Abstract
Background
Rhabdomyosarcoma (RMS) is divided into two major histological subtypes: alveolar (ARMS) and embryonal (ERMS), with most ARMS expressing one of two oncogenic genes fusing PAX3 or PAX7 with FOXO1 (P3F and P7F, respectively). The Children’s Oncology Group (COG) carried out a multi-institutional clinical trial to evaluate the prognostic value of PAX-FOXO1 fusion status.
Methods
Study participants were treated on COG protocol D9803 for intermediate risk ARMS or ERMS using multi-agent chemotherapy, radiotherapy, and surgery. Central diagnostic pathology review and molecular testing for fusion genes were carried out on prospectively collected specimens. Event-free (EFS) and overall survival (OS) at 5 years were correlated with histological subtype and PAX-FOXO1 status.
Results
Of 616 eligible D9803 enrollees, 434 cases had adequate clinical, molecular, and pathology data for definitive classification as ERMS, ARMS P3F+ or P7F+, or ARMSn (without detectable fusion). EFS was worse for those with ARMS P3F+ (54%) and P7F+ (65%) than those with ERMS (77%; P < 0.001). EFS for ARMSn and ERMS were not statistically different (90% vs. 77%, P = 0.15). ARMS P3F+had poorer OS (64%) than ARMS P7F+ (87%), ARMSn (89%), and ERMS (82%; P = 0.006).
Conclusions
ARMSn has an outcome similar to ERMS and superior EFS compared to ARMS with either P3F or P7F, when given therapy designed for children with intermediate risk RMS. This prospective analysis supports incorporation of PAX-FOXO1 fusion status into risk stratification and treatment allocation.
Keywords: PAX-FOXO1, rhabdomyosarcoma, survival
INTRODUCTION
Rhabdomyosarcoma (RMS), the most common soft tissue sarcoma in children, comprises two major histological categories with alveolar (ARMS) and embryonal (ERMS) histological features [1,2]. ARMS is usually associated with a worse outcome [3–5]. For example, the current risk stratification scheme used by the Children’s Oncology Group (COG) excludes ARMS from the low risk stratum regardless of other clinical features. Nonetheless, in the most recent COG D9803 study for children with intermediate risk RMS (D9803), 4-year failure-free survival for those with non-metastatic ARMS ranged between 60% (for unfavorable primary sites with incomplete resection at study entry) and 83% (for favorable primary sites or complete resection at study entry) [4]. This finding demonstrates that the outcome for those with intermediate risk ARMS is heterogeneous and may be amenable to more precise risk-stratification.
ARMS is usually associated with a balanced chromosomal translocation generating novel proteins fusing the DNA binding domain of either PAX3 or PAX7 from chromosomes 2 and 1, respectively, with the carboxyl terminus of FOXO1, encoded by the FOXO1 gene on chromosome 13 [6–8]. Detection of the fusion transcript by RT-PCR or the t(2;13) or t(1;13) translocation by fluorescence in situ hybridization (FISH) is routinely used for molecular diagnosis because PAX3-FOXO1 (P3F) or PAX7-FOXO1 (P7F) fusions are present in approximately 80% of ARMS and not in other cancers (reviewed in Slater and Shipley [9]). Perhaps surprisingly given the distinct histological differences between typical ARMS and ERMS, microarray-based gene expression profiling demonstrates that fusion gene-positive ARMS cases cluster together while fusion gene-negative ARMS (ARMSn) more closely resembles ERMS [10–13]. Hence, fusion gene expression confers distinct biological properties to a subset of ARMS cases, despite similar histological appearance.
Whether PAX-FOXO1 fusion status also influences clinical outcome is less clear. Prior studies investigating its prognostic significance have been retrospective analyses of selected cases available for study. The reports usually represent relatively small sample sizes, including patients with localized and metastatic disease, and often combining multiple clinical trials spanning what could be many years [10,11,14–18]. We have previously reported that these convenience cohorts may not be representative of less selected populations [14,19], making extrapolation of the results to all patients hazardous. Further, the histological definition of ARMS histology evolved since 1995, raising the possibility that some ARMSn cases actually represent ERMS based on current pathology definitions.
Definitively elucidating the prognostic importance of PAX-FOXO1 fusion status in ARMS and determining whether outcome for ARMSn is similar to ERMS (as predicted by the gene expression data and one recent report [18]) requires a prospective trial using a homogenous study population in which there was uniform evaluation of pathology and PAX-FOXO1 fusion status. In this manuscript, we report such an analysis from the COG D9803 trial for intermediate risk RMS.
METHODS
Patients and Therapy on D9803 Clinical Trial
Analyses were performed on pathology material from children and young adults enrolled on COG D9803 between 1999 and 2005 [4]. As previously described, the D9803 study enrolled participants (n = 616) with intermediate risk clinical features, including those with (a) Stages 2 and 3, Group III ERMS, and (b) non-metastatic ARMS [5]. Participants with undifferentiated sarcoma and others less than 10 years of age with Stage 4, Group IV ERMS were also eligible for D9803, but they were excluded from this analysis. Participants were randomly assigned to receive either standard chemotherapy using vincristine, actinomycin D, and cyclophosphamide (VAC) or VAC alternating with vincristine, topotecan, and cyclophosphamide (VTC). Participants with parameningeal RMS and intracranial extension were non-randomly assigned to VAC therapy. Randomized participants had similar clinical and demographic parameters, and they all received radiation; outcome was equivalent on both treatment arms [4]. Because treatment regimen did not influence outcome, all participants were combined for the purposes of PAX-FOXO1 fusion status analysis.
