Abstract
Canine parvovirus (CPV) infection induces reorganization of nuclear structures. Our studies indicated that late-stage infection induces accumulation of nuclear pore complexes (NPCs) and lamin B1 concomitantly with a decrease of lamin A/C levels on the apical side of the nucleus. Newly formed CPV capsids are located in close proximity to NPCs on the apical side. These results suggest that parvoviruses cause apical enrichment of NPCs and reorganization of nuclear lamina, presumably to facilitate the late-stage infection.
TEXT
The nuclear lamina is a protein-rich structural scaffold, the main components of which are the dynamic type V intermediate filament proteins called lamins. Lamin proteins comprise two subtypes, type A (lamin A, A10, C, and C2) and type B (B1, B2, and B3). The former are alternative splice products of the LMNA gene, and the latter are encoded by LMNB1 (B1) and LMNB2 (B2 and germ line-specific B3) genes (1). In structure, the lamins of both subtypes contain a central α-helical rod with globular head (N) and tail (C) domains. In vitro, the lamins dimerize in a parallel fashion followed by filament assembly (2, 3). The lamina is connected to the cytoskeleton via the nuclear envelope (NE)-spanning linker of nucleoskeleton and cytoskeleton (LINC) complex (4). Lamina also interacts with nuclear pore complexes (NPCs) consisting of ∼30 nucleoporins (Nups) and regulating bidirectional transport of molecules over the NE (5). Together, the NPCs and lamina define a dynamic barrier that mediates signals in response to cellular stress conditions such as virus infections and limits release of viral progeny after nuclear assembly (6–9). To overcome this barrier, viruses are known to alter the architecture of all aforementioned subnuclear structures, which in turn contribute to the function of the nucleus (10). Nuclear egress of parvovirus (PV) capsids has been suggested to occur by active export via the NPC pathway prior to cell lysis (11–13). However, the influence of parvovirus egress on the organization of NPCs and nuclear lamina is unknown. Here, we examined the distribution and organization of NPCs as well as A and B type lamins in canine parvovirus (CPV)-infected cells in late infection in comparison with those in noninfected S- and G-phase (G1/G2) cells.
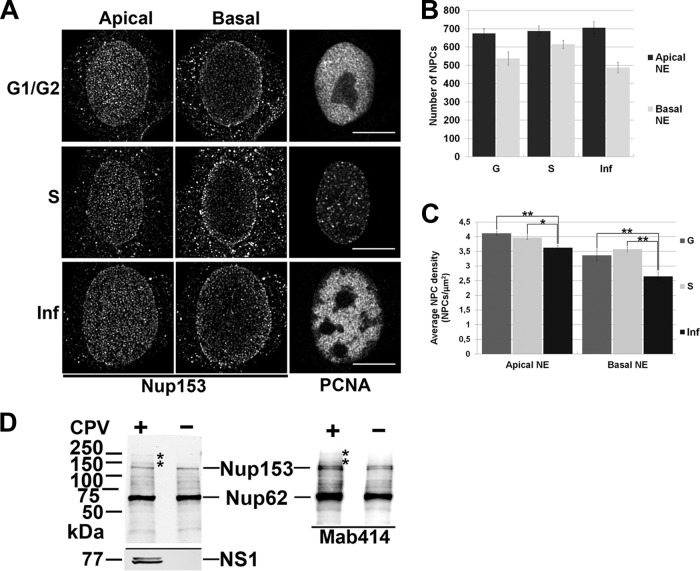
The NPCs are dynamic structures capable of assembly, disassembly, and redistribution during the cell cycle (14, 15). The nucleus has a spatially polarized architecture (16); however, the NPC distributions between the apical and basal sides of the nucleus have not been comparatively determined. Here, we examined the spatial distributions of NPCs at the apical and basal sides of nuclei in CPV-infected (multiplicity of infection [MOI] of 1 at 24 h postinfection [p.i.]) and mock-infected S- or G1/G2-phase Norden laboratory feline kidney (NLFK) cells. The NPCs were immunostained with a nucleoporin 153 (Nup153) antibody (Ab) (ab24700; Abcam, Cambridge, United Kingdom) also recognizing Nup62. A proliferating cell nuclear antigen (PCNA) Ab (ab18197; Abcam, Cambridge, United Kingdom) was used as a marker for cell cycle phases and the presence of parvoviral replication bodies (17, 18) (Fig. 1A). In confocal microscopy, z stacks consisting of an average of 30 z planes spaced by 0.15 μm were collected with the z axis corresponding to the apical-basal axis of the cell nucleus. Nuclei were scanned over a range of 4 to 6 μm. The middle z plane was applied to define the positions of the basal and apical surfaces. Confocal microscopy of infected