Background: Maintaining the mitochondrial NAD+ pool is important, whereas its generation in mammalian cells is not understood.
Results: A plant transporter expressed in human cells increases mitochondrial NAD+ but shifts metabolism from respiration to glycolysis.
Conclusion: In human cells, NAD+ is synthesized in mitochondria rather than imported from the cytosol.
Significance: Separation of subcellular NAD+ pools may be critical for metabolism in mammalian cells.
Keywords: cell compartmentalization, glycolysis, metabolism, mitochondria, respiration, NAD transport, NMNAT3
Abstract
The mitochondrial NAD pool is particularly important for the maintenance of vital cellular functions. Although at least in some fungi and plants, mitochondrial NAD is imported from the cytosol by carrier proteins, in mammals, the mechanism of how this organellar pool is generated has remained obscure. A transporter mediating NAD import into mammalian mitochondria has not been identified. In contrast, human recombinant NMNAT3 localizes to the mitochondrial matrix and is able to catalyze NAD+ biosynthesis in vitro. However, whether the endogenous NMNAT3 protein is functionally effective at generating NAD+ in mitochondria of intact human cells still remains to be demonstrated. To modulate mitochondrial NAD+ content, we have expressed plant and yeast mitochondrial NAD+ carriers in human cells and observed a profound increase in mitochondrial NAD+. None of the closest human homologs of these carriers had any detectable effect on mitochondrial NAD+ content. Surprisingly, constitutive redistribution of NAD+ from the cytosol to the mitochondria by stable expression of the Arabidopsis thaliana mitochondrial NAD+ transporter NDT2 in HEK293 cells resulted in dramatic growth retardation and a metabolic shift from oxidative phosphorylation to glycolysis, despite the elevated mitochondrial NAD+ levels. These results suggest that a mitochondrial NAD+ transporter, similar to the known one from A. thaliana, is likely absent and could even be harmful in human cells. We provide further support for the alternative possibility, namely intramitochondrial NAD+ synthesis, by demonstrating the presence of endogenous NMNAT3 in the mitochondria of human cells.
Introduction
Mitochondria constitute a unique type of organelle. They are enclosed by a double membrane system and contain their own genome and protein translation machinery. Moreover, mitochondria are essential for oxidative metabolism in eukaryotic cells and represent the major contributor to energy transduction and ATP production. Not surprisingly, mitochondrial dysfunction has been associated with various diseases including metabolic disorders, neurodegenerative diseases, and cancer (1–7). In mitochondria, virtually all metabolic processes depend on NAD. Accordingly, the NAD concentration in these organelles is high and constitutes up to 70% of the total cellular pool (8–10). All major metabolic pathways, including fatty acid catabolism, TCA cycle, and oxidative phosphorylation via the respiratory chain, involve NAD-dependent redox reactions. In addition, mitochondrial NAD+-dependent signaling reactions, most prominently mediated by the protein deacetylase sirtuin 3 and other representatives of the Sirtuin family, play fundamental roles in the regulation of global metabolism (11). Given the degradation of NAD+ in the course of these signaling reactions, the mitochondrial NAD+ pool needs to be constantly replenished. Even though this organellar NAD+ pool is regarded to be largely autonomous, its maintenance seems to be critical not only for mitochondrial homeostasis but also for most other cellular activities (12, 13). It has been reported that depletion of mitochondrial NAD+ may cause cell death. For example, upon activation of the mitochondrial permeability transition pore, mitochondrial NAD+ is depleted and degraded by activated NAD+ catabolic enzymes, resulting in the loss of cellular metabolic integrity and potentially cell death (14–18).
Surprisingly, despite the growing knowledge about NAD-dependent processes in mitochondria, the mechanisms underlying the establishment and maintenance of the mitochondrial NAD pool in mammals have remained largely unknown. In yeast and plants, membrane carriers mediating NAD+ transport across the inner mitochondrial membrane have been identified. In the plant Arabidopsis thaliana, two transporter homologs, AtNDT1 and AtNDT2,3 were shown to exhibit NAD+ transport activity (19). Although AtNDT1 is expressed in chloroplasts, AtNDT2 localizes to mitochondria. Similarly, in the yeast Saccharomyces cerevisiae, the proteins ScNDT1 and ScNDT2 constitute mitochondrial NAD+ transporters (20).
In contrast to yeast and plants, no mitochondrial NAD+ transporter has been identified in humans or other mammals (12, 21, 22). On the other hand, a mammalian isoform of the essential NAD biosynthetic enzyme NMN adenylyl transferase (NMNAT), NMNAT3, has been shown to be targeted to the mitochondrial matrix when expressed from recombinant DNA. Interestingly, such a mitochondrial NMNAT isoform is lacking in S. cerevisiae and A. thaliana. Based on these observations, a mechanism of intramitochondrial NAD+ generation was proposed that involves precursor uptake in form of NMN and further conversion to NAD+ by NMNAT3 (23, 24). However, the presence of endogenous NMNAT3 protein in human mitochondria is still a matter of debate (25). Moreover, how NMN would be imported into mitochondria is also not known, and the possibility that a yet unidentified transporter catalyzes mitochondrial NAD+ import cannot be ruled out. For example, several homologs of the established plant and yeast NAD+ transporters exist in humans. SLC25A32, a member of the mitochondrial solute carrier family SLC25 (22) has recently been described as the mitochondrial folate carrier and is the closest homolog of AtNDT2 and ScNDT1, exhibiting 30 and 35% sequence identity with AtNDT2 and ScNDT1, respectively (12, 21). Two more members of the SLC25 family, SLC25A33 and SLC25A36, display high sequence similarity with SLC25A32 (22). The strong conservation of the SLC25 proteins from yeast to humans and the fact that the predicted substrate binding sites were found almost identical still indicate that a mammalian mitochondrial transporter for a NAD-like substrate might exist (21). Furthermore, SLC25A32, SLC25A33, and SLC25A36 are the only mitochondrial carriers to share a peculiar feature (i.e. the presence of a tryptophan instead of an acidic residue in the second signature motif PX(D/E)XX(K/R) localized at the end of the third transmembrane α-helix) with yeast FLX1, NDT1, NDT2, and RIM2 and A. thaliana carriers AtNDT1 and AtNDT2 (27).
Selective modulation of the mitochondrial NAD+ pool within the organelles is experimentally challenging. For example, pharmacological inhibitors are likely to affect other cellular NAD+ pools as well and may induce acute responses independent of long term alterations of the mitochondrial NAD content (28). Using a transgenic cell-based approach, we have previously shown that a diminished mitochondrial NAD+ pool affects mitochondrial metabolism and leads to accelerate lactate production (29).
We report here the functional heterologous expression of plant and yeast mitochondrial NAD+ transporter in human cells. Both transporters mediated a strong elevation of mitochondrial NAD+ levels, as conveniently monitored by the PARAPLAY-based detection system, which exploits intramitochondrial poly(ADP-ribose) (PAR) formation as readout (23, 30). None of the human SLC25 proteins tested had such an effect. Unexpectedly, constitutive redistribution of NAD+ from the cytosol to mitochondria, established by stable expression of the AtNDT2 NAD+ transporter, not only negatively affected cell proliferation, but also changed the metabolic profile of cells and caused a switch from oxidative metabolism to glycolytic fermentation. Together with the fact that the human candidates most closely related to the plant and yeast NAD+ transporters failed to support mitochondrial NAD+ import, these findings suggest that direct NAD+ import is likely to be absent from human mitochondria. In support of this notion, we provide evidence for intramitochondrial NAD+ biosynthesis in human cells by demonstrating the presence of endogenous mitochondrial NMNAT3.
Experimental Procedures
Chemicals, Reagents, and Media
Unless otherwise specified, all chemicals and reagents were of analytical grade and purchased from Sigma and Merck. Cell culture reagents were from Merck Millipore, Lonza, and ThermoFisher Scientific. FK866 was obtained from the National Institute of Mental Health Chemical Synthesis and Drug Supply Program and Sigma. Monoclonal antibodies mouse anti-FLAG (M2), and mouse anti-β-tubulin were from Sigma, mouse anti-NMNAT3 (D-10) and mouse anti-superoxide dismutase 2 were from Santa Cruz Biotechnology, and mouse anti-PAR (10H) was from Enzo Lifescience. Monoclonal anti-GFP (JL-8) antibody was from Clontech (Takara), and fluorescent-conjugated (Alexa Fluor) secondary antibodies were from Life Technologies. HRP-conjugated goat anti-mouse/goat anti-rabbit antibodies were from Pierce/ThermoFisher Scientific. DNA-modifying and restriction enzymes were purchased from Fermentas and New England Biolabs, and oligonucleotide synthesis was done by Sigma.
Generation of Eukaryotic Expression Vectors
For transient expression of the proteins of interest, the ORFs were amplified from pre-existing plasmids or 293 cDNA and inserted into the pFLAG-CMV-5a plasmid (Sigma) via the EcoRI/BamHI restriction sites. For stable transfection of 293 cells, the ORFs of human SLC25A32 and AtNDT2 were cloned into pcDNA3.1(+) (Life Technologies) via the EcoRI/XbaI restriction sites and with the DNA sequence encoding a C-terminal FLAG epitope included in the primer sequences.
Cell Culture
293 cells were cultivated in Dulbecco's modified Eagle's medium supplemented with 10% (v/v) FCS, 2 mm glutamine, and penicillin/streptomycin. HeLa S3 cells were cultivated in Ham's F12 medium supplemented with 10% FCS, 2 mm glutamine, and penicillin/streptomycin. The cells were cultured at 37 °C in a humidified atmosphere of 5% CO2 (standard culture conditions). Transient transfection of HeLa S3 was performed for 24–48 h using Effectene reagent (Qiagen) according to the manufacturer's recommendations. For the generation of stably transfected cell lines, 293 cells were transfected by the calcium phosphate precipitation method. Monoclonal cell lines were generated after two rounds of selection in the presence of 550 μg/ml G418 and were maintained in complete 293 medium supplemented with 100 μg/ml G418.
Subcellular Fractionation
Mitochondrial and cytosolic fractions of 293 cells were prepared using a mitochondria isolation kit for cultured cells (Thermo Scientific) according to the manufacturer's protocol.
Sequence Analysis
Human relatives of AtNDT2 (Uniprot number Q8RWA5) were identified by BLAST search (Uniprot) configured with the default parameters. Multiple sequence alignment of amino acid sequences was obtained using Jalview 2.8.2 (31).
