Abstract
Background
Delineating the cascades of growth and transcription factor expression that shape the developing nervous system will improve our understanding of its molecular histogenesis and suggest strategies for cell replacement therapies. In the current investigation, we examined the ability of the proneural gene, Neurogenin1 (Neurog1; also Ngn1, Neurod3), to drive differentiation of pluripotent embryonic stem cells (ESC).
Results
Transient expression of Neurog1 in ESC was sufficient to initiate neuronal differentiation, and produced neuronal subtypes reflecting its expression pattern in vivo. To begin to address the molecular mechanisms involved, we employed microarray analysis to identify potential down-stream targets of Neurog1 expressed at sequential stages of neuronal differentiation.
Conclusions
ESC expressing Neurogenin1 begin to withdraw from cycle and form precursors that differentiate exclusively into neurons. This work identifies unique patterns of gene expression following expression of Neurog1, including genes and signaling pathways involved in process outgrowth and cell migration, regional differentiation of the nervous system, and cell cycle.
Keywords: bHLH, BMP4, ES cell, FGF, microarray, neural crest cell, Neurod3, neural induction, neuronal differentiation, Ngn1, noggin, sonic hedgehog, Tet-on
INTRODUCTION
Vertebrate neurogenesis is a stepwise process in which the primitive ectoderm is first induced to form the neural ectoderm, which folds forming the neural tube. The neural tube is then patterned along the anterior-posterior (AP) and dorsal-ventral (DV) axes by morphogens that coordinate the expression of transcription factors that confer positional identity and/or promote neural differentiation. Proneural basic helix-loop-helix (bHLH) proteins comprise one such class of transcription factors essential during early neurogenesis (review, Bertrand et al., 2002). In Drosophila, the bHLH factors achaete-scute and atonal act as generic promoters of neuronal differentiation and neuronal subtype specification (Chien et al., 1996; Jarman and Ahmed, 1998). Vertebrate atonal homologs such as Neurogenin 1 (Neurog1), Neurogenin 2 (Neurog2; Gradwohl et al., 1996; Ma et al., 1996; Sommer et al., 1996) and achaete-scute homologs such as Atoh1 (Mash1; Johnson, et al., 1990) and Ascl1 (Math1; Guillemot and Joyner, 1993) also promote neuronal differentiation both in vivo (Turner and Weintraub, 1994; Lee et al., 1995; Ma et al., 1996; Chung et al., 2002; Kim et al., 2004) and in vitro (Lo et al., 1998; Farah et al., 2000; Sun et al., 2001; Kanda et al., 2004; Satoh et al., 2010).
The expression of mammalian achaete-scute and atonal homologues within specific-–largely non-overlapping—regions of the developing central and peripheral nervous systems (CNS and PNS) suggests roles in neuronal subtype specification that have been confirmed by loss- and gain-of-function studies. For example, Neurog1 is expressed in the dorsal telecephalon where it appears to promote glutaminergic neuronal fates, Atoh1 is expressed in the ventral telencephalon specifying GABAergic neurons (Fode et al., 2000; Parras et al., 2002; Kim et al., 2011), while Neurog1 is expressed in the caudal ventricular zone of the rhombic lip, where it defines multiple GABAergic lineages (Dalgard et al., 2011). In the spinal cord, Ascl1 is expressed in a dorsal stripe near the roof plate (Gowan et al., 2001), Neurog1 is expressed in the ventral half and in a small region just below the roof plate, whereas Atoh1 is found in the intervening domain (Sommer et al., 1996; Ma, et al., 1997), where these transcription factors are thought to regulate neuronal phenotype by cross inhibition (Briscoe et al., 2000; Gowan et al., 2001; Helms et al., 2005). Loss-of-function studies have shown that Neurog1 is required for the development of dI2 dorsal spinal neurons, trigeminal and otic cranial sensory ganglia, and TrkA neurons of dorsal root ganglia (DRG) (Ma et al., 1997; Fode et al., 1998; Gowan et al., 2001). Gain-of-function studies have demonstrated that over-expression of Neurog1 biases the migration of neural crest stem cells toward dorsal root sensory ganglia in vivo (Perez et al., 1999), whereas forced expression of Neurog1 in dorsal neural tube progenitors and neural crest cells promotes their differentiation into sensory lineages (Lo et al., 2002). These data indicate that Neurog1 is required for the development of sensory neuronal lineages in both the PNS and CNS; however, it is not clear whether Neurog1 is itself sufficient to induce these lineages since the gain-of-function studies were conducted either in the embryo or in neural progenitors where the effects of morphogens and other instructive signals cannot be separated. While mis-expression of proneural genes can produce ectopic neurogenesis in a variety of species (Quan and Hassan, 2005), relatively little is known regarding the molecular mechanisms involved or down-stream gene expression following bHLH gene expression. Since bHLH transcription factor expression is strongly affected by spatial and temporal context (Powell and Jarman, 2008), we employed a gain-of-function approach in pluripotent embryonic stem (ES) cells to determine the role of Neurog1 in cell fate specification. ES cells may be a particularly informative starting material since they have a bivalent chromatin structure with promoters poised for both lineage differentiation as well as for self-renewal (e.g., Boyer et al., 2006). Lineage specifying genes such as bHLH and paired-box family members may therefore control differentiation programs by directly affecting transcription and by narrowing differentiation choices by controlling chromatin.
The current investigation identifies potential down-stream targets of Neurog1 including genes involved in cell cycle, cell migration and process outgrowth, and provides a source of neuronal precursor cells that remain sensitive to patterning molecules. Consistent with observations that Neurog1 is present in cells about to withdraw from cycle and differentiate into layer-specific neurons (Kim et al., 2011), forced expression of Neurog1 in ES cells alters their cell cycle characteristics and is sufficient to initiate neuronal differentiation in the absence of other inducing factors. In fact, Neurog1 expression was sufficient to overcome the inhibitory effects of LIF and serum proteins on ES cell differentiation (Williams et al., 1988). In addition, Neurog1 expression was also sufficient to generate both CNS and PNS neuronal subtypes typical of those dependent on Neurog1 in vivo. Yet, the positional identity and neuronal differentiation potential of induced cells could be influenced by early acting patterning factors, suggesting that Neurog1 promotes differentiation of neuronal precursors that can be influenced by the local microenvironment to subsequent regional and/or subtype specific differentiation.
RESULTS
Inducible expression of Neurog1 in ES cells
In the current investigation, we employed the Ainv15 ES cell line (Kyba et al., 2002) that expresses a “Tet-on” reverse tetracycline transactivator (rtTA) from the constitutively active ROSA26 locus, and a tet-inducible element with a LoxP targeting site and a truncated neomycin resistance cassette placed upstream of the HPRT locus. Site-specific integration of the targeting vector carrying cDNA for a gene of interest places the transgene downstream of the tet-inducible element and confers neomycin resistance, thereby allowing efficient selection of targeted cell lines. Once selected, cells are maintained in neomycin to ensure that the transgene does not undergo methylation or rearrangement, although this is unlikely at the HPRT locus (Wutz et al., 2002).
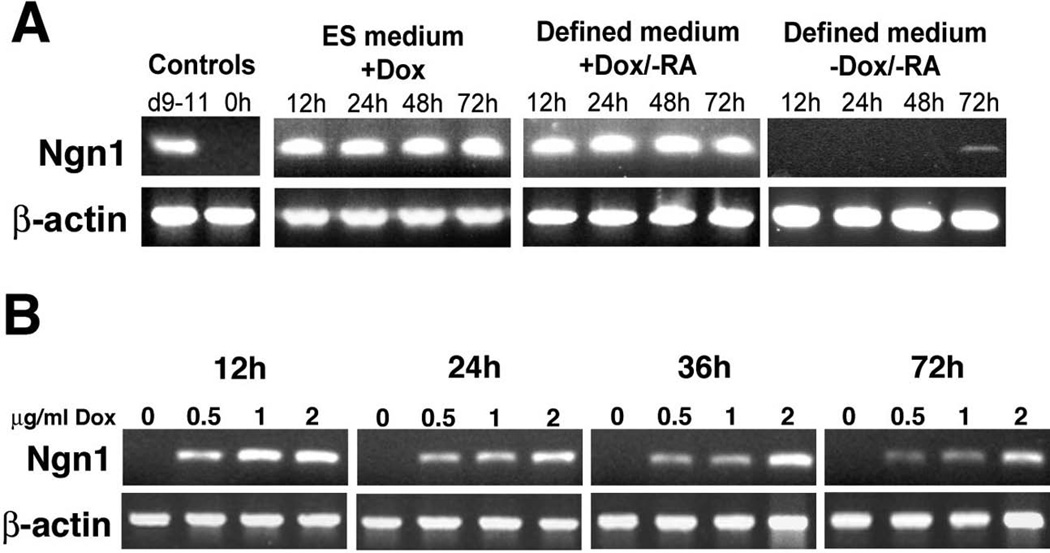
We targeted Neurog1 to the inducible locus and obtained several G418 resistant colonies which were expanded to cell lines. Based on the reliability of this inducible system (Kyba et al., 2002; M. Kyba and G. Keller, personal communication) and our own analyses in which we observed no differences between the ESC lines in their differentiation capacity or response to the tetracycline analog doxycycline (data not shown), one line (N7) was selected for most subsequent experiments. As indicated in Fig. 1A, robust Neurog1 expression in N7 cells was induced within 12 hours of addition of doxycycline (dox), to monolayer cultures grown in either complete ES medium, which inhibits spontaneous differentiation (Williams et al., 1988) or in a defined neural medium, which supports neural differentiation of ES cells in monolayer cultures. We did not observe any evidence of transgene silencing as Neurog1 expression was maintained throughout the culture period. N7 cells cultured in complete (ES) medium without dox did not express Neurog1 at 12, 24, 48, or 72h (Fig. 1A). N7 cells cultured in defined neural medium without dox were intolerant of these culture conditions often undergoing cell death by 72h (not shown). However, in cells that survived to 72h it was possible to detect slight expression of Neurog1 (Fig. 1A), likely due to stochastic neural differentiation and activation of endogenous Neurog1 elicited by culture in these conditions. When cultured in a different serum-free medium containing 5% knock-out serum replacer (Fig. 1B), we observed no expression of Neurog1 in the absence of dox, but, in the dox-treated groups, we could consistently induce expression of the Neurog1 transgene in a dose-dependent manner. Overall, these data suggest that the N7 cell line provides a consistent and tightly regulatable means to assess the sequelae of Neurog1 expression in ES cells.
Figure 1. Characterization of dox-inducible Neurog1 ES cells.
A. Cells cultured in either complete ES medium or in defined neural medium with 1 µg/ml doxycycline express Neurog1 by RT-PCR within 12 hours of treatment -- which was maintained 72h in culture. Cells cultured in defined neural medium without the addition of dox showed a very slight up-regulation of Neurog1 after 72h. Controls include pooled cDNA from day 9–11 mouse embryos (d9–11) and N7 cells cultured in ES medium without dox (0h).
B. N7 cells grown in serum free minimal medium (DMEM with 5% knockout serum replacement) with 0, 0.5, 1, or 2 µg/ml doxycycline express Neurog1 in a dose-dependent manner. After 72h, cells not exposed to doxycycline show no expression of Neurog1.
Expression of Neurog1 in ES cells induces neuronal differentiation
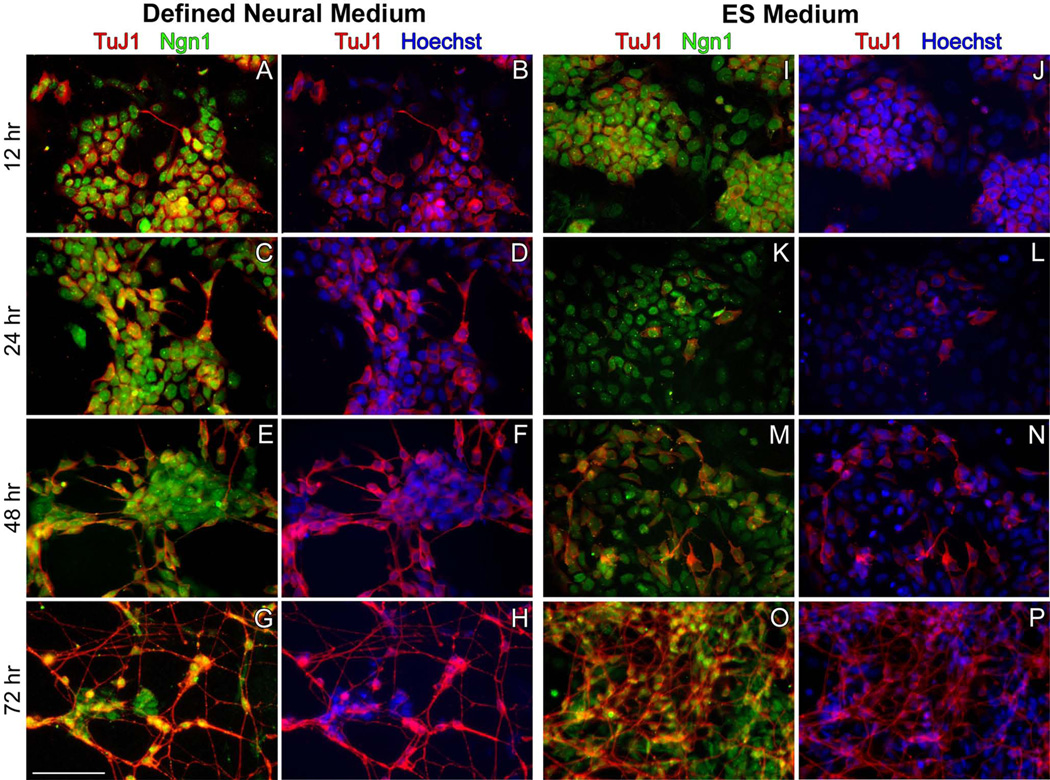
Within 24 hours of inducing the Neurog1 transgene, N7 cells exhibited overt neuronal morphologies including round, phase-bright somata and one or more long cytoplasmic processes when viewed with phase contrast microscopy. Cells cultured in complete (ES) medium or in defined neural medium (DM) with dox were fixed after 12, 24, 48, and 72 hours in culture followed by immunohistochemical localization of Neurog1 and neuronal tubulin (TuJ1 antibody). Virtually all of the cells cultured in the presence of dox were Neurog1+ at each timepoint surveyed (Fig. 2), whereas those cultured without dox were largely Neurog1− (not shown), confirming the integrity of the tet-inducible locus. TuJ1 immunostaining demonstrated weakly reactive cells in both culture conditions as early as 12 hours following transgene induction, indicating the initiation of neuronal differentiation (Fig. 2A,B,I,J), although in defined medium there were occasionally cells that exhibited a more mature neuronal phenotype (Fig. 2A,B). In general, neuronal differentiation was robust in either culture condition after 72 hours of transgene induction, but differentiation progressed more rapidly in defined medium. After 72h, cell density was also considerably greater in cultures grown in complete medium. Overall, forced expression of Neurog1 was sufficient to induce widespread neuronal differentiation.
