Abstract
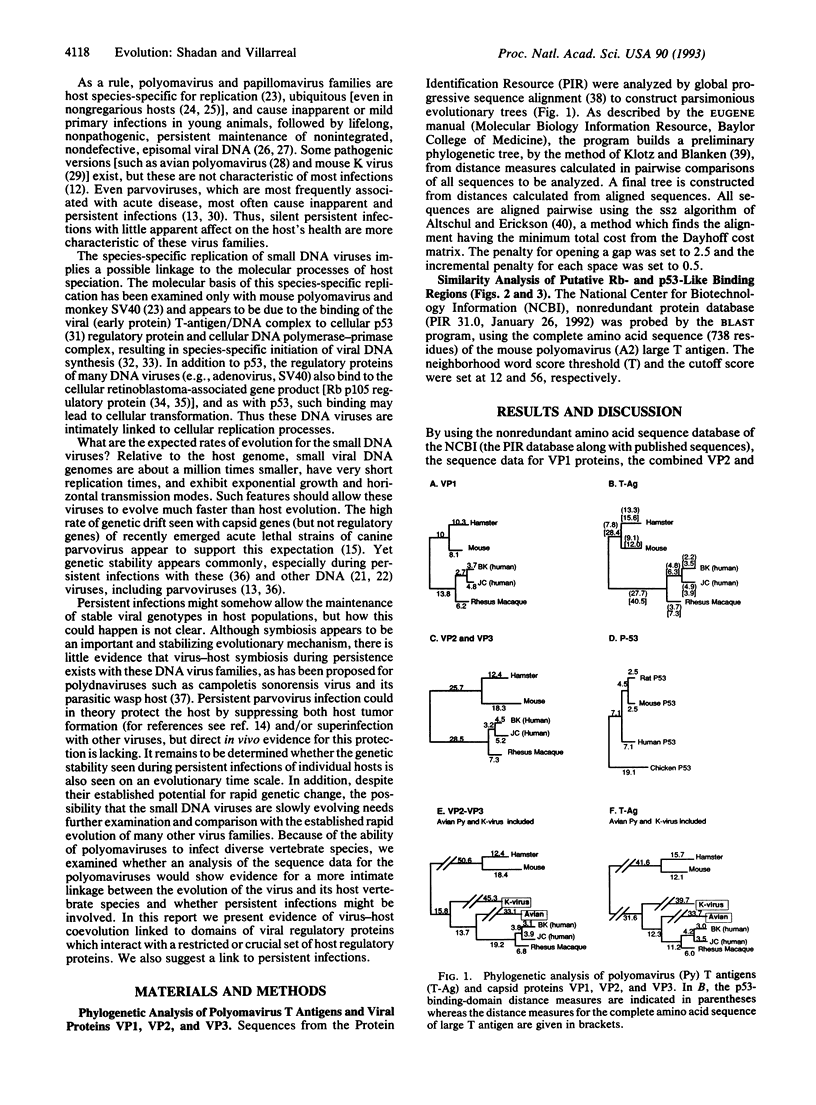
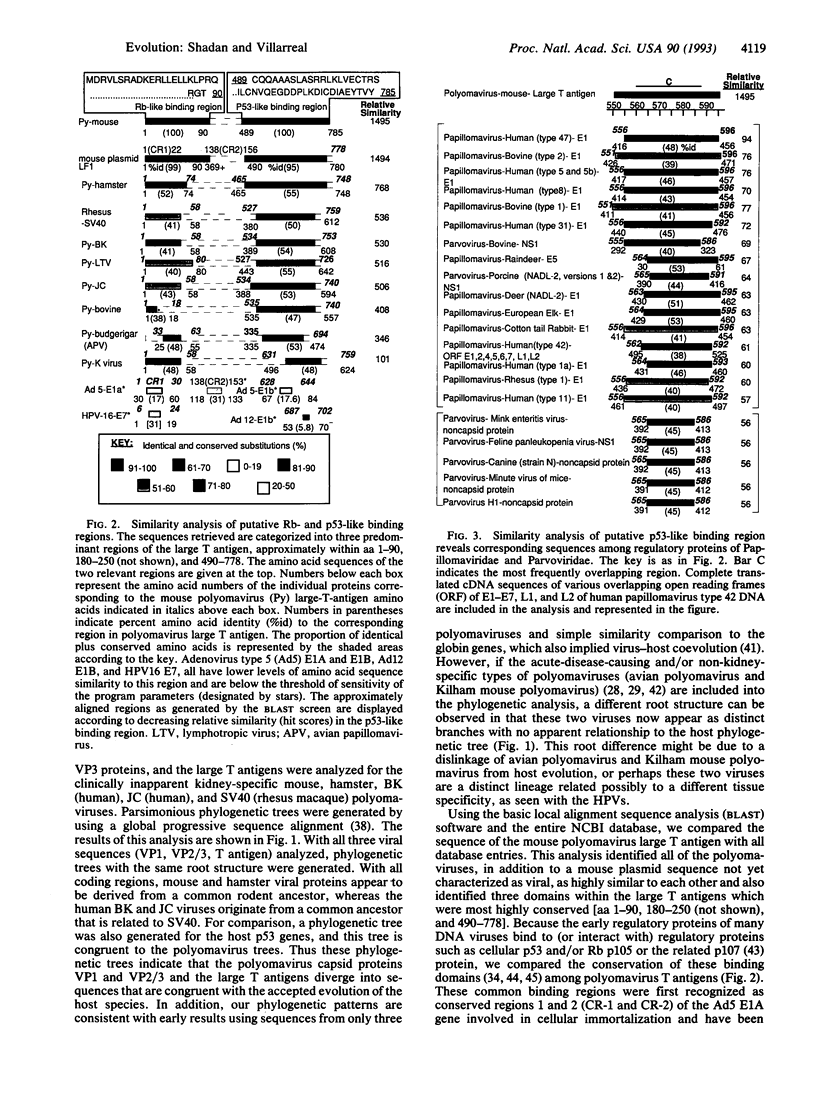
Although most RNA viral genomes (and related cellular retroposons) can evolve at rates a millionfold greater than that of their host genomes, some of the small DNA viruses (polyomaviruses and papillomaviruses) appear to evolve at much slower rates. These DNA viruses generally cause host species-specific inapparent primary infections followed by life-long, benign persistent infections. Using global progressive sequence alignments for kidney-specific Polyomaviridae (mouse, hamster, primate, human), we have constructed parsimonious evolutionary trees for the viral capsid proteins (VP1, VP2/VP3) and the large tumor (T) antigen. We show that these three coding sequences can yield phylogenetic trees similar to each other and to that of their host species. Such virus-host "co-speciation" appears incongruent with some prevailing views of viral evolution, and we suggest that inapparent persistent infections may link virus and most host evolution. Similarity analysis identified three specific regions of polyoma regulatory gene products (T antigens) as highly conserved, and two of these regions correspond to binding sites for host regulatory proteins (p53, the retinoblastoma gene product p105, and the related protein p107). The p53 site overlaps with a conserved ATPase domain and the retinoblastoma site corresponds to conserved region 1 of E1A protein of adenovirus type 5. We examined the local conservation of these binding sequences and show that the conserved retinoblastoma binding domain is characteristic and inclusive of the entire polyomavirus family, but the conserved p53-like binding domain is characteristic and inclusive of three entire families of small DNA viruses: polyomaviruses, papillomaviruses, and parvoviruses. The evolution of small-DNA-virus families may thus be tightly linked to host evolution and speciation by interaction with a subset of host regulatory proteins.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ackrill A. M., Foster G. R., Laxton C. D., Flavell D. M., Stark G. R., Kerr I. M. Inhibition of the cellular response to interferons by products of the adenovirus type 5 E1A oncogene. Nucleic Acids Res. 1991 Aug 25;19(16):4387–4393. doi: 10.1093/nar/19.16.4387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S. F., Erickson B. W. Optimal sequence alignment using affine gap costs. Bull Math Biol. 1986;48(5-6):603–616. doi: 10.1007/BF02462326. [DOI] [PubMed] [Google Scholar]
- Anderson R. M., May R. M. Population biology of infectious diseases: Part I. Nature. 1979 Aug 2;280(5721):361–367. doi: 10.1038/280361a0. [DOI] [PubMed] [Google Scholar]
