Abstract
A peptide fragment of the human tau protein which stacks to form neat cross β-sheet fibrils, resembling that found in pathological aggregation, 273GKVQIINKKLDL284 (here “R2/WT”), was modified with a spin-label at the N-terminus. With the resulting peptide, R2/G273C-SL, we probed events at time scales spanning seconds to hours after aggregation is initiated using transmission electron microscopy (TEM), thioflavin T (THT) fluorescence, ion mobility mass spectrometry (IMMS), electron paramagnetic resonance (EPR), and Overhauser dynamic nuclear polarization (ODNP) to determine if deliberate changes to its conformational states and population in solution influence downstream propensity to form fibrillar aggregates. We find varying solution conditions by adding the osmolyte urea or TMAO, or simply using different buffers (acetate buffer, phosphate buffer, or water), produces significant differences in early monomer/dimer populations and conformations. Crucially, these characteristics of the peptide in solution state before aggregation is initiated dictate the fibril formation propensity after aggregation. We conclude the driving forces that accelerate aggregation, when heparin is added, do not override the subtle intra- or interprotein interactions induced by the initial solvent conditions. In other words, the balance of protein–protein vs protein–solvent interactions present in the initial solution conditions is a critical driving force for fibril formation.
Introduction
Neurodegeneration is often associated with a class of diseases known as proteopathies or foldopathies, in which specific proteins oligomerize and aggregate to form fibrils.1−4 Examples of these proteins include α-synuclein,5 huntingtin,6 amyloid-β,3,7 and tau.8−10 Hereof, tau is an intrinsically disordered protein found in neuronal cells that plays an important role in the formation and stabilization of the microtubule cytoskeleton. The carboxyl region of adult-specific human tau contains four imperfect repeats (Figure 1A). This repeat region possesses inherent microtubule binding and microtubule stabilizing activities. Interestingly, this region also contains sequences that form the intertau cross β-sheets that are essential for the pathological tau aggregation process to fibrils.
Figure 1.
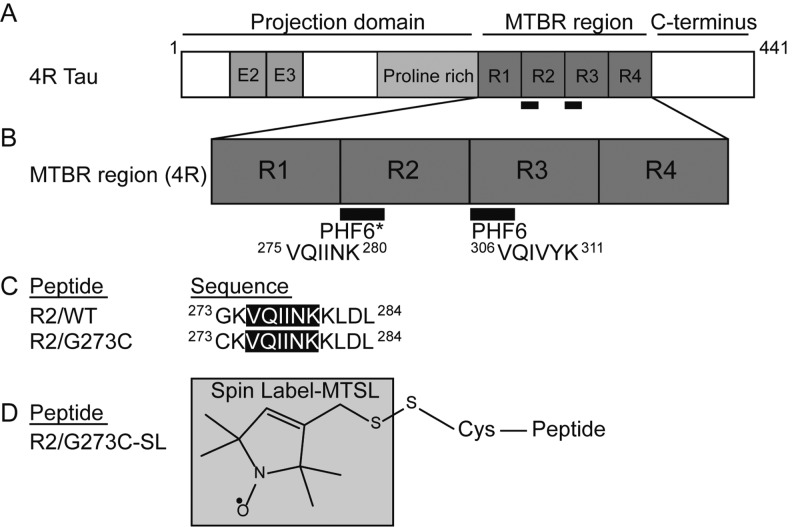
(A) Diagram of full-length four-repeat (“4R”) tau consisting of 441 amino acids, the longest isoform in the human central nervous system. This isoform has a projection domain containing two alternatively spliced exons (E2 and E3) as well as a proline-rich region and four pseudorepeats in the microtubule binding repeat region (MTBR). (B) Diagram of the MTBR showing the location and sequence of the two hexapeptide units (PHF6* and PHF6). (C) Sequence of peptides used in this study including the wild type (WT) and singly mutated versions. (D) Structure of spin-label (MTSL), which is attached to R2/G273C via a disulfide bond on the cysteine. Adapted in part from ref (28).
It is widely held that aberrant tau dissociates from microtubules and then self-aggregates10−12 to form oligomers, intermediate aggregates, and mature fibrillar pathological structures packed into cross-β conformations. However, the nature of the aggregation seeding mechanism is unclear. Is it a shift in monomer/dimer equilibrium toward an aggregation prone conformation as suggested in the nucleation conformation transition model?13 Or is it an increased concentration of oligomers/fibrils that template monomers as suggested by the nucleation polymerization model?14,15 Remarkable recent work demonstrates that pathological tau (of ill-defined structure) is transferred from one neuron to the next along synaptic circuits, leading to the trans-synaptic progression of tau pathology and the disease state.16−18 Once pathological tau is taken up by the naive postsynaptic neuron, it serves as a seed to promote aggregation of the endogenous, nonpathological tau. This newly formed pathological tau may be itself toxic or may cause toxicity through loss-of-function effects as it converts endogenous normal tau to a nonfunctional state. Whatever the exact mechanism of tau toxicity, there is increasing evidence that protein aggregation to fibrils and their specific cross β-sheet structure are reflective of tau pathology.19,20 Taken together, tau misfolding, oligomerization, and aggregation are obligatory components of tau-mediated pathogenesis. It follows that acquiring a better understanding of the driving forces for aggregation, starting from monomers in solution—of whichever tau protein or fragment that aggregates to form cross β-sheet structured fibrils—is of fundamental importance.
While it would be preferable to study tau oligomerization and aggregation using full-length tau, its large size and disordered nature exclude access to many biophysical techniques that offer molecular-level insight. Hence, we have chosen to examine a tau fragment known to be important for the aggregation process, 273GKVQIINKKLDL284 (herein referred to as “R2/WT”).21,22 This peptide contains 275VQIINK280 (also known in the literature as “PHF6*”), one of two related hexapeptide units located in the microtubule binding region (Figure 1B) and known to form interprotein β-sheet contacts that are essential to the tau aggregation/fibrillization process.21,23,24 Previous work has shown that 275VQIINK280 can form fibrils when aggregation is triggered by heparin, with morphology indistinguishable from the fibrils formed by full-length human tau.23,25,26
It was verified that R2/WT peptide, like its full length counterpart, is intrinsically disordered, populating a range of conformations from compact to extended.22 The same study proposes that the compact structures of R2/WT peptides are stabilized by hydrogen bonds or by salt bridges between the K and D residues.22 Importantly, it has been suggested in the literature that intrinsically disordered peptides can adopt aggregation-prone conformations (N* structures) and that shifting the population toward N* conformations increases aggregation rates.27−30 Most studies to date in which populations of monomers are directed toward aggregation-prone states have focused on the effect of point mutations or chemical linkers.31−33 In this paper, we test the hypothesis that similar effects can be achieved by tuning the initial solution conditions of the sample. We demonstrate on model tau fragments R2/WT and R2/G273C that subtle changes in solution conditions, such as the addition of osmolytes or changes in buffer type, influence the aggregation pathways and end products.
Our experimental strategy is to elucidate the conformational state, population, and aggregation process by probing events at time scales spanning seconds to hours after the initiation of aggregation, with assembly states covering the monomer, dimer, and fibril dimensions. Replacement of the terminal glycine residue of R2/WT with cysteine to form 273CKVQIINKKLDL284 (Figure 1C) allows the formation of nitroxide spin-labeled peptide G273C-SL with the structure shown in Figure 1D. As we will show, this modification does not significantly alter the properties of the peptide compared to R2/WT but allows us to use electron paramagnetic resonance (EPR) line shape analysis to probe the solvent accessibility, packing, and mobility of the spin-label as well as to quantify the % populations of mobile vs β-sheet embedded spin-labeled peptides, before and after the initiation of aggregation. This decomposition into mobile vs β-sheet embedded populations is made possible by the clear appearance of a spin-exchanged single line feature in the EPR spectrum of G273C-SL when subject to aggregating conditions, whose population dramatically increases with aggregation time. Such single-line EPR features, observed with singly spin-labeled proteins or peptides subject to fibrillization, have been unambiguously attributed to β-sheet packing of the spin-labeled sites in the literature.34 The same spin-labeled peptide is concurrently used to carry out Overhauser dynamic nuclear polarization (ODNP) relaxometry to monitor the surface water diffusivity within 10 Å of the spin-label. The translational diffusion of surface water necessarily involves hydrogen bond breaking, spatial displacement, and hydrogen bond reforming of water and thus reports on the strength of the dynamic surface water network that is governed by both the adhesive energy between the peptide surface and water and the cohesive energy of water lining the peptide surface. Crucially, this implies that ODNP is sensitive to detecting conformational changes of the peptide (or shifts in the population of different conformations) that alter the balance between peptide–water and peptide–peptide attraction, even in the absence of significant conformational changes or aggregation that physically buries the spin-label. Such studies are consistent with recent neutron scattering experiments on full length tau which have shown that translational water diffusivity is sensitive to the aggregation state of tau.35 Ion mobility mass spectrometry (IM-MS) is used to measure the relative populations of hydrated tau monomers, dimers, and higher order assemblies.36 And finally, transmission electron microscopy (TEM) and thioflavin T (ThT) fluorescence spectroscopy are used to assess β-sheet-rich aggregate content under various solution conditions, i.e., in the presence of osmolytes such as urea vs TMAO, or when different buffers such as acetate or phosphate vs salt-free water solvate the peptide. The question we address is whether or not the relatively subtle intra- or interprotein interactions that are present under the initial solvent conditions have a significant influence over the subsequent dramatic aggregation process induced by heparin. Our results follow.
