ABSTRACT
The risk of liver cancer in patients infected with the hepatitis B virus (HBV) and their clinical response to interferon alpha therapy vary based on the HBV genotype. The mechanisms underlying these differences in HBV pathogenesis remain unclear. In HepG2 cells transfected with a mutant HBVG2335A expression plasmid that does not transcribe the 2.2-kb doubly spliced RNA (2.2DS-RNA) expressed by wild-type HBV genotype A, the level of HBV pregenomic RNA (pgRNA) was higher than that in cells transfected with an HBV genotype A expression plasmid. By using cotransfection with HBV genotype D and 2.2DS-RNA expression plasmids, we found that a reduction of pgRNA was observed in the cells even in the presence of small amounts of the 2.2DS-RNA plasmid. Moreover, ectopic expression of 2.2DS-RNA in the HBV-producing cell line 1.3ES2 reduced the expression of pgRNA. Further analysis showed that exogenously transcribed 2.2DS-RNA inhibited a reconstituted transcription in vitro. In Huh7 cells ectopically expressing 2.2DS-RNA, RNA immunoprecipitation revealed that 2.2DS-RNA interacted with the TATA-binding protein (TBP) and that nucleotides 432 to 832 of 2.2DS-RNA were required for efficient TBP binding. Immunofluorescence experiments showed that 2.2DS-RNA colocalized with cytoplasmic TBP and the stress granule components, G3BP and poly(A)-binding protein 1 (PABP1), in Huh7 cells. In conclusion, our study reveals that 2.2DS-RNA acts as a repressor of HBV transcription through an interaction with TBP that induces stress granule formation. The expression of 2.2DS-RNA may be one of the viral factors involved in viral replication, which may underlie differences in clinical outcomes of liver disease and responses to interferon alpha therapy between patients infected with different HBV genotypes.
IMPORTANCE Patients infected with certain genotypes of HBV have a lower risk of hepatocellular carcinoma and exhibit a more favorable response to antiviral therapy than patients infected with other HBV genotypes. Using cultured human hepatoma cells as a model of HBV infection, we found that the expression of 2.2DS-RNA caused a decrease in HBV replication. In cultured cells, the ectopic expression of 2.2DS-RNA obviously reduced the intracellular levels of HBV mRNAs. Our analysis of the 2.2DS-RNA-mediated suppression of viral RNA expression showed that 2.2DS-RNA inhibited transcription via binding to the TATA-binding protein and stress granule proteins. Our findings suggest that the 2.2DS-RNA acts as a suppressive noncoding RNA that modulates HBV replication, which may in turn influence the development of chronic hepatitis B.
INTRODUCTION
The hepatitis B virus (HBV) causes acute and chronic liver diseases in humans. Millions of people worldwide suffer from HBV-induced liver disorders, and HBV infection increases the risk of hepatic cirrhosis and hepatocellular carcinoma (1, 2). Type I interferons (IFNs), including IFN-α and IFN-β, exert antiviral activity and important immunomodulatory effects in the innate immune response against HBV infection (3–5). However, the mechanism through which HBV evades the host immune response in chronically infected patients has not been fully elucidated.
In addition to the factors such as viral load and naturally occurring mutants, the HBV genotype, classified as A through J based on genomic sequence, has been shown to be associated with disease progression and responses to IFN-based therapy (6). Previous studies have shown that chronic hepatitis B patients infected with genotype A or B exhibit higher rates of seroclearance of HBV e antigen (HBeAg) and viral DNA in response to IFN-α therapy than patients with genotype C or D infection (7–9) and that chronically infected children with HBV genotype A infection have lower viral DNA loads and exhibit less severe symptoms than patients with genotype D infection (10, 11). Clinical studies have also shown that differences between HBV genotypes correlate with deoxycytidine analog resistance (12) and hepatic pathogenesis (13). However, the biological mechanism underlying these differences in HBV pathogenesis and responsiveness to IFN-α therapy have not been identified.
A diverse set of mRNAs are transcribed by HBV, including a 3.5-kb precore mRNA, a 3.5-kb pregenomic mRNA (pgRNA), 2.4-kb and 2.1-kb (pre-S/L and S) mRNAs, and a 0.9-kb (X) mRNA (14). In addition, numerous spliced mRNA variants derived from the 3.5-kb precore mRNA and pgRNA have been identified in HBV-transfected hepatoma cells, HBV transgenic mice, and the liver tissues and virions isolated from serum of chronic hepatitis B patients (15–19). Recent studies have shown that the expression of proteins encoded by certain HBV spliced transcripts is associated with viral replication, liver fibrosis, and immune surveillance (20–22). Differences in the various species and proportions of spliced RNAs occur between different HBV genotypes (23). A 2.2-kb doubly spliced RNA (2.2DS-RNA) which lacks splice donor and acceptor sites, respectively, at nucleotides 2067 and 2350 and at nucleotides 2447 and 282 is produced by HBV genotypes A and B only (19, 24). Our prior study found that the expression levels of 2.2DS-RNAs in chronic hepatitis B (CHB) patients are very different, with relatively high levels of 2.2DS-RNAs present in a portion of liver biopsy specimens from the CHB patients of genotype A (25). The detailed mechanisms that modulate the level of 2.2DS-RNA in the CHB patients have not yet been elucidated. Furthermore, it is still unclear whether 2.2DS-RNA is biologically relevant to HBV replication and pathogenic differences among HBV genotypes.
Like other environmental stressors, such as heat shock and oxidative stress, viral infection is associated with the rapid disassembly of polysomes and the formation of mRNA-ribonucleoprotein complexes, known as stress granules (SGs) and processing bodies (26). The assembly of SGs modulates mRNA function and metabolism and recruits a diverse group of mRNAs and SG-associated proteins, including the Ras-GTPase-activating protein SH3-domain-binding protein (G3BP), poly(A)-binding protein 1 (PABP1), and T-cell-restricted intracellular antigen 1, which are linked to mRNA silencing or stability (26–28). Previous studies have shown that the cleavage of G3BP by the viral protease suppresses SG formation in poliovirus-infected cells (29) and that G3BP contributes to the replication of the hepatitis C virus genome in cultured cells through interactions with viral nonstructural proteins and the 3′ untranslated regions of viral replication complexes (30). In addition, PABP1 has been shown to colocalize with viral proteins involved in viral replication in hepatitis C virus-infected cells (31). These findings suggest that viral replication modulates SG formation, which contributes to a cellular environment that is favorable for virus production. However, whether SG formation in hepatocytes is altered during HBV infection is unknown.
In our current study, we found that the expression of 2.2DS-RNA reduced the level of HBV pgRNA in human hepatoma cells transfected with a genotype A HBV expression plasmid and that the level of pgRNA was increased when the expression of 2.2DS-RNA was inhibited. The ectopic expression of 2.2DS-RNA also reduced the level of HBV pgRNA in HBV transgenic mice. We found that 2.2DS-RNA acted as a repressor of viral transcription through an interaction with the TATA-binding protein (TBP) and that the formation of the 2.2DS-RNA-TBP complex was involved in SG assembly.
MATERIALS AND METHODS
Cell culture and transfection.
The 1.3ES2 cells have an integrated copy of a 1.3-fold HBV genome and are derived from the HepG2 cell line (32). The human hepatoma cell lines, Huh7 and HepG2, the human embryonic kidney cell line, HEK293, and the mouse embryonic fibroblast cell line, NIH 3T3, were maintained under previously described conditions (33). Transfections were performed using Lipofectamine 2000 transfection reagent (Invitrogen, USA), according to the manufacturer's instructions. The transfection reagent, plasmids, and RNAs were individually diluted in Opti-MEM (Invitrogen). The diluted mixtures were combined and added to the cells in fresh Opti-MEM. The culture medium was replaced with fresh medium after an overnight incubation.
Plasmid construction.
A plasmid expressing HBV with a G2335A mutation (HBVG2335A) plasmid was generated from an HBV genotype A plasmid (34) by site-directed mutagenesis, as described previously (25). The mutation in HBVG2335A causes a nucleotide substitution at position 2335 within the polypyrimidine tract of a splice acceptor site, which alters the splicing pattern of genotype A to that of genotype D, rendering it 2.2DS-RNA deficient. The p1.3HBcl/Hyg plasmid, which contains a 1.3-fold HBV genome of genotype D (32), was used to study wild-type HBV replication. The cDNAs of the 2.2DS-RNA (24), human beta-actin (β-actin) RNA, β-galactosidase (β-Gal) RNA, and the truncated variants of 2.2DS-RNA (ΔDS-1373, ΔDS-832, ΔDS-632, and ΔDS-432) (see Fig. 4G for a schematic of the truncations) were subcloned into the pShuttle/R vector (Clontech, USA), from which transcription in mammalian cells is driven by the cytomegalovirus immediate early (CMVie) promoter. For the HBV promoter analysis, the cDNAs of the core protein promoter (nucleotides 1636 to 1851), X promoter (nucleotides 1177 to 1376), I/X enhancer and core promoter (EnhI/C, nucleotides 947 to 1851), pre-S1 promoter (nucleotides 2710 to 2800), and pre-S2/S promoter (nucleotides 2960 to 3180) were subcloned into the pGL3-Basic luciferase expression plasmid (Promega, USA) using the MluI and HindIII restriction sites (35). The cDNAs of the simian virus 40 (SV40) early enhancer/promoter and the herpes simplex virus thymidine kinase (TK) promoter were also cloned into the pGL3-Basic expression plasmid to investigate the effects of 2.2DS-RNA on the transcriptional activity of non-HBV viral promoters. The reporter plasmids used to examine transcription mediated by RNA polymerase I (RNAP-I) and RNAP-II used the rRNA promoter and the hsp70 minimal (TATA) promoter, respectively, to drive luciferase expression (36).
