Abstract
Background:
Amelogenins are the major components of enamel matrix proteins. Enamel matrix derivatives (EMD) can be used in periodontal diseases to regenerate periodontal tissues. The main aim of this study was to evaluate expression of full-length functional recombinant human amelogenin (rhAm) in Iranian lizard Leishmania (I.L.L.) as an alternative eukaryotic expression system.
Methods:
Human cDNA encoding a 175-amino acid amelogenin expression cassette was sub cloned into a pLEXSY vector. The construct was transferred into Leishmania cells by electroporation. The protein production was surveyed in the transcription and the translation levels. The expressed protein was purified and some of its biological properties were investigated in comparison to EMD and negative control.
Results:
Expression of rhAm was confirmed by RT-PCR and western blot test in Leishmania cells. Purified rhAm significantly inhibited the formation of tartrate-resistant acid phosphatase positive (TRAP+) multinuclear cells in calcitriol stimulated mouse marrow cultures. Moreover, it significantly promoted proliferation and DNA synthesis in L929 mouse fibroblast cells.
Conclusion:
Functional rhAm was successfully expressed in I.L.L. Easy handling and post translation modification were the main advantages of this expression system. It is suggested to investigate molecular properties of this rhAm in the future.
Keywords: Amelogenin, Eukaryotic expression system, Osteoclastogenesis, Lizard, Leishmania, Recombinant protein, Iran
Introduction
Amelogenins play an important role in the mineralization and organization of enamel structure (1). These proteins constitute 90% of the enamel extracellular proteins (2). The human amelogenin gene has been localized to both X and Y-chromosomes with 90% of the transcripts expressed from the X (AMELX) (3, 4). AMELEX contains 7 exons, which undergo alternative mRNA splicing. The most abundant isoform of the native lacks the internal region encoded by exon 4, related cDNA (Gen-Bank Accession No. M86932) encodes for a 175 amino acid protein (5–7). Periodontal disease is a chronic and progressive destruction of the periodontal tissues (8).
Previous studies revealed that amelogenins have a role in the regeneration of all the periodontal tissues: cementum, periodontal ligament, and bone (9–12). Amelogenin inhibits osteoclastogenesis via down regulation of RANKL, M-CSF and fibronectin expression in osteoblasts (13). It also promotes proliferation and migration of fibroblast cells (14). Emdogain® is an acidic extract of extracellular enamel matrix that can be used in the treatment of periodontal disease in human (15). Due to heterogeneity of EMD and protein purification difficulties, many researches were done to reach a large quantity of isoform recombinant amelogenin in the last twenty years (12, 16). The amelogenin molecule is phosphorylated and has many folding in its structure, providing a eukaryotic expression system for reaching an activated recombinant human amelogenin (rhAm).
Leishmania tarentolae with an expression vector, pLEXSY, has been used as a eukaryotic protein expression system previously (17, 18). Leishmania sp. protozoa are from the family of Trypanosomatidae. Regulation of protein expression in these species occurs mainly on the level of RNA and may be influenced by the structure of the inter-genic regions. They have high growth rates and easy handling like E. coli and yeast expression systems (19, 20).The post translation modification in these cells is very similar to mammalian cells in comparison to E. coli and yeast expression systems (21).
We designed this study to express functional rhAm in Iranian lizard Leishmania (I.L.L.) (22) as a novel expression system to obtain functional rhAm.
Materials and Methods
Plasmid construction and gene cloning
The human amelogenin gene sequence (Gen Bank Accession Number: M86932 variant: 2), encoding mature amelogenin protein (175 amino acid), was synthesized into pGH vector and added SalI and NheI restriction sites at up and downstream relatively. The synthesized gene was amplified by M13 primers (Table 1) and digested by using SalI and NheI restriction enzymes then sub cloned into pLEXSY-hyg2 plasmid that encoded 6His tag gene (EGE-232, Jena Bioscience, Germany). Recombinant pLEXSY-hyg-2-hAm was transformed into E. coli XL1-Blue (stratagene, UK). Transformed colony was selected via colony PCR by using P1442 and A264 primers (Table 1) which is flanking the multiple cloning site of pLEXSY-hyg2 plasmid. Final confirmation performed by sequencing the PCR product by using hAm primers (Table 1). The study was approved by the Ethical Committee of the university.
Table 1:
Primers sequence used in this study
| Name | Sequence (5′ to 3′) |
|---|---|
| M13 F | GTAAAACGACGGCCAGTG |
| M13R | GGAAACAGCTATGACCATG |
| hAm F | GTCGACATGGGGACCTGGATT |
| hAm R | GCTAGCATCCACTTCCTCCCG |
| P 1442 | CCGACTGCAACAAGGTGTAG |
| A 264 | CATCTATAGAGAAGTACACGTAAAAG |
| F 3001 | GATCTGGTTGATTCTGCCAGTAG |
Cultivation of Iranian Lizard Leishmania (I.L.L.)
The I.L.L. cells were cultivated at 26°C in LB broth medium (Himedia, India) containing 100U/ml penicillin, 100mg/mlstreptomycin (GIBCO, USA) under shaking condition (110 rpm) and passaged by diluting suspension 5–10 fold in fresh media every 3–5 days (23).