Experimental Methods
Materials
Peptides were synthesized by Genscript Corp. (Piscataway, NJ) with N-terminal acetylation and C-terminal amidation in order to remove the electric charges on the ends of the peptide. The nitroxide probe (1-oxyl-2,2,5,5-tetramethylpyrroline-3-methyl)methanethiosulfonate spin-label (MTSL) was purchased from Santa Cruz Biotechnology (Dallas, TX). Urea was purchased from Affymetrix, Inc. (Cleveland, OH). Tetramethylamine N-oxide (TMAO) was purchased from Acros Organics (Fair Lawn, NJ). Heparin (6 kDa average molecular weight) was purchased from Sigma-Aldrich (St. Louis, MO). PD MidiTrap G-10 desalting columns were purchased from GE Healthcare (Wauwatosa, WI). Thioflavin-T (ThT) was purchased from AnaSpec Inc. (Fremont, CA).
Peptide Spin-Labeling
To prepare R2/G273C-SL, R2/G273C was dissolved in 6 M guanidine hydrochloride in order to prevent aggregation during the labeling process. MTSL, dissolved in dimethyl sulfoxide (DMSO), was added in excess to the peptide (×10 molar concentration). The labeling process was allowed to occur overnight at 4 °C. Excess spin-label and guanidine hydrochloride were then removed using a PD MidiTrap G-10 desalting column, while exchanging the peptide into pure water. Finally, the spin-labeled peptide was lyophilized and stored at −20 °C until ready to use. The peptide was then dissolved in the desired buffer.
Overhauser Dynamic Nuclear Polarization (ODNP)
ODNP measures the local water dynamics with site-specific resolution of 5–10 Å (2–4 hydration layers) around a spin-label which is attached to a single cysteine of a peptide or protein.37 ODNP measurements of R2/G273C-SL were carried out at 0.35 T corresponding to a proton Larmor frequency of 14.8 MHz and electron Larmor frequency of 9.8 GHz. A 3.5 μL sample was loaded into a 0.6 mm i.d., 0.84 mm o.d. quartz capillary tube (VitroCom), sealed at one end with beeswax and the other end with critoseal, and attached to a home-built NMR probe. The probe was connected to a Bruker Avance spectrometer and held inside the dielectric (ER 4123D) EPR resonator. In order to avoid heating caused by microwave irradiation, room-temperature air was blown over the sample. ODNP was performed by continuously pumping microwave irradiation at 9.8 GHz, thus saturating the nitroxide central EPR transition, while simultaneously recording proton NMR signal at 14.8 MHz. Proton spin–lattice relaxation times, T1, were obtained using an inversion–recovery pulse sequence.
An in-depth description of ODNP theory can be found in the literature, and so only a brief overview is given here.38,39 The general idea of ODNP is to transfer the high spin polarization of an unpaired electron, supplied by a site-specific nitroxide spin-label, to the protons on water which are in close proximity upon saturation of the EPR transition (i.e., the Overhauser effect), thus greatly amplifying the NMR signal of water. The negative NMR signal enhancement can only be observed when the translational diffusion of water is rapid enough to induce dipolar relaxation with the electron spin of nitroxide spin-labels. This enhancement in NMR signal at maximum saturation, Emax, can be written as
| 1 |
The value of interest from this equation is the coupling factor, ξ, which is used to determine dynamics and subsequently the diffusion coefficient. The saturation factor of the electron spin is described by s. We have previously found that smax = 1 is valid when nitroxide spin-labels are tethered to slow tumbling molecules or assemblies, such as proteins.38 The leakage factor, f, describes the efficiency with which the electron spin facilitates the proton nuclear spin relaxation and can be described by f = 1 – T1/T10, where T1 and T10 are the spin–lattice relaxation times in the presence and absence of the spin-label, respectively. Finally, the parameter γe/γH is the ratio of the electron and proton larmor frequencies which is equal to 658.
One important challenge of quantifying the coupling factor is that determination of the leakage factor is highly error prone if the T1 relaxation time is dominated by the contribution of T10, as found in samples with low spin-label concentrations. This error can lead to inaccurate measurements of the coupling factor and therefore also the local diffusion coefficient. Thus, in this study we instead use a new ODNP analysis method which has been previously described to calculate the relaxivity constant, kσ, which reports on the fast (picoseconds) time-scale dynamics around the spin-label.39
| 2 |
where CSL is the spin-label concentration. It has been shown previously how kσ relates to the translational correlation time of water, τc, by writing it in terms of the spectral density function, J(ω,τc), of the dipole–dipole Hamiltonian.40
| 3 |
where B0 is the magnetic field. Thus, changes in kσ can be unambiguously assigned to changes in the diffusion dynamics of loosely bound hydration water which reports on protein conformational changes and interprotein interactions. Even though the analysis of kσ does not yield the diffusion coefficient, it is a very useful scale with high values of kσ indicating faster and low values indicating slower diffusing local hydration water around the spin-labels.
Lyophilized samples of R2/G273C-SL were dissolved in 20 mM ammonium acetate buffer (pH = 7.0), sodium phosphate buffer (pH = 7.0), or water to a concentration of 400 μM. For samples after aggregation, heparin is added at a molar ratio of 4:1 (peptide to heparin), which is a widely used practice to induce aggregation and fibrillization of tau peptides and proteins.41 For the study involving urea or TMAO, these osmolytes were added to the solution of tau peptides at 4.4 and 2 M concentrations, respectively.
Electron Paramagnetic Resonance (EPR)
Continuous wave EPR is capable of probing mobility of the spin-label as well as its accessibility to solvent. All EPR spectra were acquired using a Bruker EMX X-band spectrometer and dielectric (ER4123D) cavity. A 3.5 μL sample was loaded into a capillary and sealed at one end with beewax and the other with critoseal. The sample was then irradiated with 6 mW of microwave power at 9.8 GHz using a 0.3 G modulation amplitude and a sweep width of 150 G. Sample preparation and conditions were the same as for the ODNP experiment.
EPR Simulation
EPR simulation is able to fit experimental EPR data to either single or multiple components and was used to determine percent β-sheet content. EPR simulation was completed using the MultiComponent software of Dr. Christian Altenbach. In each spectra, the A tensors were fixed at Axx = 6.2, Ayy = 5.9, and Azz = 37 while the g tensors were fixed at gxx = 2.0078, gyy = 2.0058, and gzz = 2.0022 which have been previously determined for MTSL.42 The rotational correlation time, R, and the order parameter S were used as fit parameters. In the case of a two-component fit, the Heisenberg spin exchange frequency (ω) was allowed to vary in order to obtain a single line component.
Ion Mobility Mass Spectrometry (IM-MS)
Nanoelectrosprayed ionization (n-ESI) IM-MS is a method capable of detecting multiple conformations of the same species and oligomers at different sizes having the same mass to charge (m/z) ratios. The ions are generated by ESI, captured, and stored in an ion funnel and subsequently pulsed into a drift cell filled with He gas at high pressure (∼12–13 Torr). The ions experience collisions with the buffer gas molecules and additional force due to a weak electrical field. These combined effects allow the ions to drift with a constant velocity which can then be related to the reduced mobility K0 independent from instrumental parameters. This value can be used to calculate the absolute collision cross sections σ given the size n and charge z of the species.43,44
| 4 |
where m and mb are the molecular weights of the ions and buffer gas molecules, respectively, ze is the charge of the ion, N is the buffer gas density, and Ωavg is the average collisional cross section integral, which approximates to be the same as the average collision cross section σ. The IM-MS instrument was built in-house and consists of a nano-ESI source, an ion funnel, a 2 m long drift cell, and a quadrupole mass filter.45
Lyophilized samples of R2/WT and R2/G273C-SL were dissolved in 20 mM ammonium acetate buffer (pH = 7.0), sodium phosphate buffer (pH = 7.0), or water, depending on the sample of study, to a concentration of 100 μM. For the experiments involving heparin or urea, the concentration of protein samples was reduced to 50 μM. When urea was present, the concentration of urea was kept constant at 1.1 M, so that the ratio of protein to urea is the same 4:1 for all experiments including ODNP, EPR, and TEM.
Transmission Electron Microscopy (TEM)
TEM was used to visualize aggregates at nanometer scales. Images were obtained using a JEOL-1230 model microscope coupled to an ORCA camera with AMT Image Capture Software Version 5.24 (Advanced Microscopy Techniques, Woburn, MA). Unless otherwise noted in the text, aggregation mixtures contained 100 μM peptide and 25 μM heparin in 20 mM ammonium acetate buffer (pH = 7.0), 20 mM sodium phosphate buffer (pH = 7.0), or water, depending on the sample of study. Samples of these mixtures were removed after some amount of time (generally 24 h) and fixed for 5 min in 1.6% glutaraldehyde (Ted Pella, Inc.). Fixed samples were placed on TEM grids (300 mesh, Formvar/copper, Electron Microscopy Sciences) for 1.5 min, rinsed with water, and then stained for 20 s with 2% uranyl acetate (Ted Pella, Inc.). The physical dimensions of aggregates were measured using unaltered images and ImageJ software (version 1.47b, NIH). Statistics were carried out using the GraphPad Prism software.
Thioflavin T Assay
Thioflavin T is a fluorescent dye that undergoes a shift in excitation and emission maxima upon bind to and detect β-sheet amyloid structures and is therefore useful for monitoring the kinetics of tau aggregation. A ThT stock solution (3 mM) was prepared in water and stored in the dark. Mixtures of peptide (50 μM) with and without heparin (12.5 μM) were prepared in either 20 mM ammonium acetate buffer (pH = 7.0) with or without urea (4.4 M) or TMAO (2 or 0.2 M), 20 mM sodium phosphate buffer (pH 7.0), or water containing ThT (2.2 μM). ThT fluorescence was measured at 450ex/488em emission in a Tecan M220 Infinite Pro microplate reader. Data presented are the average of five independent experiments. Although the data are presented in two separate figures for organizational purposes, all experimental conditions were included in each experiment. To account for any differences in baseline signal arising from the various buffers and osmolytes, as well as for experiment-to-experiment variability in the strength of fluorescence signal, the t = 0 value of the appropriate “no heparin” condition was subtracted from each time point (both + and – heparin).
