upon YB-1 silencing, colocalization of dsRNA and YB-1 was first examined (Fig. 7B). The percentage of dsRNA that colocalized with YB-1 was severely reduced, from 60% to 8%, by YB-1 silencing (Fig. 7C, left graph), confirming siRNA-mediated knockdown of YB-1 in the HCV replication complex. The formation of HCV replication complex was also evaluated by analyzing the colocalization of dsRNA with NS5A. Notably, different from that of YB-1 silencing before HCV infection (Fig. 6B), knockdown of YB-1 in HCV-infected cells did not affect the colocalization of dsRNA with NS5A (Fig. 7B and C, right graph), suggesting that eliminating YB-1 from the assembled HCV replication complex does not disrupt the replication complex. The effect of YB-1 silencing in HCV-infected cells on HCV propagation was further investigated. The YB-1 interacting partner, DDX3, which is also an NS5A-interacting protein (Fig. 1B) involved in HCV RNA replication (34), was silenced in parallel. siRNA-mediated individual knockdown of YB-1 and DDX3 resulted in comparable reductions, 74% and 78%, respectively, by immunoblotting.
FIG 7.
Knockdown of YB-1 does not affect steady-state HCV RNA replication but represses production of infectious particles. (A) The process of YB-1 or DDX3 silencing on HCV infection. Huh-7.5.1 cells infected with J6/JFH (genotype 2a) at a high MOI (5 to 7.5) were transfected with control (siCtrl), YB-1 (siYB-1), or DDX3 (siDDX3) siRNA at 8 h postinfection. Three days posttransfection, cells and culture media were fixed or harvested. (B) Immunostaining for YB-1 (red), dsRNA (green), and NS5A (blue) was observed by confocal microscopy. The colocalization images show the results as analyzed by Zen 2009 software (Zeiss); colocalization sites of dsRNA with YB-1 or NS5A are indicated in white. (C) Quantitative results from panel B are shown as means ± SDs relative to those for control siRNA-treated cells (siCtrl, n = 128; siYB-1, n = 153). For panels D to G, results are presented as means ± SDs from three independent experiments relative to those for control siRNA-transfected cells. (D) Intracellular HCV RNA levels were quantified by real-time RT-PCR normalized to the GAPDH data. (E) Infectious virus titers in culture media were determined by focus-forming unit analysis. (F) Intracellular protein levels were detected by Western blotting for NS5A, NS3, and core, and GAPDH was used as an internal control. NS5A, NS3, and core protein expression levels were quantified by ImageJ and normalized to GAPDH levels. A representative immunoblot is shown on the right. YB-1 and DDX3 were detected to confirm knockdown efficiency. (G) Quantitation of the p58/p56 ratios of NS5A. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Interestingly, in contrast to the effects of silencing of YB-1 before reporter genome transfection (Fig. 4B and C), knockdown of YB-1 in HCV-infected cells had no significant impact on the intracellular HCV RNA levels (Fig. 7D, left graph), while silencing of DDX3 had a pronounced (30%) reduction in the amount of HCV RNA (Fig. 7D, right graph). However, the infectious virus production was more severely repressed by YB-1 silencing (49%) (Fig. 7E, left graph) than by DDX3 silencing (37%) (Fig. 7E, right graph). As the intracellular HCV RNA level was unaltered, the YB-1 silencing-mediated reduction of infectivity suggests a regulatory role for YB-1 in infectious virus production. Notably, knockdown of YB-1 resulted in a marked (62%) reduction of the NS5A level (Fig. 7F), while effects were absent on the core and modest on NS3 (32% reduction) (Fig. 7F). Preferential reduction of the NS5A level was also observed in similar DDX3-silencing experiments, albeit less severely (Fig. 7F).
NS5A exists in two phosphorylation forms, hyperphosphorylated (p58) and hypophosphorylated (p56) (62). The phosphorylation status of NS5A has been proposed to regulate the equilibrium between HCV RNA replication and virus assembly (63). Interestingly, our results showed that knockdown of YB-1 in HCV-infected Huh-7.5.1 cells preferentially downregulated p58 over p56, resulting in a 44% decline in the p58/p56 ratio (Fig. 7G, left graph). In contrast, DDX3 silencing-mediated reduction of NS5A had no apparent preference for either NS5A phosphorylation form and hence did not affect the p58/p56 ratio (Fig. 7G, right graph). These results indicate that YB-1 knockdown not only reduces the NS5A level but also alters the ratio of p58 to p56, which is different from that of DDX3.
To validate that YB-1 functions in the initial stage of HCV RNA replication rather than in steady-state HCV RNA replication, YB-1 was silenced with different siRNA doses in the replicon-containing cells supporting persistent HCV RNA replication (Ava5, genotype 1b), and the replicon RNA levels were analyzed. The different degrees of YB-1 silencing were verified by immunoblotting (79% and 90% reduction) (Fig. 8B). Consistent with that observed in the HCV-infected cells (Fig. 7D), knockdown of YB-1 in replicon cells had no impact on the HCV RNA levels (Fig. 8A), even at the dose of siRNA inhibiting HCV life cycle when administered before HCV infection (Fig. 2). Remarkably, while the NS3 level was almost not affected by YB-1 knockdown, the amount of NS5A was reduced by YB-1 silencing in a dose-dependent manner from 22% to 62% (Fig. 8B), in accordance with that observed in HCV-infected cells (Fig. 7F). Thus, our results suggest that YB-1 is not involved in the steady-state HCV RNA replication but participates in the production of infectious particles.
FIG 8.

YB-1 silencing has no effect on steady-state HCV RNA replication in replicon cells but specifically reduces NS5A levels. Ava5 cells harboring the subgenomic replicon (genotype 1b) were transfected with control siRNA (siCtrl; 100 nM) or 50 or 100 nM YB-1 siRNA (siYB-1) and harvested 72 h later. (A) Intracellular HCV replicon RNA levels were analyzed by quantitative RT-PCR and normalized to GAPDH RNA. Means ± SDs from three independent experiments are shown as amount relative to that of control siRNA-transfected cells. (B) Intracellular protein levels were detected by Western blotting for NS3, NS5A, YB-1, and GAPDH, quantified by ImageJ, and normalized to GAPDH. Results are represented as percentages relative to that of control siRNA-transfected cells. A representative immunoblot of three independent experiments is shown.
YB-1 knockdown promotes NS5A degradation.