Figure 2. Expression of Neurog1 induces widespread neuronal differentiation of ES cells.
Cells were cultured in defined neural medium (A–H) or in complete ES maintenance medium (I–P) supplemented with 1 µg/ml doxycycline. In both media, as early as 12h after dox induction, ESC expressed Neurog1 (green) (A,I). Expression of neuronal tubulin (TuJ1, red) illustrates the initiation of neuronal differentiation within 12 hours of dox treatment in both culture conditions (B,J). Cells grown in defined neural medium demonstrate overt neuronal differentiation (evidenced by neurite extension) as early as 12 hours after treatment whereas cells cultured in ES Medium do not exhibit morphological characteristics of mature neurons until 48 hours of treatment with doxycycline (M,N). These differences were maintained at 48h (C,D, K,L). By 72 hours, neuronal differentiation was extensive in both culture conditions. Cells grown in defined neural medium were highly enriched in neurons (G,H), whereas cultures grown in complete medium show the persistence of non-neuronal cells (O,P). Scale bar = 100 µm.
Neurog1 expression reduces proliferation and increases G1 phase of the cell cycle
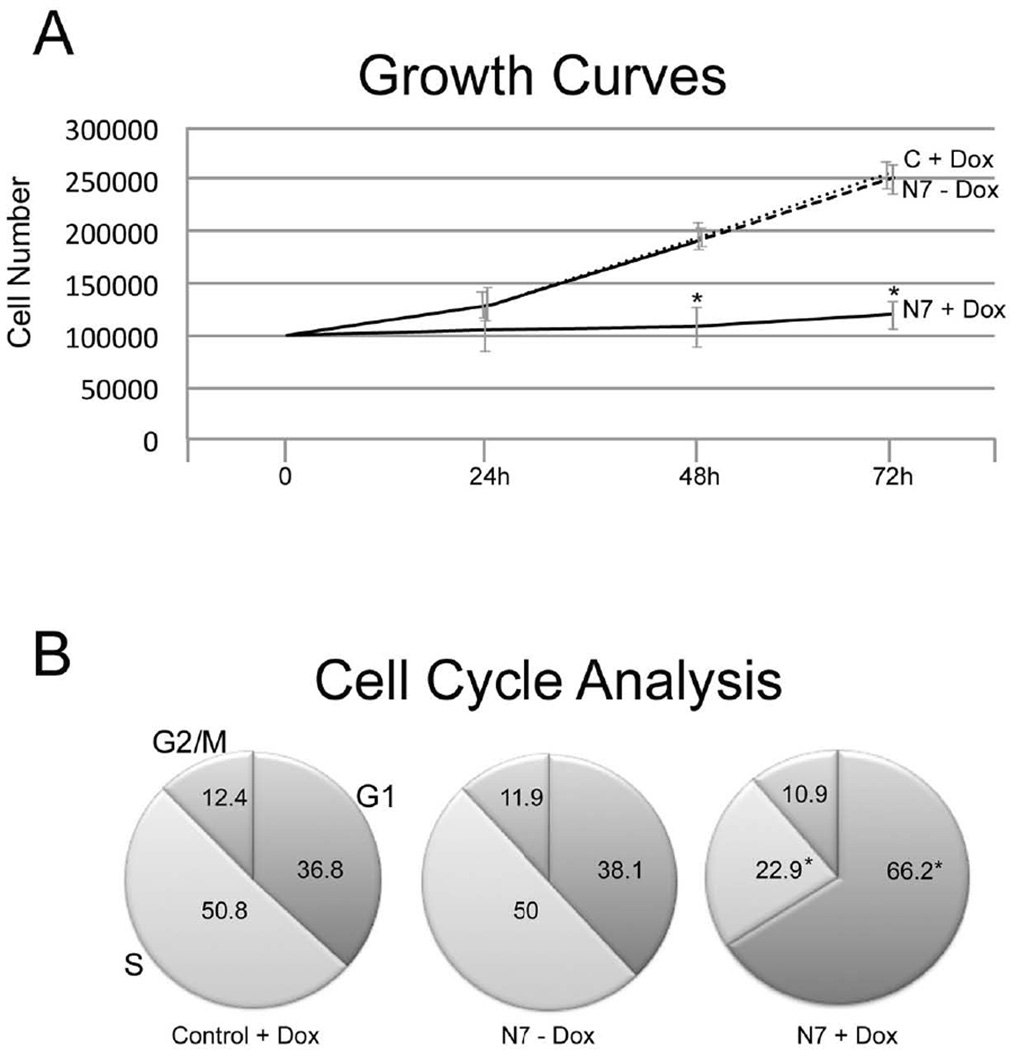
Neurog1 has previously been reported to promote cell cycle exit (Farah et al., 2000), to increase G1/G0 and decrease S phase (Piao et al., 2012), but the kinetics and target genes are not known. To examine this directly, we first determined the growth characteristics of control Ainv15 and N7 cells in the presence and absence of doxycycline at 24h intervals. There was no significant difference in cell number between control Ainv15 ES cells exposed to doxycycline and N7 cells grown without dox at any timepoint (Fig. 3A). Although there was no significant change in cell number at 24h of culture, by 48 and 72h of transgene induction, there was a significant decrease in the number of Neurog1 expressing N7 cells compared with uninduced N7 or control Ainv15 ES cells (p ≤ 0.001, Student’s t test).
Figure 3. Expression of Neurogenin1 decreases proliferation and alters cell cycle characteristics.
A. At 24h, there was no significant effect of transgene expression on cell number, but at 48 and 72h there were significantly fewer N7 cells grown with Dox, compared with either N7 cells alone (N7 - Dox), or control Ainv15 cells with Dox (C + Dox). Means represent cell number from four biological replicates, 12 wells each. * = p ≤ 0.001, Students t test.
B. N7 (N7 - Dox) and control Ainv15 (Control + Dox) cells were grown for 72h ± Doxycycline, then subjected to cell cycle analysis. Induction of Neurog1 expression (N7 + Dox) significantly increased G1 and decreased S phase compared with cells absent transgene induction. * = p ≤ 0.001, Student’s t test.
To determine where in the cell cycle Neurog1 expression affects proliferation, we cultured control Ainv15 and N7 cells ± dox for 72h, labeled the cells with propidium iodide and used FACS to analyze cell cycle characteristics. Consistent with the proliferation data, there was little difference in the cycle characteristics of control Ainv15 ES cells grown in the presence of doxycycline and N7 cells grown without dox—36.8 and 38.1% of the cells were in G1, 50.8 and 50% in S, and 12.4 vs 11.9% in G2/M in control + dox, and N7 – dox cells, respectively (Fig. 3B). Induction of Neurog1 expression had a significant effect on the number of cells in G1 phase; with 66.2% in G1, 22.9% in S, and 10.9% in G2/M. Cell death was similar in N7 ESC exposed to doxycycline (8.8 ± 2.7%) and N7 not exposed to dox (8.2 ± 0.5%), but only 4.0 ± 0.8% in control Ainv15 ES cells grown with doxycycline. These data demonstrate that induction of Neurog1 expression in ES cells slows proliferation by significantly increasing G1 and decreasing the proportion of cells in the S phase of the mitotic cycle.
Treatment with retinoic acid posteriorizes induced N7 cells
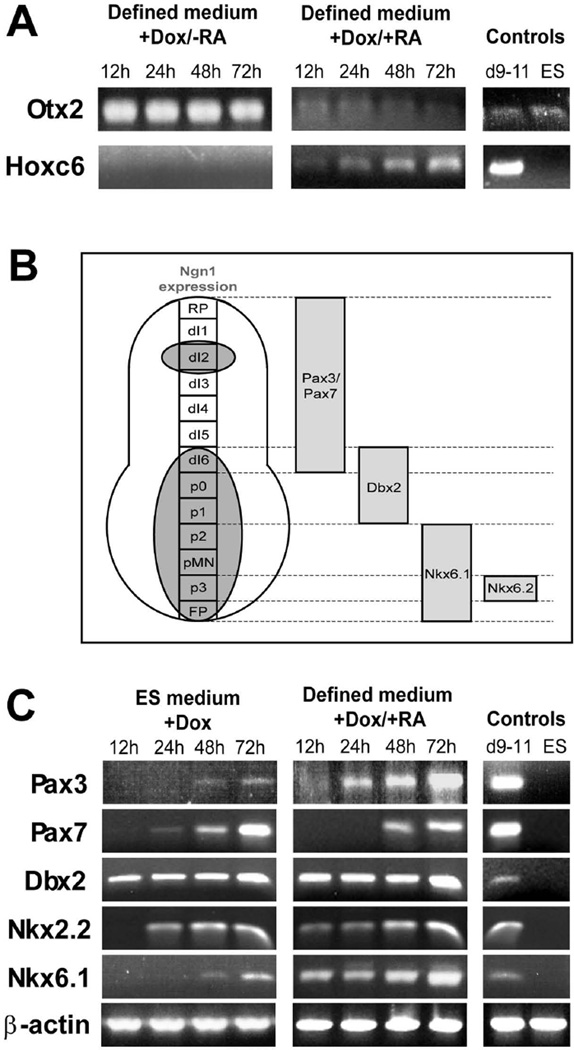
To determine the anterior-posterior characteristics of the differentiating cells, and to determine their responsiveness to patterning factors, cells were grown in defined neural medium with and without 1 µm retinoic acid (RA). Expression of Otx2, a forebrain marker (Mallamaci et al., 1996), and Hoxc6, expressed in the rostral cervical spinal cord, were examined. In both culture media, cells expressed Otx2 but not Hoxc6 at all time points assayed, indicative of a forebrain phenotype (Fig. 4A). When N7 cells were cultured for 3 days in defined medium supplemented with 1 µM RA and dox, there was a significant reduction in Otx2 expression and concomitant up-regulation of Hoxc6 (Fig. 4A). Like previous studies of ESC (Wichterle et al., 2002), these findings suggest that N7 cells neuralized by forced Neurog1 expression have an initial anterior (forebrain) phenotype that can be posteriorized by exposure to RA.
Figure 4. Expression of dorsal and ventral markers reflects Neurog1 expression in vivo.
A. Cells induced in the absence of retinoic acid (RA) demonstrate an anterior neural phenotype (expression of Otx2), whereas a marker of spinal cord (Hoxc6) was not expressed. Cells treated with 1 µM RA show diminished Otx2 expression and increased expression of Hoxc6 indicating that the anterior-posterior identity of the neuronal precursors can be modulated by treatment with RA.
B. Neurog1 is expressed in the ventral half of the developing neural tube, in the roof plate and neural crest-associated structures such as dorsal root ganglia. Pax3 and Pax7 are expressed in the dorsal half of the cord, Dbx2 in the middle third, Nkx6.1 and Nkx2.2 in the ventral spinal cord.
C. With differentiation in either complete ES medium (in which some RA is likely present in the serum), or in defined neural medium supplemented with 1 µM RA, expression of dorsal (Pax3 and Pax7), intermediate (Dbx2), and ventral (Nkx2.2 and 6.1) markers were observed in RT-PCR.
Expression of dorsal and ventral markers reflects Neurog1 expression in vivo
Molecular markers of dorsal-ventral identity in the posterior neural tube have been well-characterized (reviewed in Jessell, 2000; Helms and Johnson 2003; Helms et al., 2005; Fig. 4B); Neurog1 is expressed in the ventral half and near the roof plate of the developing posterior neural tube (Sommer et al., 1996; Ma, et al., 1997, Fig. 4B). We therefore sought to determine if induction of the Neurog1 transgene could produce a similarly mixed population of dorsal and ventral subtypes following treatment with retinoic acid. We examined induced cell cultures grown in ES cell medium, in which an indeterminate amount of RA was likely present in the fetal bovine serum constituent (Napoli, 1986), and in defined neural medium supplemented with 1 µM RA. When assayed by RT-PCR, induced cultures expressed markers associated with the dorsal (Pax3, Pax7), ventral (Nkx2.2, Nkx6.1) and middle (Dbx2) thirds of the developing neural tube (Fig. 4C), which generally corresponds to regions of Neurog1 expression in vivo. Differences in the timing of expression were observed between the proteins assayed; e.g., Dbx2 expression was evident within 12h of inducing the Neurog1 transgene, although it was not expressed by undifferentiated ESC, while robust Pax7 expression was not observed until 48h after induction. Interestingly, in subsequent experiments, Pax7 and Nkx6.1 were rarely co-expressed (Fig. 7), indicating the presence of distinct dorsal and ventral subtypes. Thus, forced expression of Neurog1 in ES cells produces a population of precursors that approximately recapitulates the in vivo pattern of Neurog1 expression.
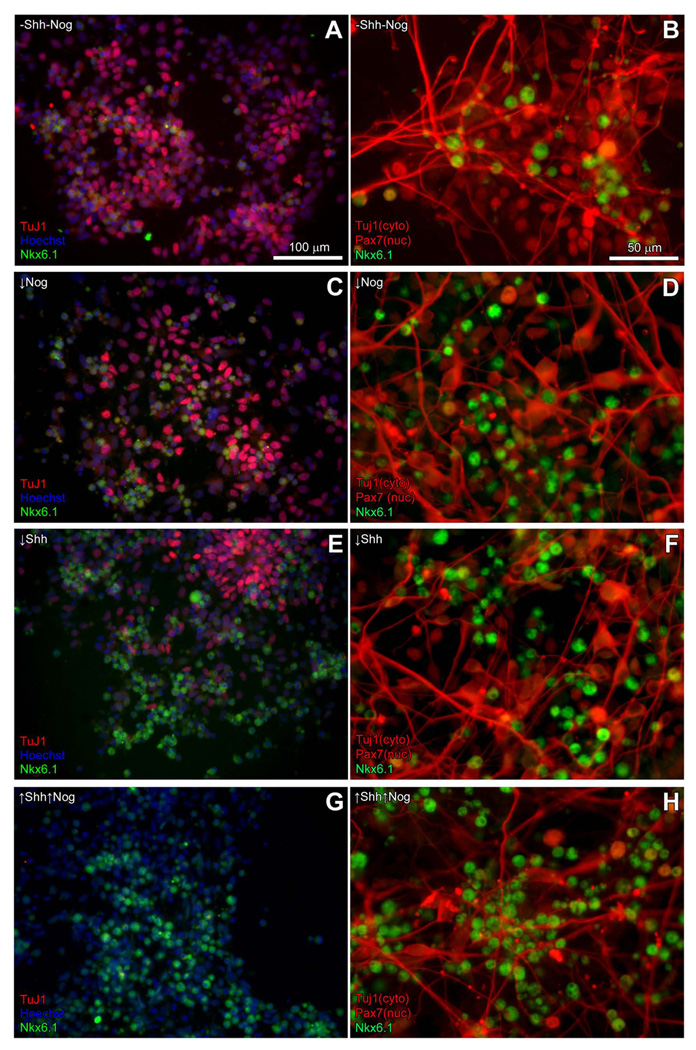
Figure 7. Shift in dorsal-ventral phenotypes with Shh and Nog treatment.
