Background: Rep68 catalyzes site-specific integration into chromosome 19 AAVS1 site.
Results: Rep68 forms a heptameric complex on the AAVS1 site.
Conclusion: Assembly requires the cooperative interaction of all functional domains and requires a minimum of two GCTC repeats.
Significance: These results provide insights into the first step of the site-specific integration reaction.
Keywords: analytical ultracentrifugation, crystal structure, DNA helicase, DNA viruses, DNA-protein interaction, electron microscopy (EM), virology, adeno-associated virus
Abstract
Adeno-associated virus (AAV) is the only eukaryotic virus with the property of establishing latency by integrating site-specifically into the human genome. The integration site known as AAVS1 is located in chromosome 19 and contains multiple GCTC repeats that are recognized by the AAV non-structural Rep proteins. These proteins are multifunctional, with an N-terminal origin-binding domain (OBD) and a helicase domain joined together by a short linker. As a first step to understand the process of site-specific integration, we proceeded to characterize the recognition and assembly of Rep68 onto the AAVS1 site. We first determined the x-ray structure of AAV-2 Rep68 OBD in complex with the AAVS1 DNA site. Specificity is achieved through the interaction of a glycine-rich loop that binds the major groove and an α-helix that interacts with a downstream minor groove on the same face of the DNA. Although the structure shows a complex with three OBD molecules bound to the AAVS1 site, we show by using analytical centrifugation and electron microscopy that the full-length Rep68 forms a heptameric complex. Moreover, we determined that a minimum of two direct repeats is required to form a stable complex and to melt DNA. Finally, we show that although the individual domains bind DNA poorly, complex assembly requires oligomerization and cooperation between its OBD, helicase, and the linker domains.
Introduction
Adeno-associated virus (AAV)3 is the only known eukaryotic virus that can establish latency by integrating its genome site specifically into the host genome (1–4). The integration site known as AAVS1 is located within a 4-kb region in human chromosome 19 at 19q13.4 (3–5). This site is within a regulatory region that controls the myosin-binding subunit 85 (MBS85) also known as protein phosphatase 1 regulatory protein (PPP1R12C) (6). Latency can also arise by episomal persistence, and both mechanisms happen in the absence of a helper virus such as adenovirus (7, 8). The AAVS1 site resembles a specific region found in the palindromic sequences known as inverted terminal repeats (ITR) at both ends of the AAV genome (9–13). The ITRs span a region of 146 nucleotides that fold into a T-shaped hairpin structure with the stem of the hairpin containing the Rep-binding site (RBS) and the terminal resolution sequence (trs); the latter undergoes a strand-specific nicking reaction that is essential for DNA replication and integration (14–17). The RBS is made up of multiple repeats of the tetranucleotide sequence 5′-GCTC-3′ or small variations of this sequence. Interestingly, most of the AAV serotypes have three contiguous repeats flanked by pseudo-repeats, a feature that is shared with AAVS1 (18). The site-specific integration process is contingent on the presence of the large regulatory proteins Rep78/Rep68, the AAVS1 site, and a cis-acting viral DNA sequence (1, 17, 19, 20). Earlier studies reported that for the AAVS1 region a minimum sequence of 33 bp containing the RBS and trs sequences is essential for site-specific integration (16, 17, 21). Furthermore, a 16-bp sequence from the p5 promoter known as the P5 integration efficiency element (P5IEE) was identified as the minimal viral cis-acting element able to mediate site-specific integration (22, 23). However, the mechanistic details of how AAV Rep78/Rep68 proteins drive this mechanism are not known. The requirement of site-specific nicking of the AAVS1 trs site suggests that some of the steps resemble the terminal resolution reaction during AAV DNA replication. This process is a variation of the mechanism used by DNA relaxases during DNA conjugation (20, 24, 25). The first step in site-specific integration is the assembly of a Rep78/Rep68-DNA complex guided by the recognition of GCTC repeats through the N-terminal origin-binding domain (OBD). The crystal structure of AAV5 OBD bound to a 26-bp AAV5 RBS illustrated for the first time the structural determinants of repeat recognition (26). Each OBD molecule interacts with two contiguous repeats using two secondary structure motifs as follows: a loop contacts the major groove bases of one repeat while an α-helix interacts with a downstream repeat through the minor groove. Thus, two contiguous OBD molecules share a repeat, with one molecule interacting through the major groove and the second molecule binding in the minor groove on the opposite face of the DNA. Whether this DNA recognition mode is conserved in other AAV Rep serotypes and in the AAVS1 site is not known due to the lack of structural information of a more representative member, such as AAV2. Intriguingly, differences in serotype specificity for ITRs have been reported such that Rep proteins are functional only when binding to their own serotype ITR (27, 28). Whether the difference in specificity is due to the number and/or arrangement of repeats found in the origin of replication of the different AAV serotypes has not been established. In addition, the effect of the helicase domain in the overall formation of the initial complex in the context of Rep78/Rep68 proteins is not known. To understand the mechanism of Rep68 assembly on the AAVS1 site and the role that each of the AAV Rep68 functional domains plays in this process, we first determined the crystal structures of AAV2 OBD in complex with integration site AAVS1. Next, we characterized the complex of Rep68 bound to a 41-mer AAVS1 site and performed binding studies with each of the Rep68 individual functional domains. Our results show that AAV2 OBD has a similar binding mode to AAV5; however, the longer recognition loop in AAV2 has more extended contacts with the major group due to its inherent flexibility by several glycine residues. We determined that Rep68 forms a heptameric complex with the AAVS1 site and that high binding affinity for AAVS1 is due to the cooperative binding and assembly of the OBD, linker, and helicase domains.
Materials and Methods
Protein Expression and Purification
OBD Proteins
The DNA region encoding amino acids 1–208 and 1–224 from adeno-associated virus type 2 (AAV2) (GenBankTM AAC03774.1) was cloned into pET-15b (Novagen) using restriction sites NdeI and XhoI. The residue Cys-151 was mutated to serine as it was found to produce disulfide bonds and inhibit crystallization. Residue Tyr-156 was mutated to phenylalanine to eliminate any potential nuclease activity. The OBDN208 was overexpressed by growth in Escherichia coli strain BL21 pLysS at 37 °C in Luria-Bertani (LB) broth until reaching an absorbance of 0.6. Isopropyl β-d-thiogalactopyranoside was added to a final concentration of 1 mm. Cells were harvested after 5 h and stored at −80 °C. The cell pellet was resuspended in binding buffer (25 mm Tris-HCl, 500 mm NaCl, 10 mm imidazole, 10% glycerol, 1 mm TCEP, pH 7.9) and lysed by sonication. OBD was purified with nickel-nitrilotriacetic acid column (Qiagen) using step gradients of 75 and 125 mm imidazole to wash nonspecific proteins binding to the column and was eluted with 300 mm imidazole. Protein was loaded onto a Hi-Load desalting column (GE Healthcare) to change into thrombin buffer (25 mm Tris-HCl, 200 mm NaCl, 10% glycerol, pH 8.0). Hexa-histidine tag was cut by addition of thrombin (1 unit/mg) and removed by passing through a nickel-nitrilotriacetic acid column. Untagged OBD was collected from the flow-through, concentrated, and further purified by gel filtration on a Hi-Load 16/60 Superdex 75 column (GE Healthcare) previously equilibrated with GF buffer (30 mm Tris-HCl, pH 7.5, 150 mm NaCl). The protein was concentrated to ∼12 mg/ml using Millipore Centricon (10-kDa cutoff) prior to crystallization.
Rep40 (Helicase Domain)
AAV-2 Rep40 (residues 225–534) were expressed and purified using pET-15b as described elsewhere (29).
Rep68 Proteins
All mutant proteins were generated using the pHisRep68/15b plasmid, which contains the AAV2 Rep68 ORF subcloned in vector pET-15b (Novagen). The Rep68octlink construct was generated by substitution of residues 206–224 of AAV2 Rep68 with the mouse Oct-1 linker residues 328–346 (GenBankTM CAA49791) using the gene synthesis services from GeneScript. Proteins were expressed in E. coli BL21(DE3) cells (Novagen) and purified as described previously (30). The size exclusion buffer contains (25 mm Tris-HCl, pH 8.0, 200 mm NaCl, and 2 mm TCEP). In brief, cell pellets were lysed in Ni-Buffer A (20 mm Tris-HCl, pH 7.9 at 4 °C, 500 mm NaCl, 5 mm imidazole, 10% glycerol, 0.2% CHAPS, and 1 mm TCEP) and purified using a nickel column. The hexa-histidine tag was removed by PreScission protease, and Rep68 was further purified by gel filtration chromatography using a HiLoad Superdex 200 16/60 column (GE Healthcare) and Size Exclusion buffer. Rep68 WT and mutant proteins were concentrated to 10 mg/ml, flash-frozen in liquid N2, and kept at −80 °C.