Cell Viability
Cell proliferation was determined by a resazurin-based viability assay (Sigma). A total of 10.000 cells/well was seeded in the cavities of 96-well plates in 100 μl of medium. The next day, medium was exchanged for fresh medium supplemented with the desired treatment (FK866 (2 μm), NA (100 μm), or respective solvents for controls). After the indicated time points, the medium was exchanged for medium supplemented with 10% (v/v) resazurin and incubated for 2 h at 37 °C. Following incubation, samples were measured fluorometrically (excitation, 540 nm; emission, 590 nm). Samples were measured in triplicate and corrected for background signals from blanks (medium). The data obtained for the first measurement (time point 0 h) were set as 100%, and all subsequent values are reported as relative percentages.
Determination of Medium pH and Lactate Concentration
To measure pH values and lactate concentrations, 225,000 cells were seeded in the cavities of a 6-well plate in 2 ml of medium. After indicated time points, the medium was removed, and the pH was measured with a pH meter. Lactate measurements were performed in triplicate on medium as previously described by Vahsen et al. (32). Prior to measurement, samples were diluted 100 times in water. In parallel, cells from the same samples were washed with PBS and harvested in 200 μl of lysis buffer, and protein determination was carried out to normalize lactate concentration to protein amount.
Protein Determination, SDS-PAGE, and Western Blot Analysis
Cells were washed with PBS and lysed in 20 mm Tris/HCl (pH 7.4), 1 mm EDTA, 2% SDS, and 150 mm NaCl. Genomic DNA was sheared by passage through a syringe with a 23-gauge needle. Protein concentration was determined by BCA protein kit (Pierce). SDS-PAGE and immunoblotting analyses were performed according to standard procedures. Enhanced chemiluminescence (SuperSignal; Pierce) was used for immunodetection. Pictures were taken using the ChemiDoc XRS+ imager (Bio-Rad).
Immunocytochemistry
Cells grown on coverslips were fixed for 45 min with ice-cold 4% (v/v) formaldehyde in PBS and permeabilized for 15 min using 0.5% (v/v) Triton X-100 in PBS. After a blocking step with complete medium for 1 h at room temperature, primary antibody in complete medium was added to cells and incubated overnight at 4 °C. Cells were then washed three times with PBS and once with PBS containing 0.1% (v/v) Triton X-100 before addition of secondary antibody diluted in complete medium and incubation for 1 h at room temperature. Nuclei were subsequently stained with DAPI, and cells were washed once with PBS containing 0.1% (v/v) Triton X-100 and twice with PBS before mounting onto slides. Images were taken using a Leica DMI6000B epifluorescence microscope (Leica Microsystems).
siRNA Knockdown Experiments
Silencer Select NMNAT3 siRNA and control siRNA and transfection reagent Lipofectamine 2000 were purchased from ThermoFisher Scientific. Knockdown efficiency of NMNAT3 siRNA was determined by 1) QRT-PCR analysis and 2) co-transfection of NMNAT3 siRNA along with plasmid encoding FLAG-tagged NMNAT3 followed by FLAG immunoblot analysis. For QRT-PCR analyses, 5 × 105 293 cells were seeded in 6-well plates 24 h before transfection with 100 pmol of siRNA. After 48 h, 5 μg of total RNA, isolated using RNeasy mini kit (Qiagen), were reversely transcribed into cDNA using RevertAid reverse transcriptase (ThermoFisher Scientific). QRT-PCR analyses were performed with a LightCycler® 480 system (Roche) using LightCycler® 480 probes Master Mix (Roche) and predesigned TaqMan gene expression assays for human NMNAT3 and β-actin (ThermoFisher Scientific). For co-transfection experiments, 3 × 105 293 cells were seeded in 12-well plates 1 day before co-transfection with 300 ng of plasmid DNA and 9 pmol of siRNA. After 24 h, cells were lysed and subjected to FLAG immunoblot analysis using 25 μg of total protein. For analyzing the metabolic consequences of down-regulated NMNAT3 gene expression, 1.3 × 106 293 cells were seeded in 6-cm dishes 24 h before transfection with 240 pmol of siRNA. After 2, 4, and 6 days, 1.5 × 106 cells were passaged and transfected with 240 pmol of siRNA upon seeding. One day after the last siRNA transfection, cells were transferred into a 96-well plate and incubated for 24 h prior to oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) determination with a Seahorse XF96 Analyzer.
Measurement of Cellular Glycolytic and Oxygen Consumption Rate
The OCR and ECAR in cultured cells were monitored in a Seahorse XF96 Analyzer (Seahorse Biosciences). Here, the OCR is initially measured under normal conditions to determine the “basal respiration.” The addition of ATP synthase inhibitor oligomycin shows oxygen consumption independent of oxidative phosphorylation (“leak activity”). Maximal respiration (also referred to as “respiratory capacity”) is measured upon addition of the uncoupler CCCP. The “respiratory reserve” of cells is the difference between basal and maximal respiration. Finally, the addition of respiratory chaincomplex I inhibitor rotenone yields complex I-independent respiration, whereas the addition of antimycin A, an inhibitor of respiratory chain complex III, reveals oxygen consumption independent of mitochondrial respiration (nonmitochondrial respiration). The ECAR is measured as basal acidification rate before addition of glucose, which determines glycolysis-dependent ECAR. Oligomycin is added to measure maximal glycolytic activity (“glycolytic capacity”), whereas 2-deoxyglucose is added to inhibit glycolysis and determine nonglycolytic ECAR.
Preparatory analyses were performed to optimize cell number and concentrations of coating agent (poly-l-lysine), carbonyl cyanide m-chlorophenyl hydrazone (CCCP), and oligomycin. The cell culture microplates were coated with 0.01% poly-l-lysine, washed with PBS, and air-dried overnight. The day before analysis, the coated microplates were sterilized by 30 min of exposure to UV light before seeding 30,000 cells/well in 80 μl of growth medium. The cells were incubated overnight at standard culture conditions. On the day of analysis, the growth medium was replaced with sterile filtered unbuffered assay medium (supplemented with 1.85 g/liter NaCl). Mitochondrial respiration and glycolysis were studied in two different assays. For analysis of mitochondrial respiratory function, the assay medium (pH 7.4, unbuffered) was supplemented with 2 mm l-glutamine, 2 mm sodium pyruvate, and 10 mm glucose, whereas the medium (pH 7.35, unbuffered) for glycolysis function measurements was supplied with 2 mm l-glutamine only. Following the addition of assay medium, the microplate was incubated for 1–2 h in a CO2-free incubator (XF Prep Station, Seahorse Biosciences) at 37 °C, to remove CO2 from the plate and medium. The assay reagents were diluted in assay medium. For mitochondrial respiration analysis, the final concentrations were 3 μm oligomycin, 0.5–1 μm CCCP, 1 μm rotenone, and 1 μm antimycin A. For analysis of glycolysis, we used 10 mm glucose, 3 μm oligomycin, and 100 mm 2-deoxyglucose (Seahorse Biosciences). To assess the impact of pyruvate on cellular metabolism, cells were starved for pyruvate for two passages before performing a basic OCR with ranging concentrations of pyruvate or methyl pyruvate (0, 2, or 10 mm in the assay medium.
Following analysis, for normalization to cell number, the cells were fixed in 4% (v/v) formaldehyde solution with 0.5 μg/ml Hoechst 33342. The microplates were stored at 4 °C, before the nuclei were imaged on a BD Pathway 855 Bioimager (BD Biosciences, 3 × 3 montage, 10× objective, excitation filter 380/10 and emission filter 435LP). To count the cells, the images were processed and segmented using the CellProfiler Software. The nuclei were counted in a fixed subregion of each well. Thereafter the nuclei count for each well was divided by the average number from untreated control wells, giving a scaling factor applied to the XF data to correct for the differences in cell number.
HPLC Analysis
Nucleotides were extracted from cell suspensions by the addition of 1 m ice-cold perchloric acid followed by neutralization with 3 m KHCO3. After centrifugation and filtration, supernatants were subjected to HPLC analysis on a Prominence HPLC system (Shimadzu) using a Nucleosil 300–5 C18 column (250 × 4.6 mm, 5mkn, Macherey-Nagel). Solvent A was composed of potassium phosphate buffer (50 mm, pH 5.8) and tetra-n-butyl-ammonium bromide (2 mm). Solvent B consisted of 50% acetonitrile (v/v), potassium phosphate buffer (100 mm, pH 5.8), and 2 mm of tetra-n-butyl-ammonium bromide. Chromatograms were recorded at 258 and 338 nm.
Results
Heterologous Expression of Plant or Yeast Mitochondrial NAD+ Transporters Increases Mitochondrial NAD+ Availability in Human Cells
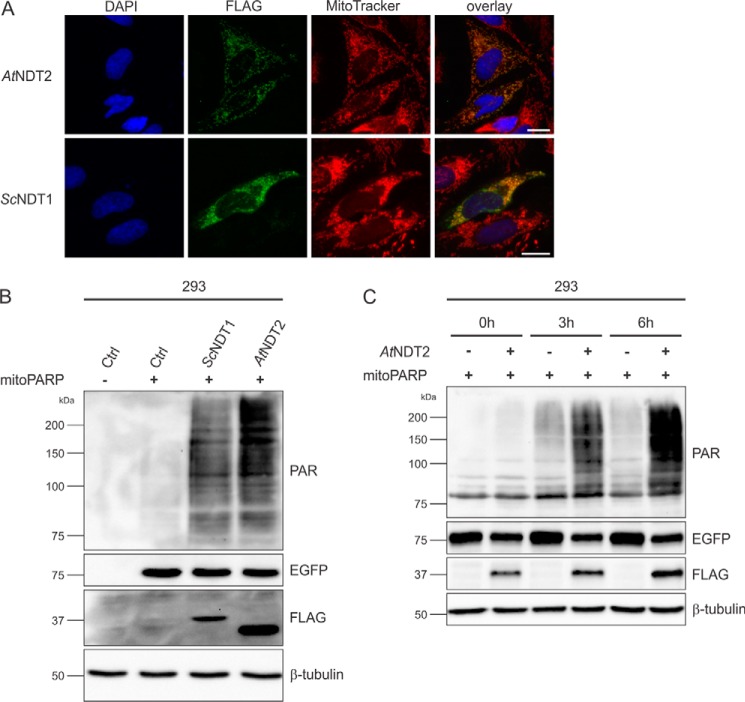
Because the exact mechanism of mitochondrial NAD generation has remained unknown, specific modulation of this pool through influencing NAD biosynthetic pathways is currently not feasible. Based on the recent identification of yeast and plant mitochondrial NAD+ transporters (19, 20), we reasoned that their heterologous expression in human cells could mediate increased NAD+ contents in these organelles. To test this hypothesis, the ORFs encoding the plant AtNDT2 or yeast ScNDT1 NAD+ transporters were expressed in human cells. As shown in Fig. 1A, expression of these transporters resulted in the expected localization of the proteins, as indicated by their co-localization with the mitochondria-specific dye MitoTracker. To determine whether overexpression of these transporters in human cells had an influence on mitochondrial NAD+ contents, we made use of the previously established PARAPLAY system (23, 30). This system is based on the accumulation of immunodetectable PAR mediated by mitoPARP, a mitochondrially targeted fusion protein consisting of an enhanced GFP (EGFP) portion and the catalytic domain of PARP1, which converts mitochondrial NAD+ to PAR in a concentration-dependent manner (23, 29). In HEK293 cells (subsequently referred to as 293 cells) transiently expressing mitoPARP, the PAR signal is readily detectable (Fig. 1B). Importantly, the signal was strongly increased in cells transiently co-expressing either ScNDT1 or AtNDT2 along with mitoPARP (Fig. 1B, two right lanes). These observations established that both NAD+ transporters were expressed as functional proteins and were able to substantially enhance NAD+ availability in mitochondria.
