ABSTRACT
Insect-borne plant viruses cause significant agricultural losses and jeopardize sustainable global food production. Although blocking plant virus transmission would allow for crop protection, virus receptors in insect vectors are unknown. Here we identify membrane alanyl aminopeptidase N (APN) as a receptor for pea enation mosaic virus (PEMV) coat protein (CP) in the gut of the pea aphid, Acyrthosiphon pisum, using a far-Western blot method. Pulldown and immunofluorescence binding assays and surface plasmon resonance were used to confirm and characterize CP-APN interaction. PEMV virions and a peptide comprised of PEMV CP fused to a proline-rich hinge (-P-) and green fluorescent protein (CP-P-GFP) specifically bound to APN. Recombinant APN expressed in Sf9 cells resulted in internalization of CP-P-GFP, which was visualized by confocal microscopy; such internalization is an expected hallmark of a functional gut receptor. Finally, in assays with aphid gut-derived brush border membrane vesicles, binding of CP-P-GFP competed with binding of GBP3.1, a peptide previously demonstrated to bind to APN in the aphid gut and to impede PEMV uptake into the hemocoel; this finding supports the hypothesis that GBP3.1 and PEMV bind to and compete for the same APN receptor. These in vitro data combined with previously published in vivo experiments (S. Liu, S. Sivakumar, W. O. Sparks, W. A. Miller, and B. C. Bonning, Virology 401:107–116, 2010, http://dx.doi.org/10.1016/j.virol.2010.02.009) support the identification of APN as the first receptor in a plant virus vector. Knowledge of this receptor will provide for technologies based on PEMV-APN interaction designed to block plant virus transmission and to suppress aphid populations.
IMPORTANCE A significant proportion of global food production is lost to insect pests. Aphids, in addition to weakening plants by feeding on their sap, are responsible for transmitting about half of the plant viruses vectored by insects. Growers rely heavily on the application of chemical insecticides to manage both aphids and aphid-vectored plant viral disease. To increase our understanding of plant virus-aphid vector interaction, we provide in vitro evidence supporting earlier in vivo work for identification of a receptor protein in the aphid gut called aminopeptidase N, which is responsible for entry of the plant virus pea enation mosaic virus into the pea aphid vector. Enrichment of proteins found on the surface of the aphid gut epithelium resulted in identification of this first aphid gut receptor for a plant virus. This discovery is particularly important since the disruption of plant virus binding to such a receptor may enable the development of a nonchemical strategy for controlling aphid-vectored plant viruses to maximize food production.
INTRODUCTION
Viruses that infect crop plants restrict our ability to consistently produce high yields from agricultural crops. Many of these viruses are transmitted to plants by pestiferous insects, with aphids transmitting nearly half of the 600 insect-borne plant viruses. Aphids therefore represent a significant threat to global agriculture (1). Viruses in the family Luteoviridae are phloem-restricted RNA viruses transmitted exclusively by aphids and cause disease in multiple food crops (2). Luteovirids are transmitted in a circulative and persistent manner which involves specific molecular interactions between the virus and the aphid (3). For this type of transmission, ingested virions cross the aphid gut and salivary gland epithelial barriers for transmission to additional plant hosts.
Luteovirus-aphid interactions are mediated by the viral capsid proteins consisting of a major coat protein (CP; 22 kDa) and one minor coat protein readthrough domain (CP-RTD; 35 to 55 kDa) (3). The RTD is not required for virus particle assembly or for uptake of virus from the gut into the aphid hemocoel, but both CP and RTD are essential for aphid transmission and are the sole determinants of vector specificity (3). The virus binds to a receptor in either the midgut, hindgut, or both for transcytosis across the aphid gut epithelium and release into the hemocoel (4). A second receptor-mediated transcytosis event occurs at the accessory salivary glands (ASG) from which virus particles are secreted with saliva to inoculate the plant phloem during subsequent feedings (5). Only a fraction of the virions present in the aphid hemolymph cross into the ASG, and a threshold amount of virus in the hemocoel is required before transmission via the ASG can occur (3). Hence, impeding the binding of a plant virus to the aphid gut receptor could reduce the amount of virus present in the hemocoel to less-than-threshold levels, thereby reducing plant virus transmission.
By screening a phage display library for peptides that bind to the gut epithelium of the pea aphid, Liu et al. (6) isolated the peptide GBP3.1 (amino acid sequence TCSKKYPRSPCM). Feeding pea aphids on the phage expressing this peptide, or on a GBP3.1-GFP fusion protein prior to transfer to a pea enation mosaic virus (PEMV)-infected plant, significantly reduced the amount of virus present in the hemocoel of the aphids relative to control treatments. In subsequent work, the peptide GBP3.1 was shown by UV-cross-linking to bind to pea aphid APN (7).
Although many studies have investigated the specific molecular interactions that facilitate virus movement in the aphid (8–11), the receptors involved in virus recognition are unknown. Identification of virus receptors in their insect vectors has been unsuccessful for circulative viruses as a whole. However, many insect proteins that bind virus particles in vitro have been identified primarily using far-Western blotting (8, 9, 12, 13). Two proteins isolated from the head of the aphid Sitobion avenae, SaM35 and SaM50, bound to barley yellow dwarf virus-MAV (BYDV-MAV) particles and were thought to be potential receptors in the ASG (8). In the aphid Myzus persicae, Rack-1, GAPDH3, and actin were shown to bind to Beet western yellows virus (9). A similar study of the small brown planthopper vector (Laodelphax striatellus) also identified Rack-1 and GAPDH3, along with three ribosomal proteins capable of binding rice stripe virus (12). The authors of that suggested that Rack-1, GAPDH3, and actin function in endocystosis and intracellular transport but are not actual virus receptors. Yang et al. (10) used a different approach based on two-dimensional (2D) gel electrophoresis comparisons of proteins from the greenbug, Schizaphis graminum, differing in the ability to transmit cereal yellow dwarf virus-RPV. Two proteins that were differentially expressed and also confirmed to bind virions were luciferase and cyclophilin (10). However, as in the previous studies, the authors could only link these proteins to the endocytosis pathway. Identification of the plant virus receptors used in their insect vectors is crucial for the development of novel strategies to block plant virus transmission.
To further investigate luteovirus-aphid interactions, we used PEMV and the pea aphid, Acyrthosiphon pisum. PEMV provides an ideal model virus as it is the only luteovirid that is not phloem-restricted and is thus mechanically transmissible to plants without reliance on aphids for transmission to the plant (14). PEMV consists of two taxonomically distinct positive-sense RNAs. PEMV-1 is the sole member of the genus Enamovirus (Luteoviridae) with genome organization similar to that of the Poleroviruses (15), whereas PEMV-2 belongs to the genus Umbravirus (14). In addition, the genome of the pea aphid has been sequenced (16), facilitating identification of putative receptor proteins, and a peptide that interferes with PEMV uptake into the aphid hemocoel has been identified (6). While PEMV is transmitted by at least 10 aphid species, the pea aphid and the green peach aphid, Myzus persicae, are the most important vectors of this virus resulting in the most severe impacts on crop yield (17).
The goal of our research was to provide in vitro support for identification of the receptor for PEMV in the pea aphid gut. In the present study, we (i) demonstrate that PEMV binds to membrane alanyl aminopeptidase N (APN) in the pea aphid (18) using a far-Western blot method, (ii) confirm that PEMV binds APN in pulldown assays, (iii) characterize the interaction between PEMV CP fused to a proline-rich hinge (-P-) and green fluorescent protein (CP-P-GFP) and APN by pulldown and immunofluorescence binding assays, and surface plasmon resonance (SPR), (iv) demonstrate that expression of pea aphid APN in Sf9 cells provides a functional receptor for internalization of CP-P-GFP into these cells, and (v) show that GBP3.1, a peptide previously shown to bind to APN in the pea aphid gut and impede uptake of PEMV into the aphid hemocoel (6, 7), competes with CP-P-GFP for binding to pea aphid gut-derived brush border membrane vesicles (BBMV). Competition between CP-P-GFP and GBP3.1 supports the hypothesis that GBP3.1 competes with PEMV for receptor binding. Taken together, these findings support the hypothesis that APN functions as a gut receptor for PEMV in the pea aphid.