Histology Review and Fusion Gene Testing
We restricted our survival analysis to the 434 cases with either ARMS or ERMS, proven PAX-FOXO1 fusion status for the ARMS cases, and adequate clinical data. During the conduct of D9803, we observed an increase from 30% to 41% in the frequency of ARMS as classified by COG central pathology, which led us to conduct a systematic pathologic re-review. For the current analysis, the diagnosis of ARMS was based upon central pathology re-review using more strict criteria for ARMS (Rudzinski et al., unpublished COG data; manuscript forthcoming). D9803 cases with a prior diagnosis of ARMS (n = 255) and a randomly selected subset with ERMS (n = 38) were jointly reviewed by two pathologists (DMP and ER). Review material included H&E slides from all patients and myogenin stains from 250 patients. The reviewers were blinded to tumor stage, anatomic site, clinical outcome, and fusion status. ARMS is recognized by classical and solid patterns. The classical pattern contains anastomosing fibrovascular connective tissue septa forming an alveolar pattern lined by neoplastic cells. The “solid” variant of ARMS grows as a solid mass of closely aggregated cells with no or scarcely discernible alveolar patterns. The cytology of solid ARMS is identical to the one encountered in the classic form. The densely packed cells are round, often similar to each other, and usually exhibiting a high nuclear:cytoplasmic ratio. We also relied on immunohistochemical staining for myogenin, which was strong and diffusely positive. Lesions without clear-cut ARMS morphology that lacked strong, diffuse myogenin expression were considered to be ERMS.
PAX-FOXO1 analysis was accomplished by real-time RT-PCR and/or fluorescence in situ hybridization (FISH) as previously described [14,20] on all ARMS cases on the D9803 study. The approximately 20% of ARMS cases negative for these gene rearrangements, a percentage consistent with other reports [14,15], were coded as ARMSn, as opposed to the ARMS P3F+ or P7F+ cases.
Statistical Analyses
Event-free survival (EFS) was defined as the time from study enrollment to the first occurrence of progression, relapse after response, or death as a first event from any cause. Overall survival (OS) was defined as the time from study enrollment to death from any cause. Follow-up for patients not experiencing an event of interest was censored at their time of last follow-up. Time-to-event distributions were estimated using the Kaplan–Meier method and they were compared among subsets of patients using the log-rank test. Statistical analyses were performed using SAS version 9.2 (Cary, NC).
RESULTS
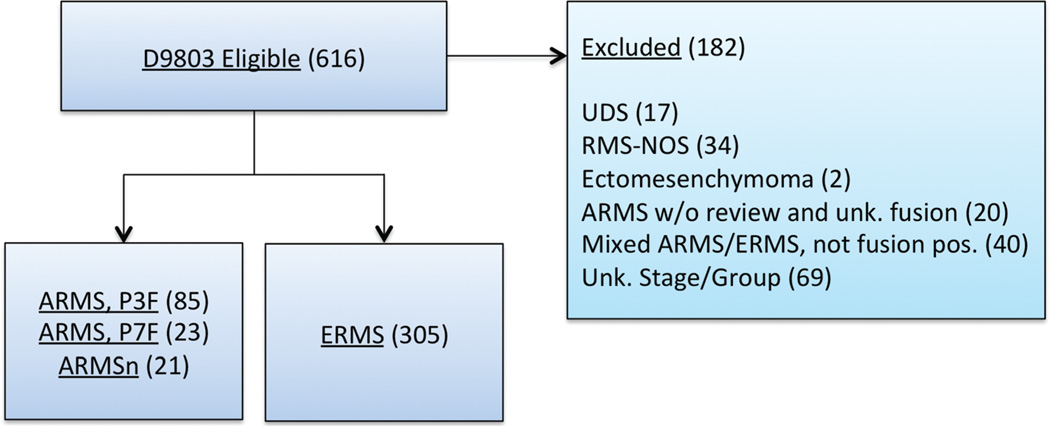
This survival analysis of children with intermediate risk RMS treated on the COG D9803 study includes 129 with confirmed ARMS and known fusion status (PAX3-FOXO1 positive [P3F+], n = 85; PAX7-FOXO1 positive [P7F+], n = 23 and translocation negative [ARMSn], n = 21) and 305 ERMS cases that included botryoid and spindle cell morphology (Fig. 1).
Fig. 1.
CONSORT diagram depicting the subsets of the COG D9803 study participants utilized for the current analysis. UDS, undifferentiated sarcoma; NOS, not otherwise specified; Unk, unknown.