FIGURE 1.
Heterologous expression of mitochondrial NAD+ transporters in human cells increases mitochondrial NAD+ availability. A, established mitochondrial NAD+ transporters from A. thaliana (AtNDT2) and S. cerevisiae (ScNDT1) were expressed with a C-terminal FLAG epitope in HeLa S3 cells and detected by FLAG immunocytochemistry. The images show the nuclei (DAPI) and mitochondrial structures (MitoTracker) and the overexpressed proteins (FLAG). The merged images reveal co-localization of the recombinant NAD+ transporters with mitochondria. Scale bar, 10 μm. B, 293 cells were co-transfected with a vector encoding mitoPARP and plasmids encoding the indicated mitochondrial NAD+ transporters or a control plasmid. Cell lysates were subjected to PAR immunoblot analyses. The intensity of the PAR immunoreactivity reflects the mitochondrial NAD+ content. The expression of the transporters (FLAG) and mitoPARP (EGFP) was confirmed, and β-tubulin served as a loading control. C, 293 cells were co-transfected with a vector encoding mitoPARP and either a vector encoding AtNDT2 or a control plasmid in the presence of the PARP inhibitor 3-aminobenzamide (1 mm). 24 h after transfection, PARP inhibition was released by medium exchange. At the indicated time points, cells were lysed and subjected to immunoblot analysis. The intensity of the PAR immunoreactivity reflects the mitochondrial NAD+ content. The expression of the transporter (FLAG) and mitoPARP (EGFP) was confirmed, and β-tubulin served as a loading control. Ctrl, control.
Notably, the presence of the transporters increased mitochondrial NAD+ contents, even though the NAD+ concentration is already constitutively higher in mitochondria than in the cytosol (8). This is in accordance with findings in yeast cells overexpressing the mitochondrial NAD+ carrier protein ScNDT1, yielding an increased mitochondrial NAD+ content (33). Preincubation of 293 cells, overexpressing mitoPARP and AtNDT2, with the PARP inhibitor 3-aminobenzamide further substantiated that the increased mitochondrial NAD+ influx was dependent on the transporter activity. Upon release of the inhibition, PAR was produced far more readily in cells co-expressing the transporter compared with the cells expressing mitoPARP alone (Fig. 1C). This result indicated that the mitoPARP construct had access to more NAD+ when the plant NAD+ carrier AtNDT2 was expressed. That is, mitochondrial NAD+ content was considerably increased upon overexpression of the carrier.
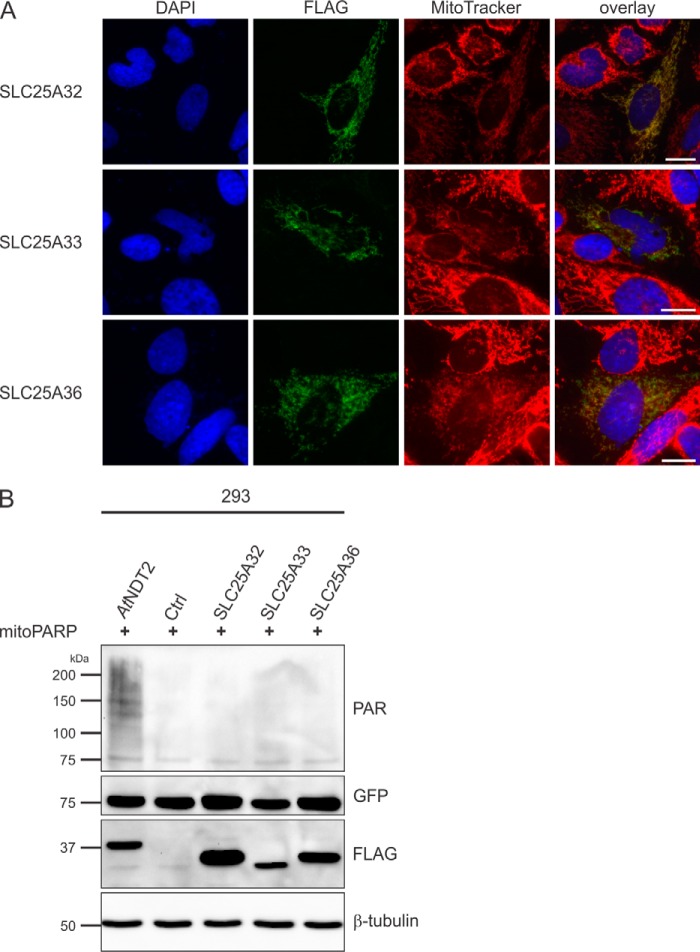
In view of the highly sensitive detection of mitochondrial NAD+ import using the PARAPLAY system, we decided to test whether known or predicted human mitochondrial transporters might mediate mitochondrial NAD+ transport. First, we considered the mitochondrial carrier SLC25A32 (Uniprot number Q9H2D1), which is the closest human relative to ScNDT1 and AtNDT2 (12, 22). SLC25A32 shares ∼30% sequence identity with AtNDT2 (Fig. 2) and was previously described as a human mitochondrial folate transporter (34). According to a recent report, SLC25A32 does not mediate mitochondrial NAD+ import (12). Indeed, overexpressed SLC25A32 localized to the mitochondria (Fig. 3A) but did not affect mitochondrial PAR formation, indicating no influence on mitochondrial NAD+ levels (Fig. 3B). Given this result, we also tested SLC25A33 (Uniprot number Q9BSK2) and SLC25A36 (Uniprot number Q96CQ1), the next two human proteins most closely related to the mitochondrial NAD+ carriers from yeast and plants (Fig. 2). Although SLC25A36 shares ∼30% identity with SL25A32, SLC25A33 in turn is ∼61% identical to SLC25A36 (21). SLC25A33 has previously been described as pyrimidine nucleotide carrier (PNC1) involved in mitochondrial pyrimidine nucleotide import (26), and for SLC25A36 a similar function has been suggested (22). When overexpressed in HeLa S3 cells, the two proteins co-localized with MitoTracker (Fig. 3A). However, when co-expressed with mitoPARP, PAR levels were unchanged compared with control cells, indicating that none of these proteins influenced mitochondrial NAD+ levels (Fig. 3B).
FIGURE 2.
Protein sequence alignment of ScNDT1 and AtNDT2 with closest human relatives. The alignment was carried out using JALVIEW (31). The level of conservation is indicated by shades of blue, with darker blue corresponding to a higher level of conservation.
FIGURE 3.
Human SLC25A32, SLC25A33, and SLC25A36 proteins do not exhibit mitochondrial NAD+ transporter activity. A, HeLa S3 cells were transiently transfected with plasmids encoding C-terminally FLAG-tagged human SLC25A32, SLC25A33, and SLC25A36 proteins. The images show the nuclei (DAPI) and mitochondrial structures (MitoTracker) and the overexpressed proteins (FLAG). As revealed by the merged images, the recombinant proteins co-localize with mitochondria. Scale bar, 10 μm. B, 293 cells were transiently co-transfected with a vector encoding mitoPARP and either control vectors or vectors encoding the indicated transporter. Cell lysates were subjected to PAR immunoblot analyses. The presence of AtNDT2 strongly increases polymer (PAR) formation indicative of elevated mitochondrial NAD+. The expression of the transporters (FLAG) and mitoPARP (EGFP) was confirmed, and β-tubulin served as a loading control. Ctrl, control.
Stable Expression of the Mitochondrial NAD+ Transporter AtNDT2 in 293 Cells Results in Constitutively Increased Mitochondrial NAD+ Availability and Reduced Cell Proliferation
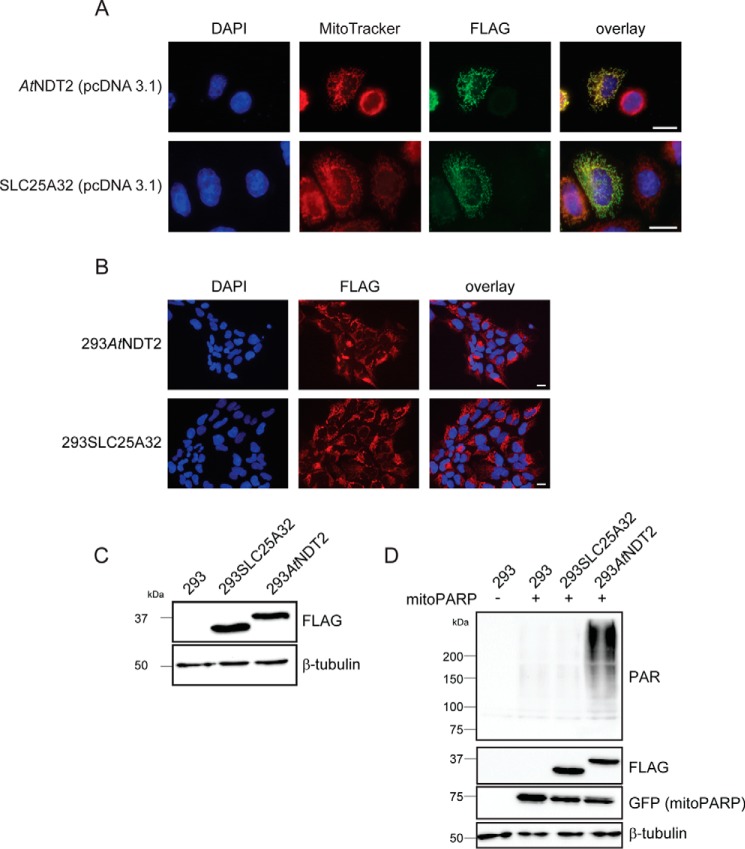
Having established a tool to redistribute cellular NAD+ such that mitochondrial NAD+ levels are elevated, we wished to test whether this change may also have physiological consequences for the cell. We generated stably transfected 293 cells overexpressing the mitochondrial NAD+ transporter AtNDT2 or, as a control cell line, the human SLC25A32 protein, designated 293AtNDT2 and 293SLC25A32 cells, respectively. The ORFs of the two proteins were subcloned into a suitable vector system for stable transfection (see “Experimental Procedures”), which did not affect the subcellular localization of the expressed proteins (Fig. 4A). Monoclonal, stably transfected cell lines were selected, and homogenous expression of the recombinant protein in all cells was confirmed by immunocytochemistry and Western blot analysis (Fig. 4, B and C). To verify that mitochondrial NAD+ was also elevated in stably transfected 293AtNDT2 cells, as would be expected from the transient overexpression (Fig. 1B), the generated cell lines were transfected with the vector encoding mitoPARP. MitoPARP expression led to a strongly increased PAR signal in 293AtNDT2 cells compared with control 293 cells (Fig. 4D), indicating that these cells had a constitutively elevated mitochondrial NAD+ content. On the other hand, PAR formation in mitoPARP-transfected 293SLC25A32 cells was indistinguishable from that observed in untransfected cells (Fig. 4D).