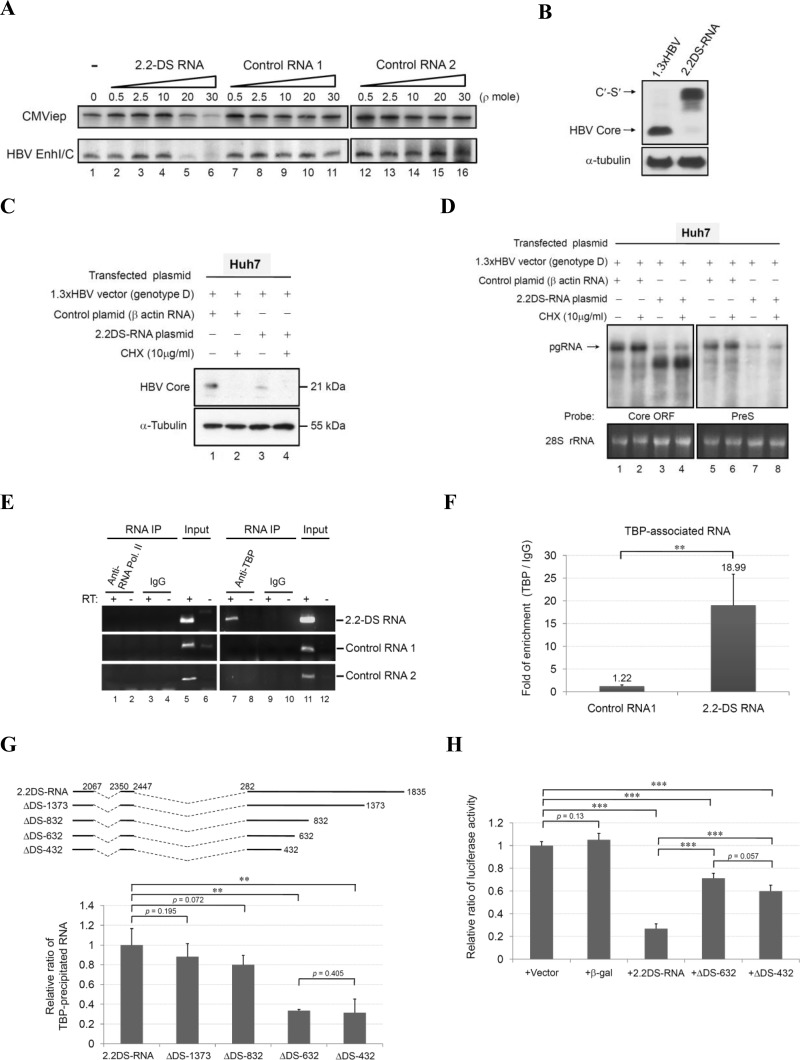
FIG 4.
The binding of HBV 2.2DS-RNA to TBP suppresses the transcriptional activity of the CMVie and HBV EnhI/C promoters. (A) Suppression of transcription by 2.2DS-RNA in vitro. Reporter DNA templates containing the CMVie promoter or the EnhI/C promoter were added to nuclear extracts, and transcription was quantified in the absence (lane 1) or presence of 2.2DS-RNA (lanes 2 to 6), negative-control RNA 1 (Renilla luciferase RNA; lanes 7 to 11), or negative-control RNA 2 (β-actin RNA; lanes 12 to 16). (B) Expression of the C′-S′ fusion protein in Huh7 cells transfected with 2.2DS-RNA expression plasmid. The cell extracts at 3 days were analyzed by Western blotting to detect the HBV core and cellular α-tubulin proteins. (C) Huh7 cells were cotransfected with the HBV genotype D expression plasmid and the control RNA or 2.2DS-RNA expression plasmid for 6 h before treatment with 10 μg/ml cycloheximide (CHX) for 2 days. The cell extracts were analyzed by Western blotting to detect the HBV core protein and cellular α-tubulin protein. (D) Total RNA was analyzed by Northern blotting using the core ORF and pre-S radiolabeled probes. The β-actin expression plasmid was used as a negative control, and the α-tubulin protein and 28S rRNA were used as loading controls for Western and Northern blotting, respectively. (E and F) The interaction between 2.2DS-RNA and TBP in Huh7 cells. The cells were transfected with plasmids that expressed the 2.2DS-RNA, negative-control RNA 1 (Renilla luciferase RNA), and negative-control RNA 2 (β-Gal RNA), and the cell lysates were subjected to immunoprecipitation (IP) using an anti-RNAP-II or anti-TBP antibody (E). The precipitated RNAs were analyzed by conventional RT-PCR using sequence-specific primer sets complementary to the RNAs indicated, and the relative amounts of TBP-associated RNA were quantified using qRT-PCR (F). (G) The cells were transfected with plasmids that expressed the truncated mutants or full-length 2.2DS-RNA, and the cell lysates were analyzed by RIP assay, as described above. Relative enrichment is reported as the relative ratios ± standard deviations of two independent experiments. (H) The cells cotransfected with the EnhI/C reporter plasmid and the indicated plasmid were prepared for the analysis of EnhI/C activity. The results are reported as the relative ratios ± standard deviations of three independent experiments (**, P < 0.01; ***, P < 0.001).
Northern blot analysis.
Total RNA was extracted from cells transfected with the 2.2DS-RNA expression plasmid and the liver tissue of HBV transgenic mice injected with the 2.2DS-RNA expression plasmid using TRIzol reagent (Invitrogen). Following electrophoresis in a formaldehyde-denaturing agarose gel, the resolved RNAs were transferred to a nylon membrane and subjected to UV-induced cross-linking. The membrane-bound RNA was probed using a radiolabeled oligonucleotide complementary to sequences in the core mRNA (nucleotides 1903 to 2449), the X mRNA (nucleotides 1374 to 1835), or the pre-S mRNA (nucleotides 2848 to 222) of HBV. The radioisotope-labeled probes were generated using Klenow fragment DNA polymerase (Pol) (Promega) and [α-32P]dCTP (PerkinElmer, USA), according to the manufacturer's instructions.
In vitro transcription.
We examined the effects of the exogenously transcribed 2.2DS-RNA on transcription in vitro using a HeLaScribe Nuclear Extract In Vitro Transcription System (Promega). Recombinant 2.2DS-RNA was transcribed in vitro using a T7 RiboMAX Express Large Scale RNA Production System (Promega), according to the manufacturer's instructions. The EnhI/C reporter DNA templates were generated by subcloning the EnhI/C promoter into the control DNA template (CMVie promoter) provided in the HeLaScribe kit. The DNA templates were linearized and added to a solution containing the nuclear extract and 10 μCi of 3,000 Ci/mmol [α-32P]UTP (PerkinElmer), according to the instructions for the HeLaScribe kit. After incubation at 30°C for 60 min, the DNA template was digested using RNase-free DNase I (Promega), and the reaction was terminated by the addition of stop solution. The transcripts were isolated by phenol-chloroform extraction and subjected to electrophoresis.
Western blot analysis.
Samples were homogenized in radioimmunoprecipitation assay (RIPA) buffer containing 1% phenylmethylsulfonyl fluoride (PMSF), and the concentration of total protein in the soluble fraction was quantified using a Pierce bicinchoninic acid (BCA) protein assay kit (Thermo Scientific, USA). The proteins were subjected to SDS-PAGE, and the resolved protein bands were transferred to a polyvinylidene difluoride membrane. The membrane was probed using a rabbit anti-HBV core antibody (Dako, USA) or mouse anti-α-tubulin antibody (GeneTex, USA). Primary antibody reactivity was visualized using a horseradish peroxidase-conjugated secondary antibody (Jackson Laboratory, USA).
Quantification of HBeAg and HBsAg.
The levels of HBV surface antigen (HBsAg) and HBeAg in the cell culture medium were measured using an enzyme-linked immunosorbent assay (ELISA) kit (General Biological, Taiwan) and a formazan substrate, according to the manufacturer's instructions.
Recombinant adenovirus production and infection in cell culture.
The cDNA of the 2.2DS-RNA was subcloned into the pAdeno-X plasmid (Clontech), in which expression is controlled using a doxycycline-inducible promoter. Adeno-X 293 cells (Clontech) were transfected with the recombinant pAdeno-X plasmid to propagate the recombinant Ad2.2-DS adenovirus, according to the manufacturer's instructions. The cells were harvested by centrifugation and lysed using a freeze-thaw method. The Ad2.2-DS virus was purified by CsCl ultracentrifugation. The 1.3ES2 cells were infected with Ad2.2-DS at a multiplicity of infection of 50. At 2 h postinfection, the virus-containing medium was removed, and the virus-infected cells were washed with phosphate-buffered saline (PBS). The cells were incubated in fresh, virus-free culture medium containing 100 ng/ml doxycycline to induce ectopic 2.2DS-RNA expression.
Luciferase reporter assay.
The Huh7 cells were cotransfected with the 2.2DS-RNA expression plasmid and a luciferase reporter plasmid. On day 2 posttransfection, the cells were lysed using Glo Lysis Buffer (Promega). The luminescence of the cell lysates was analyzed immediately in a luminometer using a Bright-Glo luciferase assay system (Promega), according to the manufacturer's instructions.
RIP assay.
Huh7 cells were transfected with the 2.2DS-RNA expression plasmid or a control RNA expression plasmid, and RNA was isolated from the cells using an EZ-Magna RNA immunoprecipitation (RIP) RNA-binding protein immunoprecipitation kit (EMD Millipore, USA). The RIP was performed using a mouse anti-RNAP-II antibody (EMD Millipore) or a mouse anti-TBP antibody (EMD Millipore). The RNAs in the precipitates were analyzed using reverse transcription and conventional PCR (RT-PCR) and quantitative reverse transcription and real-time PCR (qRT-PCR).
RT-PCR and qRT-PCR.
Using total RNA or the precipitated RNAs obtained in the RIP assays as templates, cDNAs were synthesized by reverse transcription using Superscript II reverse transcriptase (Invitrogen) and oligo(dT) primer, according to the manufacturer's instructions. Conventional PCR was performed using DreamTaq DNA polymerase (Thermo Scientific) and the following primer sets: HBV1906-1926 (5′-CATTGACCCGTATAAAGAATT-3′) and HBV310-331 (5′-ATTGGAGGTTGGGGACTGCGA-3′); HBV2097-2114 (5′-GACTCTAGCTACCTGGGT-3′) and HBV610-631 (5′-TGCGAAAGCCCAGGACGATGG-3′); 2.2DS-RNA forward (5′-ATGCAACTTTTTCACCTCTGC-3′) and 2.2DS-RNA reverse (5′-ATTGAGATCCCCGAGATTGAG-3′); control RNA 1 forward (5′-TTGAACCATTCAAAGAGAAAG-3′) and control RNA 1 reverse (5′-TTATTGTTCATTTTTGAGAACTCG-3′); and control RNA 2 forward (5′-GCCAGCTGGCGCAGGTAGCAG-3′) and control RNA 2 reverse (5′-ATTGAGATCCCCGAGATTGAG-3′). The antibody-precipitated RNAs in the RIP assay were quantified using qRT-PCR. Quantitative analysis was performed using TaqMan master mix (Roche, USA), a Universal Probe Library (Roche), and the following primers in each reaction mixture: Q-2.2DS-RNA forward primer, 5′-GTCCTACTGTTCAAGCCTCCA-3′; Q-2.2DS-RNA reverse primer, 5′-ACGGGTCAATGTCCATGC-3′; Q-Control RNA 1 forward primer, 5′-CCAGGATTCTTTTCCAATGC-3′; and Q-Control RNA 1 reverse primer, 5′-CTTGCGAAAAATGAAGACCTTT-3′. The fold enrichment of the antibody-precipitated RNAs was quantified by using the comparative threshold cycle (CT) method (TBP/IgG).
