SUMMARY
The physiological function of cell wall amidases has been investigated in several proteobacterial species. In all cases, they have been implicated in the cleavage of cell wall material synthesized by the cytokinetic ring. Although typically non-essential, this activity is critical for daughter cell separation and outer membrane invagination during division. In Escherichia coli, proteins with LytM domains also participate in cell separation by stimulating amidase activity. Here, we investigated the function of amidases and LytM proteins in the opportunistic pathogen Pseudomonas aeruginosa. In agreement with studies in other organisms, PaAmiB and three LytM proteins were found to play crucial roles in P. aeruginosa cell separation, envelope integrity, and antibiotic resistance. Importantly, the phenotype of amidase-defective P. aeruginosa cells also differed in informative ways from the E. coli paradigm; PaAmiB was found to be essential for viability and the successful completion of cell constriction. Our results thus reveal a key role for amidase activity in cytokinetic ring contraction. Furthermore, we show that the essential function of PaAmiB can be bypassed in mutants activated for a Cpx-like envelope stress response, suggesting that this signaling system may elicit the repair of division machinery defects in addition to general envelope damage.
Keywords: cell wall, peptidoglycan, cell division, antibiotic, cell envelope, amidase
INTRODUCTION
During the process of cell division, bacteria remodel their cellular envelope to construct new poles for the incipient daughter cells (Typas et al., 2011). This dramatic transformation is carried out by the cytokinetic ring apparatus called the divisome, which assembles at the nascent division site (de Boer, 2010; Lutkenhaus et al., 2012). Once formed, the divisome initiates the invagination of the inner membrane concomitantly with the synthesis of a new peptidoglycan (PG) cell wall structure called septal PG (de Pedro et al., 1997; de Boer, 2010). Gram-negative bacteria also possess an outer membrane, which surrounds the PG layer. Its constriction is coordinated with that of the inner membrane (Burdett and Murray, 1974a; Burdett and Murray, 1974b; de Boer, 2010). To allow for outer membrane invagination, degradative enzymes must split the septal PG material shared by the nascent daughter cells (Heidrich et al., 2001; de Boer, 2010). The septal PG splitting process has been most extensively studied in Escherichia coli, where enzymes called LytC-type PG amidases play a major role (Heidrich et al., 2001; Heidrich et al., 2002; Priyadarshini et al., 2007). E. coli encodes three LytC-type PG amidases, AmiA, AmiB, and AmiC (Heidrich et al., 2001). A triple mutant lacking all three of these proteins is viable, but fails to complete cell division and forms chains of daughter cells connected by what appear to be unsplit septal PG and a shared outer membrane (Heidrich et al., 2001; Priyadarshini et al., 2007).
To control amidase activity and prevent cell lysis due to unwanted PG degradation, cell separation amidases are equipped with autoinhibitory helices that occlude their active sites (Yang et al., 2012). Therefore, these enzymes require activation by the divisome proteins EnvC and NlpD, which employ LytM domains to relieve amidase autoinhibition (Uehara et al., 2009; Uehara et al., 2010). LytM factors typically function as metalloendopeptidases and possess intrinsic PG hydrolytic activity (Firczuk et al., 2005; Lu et al., 2006; Firczuk and Bochtler, 2007; Cohen et al., 2009; Singh et al., 2012). However, EnvC and NlpD represent a subset of LytM factors with degenerate active sites (referred to here as dLytM factors) that lack the residues for Zn2+ ion coordination and instead serve to temporally and spatially regulate the activation of amidase activity (Uehara et al., 2010; Peters et al., 2013). In E. coli, the regulatory activity of the dLytM factors is specific, with EnvC activating AmiA and AmiB, but not AmiC, and NlpD activating AmiC, but not AmiA or AmiB (Uehara et al., 2010).
Defects in cell separation, including loss of function of the amidases, have been associated with increased sensitivity of gram-negative bacteria to antibiotics, detergents, and defensins (Heidrich et al., 2002; Ize et al., 2003; Weatherspoon-Griffin et al., 2011; Craig et al., 2013). Hypersensitivity to these agents suggests that failure to complete division leads to a perturbation of the outer membrane permeability barrier. Disruption of the outer membrane barrier function is often associated with membrane biogenesis problems, which typically result in the induction of envelope stress responses in an attempt to ameliorate the defect. Several envelope stress responses have been identified in E. coli, including the Cpx, Rcs, and σE systems (Raivio and Silhavy, 2001; Majdalani and Gottesman, 2005; Vogt and Raivio, 2011). The Cpx two-component regulatory system utilizes the histidine kinase CpxA to regulate the activity of the response regulator CpxR, which when activated by phosphorylation, controls the expression of >50 genes (Raivio and Silhavy, 1997; De Wulf and Lin, 2000; Bury-Moné et al., 2009). Transcription of the amidase genes amiA and amiC is stimulated upon induction of the Cpx response (Weatherspoon-Griffin et al., 2011), suggesting that amidase activity may play a role in counteracting envelope stress and maintaining envelope homeostasis.
Pseudomonas aeruginosa is a ubiquitously occurring opportunistic gram-negative pathogen responsible for 8% of hospital-acquired infections (Center for Disease Control, 2014). P. aeruginosa infections are difficult to treat due to the intrinsic resistance of this bacterium to a variety of different antibiotics (Strateva and Yordanov, 2009). The resistance mechanisms are diverse and include multiple efflux pumps, a chromosomally encoded β-lactamase, and biofilm formation (Breidenstein et al., 2011). However, one of the greatest contributors to the intrinsic antibiotic resistance of P. aeruginosa is the low permeability of its outer membrane, which blocks the access of drug molecules to their intracellular targets (Yoshimura and Nikaido, 1982; Hancock and Bell, 1988; MacLeod et al., 2000). Therefore, development of new antimicrobial potentiation drugs that disrupt the outer-membrane permeability barrier and restore antibiotic sensitivity is a promising strategy for overcoming multidrug-resistant P. aeruginosa infections (Tängdén, 2014). Given that cell separation defects are known to compromise outer membrane integrity in other gram-negative bacteria (Heidrich et al., 2002; Ize et al., 2003; Weatherspoon-Griffin et al., 2011), amidase activity represents an attractive target for such potentiator drugs. P. aeruginosa encodes two LytC-type PG amidases, PaAmiA and PaAmiB (Scheurwater et al., 2007). Although AmiB has been shown to localize to the division plane (Jorgenson et al., 2014) and possess the expected PG amidase enzymatic activity (Scheurwater et al., 2007), little is known about the role of the amidases in cell division or drug resistance in P. aeruginosa.
In this report, we show that PaAmiB is required for cell separation of P. aeruginosa. However, unlike the amidases studied in other proteobacteria, this protein was additionally found to be essential for growth in rich medium and critical for efficient completion of cell constriction. Importantly, loss of PaAmiB function was indeed shown to compromise the outer membrane permeability barrier of strain PAO1 and hypersensitize it to antibiotics, including aminoglycosides and vancomycin. Inactivation of three dLytM proteins resulted in similar cell division, viability, and permeability defects, which is consistent with these factors playing partially redundant roles in PaAmiB activation. Conversely, loss of PaAmiA had no observable phenotype on its own, nor did it significantly exacerbate the PaAmiB defect. Finally, we identified mutations that suppress the viability defect and drug sensitivity of a ΔamiA ΔamiB double mutant, mapping them to a cpxA ortholog of P. aeruginosa. The suppressor mutations resemble cpxA* alleles from E. coli (Raivio and Silhavy, 1997) and likely result in the constitutive induction of a Cpx-like envelope stress response in P. aeruginosa. Our results, therefore, highlight a critical role for amidase activity in the cell constriction function of the divisome and validate PaAmiB as an attractive target for antibiotic potentiators in P. aeruginosa. Additionally, our suppressor analysis implicates a Cpx-like response in rescuing cells from the lethal effects of divisome malfunction.
RESULTS
Pseudomonas aeruginosa AmiB is essential for cell growth and division
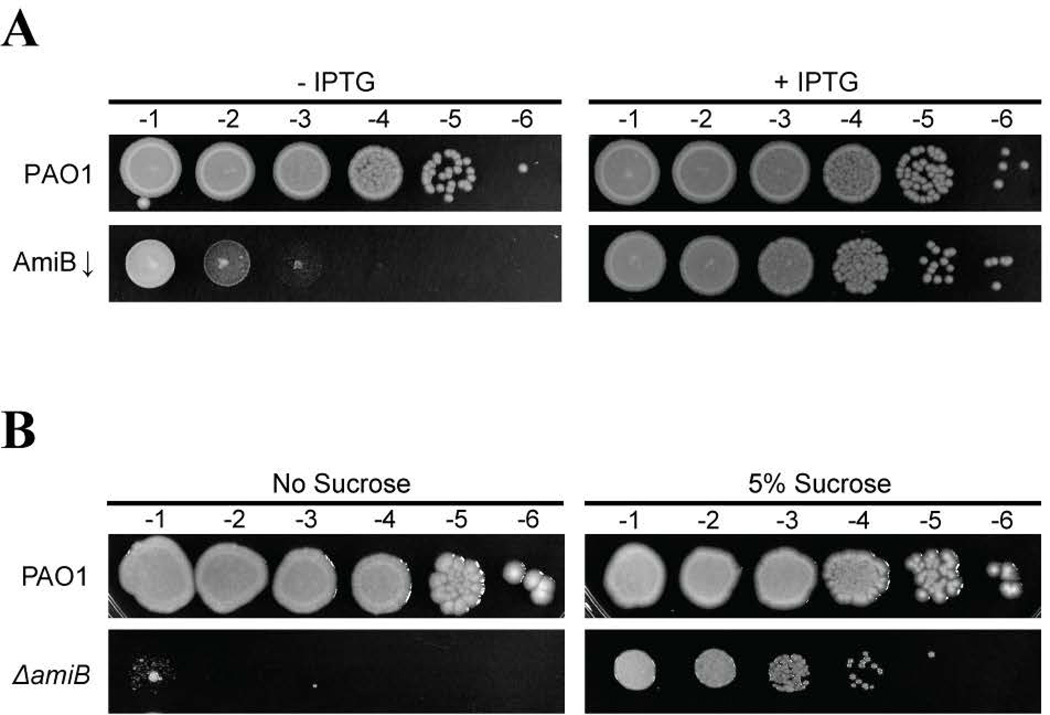
To examine the in vivo function of Pseudomonas PG amidases, we set out to disrupt the genes encoding these proteins in the PAO1 genetic background. Despite multiple attempts, we were unable to construct an amiB deletion strain that would propagate on LB medium, suggesting that amiB is essential. Consistent with this observation, the amiB mutants present in the two-allele PAO1 transposon library are unlikely to be nulls, harboring insertions at the extreme 3’ end of the gene, downstream of the catalytic domain (Held et al., 2012). To formally test amiB essentiality, we placed an IPTG-inducible version of amiB (PTOPLAC::amiB) at the Tn7 integration locus and deleted the native copy of the gene. The resulting AmiB depletion strain [ΔamiB (PTOPLAC::amiB)] was viable in the presence of IPTG, but exhibited a severe plating defect in the absence of the inducer, confirming the essentiality of AmiB for Pseudomonas growth (Fig. 1A). During the construction of the depletion strain, we serendipitously discovered that cells defective for AmiB can be propagated on LB medium supplemented with 5% sucrose, which likely functions as an osmoprotectant to stabilize the cell envelope (Fig. 1B). This phenotype is not strain-dependent, since deletion of amiB in PA14, a genetically divergent strain of P. aeruginosa, was also lethal, unless cells were grown in the presence of sucrose (Fig. S1). The amiB gene is just upstream of mutL, which encodes a component of the DNA mismatch repair system (Oliver et al., 2002) (Fig. S2A). Therefore, we assessed the potential polarity of the amiB deletion on mutL expression by comparing the frequency of spontaneous streptomycin-resistant mutants in the PaAmiB depletion strain relative an amiB+ control. Resistant mutants arose at approximately the same rate in both strains (Fig. S2B), indicating that the expression of mutL is unlikely to be affected by the deletion of amiB.