FIGURE 4.
Stable expression of plant NAD+ transporter AtNDT2 in 293 cells constitutively increases the mitochondrial NAD+ content. A, subcellular localization of AtNDT2 and human SCL25A32 in HeLa S3 cells transiently transfected with the vector for stable transfection. AtNDT2 and SLC25A32 are detected by their FLAG epitope, nuclei are stained with DAPI, and mitochondrial structures are stained with MitoTracker. Scale bar, 10 μm. B, fluorescence micrographs of stably transfected monoclonal 293 cells expressing C-terminally FLAG-tagged AtNDT2 (293AtNDT2) or SLC25A32 (293SLC25A32). AtNDT2 and SLC25A32 are detected by their FLAG epitope, and nuclei are stained with DAPI. All cells exhibit immunoreactivity toward the FLAG antibody. Scale bar, 10 μm. C, FLAG immunoblot analysis of 293AtNDT2 and 293SLC25A32 cell lysates. β-Tubulin served as a loading control. D, lysates from parental 293 cells and monoclonal 293AtNDT2 and 293SLC25A32 cells transiently transfected with the vector encoding mitoPARP (detected via its EGFP portion) were subjected to PAR immunoblot analyses. The level of PAR immunoreactivity reflects the mitochondrial NAD+ content. β-Tubulin served as a loading control.
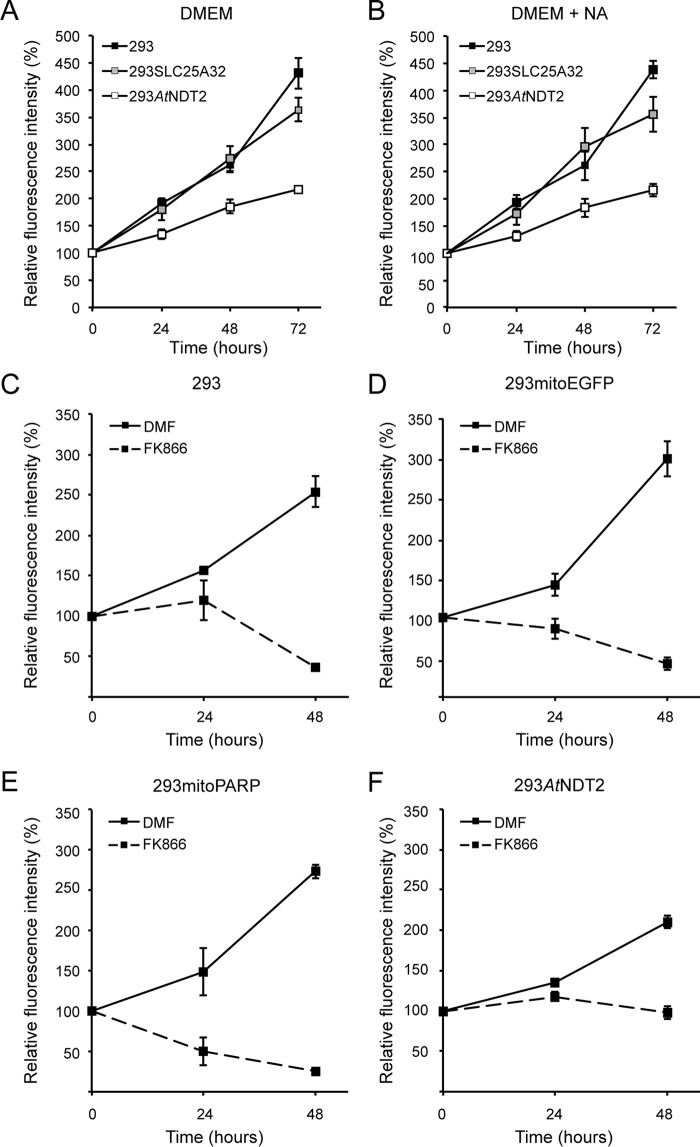
We noticed that 293AtNDT2 cells appeared to proliferate at a slower rate than the control 293 and 293SLC25A32 cells. Quantitative measurement of the cellular growth revealed that the proliferation rate of 293AtNDT2 cells was reduced by as much as ∼50% compared with the control cells (Fig. 5A). This finding was rather surprising, because one would expect a beneficial effect from a robustly maintained mitochondrial NAD+ pool.
FIGURE 5.
Stable expression of AtNDT2 reduces proliferation of 293 cells and attenuates cell death induced by inhibition of NAD biosynthesis. A and B, cell proliferation of 293AtNDT2, 293SLC25A32 and 293 parental cells was measured in the absence (A) and presence (B) of 100 μm NA. The results show the averages of three independent experiments (means ± S.D.). C–F, cell proliferation was measured in the indicated cell lines in the absence and presence of the NamPRT inhibitor FK866 (2 μm) or its solvent DMF. The results show the averages of three independent experiments (means ± S.D.).
293AtNDT2 Cells Are Desensitized toward Inhibition of NAD Biosynthesis
To rule out that the cause for the reduced cell growth of 293AtNDT2 cells was related to a deficiency of the overall cellular NAD supply, we repeated the proliferation experiment in the presence of an alternative NAD precursor, NA. Nicotinamide, present in all standard culture media, is converted to NAD+ via NMN, whereas NA enters the Preiss-Handler pathway (35) and is transformed into NAD+ via nicotinic acid mononucleotide and nicotinic acid adenine dinucleotide. That is, except for the dinucleotide formation step, both pathways proceed independently from one another. Even though NA alone is sufficient to maintain NAD+ levels and cell survival in 293 cells (23, 36, 37), treatment with NA did not improve proliferation of 293AtNDT2 cells, indicating that the low growth rate was not due to insufficient cellular NAD availability (Fig. 5B). We then tested the sensitivity of the cells to FK866, a potent inhibitor of NamPRT, the rate-limiting enzyme of NAD+ biosynthesis from nicotinamide in mammalian cells. Inhibition of NamPRT leads to NAD depletion and subsequent cell death of 293 cells (Fig. 5, C–F) (23). Strikingly, 293AtNDT2 cells were far less sensitive to FK866 treatment than control cells (Fig. 5, C, D, and F). The difference was even more dramatic in comparison to 293mitoPARP cells, which were most sensitive to the induced NAD depletion (Fig. 5E). Taken together, these data suggest that the cellular NAD supply is not impaired in 293AtNDT2 cells. Moreover, the increased mitochondrial NAD+ availability in these cells appears to provide remarkable protection toward FK866-mediated cell death.
Altering Mitochondrial NAD+ Levels Evokes a Metabolic Shift from Oxidative Phosphorylation to Glycolysis
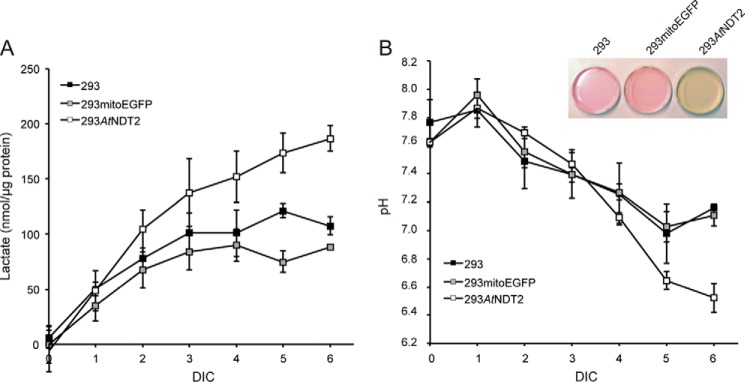
293mitoPARP cells were previously found to display increased medium acidification compared with control 293 cells (29). Unexpectedly, when culturing 293AtNDT2 cells, we also observed an accelerated color shift of the pH indicator phenol red, indicating that medium acidification occurred faster in these cells compared with controls and prompted us to monitor lactate concentration in the cell culture medium. Fig. 6A shows that lactate production in 293AtNDT2 cells was almost two times faster compared with control cells. Accordingly, despite the presence of a buffer, the pH value of the culture medium from 293AtNDT2 cells dropped faster than in control cells (Fig. 6B).
FIGURE 6.
Redistribution of NAD+ to mitochondria of 293AtNDT2 cells leads to increased acidification of the culture medium because of enhanced lactate production. A, the lactate concentration in cell culture supernatants of the indicated cell lines was determined during growth for 6 days in culture (DIC) without medium exchange. The results were normalized to cellular protein concentration and are shown as the averages of three independent experiments (means ± S.D.). B, the pH was measured in the medium collected in A. The results are shown as the averages of three independent experiments (means ± S.D.). The inset shows representative culture dishes of the indicated cell lines after growth for 6 days in culture without medium exchange.