FISH.
The fluorescence in situ hybridization (FISH) analysis was performed, as described previously (37, 38), with slight modification. The Huh7 cells were transfected with the expression plasmids for 24 h and fixed in 4% paraformaldehyde, followed by an overnight incubation in 70% ethanol at 4°C. The fixed cells were permeabilized and incubated in hybridization buffer containing 4× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 25% formamide, 3× Denhardt's solution, 2% RNase-free bovine serum albumin, 250 ng/ml salmon sperm DNA, 250 ng/ml yeast tRNA, 1 mM vanadyl ribonucleoside complex, and 10% dextran sulfate at room temperature for 30 min. The cells were probed using a digoxigenin (DIG)-labeled oligonucleotide (Integrated DNA Technologies, USA) complementary to nucleotides 1967 to 1988 of HBV (5′-TCTCGAATAGAAGGAAAGAAGT-DIG) at 23°C below the melting temperature of the probe. The labeled RNA was detected using a sheep anti-DIG antibody (Thermo Scientific), and TBP was detected using a rabbit anti-TBP antibody (Thermo Scientific). A mouse anti-G3BP antibody (Abcam, USA) was used for G3BP detection, and PABP1 was detected using a mouse anti-PABP1 antibody (Santa Cruz Biotechnology, USA). After cells were washed with 1× PBS, they were incubated with donkey anti-sheep Alexa Fluor 594, goat anti-rabbit Alexa Fluor 488, and goat anti-mouse Alexa Fluor 647 secondary antibodies (Invitrogen) for 1 h. After cells were washed with 1× PBS, they were immobilized in mounting medium (Vector Laboratories, USA) supplemented with 4′-6-diamidino-2-phenylindole (DAPI) for nuclear staining. The cells were examined using a Leica TCS SP5-II confocal fluorescence microscope.
RESULTS
2.2DS-RNA suppresses HBV transcription in vitro and in vivo.
To examine the effects of 2.2DS-RNA on HBV replication, we first investigated the expression of 2.2DS-RNA on HBV transcription in HepG2 cells. The cells were transfected with a plasmid encoding the genome of HBV genotype A, from which the 2.2DS-RNA is transcribed. We also transfected cells with a plasmid encoding the mutant HBVG2335A genome, which is 2.2DS-RNA deficient (25). The results of the Northern blotting of the cells transfected with the HBV genotype A expression plasmid showed that the level of pgRNA was lower than that in the cells transfected with the HBVG2335A expression plasmid (Fig. 1A and B). The results of the Western blotting showed that the level of the HBV core protein in the cells transfected with the wild-type HBV genotype A expression plasmid was lower than that in the cells transfected with the HBVG2335A plasmid.
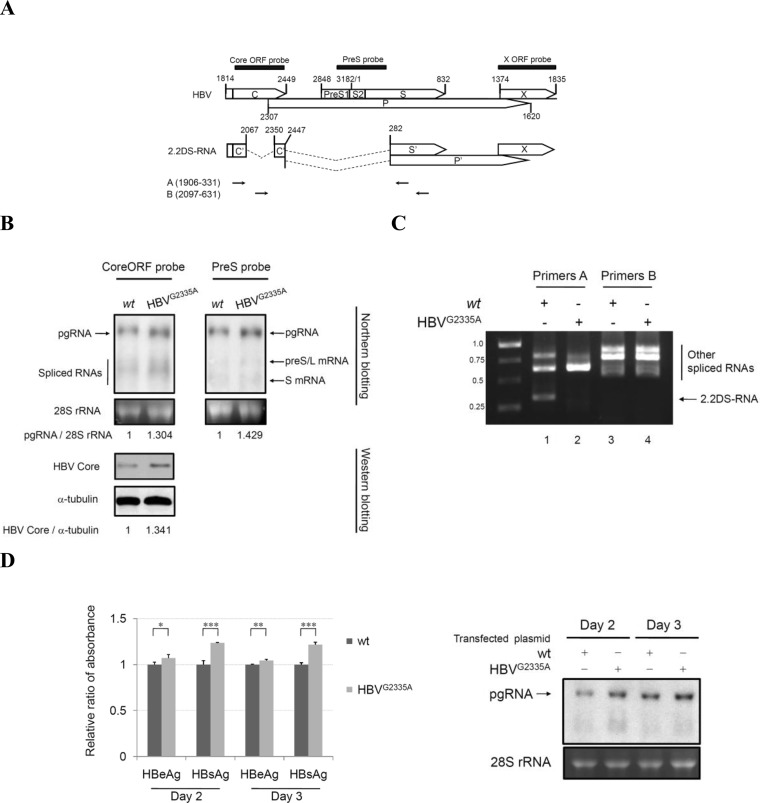
FIG 1.
The HBV 2.2DS-RNA suppresses HBV pgRNA expression in HepG2 cells. (A) Diagrammatic representation of the 2.2DS-RNA splicing variant derived from the HBV precore mRNA and pgRNA. The numbers correspond to the nucleotide positions in the wild-type HBV genome, in which the EcoRI cleavage site is defined as nucleotide position 1. The capital letters within the open boxes indicate the conventional ORFs of HBV, whereas the prime symbol (′) indicates truncated ORFs in the 2.2DS-RNA. The filled boxes represent complementary regions of the specific HBV probes used for the Northern blot analysis. The arrows indicate the primer annealing sites used for the RT-PCR amplification. (B) HepG2 cells were transfected with the HBV genotype A (wild type, wt) or HBVG2335A expression plasmid. Total RNA was analyzed by Northern blotting using two radiolabeled probes complementary to the core protein ORF or pre-S sequences. Cell extracts were analyzed by immunoblotting using an anti-HBV core antibody. The 28S rRNA and α-tubulin protein were used as loading controls for Northern and Western blotting, respectively. (C) Total RNA was subjected to RT-PCR using the HBV-specific primers A and B, and the cDNAs were subjected to electrophoresis for Northern blotting using the core ORF probe. The arrow indicates the cDNA of 2.2DS-RNA (approximately 0.3 kb). (D) The levels of HBeAg and HBsAg in the cultured medium were analyzed using an ELISA, and total RNA was analyzed by Northern blotting using the core ORF probe. The levels of HBeAg and HBsAg are reported as the relative ratios ± standard deviations of three replicates in one representative experiment (*, P < 0.05; **, P < 0.001; ***, P < 0.001).
To confirm that 2.2DS-RNA was not expressed in the cells transfected with the HBVG2335A expression plasmid, we compared the spliced RNA profile of these cells with that of the cells transfected with the wild-type HBV genotype A plasmid using RT-PCR. Primer sets A (HBV1906-1926/HBV310-331) and B (HBV2097-2114/HBV610-631) were used to detect the spliced RNAs. We found that 2.2DS-RNA was absent in the cells transfected with the HBVG2335A plasmid (Fig. 1C). We also compared viral replication in cells transfected with the HBV genotype A plasmid with that of cells transfected with the HBVG2335A plasmid by measuring the levels of HBeAg and HBsAg, which are used as serological indicators of chronic hepatitis B (39), as well as the level of pgRNA. As shown in Fig. 1D, the levels of HBeAg and HBsAg were higher in the cells transfected with the HBVG2335A plasmid than in the cells transfected with the wild-type HBV genotype A expression plasmid. Northern blot analysis also showed that the level of pgRNA was higher in the cells transfected with the HBVG2335A plasmid (Fig. 1D).
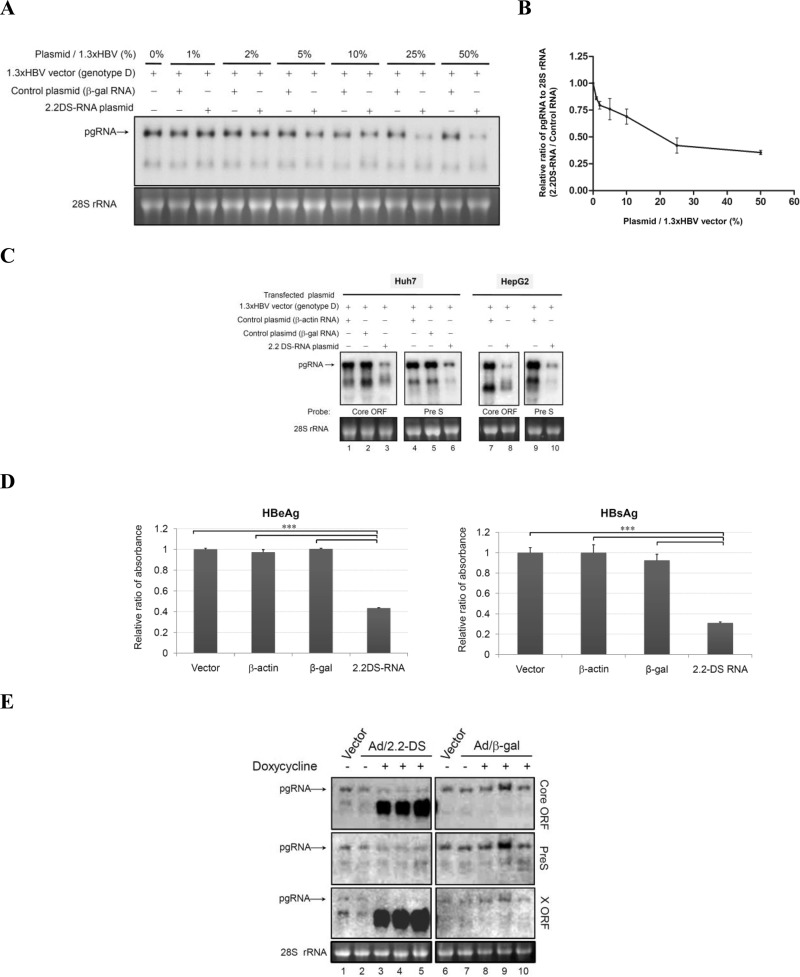
In gain-of-function experiments, we cotransfected HepG2 cells with various amounts of the 2.2DS-RNA expression plasmid and the HBV genotype D expression vector, which is 2.2DS-RNA deficient. We found that a reduction of pgRNA was observed even in the presence of a small amount (1 to 2%) of the 2.2DS-RNA plasmid compared with the amount of pgRNA in the presence of the control plasmid (Fig. 2A). Further statistical analysis showed that the level of pgRNA was suppressed by 2.2DS-RNA in a dose-dependent manner (Fig. 2B). Further experiment showed that the levels of the pgRNA and subgenomic RNAs in the Huh7 cells were reduced to 40% and 25% of those in the control cells with a β-actin expression plasmid, respectively (Fig. 2C). The levels of pgRNA and the subgenomic RNAs in the HepG2 cells were also lower than those in the control cells (Fig. 2C, lanes 7 to 10). We also investigated whether 2.2DS-RNA expression suppresses the secretion of HBeAg and HBsAg. In the Huh7 cells transfected with the 2.2DS-RNA expression plasmid, the levels of HBeAg and HBsAg were reduced to 40% and 30% of those in the vector control cells, respectively (Fig. 2D). To investigate the effect of 2.2DS-RNA expression on HBV replication, we used a doxycycline-inducible adenovirus expression system to drive the ectopic expression of 2.2DS-RNA in 1.3ES2 cells, which have a chromosomally integrated copy of the viral genome. The level of pgRNA transcribed from the integrated HBV genome in the cells in which 2.2DS-RNA was ectopically expressed was lower (Fig. 2E, lanes 3 to 5) than that in the cells expressing the control RNA (Fig. 2E, lanes 8 to 10). These results collectively showed that 2.2DS-RNA suppressed the expression of pgRNA in the HepG2, Huh7, and 1.3ES2 cells expressing HBV genotype D mRNAs.