Results and Discussion
Aggregation Propensity of R2/WT and R2/G273C-SL
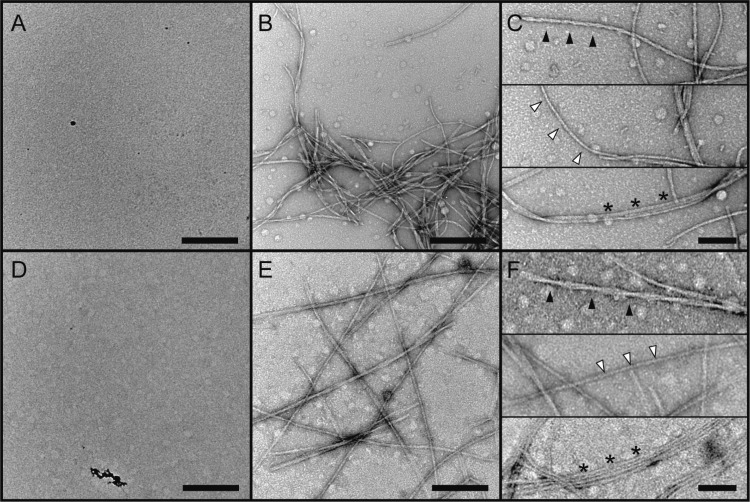
We first used TEM to assess the relative abilities of R2/G273C-SL and R2/WT peptides to form fibrillar aggregates in the presence of heparin and to compare the morphologies of the fibers produced. Without heparin, neither peptide forms substantial aggregates or fibrils during the time course of the experiment (Figure 2, panels A and D). Upon heparin addition and subsequent incubation for ∼24 h, both peptides form fibrillar aggregates (Figure 2, panels B and E). Higher magnification reveals that each peptide forms a combination of twisted, “paired helical filament”-like46 fibers, individual straight fibers, and bundled straight fibers that run parallel to each other (Figure 2, panels C and F). For a more quantitative comparison of fiber morphology, we compared the widths of individual straight fibers made of R2/G273C-SL vs R2/WT. The fiber widths were nearly identical: R2/G273C-SL fibers had an average width of 9.9 nm ± 0.17 SEM (N = 71), while R2/WT fibers had an average width of 9.7 nm ± 0.15 SEM (N = 129). This difference is not statistically significant. Notably, these dimensions fall within the prototypical range found for a variety of amyloid fibrils.47 Taken together, the TEM data indicate that at the macroscopic length scale R2/G273C-SL and R2/WT derived fibrils are indistinguishable.
Figure 2.
TEM comparison of R2/G273C-SL and R2/WT fiber formation in the absence of heparin (A and D, respectively) and in the presence of heparin (B and E, respectively). A comparison of fiber morphology is shown in (C and F) for R2/G273C-SL and R2/WT, respectively, with black arrows showing PHF-like fibers, white arrows showing individual straight fibers, and stars signifying bundled fibers. All peptides were dissolved at 100 μM in 20 mM ammonium acetate buffer, pH 7.0, and 25 μM heparin (6 kDa) was used at a 1:4 molar ratio with respect to peptide. (A, B, D, and E) scale bar = 250 nm; (C and F) scale bar = 100 nm.
Next, we compared IM-MS data of the two peptides. In the absence of heparin, the mass spectra indicate that both peptides exist predominantly in two charge states: n/z = 1/3 and 1/2.
n/z = 1/2
The arrival time distribution (ATD) of R2/G273C-SL peptide contains two features (Figure 3D). The ATD features are similar to those of the R2/G273C peptide (containing no MTSL probe, data not shown), suggesting that the short arrival time is a dimer and the other feature is a monomer. The dimer species with n/z = 2/4 is more abundant with R2/G273C-SL compared to R2/WT (see Figure 3D vs 3B, respectively). The cross sections of the spin-labeled peptide’s monomer and dimer (n/z = 1/2 and 2/4) are both ∼12.5% larger than those of the WT reflecting the presence of the spin-label.
Figure 3.
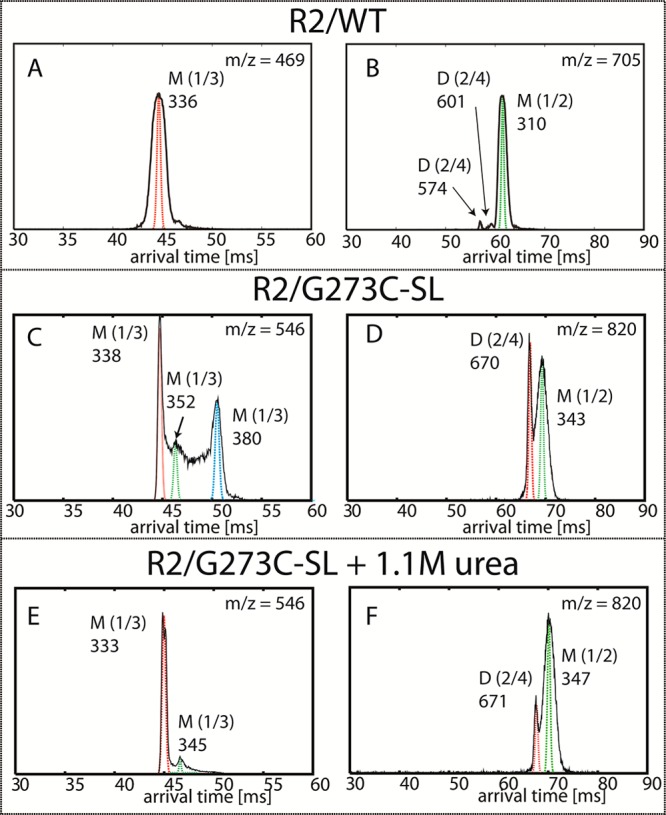
Representative ATDs at n/z = 1/3 and 1/2 of 12-mer R2/WT (m/z = 469 for panel A and 705 for panel B), R2/G273C-SL (m/z = 546 for panel C and 820 for panel D) peptides, and R2/G273C-SL with 1.1 M urea (m/z = 546 for panel E and 820 for panel F). The peptides are dissolved in 20 mM ammonium acetate buffer at pH = 7.0. Each feature is labeled with oligomer size (M = monomer, D = dimer) and experimental cross section (σ, Å2).
n/z = 1/3
The ATD of the R2/WT contains a single peak that is assigned as a monomer (Figure 3A). However, the ATD of the R2/G273C-SL peptide is more complex. The possible reason for this complexity is discussed in the Supporting Information. The important point is that the dominant peak at the shortest arrival time is a monomer. Its cross section is comparable to that of the R2/WT monomer, which is somewhat surprising given the results of the n/z = 1/2 mass peak, and indicates a more compact fold than R2/WT. The feature with the longest arrival time has a cross section of σ = 380 Å2. This is likely an extended conformation of the fully protonated species. Another possibility is that this feature corresponds to a water bound monomer in which the water molecules were dissociated before reaching the detector, as previous observed in another system.48
Importantly, the IM-MS data show the predominant population of both peptides is monomeric with comparable conformations, while TEM data show the fibrillar end point of R2/WT aggregation is preserved upon mutation and spin-labeling. On the basis of the above-described comparison of R2/WT and R2/G273C-SL, we conclude they are sufficiently similar so that conclusions based on biophysical analysis of R2/G273C-SL can be extrapolated to R2/WT.