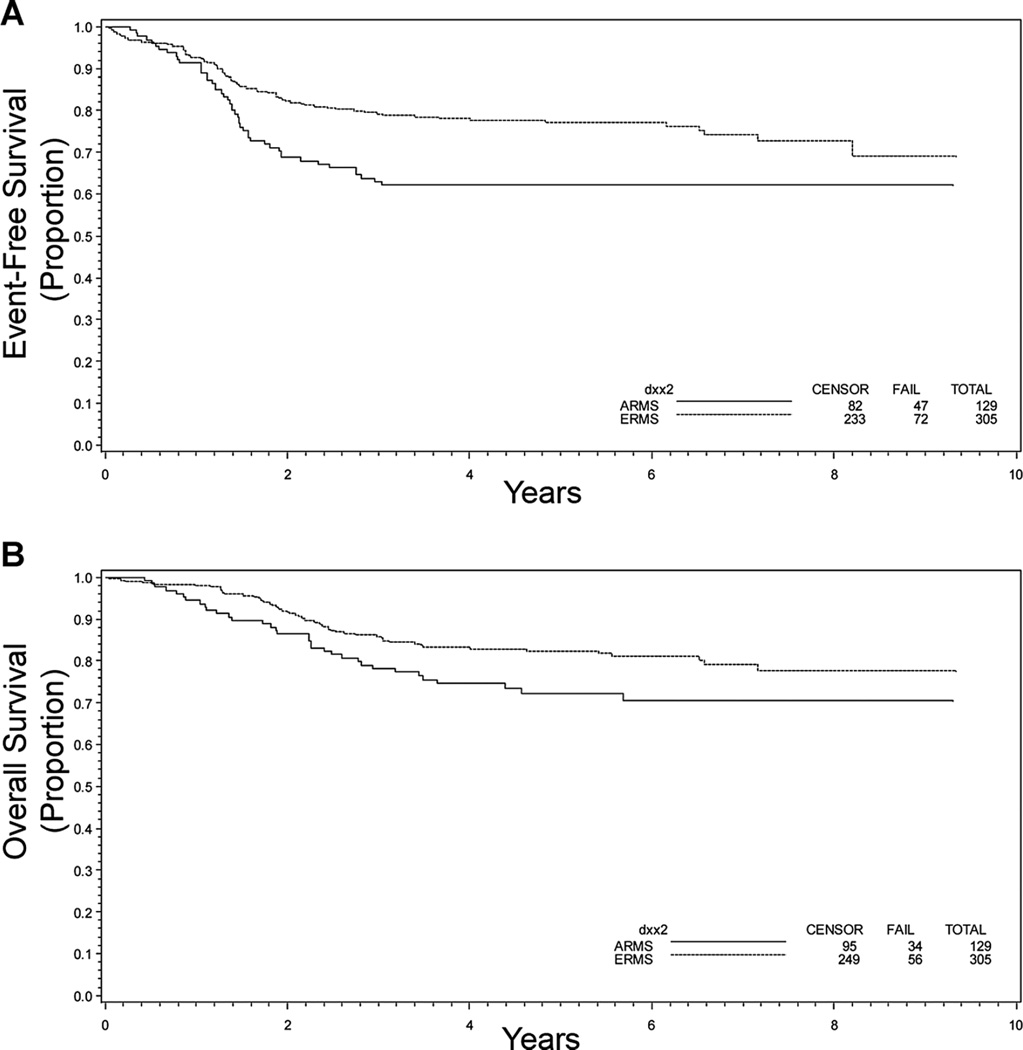
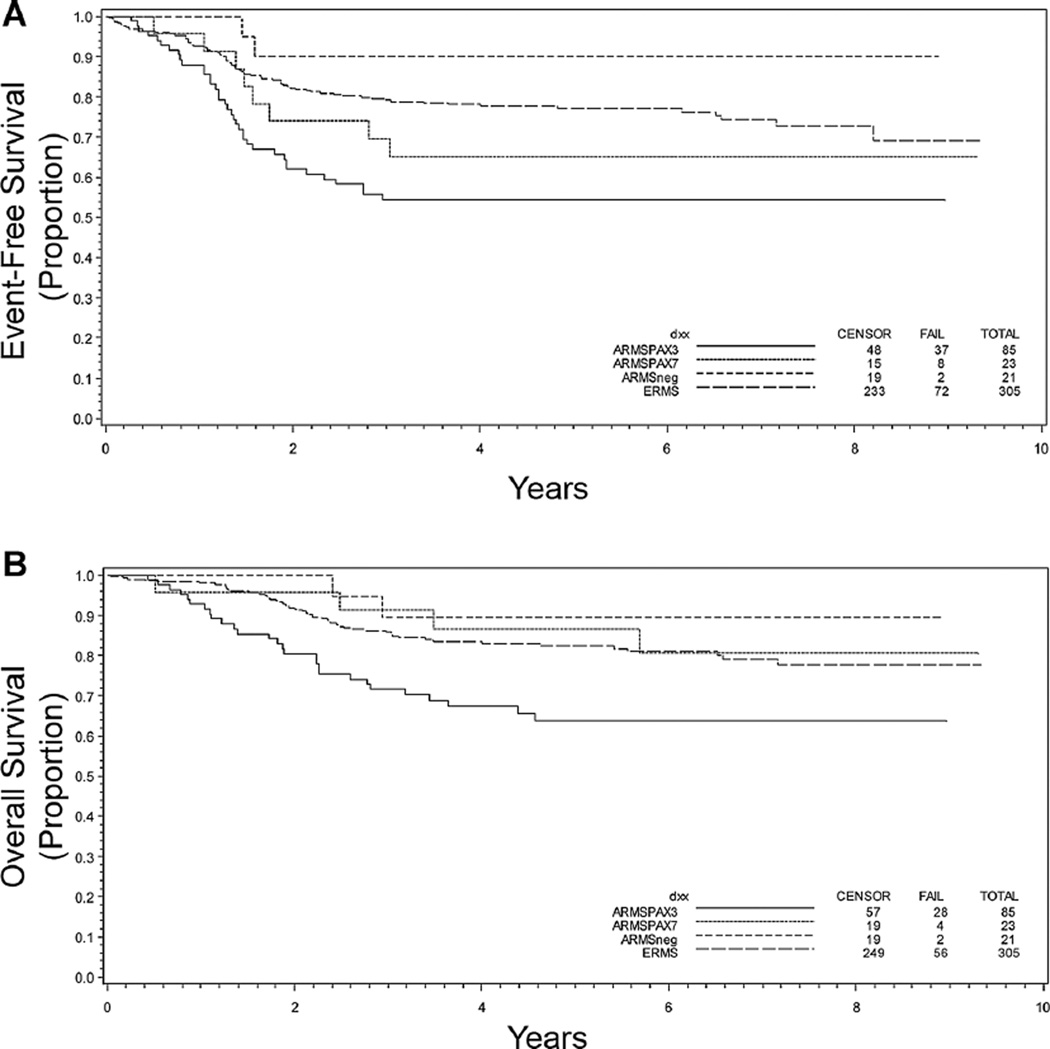
Outcome at 5 years for these patients was evaluated to determine how histology or PAX-FOXO1 status contributed to the clinical outcome. Both the EFS and OS at 5 years were worse for those with ARMS compared to ERMS (Fig. 2A and B). PAX-FOXO1 fusion status provided additional prognostic information within the ARMS population: those with P3F+ and P7F+ tumors had an inferior EFS compared to those with either ARMSn or ERMS (Table I and Fig. 3A). OS was also worse for those with tumors that were P3F+ as compared to patients withP7F+, ARMSn, and ERMS disease, all of whom had similar outcomes (Table I and Fig. 3B).
Fig. 2.
Kaplan–Meier curves depicting (A) event-free survival and (B) overall survival in years following diagnosis for ARMS (n = 129) and ERMS (n = 305) based on re-reviewed histologic diagnosis. P = 0.0035 (A) and 0.040 (B).
TABLE I.
Five-Year EFS and OS in D9803 Study
