Abstract
Background:
Limited studies have focused on the association between the protein tyrosine phosphates non-receptor type 22 (PTPN22) genetic polymorphisms and Juvenile idiopathic arthritis (JIA) susceptibility in different populations, but the results were inconclusive. Therefore, this meta-analysis of PTPN22 polymorphism (1858 C>T) was performed to get a precise systematic estimation. The “rs” number of the PTPN22 polymorphism (1858 C>T) is 4.
Methods:
A systematic literature search strategy was carried out using English databases (PubMed, Embase.) for the eligible studies. We ultimately identified 11 records from 10 articles involving the relationship between PTPN22 genetic polymorphisms and JIA risk from PubMed and Embase databases. Overall, 4552 cases and 10161 controls were investigated in this study to evaluate the association between PTPN22 (C allele vs. T allele) genotype and JIA susceptibility.
Results:
Analysis using random effects model showed an increased risk of JIA with T allele of rs2476601 vs. A allele (P<0.001). Subgroup analysis suggested that the PTPN22 polymorphism (1858C>T) was significantly associated with JIA risk in America population (OR=1.52, 95% CI:1.30–1.78). Additionally, the subgroup analysis also showed that the associations were still significant in case number more than 500 (OR=1.38, 95% CI: 1.04–1.83), while in the case number less than 500 was OR=1.55, 95% CI: 1.39–1.72.
Conclusions:
SNPs of PTPN22 (1858C>T) showed an increased risk of developing JIA.
Keywords: Phosphates non-receptor type 22, PTPN22, Polymorphism, Juvenile idiopathic arthritis, JIA, Meta-analysis
Introduction
Juvenile idiopathic arthritis (JIA) is one of the most common autoimmune diseases under the age of 16, which had been influenced by both genetic and environmental factors (1). The prevalence of JIA was 15 per 100,000 children/year (95% CI: 13–17) in the Nordic countries (2). Nevertheless, fewer studies have shown in Asian population. JIA has many complications such as macrophage activation syndrome, nodular regenerative hyperplasia of the liver and Uveitis (3). Among them, the boy can exist a more unfavorable prognosis of uveitis than in girls under the clinical course, although it was more susceptible to girls.
Several viruses such as Epstein-Barr virus could induce the appearance of JIA (4, 5). Meanwhile, familial aggregation studies, case-control studies yet had provided clues of the relationship between genetic variants such as PTPN22 (1858C>T), V-set Domain-containing T cell Activation Inhibitor 1 (VTCN1), methyl-CpG-binding protein 2 (MeCp2) and autoimmune diseases risk. However, eligible studies of the association between PTPN22 genetic polymorphism and JIA risk were controversial.
Protein tyrosine phosphates non-receptor type 22 (PTPN22 gene), located on chromosome 1p13, encodes specific lymphoid protein tyrosine phosphatase (LYP), is vital in negative regulation of T lymphocyte activation (6). The R620W polymorphism in PTPN22 gene at the nucleotide 1858 (1858C>T) in codon 620 (620Arg>Trp) has been associated with various autoimmune diseases. The disease-associated LYP variant Trp 620 could inhibt the interaction function of LYP with CSK. Consequently, an imbalanced regulation of T cell induction might generate by activiting the T cell receptor-associated kinases, and this process might be contributed to overactiving immune responses (7). For instance, PTPN22 (1858C>T) gene as a susceptible locus exists the potential relationship with Graves’ disease (GD) risk in European population (8) and Systemic Lupus Erythematosus (SLE) susceptiblity in Chinese population (9).
Considering available studies of PTPN22 polymorphism and JIA risk is not conclusive, we carried out an updated meta-analysis to discern the truly relationship.
Methods
Literature search
Records were screened from different databases including both PubMed and Embase database (all the studies were retrospected from February 2000 to July 2015). The keywords in Pubmed terms including “PTPN22 1858C/T” or “rs2476601” or “PTPN22 R620W” together with “Juvenile idiopathic arthritis” or “JIA” or “Juvenile rheumatoid arthritis” or “JRA” or “Juvenile chronic arthritis” or “JCA” or “Juvenile arthritis” or “JA”. The same retrieve strategy was also performed in Embase database. Meanwhile, scholar website such as http://scholar.google.com/ used to find full eligible records. The selective studies in our meta-analysis were abided by the criteria as follows: 1) case-control studies or cohort studies in population; 2) selected studies provided sufficient data to calculate pooled ORs (odds ratios) with the corresponding 95% CIs (Confidence Interval), which were used to evaluate the association of PTPN22 polymorphisms and JIA risk; 3) control population did not contain malignant tumor patients; 4) supplements, letters, case reports, review articles, conference papers and other meta-analysis were all excluded.