cells showed unequal distributions of NPCs on the apical and the basal sides of NE. First, the number of NPCs at the apical side was ∼31% higher than that at the basal side (Fig. 1B). In G1/G2 cells, the distribution of NPCs was also asymmetric, with ∼20% more NPCs at the apical than the basal side. In the S phase, NPCs were more equally distributed, with only ∼10% more NPCs localized to the apical side. Second, the overall NPC densities on both the apical and basal sides were significantly decreased in infection (Fig. 1C). In the infected cells, the apical NPC density (number ± standard deviation [SD], 3.6 ± 0.51 NPC/μm2, n = 22) was lower than in the S-phase cells (4.0 ± 0.42 NPC/μm2, n = 21, Student′s t test P < 0.05) or the G-phase control cells (4.12 ± 0.48 NPC/μm2, n = 22, P < 0.01) (Fig. 1A and C). An even more prominent decrease was seen at the basal side of infected-cell nuclei, where the NPC density (2.51 ± 0.65 NPC/μm2, n = 22) was ∼25% lower than in the mock-infected G-phase (G1/G2) cells and ∼30% lower than in the S-phase cells (3.36 ± 0.88 NPC/μm2 [n = 21, P < 0.01] and 3.57 ± 0.31 NPC/μm2 [n = 22, P < 0.01], respectively) (Fig. 1A and C). Our results showed that infection was accompanied by a profound modification of the NPC network, including a significant reduction in the density of NPCs at the basal side, resembling the overall NPC distribution in G1/G2 cells. Earlier studies showed that cell cycle-dependent increases in the amount of NPCs and the nuclear volume occur simultaneously but do so with different regulation mechanisms (15, 19). The frequency of NPC biogenesis fluctuates during cell cycle progression, being highest in the S and G2 phases (19–21). CPV infection is accompanied by cell cycle arrest in the S phase (22–24). Notably, in contrast with the high density of NPCs seen in S-phase cells, we observed significantly decreased density in infected cells. To exclude the possibility that the decrease in NPC density was due to infection-induced degradation, the structural integrity of Nup153 in the infected cells at 24 h p.i. was analyzed by Western blotting (4.2 × 104 cells per well). The analysis of FG-repeated Nup153 and Nup62 (Nup153 Ab, monoclonal antibody 414 [Mab414], and ab24609; Abcam) in infected and mock-infected cells showed no major differences in abundance or integrity (Fig. 1D). For comparison, actinomycin D (Act D)-treated (0.5 to 1 μg/ml, 24 h) apoptotic cells showed cleavage of Nup153 (data not shown). However, in the infected cells, two additional bands with lower electrophoretic mobility were seen. The change may have reflected a posttranslational modification of Nup153, such as increased phosphorylation. With many viruses, Nup153 undergoes structural modification to support viral replication and spread. As an example, viruses use phosphorylation of Nups to alter the nucleocytoplasmic transport of the host (25). Furthermore, phosphorylation of Nups can occur in response to DNA damage, commonly detected in parvovirus infections (26, 27), and can indicate an infection-induced functional change of Nup153 (28, 29). Our analyses do not exclude the possibility of Nup153 becoming detached from the NPCs in infection. However, the amount of homogenously distributed Nup153 in the cytoplasm seemed to remain unaltered as judged by confocal microscopy (Fig. 1A). Taken together, these results demonstrated that CPV infection is accompanied by accumulation of NPCs at the apical side of the nucleus along with a decrease in their overall density concomitantly with structural modification of Nup153.
FIG 1.