Effects of Osmolytes on Oligomeric State and Aggregation Propensity
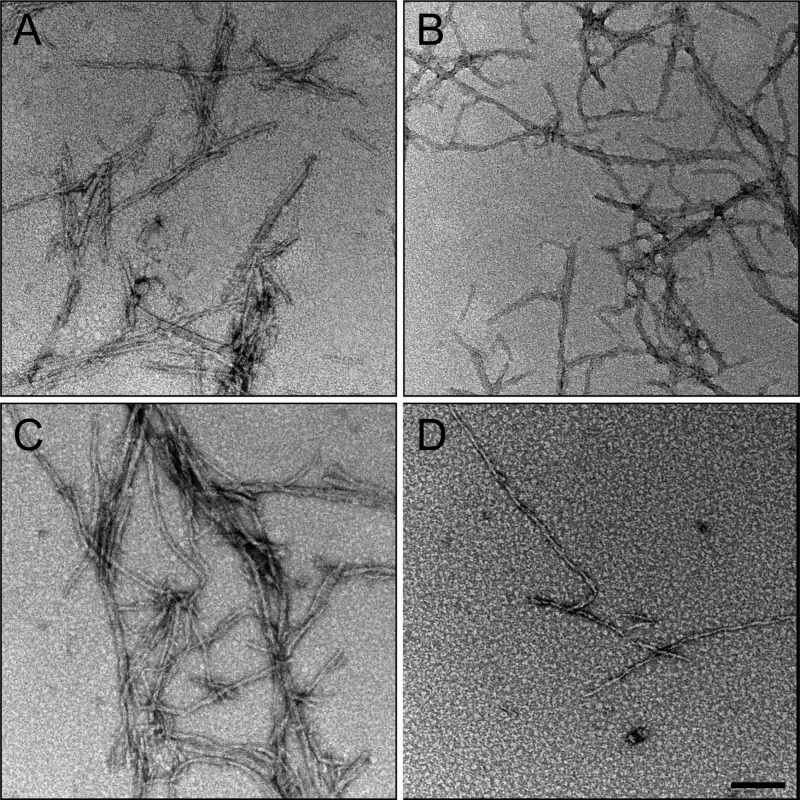
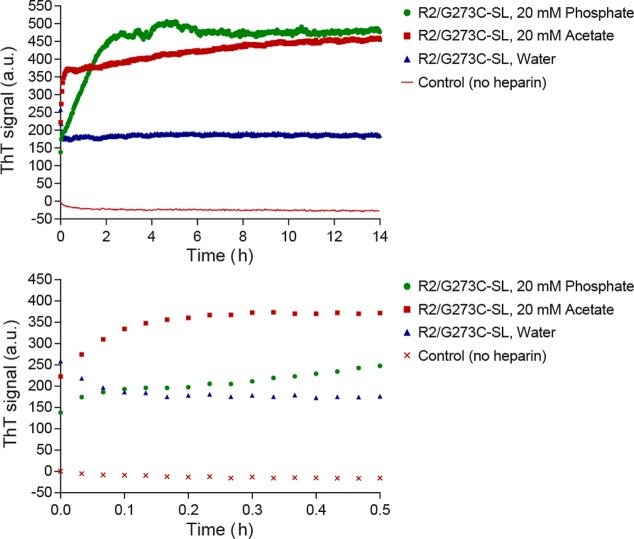
We first examine the changes in R2/G273C-SL aggregation at the macroscopic scale in the presence of the osmolyte urea (a standard chaotrope) or TMAO (a standard kosmotrope). In the absence of osmolytes, R2/G273C-SL in acetate buffer forms abundant fibrillar aggregates upon heparin addition (Figure 4A). The addition of 2 M TMAO (Figure 4B) resulted in qualitatively more fibrils than 0.2 M TMAO (Figure 4C) or buffer alone. In the presence of urea (4.4 M), fibril formation is largely prevented, with smaller quantities of fibrillar species observed (Figure 4D). Consistent with the TEM results, ThT assays demonstrate that R2/G273C-SL forms β-sheet-rich aggregates in the presence of heparin, that urea significantly slows aggregation, and that TMAO promotes aggregation in a concentration-dependent manner (Figure 5). The initial drop in signal for R2/G273C-SL in 2 M TMAO is attributed to aggressive rapid clumping, initially upon mixing, thereby excluding ThT from binding. In the absence of heparin, the peptide in all buffer conditions shows no increase in ThT signal over 14 h.
Figure 4.
TEM showing the effect of osmolytes on R2/G273C-SL fiber formation. TEM of fibers formed after 24 h of incubation of peptide (100 μM) with 6 kDa heparin (25 μM) in 20 mM ammonium acetate buffer alone (panel A), plus 2 M TMAO (panel B), 0.2 M TMAO (panel C), or 4.4 M urea (panel D). Scale bar = 100 nm.
Figure 5.
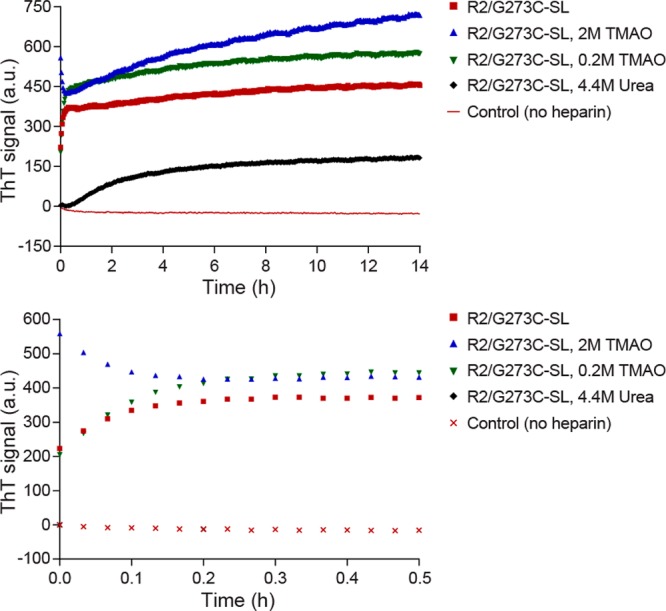
Thioflavin T fluorescence reveals effects of heparin and osmolytes on tau aggregation. Top panel is full scale while the bottom panel is zoomed in to show the first 30 min. The spin-labeled tau peptide R2/G273C-SL (25 μM) was incubated in 20 mM ammonium acetate buffer, pH 7.0, in the presence or absence of 6 kDa heparin (6.25 μM), TMAO (2 M), TMAO (0.2 M), and urea (4.4 M). All four non-heparin conditions generate identical and overlapping ThT signals, and so only one is shown for simplicity. Data shown are averaged from five separate experiments.
Next, using IM-MS, we focus on the osmolyte-induced differences in tau peptide conformations and populations prior to the addition of heparin. IM-MS analysis in the presence of TMAO was unsuccessful, as even at low TMAO concentrations and short time scales we consistently encountered technical problems due to clogging of the capillary electrospray tips, consistent with the rapid formation of tau oligomers or aggregates. In contrast, in the presence of urea, the ATDs at m/z = 820 indicate the population of dimers, which was ∼30% of the total population to begin with is further reduced to approximately 15%, as determined from the reduced signal intensity of the dimer feature relative to the monomer feature (Figure 3, panels F vs D). This suggests that urea breaks apart interprotein contacts within dimers, thereby shifting the population toward monomers. These results are consistent with computational studies showing that aggregates are destabilized in the presence of urea.49−51 MD simulations of the R2/WT peptide in urea have shown that urea binds to the peptide backbone and side chains, impeding the peptide’s ability to form compact intra- or interpeptide hydrogen bonded structures, leading primarily to extended conformations and preventing inter-tau bond formation.51
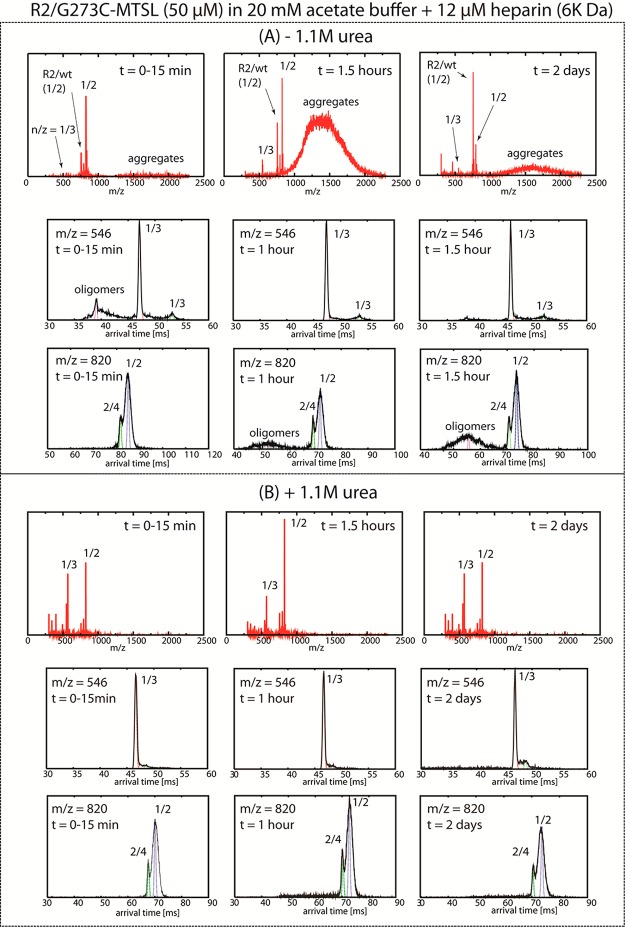
Upon addition of heparin to the solution, IM-MS data show that unresolved tau/aggregate peaks appear at time t = 0–15 min in the high m/z region of the mass spectrum around 1000–2000 (Figure 6A). At 1.5 h, the high m/z signal increases significantly, representing vigorous aggregation to form soluble oligomers. When the peptide has been allowed to aggregate for 2 days, the intensities decrease significantly, suggesting that the majority of the peptide monomer and oligomers have been converted to aggregates that can no longer be detected by MS (Figure 6A, top row), most likely due to the formation of insoluble fibrils. The ATD data in the second and third rows of Figure 6A show that at time t = 0–15 min peptide oligomers of intermediate sizes can be found at m/z = 546 (n/z = 1/3), which quickly deplete to transform into larger size oligomers with lower charges at m/z = 820 (n/z = 1/2). Oligomers gradually continue to form over the entire time scale of the experiment, presumably due to their lower charge states. In contrast, the mass spectral data in the presence of urea (Figure 6B) show no fingerprints of larger aggregates, even 2 days after heparin addition, while the ATD data show only a dimer peak at m/z = 820 across all time points. A small trace of oligomers larger than a dimer appears at t = 1 h in the presence of urea but does not increase in intensity with additional incubation time. Here, the IM-MS data together with TEM and ThT data confirm that adding urea shifts the tau peptide population toward monomer in the presence of heparin and strongly disrupts inter-tau interactions and reduces β-sheet formation. Apparently, the solution conditions that drive tau conformation toward monomer populations also hinder the formation of higher order oligomers and aggregates, even upon heparin addition. While the addition of heparin facilitates aggregate formation, this effector does not override the common molecular level interactions engrained in the solution conditions that appear to dictate interprotein interactions at the monomer, dimer, and higher order oligomer level.
Figure 6.
