Background: Epitopes of pneumococcal virulence proteins (PnVPs) are valuable for vaccine development.
Results: Novel immunodominant epitopes were mapped on six surface PnVPs.
Conclusion: These epitopes were highly conserved, specific for invasive pneumococcal disease, and localized in functional domains of PnVPs.
Significance: We identified epitopes reactive on the surface of intact encapsulated bacterial cells that could be suitable for vaccine development.
Keywords: epitope mapping, pneumonia, Streptococcus, vaccine development, zinc finger
Abstract
The identification of immunodominant B cell epitopes within surface pneumococcal virulence proteins in pediatric patients with invasive pneumococcal disease (IPD) is a valuable approach to define novel vaccine candidates. To this aim, we evaluated sera from children with IPD and age-matched controls against 141 20-mer synthetic peptides covering the entire sequence of major antigenic fragments within pneumococcal virulence proteins; namely, choline-binding protein D (CbpD), pneumococcal histidine triad proteins (PhtD and PhtE), pneumococcal surface protein A (PspA), plasminogen and fibronectin binding protein B (PfbB), and zinc metalloproteinase B (ZmpB). Ten immunodominant B cell epitopes were identified: CbpD-pep4 (amino acids (aa) 291–310), PhtD-pep11 (aa 88–107), PhtD-pep17 (aa 172–191), PhtD-pep19 (aa 200–219), PhtE-pep32 (aa 300–319), PhtE-pep40 (aa 79–98), PfbB-pep76 (aa 180–199), PfbB-pep79 (aa 222–241), PfbB-pep90 (aa 484–503), and ZmpB-pep125 (aa 431–450). All epitopes were highly conserved among different pneumococcal serotypes, and four of them were located within the functional zinc-binding domain of the histidine triad proteins PhtD and PhtE. Peptides CbpD-pep4, PhtD-pep19, and PhtE-pep40 were broadly recognized by IPD patient sera with prevalences of 96.4%, 92.9%, and 71.4%, respectively, whereas control sera exhibited only minor reactivities (<10.7%). Their specificities for IPD were 93.3%, 95%, and 96.7%; their sensitivities were 96.4%, 92.9%, and 71.4% and their positivity likelihood ratios for IPD were 14.5, 18.6, and 21.4, respectively. Furthermore, purified antibodies against CbpD-pep4, PhtD-pep19, and PhtE-pep40 readily bound on the surfaces of different pneumococcal serotypes, as assessed by FACS and immunofluorescence analysis. The identified immunodominant B cell epitopes provide a better understanding of immune response in IPD and are worth evaluation in additional studies as potential vaccine candidates.
Introduction
Streptococcus pneumoniae is a leading cause of invasive diseases with considerable morbidity and mortality worldwide, including pneumonia, meningitis, and sepsis, predominantly in infants, the elderly, and immunocompromised individuals (1, 2). Currently available vaccines for pneumococcal disease prevention are on the basis of bacterial polysaccharides, either unconjugated (as pneumococcal polysaccharide vaccine) or conjugated with a protein carrier (as pneumococcal conjugate vaccine) (3, 4). However, polysaccharides induce serotype-specific opsonophagocytic antibodies, limiting the clinical effectiveness of these vaccines strictly to vaccine serotypes. Moreover, the widespread use of pneumococcal conjugate vaccines in the last decade has been associated with the emergence of non-vaccine serotypes with significant pathogenic potential, indicating that serotype replacement has already occurred (5, 6).
A promising alternative to polysaccharide-based vaccines is the use of pneumococcal virulence proteins (PnVPs),2 which are well conserved among the majority of pneumococcal serotypes (7). Therefore, antibodies raised against them are expected to be effective against the vast majority of serotypes. Previous studies using genomic surface display libraries identified several large antigenic regions within PnVPs that are consistently recognized by adult patients with invasive pneumococcal disease (IPD) (8, 9). Beghetto et al. (9) identified a panel of large antigenic fragments (with an average length of >200 aa) within surface PnVPs; namely, the choline-binding protein D (CbpD), the pneumococcal histidine triad proteins PhtD and PhtE, the pneumococcal surface protein A (PspA), the plasminogen and fibronectin binding protein B (PfbB), and the zinc metalloproteinase B (ZmpB). However, the total length of these antigenic fragments (1702 aa) hampers the construction of synthetic analogues that could be used as stable vaccine components.
Our study aimed to map, in high resolution, the exact location of immunodominant B cell epitopes within these previously identified large antigenic regions using a pediatric patient cohort with IPD. We further characterized them regarding their sequence homology among different pneumococcal serotypes, their immunoreactivity, and their surface accessibility among different pneumococcal serotypes. Such information could be crucial to shed light on the nature of antigenic epitopes recognized by pediatric patients during IPD and is an essential first step for the development of an effective epitope-based, serotype-independent pneumococcal vaccine.
Experimental Procedures
Study Population
Twenty-eight patients (13 male) aged 2–16 years (median age, 7.5 years) convalescing from IPD and 60 age-matched controls (27 male) with no evidence of acute infection or known history of IPD were enrolled in the study. IPD patients had radiologically confirmed lobar pneumonia, and S. pneumoniae was isolated in the blood/pleural fluid. IPD patient sera were obtained during the convalescent phase (21 ± 7 days after hospital admission). Children with chronic underlying disease or immunosuppression were excluded. The institutional hospital ethics review board approved the study protocol, and written informed consent was obtained from the guardians of all subjects.
Peptide Synthesis
Pin-bound Peptides
141 20-mer synthetic peptides overlapping by six aa residues were prepared, covering the previously identified antigenic fragments of a total length of 1702 aa: CbpD (aa 248–338), PhtD (aa 38–316), PhtE (aa 55–178 and 235–387), PspA (aa 90–195 and 76–432), PfbB (aa 133–334, 443–606, and 851–1133), and ZmpB (aa 346–654) (9). Synthetic peptides were synthesized on derivatized polystyrene pins that form part of the holders that fit into microtiter plates (Mimotopes UK, Ltd).
Soluble Peptides
The 10 most antigenic peptides identified by epitope mapping using peptides covalently attached to polystyrene pins, were resynthesized in their free soluble form. The synthesis was performed using automated Fmoc (N-[9 fluorenyl] methoxycarbonyl) solid-phase synthesis (Biosynthesis Inc.). Each peptide was purified by reversed-phase high performance liquid chromatography and exhibited a single peak at its predicted molecular weight by mass spectrometry.
ELISA
Epitope Mapping
Pins were immersed in 96-well microtiter polystyrene plates (Nunc) containing PBS (pH 7.4) with 2% BSA (PBS-BSA) for 1 h at 37 °C. Sera (1/200) in PBS-BSA were added and incubated overnight at 4 °C. Plates were washed with PBS and incubated with goat anti-human IgG conjugated to horseradish peroxidase (Jackson ImmunoResearch Laboratories, 1:1000) for 1 h at 37 °C. Antibody binding was detected using a substrate solution (2,2′-azino-cis-3-ethylbenzthiazoline-6-sulfonic acid (Sigma Chemicals) and read at 405 nm using a Chromate reader (Awareness Technology). For subsequent experiments, bound antibodies were removed by sonication in a water bath containing PBS with 1% sodium dodecyl sulfate and 0.1% 2-mercaptoethanol at 60 °C.