MATERIALS AND METHODS
Insects.
Pea aphids, Acyrthosiphon pisum Harris (Aphidinae: Macrosiphini), were obtained from Berkshire Biological Supply Company (Westhampton, MA) and reared on the broad bean, Vicia faba. Aphid colonies were maintained in growth chambers at 24°C with a 12-h light/12-h dark cycle.
Preparation of aphid brush border membrane vesicles.
BBMV of adult A. pisum were prepared from whole aphids as described previously (19). The final BBMV pellets was resuspended in ice-cold buffer (0.3 M mannitol, 5 mM EGTA, 17 mM Tris-HCl [pH 7.5]) diluted 1:2 in water. Protease inhibitor cocktail (Sigma-P8340) was added to a 1:100 dilution, and samples were snap-frozen in liquid nitrogen and stored at −80°C. The protein concentration was determined (20). Aminopeptidase activity was measured (21) and was typically enriched 10- to 15-fold in the final BBMV suspensions relative to the initial homogenates.
Purification of pea enation mosaic virus.
Seven-day-old pea plants (Pisum sativum) were mechanically infected with wild-type PEMV or PEMV RNA1Δ (22) and harvested at 10 to 14 days postinfection. PEMV was purified from plants as described previously (22). Virus was then centrifuged at 147,000 × g through a 30% sucrose cushion made with 0.2 M sodium acetate (pH 7). The final pellet was washed three times in 0.2 M sodium acetate buffer and resuspended in the same buffer. Sample purity was assessed by SDS-PAGE analysis. The protein concentration of PEMV was determined by densitometric analysis with ImageJ software (23) of the Coomassie blue-stained bands with reference to known bovine serum albumin (BSA) concentrations resolved by SDS-PAGE.
Production of GFP and GFP fusion peptides.
The pBAD/His B (Invitrogen, Carlsbad, CA) vector was used for the expression of GFP, R4-GFP (R4 peptide, amino acid sequence WCDQLLQQMQCW), and GFP-RTD (Fig. 1). Three oligonucleotides were used for integrating the RTD and GFP DNA sequences: (i) a forward primer containing a PstI site and the 5′ end of the GFP sequence, (ii) an oligonucleotide connecting the 3′end of the GFP and the 5′end of the RTD sequence, and (iii) a reverse primer complementary to the 3′ end of the RTD sequence. The primers used for the construction of the GFP-RTD fusion sequence were GFPPstI and GFPRTD, as well as the reverse primer RTDHindIII (see Table 1 for the primer sequences). The cDNA encoding the R4-GFP protein was generated by PCR using the forward primers R4SacIF and shGFP-R (Table 1), with GFP cDNA as a template. The GFP-PCR products were purified by using a QIAquick gel extraction kit (Qiagen, Valencia, CA), digested with the respective restriction enzymes, cleaned by using a QIAquick nucleotide removal kit (Qiagen), and ligated into pBAD/His B. Top10 Competent Cells (Life Technologies, Carlsbad, CA) were used for the expression of GFP and the GFP fusion proteins using standard procedures. His-tagged GFP and fusion proteins were purified as described previously (6).
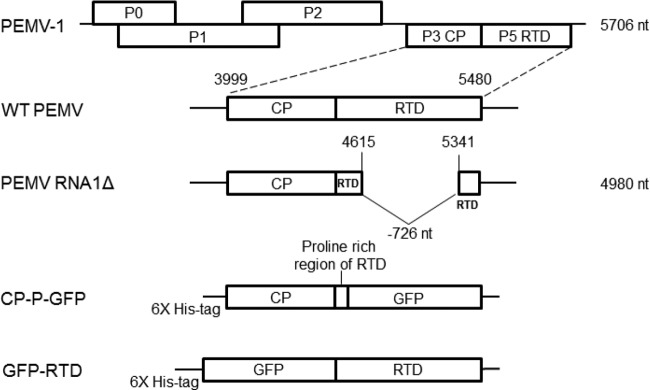
FIG 1.
Viruses and fusion protein constructs. Schematic diagrams are shown for PEMV-1 RNA (at the top) and for wild-type (WT) PEMV and PEMV RNA1Δ and the fusion proteins CP-P-GFP and GFP-RTD. The length of the genomic RNA is indicated on the right. The short remaining 5′ sequence of the RTD in PEMV RNA1Δ is in a different reading frame and does not produce a translated protein product (22).
TABLE 1.
Primers used for constructs described in this study
| Primer | Sequence (5′-3′)a | Restriction enzyme |
|---|---|---|
| GFPPstI | GCGCTGCAGCTgtgagcaagggcgaggagctg | PstI |
| GFPRTD | GGACGAGCTGTACAAGGGGGACGACGCTCCCCCG | |
| RTDHindIII | GGAAGCTTTTAatctaagggacttctgg | HindIII |
| R4SacIF | ACGGAGCTCGgcgtggtgcgatcagctgctgcagcagatgcagtgctgggcggtgagcaagggcgag | SacI |
| shGFP-R | CCGAAGCTTTTAgccgctttacttgtacagctcg | HindIII |
Lowercase text indicates coding region of the fusion protein. Restriction sites are underlined.
The CP-P-GFP fusion construct (Fig. 1) was expressed in the baculovirus expression system, as described previously (24). CP-P-GFP was purified by using the Ni-NTA agarose resin (Qiagen) as described below.
2D gel electrophoresis and far-Western blotting.
Whole aphid BBMV protein derived from A. pisum was subjected to a 2D-Clean-Up kit according to the manufacturer's directions (GE Health Sciences). The final pellets from 300 μg of initial protein were resuspended in rehydration buffer (GE Healthcare) and a trace amount of bromophenol blue. A sonicating water bath was used to increase protein solubilization. Reconstituted proteins were centrifuged in a microcentrifuge at 15,200 × g for 5 min to remove insoluble material. The protein was then applied to 7-cm Immobiline DryStrip gels (GE Healthcare) for overnight rehydration. Both pH 3 to 10 and pH 4 to 7 strips were used. Focusing was performed using the IPGphor (Bio-Rad) at 50 V for 10 h, 300 V for 1 h, 1,000 V 1 h (gradient), 5,000 V for 1.5 h (gradient), and 5,000 V for 1 h. IPG strips were then equilibrated for 15 min in equilibration buffer I (EB I; Bio-Rad), followed by 15 min in EB II. For the second dimension, the IPG strips were loaded and run on SDS–10% PAGE gels at 100 V. Gels were either stained with Coomassie blue or equilibrated in transfer buffer prior to overnight transfer to a polyvinylidene difluoride (PVDF) membrane at 4°C at 30 V.
Membranes were blocked in 5% nonfat dry milk in phosphate-buffered saline with 0.1% Tween 20 (PBS-T) overnight at 4°C. Membranes were then incubated with PEMV or PEMV RNA1Δ (20 μg/ml) or GFP fusion proteins (10 μg/ml) in 1% nonfat dry milk in PBS-T overnight at 4°C. Bound ligand was detected using affinity-purified PEMV coat protein antiserum (1:100) or GFP antiserum (Sigma; 1:5,000), followed by a horseradish peroxidase (HRP)-conjugated anti-rabbit IgG (1:5,000). The PVDF membranes were incubated in HyGlo chemiluminescent HRP detection reagent for 1 min, with luminescence detected on X-ray film according to standard procedures. Antibody-only controls were run in parallel by eliminating incubation with the ligand (virus or GFP fusion) to identify nonspecific binding of the antibodies to aphid BBMV. The experiments were replicated at least four times with each ligand.
Protein identification.
Protein spots bound by ligands were picked from Coomassie stained gels with reference to probed membranes and submitted to the Iowa State University Protein Facility for identification using matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) tandem mass spectrometry (MS/MS). All spectra were processed by using a MASCOT (MatrixScience, London, United Kingdom) database search. Peak lists were generated by Analyst QS (AB/MDS Sciex, Toronto, Canada) and were used for MS/MS ion searches. Protein identification was based on the probability based Mowse Score. The significance threshold P value was set to <0.05.
Expression of pea aphid APN in Escherichia coli for the production of polyclonal antibodies.