FIG 2.
The HBV 2.2DS-RNA suppresses viral mRNA expression in cultured hepatoma cells. (A) The dosage effect of 2.2DS-RNA in modulation of HBV pgRNA expression. HepG2 cells were cotransfected with 5 μg of the HBV genotype D and various amounts of the 2.2DS-RNA expression plasmids (1% to 50% of 2.2DS-RNA plasmid to 1.3× HBV vector). Total RNA was extracted on day 3 posttransfection and analyzed by Northern blotting using the core ORF probe. The β-Gal expression plasmid expressed the negative control RNA, and the 28S rRNA was used as a loading control. (B) The ratios of the pgRNA to 28S rRNA band intensities are shown. The results are reported as relative ratios (2.2DS-RNA to control RNA) ± standard deviations of two representative experiments. (C) 2.2DS-RNA expression suppresses the level of HBV mRNAs in Huh7 and HepG2 cells. Total RNA was analyzed by Northern blotting using various probes, as indicated. The β-actin expression plasmid expressed the negative control RNA. (D) The levels of HBeAg and HBsAg in the culture medium were analyzed using an ELISA. The results are reported as relative ratios ± standard deviations of three replicates in one representative experiment (***, P < 0.001). (E) 1.3ES2 cells were simultaneously infected with the Tet transactivator-expressing adenovirus and the adenovirus that included the Tet-regulated control (Vector; lanes 1 and 6, respectively), the Tet-regulated 2.2DS-RNA (Ad/2.2-DS; lanes 2 to 5), or the Tet-regulated β-Gal mRNA adenovirus (Ad/β-gal; lanes 7 to 10) using a multiplicity of infection of 50. The expression of 2.2DS-RNA was induced using 100 ng/ml doxycycline for 2 days, and total RNA was analyzed by Northern blotting using a radiolabeled probe complementary to the core protein ORF, pre-S, or the X protein ORF. The adenovirus expressing the Tet-regulated β-Gal vector was used as a negative RNA control.
2.2DS-RNA-induced suppression of viral and cellular promoter activity in vitro.
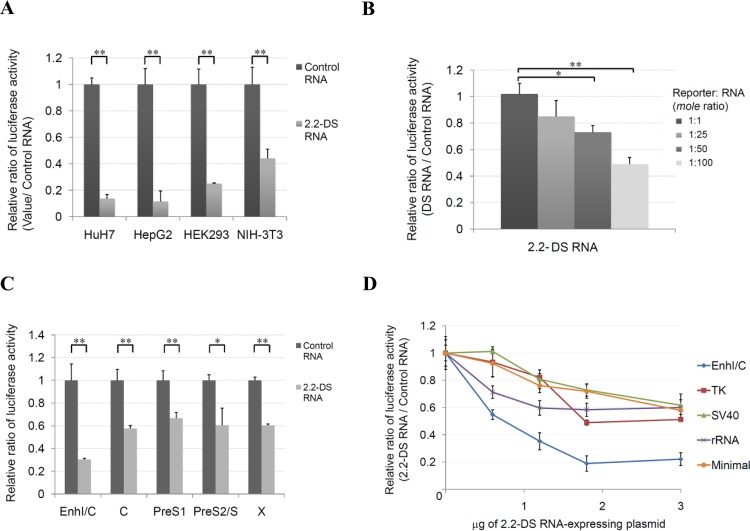
We hypothesized that 2.2DS-RNA repressed the activity of the HBV promoter responsible for transcribing the pgRNA. To test our hypothesis, we transfected Huh7 and HepG2 cells and the nonhepatoma cell lines, HEK293 and NIH 3T3, with a luciferase reporter plasmid which used the HBV EnhI/C promoter to drive luciferase expression in the presence or absence of 2.2DS-RNA expression. We found that the transcriptional activity of the EnhI/C promoter in the cells cotransfected with the 2.2DS-RNA expression plasmid was substantially lower than that in the cells cotransfected with the control RNA plasmid, regardless of cell type (Fig. 3A). These results showed that the 2.2DS-RNA-induced suppression of pgRNA expression was not tissue specific.
FIG 3.
The HBV 2.2DS-RNA suppresses the transcriptional activity of viral and cellular promoters in Huh7 cells. (A) The cells were cotransfected with the EnhI/C reporter plasmid and the control plasmid or the 2.2DS-RNA expression plasmid. Total cell lysates were prepared for the analysis of EnhI/C activity on day 2 posttransfection. (B) Huh7 cells were cotransfected with a 1:1, 1:25, 1:50, or 1:100 molar ratio of the EnhI/C reporter and the exogenously recombinant 2.2DS-RNA. (C) Suppression of the transcriptional activity of HBV promoters. Cells were cotransfected with the control plasmid or the 2.2DS-RNA expression plasmid and a reporter plasmid containing one of the following HBV promoters: EnhI/C, C (basal core promoter), pre-S1, pre-S2, or X. (D) The cells were cotransfected with 0, 0.15, 0.6, 1.2, 1.8, or 3 μg of the control or 2.2DS-RNA expression plasmid and a reporter plasmid containing one of the following promoters: HBV EnhI/C, herpes simplex virus thymidine kinase (TK), simian virus 40 early enhancer/promoter (SV40), rRNA, or the minimal promoter. Total cell lysates were prepared for the analysis of promoter activity on day 2 posttransfection. If necessary, vector DNA and/or tRNA was added to make the total amounts of RNA and DNA equivalent to those used in the transfections described in panel B or D above. The results show the relative ratios ± standard deviations of three independent experiments (**, P < 0.01; *, P < 0.05).
To exclude the possibility that the 2.2DS-RNA-induced suppression of pgRNA expression was related to the use of the CMVie promoter for the ectopic expression of 2.2DS-RNA, we transfected Huh7 cells with the luciferase reporter plasmid and various amounts of exogenously transcribed 2.2DS-RNA. We found that the transcriptional activity of the EnhI/C promoter was suppressed by the exogenous 2.2DS-RNA in a dose-dependent manner (Fig. 3B). We further investigated whether the promoters responsible for transcribing HBV mRNAs were also affected by 2.2DS-RNA expression. The results showed that the transcriptional activities of the core, X, EnhI/C, pre-S1, and pre-S2/S promoters were suppressed by 2.2DS-RNA, with the EnhI/C promoter demonstrating greatest sensitivity to the inhibitory effects of 2.2DS-RNA (Fig. 3C). High-level 2.2DS-RNA expression reduced the transcriptional activity of the EnhI/C promoter to approximately 22% of that observed in the absence of 2.2DS-RNA. These results showed that 2.2DS-RNA expression correlated with global suppression of HBV transcription in Huh7 cells.
To investigate the effects of 2.2DS-RNA on the transcriptional activity of non-HBV promoters, we performed luciferase reporter assays in Huh7 cells using the SV40, TK, rRNA, and hsp70 minimal promoters. We found that the expression of 2.2DS-RNA suppressed the transcriptional activity of all of these non-HBV promoters in a dose-dependent manner (Fig. 3D), reducing their activities to approximately 60% of those observed in the absence of 2.2DS-RNA. These results showed that 2.2DS-RNA functioned as a nonspecific suppressor of transcriptional activity.
2.2DS-RNA suppresses transcription in vitro.
Previous studies have shown that certain host and viral noncoding RNAs (ncRNAs) modulate RNAP-II-mediated transcription in trans by targeting RNAP-II or associated transcription factors (40, 41). We investigated whether 2.2DS-RNA suppresses HBV transcription through a similar mechanism in which it interacts with essential transcription factors. We performed an in vitro analysis of the transcriptional activities of the CMVie and HBV EnhI/C promoters in the presence of exogenously transcribed 2.2DS-RNA, and we observed an obvious reduction in the transcriptional activity of both promoters in the presence of 20 to 30 pmol of the 2.2DS-RNA (Fig. 4A, lanes 5 and 6) compared with that in the presence of the control RNAs (Fig. 4A, lanes 10, 11, 15, and 16). These results showed that 2.2DS-RNA might function as an ncRNA that suppressed the transcriptional activity of both HBV and non-HBV promoters in vitro.
