Background: The conserved cytomegalovirus proteins pUL50 and pUL53 heterodimerize and form a core nuclear egress complex.
Results: The crystal structure of pUL50-pUL53 was solved at 2.44 Å resolution, revealing an N-terminal hook-like extension of pUL53.
Conclusion: Data unravel the core NEC architecture, providing a scaffold for viral-cellular NEC protein interactions.
Significance: The identified NEC structure will stimulate the development of novel antiviral strategies.
Keywords: dimerization, herpesvirus, nucleus, protein complex, scaffold protein, dimer, nuclear egress, human cytomegalovirus, nuclear capsid egress, core nuclear egress complex, heterodimer pUL50-pUL53, crystal structure, scaffold for virus-host protein interactions
Abstract
Nuclear replication of cytomegalovirus relies on elaborate mechanisms of nucleocytoplasmic egress of viral particles. Thus, the role of two essential and conserved viral nuclear egress proteins, pUL50 and pUL53, is pivotal. pUL50 and pUL53 heterodimerize and form a core nuclear egress complex (NEC), which is anchored to the inner nuclear membrane and provides a scaffold for the assembly of a multimeric viral-cellular NEC. Here, we report the crystal structure of the pUL50-pUL53 heterodimer (amino acids 1–175 and 50–292, respectively) at 2.44 Å resolution. Both proteins adopt a globular fold with mixed α and β secondary structure elements. pUL53-specific features include a zinc-binding site and a hook-like N-terminal extension, the latter representing a hallmark element of the pUL50-pUL53 interaction. The hook-like extension (amino acids 59–87) embraces pUL50 and contributes 1510 Å2 to the total interface area (1880 Å2). The pUL50 structure overall resembles the recently published NMR structure of the murine cytomegalovirus homolog pM50 but reveals a considerable repositioning of the very C-terminal α-helix of pUL50 upon pUL53 binding. pUL53 shows structural resemblance with the GHKL domain of bacterial sensory histidine kinases. A close examination of the crystal structure indicates partial assembly of pUL50-pUL53 heterodimers to hexameric ring-like structures possibly providing additional scaffolding opportunities for NEC. In combination, the structural information on pUL50-pUL53 considerably improves our understanding of the mechanism of HCMV nuclear egress. It may also accelerate the validation of the NEC as a unique target for developing a novel type of antiviral drug and improved options of broad-spectrum antiherpesviral therapy.
Introduction
Human cytomegalovirus (HCMV,4 family Herpesviridae) is a major human pathogen showing a worldwide distribution. Its clinical importance has occasionally been underestimated, as infection of the immunocompetent host may be limited to mild forms of symptoms (1). The main pathogenesis of HCMV is manifested by severe systemic or even life-threatening disease in immunosuppressed hosts and upon congenital infection of neonates (2, 3). HCMV pathogenesis is determined by various parameters of immune control, viral productivity, viremia, tissue tropism, and organ damage, as well as manifold regulatory events of virus-host interaction (1). Hence, the viral productive replication cycle is largely coregulated by the interaction between viral and cellular proteins and by the formation of virus-host multiprotein complexes.
Recently, the viral nuclear egress complex (NEC) has attracted the deep interest of researchers because it represents a regulatory key position of viral replication and a putative target for novel antiviral strategies (4–9). As a characteristic feature of most DNA viruses, HCMV starts genomic replication in the host cell nucleus, where preformed capsids are packaged and exported to the cytoplasm for further virion maturation. The transition of capsids through the nuclear envelope (nuclear egress) is a multistep regulatory process that involves a phosphorylation-triggered distortion of the nuclear lamina (10–17). The HCMV-encoded protein kinase pUL97 was identified as the first herpesviral kinase with lamin-phosphorylating activity (11, 16, 17). Importantly, the recruitment of lamin-phosphorylating viral and cellular kinases as well as further lamin-modifying host factors (such as prolyl cis/trans-isomerase Pin1 (13))5 is accomplished by two conserved viral nuclear egress proteins, pUL50 and pUL53. As an essential step in HCMV replication, pUL50 and pUL53 heterodimerize and form the core of the NEC that serves as a scaffold for the recruitment of a group of viral and cellular NEC-associated proteins. The composition of the multimeric NEC of human and murine CMVs has recently been defined by proteomic analyses (15, 18).
Here, we report the first crystal structure of the HCMV core NEC. Functionally relevant fragments of pUL50 and pUL53 were coproduced in Escherichia coli, copurified, and crystallized to determine the three-dimensional structure at 2.44 Å resolution. Implications for the assembly of the multimeric NEC are discussed in view of the observed hook-like interaction between pUL53 and pUL50 and the larger assembly of pUL50-pUL53 heterodimers into hexameric ring-like structures in the crystals.
Experimental Procedures
Protein Expression and Purification
PCR amplification of HCMV ORFs UL50 and UL53 was performed using Vent polymerase reactions as described elsewhere (19). PCR primers used were: 5-UL50-BsaI-ACCT, TGAGGTCTCAAGGTATGGAGATGAACAAGGTTCTCC; 3-UL50(175 STOP)-XbaI, TAGTCTAGATCACGACGTCGACGCGGCTTTC; 5-UL53(50–292)-NheI, TAGGCTAGCCCGTCGCCGGCCGACGCG; 3-UL53(50–292 STOP)-EcoRI, TAGGAATTCTCAGCCGCTGGACTGACACAGCTCC.