Both 28-kDa (amino acids 385 to 633) and 67-kDa (amino acids 216 to 794) fragments of full-length pea aphid APN (NP_001119606) were generated by PCR and cloned into pBAD/His B. Proteins were expressed as described above. Due to low binding of His-tagged APN fragments to the nickel resin under native conditions, the proteins were purified under denaturing conditions using 6 M guanidine hydrochloride. The recombinant APN was then refolded by dialysis in PBS with two buffer exchanges, using Slide-A-Lyzer dialysis cassettes (Life Technologies). For the production of antiserum, a mixture of the refolded soluble 28-kDa fragment and PAGE gel slices containing the 67-kDa fragment were used for inoculation of rabbits at the Iowa State University Hybridoma Facility.
Baculovirus expression of pea aphid APN in Sf9 cells.
A Bac-to-Bac baculovirus expression system (Invitrogen) was used to generate recombinant baculoviruses. To clone pea aphid APN (NP_001119606) into the baculovirus transfer vector pFastBac1(Invitrogen), the cDNA was first amplified using gene-specific primers containing NotI and HindIII restriction sites and ligated into pGEM T-Easy vector (Promega). There were 10 amino acid differences between the published sequence and the protein obtained, none of which fell within the catalytic domain. These differences were D22A, N51S, K234N, K268E, S303Y, I322V, A352E, E376Q, M874T, and I911T, with the first amino acid named from the published sequence. A version of APN lacking the glycosylphosphatidylinositol (GPI) anchor and containing a C-terminal histidine tag was also constructed to facilitate purification of APN by adding the 6× His tag and a stop codon prior to the GPI signal sequence. The sequences were excised from the vector and ligated into NotI/HindIII-digested pFastBac1. The recombinant bacmids [vAPN-GPI(−) and vAPN-GPI(+)] were generated according to the manufacturer's directions, and PCR was used to confirm the presence of inserts. Sf9 cells grown in a monolayer and maintained in Sf900 medium at 28°C were transfected with recombinant bacmids using Cellfectin II reagent (Invitrogen). Supernatant was harvested 4 days postinfection. Recombinant baculoviruses were plaque purified, and recombinant APN was expressed in baculovirus-infected cells according to standard procedures. Untransformed bacmid retaining the lacZ insert (vLacZα) was used as a negative-control virus.
Purification of His-tagged APN.
Sf9 cells were infected with recombinant baculovirus expressing APN and harvested at 72 h postinfection. APN-GPI(−) was purified from both the Sf900 medium and the cell lysates. To purify from the medium, sodium phosphate and imidazole were added to final concentrations of 50 and 10 mM, respectively. The pH was not adjusted in order to avoid precipitation of some components of the medium. The medium was incubated with the Ni-NTA agarose resin (Qiagen) overnight at 4°C. The resin was washed, and the protein was eluted using standard buffers described by the manufacturer. APN purification from the cells was conducted under native conditions using a batch purification method with the Ni-NTA resin as described by the manufacturer. Purified vAPN-GPI(−) was used for SPR analysis.
Immunofluorescence detection of pea aphid APN expressed in Sf9 cells.
Two methods were used for the immunofluorescence assays. For the first method, Sf9 cells were infected with the recombinant baculoviruses expressing APN. At 48 h postinfection, cells were seeded on microscope slides, washed once in PBS, and fixed with 3.7% formaldehyde in PBS for 30 min. All washes and incubations were done using a Coplin jar. After fixation, the cells were permeabilized with PBS containing 0.3% Triton X-100 (PT buffer) for 1 h at 37°C, followed by blocking for 1 h in PT buffer containing 3% BSA (PTB buffer). The cells were then incubated with a 1:250 dilution of anti-APN antiserum in PTB buffer containing 0.5% BSA overnight at 4°C. The following day, the cells were washed three times in PT buffer and then incubated in 1:500 Alexa 488-conjugated goat anti-rabbit secondary antibody (Invitrogen) in PTB containing 0.5% BSA for 1 h at 37°C. The cells were then washed three times in PT buffer, incubated in 1:1,000 DAPI (4′,6′-diamidino-2-phenylindole; Kirkegaard & Perry Laboratories, Inc.) in PT buffer to stain the nuclei, and then washed three times in PT buffer. The slides were mounted in 50% glycerol in PBS. Alexa 488 fluorescence was detected using standard epifluorescence microscopy using a fluorescein isothiocyanate (FITC) HYQ filter (Nikon) with 460- to 500-nm excitation and 510- to 560-nm emission. DAPI fluorescence was visualized with a UV-2E/C filter (Nikon) with a 340- to 380-nm excitation and 435- to 485-nm emission. Images were also taken under a bright field. Uninfected cells and cells infected with vLacZα served as negative controls.
For the second method, the cells were seeded and infected on poly-l-lysine-coated 12-mm glass coverslips (BD Biosciences) in 24-well plates. At 48 h postinfection, the cells were washed three times in insect PBS (1 mM sodium phosphate, 10.5 mM potassium phosphate, 140 mM sodium chloride, 40 mM potassium chloride [pH 6.2]) on a shaker. The cells were fixed in 2% formaldehyde in PBS for 30 min at room temperature. After a brief rinse in PBS, cells were permeabilized in PT buffer for 1 h at 28°C, followed by blocking in 3% PTB for 1 h. Antibody and DAPI incubations were conducted as described above, but on a shaker. To investigate the binding of CP-P-GFP to APN expressed in Sf9 cells, the same procedure was followed except that the antibody steps were replaced by incubation with 10 μg of CP-P-GFP/ml. The Sf9 cells were incubated with the CP-P-GFP either 1 h at 28°C or overnight at 4°C. Uninfected cells and cells infected with vLacZα served as controls. Experiments for antibody detection of APN were repeated four times between the two methods. Experiments for CP-P-GFP binding were repeated six times, with CP-P-GFP binding observed in all experiments.
Confocal microscopy.
Sf9 cells were seeded on the glass coverslips and infected with baculovirus expressing APN as described above. At 48 h postinfection, cells were washed once in PBS and incubated with 10 μg of CP-P-GFP or E. coli-expressed GFP/ml for 1 h at room temperature with gentle rocking. The cells were then washed three times in PBS for 5 min per wash, fixed in 2% formaldehyde, and then washed once in PBS for 5 min. The cells were stained with 1:1,000 DAPI for 5 min and washed three times in PBS for 5 min per wash. Controls included uninfected Sf9 cells and cells infected with vLacZα. Glass coverslips were mounted on microscope slides in a 1:1 solution of glycerol and PBS and visualized using a Leica SP5 X MP microscope (Exton, PA). A 63×/1.4 oil objective lens was used with a 5× zoom. The DAPI excitation was 405 nm, and the emission range was 416 to 464 nm. GFP excitation was 488 nm, and the emission range was 504 to 581 nm. A total of 50 to 100 cells for each of three dishes were examined for fluorescence per treatment, and images were generated for representative cells. Confocal experiments were conducted twice, with a total of >300 cells examined per treatment.
Cross-linking pulldown assays with PEMV.
Purified PEMV was labeled with a Sulfo-SBED biotin label transfer reagent (Thermo Scientific). All steps were performed in the dark to avoid activation of the aryl azide group prior to cross-linking. The Sulfo-SBED reagent was dissolved in dimethylformamide (DMF) and added to 250 μg of PEMV in 5 ml of 0.2 M sodium phosphate at pH 7. A final concentration of 8% DMF was maintained in the solution to avoid precipitation of the labeled virus. The final concentration of the Sulfo-SBED in the reaction was 110 μM. The reaction mixture was incubated at room temperature for 30 min with occasional mixing. To remove unbound Sulfo-SBED reagent, virus solution was passed through a Sephadex G-25 column (GE Healthcare). Labeled virus was stored at 4°C in the dark.