As shown in Fig. 1A, the 2.2DS-RNA contains three potential open reading frames (ORFs) that could encode a fusion C′-S′ protein, a truncated polymerase, and the X protein. Western blotting showed that the C′-S′ proteins were expressed in Huh7 cells transfected with the 2.2DS-RNA expression plasmid (Fig. 4B). To determine whether these proteins are required for the 2.2DS-RNA-induced suppression of HBV pgRNA expression, we examined the effect of 2.2DS-RNA on viral transcription in cells transfected with the HBV genotype D expression plasmid and treated with cycloheximide. Western blotting showed that the de novo synthesis of the plasmid-encoded HBV core protein was completely arrested by cycloheximide (Fig. 4C), whereas the Northern blotting showed that the inhibition of the de novo expression of viral proteins had no valid effect on the 2.2DS-RNA-induced suppression of HBV mRNA expression (Fig. 4D, lanes 4 and 8) compared with that in the cells cotransfected with the control RNA expression plasmid (Fig. 4D, lanes 2 and 6). The results suggested that the 2.2DS-RNA-induced suppression of HBV transcription might be independent of the 2.2DS-RNA-encoded proteins and might involve the inhibition of essential transcription factors.
Binding of TBP to 2.2DS-RNA initiates accumulation of cytoplasmic TBP.
Both RNAP-II and TBP are crucial components of the basal transcription machinery (42, 43). To gain further insight into the mechanism through which cellular transcription factors may be modulated by 2.2DS-RNA, we immunoprecipitated RNAP-II and TBP in Huh7 cells transfected with a 2.2DS-RNA expression plasmid and analyzed the immunoprecipitates using conventional RT-PCR and qRT-PCR. The data showed that neither 2.2DS-RNA nor the control RNAs were associated with RNAP-II (Fig. 4E), and the control RNAs were essentially undetectable in the anti-TBP immunoprecipitates. However, we found that a specific association occurred between the 2.2DS-RNA and TBP (Fig. 4E, lane 7). Using qRT-PCR, we determined that the amount of TBP associated with 2.2DS-RNA was approximately 15.6-fold higher (18.99 ± 6.86) than that associated with the control RNA 1 (1.22 ± 0.3) (Fig. 4F).
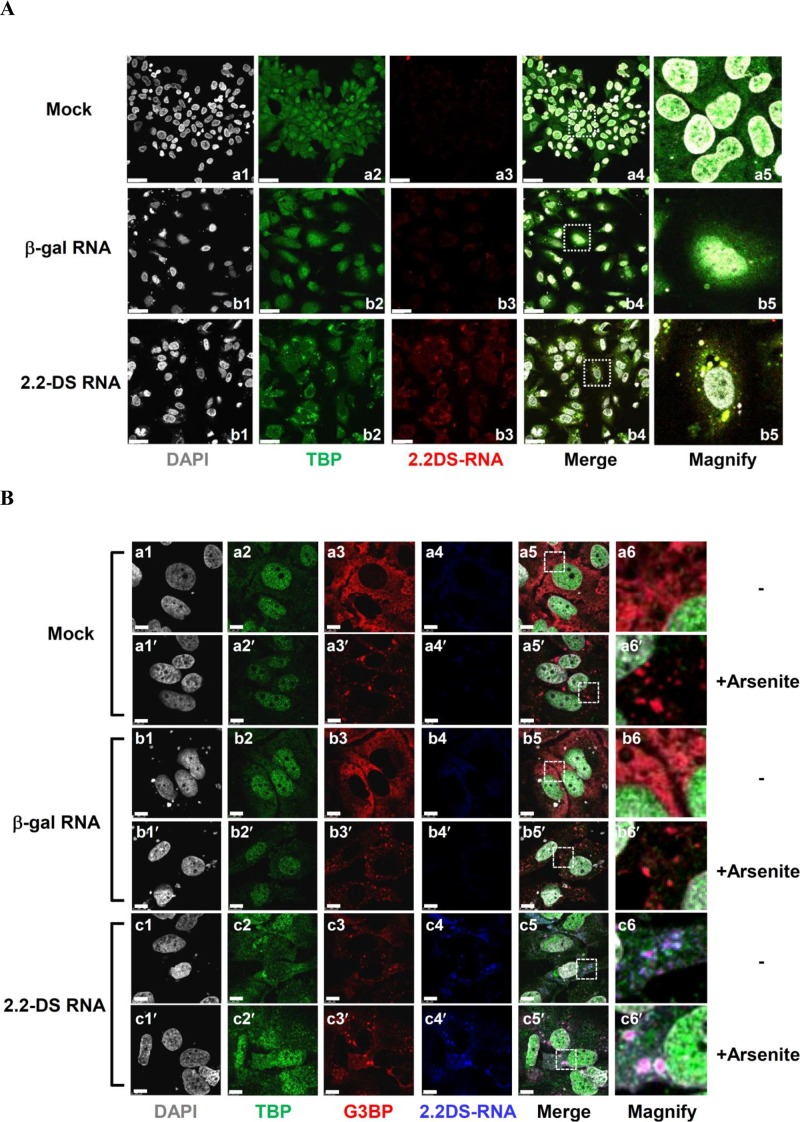
The 2.2DS-RNA has deletions in the central and 3′ terminal regions of the core ORF, the internal region of the Pol ORF, or within the pre-S/S region in HBV-infected cells (19, 24). To delimit the region within 2.2DS-RNA that is required for TBP binding, we constructed four truncated versions of 2.2DS-RNA, designated ΔDS-1373, ΔDS-832, ΔDS-632, and ΔDS-432 (Fig. 4G), in which progressively larger deletions were made at the 3′ terminus of the 2.2DS-RNA, and used them in RIP assays in which TBP was precipitated. As shown in Fig. 4G, similar levels of ΔDS-1373, ΔDS-832, and 2.2DS-RNA were bound by TBP (0.88 ± 0.13, 0.80 ± 0.09, and 1.00 ± 0.16, respectively), whereas the relative levels of ΔDS-632 (0.34 ± 0.01) and ΔDS-432 (0.32 ± 0.17) bound by TBP were lower than the level of 2.2DS-RNA. When ΔDS-632 and ΔDS-432 were individually tested for the ability to repress transcriptional activity of the HBV EnhI/C promoter, we found that the levels of luciferase activity were higher in the cells transfected with the ΔDS-632 (0.71 ± 0.04) and ΔDS-432 (0.60 ± 0.05) expression plasmids than the level in cells transfected with the 2.2DS-RNA expression plasmid (0.27 ± 0.04) (Fig. 4H). The results showed that TBP bound 2.2DS-RNA in Huh7 cells and that sequences within nucleotide positions 432 to 832 of 2.2DS-RNA were involved in TBP binding. Moreover, these results suggested that the binding of 2.2DS-RNA to TBP contributed to the transcriptional repression of the HBV EnhI/C promoter. We performed a FISH analysis to determine the cellular location at which 2.2DS-RNA interacts with TBP. In the cells transfected with the control RNA expression plasmid and the mock-transfected cells, TBP was detected throughout the cell, but the most intense fluorescent staining of TBP occurred in the nucleus (Fig. 5A). In the cells transfected with the 2.2DS-RNA expression plasmid, diffuse TBP staining was observed in the nucleus, but intensely fluorescent TBP foci were also observed in the cytoplasm (Fig. 5A, bottom panel). These results collectively indicated that a specific interaction occurred between 2.2DS-RNA and TBP in the cytoplasm, which resulted in the accumulation of cytoplasmic 2.2DS-RNA-TBP complexes.
FIG 5.
Cytoplasmic 2.2DS-RNA-TBP complexes colocalize with SG proteins. (A) The HBV 2.2DS-RNA induces the formation of cytoplasmic TBP foci in Huh7 cells. The cells were transfected with the control (empty) vector (mock), the β-Gal expression plasmid (negative-control RNA), or the 2.2DS-RNA expression plasmid. The nuclei were stained using DAPI (gray). An anti-TBP primary antibody was used to detect TBP (green). The 2.2DS-RNA was visualized by hybridization with a DIG-labeled probe and immunostaining using an anti-DIG primary antibody (red). The colocalization of cytoplasmic TBP and 2.2DS-RNA appears as yellow foci in the merged panels (scale bar, 25 μm). (B) Colocalization of TBP-2.2DS-RNA complexes with SG proteins. The cells were transfected as described above and treated with 500 μM arsenite (+Arsenite) or vehicle control (−) at 37°C for 30 min. The cellular distribution of G3BP was examined using an anti-G3BP antibody (red). The nuclei (gray) and TBP (green) were detected, as described above, but 2.2DS-RNA was detected based on blue fluorescence. The colocalization of 2.2DS-RNA, TBP, and G3BP is visible as white foci in the merged panels. The boxed regions in the merged images were magnified to highlight the results.
Binding of 2.2DS-RNA by TBP coincides with SG assembly.
Viral infection is associated with the formation of cytoplasmic ribonucleoprotein complexes, known as SGs (26). To determine whether cytoplasmic 2.2DS-RNA-TBP complexes interact with components of SGs, we investigated whether the 2.2DS-RNA-TBP complexes associated with G3BP, a known SG component (30), using FISH. We found that intensely fluorescent 2.2DS-RNA foci colocalized with intensely fluorescent G3BP foci in the cytoplasm of Huh7 cells (Fig. 5B, frame c5). A portion of the cytoplasmic G3BP foci also colocalized with intensely fluorescent TBP foci in the cells transfected with the 2.2DS-RNA expression plasmid, whereas G3BP and TBP did not colocalize in the cells transfected with the control RNA expression plasmid or the mock-transfected cells (Fig. 5B, frames a5 and b5).