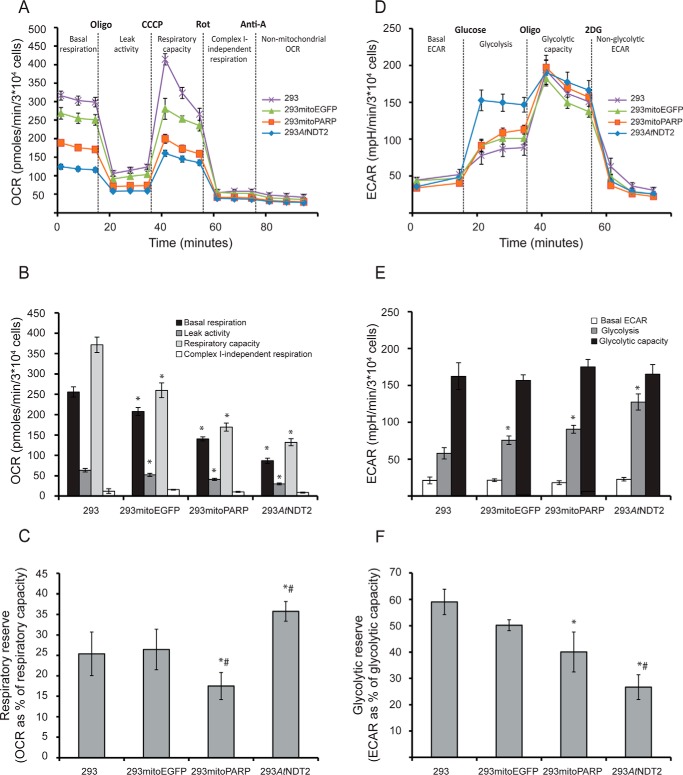
Next, we assessed mitochondrial respiration and glycolytic rate using a Seahorse XF analyzer (see “Experimental Procedures”). Strikingly, the OCR during both basal and maximal (upon the addition of the uncoupler CCCP) respiration was much lower in 293AtNDT2 compared with control cells (Fig. 7, A and B). 293mitoPARP cells showed a similar, but less drastic shift in the OCR. Interestingly, the comparison of basal respiration and respiratory capacity revealed that 293AtNDT2 cells have a slightly higher respiratory reserve (Fig. 7C). As indicated by the oligomycin sensitivity, all cell lines tested appeared to dedicate a similar proportion of basal respiration to ATP production, suggesting that ATP synthase activity is not impaired in 293mitoPARP or 293AtNDT2 cells. However, the differences in oligomycin-sensitive basal respiration indicated that absolute ATP production from oxidative phosphorylation was apparently lower in 293AtNDT2 cells. In addition, measurements of OCR after addition of the complex I inhibitor rotenone was similar in all cell lines, suggesting that complex II did not compensate for reduced respiration. Conversely, measurements of the ECAR, an indicator of the cellular glycolytic rate, showed that 293AtNDT2 cells had a strongly elevated glycolytic rate in comparison with the other investigated cells (Fig. 7, D and E). Because the maximum glycolytic capacity was not altered, these cells used a higher proportion of their glycolytic capacity under basal conditions, leading to a reduced glycolytic reserve (Fig. 7F). Again, 293mitoPARP cells showed a similar trend in these experiments, but to a lesser extent. These results indicate that alterations of mitochondrial NAD+ availability lead to a metabolic shift that favors glycolysis and reduces respiration. Surprisingly, this shift was detected in the two cell lines with opposite alterations of the mitochondrial NAD+ pool, namely increased (293AtNDT2) or decreased (293mitoPARP) mitochondrial NAD+ content. This metabolic shift was also reflected in the nucleotide content of the stably transfected cells (Table 1). Although the overall cellular NAD+ content of 293AtNDT2 cells was increased by ∼50% compared with parental 293 cells, the ATP levels were dramatically reduced to ∼25%. ADP levels were also reduced, although to a lesser extent. Interestingly, NADP+, which is synthesized from NAD+ mainly in the cytosol, was decreased as well. In contrast, AMP levels in these cells were increased. These results further support the shift from a highly efficient ATP-producing metabolic pathway such as respiration to a much less effective one, glycolysis.
FIGURE 7.
Subcellular distribution of cellular NAD+ determines the metabolic fate of cells. The activity and integrity of glycolysis and mitochondrial respiration in 293AtNDT2 cells were determined by extracellular flux analysis and compared with parental 293 cells and 293 cells stably expressing mitochondrial EGFP (293mitoEGFP) or the mitoPARP construct (293mitoPARP). A, mitochondrial respiration was assessed by measuring OCR after sequential addition of oligomycin (Oligo, 3 μm), CCCP (0.5–1 μm), rotenone (Rot, 1 μm), and antimycin A (Anti-A, 1 μm). The figure shows representative data from one of four experiments. B, quantification of the mitochondrial respiration data obtained in A. The rate determined after the addition of antimycin A was subtracted as background. The results are shown as the averages of four independent experiments (means ± S.D.). *, p ≤ 0.001 versus 293. C, respiratory reserve capacity (respiratory capacity − basal respiration) calculated as percentages of respiratory capacity. The results are shown as averages from four independent experiments (means ± S.D.). *, p < 0.05 versus 293; #, p < 0.05 versus 293mitoEGFP. D, glycolysis was studied by analyzing ECAR after sequential additions of glucose (10 mm), oligomycin (Oligo, 3 μm), and 2-deoxyglucose (2DG, 100 mm). The figure shows representative data from one of four experiments. E, quantification of the glycolysis data obtained in C (means ± S.D.). The rate determined after addition of 2-deoxyglucose was subtracted as background. *, p ≤ 0.001 versus 293. F, the glycolytic reserve capacity (glycolytic capacity − basal glycolysis) was calculated as percentage of glycolytic capacity. The figure shows data (means ± S.D.) from three independent experiments. *, p < 0.05 versus 293; #, p < 0.05 versus mitoEGFP.
TABLE 1.
Relative nucleotide content of 293 parental and 293AtNDT2 cells
| Relative nucleotide content |
|||||
|---|---|---|---|---|---|
| NAD+ | ATP | ADP | AMP | NADP+ | |
| % | |||||
| 293 | 100 ± 1.1 | 100 ± 3.3 | 100 ± 3.4 | 100 ± 4.6 | 100 ± 6.9 |
| 293AtNDT2 | 151.7 ± 5.5 | 25.7 ± 12.8 | 74.8 ± 4.5 | 125.9 ± 3.8 | 30.4 ± 7.6 |
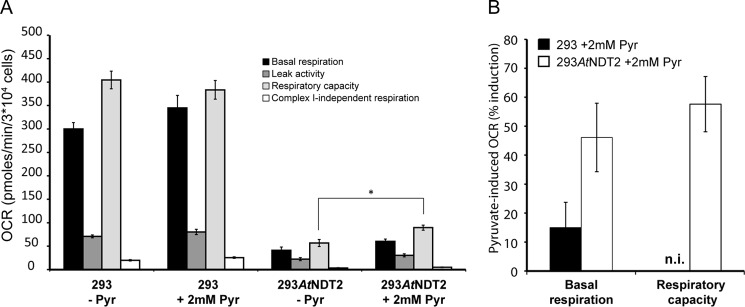
One possible reason for this metabolic shift in 293AtNDT2 cells could be the lack of supply of glycolytic product pyruvate for mitochondrial respiration. We therefore repeated the ECAR and OCR analyses in the absence and presence of pyruvate. Indeed, supplementation with 2 mm pyruvate moderately but significantly increased mitochondrial respiration in 293AtNDT2 cells (Fig. 8A). Both basal respiration and respiratory capacity (maximal respiration) were increased in 293AtNDT2 cells, whereas 293 cells only showed a slightly elevated basal respiration rate (Fig. 8B). Similar results were also obtained by supplementation with higher concentrations of pyruvate (up to 10 mm) or methyl pyruvate (data not shown).
FIGURE 8.
Pyruvate supplementation partially restores mitochondrial respiration in 293AtNDT2 cells. A, mitochondrial respiration was assessed by measuring OCR in 293 and 293AtNDT2 cells incubated in the absence (−Pyr) or presence (+2 mm Pyr) of pyruvate, after pyruvate starvation. The figure shows results from a representative experiment. *, p < 0.05. B, quantification of the relative pyruvate-dependent induction of basal and maximal respiration (respiratory capacity) in 293 and 293AtNDT2 cells. n.i., no induction.
The Presence of NMNAT3 in Mammalian Mitochondria Suggests Autonomous, NAD+ Carrier-independent Generation of the Mitochondrial NAD+ Pool
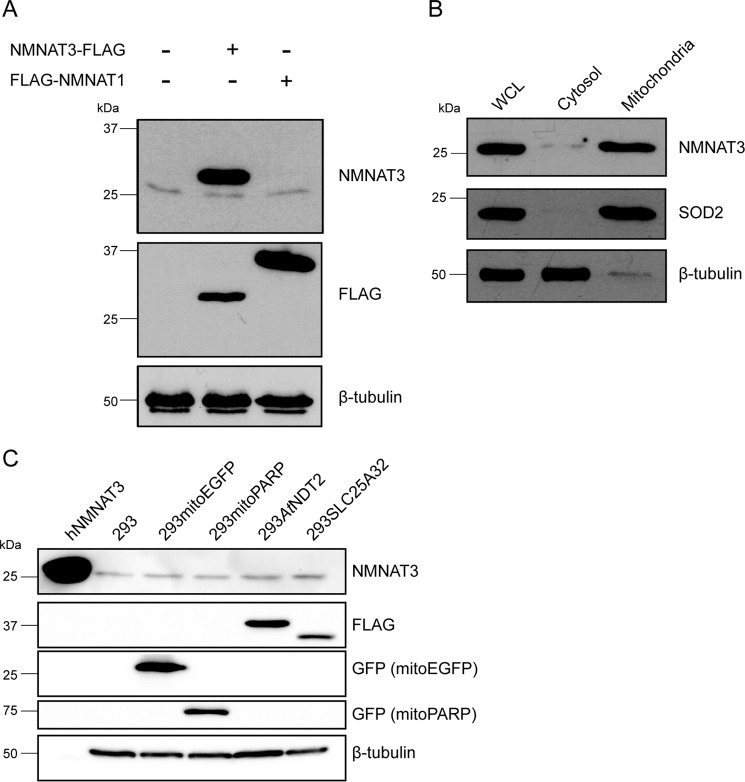
A mammalian mitochondrial NAD+ transporter has not been identified (12, 21, 22). In fact, the data presented here indicated that the presence of a functional mitochondrial NAD+ transporter, similar to those found in yeast and plants, could actually be harmful for human cells. Nonetheless, given that in mammalian mitochondria virtually all processes are dependent on, associated with, or regulated by NAD, the nucleotide needs to be continuously supplied to these organelles. Therefore, we wished to verify whether intramitochondrial biosynthesis of NAD might be mediated by endogenous NMNAT3. We decided to apply a newly available monoclonal NMNAT3 antibody (D-10; Santa Cruz Biotechnology). The immunoblot analysis revealed a single band, which migrated at a position corresponding to a molecular mass of ∼28 kDa (Fig. 9A), a value expected from the calculation of the theoretical molecular mass of endogenous NMNAT3 residing in the mitochondrial matrix.
FIGURE 9.