Transfection of I.L.L. cells
108 cells of I.L.L. (22) were washed by electroporation buffer (eppendorf, Germany) and suspended in 1 ml electroporation buffer. The recombinant pLEXSY-hyg2-hAm linearized by SwaI (Ferments, Lithuania). Then 50μl of linearized DNA containing 5μgr DNA was added to 450 μl of cell suspension containing 108 cell/min a 4 mm cuvette. The I.L.L. cells were transfected by using two pulse 2000V (interval: 10S) in a Multiporator (eppendorf, Germany) (24). Stable transfectants were selected on LB broth media containing 100 μg/ml hygromycin (sigma). Integration of the expression cassette into the small subunit rRNA (ssu) locus of I.L.L. was confirmed by diagnostic PCR reaction by using F3001 forward primer (located on ssu locus of I.L.L. genome) and hAm reverse primer (Table 1).
Expression of rhAm in the I.L.L. cells
Expression of rhAm in the I.L.L. cells was evaluated by RT-PCR, SDS-PAGE and western blot tests. Total RNA was extracted from the cells by using RNeasy mini kit (QIAGEN). Gene cDNA was synthesized by random hexamers and amplified by PCR with hAm specific primers (Table 1). To determine rhAm protein in cells, transfected promastigotes were analyzed on 12% SDS-PAGE gel. For detection secretory rhAm, supernatant of cultures was filtered (0.2 μm). Then all proteins in supernatant were precipitated by ice-cold trichloroacetic acid 50%. The pellet was prepared and analyzedon 12% SDS-PAGE gel (25). The same procedure was carried out by I.L.L. wild type of Leishmania and compared with transformed one.
For Western blot analysis, protein bands were blotted on the nitrocellulose membrane then it was blocked by 3% skimmed milk in TBS buffer (20 mM Tris and 150 mM NaCl, pH 7.6) for 1 hour. Then membrane was washed and incubated with 1/1000 diluted rabbit polyclonal antibody to hAm (Abcam, UK) for 2 hours at room temperature. The washing process repeated again and incubated with 1/2000 diluted alkaline phosphatase conjugated goat polyclonal anti-rabbit antibody (Abcam, UK) for an additional 2 hours at room temperature. Finally, alkaline phosphatase substrate (NBT-BCIP) was added to membrane pre-soaked to alkaline phosphates buffer. Emdogain® (Straumann, USA) was used as positive control for western blotting test.
Purification of rhAm by Affinity Chromatography
The rhAm fused to his-tag was purified from cell lysate by using Ni-NTA His•Bind® resins (Novagen, Germany) under nature condition. The purified protein was concentrated and desalted by using Amicon Ultra-15 units with a cut-off of 10 kDa (Millipore, Germany).
Osteoclast differentiation Assay
Osteoclasts were generated from mice bone marrow nucleated cells. 6-week-old Swiss-Webster male mice were sacrificed by cervical dislocation. The procedures were reviewed and approved by the Shahid Beheshti University of Medical Sciences Ethics Committee. These cells were collected from the femurs and tibias of mice, as described by Holliday (26). The marrow cells were suspended in α-MEM that contained 10% FBS and 10−8 M active metabolite of vitamin D3, 1, 25-dihydroxy-cholecalciferol [1,25 (OH)2D3]. Cell suspension was plated in 24-well plate at a density of 500 000 nucleated cells/cm2. The cultures were maintained at 37 °C in a humidified atmosphere of 98% air and 5% CO2. After 6 days, the media was removed and adherent cells were fixed with 2% paraformaldehyde. Tartrate-resistant acid phosphatase (TRAP) staining was performed according to the kit instruction (sigma, USA). TRAP+ mononuclear cells (less than three nuclei) and TRAP+ multinucleated cells (three or more nuclei) were counted under a 40X microscope from 3 wells in 10 random areas per well in each group (27).
Proliferation Assay
L929 mouse fibroblast cell line was obtained from National Cell Bank of Iran (Pasteur Institute of Iran). The L929 cell line was sub-cultured and then cell suspension was prepared in DMEM media. The cells were seeded in 96-well cell culture plate at a density 5×103 per well. After 24 h, it was exchanged with fresh serum free media. EMD and rhAm were added to related groups at the 1μg/ml and 10μg/ml concentrations but not into control group. The cultures were maintained at 37 °C in a humidified atmosphere of 98% air and 5% CO2 for 4 days. Then 20 μl of MTT solution (0.5mg/ml) (Sigma, USA) was added per well and incubated for 4h to form formazan crystals in viable cells. The crystals were dissolved in acidic isopropanol and absorbance was measured at 570nm with reference filter 620 (16).
DNA synthesis
DNA synthesis was measured by BrdU colorimetric kit (Roch, Germany). The L929 cell culture was prepared and exposed as the same as MTT assay. The cells were incubated for 2 hours with BrdU and then BrdU incorporation was measured in accordance with the manufacturer’s instruction BrdU ELISA kit (16).
Statistical analysis
All experiments were performed at least three times. Descriptive statistics (means and standard deviations) were calculated. Shapiro-Wilk and Levene tests were used to check normality and homogeneity of variance assumptions respectively. One way ANOVA was used for comparison among groups. Pairwise comparisons between groups of interest were performed using Benjamini-Hochberg post hoc test. The P<0.05 was considered statistically significant.