Figure 1. AmiB is conditionally essential in Pseudomonas.
A. Overnight cultures of PAO1 [WT] and BPA46 [ΔamiB (PTOPLAC::amiB)] were subcultured 1:10 into LB with 1mM IPTG and allowed to grow for 2h at 30°C. They were then normalized to OD600 of 0.5, washed with fresh LB, and subjected to serial dilution. 5µL of each dilution were spotted onto LB plates with or without 1mM IPTG as indicated.
B. Overnight cultures of PAO1 [WT] and BPA157 [ΔamiB] grown in LB supplemented with 5% sucrose were normalized to OD600 of 0.5 and subjected to serial dilution, before 5µL of each dilution were spotted onto LB plates either containing or lacking 5% sucrose. Plates were incubated at 30°C for 2d prior to imaging.
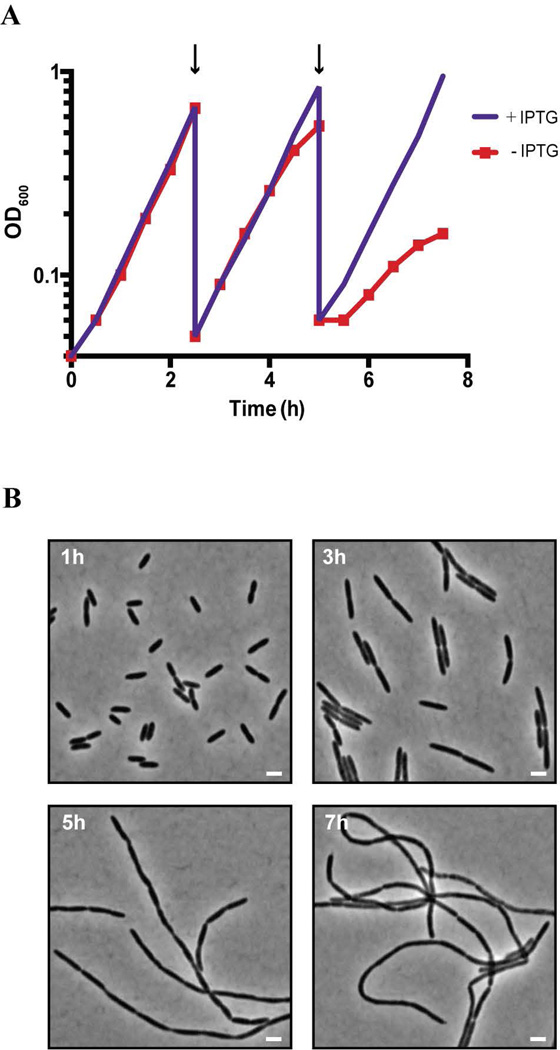
To determine the phenotypic consequences of PaAmiB inactivation, we monitored growth and cellular morphology for several generations following PaAmiB depletion (Fig. 2). Cells of the PaAmiB depletion strain [ΔamiB (PTOPLAC::amiB)] grown with or without IPTG displayed similar growth rates for approximately 5 hours following the initiation of PaAmiB depletion, after which the growth rate of the uninduced culture markedly declined (Fig. 2A). Cell morphology remained unchanged at the early stages of depletion, but following three hours of growth in the absence of the inducer, PaAmiB-depleted cells began to form elongated cell pairs (Fig. 2B). This apparent cell separation defect became progressively worse with time. After five hours of PaAmiB depletion, long cell filaments were observed (Fig. 2B). Some of these filamentous cells had regularly spaced constrictions indicative of a cell separation defect. However, many filaments with irregularly positioned, shallow constrictions were also observed and increased in abundance during continued growth in depletion conditions (Fig. 2B and Fig. S3A).
Figure 2. Depletion of AmiB leads to inhibition of growth and aberrant cell morphology.
A. Growth curve of BPA46 [ΔamiB (PTOPLAC::amiB)] with and without 1mM IPTG. An overnight cell culture was washed with fresh LB and diluted to an OD600 of 0.04 in LB either containing or lacking 1mM IPTG. Cells were then grown with agitation at 37°C, with OD600 readings taken every 30min. At 2.5h and 5h time points (marked with arrows), the cultures were diluted to prevent entry into stationary phase.
B. At the indicated time points of the growth curve presented in (A), samples of cells grown without IPTG were chemically fixed and preserved on ice. These cells were subsequently imaged using phase contrast optics. The bars equal 2µm.
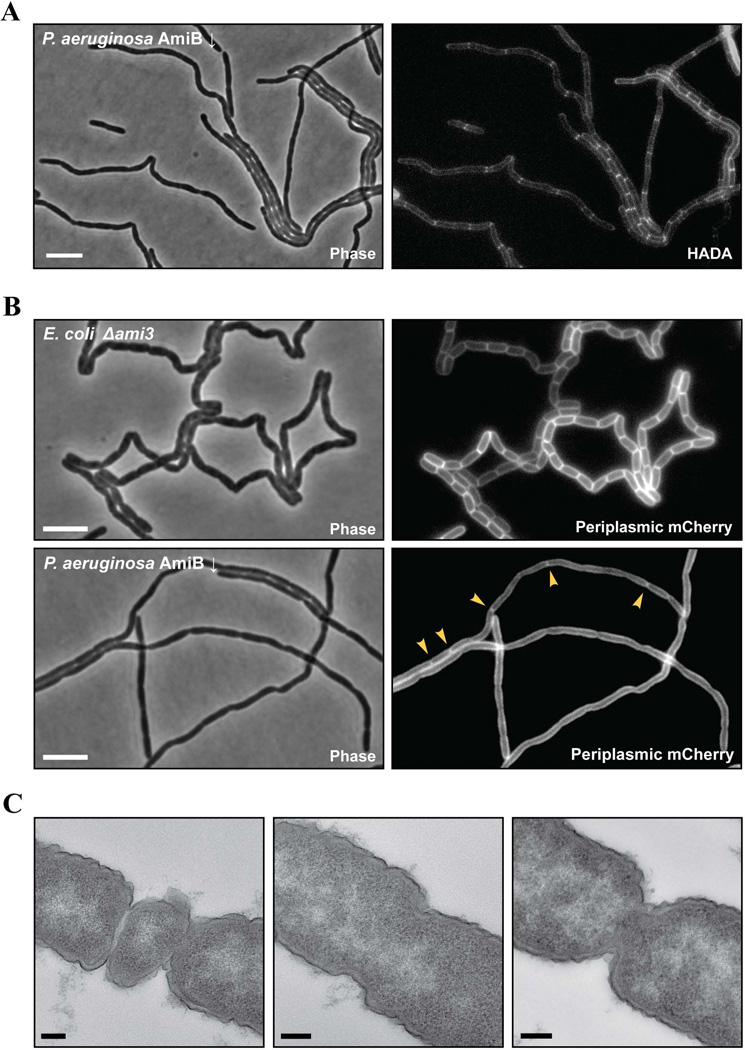
We further investigated the state of cell constriction in the PaAmiB-depleted cells by visualizing septal PG biogenesis with fluorescent D-amino acid (HADA) staining (Kuru et al., 2012) and the progress of inner membrane invagination with a periplasmic mCherry marker. Most areas of constriction along the length of the cell filaments were associated with a transverse band of HADA staining, indicating that septal PG biogenesis has been successfully initiated at these sites (Fig. 3A). However, very few constrictions in the filaments displayed a band of periplasmic fluorescence, suggesting that inner membrane invagination and fusion were largely incomplete (Fig. 3B). This observation is in striking contrast to the phenotype of E. coli triple amidase mutant, which shows very regular bands of periplasmic mCherry fluorescence indicative of advanced inner membrane invagination (Fig. 3B). Similar results were obtained by monitoring inner membrane invagination directly with a membrane-anchored mCherry fusion (data not shown). Transmission electron microscopy of thin sections of the PaAmiB-depleted filaments confirmed that completed septa showing the hallmarks of a cell separation defect were rare, and the majority of the observed constrictions in the micrographs indeed appeared to be incomplete (Fig. 3C).
Figure 3. Loss of AmiB interferes with cytokinesis.
A. Phase contrast and HADA fluorescence images of BPA46 [ΔamiB (PTOPLAC::amiB)]. Overnight culture of cells grown in LB supplemented with 1mM IPTG was washed, diluted 1:2000 in LB lacking the inducer, and allowed to grow for 5h at 37°C. Then, HADA was added to the final concentration of 30µM, and the sample was incubated at 37°C for 30min. Finally, the cells were washed to eliminate unincorporated label and imaged with phase contrast and DAPI epifluorescence optics. The space bar is 5µm.
B. Phase contrast and mCherry fluorescence images of P. aeruginosa BPA46/pAAY65 [ΔamiB (PTOPLAC::amiB) / Psyn135::ssdsbA-mCherry] and E. coli TB171/pAAY65 [ΔamiA ΔamiB ΔamiC / Psyn135ssdsbA-mCherry]. Overnight cultures grown in LB with gentamicin (and IPTG for BPA46) were washed to eliminate the antibiotic and the inducer, diluted 1:2000 (BPA46) or 1:5000 (TB171), and allowed to grow for 5h at 37°C prior to imaging. Arrowheads indicate P. aeruginosa division sites with periplasmic mCherry accumulation. The space bars are 5µm.
C. Electron micrographs of BPA46 [ΔamiB (PTOPLAC::amiB)]. Cells were grown as in (B), but after the 5h incubation, they were chemically fixed, used to prepare thin sections, and imaged with a transmission electron microscopy. The space bars are 100nm.
In contrast to the loss of PaAmiB function, deletion of amiA had no effect on either cell growth or morphology (Fig. S3B) and did not exacerbate the phenotypes associated with PaAmiB depletion (data not shown). We conclude that, as in E. coli, PaAmiB function contributes to the completion of cell division in P. aeruginosa, which is consistent with the septal localization of a native-site PaAmiB-mCherry fusion (Jorgenson et al., 2014) (Fig. S4). However, although in E. coli, amidase inactivation largely manifests as a defect in septal PG splitting, the phenotype of PaAmiB suggests that it also plays an important role in promoting cell constriction (see Discussion).
Three partially redundant dLytM proteins are required for daughter cell separation
To promote cell separation in E. coli, PG amidases require activation by partner proteins with LytM (Peptidase M23) domains (Firczuk and Bochtler, 2007; Uehara et al., 2010). E. coli encodes four proteins with LytM domains: EnvC, NlpD, YgeR, and MepM (YebA) (Uehara et al., 2009). EnvC, NlpD, and YgeR are dLytM proteins without critical endopeptidase active site residues. Of these, EnvC and NlpD play the main roles in cell separation, activating either AmiA and AmiB, or AmiC, respectively (Uehara et al., 2010). MepM, on the other hand, is an active endopeptidase involved in cell elongation (Singh et al., 2012). P. aeruginosa encodes nine different proteins with recognizable LytM domains, three of which (PA3623, PA4924, and PA5133) are predicted to produce dLytM factors that may play a role in amidase activation and cell separation. A phylogenetic tree generated using multiple sequence alignments of the E. coli and P. aeruginosa LytM proteins revealed that PA3623 and PA4924 closely cluster with E. coli NlpD and YgeR (Fig. S5A). These factors also have similar domain architecture, carrying a lipoprotein signal sequence at the N terminus, a PG-binding LysM domain, and the degenerate LytM domain at the C terminus (Fig. S5B). Finally, PA3623 shares 41% identity with E. coli NlpD, and its gene is encoded at a genomic locus (between pcm and rpoS genes) similar to nlpD in E. coli (Fig. S2C). Hereafter, we refer to PA3623 and PA4924 as PaNlpD and NlcS (NlpD-like cell separation factor), respectively. Another grouping of proteins involves PA5133 and E. coli EnvC, which share 32% identity and are both predicted to have a periplasm-targeted signal sequence, two coiled-coil domains, and a dLytM domain (Fig. S5). E. coli envC and PA5133 are also predicted to be co-transcribed with orthologous putative polysaccharide deacetylase-encoding genes, yibQ and PA5135, respectively. Therefore, we will henceforth refer to PA5133 as PaEnvC. The remaining LytM domain factors in Pseudomonas show strong divergence from the dLytM proteins of E. coli, making their participation in amidase activation less likely (Fig. S5A).