| Biologic subset | Patients (#) | EFS (95% CI) | OS |
|---|---|---|---|
| Molecular subtype | |||
| ERMS | 305 | 77% (72%, 82%) | 82% (77%, 86%) |
| ARMS PAX3 | 85 | 54% (43%, 65%) | 64% (52%, 74%) |
| ARMS PAX7 | 23 | 65% (42%, 81%) | 87% (64%, 96%) |
| ARMS negative | 21 | 90% (65%, 97%) | 89% (64%, 97%) |
| Stage 1 or Stage 2, 3/Group I/II | |||
| ARMS PAX3 | 28 | 65% (44%, 80%) | 70% (47%, 85%) |
| ARMS PAX7 | 12 | 75% (41%, 91%) | 92% (54%, 99%) |
| ARMS negative | 10 | 100% | 100% |
| Stage 2, 3/Group III | |||
| ERMS | 261 | 77% (71%, 81%) | 82% (77%, 86%) |
| ARMS PAX3 | 57 | 49% (36%, 62%) | 61% (47%, 73%) |
| ARMS PAX7 | 11 | 55% (23%, 78%) | 82% (45%, 95%) |
| ARMS negative | 11 | 82% (45%, 95%) | 80% (41%, 95%) |
Fig. 3.
Kaplan–Meier curves depicting (A) event-free and (B) overall survival in years following diagnosis based on molecular classification of disease as being ERMS (n = 305) or ARMSn (n = 21), ARMS PF3+ (n = 85), or ARMS PF7+ (n = 23). P < 0.001 (A) and 0.0035 (B).