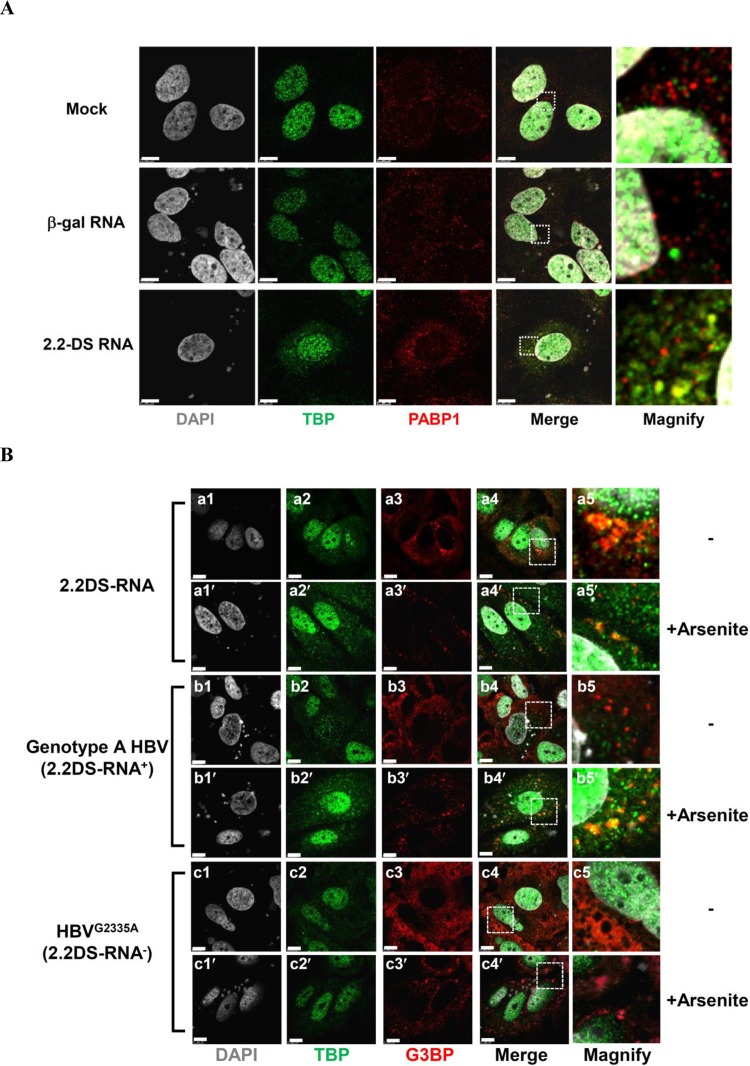
Arsenite inhibits translation and activates multiple stress signaling pathways, inducing the formation of large stress granules in the cytoplasm of cells (44). Therefore, we investigated the effect of arsenite on the colocalization of SG proteins and TBP in the presence of 2.2DS-RNA. Arsenite treatment caused the assembly of SGs in Huh7 cells, as evidenced by the formation of numerous intensely fluorescent G3BP foci in the cytoplasm, a small portion of which colocalized with TBP in the cells transfected with the 2.2DS-RNA expression plasmid (Fig. 5B, frame c5′). In contrast, arsenite treatment did not cause TBP to colocalize with G3BP foci in the cytoplasm of cells transfected with the control RNA expression plasmid or the mock-transfected cells (Fig. 5B, frames a5′ and b5′). We also examined the cellular distribution of PABP1, another SG marker (44), in cells transfected with the 2.2DS-RNA expression plasmid and found that intensely fluorescent PABP1 foci also colocalized with TBP in the cytoplasm (Fig. 6A).
FIG 6.
The 2.2DS-RNA-induced formation of SGs coincides with the accumulation of TBP in the cytoplasm of Huh7 cells. (A) The cells were transfected with the vehicle control (mock), β-Gal expression plasmid (negative-control RNA), or 2.2DS-RNA expression plasmid. The nuclei were stained using DAPI (gray). TBP and PABP1 were detected using anti-TBP (green) and anti-PABP1 (red) antibodies, respectively. In the merged panels, the colocalization of TBP and PABP1 is visible as yellow foci in the cells expressing 2.2DS-RNA. (B) The cells were transfected with the HBV 2.2DS-RNA, genotype A (2.2DS-RNA+), or HBVG2335A (2.2DS-RNA−) expression plasmids in the presence (+Arsenite) or absence (−) of 500 μM arsenite. The cells were fixed, and the nuclei were stained using DAPI (gray). TBP and G3BP were detected using anti-TBP (green) and anti-G3BP (red) antibodies, respectively. The colocalization of TBP and G3BP foci is visible as yellow foci in the cells transfected with the HBV genotype A or 2.2DS-RNA expression plasmid (merged panels). The boxed regions in the merged images are magnified to highlight the results (scale bar, 7.5 μm).
To investigate whether the expression of 2.2DS-RNA induces SG formation, we compared the cellular distribution of G3BP between cells transfected with the 2.2DS-RNA, HBV genotype A, and HBVG2335A (2.2DS-RNA-deficient) expression plasmids. We found that, in the cells transfected with the 2.2DS-RNA expression plasmid, SGs formed in both the presence and absence of arsenite (Fig. 6B, frames a4 and a4′). However, although SGs formed in the genotype A-transfected cells following arsenite treatment, TBP did not colocalize obviously with the aggregated G3BP in the absence of arsenite (Fig. 6B, frame b4). In contrast, SGs did not form in the cytoplasm of the HBVG2335A-transfected cells (Fig. 6B, frame c4). Following arsenite treatment, the level of TBP foci associated with the G3BP aggregates in the genotype A-transfected cells was relatively greater (Fig. 6B, frame b4′) than that in the HBVG2335A-transfected cells (Fig. 6B, frame c4′). These results suggested that 2.2DS-RNA contributed to SG formation in the Huh7 cells and that the association of TBP foci with G3BP aggregates might be dependent on the level of 2.2DS-RNA expression.
DISCUSSION
In our investigation of the effects of 2.2DS-RNA on HBV transcription, we found that 2.2DS-RNA overexpression correlated with reduced levels of HBV transcripts and the reduced transcriptional activity of HBV promoters, certain cellular promoters, and certain promoters of other viruses. In addition, 2.2DS-RNA suppressed transcription in vitro through a mechanism that involves 2.2DS-RNA-TBP binding, in which certain sequences located within nucleotide positions 432 to 832 of 2.2DS-RNA are required for efficient TBP binding. We also found that 2.2DS-RNA sequesters TBP in the cytoplasm and that the interaction of 2.2DS-RNA and TBP induces SG formation. Various highly conserved ncRNAs have been identified that serve regulatory roles in a variety of cellular processes through interactions with cellular factors (41, 45–48). Future studies of the sequences and structure of 2.2DS-RNA are warranted to identify the molecular basis of the interaction between TBP and 2.2DS-RNA, in which sequences within nucleotide positions 432 to 832 of 2.2DS-RNA contribute to the repression of HBV transcription.
In the initiation of RNAP-II-mediated transcription, RNAP-II binds TBP and various other general transcription factors at the promoter region of the gene, forming the RNAP-II transcription complex (49). Although we identified the interaction of 2.2DS-RNA and TBP in vivo, no interaction between 2.2DS-RNA and RNAP-II was detected (Fig. 4E). Our current data provide no explanation for the exclusion of RNAP-II from the 2.2DS-RNA-TBP complex. Future studies are required to determine whether the interaction of 2.2DS-RNA with TBP in the cytoplasm suppresses transcription by disrupting RNAP-II binding to TBP in the nucleus.
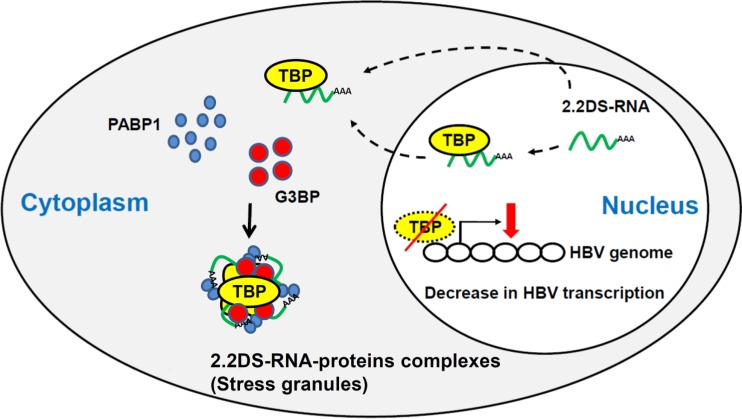
We propose a model of HBV infection in hepatocytes in which 2.2DS-RNA suppresses HBV transcription through a pathway that involves the formation of the 2.2DS-RNA-TBP complex and SG assembly (Fig. 7). In our model, 2.2DS-RNA interacts with TBP in the nucleus before translocation to the cytoplasm and/or binds cytoplasmic and newly translated TBP, forming TBP-2.2DS-RNA complexes in the cytoplasm. A portion of the cytoplasmic TBP-2.2DS-RNA complexes associates with G3BP and PABP1, inducing SG formation and the association of 2.2DS-RNA, TBP, and SGs. The formation of TBP-2.2DS-RNA complexes in the cytoplasm reduces the level of TBP in the nucleus, and the depletion of nuclear TBP suppresses viral transcription, which in turn inhibits HBV replication. The 2.2DS-RNA might also affect SG-mediated pathways involved in the host cell response to viral infection.
FIG 7.
Model for 2.2DS-RNA-mediated suppression of HBV transcription in hepatocytes. The dotted lines indicate that 2.2DS-RNA (green) may interact with TBP (yellow) in the nucleus and/or cytoplasm. The 2.2DS-RNA-TBP complexes accumulate in the cytoplasm, and a portion of the complexes associates with PABP1 (blue) and G3BP (red), which induces SG assembly. The accumulation of TBP in the cytoplasm depletes the nucleus of TBP, which in turn inhibits HBV transcription.
The TBP is a crucial component of the basal transcriptional machinery in all eukaryotes (50). Therefore, changes in the level of nuclear TBP will affect both viral and cellular TBP-mediated transcription. With the exception of the pre-S1 promoter, HBV promoters do not contain a canonical TATA box (51). However, HBV promoters do contain putative TATA-like sequences that are bound by TBP in a sequence-specific manner (52–54). We confirmed that the transcriptional activities of HBV promoters were statistically suppressed by 2.2DS-RNA, as were the activities of other viral promoters (Fig. 3C). The suppression of the transcriptional activity of the X promoter provides additional evidence supporting our model of the 2.2DS-RNA-mediated suppression of HBV replication.
The X protein of HBV activates the transcription of HBV genes, as well as a repertoire of host genes, including TBP (55, 56). The X protein has also been shown to bind TBP in vitro and to increase TBP expression in cells (57, 58). Therefore, it is possible that the 2.2DS-RNA-mediated suppression of X protein expression inhibits both HBV and cellular transcription as both require TBP. However, although 2.2DS-RNA suppressed the transcriptional activity of the viral and cellular promoters that were used in the expression plasmids in our study, transcription from the integrated HBV genome was not strongly inhibited by 2.2DS-RNA expression (Fig. 2E). These results suggest that the transcriptional activity of promoters in episomal DNA, such as the covalently closed circular genome (cccDNA) of HBV, might be more sensitive to the inhibitory effects of 2.2DS-RNA than promoters in chromosomal DNA. In the future, we will compare the inhibitory effect of 2.2DS-RNA on transcription from the HBV cccDNA with that from the chromosomally integrated HBV genome in 1.3ES2 cells.