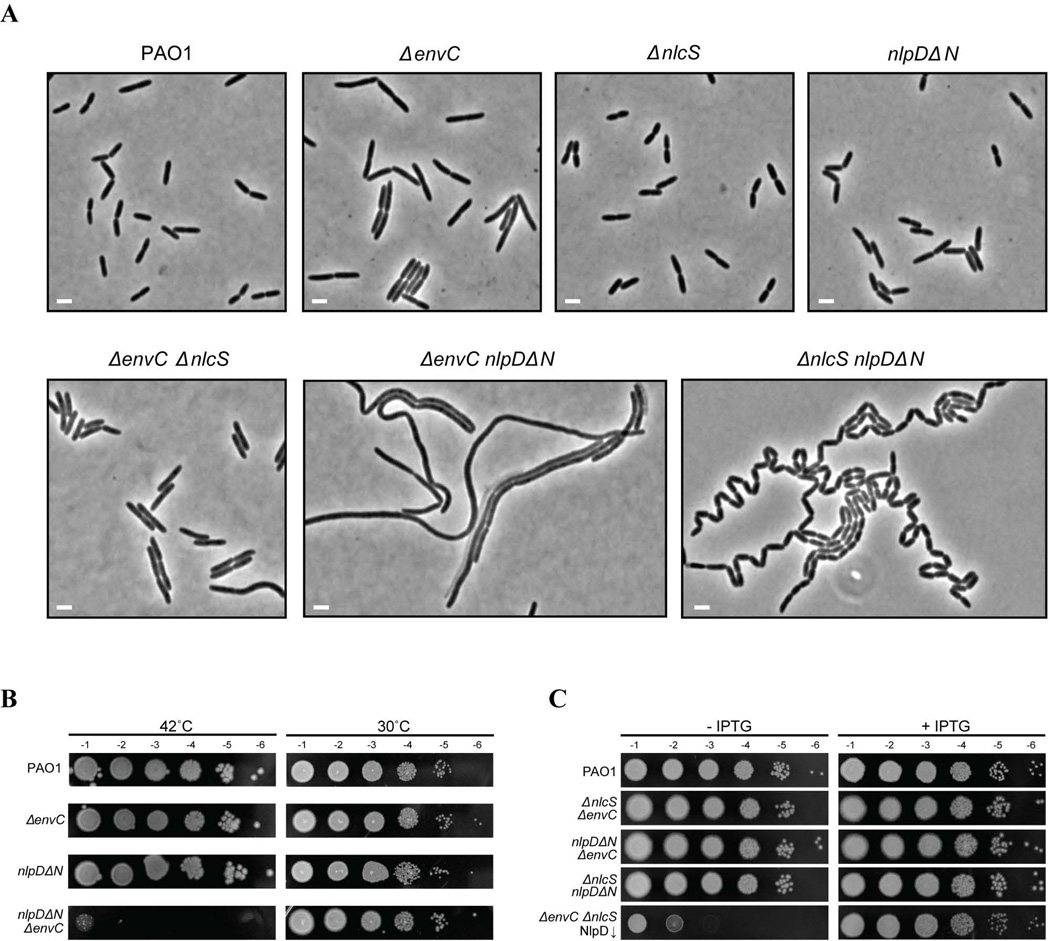
We investigated the involvement of PaNlpD, NlcS, and PaEnvC in cell separation by assessing their loss-of-function phenotypes. Derivatives of PAO1 with single deletions of nlpD, nlcS, or envC were readily constructed, indicating that these genes are not essential. None of the single deletion mutants displayed a significant division phenotype, with only ΔenvC cells appearing slightly elongated at 42°C relative to the wild-type PAO1 control (Figs. 4A and S6). One minor complication with the ΔnlpD strain is the likely presence of a promoter for the downstream rpoS gene within the nlpD coding sequence (Fig. S2C). rpoS encodes an alternative sigma factor that alters the expression of a large number of genes in response to a variety of stresses (Venturi, 2003; Schuster et al., 2003). Accordingly, we found that the ΔnlpD mutation induced overproduction of the pigment pyocyanin (Fig. S2D), a phenotype associated with an RpoS defect in P. aeruginosa (Suh et al., 1999). Therefore, to study the effect of NlpD inactivation without the complications of additional rpoS phenotypes, we constructed a small in-frame deletion in the nlpD gene (nlpDΔN) that removes the coding sequence for the N-terminal lipoprotein signal sequence from NlpD and thus prevents its export to the periplasm, where it normally functions. Importantly, nlpDΔN and ΔnlpD mutants displayed the same morphological phenotypes when combined with other dLytM factor mutants, but the rpoS-related pyocyanin phenotype was only observed in the ΔnlpD the background (Fig. S2D and data not shown). Thus, a strain with the nlpDΔN mutation is NlpD⍰, but remains RpoS+.
Figure 4. Phenotypic analysis of dLytM factor mutants.
A. Phase contrast images of PAO1 [WT], BPA107 [ΔenvC], BPA14 [ΔlytM2], BPA70 [nlpDΔN], BPA57 [ΔenvC ΔlytM2], BPA109 [ΔenvC nlpDΔN], and BPA72 [ΔlytM2 nlpDΔN]. Overnight cultures of cells grown in LB at 30°C were diluted 1:2000 in LB and grown for 4h at 42°C. Cells were then placed on agarose pads and imaged with phase contrast optics. All space bars are 2µm.
B. Viability of PAO1 [WT], BPA107 [ΔenvC], BPA70 [nlpDΔN], and BPA109 [ΔenvC nlpDΔN] cells. Overnight cultures were subcultured 1:10 into LB and allowed to grow for 2h at 30°C. They were then normalized to OD600 of 0.5 and subjected to serial dilution. 5µL aliquots of the dilutions were spotted onto LB plates, which were then incubated either at 42°C or 30°C.
C. Viability of PAO1/pPSV38 [WT], BPA57/pPSV38 [ΔenvC ΔlytM2], BPA109 /pPSV38 [ΔenvC nlpDΔN], and BPA72/pPSV38 [ΔlytM2 nlpDΔN], and BPA204/pPSV38 [ΔenvC ΔlytM2 (attTn7::lacIq) (PTOPLAC::nlpD)]. Overnight cultures grown in LB supplemented with 15µg/mL gentamicin and 1mM IPTG were washed, diluted 1:10 in LB with 1mM IPTG, and allowed to grow for 2h at 30°C. The cells were then washed with fresh LB, normalized to OD600 of 0.5, and subjected to serial dilution, spotting 5µL of each dilution on LB plates either containing or lacking 1mM IPTG. The plates were incubated at 30°C. The plasmid pPSV38 encodes lacIq and was needed for effective depletion of NlpD in strain BPA204/pPSV38 [ΔenvC ΔlytM2 (attTn7::lacIq) (PTOPLAC::nlpD)].
Only double dLytM mutants with nlpD disruptions were found to display significant morphological phenotypes (Fig. 4A). A severe cell separation defect was observed with the nlpDΔN ΔnlcS mutant, which formed long chains of deeply constricted cells at both 30°C and 42°C (Figs. 4A and S6). The nlpDΔN ΔenvC mutant, on the other hand, grew and divided normally at 30°C, but displayed a lethal division defect and formed relatively smooth filaments at 42°C (Fig. 4A–B). This phenotype is reminiscent of the division problems observed for a single ΔenvC mutant in E. coli grown in low salt medium at 42°C (Ichimura et al., 2002; Hara et al., 2002). Thus, as in E. coli, dLytM factors of P. aeruginosa play an important role in the cell separation process, and like PaAmiB, in some instances appear to also function in cell constriction.
By analogy with E. coli, the most likely function of the dLytM factors in P. aeruginosa is the activation of the cell separation amidases. Given that PaAmiB is essential in this organism, we expected that some combination of its potential activators should also be essential for growth. The viability of all mutant combinations lacking two of the three recognizable dLytM factors suggested the possibility that all three may have at least some ability to stimulate PaAmiB activity in vivo. We, therefore, constructed a PaNlpD depletion strain in which the sole copy of nlpD was placed under the control of an IPTG-inducible promoter in a mutant deleted for envC and nlcS [ΔnlcS ΔenvC (PTOPLAC::nlpD)]. This strain displayed an IPTG-dependent growth phenotype and a cell division defect similar to PaAmiB depletion, which is consistent with the possibility that all three P. aeruginosa dLytM proteins play partially redundant roles in PaAmiB activation (Fig. 4C and Fig. S7A). We conclude that the activity of at least one dLytM factor is required for proper cell division and viability of P. aeruginosa.