- Bando H., Kusuda J., Gojobori T., Maruyama T., Kawase S. Organization and nucleotide sequence of a densovirus genome imply a host-dependent evolution of the parvoviruses. J Virol. 1987 Feb;61(2):553–560. doi: 10.1128/jvi.61.2.553-560.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berns K. I. Parvovirus replication. Microbiol Rev. 1990 Sep;54(3):316–329. doi: 10.1128/mr.54.3.316-329.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black F. L. Infectious diseases in primitive societies. Science. 1975 Feb 14;187(4176):515–518. doi: 10.1126/science.163483. [DOI] [PubMed] [Google Scholar]
- Blissard G. W., Theilmann D. A., Summers M. D. Segment W of Campoletis sonorensis virus: expression, gene products, and organization. Virology. 1989 Mar;169(1):78–89. doi: 10.1016/0042-6822(89)90043-3. [DOI] [PubMed] [Google Scholar]
- Bradley M. K., Smith T. F., Lathrop R. H., Livingston D. M., Webster T. A. Consensus topography in the ATP binding site of the simian virus 40 and polyomavirus large tumor antigens. Proc Natl Acad Sci U S A. 1987 Jun;84(12):4026–4030. doi: 10.1073/pnas.84.12.4026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell B. A., Villarreal L. P. Host species specificity of polyomavirus DNA replication is not altered by simian virus 40 72-base-pair repeats. Mol Cell Biol. 1985 Jun;5(6):1534–1537. doi: 10.1128/mcb.5.6.1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L., Faha B., Dembski M., Tsai L. H., Harlow E., Dyson N. Independent binding of the retinoblastoma protein and p107 to the transcription factor E2F. Nature. 1992 Jan 9;355(6356):176–179. doi: 10.1038/355176a0. [DOI] [PubMed] [Google Scholar]
- Chan S. Y., Ho L., Ong C. K., Chow V., Drescher B., Dürst M., ter Meulen J., Villa L., Luande J., Mgaya H. N. Molecular variants of human papillomavirus type 16 from four continents suggest ancient pandemic spread of the virus and its coevolution with humankind. J Virol. 1992 Apr;66(4):2057–2066. doi: 10.1128/jvi.66.4.2057-2066.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeCaprio J. A., Ludlow J. W., Figge J., Shew J. Y., Huang C. M., Lee W. H., Marsilio E., Paucha E., Livingston D. M. SV40 large tumor antigen forms a specific complex with the product of the retinoblastoma susceptibility gene. Cell. 1988 Jul 15;54(2):275–283. doi: 10.1016/0092-8674(88)90559-4. [DOI] [PubMed] [Google Scholar]
- Deppert W., Steinmayer T. Metabolic stabilization of p53 in SV40-transformed cells correlates with expression of the transformed phenotype but is independent from complex formation with SV40 large T antigen. Curr Top Microbiol Immunol. 1989;144:77–83. doi: 10.1007/978-3-642-74578-2_9. [DOI] [PubMed] [Google Scholar]