We observed SG assembly in the cells ectopically expressing 2.2DS-RNA and in those transfected with the HBV genotype A expression plasmid, whereas SG assembly was not observed in the cells transfected with the 2.2DS-RNA-deficient HBVG2335A expression plasmid (Fig. 6B). However, the results of our current study do not fully clarify the causal relationship between the interaction of 2.2DS-RNA with TBP and the formation of SGs. Future investigations of the molecular basis of the colocalization of 2.2DS-RNA and TBP with SG proteins in HBV-transfected cells are warranted. Previous studies have shown that transcription factors, such as the Rpb4p subunit of RNAP-II and the steroid receptor coactivator-3, are involved in SG-mediated processes (26, 59, 60), which suggests that other transcription factors, such as TBP, might also be recruited to SGs. Furthermore, whether 2.2DS-RNA-mediated SG formation induces known SG-related processes, such as mRNA degradation, and whether SG formation is biologically relevant to HBV pathogenesis require further study.
Recent clinical studies have shown that patients infected with HBV genotype A or B, each of which transcribes 2.2DS-RNA, have less severe symptoms and liver pathology and higher rates of viral clearance in response to IFN-α therapy than patients infected with HBV genotype C or D, each of which is 2.2DS-RNA deficient (7, 9, 61, 62). Our prior study found that relatively high levels of 2.2DS-RNAs were present in CHB patients with genotype A infection but not in those with genotype D infection (25). These results indicated that the expression of 2.2DS-RNA could be induced and regulated differently in CHB patients than it is in tissue culture. Based on our current findings that the level of pgRNA was suppressed by 2.2DS-RNA even in the presence of small amounts of the 2.2DS-RNA plasmids (Fig. 2A and B), it is conceivable that the level of pgRNA and viral transcription may be suppressed in CHB patients who highly express 2.2DS-RNA. Moreover, the 2.2DS-RNA-induced suppression of pgRNA may be persistent in the CHB patients of genotype A, which is not the case for patients infected with other genotypes that are 2.2DS-RNA deficient. The difference may raise questions about whether the long-term expression of 2.2DS-RNA in CHB patients of genotype A could influence outcomes of chronic HBV infection and IFN-based therapy. Further experiments are still needed to understand the correlation of viral replication with the expression of 2.2DS-RNA in CHB patients, as well as the factors modulating the level of 2.2DS-RNA.
In conclusion, our results collectively indicate that the 2.2DS-RNA of HBV correlates with the inhibition of expression of HBV pgRNA at the transcriptional level through a mechanism that involves TBP-2.2DS-RNA binding and induces SG assembly. Our findings offer experimental evidence unveiling a previously unknown association between the interaction of TBP with 2.2DS-RNA and suppression of viral transcription. Moreover, the differential expression of 2.2DS-RNA by the various HBV genotypes may be one factor that influences differences in long-term outcomes of CHB disease and responses to antiviral therapy.
ACKNOWLEDGMENTS
This work was supported by research grants from the Ministry of Science and Technology and the National Health Research Institutes (NHRI), Taiwan (grants 02D2-MM-NSC13 and MGPP07-014, respectively). K.-N.T. performed his thesis research under the auspices of the Graduate Program of Biotechnology in Medicine at the National Tsing Hua University and the NHRI.
The funding agencies had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
We thank Shu-Hsia Chen, Jui-Chou Hsu, Guann-Yi Yu, and Li-Rung Huang for kindly reviewing the manuscript. We thank Shu-Fen Hu of the National Health Research Institutes Optical Biology Core for technical assistance with the confocal microscopy, and we thank Wen-Ling Chen for assistance with the FISH analysis. We also thank Y. H. W. Lee for kindly providing the hsp70 minimal and rRNA reporter plasmids.
REFERENCES
- 1.Iloeje UH, Yang HI, Su J, Jen CL, You SL, Chen CJ. 2006. Predicting cirrhosis risk based on the level of circulating hepatitis B viral load. Gastroenterology 130:678–686. doi: 10.1053/j.gastro.2005.11.016. [DOI] [PubMed] [Google Scholar]
- 2.Kao JH, Chen DS. 2002. Global control of hepatitis B virus infection. Lancet Infect Dis 2:395–403. doi: 10.1016/S1473-3099(02)00315-8. [DOI] [PubMed] [Google Scholar]
- 3.Bertoletti A, Ferrari C. 2003. Kinetics of the immune response during HBV and HCV infection. Hepatology 38:4–13. doi: 10.1053/jhep.2003.50310. [DOI] [PubMed] [Google Scholar]
- 4.Biron CA. 2001. Interferons alpha and beta as immune regulators—a new look. Immunity 14:661–664. doi: 10.1016/S1074-7613(01)00154-6. [DOI] [PubMed] [Google Scholar]
- 5.Akira S, Uematsu S, Takeuchi O. 2006. Pathogen recognition and innate immunity. Cell 124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 6.Lin CL, Kao JH. 2011. The clinical implications of hepatitis B virus genotype: recent advances. J Gastroenterol Hepatol 26(Suppl 1):S123–S130. doi: 10.1111/j.1440-1746.2010.06541.x. [DOI] [PubMed] [Google Scholar]
- 7.Erhardt A, Blondin D, Hauck K, Sagir A, Kohnle T, Heintges T, Haussinger D. 2005. Response to interferon alfa is hepatitis B virus genotype dependent: genotype A is more sensitive to interferon than genotype D. Gut 54:1009–1013. doi: 10.1136/gut.2004.060327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kao JH, Wu NH, Chen PJ, Lai MY, Chen DS. 2000. Hepatitis B genotypes and the response to interferon therapy. J Hepatol 33:998–1002. doi: 10.1016/S0168-8278(00)80135-X. [DOI] [PubMed] [Google Scholar]
- 9.Janssen HL, van Zonneveld M, Senturk H, Zeuzem S, Akarca US, Cakaloglu Y, Simon C, So TM, Gerken G, de Man RA, Niesters HG, Zondervan P, Hansen B, Schalm SW, HBV 99-01 Study Group, Rotterdam Foundation for Liver R. 2005. Pegylated interferon alfa-2b alone or in combination with lamivudine for HBeAg-positive chronic hepatitis B: a randomised trial. Lancet 365:123–129. doi: 10.1016/S0140-6736(05)17701-0. [DOI] [PubMed] [Google Scholar]
- 10.Oommen PT, Wirth S, Wintermeyer P, Gerner P. 2006. Relationship between viral load and genotypes of hepatitis B virus in children with chronic hepatitis B. J Pediatr Gastroenterol Nutr 43:342–347. doi: 10.1097/01.mpg.0000233191.95447.1e. [DOI] [PubMed] [Google Scholar]
- 11.Thakur V, Guptan RC, Kazim SN, Malhotra V, Sarin SK. 2002. Profile, spectrum and significance of HBV genotypes in chronic liver disease patients in the Indian subcontinent. J Gastroenterol Hepatol 17:165–170. doi: 10.1046/j.1440-1746.2002.02605.x. [DOI] [PubMed] [Google Scholar]
- 12.Zollner B, Petersen J, Puchhammer-Stockl E, Kletzmayr J, Sterneck M, Fischer L, Schroter M, Laufs R, Feucht HH. 2004. Viral features of lamivudine resistant hepatitis B genotypes A and D. Hepatology 39:42–50. doi: 10.1002/hep.20016. [DOI] [PubMed] [Google Scholar]
- 13.Yang HI, Yeh SH, Chen PJ, Iloeje UH, Jen CL, Su J, Wang LY, Lu SN, You SL, Chen DS, Liaw YF, Chen CJ. 2008. Associations between hepatitis B virus genotype and mutants and the risk of hepatocellular carcinoma. J Natl Cancer Inst 100:1134–1143. doi: 10.1093/jnci/djn243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tiollais P, Pourcel C, Dejean A. 1985. The hepatitis B virus. Nature 317:489–495. doi: 10.1038/317489a0. [DOI] [PubMed] [Google Scholar]
- 15.Suzuki T, Kajino K, Masui N, Saito I, Miyamura T. 1990. Alternative splicing of hepatitis B virus RNAs in HepG2 cells transfected with the viral DNA. Virology 179:881–885. doi: 10.1016/0042-6822(90)90160-S. [DOI] [PubMed] [Google Scholar]
- 16.Su TS, Lai CJ, Huang JL, Lin LH, Yauk YK, Chang CM, Lo SJ, Han SH. 1989. Hepatitis B virus transcript produced by RNA splicing. J Virol 63:4011–4018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Choo KB, Liew LN, Chong KY, Lu RH, Cheng WT. 1991. Transgenome transcription and replication in the liver and extrahepatic tissues of a human hepatitis B virus transgenic mouse. Virology 182:785–792. doi: 10.1016/0042-6822(91)90619-M. [DOI] [PubMed] [Google Scholar]
- 18.Chen PJ, Chen CR, Sung JL, Chen DS. 1989. Identification of a doubly spliced viral transcript joining the separated domains for putative protease and reverse transcriptase of hepatitis B virus. J Virol 63:4165–4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gunther S, Sommer G, Iwanska A, Will H. 1997. Heterogeneity and common features of defective hepatitis B virus genomes derived from spliced pregenomic RNA. Virology 238:363–371. doi: 10.1006/viro.1997.8863. [DOI] [PubMed] [Google Scholar]
- 20.Wang YL, Liou GG, Lin CH, Chen ML, Kuo TM, Tsai KN, Huang CC, Chen YL, Huang LR, Chou YC, Chang C. 2015. The inhibitory effect of the hepatitis B virus singly spliced RNA-encoded p21.5 protein on HBV nucleocapsid formation. PLoS One 10:e0119625. doi: 10.1371/journal.pone.0119625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mancini-Bourgine M, Bayard F, Soussan P, Deng Q, Lone YC, Kremsdorf D, Michel ML. 2007. Hepatitis B virus splice-generated protein induces T-cell responses in HLA-transgenic mice and hepatitis B virus-infected patients. J Virol 81:4963–4972. doi: 10.1128/JVI.02619-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Soussan P, Tuveri R, Nalpas B, Garreau F, Zavala F, Masson A, Pol S, Brechot C, Kremsdorf D. 2003. The expression of hepatitis B spliced protein (HBSP) encoded by a spliced hepatitis B virus RNA is associated with viral replication and liver fibrosis. J Hepatol 38:343–348. doi: 10.1016/S0168-8278(02)00422-1. [DOI] [PubMed] [Google Scholar]