Soluble Peptide ELISA
Microtiter plates (Nunc) were coated in triplicate with peptides (3 or 15 μg/ml) in PBS or carbonate bicarbonate (pH 9.6) for 2 h at 4 °C. After blocking with PBS containing 2% bovine serum albumin, sera (1/50) were incubated overnight at 4 °C. Goat anti-human alkaline phosphatase-conjugated IgG antibody (Jackson ImmunoResearch Laboratories) was added (1:3000), and antibody binding was detected using the substrate 4-nitrophenyl-phosphate-disodium salt hexahydrate (Sigma Chemicals) at 405 nm (Chromate reader, Awareness Technology). ELISA results were converted to ELISA binding units [(100* absorbance at 405 nm) / cutoff value]. The cutoff value for each peptide assay was determined using the mean optical density plus three standard deviations of normal controls.
Computer Predictions and Homology Search
Peptide sequences were compared against the UniProtKB database (version 2015_01). A similarity search was performed using the Expacy SIB Blast network server using PAM30 matrix.
Purification of Specific Anti-peptide Antibodies
IgGs from IPD sera that exhibited high anti-peptide reactivity in ELISA were purified using a protein A-Sepharose 4B column (Sigma), concentrated, and dialyzed against PBS. Specific immunoaffinity columns of CNBr-activated Sepharose 4B (GE Healthcare Bio-Sciences) were generated by standard methods by coupling of 7 mg of each peptide. IgG fractions from IPD sera were passed slowly through the columns, and bound specific anti-peptide antibodies were eluted using 0.1 m HCl-glycine (pH 2.7). The pH value was neutralized immediately after elution. The eluates were concentrated using polyethylene glycol and dialyzed against PBS (pH 7.3). The final concentration of specific antibody was determined by UV absorbance at 280 nm (UNICAM γ-Helios) and Bradford assays. Anti-peptide antibodies specifically recognized the peptide against which they were purified in homologous ELISA but not the other two peptides in heterologous assays. On the contrary, flowthrough fractions from the immunoaffinity columns exhibited only a minimal reactivity against the peptides, suggesting that almost all specific anti-peptide IgGs for each peptide had been transferred to the eluate of the corresponding CNBr column. SDS-PAGE of the purified antibodies revealed two bands at 50 and 22 kD (and a weak band at 73 kD), as expected, confirming the purity of the IgG antibodies.
Bacterial Cells
Clinical isolates of serotypes 1, 3, 6B, 18C, 19A, 22F, and 23B were isolated from patients with confirmed bacteremia and provided by the Microbiology Department of “Aghia Sophia” Children's Hospital. Serogrouping was performed by latex agglutination test and serotyping by Quellung reaction.
Western Blot Analysis
Pneumococcal cells were grown in Todd-Hewitt broth (Laboratorios Conda, S.A.) for 1–2 days at 37 °C. Cells were collected by centrifugation (3000 × g, 15 min). Bacteria were lysed using buffer containing 0.01% sodium dodecyl sulfate, 0.2% sodium deoxycholate, and 0.15 m sodium citrate. Protein extracts were obtained after repeated freeze-thaw cycles, followed by sonication, and pelleted by centrifugation. Protein extracts were applied to 12% SDS-polyacrylamide gel, followed by electrotransfer onto nitrocellulose, and analyzed for reactivity against affinity-purified anti-peptide antibodies (8 μg/ml) and whole IPD serum (1/75) in PBS-BSA by immunoblotting.
Flow Cytometric Analysis
Pneumococcal cells derived from the early logarithmic growth phase of cultures (A600 = 0.2–0.26) were harvested by centrifugation (1500 × g, 15 min) and blocked using PBS containing 2% FCS (PBS-FCS) for 30 min at room temperature. Purified antibodies diluted in PBS-FCS (150–200 μg/ml) were added and reacted for 2vh at 37 °C. After washing with PBS, cells were incubated with goat anti-human FITC-conjugated IgG (Jackson ImmunoResearch Laboratories) (1/80) for 30 min at room temperature. The samples were resuspended in PBS and analyzed with a fluorescence-activated cell sorter (FACSCalibur, BD Biosciences) and analyzed using CellQuest software (BD Biosciences).
Immunofluorescence Microscopy
Pneumococcal cells were treated as in flow cytometry assay apart from the bacterial staining that was performed using goat anti-human IgG Alexa Fluor 488-conjugated (1/200) and analyzed with an immunofluorescence microscope (Axiophot, Zeiss).
Statistical Analysis
In the initial epitope mapping, immunoreactivities were analyzed using one-way analysis of variance followed by Tukey post hoc test for pair-wise comparison of samples. In soluble peptide assays, the sensitivity and specificity of selected epitopes was estimated using receiver operating characteristic curves on the basis of their immunoreactivities in IPD patient and control sera. The overall performance of the receiver operating characteristic analysis was quantified by computing the area under the curve. An area of 1 indicated perfect performance, whereas 0.5 indicated a performance that was not different from chance. Logistic regression models were used to derive linear predictors and compare the areas under the curve. All reported p values are two-tailed. Statistical significance was set at p < 0.05, and analyses were conducted using Minitab v16.0 and GraphPad Prism v5.03 software.
Results
Fine Specificity of Immunodominant B Cell Epitopes
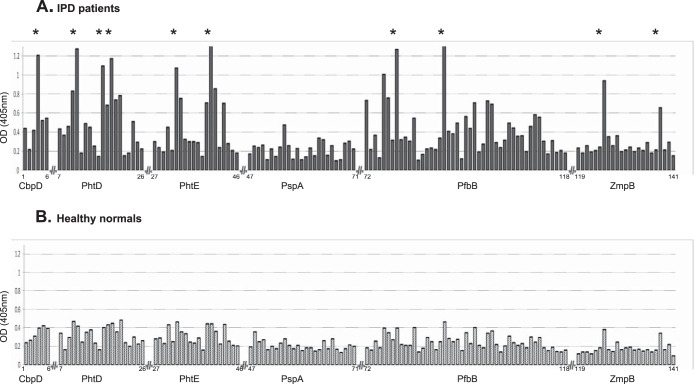
Pin-bound peptides are regenerated after each use, and, therefore, they have a use limit (of about 15 times) to give reliable and reproducible results. In this regard, we selected the sera of 10 IPD patients that displayed the clearest binding profile (with low background and high signal-to-noise ratio) in immunoblotting against pneumococcal cell wall extracts. Five age-matched controls were also used. IPD sera reacted preferentially with 10 distinct epitopes, whereas control sera displayed low absorbance levels for all peptides (Fig. 1). CbpD-pep4, PhtE-pep32, and ZmpB-pep125 reacted with 90% of the sera, whereas PhtD-pep11, PhtD-pep17, PhtD-pep19, PhtE-pep40, PfbB-pep76, PfbB-pep79 and PfbB-pep90 were recognized by all sera (100%). Importantly, the sequence of three of the epitopes identified in our study were found to exist in additional repeats within their parent protein. In detail, CbpD-pep4 sequence appeared twice within CbpD, PfbB-pep76 appeared in six copies within PfbB, whereas the PfbB-pep90 sequence existed in four repeats within PfbB (Fig. 2). Therefore, enhanced immunoreactivity of the parent protein with anti-peptide antibodies is expected compared with the reactivity of the single peptide. Analysis of signal intensities of the pin-bound peptides in patient sera shows a clear accumulation of signals corresponding to 10 distinct epitopes (analysis of variance with post hoc Tukey test, F = 147.3, p < 0.0001; Fig. 1A). Control sera displayed low absorbance levels with no distinct signal peaks among individual peptides (p > 0.05, Fig. 1B).
FIGURE 1.