Whole guts from pea aphids were dissected in PBS containing protease inhibitor cocktail at a dilution of 1:100 (Sigma). Protease inhibitors were present throughout the following procedure. Guts were washed three times in PBS and then incubated with 10 μg of labeled PEMV for 1 h at 4°C in the dark. The guts were gently mixed every 5 to 10 min. The guts were then washed seven times in PBS to remove unbound PEMV. The guts were exposed to UV for 15 min (PEMV) to cross-link proteins interacting with the virus. The guts were then briefly centrifuged and resuspended in lysis buffer (250 mM potassium acetate, 10 mM magnesium acetate, 50 mM HEPES [pH 7.4], 0.1% NP-40) and thoroughly homogenized. The suspension was centrifuged at 13,300 × g to pellet the insoluble material. The supernatants were added to 100 μl of streptavidin-agarose beads (Invitrogen) preequilibrated in lysis buffer without NP-40. The beads were rotated at room temperature for 1 h. The beads were then washed seven times in lysis buffer without NP-40 and boiled in SDS loading buffer for 5 min, and the supernatants were loaded for SDS-PAGE and Western blot detection with purified APN antiserum. APN pulled down with PEMV in test and control treatments and visualized by Western blotting was quantified using ImageJ analysis (23). Statistical differences were determined by using the Student t test. Controls for PEMV pulldown assays included streptavidin beads incubated with PEMV alone, beads incubated with aphid guts alone, and beads incubated with buffer only.
SPR binding assays.
Surface plasmon resonance (SPR) assays using a BIAcore T100 (BIAcore, Uppsala, Sweden) were used for quantification of the relative binding of CP-P-GFP, GFP, and RTD-GFP to APN-GPI(−). The buffer HBS-N (BIAcore) was used for all experiments. Preparations of CP-P-GFP, GFP, and RTD-GFP were dialyzed in HBS-N to a final concentration of 6 μM. APN-GPI(−) was immobilized on to the carboxymethylated dextran (CM5) sensor chip surface (BIAcore) through standard amine coupling method. Carboxyl groups along the carboxymethylated dextran chains of the sensor chip surface were activated by injecting a mixture of NHS (0.1 M N-hydroxysuccinimide) and EDC [0.1 M 1-ethyl-3-(3-dimethylamino-propyl)carbodiimide hydrochloride; 1:1 (vol/vol)]. APN-GPI(−) in coupling buffer (10 mM sodium acetate [pH 4.5]) was injected over the chip surface at 0.1 μg/μl to obtain an immobilization target level of 2,000 resonance units (RU). After coupling, unreacted surface ester groups were blocked by injecting 1 M ethanolamine (pH 8.5) onto the chip surface. Analysis of the interaction of APN-GPI(−) with CP-P-GFP, GFP, and RTD-GFP was performed by injecting proteins (6 μM in HBS-N) at 30 μl/min for 60 s, dissociation for 60 s, regeneration with 50 mM NaOH for 60 s, and stabilization for 120 s. Reference flow cells without immobilization of APN-GPI(−) were included in the experiments. SPR assays were conducted at least twice, and the data were analyzed by one-way analysis of variance.
BBMV pulldown competition assays.
Pulldown binding assays using pea aphid BBMV were carried out as described previously (7). Briefly, 10-μg portions of BBMV were incubated with ligands (CP-P-GFP with or without test peptide-GFP fusion proteins) for 1 h at room temperature. The BBMV were washed three times, the membrane fraction was pelleted by centrifugation, and the total proteins associated with the membrane in the pelleted fraction were examined by Western blotting. For the competition assays, CP-P-GFP (50 nM) was incubated with BBMV in the presence of a 1,000-fold molar excess of GBP3.1-GFP (50 μM). As a negative control, the same reaction was performed using 1,000-fold molar excess of a nonbinding control peptide R4-GFP instead of GBP3.1-GFP. GBP3.1-GFP, CP-P-GFP, R4-GFP, and GFP were run alone with BBMV as controls. This experiment was repeated twice.
RESULTS
PEMV binds to pea aphid APN.
To identify membrane-associated proteins that interact with PEMV in the aphid gut, brush border membrane vesicles (BBMV) rather than whole aphids were used for 2D ligand blot analyses. BBMV were separated by 2D gel electrophoresis, blotted to PVDF membranes, and overlaid with purified PEMV, coat protein-GFP fusion (CP-P-GFP), or the readthrough domain-GFP fusion (GFP-RTD; Fig. 1). Bound PEMV or GFP fusion protein was detected with antiserum specific to the PEMV coat protein or GFP, respectively. Wild-type PEMV, PEMV lacking the RTD (PEMV RNA1Δ), and CP-P-GFP all bound specifically to an aphid protein of ∼150 kDa which appeared on some blots as multiple spots (Fig. 2A and B). This protein was isolated from Coomassie blue-stained gels with reference to probed membranes and identified as membrane alanyl aminopeptidase N (APN; gi|187179337) by MALDI-TOF MS/MS. Although the pea aphid has six full-length isoforms of APN (25), four of the five hits identified by Mascot Search, were to the same APN sequence (accession no. NP_001119606). The additional binding observed in both CP-P-GFP and GFP blots was attributed to binding of the GFP antibody to aphid BBMV proteins. In contrast to the specific binding of PEMV and CP-P-GFP, binding of the GFP-RTD fusion lacked specificity and bound many aphid proteins, including APN (Fig. 2B).
FIG 2.
PEMV CP and RTD bind aminopeptidase N. (A) Binding of WT PEMV and PEMV RNA1Δ to pea aphid BBMV. No binding was detected for the antibody (no ligand) control. (B) Binding of CP-P-GFP and GFP-RTD to pea aphid BBMV. Nonspecific binding of the GFP antibody to aphid BBMV proteins was detected in blots with CP-P-GFP or GFP as ligand. In panels A and B, arrows indicate consistent binding. Far-Western blotting conducted using pH 3 to 10 and pH 4 to 7 was replicated at least four times per ligand. (C) PEMV binding of APN using a pulldown assay. PEMV was labeled with a biotin transfer reagent to UV cross-link proteins bound to PEMV. The PEMV-APN complex was pulled down using streptavidin-agarose beads, and APN was detected by Western blotting with APN antiserum (at left). The amount of APN pulled down by PEMV averaged 4.5-fold more than the control lacking PEMV across four replicates (Student t test P = 0.025; as determined by the ImageJ program [at right]). In panels A to C, the positions of molecular mass standards are indicated on the left. (D) BIAcore SPR analysis of CP-P-GFP, GFP-RTD, GFP interactions with APN. Sensorgram showing real-time interactions between 6 μM concentrations of GFP and the GFP fusion proteins with immobilized APN-GPI(−) are shown. L1 chip surfaces were prepared with 2,000 RU of APN-GPI(−). The data shown are representative of two independent experiments.
Pulldown assays with pea aphid guts were used to confirm binding of PEMV to APN. PEMV was labeled with a biotin transfer reagent to UV cross-link proteins bound to PEMV. The biotin label was used to pull down the protein complex with streptavidin-linked agarose beads. Purified anti-APN IgG was used to detect the coprecipitated APN by Western blotting (Fig. 2C). Although some APN associated nonspecifically with the streptavidin-agarose beads, the amount of APN pulled down in the presence of PEMV was significantly greater than in the absence of PEMV across four replicate experiments, with band intensities of APN measured by densitometric analysis using the ImageJ program (Fig. 2C; Student t test, P = 0.025). The association of pea aphid APN with biotin may account for this background binding. The association of pea aphid APN with PEMV, as shown by UV cross-linking, confirms colocalization of PEMV with APN in the pea aphid gut.
Binding of PEMV CP and RTD to APN.
The real-time binding of baculovirus-expressed APN without a GPI anchor, APN-GPI(−), to CP-P-GFP, GFP-RTD, and GFP was quantified using SPR (Fig. 2D). The relative binding units ± the standard errors of the mean to APN were as follows: GFP, 0.00 ± 0.00; GFP-RTD, 3.079 ± 0.37 (P = 0.021399); and CP-P-GFP, 100.38 ± 0.51 (P = 3.91E–05).
PEMV CP binds recombinant APN.
The binding of the PEMV coat protein with the proline rich hinge to Sf9 cells with or without baculovirus-expressed recombinant pea aphid APN-GPI(+) (Fig. 3A) was visualized by incubation with CP-P-GFP. The localization of APN to the membrane of baculovirus-infected Sf9 cells was confirmed by Western blotting of membrane and soluble fractions (data not shown), a finding consistent with previous reports for Spodoptera litura GPI-anchored APN in Sf21 cells (26) and localization to the pea aphid midgut epithelium membrane (18).