- 23.Sommer G, van Bommel F, Will H. 2000. Genotype-specific synthesis and secretion of spliced hepatitis B virus genomes in hepatoma cells. Virology 271:371–381. doi: 10.1006/viro.2000.0331. [DOI] [PubMed] [Google Scholar]
- 24.Wu HL, Chen PJ, Tu SJ, Lin MH, Lai MY, Chen DS. 1991. Characterization and genetic analysis of alternatively spliced transcripts of hepatitis B virus in infected human liver tissues and transfected HepG2 cells. J Virol 65:1680–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang CC, Kuo TM, Yeh CT, Hu CP, Chen YL, Tsai YL, Chen ML, Chou YC, Chang C. 2013. One single nucleotide difference alters the differential expression of spliced RNAs between HBV genotypes A and D. Virus Res 174:18–26. doi: 10.1016/j.virusres.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 26.Anderson P, Kedersha N. 2008. Stress granules: the Tao of RNA triage. Trends Biochem Sci 33:141–150. doi: 10.1016/j.tibs.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 27.Kedersha N, Stoecklin G, Ayodele M, Yacono P, Lykke-Andersen J, Fritzler MJ, Scheuner D, Kaufman RJ, Golan DE, Anderson P. 2005. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J Cell Biol 169:871–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kedersha N, Cho MR, Li W, Yacono PW, Chen S, Gilks N, Golan DE, Anderson P. 2000. Dynamic shuttling of TIA-1 accompanies the recruitment of mRNA to mammalian stress granules. J Cell Biol 151:1257–1268. doi: 10.1083/jcb.151.6.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.White JP, Cardenas AM, Marissen WE, Lloyd RE. 2007. Inhibition of cytoplasmic mRNA stress granule formation by a viral proteinase. Cell Host Microbe 2:295–305. doi: 10.1016/j.chom.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 30.Yi Z, Pan T, Wu X, Song W, Wang S, Xu Y, Rice CM, Macdonald MR, Yuan Z. 2011. Hepatitis C virus co-opts Ras-GTPase-activating protein-binding protein 1 for its genome replication. J Virol 85:6996–7004. doi: 10.1128/JVI.00013-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ariumi Y, Kuroki M, Kushima Y, Osugi K, Hijikata M, Maki M, Ikeda M, Kato N. 2011. Hepatitis C virus hijacks P-body and stress granule components around lipid droplets. J Virol 85:6882–6892. doi: 10.1128/JVI.02418-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chou YC, Jeng KS, Chen ML, Liu HH, Liu TL, Chen YL, Liu YC, Hu CP, Chang C. 2005. Evaluation of transcriptional efficiency of hepatitis B virus covalently closed circular DNA by reverse transcription-PCR combined with the restriction enzyme digestion method. J Virol 79:1813–1823. doi: 10.1128/JVI.79.3.1813-1823.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chong CL, Chen ML, Wu YC, Tsai KN, Huang CC, Hu CP, Jeng KS, Chou YC, Chang C. 2011. Dynamics of HBV cccDNA expression and transcription in different cell growth phase. J Biomed Sci 18:96. doi: 10.1186/1423-0127-18-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yuh CH, Chang YL, Ting LP. 1992. Transcriptional regulation of precore and pregenomic RNAs of hepatitis B virus. J Virol 66:4073–4084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hong MH, Chou YC, Wu YC, Tsai KN, Hu CP, Jeng KS, Chen ML, Chang C. 2012. Transforming growth factor-β1 suppresses hepatitis B virus replication by the reduction of hepatocyte nuclear factor-4α expression. PLoS One 7:e30360. doi: 10.1371/journal.pone.0030360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kao CF, Chen SY, Lee YH. 2004. Activation of RNA polymerase I transcription by hepatitis C virus core protein. J Biomed Sci 11:72–94. doi: 10.1007/BF02256551. [DOI] [PubMed] [Google Scholar]
- 37.de Planell-Saguer M, Rodicio MC, Mourelatos Z. 2010. Rapid in situ codetection of noncoding RNAs and proteins in cells and formalin-fixed paraffin-embedded tissue sections without protease treatment. Nat Protoc 5:1061–1073. doi: 10.1038/nprot.2010.62. [DOI] [PubMed] [Google Scholar]
- 38.Li ZF, Liang YM, Lau PN, Shen W, Wang DK, Cheung WT, Xue CJ, Poon LM, Lam YW. 2013. Dynamic localisation of mature microRNAs in human nucleoli is influenced by exogenous genetic materials. PLoS One 8:e70869. doi: 10.1371/journal.pone.0070869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lok AS, McMahon BJ. 2001. Chronic hepatitis B. Hepatology 34:1225–1241. doi: 10.1053/jhep.2001.29401. [DOI] [PubMed] [Google Scholar]
- 40.Goodrich JA, Kugel JF. 2006. Non-coding-RNA regulators of RNA polymerase II transcription. Nat Rev Mol Cell Biol 7:612–616. doi: 10.1038/nrm1946. [DOI] [PubMed] [Google Scholar]
- 41.Yamamoto R, Koseki S, Ohkawa J, Murakami K, Nishikawa S, Taira K, Kumar PK. 1997. Inhibition of transcription by the TAR RNA of HIV-1 in a nuclear extract of HeLa cells. Nucleic Acids Res 25:3445–3450. doi: 10.1093/nar/25.17.3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sharp PA. 1992. TATA-binding protein is a classless factor. Cell 68:819–821. doi: 10.1016/0092-8674(92)90023-6. [DOI] [PubMed] [Google Scholar]
- 43.Hampsey M. 1998. Molecular genetics of the RNA polymerase II general transcriptional machinery. Microbiol Mol Biol Rev 62:465–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Burgess HM, Richardson WA, Anderson RC, Salaun C, Graham SV, Gray NK. 2011. Nuclear relocalisation of cytoplasmic poly(A)-binding proteins PABP1 and PABP4 in response to UV irradiation reveals mRNA-dependent export of metazoan PABPs. J Cell Sci 124:3344–3355. doi: 10.1242/jcs.087692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Martianov I, Ramadass A, Serra Barros A, Chow N, Akoulitchev A. 2007. Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature 445:666–670. doi: 10.1038/nature05519. [DOI] [PubMed] [Google Scholar]
- 46.Mariner PD, Walters RD, Espinoza CA, Drullinger LF, Wagner SD, Kugel JF, Goodrich JA. 2008. Human Alu RNA is a modular transacting repressor of mRNA transcription during heat shock. Mol Cell 29:499–509. doi: 10.1016/j.molcel.2007.12.013. [DOI] [PubMed] [Google Scholar]
- 47.Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA, Blencowe BJ, Prasanth SG, Prasanth KV. 2010. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell 39:925–938. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Borah S, Darricarrere N, Darnell A, Myoung J, Steitz JA. 2011. A viral nuclear noncoding RNA binds re-localized poly(A) binding protein and is required for late KSHV gene expression. PLoS Pathog 7:e1002300. doi: 10.1371/journal.ppat.1002300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thomas MC, Chiang CM. 2006. The general transcription machinery and general cofactors. Crit Rev Biochem Mol Biol 41:105–178. doi: 10.1080/10409230600648736. [DOI] [PubMed] [Google Scholar]
- 50.Hernandez N. 1993. TBP, a universal eukaryotic transcription factor? Genes Dev 7:1291–1308. doi: 10.1101/gad.7.7b.1291. [DOI] [PubMed] [Google Scholar]
- 51.Paran N, Ori A, Haviv I, Shaul Y. 2000. A composite polyadenylation signal with TATA box function. Mol Cell Biol 20:834–841. doi: 10.1128/MCB.20.3.834-841.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bogomolski-Yahalom V, Klein A, Greenblat I, Haviv Y, Tur-Kaspa R. 1997. The TATA-less promoter of hepatitis B virus S gene contains a TBP binding site and an active initiator. Virus Res 49:1–7. doi: 10.1016/S0168-1702(96)01429-3. [DOI] [PubMed] [Google Scholar]
- 53.Quarleri J. 2014. Core promoter: a critical region where the hepatitis B virus makes decisions. World J Gastroenterol 20:425–435. doi: 10.3748/wjg.v20.i2.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen IH, Huang CJ, Ting LP. 1995. Overlapping initiator and TATA box functions in the basal core promoter of hepatitis B virus. J Virol 69:3647–3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bouchard MJ, Schneider RJ. 2004. The enigmatic X gene of hepatitis B virus. J Virol 78:12725–12734. doi: 10.1128/JVI.78.23.12725-12734.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.van Breugel PC, Robert EI, Mueller H, Decorsiere A, Zoulim F, Hantz O, Strubin M. 2012. Hepatitis B virus X protein stimulates gene expression selectively from extrachromosomal DNA templates. Hepatology 56:2116–2124. doi: 10.1002/hep.25928. [DOI] [PubMed] [Google Scholar]
- 57.Johnson SA, Mandavia N, Wang HD, Johnson DL. 2000. Transcriptional regulation of the TATA-binding protein by Ras cellular signaling. Mol Cell Biol 20:5000–5009. doi: 10.1128/MCB.20.14.5000-5009.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Qadri I, Maguire HF, Siddiqui A. 1995. Hepatitis B virus transactivator protein X interacts with the TATA-binding protein. Proc Natl Acad Sci U S A 92:1003–1007. doi: 10.1073/pnas.92.4.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lotan R, Bar-On VG, Harel-Sharvit L, Duek L, Melamed D, Choder M. 2005. The RNA polymerase II subunit Rpb4p mediates decay of a specific class of mRNAs. Genes Dev 19:3004–3016. doi: 10.1101/gad.353205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yu C, York B, Wang S, Feng Q, Xu J, O'Malley BW. 2007. An essential function of the SRC-3 coactivator in suppression of cytokine mRNA translation and inflammatory response. Mol Cell 25:765–778. doi: 10.1016/j.molcel.2007.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hou J, Schilling R, Janssen HL, Hansen BE, Heijtink R, Sablon E, Williams R, Lau GK, Schalm SW, Naoumov NV. 2007. Genetic characteristics of hepatitis B virus genotypes as a factor for interferon-induced HBeAg clearance. J Med Virol 79:1055–1063. doi: 10.1002/jmv.20935. [DOI] [PubMed] [Google Scholar]
- 62.Wai CT, Chu CJ, Hussain M, Lok AS. 2002. HBV genotype B is associated with better response to interferon therapy in HBeAg(+) chronic hepatitis than genotype C. Hepatology 36:1425–1430. doi: 10.1002/hep.1840360619. [DOI] [PubMed] [Google Scholar]