A, epitope mapping of the antigenic fragments CbpD (aa 248–338), PhtD (aa 38–316), PhtE (aa 55–178 and 235–387), PspA (aa 90–195 and aa 76–432), PfbB (aa 133–334, 443–606, and 851–1133), and ZmpB (aa 346–654) using 141 20-mer synthetic peptides overlapping by six residues. The peptides are numbered on the x axis according to their cognate protein. The y axis denotes the mean optical density (OD) at 405 nm in 10 sera from patients convalescing from IPD after background subtraction. The asterisks denote peptides with a clear accumulation of signals (analysis of variance with post hoc Tukey test, F = 147.3, p < 0.0001); namely, CbpD-pep4, PhtD-pep11, PhtD-pep17, PhtD-pep19, PhtE-pep32, PhtE-pep40, PfbB-pep76, PfbB-pep79, PfbB-pep90, and ZmpB-pep125. B, control sera (n = 5) displayed low absorbance levels with no discernible signal peaks among individual peptides (analysis of variance with post hoc Tukey test, p > 0.05).
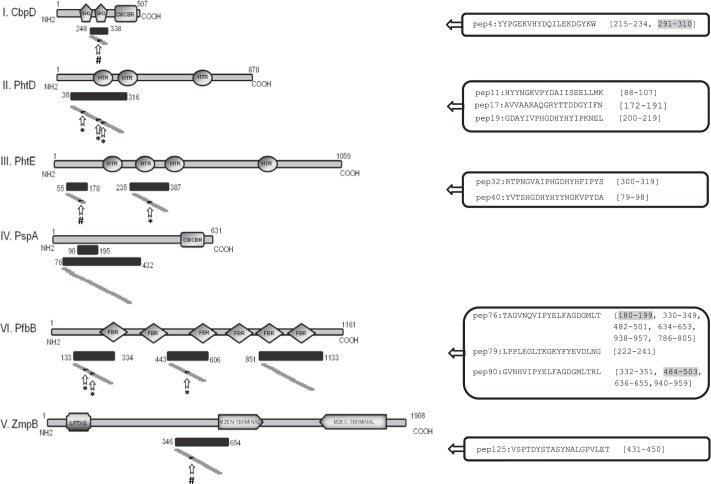
FIGURE 2.
Schematic representation of the six surface pneumococcal proteins mapped in our study. Regions previously identified as antigenic are presented with their localization within cognate protein (black bars underneath each protein). Small gray boxes represent the synthetic 20-mer peptides spanning the entire length of the antigenic regions. Arrows indicate the 10 immunodominant B cell epitopes identified by epitope mapping. Their amino acid sequences are also shown. For epitopes existing in multiple repeats within cognate protein, the amino acid positions of all repeats are presented (the original one is highlighted with a gray background). Asterisks indicate recognition by the 100% of the sera tested, and # indicates recognition by the 90% of the sera. SH3, SH3-like domain; CWCBD, cell wall choline binding domain; HTR, histidine triad domain; LPTXG, LPTXG motif cell wall anchor domain; M26 N-TERMINAL, peptidase M26 N-terminal domain; M26 C-TERMINAL, peptidase M26 C-terminal domain; FBR, fibronectin and binding repeat SSURE domain.
Sequence Homologies
A similarity search revealed that all immunodominant epitopes are well conserved (95–100%) among all pneumococcal serotypes. Only a very limited number of other streptococcal strains (belonging to Streptococcus mitis, Streptococcus oralis, and Streptococcus pseudopneumoniae) exhibit some sequence homology (about 80%) to these peptides.
Three of the epitopes (CbpD-pep4, PfbB-pep76, and PfbB-pep90) were found to exist in multiple repeats within their parent protein (two, six, and four repeats, respectively). Remarkably, 50% of the epitopes were found to be located in the pneumococcal histidine triad proteins (Phts) PhtD and PhtE. These proteins belong to a family of highly homologous proteins containing multiple copies of the eponymous His triad motif (HXXHXH); namely, the PhtA, PhtB, PhtD, and PhtE proteins (10). Because they are closely related regarding their amino acid sequences (possessing about 87% identity in their N-terminal regions), the epitopes identified in the PhtD and PhtE proteins share highly homologous sequences with the other members of the family. Therefore, PhtD-pep11, PhtD-pep17, and PhtD-pep19 are 100% homologous with regions of PhtA and PhtB and 90% homologous with regions in the PhtE protein. Similarly, PhtE-pep32 and PhtE-pep40 are 100% homologous with regions in the PhtA, PhtB, and PhtE proteins. Surprisingly, 4 of 10 immunodominant epitopes were found to reside exactly within the zinc finger domain of the PhtD and PhtE proteins (11). Given that PhtD and PhtE proteins possess multiple repeats of the zinc finger domain, additional high homologous regions to the epitopes exist in each of them. These regions are of a total length of 11–18 aa, and their homology to the PhtD and PhtE epitopes ranges from 72% to 92%.
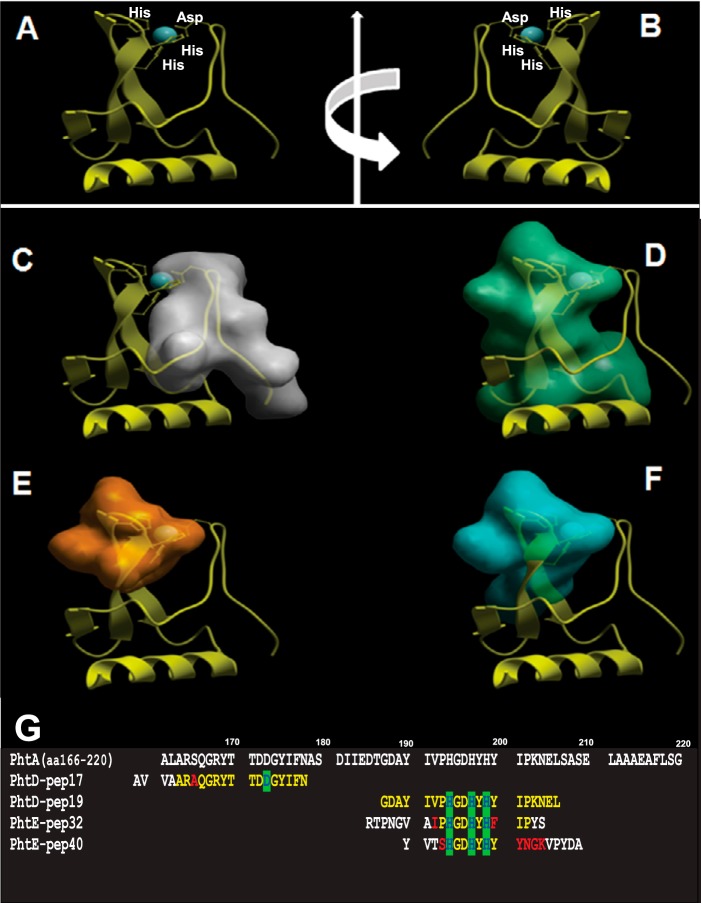
On the basis of the solved three-dimensional structure of PhtA (PDB code 2CS7), we explored the conformational properties of the histidine triad epitopes (Fig. 3) (11). In the parent structure, the zinc binding site is assembled by three histidine residues (His-194, His-197, and His-199; motif HXXHXH) forming the base of a triangle, with the zinc ion sitting above the plane of these residues and an aspartic acid residue (Asp-173) forming the apex of the triangle. PhtD-pep19, PhtE-pep32, and PhtE-pep40 epitopes possess the three zinc-bound histidine residues and are located within the two β strands of the structure. On the other hand, the PhtD-pep17 epitope, holding the zinc-bound aspartic acid residue, is located within the loop opposite to β strands that compromise the zinc binding site (Fig. 3).