Outer membrane function is compromised upon AmiB inactivation
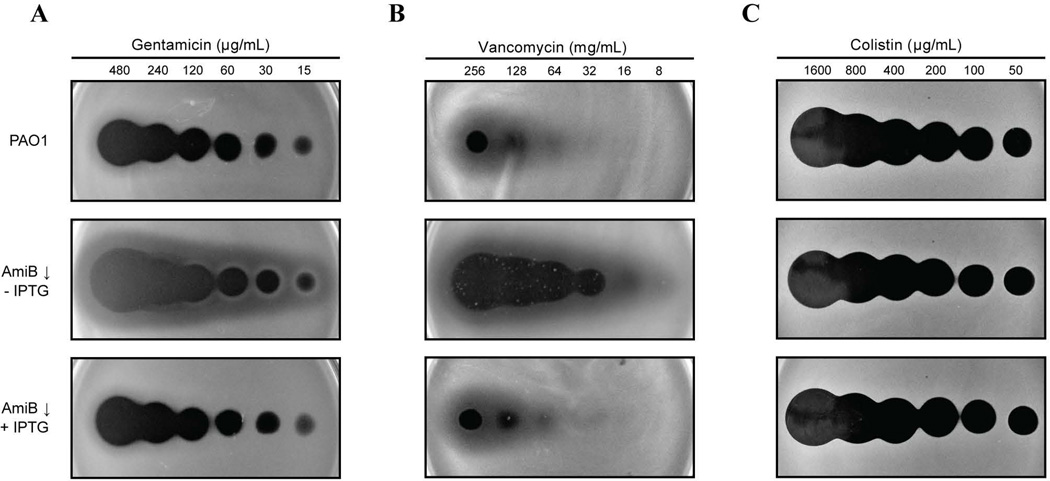
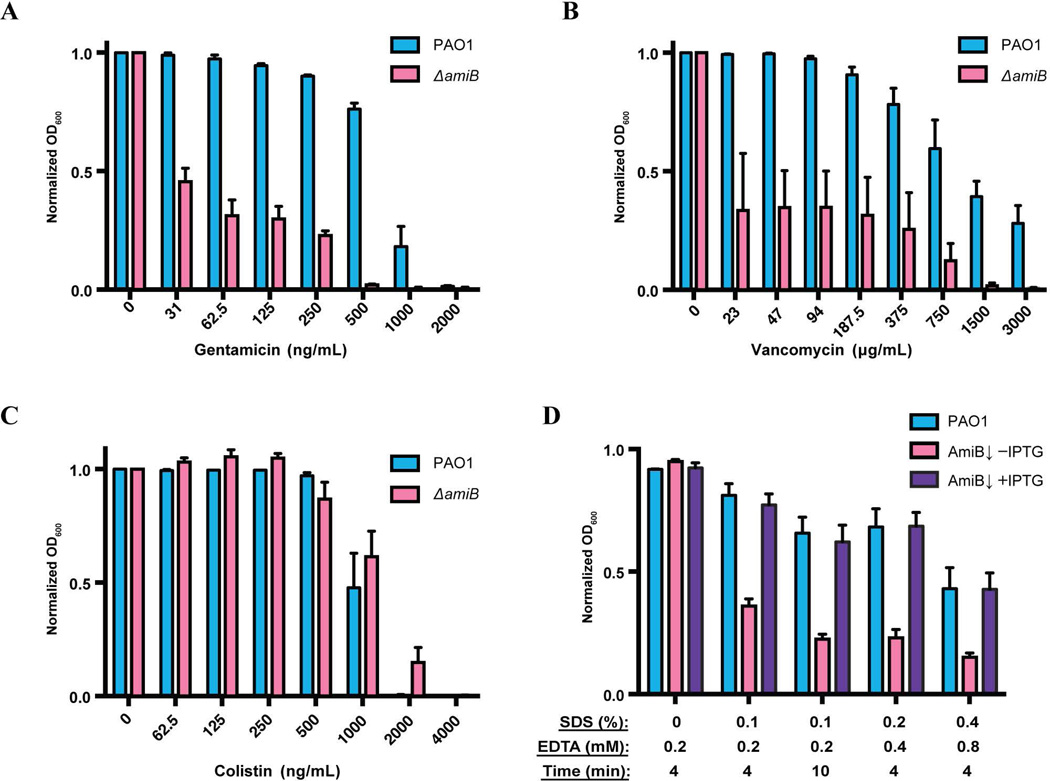
The outer membrane permeability barrier of P. aeruginosa is exceedingly robust, resulting in a high intrinsic resistance to antibiotics (Yoshimura and Nikaido, 1982; Hancock and Bell, 1988). Because the loss of PG amidase activity has been associated with outer membrane defects and antibiotic hypersensitivity in other gram-negative organisms (Heidrich et al., 2002; Ize et al., 2003; Weatherspoon-Griffin et al., 2011), we wondered whether PaAmiB inactivation would also be an attractive means of overcoming drug resistance in P. aeruginosa. In order to rapidly compare the sensitivity of PAO1 [WT] with the PaAmiB depletion strain BPA46 [ΔamiB (PTOPLAC::amiB)] to a range of different drugs, an antibiotic spotting assay was used. Soft agar was inoculated with either PAO1 or BPA46 and distributed over LB agar plates. In the case of BPA46, soft agar was prepared either with or without addition of IPTG. Once the top agar solidified, a fixed volume of an antibiotic dilution series was spotted onto the agar, and the plates were incubated for 2 days at room temperature (~22°C). Antibiotic sensitivity was then assessed by comparing the resulting zones of clearing on the bacterial lawns. Notably, although BPA46 could not form colonies without IPTG, it was able to undergo a sufficient number of mass doublings to form a relatively robust lawn in soft-agar even in the absence of the inducer. The spotting assay using these lawns revealed that cells depleted for PaAmiB were hypersensitive to gentamicin, tobramycin, vancomycin, meropenem, ertapenem, and the LpxC inhibitor PF-5081090, as indicated by the larger zones of clearing observed for BPA46 in the absence of inducer relative to PAO1 or BPA46 grown with IPTG (Fig. 5, data not shown). We also showed that the inactivation of the three dLytM factors that likely stimulate PaAmiB in vivo leads to a similar vancomycin hypersensitivity phenotype (Fig. S7B). Conversely, AmiB-depleted cells showed normal sensitivity to colistin, imipenem, rifampicin, erythromycin, and nalidixic acid (Fig. 5, data not shown), indicating that the observed hypersensitivities are not simply a consequence of the growth defect associated with loss of PaAmiB function. Nevertheless, to avoid the possibility of PaAmiB essentiality confounding the interpretation of the results, we also investigated the drug sensitivity of ΔamiB cells grown under permissive conditions (LB with 5% sucrose). Cells of PAO1 or its ΔamiB derivative were grown overnight in liquid medium in the presence of a range of different gentamicin, vancomycin, or colistin concentrations, and the OD600 of the resulting cultures was measured the following morning. The results mirrored those for the antibiotic spotting assay, clearly indicating that PaAmiB inactivation results in hypersensitivity to gentamicin and vancomycin, but not colistin (Fig. 6A–C).
Figure 5. Antibiotic susceptibility of AmiB-depleted cells.
A. Gentamicin spotting assay. Overnight cultures of PAO1 [WT] and BPA46 [ΔamiB (PTOPLAC::amiB)] were diluted 1:100 in LB supplemented with 1mM IPTG and allowed to grow at 30°C for 3h. Following two washes with fresh LB, the cells were resuspended in LB lacking IPTG and allowed to grow for another 2h. They were then normalized to OD600 of 0.4, diluted 1:40 in molten LB 0.8% agar either containing or lacking 1mM IPTG as indicated, and distributed over LB plates. After allowing 30min at room temperature for the agar to solidify, 5µL aliquots of gentamicin solutions at the indicated concentrations were spotted onto the surface of the plates. Plates were incubated at room temperature (~22°C) for 2d and then photographed.
B. Vancomycin spotting assay. Cells were treated the same as in (A), except that 10µL aliquots of vancomycin solutions at indicated concentrations were spotted onto the plates.
C. Colistin spotting assay. Cells were treated the same as in (A), except that 5µL aliquots of colistin solutions at indicated concentrations were spotted onto the plates.
Figure 6. Drug and detergent sensitivity determinations in liquid culture.
A–C. Gentamicin (A), vancomycin (B) and colistin (C) MIC assays. Overnight cultures of PAO1 [WT] and BPA157 [ΔamiB] grown from isolated colonies in LB with 5% sucrose were subcultured 1:5 and allowed to grow for 2h at 30°C. The cells were then normalized to OD600 of 0.005 and diluted in LB, which was supplemented with 5% sucrose and the indicated concentrations of antibiotics. Optical density readings were taken following ~24 h of growth at 30°C. The experiment was performed three times, with two biological replicates of BPA157 and one biological replicate of PAO1 each time. The values are normalized to the no-antibiotic controls, and the error bars represent standard error. The vancomycin experiment includes two samples of BPA157 that showed dramatically higher OD readings compared to the other four samples and likely contained AmiB-bypass suppressor mutants.
D. Susceptibility to lysis by SDS. Overnight cultures of PAO1 [WT] and BPA46 [ΔamiB (PTOPLAC::amiB)] were washed, diluted 1:1000 in LB either containing or lacking 1mM IPTG, and allowed to grow for 5h at 37°C. The cells were then normalized to OD600 of 0.5, mixed with SDS and/or EDTA at the indicated final concentrations, and incubated for 4–10min before measuring final optical density. The experiment was performed in triplicate, each time with three technical replicates per biological sample. The chart presents values normalized to the initial optical density prior to the addition of SDS/EDTA, and the bars represent standard error.
The hypersensitivity of cells lacking PaAmiB to a variety of antibiotics with different modes of action is indicative of an outer membrane permeability barrier defect. Consistent with this possibility, cells depleted for PaAmiB were also hypersensitive to treatment with a detergent (Fig. 6D). The hypersensitivity to vancomycin, which is normally excluded from gram-negative cells due to its large size and inability to pass through the outer membrane (Vaara, 1993; Mori et al., 2012), also suggests that the permeability barrier problems extend beyond a malfunctioning of the efflux pumps. Further support for this possibility comes from the antibiotic spotting assay, which indicates that a PAO1 derivative lacking the five major efflux systems (Chuanchuen et al., 2005) still displays antibiotic hypersensitivity upon AmiB depletion (Fig. S8).
In addition to examining the intrinsic resistance of P. aeruginosa to antibiotics, we also investigated the effect of PaAmiB inactivation on an acquired drug resistance mechanism. Carbapenem antibiotics like meropenem are imporant therapeutics used clinically to treat P. aeruginosa infections, but loss-of-function mutations in oprD, which encodes the major outer membrane porin, block carbapenem entry and are a common cause of carbapenem resistance (Köhler et al., 1999; Naenna et al., 2010). Importantly, we found that the loss of PaAmiB function could restore meropenem sensitivity to a PAO1 strain deficient in oprD (Fig. S9). We, therefore, conclude that PaAmiB inactivation results in an outer membrane permeability defect in P. aeruginosa and overcomes both intrinsic and acquired resistance mechanisms in this highly drug-resistant organism.
Amidase essentiality is suppressed by the activation of envelope stress responses
Given that the E. coli triple amidase mutant (ΔamiA ΔamiB ΔamiC) is viable (Heidrich et al., 2001), we were surprised to find that PaAmiB is essential for the growth of P. aeruginosa. We reasoned that suppressor mutants that bypass PaAmiB essentiality in P. aeruginosa might reveal why amidase activity is essential in this organism, but not in E. coli. For the bypass suppressor selection, we constructed a P. aeruginosa strain deleted for both LytC-type PG amidases (ΔamiA ΔamiB) to avoid the possible isolation of ΔamiB suppressors that overproduce PaAmiA. The double mutant was maintained on LB supplemented with 5% sucrose and plated on standard LB medium to select for spontaneous suppressors. The suppressor mutants were purified on LB plates without sucrose to confirm their ability to grow under restrictive conditions, and whole-genome Illumina sequencing was used to identify the mutations responsible for suppression.
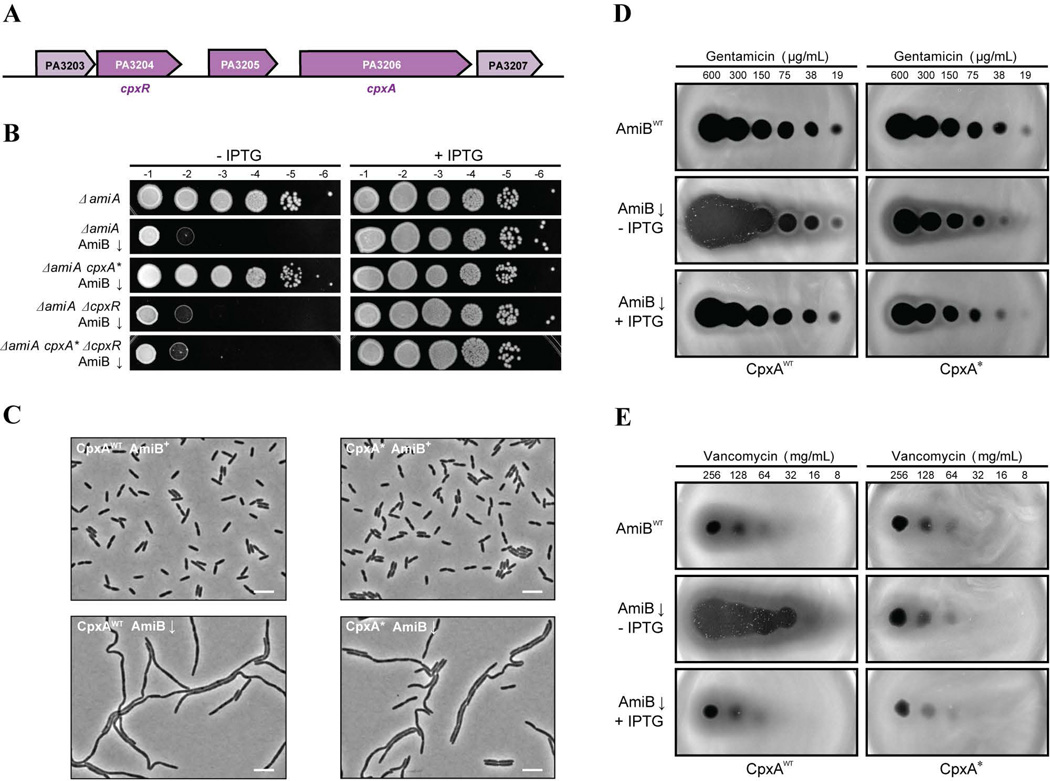
Mutations bypassing AmiB-dependent growth were identified in several different loci. The two independent suppressors we focused on for this study had missense mutations in the PA3206 gene (Fig. 7A), which encodes a putative sensor histidine kinase. One allele results in an A154S substitution in the second predicted transmembrane domain, while the other allele corresponds to an I273T change in the cytoplasmic kinase domain. To confirm the suppression phenotype of mutations in PA3206, the allele generating the A154S substitution was reconstructed in a PaAmiB depletion strain that also lacked PaAmiA [ΔamiA ΔamiB (PTOPLAC::amiB)]. The resulting strain was indeed able to grow in the absence of AmiB induction, indicating that the PA3206 mutation restores viability to AmiB-deficient cells (Fig. 7B). The closest homolog of PA3206 in E. coli is the histidine kinase CpxA (31% identity; 53% similarity). In E. coli, CpxA is a component of a well-characterized envelope stress response that regulates the expression of over fifty genes in response to a variety of membrane and PG perturbations (DiGiuseppe and Silhavy, 2003; Bury-Moné et al., 2009; Evans et al., 2013). In addition to CpxA, the Cpx system includes the response regulator CpxR, which is regulated by CpxA through phosphorylation and dephosphorylation (Raivio and Silhavy, 1997), and the periplasmic protein CpxP, which inhibits the Cpx response when overexpressed (Raivio et al., 1999). Consistent with the possibility that PA3206 is the P. aeruginosa ortholog of CpxA, the adjacent genes at its genomic locus, PA3205 and PA3204, encode homologs of CpxP (31% identity; 42% similarity) and CpxR (46% identity; 64% similarity), respectively (Fig. 7A). However, the closest ortholog of PA3205 in E. coli is actually the periplasmic chaperone Spy, whose gene is also a part of the Cpx regulon (Raivio et al., 2000). Nevertheless, based on the observed similarities, we henceforth refer to PA3206 as PaCpxA and PA3204 as PaCpxR.