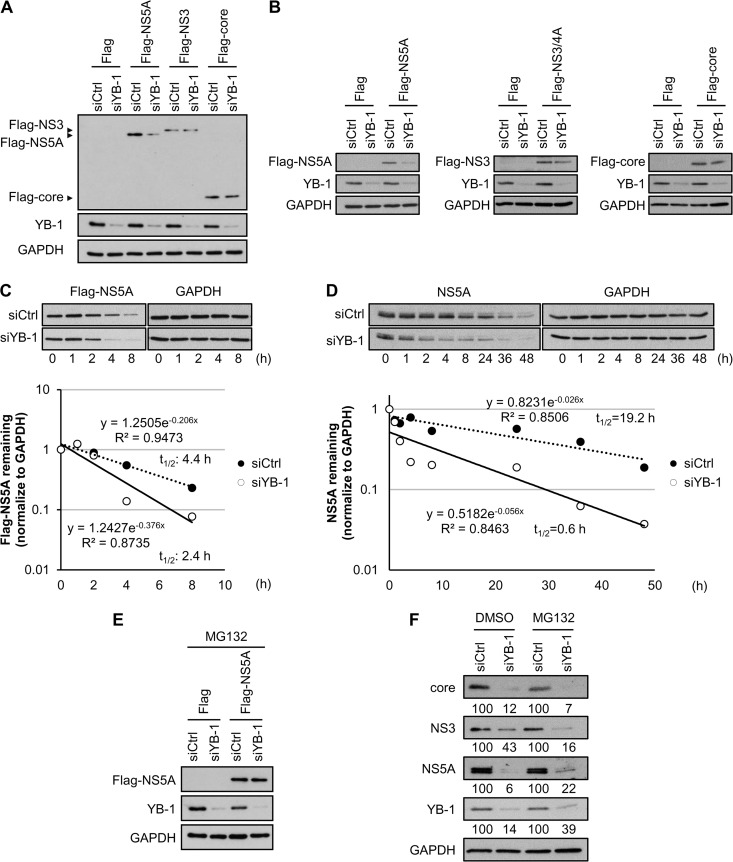
As the NS5A protein level was shown to be more susceptible to YB-1 silencing than those of NS3 and core (Fig. 2A, 7F, and 8B), we further investigated whether YB-1 regulates the NS5A protein level. YB-1- or control siRNA-transfected Huh-7.5.1 cells were transfected individually with Flag-tagged NS5A, NS3, or core of HCV genotype 2a. Knockdown of YB-1 markedly reduced the Flag-NS5A protein level but had no discernible effects on the expression of Flag-NS3 and Flag-core, even in the context of the absence of HCV replication (Fig. 9A). Similar results were also obtained with Flag-tagged NS5A, NS3, and core from genotype 1b (Fig. 9B), indicating that among the three interacting viral proteins, YB-1 specifically and positively modulates the NS5A expression level.
FIG 9.
YB-1 knockdown promotes NS5A degradation. Control (siCtrl) or YB-1 (siYB-1) siRNA-transfected Huh-7.5.1 cells were transfected with expression plasmid for Flag-NS5A, Flag-NS3, or Flag-core of HCV genotype 2a (A) or Flag-NS5A, Flag-NS3/4A, or Flag-core of HCV genotype 1b (B). Twenty-four hours (A) or 16 h (B) posttransfection, protein levels were detected by Western blotting for viral proteins with anti-Flag antibody, GAPDH as an internal control, and YB-1 to confirm silencing efficiency. (C) (Top) Control- or YB-1 siRNA-transfected Huh-7.5.1 cells were transfected with Flag-NS5A (genotype 2a) expression plasmid. Forty-eight hours later, cells were treated with cycloheximide at 100 μg/ml. Cells were harvested and analyzed for Flag-NS5A levels by Western blotting with an anti-Flag antibody at the indicated time points after cycloheximide addition. GAPDH was used as an internal control. (Bottom) Flag-NS5A levels were quantified by ImageJ and normalized to GAPDH levels. The half-life of Flag-NS5A was determined by regression analysis. Protein levels of Flag-NS5A without cycloheximide treatment (0 h) are set as 1. (D) (Top) Ava5 replicon cells (genotype 1b) were transfected with control or YB-1 siRNA. At 72 h posttransfection, cells were treated with cycloheximide at 100 μg/ml. Cells were harvested and analyzed for NS5A levels by Western blotting at the indicated time points after cycloheximide addition. GAPDH was used as an internal control. (Bottom) NS5A levels were quantified by ImageJ and normalized to GAPDH levels, and the half-life of NS5A was determined by regression analysis. Protein levels of NS5A without cycloheximide treatment (0 h) are set as 1. (E) Control or YB-1 siRNA-transfected Huh-7.5.1 cells were transfected with Flag or Flag-NS5A (genotype 2a) expression plasmid. At 20 h posttransfection, cells were treated with MG132 at 10 μM for 4 h to inhibit proteasome activities. Protein levels were detected by Western blotting for NS5A with an anti-Flag antibody; GAPDH was used as an internal control, and YB-1 was included for confirming knockdown efficiency. (F) Huh-7.5.1 cells transfected with control or YB-1 siRNA were infected with J6/JFH (genotype 2a) at an MOI of 0.5 as described in the legend to Fig. 2. Four hours before harvest, infected cells were treated with DMSO or MG132 (10 μM). Intracellular protein levels were detected by Western blotting for core, NS3, NS5A, YB-1, and GAPDH, quantified by ImageJ, and normalized to GAPDH. Results are represented as percentages relative to that of control siRNA-transfected cells.
To clarify whether the decrease of Flag-NS5A protein level in YB-1 silencing cells is due to protein degradation, the half-life of Flag-NS5A was assessed with the translation inhibitor cycloheximide. In the presence of cycloheximide, knockdown of YB-1 accelerated the degradation of Flag-NS5A compared with that of control cells, as demonstrated by the reduction of NS5A half-life from 4.4 h to 2.4 h (Fig. 9C). The positive regulatory effect of YB-1 on NS5A protein stability was then examined in the context of HCV RNA replication. To this end, replicon cells (Ava5, genotype 1b) transfected with YB-1 or control siRNA were treated with cycloheximide; the NS5A half-life was found to be dramatically reduced, from 19.2 h to 0.6 h, by YB-1 silencing (Fig. 9D). The results suggest that YB-1 protects NS5A from protein degradation. Variations between the half-life of NS5A expressed ectopically and in replicon cells were similarly observed in previous studies of core and NS5B (64–67).
We next investigated whether the rapid degradation of NS5A evoked by YB-1 silencing is proteasome dependent by treating YB-1 knockdown or control cells expressing Flag-NS5A with a proteasome inhibitor, MG132. MG132 clearly abolished YB-1 silencing-mediated Flag-NS5A reduction (Fig. 9E). To confirm this effect in the context of HCV infection, control or YB-1 knockdown cells were infected with J6/JFH (MOI of 0.5) for 48 h and treated with MG132 for 4 h before analysis. MG132 partially restored the NS5A protein level, while recovery of NS3 and core protein levels was not observed (Fig. 9F). Taken together, our results indicate that knockdown of YB-1 leads to NS5A instability.
Phosphorylation of YB-1 at serine 102 (S102) modulates NS5A–YB-1 interaction and NS5A stability.
We were also interested to know whether NS5A–YB-1 interaction is crucial for maintaining NS5A stability and whether this interaction is elicited by HCV-induced signaling. Along this line, it is noted that both HCV infection and NS5A alone activate the PI3K/Akt pathway (23, 68), and phosphorylation at the YB-1 S102 residue by Akt is known to modulate multiple functions of YB-1 (69). To examine if the YB-1–NS5A interaction is dependent on the phosphorylation of YB-1 at the S102 residue, coimmunoprecipitation was performed with Huh-7.5.1 cell extracts coexpressing Flag-NS5A and either HA-YB-1, the nonphosphorylatable mutant HA-YB-1(S102A), or the phospho-mimic mutant HA-YB-1(S102D). Interactions between the YB-1(S102A) mutant and NS5A were disturbed, resulting in a 62% reduction compared to that with wild-type YB-1, while binding between the YB-1(S102D) mutant and NS5A was comparable to wild-type YB-1–NS5A interaction (Fig. 10A).
FIG 10.