Results
Plasmid Construction and gene Cloning
The hAm gene (531 bp) was sub cloned into SalI and NheI sites of pLEXSY-hyg 2 (Fig. 1).
Fig. 1:
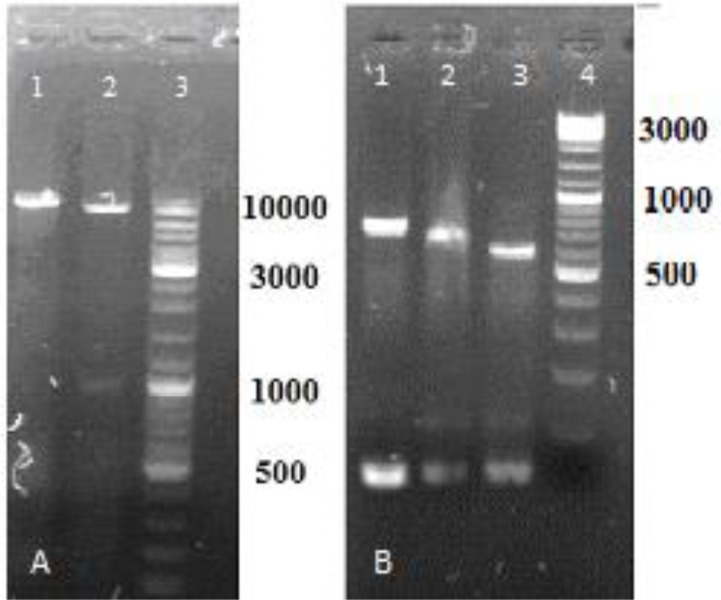
Digestion by NheI and SalI restriction enzymes, (A) pLexyhyg2; lane1, digestion by NheI; lane2, digestion of stuffer fragment by NheI and SalI (1000bp); lane3, DNA size marker 100–100000 bp (Fermentas, Lithuania). (B) PCR product digestion; lane1 PCR product 717bp.; lane2 digestion by NheI (618 bp); lane3 digesestion by NheI and SalI 531bp. fragment; lane4, DNA size marker 100–3000 bp (Fermentas, Lithuania)
The Fig. 2 shows PCR product of recombinant pLEXSY-hyg 2-hAm plasmid (830 bp fragment). Sequencing results confirmed the presence of hAm gene in recombinant construct in an accuracy position.
Fig. 2:
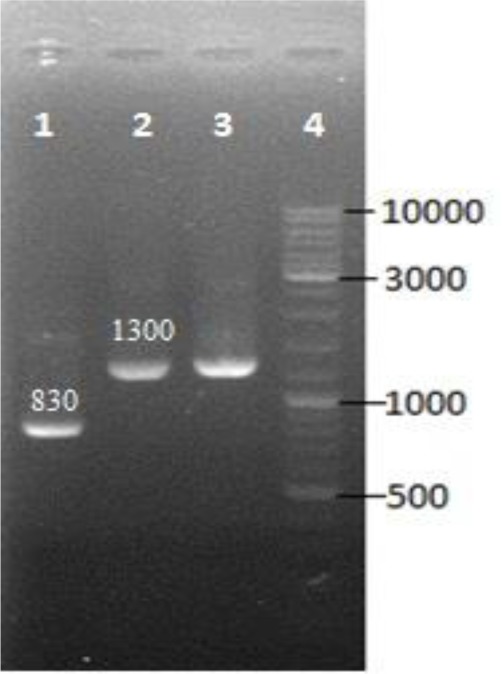
Colony PCR by using P1442 and A264 primers flanking the multiple cloning site of pLEXSY-hyg2 plasmid was performed.; lane 1, desired colony (830 bp fragment) lane 2,3 colony with re-ligation of the stuffer segment of vector with the 1300 bp fragment and lane4, DNA size marker
Transfection and Selection of Recombinant I.L.L. cells
Recombinant pLEXSY-hyg2 plasmid was transfected into I.L.L. promastigote by electroporation. They were cultured in the presence of hygromycin (up to100 μg/ml). Resistant transfected promastigotes were surveyed by PCR reaction. The PCR products of transfected promastigotes 1828 bp and non-transfected promastigotes are shown in Fig. 3.