Fluorescence Anisotropy DNA Binding Assay
Binding assays were performed using 5 nm fluorescein-labeled 41-mer AAVS1 DNA. Rep68 constructs at different concentrations were mixed with DNA at a final volume of 200 μl using the following buffer: 25 mm HEPES, pH 7.0, 200 mm NaCl, 1 mm TCEP. Fluorescence readings were taken on a PC1 fluorimeter (ISS, Inc.) with excitation and emission filters at 492 and 528 nm, respectively. Tubes were equilibrated at 20 °C for 20 min before measurement. Each anisotropy point is the average of 10 measurements. Anisotropy is calculated as the ratio of the difference between vertical and horizontal emission intensities over the total normalized intensity. The fraction of DNA bound (B) was calculated using Equation 1,
where [A]x represents the anisotropy measured at protein concentration X; [A]DNA is the anisotropy of free fluorescence DNA, and [A]final is the anisotropy at saturation. To obtain overall apparent binding constants that could be used to compare different mutants, data were fitted to a single binding site model with a Hill coefficient. This model was selected because at this time sufficient details of the assembly process are not known. Fitting was performed using the program Prism 6TM (GraphPad). Each experiment was done in triplicate.
Analytical Ultracentrifugation
Sedimentation velocity experiments were carried out using a Beckman Optima XL-I analytical ultracentrifuge (Beckman Coulter, Inc.) equipped with both four- and eight-hole rotors. Samples (2, 5, 10, 20, and 40 μm) were loaded in the cells, using in all cases size exclusion buffer. Samples were centrifuged in two-sector carbon-filled Epon centerpieces at 20 °C. Sectors were loaded with 420 μl of sample volume. Typically, 200 or more scans were collected at 5-min intervals at 25,000 rpm. Sedimentation profiles were collected using both UV absorption (280 nm) and Rayleigh interference optical systems. Results were analyzed using both SEDFIT and SEDPHAT (31, 32).
Crystallization and X-ray Structure Determination
The oligonucleotides used for crystallization were purchased from Integrated DNA Technologies, Inc. as follows: site A, 5′-CTCGGCGCTCGCTCGCTCGCT-3′ and 5′-GAGCGAGCGAGCGAGCGCCGA-3′; site B, 5′-GCGCTCGCTCGCTCGCTGGCC-3′ and 5′-CGCCCAGCGAGCGAGCGAGCG-3′. DNA was purified on a Mono Q-5/50GL column. The purified DNA was desalted, lyophilized, and resuspended in TE buffer (10 mm Tris-HCl, pH 8.0, 40 mm NaCl, 1 mm EDTA). The oligonucleotides were mixed in 1:1 molar ratio, heated to 90 °C for 5 min, and cooled slowly to room temperature. OBD and double-stranded DNA were mixed in 1:2 (complex A) and 1:3 (complex B) molar ratios, respectively. These complexes were concentrated to a final concentration of protein of about 18 mg/ml. The buffer concentration was exchanged during the concentration process to 30 mm Tris-HCl, pH 7.3, and 40 mm NaCl. All crystallization experiments were carried out using the hanging and sitting-drop methods with commercially available screening kits at 4 °C. Crystals of complex A grew from 3-μl hanging drop after 2–3 weeks. The best crystals were obtained from reservoir solution containing 50 mm sodium citrate, pH 4.3, 5.75–7.25% PEG-6K, and 0.2 m LiCl. Crystals of complex B grew from 6-μl sitting drop where reservoir solution was 100 mm sodium phosphate/citrate buffer, pH 4.2, 5–7.5% PEG-3K and 0.2 m NaCl. Both crystal forms were cryo-protected in their corresponding reservoir solution by adding ethylene glycol to 25% before flash-freezing in liquid nitrogen. Diffraction data were collected at the National Synchrotron Light Source at Brookhaven National Laboratory beamline X6a. The data were processed with the program HKL2000 (33), and the structure was solved by molecular replacement using the program PHENIX. We used the structure of the AAV2 OBD as a search model (Protein Data Bank code 4ZO0). Model building was carried out using PHENIX (34) and manual building using the program COOT (35). Analysis of the buried surface area was determined by calculating the difference in a solvent-accessible area (ASA) as shown in Equation 2,
The individual solvent-accessible areas were calculated using Chimera (36). Figures were generated by PyMOL (37), DOG (38), and Adobe Photoshop.
Transmission Electron Microscopy Analysis
Protein samples at 0.1 mg/ml were adsorbed directly onto carbon-coated copper grids. Following negative staining with 0.75% (w/v) uranyl formate, samples were visualized in an electron microscope Tecnai F20 operated at 200 kV, and images were collected at a magnification of ×50,000 under low dose conditions on a Gatan 4k x 4k CCD camera. Particle windowing, two-dimensional alignment, and classification reconstruction were carried out with EMAN2. The entire process followed the default settings of this image-processing software for eight iterative alignments. The two-dimensional averages were obtained from a final set of 520 particles.
Results
To characterize the assembly of Rep68 on the integration site AAVS1, we utilized a three-pronged strategy. First, we determined the x-ray structure of AAV-2 OBD bound to AAVS1 DNA to define the structural details of specific repeat recognition. Next, we determined the stoichiometry of the Rep68-AAVS1 complex by sedimentation velocity and electron microscopy. Finally, we carried out a detailed analysis of the role of each domain in DNA binding and the minimal number of repeats that are needed to form a stable complex.
Overview of Structure Determination
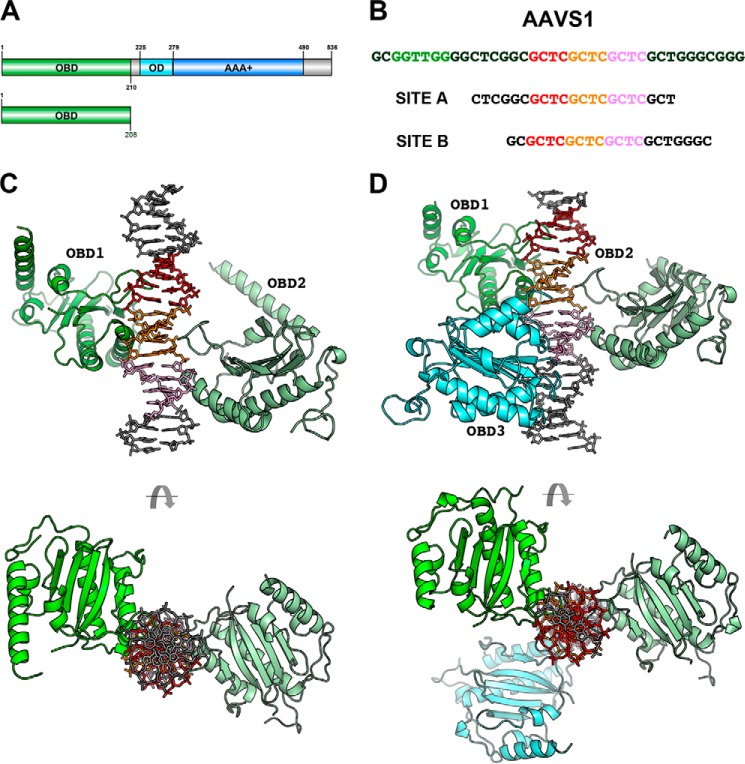
We have determined the structure of two AAV2 OBD-AAVS1 complexes with different stoichiometries. Complex A has a 2:1 OBD/AAVS1 ratio, and complex B has three molecules of OBD per AAVS1 site. The AAV2 OBD used in crystallization spans amino acids 1–208 of Rep78/Rep68 (Fig. 1A). Both were crystallized with 21-mer AAVS1 DNA sites containing a core of three direct GCTC repeats but differing in the number of residues upstream and downstream of this core (Fig. 1B). The stoichiometry of the complexes parallel the protein-DNA ratios used during crystallization. Complex A crystallized in space group C2 with two complexes per asymmetric unit, and complex B crystals belong to P21 with one complex in the asymmetric unit. The structures were solved by molecular replacement using the structure of AAV2-OBD as a search model (Protein Data Bank code 4ZO0). Structures of the complexes were refined to 2.5 and 2.6 Å, respectively; refinement statistics are shown in Table 1. These structures may represent snapshots of consecutive binding events as the DNA is titrated with higher concentrations of the protein (Fig. 1, C and D).
FIGURE 1.
Overall x-ray structures of AAV2 OBD-AAVS1 complexes. A, domain representation of Rep68 and OBD construct used in crystallization. B, sequence of AAVS1 integration site and DNA sites used for crystallization. GCTC repeat 1 is colored red, and repeats 2 and 3 are orange and pink, respectively. Green sequence represents the trs site. C, structure of complex A with a 2:1 stoichiometry with OBD1 (green) binding to repeats 1 and 2; OBD2 (pale green) binding to repeats 2 and 3. D, structure of complex B with a 3:1 stoichiometry showing OBD1 (green) binding to repeats 1 and 2; OBD2 (pale green) binding to repeats 2 and 3, and OBD3 (cyan) binding to repeat 3 and pseudo-repeat.
TABLE 1.