YB-1 stabilizes NS5A through its serine 102 phosphorylation-mediated interaction with NS5A. (A) Coimmunoprecipitation was performed with Huh-7.5.1 cells cotransfected with a Flag vector (−) or expression plasmids for Flag-NS5A (+) (genotype 2a) and either an HA vector, HA-YB-1, HA-YB-1(S102A), or HA-YB-1(S102D) in the presence of RNase A with anti-Flag beads. For detection of HA-YB-1, 20-μg cell lysates (1.67% of total input protein) and four-ninths of the precipitates (IP) were subjected to Western blotting with an anti-HA antibody. To detect Flag-NS5A, 20-μg cell lysates (1.67% of total input protein) and two-ninths of the precipitates were subjected to Western blotting with an anti-Flag antibody. GAPDH was included as an internal control. Levels of immunoprecipitated HA-YB-1 variants were quantified by ImageJ and are shown as percentages of wild-type HA-YB-1. (B) YB-1 siRNA-transfected Huh-7.5.1 cells were cotransfected with plasmids encoding Flag-NS5A (genotype 2a) and either siRNA-resistant GFP-YB-1 (GFP-YB-1R), GFP-YB-1(S102A) [GFP-YB-1(S102A)R], or GFP-YB-1(S102D) [GFP-YB-1(S102D)R]. Twenty-four hours posttransfection, levels of Flag-NS5A and mutants for GFP-YB-1 or GFP-YB-1 were analyzed by Western blotting with anti-Flag and anti-GFP antibodies, respectively. GAPDH was used as an internal control. Flag-NS5A levels were quantified by ImageJ and normalized to GAPDH levels and are shown as percentages of that of YB-1 knockdown cells cotransfected with Flag-NS5A and wild-type GFP-YB-1. (C) (Top) YB-1 siRNA-transfected Huh-7.5.1 cells were cotransfected with plasmids encoding Flag-NS5A (genotype 2a) and either siRNA-resistant GFP-YB-1 (GFP-YB-1R), GFP-YB-1(S102A) [GFP-YB-1(S102A)R], or GFP-YB-1(S102D) [GFP-YB-1(S102D)R]. Forty-eight hours later, cells were treated with cycloheximide at 100 μg/ml. Cell lysates were prepared and analyzed for Flag-NS5A levels by immunoblotting with an anti-Flag antibody at the indicated time points after cycloheximide addition. GAPDH was used as an internal control. (Bottom) Flag-NS5A levels were quantified by ImageJ and normalized to GAPDH levels, and the half-life of Flag-NS5A was determined by regression analysis. Protein levels of Flag-NS5A without cycloheximide treatment (0 h) are set as 1. (D) Huh-7.5.1 cells transfected with control siRNA (siCtrl) or YB-1 siRNA (siYB-1) for 24 h were transfected with the HA vector or plasmid encoding either siRNA-resistant HA-YB-1 (HA-YB-1R), HA-YB-1(S102A) [HA-YB-1(S102A)R], or HA-YB-1(S102D) [HA-YB-1(S102D)R] and infected with J6/JFH (genotype 2a) at an MOI of 0.5 24 h later. Cells lysates prepared at 48 h postinfection were subjected to Western blotting for NS5A, endogenous YB-1, and HA-tagged YB-1 variants, and GAPDH was used as an internal control. NS5A levels were quantified by ImageJ and normalized to GAPDH levels, and are shown as percentages of that of cells transfected with control siRNA and HA vector.
Next, we investigated the importance of the phosphorylation-mediated NS5A–YB-1 interaction for YB-1 to stabilize NS5A. NS5A protein levels were examined in YB-1 knockdown Huh-7.5.1 cells transfected with Flag-NS5A and either siRNA-resistant GFP-YB-1, the GFP-YB-1(S102A) mutant, or the GFP-YB-1(S102D) mutant. Compared with wild-type YB-1, the YB-1(S102A) mutant, which only partially interacted with NS5A, failed to maintain the NS5A level in showing a 67% reduction compared to that with the wild type, while the phospho-mimic YB-1(S102D) mutant not only interacted with NS5A but also maintained the NS5A level in YB-1 knockdown cells (Fig. 10B). The diverse NS5A levels in YB-1 knockdown cells expressing different YB-1 variants were then verified to reflect the altered NS5A stability. A decreased NS5A half-life from 2.2 h to 0.8 h was observed in YB-1 knockdown cells expressing the siRNA-resistant GFP-YB-1(S102A) mutant compared to that in cells transfected with wild-type GFP-YB-1, while a comparable half-life of NS5A was shown with expression of the GFP-YB-1(S102D) mutant (1.7 h) and wild-type GFP-YB-1 (Fig. 10C). Thus, NS5A–YB-1 interaction is mediated by phosphorylation of YB-1 at the S102 residue, which is important for NS5A stabilization.
To validate the role of YB-1 S102 phosphorylation in HCV infection, Huh-7.5.1 cells coexpressing YB-1 siRNA and either siRNA-resistant wild-type HA-YB-1, HA-YB-1(S102A), or HA-YB-1(S102D) were infected with J6/JFH virus. Consistent with their NS5A-interacting activities and their abilities to stabilize ectopically expressed NS5A, both wild-type YB-1 and the YB-1(S102D) mutant rescued the NS5A levels from 5% to 64% and 52%, respectively, compared to control cells, in HCV infection (Fig. 10D). However, the phosphorylation-null HA-YB-1(S102A) mutant led to 24% and, therefore, a much lower restoration of NS5A expression (Fig. 10D). Taken together, our findings indicate that YB-1 stabilizes the pivotal viral protein NS5A via phosphorylation-dependent interaction with NS5A through which YB-1 plays a role in HCV infection and HCV RNA replication.
DISCUSSION
NS5A is a pivotal viral protein involved in multiple steps in the HCV life cycle, including HCV RNA replication (14, 63), infectious virus production (12, 13, 15), and the recently suggested early stage of HCV RNA replication (19–22). In this study, we revealed that YB-1 is a novel NS5A-interacting cellular factor (Fig. 1) and that such an interaction stabilizes NS5A (Fig. 10). The colocalization of YB-1 and NS5A in the HCV replication complexes and also in the cytoplasm in HCV-infected cells (Fig. 5) implies that YB-1 may modulate various steps of the HCV life cycle via maintaining the cellular NS5A level. Supporting this notion, knockdown of YB-1 before HCV infection and reporter genome transfection reduced HCV replication complex formation and RNA replication (Fig. 4 and 6), while YB-1 silencing in HCV-infected and replicon cells did not affect intracellular HCV RNA levels (Fig. 7D, left graph, and 8A), indicating YB-1 involvement in the establishment of HCV replication complex formation. Moreover, as the intracellular HCV RNA level was not altered by YB-1 knockdown in HCV-infected cells (Fig. 7D), reduced infectivity in the culture medium (Fig. 7E) conceivably suggests that YB-1 is important for the production of infectious HCV particles. Interestingly, by using a plasmid-derived JFH-1-expressing system, Chatel-Chaix et al. showed that knockdown of YB-1 promoted infectious virus production (43, 44). Notably, the kinetics of the plasmid-derived JFH-1 expression is much slower than that of cell culture-derived HCV (HCVcc) infection adopted in our study (22, 43). While Chatel-Chaix et al. assessed the role of YB-1 silencing in virus production at a defined early stage of the plasmid-derived infection system (43), we detected the inhibitory effect of YB-1 knockdown on the production of HCV infectious virus at the late stage of the HCV life cycle (Fig. 2C, 2E, and 7E and reference 22). Results from the use of different infection systems and at different time windows have hence revealed distinctive roles for YB-1 in virus production at different stages of the HCV life cycle.