FIG 3.
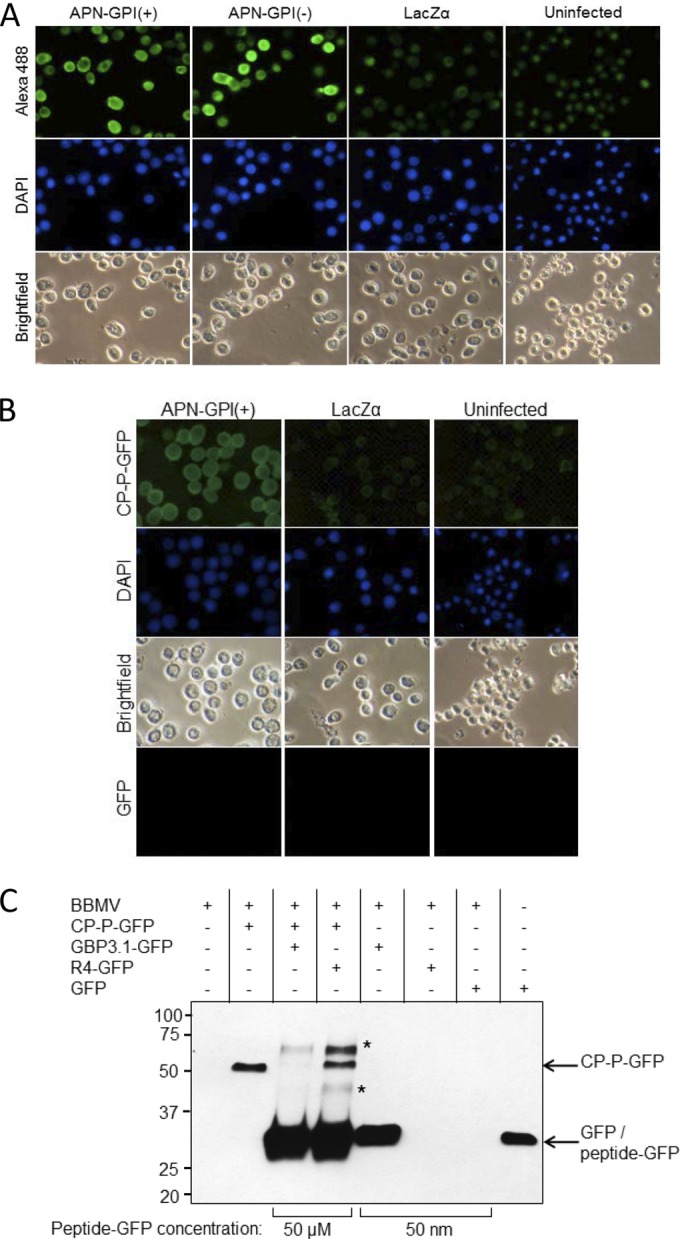
PEMV binding to APN is specific. (A) Baculovirus expression of recombinant APN in Sf9 cells. Sf9 cells were infected with the recombinant baculoviruses vAPN-GPI(+), vAPN-GPI(−), or vLacZα as a control, and expression of APN at 48 h was detected using an anti-APN primary antibody and an Alexa 488-conjugated secondary antibody. Nuclei were stained with DAPI. Uninfected cells served as a control. Alexa 488 (top row) and DAPI fluorescence (middle row) were visualized by epifluorescence microscopy. Bright-field images are also shown. The images are representative of four experiments. (B) Binding of CP-P-GFP to Sf9 cells expressing recombinant APN. Sf9 cells were infected with vAPN-GPI(+) or negative-control baculovirus, vLacZα, and at 48 h postinfection the cells were incubated with CP-P-GFP or GFP, followed by DAPI to stain the nuclei. Uninfected Sf9 cells served as an additional control. Increased fluorescence was observed in Sf9 cells expressing recombinant APN. No fluorescence was observed when cells were incubated with GFP. The images are representative of six experiments. Note that baculovirus infection results in enlargement of the nucleus, as seen for DAPI-stained, infected cells. All images were taken at the same magnification. (C) GBP3.1 competes with CP-P-GFP for binding to aphid BBMV. Shown is a Western blot with anti-GFP antiserum for the detection of GFP fusion proteins bound to BBMV in pulldown assays. The binding of CP-P-GFP (50 nM) was outcompeted by the addition of a 1,000-fold molar excess of GBP3.1-GFP (50 μM) but not by excess of the nonbinding control peptide R4-GFP. At 50 nM, GBP3.1-GFP showed strong binding to the BBMV compared to no observed binding of R4-GFP and GFP at the same concentration. R4-GFP was detected when used in excess (50 μM) but did not compete with the binding of CP-P-GFP (∼52 kDa). A total of 50 ng of GFP protein was used as positive control for the GFP antiserum. *, Proteins of ∼60 and 40 kDa detected after the addition of 50 μM peptide are hypothesized to be a dimer (∼57 kDa) and a degradation product of the peptide-GFP proteins, respectively. Conserved cysteines at positions 2 and 11 of the peptides provide for disulfide bonds which mediate dimerization at high concentration.
An increase in fluorescence was detected in Sf9 cells expressing APN compared to the controls, indicating increased binding of CP-P-GFP (Fig. 3B). Low levels of background fluorescence in cells infected with a control virus (vLacZα) and in uninfected cells may result from weak CP binding to Spodoptera frugiperda APN in the Sf9 cells. Fluorescence was not observed in Sf9 cells incubated with GFP supporting weak binding of CP to S. frugiperda APN and indicating that the observed fluorescence from CP-P-GFP did not result from GFP binding.
The gut binding peptide GPB3.1 competes with CP-P-GFP for binding to BBMV.
We previously identified the peptide GBP3.1 that impedes uptake of PEMV into the aphid hemocoel (6) and binds to pea aphid APN (7). To better mimic binding under in vivo conditions, BBMV, rather than recombinant APN, were used for competition assays. Using a BBMV pulldown assay, CP-P-GFP binding was out-competed by addition of excess GBP3.1-GFP (Fig. 3C). Excess nonbinding peptide R4-GFP did not compete with CP-P-GFP for binding. These in vitro pulldown assay results showing that GBP3.1-GFP specifically competes with CP-P-GFP for binding to aphid BBMV support the in vivo competition between GBP3.1 and PEMV for binding to the receptor, APN (6, 7).
CP-mediated entry into Sf9 cells expressing APN.
To address whether CP-P-GFP is internalized into Sf9 cells expressing pea aphid APN-GPI(+), cells were incubated with CP-P-GFP, and fluorescence in living cells was observed by confocal microscopy. Fluorescence was observed inside the Sf9 cells, confirming that pea aphid APN provides a functional receptor for uptake of CP-P-GFP into the cell (Fig. 4). Minimal fluorescence was observed in the control treatments of Sf9 cells infected with a baculovirus expressing LacZ and uninfected Sf9 cells.
FIG 4.
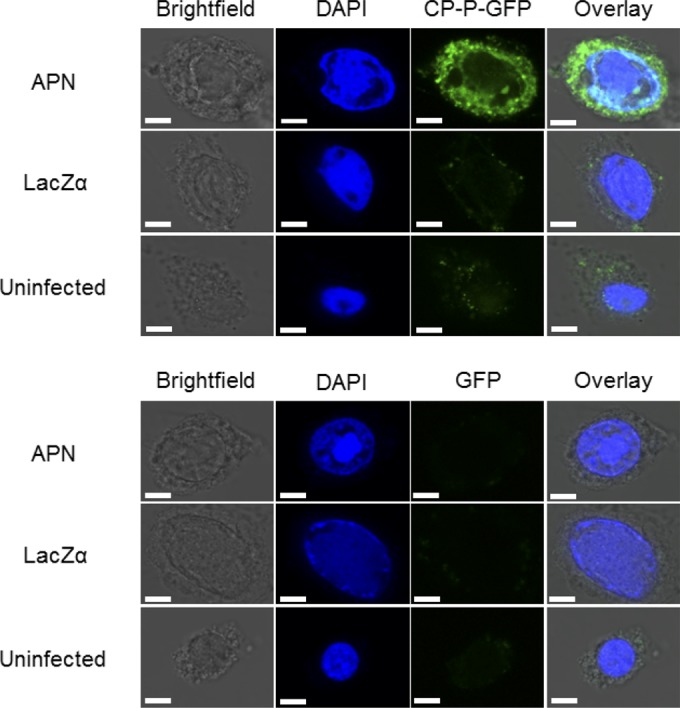
Aminopeptidase N is necessary for internalization of CP-P-GFP. Visualization of CP-P-GFP endocytosed into APN-expressing Sf9 cells using confocal microscopy. Sf9 cells were infected with vAPN-GPI(+) or vLacZα. At 48 h postinfection, the cells were incubated with CP-P-GFP or GFP, followed by DAPI to stain the nuclei. Uninfected Sf9 cells served as an additional control. DAPI and intracellular GFP fluorescence were detected by the examination of multiple cell layers using a confocal microscope. Increased fluorescence was observed in Sf9 cells expressing recombinant APN. No fluorescence was observed when cells were incubated with GFP. The images are representative of two replicate experiments, with fluorescence seen in approximately half of the cells in the CP-P-GFP treatment. Scale bars, 5 μm.