Infection- and cell cycle-dependent distribution of nuclear pore complexes. (A) Confocal microscopy images of the infected cells (Inf) at 24 h p.i. and the mock-infected cells in S and G1/G2 phases. NPCs and PCNA were visualized with Nup153 (left and middle panels) and PCNA (right panel) antibodies. (B) The amount of NPCs calculated from the apical and basal sides of NE. (C) Average density of NPCs in apical and basal side of NE. The average values of the results of triplicate experiments ± SDs are shown. (D) Western blot analysis of Nup153 and Nup62 proteins harvested from the infected and mock-infected cells and their structural integrity. Nups were detected with Nup153-specific antibody and Mab414 antibody recognizing FG-repeated Nup62 and Nup153 in feline cells. Statistical significance in comparison to the mock-infected cells is shown (Student's t test P values: *, P < 0.05; **, P < 0.01). Bars, 10 μm. Error bars represent the 95% confidence interval.
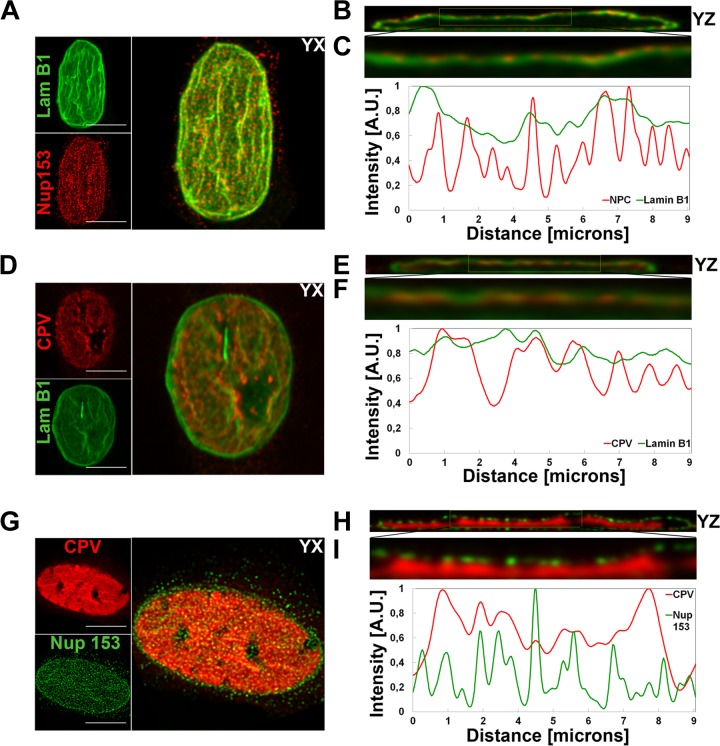
NPCs are anchored into the nuclear lamina (30, 31). B-type lamins concentrate in pore-rich regions, whereas A-type lamins are found in pore-free islands (32). Accordingly, changes in the distribution of NPCs correlate with nuclear lamina reorganization (19, 33). As CPV infection was accompanied by significant changes in the distribution of NPCs, we next analyzed the distributions of lamins at the apical and basal sides of the nucleus. Immunofluorescence analysis was carried out with antibodies recognizing lamins A/C and B1 (NCL-LAM-A/C [monoclonal; Leica Biosystems, Newcastle, United Kingdom] and ab16048 [polyclonal; Abcam, Cambridge, United Kingdom]). In virus-infected cells, similarly to NPCs, lamin B1 was enriched in clusters along the apical side of the NE (Fig. 2A), whereas in mock-infected cells, lamin B1 was distributed more equally between the two sides. These data suggested that infection affected the composition of the lamina or the lamin epitope. To study the distributions of lamins A/C and B1 in more detail, we first compared their intensities at the apical and basal sides (n = 24) (Fig. 2B) and then determined the surface intensity ratios (apical intensity divided by basal intensity) for both lamins (n = 24) (Fig. 2C). At the apical side of mock-infected cells, the intensity of lamin AC was ∼16% higher than that of lamin B1. In infection, the intensity of lamin A/C at the apical side was significantly (P < 0.05) decreased and was ∼6% lower than that of lamin B1 (Fig. 2B). At the basal side, the average intensities of both lamins were significantly (P < 0.05) decreased in infection in comparison with those in mock-infected