Left panels: Immunostaining with Nkx6.1 (green nuclei) and Pax7 antibodies (red nuclei) indicate that more Pax7+ precursors are present in the absence of or at low doses of Sonic Hedgehog (Shh) and/or Noggin (Nog). Higher doses of Shh and/or Nog reduce the number of Pax7+ cells observed and generate more Nkx6.1+ cells (not shown), and the application of high doses of Shh and Nog together (↑Shh↑Nog) resulted in a dramatic increase in Nkx6.1+ cells and a concomitant decrease in the number of Pax7+ cells. Total cell number in each field was determined by Hoechst nuclear staining (blue). Scale bar = 100 micrometers. Right panels: After cell counts, cultures were stained with Tuj1 to distinguish neurons (red cytoplasm), which are largely Pax7- (red nuclei) and Nkx6.1-(green nuclei) as might be expected for markers that are normally down-regulated with differentiation. Scale bar = 50 micrometers.
Forced expression of Neurog1 produces CNS and PNS neurons
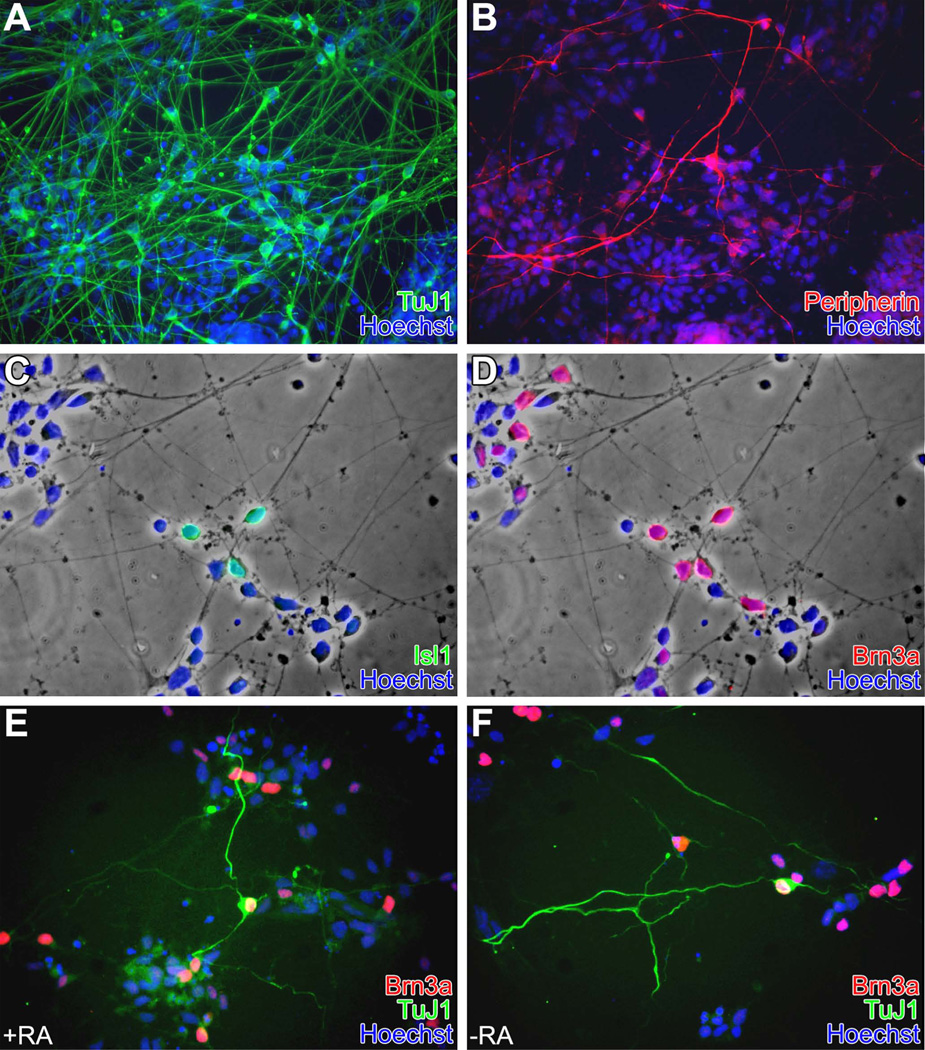
The distribution of CNS and PNS phenotypes was determined after 5 days of culture with and without the addition of RA. Cells were grown in defined medium plus dox to induce the Neurog1 transgene for 72h then switched to a medium supplemented with 5% knockout serum replacer (SRM5) without dox for 48h, as these conditions increased cell survival. After 5 days, there was extensive neuronal differentiation as evidenced by TuJ1 immunoreactivity (Fig. 5A); a subset of these cells (Fig. 5B) was also positive for peripherin, with a Tuj1+:peripherin+ ratio of approximately 21:1 (Table 1). To obtain quantitative data, we scored only peripherin+ somata, so these data likely underestimate the total, as there were many peripherin+ fibers whose cell bodies were outside the field of view. Some neurons expressed Islet1 (Fig. 5C), and, invariably, these cells were also Brn3a+ (Fig. 5D), an expression pattern typical of sensory ganglia (SG) of the PNS as well as of dI3 dorsal interneurons in the CNS (Gowan et al., 2001). There were also many Brn3a+/Isl1-cells (Fig. 5 C,D); an expression pattern that characterizes dI-1, 2, or 5 interneurons in the CNS (reviewed in Helms and Johnson, 2003). Nearly all (about 97%) of the peripherin+ cells were also Brn3a+ and had long, well-developed processes and unipolar or bipolar morphologies (Fig. 5 E,F) – a pattern most consistent with a SG neuronal phenotype. Interestingly, there was little statistically significant difference in marker expression in groups exposed to RA versus those not exposed to RA, although the overall number of neurons present as evidenced by TuJ1 immunoreactivity was slightly higher in the RA-treated cultures as was the number of peripherin+ neurons (Table 1).
Figure 5. Induced N7 cells form CNS and PNS phenotypes.
N7 cells were grown for three days in defined neural medium in the presence of Dox, followed by two days without Dox ± 1 µM RA.
A&B: N7 cultures grown with RA for 5 days exhibit widespread neurogenesis. Some TuJ1+ neurons were also Peri+. Similar results were observed in RA-free cultures (not shown), although there was a slight attenuation in the overall number of neurons observed (see text and Table 1).
C&D: Cells cultured in defined medium with RA for 5 days illustrating the presence of Isl1+ and Brn3a+ neurons. Isl1+/Brn3a+ are co-expressed in dorsal interneurons (lamina 3), and in sensory ganglia; Isl1-Brn3a+ expression is typical of dI-1, 2, or 5 in CNS (dI-2 precursors are normally Neurog1+); Isl1+/Brn3a-expression is typical of CNS motor neurons. Most Isl1+ cells were Brn3a+, but not all Brn3a+ cells were Isl1+.
E&F: Cells cultured +RA (E) or −RA (F) for 5 days illustrating Peri+/Brn3a+ (SG phenotype) cells. The overall frequency was similar in +RA and –RA treated cultures. Nearly all cells that were Peri+ were also Brn3a+.
Table 1. Distribution of neuronal markers.
N7 cells were grown for three days in defined neural medium in the presence of Dox, followed by two days without Dox in defined neural medium ± 1 µM retinoic acid (RA). At least 1000 cells from 10 randomly selected fields in 3 replicate dishes were scored for immunoreactivity to the markers indicated and divided by the total cell number in each field determined by Hoechst nuclear staining. The data presented are the overall means for each treatment group ± s.e.m. across the 3 replicates. P values are based on results of ANOVA, followed by pairwise Student’s t-test.
| TuJ1/Peripherin staining | +RA | −RA | +RA vs. −RA p= |
| %TuJ1+ | 69.2±1.2 | 63.7±1.5 | <0.001 |
| %Peripherin+ somata | 3.5±0.3 | 2.9±0.2 | 0.022 |
| TuJ1+ Peripherin+ | 20.9±1.6 : 1 | 22.7±1.3 : 1 | 0.143 |
| Brn3a/Peripherin staining | +RA | −RA | +RA vs. −RA p= |
| %Peripherin+ somata | 3.4±0.4 | 2.9±0.2 | 0.032 |
| %Brn3a+ | 17.8±1.0 | 16.1±1.0 | 0.063 |
| % of Peri+ also Brn3a+ | 96.9±2.4 | 96.8±2.5 | 0.4755 |
| Brn3a/Isl1 staining | +RA | -RA | +RA vs. −RA p= |
| %Brn3a (total) | 18.1±0.9 | 16.7±1.1 | 0.067 |
| Brn3a+/Isl1+ | 7.4±0.7 | 6.5±0.5 | 0.065 |
| Brn3a+/Isl1− | 11.3±0.8 | 10.3±1.2 | 0.168 |
| %Isl1+ (total) | 7.4±0.6 | 6.5±0.5 | 0.095 |
| Isl1+/Brn3a− | 0.2±0.1 | 0.1±0.1 | 0.191 |
Induced cells respond to dorsal-ventral patterning factors
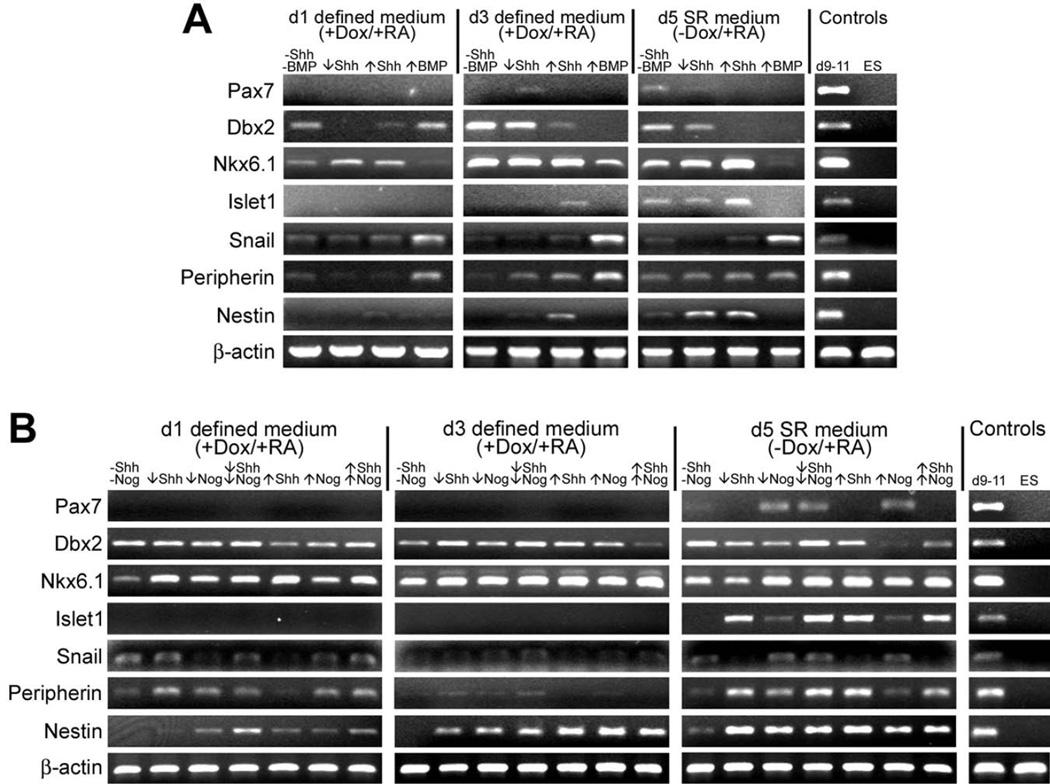
Since forced expression of Neurog1 in ES cells is sufficient to promote both dorsal and ventral neural phenotypes, we wished to determine if the resulting neuronal cells could be patterned by exogenous factors. We cultured N7 cells in defined medium with dox and RA for the first three days and then changed the culture media to a serum replacement medium (SRM) with RA but not dox, to enhance cell survival. To test the effects of exogenous factors, media were supplemented with Sonic hedgehog (Shh), Bone Morphogentic Protein-4 (BMP4), or Noggin (Nog) recombinant proteins from the beginning of each experiment. RT-PCR analysis indicated that Shh treatment induced expression of the ventral marker, Nkx6.1, and suppressed expression of the dorsal marker, Pax7 (Fig. 6A). Exposure of cells to either high (25 nM) or low (2.5 nM) concentrations of Shh, resulted in a dose-dependent expression of ventral markers. On day 1, cultures grown in either high or low levels of Shh expressed higher levels of Nkx6.1 compared to those without Shh. At day 3, Nkx6.1 expression levels were similar between Shh-treated and untreated cultures, but by day 5, cells grown in the high Shh condition expressed higher levels of Nkx6.1. Expression of the intermediate dorsal-ventral marker, Dbx2, was reduced by treatment with Shh in a dose-dependent manner. Treatment with Shh also resulted in the expression of Islet1, which is associated with motor neurons in the ventral spinal cord, and nestin, expressed by neural ectoderm and neural stem cells.
Figure 6. Induced N7 cells respond to patterning factors.
N7 cells were cultured for 3 days in defined neural medium with 1 µg/ml doxycycline (+ Dox) and then switched to serum replacement (SR) medium without Dox for 2 additional days of culture. Media were supplemented with 1 µM retinoic acid (RA) and growth factors as indicated for the entire time in culture. Samples from non-induced N7 cells grown in complete medium (ES) and pooled RNA from d9, 10, and 11 embryos (d9–11) show expected expression of all markers assayed. β-actin is included as a positive control. ↓Shh = 2.5 nM, ↑Shh = 25 nM, ↑BMP = 5 nM BMP4, ↓Nog = 2 nM, ↑Nog = 5 nM
A. Shh and BMP4 treatment: Cells induced in the presence of either low or high doses of Shh show reduced expression of both dorsal (Pax7) and intermediate (Dbx2) neural tube markers and increased expression of a ventral marker (Nkx6.1) compared to untreated cells. In Shh-treated cultures, down-regulation of Dbx2 and up-regulation of Nkx6.1 and Islet1, were dose-dependent. Shh treatment also resulted in up-regulation of the neural stem cell-associated intermediate filament protein, Nestin, by d5 of culture. BMP4-treated cultures show up-regulation of Snail, a marker of pre-migratory neural crest, and Peripherin, a peripheral neuronal marker, and abrogation of all other neural markers assayed.