DISCUSSION
Luteovirus receptors.
Luteovirids are recognized by specific receptors in either the midgut or hindgut of the aphid vector. Viruses in the genus Luteovirus tend to be acquired through the hindgut, whereas viruses in the genus Polerovirus use either the mid- or the hindgut (4). Different viruses use different receptor proteins in a given aphid vector. For example, Soybean dwarf virus is acquired through the hindgut epithelium of the green peach aphid, Myzus persicae (27), while potato leafroll virus and beet western yellow virus are acquired through the midgut epithelium of M. persicae (28, 29). Moreover, there is evidence that related viruses use different capsid protein domains and different receptors in the same vector (e.g., Barley yellow dwarf virus, BYDV-RPV, and BYDV-PAV in the bird cherry-oat aphid, Rhopalosiphum padi) (30).
Although luteovirus-aphid interactions have long been the focus of investigation, the specific aphid receptors involved in luteovirus transmission have not previously been identified. In the present study, we demonstrated that PEMV binds to membrane alanyl aminopeptidase N (APN) using a 2D far-Western blot method with BBMV from whole pea aphids. In contrast to previous studies, we used BBMV to increase the probability of identifying a receptor under in vitro conditions by enriching for membrane proteins. Although we used whole aphid BBMV rather than pea aphid gut-derived BBMV, the majority of the protein from whole aphid BBMV preparations would be derived from the gut and APN is localized to the gut (18). Difficulties encountered in detecting PEMV-APN interaction without a cross-linking step with biotin-labeled PEMV suggest that PEMV-receptor binding in the aphid gut is transient. Such transient binding of luteoviruses to aphid receptors may explain why previous attempts to identify receptors were unsuccessful (8–10).
APN as a luteovirus receptor.
Collectively, our data combined with previously published data support the hypothesis that APN serves as a functional receptor for PEMV in the pea aphid vector (Fig. 5). APN is commonly a major component in the insect gut epithelial membrane with a primary function to cleave N-terminal amino acids. APN is estimated to comprise 15% of the total midgut protein in the pea aphid (18). This midgut localization is consistent with the role of APN as a gut receptor given that the midgut is the site for PEMV uptake (4) and that, based on activity levels, APN expression is relatively low in the hindgut (31). Consistent with the high molecular mass relative to the predicted molecular mass, APN is heavily glycosylated and known to be a target for aphicidal lectins (32–35). The role glycans play, if any, in luteovirus-aphid interactions is unclear (36, 37). APN is also one of the receptors for Bacillus thuringiensis δ-endotoxins in insects, although these toxins are relatively ineffective against aphids (19, 38, 39). There is a precedent for APN functioning as a virus receptor as some group I coronaviruses such as human coronavirus 229E use APN for cell entry (40).
FIG 5.
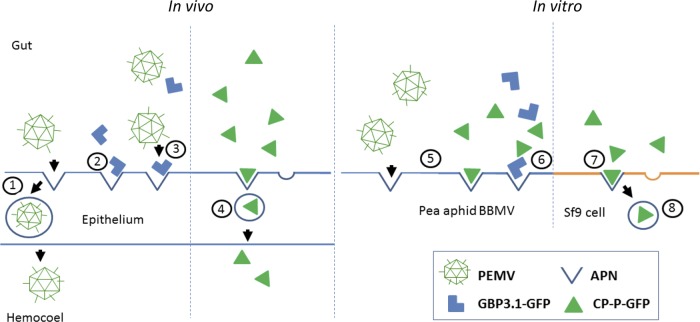
Schematic summary of evidence for APN as the receptor for PEMV. Shown are interactions between the virus PEMV, the peptide GBP3.1, the PEMV CP-P-GFP fusion protein, and APN based on published data (in vivo) and data presented here (in vitro). Numbered circles: 1, PEMV binds to a putative receptor and transcytoses across the gut epithelium of the aphid vector (3, 6); 2, the pea aphid gut binding peptide GBP3.1 binds to APN (7); 3, GBP3.1-GFP competes with PEMV in vivo reducing the amount of virus in the aphid hemocoel (6); 4, CP-P-GFP transcytoses across the aphid gut epithelium into the aphid hemocoel similar to PEMV (24); 5, both PEMV and CP-P-GFP bind APN in vitro (Fig. 2A and B); 6, GBP3.1-GFP competes with CP-P-GFP for binding in vitro (Fig. 3C); 7, CP-P-GFP binds recombinant pea aphid APN on Sf9 cells (Fig. 2A); 8, Pea aphid APN mediates internalization of CP-P-GFP into Sf9 cells (Fig. 4).
As evident from the far Western blots in Fig. 2, APN has multiple isoforms. These isoforms likely result from differential posttranslational modification since four of the five hits from MALDI-TOF analysis were to the same APN, with the fifth hit to an APN-like, partial sequence (accession no. XP_001944286). The biological significance of the isoforms, if any, is unknown.
The function of APN as the receptor for PEMV in vivo has been demonstrated: GBP3.1 directly competes in vivo with PEMV virions for receptor binding and impedes the uptake of PEMV into the aphid hemocoel (6). The downstream effect of this would be reduced virus titer in the aphid hemocoel and consequent reduced transmission of the plant virus (3). We have since shown that both GBP3.1 and PEMV bind to APN. Hence, the in vitro binding of PEMV to pea aphid APN shown in the present study has been confirmed under in vivo conditions (Fig. 5).
Although our attempts to visualize PEMV entry into Sf9 cells infected with the APN-expressing recombinant baculovirus (either by transmission electron microscopy or by use of FITC-labeled PEMV) were not successful, the following factors (depicted in Fig. 5) taken together support the role of APN as a functional receptor for PEMV in the pea aphid: APN is a membrane-associated protein (18) and is bound specifically by CP-P-GFP and PEMV (Fig. 2 and 3B), and (ii) the binding of both CP-P-GFP and PEMV is impeded by GBP3.1-GFP (Fig. 3C) (6) that also binds APN (7). Hence, CP-P-GFP and PEMV compete with GBP3.1-GFP for receptor binding; recombinant APN on Sf9 cells mediates internalization of CP-P-GFP. The low efficacy and inconsistency of RNA interference in aphids prevented functional confirmation of APN as the receptor for PEMV by silencing in the pea aphid (41).
The readthrough domain binds nonspecifically.
In contrast to the specific binding of PEMV and CP-P-GFP to APN, the GFP-RTD fusion protein bound multiple BBMV proteins, including APN. The relatively weak binding of GFP-RTD compared to CP-P-GFP (Fig. 2D), combined with the lack of specific binding of GFP-RTD (Fig. 2B), supports the hypothesis that the viral RTD functions in initial interactions between the virus and the gut epithelial cells, providing the virus with a catch-hold from which it binds specifically to receptors that allow for entry into the cell (42). Entry of CP-P-GFP into Sf9 cells expressing APN-GPI(+) is consistent with the observation that luteovirids lacking the RTD can cross the gut barrier and enter the aphid hemocoel (28, 43–45). The RTD is required for aphid transmission (46–48), although its role is not fully understood. While GFP-RTD bound multiple BBMV proteins as observed in 2D ligand blots, the binding of wild-type PEMV (with virions composed of both CP and RTD) was specific in ligand blots for APN (Fig. 2A and B). This apparent anomaly may result from the relative abundance of the CP and CP-RTD proteins in the PEMV virion: readthrough of the CP stop codon occurs at a relatively low frequency of ∼10% (49). We expect the RTD to contribute little to the strength of virion binding to APN, similar to the relative binding strengths seen for CP-P-GFP and GFP-RTD by SPR (Fig. 2D).