Data extraction and synthesis
All retrieved records from the databases inclusive or not were examined by two independent reviewers (ShiLing Zhong and Nan Sun), and disagreements were solved by a third researcher (YunYan Li). For each eligible study, the following characteristics were collected: first author, year of publication, ethnicity and region of the population being studied, polymorphisms examined, study time, pathologic, source of control, characteristics of cases and controls.
Statistical Analysis
All analysis were performed in stata version 12.0 (StataCorp Lp, College station, Texas, USA). The combined ORs and the corresponding 95% CIs were calculated and demonstrated in the forest plots by using the fixed or the random effects model. We used random effects model when P value of heterogenous test was no more than 0.1 (P≤ 0.1). When P value of heterogenous test was more than 0.1 (P>0.1), fixed effects model were used. Besides, heterogeneity was also measured in our meta-analysis through using Cochran’s Q and the inconsistency index (I2) statistic. Cumulative meta-analysis, sensitivity analysis and Galbraith plot were also used to find the potential information of meta-analysis. Subgroup analysis was used to investigate better possible reasons of between-study heterogeneity. The subgroups are as follows: geographical locations (European and Amecria, and other regions), number of case (<500 vs. ≥500), source of control (population-based vs. hospital-based). Meanwhile, publication bias was investigated by Begg’s funnel plot, in which the standard error of log OR of each study was plotted against its OR; Funnel-plot asymmetry was further assessed by the method of Egger’s linear regression test, which could assess the relationship between effect size, and variance differs between large and small studies.
Results
Study Selection and Study Characteristics
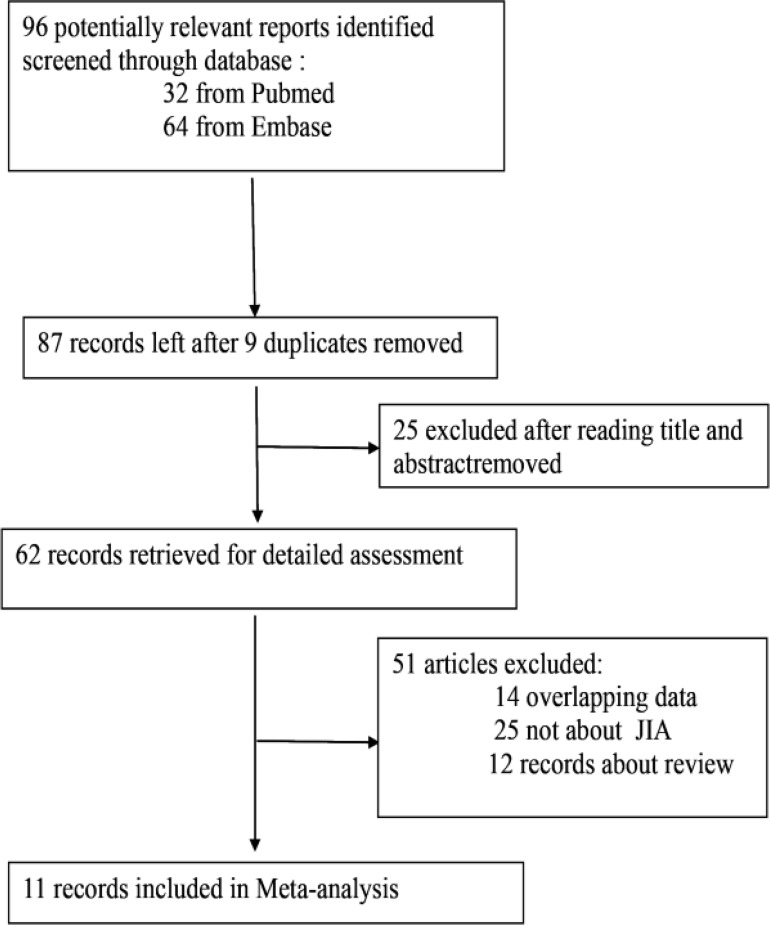
Based on search strategy above, 96 individual records were identified. However, only 62 full-texts were available for the further estimation. According to the inclusive and exclusive criteria, 51 articles were excluded including 14 overlapping articles, 25 uncorrelated articles and 12-review paper about JIA. We ultimately identified 11 records from 10 articles involving the relationship between PTPN22 genetic polymorphisms and JIA risk from Pubmed and Embase databases (10) in Fig. 1.