B. Shh and Noggin treatment: Cells were induced in the presence of combinations of Shh and a BMP antagonist, Noggin (Nog). In general, all treatment groups showed increased expression of Nkx6.1 compared to untreated cells. Expression of Pax7 increased in Nog-treated groups, likely due to an overall increase in the number of neural progenitors as evidenced by increased expression of Nestin. Cultures treated with high doses of Shh and Nog show no expression of Pax7, indicating that these cultures are ventralized by Shh treatment. Shh-treated cells also show increased expression of Islet1 and Peripherin, markers both present in ventral motor neurons, and diminished expression of Snail. High doses of Nog also reduced expression of the intermediate neural tube marker Dbx2, suggesting a role for BMP signaling in establishing this lineage.
Treatment with BMP4 (5 nM) drastically reduced CNS neural differentiation as evidenced by down-regulation of all three CNS neural progenitor markers (Pax7, Dbx2, and Nkx6.1) (Fig. 6A). RT-PCR analysis confirmed expression in BMP4 exposed cultures of GATA-4, Brachyury, and Claudin6, markers of endoderm, mesoderm, and epidermal ectoderm respectively (not shown). BMP4 treated cultures also showed a marked increase in the expression of Snail, a marker of EMT and early neural crest cells and of peripherin, expressed in the PNS and motor neurons (Fig. 6A), although we did not observe many overtly neuronal cells in these cultures. Taken together, these data support the recognized role of BMPs as neural antagonists in the early gastrula stage embryo as well as during the early stages of mouse ES cell differentiation. However, the expression of neural crest and PNS markers suggests that, in the context of forced Neurog1 expression, BMPs can act instructively to promote neural crest and PNS phenotypes (e.g., Aihara et al., 2010).
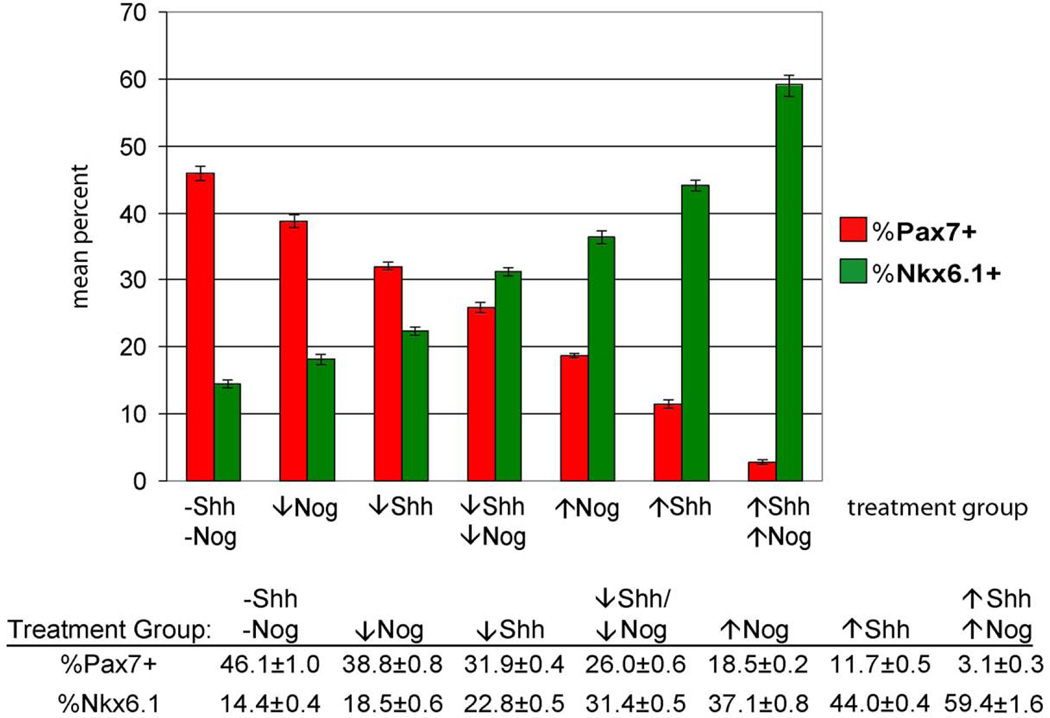
To address the possibility that endogenous BMPs might influence the neuronal differentiation observed in our cultures, we treated cell cultures with low (2 nM) or high (5 nM) levels of the BMP antagonist, Noggin (Nog), in combination with low or high Shh treatment. Consistent with previous observations, Shh-treated cultures expressed low levels of the dorsal marker, Pax7 (Fig. 6B). Interestingly, Pax7 expression in Nog-treated cultures was higher than untreated controls. However, as we observed that Nog treatment appeared to enhance neuronal differentiation and/or survival such that there were many more neurons and primitive neural “rosettes” present in Nog-treated groups compared either to controls or cultures treated with Shh alone (not shown). Since the enhanced Pax7 signal could be due to the increase in neuronal differentiation, we examined the expression of Nkx6.1 and Pax7 using immunohistochemistry (Fig. 7) after 5 days of differentiation. Very few cells co-expressed Pax7 and Nkx6.1, indicating that the cultures contained a mixed population of precursor cells that could be characterized as distinctly "dorsal" (i.e., Pax7+) or "ventral" (i.e., Nkx6.1+). After quantifying the Pax7 and Nkx6.1-positive populations, we then applied the Tuj1 antibody to assess overall neuronal differentiation and found little difference between the treatment groups. Moreover, the Tuj1+ neurons were generally neither Pax7+ nor Nkx6.1+, further confirming that these markers labeled precursors rather than more mature neurons (Fig. 7 right panel). In cultures not treated with Shh or Nog, 46.1% (± 1.0) of cells were Pax7 positive, while 14.4% (± 0.4) were Nkx6.1 positive (Fig. 8). Treatment with either Shh or Nog at low or high levels resulted in a significant reduction in Pax7 immunoreactivity and a corresponding increase in the number of Nkx6.1 positive cells. We also observed more ventral phenotypes in Shh treated groups compared with Nog treatment, and the ventralizing effect appeared to be dose-dependent. Interestingly, co-application of Shh and Nog at either dose produced more ventral phenotypes than either factor alone, suggesting that these two molecules may act synergistically to promote ventral phenotypes. Altogether, the data suggest that forced expression of Neurog1 in ES cells produces a population of neuronal precursors that remain responsive to extrinsic patterning signals.
Figure 8. Quantification of dorsal-ventral phenotypes with Shh and Nog treatment.
There was a dose-dependent decrease in the percentage of Pax7 + cells and an increase in the percentage of Nkx6.1 positive cells following Nog and Shh treatment. Differences observed between all treatment groups were statistically significant (p ≤ 0.05, ANOVA, followed by pairwise Student’s t-test). n = 1000 cells from 10 random fields from two replicate cultures. ↓Shh = 2.5 nM, ↑Shh = 25 nM, ↓Nog = 2 nM, ↑Nog = 5 nM
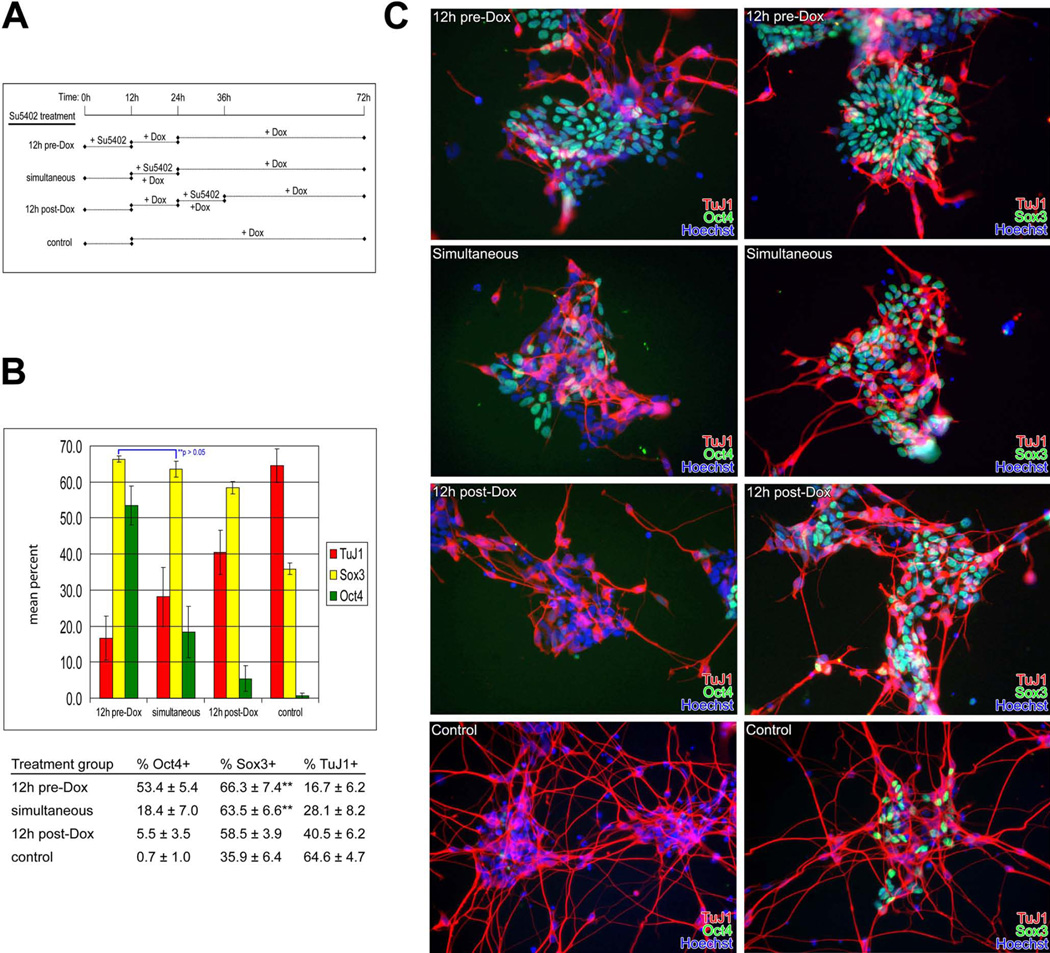
Neural induction via forced Neurog1 expression can be influenced by FGF signaling
The role of FGF signaling in neural induction and ESC differentiation is unresolved, with some studies suggesting that FGF signaling is indispensable in neural induction both in vivo (Streit et al., 2000) and in ES cells in vitro (Ying et al., 2003a). To assess the role of FGF signaling during neuronal differentiation of ES cells in the context of Neurog1 expression, we exposed N7 cells in defined medium to a 12-hour pulse of 5 nM SU5402, which targets the kinase domain of FGFR1 and therefore abrogates all signaling via this receptor (Mohammadi et al., 1997). We treated cells at three intervals: 12 hours prior to the addition of dox (pre-dox), concurrent with the addition of dox (simultaneous), and 12 hours following the addition of dox (post-dox); the control group received dox 12h after plating (Fig. 9A). Cells were then cultured for 72h in defined neural medium containing dox then assayed for the expression of Sox3, a marker of neural ectoderm and neuronal precursors, TuJ1 labeling early neurons, or Oct4, expressed in undifferentiated ES cells. Media changes and monitoring via phase microscopy occurred at 12, 24, 36, and 48 hours at which time we observed no differences in cell death (as evidenced by non-adherent cells observed in the plates and trypan blue staining of withdrawn media) across any of the treatment groups compared to controls. Consistent with either a requirement for Fgf/ERK signaling for ESC differentiation (Kunath et al., 2007), or alternatively, that Fgf/ERK signaling prevents de-differentiation to a more primitive ES-like state (Greber et al., 2010), there were considerably more Oct4 expressing cells (53.4 ± 5.4%) in the pre-dox group (Fig. 9B). Neuronal differentiation as evidenced by Tuj1 immunoreactivity was strikingly reduced in the pre-dox group (16.7 ± 6.2%) and the number of Sox3+ precursors (66.3 ± 7.4%) was higher compared to the other treatment groups. Simultaneous inhibition of FGF signaling with Neurog1 induction resulted in many fewer Oct4+ ES cells (18.4 ± 2.3%), and slightly more TuJ1+ neurons (28.1 ± 1.9%) compared with the pre-dox group. The number of Sox3+ precursors was slightly reduced in the simultaneous group (63.5 ± 6.6%) compared to the pre-dox group, but the difference only approached statistical significance (p=0.06). In the post-dox group, very few Oct4+ ES cells remained (5.5 ± 3.5%). There was also a slight, but statistically significant (p < 0.05), reduction in the number of Sox3+ precursors (58.8 ± 3.9%) in the post-dox group compared to either the pre-dox or simultaneous treatment groups, and a concomitant increase in the number of mature neurons (40.5 ± 6.2%). Controls receiving no SU5402 treatment showed a dramatic overall increase in differentiation as there were few Oct4+ cells remaining (0.7 ± 1.0%), and a significant reduction in the number of Sox3+ precursors (35.9 ± 6.4%). The number of TuJ1+ neurons in the control group was much higher than any of the treatment groups (64.6 ± 4.7%), suggesting that the reduction in Sox3+ cell numbers was likely the result of increased differentiation of the Sox3+ precursor pool into (Tuj1+/Sox3-) neurons. Overall, these data indicate that the induction of Sox3+ neuronal precursors in the context of Neurog1 expression is largely Fgf-independent, but there is a temporal window within which neuronal differentiation can be held in check via Fgf-mediated persistence of the Oct4 pluripotency pathway.
Figure 9. Neuronal differentiation via forced Neurog1 expression is affected by FGF signaling.
A. Treatment groups. N7 cells were plated in defined medium (time = 0h) and exposed to a 12h pulse of 5 nM SU5402. One group (pre-induction) was treated with SU5402 at the time of plating, after which the medium was exchanged for fresh medium supplemented with 1 µg/ml doxycycline (+Dox) for the remainder of the culture period. The second group (simultaneous) was plated in defined medium for 12h followed by addition of SU5402 and Dox. At 24h, the medium was exchanged for fresh medium supplemented with Dox for the remainder of the culture period. The final group (post-induction) was plated in defined medium for 12h followed by addition of Dox. At 24h, a 12-hour pulse of SU5402 was administered, and, at 36h, the medium was exchanged for fresh medium supplemented with Dox for the remainder of the culture period.
B. Oct4/Sox3/TuJ1 cell counts. There was a decrease in the number of Sox3+ cells between the pre-induction and simultaneous treatment groups that was not significant (**, p = 0.058), while the decrease in the number of Sox3+ cells in the post-dox group was slight but significant compared with the other treatment groups. There was a striking increase in TuJ1 expression with delayed SU5402 treatment and a corresponding decrease in the number of Oct4+ cells, while the number of Sox3+ cells observed dropped only slightly with delayed treatment. Untreated controls demonstrated markedly increased neuronal differentiation, a concomitant reduction in neuronal precursor cells, with very few cells expressing Oct4. At least 1000 cells from 10 randomly selected fields in two replicate dishes were scored for Oct4, Sox3, and TuJ1, and analyzed by pairwise Student’s t-test.