The N-terminal fragment of pUL50 (residues 1–175 of the ORF-UL50 1–397) and the central fragment of pUL53 (residues 50–292 of the ORF-UL53 1–376) were cloned into vectors pE-SUMO(Amp) (LifeSensors Inc.) or pET-28b(+) (Novagen), respectively. The proteins SUMO-pUL50(1–175) and His-pUL53(50–292) were coproduced in E. coli BL21(DE3). Bacterial cells were grown in LB medium in the presence of 100 μg/ml ampicillin and 32 μg/ml kanamycin at 33 °C to an A600 of 0.4 before the temperature was lowered to 20 °C, and protein expression was induced with 0.25 mm isopropyl β-d-thiogalactopyranoside overnight. For selenomethionine labeling, the transformed E. coli BL21(DE3) cells were grown in auto-inducing PASM-5052 medium supplemented with selenomethionine as described before (20). The main cultures were grown at 37 °C to an A600 of 0.4, and protein expression continued at 20 °C overnight. Bacterial cells were harvested by centrifugation, resuspended in HisTrap binding buffer (50 mm phosphate buffer, pH 7.4, 300 mm NaCl, 30 mm imidazole) containing protease inhibitors, lysozyme, and DNase, and disrupted by sonication. Purification was performed by immobilized metal ion affinity chromatography, followed by cleavage of both SUMO and His tags with SUMO protease and thrombin, respectively. The cleaved tags were removed by a second ion affinity chromatography, and the subsequent size exclusion chromatography step was performed as described before using a 50 mm Tris/HCl, pH 8.0, 150 mm NaCl, 2 mm tris(2-carboxyethyl)phosphine (TCEP) buffer (14).
Crystallization of pUL50(1–175)-pUL53(50–292)
Native unlabeled crystals were obtained by sitting drop vapor diffusion in CrystalQuick 96-well plates (Greiner Bio-One). Crystals grew after 1 week at 20 °C after mixing equal volumes of protein (10–13 mg/ml in gel filtration buffer) and reservoir solution (0.2 m CaCl2, 20% w/v PEG 3350). Selenomethionine-derivatized crystals were obtained via the batch method by mixing 10 μl of protein solution (9–10 mg/ml in gel filtration buffer) with 10 μl of reservoir solution (0.2 m CaCl2, 20% w/v PEG 3350) in round-bottom PCR plates (Porvair Sciences) after 10 days at 20 °C. Native and selenomethionine-labeled crystals were soaked in cryo-protectant (80% mother liquor, 20% w/v PEG 400) and subsequently in perfluoropolyether cryo-oil (Hampton Research) prior to flash-cooling in liquid nitrogen.
Structure Determination
Datasets were collected at BESSY synchrotron Berlin, Germany and PETRA in Hamburg, Germany and processed with the program XDS (Table 1) (21, 22). The phase problem was solved via single anomalous dispersion phasing using the program HKL2MAP and the SHELX program suite (23, 24). A clear anomalous signal (|F(+) − F(−)|/σF > 1) was detectable up to a resolution of 3.4 Å in the selenomethionine dataset collected at the peak wavelength. Upon identification of the selenium positions with SHELXD (correlation coefficient = 49/24% for all/weak data) (25), a partial model containing 255 residues could be traced with SHELXE (correlation coefficient = 31% between partial structure and native dataset). The model was further completed with module AUTOBUILD in program PHENIX (26), manually inspected and corrected with the program COOT (27), and refined with the program REFMAC (28). All structure illustrations presented below were prepared with the program PyMOL (29).
TABLE 1.
Crystallographic data and refinement statistics
| Native pUL50-pUL53 | Se-Met pUL50-pUL53 | |
|---|---|---|
| Data collection | BESSY 14.1 | PETRA III P14 |
| Space group | P6 | P6 |
| Cell dimensions | ||
| a, b, c (Å) | 118.16 118.16 73.62 | 119.67 119.67 74.13 |
| α, β, γ (°) | 90 90 120 | 90 90 120 |
| Resolution (Å) | 50–2.44 (2.50–2.44)a | 50–3.00 (3.08–3.00) |
| No. of reflections | 21,930 | 23,546 |
| Wavelength (Å) | 0.9184 | 0.9794 |
| Mean I/σI | 13.5 (1.3) | 18.2 (1.4) |
| CC ½ (%) | 99.9 (79.0) | 99.9 (58.3) |
| Crystal mosaicity (°) | 0.08 | 0.17 |
| Completeness (%) | 99.9 (99.9) | 100 (99.9) |
| Rsym (%) | 8.8 (129.8) | 12.0 (193.9) |
| Redundancy | 6.8 (6.6) | 20.00 (10.1) |
| Wilson B-factor (Å2) | 68.3 | 98.2 |
| Mean (|F(+) − F(−)|/σF) | 0.8 | 1.8 |
| Refinement | ||
| Rwork/Rfree (%) | 22.1/28.7 | |
| No. of protein atoms | 3081 | |
| No. of non-protein atoms | 1 Zn2+, 20 solvent molecules | |
| r.m.s.d. valuesb | ||
| Bond lengths (Å) | 0.010 | |
| Angles (°) | 1.413 | |
| Φ/Ψ plotc | ||
| Most favored (%) | 92.0 | |
| Allowed (%) | 7.5 | |
| Generally allowed (%) | 0.6 | |
| Disallowed (%) | 0.0 | |
| Average B value (Å2) | ||
| Protein | 78.9 | |
| Zn2+ | 62.2 | |
| Solvent molecules | 54.0 | |
aValues for the highest resolution shell are provided in parentheses.