ESI-q-mass spectra of 50 μM G273C-SL in 12.5 μM 6 kDa heparin with and without 1.1 M urea dissolved in 20 mM ammonium acetate buffer. (A) In the absence of urea, large oligomers are detected at both n/z = 1/2 and 1/3 ATDs. The mass spectra show high signal-to-noise at high m/z indicating the prevalence of large and unresolved aggregates. Note the presence of a minor peak at n/z = 1/2, which likely represents an R2/WT impurity. (B) The presence of urea suppresses the aggregation process, as indicated by no significant changes in the mass spectra or ATDs after 2 days.
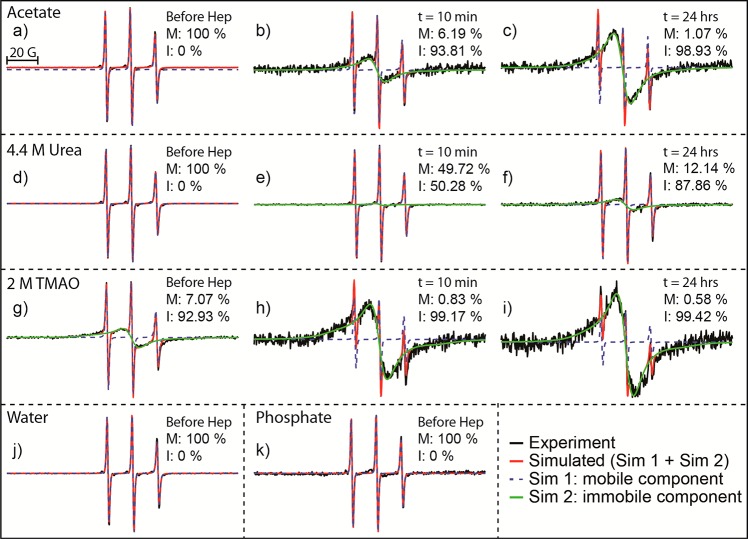
Finally, we seek to measure the peptide surface’s local environment by ODNP relaxometry,38 which reports on the translational diffusivity of surface water near the spin-labeled site of the R2/G273C-SL peptide by means of an electron–1H cross-relaxivity parameter, kσ, as defined in the literature.52,53 The reference value is the surface water diffusivity of the R2/G273C-SL peptide in bulk solution state before the addition of heparin and in the absence of osmolytes. Under these conditions, we know from IM-MS data that R2/G273C-SL predominantly exists as monomers, as previously discussed (Figure 3C,D), while quantitative EPR line shape analysis verifies the spin-label is mobile and fully solvent-exposed. Most importantly, EPR finds the spin-label is represented by a single mobile population (see Figure 7a and Supporting Information Figure S4), warranting the interpretation of the ODNP parameter kσ in terms of water dynamics. The kσ value of the R2/G273C-SL surface in acetate buffer, without added heparin or osmolytes, is 20.8 ± 1.0 s–1 M–1 (Figure 8). When heparin is added, the peptide surface water diffusivity or accessibility dramatically slows within the experimental dead time (∼ minutes), as reflected in the lowering of kσ to 2.5 ± 2.3 s–1 M–1. This value remains unchanged throughout the 24 h time course of the experiment during which aggregation proceeds. This implies that upon addition of heparin the peptide assumes a local conformation in which the spin-label is partially buried from the solvent. Hence, the local environment of the spin-label does not further change with macroscopic aggregate formation.
Figure 7.
Experimental EPR spectra together with quantitative line shape simulation and analysis of 400 μM R2/G273C-SL in 20 mM ammonium acetate buffer, pH 7.0, and 100 μM 6 kDa heparin with no osmolytes (a–c), 4.4 M urea (d–f), or 2 M TMAO (g–i) before heparin addition (a, d, and g, respectively), 10 min after heparin addition (b, e, and h, respectively), and 24 h after heparin addition (c, f, and i, respectively). Also shown is the peptide dissolved in water and 20 mM phosphate buffer, pH 7.0, before heparin addition (j and k, respectively). Experimental EPR data are shown in black, the total simulated EPR spectrum is shown in red, the mobile component of the simulation is shown as a blue dotted line, and the immobile component is shown in green. For each spectrum, the derived % population of the mobile (M) and immobile (I) components is listed.
Figure 8.
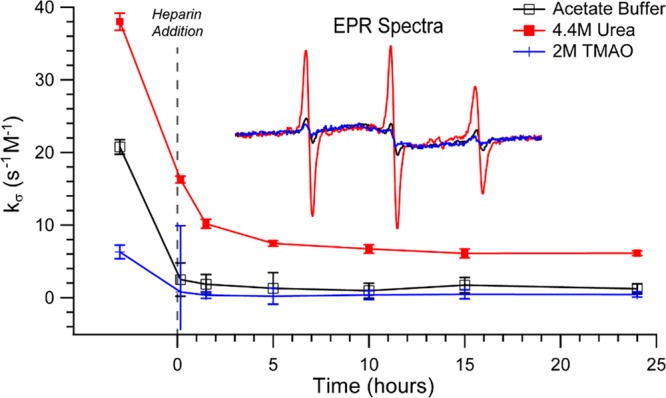
ODNP time course of 400 μM R2/G273C-SL in 20 mM acetate buffer, pH 7.0, and 100 μM 6 kDa heparin with either no osmolytes, 4.4 M urea, or 2 M TMAO. Note the connecting line from before to after heparin addition is meant as a guide. Inset: EPR spectra taken of all three conditions after 24 h and normalized by their respective second integrals.
In contrast, when urea is present at 4.4 M, the hydration dynamics of the R2/G273C-SL surface are significantly more active even before heparin is added, as reflected in kσ = 38.0 ± 1.2 s–1 M–1 (Figure 8). This increase in surface water dynamics in the presence of urea can only be in part due to the breaking apart of dimer populations that result in the spin-label being more exposed to the solvent. As previously described, IM-MS results show that the dimer population in the presence of urea is reduced to half of its initial, already small, dimer population. MD simulations of the wild-type peptide have shown that monomers in urea are more extended than in the absence of urea.51 Quantitative EPR analysis further verifies that the narrow line shape remains unaltered and that the EPR spectrum of G273C-SL before heparin addition consists of a single mobile population, with or without urea (compare Figure 7, panel a vs d; also see Supporting Information Figure S4). Thus, the observed change in kσ (Figure 8) cannot be simply due to the increased exposure of spin-label in the presence of urea. Instead, the dramatic increase in the tau peptide’s surface water diffusivity in the presence of urea is attributed to a weakened interaction between the peptide surface and the water hydrating the peptide surface and the weakened cohesion between water molecules of the dynamic hydration network near the peptide surface. The interpretation that urea weakens the peptide’s surface water network is further corroborated by reference measurements of the spin probe in solution alone (in the absence of peptide), where local water diffusivity slows in the presence of urea, as verified with a decrease in the value of kσ (see Table S1; from kσ = 56.6 ± 0.5 s–1 M–1 for acetate buffer to kσ = 33.4 ± 0.4 s–1 M–1 for 4.4 M urea dissolved in acetate buffer). The measured effect of urea decreasing the local water diffusivity near the free spin probe in bulk aqueous solution is expected, as urea is known to increase the bulk water viscosity by more than 20% at 4.4 M.54
When heparin is added to the solution of R2/G273C-SL in the presence of urea, the surface hydration dynamics slow dramatically over a period of several hours to yield kσ of 6.1 ± 0.3 s–1 M–1. Again, this change suggests that even in the presence of urea the peptide assumes an altered local conformation in which the spin-label is buried from the solvent and/or experiences an altered topological environment once aggregation is initiated. It is interesting that the changes do not result in significant aggregate formation as detectable by TEM, ThT, and IM-MS. Quantitative EPR line shape analysis finds that the characteristic single-line EPR feature signifying immobilized spin-labels embedded in parallel β-sheet structures increases from 0 to ∼50% within 15 min of adding heparin, even in the presence of urea (see Figure 7, panels d–f). This means a significant population of the tau fragments stack in β-sheet arrangements in the presence of urea when heparin is added, even though urea largely hinders the growth to larger fibrillar assemblies detectable by TEM or ThT staining. Consistently, the ATD data in the presence of urea hint at the formation of small populations of higher order assemblies (Figure 6B). Notably, the kσ value in the presence of urea is higher across all aggregation times compared to without, implying a higher surface water diffusivity experienced by the spin-label regardless of the tau conformation or oligomer distribution in the presence of heparin. This suggests that urea nonspecifically weakens the strength of the hydration water structure near the surface of the peptide.
TMAO (2 M) has the opposite effect on surface water diffusivity near the terminal site of R2/G273C-SL. Even before the addition of heparin, the hydration dynamics in the presence of TMAO are slowed significantly, as represented by a kσ value of 6.3 ± 0.9 s–1 M–1 (Figure 8). The reference measurements of free spin probes in acetate buffer containing the same TMAO concentration show that kσ changes from 56.6 ± 0.5 to 31.5 ± 0.4 s–1 M–1, which is consistent with an increase in bulk water viscosity by TMAO, similar to the changes observed when 4.4 M urea was added. Thus, the contrasting effects of TMAO and urea at the peptide surface are even more striking, especially given the much more dramatic decrease in the kσ value at the peptide surface when TMAO is added to the solution of the peptide, compared to in bulk solution. The effects of TMAO vs urea are therefore not that of altering bulk solution properties, but rather a result of their distinct interactions with the peptide surface and the peptide surface’s hydration water. Specifically, the significantly depressed kσ value even in the absence of heparin suggests that TMAO facilitates partial intraprotein folding and/or protein–protein interaction, thereby limiting the solvent accessibility of water to the spin-label tethered to the peptide surface. In fact, quantitative EPR line shape analysis finds that G273C-SL before heparin addition already contains a significant, single-line, immobile component (Figure 7, panel g). This population further increases to consume nearly 100% of G273C-SL, after a few hours upon heparin addition. This is consistent with the observation that the peptides in the presence of TMAO, even before the addition of heparin, could not be studied by IM-MS due to formation of assemblies that clogged the IM-MS tip. Within minutes of adding heparin, the value of kσ that reflects on surface water diffusion further drops to around the same value as that for the peptide in its fully aggregated state (kσ = 0.4 ± 0.1 s–1 M–1). This illustrates that the spin-label, within minutes of adding heparin, quickly experiences a local packing and topological environment indistinguishable from a mature fibril.