Practical applications.
The identification of receptors for plant viruses in their insect vectors has important implications for agriculture. Current management of both aphids and aphid-transmitted plant viruses relies primarily on the application of chemical insecticides. Knowledge of plant virus-vector molecular interactions allows for technologies to both disrupt plant virus transmission and to manage the aphid vector: plant expression of peptides, such as GBP3.1, that bind specifically to plant virus receptors could be used to block plant virus transmission (6). Upon aphid feeding, these peptides would interfere with the uptake of virions into the aphid hemocoel, thereby reducing virus transmission in the field. In addition, plant expression of PEMV CP fused to an insect-specific neurotoxin confers oral toxicity via APN-mediated delivery of the toxin to the aphid hemocoel for transgenic plant resistance against aphids (24, 50). Knowledge of the receptor may allow for adaptation of this technology to target other insect pests of agricultural importance.
ACKNOWLEDGMENTS
We thank S. Sivakumar for construction of the recombinant baculovirus expressing CP-P-GFP, and we thank W. Allen Miller for helpful discussions.
This study was funded by the USDA grant AFRI 2008-03996, by the Iowa State University Plant Sciences Institute Virus-Insect Interactions Initiative, and by Hatch Act and State of Iowa funds.
REFERENCES
- 1.Dedryver CA, Le Ralec A, Fabre F. 2010. The conflicting relationships between aphids and men: a review of aphid damage and control strategies. C R Biol 333:539–553. doi: 10.1016/j.crvi.2010.03.009. [DOI] [PubMed] [Google Scholar]
- 2.Hogenhout SA, Ammar E-D, Whitfield AE, Redinbaugh MG. 2008. Insect vector interactions with persistently transmitted viruses. Annu Rev Phytopathol 46:327–359. doi: 10.1146/annurev.phyto.022508.092135. [DOI] [PubMed] [Google Scholar]
- 3.Gray S, Gildow FE. 2003. Luteovirus-aphid interactions. Annu Rev Phytopathol 41:539–566. doi: 10.1146/annurev.phyto.41.012203.105815. [DOI] [PubMed] [Google Scholar]
- 4.Brault V, É Herrbach Reinbold C. 2007. Electron microscopy studies on luteovirid transmission by aphids. Micron 38:302–312. doi: 10.1016/j.micron.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 5.Brault V, Uzest M, Monsion B, Jacquot E, Blanc S. 2010. Aphids as transport devices for plant viruses. C R Biol 333:524–538. doi: 10.1016/j.crvi.2010.04.001. [DOI] [PubMed] [Google Scholar]
- 6.Liu S, Sivakumar S, Sparks WO, Miller WA, Bonning BC. 2010. A peptide that binds the pea aphid gut impedes entry of Pea enation mosaic virus into the aphid hemocoel. Virology 401:107–116. doi: 10.1016/j.virol.2010.02.009. [DOI] [PubMed] [Google Scholar]
- 7.Chougule NP, Li H, Liu S, Linz LB, Narva KE, Meade T, Bonning BC. 2013. Retargeting of the Bacillus thuringiensis toxin Cyt2Aa against hemipteran insect pests. Proc Natl Acad Sci U S A 110:8465–8470. doi: 10.1073/pnas.1222144110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li C, Cox-Foster D, Gray SM, Gildow F. 2001. Vector specificity of barley yellow dwarf virus (BYDV) transmission: identification of potential cellular receptors binding BYDV-MAV in the aphid, Sitobion avenae. Virology 286:125–133. doi: 10.1006/viro.2001.0929. [DOI] [PubMed] [Google Scholar]
- 9.Seddas P, Boissinot S, Strub JM, Van Dorsselaer A, Van Regenmortel MH, Pattus F. 2004. Rack-1, GAPDH3, and actin: proteins of Myzus persicae potentially involved in the transcytosis of beet western yellows virus particles in the aphid. Virology 325:399–412. doi: 10.1016/j.virol.2004.05.014. [DOI] [PubMed] [Google Scholar]
- 10.Yang X, Thannhauser TW, Burrows M, Cox-Foster D, Gildow FE, Gray SM. 2008. Coupling genetics and proteomics to identify aphid proteins associated with vector-specific transmission of polerovirus (Luteoviridae). J Virol 82:291–299. doi: 10.1128/JVI.01736-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cilia M, Tamborindeguy C, Fish T, Howe K, Thannhauser TW, Gray S. 2011. Genetics coupled to quantitative intact proteomics links heritable aphid and endosymbiont protein expression to circulative polerovirus transmission. J Virol 85:2148–2166. doi: 10.1128/JVI.01504-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li S, Xiong R, Wang X, Zhou Y. 2011. Five proteins of Laodelphax striatellus are potentially involved in the interactions between rice stripe virus and vector. PLoS One 6:e26585. doi: 10.1371/journal.pone.0026585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kikkert M, Meurs C, F van de Wetering Dorfmuller S, Peters D, Kormelink R, Goldbach R. 1998. Binding of tomato spotted wilt virus to a 94-kDa thrips protein. Phytopathol 88:63–69. doi: 10.1094/PHYTO.1998.88.1.63. [DOI] [PubMed] [Google Scholar]
- 14.Demler SA, de Zoeten GA, Adam G, Harris KF. 1996. Pea enation mosaic enamovirus: properties and aphid transmission, p 303–344. In Harrison BD, Murant AF (ed), The plant viruses, vol 5: polyhedral virions and bipartite RNA genomes Springer Science and Business Media, New York, NY. [Google Scholar]
- 15.Demler SA, de Zoeten GA. 1991. The nucleotide sequence and luteovirus-like nature of RNA 1 of an aphid non-transmissible strain of pea enation mosaic virus. J Gen Virol 72(Pt 8):1819–1834. doi: 10.1099/0022-1317-72-8-1819. [DOI] [PubMed] [Google Scholar]
- 16.The International Aphid Genomics C. 2010. Genome sequence of the pea aphid, Acyrthosiphon pisum. PLoS Biol 8:e1000313. doi: 10.1371/journal.pbio.1000313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Zoeten GA, Skaf JS. 2001. Pea enation mosaic and the vagaries of a plant virus. Adv Virus Res 57:323–350. doi: 10.1016/S0065-3527(01)57007-4. [DOI] [PubMed] [Google Scholar]
- 18.Cristofoletti PT, de Sousa FA, Rahbe Y, Terra WR. 2006. Characterization of a membrane-bound aminopeptidase purified from Acyrthosiphon pisum midgut cells: a major binding site for toxic mannose lectins. FEBS J 273:5574–5588. doi: 10.1111/j.1742-4658.2006.05547.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li H, Chougule NP, Bonning BC. 2011. Interaction of the Bacillus thuringiensis delta endotoxins Cry1Ac and Cry3Aa with the gut of the pea aphid, Acyrthosiphon pisum (Harris). J Invertebr Pathol 107:69–78. doi: 10.1016/j.jip.2011.02.001. [DOI] [PubMed] [Google Scholar]
- 20.Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. [DOI] [PubMed] [Google Scholar]
- 21.Garczynski SF, Adang MJ. 1995. Bacillus thuringiensis Cry1A(C) delta-endotoxin binding aminopeptidase in the Manduca sexta midgut has a glycosyphosphatidyinositol anchor. Insect Biochem Mol Biol 25:409–415. doi: 10.1016/0965-1748(94)00072-7. [DOI] [Google Scholar]
- 22.Liu S, Sivakumar S, Wang ZH, Bonning BC, Miller W. 2009. The readthrough domain of pea enation mosaic virus coat protein is not essential for virus stability in the hemolymph of the pea aphid. Arch Virol 154:469–479. doi: 10.1007/s00705-009-0327-7. [DOI] [PubMed] [Google Scholar]