br.m.s.d. values, root mean square deviation values.
cAs calculated with the program PROCHECK (21).
Results and Discussion
Crystal structure of the HCMV pUL50-pUL53 Core NEC
The formation of a core NEC on the basis of an inner nuclear membrane-anchored heterodimer pUL50-pUL53 is pivotal for the nuclear steps of HCMV replication and the subsequent morphogenesis of viral particles. Previous analyses of biochemical properties pointed to a tightly interlocked high-affinity heterodimer formed between pUL50 and pUL53, similar to the respective homologs of other herpesviruses (9, 19, 30, 31). A mutational analysis of the protein segments that participate in the pUL50-pUL53 interaction supported this notion, namely that a unique type of interlock is provided by N-terminal α-helical segments of pUL53 that become tightly hooked into a platform provided by pUL50.
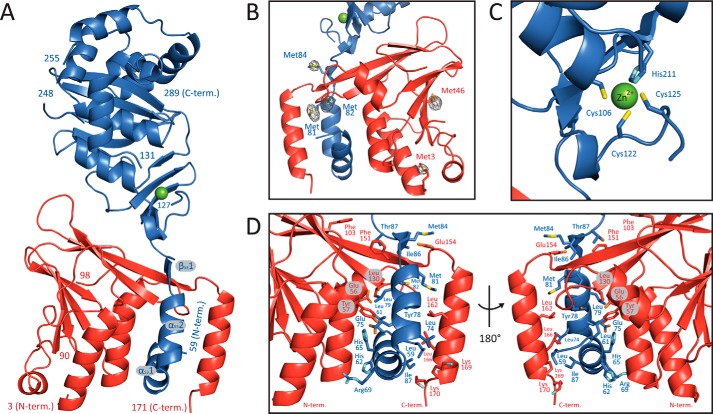
In the 2.44 Å crystal structure, the segments 3–171 of pUL50 and 59–289 of pUL53 could be traced almost contiguously in the electron density maps (Fig. 1). In comparison with the fragments that were present in the crystallization setup (residues 1–175 of pUL50 and 50–292 of pUL53), only a few terminal residues are not resolved in the current model. Three additional segments, namely 91–97 in pUL50 and 128–130 and 249–254 in pUL53, could not be located with confidence and were therefore omitted from the crystal structure (Table 1) (21). pUL50 and pUL53 adopt highly globular folds and consist of mixed α and β secondary structure elements (Fig. 1, A and B). Each protein contains two all-antiparallel β-sheets. Although pUL50 contains a five-stranded and three-stranded β-sheet, pUL53 displays two four-stranded sheets. In comparison with pUL50, pUL53 displays two remarkable features, namely a zinc-binding site (Fig. 1C) and a hook-like N-terminal extension (residues 59–87), which embraces pUL50 and represents a hallmark of the pUL50-pUL53 interaction (Fig. 1D and see below).
FIGURE 1.
Crystal structure of the pUL50-pUL53 complex. A, ribbon representation of the complex. pUL50 is shown in red, and pUL53 is in blue. The secondary structure elements of the hook-like N-terminal extension of pUL53 are designated as α531, α532, and β531. B, display of a portion of the phased anomalous difference electron density map that validates the location of the methionine residues in the final model. The map is displayed at a 5 σ cut-off and is calculated with the anomalous differences observed in the Se-Met peak dataset (Table 1). C, close-up view of the zinc-binding site. D, detailed representation of the interaction of the hook-like extension of pUL53 with pUL50 shown in two different orientations. Residues displayed in a stick representation contribute in excess of 50 Å2 of their surface area to the interaction.
The structure of pUL53 comprises an unexpected zinc-binding site (Fig. 1C). Zinc binds to three cysteine residues (Cys106, Cys122, and Cys125) and a histidine (His211). These residues are displayed from the four-stranded β-sheet of pUL53 that is not part of the GHKL-like fold of pUL53 (see below). Zinc is tetrahedrally coordinated, and its binding mode is in full agreement with that typically observed in zinc-binding proteins (32). All zinc-coordinating residues are fully conserved among pUL53 homologous proteins (supplemental Fig. 1) (14, 33, 34). This notwithstanding, the exact function of the zinc-binding site currently remains elusive.