FIGURE 3.
Structural analysis of the zinc finger-related epitopes. A, overall three-dimensional structure histidine triad motif corresponding to aa 166–220 of the PhtA structure (PDB code 2CS7). B, the same structure after 180° rotation. C, localization of the PhtD-pep17 epitope (gray) within the loop that contains the Asp residue that compromises the zinc-binding site. D, localization of the PhtD-pep19 epitope (green). E, localization of the PhtE-pep32 epitope (orange). F, localization of the PhtD-pep40 epitope (blue). The latter three epitopes reside within the two β-strands that contain the three histidine residues of the Pht motif. G, multiple sequence alignment of the PhtA fragment (aa 166–220) and the sequences of the PhtD-pep17, PhtD-pep19, PhtE-pep32, and PhtE-pep40 epitopes. Identical residues are shown in yellow and conservative substitutions in red. The histidine and aspartic acid residues are highlighted in green.
Immunoreactivity of Immunodominant Epitopes in IPD Patients and Controls
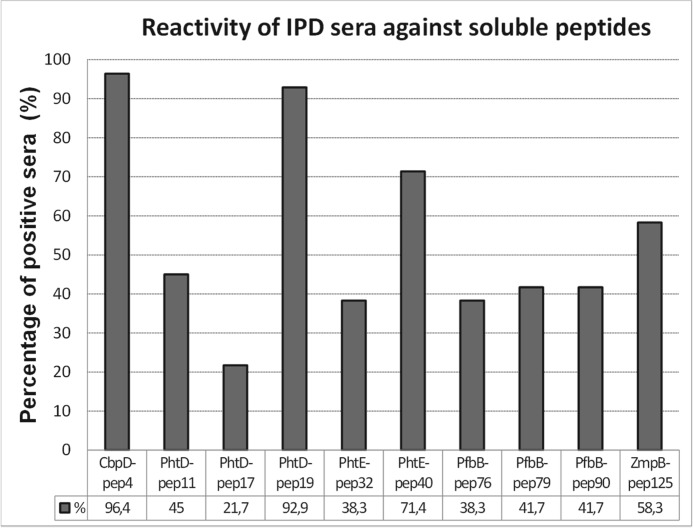
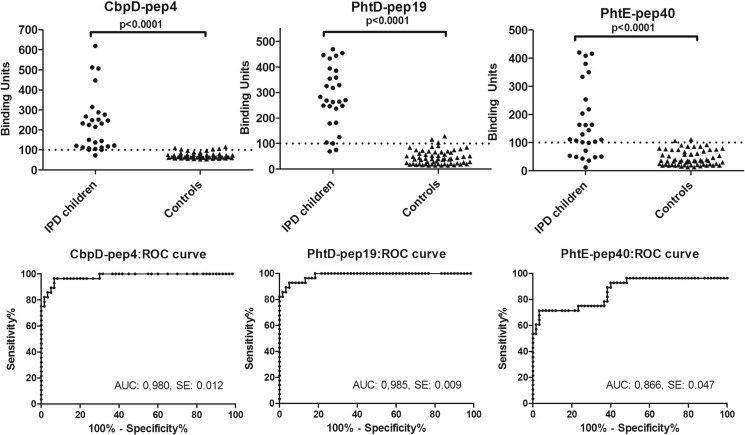
Immunoreactivity against the 10 initially identified epitopes by epitope mapping was further evaluated in all IPD patients (n = 28) using immunosorbent assays on the basis of soluble (free) peptide analogues. These assays have no use limit and, therefore, can be applied to any number of samples. CbpD-pep4, PhtD-pep19, and PhtE-pep40 exhibited significant reactivity against the vast majority of IPD sera, with prevalences of 96.4%, 92.9%, and 71.4% respectively, whereas they displayed low reactivity against control sera (with prevalences of 10.7%, 7.1%, and 7.1%, respectively). The remaining seven soluble peptide epitopes exhibited lower reactivity against IPD sera, ranging from 21.7% to 58.3% (and no reactivity against control sera), most likely because of their inability to coat the plates efficiently and/or at the same time react with patient antibodies (Fig. 4). Each of the three immunodominant epitopes (as confirmed in both immobilized and soluble peptide assays) reacted with the vast majority of patient sera and significantly discriminated patients from controls (p < 0.0001) (Fig. 5). The sensitivities for the CbpD-pep4-, PhtD-pep19-, and PhtE-pep40-based assays were 96.4%, 92.9%, and 71.4%, respectively, whereas their specificities were 93.3%, 95%, and 96.7%, respectively. Remarkably, each IPD patient recognized at least one of the three most antigenic epitopes. Therefore, the combination of all three peptides increased the sensitivity for IPD to 100% with a specificity of 88.3%. The area under the curve was very high for the assays of CbpD-pep4, PhtD-pep19, and PhtE-pep40, reaching 0.98, 0.99, and 0.87, respectively, whereas their positivity likelihood ratios for IPD were 14.5, 18.6, and 21.4, respectively (Fig. 5).
FIGURE 4.
Prevalence of antibodies against the 10 immunoreactive epitopes (identified by epitope mapping) in sera from 28 patients with IPD.
FIGURE 5.
A, prevalence of antibodies against the three most immunoreactive epitopes in sera from 28 patients with IPD and 60 controls. Statistical significant differences were observed between the two groups for all peptides (p < 0.0001). Dotted lines represent the cutoff value for each peptide. B, receiver operating characteristic curves (ROC) showing the sensitivity and specificity of relevant ELISA results for each peptide. The values of area under curve (AUC) and standard error are also presented (above the x axis) for each peptide.
Evaluation of Reactivity of Affinity-purified Anti-peptide Antibodies against Whole-cell Lysates and Live Intact Pneumococcal Cells
We assessed the specificity of the recognition of immunodominant epitopes using affinity-purified specific antibodies against CbpD-pep4, PhtD-pep19, and PhtE-pep40 peptides from IPD sera. Immunoblotting experiments demonstrated the ability of purified antibodies to recognize, besides the peptides, their corresponding parent protein on whole-cell pneumococcal lysates. Specific antibodies against CbpD-pep4 recognized a low molecular mass protein band consistent with the molecular mass of CbpD (41 kDa), whereas specific antibodies against PhtD-pep19 and PhtE-pep40 recognized two major proteins with molecular masses of ∼93 and 107 kDa, matching the molecular mass of PhtD (93.5 kDa) and PhtE (114.6 kDa), respectively (Fig. 6) (12). Because of the high sequence homology between Phts, antibodies against PhtD-pep19 and PhtE-pep40 recognized both bands assigned to PhtD and PhtE proteins, and they also reacted with a slightly weaker band possibly corresponding to PhtA (91.5 kDa) or PhtB (92.1 kDa) protein. This multiple Phts recognition pattern is typical for antibodies targeting the amino-terminal region of the proteins (12). Some reactivity was also detected with proteins of smaller molecular weight, most likely attributed to protein degradation (Fig. 6). Regarding CbpD, the corresponding band (∼41 kDa) was rather faint. This can be attributed to a lower extent of expression of CbpD in the particular S. pneumoniae strain or to a low recovery of this particular protein in the extract. Another explanation is that affinity-purified anti-CbpDpep4 antibodies used in the current lane may be in suboptimal concentration for immunoblotting. However, the aim of this experiment was to confirm the specificity of the affinity-purified anti-peptide antibodies (represented by their potential to recognize, besides the peptides, their corresponding parent protein) and not to quantify their immunoreactivity (reflected by the intensity of each band).