HCV co-opts ribonucleoprotein complexes via virus-host protein-protein and RNA-protein interactions to facilitate its propagation (70). Several studies, including our own, have shown that host factors YB-1 and DDX3 not only participate in the HCV life cycle (Fig. 2 and 7 and references 34, 35, and 43) but also associate with HCV RNA (42, 43) and interact with HCV core, NS3, and NS5A (Fig. 1 and references 31 to 33, 43, and 44). Although sharing the same viral interacting partners, YB-1 and DDX3 seem to play several different roles in the HCV life cycle. While DDX3 is required for HCV IRES-mediated translation (29, 71), YB-1 plays either a moderate or a minor role that suppresses or promotes HCV translation under normal or stress conditions, respectively (Fig. 3C). The regulatory effects of DDX3 on HCV IRES are independent of the HCV 3′ NTR (71). However, the effects of YB-1 on HCV IRES-mediated translation are dependent on the presence of the HCV 3′ NTR (Fig. 3). While YB-1 is a positive regulator required in HCV RNA replication but dispensable in maintaining the steady-state HCV RNA replication (Fig. 4, 7D, left side, and 8A), DDX3 participates in steady-state HCV RNA replication (Fig. 7D, right graph, and reference 34). Moreover, unlike YB-1, DDX3 is not involved the equilibrium between HCV RNA replication and infectious virus production under defined conditions (44). The comparable reductions of HCV RNA levels and infectivity after DDX3 knockdown in HCV-infected cells (Fig. 7D, right graph, and 7E, right graph) indicate that such infectivity decreases by DDX3 silencing is a consequence of impaired HCV RNA replication, which might be different from the case with YB-1.
Besides participating in HCV RNA translation, RNA replication, and virus production of the HCV life cycle, NS5A has further been proposed to regulate switches between these life cycle stages (13, 72). The HCV genome RNA is used as the template for both IRES-mediated translation and negative-strand RNA synthesis, two essential but opposing processes (3). To avoid functional conflicts, a mechanism which coordinates these two processes for efficient HCV replication is necessary and is believed to be accomplished by cooperation between viral and host proteins (70, 73, 74). NS5A interacts with both the HCV 5′ and 3′ NTRs and hence has been proposed to regulate the switch from HCV translation to RNA replication (72). Given that YB-1 associates with NS5A to sustain the NS5A level (Fig. 10), and that YB-1 has been identified as the HCV 5′ and 3′NTR-interacing protein (42, 75, 76), it is likely that YB-1 regulates the translation-replication switch in the HCV life cycle through the NS5A–YB-1 complex. Interestingly, in the reporter genome assay, the observation that YB-1 moderately inhibits primary translation (Fig. 3C, normal condition [i.e., without sodium arsenite], and Fig. 4) while playing a critical role in HCV RNA replication may further support this notion. Recently, NS3, another YB-1-interacting viral protein, has been reported to regulate the switch from translation to replication of HCV RNA via replacing the human La protein, an HCV IRES trans-acting factor, from the HCV IRES (73). It is thus possible that YB-1 may interexchange with viral proteins, such as NS3 and NS5A, to participate in the translation-replication switch in the HCV life cycle.
Phosphorylation of NS5A has been suggested to modulate HCV RNA replication. A study based on HCV genotype 1b has revealed that hyperphosphorylation of NS5A represses HCV RNA replication (77). Nevertheless, hyperphosphorylation of genotype 2a NS5A either enhances or inhibits HCV RNA replication, depending on the serine residues examined (13, 78, 79). Since NS5A phosphorylation also regulates virion assembly (13), phosphorylation of NS5A has been suggested to control the switch from HCV RNA replication to virus production (13), and the ratio of the hyperphosphorylated (p58) to hypophosphorylated (p56) forms of NS5A may be important for both events (63). In this study, our observation revealed that knockdown of YB-1 in HCV-infected cells not only reduced the total NS5A level but also preferentially downregulated p58 rather than p56 (Fig. 7G, left graph). However, DDX3 knockdown-mediated NS5A reduction had no preference for either forms of NS5A (Fig. 7G, right graph). Interestingly, YB-1 has been shown to play a role in the equilibrium between HCV RNA replication and virus release in an early stage of a plasmid-driven JFH1 infection system (43, 44). Therefore, in view of the fact that p58 is more labile than p56 (66), it will be interesting to investigate whether YB-1 regulates the switch from HCV RNA replication to virion production via differential modulation of the stability of NS5A phosphorylation forms.
In this study, we also found that YB-1 interacts with NS5A and protects it from degradation, which is dependent on the phosphorylation of YB-1 at the S102 residue (Fig. 10). Interestingly, among the known signaling pathways inducing phosphorylation of YB-1 at S102 (41), PI3K/Akt signaling was previously demonstrated to be activated by NS5A (23). Hence, through activating the PI3K/Akt pathway, NS5A may induce phosphorylation of YB-1 at S102, which, in turn, promotes NS5A–YB-1 interaction and NS5A stabilization, thereby resulting in NS5A maintaining its level during HCV infection. Positive-stranded RNA viruses adopt cellular degradation pathways to maintain appropriate viral protein levels that avoid inhibitory effects of high-level RNA-dependent RNA polymerases (RdRps) on virus assembly, or premature apoptosis induced by excess viral proteases, thus supporting productive viral propagation (80). Accordingly, the protein levels of HCV core, NS2, NS5B, and NS5A are all tightly regulated by cellular protein degradation pathways (24, 80). Moreover, studies of positive-stranded RNA viruses, including encephalomyocarditis virus (EMCV) and hepatitis A virus (HAV), have implied that the degradation-mediated regulation is fine-tuned dynamically at different viral life cycle stages (80). Our finding of the regulation of NS5A stability by cellular signaling-induced YB-1 phosphorylation suggests that HCV can temporally regulate NS5A stability and likely the p58/p56 ratio of NS5A, facilitating efficient viral propagation.
Besides stabilizing NS5A, it is very likely that the NS5A–YB-1 interaction may convert NS5A conformation and the NS5A interactome, which hence fine-tunes the roles of NS5A during HCV infection. Both positive and negative roles for NS5A in HCV IRES-mediated translation have been reported with different assay systems and cell lines (17, 18, 81), implying that specific NS5A interactions with other proteins may determine the effects of NS5A on HCV translation. Given the diverse implication of YB-1, DDX3, NS3, and core in HCV translation (Fig. 3C and references 29, 71, 73, and 82), interconnection among these host and viral interacting partners may synergistically or counteractively promote or inhibit HCV translation to temporally fulfill the needs of the virus. Moreover, the different roles of YB-1 and DDX3 in steady-state HCV RNA replication (Fig. 7) suggest that YB-1 and DDX3 may not function as a complex but associate with NS5A sequentially in HCV RNA replication. It is possible that NS5A associates with YB-1 and other cellular cofactors early after infection to facilitate the establishment of HCV RNA replication environment, while part of NS5A subsequently dissociates from YB-1 to cooperate with DDX3-containing ribonucleoprotein complex as part of the replication complex. The markedly reduced colocalization of dsRNA with YB-1, but not with NS5A (Fig. 7B and C), by knockdown of YB-1 in HCV-infected cells further substantiates that NS5A and YB-1 may reside in different microenvironments of the HCV replication complex.
The NS3/YB-1 interaction (43, 44) and the proposed transient role of YB-1 in inhibiting NS3-dependent HCV release under defined conditions (44) reveal another role of YB-1 in HCV production. Moreover, YB-1 translocates to lipid droplets in a core-dependent but NS5A-independent way in HCV-producing cells (43). Given that YB-1 interacts with both core and NS5A (Fig. 1), YB-1 may promote the association of NS5A with lipid droplets, as that of Rab18 (83), via the core–YB-1 interaction, to enhance HCV infectious virus production. It is noteworthy that the HCV replication complex, NS5A and core, is transported to lipid droplets via microtubules (84). As YB-1 is known to promote microtubule assembly in vitro (85), it is likely that YB-1 may also directly regulate HCV production through modulating the microtubule cytoskeleton. It is also possible that YB-1 may participate in other steps in HCV life cycle, including the entry step. Further studies will be needed to investigate how HCV regulates the interactome of NS5A spatiotemporally by interchange YB-1 with other NS5A interacting partners and how YB-1 switches its viral partners during the infection process to achieve efficient HCV propagation.