Effects of Solution Buffer on Oligomeric State and Aggregation Propensity
Next, we test the effects of more subtle modulators of protein conformation by varying the buffer composition. Surprisingly, when dissolving the peptide in solvents of 20 mM phosphate buffer (pH = 7), 20 mM acetate buffer (pH = 7) vs water, stark differences were observed at all stages of aggregation, from the early population of tau in solution before heparin addition to the quantity of macroscopic fibrils. ThT assays show that R2/G273C-SL forms β-sheet aggregates in the presence of heparin under all three solvent conditions; however, clear differences are observed in the amount of ThT active units formed, as seen in Figure 9. A substantial amount of ThT activity is observed when the peptide is dissolved in phosphate buffer and acetate buffer, and much less activity is seen when the peptide is dissolved in only water. While the ThT activity of aggregating tau in acetate buffer was highly reproducible, ThT data of the peptide dissolved in phosphate buffer were found to be variable, making it difficult to compare the extent of aggregation in the two buffers. Often, an initial drop was observed in the ThT signal of G273C-SL samples in phosphate buffer or an initially slow slope of increase in ThT amplitude observed as shown in Figure 9 (green trace), followed typically by a higher ThT amplitude in phosphate compared to acetate buffer, once aggregation is mature. This may be attributed to kinetically rapid aggregation in phosphate buffer, initially hampering the access of ThT dye to the β-sheets of tau peptide aggregates, similar to what we observed with R2/G273C-SL aggregation in the presence of 2 M TMAO (Figure 5), or to the generation of non-β-sheet aggregates.
Figure 9.
Thioflavin T fluorescence reveals effects of heparin and solvent on tau aggregation. Top panel is full scale while the bottom panel is zoomed in to show the first 30 min. The spin-labeled tau peptide R2/G273C-SL (25 μM) was incubated in 20 mM phosphate buffer (pH = 7.0), 20 mM ammonium acetate buffer (pH = 7.0), or water in the presence or absence of 6 kDa heparin (6.25 μM). All four non-heparin conditions generate identical and overlapping ThT signals, and so only one is shown for simplicity. Data shown are averaged from five separate experiments.
TEM images (Figure 10 inset) show that R2/G273C-SL aggregates and forms fibrils in all three solvent conditions. Still in water, the fibril formation is not as vigorous as in phosphate and acetate buffer. This is consistent with ThT assays which show a higher increase in aggregation when R2/G273C-SL is incubated with heparin in either acetate or phosphate buffer compared to water. Interestingly, fibrils formed in phosphate buffer tend to appear clumped, while fibrils formed in acetate buffer are more evenly dispersed and highly reproducible. This is a possible cause for the variability seen in the ThT data for the peptide in phosphate buffer, as fibril clumping could interfere with the access of the ThT to the fibrils or off-pathway, non-β-sheet, aggregates may be not ThT-active.
Figure 10.
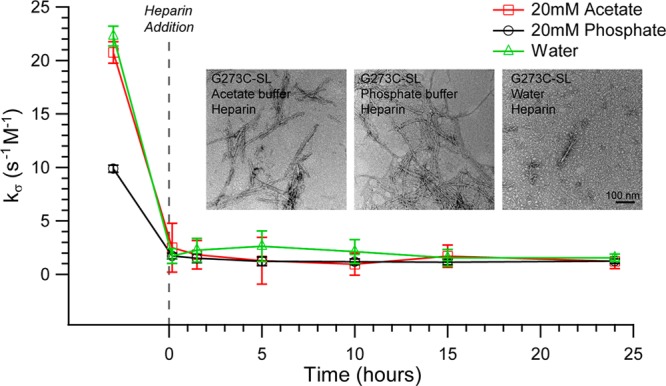
ODNP time course of 400 μM R2/G273C-SL in 20 mM ammonium acetate buffer, 20 mM phosphate buffer, or water along with 100 μM 6 kDa heparin where applicable. Inset: TEM of 100 μM R2/G273C-SL in 20 mM ammonium acetate buffer, 20 mM phosphate buffer, or water incubated with 25 μM 6 kDa heparin.
The IM-MS data for R2/G273C-SL are consistent with the TEM results. Monomers occur at n/z = 1/3 and n/z = 1/2 while dimers only occur at n/z = 1/2 in all solvents. An analysis of the ATDs indicates that monomer is dominant for water solvent, preferred for ammonium acetate solvent and roughly equivalent to dimer in phosphate solvent. This trend suggests that in phosphate buffer interactions between the peptides and aggregation is facilitated even before heparin is added.
In the absence of heparin, ODNP-derived surface water diffusivity (Figure 10) is comparably high in acetate buffer or water, as signified with a kσ of 20.8 ± 1.0 and 22.3 ± 0.9 s–1 M–1, respectively, but much slower in phosphate buffer, represented by a kσ of 9.9 ± 0.3 s–1 M–1. This correlates well with the IM-MS data trend that shows that the phosphate buffer shifts the conformation toward higher dimer populations. However, upon heparin addition, the water dynamics collapse to the same value in all three solutions (kσ ∼ 1.2 s–1 M–1), corresponding to the local solvent environment of the R2/G273C-SL peptide surface topology in its aggregated state. The control experiment of measuring local water dynamics near a free spin-label, not tethered to a tau peptide, showed no meaningful differences between phosphate buffer (kσ = 67.2 ± 0.3 s–1 M–1), acetate buffer (kσ = 56.6 ± 0.5 s–1 M–1), and water (kσ = 63.9 ± 0.4 s–1 M–1). Even though there is an ∼16% difference in kσ for the free spin-label between the acetate buffer and phosphate buffer, this is a small difference compared to the ∼52% difference in kσ between the two buffers on the surface of the tau peptide. Furthermore, the small change goes in the opposite direction between these two buffers for the free spin-label vs the spin-label on the tau peptide surface. This supports the conclusion that the differences in kσ are due to differences in the conformational population of tau peptides established in different buffer solutions prior to the initiation of aggregation. While IM-MS and ODNP data display clear contrast in the conformational equilibrium of G273C-SL in the different buffers prior to heparin addition, EPR line shape analysis finds that G273C-SL is represented with a single, high-mobility, population in acetate and phosphate buffer and water (Figure 7, panels a, j, and k). Importantly, this demonstrates that it is not the formation of higher order oligomers or aggregates under different buffer conditions that seeds or facilitates fibril growth, but indeed the shift in equilibrium toward aggregation-prone solution species as reflected in increased dimer population, still harboring highly mobile spin-labels, that is modulated by the choice of buffers.
Overall, we find that downstream aggregation events are highly sensitive to changes in early protein conformations and oligomer populations, not only when adding osmolytes (i.e., urea and TMAO) with known effects on protein conformation or protein–protein interactions55,56 but also when altering the type of buffer, which is usually regarded a subtle or even inconsequential detail in sample preparation for protein aggregation studies. We conclude that the effect of the respective solution conditions (i.e., buffers, osmolytes, etc.) on the initial protein conformations and populations is a critical factor, as they significantly and consistently affect the oligomerization and aggregation properties of tau peptide systems downstream. While we cannot exclude the possibility that differing solution conditions may alter the interaction of heparin with the peptide itself, this is believed to be a minor factor given that the tau–heparin interaction is thought to be mediated by dominant electrostatic interactions. Crucially the main changes in dimer population or peptide–solvent interaction with altered solution conditions occur before heparin addition.
Conclusions
The aggregation process and accompanying transient changes of tau are challenging to study experimentally, given that tau is an intrinsically disordered protein with multiple monomeric and small oligomeric populations coexisting in solution. Effects that shift these populations, however, can have significant consequences for tau aggregation and fibrillization. Here we report on the study of tau peptide aggregation using a set of techniques including TEM and ThT assays to characterize amyloid fibril formation, IM-MS to derive tau populations in solution and their cross sections, EPR line shape analysis to quantify the population of mobile spin-labels vs immobile spin-labels embedded in β-sheet structured interfaces, and ODNP-derived surface water diffusivity/accessibility near the spin-label of the tau peptide in situ as aggregation proceeds. Specifically, we find that urea and TMAO both dramatically affect oligomer formation and aggregation propensity of the short peptide R2/G273C-SL, shifting the tau conformation and/or population in opposite directions with regards to aggregation propensities. IM-MS data and ODNP measurements find consistent results that urea extends the tau conformation and breaks apart inter-tau contacts and confirm that urea significantly weakens the cohesive and dynamic water network hydrating the R2/G273C-SL surface. Urea not only shifts the population toward monomers but also persistently disrupts the surface water network on the tau peptide and clusters near the tau surface and so prevents inter-tau dimerization, as has been also recently shown by a computational study.51 This is consistent with the ODNP results that display weakened, and thus fast diffusing, hydration water near the tau peptide surface, before as well as after aggregation is initiated. The opposite effect is apparent in the presence of TMAO. The presence of acetate and phosphate buffers also shifts the tau peptide toward more aggregation prone populations compared to water which, in turn, was shown to affect the ability of the peptide to aggregate after the addition of heparin. The observation that the initial conformations induced via osmolytes or buffers affect aggregation all the way through to the fibrillar state of the peptide suggests that the factors modulating the competition between solvent–protein and protein–protein interactions in solution state are essential factors that not only influence dimer formation propensity but also downstream protein aggregation and fibril formation propensities. This suggests that it is not the concentration of higher order oligomers or aggregates that catalyzes or modulates aggregation and fibril growth as proposed by the nucleation polymerization model,14,15 at least in the case of the here-studied tau fragment. Even though heparin drives aggregation kinetics dramatically toward fibril formation, intrinsic interprotein interactions that are differentially modulated by solution conditions persist throughout the aggregation process.