C. Oct4/Sox3/TuJ1 immunohistochemistry. The cell counts described above were obtained from cell cultures that were fixed after 72h and co-stained with TuJ1 and Oct4 antibodies (left panels) or TuJ1 and Sox3 antibodies (right panels). Nuclei are labeled with Hoechst 33258. Blocking FGF signaling resulted in fewer neurons (TuJ1+) in a manner depending on the timing of treatment. Control cells receiving no SU5402 show robust neuronal differentiation.
Microarray analysis
After 24 hours of transgene induction, 1384 genes were significantly differentially expressed; 951 up-regulated. At 48h, 5894 genes were significantly altered (2856 up-regulated); increasing to 7205 (3263 up-regulated) after 72h of doxycycline exposure. Gene ontology classification analysis identified significant alterations in the expression of genes associated with: nervous system development (48%), development (24%), metabolism (4%), cell cycle (4%), signaling (4%), and cellular organization (4%). Supplemental Table 1 summarizes the results of functional annotation clustering of genes significantly up- and down-regulated at 24, 48 and 72 hours of transgene induction. There were significant changes in the expression of genes involved in neurogenesis, in process outgrowth and cell migration, those involved in the regional development of the nervous system, in cell cycle, as well as genes involved in ESC homeostasis.
We carried out KEGG pathway analysis to map alterations in signaling pathways and molecular functions and identified sequential induction of genes in pathways involved in axon guidance, cancer and melanogenesis (Supplemental Table 2). By 72h of transgene induction, 19 pathways were activated, including: ErbB, Neurotrophin, Wnt, Mapk, Focal adhesion, Calcium, and Insulin signaling pathways. After 72h, only one pathway, Cysteine and methionine metabolism, was significantly down-regulated.
Consistent with our RT-PCR results, Neurog1 was up-regulated 74, 74 and 42.7 fold at 24h intervals compared with uninduced cells. Other bHLH genes were also induced: Neurod4 increased by 33, 206 and 127.8 fold; Neurod1 increased 9.7, 11.5 and 10.2 fold, while Neurog2 was up-regulated by: 0, 4.4 and 3.8 fold (Table 2). Pluripotency factors expressed by undifferentiated ES cells: Eed, Eras, Fbxo15, Foxd3, Klf2, Klf4, Nanog, Pou5f1 (Oct4), Sox2, Zic3, were significantly down-regulated with differentiation. Genes associated with neurogenesis including: Ebf2, Ebf3, Elavl3, Elavl4, Fabp7, Hes5, Lhx2, Ncam1, Nhlh1, Nhlh4, Pax3, Pax6, Reln, Zic1, were induced, while the negative regulator of neurogenesis Rest (RE1-silencing factor), was down-regulated. Many genes expressed in the ventricular zone (Fu et al., 2009) and by neural stem cells including Dll1, Dcx, Prom1, Reln, Coup-TFI and II (Naka et al., 2008), and Hes5 (Basak and Taylor 2007; Hatakeyama et al., 2004) were expressed, as were genes associated with cell cycle exit and differentiation including Ebf2,3 (Pozzoli 2001; Garcia-Dominguez et al., 2003). Id4, Pax3, Pax6, Sox3, Sox4 and Sox11 (Bergsland et al., 2006) were also increased. Factors involved in process outgrowth and cell migration including ephrins and their Eph receptors, semaphorins and plexins were induced by Neurog1. Interestingly, epithelial genes including junctional proteins and cell-cell adhesion molecules such as E-Cadherin (Cdh1) were down-regulated whereas N-Cadherin (Cdh2) expression significantly increased.
TABLE 2. Microarray expression analysis.
Fold change in gene expression following 24h, 48h and 72h of transgene induction
| Gene | UniGene# | 24h | 43h | 72h | Gene | UniGene# | 24h | 43h | 72h | Gene | UniGene# | 24h | 43h | 72h | Gene | UniGene# | 24h | 43h | 72h |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Factors associated with pluripotency | Neural Patterning | Cell Surface | Cell Cycle | ||||||||||||||||
| Eed | Mm.380914 | - | −3.6 | −4 | Eya1 | Mm.250185 | - | 7 | 11.6 | Slit1 | Mm.40322 | - | 8.6 | - | G1/S checkpoint | ||||
| Eras | Mm.250895 | −4.9 | −33.9 | −65.8 | Foxa2 | Mm.938 | - | 21.2 | 13.7 | Slit2 | Mm.289739 | - | 3.5 | 8.1 | Cdkn1a | Mm.195663 | - | 5.0 | 3.0 |
| Fbxo15 | Mm.28369 | - | - | −14.4 | Foxd1 | Mm.347441 | - | 11.1 | 23.9 | Synpr | Mm.37515 | - | 47.5 | 144.9 | Suv39h1 | Mm.9244 | 2.9 | −35.7 | −17.6 |
| Foxd3 | Mm.4758 | - | −12.4 | - | Foxg1 | Mm.390496 | - | - | 13 | Unc5c | Mm.24430 | - | 12.2 | 31.8 | Trp53 | Mm.222 | - | −3.3 | −5.5 |
| Klf2 | Mm.26938 | - | −27.8 | −40.7 | Foxp2 | Mm.332919 | - | 46.7 | 438 | Signaling | |||||||||
| Klf4 | Mm.4325 | −8.4 | −15.5 | −22.9 | Hoxa1 | Mm.l97 | 24.3 | - | - | Bmp1 | Mm.27757 | 2.7 | 5.8 | 4.7 | G1/S | ||||
| Nanog | Mm.440503 | −4.7 | −22.9 | −33.1 | Hoxa2 | Mm.131 | - | 18 | 24.4 | Bmp4 | Mm.6813 | −6.2 | - | −8.1 | Btg2 | Mm.392646 | 3.6 | 4.0 | - |
| Pou5f1 | Mm.17031 | −4.8 | −27.1 | −38.2 | Hoxa4 | Mm.439647 | - | 21.9 | - | Dll1 | Mm.4875 | 15.8 | 8.3 | 4.2 | Cables1 | Mm.40717 | - | 2.3 | 2.7 |
| Sox2 | Mm.65396 | - | - | −2.7 | Hoxb1 | Mm.890 | 10.7 | 16.5 | - | Dll3 | Mm.12896 | 19.9 | 20.5 | 5.5 | Ccnd1 | Mm.l6110 | −2.2 | −6.1 | −6.9 |
| Tert | Mm.10109 | - | - | −3.5 | Hoxb2 | Mm.281153 | 33.5 | 92.7 | 73.4 | Dll4 | Mm.143719 | 58.2 | 56 | 18.8 | Ccne1 | Mm.l6110 | −6.1 | −6.9 | |
| Zic3 | Mm.255890 | −2.2 | −5.5 | −10.3 | Hoxb4 | Mm.3546 | 22.6 | 43.3 | 45.6 | Dner | Mm.292560 | - | 12 | 23 | Cdca5 | Mm.23526 | - | −3.1 | −3.7 |
| Hoxd9 | Mm.26544 | - | - | 10.6 | Fgf4 | Mm.4956 | −4.7 | −48.3 | −8.1 | Cdkn1a | Mm.195663 | - | 5.0 | 3.0 | |||||
| lrx1 | Mm.316056 | - | 4.2 | 6.5 | Fgf5 | Mm.5505 | - | - | - | Ccnd2 | Mm.333406 | - | 3.0 | 6.0 | |||||
| lrx3 | Mm.238044 | 8.5 | 39.9 | 42.6 | Fgf12 | Mm.7996 | - | 20.9 | 30.6 | Gspt1 | Mm.325827 | - | 2.1 | 2.1 | |||||
| lsl1 | Mm.42242 | 10.1 | 89.3 | 197 | Fgf13 | Mm.7995 | - | - | 5.2 | ||||||||||
| Neural Differentiation | Lhx2 | Mm.142856 | 54.8 | 144.7 | 192.1 | Fgf15 | Mm.3904 | - | 3.6 | - | S | ||||||||
| Ascl1 | Mm.136217 | - | - | - | Onecut1 | Mm.303355 | 18.3 | 13 | 27.3 | Fgf17 | Mm.12814 | −12.9 | −91.7 | −55.6 | Brca2 | Mm.236256 | - | −2.8 | −3.4 |
| Brn2 (Pou3f2) | Mm.l29387 | - | - | 7.2 | Onecut2 | Mm.234723 | 8 | 120.6 | 184.5 | Frzb | Mm.427436 | - | 46.3 | 35.9 | Cdt1 | Mm.21873 | - | −2.8 | −3.7 |
| Dclk1 | Mm.393242 | - | 41 | 119.9 | Six1 | Mm.4645 | - | 5.2 | 15.5 | Gli1 | Mm.391450 | - | - | −14.6 | Pola1 | Mm.l923 | - | −2.5 | −3.0 |
| Dcx | Mm.l2871 | 8.2 | 193.8 | 359.4 | Gli2 | Mm.273292 | −2.9 | - | −3.8 | Skp2 | Mm.35584 | −2.5 | −2.2 | −2.3 | |||||
| Ebf2 | Mm.319947 | 95.7 | 158.5 | 96.3 | Gmnn | Mm.12239 | - | −2.7 | −3 | ||||||||||
| Ebf3 | Mm.258708 | 47.7 | 186.2 | 281.9 | Cell Surface | Hes5 | Mm.137268 | 21 | 384.7 | 203.2 | G2/M Checkpoint | ||||||||
| Ednrb | Mm.229532 | - | - | 194.6 | Cdh1 | Mm.35605 | - | −8.9 | −10.7 | Hes6 | Mm.280029 | 17.9 | 7.5 | 2.6 | cdk1 | Mm.281367 | - | - | - |
| Elavl3 | Mm.390167 | - | 30.9 | 35 | Cdh2 | Mm.257437 | - | 18.3 | 31.8 | Hhat | Mm.145857 | 3.6 | 5.2 | 4.7 | Cdt1 | Mm.21873 | - | −2.8 | −3.7 |
| Elavl4 | Mm.3970 | - | 75 | 143.6 | Dab1 | Mm.289682 | - | 2.9 | 5.4 | Id2 | Mm.34871 | - | 2.9 | - | Pola1 | Mm.l923 | - | −2.5 | −3.0 |
| Fabp7 | Mm.3644 | 49.4 | 201.1 | 443.8 | Dcc | Mm.l67882 | - | 61.5 | 114 | Id3 | Mm.110 | −10.9 | - | - | Tipin | Mm.l96219 | - | −2.6 | −3.4 |
| Gfap | Mm.1239 | - | - | - | Efna1 | Mm.15675 | - | - | - | Id4 | Mm.458006 | 5.8 | 20.7 | 14.8 | |||||
| Isl1 | Mm.42242 | 10.1 | 89.3 | 197 | Efna5 | Mm.7978 | - | 5.2 | 6.6 | Igf1r | Mm.275742 | - | 3.1 | - | |||||
| Lhx2 | Mm.142856 | 54.8 | 144.7 | 192.1 | Efnb1 | Mm.3374 | 12.7 | 2.9 | - | Igf2bp3 | Mm.281018 | - | 3.4 | 2.3 | G2/M | ||||
| Myt1 | Mm.458718 | - | 24 | 66.2 | Efnb2 | Mm.209813 | - | 8.3 | 9.6 | Igfbp2 | Mm.141936 | - | −2.6 | −4.9 | Birc5 | Mm.8552 | - | −3.4 | −5.0 |
| Ncam1 | Mm.4974 | - | - | 6.8 | Epha1 | Mm.133330 | - | - | −33.6 | Igfbp4 | Mm.233799 | - | 25.4 | 13.5 | Ccnb2 | Mm.22592 | - | - | −3.1 |
| Nefh | Mm.298283 | −2.3 | −2.4 | −2.6 | Epha2 | Mm.2581 | - | −6 | −6 | Igfbp5 | Mm.405761 | - | 18.1 | 25.3 | Cdc25b | Mm.38444 | 4.6 | - | - |
| Nefl | Mm.1956 | −2.7 | 4.2 | 23.4 | Epha3 | Mm.1977 | - | −4.3 | - | Igfbp7 | Mm.233470 | −3.2 | - | −4.9 | Cdkn1a | Mm.l95663 | - | 5.0 | 3.0 |
| Nefm | Mm.390700 | - | - | 257.7 | Epha4 | Mm.400747 | −2.2 | 2.2 | 2 | Igfbpl1 | Mm.3919 | 9.4 | 67.1 | 58 | Gadd45g | Mm.281298 | 9.6 | - | - |
| Neurod1 | Mm.4636 | 9.7 | 11.5 | 10.2 | Epha5 | Mm.137991 | - | 18 | 43.7 | Inhbb | Mm.3092 | −11.5 | −43.7 | −131 | Gpc1 | Mm.297976 | - | 3.6 | 3.9 |
| Neurod4 | Mm.10695 | 33 | 206 | 127.8 | Ephb1 | Mm.22897 | - | - | - | Jag1 | Mm.22398 | - | 6.2 | 3.8 | Hmga2 | Mm.l57190 | - | - | 3.0 |
| Neurog1 | Mm.266665 | 74.4 | 74 | 42.7 | Ephb2 | Mm.250981 | - | 2.6 | 3.7 | Nlk | Mm.9001 | - | - | 2.1 | |||||
| Neurog2 | Mm.42017 | - | 4.4 | 3.8 | Esrrg | Mm.89989 | - | - | 98 | Nodal | Mm.57195 | −2.2 | −14 | −21.5 | |||||
| Nhlh1 | Mm.2474 | 27.4 | 101.5 | 105.4 | Itga1 | Mm.317280 | - | −4.8 | - | Notch1 | Mm.290610 | - | 2.8 | - | M | ||||
| Nhlh2 | Mm.137286 | - | 109.9 | 204.3 | Itga5 | Mm.16234 | - | - | −3.1 | Notch2 | Mm.254017 | 3 | 2.7 | - | Aurka | Mm.249363 | - | −2.9 | −3.7 |
| Nr2f1 | Mm.439653 | - | 90.1 | 197.9 | Itga6 | Mm.225096 | - | −2.5 | 4.6 | Notch4 | Mm.l73813 | - | −4 | - | Birc5 | Mm.8552 | - | −3.4 | −5.0 |
| Nr2f2 | Mm.l58143 | - | 74.9 | 106.2 | Netrin1 | Mm.39095 | −3.2 | 14.2 | - | Ntrk1 | Mm.80682 | - | 11.4 | - | Bub1b | Mm.29133 | - | −4.2 | −4.8 |
| Nrxn1 | Mm.480021 | - | 76.3 | 227.6 | Ntrk1 | Mm.80682 | - | 11.4 | - | Ntrk2 | Mm.l30054 | - | - | - | Ccdc99 | Mm.l94368 | - | −2.4 | −3.0 |
| Olig1 | Mm.39300 | - | - | - | Ntrk3 | Mm.421361 | - | 6.2 | 22 | Ntrk3 | Mm.33496 | - | 6.2 | 22 | Ccnb1 | Mm.260114 | - | −3.3 | −4.5 |
| Olig2 | Mm.37289 | - | - | - | Pcdh8 | Mm.390715 | 10.5 | 155.1 | 108 | Sfrp2 | Mm.19155 | - | - | 12.4 | Ccnb2 | Mm.22592 | - | - | −3.1 |
| Olig3 | Mm.156946 | 4.5 | 19.6 | - | Plxna1 | Mm.3789 | - | 3 | 2.3 | Smad1 | Mm.223717 | - | - | 4.2 | Ccng2 | Mm.3527 | 3.7 | 6.2 | 5.6 |
| Pax3 | Mm.1371 | 20.2 | 177 | 98.3 | Plxna2 | Mm.2251 | 7.1 | 64.1 | 58.1 | Smad2 | Mm.391091 | - | - | 2.1 | Cdc20 | Mm.289747 | - | −3.0 | −4.2 |
| Pax6 | Mm.33870 | - | 10.8 | 23.1 | Plxnb1 | Mm.53862 | - | 2.8 | 2.5 | Smad3 | Mm.7320 | 2.8 | 2.8 | - | Cep55 | Mm.9916 | - | −6.1 | −6.3 |
| Prom1 | Mm.6250 | - | 10 | 0 | Plxnc1 | Mm.256712 | - | 10 | 11.6 | Smad4 | Mm.100399 | - | - | - | Ercc6l | Mm.31911 | - | −4.9 | −6.6 |
| Reln | Mm.425236 | - | 40.8 | 43.3 | Plxnd1 | Mm.3085 | - | 5.9 | 6.9 | Smad7 | Mm.34407 | −5.1 | −8.7 | −6.6 | F630043A04Rik | Mm.30173 | - | −2.3 | −3.9 |
| Rest | Mm.28840 | −4.5 | −6.4 | −15.6 | Robo1 | Mm.310772 | - | 4.6 | 6.3 | Socs2 | Mm.4132 | −2.1 | −4.4 | −17.2 | Fbxo5 | Mm.197520 | - | −2.9 | −4.0 |
| Ret | Mm.57199 | - | 96.8 | 251.1 | Sema3c | Mn.5071 | - | - | 12.8 | Socs6 | Mm.91920 | - | 4.7 | 5.2 | Hells | Mm.57223 | - | −3.5 | −4.0 |
| Runx1t1 | Mm.4909 | - | 5.6 | 19.3 | Sema3d | Mm.89313 | - | - | 13.8 | Stat1 | Mm.277406 | 3.9 | - | - | Incenp | Mm.29755 | - | −2.7 | −3.1 |
| Sox11 | Mm.41702 | 4.2 | 34.7 | 43.1 | Sema4a | Mm.439752 | - | −5.4 | −22.2 | Stat3 | Mm.249934 | - | −2 | −2 | Ndc80 | Mm.225956 | - | −2.6 | −3.3 |
| Sox3 | Mm.35784 | 9.9 | 15.7 | 11.9 | Sema4b | Mm.275909 | - | - | −2.5 | Stat6 | Mm.121721 | - | −5.3 | −11.4 | Nsk2 | Mm.33773 | - | −2.8 | −4.0 |
| Sox4 | Mm.240627 | 6.1 | 13.2 | 18 | Sema4c | Mm.29558 | - | - | 9.9 | Tcf3 | Mm.440067 | - | −2.3 | - | Nup43 | Mm.299887 | - | −3.3 | −3.7 |