Data collection and refinement statistics
| Complex A | Complex B | |
|---|---|---|
| Data collection | ||
| Space group | C2 | P21 |
| Cell dimensions | ||
| a, b, c (Å) | 183.1, 79.6, 139.5 | 71.0, 137.26, 79.25 |
| α, β, γ (°) | 90, 98.1, 90 | 90, 111.7, 90 |
| Wavelength (Å) | 0.979 | 0.979 |
| Resolution (Å) | 30–2.50 (2.54–2.50) | 30–2.6 |
| No. of measured | 246,245 | 426,030 |
| No. of unique | 65,548 | 105,074 |
| Data coverage (%)a | 95.2 (77.4) | 99.1 (98.9) |
| Rmerge(%)a,b | 0.07 (0.572) | 0.075 (0.311) |
| I/σa | 18.7 (1.5) | 12.4 (4.1) |
| Refinement statistics | ||
| Resolution range | 30.0–2.5 | 33.0–2.6 |
| Reflections | 65,509 | 36,889 |
| Rcryst(%)c | 18.3 | 20.46 |
| Rfree(%)d | 22.3 | 22.95 |
| Non-hydrogen atoms | 8020 | 5496 |
| Protein | 6025 | 4581 |
| Water | 302 | 58 |
| Average B-factors(Å2) | 91.2 | 38.6 |
| Protein | 91.9 | 38.6 |
| Metal | 98.7 | 105.1 |
| Water | 74.1 | 36.6 |
| Root mean square deviations | ||
| Bonds (Å) | 0.008 | 0.013 |
| Angles (Å) | 1.09 | 1.68 |
| Ramachandran plot quality | ||
| Most favored (%) | 96.8 | 97 |
| Additional allowed (%) | 2.7 | 2.3 |
| Generously allowed(%) | 0.41 | 0.7 |
| Disallowed (%) | 0 | 0 |
aValues for the outmost shells are given in parentheses.
b Rmerge = Σ|I − 〈I〉|/Σ I, where I is the integrated intensity of a given reflection.
cRcryst = Σ ‖Fo| − |Fc‖ Σ |Fo|.
dFor Rfree calculations, 5% of data was excluded from refinement.
Overall Structure of OBD-AAVS1 Complex
Complex A can be superimposed onto complex B with an overall root mean square deviation of 1.14 Å for two OBD molecules and 17 bp of DNA. Although both complexes crystallize in different space groups, the overall interactions with DNA are identical thus confirming the validity of the specific contacts between OBD and DNA. Each OBD molecule interacts with two consecutive GCTC repeats where the DNA recognition loop (LDB) interacts with the major groove of the first repeat, and α-helix D interacts with the second repeat through the minor groove (Fig. 2). This particular binding mode establishes that multiple OBD molecules will share repeats such that an OBD molecule interacting through the major groove will face a second OBD molecule binding upstream through the minor groove in the opposite face of DNA (Fig. 2). The OBDs dock onto DNA in a head to tail orientation spiraling around the DNA axis by ∼144°. The AAVS1 site contains an RBS with three perfect 5′-GCTC-3′ repeats and two “imperfect” repeats with the sequence 5′-CGGC-3′ and 5′-GCTG-3′ located upstream and downstream of the perfect repeats, respectively (Fig. 1B). Two of the OBDs in complexes A and B bind the same repeats, and the third OBD in complex B binds the final GCTC repeat and the downstream pseudo-repeat. This type of binding mode positions the C-terminal ends of the OBD molecules pointing upstream of the RBS site where the helicase domains would be positioned in the full-length Rep68/Rep78 proteins.
FIGURE 2.
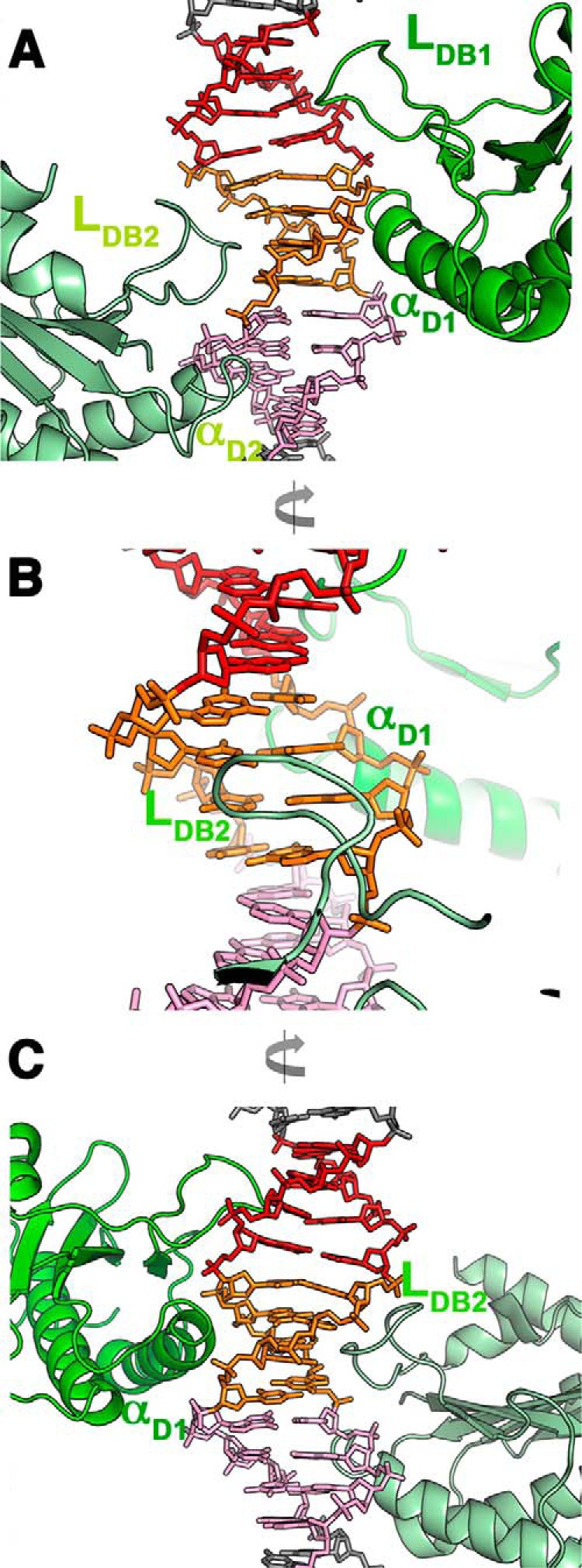
Docking of OBD into major and minor grooves. A, details of complex B showing OBD1 (green) and OBD2 (pale green) binding mode with loops LDB docking into contiguous major grooves and α-helix D docking into minor grooves. OBD1 binds to repeats 1 (red) and 2 (orange), and OBD2 binds to repeats 2 and 3 (pink). B, view 90° from A showing that although the OBD2 loop LDB docks into major groove, helix D from OBD1 binds the minor groove in the opposite face of DNA. C, view 180° from A, showing that only the N-terminal tip of helix D docks into the minor groove.
Sequence-specific Recognition of GCTC Repeats in AAVS1
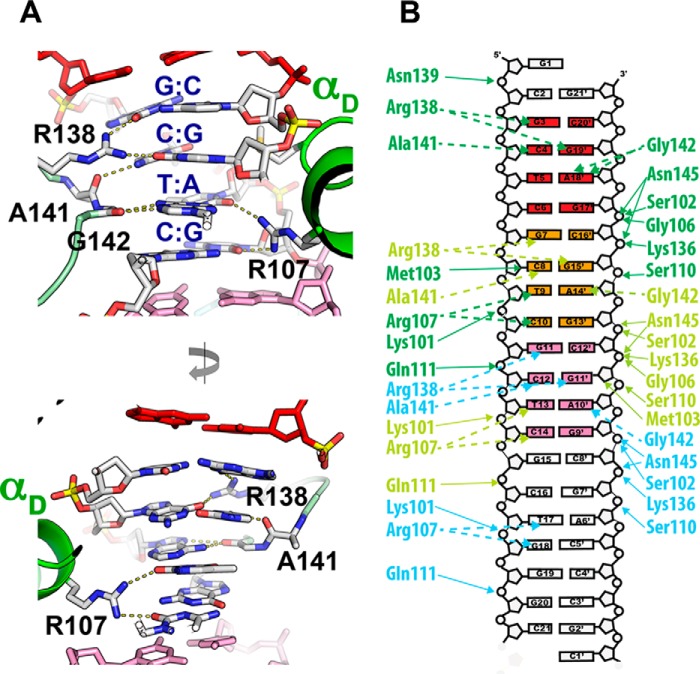
Two OBD molecules binding to contiguous repeats cooperate in recognizing a full GCTC repeat sequence (Fig. 3). Four residues generate all specific contacts with the bases as follows: three are located in the DNA recognition loop LDB of one OBD molecule (Arg-138, Ala-141, and Gly-142), and the upstream OBD molecule contacts DNA via Arg-107 (Fig. 3A). For instance, for the recognition of the central repeat (Fig. 1B, repeat 2), residues from the LDB loop specify the sequence GCT with Arg-138 making hydrogen bonds with guanine 7 via O6 and also with the guanine 15′ of the second base pair (Fig. 3, A and B). Cytosine 8 complementary to guanine 15′ is recognized by the main chain carbonyl of Ala-141 (Fig. 3A, bottom panel). The recognition of the third base pair (T:A) is provided via Gly-142 whose main chain carbonyl makes a bidentate interaction with adenine 14′. Finally, Arg-107 of the upstream OBD makes a bidentate interaction with thymine 9 and cytosine 10. Thus, of the eight possible nucleotides in the GCTC sequence, only two do not directly contact the protein and are located at the ends of the tetranucleotide repeat (Fig. 3). The OBD-DNA-specific contacts are stabilized by 6–8 phosphate-backbone contacts per OBD molecule. Residues that provide these interactions are also located in loop LDB and αD elements. There are no direct contacts between the different OBD molecules, and whether the binding of multiple molecules is a cooperative event mediated by DNA, remains to be determined. The DNA is essentially B-form with no major distortions in any of its geometric parameters as determined by the program 3DNA (39, 40).