Figure 7. Suppression of AmiB disruption phenotypes by a CpxA*-like mutation.
A. Genomic context of cpxA in PAO1 genome. cpxA (PA3206) is located adjacently to PA3205 and PA3204, which encode homologs of cpxP and cpxR, respectively.
B. Viability of PAO1 [WT], BPA60 [ΔamiA], BPA220 [ΔamiB ΔamiA (PTOPLAC::amiB)], BPA231 [ΔamiB ΔamiA cpxA(A154S) (PTOPLAC::amiB)], BPA245 [ΔamiB ΔamiA ΔcpxR (PTOPLAC::amiB)], and BPA247 [ΔamiB ΔamiA cpxA(A154S) ΔcpxR (PTOPLAC::amiB)] cells. Cells were treated as for Fig. 1A.
C. Phase images of BPA220 [ΔamiB ΔamiA (PTOPLAC::amiB)] and BPA231 [ΔamiB ΔamiA cpxA(A154S) ΔcpxR (PTOPLAC::amiB)]. Overnight cultures grown in LB supplemented with 1mM IPTG were washed and diluted 1:2000 in LB either containing or lacking 1mM IPTG. The cells were grown at 37°C for 5h prior to imaging on agarose pads with phase contrast optics. Bars equal 5µm.
D–E. Gentamicin (D) and Vancomycin (E) spotting assays for PAO1 [WT], BPA46 [ΔamiB (PTOPLAC::amiB)], BPA237 [cpxA(A154S)], and BPA294 [ΔamiB cpxA(A154S) (PTOPLAC::amiB)]. The assays were performed following the same protocol as in Fig. 5.
Our analysis revealed that loss of PaAmiB function results in severe defects in cell division and the barrier function of the outer membrane. We, therefore, suspected that the suppressor mutations in cpxA might promote constitutive induction of a Cpx-like response in P. aeruginosa, allowing cells to cope with the division defect and envelope damage that results from PaAmiB inactivation. In support of this hypothesis, many alleles of E. coli cpxA that result in constitutive Cpx induction (known as cpxA*) possess amino acid substitutions in the second transmembrane helix of CpxA (Raivio and Silhavy, 1997), the location of the A154S substitution in our PaAmiB suppressor. If the cpxA(A154S) mutation behaves like a cpxA* allele, its ability to suppress the AmiB defect should depend on PaCpxR. This possibility proved to be the case, as AmiB depletion was lethal in cells lacking cpxR, whether or not they also harbored the cpxA(A154S) allele (Fig. 7B).
In addition to the suppression of PaAmiB essentiality, we investigated the ability of the cpxA(A154S) allele to suppress the cell division and antibiotic hypersensitivity phenotypes associated with the loss of PaAmiB function. The PaAmiB depletion strain carrying the cpxA(A154S) allele formed cell chains that appeared shorter than the corresponding CpxA(WT) strain. However, the mutant PaCpxA did not fully suppress the cell division defect observed upon PaAmiB depletion, and many long chains of cells were still present after growth without induction (Fig. 7C). In contrast to the division phenotype, the cpxA(A154S) mutation dramatically reduced the hypersensitivity of the PaAmiB depletion strain to gentamicin and vancomycin (Fig. 7D–E).
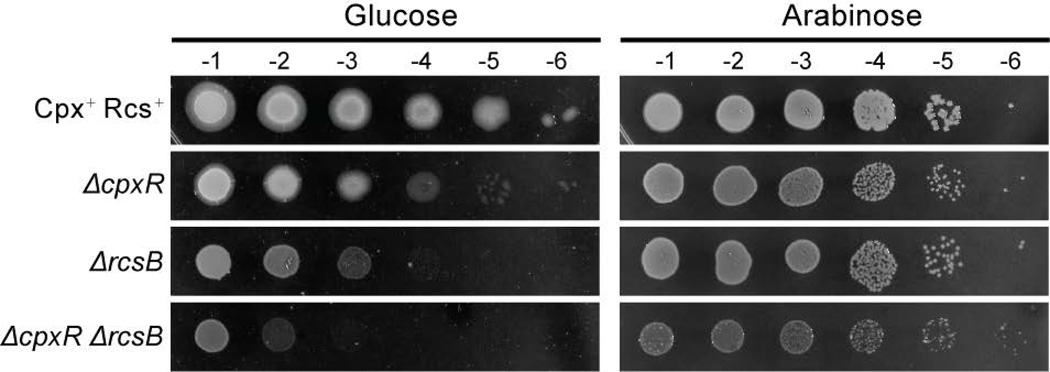
The finding that activation of a putative Cpx-like envelope stress response is capable of suppressing the essentiality of PaAmiB in P. aeruginosa led us to wonder whether a similar response promotes the viability of the triple amidase mutant in E. coli. To test this possibility, we constructed an E. coli amidase depletion strain in which all three of the amidase genes were disrupted and an ectopic copy of amiA was placed under control of the arabinose promoter at the phage λ attachment site [ΔamiA ΔamiB amiC::KanR (PBAD::amiA)]. As expected from previous results, this strain was viable with or without amiA induction (Fig. 8). The isogenic strain with cpxR deletion also remained viable upon AmiA depletion, but formed smaller colonies than its Cpx+ parental strain, suggesting that the Cpx response is ameliorating some of the detrimental effects of amidase inactivation. Furthermore, both ΔcpxR and Cpx+ colonies grown without induction had a mucoid appearance (Fig. 8), which is a phenotype associated with the activation of another major envelope stress response pathway in E. coli, the Rcs system (Majdalani and Gottesman, 2005). The Rcs response involves a complex phospho-relay, with the response regulator RcsB ultimately mediating the changes in gene expression (Majdalani and Gottesman, 2005). Deletion of rcsB in the E. coli amidase depletion strain led to a plating defect in the absence of inducer that was more severe than that observed upon cpxR disruption (Fig. 8). However, small colonies were formed and the strain remained viable without arabinose (Fig. 8). We, therefore, deleted both cpxR and rcsB in the E. coli amidase depletion strain. As opposed to the individual mutants, inactivation of both stress responses resulted in a severe plating defect and the loss of viability upon amidase depletion (Fig. 8). We conclude that Cpx and Rcs stress responses in E. coli play partially redundant roles in supporting the growth of cells that lack cell separation amidase activity. In combination with the PaAmiB suppressors characterized in P. aeruginosa, this finding suggests that the difference in amidase essentiality in E. coli and P. aeruginosa may stem, at least in part, from differing levels of Cpx- and/or Rcs-mediated gene expression in these organisms. Furthermore, the results also suggest the possibility that a factor or a collection of factors induced by these stress response systems is capable of correcting the cell division problems and/or repairing the cell envelope damage resulting from amidase inactivation.
Figure 8. Viability of E. coli triple amidase mutant is dependent on the Cpx and Rcs stress responses.
Cultures of AAY14 [ΔamiA ΔamiB ΔamiC (PBAD::ssdsbA-amiA)], and its derivatives lacking cpxR, rcsB, or both of these genes were grown overnight in LB supplemented with 0.2% arabinose, washed with 1x M9 salts, diluted 1:10 in M9 minimal medium supplemented with 0.2% maltose and 0.002% arabinose, and allowed to grow for 2h at 30°C. Cells were then normalized to OD600 of 0.5 and serial dilutions were prepared, 5µL aliquots of which were spotted onto M9 plates supplemented either with 0.2% glucose or 0.2% arabinose. Plates were incubated at 30°C for 2d and then photographed.
DISCUSSION
The physiological function of LytC-type amidases has been investigated in several different proteobacteria, including E. coli, Salmonella typhimurium, Vibrio cholerae, and Neisseria gonorrhoeae (Heidrich et al., 2001; Garcia and Dillard, 2006; Priyadarshini et al., 2007; Weatherspoon-Griffin et al., 2011; Moll et al., 2014). In all cases, these enzymes have been found to play a significant role in the process of septal PG splitting and daughter cell separation. In E. coli, proteins with dLytM domains were also implicated in cell separation and found to be critical activators of the amidases at the cytokinetic ring (Uehara et al., 2009; Uehara et al., 2010; Yang et al., 2012). Subsequently, proteins with dLytM domains were identified as cell separation factors in other gram-negative bacteria, presumably functioning via the activation of amidases or other potential cell separation enzymes (Goley et al., 2010; Möll et al., 2010; Poggio et al., 2010; Moll et al., 2014; Ercoli et al., 2015). Here, we investigated the function of amidases and dLytM proteins in the opportunistic pathogen P. aeruginosa. In agreement with the studies in other organisms, we find that PaAmiB, along with three dLytM proteins, plays a critical role in P. aeruginosa cell division. Importantly, however, our results differed from the E. coli paradigm, revealing a critical contribution of amidase activity to the success of cell constriction by the divisome.
Amidase activity and cell constriction
It has been proposed that PG amidases in E. coli participate in a positive feedback loop of cell wall synthesis and hydrolysis that stimulates divisome contraction during cytokinesis (Gerding et al., 2009). This model is based on studies of the essential cell division protein FtsN and its localization to the divisome. FtsN is a single-pass membrane protein with an N-terminal transmembrane domain and a periplasmic C-terminal domain consisting of a small, membrane-proximal essential domain (EFtsN) required for cell division and a non-essential, PG-binding SPOR domain (SFtsN) at the extreme C-terminus (Yang et al., 2004; Ursinus et al., 2004; Möll and Thanbichler, 2009; Gerding et al., 2009; Arends et al., 2009). Although the details of constriction initiation are still being elucidated (Liu et al., 2015; Tsang and Bernhardt, 2015a; Weiss, 2015; Tsang and Bernhardt, 2015b), the midcell localization of a small number of FtsN molecules is thought to start the process by stimulating septal PG synthesis by cell wall synthases of the divisome. According to the current model (Gerding et al., 2009; Lutkenhaus, 2009), it is EFtsN that functions to activate cell wall synthesis, and the resulting nascent septal PG is then rapidly processed by the amidases. The peptide-free glycan chains produced by the amidases serve as ligands for binding of SFtsN (Ursinus et al., 2004) and, therefore, are thought to promote the recruitment of more FtsN to the division site. Additional rounds of PG synthesis and remodeling are then promoted by the newly recruited EFtsN domains, which attract more FtsN to the septum via the SPOR domain in a self-enhancing cycle that drives cell constriction (Gerding et al., 2009).
According to the proposed FtsN-stimulated constriction cycle, amidase activity plays a key role in the contraction of the divisome. However, in this regard, the experimental results are mixed. On the one hand, the recruitment of SFtsN to the division site is known to require at least one of the three E. coli amidases (Gerding et al., 2009). Additionally, electron micrographs of PG sacculi purified from an E. coli triple amidase mutant display rings of dark staining that appear to be unprocessed, incompletely closed septa (Heidrich et al., 2001; Priyadarshini et al., 2007), suggesting potential problems with the constriction process. On the other hand, amidase-defective cell chains seem largely capable of completing inner membrane constriction and fusion (Heidrich et al., 2001; Priyadarshini et al., 2007), implying minimal contribution of amidase activity to cytokinetic ring closure.