Fig. 3:
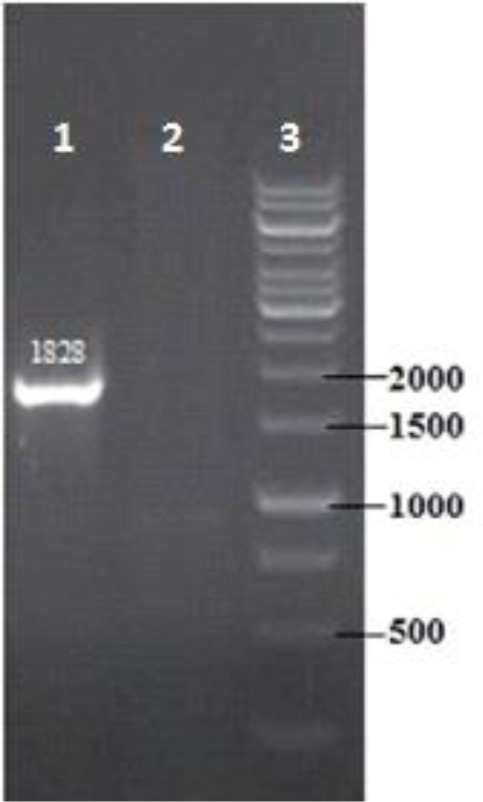
Confirmation of genomic integration by PCR. Diagnostic PCR was performed by using forward primer, hybridizing to the ssu sequence (F3001) and reveres primer of hAM within expression cassette. Lan 1, transfected I.L.L; lane 2 wildtype control, lane 3, DNA size marker
Analysis of Expression Recombinant hAm
RT-PCR was performed on transfected and non-transfected I.L.L. promastigotes by using hAm specific primers. The length of desired product should be 537 bp (Fig. 4). Expressed hAm protein was electrophoresed on SDS-PAGE, transferred on nitrocellulose membrane and analyzed by western blot compared to cell culture supernatant. SDS-PAGE gel staining results show that two bands 19kDa and 50kDa in recombinant cell lysate were stronger than the control and there is no obvious difference between case and control groups in cell culture supernatant.
Fig. 4:
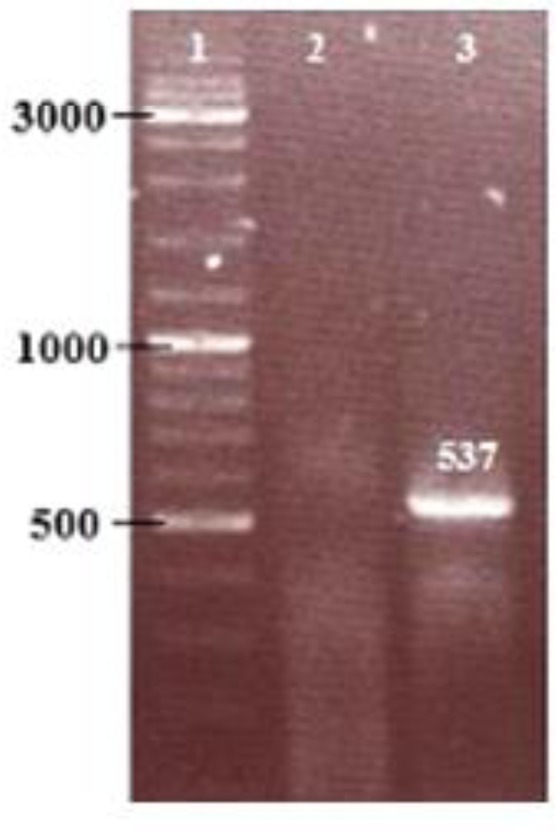
RT - PCR analysis of gene expression. PCR was performed by using hAm forward and reverse primers. lane 1, DNA size marker; lane 2, non-transfected control cDNA; lane 2, transfected DNA
Theoretical molecular weight of rhAm include signal peptide and His-tag is about 23 kDa and without them is about 20 kDa. In western blot test 19 kDa, 30 kDa, 50 kDa, 52kDa and 70 kDa isoforms were observed in recombinant cell lysate but not in culture supernatant comparison to control group (Fig. 5).
Fig. 5:

Analysis of recombinant hAm expression by using the specific polyclonal antibody to hAmin western blot test. lane 1, non-transfected cell lysate; lane 2, transfected cell lysate; lane 3, protein size marker in kDa.; lane 4, purified rhAm from cell lysate by using Ni-NTA His•Bind® resins, lane 5, positive control (Emdogaine) for western blot test; lane 6, supernatant of cell culture related to transfected I.L.L.; lane 7, supernatant of cell culture related to non-transfected I.L.L
Purification of rhAM
Purification of rhAm from cell lysate was performed by using Ni-NTA His•Bind® resins. Western blot test showed three bands in 19 kDa, 30 kDa, 37 kDa and a weak band in 57 kDa (Fig. 5).
Osteoclast differentiation assay
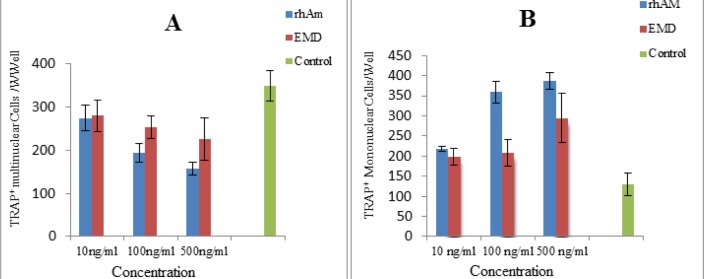
Osteoclast precursors in mouse bone marrow can be induced to form multinucleated osteoclast-like cells in the presence of 1, 25 (OH)2D3 (28). Formation of TRAP+ multinuclear cells was inhibited significantly in presence of rhAm in all doses [10ng/ml, 100ng/ml and 500ng/ml (P=0.0001)] in comparison to control group. Similarly EMD was inhibited multinuclear formation in comparison to control group [10 ng/ml (P= 0.039), 100 ng/ml (P=0.009) and 500ng/ml (P=0.0001)]. Counting of TRAP+ mononuclear was increased significantly in presence of rhAm in all doses in comparison to control group [10ng/ml (P=0.006), 100ng/ml (P=0.0001) and 500ng/ml (P=0.0001)]. Also number of the TRAP+ mononuclear was increased significantly in the presence of EMD in all doses in comparison to control group. [10ng/ml (P= 0.023), 100ng/ml (P=0.014) and 500ng/ml (P=0.0001)] (Fig. 6).