cells. Determination of surface intensity ratios showed that, in general, lamin intensities were higher at the apical side than at the basal side (Fig. 2C) as reported earlier (16). In infection, the surface intensity ratio of lamin B1 was slightly increased whereas that of lamin A/C was similar to that in the mock-infected cells (Fig. 2C). These results showed that infection was accompanied by a decrease in the abundance of lamin A/C in the lamina concomitantly with enrichment of lamin B1 at the apical side. We then analyzed whether the infection-induced changes in distributions of lamins were due to their degradation. Western blot analysis (4.2 × 104 cells per well) indicated that lamins A/C and B1 (ab8984 and ab16048; Abcam, Cambridge, United Kingdom) remained intact in infection (Fig. 3A) but disintegrated in Act D-induced (1 and 5 μg/ml, 24 h) apoptotic cells (Fig. 3B). This agrees with earlier data showing that parvoviral nuclear egress does not induce degradation of lamins (34). Importantly, the expression levels of lamin A/C and B1 in the infected and mock-infected cells were comparable (Fig. 3A). In parallel, confocal imaging showed no marked discontinuity in lamin A/C or B1 staining in the infected cells; i.e., the lamin A/C and lamin B1 layers remained continuous even when viral capsids accumulated in the nuclear periphery at late-stage infection (Fig. 3C). Finally, the apical distributions of lamin B1, Nup153, and newly formed CPV capsids in infection were compared using deconvoluted confocal yz cross sections and analyzed with the program ImageJ. The apical distribution of lamin B1 was found to be similar to those of Nup153 (Fig. 4A, B, and C) and virus capsids (Fig. 4D, E, and F). Importantly, virus capsids were concentrated beneath apical NPCs (Fig. 4G, H, and I). In summary, our results indicated that, in CPV infection, lamin B1 was enriched in the apical side concomitantly with an overall decrease of lamin A/C levels. Instead of inducing degradation, parvovirus infection might influence the organizational and/or functional status of nuclear lamins.
FIG 2.
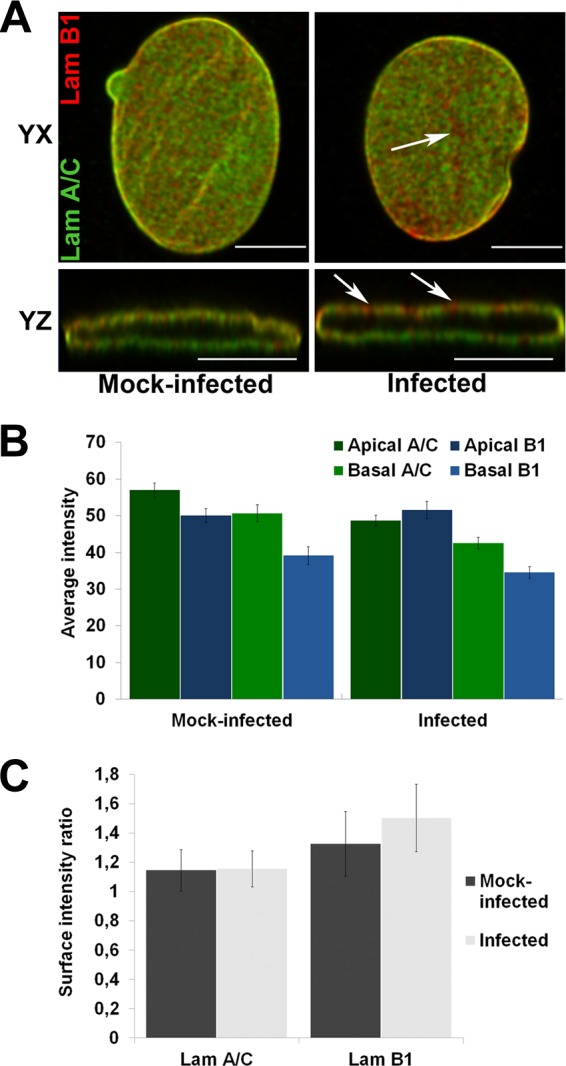
Distributions of lamin A/C and lamin B1. (A) Confocal yx and yz cross sections taken through the nucleus show localization of lamin A/C (Lam A/C) (green) and lamin B1 (Lam B1) (red) Abs in mock-infected (left) and infected (right) cells. Arrowheads show lamin B1-enriched areas. (B) Average intensities of lamin A/C and B in apical and basal sides of NE. (C) Surface intensity ratio between the apical and basal sides for both lamins individually per ImageJ analysis. Error bars represent the 95% confidence intervals. Bars, 10 μm.