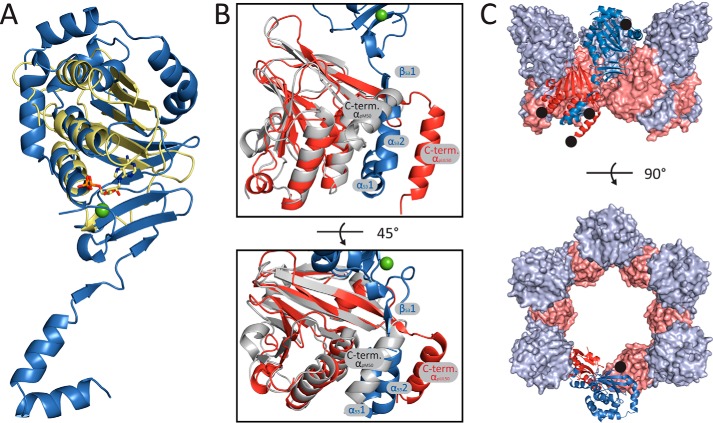
As noted in the recently determined NMR structure of pUL50 homolog, pM50, of murine CMV (MCMV), the protein fold displays only very limited local similarities to other structures currently available from the Protein Data Bank and can therefore not readily be assigned to a known protein fold (9, 35). However, this is not the case for pUL53. A search for structurally related proteins using PDBeFold (36) revealed a significant similarity between the pUL53 fold and the GHKL domain of bacterial sensory histidine kinases despite a low sequence similarity of <15%. The GHKL domain is found in a number of functionally diverse ATP-binding proteins including gyrases, Hsp90, histidine kinases, and MutL that all contain an unconventional Bergerat ATP-binding fold (37). All four strands from the second antiparallel β-sheet of pUL53 (Ile188–Glu195, Arg198–Phe205, Tyr228–Val235, His238–Cys245) and two α-helices (Gln171–Asn185, Pro213–Ala223) have structural equivalent elements in the GHKL domain, whereas additional N-terminal and C-terminal stretches of pUL53 adopt a deviating topology (Fig. 2A). Interestingly, the ATP-binding site of the GHKL domain is not conserved in pUL53, and instead a zinc-binding site is found at a topologically equivalent position (Fig. 2A and see above). A structural comparison between pUL53 and pUL50 revealed that those elements conserved between pUL53 and GHKL domains are also partially present in pUL50. However, the structural similarity between pUL50 and GHKL domains is significantly less pronounced than that found between pUL53 and GHKL domains.
FIGURE 2.
Implications of the pUL50-pUL53 crystal structure. A, HCMV pUL53 shows a structural resemblance to the GHKL domain, as depicted by an overlay of pUL53 (blue) and the GHKL domain from Staphylococcus aureus VraS (yellow; PDB code 4GT8). The ADP molecule bound to the VraS protein is shown in sticks and colored according to the atom types. The zinc ion bound to pUL53 is highlighted as a green sphere. The search for structural homologs and the overlay were both performed with PDBeFold (36). B, superposition of the crystal structure of pUL50 and the NMR structure of pM50 (PDB code 5A3G) (9). pUL50, pUL53, and pM50 are shown in red, blue, and gray, respectively. In comparison with the structure of pM50, the C-terminal helix of pUL50 is displaced by helices α531 and α532 from pUL53 that form a hook-like extension. C, hexameric ring-like structure formed by the pUL50-pUL53 complex in the crystals. Ribbon and surface representations of pUL50 are shown in red and light red, and representations of pUL53 are shown in blue and light blue, respectively. Although the pUL50 molecules form a contiguous ring, the pUL53 molecules are displayed individually and form the rim of the ring. The locations of the protein termini are marked with black dots.
Specific Structural Determinants Accomplish a Tight Interlocking of the pUL50-pUL53 Heterodimer
pUL50 and pUL53 readily form a complex when mixed in solution (38). The crystal structure reveals the hallmarks of the interaction and at the same time corroborates previous findings from mutagenesis experiments (14, 38). In the pUL50-pUL53 complex, each protein contributes as much as 1880 Å2 of solvent-accessible surface area to the protein interface. Of these, the N-terminal pUL53 hook (residues 59–87) contributes by far the largest portion, namely 1510 Å2 (80%) to the interface area. The structure of the complex shows that the pUL53 hook is not formed by a single secondary structure element but is formed by two α-helices (from here on termed α531 and α532) that are followed by a short β-strand β531 (Fig. 1A). α531 and α532 become embedded between three α-helices of pUL50, and the short β-strand β531 is added via a total of three main-chain hydrogen bonds to the side of the extended middle strand of the three-stranded β-sheet of pUL50. A conserved proline residue (Pro72 in pUL53, supplemental Fig. 1) appears to be responsible for the disruption of the N-terminal segment of pUL53 into two separate helices, α531 and α532, and thus for its hook-like appearance.
The interaction between pUL50 and the hook region of pUL53 is based on mixed physicochemical properties. The hook structure itself appears to be stabilized by a small hydrophobic core, which in the complex becomes augmented by the contribution of hydrophobic residues provided by the three pUL50 α-helices that interact with the pUL53 hook. This is particularly obvious when considering the amphipathic polarity of one of these helices, namely the very C-terminal α-helix of pUL50. Here, all hydrophobic residues that are displayed on one face of the helix become buried in the interface between pUL50 and pUL53, whereas all polar residues remain accessible from the surface of the complex.