FIGURE 6.
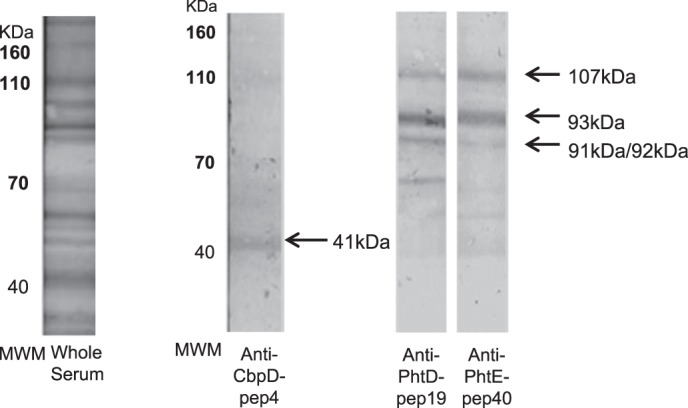
Affinity-purified anti-peptide antibodies recognized their cognate protein on whole-cell lysates by immunoblotting. The binding profile of the whole serum of a patient with IPD before the purification procedure is depicted in the left panel. The binding profiles of the eluates of the three immunoaffinity columns (CbpD-pep4, PhtD-pep19, and PhtE-pep40) are depicted in the right panels. Apparent molecular size markers in kilodaltons are shown (MWM). Arrows indicate the reactive bands and their estimated molecular mass nearly consistent with the molecular mass of cognate protein, respectively: CbpD (41 kDa), PhtD (93.5 kDa) and PhtE (114.6 kDa), and PhtA (91.5 kDa) and PhtB (92.1 kDa).
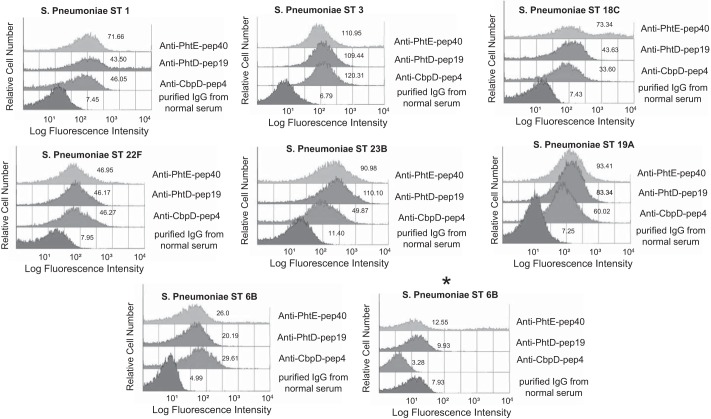
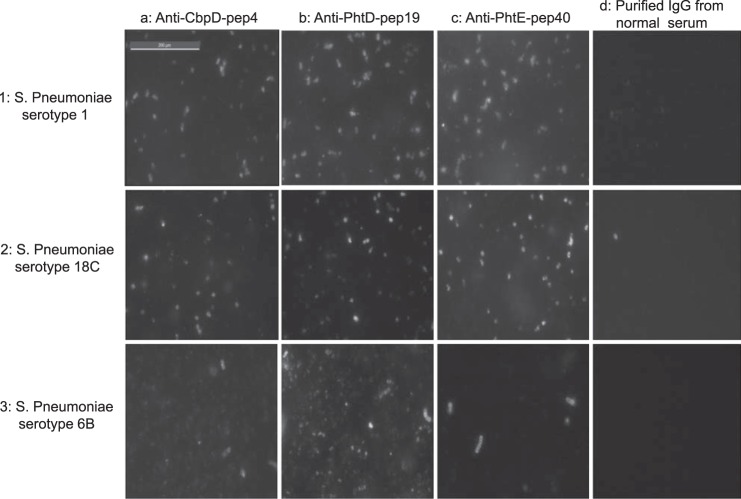
Surface binding of affinity-purified antibodies against CbpD-pep4, PhtD-pep19, and PhtE-pep40 onto the surface of live intact pneumococcal cells of seven different serotypes was detected by flow cytometry (Fig. 7). Purified antibodies readily bound on the surface of seven of eight encapsulated clinical isolates tested, as shown by geometric mean fluorescence intensities. Because antibodies against PhtD-pep19 and PhtE-pep40 were cross-reacting with all Pht family members, it is not possible to determine whether surface labeling was due to each one separately or to a synergistic binding effect. Interestingly, two 6B clinical isolates displayed different surface binding reactivity. Although one 6B isolate was bound by all anti-peptide antibodies, a different 6B isolate displayed no or low surface binding reactivity. Purified IgG from control serum exhibited a consistently low reactivity against all tested pneumococcal serotypes. Similar results were obtained by immunofluorescence microscopy (Fig. 8).
FIGURE 7.
Specific affinity-purified anti-peptide antibodies readily bound onto live pneumococcal cells of different serotypes. The asterisk corresponds to a different strain (clinical isolate) of S. pneumoniae of serotype 6B that exhibited no surface staining with specific anti-peptide antibodies. Purified IgG from normal serum was used as a negative control. The numbers next to the histograms reflect the geometric mean fluorescence intensity of the population.
FIGURE 8.
Stained S. pneumoniae surface structures were observed using a immunofluorescence microscope (Zeiss LSM 510). Affinity-purified specific antibodies against CbpD-pep4 (column a), PhtD-pep19 (column b), and PhtE-pep40 (column c) reacted with intact live S. pneumoniae cell serotypes 1 (row 1) and18C (row 2). Weaker surface staining was observed onto a strain of live S. pneumoniae serotype 6B (row 3). Row 4 represents S. pneumoniae cells of the three serotypes incubated with purified IgG from normal serum (negative control). Representative panels of one of three individual experiments are shown.
Discussion
To our knowledge, this is the first study to unveil the fine specificity of immunodominant highly conserved B cell epitopes targeted by immune response in pediatric patients with IPD. Importantly, all identified epitopes are located within functional surface proteins, implying that antibodies against these epitopes could inhibit major functions of the bacterium associated with impaired invasive capacity and virulence. One epitope was identified within CbpD that is involved in adherence, complement inhibition, and competence-induced cell lysis (13). Two epitopes have been identified within PhtE and three epitopes within PhtD. Pht proteins inhibit surface complement deposition and mediate bacterial adherence through zinc binding (14–16). One epitope has been identified within ZmpB and three epitopes within PfbB. Both of these proteins mediate bacterial adherence to epithelial cells, which is considered an essential early step in colonization and disease (17, 18). Furthermore, the fact that all epitopes reside within proteins that fulfill important non-redundant roles in bacterial virulence minimizes the possibility of negative selection or strain replacement under immune pressure (19). No immunodominant epitope was identified within the extensively studied PspA, a finding most likely explained by the existence of conformational epitopes instead of linear epitopes that could not be detected by the epitope mapping method used here or, alternatively, by the high primary sequence variability of PspA (20, 21). All identified epitopes in our study were included within the large antigenic fragments defined previously by Giefing et al. (8), who screened the whole pneumococcal genome against sera from adults following IPD. Notably, in the study by Giefing et al. (8), 193 fragments were identified as antigenic, having a total length of 13494 aa (8).