FIG 3.
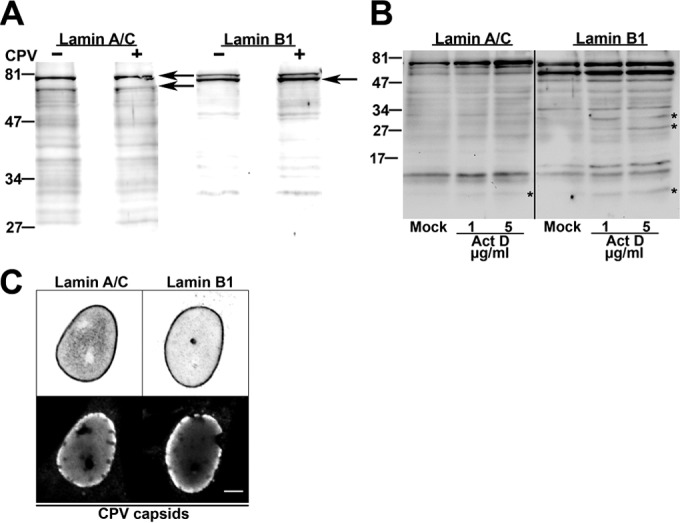
Structural integrity of lamins. (A and B) Western blot analysis of structural integrity of lamins A/C and B1 in infected, mock-infected cells (A) and actinomycin D (1 to 5 μg/ml)-treated cells (B). Asterisks indicate disintegration products of lamins. (C) Confocal microscopy sections of infected cells showing distributions of lamin A/C and lamin B1 and accumulation of viral capsids at the nuclear periphery at 24 h p.i. Bar, 5 μm.
FIG 4.
Intranuclear localization of lamin B1, NPCs, and viral capsids. Confocal microscopy-derived apical maximum-intensity projections are visualized with yz cross sections showing intranuclear distribution of NPCs (red) and lamin B1 (green) (A and B), virus capsids (red) and lamin B1 (green) (D and F), and virus capsids (red) and NPCs (green) (G and H). Capsids, NPC, and lamin B1 were visualized with capsid protein, Nup153, and lamin B1 antibodies. (C, F, and I) Normalized correlative intensity profiles from yz cross-section closeups are shown. Results of fluorescence line profile analysis of the intensity of capsids (red), NPCs (red/green), and lamin B1 (green) in a single optical section through the center of each nucleus are shown beside each image. Analysis was performed with ImageJ and the Plot RGB Profile plugin. A.U., arbitrary units. Bars, 10 μm.
To conclude, we observed that, in late-stage parvovirus infection, significant relocation of NPCs and reorganization of nuclear lamina occurred at the apical side of the nucleus. These changes were associated with the location of viral capsids in close proximity to apical NPCs. These results suggest that reorganization of the nuclear envelope might be important for viral egress. This report extends knowledge on parvovirus nuclear egress and the accompanying virus-induced changes in organization of the NPCs and the nuclear lamina.
ACKNOWLEDGMENTS
We thank Colin Parrish for the infectious CPV clone and CPV antibodies. We are grateful to Klaus Hedman for comments on the manuscript.
This work was financed by the Academy of Finland under the award numbers 135609 (E.A.N.), 267471 (T.O.I.), and 138388 (M.V.-R.).