Among those residues that contribute in excess of 50 Å2 to the protein interface (11 residues from pUL50 and 18 residues from pUL53), an equally high portion of polar and apolar residues was detected (15 versus 14 residues, respectively) (Fig. 1D). However, most strikingly, an inspection of the sequence conservation between homologs of pUL50 and pUL53 indicated that the interacting residues do not show a specifically high degree of conservation (supplemental Fig. 1). This holds particularly true for pUL50 and stands in striking contrast to the pUL53 zinc-binding residues that are fully conserved.
The previous biochemical mapping of the pUL50 regions responsible for pUL53 interaction was mostly deduced from serial N-terminal and C-terminal truncation constructs analyzed by coimmunoprecipitation experiments (12, 14). This led to the definition of functionally important protein segments. Recently, these segments could be confined to the N-terminal half of pUL50, i.e. residues 1–209. Within this segment, residues 1–181 proved to be responsible for an optimal interaction with pUL53 whereby residues 10–169 were sufficient for a basal level of interaction (14). The crystal structure and an alignment of the pUL50 residues involved in pUL53 interaction indicate that these residues are entirely contained within the biochemically mapped region 10–169. The interaction-conferring N-terminal half of pUL50 also covers two regions conserved among herpesviruses, i.e. CR1 and CR2 (supplemental Fig. 1).
As far as pUL53 is concerned, previous mapping analyses performed by Sam et al. (38) stressed the importance of specific residues contained in the central region of pUL53, i.e. residues 58–313. In addition, fragment 50–84 was proposed to form an α-helix that would be required for pUL50 interaction. With the caveat that the crystal structure reveals two separate α-helices (α531 and α532) arranged in a hook-like fashion, the previously reported interaction mapping is fully consistent with the crystal structure. Interestingly, this stretch is entirely and exclusively covered by CR1, one of the four regions CR1–CR4 conserved among herpesviruses.
Concerning structural NEC data, only one pUL50-like structure is available in the Protein Data Bank (PDB) so far (35). The respective NMR structure of the MCMV homolog pM50 closely resembles the structure of pUL50 observed in the crystal-derived complex (Fig. 2B) (9). Titration experiments in combination with heteronuclear single quantum correlation (HSQC) measurements allowed mapping of the pM53-binding site on the surface of pM50 (9). When assuming that the MCMV and HCMV homologs behave strongly similar, then a comparison of these structures suggests that binding of pUL53 to pUL50 causes a considerable repositioning of the very C-terminal α-helix of pUL50. As a consequence of the displacement of this C-terminal helix, the hook-like N-terminal extension of pUL53 becomes trapped between the shifted C-terminal pUL50 helix and two additional pUL50 helices (Fig. 2B).
Tertiary and Quaternary Arrangements of the pUL50-pUL53 Heterodimers Are Important for Providing a Scaffold Structure for Multimeric NEC Assembly
The fragments of pUL50 and pUL53 used in this study readily form heterodimers in vitro (data not shown) (38). In the crystals, however, we observe even larger symmetric assemblies, namely hexameric ring-like structures that result from the application of the P6 crystal symmetry to the pUL50-pUL53 heterodimer present in the crystallographic asymmetric unit (Fig. 2C). In these oligomeric assemblies, the pUL50 molecules contact each other and form hexameric ring-like structures, whereas the pUL53 molecules appear to be displayed individually from the rim of the pUL50 rings (Fig. 2C). In these oligomers, each individual pUL53 molecule is in contact with two pUL50 molecules without contacting other pUL53 molecules.
These hexameric assemblies are considered remarkable for several reasons. Although they are generated by mere crystal packing contacts, it is notable that space group P6 is only rarely observed in protein crystallography. Less than 0.25% of all protein crystals display this symmetry (PDB statistics, by summer 2015) (35). Cases in which this space group is observed appear not to be fortuitous but rather reflect a preferred functional assembly of biological relevance. For example, space group P6 has been detected in viral capsid proteins such as the hexameric assembly of the human immunodeficiency virus type 1 (HIV-1) capsid protein (39) and in proteins that are part of highly symmetric complexes of bacterial secretion systems (40). For these reasons, the possibility arises that such a hexameric arrangement of the relevant full-length proteins might correspond to a preferred oligomeric state of pUL50-pUL53 readily assembled under specific conditions of viral replication. This might occur under molecular crowding during steps of viral nuclear replication, specifically the nuclear rim recruitment of pUL53 by pUL50.
Moreover, the hexameric arrangement of pUL50-pUL53 matches the honeycomb pattern observed for the homologous herpes simplex virus (HSV-1) NEC core proteins, pUL34-pUL31, in cryo-electron micrographs (41). Also the observed diameter of ∼11 nm of the pores of the ring-like arrays of pUL34-pUL31 is consistent with our findings (Fig. 2C). These viral NEC proteins pUL34 and pUL31 can initiate membrane budding in vitro through the formation of ordered coats on the inner surface of vesicles. The inward topology of the observed in vitro budding of the core NEC resembles membrane budding of viral capsids, and suggests the existence of a minimal virus-encoded membrane-budding machinery. The ring-like structure is also of interest when considering further collective functions of the NEC. In general, the HCMV-specific NEC not only clusters proteins to distinct membrane sites and induces the formation of locally restricted lamina-depleted areas (13), but also offers multiple attachment points for additional quaternary protein-protein interactions. Thus, the highly ordered ring-like arrangement might promote the controlled association of further NEC proteins. The ring-like structures may also suggest a high degree of structural conservation between herpesviral core NECs, and might suggest that similar mechanisms apply for NEC-induced effects on the nuclear lamina and host membranes during nuclear egress.