Homology searches revealed that the identified epitopes are almost completely conserved among different pneumococcal serotypes, breaking ground for serotype-independent vaccine development. Moreover, the identified epitopes share some sequence homology to a very limited number of other streptococcal strains of the nasopharyngeal flora (22). Interestingly, 80% of the epitopes identified in our study exist in multiple repeats within pneumococcal surface proteins. Epitopes CbpD-pep4, PfbB-pep76, and PfbB-pep90 exist in two, six, and four repeats respectively within their cognate protein, whereas epitopes PhtD-pep11, PhtD-pep17, PhtD-pep19, PhtE-pep32, and PhtE-pep40 have identical (or 90% homologous) sequences in all four members of Pht proteins and share highly homologous regions within their cognate Pht protein. This finding may imply a greater avidity of anti-peptide antibodies (through divalent interactions), leading to more effective protection (23). The three-dimensional structural analysis revealed that PhtD-pep17, PhtD-pep19, PhtE-pep32, and PhtE-pep40 reside within the zinc-binding domain of cognate protein and possess the residues that are critical for zinc binding, i.e. HXXHXH or Asp (11). This finding suggests that binding of anti-peptide antibodies to this exact region may block the Pht protein-zinc interaction and, therefore, hamper Pht proteins function, including zinc concentration homeostasis in the bacterial environment, adherence to epithelial cells, and C3b deposition inhibition, leading to impaired opsonophagocytosis and, therefore, attenuated bacterial virulence (14–16, 24). In this regard, previous studies have shown that a quadruple S. pneumoniae mutant in which all Pht genes are deleted is completely avirulent (25), whereas antibodies against PhtD and PhtE prevent bacterial adhesion to the human respiratory epithelium (24). We are currently undertaking further studies to investigate how antibody binding may affect the zinc binding potential of zinc-finger B cell epitopes as well as their changes in conformation and function, as we have described previously in a different clinical setting (26).
Three of 10 initially selected epitopes, i.e. CbpD-pep4, PhtD-pep19, and PhtE-pep40, displaying the highest immunoreactivity in soluble peptides ELISA against IPD patients' sera, were further confirmed as specific targets of the humoral immune response to IPD. Understanding the differences in immune response between subjects who are exposed to S. pneumoniae and those that finally develop IPD may be the key for the development of effective vaccines against IPD. Although a history of pneumococcal exposure through nasopharyngeal carriage or mucosal infection was not known in patients and controls, we presume that such events would have occurred equally between the two study groups. It has been reported previously that pneumococcal exposure or cross-reactive commensal bacteria may elicit antibodies against surface pneumococcal proteins in healthy controls (27, 28). Nevertheless, the identified epitopes were found to be disease-specific, most likely because they are exposed and recognized at the time of epithelial adhesion, bloodstream invasion, and massive inflammation, which is observed during IPD. Alternatively, the identified epitopes may represent targets of high-avidity antibodies that are generated through somatic hypermutation and/or gene conversion of the antibody-variable region following their affinity maturation in IPD (29). Notably, all IPD sera were found to be immunoreactive against at least one epitope, suggesting that a novel synthetic analogue combining all three peptides could broaden its immunogenic potential, as described previously (30–32).
At this point, one could wonder why antibodies against the CbpD-pep4, PhtD-pep19, and PhtE-pep40 peptides did not actually protect our patients from IPD. In our study, antibodies against these peptides were detected in the sera of the vast majority of IPD patients only in the convalescent phase, taken at 21 ± 7 days after hospital admission (plus a short incubation period after S. pneumoniae infection and before hospital admission). These antibodies did not pre-exist, as implied by the finding that control sera from children with no known history of IPD exhibited only a minor reactivity against these peptides. In addition, ongoing work in our laboratory has shown promising results for their protective capacity against different pneumococcal strains (33, 34). More specifically, affinity-purified antibodies against the peptide epitopes exhibited significant in vitro opsonophagocytic activity (33), and anti-peptide antibodies developed in mice significantly increased the survival of the experimental animals in passive immunization experiments followed by S. pneumoniae-induced lethal sepsis (34). Similar approaches in which convalescent sera were screened for detection of novel antigens have been applied previously for other pathogens, including Staphylococcus aureus and Streptococcus pyogenes, and successfully identified promising peptide vaccines that are currently in phase II clinical studies (35, 36).
One limitation of our study is that, because we mapped previously identified antigenic fragments of PnPs, other immunogenic regions (with a lower reactivity) may also exist. However, the development of a successful epitope-based vaccine does not require the identification of all antigenic epitopes but a sufficient number of immunodominant ones. Another limitation is that epitope mapping was held using peptides covalently attached by their C terminus to derivatized polystyrene pins, whereas the immunoreactivity of IPD sera was evaluated using soluble peptide ELISAs. The latter was necessary because of the reuse limit of pin-bound peptides for reliable results (about 15 repeats). As a result, among the 10 initially identified epitopes through pin-bound peptide ELISAs, three epitopes exhibited a high reactivity against IPD sera in soluble peptide ELISAs. However, we cannot exclude that some of the remaining soluble peptides displayed limited immunoreactivity because they could not meet the requirements for an efficient immunosorbent assay setup; i.e. to adsorb efficiently to the plastic surface or to use their limited set of side chains for simultaneous interaction with the plastic surface and the antibody molecule (37).
Affinity-purified anti-peptide antibodies from IPD sera specifically recognized their cognate protein in pneumococcal whole-cell lysates. This finding excludes a nonspecific binding because of the high conformational freedom of small peptides or a cross-reactivity of unrelated human antibodies. As expected, anti-PhtD-pep19 and anti-PhtE-pep40 antibodies cross-recognized all Phts in immunoblotting because these two peptides appear with a very high sequence homology within these proteins (12). Moreover, affinity-purified antibodies against the three immunodominant epitopes exhibited significant binding on encapsulated live intact pneumococcal cells of different serotypes. Therefore, antibodies against the peptides are capable to bind their cognate protein in its natural conformation on the surface of intact encapsulated bacterial cells. This finding suggests that the capsule of the bacterial cells cannot mask the anti-peptide antibody recognition for the majority of clinical isolates. Purified antibodies recognized pneumococcal cells of seven different serotypes to the same extent, confirming the epitope conservation among different serotypes. Our finding that one of the two 6B isolates displayed an absence of surface binding could be attributed to the known variation in bacterial capsule thickness or to a lower expression of PhtE, PhtD, and CbpD proteins in this particular strain (38, 39). Previous reports have demonstrated that surface-accessible epitopes may elicit antibody-mediated protection against pneumococcal disease (21, 40, 41), underlining the potential of our epitopes for the development of future vaccines. Additional studies evaluating the in vitro opsonophagocytic and in vivo protective efficacy of anti-peptide antibodies are required.
In conclusion, our work unveiled the fine specificity of major linear B cell epitopes within surface PnVPs that are involved in host-pathogen interaction and share interesting functional and immunological characteristics. It remains to evaluate the protective efficacy of the identified synthetic peptide epitopes against pneumococcal disease in additional animal studies. However, their short length, ease of production, cost-effectiveness, high degree of conservation among different serotypes, specificity for IPD, localization in functional protein domains, and opsonizing ability strongly suggest that they could be of great interest regarding their implications in the development of novel immunogens for new-generation epitope-based vaccines. Most importantly, such information was obtained by screening pediatric sera because children are at high risk for IPD and consist a main target population for optimal protection.
Author Contributions
T. L. performed the experiments, data acquisition, and data analysis and drafted the article. J. G. R. designed the study, performed the experiments, analyzed the data, and revised the article. C. P. performed the FACS analysis. A. Z. T., G. C., and M. T. supervised the study and revised the article. V. S. designed the study, had overall supervision, and revised the article.