Fig. 6:
Bar chart of counting TRAP+ multinuclear cells, part A, and TRAP+ mononuclear cells, part B, in expose to different doses of rhAm and EMD in comparison to control group. Data are means ± SD of result from three determinations.
A) Formation of the TRAP+ multinuclear was inhibited in all concentration of rhAm and EMD groups in comparison to control group. There was significant difference between rhAm and EMD groups in 500 ng/ml in number of TRAP+ multinuclear (P=0.04) but not in10 ng/ml and 100 ng/ml doses. B) There was an increase in the number of TRAP+ mononuclear cells in all groups in comparison to control group. There was no significant difference between rhAm and EMD group in 10 ng/ml dose, but there was in 100 ng/ml (P=0.0001) and 500 ng/ml (P=0.005) in the number of mononuclear cells. Bars represent standard deviations (SD)
Proliferation Assay
L929 fibroblast cell line was incubated in serum free DMEM with rhAm or EMD for 4days. MTT assay showed that exposed cells with rhAM were proliferated in a dose dependent manner in comparison to control group (Fig. 7).
Fig. 7:
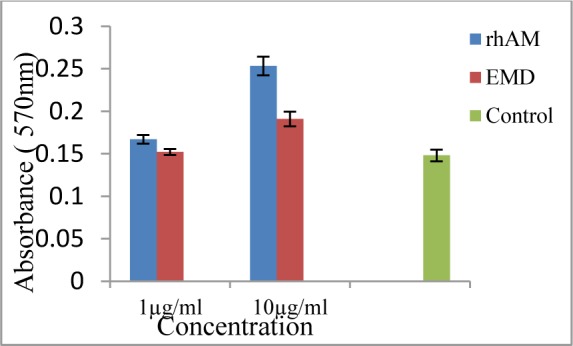
Proliferation assay was performed by using L929 mouse fibroblast cell line and MTT salt. Cell proliferation significantly increased in the presence of 1μg/ml rhAm (P=0.015), 10μg/ml rhAm (P=0.0001) and10μg/ml EMD (P=0.0001) but not in1μg/ml EMD in compare to control group. There was a significant difference between 1μg/ml rhAm treated group and 1μg/ml EMD treated group (P=0.04). Also there was a significant difference between 10μg/ml rhAm treated group and 10μg/ml EMD treated group (P=0.0001). Bars represent standard deviations (SD)
DNA synthesis
Incubation of L929 fibroblast cell line was performed with rhAm or EMD in a serum free media for 4 days. BrdU test revealed that DNA synthesis was increased in rhAm group in a dose dependent manner in comparison to control group (Fig. 8).
Fig. 8:
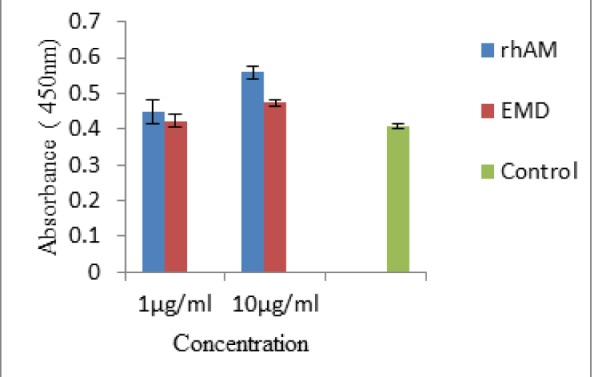
Brdu incorporation in L929 cell line was significantly increased in the presence of 1μg/mlrhAm (P=0.041), 10μg/ml rhAm (P=0.0001) and10μg/ml EMD (P=0.003) but not in1μg/ml EMD in comparison to the control group. There was a significant difference between 10μg/ml rhAm treated group and 10μg/ml EMD treated group (P=0.0001) but not in 1μg/ml concentration. Bars represent standard deviation (SD)
Discussion
Amelogenins are the major components of enamel matrix proteins. These proteins are translated from mRNA splicing variants then undergo post-translational modification and they play an important role in the mineralization and organization of enamel structure (29–31). These highly conserved proteins have tendency to the unique physiological characteristic of self-assemble into spherical mono dispersed aggregates (15).