Specific residues within the interaction surfaces of the core NEC proteins (in particular Glu56 and Tyr57 in pUL50) seem to be essential not only for heterodimerization but also for subsequent scaffolding steps during multimeric NEC formation (9, 15). Although intracellular membrane trafficking of pUL50 is determined by residues and sequential domains other than Glu56/Tyr57 (19), the two residues were both found to be essential not only for pUL50-pUL53 complex formation but also for the recruitment of pUL53 to the nuclear rim promoting viral replication efficiency (14, 15). As depicted in Fig. 1, Tyr57 is an integral part of the pUL50-pUL53 binding interface and becomes significantly buried upon complex formation. Glu56 enhances the stability of the pUL50 interaction helix by forming an intramolecular salt bridge with Lys123. Both residues are strictly conserved among the pUL50 homologs (supplemental Fig. 1). A role of Glu56 and Tyr57 in assuring the structural integrity of the complex may explain that mutation of either of the two residues results in severe functional impairment of the core NEC. As highlighted by our recent investigation of pUL50-pUL53 interaction using truncation and replacement mutants, the biochemical and structural relevance of an N-terminal amphipathic α-helix in pUL50 stretching from amino acids 3–20 (12) has now been confirmed (Fig. 1). Our previous experimentation demonstrated that any deletion of N-terminal residues led to a reduction of signals in interaction assays (e.g. coimmunoprecipitation) and that truncations downstream of Asp10 resulted in a strict loss of both protein stability and capability of pUL53 binding (14).
Recently published proteomic data provided for the first time a candidate list of the associated viral and cellular constituents of the multimeric NEC for both HCMV and MCMV (15, 18). For HCMV, at least four proteins were identified that directly interact with pUL50, i.e. pUL53, p32/gC1qR, emerin, and PKCα (12, 14, 15) in addition to the regulatory interaction with viral kinase pUL97 that phosphorylates pUL50 at specific sites (42). A number of additional NEC-associated proteins possibly providing accessory functions are still under investigation. Thus, it is of no surprise that the NEC of HCMV and other herpesviruses is currently considered a promising target for the development of novel antiviral strategies. Such strategies might aim either at a direct block of protein-protein interactions via in silico-designed small molecule inhibitors or at an interference with regulatory mechanisms that control NEC assembly, such as the inhibition of NEC protein phosphorylation through kinase inhibitors (6, 43). We anticipate that the structural information presented here for the pUL50-pUL53 complex will accelerate the validation of the NEC as a unique antiherpesviral target and might promote the development of a novel type of NEC-directed drugs.
Author Contributions
M. M designed the study. M. M, Y. A. M., H. S, S. A. W., and J. M. wrote the paper. M. M constructed vectors for expression of proteins. S. A. W., C. E. S., M. S., and K. H. purified and crystallized the proteins. Y. A. M. and S. A. W. determined the x-ray structure shown in Figures 1 and 2 and Table 1. J. M., M. M., and H. S. performed and analyzed additional experiments shown in supplemental Fig. 1 and Fig. 2A, respectively. S. A. W. contributed to the preparation of the figures and Table 1. All authors analyzed the results and approved the final version of the manuscript.
Supplementary Material
Acknowledgments
We thank Karine Sparta (HZB Berlin) and Johanna Kallio and Thomas Schneider (European Molecular Biology Laboratory (EMBL) Hamburg) for help with data collection and data reduction at the respective synchrotron radiation sources. We further thank Michael Weyand and Clemens Steegborn (University of Bayreuth) for providing us with the opportunity to test the diffraction of crystals. We greatly acknowledge the scientific discussion and support by Corina Hutterer and Eric Sonntag as well as excellent technical assistance by Sabrina Wagner (Institute for Clinical and Molecular Virology, Friedrich-Alexander University of Erlangen-Nürnberg).
This work was supported by Deutsche Forschungsgemeinschaft via SFB796 (A2, A3/Z1, C3) and MA1289/8-1 and by Wilhelm Sander-Stiftung (2011.085.1-2) and Bayerische Forschungsstiftung (ForBIMed–Biomarker in der Infektionsmedizin/I1). The authors declare that they have no conflicts of interest with the contents of this article.
This article was selected as a Paper of the Week.

This article contains supplemental Fig. 1.
The atomic coordinates and structure factors (code 5D5N) have been deposited in the Protein Data Bank (http://wwpdb.org/).
J. Milbradt, H. Sticht, and M. Marschall, unpublished data.
- HCMV
- human cytomegalovirus
- MCMV
- murine cytomegalovirus
- CR
- conserved region
- NEC
- nuclear egress complex
- SUMO
- small ubiquitin-like modifier.