Acknowledgments
We thank Prof. David Greenberg for sera from IPD children for the initial screening. We also thank Maria Mavrouli, Christos Adamopoulos, and Haris Alexopoulos for technical assistance and all parents and children who volunteered to participate in our study.
This work was supported by Special Account for Research Funds (ELKE) Grants 70/3/9687 and 70/3/9473. The authors declare that they have no conflicts of interest with the contents of this article.
- PnVP
- pneumococcal virulence protein
- IPD
- invasive pneumococcal disease
- aa
- amino acids.
References
- 1.O'Brien K. L., Wolfson L. J., Watt J. P., Henkle E., Deloria-Knoll M., McCall N., Lee E., Mulholland K., Levine O. S., Cherian T., and Hib and Pneumococcal Global Burden of Disease Study Team (2009) Burden of disease caused by Streptococcus pneumoniae in children younger than 5 years: global estimates. Lancet 374, 893–902 [DOI] [PubMed] [Google Scholar]
- 2.Ciapponi A., Elorriaga N., Rojas J. I., Romano M., Martí S. G., Bardach A., and Ruvinsky S. (2014) Epidemiology of pediatric pneumococcal meningitis and bacteremia in Latin America and the Caribbean: a systematic review and meta-analysis. Pediatr. Infect. Dis. J. 33, 971–978 [DOI] [PubMed] [Google Scholar]
- 3.Hansen J., Black S., Shinefield H., Cherian T., Benson J., Fireman B., Lewis E., Ray P., and Lee J. (2006) Effectiveness of heptavalent pneumococcal conjugate vaccine in children younger than 5 years of age for prevention of pneumonia: updated analysis using World Health Organization standardized interpretation of chest radiographs. Pediatr. Infect. Dis. J. 25, 779–781 [DOI] [PubMed] [Google Scholar]
- 4.Pilishvili T., Lexau C., Farley M. M., Hadler J., Harrison L. H., Bennett N. M., Reingold A., Thomas A., Schaffner W., Craig A. S., Smith P. J., Beall B. W., Whitney C. G., Moore M. R., and Active Bacterial Core Surveillance/Emerging Infections Program Network (2010) Sustained reductions in invasive pneumococcal disease in the era of conjugate vaccine. J. Infect. Dis. 201, 32–41 [DOI] [PubMed] [Google Scholar]
- 5.Chibuk T. K., Robinson J. L., and Hartfield D. S. (2010) Paediatric complicated pneumonia and pneumococcal serotype replacement: trends in hospitalized children pre and post introduction of routine vaccination with pneumococcal conjugate vaccine (PCV7). Eur. J. Pediatr. 169, 1123–1128 [DOI] [PubMed] [Google Scholar]
- 6.Scott J. R., Millar E. V., Lipsitch M., Moulton L. H., Weatherholtz R., Perilla M. J., Jackson D. M., Beall B., Craig M. J., Reid R., Santosham M., and O'Brien K. L. (2012) Impact of more than a decade of pneumococcal conjugate vaccine use on carriage and invasive potential in Native American communities. J. Infect. Dis. 205, 280–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tai S. S. (2006) Streptococcus pneumoniae protein vaccine candidates: properties, activities and animal studies. Crit. Rev. Microbiol. 32, 139–153 [DOI] [PubMed] [Google Scholar]
- 8.Giefing C., Meinke A. L., Hanner M., Henics T., Bui M. D., Gelbmann D., Lundberg U., Senn B. M., Schunn M., Habel A., Henriques-Normark B., Ortqvist A., Kalin M., von Gabain A., and Nagy E. (2008) Discovery of a novel class of highly conserved vaccine antigens using genomic scale antigenic fingerprinting of pneumococcus with human antibodies. J. Exp. Med. 205, 117–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beghetto E., Gargano N., Ricci S., Garufi G., Peppoloni S., Montagnani F., Oggioni M., Pozzi G., and Felici F. (2006) Discovery of novel Streptococcus pneumoniae antigens by screening a whole-genome λ-display library. FEMS Microbiol. Lett. 262, 14–21 [DOI] [PubMed] [Google Scholar]
- 10.Plumptre C. D., Ogunniyi A. D., and Paton J. C. (2012) Polyhistidine triad proteins of pathogenic streptococci. Trends Microbiol. 20, 485–493 [DOI] [PubMed] [Google Scholar]
- 11.Riboldi-Tunnicliffe A., Isaacs N. W., and Mitchell T. J. (2005) 1.2 Angstroms crystal structure of the Pneumococcus PhtA histidine triad domain a novel zinc binding fold. FEBS Lett. 579, 5353–5360 [DOI] [PubMed] [Google Scholar]
- 12.Adamou J. E., Heinrichs J. H., Erwin A. L., Walsh W., Gayle T., Dormitzer M., Dagan R., Brewah Y. A., Barren P., Lathigra R., Langermann S., Koenig S., and Johnson S. (2001) Identification and characterization of a novel family of pneumococcal proteins that are protective against sepsis. Infect. Immun. 69, 949–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kausmally L., Johnsborg O., Lunde M., Knutsen E., and Håvarstein L. S. (2005) Choline-binding protein D (CbpD) in Streptococcus pneumoniae is essential for competence-induced cell lysis. J. Bacteriol. 187, 4338–4345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kallio A., Sepponen K., Hermand P., Denoël P., Godfroid F., and Melin M. (2014) The role of pneumococcal histidine triad (Pht) proteins in the attachment of Streptococcus pneumoniae to respiratory epithelial cells. Infect. Immun. 82, 1683–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Godfroid F., Hermand P., Verlant V., Denoël P., and Poolman J. T. (2011) Preclinical evaluation of the Pht proteins as potential cross-protective pneumococcal vaccine antigens. Infect. Immun. 79, 238–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rioux S., Neyt C., Di Paolo E., Turpin L., Charland N., Labbé S., Mortier M. C., Mitchell T. J., Feron C., Martin D., and Poolman J. T. (2011) Transcriptional regulation, occurrence and putative role of the Pht family of Streptococcus pneumoniae. Microbiology 157, 336–348 [DOI] [PubMed] [Google Scholar]
- 17.Gong Y., Xu W., Cui Y., Zhang X., Yao R., Li D., Wang H., He Y., Cao J., and Yin Y. (2011) Immunization with a ZmpB-based protein vaccine could protect against pneumococcal diseases in mice. Infect. Immun. 79, 867–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Papasergi S., Garibaldi M., Tuscano G., Signorino G., Ricci S., Peppoloni S., Pernice I., Lo Passo C., Teti G., Felici F., and Beninati C. (2010) Plasminogen- and fibronectin-binding protein B is involved in the adherence of Streptococcus pneumoniae to human epithelial cells. J. Biol. Chem. 285, 7517–7524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Croucher N. J., Harris S. R., Fraser C., Quail M. A., Burton J., van der Linden M., McGee L., von Gottberg A., Song J. H., Ko K. S., Pichon B., Baker S., Parry C. M., Lambertsen L. M., Shahinas D., Pillai D. R., Mitchell T. J., Dougan G., Tomasz A., Klugman K. P., Parkhill J., Hanage W. P., and Bentley S. D. (2011) Rapid pneumococcal evolution in response to clinical interventions. Science 331, 430–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hollingshead S. K., Becker R., and Briles D. E. (2000) Diversity of PspA: mosaic genes and evidence for past recombination in Streptococcus pneumoniae. Infect. Immun. 68, 5889–5900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Daniels C. C., Coan P., King J., Hale J., Benton K. A., Briles D. E., and Hollingshead S. K. (2010) The proline-rich region of pneumococcal surface proteins A and C contains surface-accessible epitopes common to all pneumococci and elicits antibody-mediated protection against sepsis. Infect. Immun. 78, 2163–2172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nobbs A. H., Lamont R. J., and Jenkinson H.F. (2009) Streptococcus adherence and colonization. Microbiol. Mol. Biol. Rev. 73, 407–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fried A. J., Altrich M. L., Liu H., Halsey J. F., and Bonilla F. (2013) Correlation of pneumococcal antibody concentration and avidity with patient clinical and immunologic characteristics. J. Clin. Immunol. 33, 847–856 [DOI] [PubMed] [Google Scholar]