FIGURE 3.
AAV2 OBD-AAVS1 contacts. A, recognition of GCTC repeats by loop LDB and helix D. Upper panel, Arg-138 makes bidentate hydrogen bond contacts with two guanines in GC pairs in opposite strands of DNA. Ala-141 backbone carbonyl makes hydrogen bond contact with cytosine in the second base pair, and Gly-142 interacts with the adenine in the third base pair. Upstream OBD1 molecule interacts with the minor groove in the opposite DNA face through Arg-107. Bottom panel, details of the minor groove interactions, where Arg-107 makes hydrogen bonds with thymine and cytosine in the third and fourth base pairs. B, schematic diagram of protein-DNA contacts. Hydrogen bonds are represented by dotted lines, and phosphate contacts are represented as solid lines. Line coloring is according to OBD molecule with OBD1(green), OBD2 (pale green), and OBD3 (cyan).
Recognition of GCTC Repeats by Different AAV Rep Serotypes
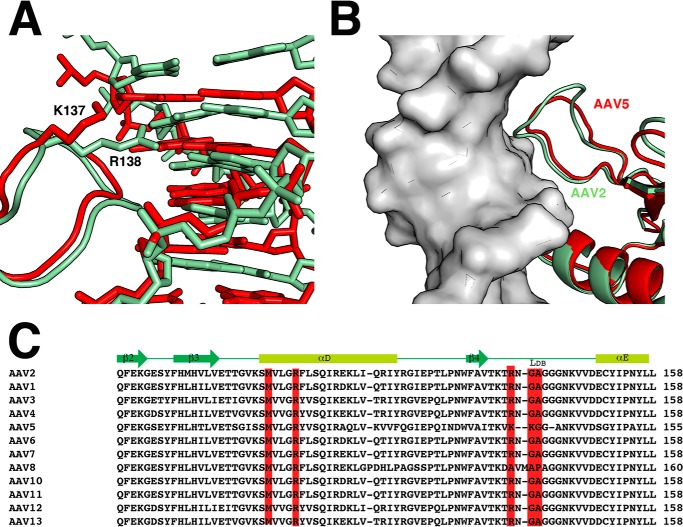
Although the principles of repeat recognition were shown by the structure of the AAV5 OBD-RBS complex, there are significant differences between AAV5 and the majority of the other AAV serotypes, particularly in the recognition loop LDB (Fig. 4C). In AAV2 and the majority of AAV Rep serotypes, this region spans ∼10 residues and contains the sequence RNGAGGG. In contrast, AAV5 has a shorter recognition loop with the sequence K–KGGA. The underlined residues represent amino acids that interact with DNA bases as discussed above (Fig. 3B). Using the structure of AAV5 OBD bound to the RBS element from the AAV5 ITR, we compare its recognition of the GCTC repeats with AAV2 OBD (26). Although the two structures superimpose with a root mean square deviation of ∼1 Å, their respective repeat DNAs “sit” differently with respect to the superimposed OBD domains. The AAV5 RBS is translated up the helical axis by ∼1–1.5 Å, and the conformation of the recognition loops moves up accordingly (Fig. 4A). The fact that both OBDs make similar specific interactions with DNA despite differences in their relative positions can be attributed to the flexibility of the glycine-rich recognition loop that can accommodate the conformational requirements needed to fit into the major groove (Fig. 4B). The conformation of the recognition loop follows the DNA backbone making significant van der Waal contacts with its ribose groups. This is more pronounced in the larger AAV2 loop (Fig. 4B). In AAV5, Lys-137 plays the equivalent role of residue Arg-138 in AAV2. It interacts with the first guanine of the repeat. However, in contrast to Arg-138, Lys-137 does not make any interactions with the guanine in the second base pair of the repeat. In AAV5, residue Lys-138 interacts with cytosine through its main chain carbonyl similarly as Ala-141 in AAV2. Finally, the main chain carbonyl of AAV5 Gly-139 and AAV2 Gly-142 plays equivalent roles in recognizing adenine in the third base pair of the repeat (Fig. 4A). Thus, with the exception of Lys-137 and Arg-138, most of the interactions are made through main chain atoms. Consequently, sequence variations in the loop that allow sufficient conformational flexibility to dock into the major groove are one of the main requirements to make specific contacts (Fig. 4C).
FIGURE 4.
Comparison of AAV2 and AAV5 binding to GCTC repeats. A, superposition of structurally aligned structures of AAV2-OBD (pale green) and AAV5 OBD (red) bound to AAVS1 and AAV5 origin RBS. Equivalent residues Arg-138 (AAV2) and Lys-137 (AAV5) are represented as sticks. B, differences in the conformation of loops LDB between AAV2 (pale green) and AAV5 (red) OBDs. AAV2 loop LDB makes more extensive contacts with the DNA backbone due to its larger size; however, the conformation of the second half of the loops is very similar. C, sequence and structural alignment of AAV serotypes from β2 to αE that include residues involved in specific DNA contacts (red) and backbone DNA contacts (blue).
Rep68 Forms a Heptameric Complex on AAVS1
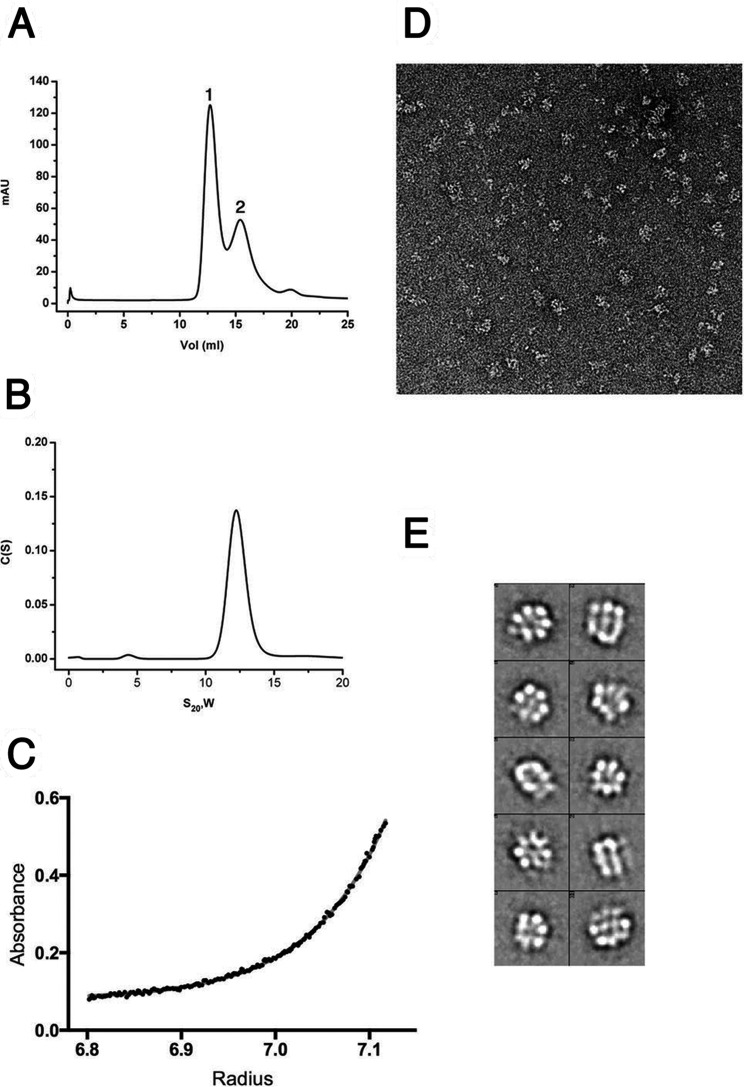
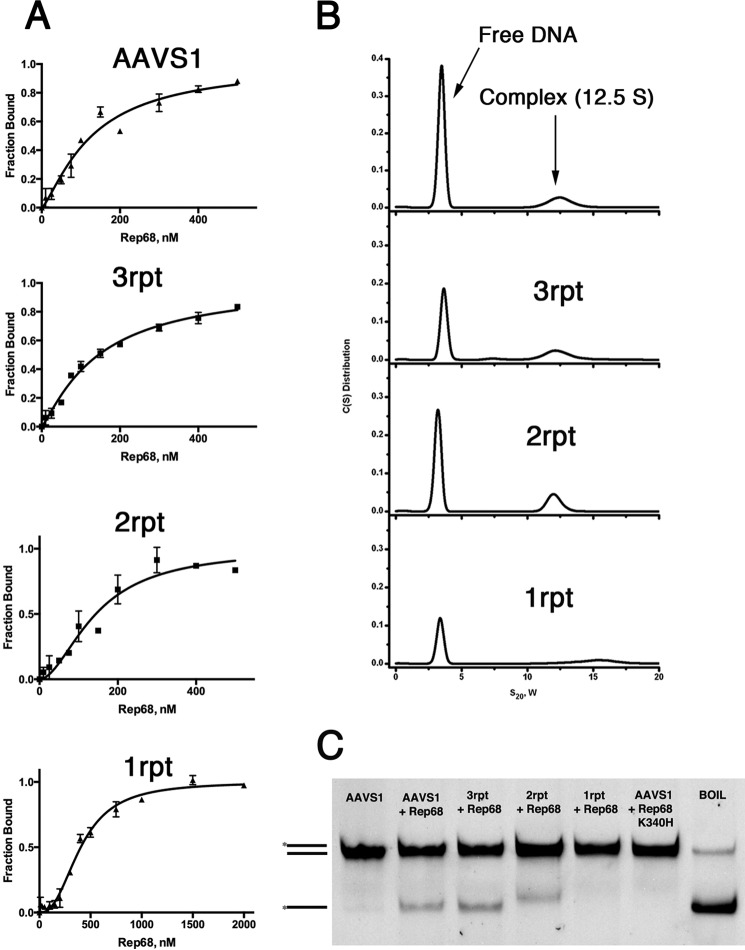
Recognition of the GCTC repeats is only one of the events required for the assembly of Rep68 on the AAVS1 DNA site; however, not much is known about the nature of the final complex. To start the characterization of the Rep68-AAVS1 complex, we first used analytical gel filtration. The elution profile shows two peaks as follows: one eluting at ∼12.5 ml that represents a Rep68-AAVS1 complex, and the second eluting peak represents excess DNA (Fig. 5A). The purified Rep68-AAVS1 complex is stable even at a concentration of 500 mm NaCl (data not shown). The elution profile of the complex differs drastically from that of apo-Rep68, which shows the presence of multiple species (41). To further characterize this complex, we used sedimentation velocity. Experiments were performed at 20 °C using the