References
- 1.Griffiths P., Baraniak I., and Reeves M. (2015) The pathogenesis of human cytomegalovirus. J. Pathol. 235, 288–297 [DOI] [PubMed] [Google Scholar]
- 2.Mocarski E. S. Jr., Shenk T., Griffiths P.D., and Pass R. F. (2013) Cytomegaloviruses. in Fields Virology, (Knipe D. M., and Howley P. M., eds), 6th Ed., pp. 1960–2014, Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 3.Hamilton S. T., van Zuylen W., Shand A., Scott G. M., Naing Z., Hall B., Craig M. E., and Rawlinson W. D. (2014) Prevention of congenital cytomegalovirus complications by maternal and neonatal treatments: a systematic review. Rev. Med. Virol. 24, 420–433 [DOI] [PubMed] [Google Scholar]
- 4.Marschall M., and Stamminger T. (2009) Molecular targets for antiviral therapy of cytomegalovirus infections. Future Microbiol. 4, 731–742 [DOI] [PubMed] [Google Scholar]
- 5.Lee C. P., and Chen M. R. (2010) Escape of herpesviruses from the nucleus. Rev. Med. Virol. 20, 214–230 [DOI] [PubMed] [Google Scholar]
- 6.Marschall M., Feichtinger S., and Milbradt J. (2011) Regulatory roles of protein kinases in cytomegalovirus replication. Adv. Virus Res. 80, 69–101 [DOI] [PubMed] [Google Scholar]
- 7.Tandon R., and Mocarski E. S. (2012) Viral and host control of cytomegalovirus maturation. Trends Microbiol. 20, 392–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mettenleiter T. C., Müller F., Granzow H., and Klupp B. G. (2013) The way out: what we know and do not know about herpesvirus nuclear egress. Cell. Microbiol. 15, 170–178 [DOI] [PubMed] [Google Scholar]
- 9.Leigh K. E., Sharma M., Mansueto M. S., Boeszoermenyi A., Filman D. J., Hogle J. M., Wagner G., Coen D. M., and Arthanari H. (2015) Structure of a herpesvirus nuclear egress complex subunit reveals an interaction groove that is essential for viral replication. Proc. Natl. Acad. Sci. U.S.A. 112, 9010–9015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Muranyi W., Haas J., Wagner M., Krohne G., and Koszinowski U. H. (2002) Cytomegalovirus recruitment of cellular kinases to dissolve the nuclear lamina. Science 297, 854–857 [DOI] [PubMed] [Google Scholar]
- 11.Hamirally S., Kamil J. P., Ndassa-Colday Y. M., Lin A. J., Jahng W. J., Baek M. C., Noton S., Silva L. A., Simpson-Holley M., Knipe D. M., Golan D. E., Marto J. A., and Coen D. M. (2009) Viral mimicry of Cdc2/cyclin-dependent kinase 1 mediates disruption of nuclear lamina during human cytomegalovirus nuclear egress. PLoS Pathog. 5, e1000275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Milbradt J., Auerochs S., Sticht H., and Marschall M. (2009) Cytomegaloviral proteins that associate with the nuclear lamina: components of a postulated nuclear egress complex. J. Gen. Virol. 90, 579–590 [DOI] [PubMed] [Google Scholar]
- 13.Milbradt J., Webel R., Auerochs S., Sticht H., and Marschall M. (2010) Novel mode of phosphorylation-triggered reorganization of the nuclear lamina during nuclear egress of human cytomegalovirus. J. Biol. Chem. 285, 13979–13989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Milbradt J., Auerochs S., Sevvana M., Muller Y. A., Sticht H., and Marschall M. (2012) Specific residues of a conserved domain in the N terminus of human cytomegalovirus pUL50 protein determine its intranuclear interaction with pUL53. J. Biol. Chem. 287, 24004–24016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Milbradt J., Kraut A., Hutterer C., Sonntag E., Schmeiser C., Ferro M., Wagner S., Lenac T., Claus C., Pinkert S., Hamilton S. T., Rawlinson W. D., Sticht H., Couté Y., and Marschall M. (2014) Proteomic analysis of the multimeric nuclear egress complex of human cytomegalovirus. Mol. Cell. Proteomics 13, 2132–2146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marschall M., Marzi A., aus dem Siepen P., Jochmann R., Kalmer M., Auerochs S., Lischka P., Leis M., and Stamminger T. (2005) Cellular p32 recruits cytomegalovirus kinase pUL97 to redistribute the nuclear lamina. J. Biol. Chem. 280, 33357–33367 [DOI] [PubMed] [Google Scholar]
- 17.Kuny C. V., Chinchilla K., Culbertson M. R., and Kalejta R. F. (2010) Cyclin-dependent kinase-like function is shared by the β- and γ-subset of the conserved herpesvirus protein kinases. PLoS Pathog. 6, e1001092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lemnitzer F., Raschbichler V., Kolodziejczak D., Israel L., Imhof A., Bailer S. M., Koszinowski U., and Ruzsics Z. (2013) Mouse cytomegalovirus egress protein pM50 interacts with cellular endophilin-A2. Cell. Microbiol. 15, 335–351 [DOI] [PubMed] [Google Scholar]
- 19.Schmeiser C., Borst E., Sticht H., Marschall M., and Milbradt J. (2013) The cytomegalovirus egress proteins pUL50 and pUL53 are translocated to the nuclear envelope through two distinct modes of nuclear import. J. Gen. Virol. 94, 2056–2069 [DOI] [PubMed] [Google Scholar]
- 20.Studier F. W. (2005) Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 41, 207–234 [DOI] [PubMed] [Google Scholar]