In this study, we found that YB-1 stabilizes and interacts with NS5A of both genotype 1b and 2a (Fig. 1 and 9), suggesting a genotype independence of this regulatory mechanism. Moreover, our observations that both NS5A stabilization and NS5A–YB-1 interaction are dependent on the phosphorylation of YB-1 serine 102 (Fig. 10) suggest that phosphorylation of the YB-1 S102 residue may be used as a novel target for anti-HCV therapy. Accordingly, the preclinical PI3K inhibitor LY294002 moderately inhibits HCV infection at a dose specifically inhibiting PI3K (86, 87). Inhibitors targeting the PI3K/Akt pathway, one of the signaling pathways activated by HCV infection and NS5A (23, 68) and inducing YB-1 phosphorylation at serine 102 (41), are now in clinical development for anticancer therapy (88). Moreover, a cell-permeative peptide has been designed and proved to antagonize YB-1 phosphorylation at serine 102 in cultured cancer cells (89). It would be interesting to elucidate the anti-HCV effects of these developing anticancer drugs. These potential host-targeting antiviral drugs may theoretically have a high genetic barrier to drug resistance. Taken together, our results demonstrate how NS5A, and possibly its p58-to-p56 ratio, are maintained in the HCV life cycle to support efficient HCV infection and further provide a potential aspect for the development of novel strategies for anti-HCV therapy.
ACKNOWLEDGMENTS
This study was supported by grants from the National Health Research Institute (NHRI-EX99-9501BI, NHRI-EX100-10014BI, NHRI-EX101-10014BI, and NHRI-EX102-10014BI), the Ministry of Science and Technology (NSC97-2320-B-010-014-MY3, NSC98-2320-B-009-003-MY3, NSC101-2320-B-009-001-MY3, and MOST103-2320-B-009-006), the UST-UCSD International Center of Excellence in Advanced Bio-engineering sponsored by the Taiwan Ministry of Science and Technology I-RiCE Program (MOST102-2911-I-009-101 and MOST103-2911-I-009-101), and the Aiming for the Top University Program of the National Chiao-Tung University and the Ministry of Education (103W963 and 104W963), Taiwan, Republic of China, to Yan-Hwa Wu Lee.
We thank Tsung-Sheng Su, Tzu-Hao Cheng, Li-Ru You, and Chun-Ming Chen for helpful discussions and comments. We also thank Kong-Bung Choo for critical readings of the manuscript and constructive suggestions. We are grateful to Charles M. Rice (Rockefeller University) for kindly providing Ava5 replicon cells as well as the pFL-J6/JFH, J6/JFH(p7-Rluc2A), J6/JFH(p7-Rluc2A)GNN, and J6/JFH(p7-Rluc2A)K33A/R35A plasmids. We are also grateful to Francis V. Chisari (Scripps Research Institute) for kindly offering Huh-7.5.1 cells and to Lih-Hwa Hwang (National Yang-Ming University) for kindly providing the pFlag-NS5A, pFlag-NS3/4A, and pFlag-core plasmids. We also thank the Imaging Core Facility of Nanotechnology of the University System of Taiwan-National Yang-Ming University (UST-YMU) for confocal microscopy technical services.
REFERENCES
- 1.Lavanchy D. 2009. The global burden of hepatitis C. Liver Int 29(Suppl 1):S74–S81. [DOI] [PubMed] [Google Scholar]
- 2.Bartenschlager R, Lohmann V, Penin F. 2013. The molecular and structural basis of advanced antiviral therapy for hepatitis C virus infection. Nat Rev Microbiol 11:482–496. doi: 10.1038/nrmicro3046. [DOI] [PubMed] [Google Scholar]
- 3.Moradpour D, Penin F, Rice CM. 2007. Replication of hepatitis C virus. Nat Rev Microbiol 5:453–463. doi: 10.1038/nrmicro1645. [DOI] [PubMed] [Google Scholar]
- 4.Bartenschlager R, Cosset FL, Lohmann V. 2010. Hepatitis C virus replication cycle. J Hepatol 53:583–585. doi: 10.1016/j.jhep.2010.04.015. [DOI] [PubMed] [Google Scholar]
- 5.Scheel TK, Prentoe J, Carlsen TH, Mikkelsen LS, Gottwein JM, Bukh J. 2012. Analysis of functional differences between hepatitis C virus NS5A of genotypes 1–7 in infectious cell culture systems. PLoS Pathog 8:e1002696. doi: 10.1371/journal.ppat.1002696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tellinghuisen TL, Marcotrigiano J, Gorbalenya AE, Rice CM. 2004. The NS5A protein of hepatitis C virus is a zinc metalloprotein. J Biol Chem 279:48576–48587. doi: 10.1074/jbc.M407787200. [DOI] [PubMed] [Google Scholar]
- 7.Gosert R, Egger D, Lohmann V, Bartenschlager R, Blum HE, Bienz K, Moradpour D. 2003. Identification of the hepatitis C virus RNA replication complex in Huh-7 cells harboring subgenomic replicons. J Virol 77:5487–5492. doi: 10.1128/JVI.77.9.5487-5492.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miyanari Y, Hijikata M, Yamaji M, Hosaka M, Takahashi H, Shimotohno K. 2003. Hepatitis C virus non-structural proteins in the probable membranous compartment function in viral genome replication. J Biol Chem 278:50301–50308. doi: 10.1074/jbc.M305684200. [DOI] [PubMed] [Google Scholar]
- 9.Egger D, Wölk B, Gosert R, Bianchi L, Blum HE, Moradpour D, Bienz K. 2002. Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. J Virol 76:5974–5984. doi: 10.1128/JVI.76.12.5974-5984.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shi ST, Lee KJ, Aizaki H, Hwang SB, Lai MM. 2003. Hepatitis C virus RNA replication occurs on a detergent-resistant membrane that cofractionates with caveolin-2. J Virol 77:4160–4168. doi: 10.1128/JVI.77.7.4160-4168.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miyanari Y, Atsuzawa K, Usuda N, Watashi K, Hishiki T, Zayas M, Bartenschlager R, Wakita T, Hijikata M, Shimotohno K. 2007. The lipid droplet is an important organelle for hepatitis C virus production. Nat Cell Biol 9:1089–1097. doi: 10.1038/ncb1631. [DOI] [PubMed] [Google Scholar]
- 12.Appel N, Zayas M, Miller S, Krijnse-Locker J, Schaller T, Friebe P, Kallis S, Engel U, Bartenschlager R. 2008. Essential role of domain III of nonstructural protein 5A for hepatitis C virus infectious particle assembly. PLoS Pathog 4:e1000035. doi: 10.1371/journal.ppat.1000035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tellinghuisen TL, Foss KL, Treadaway J. 2008. Regulation of hepatitis C virion production via phosphorylation of the NS5A protein. PLoS Pathog 4:e1000032. doi: 10.1371/journal.ppat.1000032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tellinghuisen TL, Marcotrigiano J, Rice CM. 2005. Structure of the zinc-binding domain of an essential component of the hepatitis C virus replicase. Nature 435:374–379. doi: 10.1038/nature03580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Masaki T, Suzuki R, Murakami K, Aizaki H, Ishii K, Murayama A, Date T, Matsuura Y, Miyamura T, Wakita T, Suzuki T. 2008. Interaction of hepatitis C virus nonstructural protein 5A with core protein is critical for the production of infectious virus particles. J Virol 82:7964–7976. doi: 10.1128/JVI.00826-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He Y, Yan W, Coito C, Li Y, Gale M Jr, Katze MG. 2003. The regulation of hepatitis C virus (HCV) internal ribosome-entry site-mediated translation by HCV replicons and nonstructural proteins. J Gen Virol 84:535–543. doi: 10.1099/vir.0.18658-0. [DOI] [PubMed] [Google Scholar]