- Deppert W., Steinmayer T., Richter W. Cooperation of SV40 large T antigen and the cellular protein p53 in maintenance of cell transformation. Oncogene. 1989 Sep;4(9):1103–1110. [PubMed] [Google Scholar]
- Domingo E., Martínez-Salas E., Sobrino F., de la Torre J. C., Portela A., Ortín J., López-Galindez C., Pérez-Breña P., Villanueva N., Nájera R. The quasispecies (extremely heterogeneous) nature of viral RNA genome populations: biological relevance--a review. Gene. 1985;40(1):1–8. doi: 10.1016/0378-1119(85)90017-4. [DOI] [PubMed] [Google Scholar]
- Domingo E. RNA virus evolution and the control of viral disease. Prog Drug Res. 1989;33:93–133. doi: 10.1007/978-3-0348-9146-2_5. [DOI] [PubMed] [Google Scholar]
- Doolittle R. F., Feng D. F., Johnson M. S., McClure M. A. Origins and evolutionary relationships of retroviruses. Q Rev Biol. 1989 Mar;64(1):1–30. doi: 10.1086/416128. [DOI] [PubMed] [Google Scholar]
- Dubensky T. W., Villarreal L. P. The primary site of replication alters the eventual site of persistent infection by polyomavirus in mice. J Virol. 1984 May;50(2):541–546. doi: 10.1128/jvi.50.2.541-546.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyson N., Bernards R., Friend S. H., Gooding L. R., Hassell J. A., Major E. O., Pipas J. M., Vandyke T., Harlow E. Large T antigens of many polyomaviruses are able to form complexes with the retinoblastoma protein. J Virol. 1990 Mar;64(3):1353–1356. doi: 10.1128/jvi.64.3.1353-1356.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyson N., Guida P., McCall C., Harlow E. Adenovirus E1A makes two distinct contacts with the retinoblastoma protein. J Virol. 1992 Jul;66(7):4606–4611. doi: 10.1128/jvi.66.7.4606-4611.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dörries K., Loeber G., Meixensberger J. Association of polyomaviruses JC, SV40, and BK with human brain tumors. Virology. 1987 Sep;160(1):268–270. doi: 10.1016/0042-6822(87)90071-7. [DOI] [PubMed] [Google Scholar]
- Eki T., Enomoto T., Masutani C., Miyajima A., Takada R., Murakami Y., Ohno T., Hanaoka F., Ui M. Mouse DNA primase plays the principal role in determination of permissiveness for polyomavirus DNA replication. J Virol. 1991 Sep;65(9):4874–4881. doi: 10.1128/jvi.65.9.4874-4881.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewen M. E., Xing Y. G., Lawrence J. B., Livingston D. M. Molecular cloning, chromosomal mapping, and expression of the cDNA for p107, a retinoblastoma gene product-related protein. Cell. 1991 Sep 20;66(6):1155–1164. doi: 10.1016/0092-8674(91)90038-z. [DOI] [PubMed] [Google Scholar]
- Feng D. F., Doolittle R. F. Progressive sequence alignment as a prerequisite to correct phylogenetic trees. J Mol Evol. 1987;25(4):351–360. doi: 10.1007/BF02603120. [DOI] [PubMed] [Google Scholar]