- 21.Laskowski R. A., MacArthur M. W., Moss D. S., and Thornton J. M. (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 22.Kabsch W. (2010) XDS. Acta Crystallogr. D. Biol. Crystallogr. 66, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pape T., and Schneider T. R. (2004) HKL2MAP: a graphical user interface for phasing with SHELX programs. J. Appl. Crystallogr. 37, 843–844 [Google Scholar]
- 24.Sheldrick G. M. (2010) Experimental phasing with SHELXC/D/E: combining chain tracing with density modification. Acta Crystallogr. D. Biol. Crystallogr. 66, 479–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schneider T. R., and Sheldrick G. M. (2002) Substructure solution with SHELXD. Acta Crystallogr. D. Biol. Crystallogr. 58, 1772–1779 [DOI] [PubMed] [Google Scholar]
- 26.Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L.-W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., and Zwart P. H. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D. Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Emsley P., Lohkamp B., Scott W. G., and Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D. Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G. W., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., Read R. J., Vagin A., and Wilson K. S. (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr. D. Biol. Crystallogr. 67, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.DeLano W. L. (2010) The PyMOL Molecular Graphics System, version 1.3r1, Schrödinger, LLC, New York [Google Scholar]
- 30.Milbradt J., Auerochs S., and Marschall M. (2007) The cytomegaloviral proteins pUL50 and pUL53 are associated with the nuclear lamina and interact with cellular protein kinase C. J. Gen. Virol. 88, 2642–2650 [DOI] [PubMed] [Google Scholar]
- 31.Funk C., Ott M., Raschbichler V., Nagel C. H., Binz A., Sodeik B., Bauerfeind R., and Bailer S. M. (2015) The herpes simplex virus protein pUL31 escorts nucleocapsids to sites of nuclear egress, a process coordinated by its N-terminal domain. PLoS Pathog. 11, e1004957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harding M. M. (2004) The architecture of metal coordination groups in proteins. Acta Crystallogr. D. Biol. Crystallogr. 60, 849–859 [DOI] [PubMed] [Google Scholar]
- 33.Thompson J. D., Higgins D. G., and Gibson T. J. (1994) ClustalW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties, and weight matrix choice. Nucleic Acids Res. 22, 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lötzerich M., Ruzsics Z., and Koszinowski U. H. (2006) Functional domains of murine cytomegalovirus nuclear egress protein M53/p38. J. Virol. 80, 73–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rose P. W., Prlić A., Bi C., Bluhm W. F., Christie C. H., Dutta S., Green R. K., Goodsell D. S., Westbrook J. D., Woo J., Young J., Zardecki C., Berman H. M., Bourne P. E., and Burley S. K. (2015) The RCSB Protein Data Bank: views of structural biology for basic and applied research and education. Nucleic Acids Res. 43, D345–D356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krissinel E., and Henrick K. (2004) Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D. Biol. Crystallogr. 60, 2256–2268 [DOI] [PubMed] [Google Scholar]
- 37.Dutta R., and Inouye M. (2000) GHKL, an emergent ATPase/kinase superfamily. Trends Biochem. Sci. 25, 24–28 [DOI] [PubMed] [Google Scholar]
- 38.Sam M. D., Evans B. T., Coen D. M., and Hogle J. M. (2009) Biochemical, biophysical, and mutational analyses of subunit interactions of the human cytomegalovirus nuclear egress complex. J. Virol. 83, 2996–3006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gres A. T., Kirby K. A., KewalRamani V. N., Tanner J. J., Pornillos O., and Sarafianos S. G. (2015) X-ray crystal structures of native HIV-1 capsid protein reveal conformational variability. Science 349, 99–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ruiz F.M., Santillana E., Spínola-Amilibia M., Torreira E., Culebras E., and Romero A. (2015) Crystal structure of Hcp from Acinetobacter baumannii: a component of the type VI secretion system. PLoS ONE 10, e0129691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bigalke J. M., Heuser T., Nicastro D., and Heldwein E. E. (2014) Membrane deformation and scission by the HSV-1 nuclear egress complex. Nat. Commun. 5, 4131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sharma M., and Coen D. M. (2014) Comparison of effects of inhibitors of viral and cellular protein kinases on human cytomegalovirus disruption of nuclear lamina and nuclear egress. J. Virol. 88, 10982–10985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sharma M., Bender B. J., Kamil J. P., Lye M. F., Pesola J. M., Reim N. I., Hogle J. M., and Coen D. M. (2015) Human cytomegalovirus UL97 phosphorylates the viral nuclear egress complex. J. Virol. 89, 523–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.