Similar to recent studies with the cell separation protein RlpA (Jorgenson et al., 2014), our results in P. aeruginosa showcase the utility of studying cell biological processes in related genera to enhance our understanding of the underlying mechanisms. At the first glance, P. aeruginosa cells depleted for PaAmiB looked similar to the cell chains formed by E. coli mutants lacking amidase activity. However, closer inspection revealed that the cell constrictions in the P. aeruginosa chains were more shallow and irregularly positioned compared to those seen in the E. coli chains. Moreover, fluorescence microscopy with membrane and periplasmic markers, as well as electron microscopy, indicated that the observed sites of membrane invagination typically fail to complete inner membrane constriction and fusion. These observations strongly support the notion (Gerding et al., 2009) that septal PG processing by amidases positively influences the activity and/or the stability of the divisome, allowing the machinery to successfully complete cell constriction once the process is initiated. This role for the amidases appears to be more pronounced in P. aeruginosa than in E. coli, which might lead to the observed essentiality of PaAmiB. Nevertheless, based on these results and the prior studies discussed above, it is likely that amidase activity contributes similarly to the divisome function in E. coli and other proteobacteria. In support of this possibility, E. coli mutants deleted for envC or ftsEX display extreme defects in cell constriction when grown at high temperatures in low-salt media (de Leeuw et al., 1999; Ichimura et al., 2002; Hara et al., 2002; Schmidt et al., 2004; Reddy, 2006). These phenotypes have been previously interpreted to suggest that EnvC and its partner complex FtsEX have roles in divisome function/stability that are distinct from their involvement in amidase activation (Uehara et al., 2010; Yang et al., 2011). However, our finding that PaAmiB is critical for the completion of cell constriction in P. aeruginosa raises the distinct possibility that the division phenotypes of E. coli envC and ftsEX mutants are at least partially due to a defect in amidase activation. Further work will be required to test this model and to determine whether the function of the amidases in promoting cell constriction is primarily mediated through the proposed FtsN-stimulated constriction cycle (Gerding et al., 2009).
Envelope stress responses and the suppression of division defects
Clues as to why amidase activity is more important for division in P. aeruginosa compared to E. coli were provided by the suppressor mutations that render PaAmiB non-essential. Two independent suppressors mapped to a gene that appears to encode the P. aeruginosa ortholog of E. coli CpxA. The suppression activity of the cpxA alleles required the presence of the P. aeruginosa CpxR response regulator ortholog, indicating that they likely result in the constitutive activation of a Cpx-like envelope stress response. Variants of E. coli CpxA that elevate Cpx signaling levels are called CpxA*, and since the PaCpxA mutations that suppress PaAmiB essentiality map to a region of the protein that is modified in many E. coli CpxA* isolates (Raivio and Silhavy, 1997), we suspect that our suppressor alleles represent P. aeruginosa CpxA* equivalents. However, it is also possible that the suppressor cpxA alleles are null mutants that lead to higher basal levels of the Cpx-like response by inactivating the phosphatase activity of PaCpxA and thus increasing phospho-PaCpxR concentration. Regardless, our results implicate a Cpx-like response of P. aeruginosa in amelioration of cell division problems and/or their lethal consequences. Furthermore, our finding that E. coli mutants lacking amidase activity require functional Cpx and Rcs systems for growth indicates that envelope stress responses play a greater role in allowing gram-negative bacteria to survive division defects than previously appreciated. Consistent with this idea, the Rcs system in E. coli is known to stimulate the transcription of the key cell division genes ftsA and ftsZ (Carballès et al., 1999), while the Cpx system activates the expression of amidase genes amiA and amiC (Weatherspoon-Griffin et al., 2011). Further investigation is required to determine the mechanism by which the Cpx-like response in P. aeruginosa bypasses the amidase requirement for cell division and viability. The identification of what appear to be CpxA* equivalents in this organism should prove to be valuable tools both for addressing this question and for characterizing the Cpx regulon in this important pathogen.
dLytM factors and redundancy in amidase activation
The essentiality of PaAmiB predicted that dLytM factors that activate its enzymatic activity would also be essential. Indeed, we found that the simultaneous inactivation of PaEnvC, PaNlpD, and NlcS was lethal. However, none of the single disruptions of these factors produced a strong phenotype. Double mutants lacking pairs of dLytM proteins were also viable and exhibited a range of division phenotypes. Therefore, in contrast to the E. coli system, where each amidase only appears to be activated by a single dLytM protein, our genetic results suggest that PaAmiB is activated by three different and partially redundant dLytM proteins. Similar results in V. cholerae suggest that its lone amidase is activated by both of its major dLytM proteins VcEnvC and VcNlpD (Moll et al., 2014). Thus, while the general theme of amidase activation by dLytM proteins may be conserved in gamma-proteobacteria, the regulatory interactions appear to differ, perhaps due to niche-specific adaptations.
The double dLytM mutant lacking the paralogous proteins PaNlpD and NlcS displayed a severe division defect, forming chains of deeply constricted cells at all temperatures tested. Thus, it appears that PaEnvC is sufficient for the early stages of septal splitting and the cell constriction process. However, the remaining two dLytM factors look to be required for the final stage(s) of cell separation. This phenotype is consistent with the observation that E. coli AmiC, the amidase activated by NlpD, appears to localize to the division site as a focus very late in the constriction process, suggesting that it also normally participates only at the end of the separation process (Peters et al., 2011). Thus, the different amidases and/or dLytM activators of gamma-proteobacteria are unlikely to be performing completely redundant functions, but rather participate in overlapping, yet distinct aspects of the division process.
PG amidases as targets for antibiotic potentiators
Inactivation of cell separation enzymes has been associated with defects in the outer membrane permeability barrier of E. coli and other gram-negative bacteria (Heidrich et al., 2002; Ize et al., 2003; Weatherspoon-Griffin et al., 2011; Craig et al., 2013; Moll et al., 2014). The causative link between the failure to split septal PG and the perturbation of the outer membrane is unclear. Nevertheless, this phenotype has led to the proposal that septum-cleaving enzymes in general, and LytC-type amidases in particular, may be good targets for agents that sensitize gram-negative bacteria to antibiotics that are normally excluded by the outer membrane (Heidrich et al., 2002). Here, we show that inactivating PaAmiB or the dLytM factors also increases the sensitivity of P. aeruginosa to antibiotics, including gentamicin and vancomycin. Moreover, defects in PaAmiB were found to overcome the acquired resistance of oprD mutants to the β-lactam meropenem. Thus, targeting amidase activity may also be a valid approach for combating the intrinsic and acquired drug resistance mechanisms of P. aeruginosa. However, our isolation of suppessors of PaAmiB revealed a potential complication that may be encounted with this strategy. The cpxA alleles identified in the selection not only restore the viability of the PaAmiB depletion strain, but also mend the envelope permeability barrier. Further work will be required to determine how this Cpx-like stress response repairs the envelope in cells lacking PaAmiB and whether mutants that (hyper)activate the system will present problems for the development of antibiotic potentiators that overcome the intrinsic resistance of gram-negative bacteria.
EXPERIMENTAL PROCEDURES
Strains, media, and strain manipulation
P. aeruginosa cells were grown in LB (1% tryptone, 0.5% yeast extract, 0.5% NaCl). When necessary, the medium was supplemented with 1mM IPTG or 5% sucrose. For plasmid maintenance or integration, gentamicin was used the concentration of 30 µg/mL (plates) or 15µg/mL (liquid). E. coli cells were grown in LB or M9 minimal medium supplemented with 0.2% casamino acids and 0.2% sugar (arabinose, glucose, or maltose) unless indicated otherwise. For E. coli, antibiotics were used at the concentrations of 20 µg/mL for kanamycin, 20µg/mL for ampicillin, or 15µg/mL for gentamicin. During P. aeruginosa strain construction, plasmids were conjugated into P. aeruginosa from an E. coli donor on LB plates, and counter-selection against E. coli was accomplished on Pseudomonas Isolation Agar (PIA, Difco) or Voger-Bonner minimal medium (VBMM) supplemented with 60µg/mL gentamicin. Replicating plasmids were introduced into P. aeruginosa by electroporation. P. aeruginosa strains, E. coli strains, and all plasmids used in this study are listed in Tables S1, S2, and S3, respectively. Detailed descriptions of the strain and plasmid construction procedures can be found in Supplemental Material.
Growth assays and microscopy
For spot dilution assays with P. aeruginosa, overnight cell cultures were typically subcultured 1:10 in LB medium (supplemented with IPTG for the strains with PTOPLAC-directed gene expression) and allowed to grow at 30°C for 2h. The cells were then washed to eliminate the inducer, normalized to OD600 of 0.5, and subjected to serial 10-fold dilutions. 5µL of the 10−1 to 10−6 dilutions were then spotted onto the plates indicated in the text and incubated at 30°C for ~24h prior to imaging (unless noted otherwise). For the E. coli spot dilution, overnight cell cultures grown in LB with 0.2% arabinose were washed twice with 1x M9 salts, subcultured 1:10 in M9 medium supplemented with 0.2% maltose and 0.002% arabinose, and allowed to grow at 30°C for 2h. The cells were again washed twice with 1x M9 salts, normalized to OD600 of 0.5, and subjected to serial 10-fold dilutions, with 5µL of each dilution spotted onto the plates. The plates were incubated at 30°C for 2d.
For the growth curve assay, an overnight culture of BPA46 grown in LB supplemented with 1mM IPTG was washed to eliminate the inducer, diluted in LB or LB supplemented with IPTG to OD600 of 0.04 and allowed to grow in a 37°C water bath with 250rpm shaking. Growth was monitored via optical density readings taken every 30min. Additionally, starting at the 1h time point, 500µL aliquots of cells were taken out every 1h, fixed with 2.4% formaldehyde/0.04% glutaraldehyde in 33 mM sodium phosphate (pH 7.4), and stored on ice. Cells were imaged on 1.5% agarose pads, using Nikon TE2000 inverted microscope outfitted with Nikon Intensilight illuminator, a Coolsnap HQ2 charge-coupled device camera from Photometrics, and a CFI Plan Apo DM x100 objective lens (1.4 NA). For imaging live cells, exponentially growing cultures were spotted onto LB agarose pads. mCherry and HADA fluorescence images were taken with the ET-mCherry (Chroma 49008) and ET-DAPI (Chroma 49000) filter sets, respectively. For imaging with HADA (HCC-amino-D-alanine), exponentially growing BPA46 cells were incubated with 30µM HADA in LB at 37°C for 30min and then washed to eliminate unincorporated label prior to imaging. For electron microscopy, cells were grown as for light microscopy, but then chemically fixed overnight in FGP fixative (2.5% paraformaldehyde, 5% glutaraldehyde, 0.06% picric acid, 0.2 M cacodylate buffer (EMS)). The samples were treated with 1% osmium tetroxide/1.5% potassium ferrocyanide and then with 1% uranyl acetate, dehydrated, embedded in Epon resin, sectioned, and imaged with JEOL 1200EX transmission electron microscope equipped with an AMT 2k CCD camera.
Antibiotic and detergent sensitivity assays
For the antibiotic spotting assay, overnight cell cultures were diluted 1:100 in LB supplemented with IPTG and allowed to grow for 3h at 30°C. These cultures were washed 1x with LB, resuspended in fresh LB, and allowed to grow for another 2h at 30°C without the inducer. Then, the cells were again washed 1x with LB and normalized to OD600 of 0.4. 100µL cell aliquots were mixed with 4mL samples of 0.8% LB top agar (either with or without 1mM IPTG, as indicated), which was pre-melted and stored at 50°C. The agar was then poured over LB plates and allowed to solidify for 30min at room temperature. Either 5µL or 10µL of antibiotics at the concentrations indicated in the text were spotted onto the surface of the top agar, and the plates were incubated upright for 2d at room temperature (~22°C) prior to imaging.