fluorescein-labeled 42-mer AAVS1 DNA site at a concentration of 2 μm. Data were collected at 492 nm to detect species containing DNA. The sedimentation profile shows a single peak sedimenting at 12.5 s20,w. The presence of a single peak allowed us to fit the data to a single ideal species model to estimate the molecular mass. We obtained a value of 438 kDa, close to the theoretical value for a heptameric complex (453 kDa) (Fig. 5B). A similar estimate was obtained by sedimentation equilibrium (Fig. 5C). To further confirm these results, we used negative-stain transmission electron microscopy to obtain structural information by single-particle reconstruction. Fig. 5D shows a representative electron microscopic image of a negatively stained Rep68-AAVS1 complex. A reference-free two-dimensional alignment without imposing any symmetry was carried out that clearly shows the presence of seven-member ring structures (Fig. 5E). Thus, Rep68 forms a heptameric complex when bound to the AAVS1 site.
FIGURE 5.
Solution studies of Rep68-AAVS1 complex. A, size exclusion chromatography profile of the Rep68-AAVS1 complex. The DNA site used is a 41-mer DNA shown in Fig. 1B. The profile shows two peaks with the faster eluting peak corresponding to the Rep68-DNA complex, and the second peak corresponding to the excess free-DNA. mAU, milliabsorbance units. B, purified complex was analyzed by sedimentation velocity using a fluorescein-labeled AAVS1 DNA. Data were collected at 492 nm, and data were analyzed using the program Sedfit. Estimated molecular weight corresponds to a heptameric complex. C, equilibrium sedimentation of Rep68-AAVS1 was carried out in the same conditions as B and was fitted as a single sedimenting species model. The distribution of the residuals of the fit is shown. D, EM image of negatively stained Rep68-AAVS1 complex. The rings are easily recognized in the raw images. E, representative two-dimensional averages showing the heptameric complex.
Minimum of Two Direct Repeats Is Required for Stable Rep68-RBS Complex
To determine the minimal number of repeats that are required to obtain a stable complex, we calculated the binding affinity and stoichiometry of Rep68 to DNA sites with one, two, and three GCTC repeats. Binding isotherms were determined using fluorescence anisotropy measurements on 41-mer DNA sites labeled at the 5′ end with carboxyfluorescein. Data were fitted using a single site model with a Hill coefficient. Fig. 6 shows that Rep68 binds to the AAVS1 with a KD, app of 128 nm (Fig. 6A). Sites with two and three repeats have affinities of ∼140 nm, which is close to the binding constant for the AAVS1 site. Moreover, the h values obtained from the fitting range from 1.2 to 1.7 imply positive cooperativity during complex formation. In contrast, the single repeat site increases the binding constant to about ∼400 nm. To determine whether Rep68 forms the same complex on these sites, we performed sedimentation velocity. Fig. 6B shows the comparison of the sedimentation velocity profiles of the mutant sites with the AAVS1 complex. Whereas 3rpt and 2rpt form complexes that sediment at 12.5 S as the AAVS1 site, the 1rpt site does not. Thus, Rep68 needs at least two GCTC repeats to assemble into a stable heptameric complex.
FIGURE 6.
Minimum number of repeats required for stable Rep68-RBS complex. A, equilibrium binding studies using fluorescent anisotropy with 41-mer DNA sites labeled with carboxyfluorescein at 5 nm concentration. Binding was done in 25 mm HEPES, pH 7.0, 200 mm NaCl, 1 mm TCEP at room temperature. Data were fitted with a single site model with Hill coefficient. From top to bottom AAVS1 site is as follows: TGGCGGCGGTTGGGGCTCGGCGCTCGCTCGCTCGCTGGGCG; 3rpt, TGGCGGCGGTTGGGGCTCGGCGCTCGCTCGCTCAAAGGGCG; 2rpt, TGGCGGCGGTTGGGGCTCGGCAAAGGCTCGCTCGCTGGGCG; 1rpt, TGGCGGCGGTTGGGGCTCGGCGCTCAAAGAAAGAAAGGGCG. B, sedimentation velocity profiles of Rep68 (6 μm) with fluorescein-labeled DNA (2 μm). Samples were centrifuged at 25,000 rpm and 20 °C, and scans were collected at 492 nm. Data were analyzed using the program SEDFIT (32. Plots show sedimentation profiles of Rep68 in complex with the following DNA sites (top to bottom): AAVS1, 3rpt, 2rpt, and 1rpt. C, strand displacement melting assay on AAVS1 41-mer fluorescein-labeled dsDNA and AAVS1 mutant DNA sites 3rpt, 2rpt, and 1rpt. Rep68-K340H ATPase dead mutant is used as a negative control.
To determine whether the formation of a stable complex translates into efficient DNA melting, we tested the ability of Rep68 to melt the AAVS1 mutant DNA sites with varying numbers of GCTC repeats using the strand-displacement assay (42). The method consists of the generation of fluorescein-labeled single-stranded DNA upon addition of ATP and magnesium. The results show that although AAVS1, 3rpt, and 2rpt sites are melted, the Rep68 is not able to melt the 1rpt site (Fig. 6C).