Fig. 1:
Flow chart of study selection
According to the criteria, all articles were screened carefully to assess the eligibility and reliability. The characteristics of the studies were showed (4552 cases and 10161 controls) in Table 1.
Table 1:
Characteristics of the studies related with the effects of PTPN22 genetic polymorphisms and JIA risk
| Ref | Ethnicity study time | Pathologic diagnosis | source of controls | case group | control group | OR | |
|---|---|---|---|---|---|---|---|
| 1 | Eurpean (Norway) | NA | ILAR | Population | 230 cases | 1400 controls | 1.17 (0.9–1.5) |
| 2 | European (British) | NA | ILAR | Population | 661 cases from the British | 595 controls from general practitioners | 1.53 (1.2–2.0) |
| 3 | European (Norway) | NA | ILAR | Hospital | 320 cases | 555 controls | 1.41 (1.01–1.96) |
| 4 | European (Czech) | 2006 | ILAR | Population | 130 cases | 400 controls | 2.7 (1.8–4.2) |
| 5 | European (Hungarian) | NA | ILAR | Population | 150 cases | 200 controls | 1.13 (0.66–1.95) |
| 6 | America | NA | ILAR * | Population | 809 cases from US and German | 2990 controls from same place | 1.65 (1.38–1.98) |
| 7 | America | NA | ILAR | Population | 809 cases from US and German | 2990 controls from same place | 1.64 (1.37–1.97) |
| 8 | Northern European | NA | ILAR | Population | 636 cases from Pediatric Rheumatology clinics at the University | 733 healthy adults (59% female) | 1.29 (1.02–1.62) |
| 9 | European (Greece) | NA | ILAR | Hospital | 128 cases(70.31% femal,29.69% male) | 221 controls were from Thessaloniki | 0.44 (0.21–0.97) |
| 10 | Australia | NA | ILAR | Hospital | 318 cases | 556 controls | 1.62 (1.15–2.3) |
| 11 | America(Utah) | NA | ILAR | population | 155 cases (85% femal,15% male) | 411 controls (66% female) | 1.61 (1.11–2.31) |
NA: not available; ILAR: International League of Associations for Rheumatology
Results
PTPN22 genotype with JIA susceptibility:
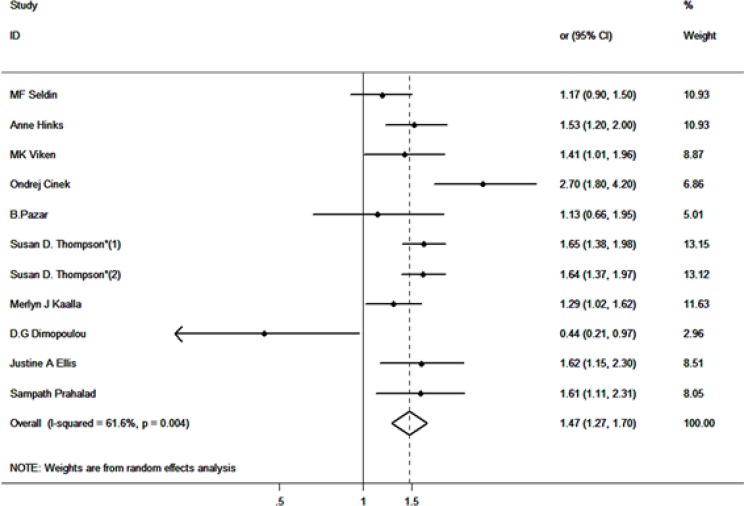
Overall, 4552 cases and 10161 controls were investigated in this study to evaluate the association between PTPN22 (C allele vs. T allele) genotype and JIA susceptibility. Four articles were carried out in America population and 6 in European population. The random effects model showed that an increased susceptibility was associated between the PTPN22 genotype and JIA risk (OR=1.47, 95% CI: 1.27–1.70, P<0.001). The forest plot is showed in Fig. 2.