- 24.Khan M. N., and Pichichero M. E. (2012) Vaccine candidates PhtD and PhtE of Streptococcus pneumoniae are adhesins that elicit functional antibodies in humans. Vaccine 30, 2900–2907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ogunniyi A. D., Grabowicz M., Mahdi L. K., Cook J., Gordon D. L., Sadlon T. A., and Paton J. C. (2009) Pneumococcal histidine triad proteins are regulated by the Zn2+-dependent repressor AdcR and inhibit complement deposition through the recruitment of complement factor H. FASEB J. 23, 731–738 [DOI] [PubMed] [Google Scholar]
- 26.Routsias J. G., Kosmopoulou A., Makri A., Panou-Pomonis E., Sakarellos C., Sakarellos-Daitsiotis M., Moutsopoulos H. M., and Tzioufas A. G. (2004) Zinc ion dependent B-cell epitope, associated with primary Sjogren's syndrome, resides within the putative zinc finger domain of Ro60kD autoantigen: physical and immunologic properties. J. Med. Chem. 47, 4327–4334 [DOI] [PubMed] [Google Scholar]
- 27.Adrian P. V., Bogaert D., Oprins M., Rapola S., Lahdenkari M., Kilpi T., de Groot R., Käyhty H., and Hermans P. W. (2004) Development of antibodies against pneumococcal proteins α-enolase, immunoglobulin A1 protease, streptococcal lipoprotein rotamase A, and putative proteinase maturation protein A in relation to pneumococcal carriage and otitis media. Vaccine 22, 2737–2742 [DOI] [PubMed] [Google Scholar]
- 28.Simell B., Ahokas P., Lahdenkari M., Poolman J., Henckaerts I., Kilpi T. M., and Käyhty H. (2009) Pneumococcal carriage and acute otitis media induce serum antibodies to pneumococcal surface proteins CbpA and PhtD in children. Vaccine 27, 4615–4621 [DOI] [PubMed] [Google Scholar]
- 29.Zhou J., Lottenbach K. R., Barenkamp S. J., and Reason D. C. (2004) Somatic hypermutation and diverse immunoglobulin gene usage in the human antibody response to the capsular polysaccharide of Streptococcus pneumoniae Type 6B. Infect. Immun. 72, 3505–3514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Posfay-Barbe K. M., Galetto-Lacour A., Grillet S., Ochs M. M., Brookes R. H., Kraehenbuhl J. D., Cevey-Macherel M., Gehri M., Gervaix A., and Siegrist C. A. (2011) Immunity to pneumococcal surface proteins in children with community-acquired pneumonia: a distinct pattern of responses to pneumococcal choline-binding protein A. Clin. Microbiol. Infect. 17, 1232–1238 [DOI] [PubMed] [Google Scholar]
- 31.Verhoeven D., Xu Q., and Pichichero M. E. (2014) Vaccination with a Streptococcus pneumoniae trivalent recombinant PcpA, PhtD and PlyD1 protein vaccine candidate protects against lethal pneumonia in an infant murine model. Vaccine 32, 3205–3210 [DOI] [PubMed] [Google Scholar]
- 32.Cao J., Chen D., Xu W., Chen T., Xu S., Luo J., Zhao Q., Liu B., Wang D., Zhang X., Shan Y., and Yin Y. (2007) Enhanced protection against pneumococcal infection elicited by immunization with the combination of PspA, PspC, and ClpP. Vaccine 25, 4996–5005 [DOI] [PubMed] [Google Scholar]
- 33.Papastamatiou T., Routsias J. G., Lagousi T., Tsakris A., and Spoulou V. (2015) 33rd Annual Meeting of European Society for Paediatric Infectious Diseases, May 12–16, 2015, p. 80, European Society for Paediatric Infectious Diseases, Leipzig, Germany [Google Scholar]
- 34.Papastamatiou T., Routsias J. G., Lagousi T., Tsakris A., and Spoulou V. (2015) 33rd Annual Meeting of European Society for Paediatric Infectious Diseases, May 12–16, 2015, p. 31, European Society for Paediatric Infectious Diseases, Leipzig, Germany [Google Scholar]
- 35.Meinke A., Henics T., Hanner M., Minh D. B., and Nagy E. (2005) Antigenome technology: a novel approach for the selection of bacterial vaccine candidate antigens. Vaccine 23, 2035–2041 [DOI] [PubMed] [Google Scholar]
- 36.Kuklin N. A., Clark D. J., Secore S., Cook J., Cope L. D., McNeely T., Noble L., Brown M. J., Zorman J. K., Wang X. M., Pancari G., Fan H., Isett K., Burgess B., Bryan J., Brownlow M., George H., Meinz M., Liddell M. E., Kelly R., Schultz L., Montgomery D., Onishi J., Losada M., Martin M., Ebert T., Tan C. Y., Schofield T. L., Nagy E., Meineke A., Joyce J. G., Kurtz M. B., Caulfield M. J., Jansen K. U., McClements W., and Anderson A. S. (2006) A novel Staphylococcus aureus vaccine: iron surface determinant B induces rapid antibody responses in rhesus macaques and specific increased survival in a murine S. aureus sepsis model. Infect. Immun. 74, 2215–2223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ivanov V. S., Suvorova Z. K., Tchikin L. D., Kozhich A. T., and Ivanov V. T. (1992) Effective method for synthetic peptide immobilization that increases the sensitivity and specificity of ELISA procedures. J. Immunol. Methods 153, 229–233 [DOI] [PubMed] [Google Scholar]
- 38.Weiser J. N., Austrian R., Sreenivasan P. K., and Masure H. R. (1994) Phase variation in pneumococcal opacity: relationship between colonial morphology and nasopharyngeal colonization. Infect. Immun. 62, 2582–2589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Browall S., Norman M., Tångrot J., Galanis I., Sjöström K., Dagerhamn J., Hellberg C., Pathak A., Spadafina T., Sandgren A., Bättig P., Franzén O., Andersson B., Örtqvist Å., Normark S., and Henriques-Normark B. (2014) Intraclonal variations among Streptococcus pneumoniae isolates influence the likelihood of invasive disease in children. J. Infect. Dis. 209, 377–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hamel J., Charland N., Pineau I., Ouellet C., Rioux S., Martin D., and Brodeur B. R. (2004) Prevention of pneumococcal disease in mice immunized with conserved surface-accessible proteins. Infect. Immun. 72, 2659–2670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gor D. O., Ding X., Briles D. E., Jacobs M. R., and Greenspan N. S. (2005) Relationship between surface accessibility for PpmA, PsaA, and PspA and antibody-mediated immunity to systemic infection by Streptococcus pneumoniae. Infect. Immun. 73, 1304–1312 [DOI] [PMC free article] [PubMed] [Google Scholar]