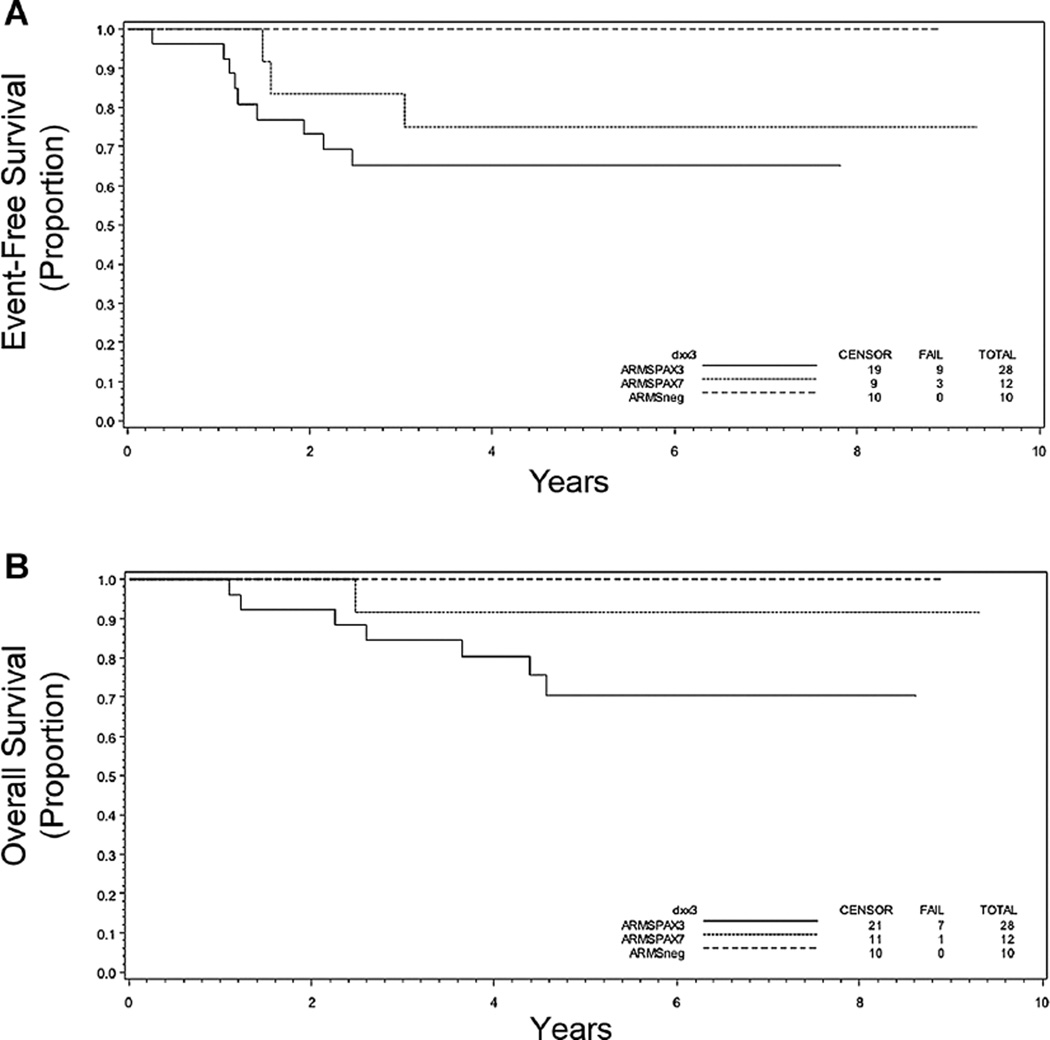
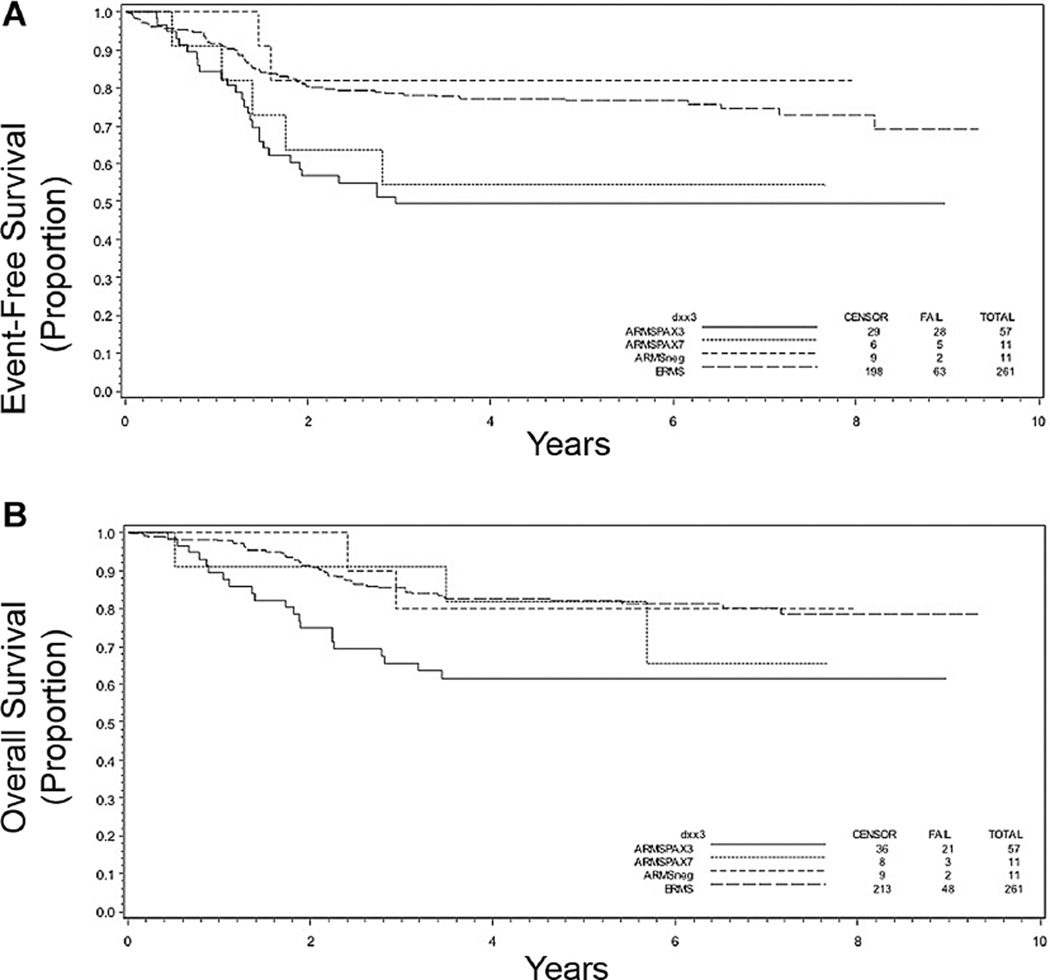
Fusion status also influenced outcome within distinct clinical subgroups. Focusing on those with the more favorable Stage 1 or Stage 2, 3, and Group I/II disease, the 5-year EFS for those with P3F+ and P7F+ ARMS was similar (65% and 75%, respectively) with a trend toward EFS being inferior when compared to those with ARMSn (100%) disease (P = 0.13; Table I and Fig. 4A). The 5-year OS for P3F+ tumors was somewhat worse than that for P7F+ and ARMSn+ tumors (P = 0.21; Table I and Fig. 4B). It should be emphasized that these data are based on a very small number of cases. For the subset with Stage 2, 3, Group III RMS, the presence of either P3F+ or P7F+ portended worse EFS at 5 years (P < 0.001; Table I and Fig. 5A). OS was significantly worse only in patients with P3F+ disease (P = 0.015; Table I and Fig. 5B).