Fig. 2:
Association between PTPN22 polymorphism and JIA risk analyzed by forest plot of meta-analysis. The forest plots of pooled OR with 95% CI (C allele vs. T allele; OR=1.47, 95% CI: 1.27–1.70; random effects model, P<0.001)
Subgroup analysis
Considering the comparable heterogeneity in light of the I2 and P value (I2=61.6%, P=0.004), subgroup analysis was performed to recognize substantial between-study heterogeneity. We classified Greece, British and Norway as European regions while identified Utah as America region. Additionally, remaining places were defined as other regions. Meanwhile, according to the difference of case number and population resource, stratification analysis was carried out, respectively.
The subgroup analysis results are shown in Table 2.
Table 2:
Subgroup analysis of the association between PTPN22 polymorphisms and JIA
| Polymorphism | Allele C vs. Allete T | No. of studies (cases/controls) | Odds ratio | M | Heterogeneity | PE | ||
|---|---|---|---|---|---|---|---|---|
| OR(95%CI) | POR | I2(%) | PH | |||||
| PTPN22(rs2476601) | All studies | 11 (4552 / 10161) | 1.42(1.20, 1.68) | <0.001 | R | 61.6 | 0.004 | 0.303 |
| subgroup analysis by number of case | ||||||||
| <500 | 7 (1431 / 3743) | 1.38(1.04, 1.83) | 0.025 | R | 72.4% | 0.001 | 0.619 | |
| ≥500 | 4 (3121 / 6418) | 1.55(1.39, 1.72) | <0.001 | R | 8.0% | 0.353 | 0.309 | |
| subgroup analysis by number of control | ||||||||
| population-based | 8 (3786 / 8829) | 1.52(1.32, 1.76) | <0.001 | R | 55.5% | 0.028 | 0.979 | |
| hospital-based | 3 (766 / 1332) | 1.36(1.15, 1.60) | <0.001 | R | 8.8% | 0.334 | 0.882 | |
| subgroup analysis by region | ||||||||
| America | 4 (2209 / 6901) | 1.52(1.30, 1.78) | <0.001 | F | 45.7% | 0.137 | 0.585 | |
| Augean | 6 (2025 / 2704) | 1.36(1.01, 1.83) | 0.041 | R | 74.4% | 0.002 | 0.555 | |
| Adjusted result | Adjusted studies | 9 (4294 / 9540) | 1.48(1.36, 1.62) | <0.001 | F | 8.8% | 0.362 | 0.268 |
Sensitivity analysis and Galbraith plot
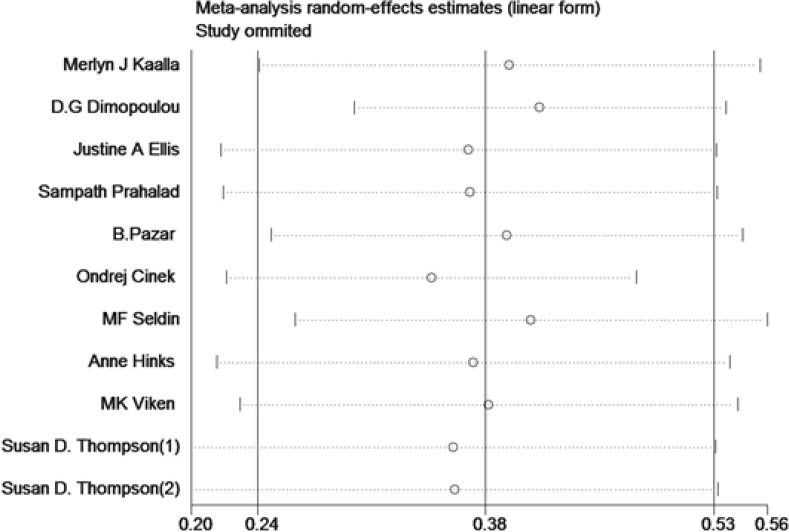
Sensitivity analysis could help to examine the possible source of heterogeneous articles by sequentially excluding one article at a time. The sensitivity analysis showed that article of Number 4 might be the main source of heterogenity (Fig. 3). Additionally, we used the Galbraith plot to recognize the sensitive records from eligible studies. The results are shown that records from Number 9 and Number 4 might be the main heterogeneous studies.