NMNAT3 is present in mitochondria of human cells, and its expression level is unaffected by changes in the mitochondrial NAD+ content. A, Western blot detection of endogenous and overexpressed NMNAT3 in 293 cells with a monoclonal NMNAT3-specific antibody. The monoclonal antibody detected both the endogenous protein and the overexpressed FLAG-tagged recombinant NMNAT3. No cross-reactivity of the monoclonal antibody with NMNAT1 was observed. β-Tubulin served as a loading control. B, whole cell lysate (WCL), cytosolic and mitochondrial fractions from 293 cells were subjected to immunoblot analysis using the monoclonal NMNAT3-specific antibody. Superoxide dismutase (SOD2) served as a control for the purity of mitochondrial fraction, whereas β-tubulin served as a control for the cytosolic fraction. C, detection of endogenous NMNAT3 in lysates from 293 cell lines with altered mitochondrial NAD+ availability (293mitoPARP and 293AtNDT2), as well as control cells (293mitoEGFP and 293SLC25A32 and parental 293 cells). Lysates were subjected to immunoblot analysis using the NMNAT3-specific antibody. β-Tubulin served as a loading control. hNMNAT3, bacterially expressed, purified His6-tagged human NMNAT3 (47).
To assess the specificity of the antibody, we performed immunoblot analyses using recombinant FLAG-tagged human NMNAT3 and NMNAT1. As shown in Fig. 9A, the antibody was highly selective for NMNAT3 and did not recognize NMNAT1. Subcellular fractionation experiments confirmed that endogenous NMNAT3 was exclusively present in the mitochondrial fraction (Fig. 9B). Interestingly, the expression level of NMNAT3 protein seemed unaffected by changes in the mitochondrial NAD+ content. Cell lysates from both 293AtNDT2 (elevated mitochondrial NAD+ levels) and 293mitoPARP cells (lowered mitochondrial NAD+ levels) showed protein levels for NMNAT3 similar to those found in the lysates from control cell lines (Fig. 9C).
We reasoned that should the mitochondrial NAD pool depend on NMNAT3 activity, a reduction of NMNAT3 expression may affect the metabolic phenotype of the cells. To test this, we reduced NMNAT3 expression by a siRNA-mediated knockdown approach.
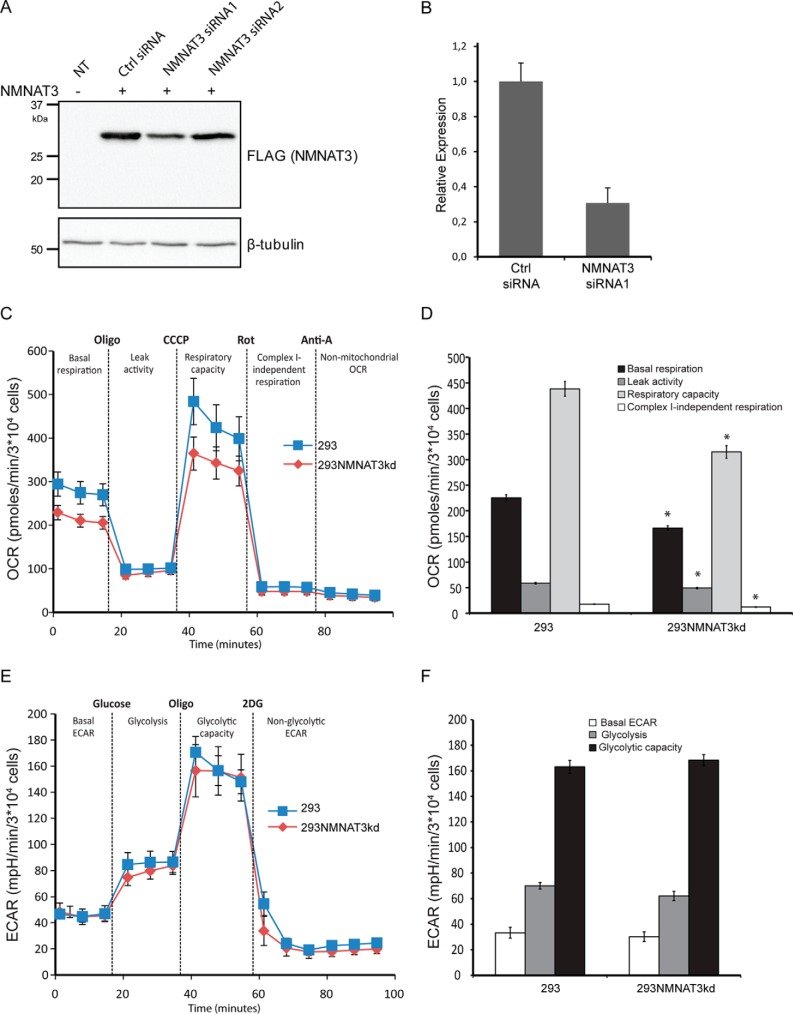
The knockdown capacity of two NMNAT3 siRNAs was verified by FLAG immunoblot analysis 24 h after transfection of siRNA along with plasmid encoding FLAG-tagged NMNAT3 (Fig. 10A) and by the relative quantification of endogenous NMNAT3 transcript levels by QRT-PCR in 293 cells after 48 h of siRNA transfection (Fig. 10B). The presence of NMNAT3 siRNA1 reduced NMNAT3 expression to ∼40% (Fig. 10A) and 30% (Fig. 10B), whereas siRNA2 was not effective. Next, to ensure maximal efficiency of the knockdown, NMNAT3 siRNA1 was used to transiently transfect parental 293 cells four consecutive times over 8 days with 48-h intervals. Subsequent metabolic analysis of these NMNAT3 knockdown cells using the Seahorse Analyzer showed a reduction in mitochondrial respiration similar to that observed in 293mitoPARP cells (Fig. 10, C and D). There was, however, no significant effect on the glycolytic rate of these cells (Fig. 10, E and F).
FIGURE 10.
Down-regulation of NMNAT3 gene expression in 293 cells reduces mitochondrial respiration. A, FLAG immunoblot analysis of 293 cell lysates prepared after 24 h of expression of plasmid encoding C-terminally FLAG-tagged NMNAT3 in the presence of two distinct NMNAT3 siRNAs and control siRNA. NT, lysates from untransfected 293 cells. B, relative NMNAT3 gene expression in 293 cells after 48 h of transfection of NMNAT3 siRNA1 and control siRNA (Ctrl) as revealed by QRT-PCR. The NMNAT3 transcript level was normalized to β-actin. C, mitochondrial respiration assessed by measuring OCR in 293 cells transfected with NMNAT3 siRNA1 (293NMNAT3kd) and control siRNA (293). D, quantification of the mitochondrial respiration data obtained in C. *, p < 0.05 E, glycolysis was analyzed by ECAR in 293 cells transfected with NMNAT3 siRNA1 (293NMNATkd) and control siRNA (293). F, quantification of the glycolysis data obtained in E.
Taken together, these observations demonstrate that NMNAT3 is present within mitochondria of all human cell lines used in this study and that under physiological conditions, the mitochondrial NAD+ pool is likely to be maintained by autonomous NAD+ generation rather than import from the cytosol.
Discussion
In the present study, we demonstrate the possibility of redistributing cellular NAD+ from the cytosol into mitochondria by heterologous overexpression of yeast or plant NAD+ transporters in human cells. Constitutive redistribution of the cellular NAD+ pool from the cytosol to mitochondria, achieved by stable expression of the plant NAD+ transporter AtNDT2, results in lower sensitivity toward NAD+ biosynthesis inhibition, but surprisingly, also in slower cell proliferation and a metabolic shift from respiration toward glycolytic fermentation. Overexpression of human proteins most closely related to the yeast and plant NAD+ transporters has no detectable effect on mitochondrial NAD+ content. We further substantiate the hypothesis of intramitochondrial NAD+ biosynthesis upon uptake of cytosolic NMN, rather than NAD+ import, by providing evidence of the presence of NMNAT3 in human mitochondria, a protein known to catalyze NAD+ synthesis from NMN and ATP in vitro.
Modulation of Mitochondrial NAD+ Levels
In a previous study, it was noted that 293mitoPARP cells have a constitutively lower NAD+ content, because of the mitochondrial NAD+ consumption introduced by the mitoPARP construct (29). 293mitoPARP cells exhibited a higher lactate production rate and lowered mitochondrial membrane potential but did not have any obvious deficiencies. Here, we succeeded in establishing a model system that is characterized by a considerable increase of mitochondrial NAD+ levels. Expression of the yeast and plant NAD+ transporters led to strongly increased mitochondrial PAR formation as detected by the PARAPLAY reporter assay visualizing the availability of intramitochondrial NAD+ far beyond the level of control cells. The fact that the transporters presumably work against a concentration gradient raises the question of the driving force, such as the possibility of a counter exchange with a gradient in the opposite direction. Segregated NAD+ pools have also been demonstrated in other organelles such as peroxisomes, the Golgi apparatus, and endoplasmic reticulum (30). So far, only for human peroxisomes, a membrane protein with described NAD+ transporter activity could be identified (38). However, whether modulations of NAD+ within these organelles by similar mechanisms are possible remains to be investigated.
The Presence of NMNAT3 in Mitochondria Suggests Autonomous NAD+ Synthesis within These Organelles
Given that previous studies have failed to identify and describe a mitochondrial transporter for NAD+ or its precursor NMN in mammalian cells, the question of how the mitochondrial NAD+ pool is established has remained unanswered. When we overexpressed the closest human relatives to the plant and yeast NAD+ transporter proteins, SLC25A32, SLC25A33, and SLC25A36, we did not detect any increase in mitochondrial PAR formation, which indicated that these proteins do not exhibit NAD+ transporter activity. These observations are in accordance with a previous study showing that SLC25A32 does not influence mitochondrial NAD+ (12). Moreover, during the preparation of this manuscript, an in-depth characterization of SLC25A33 and SLC25A36 was published that identified these two proteins as (deoxy-) pyrimidine nucleotide transporters essential for mitochondrial DNA homeostasis and also confirmed their lack of NAD+ transport activity (39).
Although all these observations seem to argue against a direct import of NAD+ into mammalian mitochondria, autonomous intramitochondrial NAD+ biosynthesis would at least require NMNAT activity to form the dinucleotide from NMN and ATP. Recombinantly expressed human NMNAT3 has been localized to the mitochondrial matrix (23). However, whether endogenous NMNAT3 is indeed present in human cells has been disputed because of the lack of experimental evidence. So far, the presence of endogenous NMNAT3 protein has only been verified in human and mouse red blood cells (40, 41), as well as in rat neurons (42). In this regard, our demonstration that endogenous NMNAT3 is indeed present in mitochondria of human cells represents an important step toward the mechanistic understanding of human NAD metabolism. Moreover, NMNAT3 knockdown in 293 cells led to a reduction of mitochondrial respiration, indicating that NMNAT3 is not only present but functional in human mitochondria. Collectively, these results support the conclusion that the mitochondrial NAD+ pool in mammalian cells is likely established by autonomous synthesis, which involves NMNAT3.