- 17.Kalliampakou KI, Kalamvoki M, Mavromara P. 2005. Hepatitis C virus (HCV) NS5A protein downregulates HCV IRES-dependent translation. J Gen Virol 86:1015–1025. doi: 10.1099/vir.0.80728-0. [DOI] [PubMed] [Google Scholar]
- 18.Khachatoorian R, Arumugaswami V, Ruchala P, Raychaudhuri S, Maloney EM, Miao E, Dasgupta A, French SW. 2012. A cell-permeable hairpin peptide inhibits hepatitis C viral nonstructural protein 5A-mediated translation and virus production. Hepatology 55:1662–1672. doi: 10.1002/hep.25533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reiss S, Rebhan I, Backes P, Romero-Brey I, Erfle H, Matula P, Kaderali L, Poenisch M, Blankenburg H, Hiet MS, Longerich T, Diehl S, Ramirez F, Balla T, Rohr K, Kaul A, Bühler S, Pepperkok R, Lengauer T, Albrecht M, Eils R, Schirmacher P, Lohmann V, Bartenschlager R. 2011. Recruitment and activation of a lipid kinase by hepatitis C virus NS5A is essential for integrity of the membranous replication compartment. Cell Host Microbe 9:32–45. doi: 10.1016/j.chom.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berger KL, Kelly SM, Jordan TX, Tartell MA, Randall G. 2011. Hepatitis C virus stimulates the phosphatidylinositol 4-kinase III alpha-dependent phosphatidylinositol 4-phosphate production that is essential for its replication. J Virol 85:8870–8883. doi: 10.1128/JVI.00059-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gastaminza P, Pitram SM, Dreux M, Krasnova LB, Whitten-Bauer C, Dong J, Chung J, Fokin VV, Sharpless KB, Chisari FV. 2011. Antiviral stilbene 1,2-diamines prevent initiation of hepatitis C virus RNA replication at the outset of infection. J Virol 85:5513–5523. doi: 10.1128/JVI.02116-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shulla A, Randall G. 2015. Spatiotemporal analysis of hepatitis C virus infection. PLoS Pathog 11:e1004758. doi: 10.1371/journal.ppat.1004758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Street A, Macdonald A, Crowder K, Harris M. 2004. The hepatitis C virus NS5A protein activates a phosphoinositide 3-kinase-dependent survival signaling cascade. J Biol Chem 279:12232–12241. doi: 10.1074/jbc.M312245200. [DOI] [PubMed] [Google Scholar]
- 24.Hou W, Tian Q, Zheng J, Bonkovsky HL. 2010. Zinc mesoporphyrin induces rapid proteasomal degradation of hepatitis C nonstructural 5A protein in human hepatoma cells. Gastroenterology 138:1909–1919. doi: 10.1053/j.gastro.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schröder M. 2010. Human DEAD-box protein 3 has multiple functions in gene regulation and cell cycle control and is a prime target for viral manipulation. Biochem Pharmacol 79:297–306. doi: 10.1016/j.bcp.2009.08.032. [DOI] [PubMed] [Google Scholar]
- 26.Chao CH, Chen CM, Cheng PL, Shih JW, Tsou AP, Wu Lee YH. 2006. DDX3, a DEAD box RNA helicase with tumor growth-suppressive property and transcriptional regulation activity of the p21waf1/cip1 promoter, is a candidate tumor suppressor. Cancer Res 66:6579–6588. doi: 10.1158/0008-5472.CAN-05-2415. [DOI] [PubMed] [Google Scholar]
- 27.Chang PC, Chi CW, Chau GY, Li FY, Tsai YH, Wu JC, Wu Lee YH. 2006. DDX3, a DEAD box RNA helicase, is deregulated in hepatitis virus-associated hepatocellular carcinoma and is involved in cell growth control. Oncogene 25:1991–2003. doi: 10.1038/sj.onc.1209239. [DOI] [PubMed] [Google Scholar]
- 28.Botlagunta M, Vesuna F, Mironchik Y, Raman A, Lisok A, Winnard P Jr, Mukadam S, Van Diest P, Chen JH, Farabaugh P, Patel AH, Raman V. 2008. Oncogenic role of DDX3 in breast cancer biogenesis. Oncogene 27:3912–3922. doi: 10.1038/onc.2008.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shih JW, Tsai TY, Chao CH, Wu Lee YH. 2008. Candidate tumor suppressor DDX3 RNA helicase specifically represses cap-dependent translation by acting as an eIF4E inhibitory protein. Oncogene 27:700–714. doi: 10.1038/sj.onc.1210687. [DOI] [PubMed] [Google Scholar]
- 30.Shih JW, Wang WT, Tsai TY, Kuo CY, Li HK, Wu Lee YH. 2012. Critical roles of RNA helicase DDX3 and its interactions with eIF4E/PABP1 in stress granule assembly and stress response. Biochem J 441:119–129. doi: 10.1042/BJ20110739. [DOI] [PubMed] [Google Scholar]
- 31.Mamiya N, Worman HJ. 1999. Hepatitis C virus core protein binds to a DEAD box RNA helicase. J Biol Chem 274:15751–15756. doi: 10.1074/jbc.274.22.15751. [DOI] [PubMed] [Google Scholar]
- 32.Owsianka AM, Patel AH. 1999. Hepatitis C virus core protein interacts with a human DEAD box protein DDX3. Virology 257:330–340. doi: 10.1006/viro.1999.9659. [DOI] [PubMed] [Google Scholar]
- 33.You LR, Chen CM, Yeh TS, Tsai TY, Mai RT, Lin CH, Wu Lee YH. 1999. Hepatitis C virus core protein interacts with cellular putative RNA helicase. J Virol 73:2841–2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ariumi Y, Kuroki M, Abe K, Dansako H, Ikeda M, Wakita T, Kato N. 2007. DDX3 DEAD-box RNA helicase is required for hepatitis C virus RNA replication. J Virol 81:13922–13926. doi: 10.1128/JVI.01517-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Randall G, Panis M, Cooper JD, Tellinghuisen TL, Sukhodolets KE, Pfeffer S, Landthaler M, Landgraf P, Kan S, Lindenbach BD, Chien M, Weir DB, Russo JJ, Ju J, Brownstein MJ, Sheridan R, Sander C, Zavolan M, Tuschl T, Rice CM. 2007. Cellular cofactors affecting hepatitis C virus infection and replication. Proc Natl Acad Sci U S A 104:12884–12889. doi: 10.1073/pnas.0704894104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shih JW, Wu Lee YH. 2014. Human DExD/H RNA helicases: emerging roles in stress survival regulation. Clin Chim Acta 436:45–58. doi: 10.1016/j.cca.2014.05.003. [DOI] [PubMed] [Google Scholar]
- 37.Eliseeva IA, Kim ER, Guryanov SG, Ovchinnikov LP, Lyabin DN. 2011. Y-box-binding protein 1 (YB-1) and its functions. Biochemistry (Mosc) 76:1402–1433. doi: 10.1134/S0006297911130049. [DOI] [PubMed] [Google Scholar]
- 38.Chang YW, Mai RT, Fang WH, Lin CC, Chiu CC, Wu Lee YH. 2014. YB-1 disrupts mismatch repair complex formation, interferes with MutSalpha recruitment on mismatch and inhibits mismatch repair through interacting with PCNA. Oncogene 33:5065–5077. doi: 10.1038/onc.2013.450. [DOI] [PubMed] [Google Scholar]
- 39.Cobbold LC, Spriggs KA, Haines SJ, Dobbyn HC, Hayes C, de Moor CH, Lilley KS, Bushell M, Willis AE. 2008. Identification of internal ribosome entry segment (IRES)-trans-acting factors for the Myc family of IRESs. Mol Cell Biol 28:40–49. doi: 10.1128/MCB.01298-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cobbold LC, Wilson LA, Sawicka K, King HA, Kondrashov AV, Spriggs KA, Bushell M, Willis AE. 2010. Upregulated c-myc expression in multiple myeloma by internal ribosome entry results from increased interactions with and expression of PTB-1 and YB-1. Oncogene 29:2884–2891. doi: 10.1038/onc.2010.31. [DOI] [PubMed] [Google Scholar]