For the antibiotic MIC assays, overnight cell cultures were recovered from stationary phase by 1:10 dilution in LB supplemented with 5% sucrose and growth at 30°C for 2h. The cells were then normalized to OD600 of 0.005 in LB with sucrose and the indicated concentrations of gentamicin, vancomycin, or colistin and grown for ~24h at 30°C. When present, the biofilm formed on the walls of the test tubes was resuspended prior to taking optical density readings.
The SDS sensitivity assay was loosely based on the protocol described in (Helander et al., 1997). Overnight cultures of PAO1 and BPA46 were washed 1x with LB, subcultured 1:1000 in LB either with IPTG (BPA46) or without IPTG (PAO1 and BPA46), and allowed to grow at 37°C for 5h. The cells were then normalized to OD600 of 0.5 in the HEPES buffer (50mM NaCl; 10mM HEPES, pH 7). For each sample, the initial optical density was measured prior to the addition of SDS and/or EDTA at the indicated concentrations. The sample was then mixed, incubated for 4–10 min, and the final optical density was measured.
Suppressor mutant selection and mutation rate experiment
In order to isolate suppressor mutations that allow growth on LB in the absence of PaAmiB, isolated colonies of BPA158 [ΔamiA ΔamiB] cells were grown overnight at 30°C in separate cultures of liquid LB medium supplemented with 5% sucrose. The cells were diluted 1:10 with LB, 100µL aliquots of each dilution were plated onto LB plates without sucrose, and the plates were incubated at 30°C for 2d. Surviving colonies were isolated, and suppression was confirmed by re-streaking isolated colonies on plain LB plates. To prevent isolation of clonal mutants, only one mutant colony per plate was submitted for whole-genome Illumina sequencing performed using standard procedures by the Infection IMED group at AstraZeneca in Waltham, MA.
To assess the mutation rate of the ΔamiB mutant without antibiotic hypersensitivity confounding the results, BPA46 [ΔamiB (PTOPLAC::amiB)] and its parental strain with wild-type amiB at the native locus, BPA35 [amiB+ (PTOPLAC::amiB)], were grown overnight from isolated colonies in LB supplemented with 1mM IPTG. The cells were then centrifuged for 20min at 3000g, resuspended in 1/100th of the original volume, and 100µL of the suspension were plated on LB plates containing 500µg/mL streptomycin and 1mM IPTG. The remaining cells were subjected to serial 10-fold dilutions, and 100µL samples of the 10−8 and 10−9 factor dilutions were plated on LB supplemented with 1mM IPTG only. The plates were incubated at 30°C, and the colonies were counted after 2d of growth.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank all members of the Bernhardt and Rudner labs for advice and helpful discussions, particularly Nick Peters for constructing the AmiA depletion strain of E. coli. We would also like to thank Sarah McLeod, Alita Miller, Scott Mills, and the rest of the Infection IMED team at AstraZeneca for advice and input. Whole genome sequencing was carried out by Vincent Isabella and Robert McLaughlin at AstraZeneca in Waltham, MA. We also thank Maria Ericsson at the HMS EM facility for her help preparing and visualizing thin sections of PAAmiB-depleted cells. Special thanks to Stephen Lory and Simon Dove for help with P. aeruginosa methods and for providing strains and expert advice. Finally, we would like to thank Herbert Schweizer for sharing the PAO1 efflux mutant with us. This work was supported by the National Institute of Allergy and Infections Diseases of the National Institutes of Health (R21 AI111713 and R01 AI083365), and funds from AstraZeneca. A.Y. was supported in part by a postdoctoral fellowship from the Cystic Fibrosis Foundation (YAKHNI14F0).
REFERENCES
- Arends SJR, Williams K, Scott RJ, Rolong S, Popham DL, Weiss DS. Discovery and characterization of three new Escherichia coli septal ring proteins that contain a SPOR domain: DamX, DedD, and RlpA. Journal of Bacteriology. 2009;192:242–255. doi: 10.1128/JB.01244-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breidenstein EBM, la Fuente-Núñez de C, Hancock REW. Pseudomonas aeruginosa: all roadslead to resistance. Trends in Microbiology. 2011;19:419–426. doi: 10.1016/j.tim.2011.04.005. [DOI] [PubMed] [Google Scholar]
- Burdett ID, Murray RG. Septum formation in Escherichia coli: characterization of septal structure and the effects of antibiotics on cell division. Journal of Bacteriology. 1974a;119:303–324. doi: 10.1128/jb.119.1.303-324.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdett ID, Murray RG. Electron microscope study of septum formation in Escherichia coli strains B and B-r during synchronous growth. Journal of Bacteriology. 1974b;119:1039–1056. doi: 10.1128/jb.119.3.1039-1056.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bury-Moné S, Nomane Y, Reymond N, Barbet R, Jacquet E, Imbeaud S, et al. Global analysis of extracytoplasmic stress signaling in Escherichia coli . PLoS Genet. 2009;5:e1000651. doi: 10.1371/journal.pgen.1000651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carballès F, Bertrand C, Bouché JP, Cam K. Regulation of Escherichia coli cell division genes ftsA and ftsZ by the two-component system rcsC-rcsB . Molecular Microbiology. 1999;34:442–450. doi: 10.1046/j.1365-2958.1999.01605.x. [DOI] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention. Antibiotic Resistance Threats in the United States, 2013. 2014 http://www.cdc.gov/drugresistance/pdf/ar-threats-2013-508.pdf.
- Chuanchuen R, Murata T, Gotoh N, Schweizer HP. Substrate-dependent utilization of OprM or OpmH by the Pseudomonas aeruginosa MexJK efflux pump. Antimicrobial Agents and Chemotherapy. 2005;49:2133–2136. doi: 10.1128/AAC.49.5.2133-2136.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen DN, Sham YY, Haugstad GD, Xiang Y, Rossmann MG, Anderson DL, Popham DL. Shared catalysis in virus entry and bacterial cell wall depolymerization. Journal of Molecular Biology. 2009;387:607–618. doi: 10.1016/j.jmb.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig M, Sadik AY, Golubeva YA, Tidhar A, Slauch JM. Twin-arginine translocation system (tat) mutants of Salmonella are attenuated due to envelope defects, not respiratory defects. Molecular Microbiology. 2013;89:887–902. doi: 10.1111/mmi.12318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer PA. Advances in understanding E. coli cell fission. Current Opinion in Microbiology. 2010;13:730–737. doi: 10.1016/j.mib.2010.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Leeuw E, Graham B, Phillips GJ, Hagen-Jongman ten CM, Oudega B, Luirink J. Molecular characterization of Escherichia coli FtsE and FtsX. Molecular Microbiology. 1999;31:983–993. doi: 10.1046/j.1365-2958.1999.01245.x. [DOI] [PubMed] [Google Scholar]
- de Pedro MA, Quintela JC, Höltje JV, Schwarz H. Murein segregation in Escherichia coli . Journal of Bacteriology. 1997;179:2823–2834. doi: 10.1128/jb.179.9.2823-2834.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Wulf P, Lin ECC. Cpx two-component signal transduction in Escherichia coli: excessive CpxR-P levels underlie CpxA* phenotypes. Journal of Bacteriology. 2000;182:1423–1426. doi: 10.1128/jb.182.5.1423-1426.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiGiuseppe PA, Silhavy TJ. Signal detection and target gene induction by the CpxRA two-component system. Journal of Bacteriology. 2003;185:2432–2440. doi: 10.1128/JB.185.8.2432-2440.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ercoli G, Tani C, Pezzicoli A, Vacca I, Martinelli M, Pecetta S, et al. LytM proteins play a crucial role in cell separation, outer membrane composition, and pathogenesis in nontypeable Haemophilus influenzae . mBio. 2015;6:e02575–e02514. doi: 10.1128/mBio.02575-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans KL, Kannan S, Li G, de Pedro MA, Young KD. Eliminating a set of four Penicillin Binding Proteins triggers the Rcs phosphorelay and Cpx stress responses in Escherichia coli . Journal of Bacteriology. 2013;195:4415–4424. doi: 10.1128/JB.00596-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firczuk M, Mucha A, Bochtler M. Crystal structures of active LytM. Journal of Molecular Biology. 2005;354:578–590. doi: 10.1016/j.jmb.2005.09.082. [DOI] [PubMed] [Google Scholar]
- Firczuk MG, Bochtler M. Folds and activities of peptidoglycan amidases. FEMS Microbiology Reviews. 2007;31:676–691. doi: 10.1111/j.1574-6976.2007.00084.x. [DOI] [PubMed] [Google Scholar]
- Garcia DL, Dillard JP. AmiC functions as an N-acetylmuramyl-L-alanine amidase necessary for cell separation and can promote autolysis in Neisseria gonorrhoeae . Journal of Bacteriology. 2006;188:7211–7221. doi: 10.1128/JB.00724-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerding MA, Liu B, Bendezu FO, Hale CA, Bernhardt TG, de Boer PAJ. Self-enhanced accumulation of FtsN at division sites and roles for other proteins with a SPOR domain (DamX, DedD, and RlpA) in Escherichia coli cell constriction. Journal of Bacteriology. 2009;191:7383–7401. doi: 10.1128/JB.00811-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goley ED, Comolli LR, Fero KE, Downing KH, Shapiro L. DipM links peptidoglycan remodelling to outer membrane organization in Caulobacter . Molecular Microbiology. 2010;77:56–73. doi: 10.1111/j.1365-2958.2010.07222.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock RE, Bell A. Antibiotic uptake into gram-negative bacteria. Eur J Clin Microbiol Infect Dis. 1988;7:713–720. doi: 10.1007/BF01975036. [DOI] [PubMed] [Google Scholar]
- Hara H, Narita S, Karibian D, Park JT, Yamamoto Y, Nishimura Y. Identification and characterization of the Escherichia coli envC gene encoding a periplasmic coiled-coil protein with putative peptidase activity. FEMS Microbiol Lett. 2002;212:229–236. doi: 10.1111/j.1574-6968.2002.tb11271.x. [DOI] [PubMed] [Google Scholar]
- Heidrich C, Templin MF, Ursinus A, Merdanovic M, Berger J, Schwarz H, et al. Involvement of N-acetylmuramyl-L-alanine amidases in cell separation and antibiotic-induced autolysis of Escherichia coli . Molecular Microbiology. 2001;41:167–178. doi: 10.1046/j.1365-2958.2001.02499.x. [DOI] [PubMed] [Google Scholar]
- Heidrich C, Ursinus A, Berger J, Schwarz H, Höltje JV. Effects of multiple deletions of murein hydrolases on viability, septum cleavage, and sensitivity to large toxic molecules in Escherichia coli . Journal of Bacteriology. 2002;184:6093–6099. doi: 10.1128/JB.184.22.6093-6099.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helander I, Alakomi HL, Latva-Kala K. Polyethyleneimine is an effective permeabilizer of gram-negative bacteria. Microbiology. 1997 doi: 10.1099/00221287-143-10-3193. [DOI] [PubMed] [Google Scholar]
- Held K, Ramage E, Jacobs M, Gallagher L, Manoil C. Sequence-verified two-allele transposon mutant library for Pseudomonas aeruginosa PAO1. Journal of Bacteriology. 2012;194:6387–6389. doi: 10.1128/JB.01479-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichimura T, Yamazoe M, Maeda M, Wada C, Hiraga S. Proteolytic activity of YibP protein in Escherichia coli . Journal of Bacteriology. 2002;184:2595–2602. doi: 10.1128/JB.184.10.2595-2602.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ize B, Stanley NR, Buchanan G, Palmer T. Role of the Escherichia coli Tat pathway in outer membrane integrity. Molecular Microbiology. 2003;48:1183–1193. doi: 10.1046/j.1365-2958.2003.03504.x. [DOI] [PubMed] [Google Scholar]