Rep68 DNA Binding Requires the Cooperative Interaction of OBD, Helicase, and Linker
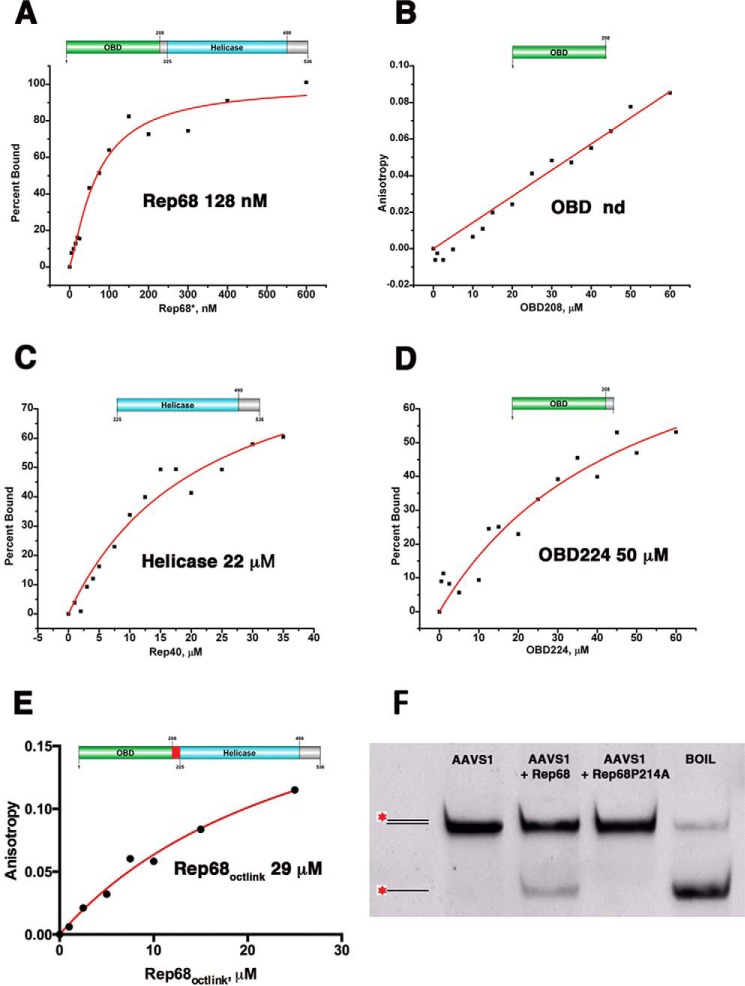
AAV Rep68 has two DNA binding domains that play a role in the overall affinity for the AAVS1 site. The helicase domain binds DNA nonspecifically, whereas the specificity for RBS is provided by the OBD. To determine the contributions of each of the domains for binding the AAVS1 site, we determined the binding affinities of Rep68, OBD, and the helicase domain (Rep40). At the experimental conditions tested, we could not measure the binding constant of OBD because it never reached saturation (Fig. 7B). Under the same conditions, Rep40 binds with an apparent dissociation constant of ∼ 22 μm (Fig. 7C). In previous studies, we and others determined that the linker is important for the oligomerization of Rep68 (30, 43). To determine the role of the linker in DNA binding, we generated the construct OBDL spanning residues 1–224 that includes the OBD and the entire linker region. We calculated the binding affinity for this construct and obtained a KD, app of ∼50 μm (Fig. 7D). These results suggest that the linker provides additional contacts with DNA increasing the affinity of the minimal OBD domain. To determine the role of the linker region in the context of Rep68, we determined the affinity of Rep68octlink, a mutant that has 18 residues from the Oct-1 protein instead of Rep68 residues 206–224. We determined previously that this mutant protein behaves as a monomer in solution (30). Rep68octlink has a KD, app of 29 μm (Fig. 7E). Moreover, in a recent study, we determined that mutation of linker residue proline 214 to alanine affects the ability of Rep68 to oligomerize and bind DNA (44). To test the effect of this mutation on the melting activity of Rep68, we performed the strand displacement reaction and showed that this mutant is unable to melt DNA (Fig. 7F). Taken together, our data show that the two functional domains plus the linker participate in DNA binding and act cooperatively to bind DNA. Moreover, our data suggest that oligomerization of Rep68 on DNA drives much of the binding affinity.
FIGURE 7.
Binding isotherms of Rep68 functional domains. Fluorescent anisotropy binding assays were carried out with a 41-mer AAVS1 DNA labeled with fluorescein. DNA concentration was 5 nm, and experiments were carried at room temperature as described under “Materials and Methods.” Binding isotherms are as follows: A, Rep68; B, OBD domain; C, helicase domain, Rep40; D, OBDL(1–224); and E, Rep68octlink. Apparent dissociation constants were determined from triplicate experiments and fitted to a one binding site-specific binding model with Hill coefficient; F, strand displacement melting assay on AAVS1 41-mer fluorescein-labeled dsDNA of Rep68 and Rep68-P214A mutant. nd, not determined.
Discussion
Our studies illustrate the binding mode that AAV Rep68 uses to assemble at the AAVS1 integration site. The x-ray structure of AAV2 OBD bound to the AAVS1 site expands our knowledge of the structural determinants that determine binding of AAV Rep proteins to GCTC repeats. Specific recognition is achieved through the interaction of residues in flexible loop LDB and α-helix D docking into contiguous major and minor grooves, respectively. The residues in helix D contacting the DNA bases are conserved in all AAV serotypes. In contrast, the nature of loop LDB varies only in AAV5 and AAV8 both in sequence and length (Fig. 4C). However, because many of the interactions of loop LDB with the DNA bases involved backbone atoms, it appears that the only prerequisite is to have enough backbone flexibility to dock into the major groove. This is provided by the high content of glycine residues found in the loop (Fig. 4C). Thus, even though in AAV5 the loop is shorter by a couple of residues, its inherent flexibility allows it to dock into the major groove where main chain groups from glycine and alanine make hydrogen bond contacts with DNA. A puzzling case is the AAV8 Rep with a longer LDB loop but no basic residues to interact with DNA, particularly no equivalent residue for Arg-138. Instead, three hydrophobic residues alanine, valine, and methionine are found in the loop together with a proline (Fig. 4C). It is not clear how these changes still allow recognition of the GCTC repeat. Future structural studies are needed to answer this question.
Our results show that high affinity and specific binding of Rep68 to DNA containing RBS sites require the concerted and cooperative binding of its two domains (OBD, helicase) and the linker. Thus, although the OBD provides the specificity to bind the GCTC repeats, affinity arises from the helicase domain and oligomerization during binding. This is in agreement with previous studies showing that binding affinity to the minimal RBS sequence (28 bp) is significantly lower than larger DNA sites with additional upstream sequences (45). The low affinity of the OBD for DNA can be understood by its peculiar binding mode. Analysis of the OBD-AAVS1 interface shows that the total buried surface area upon complex formation per monomer is only 268 Å2, one of the smallest for a specific protein-DNA complex. The number of residues per base contact is also at the lower end of specific DNA-binding proteins with only four residues contacting the DNA bases. These properties resemble those of proteins that bind DNA non-specifically (46). Moreover, the number of phosphate contacts is sparse, reflecting the low DNA binding affinity of the OBD.
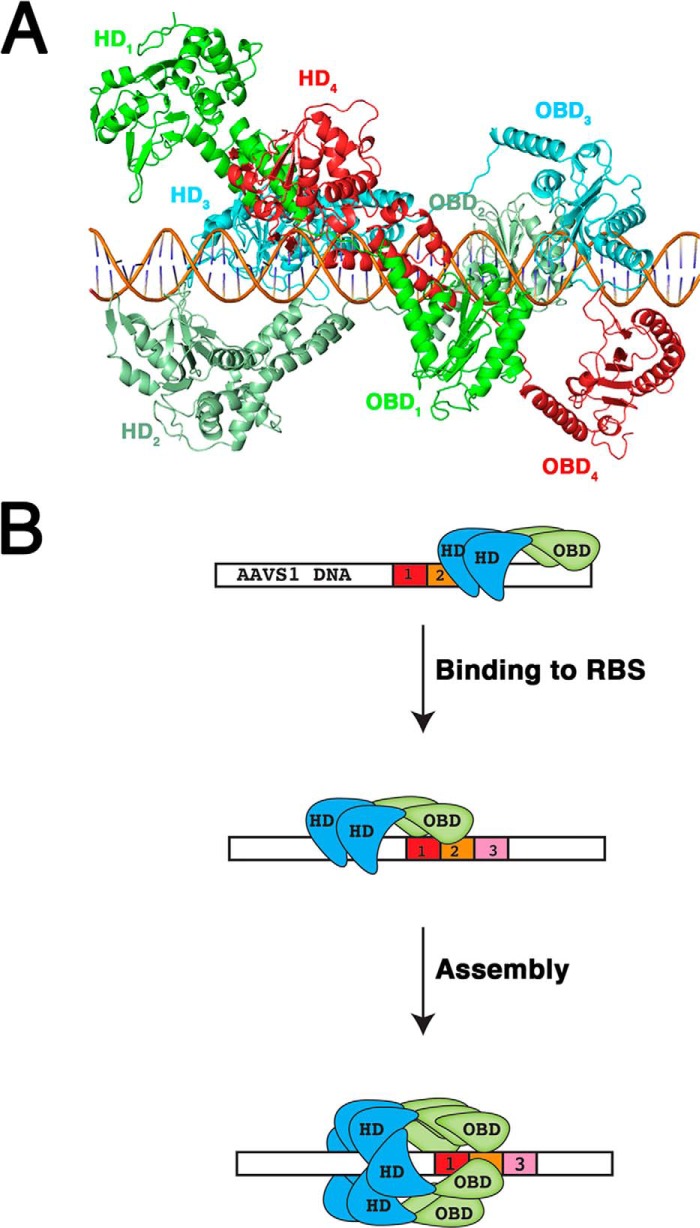
Our data suggest that oligomerization of Rep68 is a prerequisite to bind DNA with high affinity. Binding studies with the oligomerization-deficient mutant Rep68octlink show that this protein binds DNA poorly despite having all the functional side chains to make DNA contacts (Fig. 7E). The same results were obtained with a Rep68 P214A mutant that is mostly monomeric in solution (44). Nevertheless, because of steric constraints, there is a limit to the number of Rep68 molecules binding to multiple GCTC repeats at the RBS site. A model of five Rep68 molecules binding to the AAVS1 site based on the AAV5 OBD-RBS x-ray structure shows that more than three Rep68 molecules bound to the RBS simultaneously will start to clash with each other. Thus, a Rep68 molecule bound to the first repeat will clash with the helicase domain of a Rep68 molecule binding downstream in the fourth repeat (Fig. 8A). We propose a model where a Rep68 oligomer scans DNA until it recognizes the RBS site. Assembly of the final heptameric complex is accomplished by addition of further molecules guiding the helicase domain (Fig. 8B). This process may be similar to the assembly of the simian virus T large antigen 40 protein where only one direct repeat is needed to assemble the final complex (47). These facts bring the question of why there are multiple repeats at the origin of replication in all AAV serotypes and the AAVS1 site. Part of the answer lies in the binding mode of the AAV OBD domain that requires two repeats to bind DNA specifically. However, the presence of multiple identical binding sites in regulatory DNA sequences such as enhancers and promoters is a general feature known as homotypic site clustering (48–51). Thus, in addition to promoting cooperative interactions and producing a non-linear response to protein concentration, the occurrence of multiple repeats increases the local concentration of the protein by facilitating lateral diffusion on DNA (52–55). Consequently, the number of repeats found in the RBS sites is not an indicator of the number of Rep molecules recruited to DNA.