Metabolic Consequences of Altered Mitochondrial NAD+ Levels
A surprising finding of the present study is the shift from oxidative phosphorylation to glycolysis in human cells stably expressing AtNDT2 and the ensuing suppression of cell growth. This metabolic change was evident from both monitoring medium lactate levels, the nucleotide content of the cells and, in more detail, measuring oxygen consumption and glycolytic rates.
In principle, these alterations could suggest an impairment of the oxidative phosphorylation system. For example, the lowered NADP content might cause a decreased capacity to counteract oxidative damage to the respiratory chain. However, the presence of the transporter AtNDT2 is also likely to result in a decrease of cytosolic NAD+, because of the redistribution of the dinucleotide from the cytosol into the mitochondria. Under conditions of a diminished cytosolic NAD+ pool (note that AtNDT2 transports NAD+ rather than NADH (19)), maintenance of glycolysis would require a higher rate of NADH reoxidation. This regeneration of NAD+ is catalyzed by lactate dehydrogenase and would lead to increased lactate production as measured in these cells. Another intriguing aspect of such a mechanism would be the diversion of pyruvate into lactate production rather than being used as substrate for mitochondrial respiration. In line with such a potential mechanism, recent findings in mammalian cells showed that DNA damage-mediated activation of PARP1 leads to inhibition of glycolysis and subsequent mitochondrial respiratory failure (43, 44). Importantly, this phenotype could be reversed by the addition of pyruvate to the culture medium, indicating that impaired mitochondrial function was caused by stalling glycolysis and reduced availability of pyruvate produced by glycolysis. High supplement of pyruvate increased respiration in 293AtNDT2 cells (Fig. 8), indicating that such a mechanism contributes to the metabolic phenotype in our system as well. However, because no full restoration of mitochondrial respiration was achieved, there may be other factors involved. For example, the countersubstrates for NAD+ import, which are preferably ADP and AMP for the AtNDT2 transporter, could accumulate in the cytosol and mediate an increase in glycolysis. However, this would not explain the dramatic changes of mitochondrial respiration, especially for the maximal respiration in the presence of the uncoupler CCCP.
Moreover, increased mitochondrial NAD+ could potentially change acetylation patterns because of altered activity of mitochondrial sirtuins and thus influence reactions for example involved in the Krebs cycle. Preliminary results indicate that the global acetylation state of 293AtNDT2 cells is not changed (data not shown). Whether specific sirtuin targets, both mitochondrial and cytosolic/nuclear, are affected by the NAD+ redistribution remains to be elucidated.
Strikingly, 293mitoPARP cells, although seemingly opposite in terms of mitochondrial NAD+ modulation displayed the same metabolic shift. A plausible explanation for the reduced oxidative phosphorylation in this cell line appears to be the limited availability of mitochondrial NAD+. This is supported by the NMNAT3 knockdown, which changed mitochondrial respiration of parental 293 cells similar to that of the 293mitoPARP cells (Fig. 10). In turn, the deficiency in mitochondrial respiration is compensated by the observed increase of the glycolytic rate and lactate production in 293mitoPARP cells.
Interestingly, similar observations have been made in yeast cells (33). Here, upon deletion as well as overexpression of mitochondrial NAD+ carriers, ATP production was lowered, and under conditions where cellular metabolism was fully respiratory, both mutant strains showed a low biomass yield, indicative of impaired energetic efficiency (33).
Even though both 293AtNDT2 and 293mitoPARP cells accelerate glycolysis to cope with the deficient oxidative phosphorylation, the difference in their proliferation is striking. Possibly, the permanent drainage of NAD+ from the cytosol by expression of AtNDT2 lowers the capacity to produce acetyl-CoA from pyruvate to an extent that severely affects cell growth and proliferation. The moderate but significant rescue effect of pyruvate supplementation on the respiratory rate of 293AtNDT2 cells in our experiments supports this possibility. On the other hand, the continuous mitochondrial NAD+ consumption in the 293mitoPARP cells can be sufficiently compensated by enhanced glycolysis to maintain the proliferation rate. The different consequences of opposing alterations in mitochondrial NAD+ levels evoked in 293AtNDT2 and 293mitoPARP cells are also reflected by the disparate responses to inhibition of NAD+ synthesis by treatment with the NamPRT inhibitor FK866. Although increased mitochondrial NAD+ turnover in 293mitoPARP cells seems to make the cells even more vulnerable to FK866 treatment, mitochondrial accumulation of NAD+ in 293AtNDT2 cells seems to be protective. The slow proliferation rate of these cells might be accompanied by a lower NAD+ consumption enabling longer survival in the presence of FK866. Indeed, it has been previously reported that decreased NAD+ consumption by inhibition of PARP1 resulted in prolonged cell survival in the presence of FK866 (13, 23, 45).
Taken together, our data show that dramatic changes in the distribution of cellular NAD+ have deleterious effects on major metabolic pathways. Considering our demonstration of the presence of the NAD+ biosynthetic enzyme NMNAT3 within mitochondria, we reason that a mitochondrial NAD+ transporter may not only be unnecessary but might in fact be harmful to human cells. Because of the deleterious effects observed upon expression of AtNDT2, we would predict that, if a human mitochondrial NAD+ carrier exists, it would have very different properties (e.g. structural, counter substrate, kinetic behavior, etc.), and its expression would have to be low and highly regulated to avoid a similar phenotype as observed in 293AtNDT2 cells. This would also explain the difficulties encountered in its identification. Furthermore, it is worth noting that yeast and A. thaliana express mitochondrial NAD+ transporters but lack an NMNAT3 homolog. Conversely, for the Jerusalem artichoke (Helianthus tuberosus L.) a mitochondrial NMNAT isoform has been described (46), whereas we were unable to find a gene encoding a putative mitochondrial NAD+ transporter homolog in a BLAST search. Therefore, it would appear that mitochondrial NAD+ import and autonomous NAD synthesis are two independently occurring mechanisms to maintain this important organellar pool, which needs to be accurately regulated to obtain an optimal bioenergetic efficiency.
Author Contributions
M. R. V. L. and C. D. conceived and coordinated the study, designed and carried out experiments, and wrote the manuscript. I. K. N. P., V. A. K., and M. N. carried out experiments and drafted the manuscript. G. A. and F. P. contributed to the initiation of the study and drafted the manuscript. S. E. D. carried out experiments. A. A. N. and K. J. T. contributed to drafting the manuscript. M. Z. conceived and coordinated the study, designed experiments and wrote the manuscript. All authors analyzed data and reviewed and refined the manuscript for important intellectual content.
Acknowledgments
We thank Mirko Muzzi for the contribution to preliminary experimental data and Fedor Kryuchkov for assistance with HPLC analysis.
This work was supported by Norwegian Research Council Grant NFR 214435/F20, the Norwegian Cancer Society, Russian Foundation for Basic Research Grants 14-04-01765a and 14-04-32117 mol_a, and Italian Human ProteomeNet Grant RBRN07BMCT_009. The authors declare that they have no conflicts of interest with the contents of this article.
- NDT
- nicotinamide adenine dinucleotide transporter
- NMNAT
- nicotinamide mononucleotide adenylyl transferase
- NMN
- nicotinamide mononucleotide
- NA
- nicotinic acid
- NamPRT
- nicotinamide phosphoribosyl transferase
- PAR
- poly(ADP-ribose)
- PARP
- poly(ADP-ribose) polymerase
- OCR
- oxygen consumption rate
- ECAR
- extracellular acidification rate
- qRT-PCR
- quantitative RT-PCR
- CCCP
- carbonyl cyanide m-chlorophenyl hydrazone.