Acknowledgments
S.H. thanks Profs. John Lew and Rick Dahlquist for their initial help and support in the study of tau aggregation. S.H. and N.E. acknowledge support through the NIH Director’s New Innovator award (1DP2OD007221-01) and support through the Excellence Cluster on Solvation science of the RUB. This material is based upon work supported by the National Science foundation Graduate Research Fellowship under Grant DGE 1144085. N.E.L. was supported by a grant from Santa Barbara Cottage Hospital and the University of California, Santa Barbara. M.T.B. acknowledges support from the NIH (1RO1AG047116) and the NSF (CHE-1301032). J.-E.S. acknowledges NSF Grant MCB-1158577. All authors made use of the NRI-MCDB Microscopy Facility and the Materials Research Laboratory Central Facilities supported by the NSF through the Materials Research Science and Engineering Centers (# DMR 1121053). The authors acknowledge the use of the Biological Nanostructures Laboratory within the California NanoSystems Institute, supported by the University of California, Santa Barbara and the University of California, Office of the President. All authors thank Zachary Levine for his contribution to the TOC figure. We acknowledge the use of the MultiComponent software for cw ESR spectral simulation devolped by Dr. Christian Altenbach (University of California, Los Angeles, CA). The program is written in LabVIEW (National Instruments) and can be freely downloaded from the following site: http://www.chemistry.ucla.edu/directory/hubbell-wayne-l.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jpcb.5b08092.
-
Figures showing the IM-MS comparison of R2/WT and R2/G273C-SL, IM-MS reference measurement of C-MTSL, EPR of R2/G273C-SL in various osmolytes, EPR of R2/G273C-SL in various buffers, ATDs of R2/WT and R2/G273C-SL in various buffers, IM-MS comparison of R2/G273C-SL in water and phosphate buffer; discussion on the n/z = 1/3 peak from the ATD of R2/G273C-SL; table of ODNP reference measurements in various buffers
(PDF)
Author Present Address
T.D.D.: Department of Chemistry and the Beckman Institute, University of Illinois at Urbana–Champaign, Urbana, IL 61801.
Author Contributions
N.A.E., T.D.D., and N.E.L. have equally contributed to this study.
The authors declare no competing financial interest.
Supplementary Material
References
- Walker L. C.; Levine H. III; Mattson M. P.; Jucker M. Inducible Proteopathies. Trends Neurosci. 2006, 29, 438–443 10.1016/j.tins.2006.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson C. M. Protein Misfolding, Evolution and Disease. Trends Biochem. Sci. 1999, 24, 329–332 10.1016/S0968-0004(99)01445-0. [DOI] [PubMed] [Google Scholar]
- Klein W. L.; Krafft G. A.; Finch C. E. Targeting Small Abeta Oligomers: The Solution to an Alzheimer’s Disease Conundrum?. Trends Neurosci. 2001, 24, 219–224 10.1016/S0166-2236(00)01749-5. [DOI] [PubMed] [Google Scholar]
- Laganowsky A.; Liu C.; Sawaya M. R.; Whitelegge J. P.; Park J.; Zhao M.; Pensalfini A.; Soriaga A. B.; Landau M.; Teng P. K.; et al. Atomic View of a Toxic Amyloid Small Oligomer. Science 2012, 335, 1228–1231 10.1126/science.1213151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goedert M. The Significance of Tau and Alpha-Synuclein Inclusions in Neurodegenerative Diseases. Curr. Opin. Genet. Dev. 2001, 11, 343–351 10.1016/S0959-437X(00)00200-8. [DOI] [PubMed] [Google Scholar]
- Chen S.; Ferrone F. A.; Wetzel R. Huntington’s Disease Age-of-Onset Linked to Polyglutamine Aggregation Nucleation. Proc. Natl. Acad. Sci. U. S. A. 2002, 99, 11884–11889 10.1073/pnas.182276099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haass C.; Selkoe D. J. Soluble Protein Oligomers in Neurodegeneration: Lessons from the Alzheimer’s Amyloid Beta-Peptide. Nat. Rev. Mol. Cell Biol. 2007, 8, 101–112 10.1038/nrm2101. [DOI] [PubMed] [Google Scholar]
- Binder L. I.; Guillozet-Bongaarts A. L.; Garcia-Sierra F.; Berry R. W. Tau, Tangles, and Alzheimer’s Disease. Biochim. Biophys. Acta, Mol. Basis Dis. 2005, 1739, 216–223 10.1016/j.bbadis.2004.08.014. [DOI] [PubMed] [Google Scholar]
- Ward S. M.; Himmelstein D. S.; Lancia J. K.; Binder L. I. Tau Oligomers and Tau Toxicity in Neurodegenerative Disease. Biochem. Soc. Trans. 2012, 40, 667–671 10.1042/BST20120134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee V. M.; Goedert M.; Trojanowski J. Q. Neurodegenerative Tauopathies. Annu. Rev. Neurosci. 2001, 24, 1121–1159 10.1146/annurev.neuro.24.1.1121. [DOI] [PubMed] [Google Scholar]
- Kumar S.; Tepper K.; Kaniyappan S.; Biernat J.; Wegmann S.; Mandelkow E.-M.; Müller D. J.; Mandelkow E. Stages and Conformations of the Tau Repeat Domain During Aggregation and Its Effect on Neuronal Toxicity. J. Biol. Chem. 2014, 289, 20318–20332 10.1074/jbc.M114.554725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbaum-Garfinkle S.; Cobb G.; Compton J. T.; Li X. H.; Rhoades E. Tau Mutants Bind Tubulin Heterodimers with Enhanced Affinity. Proc. Natl. Acad. Sci. U. S. A. 2014, 111, 6311–6316 10.1073/pnas.1315983111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.; Culyba E. K.; Powers E. T.; Kelly J. W. Amyloid-B Forms Fibrils by Nucleated Conformational Conversion of Oligomers. Nat. Chem. Biol. 2011, 7, 602–609 10.1038/nchembio.624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orte A.; Birkett N. R.; Clarke R. W.; Devlin G. L.; Dobson C. M.; Klenerman D. Direct Characterization of Amyloidogenic Oligomers by Single-Molecule Fluorescence. Proc. Natl. Acad. Sci. U. S. A. 2008, 105, 14424–14429 10.1073/pnas.0803086105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Simone A.; Kitchen C.; Kwan A. H.; Sunde M.; Dobson C. M.; Frenkel D. Intrinsic Disorder Modulates Protein Self-Assembly and Aggregation. Proc. Natl. Acad. Sci. U. S. A. 2012, 109, 6951–6956 10.1073/pnas.1118048109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L.; Drouet V.; Wu J. W.; Witter M. P.; Small S. A.; Clelland C.; Duff K. Trans-Synaptic Spread of Tau Pathology in Vivo. PLoS One 2012, 7, e31302. 10.1371/journal.pone.0031302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clavaguera F.; Hench J.; Goedert M.; Tolnay M. Invited Review: Prion-Like Transmission and Spreading of Tau Pathology. Neuropathol. Appl. Neurobiol. 2015, 41, 47–58 10.1111/nan.12197. [DOI] [PubMed] [Google Scholar]
- Falcon B.; Cavallini A.; Angers R.; Glover S.; Murray T. K.; Barnham L.; Jackson S.; O’Neill M. J.; Isaacs A. M.; Hutton M. L.; et al. Conformation Determines the Seeding Potencies of Native and Recombinant Tau Aggregates. J. Biol. Chem. 2015, 290, 1049–1065 10.1074/jbc.M114.589309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spillantini M. G.; Goedert M.; Crowther R. A.; Murrell J. R.; Farlow M. R.; Ghetti B. Familial Multiple System Tauopathy with Presenile Dementia: A Disease with Abundant Neuronal and Glial Tau Filaments. Proc. Natl. Acad. Sci. U. S. A. 1997, 94, 4113–4118 10.1073/pnas.94.8.4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feany M. B.; Dickson D. W. Neurodegenerative Disorders with Extensive Tau Pathology: A Comparative Study and Review. Ann. Neurol. 1996, 40, 139–148 10.1002/ana.410400204. [DOI] [PubMed] [Google Scholar]
- von Bergen M.; Barghorn S.; Li L.; Marx A.; Biernat J.; Mandelkow E. M.; Mandelkow E. Mutations of Tau Protein in Frontotemporal Dementia Promote Aggregation of Paired Helical Filaments by Enhancing Local Beta-Structure. J. Biol. Chem. 2001, 276, 48165–48174. [DOI] [PubMed] [Google Scholar]
- Larini L.; Gessel M. M.; LaPointe N. E.; Do T. D.; Bowers M. T.; Feinstein S. C.; Shea J.-E. Initiation of Assembly of Tau(273–284) and Its Δk280 Mutant: An Experimental and Computational Study. Phys. Chem. Chem. Phys. 2013, 15, 8916–8928 10.1039/c3cp00063j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W.; Lee V. M. Characterization of Two Vqixxk Motifs for Tau Fibrillization in Vitro. Biochemistry 2006, 45, 15692–15701 10.1021/bi061422+. [DOI] [PubMed] [Google Scholar]