REFERENCES
- 1.Dittmer TA, Misteli T. 2011. The lamin protein family. Genome Biol 12:222–222. doi: 10.1186/gb-2011-12-5-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gruenbaum Y, Medalia O. 2015. Lamins: the structure and protein complexes. Curr Opin Cell Biol 32:7–12. doi: 10.1016/j.ceb.2014.09.009. [DOI] [PubMed] [Google Scholar]
- 3.Köster S, Weitz DA, Goldman RD, Aebi U, Herrmann H. 2015. Intermediate filament mechanics in vitro and in the cell: from coiled coils to filaments, fibers and networks. Curr Opin Cell Biol 32:82–91. doi: 10.1016/j.ceb.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ho CY, Lammerding J. 2012. Lamins at a glance. J Cell Sci 125:2087–2093. doi: 10.1242/jcs.087288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fahrenkrog B, Köser J, Aebi U. 2004. The nuclear pore complex: a jack of all trades? Trends Biochem Sci 29:175–182. doi: 10.1016/j.tibs.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 6.Osmanagic-Myers S, Dechat T, Foisner R. 2015. Lamins at the crossroads of mechanosignaling. Genes Dev 29:225–237. doi: 10.1101/gad.255968.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pascual-Garcia P, Capelson M. 2014. Nuclear pores as versatile platforms for gene regulation. Curr Opin Genet Dev 25:110–117. doi: 10.1016/j.gde.2013.12.009. [DOI] [PubMed] [Google Scholar]
- 8.Cibulka J, Fraiberk M, Forstova J. 2012. Nuclear actin and lamins in viral infections. Viruses 4:325–347. doi: 10.3390/v4030325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Malhas AN, Lee CF, Vaux DJ. 2009. Lamin B1 controls oxidative stress responses via Oct-1. J Cell Biol 184:45–55. doi: 10.1083/jcb.200804155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zakaryan H, Stamminger T. 2011. Nuclear remodelling during viral infections. Cell Microbiol 13:806–813. doi: 10.1111/j.1462-5822.2011.01596.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cotmore SF, Tattersall P. 2014. Parvoviruses: small does not mean simple. Annu Rev Virol 1:517–537. doi: 10.1146/annurev-virology-031413-085444. [DOI] [PubMed] [Google Scholar]
- 12.Engelsma DN, Valle N, Fish A, Salomé N, Almendral JM, Fornerod M. 2008. A supraphysiological nuclear export signal is required for parvovirus nuclear export. Mol Biol Cell 19:2544–2552. doi: 10.1091/mbc.E08-01-0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maroto B, Valle N, Saffrich R, Almendral JM. 2004. Nuclear export of the nonenveloped parvovirus virion is directed by an unordered protein signal exposed on the capsid surface. J Virol 78:10685–10694. doi: 10.1128/JVI.78.19.10685-10694.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Imamoto N, Funakoshi T. 2012. Nuclear pore dynamics during the cell cycle. Curr Opin Cell Biol 24:453–459. doi: 10.1016/j.ceb.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 15.Dultz E, Ellenberg J. 2010. Live imaging of single nuclear pores reveals unique assembly kinetics and mechanism in interphase. J Cell Biol 191:15–22. doi: 10.1083/jcb.201007076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim D, Wirtz D. 2015. Cytoskeletal tension induces the polarized architecture of the nucleus. Biomaterials 48:161–172. doi: 10.1016/j.biomaterials.2015.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Essers J, Theil AF, Baldeyron C, van Cappellen WA, Houtsmuller AB, Kanaar R, Vermeulen W. 2005. Nuclear dynamics of PCNA in DNA replication and repair. Mol Cell Biol 25:9350–9359. doi: 10.1128/MCB.25.21.9350-9359.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ihalainen TO, Niskanen EA, Jylhävä J, Paloheimo O, Dross N, Smolander H, Langowski J, Timonen J, Vihinen-Ranta M. 2009. Parvovirus induced alterations in nuclear architecture and dynamics. PLoS One 4:e5948. doi: 10.1371/journal.pone.0005948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maeshima K, Iino H, Hihara S, Funakoshi T, Watanabe A, Nishimura M, Nakatomi R, Yahata K, Imamoto F, Hashikawa T, Yokota H, Imamoto N. 2010. Nuclear pore formation but not nuclear growth is governed by cyclin-dependent kinases (Cdks) during interphase. Nat Struct Mol Biol 17:1065–1071. doi: 10.1038/nsmb.1878. [DOI] [PubMed] [Google Scholar]