References
- 1.Houtkooper R. H., Cantó C., Wanders R. J., and Auwerx J. (2010) The secret life of NAD+: an old metabolite controlling new metabolic signaling pathways. Endocr. Rev. 31, 194–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Koch-Nolte F., Haag F., Guse A. H., Lund F., and Ziegler M. (2009) Emerging roles of NAD+ and its metabolites in cell signaling. Sci. Signal. 2, mr1. [DOI] [PubMed] [Google Scholar]
- 3.Reddy P. H., and Reddy T. P. (2011) Mitochondria as a therapeutic target for aging and neurodegenerative diseases. Curr. Alzheimer Res. 8, 393–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Szendroedi J., Phielix E., and Roden M. (2012) The role of mitochondria in insulin resistance and type 2 diabetes mellitus. Nat. Rev. Endocrinol. 8, 92–103 [DOI] [PubMed] [Google Scholar]
- 5.Wallace D. C. (2012) Mitochondria and cancer. Nat. Rev. Cancer 12, 685–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Palmieri F. (2008) Diseases caused by defects of mitochondrial carriers: a review. Biochim. Biophys. Acta 1777, 564–578 [DOI] [PubMed] [Google Scholar]
- 7.Palmieri F. (2014) Mitochondrial transporters of the SLC25 family and associated diseases: a review. J. Inherit Metab. Dis. 37, 565–575 [DOI] [PubMed] [Google Scholar]
- 8.Di Lisa F. (2001) Mitochondrial contribution in the progression of cardiac ischemic injury. IUBMB Life 52, 255–261 [DOI] [PubMed] [Google Scholar]
- 9.Alano C. C., Tran A., Tao R., Ying W., Karliner J. S., and Swanson R. A. (2007) Differences among cell types in NAD+ compartmentalization: a comparison of neurons, astrocytes, and cardiac myocytes. J. Neurosci. Res. 85, 3378–3385 [DOI] [PubMed] [Google Scholar]
- 10.Stein L. R., and Imai S. (2012) The dynamic regulation of NAD metabolism in mitochondria. Trends Endocrinol. Metab. 23, 420–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dölle C., Rack J. G., and Ziegler M. (2013) NAD and ADP-ribose metabolism in mitochondria. FEBS J. 280, 3530–3541 [DOI] [PubMed] [Google Scholar]
- 12.Yang H., Yang T., Baur J. A., Perez E., Matsui T., Carmona J. J., Lamming D. W., Souza-Pinto N. C., Bohr V. A., Rosenzweig A., de Cabo R., Sauve A. A., and Sinclair D. A. (2007) Nutrient-sensitive mitochondrial NAD+ levels dictate cell survival. Cell 130, 1095–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pittelli M., Formentini L., Faraco G., Lapucci A., Rapizzi E., Cialdai F., Romano G., Moneti G., Moroni F., and Chiarugi A. (2010) Inhibition of nicotinamide phosphoribosyltransferase: cellular bioenergetics reveals a mitochondrial insensitive NAD pool. J. Biol. Chem. 285, 34106–34114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pieper A. A., Verma A., Zhang J., and Snyder S. H. (1999) Poly (ADP-ribose) polymerase, nitric oxide and cell death. Trends Pharmacol. Sci. 20, 171–181 [DOI] [PubMed] [Google Scholar]
- 15.Chiarugi A., and Moskowitz M. A. (2002) Cell biology. PARP-1: a perpetrator of apoptotic cell death? Science 297, 200–201 [DOI] [PubMed] [Google Scholar]
- 16.Alano C. C., Ying W., and Swanson R. A. (2004) Poly(ADP-ribose) polymerase-1-mediated cell death in astrocytes requires NAD+ depletion and mitochondrial permeability transition. J. Biol. Chem. 279, 18895–18902 [DOI] [PubMed] [Google Scholar]
- 17.Alano C. C., Garnier P., Ying W., Higashi Y., Kauppinen T. M., and Swanson R. A. (2010) NAD+ depletion is necessary and sufficient for poly(ADP-ribose) polymerase-1-mediated neuronal death. J. Neurosci. 30, 2967–2978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kristian T., Balan I., Schuh R., and Onken M. (2011) Mitochondrial dysfunction and nicotinamide dinucleotide catabolism as mechanisms of cell death and promising targets for neuroprotection. J. Neurosci. Res. 89, 1946–1955 [DOI] [PubMed] [Google Scholar]
- 19.Palmieri F., Rieder B., Ventrella A., Blanco E., Do P. T., Nunes-Nesi A., Trauth A. U., Fiermonte G., Tjaden J., Agrimi G., Kirchberger S., Paradies E., Fernie A. R., and Neuhaus H. E. (2009) Molecular identification and functional characterization of Arabidopsis thaliana mitochondrial and chloroplastic NAD+ carrier proteins. J. Biol. Chem. 284, 31249–31259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Todisco S., Agrimi G., Castegna A., and Palmieri F. (2006) Identification of the mitochondrial NAD+ transporter in Saccharomyces cerevisiae. J. Biol. Chem. 281, 1524–1531 [DOI] [PubMed] [Google Scholar]
- 21.Haitina T., Lindblom J., Renström T., and Fredriksson R. (2006) Fourteen novel human members of mitochondrial solute carrier family 25 (SLC25) widely expressed in the central nervous system. Genomics 88, 779–790 [DOI] [PubMed] [Google Scholar]
- 22.Palmieri F. (2013) The mitochondrial transporter family SLC25: identification, properties and physiopathology. Mol. Aspects Med. 34, 465–484 [DOI] [PubMed] [Google Scholar]
- 23.Nikiforov A., Dölle C., Niere M., and Ziegler M. (2011) Pathways and subcellular compartmentation of NAD biosynthesis in human cells: from entry of extracellular precursors to mitochondrial NAD generation. J. Biol. Chem. 286, 21767–21778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barile M., Passarella S., Danese G., and Quagliariello E. (1996) Rat liver mitochondria can synthesize nicotinamide adenine dinucleotide from nicotinamide mononucleotide and ATP via a putative matrix nicotinamide mononucleotide adenylyltransferase. Biochem. Mol. Biol. Int. 38, 297–306 [PubMed] [Google Scholar]
- 25.Felici R., Lapucci A., Ramazzotti M., and Chiarugi A. (2013) Insight into molecular and functional properties of NMNAT3 reveals new hints of NAD homeostasis within human mitochondria. PLoS One 8, e76938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Floyd S., Favre C., Lasorsa F. M., Leahy M., Trigiante G., Stroebel P., Marx A., Loughran G., O'Callaghan K., Marobbio C. M., Slotboom D. J., Kunji E. R., Palmieri F., and O'Connor R. (2007) The insulin-like growth factor-I-mTOR signaling pathway induces the mitochondrial pyrimidine nucleotide carrier to promote cell growth. Mol. Biol. Cell 18, 3545–3555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cappello A. R., Miniero D. V., Curcio R., Ludovico A., Daddabbo L., Stipani I., Robinson A. J., Kunji E. R., and Palmieri F. (2007) Functional and structural role of amino acid residues in the odd-numbered transmembrane α-helices of the bovine mitochondrial oxoglutarate carrier. J. Mol. Biol. 369, 400–412 [DOI] [PubMed] [Google Scholar]
- 28.Billington R. A., Genazzani A. A., Travelli C., and Condorelli F. (2008) NAD depletion by FK866 induces autophagy. Autophagy 4, 385–387 [DOI] [PubMed] [Google Scholar]
- 29.Niere M., Kernstock S., Koch-Nolte F., and Ziegler M. (2008) Functional localization of two poly(ADP-ribose)-degrading enzymes to the mitochondrial matrix. Mol. Cell. Biol. 28, 814–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dölle C., Niere M., Lohndal E., and Ziegler M. (2010) Visualization of subcellular NAD pools and intra-organellar protein localization by poly-ADP-ribose formation. Cell. Mol. Life Sci. 67, 433–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Waterhouse A. M., Procter J. B., Martin D. M., Clamp M., and Barton G. J. (2009) Jalview version 2: a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vahsen N., Candé C., Brière J. J., Bénit P., Joza N., Larochette N., Mastroberardino P. G., Pequignot M. O., Casares N., Lazar V., Feraud O., Debili N., Wissing S., Engelhardt S., Madeo F., Piacentini M., Penninger J. M., Schägger H., Rustin P., and Kroemer G. (2004) AIF deficiency compromises oxidative phosphorylation. EMBO J. 23, 4679–4689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Agrimi G., Brambilla L., Frascotti G., Pisano I., Porro D., Vai M., and Palmieri L. (2011) Deletion or overexpression of mitochondrial NAD+ carriers in Saccharomyces cerevisiae alters cellular NAD and ATP contents and affects mitochondrial metabolism and the rate of glycolysis. Appl. Environ. Microbiol. 77, 2239–2246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Titus S. A., and Moran R. G. (2000) Retrovirally mediated complementation of the glyB phenotype: cloning of a human gene encoding the carrier for entry of folates into mitochondria. J. Biol. Chem. 275, 36811–36817 [DOI] [PubMed] [Google Scholar]
- 35.Dölle C., Skoge R. H., Vanlinden M. R., and Ziegler M. (2013) NAD biosynthesis in humans: enzymes, metabolites and therapeutic aspects. Curr. Top. Med. Chem. 13, 2907–2917 [DOI] [PubMed] [Google Scholar]
- 36.Olesen U. H., Thougaard A. V., Jensen P. B., and Sehested M. (2010) A preclinical study on the rescue of normal tissue by nicotinic acid in high-dose treatment with APO866, a specific nicotinamide phosphoribosyltransferase inhibitor. Mol. Cancer Ther. 9, 1609–1617 [DOI] [PubMed] [Google Scholar]
- 37.Tan B., Young D. A., Lu Z. H., Wang T., Meier T. I., Shepard R. L., Roth K., Zhai Y., Huss K., Kuo M. S., Gillig J., Parthasarathy S., Burkholder T. P., Smith M. C., Geeganage S., and Zhao G. (2013) Pharmacological inhibition of nicotinamide phosphoribosyltransferase (NAMPT), an enzyme essential for NAD+ biosynthesis, in human cancer cells: metabolic basis and potential clinical implications. J. Biol. Chem. 288, 3500–3511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Agrimi G., Russo A., Scarcia P., and Palmieri F. (2012) The human gene SLC25A17 encodes a peroxisomal transporter of coenzyme A, FAD and NAD+. Biochem. J. 443, 241–247 [DOI] [PubMed] [Google Scholar]
- 39.Di Noia M. A., Todisco S., Cirigliano A., Rinaldi T., Agrimi G., Iacobazzi V., and Palmieri F. (2014) The human SLC25A33 and SLC25A36 genes of solute carrier family 25 encode two mitochondrial pyrimidine nucleotide transporters. J. Biol. Chem. 289, 33137–33148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Di Stefano M., Galassi L., and Magni G. (2010) Unique expression pattern of human nicotinamide mononucleotide adenylyltransferase isozymes in red blood cells. Blood Cells Mol. Dis. 45, 33–39 [DOI] [PubMed] [Google Scholar]
- 41.Hikosaka K., Ikutani M., Shito M., Kazuma K., Gulshan M., Nagai Y., Takatsu K., Konno K., Tobe K., Kanno H., and Nakagawa T. (2014) Deficiency of nicotinamide mononucleotide adenylyltransferase 3 (nmnat3) causes hemolytic anemia by altering the glycolytic flow in mature erythrocytes. J. Biol. Chem. 289, 14796–14811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kitaoka Y., Munemasa Y., Kojima K., Hirano A., Ueno S., and Takagi H. (2013) Axonal protection by Nmnat3 overexpression with involvement of autophagy in optic nerve degeneration. Cell Death Dis. 4, e860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Andrabi S. A., Umanah G. K., Chang C., Stevens D. A., Karuppagounder S. S., Gagné J. P., Poirier G. G., Dawson V. L., and Dawson T. M. (2014) Poly(ADP-ribose) polymerase-dependent energy depletion occurs through inhibition of glycolysis. Proc. Natl. Acad. Sci. U.S.A. 111, 10209–10214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fouquerel E., Goellner E. M., Yu Z., Gagné J. P., Barbi de Moura M., Feinstein T., Wheeler D., Redpath P., Li J., Romero G., Migaud M., Van Houten B., Poirier G. G., and Sobol R. W. (2014) ARTD1/PARP1 negatively regulates glycolysis by inhibiting hexokinase 1 independent of NAD+ depletion. Cell Reports 8, 1819–1831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bruzzone S., Fruscione F., Morando S., Ferrando T., Poggi A., Garuti A., D'Urso A., Selmo M., Benvenuto F., Cea M., Zoppoli G., Moran E., Soncini D., Ballestrero A., Sordat B., Patrone F., Mostoslavsky R., Uccelli A., and Nencioni A. (2009) Catastrophic NAD+ depletion in activated T lymphocytes through Nampt inhibition reduces demyelination and disability in EAE. PLoS One 4, e7897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Di Martino C., and Pallotta M. L. (2011) Mitochondria-localized NAD biosynthesis by nicotinamide mononucleotide adenylyltransferase in Jerusalem artichoke (Helianthus tuberosus L.) heterotrophic tissues. Planta 234, 657–670 [DOI] [PubMed] [Google Scholar]
- 47.Berger F., Lau C., Dahlmann M., and Ziegler M. (2005) Subcellular compartmentation and differential catalytic properties of the three human nicotinamide mononucleotide adenylyltransferase isoforms. J. Biol. Chem. 280, 36334–36341 [DOI] [PubMed] [Google Scholar]