In this study we transfected I.L.L. cells with the pLEXSY-hyg2-hAM vector included hAm gene (Gen Bank Accession Number: M86932 variant: 2) that encoded a full-length recombinant human amelogenin fused with the his-tag in C-terminal. The western blot test confirmed that two stronger bands (19 and 50 kDa) in SDS-PAGE gel were amelogenin in transfected I.L.L. cells. Also the western blot test revealed additional bands in 30, 52 and 70 kDa. This result is due to the nature of amelogenin proteins to tendency to form aggregation (10, 16, 32). To determine exact molecular weight characterization of produced rhAM, it is suggested to use mass spectrometry. Enamel Matrix Derivative extracted from porcine enamel can be used for restoration three periodontal tissue types in periodontal diseases (33). EMD is composed of heterogenic proteins therefore many researches and efforts were done to reach a large quantity of isoform recombinant amelogenin (10, 16, 32). Expression of rhAm has been reported in different expression systems. For the first time, in 1996, Deutsch et al. prepared a human cDNA, encoding for the 175-amino-acid amelogenin, by RT-PCR and sub-cloned into pGEX-KG expression plasmid for over-expression in E. coli (32). The E. coli expression system lacked cell machinery to encourage correct folding and post translational modification of proteins. Taylor et al., expressed rhAm in the eukaryotic baculo virus expression system (10). Cheng et al. used yeast, P. pastoris, to express rhAm, and then they investigated the biological and molecular characteristics of rhAm (16).
One of the new eukaryotic expression systems Leishmania tarentolae (Jena Bioscience LEXY host P10) has been used to express many proteins (18, 34). In this study, we used Iranian lizard Leishmania (22) for the expression of rhAm for the first time. Some advantages of this system are a) easy handling like E. coli and yeast expression systems. b) Cultivation in low cost media like LBc) eukaryotic protein synthesis and post translation modification more likely to mammalian cells than yeast cells. d) High cell density in suspension culture e) human safety d) extremely stable over hundreds of generation (21). To investigate biological function of rhAm in our lab, the effect of this product on ostoclastogenesis and fibroblast proliferation was surveyed.
Mouse bone marrow cultures contain both osteoblasts and osteoclasts. By adding 1,25 (OH)2D3 to culture media, osteoblasts were stimulated to increase production of RANKL, which stimulates osteoclasts progenitors to mature into osteoclasts (26). Previously Nishiguchi demonstrated that amelogenin inhibits osteoclastogenesis via down regulation of RANKL, M-CSF and fibronectin expression inosteoblasts (13). In this study, the effect of rhAm on ostoclastogenesis was determined in three doses (10, 100 and 500 ng/ml) compared to similar doses of EMD and control. Multinucleated cell formation was inhibited by different doses of rhAm and EMD. Expectedly, the mononuclear cells’ counting was increased in the presence of different doses of rhAm and EMD in comparison to control group. Leucine-rich amelogenin peptide (LRAP) inhibited osteoclast formation in the co-cultures of osteoclast progenitor and cementoblast/periodontal ligament cells of mice. LRAP significantly reduced RANKL expression and the TRAP-positive cells in 10 and 100 ng/ml concentrations (9). Porcine recombinant amelogenin significantly decreased the number of human TRAP+ odontoclastic cells in appropriate cell culture environment (35). Our study supported previous studies and revealed that rhAm has biological activity. On the other hand, EMD promoted migration and proliferation of gingival fibroblasts (36). rhAm (expressed in yeast) and EMD increased proliferation and DNA synthesis in human PDLF (16). Amelogenin stimulate fibroblast signaling, proliferation and migration via integrin interactions (14). In our study, MTT assay and BrdU incorporation assay were performed to investigate proliferation effect of rhAm L929 fibroblast cells. MTT assay showed that exposed cells with rhAM were proliferated in a dose dependent manner in comparison to control group. Also the proliferative effect of rhAm significantly was more than EMD at the same concentration. In this regard, BrdU test revealed that DNA synthesis was increased in the presence of rhAm in a dose dependent manner in comparison to the control group. Our investigation supported previous studies and confirmed some of the biological properties of amelogenin protein. Also this study supported responsibility of amelogenin for therapeutic effect of EMD. Further, it revealed that produced rhAm in I.I.L. has biological activity. In this study, we could not indicate any secreted amelogenin in media supernatant. This may be due to the lower efficiency of the LmSAP (acid phosphatase of L. mexicana) signal peptide in recombinant I.L.L. Secreted protein may be enhanced by altering some amino acids codon in signal peptide gene (37).
The final yield of purified protein was about 4 mg/l that was low in comparison to protein expressed in L. tarentolae (Jena Bioscience LEXY host P10) (17, 21) but it was compatible to protein expressed in I.L.L. (20, 25). It is suggested to characterize I.L.L. and its proteolytic enzymes.
Conclusion
We expressed and purified rhAm in a domestic Leishmania host as a new eukaryotic expression system for the first time. The biological activity of the rhAM was corroborated. The main advantages of this expression system are its easy handling like E. coli and yeast expression systems, post translation modification and cultivation in low cost media such as LB.
Ethical considerations
Ethical issues (Including plagiarism, informed consent, misconduct, data fabrication and/or falsification, double publication and/or submission, redundancy, etc.) have been completely observed by the authors.
Acknowledgements
This project was supported by a Deputy in Research of Shahid Beheshti University of Medical Sciences through grant No. 115-7950 and it was done in Cellular and Molecular Biology Research Center and Biotechnology Department of Shahid Beheshti University of Medical Sciences, Tehran, Iran. The authors declare that there is no conflict of interests.