Fig. 4.
Kaplan–Meier curves depicting (A) event-free and (B) overall survival in years following diagnosis based on molecular classification of disease as being ARMSn (n = 10), ARMS PF3+ (n = 28), or ARMS PF7+ (n = 12) within the subset of patients with Stage 1 or Stage 2, 3 and Group I/II disease. P = 0.13 (A) and 0.13 (B).
Fig. 5.
Kaplan–Meier curves depicting (A) event-free and (B) overall survival in years following diagnosis based on molecular classification of disease as being ERMS (n = 261), ARMSn (n = 11), ARMS PF3+ (n = 57), or ARMS PF7+ (n = 11) within the subset of patients with Stage 2, 3 and Group III disease. P < 0.001 (A) and 0.0054 (B).
DISCUSSION
Two analyses of the prognostic significance of PAX-FOXO1 fusion status reached conflicting conclusions, one finding an inferior outcome in fusion positive patients [10] and another finding no association [17]. The current analysis has several strengths that make our conclusions about the prognostic significance of PAX-FOXO1 fusion status more definitive. First, our report is based on analysis of a single, prospective clinical trial using uniform treatment. This contrasts with both recent European series that were based on retrospective analyses of subsets of available cases from patients treated on or according to five different clinical trials spanning nearly 2 decades [10,17]. Second, our analysis avoids the methodological flaws inherent in the study of convenience cohorts, as illustrated in results from COG and the Cooperative Soft Tissue Sarcoma Study (CWS) group. In the previous reports, the prognostic significance of fusion status was not consistent [14,17]. For example, Barr and colleagues used a convenience cohort from the Intergroup RMS Study Group III trial to show that survival was substantially better in cases where material was available for molecular analysis [14] whereas Stegmaier et al. [17] showed the reverse in a different convenience cohort. Finally, uniform pathology re-review with a consistent and revised definition of ARMS in our analysis led to a low rate of ARMSn. We have previously reported a drift in the pathological diagnosis of ARMS, leading to an increase in its incidence and in the frequency of ARMSn [21]. The 30% ARMSn rate found by Williamson et al. [10] is higher than in our current analysis and in previous reports showing only approximately 20% frequency of ARMSn cases [14,15]. Including the more favorable ERMS cases with ARMS might have skewed an outcome analysis of fusion-negative ARMS.
Although sub-type specific fusion genes essentially define many soft tissue sarcomas [22], the presence or absence of a specific fusion gene has not yet been used to guide therapy in COG or other cooperative group trials. Based on our prospective results, we now have sufficient data to use PAX-FOXO1 status to assign therapy, as suggested by Wexler and Ladanyi [23]. The substantially better EFS suggests that children with ARMSn might be effectively treated with less intensive therapy. However, this initial step towards molecularly guided therapy must be taken carefully. The seemingly exceptional survival for children with Stage 1 or Stage 2/3, Group I/II, ARMSn disease (100% at 5 years) is based on only 10 children and in the context of more intensive intermediate risk therapy.
The prognostic relevance of P3F+ versus P7F+ ARMS is not as clear. For example, our prospective analysis shows the EFS for those with P7F+ disease to be similar to that in children with P3F+ ARMS, but the OS is substantially better. This distinction was also preserved in our analyses of subsets with low or higher stage disease, but the numbers of cases in each are small. Nonetheless, a conclusion that OS for those with P7F+ ARMS approximates survival for subjects with fusion-negative disease parallels findings from a recent retrospective analysis of pooled data from European and North American cohorts [18]. Based on their findings, these authors concluded that P7F+ ARMS could be included in a lower risk stratum with the fusion negative cases. Because EFS has been used as a primary endpoint in recent COG trials—and EFS for P3F+ and P7F+ intermediate risk disease is similar—another reasonable approach would be to stratify all fusion positive ARMS together, at least in the intermediate risk group studied here.
A molecular explanation for the similar EFS, but not OS, in children with P7F+ and P3F+ RMS is not apparent. Most assume that oncogenic transcription factors influence tumor biology by controlling gene expression. For example, Davicioni et al. [11] showed that OS is better for children with tumors expressing a “P7F-like” signature as compared to tumors with a “P3F-like” signature. It perhaps should be noted that microarray gene expression profiling is typically accomplished using RMS specimens taken at diagnosis, and these studies have not revealed robust gene expression differences from unsupervised analysis of PF3+ and PF7+ subtypes [10–13]. It is possible that similar studies of recurrent specimens could reveal gene expression differences that might help explain the more favorable OS for those with P7F+ disease. Alternatively, DNA-based studies do support fundamental differences between P7F+ and P3F+ ARMS. For example, close to 25% of P3F+ ARMS have amplification of 12q13–q15, which is associated with significantly higher expression of the cell cycle regulator CDK4 [24]. Approximately two-thirds of the P7F+ ARMS have amplification of a region of 13q31, with the minimal amplicon including MIR17HG, encoding the miR-17–92 cluster of microRNAs [25]. Hence, these molecular differences may influence in tumor biology in a manner not obviously reflected by gene expression.