- 41.Stratford AL, Fry CJ, Desilets C, Davies AH, Cho YY, Li Y, Dong Z, Berquin IM, Roux PP, Dunn SE. 2008. Y-box binding protein-1 serine 102 is a downstream target of p90 ribosomal S6 kinase in basal-like breast cancer cells. Breast Cancer Res 10:R99. doi: 10.1186/bcr2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Harris D, Zhang Z, Chaubey B, Pandey VN. 2006. Identification of cellular factors associated with the 3′-nontranslated region of the hepatitis C virus genome. Mol Cell Proteomics 5:1006–1018. doi: 10.1074/mcp.M500429-MCP200. [DOI] [PubMed] [Google Scholar]
- 43.Chatel-Chaix L, Melancon P, Racine MÈ, Baril M, Lamarre D. 2011. Y-box-binding protein 1 interacts with hepatitis C virus NS3/4A and influences the equilibrium between viral RNA replication and infectious particle production. J Virol 85:11022–11037. doi: 10.1128/JVI.00719-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chatel-Chaix L, Germain MA, Motorina A, Bonneil É, Thibault P, Baril M, Lamarre D. 2013. A host YB-1 ribonucleoprotein complex is hijacked by hepatitis C virus for the control of NS3-dependent particle production. J Virol 87:11704–11720. doi: 10.1128/JVI.01474-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhong J, Gastaminza P, Cheng G, Kapadia S, Kato T, Burton DR, Wieland SF, Uprichard SL, Wakita T, Chisari FV. 2005. Robust hepatitis C virus infection in vitro. Proc Natl Acad Sci U S A 102:9294–9299. doi: 10.1073/pnas.0503596102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Blight KJ, Kolykhalov AA, Rice CM. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972–1974. doi: 10.1126/science.290.5498.1972. [DOI] [PubMed] [Google Scholar]
- 47.Lindenbach BD, Evans MJ, Syder AJ, Wölk B, Tellinghuisen TL, Liu CC, Maruyama T, Hynes RO, Burton DR, McKeating JA, Rice CM. 2005. Complete replication of hepatitis C virus in cell culture. Science 309:623–626. doi: 10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
- 48.Jones CT, Murray CL, Eastman DK, Tassello J, Rice CM. 2007. Hepatitis C virus p7 and NS2 proteins are essential for production of infectious virus. J Virol 81:8374–8383. doi: 10.1128/JVI.00690-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen YJ, Chen YH, Chow LP, Tsai YH, Chen PH, Huang CY, Chen WT, Hwang LH. 2010. Heat shock protein 72 is associated with the hepatitis C virus replicase complex and enhances viral RNA replication. J Biol Chem 285:28183–28190. doi: 10.1074/jbc.M110.118323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Carrière M, Pène V, Breiman A, Conti F, Chouzenoux S, Meurs E, Andrieu M, Jaffray P, Grira L, Soubrane O, Sogni P, Calmus Y, Chaussade S, Rosenberg AR, Podevin P. 2007. A novel, sensitive, and specific RT-PCR technique for quantitation of hepatitis C virus replication. J Med Virol 79:155–160. doi: 10.1002/jmv.20773. [DOI] [PubMed] [Google Scholar]
- 51.de Chassey B, Navratil V, Tafforeau L, Hiet MS, Aublin-Gex A, Agaugué S, Meiffren G, Pradezynski F, Faria BF, Chantier T, Le Breton M, Pellet J, Davoust N, Mangeot PE, Chaboud A, Penin F, Jacob Y, Vidalain PO, Vidal M, André P, Rabourdin-Combe C, Lotteau V. 2008. Hepatitis C virus infection protein network. Mol Syst Biol 4:230. doi: 10.1038/msb.2008.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Song Y, Friebe P, Tzima E, Jünemann C, Bartenschlager R, Niepmann M. 2006. The hepatitis C virus RNA 3′-untranslated region strongly enhances translation directed by the internal ribosome entry site. J Virol 80:11579–11588. doi: 10.1128/JVI.00675-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bradrick SS, Walters RW, Gromeier M. 2006. The hepatitis C virus 3′-untranslated region or a poly(A) tract promote efficient translation subsequent to the initiation phase. Nucleic Acids Res 34:1293–1303. doi: 10.1093/nar/gkl019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Arnaud N, Dabo S, Maillard P, Budkowska A, Kalliampakou KI, Mavromara P, Garcin D, Hugon J, Gatignol A, Akazawa D, Wakita T, Meurs EF. 2010. Hepatitis C virus controls interferon production through PKR activation. PLoS One 5:e10575. doi: 10.1371/journal.pone.0010575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim JH, Park SM, Park JH, Keum SJ, Jang SK. 2011. eIF2A mediates translation of hepatitis C viral mRNA under stress conditions. EMBO J 30:2454–2464. doi: 10.1038/emboj.2011.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hoffman B, Liu Q. 2011. Hepatitis C viral protein translation: mechanisms and implications in developing antivirals. Liver Int 31:1449–1467. doi: 10.1111/j.1478-3231.2011.02543.x. [DOI] [PubMed] [Google Scholar]
- 57.Targett-Adams P, Boulant S, McLauchlan J. 2008. Visualization of double-stranded RNA in cells supporting hepatitis C virus RNA replication. J Virol 82:2182–2195. doi: 10.1128/JVI.01565-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.El-Hage N, Luo G. 2003. Replication of hepatitis C virus RNA occurs in a membrane-bound replication complex containing nonstructural viral proteins and RNA. J Gen Virol 84:2761–2769. doi: 10.1099/vir.0.19305-0. [DOI] [PubMed] [Google Scholar]
- 59.Aizaki H, Lee KJ, Sung VM, Ishiko H, Lai MM. 2004. Characterization of the hepatitis C virus RNA replication complex associated with lipid rafts. Virology 324:450–461. doi: 10.1016/j.virol.2004.03.034. [DOI] [PubMed] [Google Scholar]
- 60.Wölk B, Büchele B, Moradpour D, Rice CM. 2008. A dynamic view of hepatitis C virus replication complexes. J Virol 82:10519–10531. doi: 10.1128/JVI.00640-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jones DM, Gretton SN, McLauchlan J, Targett-Adams P. 2007. Mobility analysis of an NS5A-GFP fusion protein in cells actively replicating hepatitis C virus subgenomic RNA. J Gen Virol 88:470–475. doi: 10.1099/vir.0.82363-0. [DOI] [PubMed] [Google Scholar]
- 62.Tanji Y, Kaneko T, Satoh S, Shimotohno K. 1995. Phosphorylation of hepatitis C virus-encoded nonstructural protein NS5A. J Virol 69:3980–3986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huang Y, Staschke K, De Francesco R, Tan SL. 2007. Phosphorylation of hepatitis C virus NS5A nonstructural protein: a new paradigm for phosphorylation-dependent viral RNA replication? Virology 364:1–9. doi: 10.1016/j.virol.2007.01.042. [DOI] [PubMed] [Google Scholar]