| Tubb3 | Mm.40068 | - | 7.7 | 8.7 | Sema4f | Mm.270543 | - | - | 6.4 | Tcf4 | Mm.4269 | - | 6.8 | 5.1 | Plk1 | Mm.16525 | 2.0 | −2.5 | −3.5 |
| Zeb1 | Mm.3929 | - | 6.4 | 6.5 | Sema5a | Mm.260374 | −3.5 | 4.3 | 4 | Wnt5a | Mm.287544 | 20.2 | 13.4 | 6.4 | Rcc2 | Mm.253 | - | −4.4 | −5.1 |
| Zfp238 | Mm.480309 | 6.6 | 11.1 | 7.8 | Sema6a | Mm.40909 | - | - | 4.7 | Wnt8b | Mm558 | 7.9 | 8.6 | - | Spc24 | Mm.295642 | - | −3.0 | −3.3 |
| Zic1 | Mm.335350 | 53.4 | 40.7 | 31 | Sema6d | Mm.330536 | 11.3 | 37.7 | 44.9 | Zeb2 | Mm.35353 | 6 | 39.4 | 27.8 | Tipin | Mm.l96219 | - | −2.6 | −3.4 |
There were also significant increases in the expression of genes associated with regional neuronal cell types, including telencephalon-associated genes Elavl4, Fabp7, Foxd1 and Foxg1. Lhx2, which plays a critical role in cortical patterning (Mangale et al., 2008; Chou et al., 2009) was particularly strongly induced by Neurogenin1. A number of factors associated with ventral neural fates, e.g., striatal (Foxp2), midbrain (Pcdh8), and midbrain dopaminergic neurons (Foxa2, Ebf3) were also induced. Hindbrain markers Hoxa2 and Hoxb2 were significantly induced, as was Hoxd9, which is expressed in thoracic lateral motor neuron columns (Dasen et al., 2005). Other genes associated with a motor neuron phenotype such as Irex3 and Isl1 were also significantly increased. Genes expressed in cephalic placodes including Eya1, Ebf2, Fbxo2, Six1, Netrin1 and its receptors DCC and Unc5C were up-regulated as were genes associated with neural crest cells, including Ednrb and Ret. Few genes associated with glial differentiation (Fu et al., 2009) were identified, and genes that inhibit oligodendrocyte or astrocyte differentiation, e.g., Hes5 (Liu et al., 2006) and Id4 (Marin-Husstege et al., 2006) were induced.
Activation or silencing of lineage specific sets of genes during development is controlled by transcription factors as well as by epigenetic regulators of chromatin structure. Because ESC have a chromatin configuration that is “poised” for lineage differentiation (e.g., Boyer et al., 2006), selective induction of chromatin regulators associated with other lineages, or removal of repressive marks on neuronal promoters/enhancers could drive neuronal differentiation. We examined the expression of transcripts encoding proteins involved in chromatin modification including SWI/SNF, polycomb family members, HDACs, etc., and found minimal (< 3 fold) changes. There were three exceptions that are of potential interest. The first is in Forkhead box (Fox) gene expression. Foxa proteins are considered “pioneering” in that they open chromatin to allow modifiers access to chromatin, Foxp factors function as classic transcription factors recruiting enzymes to regulate gene expression, while Foxo proteins appear to do both (Lalmansingh et al., 2012). Following Neurog1 expression, Foxa2 was increased 21.2 and 13.7 fold at 48 and 72h, and Foxp2 was increased 62.4 and 53.8 fold at similar time points, suggesting a role in neuronal lineage differentiation. Second, Phc2, a polycomb group member expressed in germinal zones of the nervous system (Kim et al., 2005), was induced 7.1, 8.9 and 8.3 fold. Finally, the histone methyltransferase, Suv39h1 was down-regulated 35.7 and 17.6 fold at 48 and 72h. Suv39h proteins have been shown to oppose Ring1 to coordinate early lineage decisions in the blastocyst (Alder et al., 2010), and to interact with Smads to coordinate BMP-induced gene repression and lineage differentiation (Frontelo et al., 2004). Somewhat surprisingly, given a recent study demonstrating that Ezh2 is required to inhibit transcription of non-muscle lineage genes in satellite cells (Juan et al., 2011), we did not observe alterations in either the ubiquitous Ezh1 or in Ezh2 associated with proliferating tissues (Margueron et al., 2008). Future interrogation using ChIP and knock-down will explore these possibilities more directly.
Microarray analysis also identified a complex pattern of expression of genes involved in cell cycle control. At 24 and 48h, cluster analysis did not identify any significant clusters containing cell cycle related genes, but by 72h of transgene induction, three clusters identified down-regulated genes involved in cell cycle. Cluster 3 (Enrichment Score = 7, p ≤ 5.0 × 10−9, Benjamini-Hochbert corrected) contained genes involved in cell cycle progression, Cluster 4 (Enrichment Score = 5.5, p ≤ 3.1 × 10−9, corrected) identified genes involved in chromosome organization, sister chromatid, and centromere structure, while Cluster 7 (Enrichment Score = 4.9, p ≤ 2.3 × 10−6, corrected) identified genes involved in chromosome organization (Supplemental Table 2).
Because the ESC population is relatively homogeneous (compared to brain tissue) it is possible to examine the sequential expression/repression of transcripts involved in cycle regulation (Table 2). Positive regulators of cell cycle including: Birc5, Id3, Skp2 were down-regulated, and negative regulators: Btg2, Ccng2, Ebf2,3, Gadd45g, Gspt1, Hipk2, Prmt2 were up-regulated with transgene expression. However, other positive regulators were stimulated, including: Ccnd1, Ccnd2, Cdc25b, Cyr61, Gpc1, Hmga2, Tead2. Others were strongly changed initially by transgene induction but then were reduced by 48 or 72h, including: Cdc25b, Gadd45g, and Id3. Interestingly, the tumor suppressor Trp53/p53 was down-regulated significantly both at 48 and 72h of induction. P53 plays major roles in apoptosis, cell cycle arrest, and via its downstream targets Gadd45g and p21 (Cdkn1a) which is up-regulated at all time points, which functions as a G1 regulator to produce growth arrest.
We also compared gene expression in Neurog1 expressing cells at 24 vs 48, 48 vs 72 and 24 vs 72 hour time points. Overall, microarray analysis of changes in gene expression between 24–48h identified sets of genes expressed in neural stem cells, neural crest and placodes, comparing 24 and 72h patterns, increasing numbers of mature markers including channel genes, neurotransmitter/receptor genes were identified, consistent with our previous observations that they exhibit mature axon potentials (Reyes et al., 2008).
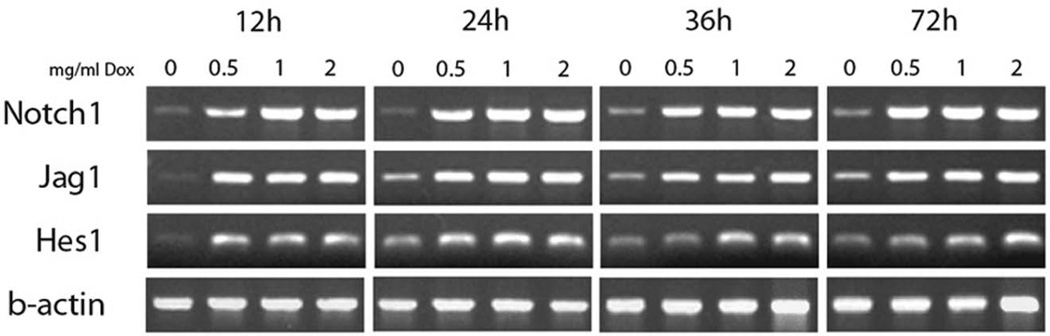
To validate the microarray data, we chose to examine Notch-Delta pathway members, as they have previously been suggested to be targets of Neurog1 (Ma et al., 1997). As early as 12h after doxycycline treatment, expression of Notch pathway members was strikingly up-regulated, remaining high during the 72 hour culture period. Over time in control cultures there was a gradual increase in expression of pathway members, Notch1, Jag1 and Hes1 as neuronal differentiation increased (Fig. 10). Consistent with these observations, at 24h array analysis identified Notch2 (3-fold), Hes5 (21) and Hes6 (17.9) as up-regulated (Table 2). By 48 hours, many pathway members including Notch1 (2.8), Notch2 (2.7), Notch4 (−4), Jag1 (6.2), Hes5 (384), Hes6 (7.5), Dner (12) were identified. At 72 h, Jag1 (3.8) and Hes5 (203.2), Hes6 (2.6), and Dner (23) were up-regulated.
Figure 10. Induced N7 Cells Express Notch-Delta Pathway Members in a dose-dependent manner.
Cells cultured in defined neural medium with 0, 0.5,1, or 2 mg/ml doxycycline up-regulate the Notch1 receptor and its ligand Jagged1, as well as the downstream target of Notch activation, Hes1. Up-regulation of genes associated with Notch-Delta signaling is correlated with the expression of Neurog1, providing further evidence of an interaction between Neurog1 and the Notch-Delta signaling pathway, as observed in microarray analysis.
Microarray data have been deposited in GEO, accession numbers: GSE42883, GSM1052734-GSM1052745.
DISCUSSION
Inducible expression of Neurog1 in ES cells provides a novel model of neurogenesis
ES cells provide a powerful model system to study the mechanisms of cell fate specification and lineage segregation at a stage in early development not readily accessible for manipulation. Studies of directed ES cell differentiation typically rely on differentiation in embryoid bodies and/or exposure to stromal cell derived factors or to non-specific morphogens such as RA. While these approaches can produce populations enriched with neurons (e.g., Zeng et al., 2011), undefined factors and endogenous cytokines present in EBs, media containing serum, or stromal cell co-culture, complicate efforts to understand the role of specific signaling pathways in lineage segregation. Differentiation of ES cells in monolayer cultures in defined media minimizes these effects (e.g., Ying et al., 2003b) and, if combined with forced expression of a lineage restricted gene, presents a unique opportunity to tease out the interactions of particular genes and growth factors in early development. An additional advantage of the inducible gene expression paradigm is the ability to maintain transgene expression following implantation (Reyes et al., 2008). Here we demonstrate that expression of Neurog1 in ES cells is sufficient to induce neuronal differentiation even in conditions that normally maintain pluripotency (adherent culture at high density with serum and LIF), in defined media where the presence of morphogens can be more precisely controlled.
Neurog1 expression in ES cells generates representative neuronal subtypes and neural crest derivatives
Consistent with the pattern of Neurog1 expression in defined domains of both the CNS and PNS (Ma et al., 1996, Sommer et al., 1996; Ma et al., 1997, Murray et al., 2000; Kim et al., 2011; Takano-Maruyama et al., 2012), expression of Neurog1 in ES cells is sufficient to generate neuronal precursors and mature cell types representative of these expression domains (Figs. 4 and 5). In fact, this paradigm may represent a reliable method to obtain sensory neurons, which have been difficult to generate from embryonic stem cells (Sasai, 2005). We observed a significant number of neurons that expressed Brn3a, a POU homeodomain transcription factor associated with sensory lineages in both the CNS and PNS. In the CNS, Brn3a+ sensory lineages arise throughout the anterior-posterior extent of the dorsal alar plate in the neural tube. In the posterior spinal cord, the alar plate generates Brn3a+ dorsal interneurons, of which the dI2 commissural lineage is specifically Neurog1-dependent (Gowan et al., 2001), whereas rostral domains of Brn3a and Isl1 expression are present in the tectum and tegmentum of the midbrain (Fedstova and Turner, 2001). A 4kb enhancer has been shown to drive reporter gene expression to the midbrain, hindbrain and spinal cord (Nakada et al., 2004), while a second 0.8 kb enhancer drives expression to cells fated to become interneurons in the ventral spinal cord (Quinones et al., 2010). The presence of peripherin+ cells in our cultures presents the intriguing possibility of having generated motor neurons which express this marker (Escurat, et al., 1990). However, these cells almost invariably co-expressed Brn3a, which has not been reported in motor neurons, but is more suggestive of PNS lineages. Regarding glial differentiation, we did not observe any up-regulation in glial markers in either our microarray analyses (Table 2) or by RT-PCR or immunohistochemistry (not shown). Moreover, our previous work similarily failed to detect GFAP+ cells in vivo or in vitro (Reyes et al., 1998), consistent with observations that Neurog1promotes neurogenesis while inhibiting gliogenesis (Sun, et al. 2001) and detailed fate mapping studies in which Neurog1 was found to be exclusively associated with neuronal lineages (Kim, et al., 2011).