FIGURE 8.
Model of Rep68 assembly on AAVS1 site. A, model of Rep68 bound to a 50-mer DNA. The model was based on the structure of the AAV5-OBD/RBS complex (Protein Data Bank code 1RZ9). The linker was modeled as an extended region to allowed interaction with DNA. Four Rep68 molecules are colored green (Rep68–1), light green (Rep68–2), blue (Rep68–3), and red (Rep68–4). Individual domains are labeled accordingly. The helicase domain (HD4) of Rep68–4 clashes with both the OBD1 and HD1 of the Rep68–1 molecule. B, oligomer of Rep68 (shown as a dimer) binds DNA using the helicase and OBD domains. Upon encountering the RBS site through specific contacts with the OBD domain, additional molecules are recruited, and the heptameric complex is assembled. Helicase domain is represented in blue, and OBD domain is green. Three direct repeats in the AAVS1 site are red, orange, and pink.
The heptameric Rep68-AAVS1 complex is different from other published reports that show a variety of Rep oligomers such as a hexamer of Rep78 on the AAV-2 ITR site (56), a Rep68 octamer bound to a single-stranded poly(dT) DNA (57), and the pentameric complex observed in the AAV5 OBD-RBS structure (26). These large varieties of complexes are the reflections of the versatility of AAV Rep proteins to be modulated depending on the DNA substrate they bind. However, the pentameric complex obtained in the x-ray structure of the AAV5 OBD-RBS complex shows the effect of the saturation of all possible repeats at high concentrations used in the crystallization conditions and does not reflect the real stoichiometry that occurs in the context of the full-length proteins. Thus, we hypothesize that in order to bind DNA with specificity and high affinity, Rep68 binds the AAVS1 site as an oligomer, at least a dimer, that subsequently assembles to form the final complex. However, it is possible that ATP binding and hydrolysis may lead to formation of a different stoichiometry reflecting the dynamic processes occurring at the time of assembly such as melting and nicking of DNA. These questions will need to be answer by future studies that are currently in progress in our laboratory.
Author Contributions
C. R. E. designed, analyzed data, and wrote the paper. F. N. M. expressed, purified the AAV2 protein, crystallized, solved, and refined the x-ray structures. F. Z. P. purified Rep68, performed analytical ultracentrifugation, collected and analyzed EM data, and performed melting assays. C. B. purified Rep68 and Rep68 mutants and performed binding assays. J. W. B.II performed analytical ultracentrifugation and analyzed the data. All authors analyzed the results and approved final version of manuscript.
Acknowledgments
We thank National Synchrotron Light Source beamline X6a members Vivian Stojanoff, Jean Jakoncic, and Edwin Lazo. We also thank Vishaka Santosh for helpful comments during the preparation of the manuscript.
This work was supported by National Institutes of Health Grant R01-GM092854. The authors declare that they have no conflicts of interest with the contents of this article.
The atomic coordinates and structure factors (codes 5BYG and 4ZQ9) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- AAV
- adeno-associated virus
- AAVS1
- adeno-associated virus integration site 1
- TCEP
- tris(2-carboxyethyl)phosphine
- OBD
- origin-binding domain
- RBS
- Rep-binding site
- ITR
- inverted terminal repeat
- trs
- terminal resolution sequence.
References
- 1.Surosky R. T., Urabe M., Godwin S. G., McQuiston S. A., Kurtzman G. J., Ozawa K., and Natsoulis G. (1997) Adeno-associated virus Rep proteins target DNA sequences to a unique locus in the human genome. J. Virol. 71, 7951–7959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Young S. M. Jr., and Samulski R. J. (2001) Adeno-associated virus (AAV) site-specific recombination does not require a Rep-dependent origin of replication within the AAV terminal repeat. Proc. Natl. Acad. Sci. U.S.A. 98, 13525–13530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kotin R. M., Siniscalco M., Samulski R. J., Zhu X. D., Hunter L., Laughlin C. A., McLaughlin S., Muzyczka N., Rocchi M., and Berns K. I. (1990) Site-specific integration by adeno-associated virus. Proc. Natl. Acad. Sci. U.S.A. 87, 2211–2215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Samulski R. J., Zhu X., Xiao X., Brook J. D., Housman D. E., Epstein N., and Hunter L. A. (1991) Targeted integration of adeno-associated virus (AAV) into human chromosome 19. EMBO J. 10, 3941–3950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kotin R. M., Menninger J. C., Ward D. C., and Berns K. I. (1991) Mapping and direct visualization of a region-specific viral DNA integration site on chromosome 19q13-qter. Genomics 10, 831–834 [DOI] [PubMed] [Google Scholar]
- 6.Tan I., Ng C. H., Lim L., and Leung T. (2001) Phosphorylation of a novel myosin binding subunit of protein phosphatase 1 reveals a conserved mechanism in the regulation of actin cytoskeleton. J. Biol. Chem. 276, 21209–21216 [DOI] [PubMed] [Google Scholar]
- 7.Schnepp B. C., Jensen R. L., Chen C. L., Johnson P. R., and Clark K. R. (2005) Characterization of adeno-associated virus genomes isolated from human tissues. J. Virol. 79, 14793–14803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berns K. I., and Giraud C. (1996) Biology of adeno-associated virus. Curr. Top. Microbiol. Immunol. 218, 1–23 [DOI] [PubMed] [Google Scholar]
- 9.Srivastava A., Lusby E. W., and Berns K. I. (1983) Nucleotide sequence and organization of the adeno-associated virus 2 genome. J. Virol. 45, 555–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berns K. I., and Linden R. M. (1995) The cryptic life style of adeno-associated virus. BioEssays 17, 237–245 [DOI] [PubMed] [Google Scholar]
- 11.Fife K. H., Berns K. I., and Murray K. (1977) Structure and nucleotide sequence of the terminal regions of adeno-associated virus DNA. Virology 78, 475–477 [DOI] [PubMed] [Google Scholar]
- 12.Hauswirth W. W., and Berns K. I. (1977) Origin and termination of adeno-associated virus DNA replication. Virology 78, 488–499 [DOI] [PubMed] [Google Scholar]
- 13.Spear I. S., Fife K. H., Hauswirth W. W., Jones C. J., and Berns K. I. (1977) Evidence for two nucleotide sequence orientations within the terminal repetition of adeno-associated virus DNA. J. Virol. 24, 627–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Snyder R. O., Samulski R. J., and Muzyczka N. (1990) In vitro resolution of covalently joined AAV chromosome ends. Cell 60, 105–113 [DOI] [PubMed] [Google Scholar]
- 15.Im D. S., and Muzyczka N. (1990) The AAV origin binding protein Rep68 is an ATP-dependent site-specific endonuclease with DNA helicase activity. Cell 61, 447–457 [DOI] [PubMed] [Google Scholar]
- 16.Linden R. M., Ward P., Giraud C., Winocour E., and Berns K. I. (1996) Site-specific integration by adeno-associated virus. Proc. Natl. Acad. Sci. U.S.A. 93, 11288–11294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Linden R. M., Winocour E., and Berns K. I. (1996) The recombination signals for adeno-associated virus site-specific integration. Proc. Natl. Acad. Sci. U.S.A. 93, 7966–7972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hüser D., Gogol-Döring A., Lutter T., Weger S., Winter K., Hammer E. M., Cathomen T., Reinert K., and Heilbronn R. (2010) Integration preferences of wild type AAV-2 for consensus rep-binding sites at numerous loci in the human genome. PLoS Pathog. 6, e1000985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ponnazhagan S., Erikson D., Kearns W. G., Zhou S. Z., Nahreini P., Wang X. S., and Srivastava A. (1997) Lack of site-specific integration of the recombinant adeno-associated virus 2 genomes in human cells. Hum. Gene Ther. 8, 275–284 [DOI] [PubMed] [Google Scholar]
- 20.Henckaerts E., Dutheil N., Zeltner N., Kattman S., Kohlbrenner E., Ward P., Clément N., Rebollo P., Kennedy M., Keller G. M., and Linden R. M. (2009) Site-specific integration of adeno-associated virus involves partial duplication of the target locus. Proc. Natl. Acad. Sci. U.S.A. 106, 7571–7576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Young S. M. Jr., McCarty D. M., Degtyareva N., and Samulski R. J. (2000) Roles of adeno-associated virus Rep protein and human chromosome 19 in site-specific recombination. J. Virol. 74, 3953–3966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feng D., Chen J., Yue Y., Zhu H., Xue J., and Jia W. W. (2006) A 16-bp Rep binding element is sufficient for mediating Rep-dependent integration into AAVS1. J. Mol. Biol. 358, 38–45 [DOI] [PubMed] [Google Scholar]