Fig. 3:
Sensitivity analysis of association between PTPN22 genetic variances and JIA
Discussion
The study showed an increased risk of JIA with PTPN22 polymorphism (1858 C>T) (OR=1.26, 95% CI: 1.27–1.70, P <0.001), especially in America population (OR=1.52, 95% CI: 1.30–1.78) through Subgroup analysis. Meta-analysis has been recognized as an important tool to define the effect of selected genetic polymorphisms and disease susceptibility, and discern the potential sources of between-study heterogeneity (11). There was a statistically significant association between PTPN22 and JIA (OR=1.44, 95% CI 1.31– 1.6, P <0.001) (12). However, two vital paper published in 2013 were not included in this pooled analysis (12). Additionally, this meta-analysis had not been performed a further stratified analysis in different population and sample size, which could influence the precision and accuracy of the conclusion.
Eleven critical individual case-control studies about the PTPN22 polymorphism and JIA risk were collected in this paper. We ultimately identified that an increased risk for JIA in crowd with T allele in locus of PTPN22 1858 (OR=1.26, 95% CI: 1.27–1.70). Subgroup analyses were mainly carried out by ethnicity, sample number and the source of case. Stratified for ethnicity, the PTPN22 polymorphism (1858 C>T) was significantly associated with JIA risk in America population (OR=1.52, 95% CI: 1.30–1.78). The subgroup analysis showed that the associations were still significant in case number more than 500 (OR=1.38, 95% CI: 1.04–1.83), while in the case number less than 500 was OR=1.55, 95% CI: 1.39–1.72. These different data among the different ethnicity might hint that different ethnic genetic backgrounds could influence the incidence of JIA. At the same time, due to the limited number of inclusive studies in this meta-analysis, the results might have insufficient statistical power to detect a true effect and generate fluctuated risk estimation. To ensure the epidemiological credibility of this meta-analysis, we further performed sensitivity analysis and Galbraith plot. After omitting some heterogeneous articles according above results, no heterogeneity had been identified. Additionally, no publication bias further proof our results reliability
PTPN22 is an intracellular phosphatase, which plays a vital function in modulating cytokine signal transduction through the JAK/STAT signaling pathways (10). As a transcription factor, STAT4 is essential for the gulation process of Th1 cell differentiation from lymphocytes, macrophages and dendritic cells (10). Th1 cells play a role in the development of JIA by producing inflammatory cytokine interferona. Several studies argued an allelic variant of PTPN22 is strongly associated with multiple autoimmune diseases including type 1 diabetes (T1D), systemic lupus erythematosus (SLE), rheumatoid arthritis (RA), Graves disease, and others. The risk allele comprises a single-nucleotide polymorphism, C1858T, resulting in an amino acid change from arginine to tryptophan at position 620 (R620W) of the encoded protein LYP. Whose functions were as a negative regulator in (TCR) signaling, our hypothesis is that the PTPN22 polymorphism might be contributed to the development of JIA through pathway above. It was interesting that mice lacking of the PTPN22 gene showed high levels of autoimmune diseases characteristic by the presence of high-titer pathogenic autoantibodies (13), this result further support our conclusion in some extent.
However, there were some possible limitations existing in this meta-analysis. First, lack of sufficient study number and limited studies scale could all influence the stability of the conclusion. At the same time, the missing of the records in Asian and African population could disturb the conclusion. Additionally, our literature searching only included English databases, which might lead to the language selection bias.
Besides, three articles recruit control subjects in the hospital-based population and that might be different from the population-based controls. Furthermore, complex environmental factors and interactive function between genes and environment in our meta-analysis were not considered. These factors above could affect the conclusion. At last, genome-wide association study (GWAS) had not been performed in available studies, which could increase statistics power. Therefore, GWAS should be considered to carry out in multi-center.
Conclusion
There was positively association between PTPN22 gene variants and developing JIA, and further studies including GWAS should be carried out in multi-center to discern more truly genetic risk factors and possible environmental factors.
Ethical considerations
Ethical issues (Including plagiarism, informed consent, misconduct, data fabrication and/or falsification, double publication and/or submission, redundancy, etc.) have been completely observed by the authors.
Acknowledgement
This paper was supported by Ningbo science and research projects (2010C50010) and Ningbo health bureau research projects (2009B03). The authors declare that there is no conflict of interests.