- 23.Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bonning BC, Pal N, Liu S, Wang Z, Sivakumar S, Dixon PM, King GF, Miller WA. 2014. Toxin delivery by the coat protein of an aphid-vectored plant virus provides plant resistance to aphids. Nat Biotechnol 32:102–105. doi: 10.1038/nbt.2753. [DOI] [PubMed] [Google Scholar]
- 25.Linz LB. 2013. Molecular interactions between Pea enation mosaic virus and its pea aphid vector. Iowa State University, Ames, IA. [Google Scholar]
- 26.Agrawal N, Malhotra P, Bhatnagar RK. 2002. Interaction of gene-cloned and insect cell-expressed aminopeptidase N of Spodoptera litura with insecticidal crystal protein Cry1C. Appl Environ Microbiol 68:4583–4592. doi: 10.1128/AEM.68.9.4583-4592.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gildow FE, Damsteegt VD, Stone AL, Smith OP, Gray SM. 2000. Virus-vector cell interactions regulating transmission specificity of soybean dwarf luteoviruses. Phytopathol 148:333–342. doi: 10.1111/j.1439-0434.2000.tb04784.x. [DOI] [Google Scholar]
- 28.Reinbold C, Gildow FE, Herrbach E, Ziegler-Graff V, Goncalves MC, JF van den Heuvel Brault V. 2001. Studies on the role of the minor capsid protein in transport of Beet western yellows virus through Myzus persicae. J Gen Virol 82:1995–2007. doi: 10.1099/0022-1317-82-8-1995. [DOI] [PubMed] [Google Scholar]
- 29.Garret A, Kerlan C, Thomas D. 1996. Ultrastructural study of acquisition and retention of potato leafroll luteovirus in the alimentary canal of its aphid vector, Myzus persicae Sulz. Arch Virol 141:1279–1292. doi: 10.1007/BF01718830. [DOI] [PubMed] [Google Scholar]
- 30.Gildow FE, Rochow WF. 1980. Transmission interference between two isolates of barley yellow dwarf virus in Macrosiphum avenae. Phytopathol 70:122–126. doi: 10.1094/Phyto-70-122. [DOI] [Google Scholar]
- 31.Cristofoletti PT, Ribeiro AF, Deraison C, Rahbe Y, Terra WR. 2003. Midgut adaptation and digestive enzyme distribution in a phloem feeding insect, the pea aphid Acyrthosiphon pisum. J Insect Physiol 49:11–24. doi: 10.1016/S0022-1910(02)00222-6. [DOI] [PubMed] [Google Scholar]
- 32.Rahbe Y, Sauvion N, Febvay G, Peumans WJ, Gatehouse AMR. 1995. Toxicity of lectins and processing of ingested proteins in the pea aphid Acyrthosiphon-Pisum. Entomol Exp Appl 76:143–155. doi: 10.1111/j.1570-7458.1995.tb01956.x. [DOI] [Google Scholar]
- 33.Sauvion N, Charles H, Febvay G, Rahbe Y. 2004. Effects of jackbean lectin (ConA) on the feeding behavior and kinetics of intoxication of the pea aphid, Acyrthosiphon pisum. Ent Exp Appl 110:31–44. doi: 10.1111/j.0013-8703.2004.00117.x. [DOI] [Google Scholar]
- 34.Fitches E, Wiles D, Douglas AE, Hinchliffe G, Audsley N, Gatehouse JA. 2008. The insecticidal activity of recombinant garlic lectins toward aphids. Insect Biochem Mol Biol 38:905–915. doi: 10.1016/j.ibmb.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 35.Majumder P, Mondal HA, Das S. 2005. Insecticidal activity of Arum maculatum tuber lectin and its binding to the glycosylated insect gut receptors. J Agric Food Chem 53:6725–6729. doi: 10.1021/jf051155z. [DOI] [PubMed] [Google Scholar]
- 36.Seddas P, Boissinot S. 2006. Glycosylation of beet western yellows virus proteins is implicated in the aphid transmission of the virus. Arch Virol 151:967–984. doi: 10.1007/s00705-005-0669-8. [DOI] [PubMed] [Google Scholar]
- 37.Revollon S, Strub JM, Fitchette AC, Wiss L, Gomord V, Van Dorsselaer A, Brault V. 2010. A reinvestigation provides no evidence for sugar residues on structural proteins of poleroviruses and argues against a role for glycosylation of virus structural proteins in aphid transmission. Virology 402:303–314. doi: 10.1016/j.virol.2010.03.037. [DOI] [PubMed] [Google Scholar]
- 38.Porcar M, Grenier AM, Federici B, Rahbe Y. 2009. Effects of Bacillus thuringiensis delta-endotoxins on the pea aphid. Appl Environ Microbiol 75:4897–4900. doi: 10.1128/AEM.00686-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chougule NP, Bonning BC. 2012. Toxins for transgenic resistance to hemipteran pests, p 405–429. In Toxins, vol 4 MDPI, Basel, Switzerland. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yeager CL, Ashmun RA, Williams RK, Cardellichio CB, Shapiro LH, Look AT, Holmes KV. 1992. Human aminopeptidase N is a receptor for human coronavirus 229E. Nature 357:420–422. doi: 10.1038/357420a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Christiaens O, Smagghe G. 2014. The challenge of RNAi-mediated control of hemipterans. Curr Opin Insect Sci 6:15–21. doi: 10.1016/j.cois.2014.09.012. [DOI] [PubMed] [Google Scholar]
- 42.Grove J, Marsh M. 2011. The cell biology of receptor-mediated virus entry. J Cell Biol 195:1071–1082. doi: 10.1083/jcb.201108131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chay CA, Gunasinge UB, Dinesh-Kumar SP, Miller WA, Gray SM. 1996. Aphid transmission and systemic plant infection determinants of barley yellow dwarf luteovirus-PAV are contained in the coat protein readthrough domain and 17-kDa protein, respectively. Virology 219:57–65. doi: 10.1006/viro.1996.0222. [DOI] [PubMed] [Google Scholar]
- 44.Gildow FE. 1999. Luteovirus transmission and mechanisms regulating vector-specificity, p 25 In Smith HG, Barker H (ed), The Luteoviridae. CAB International, Wallingford, UK. [Google Scholar]
- 45.Rouze-Jouan J, Terradot L, Pasquer F, Tanguy S, Giblot Ducray-Bourdin D. 2001. The passage of Potato leafroll virus through Myzus persicae gut membrane regulates transmission efficiency. J Gen Virol 82:17–23. doi: 10.1099/0022-1317-82-1-17. [DOI] [PubMed] [Google Scholar]
- 46.Brault V, Mutterer J, Scheidecker D, Simonis MT, Herrbach E, Richards K, Ziegler-Graff V. 2000. Effects of point mutations in the readthrough domain of the beet western yellows virus minor capsid protein on virus accumulation in planta and on transmission by aphids. J Virol 74:1140–1148. doi: 10.1128/JVI.74.3.1140-1148.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bruyere A, Brault V, Ziegler-Graff V, Simonis M-T, JF van den Heuvel Richards K, Guilley H, Jonard G, Herrbach E. 1997. Effects of mutations in the beet western yellows virus readthrough protein on its expression and packaging and on virus accumulation, symptoms and aphid transmission. Virology 230:323–334. doi: 10.1006/viro.1997.8476. [DOI] [PubMed] [Google Scholar]
- 48.Brault V, Van den Heuvel JF, Verbeek M, Ziegler-Graff V, Reutenauer A, Herrbach E, Garaud JC, Guilley H, Richards K, Jonard G. 1995. Aphid transmission of beet western yellows luteovirus requires the minor capsid read-through protein P74. EMBO J 14:650–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brown CM, Dinesh-Kumar SP, Miller WA. 1996. Local and distant sequences are required for efficient readthrough of the barley yellow dwarf virus PAV coat protein gene stop codon. J Virol 70:5884–5892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bonning BC, Chougule NP. 2014. Delivery of intrahemocoelic peptides for insect pest management. Trends Biotechnol 32:91–98. doi: 10.1016/j.tibtech.2013.08.001. [DOI] [PubMed] [Google Scholar]