In the PNS, Neurog1 has been shown to be instructive for sensory neurogenesis (Perez et al., 1999; Lo et al., 2002) and required for the generation of proximal cranial sensory ganglia and TrkA neurons of dorsal root ganglia (Ma et al., 1997; Ma et al., 2000), whereas Atoh1 is associated with development of adrenergic autonomic ganglia (Guillemot et al., 1993). Correspondingly, we failed to detect tyrosine hydroxylase, a marker of adrenergic neurons in any of our cultures (not shown) but instead found many peripherin+ neurons that invariably co-expressed Brn3a (Fig. 5E and F), a phenotype representative of both cranial and dorsal root sensory ganglia (Gowan et al., 2001). A possible caveat to this interpretation is that Brn3a (Nadal-Nicolas et al., 2009) and peripherin (Tajika et al., 2004) are also co-expressed in retinal ganglion cells, but other markers of retinal ganglion cells such as Chx10, Dlx1, Dlx2, and Brn3b (de Melo, et al., 2003) were not significantly up-regulated in our microarray analysis, making this phenotype less likely.
Derivation of neural crest from ES cells typically is a multistep process that requires sequential exposure to multiple growth and differentiation factors (Mizuseki et al., 2003; Aihara et al., 2010; Kawaguchi et al., 2010). Our findings suggest that Neurog1 expression can direct ES cells toward Ret+, Ednrb+ neural crest fates, and induces expression of Sox4 and Sox11 required in early sympathetic ganglia (Potzer et al., 2010) in the absence of exogenous instructive signals. However, our cultures contained a mixture of CNS and PNS types, and it remains to be determined what factors in addition to Neurog1 expression mediate the choice between these lineages.
ES cells expressing Neurog1 can be patterned along the anterior-posterior and dorsal-ventral axes and influenced by Fgf signaling
Expression of Neurog1 prompted early expression of forebrain (Otx2, Hoxa1) as well as midbrain (Engrailed1, not shown) markers, strong induction of Hoxb2 but not of more posterior markers including Hoxc6. However, treatment of these cultures with RA promoted expression of Hoxc6 and down-regulation of anterior markers. These data suggest that these cells possess an intrinsic anterior character that can be posteriorized via RA-mediated signals (Irioka et al., 2005). Since these cultures lack other cell types, these results clearly demonstrate that Neurog1 expressing neuronal precursors possess an anterior character in the absence of confounding ”mesendodermal” signals.
Based on previous gain-of-function studies, we expected that Neurog1 expression in ES cells would produce specific subtypes of neurons independent of any endogenous patterning molecules. However, we were surprised to find that induced cells were quite responsive to dorsal-ventral (DV) patterning molecules (Wilson and Maden, 2005) such that more ventral phenotypes were observed in cultures treated with Shh, and this effect could be augmented by addition of Noggin protein. RA treatment alone also produced ventral phenotypes. In addition, noggin treatment accelerated and BMP treatment suppressed neurogenesis in our cultures. Together, these data indicate that ES cells forced to express Neurog1 progress through a stage where they remain receptive to patterning cues.
Recent evidence indicates that FGFs may have an independent and indispensable role in the induction of neural fate in vivo (Streit et al., 2000; Wilson et al., 2000; Lanner and Rossant, 2010) and in the neuronal differentiation of ES cells (Ying et al., 2003a). Fgf signaling can maintain self-renewal of ESC (Sterneckert et al., 2010; Staviridis et al., 2010), and is required for cell cycle exit and the transition to lineage differentiation (Kunath et al., 2007). Other studies have demonstrated that Fgf signaling is essential for the progression of ES cells to an epiblast state (Stavridis, et al., 2007); preventing de-differentiation to a more primitive ICM-like state (Greber et al., 2010) and inhibiting neural differentiation (Greber et al., 2010; Greber et al., 2011; Jaeger et al., 2011). Our data suggest that expression of Neurog1 in ES cells is sufficient to generate Sox3+ progenitors independent of FGF signaling (Fig. 9B). However, the reduction in the percentage of neurons in the pre-induction and simultaneous induction treatment groups, and striking increase in the number of Oct4+ and Sox3+ cells in the earlier treatment groups, suggests that the initial specification of Sox3+ precursors in the context of Neurog1 expression does not require FGF signaling. However, there is a window where inhibition of Fgf signaling interferes with the ability of these precursors to differentiate into mature neurons. It has been suggested that inhibition of Fgf signaling can revert epiblast stem cells to an Oct4+ ES cell state (Greber et al., 2010). However, we failed to detect up-regulation of epiblast markers such as Fgf5, but we did see markers of neural ectoderm such as Pax 6 (Table 2), suggesting that neuronal differentiation via Neurog1 expression likely bypasses an epiblast state. Nonetheless, the Oct4 pathway appears to be initially intact in these cells and abrogation of Fgf signaling re-engages ES cell programs that inhibit neuronal differentiation.
ES cells neuralized via Neurog1 expression may progress through a Notch-sensitive neuronal precursor stage
Although our immunohistochemical analyses indicated that all of the cells in our cultures express Neurog1 (Fig. 2), not all matured into neurons after 3 or even 5 days in culture, similar to previous observations in embryonal carcinoma (EC) (Farah et al., 2000; Kim et al. 2004) and ES cells (Kanda et al., 2004). Although primitive neural stem cells appear relatively insensitive to Notch signaling, definitive neural stem cells may be maintained in an undifferentiated state via activation of the Notch pathway (Tropepe et al., 2001), like neuronal precursors, whose differentiation into mature neurons can be held in check by the Notch-Delta pathway (reviewed in Louvi and Artavanis-Tsakonas, 2006). We observed that induction of Neurog1 results in transient expression of the Notch ligand, Jag1 (Fig. 10 and Table 2), and of Dll1,3,4 (Table 2), as well as up-regulation of Hes1, Hes5 and Hes6, repressor-type bHLH factors that are downstream targets of the Notch signaling pathway. The Notch target gene Hes5, required for NSC maintenance (Basak and Taylor, 2007; Hatakeyama et al., 2004), was strongly induced by Neurog1. In fact, the up-regulation of these repressor-type bHLHs is more likely the effect of Jag1 expression in neighboring cells than a direct effect of Neurog1 expression. Thus, ES cells expressing Neurog1 may progress through a Notch-sensitive stage in which cells at the edge of rosettes escape inhibition to differentiate into neurons, and then repress neuronal differentiation in neighboring cells.
Gene expression patterns down-stream of Neurog1
Since Neurog1 is expressed early in the neurogenic cascade at the time of lineage commitment/determination (Blader et al., 1997), we employed microarray analysis to identify transcripts expressed down-stream of Neurog1. Many of the genes strongly induced by Neurog1 are expressed in the stratifying neural ectoderm at the time of cell cycle exit (Fu et al., 2009); coupling Neurogenin1 expression with cell cycle exit and neuronal differentiation (Kim et al., 2011). Other candidate transcription factors such as Sox4 and Sox11, which are known to play a role in the downstream effects of proneural bHLH factors, were strongly induced, while genes associated with ESC maintenance, and inhibitors of neuronal differentiation were down-regulated. In addition, genes associated with astrocyte and oligodendrocyte differentiation were not identified or were down-regulated.
Genes associated with neuronal cell migration were strongly induced by Neurogenin1, including the microtubule associated protein doublecortin (Dcx), Cdn2, reelin, CoupTF factors (Nr2f1, Nr2f2; Tripodi et al., 2004), as well as EphA5 previously identified as down-regulated in Neurog2:Neurog1 double mutant mice (Mattar et al., 2004). There was also a striking switch in expression of genes associated with an epithelial morphology including junctional proteins, as well as E-Cadherin (by ESC) to N-Cadherin by differentiating neurons. These data suggest that Neurog1 expression activates a program of neurogenesis that includes cell surface changes, induction of genes involved in cell migrations, transcription factors and cell cycle modulators.
In a screen of Ngnr1 and NeuroD targets in Xenopus, Seo et al (2007) identified similar sets of target genes: 47% of the genes induced by Ngnr1 over-expression in Xenopus were identified in our analysis, while 43% of the genes induced by NeuroD were present in our screen. Somewhat surprisingly, all of the seven core regulatory factors suggested to promote Neurogenin-mediated neurogenesis: Ebf2,3, Hes6, Myt1, Nhlh1, Neurod1, Neurod4, and Runx1t1 (Pang et al., 2011) were significantly increased in our cells. An in silico analysis to identify targets of Neurog2 (dorsal telencephalon) vs Ascl1 (ventral telencephalon) (Gohlke et al., 2008) identified only Dcx, Elavl4, Nrarp, in common with our Neurog1 down-stream targets; and one, Fzd5, was down-regulated significantly in our analysis. Interestingly, constitutive over-expression of NEUROG1 in a cell line derived from human fetal telencephalon identified 588 potential target genes; 27.7% of them were identified in this analysis at 72h. These represent a very different population as telencephalon progenitor cells expressed transcripts associated with glial cells as well as with neurons (Satoh et al., 2010). Similarly, expression of transcripts in the hindbrain neural ectoderm on E11.5 -- when Purkinje progenitors are developing -- in Neurog1 −/− embryos identified 31/117 transcripts also present in our analysis. These data suggest that ultimately, subtype specificity is likely controlled by combinations of bHLH proteins and region-restricted transcription factors (particularly homeodomain proteins), which in combination with non-coding RNAs, may regulate each other or stabilize a neurogenic program.
Other candidates that may act in concert with bHLH factors to restrict lineage progression are chromatin regulators. The ESC faces the unique problem of maintaining both self-renewal and pluripotency, which it may solve with a unique bivalent chromatin structure in which enhancers of lineage specific genes are associated with specific methylation marks that repress lineage gene expression “poised chromatin”, while pluripotency genes maintain an “active” chromatin through successive rounds of division. These modifications are lost with differentiation (Boyer et al., 2006), but many are re-set with somatic cell reprogramming (Creyghton et al., 2010). In fact, it has been suggested that since many lineage specific transcription factors belong to large families that recognize a common binding sequence, e.g., the E box, additional mechanisms, such as chromatin modification, must exist to limit the sites available for transcription factor binding (Conerly et al., 2011). Our microarray analysis identifies several transcripts encoding chromatin modifying factors as possible targets of Neurog1. These factors may function to restrict binding of non-neuronal lineage factors during Neurog1 driven differentiation, and may ultimately suggest methods for direct reprogramming of adult cells to neurons. Additional work will examine direct interactions and use shRNA to probe their roles in the context of Neurog1 expression.
Neurog1 alters cell cycle progression
Although the proneural bHLH genes Atoh1 (Math1), Asc1 (Mash1), Neurog1/2 promote cell cycle exit and expression of neuronal genes, the mechanisms involved and target genes are largely unknown. Recently two large-scale analyses of gene expression downstream of Ascl1 (Castro et al., 2011) and Atoh1 (Machold et al., 2011) identified transcripts involved in cell cycle exit and sequential phases of neural differentiation. While it is not surprising that proliferating granule cell precursors maintained Atoh1 and cell cycle genes, in the current investigation we also identified genes involved in positive regulation of the cell cycle. Of the 95 cycle-related genes expressed by Mash1+ rhombic lip precursors, 17 were also present in Neurog1 over-expressing ES cells, including: Aurka, Birc5, Brca2, Bub1b, Ccnd1, Ccnd3, Cdc20, Cdca2, Cdca5, Cdt1, E2f8, Fbxo5, Gmnn, Hells, Jub, Nek2, Trpt53/p53. Downstream of Ascl1 (Castro et al., 2011) only five were genes were common to Neurog1 cells: Ccnd1, Ccd3, Cdkn1a/p21, Plk1, Gadd45g.
Gadd45g, a strong inhibitor of cell cycle progression (e.g., Mak and Kultz, 2004) was strongly induced by Neurog1 at 24h. Ebf3 was particularly strongly up-regulated by Neurog1 (48, 162, 182-fold at 24h intervals); it has previously been reported to be downstream of NeuroD (Pozzoli et al., 2001). Ebf genes inhibit cell cycle progression at G1/S, and couple cycle exit and neural differentiation (Garcia-Domingues 2003); over-expression decreases proliferation consistent with our observations. Neuron specific activators of the cell cycle progression inhibitor Cdk5 (Cdk5r1/p35 and Cdk5r2/p39), which are responsible for producing the unique neuronal cell cycle, were also increased. Other genes such as E2f8, which is induced at G1/S and acts as a repressor of cycle progression via p53 (Christensen et al., 2005), were down-regulated in Neurogenin1 expressing cells. Cdc20, which has previously been associated with increased neural plasticity (Conway et al., 2007), was also decreased in N7 cells following transgene induction. This work demonstrates that the neurogenic bHLH genes affect genes involved in cell cycle progression, neural differentiation, migration and apoptosis, and may reflect the presence of a population of proliferative progenitors in the Neurog1 expressing cells.
ES cells, like the early epiblast, spend most of their time in S phase and have an attenuated G1 (Burdon et al., 2002; White et al., 2005), possibly to minimize exposure to differentiation factors present in the local microenvironment (Orford and Scadden, 2008). Lengthening of G1, which occurs with differentiation of the epiblast and ES cells and is characteristic of somatic cells, was also observed following expression of Neurogenin1. Whether a cell exits cycle is determined at the G1 checkpoint, and exit from cycle is required for many cell fate decisions, consistent with the presence of a population of proliferative precursors in these cultures.
The ability to induce expression of Neurog1 in ES cells constitutes a straightforward and powerful model system that provides insight into the complex interplay of signaling molecules and transcription factors that shape the 2-cell embryo into the thousands of mature cell types present in the adult PNS and CNS, and increases our understanding and eventual control of normal development, birth defects and tumor formation. Ultimately, elucidation of the molecular histogenesis of the nervous system will also improve our understanding of the factors that promote ectopic neurogenesis in a number of neurodegenerative conditions (Curtis et al., 2003). In addition, primitive precursors such as those that can be generated by the N7 cell line may be useful for cell transplantation or as a resource to identify down-stream targets involved in activation of a program of neurogenesis.