A number of questions arise as we consider how to move into an era in which molecular features are used for risk stratification and treatment assignment. First among them, perhaps, is can a multi-feature metagene signature [10,16] or the incorporation of non-coding RNAs and/or structural DNA changes provide prognostic data beyond that derived from a single molecular analyte, such as fusion gene status? Given the aforementioned difficulties with retrospective, convenience cohorts, this issue must be formally addressed in the context of a prospective trial. A second question relates to the best strategy to detect fusion genes in RMS. Our findings are based on RT-PCR or FISH assays, which yielded interpretable results in the vast majority of cases. Although RNA degradation in FFPE material can pose a challenge for an RT-PCR-based assay, emerging technologies may assuage this concern. For example, color-coding individual RNA transcripts of interest (including fusion transcripts), followed by solid-state capture and digital quantification can provide accurate and reproducible measurements using formalin-fixed, paraffin-embedded and fresh, frozen material [26]. However, even after such RNA-based tools are optimized and developed as clinical tests, FISH provides an important advantage in the ability to detect new molecular rearrangements that might have been missed by other RNA-based approaches [27]. In the context of a clinical trial, it might be acceptable to miss the small percentage of RMS cases that have variant translocations; however, central histology review and FISH-based approaches are still worthwhile steps to fuel future discovery.
ACKNOWLEDGMENTS
We gratefully acknowledge administrative and technical support from Chune Zhang (Barr laboratory) and from Natalie Beeler, Mandy Dunlevy, Julie Moore, and other staff at the COG Biopathology Center at Nationwide Children’s Hospital.
Grant sponsor: St. Baldrick’s Foundation; Grant number: #179772; Grant sponsor: NIH; Grant numbers: CA148216; CA104896 CA098413.
Additional support from Intramural Research Program, Center for Cancer Research, National Cancer Institute (FGB).
Footnotes
Conflict of interest: Nothing to declare.
REFERENCES
- 1.Womer RB, Pressey JG. Rhabdomyosarcoma and soft tissue sarcoma in childhood. Curr Opin Oncol. 2000;12:337–344. doi: 10.1097/00001622-200007000-00010. [DOI] [PubMed] [Google Scholar]
- 2.Merlino G, Helman LJ. Rhabdomyosarcoma—Working out the pathways. Oncogene. 1999;18:5340–5348. doi: 10.1038/sj.onc.1203038. [DOI] [PubMed] [Google Scholar]
- 3.Oberlin O, Rey A, Lyden E, et al. Prognostic factors in metastatic rhabdomyosarcomas: Results of a pooled analysis from United States and European cooperative groups. J Clin Oncol. 2008;26:2384–2389. doi: 10.1200/JCO.2007.14.7207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arndt CA, Stoner JA, Hawkins DS, et al. Vincristine, actinomycin, and cyclophosphamide compared with vincristine, actinomycin, and cyclophosphamide alternating with vincristine, topotecan, and cyclophosphamide for intermediate-risk rhabdomyosarcoma: Children’s oncology group study D980. J Clin Oncol. 2009;27:5182–5188. doi: 10.1200/JCO.2009.22.3768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crist WM, Anderson JR, Meza JL, et al. Intergroup rhabdomyosarcoma study-IV: Results for patients with nonmetastatic disease. J Clin Oncol. 2001;19:3091–3102. doi: 10.1200/JCO.2001.19.12.3091. [DOI] [PubMed] [Google Scholar]
- 6.Barr FG, Galili N, Holick J, et al. Rearrangement of the PAX3 paired box gene in the paediatric solid tumour alveolar rhabdomyosarcoma. Nat Genet. 1993;3:113–117. doi: 10.1038/ng0293-113. [DOI] [PubMed] [Google Scholar]
- 7.Galili N, Davis RJ, Fredericks WJ, et al. Fusion of a fork head domain gene to PAX3 in the solid tumour alveolar rhabdomyosarcoma. Nat Genet. 1993;5:230–235. doi: 10.1038/ng1193-230. [DOI] [PubMed] [Google Scholar]
- 8.Davis RJ, D’Cruz CM, Lovell MA, et al. Fusion of PAX7 to FKHR by the variant t(1;13)(p36;q14) translocation in alveolar rhabdomyosarcoma. Cancer Res. 1994;54:2869–2872. [PubMed] [Google Scholar]
- 9.Slater O, Shipley J. Clinical relevance of molecular genetics to paediatric sarcomas. J Clin Pathol. 2007;60:1187–1194. doi: 10.1136/jcp.2006.040113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Williamson D, Missiaglia E, de RA, et al. Fusion gene-negative alveolar rhabdomyosarcoma is clinically and molecularly indistinguishable from embryonal rhabdomyosarcoma. J Clin Oncol. 2010;28:2151–2158. doi: 10.1200/JCO.2009.26.3814. [DOI] [PubMed] [Google Scholar]