- Fisher A. G., Ensoli B., Looney D., Rose A., Gallo R. C., Saag M. S., Shaw G. M., Hahn B. H., Wong-Staal F. Biologically diverse molecular variants within a single HIV-1 isolate. Nature. 1988 Aug 4;334(6181):444–447. doi: 10.1038/334444a0. [DOI] [PubMed] [Google Scholar]
- Gorbalenya A. E., Koonin E. V., Wolf Y. I. A new superfamily of putative NTP-binding domains encoded by genomes of small DNA and RNA viruses. FEBS Lett. 1990 Mar 12;262(1):145–148. doi: 10.1016/0014-5793(90)80175-i. [DOI] [PubMed] [Google Scholar]
- Klotz L. C., Blanken R. L. A practical method for calculating evolutionary trees from sequence data. J Theor Biol. 1981 Jul 21;91(2):261–272. doi: 10.1016/0022-5193(81)90233-2. [DOI] [PubMed] [Google Scholar]
- Larose A., Dyson N., Sullivan M., Harlow E., Bastin M. Polyomavirus large T mutants affected in retinoblastoma protein binding are defective in immortalization. J Virol. 1991 May;65(5):2308–2313. doi: 10.1128/jvi.65.5.2308-2313.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. M., Bih F. Y., Chao Y. C., Govindarajan S., Lai M. M. Evolution of hepatitis delta virus RNA during chronic infection. Virology. 1992 May;188(1):265–273. doi: 10.1016/0042-6822(92)90756-f. [DOI] [PubMed] [Google Scholar]
- Manfredi J. J., Prives C. Binding of p53 and p105-RB is not sufficient for oncogenic transformation by a hybrid polyomavirus-simian virus 40 large T antigen. J Virol. 1990 Nov;64(11):5250–5259. doi: 10.1128/jvi.64.11.5250-5259.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manfredi J. J., Prives C. Binding of p53 and p105-RB is not sufficient for oncogenic transformation by a hybrid polyomavirus-simian virus 40 large T antigen. J Virol. 1990 Nov;64(11):5250–5259. doi: 10.1128/jvi.64.11.5250-5259.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marich J. E., Pontsler A. V., Rice S. M., McGraw K. A., Dubensky T. W. The phylogenetic relationship and complete nucleotide sequence of human papillomavirus type 35. Virology. 1992 Feb;186(2):770–776. doi: 10.1016/0042-6822(92)90045-q. [DOI] [PubMed] [Google Scholar]
- Markowitz R. B., Eaton B. A., Kubik M. F., Latorra D., McGregor J. A., Dynan W. S. BK virus and JC virus shed during pregnancy have predominantly archetypal regulatory regions. J Virol. 1991 Aug;65(8):4515–4519. doi: 10.1128/jvi.65.8.4515-4519.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May R. M., Anderson R. M. Population biology of infectious diseases: Part II. Nature. 1979 Aug 9;280(5722):455–461. doi: 10.1038/280455a0. [DOI] [PubMed] [Google Scholar]
- Mayer M., Dörries K. Nucleotide sequence and genome organization of the murine polyomavirus, Kilham strain. Virology. 1991 Apr;181(2):469–480. doi: 10.1016/0042-6822(91)90879-g. [DOI] [PubMed] [Google Scholar]