- 20.Maul GG, Maul HM, Scogna JE, Lieberman MW, Stein GS, Hsu BY, Borun TW. 1972. Time sequence of nuclear pore formation in phytohemagglutinin-stimulated lymphocytes and in HeLa cells during the cell cycle. J Cell Biol 55:433–447. doi: 10.1083/jcb.55.2.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Winey M, Yarar D, Giddings TH, Mastronarde DN. 1997. Nuclear pore complex number and distribution throughout the Saccharomyces cerevisiae cell cycle by three-dimensional reconstruction from electron micrographs of nuclear envelopes. Mol Biol Cell 8:2119–2132. doi: 10.1091/mbc.8.11.2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nykky J, Tuusa JE, Kirjavainen S, Vuento M, Gilbert L. 2010. Mechanisms of cell death in canine parvovirus-infected cells provide intuitive insights to developing nanotools for medicine. Int J Nanomedicine 5:417–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rothballer A, Kutay U. 2013. Poring over pores: nuclear pore complex insertion into the nuclear envelope. Trends Biochem Sci 38:292–301. doi: 10.1016/j.tibs.2013.04.001. [DOI] [PubMed] [Google Scholar]
- 24.Antonin W, Ellenberg J, Dultz E. 2008. Nuclear pore complex assembly through the cell cycle: Regulation and membrane organization. FEBS Lett 582:2004–2016. doi: 10.1016/j.febslet.2008.02.067. [DOI] [PubMed] [Google Scholar]
- 25.Porter FW, Palmenberg AC. 2009. Leader-induced phosphorylation of nucleoporins correlates with nuclear trafficking inhibition by cardioviruses. J Virol 83:1941–1951. doi: 10.1128/JVI.01752-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cotmore SF, Tattersall P. 2013. Parvovirus diversity and DNA damage responses. Cold Spring Harb Perspect Biol 5:a012989. doi: 10.1101/cshperspect.a012989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luo Y, Qiu J. 2013. Parvovirus infection-induced DNA damage response. Future Virol 8:245–257. doi: 10.2217/fvl.13.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Güttinger S, Laurell E, Kutay U. 2009. Orchestrating nuclear envelope disassembly and reassembly during mitosis. Nat Rev Mol Cell Biol 10:178–191. doi: 10.1038/nrm2641. [DOI] [PubMed] [Google Scholar]
- 29.Wan G, Zhang X, Langley RR, Liu Y, Hu X, Han C, Peng G, Ellis LM, Jones SN, Lu X. 2013. DNA damage-induced nuclear export of precursor microRNAs is regulated by the ATM-AKT pathway. Cell Rep 3:2100–2112. doi: 10.1016/j.celrep.2013.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hutchison CJ. 2002. Lamins: building blocks or regulators of gene expression? Nat Rev Mol Cell Biol 3:848–858. doi: 10.1038/nrm950. [DOI] [PubMed] [Google Scholar]
- 31.Guo Y, Kim Y, Shimi T, Goldman RD, Zheng Y. 2014. Concentration-dependent lamin assembly and its roles in the localization of other nuclear proteins. Mol Biol Cell 25:1287–1297. doi: 10.1091/mbc.E13-11-0644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maeshima K, Yahata K, Sasaki Y, Nakatomi R, Tachibana T, Hashikawa T, Imamoto F, Imamoto N. 2006. Cell-cycle-dependent dynamics of nuclear pores: pore-free islands and lamins. J Cell Sci 119:4442–4451. doi: 10.1242/jcs.03207. [DOI] [PubMed] [Google Scholar]
- 33.Fiserova J, Goldberg M. 2010. Relationships at the nuclear envelope: lamins and nuclear pore complexes in animals and plants. Biochem Soc Trans 38:829. doi: 10.1042/BST0380829. [DOI] [PubMed] [Google Scholar]
- 34.Nüesch JPF, Lachmann S, Rommelaere J. 2005. Selective alterations of the host cell architecture upon infection with parvovirus minute virus of mice. Virology 331:159–174. doi: 10.1016/j.virol.2004.10.019. [DOI] [PubMed] [Google Scholar]