References
- 1. Termine JD, Belcourt AB, Christner PJ, Conn KM, Nylen MU. (1980). Properties of dissociatively extracted fetal tooth matrix proteins. I. Principal molecular species in developing bovine enamel. J Biol Chem, 255( 20): 9760–9768. [PubMed] [Google Scholar]
- 2. Simmer JP, Lau EC, Hu CC, Aoba T, Lacey M, Nelson D, et al. (1994). Isolation and characterization of a mouse amelogenin expressed in Escherichia coli. Calcif Tissue Int, 54( 4): 312–319. [DOI] [PubMed] [Google Scholar]
- 3. Lau EC, Mohandas TK, Shapiro LJ, Slavkin HC, Snead ML. (1989). Human and mouse amelogenin gene loci are on the sex chromosomes. Genomics, 4( 2): 162–168. [DOI] [PubMed] [Google Scholar]
- 4. Salido EC, Yen PH, Koprivnikar K, Yu LC, Shapiro LJ. (1992). The human enamel protein gene amelogenin is expressed from both the X and the Y chromosomes. Am J Hum Genet, 50( 2): 303–316. [PMC free article] [PubMed] [Google Scholar]
- 5. Baba O, Takahashi N, Terashima T, Li W, DenBesten PK, Takano Y. (2002). Expression of alternatively spliced RNA transcripts of amelogenin gene exons 8 and 9 and its end products in the rat incisor. J Histochem Cytochem, 50( 9): 1229–1236. [DOI] [PubMed] [Google Scholar]
- 6. Li W, Mathews C, Gao C, DenBesten PK. (1998). Identification of two additional exons at the 3′ end of the amelogenin gene. Arch Oral Biol, 43( 6): 497–504. [DOI] [PubMed] [Google Scholar]
- 7. Richard B, Delgado S, Gorry P, Sire JY. (2007). A study of polymorphism in human AMELX. Arch Oral Biol, 52( 11): 1026–1031. [DOI] [PubMed] [Google Scholar]
- 8. Newman MG, Takei H, Klokkevold PR, Carranza FA. (2012). Carranza’s clinical periodontology, 11th ed W.B. Saunders Co. United States of America, pp.: 626–630. [Google Scholar]
- 9. Hatakeyama J, Philp D, Hatakeyama Y, Haruyama N, Shum L, Aragon MA, et al. (2006). Amelogenin-mediated regulation of osteoclastogenesis, and periodontal cell proliferation and migration. J Dent Res, 85( 2): 144–149. [DOI] [PubMed] [Google Scholar]
- 10. Taylor AL, Haze-Filderman A, Blumenfeld A, Shay B, Dafni L, Rosenfeld E, et al. (2006). High yield of biologically active recombinant human amelogenin using the baculovirus expression system. Protein Expression and Purification, 45( 1): 43–53. [DOI] [PubMed] [Google Scholar]
- 11. Robinson C, Brookes SJ, Shore RC, Kirkham J. (1998). The developing enamel matrix: nature and function. Eur J Oral Sci, 106: 282–291. [DOI] [PubMed] [Google Scholar]
- 12. Haze A, Taylor AL, Haegewald S, Leiser Y, Shay B, Rosenfeld E, et al. (2009). Regeneration of bone and periodontal ligament induced by recombinant amelogenin after periodontitis. J Cell Mol Med, 13( 6): 1110–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nishiguchi M, Yuasa K, Saito K, Fukumoto E, Yamada A, Hasegawa T, et al. (2007). Amelogenin is a negative regulator of osteoclastogenesis via downregulation of RANKL, M-CSF and fibronectin expression in osteoblasts. Arch Oral Biol, 52( 3): 237–243. [DOI] [PubMed] [Google Scholar]
- 14. Almqvist S, Werthen M, Johansson A, Agren MS, Thomsen P, Lyngstadaas SP. (2010). Amelogenin is phagocytized and induces changes in integrin configuration, gene expression and proliferation of cultured normal human dermal fibroblasts. J Mater Sci Mater Med, 21( 3): 947–954. [DOI] [PubMed] [Google Scholar]
- 15. Gestrelius S, Lyngstadaas S, Hammarström L. (2000). Emdogain–periodontal regeneration based on biomimicry. Clin Oral Investig, 4( 2): 120–125. [DOI] [PubMed] [Google Scholar]
- 16. Cheng L, Lin ZK, Shu R, Liu DL, Zhang XL, Liu B, et al. (2012). Analogous effects of recombinant human full-length amelogenin expressed by Pichia pastoris yeast and enamel matrix derivative in vitro. Cell Prolif, 45( 5): 456–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Breitling R, Klingner S, Callewaert N, Pietrucha R, Geyer A, Ehrlich G, et al. (2002). Nonpathogenic trypanosomatid protozoa as a platform for protein research and production. Protein Expr Purif, 25( 2): 209–218. [DOI] [PubMed] [Google Scholar]
- 18. Dortay H, Schmöckel SM, Fettke J, Mueller-Roeber B. (2011). Expression of human c-reactive protein in different systems and its purification from Leishmania tarentolae. Protein Expr Purif, 78( 1): 55–60. [DOI] [PubMed] [Google Scholar]