- Mole S. E., Gannon J. V., Ford M. J., Lane D. P. Structure and function of SV40 large-T antigen. Philos Trans R Soc Lond B Biol Sci. 1987 Dec 15;317(1187):455–469. doi: 10.1098/rstb.1987.0072. [DOI] [PubMed] [Google Scholar]
- Moreno J. P., Villarreal L. P. Analysis of cellular DNA synthesis during polyoma virus infection of mice: acute infection fails to induce cellular DNA synthesis. Virology. 1992 Feb;186(2):463–474. doi: 10.1016/0042-6822(92)90011-d. [DOI] [PubMed] [Google Scholar]
- Murakami Y., Eki T., Yamada M., Prives C., Hurwitz J. Species-specific in vitro synthesis of DNA containing the polyoma virus origin of replication. Proc Natl Acad Sci U S A. 1986 Sep;83(17):6347–6351. doi: 10.1073/pnas.83.17.6347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak M. A., May R. M., Anderson R. M. The evolutionary dynamics of HIV-1 quasispecies and the development of immunodeficiency disease. AIDS. 1990 Nov;4(11):1095–1103. doi: 10.1097/00002030-199011000-00007. [DOI] [PubMed] [Google Scholar]
- Pagano M., Dürst M., Joswig S., Draetta G., Jansen-Dürr P. Binding of the human E2F transcription factor to the retinoblastoma protein but not to cyclin A is abolished in HPV-16-immortalized cells. Oncogene. 1992 Sep;7(9):1681–1686. [PubMed] [Google Scholar]
- Parrish C. R., Aquadro C. F., Strassheim M. L., Evermann J. F., Sgro J. Y., Mohammed H. O. Rapid antigenic-type replacement and DNA sequence evolution of canine parvovirus. J Virol. 1991 Dec;65(12):6544–6552. doi: 10.1128/jvi.65.12.6544-6552.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pipas J. M. Common and unique features of T antigens encoded by the polyomavirus group. J Virol. 1992 Jul;66(7):3979–3985. doi: 10.1128/jvi.66.7.3979-3985.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochford R., Campbell B. A., Villarreal L. P. Genetic analysis of the enhancer requirements for polyomavirus DNA replication in mice. J Virol. 1990 Feb;64(2):476–485. doi: 10.1128/jvi.64.2.476-485.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochford R., Moreno J. P., Peake M. L., Villarreal L. P. Enhancer dependence of polyomavirus persistence in mouse kidneys. J Virol. 1992 Jun;66(6):3287–3297. doi: 10.1128/jvi.66.6.3287-3297.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochford R., Moreno J. P., Peake M. L., Villarreal L. P. Enhancer dependence of polyomavirus persistence in mouse kidneys. J Virol. 1992 Jun;66(6):3287–3297. doi: 10.1128/jvi.66.6.3287-3297.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rott O., Kröger M., Müller H., Hobom G. The genome of budgerigar fledgling disease virus, an avian polyomavirus. Virology. 1988 Jul;165(1):74–86. doi: 10.1016/0042-6822(88)90660-5. [DOI] [PubMed] [Google Scholar]