- 64.Shirakura M, Murakami K, Ichimura T, Suzuki R, Shimoji T, Fukuda K, Abe K, Sato S, Fukasawa M, Yamakawa Y, Nishijima M, Moriishi K, Matsuura Y, Wakita T, Suzuki T, Howley PM, Miyamura T, Shoji I. 2007. E6AP ubiquitin ligase mediates ubiquitylation and degradation of hepatitis C virus core protein. J Virol 81:1174–1185. doi: 10.1128/JVI.01684-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gao L, Tu H, Shi ST, Lee KJ, Asanaka M, Hwang SB, Lai MM. 2003. Interaction with a ubiquitin-like protein enhances the ubiquitination and degradation of hepatitis C virus RNA-dependent RNA polymerase. J Virol 77:4149–4159. doi: 10.1128/JVI.77.7.4149-4159.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pietschmann T, Lohmann V, Rutter G, Kurpanek K, Bartenschlager R. 2001. Characterization of cell lines carrying self-replicating hepatitis C virus RNAs. J Virol 75:1252–1264. doi: 10.1128/JVI.75.3.1252-1264.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Moradpour D, Kary P, Rice CM, Blum HE. 1998. Continuous human cell lines inducibly expressing hepatitis C virus structural and nonstructural proteins. Hepatology 28:192–201. [DOI] [PubMed] [Google Scholar]
- 68.Liu Z, Tian Y, Machida K, Lai MM, Luo G, Foung SK, Ou JH. 2012. Transient activation of the PI3K-AKT pathway by hepatitis C virus to enhance viral entry. J Biol Chem 287:41922–41930. doi: 10.1074/jbc.M112.414789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kosnopfel C, Sinnberg T, Schittek B. 2014. Y-box binding protein 1—a prognostic marker and target in tumour therapy. Eur J Cell Biol 93:61–70. doi: 10.1016/j.ejcb.2013.11.007. [DOI] [PubMed] [Google Scholar]
- 70.Nagy PD, Pogany J. 2012. The dependence of viral RNA replication on co-opted host factors. Nat Rev Microbiol 10:137–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Geissler R, Golbik RP, Behrens SE. 2012. The DEAD-box helicase DDX3 supports the assembly of functional 80S ribosomes. Nucleic Acids Res 40:4998–5011. doi: 10.1093/nar/gks070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Huang L, Hwang J, Sharma SD, Hargittai MR, Chen Y, Arnold JJ, Raney KD, Cameron CE. 2005. Hepatitis C virus nonstructural protein 5A (NS5A) is an RNA-binding protein. J Biol Chem 280:36417–36428. doi: 10.1074/jbc.M508175200. [DOI] [PubMed] [Google Scholar]
- 73.Ray U, Das S. 2011. Interplay between NS3 protease and human La protein regulates translation-replication switch of hepatitis C virus. Sci Rep 1:1. doi: 10.1038/srep00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gamarnik AV, Andino R. 1998. Switch from translation to RNA replication in a positive-stranded RNA virus. Genes Dev 12:2293–2304. doi: 10.1101/gad.12.15.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lu H, Li W, Noble WS, Payan D, Anderson DC. 2004. Riboproteomics of the hepatitis C virus internal ribosomal entry site. J Proteome Res 3:949–957. doi: 10.1021/pr0499592. [DOI] [PubMed] [Google Scholar]
- 76.Pacheco A, Reigadas S, Martínez-Salas E. 2008. Riboproteomic analysis of polypeptides interacting with the internal ribosome-entry site element of foot-and-mouth disease viral RNA. Proteomics 8:4782–4790. doi: 10.1002/pmic.200800338. [DOI] [PubMed] [Google Scholar]
- 77.Appel N, Pietschmann T, Bartenschlager R. 2005. Mutational analysis of hepatitis C virus nonstructural protein 5A: potential role of differential phosphorylation in RNA replication and identification of a genetically flexible domain. J Virol 79:3187–3194. doi: 10.1128/JVI.79.5.3187-3194.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fridell RA, Valera L, Qiu D, Kirk MJ, Wang C, Gao M. 2013. Intragenic complementation of hepatitis C virus NS5A RNA replication-defective alleles. J Virol 87:2320–2329. doi: 10.1128/JVI.02861-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.LeMay KL, Treadaway J, Angulo I, Tellinghuisen TL. 2013. A hepatitis C virus NS5A phosphorylation site that regulates RNA replication. J Virol 87:1255–1260. doi: 10.1128/JVI.02154-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Choi AG, Wong J, Marchant D, Luo H. 2013. The ubiquitin-proteasome system in positive-strand RNA virus infection. Rev Med Virol 23:85–96. doi: 10.1002/rmv.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hoffman B, Li Z, Liu Q. 10 April 2015. Down-regulation of viral RNA translation by hepatitis C virus nonstructural protein 5A requires the polyU/UC sequence in the 3′ untranslated region. J Gen Virol doi: 10.1099/vir.0.000141. [DOI] [PubMed] [Google Scholar]
- 82.Lourenço S, Costa F, Débarges B, Andrieu T, Cahour A. 2008. Hepatitis C virus internal ribosome entry site-mediated translation is stimulated by cis-acting RNA elements and trans-acting viral factors. FEBS J 275:4179–4197. doi: 10.1111/j.1742-4658.2008.06566.x. [DOI] [PubMed] [Google Scholar]
- 83.Salloum S, Wang H, Ferguson C, Parton RG, Tai AW. 2013. Rab18 binds to hepatitis C virus NS5A and promotes interaction between sites of viral replication and lipid droplets. PLoS Pathog 9:e1003513. doi: 10.1371/journal.ppat.1003513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lai CK, Saxena V, Tseng CH, Jeng KS, Kohara M, Lai MM. 2014. Nonstructural protein 5A is incorporated into hepatitis C virus low-density particle through interaction with core protein and microtubules during intracellular transport. PLoS One 9:e99022. doi: 10.1371/journal.pone.0099022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chernov KG, Mechulam A, Popova NV, Pastre D, Nadezhdina ES, Skabkina OV, Shanina NA, Vasiliev VD, Tarrade A, Melki J, Joshi V, Baconnais S, Toma F, Ovchinnikov LP, Curmi PA. 2008. YB-1 promotes microtubule assembly in vitro through interaction with tubulin and microtubules. BMC Biochem 9:23. doi: 10.1186/1471-2091-9-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Berger KL, Cooper JD, Heaton NS, Yoon R, Oakland TE, Jordan TX, Mateu G, Grakoui A, Randall G. 2009. Roles for endocytic trafficking and phosphatidylinositol 4-kinase III alpha in hepatitis C virus replication. Proc Natl Acad Sci U S A 106:7577–7582. doi: 10.1073/pnas.0902693106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pisonero-Vaquero S, García-Mediavilla MV, Jorquera F, Majano PL, Benet M, Jover R, González-Gallego J, Sánchez-Campos S. 2014. Modulation of PI3K-LXRalpha-dependent lipogenesis mediated by oxidative/nitrosative stress contributes to inhibition of HCV replication by quercetin. Lab Invest 94:262–274. doi: 10.1038/labinvest.2013.156. [DOI] [PubMed] [Google Scholar]
- 88.Bauer TM, Patel MR, Infante JR. 2015. Targeting PI3 kinase in cancer. Pharmacol Ther 146:53–60. doi: 10.1016/j.pharmthera.2014.09.006. [DOI] [PubMed] [Google Scholar]
- 89.Law JH, Li Y, To K, Wang M, Astanehe A, Lambie K, Dhillon J, Jones SJ, Gleave ME, Eaves CJ, Dunn SE. 2010. Molecular decoy to the Y-box binding protein-1 suppresses the growth of breast and prostate cancer cells whilst sparing normal cell viability. PLoS One 5:e12661. doi: 10.1371/journal.pone.0012661. [DOI] [PMC free article] [PubMed] [Google Scholar]