- 19. Fritsche C, Sitz M, Weiland N, Breitling R, Pohl HD. (2007). Characterization of the growth behavior of Leishmania tarentolae–a new expression system for recombinant proteins. J Basic Microbiol, 47( 5): 384–393. [DOI] [PubMed] [Google Scholar]
- 20. Mirzaahmadi S, Asaadi-Tehrani G, Bandehpour M, Davoudi N, Tahmasbi L, Hosseinzadeh N, et al. (2011). Expression of recombinant human coagulation factor VII by the Lizard Leishmania expression system. J Biomed Biotechnol, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Niimi T. (2012). Recombinant protein production in the eukaryotic protozoan parasite Leishmania tarentolae: a review. In: Recombinant Gene Expression, Reviews and protocols. Springer Protocols. 3rd ed, Humana Press; pp. 307–315. [DOI] [PubMed] [Google Scholar]
- 22. Kazemi B, Tahvildar-Bidroni GH, Hashemi Fesharaki SR, EJ (2004). Isolation a lizard Leishmania promastigote from its natural hostin Iran. J Biol Sci, 4( 5): 620–623. [Google Scholar]
- 23. Kazemi B, Heidari MH, Naderi M, Piryaei A, Nazari-Pouya M-R. (2008). Study on ultrastructure of Leishmania major and Lizard leishmania. J Cell Anim Biol, 2( 6): 129–133. [Google Scholar]
- 24. Beverley SM, Clayton CE. (1993). Transfection of Leishmania and Trypanosoma brucei by Electroporation. Protocols in molecular parasitology: Methods Mol Biol, 21: 333–348. [DOI] [PubMed] [Google Scholar]
- 25. Taromchi AH, Kazemi B, Mahmazi S, Bandehpour M. (2013). Heterologous Expression of Human IL-29 (IFN-lambda 1) in Iranian Lizard Leishmania. Iran J Biotech, 11( 3): 168–174. [Google Scholar]
- 26. Holliday LS, Dean AD, Greenwald JE, G SL. (1995). C-type natriuretic peptide increases bone resorptionin 1,25-dihydroxy vitamin D3 stimulated mouse bone marrow cultures. J Biol Chem, 270: 18983–18989. [DOI] [PubMed] [Google Scholar]
- 27. Otsuka E, Kato Y, Hirose S, Hagiwara H. (2000). Role of Ascorbic Acid in the Osteoclast Formation: Induction of Osteoclast Differentiation Factor with Formation of the Extracellular Collagen Matrix 1. Endocrinology, 141( 8): 3006–3011. [DOI] [PubMed] [Google Scholar]
- 28. Takahashi N, Yamana H, Yoshki S, Roodman GD, Mundy GR, Jones SJ, et al. (1988). Osteoclast-Like Cell Formation and its Regulation by Osteotropic Hormones in Mouse Bone Marrow Cultures. Endocrinology, 122( 4): 1373–1382. [DOI] [PubMed] [Google Scholar]
- 29. Fincham AG, Moradian-Oldak J, Simmer JP, Sarte P, Lau EC, Diekwisch T, et al. (1994). Self-assembly of a recombinant amelogenin protein generates supramolecular structures. J Struct Biol, 112( 2): 103–109. [DOI] [PubMed] [Google Scholar]
- 30. Moradian-Oldak J, Simmer JP, Lau EC, Sarte PE, Slavkin HC, Fincham AG. (1994). Detection of monodisperse aggregates of a recombinant amelogenin by dynamic light scattering. Biopolymers, 34( 10): 1339–1347. [DOI] [PubMed] [Google Scholar]
- 31. Diekwisch T, David S, Bringas P, Jr., Santos V, Slavkin HC. (1993). Antisense inhibition of AMEL translation demonstrates supramolecular controls for enamel HAP crystal growth during embryonic mouse molar development. Development, 117( 2): 471–482. [DOI] [PubMed] [Google Scholar]
- 32. Deutsch D, Chityat E, Hekmati M, Palmon A, Farkash Y, Dafni L. (1996). High expression of human amelogenin in E. coli. Adv Dent Res, 10( 2): 187–193; discussion 194. [DOI] [PubMed] [Google Scholar]
- 33. Hammarström L, Heijl L, Gestrelius S. (1997). Periodontal regeneration in a buccal dehiscence model in monkeys after application of enamel matrix proteins. J Clin Periodontol, 24( 9 Pt 2): 669. [DOI] [PubMed] [Google Scholar]
- 34. Sugino M, Niimi T. (2012). Expression of multisubunit proteins in Leishmania tarentolae. In: Recombinant Gene Expression, Reviews and protocols. Springer Protocols. 3rd ed, Humana Press; pp. 317–325. [DOI] [PubMed] [Google Scholar]
- 35. Yagi Y, Suda N, Yamakoshi Y, Baba O, Moriyama K. (2009). In vivo application of amelogenin suppresses root resorption. J Dent Res, 88( 2): 176–181. [DOI] [PubMed] [Google Scholar]
- 36. Rincon JC, Haase H, Bartold P. (2003). Effect of Emdogain® on human periodontal fibroblasts in an in vitro wound-healing model. J Periodont Res, 38( 3): 290–295. [DOI] [PubMed] [Google Scholar]
- 37. Klatt S, Konthur Z. (2012). Secretory signal peptide modification for optimized antibody-fragment expression-secretion in Leishmania tarentolae. Microb Cell Fact, 11: 97. [DOI] [PMC free article] [PubMed] [Google Scholar]