- Jorgenson MA, Chen Y, Yahashiri A, Popham DL, Weiss DS. The bacterial septal ring protein RlpA is a lytic transglycosylase that contributes to rod shape and daughter cell separation in Pseudomonas aeruginosa . Molecular Microbiology. 2014;93:113–128. doi: 10.1111/mmi.12643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler T, Michea-Hamzehpour M, Epp SF, Pechere JC. Carbapenem activities against Pseudomonas aeruginosa: respective contributions of OprD and efflux systems. Antimicrobial Agents and Chemotherapy. 1999;43:424–427. doi: 10.1128/aac.43.2.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuru E, Hughes HV, Brown PJ, Hall E, Tekkam S, Cava F, et al. In situ probing of newly synthesized peptidoglycan in live bacteria with fluorescent D-amino acids. Angew Chem Int Ed. 2012;51:12519–12523. doi: 10.1002/anie.201206749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Persons L, Lee L, de Boer PAJ. Roles for both FtsA and the FtsBLQ subcomplex in FtsN-stimulated cell constriction in Escherichia coli . Molecular Microbiology. 2015;95:945–970. doi: 10.1111/mmi.12906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu JZ, Fujiwara T, Komatsuzawa H, Sugai M, Sakon J. Cell wall-targeting domain of glycylglycine endopeptidase distinguishes among peptidoglycan cross-bridges. J Biol Chem. 2006;281:549–558. doi: 10.1074/jbc.M509691200. [DOI] [PubMed] [Google Scholar]
- Lutkenhaus J. FtsN -- trigger for septation. Journal of Bacteriology. 2009;191:7381–7382. doi: 10.1128/JB.01100-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutkenhaus J, Pichoff S, Du S. Bacterial cytokinesis: From Z ring to divisome. Cytoskeleton. 2012;69:778–790. doi: 10.1002/cm.21054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLeod DL, Nelson LE, Shawar RM, Lin BB, Lockwood LG, Dirk JE, et al. Aminoglycoside-resistance mechanisms for cystic fibrosis Pseudomonas aeruginosa isolates are unchanged by long-term, intermittent, inhaled tobramycin treatment. J Infect Dis. 2000;181:1180–1184. doi: 10.1086/315312. [DOI] [PubMed] [Google Scholar]
- Majdalani N, Gottesman S. The Rcs phosphorelay: a complex signal transduction system. Annu Rev Microbiol. 2005;59:379–405. doi: 10.1146/annurev.micro.59.050405.101230. [DOI] [PubMed] [Google Scholar]
- Moll A, Dorr T, Alvarez L, Chao MC, Davis BM, Cava F, Waldor MK. Cell separation in Vibrio cholerae is mediated by a single amidase whose action Is modulated by two nonredundant activators. Journal of Bacteriology. 2014;196:3937–3948. doi: 10.1128/JB.02094-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möll A, Thanbichler M. FtsN-like proteins are conserved components of the cell division machinery in proteobacteria. Molecular Microbiology. 2009;72:1037–1053. doi: 10.1111/j.1365-2958.2009.06706.x. [DOI] [PubMed] [Google Scholar]
- Möll A, Schlimpert S, Briegel A, Jensen GJ, Thanbichler M. DipM, a new factor required for peptidoglycan remodelling during cell division in Caulobacter crescentus . Molecular Microbiology. 2010;77:90–107. doi: 10.1111/j.1365-2958.2010.07224.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori N, Ishii Y, Tateda K, Kimura S, Kouyama Y, Inoko H, et al. A peptide based on homologous sequences of the β-barrel assembly machinery component BamD potentiates antibiotic susceptibility of Pseudomonas aeruginosa . Journal of Antimicrobial Chemotherapy. 2012;67:2173–2181. doi: 10.1093/jac/dks174. [DOI] [PubMed] [Google Scholar]
- Naenna P, Noisumdaeng P, Pongpech P, Tribuddharat C. Detection of outer membrane porin protein, an imipenem influx channel, in Pseudomonas aeruginosa clinical isolates. Southeast Asian J Trop Med Public Health. 2010;41:614–624. [PubMed] [Google Scholar]
- Oliver A, Baquero F, Blázquez J. The mismatch repair system (mutS, mutL and uvrD genes) in Pseudomonas aeruginosa: molecular characterization of naturally occurring mutants. Molecular Microbiology. 2002 doi: 10.1046/j.1365-2958.2002.02855.x. [DOI] [PubMed] [Google Scholar]
- Peters NT, Dinh T, Bernhardt TG. A fail-safe mechanism in the septal ring assembly pathway generated by the sequential recruitment of cell separation amidases and their activators. Journal of Bacteriology. 2011;193:4973–4983. doi: 10.1128/JB.00316-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters NT, Morlot C, Yang DC, Uehara T, Vernet T, Bernhardt TG. Structure-function analysis of the LytM domain of EnvC, an activator of cell wall remodelling at the Escherichia coli division site. Molecular Microbiology. 2013;89:690–701. doi: 10.1111/mmi.12304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poggio S, Takacs CN, Vollmer W, Jacobs-Wagner C. A protein critical for cell constriction in the Gram-negative bacterium Caulobacter crescentus localizes at the division site through its peptidoglycan-binding LysM domains. Molecular Microbiology. 2010;77:74–89. doi: 10.1111/j.1365-2958.2010.07223.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priyadarshini R, de Pedro MA, Young KD. Role of peptidoglycan amidases in the development and morphology of the division septum in Escherichia coli . Journal of Bacteriology. 2007;189:5334–5347. doi: 10.1128/JB.00415-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raivio TL, Silhavy TJ. Transduction of envelope stress in Escherichia coli by the Cpx two-component system. Journal of Bacteriology. 1997;179:7724–7733. doi: 10.1128/jb.179.24.7724-7733.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raivio TL, Silhavy TJ. Periplasmic stress and ECF sigma factors. Annu Rev Microbiol. 2001;55:591–624. doi: 10.1146/annurev.micro.55.1.591. [DOI] [PubMed] [Google Scholar]
- Raivio TL, Laird MW, Joly JC, Silhavy TJ. Tethering of CpxP to the inner membrane prevents spheroplast induction of the Cpx envelope stress response. Molecular Microbiology. 2000;37:1186–1197. doi: 10.1046/j.1365-2958.2000.02074.x. [DOI] [PubMed] [Google Scholar]
- Raivio TL, Popkin DL, Silhavy TJ. The Cpx envelope stress response is controlled by amplification and feedback inhibition. Journal of Bacteriology. 1999;181:5263–5272. doi: 10.1128/jb.181.17.5263-5272.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy M. Role of FtsEX in cell division of Escherichia coli: viability of ftsEX mutants is dependent on functional SufI or high osmotic strength. Journal of Bacteriology. 2006;189:98–108. doi: 10.1128/JB.01347-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheurwater EM, Pfeffer JM, Clarke AJ. Production and purification of the bacterial autolysin N-acetylmuramoyl-L-alanine amidase B from Pseudomonas aeruginosa . Protein Expression and Purification. 2007;56:128–137. doi: 10.1016/j.pep.2007.06.009. [DOI] [PubMed] [Google Scholar]
- Schmidt KL, Peterson ND, Kustusch RJ, Wissel MC, Graham B, Phillips GJ, Weiss DS. A predicted ABC transporter, FtsEX, is needed for cell division in Escherichia coli . Journal of Bacteriology. 2004;186:785–793. doi: 10.1128/JB.186.3.785-793.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster M, Hawkins AC, Harwood CS, Greenberg EP. The Pseudomonas aeruginosa RpoS regulon and its relationship to quorum sensing. Molecular Microbiology. 2003;51:973–985. doi: 10.1046/j.1365-2958.2003.03886.x. [DOI] [PubMed] [Google Scholar]
- Singh SK, SaiSree L, Amrutha RN, Reddy M. Three redundant murein endopeptidases catalyse an essential cleavage step in peptidoglycan synthesis of Escherichia coli K12. Molecular Microbiology. 2012;86:1036–1051. doi: 10.1111/mmi.12058. [DOI] [PubMed] [Google Scholar]
- Strateva T, Yordanov D. Pseudomonas aeruginosa - a phenomenon of bacterial resistance. Journal of Medical Microbiology. 2009;58:1133–1148. doi: 10.1099/jmm.0.009142-0. [DOI] [PubMed] [Google Scholar]
- Suh SJ, Silo-Suh L, Woods DE, Hassett DJ, West SE, Ohman DE. Effect of rpoS mutation on the stress response and expression of virulence factors in Pseudomonas aeruginosa . Journal of Bacteriology. 1999;181:3890–3897. doi: 10.1128/jb.181.13.3890-3897.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tängdén T. Combination antibiotic therapy for multidrug-resistant gram-negative bacteria. Upsala Journal of Medical Sciences. 2014;119:149–153. doi: 10.3109/03009734.2014.899279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang M-J, Bernhardt TG. A role for the FtsQLB complex in cytokinetic ring activation revealed by an ftsL allele that accelerates division. Molecular Microbiology. 2015a;95:925–944. doi: 10.1111/mmi.12905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang M-J, Bernhardt TG. Guiding divisome assembly and controlling its activity. Current Opinion in Microbiology. 2015b;24:60–65. doi: 10.1016/j.mib.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Typas A, Banzhaf M, Gross CA, Vollmer W. From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat Rev Micro. 2011;10:123–136. doi: 10.1038/nrmicro2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara T, Dinh T, Bernhardt TG. LytM-domain factors are required for daughter cell separation and rapid ampicillin-induced lysis in Escherichia coli . Journal of Bacteriology. 2009;191:5094–5107. doi: 10.1128/JB.00505-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara T, Parzych KR, Dinh T, Bernhardt TG. Daughter cell separation is controlled by cytokinetic ring-activated cell wall hydrolysis. The EMBO Journal. 2010:1–11. doi: 10.1038/emboj.2010.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ursinus A, van den Ent F, Brechtel S, de Pedro M, Höltje JV, Löwe J, Vollmer W. Murein (peptidoglycan) binding property of the essential cell division protein FtsN from Escherichia coli . Journal of Bacteriology. 2004;186:6728–6737. doi: 10.1128/JB.186.20.6728-6737.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaara M. Antibiotic-supersusceptible mutants of Escherichia coli and Salmonella typhimurium . Antimicrobial Agents and Chemotherapy. 1993;37:2255–2260. doi: 10.1128/aac.37.11.2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venturi V. Control of rpoS transcription in Escherichia coli and Pseudomonas: why so different? Molecular Microbiology. 2003;49:1–9. doi: 10.1046/j.1365-2958.2003.03547.x. [DOI] [PubMed] [Google Scholar]
- Vogt SL, Raivio TL. Just scratching the surface: an expanding view of the Cpx envelope stress response. FEMS Microbiol Lett. 2011;326:2–11. doi: 10.1111/j.1574-6968.2011.02406.x. [DOI] [PubMed] [Google Scholar]
- Weatherspoon-Griffin N, Zhao G, Kong W, Kong Y, Morigen Andrews-Polymenis H, et al. The CpxR/CpxA two-component system up-regulates two Tat-dependent peptidoglycan amidases to confer bacterial resistance to antimicrobial peptide. Journal of Biological Chemistry. 2011;286:5529–5539. doi: 10.1074/jbc.M110.200352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss DS. Last but not least: new insights into how FtsN triggers constriction during Escherichia coli cell division. Molecular Microbiology. 2015;95:903–909. doi: 10.1111/mmi.12925. [DOI] [PubMed] [Google Scholar]
- Yang DC, Peters NT, Parzych KR, Uehara T, Markovski M, Bernhardt TG. An ATP-binding cassette transporter-like complex governs cell-wall hydrolysis at the bacterial cytokinetic ring. Proc Natl Acad Sci USA. 2011;108:e1052–e1060. doi: 10.1073/pnas.1107780108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang DC, Tan K, Joachimiak A, Bernhardt TG. A conformational switch controls cell wall-remodelling enzymes required for bacterial cell division. Molecular Microbiology. 2012;85:768–781. doi: 10.1111/j.1365-2958.2012.08138.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J-C, Van Den Ent F, Neuhaus D, Brevier J, Lowe J. Solution structure and domain architecture of the divisome protein FtsN. Molecular Microbiology. 2004;52:651–660. doi: 10.1111/j.1365-2958.2004.03991.x. [DOI] [PubMed] [Google Scholar]
- Yoshimura F, Nikaido H. Permeability of Pseudomonas aeruginosa outer membrane to hydrophilic solutes. Journal of Bacteriology. 1982;152:636–642. doi: 10.1128/jb.152.2.636-642.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.