- Soeda E., Maruyama T., Arrand J. R., Griffin B. E. Host-dependent evolution of three papova viruses. Nature. 1980 May 15;285(5761):165–167. doi: 10.1038/285165a0. [DOI] [PubMed] [Google Scholar]
- Som T., Armstrong K. A., Volkert F. C., Broach J. R. Autoregulation of 2 micron circle gene expression provides a model for maintenance of stable plasmid copy levels. Cell. 1988 Jan 15;52(1):27–37. doi: 10.1016/0092-8674(88)90528-4. [DOI] [PubMed] [Google Scholar]
- Steinhauer D. A., Holland J. J. Rapid evolution of RNA viruses. Annu Rev Microbiol. 1987;41:409–433. doi: 10.1146/annurev.mi.41.100187.002205. [DOI] [PubMed] [Google Scholar]
- Sugimoto C., Hara K., Taguchi F., Yogo Y. Regulatory DNA sequence conserved in the course of BK virus evolution. J Mol Evol. 1990 Dec;31(6):485–492. doi: 10.1007/BF02102075. [DOI] [PubMed] [Google Scholar]
- Tack L. C., Cartwright C. A., Wright J. H., Eckhart W., Peden K. W., Srinivasan A., Pipas J. M. Properties of a simian virus 40 mutant T antigen substituted in the hydrophobic region: defective ATPase and oligomerization activities and altered phosphorylation accompany an inability to complex with cellular p53. J Virol. 1989 Aug;63(8):3362–3367. doi: 10.1128/jvi.63.8.3362-3367.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tack L. C., Wright J. H. Altered phosphorylation of free and bound forms of monkey p53 and simian virus 40 large T antigen during lytic infection. J Virol. 1992 Mar;66(3):1312–1320. doi: 10.1128/jvi.66.3.1312-1320.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tominaga T., Yogo Y., Kitamura T., Aso Y. Persistence of archetypal JC virus DNA in normal renal tissue derived from tumor-bearing patients. Virology. 1992 Feb;186(2):736–741. doi: 10.1016/0042-6822(92)90040-v. [DOI] [PubMed] [Google Scholar]
- Wang E. H., Friedman P. N., Prives C. The murine p53 protein blocks replication of SV40 DNA in vitro by inhibiting the initiation functions of SV40 large T antigen. Cell. 1989 May 5;57(3):379–392. doi: 10.1016/0092-8674(89)90913-6. [DOI] [PubMed] [Google Scholar]
- Weiner B. M., Bradley M. K. Specific mutation of a regulatory site within the ATP-binding region of simian virus 40 large T antigen. J Virol. 1991 Sep;65(9):4973–4984. doi: 10.1128/jvi.65.9.4973-4984.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte P., Williamson N. M., Harlow E. Cellular targets for transformation by the adenovirus E1A proteins. Cell. 1989 Jan 13;56(1):67–75. doi: 10.1016/0092-8674(89)90984-7. [DOI] [PubMed] [Google Scholar]
- de Villiers E. M. Heterogeneity of the human papillomavirus group. J Virol. 1989 Nov;63(11):4898–4903. doi: 10.1128/jvi.63.11.4898-4903.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]