- 11.Davicioni E, Anderson MJ, Finckenstein FG, et al. Molecular classification of rhabdomyosarcoma—Genotypic and phenotypic determinants of diagnosis. Am J Pathol. 2009;174:550–564. doi: 10.2353/ajpath.2009.080631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lae M, Ahn EH, Mercado GE, et al. Global gene expression profiling of PAX-FKHR fusion-positive alveolar and PAX-FKHR fusion-negative embryonal rhabdomyosarcomas. J Pathol. 2007;212:143–151. doi: 10.1002/path.2170. [DOI] [PubMed] [Google Scholar]
- 13.Wachtel M, Dettling M, Koscielniak E, et al. Gene expression signatures identify rhabdomyosarcoma subtypes and detect a novel t(2;2)(q35;p23) translocation fusing PAX3 to NCOA1. Cancer Res. 2004;64:5539–5545. doi: 10.1158/0008-5472.CAN-04-0844. [DOI] [PubMed] [Google Scholar]
- 14.Barr FG, Smith LM, Lynch JC, et al. Examination of gene fusion status in archival samples of alveolar rhabdomyosarcoma entered on the Intergroup Rhabdomyosarcoma Study-III trial: A report from the Children’s Oncology Group. J Mol Diagn. 2006;8:202–208. doi: 10.2353/jmoldx.2006.050124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sorensen PH, Lynch JC, Qualman SJ, et al. PAX3-FKHR and PAX7-FKHR gene fusions are prognostic indicators in alveolar rhabdomyosarcoma: A report from the children’s oncology group. J Clin Oncol. 2002;20:2672–2679. doi: 10.1200/JCO.2002.03.137. [DOI] [PubMed] [Google Scholar]
- 16.Davicioni E, Anderson JR, Buckley JD, et al. Gene expression profiling for survival prediction in pediatric rhabdomyosarcomas: A report from the children’s oncology group. J Clin Oncol. 2010;28:1240–1246. doi: 10.1200/JCO.2008.21.1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stegmaier S, Poremba C, Schaefer KL, et al. Prognostic value of PAX-FKHR fusion status in alveolar rhabdomyosarcoma: A report from the cooperative soft tissue sarcoma study group (CWS) Pediatric Blood Cancer. 2011;57:406–414. doi: 10.1002/pbc.22958. [DOI] [PubMed] [Google Scholar]
- 18.Missiaglia E, Williamson D, Chisholm J, et al. PAX3/FOXO1 fusion gene status is the key prognostic molecular marker in rhabdomyosarcoma and significantly improves current risk stratification. J Clin Oncol. 2012;30:1670–1677. doi: 10.1200/JCO.2011.38.5591. [DOI] [PubMed] [Google Scholar]
- 19.Rosenberg AR, Skapek SX, Hawkins DS. The inconvenience of convenience cohorts: Rhabdomyosarcoma and the PAX-FOXO1 biomarker. Cancer Epidemiol Biomarkers Prev. 2012;21:1012–1018. doi: 10.1158/1055-9965.EPI-12-0207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dumont SN, Lazar AJ, Bridge JA, et al. PAX3/7-FOXO1 fusion status in older rhabdomyosarcoma patient population by fluorescent in situ hybridization. J Cancer Res Clin Oncol. 2012;138:213–220. doi: 10.1007/s00432-011-1089-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anderson JR, Barr FG, Hawkins DS, et al. Fusion-negative alveolar rhabdomyosarcoma: Modification of risk stratification is premature. J Clin Oncol. 2010;28:e587–e588. doi: 10.1200/JCO.2010.30.5466. author reply e589–590. [DOI] [PubMed] [Google Scholar]
- 22.Spunt SL, Skapek SX, Coffin CM. Pediatric nonrhabdomyosarcoma soft tissue sarcomas. Oncologist. 2008;13:668–678. doi: 10.1634/theoncologist.2007-0182. [DOI] [PubMed] [Google Scholar]
- 23.Wexler LH, Ladanyi M. Diagnosing alveolar rhabdomyosarcoma: Morphology must be coupled with fusion confirmation. J Clin Oncol. 2010;28:2126–2128. doi: 10.1200/JCO.2009.27.5339. [DOI] [PubMed] [Google Scholar]
- 24.Barr FG, Duan F, Smith LM, et al. Genomic and clinical analyses of 2p24 and 12q13-q14 amplification in alveolar rhabdomyosarcoma: A report from the Children’s Oncology Group. Genes Chromosomes Cancer. 2009;48:661–672. doi: 10.1002/gcc.20673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reichek JL, Duan F, Smith LM, et al. Genomic and clinical analysis of amplification of the 13q31 chromosomal region in alveolar rhabdomyosarcoma: A report from the Children’s Oncology Group. Clin Cancer Res. 2011;17:1463–1473. doi: 10.1158/1078-0432.CCR-10-0091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Geiss GK, Bumgarner RE, Birditt B, et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol. 2008;26:317–325. doi: 10.1038/nbt1385. [DOI] [PubMed] [Google Scholar]
- 27.Sumegi J, Streblow R, Frayer RW, et al. Recurrent t(2;2) and t(2;8) translocations in rhabdomyosarcoma without the canonical PAX-FOXO1 fuse PAX3 to members of the nuclear receptor transcriptional coactivator family. Genes Chromosomes Cancer. 2010;49:224–236. doi: 10.1002/gcc.20731. [DOI] [PMC free article] [PubMed] [Google Scholar]