- 23.Xu Z. X., Chen J. Z., Yue Y. B., Zhang J. Q., Li Z. H., Feng D. M., Ruan Z. C., Tian L., Xue J. L., Wang Q. J., and Jia W. (2009) A 16-bp RBE element mediated Rep-dependent site-specific integration in AAVS1 transgenic mice for expression of hFIX. Gene Ther. 16, 589–595 [DOI] [PubMed] [Google Scholar]
- 24.González-Prieto C., Agúndez L., Linden R. M., and Llosa M. (2013) HUH site-specific recombinases for targeted modification of the human genome. Trends Biotechnol. 31, 305–312 [DOI] [PubMed] [Google Scholar]
- 25.Chandler M., de la Cruz F., Dyda F., Hickman A. B., Moncalian G., and Ton-Hoang B. (2013) Breaking and joining single-stranded DNA: the HUH endonuclease superfamily. Nat. Rev. Microbiol. 11, 525–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hickman A. B., Ronning D. R., Perez Z. N., Kotin R. M., and Dyda F. (2004) The nuclease domain of adeno-associated virus rep coordinates replication initiation using two distinct DNA recognition interfaces. Mol. Cell 13, 403–414 [DOI] [PubMed] [Google Scholar]
- 27.Chiorini J. A., Afione S., and Kotin R. M. (1999) Adeno-associated virus (AAV) type 5 Rep protein cleaves a unique terminal resolution site compared with other AAV serotypes. J. Virol. 73, 4293–4298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grimm D., Pandey K., Nakai H., Storm T. A., and Kay M. A. (2006) Liver transduction with recombinant adeno-associated virus is primarily restricted by capsid serotype not vector genotype. J. Virol. 80, 426–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.James J. A., Escalante C. R., Yoon-Robarts M., Edwards T. A., Linden R. M., and Aggarwal A. K. (2003) Crystal structure of the SF3 helicase from adeno-associated virus type 2. Structure 11, 1025–1035 [DOI] [PubMed] [Google Scholar]
- 30.Zarate-Perez F., Bardelli M., Burgner J. W. 2nd, Villamil-Jarauta M., Das K., Kekilli D., Mansilla-Soto J., Linden R. M., and Escalante C. R. (2012) The interdomain linker of AAV-2 Rep68 is an integral part of its oligomerization domain: role of a conserved SF3 helicase residue in oligomerization. PLoS Pathog. 8, e1002764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vistica J., Dam J., Balbo A., Yikilmaz E., Mariuzza R. A., Rouault T. A., and Schuck P. (2004) Sedimentation equilibrium analysis of protein interactions with global implicit mass conservation constraints and systematic noise decomposition. Anal. Biochem. 326, 234–256 [DOI] [PubMed] [Google Scholar]
- 32.Schuck P. (2003) On the analysis of protein self-association by sedimentation velocity analytical ultracentrifugation. Anal. Biochem. 320, 104–124 [DOI] [PubMed] [Google Scholar]
- 33.Otwinowski Z., and Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 34.Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., and Terwilliger T. C. (2002) PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 35.Emsley P., and Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 36.Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., and Ferrin T. E. (2004) UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 37.DeLano W. L. (2010) The PyMOL Molecular Graphics System, Version 1.3r1 Schrödinger, LLC, New York [Google Scholar]
- 38.Ren J., Wen L., Gao X., Jin C., Xue Y., and Yao X. (2009) DOG 1.0: illustrator of protein domain structures. Cell. Res. 19, 271–273 [DOI] [PubMed] [Google Scholar]
- 39.Lu X. J., and Olson W. K. (2003) 3DNA: a software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures. Nucleic Acids Res. 31, 5108–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lu X. J., and Olson W. K. (2008) 3DNA: a versatile, integrated software system for the analysis, rebuilding and visualization of three-dimensional nucleic-acid structures. Nat. Protoc. 3, 1213–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zarate-Perez F., Mansilla-Soto J., Bardelli M., Burgner J. W. 2nd, Villamil-Jarauta M., Kekilli D., Samso M., Linden R. M., and Escalante C. R. (2013) Oligomeric properties of adeno-associated virus Rep68 reflect its multifunctionality. J. Virol. 87, 1232–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Duderstadt K. E., Chuang K., and Berger J. M. (2011) DNA stretching by bacterial initiators promotes replication origin opening. Nature 478, 209–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maggin J. E., James J. A., Chappie J. S., Dyda F., and Hickman A. B. (2012) The amino acid linker between the endonuclease and helicase domains of adeno-associated virus type 5 Rep plays a critical role in DNA-dependent oligomerization. J. Virol. 86, 3337–3346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Musayev F. N., Zarate-Perez F., Bardelli M. R., Bishop C. M., Saniev E. F., Linden R. M., Henckaerts E., and Escalante C. R. (2015) Structural studies of AAV2 Rep68 reveal a partially structured linker and compact domain conformation. Biochemistry 10.1021/acs.biochem.5b00610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chiorini J. A., Wiener S. M., Owens R. A., Kyöstió S. R., Kotin R. M., and Safer B. (1994) Sequence requirements for stable binding and function of Rep68 on the adenovirus type 2 inverted terminal repeats. J. Virol. 68, 7448–7457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim R., Corona R. I., Hong B., and Guo J. T. (2011) Benchmarks for flexible and rigid transcription factor-DNA docking. BMC Struct. Biol. 11, 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chang Y. P., Xu M., Machado A. C., Yu X. J., Rohs R., and Chen X. S. (2013) Mechanism of origin DNA recognition and assembly of an initiator-helicase complex by SV40 large tumor antigen. Cell Rep. 3, 1117–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Berman B. P., Nibu Y., Pfeiffer B. D., Tomancak P., Celniker S. E., Levine M., Rubin G. M., and Eisen M. B. (2002) Exploiting transcription factor binding site clustering to identify cis-regulatory modules involved in pattern formation in the Drosophila genome. Proc. Natl. Acad. Sci. U.S.A. 99, 757–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lifanov A. P., Makeev V. J., Nazina A. G., and Papatsenko D. A. (2003) Homotypic regulatory clusters in Drosophila. Genome Res. 13, 579–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li L., Zhu Q., He X., Sinha S., and Halfon M. S. (2007) Large-scale analysis of transcriptional cis-regulatory modules reveals both common features and distinct subclasses. Genome Biol. 8, R101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gotea V., Visel A., Westlund J. M., Nobrega M. A., Pennacchio L. A., and Ovcharenko I. (2010) Homotypic clusters of transcription factor binding sites are a key component of human promoters and enhancers. Genome Res. 20, 565–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim J. G., Takeda Y., Matthews B. W., and Anderson W. F. (1987) Kinetic studies on Cro repressor-operator DNA interaction. J. Mol. Biol. 196, 149–158 [DOI] [PubMed] [Google Scholar]
- 53.Giniger E., and Ptashne M. (1988) Cooperative DNA binding of the yeast transcriptional activator GAL4. Proc. Natl. Acad. Sci. U.S.A. 85, 382–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Coleman R. A., and Pugh B. F. (1995) Evidence for functional binding and stable sliding of the TATA-binding protein on nonspecific DNA. J. Biol. Chem. 270, 13850–13859 [DOI] [PubMed] [Google Scholar]
- 55.Hertel K. J., Lynch K. W., and Maniatis T. (1997) Common themes in the function of transcription and splicing enhancers. Curr. Opin. Cell Biol. 9, 350–357 [DOI] [PubMed] [Google Scholar]
- 56.Smith R. H., Spano A. J., and Kotin R. M. (1997) The Rep78 gene product of adeno-associated virus (AAV) self-associates to form a hexameric complex in the presence of AAV ori sequences. J. Virol. 71, 4461–4471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mansilla-Soto J., Yoon-Robarts M., Rice W. J., Arya S., Escalante C. R., and Linden R. M. (2009) DNA structure modulates the oligomerization properties of the AAV initiator protein Rep68. PLoS Pathog. 5, e1000513. [DOI] [PMC free article] [PubMed] [Google Scholar]