- Sawaya M. R.; Sambashivan S.; Nelson R.; Ivanova M. I.; Sievers S. A.; Apostol M. I.; Thompson M. J.; Balbirnie M.; Wiltzius J. J.; McFarlane H. T.; et al. Atomic Structures of Amyloid Cross-Beta Spines Reveal Varied Steric Zippers. Nature 2007, 447, 453–457 10.1038/nature05695. [DOI] [PubMed] [Google Scholar]
- von Bergen M.; Friedhoff P.; Biernat J.; Heberle J.; Mandelkow E. M.; Mandelkow E. Assembly of Tau Protein into Alzheimer Paired Helical Filaments Depends on a Local Sequence Motif ((306)Vqivyk(311)) Forming Beta Structure. Proc. Natl. Acad. Sci. U. S. A. 2000, 97, 5129–5134 10.1073/pnas.97.10.5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goux W. J.; Kopplin L.; Nguyen A. D.; Leak K.; Rutkofsky M.; Shanmuganandam V. D.; Sharma D.; Inouye H.; Kirschner D. A. The Formation of Straight and Twisted Filaments from Short Tau Peptides. J. Biol. Chem. 2004, 279, 26868–26875 10.1074/jbc.M402379200. [DOI] [PubMed] [Google Scholar]
- Straub J. E.; Thirumalai D. Toward a Molecular Theory of Early and Late Events in Monomer to Amyloid Fibril Formation. Annu. Rev. Phys. Chem. 2011, 62, 437–463 10.1146/annurev-physchem-032210-103526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M. S.; Co N. T.; Reddy G.; Hu C. K.; Straub J. E.; Thirumalai D. Factors Governing Fibrillogenesis of Polypeptide Chains Revealed by Lattice Models. Phys. Rev. Lett. 2010, 105, 218101. 10.1103/PhysRevLett.105.218101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellarin R.; Caflisch A. Interpreting the Aggregation Kinetics of Amyloid Peptides. J. Mol. Biol. 2006, 360, 882–892 10.1016/j.jmb.2006.05.033. [DOI] [PubMed] [Google Scholar]
- Bellesia G.; Shea J.-E. Effect of B-Sheet Propensity on Peptide Aggregation. J. Chem. Phys. 2009, 130, 145103. 10.1063/1.3108461. [DOI] [PubMed] [Google Scholar]
- Sciarretta K. L.; Gordon D. J.; Petkova A. T.; Tycko R.; Meredith S. C. Aβ40-Lactam(D23/K28) Models a Conformation Highly Favorable for Nucleation of Amyloid†. Biochemistry 2005, 44, 6003–6014 10.1021/bi0474867. [DOI] [PubMed] [Google Scholar]
- Gessel M. M.; Bernstein S.; Kemper M.; Teplow D. B.; Bowers M. T. Familial Alzheimer’s Disease Mutations Differentially Alter Amyloid B-Protein Oligomerization. ACS Chem. Neurosci. 2012, 3, 909–918 10.1021/cn300050d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morriss-Andrews A.; Shea J.-E. Computational Studies of Protein Aggregation: Methods and Applications. Annu. Rev. Phys. Chem. 2015, 66, 643–666 10.1146/annurev-physchem-040513-103738. [DOI] [PubMed] [Google Scholar]
- Margittai M.; Langen R. Template-Assisted Filament Growth by Parallel Stacking of Tau. Proc. Natl. Acad. Sci. U. S. A. 2004, 101, 10278–10283 10.1073/pnas.0401911101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fichou Y.; Schirò G.; Gallat F.-X.; Laguri C.; Moulin M.; Combet J.; Zamponi M.; Härtlein M.; Picart C.; Mossou E.; et al. Hydration Water Mobility Is Enhanced around Tau Amyloid Fibers. Proc. Natl. Acad. Sci. U. S. A. 2015, 112, 6365–6370 10.1073/pnas.1422824112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein S. L.; Dupuis N. F.; Lazo N. D.; Wyttenbach T.; Condron M. M.; Bitan G.; Teplow D. B.; Shea J. E.; Ruotolo B. T.; Robinson C. V.; et al. Amyloid-Beta Protein Oligomerization and the Importance of Tetramers and Dodecamers in the Aetiology of Alzheimer’s Disease. Nat. Chem. 2009, 1, 326–331 10.1038/nchem.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altenbach C.; Oh K.-J.; Trabanino R. J.; Hideg K.; Hubbell W. L. Estimation of Inter-Residue Distances in Spin Labeled Proteins at Physiological Temperatures: Experimental Strategies and Practical Limitations†. Biochemistry 2001, 40, 15471–15482 10.1021/bi011544w. [DOI] [PubMed] [Google Scholar]
- Armstrong B. D.; Han S. Overhauser Dynamic Nuclear Polarization to Study Local Water Dynamics. J. Am. Chem. Soc. 2009, 131, 4641–4647 10.1021/ja809259q. [DOI] [PubMed] [Google Scholar]
- Franck J. M.; Pavlova A.; Scott J. A.; Han S. Quantitative Cw Overhauser Effect Dynamic Nuclear Polarization for the Analysis of Local Water Dynamics. Prog. Nucl. Magn. Reson. Spectrosc. 2013, 74, 33–56 10.1016/j.pnmrs.2013.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franck J. M.; Scott J. A.; Han S. Nonlinear Scaling of Surface Water Diffusion with Bulk Water Viscosity of Crowded Solutions. J. Am. Chem. Soc. 2013, 135, 4175–4178 10.1021/ja3112912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goedert M.; Jakes R.; Spillantini M. G.; Hasegawa M.; Smith M. J.; Crowther R. A. Assembly of Microtubule-Associated Protein Tau into Alzheimer-Like Filaments Induced by Sulphated Glycosaminoglycans. Nature 1996, 383, 550–553 10.1038/383550a0. [DOI] [PubMed] [Google Scholar]
- Columbus L.; Kálai T.; Jekö J.; Hideg K.; Hubbell W. L. Molecular Motion of Spin Labeled Side Chains in A-Helices: Analysis by Variation of Side Chain Structure†. Biochemistry 2001, 40, 3828–3846 10.1021/bi002645h. [DOI] [PubMed] [Google Scholar]
- Mason E. A.; McDaniel E. W.. Transport Properties of Ions in Gases; John Wiley & Sons: New York, 1988. [Google Scholar]
- Gidden J.; Ferzoco A.; Baker E. S.; Bowers M. T. Duplex Formation and the Onset of Helicity in Poly D(Cg)N Oligonucleotides in a Solvent-Free Environment. J. Am. Chem. Soc. 2004, 126, 15132–15140 10.1021/ja046433+. [DOI] [PubMed] [Google Scholar]
- Kemper P. R.; Dupuis N. F.; Bowers M. T. A New, Higher Resolution, Ion Mobility Mass Spectrometer. Int. J. Mass Spectrom. 2009, 287, 46–57 10.1016/j.ijms.2009.01.012. [DOI] [Google Scholar]
- Goedert M.; Spillantini M. G.; Cairns N. J.; Crowther R. A. Tau Proteins of Alzheimer Paired Helical Filaments: Abnormal Phosphorylation of All Six Brain Isoforms. Neuron 1992, 8, 159–168 10.1016/0896-6273(92)90117-V. [DOI] [PubMed] [Google Scholar]
- Chiti F.; Dobson C. M. Protein Misfolding, Functional Amyloid, and Human Disease. Annu. Rev. Biochem. 2006, 75, 333–366 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- Do T. D.; Bowers M. T. Diphenylalanine Self Assembly: Novel Ion Mobility Methods Showing the Essential Role of Water. Anal. Chem. 2015, 87, 4245–4252 10.1021/ac5046774. [DOI] [PubMed] [Google Scholar]
- Klimov D. K.; Straub J. E.; Thirumalai D. Aqueous Urea Solution Destabilizes Aβ16–22 Oligomers. Proc. Natl. Acad. Sci. U. S. A. 2004, 101, 14760–14765 10.1073/pnas.0404570101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Z.; Li J.; Yin C.; Yang Z.; Wu J.; Zhou R. Effect of Urea Concentration on Aggregation of Amyloidogenic Hexapeptides (Nfgail). J. Phys. Chem. B 2014, 118, 48–57 10.1021/jp407776e. [DOI] [PubMed] [Google Scholar]
- Levine Z. A.; Larini L.; LaPointe N. E.; Feinstein S. C.; Shea J. E. Regulation and Aggregation of Intrinsically Disordered Peptides. Proc. Natl. Acad. Sci. U. S. A. 2015, 112, 2758–2763 10.1073/pnas.1418155112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franck J. M.; Pavlova A.; Scott J. A.; Han S. Quantitative Cw Overhauser Effect Dynamic Nuclear Polarization for the Analysis of Local Water Dynamics. Prog. Nucl. Magn. Reson. Spectrosc. 2013, 74, 33–56 10.1016/j.pnmrs.2013.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain S.; Franck J. M.; Han S. Transmembrane Protein Activation Refined by Site-Specific Hydration Dynamics. Angew. Chem., Int. Ed. 2013, 52, 1953–1958 10.1002/anie.201206147. [DOI] [PubMed] [Google Scholar]
- Kawahara K.; Tanford C. Viscosity and Density of Aqueous Solutions of Urea and Guanidine Hydrochloride. J. Biol. Chem. 1966, 241, 3228–3232. [PubMed] [Google Scholar]
- Candotti M.; Esteban-Martin S.; Salvatella X.; Orozco M. Toward an Atomistic Description of the Urea-Denatured State of Proteins. Proc. Natl. Acad. Sci. U. S. A. 2013, 110, 5933–5938 10.1073/pnas.1216589110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello C. C.; Barrick D. Measuring the Stability of Partly Folded Proteins Using Tmao. Protein Sci. 2003, 12, 1522–1529 10.1110/ps.0372903. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.