References
- 1. Ravelli A, Martini A. (2007). Juvenile idiopathic arthritis. The Lancet, 369: 767–778. [DOI] [PubMed] [Google Scholar]
- 2. Berntson L, Gäre BA, Fasth A, Herlin T, Kristinsson J, Lahdenne P, Marhaug G, Nielsen S, Pelkonen P, Rygg M. (2003). Incidence of juvenile idiopathic arthritis in the Nordic countries. A population based study with special reference to the validity of the ILAR and EULAR criteria. J Rrheumatol, 30: 2275–2282. [PubMed] [Google Scholar]
- 3. Angeles-Han ST, McCracken C, Yeh S, Jenkins K, Stryker D, Rouster-Stevens K, Vogler LB, Lambert SR, Drews-Botsch C, Prahalad S. (2015). Characteristics of a cohort of children with Juvenile Idiopathic Arthritis and JIA-associated Uveitis. Pediatr Rheumatol Online J, 13: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kawada J-i, Kitagawa Y, Iwata N , Ito Y. (2013). Clinical characteristics of influenza virus infection in juvenile idiopathic arthritis patients treated with tocilizumab. Modern Rheumatol, 23: 972–976. [DOI] [PubMed] [Google Scholar]
- 5. Massa M, Mazzoli F, Pignatti P, De Benedetti F, Passalia M, Viola S, Samodal R, La Cava A, Giannoni F, Ollier W. (2002). Proinflammatory responses to self HLA epitopes are triggered by molecular mimicry to Epstein-Barr virus proteins in oligoarticular juvenile idiopathic arthritis. Arthritis & Rheumatism, 46: 2721–2729. [DOI] [PubMed] [Google Scholar]
- 6. Ostanek L, Ostanek-Panka M, Bobrowska-Snarska D, Binczak-Kuleta A, Fischer K, Kaczmarczyk M, Ciechanowicz A, Brzosko M. (2014). PTPN22 1858C>T gene polymorphism in patients with SLE: association with serological and clinical results. Mol Biol Rep, 41: 6195–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pradhan V, Borse V, Ghosh K. (2010). PTPN22 gene polymorphisms in autoimmunediseases with special reference to systemic lupus erythematosus disease susceptibility. J Postgraduate Med, 56: 239–242. [DOI] [PubMed] [Google Scholar]
- 8. Velaga M, Wilson V, Jennings C, Owen C, Herington S, Donaldson P, Ball S, James R, Quinton R, Perros P. (2004). The codon 620 tryptophan allele of the lymphoid tyrosine phosphatase (LYP) gene is a major determinant of Graves’ disease. J Clin Endocrinol Metabol, 89: 5862–5865. [DOI] [PubMed] [Google Scholar]
- 9. Han J-W, Zheng H-F, Cui Y, Sun L-D, Ye D-Q, Hu Z, Xu J-H, Cai Z-M, Huang W, Zhao G-P. (2009). Genome-wide association study in a Chinese Han population identifies nine new susceptibility loci for systemic lupus erythematosus. Nature Genetics, 41: 1234–1237. [DOI] [PubMed] [Google Scholar]
- 10. Stanford SM, Bottini N. (2014). PTPN22: the archetypal non-HLA autoimmunity gene. Nat Rev Rheumatol, 10: 602–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Higgins J, Thompson SG. (2002). Quantifying heterogeneity in a meta-analysis. Stat Med, 21: 1539–1558. [DOI] [PubMed] [Google Scholar]
- 12. Kaalla MJ, Broadaway KA, Rohani-Pichavant M, Conneely KN, Whiting A, Ponder L, Okou DT, Angeles-Han S, Rouster-Stevens K, Brown MR, Vogler LB, Jorde LB, Bohnsack JF, Epstein MP, Prahalad S. (2013). Meta-analysis confirms association between TNFAG238A variant and JIA, and between PTPN22-C1858T variant and oligoarticular, RF-polyarticular and RF-positive polyarticular JIA. Pediatr Rheumatol Online J, 11: 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kyogoku C, Ortmann WA, Lee A, Selby S, Carlton VE, Chang M, Ramos P, Baechler EC, Batliwalla FM, Novitzke J. (2004). Genetic association of the R620W polymorphism of protein tyrosine phosphatase PTPN22 with human SLE. Am J Human Genetics, 75: 504–507. [DOI] [PMC free article] [PubMed] [Google Scholar]