EXPERIMENTAL PROCEDURES
Targeting vector construction and generation of inducible Neurog1 cell lines
A Neurog1 insert from pCS2-ND3 (M. McCormick) was ligated with an EcoRI-NotI fragment from pI2R (Clontech, Palo Alto, CA) containing an IRES-DsRed2 cassette into the pLox targeting vector to generate the targeting construct, pLox-N1-I2R. To generate targeted cell lines, 1.0 × 106 Ainv15 ES cells (Kyba et al., 2002) were plated in a 60mm dish and co-transfected 24 hours later with 1 µg each of pLox-N1-I2R and pSalk-Cre (M. Kyba) using Lipofectamine/Plus Reagent (Invitrogen) in serum-free DMEM. After 3 hours, the transfection medium was replaced with complete ES medium. The following day, the cells were split to a 150 mm dish in complete ES medium (see below) containing 350 µg/ml G418 (Invitrogen). After 10 days in selection 12 colonies remained, from which we were able to establish 7 independent cell lines. Three of these lines were selected for additional analysis to confirm targeted integration of the Neurog1 transgene at the tet-inducible locus, stable expression of rtTA, doxycycline-inducible expression of the Neurog1, and uniform neuronal differentiation after 1 and 3 days of culture in defined neural medium (described below) with1 µg/ml doxycycline (Supplemental Fig. 1). We observed no differences between the cell lines and we therefore selected one of the lines, N7, to carry out subsequent experiments.
Cell culture and neural differentiation
Ainv15 ES cells were cultured on 0.1% gelatin coated substrates in complete ES medium (ESM) consisting of high-glucose DMEM (Invitrogen, Carlsbad, CA) supplemented with 10% ES-tested FBS (Atlanta Biologicals, Norcross, GA or Harlan Bioproducts, Indianapolis, IN), 10−4 M β-mercaptoethanol (Sigma, St. Louis, MO), 0.224 µg/ml L-glutamine (Invitrogen), 1.33 µg/ml HEPES (Invitrogen), and 1000 units/ml human recombinant LIF (Oncogene Research Products, San Diego, CA). N7 cells were maintained in ESM plus 350 µg/ml G418 (Invitrogen) and 1.5 µg/ml Puromycin (Sigma). For neural differentiation, cells were plated at 5.0 104 cells/cm2 in gelatin-coated 6- or 12-well plates in ESM or in defined medium supplemented with 1 µg/ml doxycycline (Sigma), and media changed every day. Defined neural medium consisted of 80% F-12/DMEM and 20% Neurobasal media with 10 µl/ml MEM pyruvate, 8 µl/ml N2 supplement, and 4 µl/ml RA-free B27 supplement (all media components from Invitrogen). Where indicated, media were supplemented with 1 µM all-trans-retinoic acid (Sigma). For long-term culture, defined medium was replaced after 3 days with doxycycline-free serum replacement medium (SRM5) consisting of high glucose DMEM supplemented with 5% knockout serum replacer (Invitrogen), 10−4 M β-mercaptoethanol, 0.224 µg/ml L-glutamine, and 1.33 µg/ml HEPES to enhance cell survival. Where indicated, recombinant growth factors (all from R&D Systems, Minneapolis, MN) were added to freshly prepared media at the following concentrations: 25 nM (high) or 2.5 nM (low) mouse Shh; 5 nM human BMP4; and 5 nM (high) or 2 nM (low) Noggin-Fc chimera. To examine the role of FGF signaling, cultures were exposed at intervals to 5 nM SU5402 (Calbiochem) for 12 hours to block FGFR1 signaling.
Proliferation and cell cycle analysis
To analyze the effects of transgene induction on proliferation, Ainv15 and N7 ES cells were plated at 1 × 105 cells/well in 12 well plates in triplicate (with four biological replicates) in complete ES medium ± 2 µg/ml doxycycline. After 24, 48 and 72 hours, cells were disaggregated using trypsin and counted in a Coulter counter (Becton Dickinson).
For cell cycle analysis, cells were plated in triplicate at 2 × 105 in 6 well plates in complete ES medium ± 2 µg/ml doxycycline. After 72h, cells were fixed in cold absolute ethanol, suspended in PBS and DNAse free RNase A (Sigma) at a final concentration of 20 µg/ml. Samples were incubated at 37° for 30 minutes and DNA labeled with 0.1 mg/ml propidium iodide (Sigma). Analysis was done in a FACS Calibur using Cell Quest Pro MAC 9.0 and cell cycle analysis done with Mod Fit LT Mac 3.1 SP3.
RT-PCR
RNAs were extracted from lysates of cells treated with TRIzol reagent (Invitrogen), DNAsed, and quantified by spectrophotometry. Control RNA samples were obtained from N7 cells cultured in ES medium and from pooled samples of gestation day 9, 10, and 11 mouse embryos. RNAs (1 µg each) served as templates in reverse transcription reactions with oligo-dT primers, and 1/20 of the single-strand cDNA products were used in each PCR amplification. General PCR conditions were 94°C/3 min, 94°C/30 s, 53° – 63°C/1 min, 72°C/1 min for 25–40 cycles. Specific information regarding PCR primer sequences and reaction conditions is provided in the on-line supplementary material. The PCR products (10 µl each) were electrophoresed in 1.5% agarose gels in the presence of ethidium bromide, visualized on a UV transilluminator and digitally photographed using a BioRad Gel Documentation system. Primers and conditions are summarized in Supplemental Table 3.
Immunohistochemistry
To examine cell type-specific and positional markers, cells were fixed in 2% paraformaldehyde for 15 min at 25° C, then stored in PBS at 4° prior to processing for immunohistochemistry (IHC). Briefly, cells were permeabilized and non-specific antibody binding blocked using host serum prior to incubation with primary antibodies at 4° overnight. Cells were washed, then exposed to secondary antibody conjugated to Cy3 or FITC (Jackson ImmunoResearch Laboratories) for 30 min at 25° C. Nuclear staining for total cell number counts was achieved by incubating cells with 1 µM Hoechst 33285 (Sigma) for 2 mins. Cells were washed, then mounted using Prolong Gold (Molecular Probes/Invitrogen) and 25 mm coverslips. Cells were examined, counted and photomicrographs imported into Photoshop using a Leica DMIRB inverted microscope and Olympus digital camera.
Primary antibodies included: Sox3 (Mike Klymkowsky; 1:1000) expressed by in neural ectoderm and neuronal precursors (Zhang et al., 2003), Tuj1 (Covance MMS-435P; 1:250) which recognizes a neuron-specific class III β-tubulin expressed very early in neuronal differentiation and persisting in mature neurons (Moody et al., 1987), peripherin (7C5, AbCam ab4573, 1:1000) which is expressed in the PNS, Brn3a (Eric Turner, 1:1000) expressed in sensory neurons, tyrosine hydroxylase (Chemicon AB5968, 1:1000) expressed in adrenergic neurons, Oct4 (Santa Cruz sc-8628, 1:100) expressed in undifferentiated ES cells. Dorsal-ventral markers included: Pax7 (Developmental Studies Hybridoma Bank supernatant, 1:20) labeling dorsal neural progenitors, Nkx6.1 (N15, Santa Cruz sc-15027, 1:100) expressed in ventral progenitors, and Islet 1 which recognizes Islet 1 and Islet 2 (Developmental Studies Hybridoma Bank, 39.4D5, 1:100) labeling ventral motor neurons and dorsal root (sensory) ganglia, or Dbx2 expressed in the middle zone (mid) of the neural tube (Agalliu and Schieren, 2009).
For quantitative analysis of the cell types present, cell counts were performed on 10 microscopic fields at 20× from at least two replicate cultures, counting at least 1000 cells per replicate. For each field, the number of a given cell type was divided by the total number of cells present as determined by Hoechst nuclear staining to calculate a percentage for each field which was then summed to obtain a mean percentage for each replicate. The data presented are the overall mean ± sem across all replicates. Data were analyzed using ANOVA, Students t, or Chi-square test where appropriate (with Bonferroni correction as necessary for multiple comparisons); a p value of ≤ 0.05 was considered significant.
Microarray Analysis
RNAs were extracted as described above from triplicate samples (cells differentiated in three independent experiments) of control N7 cells (−Dox) at 24h culture intervals and N7 cells exposed to doxycycline for 24, 48 and 72h. RNA was hybridized to Affymetrix Mouse 430 2.0 arrays (Santa Clara, CA, http://www.affymetrix.com) by the NIH NINDS arraying core at TGen (http://www.tgen.org). The signal intensity of each array was normalized to a mean of 150 by robust multiarray averaging (RMA), then GeneSpring (Agilent) software was employed to select genes with ≥ 2 fold change between groups. At each time point, N7 cells (−Dox) were employed as the reference for comparison with +Dox samples. Welch t-tests were then carried out to identify significant differences between controls and Neurogenin1 expressing cells. Gene expression was also compared in +Dox (and −Dox control) samples at sequential timepoints (e.g., 24 vs 48h, 48 vs 72h, and 24 vs 72 hours of transgene induction). Benjamini-Hochbert false discovery corrections were performed on each comparison to reduce the number of false positive results. Finally, Gene Ontology, Functional Annotation Clustering, and KEGG pathway analyses were carried out using DAVID V 6.7 (Huang et al., 2009).
Supplementary Material
Functional annotation clustering analysis at 24, 48 and 72h of transgene induction identifies clusters of genes that are mapped to Gene Ontology classifications (GO terms).
KEGG pathway maps at of genes up- and down-regulated at sequential stages of differentiation. n = number of genes, p = significance with Benjamini-Hochberg correction for multiple comparisons.
Forward and reverse primer pairs used in PCR, expected product size (Product), AT = annealing temperature, cycles = RT-PCR cycles employed, Denaturant ± DMSO.
Key Findings.
Inducible expression of Neurogenin1 in ESC provides a novel model of neurogenesis.
Pulsed expression of Neurogenin1 produces PNS and CNS neuronal sub-types.
Neurog1 promotes differentiation of neuronal precursors that exclusively form neurons.
This is the first description of the gene expression cascade downstream of Neurog1
ACKNOWLEDGEMENTS
This research was supported by NIH-NS 39438, JMV was supported by DE-07057. The authors are grateful to Drs. Sean Morrison, Ben Novitch, Pamela Raymond and David Turner for advice with the work, to Dr. Mary McCormick for the Neurog1 cDNA, to Dr. Mike Klymkowski for the anti-Sox3 antibody, and to Dr. Michael Kyba for constructs, Ainv15 cells and advice. The authors acknowledge the use of DAVID V. 6.7, the University of Michigan Comprehensive Cancer Center FACS Core facility, and antibodies from the Developmental Biology Hybridoma Bank.
Footnotes
APPENDIX A. SUPPLEMENTAL DATA
Supplemental Tables 1,2,3,4.
REFERENCES
- Agalliu D, Schieren I. Heterogeneity in the developmental potential of motor neuron progenitors revealed by clonal analysis of single cells in vitro. Neural Dev. 2009;4:2. doi: 10.1186/1749-8104-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aihara Y, Hayashi Y, Hirata M, Shibata S, Nagoshi N, Nakanishi M, Ohnuma K, Warashina M, Michiue T, Uchiyama H, Okano H, Asahima M, Furue MK. Induction of neural crest cells from mouse embryonic stem cells in a serum-free monolayer culture. Int J Dev Biol. 2010;54:1287–1294. doi: 10.1387/ijdb.103173ya. [DOI] [PubMed] [Google Scholar]
- Alder O, Laviall F, Helness A, Brookes E, Pinho S, Chandrashekran A, Arnaud P, Pombo A, O’Neill L, Azuara V. Ring1B and Suv39h1 delineage distinct chromatin states at bivalent genes during early mouse lineage commitment. Development. 2010;137:2483–2492. doi: 10.1242/dev.048363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basak O, Taylor V. Identification of self-replicating multipotent progenitors in the embryonic nervous system by high Notch activity and Hes5 expression. Eur J Neurosci. 2007;25:1006–1022. doi: 10.1111/j.1460-9568.2007.05370.x. [DOI] [PubMed] [Google Scholar]
- Bergsland M, Werme M, Malewicz M, Perlmann T, Muhr J. The establishment of neuronal properties is controlled by Sox4 and Sox 11. Genes Dev. 2006;20:3475–3486. doi: 10.1101/gad.403406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand N, Castro DS, Guillemot F. Proneural genes and the specification of neural cell types. Nat Rev Neurosci. 2002;3:517–530. doi: 10.1038/nrn874. [DOI] [PubMed] [Google Scholar]
- Blader P, Fischer N, Gradwohl G, Guillemot F, Strahle U. The activity of neurogenin1 is controlled by local cues in the zebrafish embryo. Develop. 1997;124:4557–4569. doi: 10.1242/dev.124.22.4557. [DOI] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlingr J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, Bell GW, Otte AP, Vidal M, Gifford DK, Young RA, Jaenisch R. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- Briscoe J, Pierani A, Jessell TM, Ericson J. A homeodomain protein code specifies progenitor cell identity and neuronal fate in the ventral neural tube. Cell. 2000;101:325–345. doi: 10.1016/s0092-8674(00)80853-3. [DOI] [PubMed] [Google Scholar]
- Burdon T, Smith A, Savatier P. Signalling, cell cycle and pluripotency in embryonic stem cells. Trends Cell Biol. 2002;12:432–438. doi: 10.1016/s0962-8924(02)02352-8. [DOI] [PubMed] [Google Scholar]
- Castro DS, Martynoga B, Parras C, Ramesh V, Pacary E, Johnston C, Drechsel D, Lebel-Patter M, Garcia LG, Hunt C, Dolle D, Bithell A, Ettwiller L, Buckley N, Guillemot F. A novel function of the proneural factor Ascl1 in progenitor proliferation identified by genome-wide characterization of its targets. Genes Dev. 2011;25:930–945. doi: 10.1101/gad.627811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien CT, Hsiao CD, Jan LY, Jan YN. Neuronal type information encoded in the basic-helix-loop-helix domain of proneural genes. Proc Natl Acad Sci U S A. 1996;93:13239–13244. doi: 10.1073/pnas.93.23.13239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou SJ, Perez-Garcia CG, Kroll TT, O’Leary DD. Lhx2 specifies regional fate in Emx1 lineage of telencephalic progenitors generating cerebral cortex. Nat. Neurosci. 2009;12:1381–1389. doi: 10.1038/nn.2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen J, Cloos P, Toftegaard U, Klinkenberg D, Bracken AP, Trinh E, Heeran M, Di Stefano L, Helin K. Characterization of E2F8, a Novel E2F-like cell-cycle regulated repressor of E2F-activated transcription. Nucleic Acids Res. 2005;33:5458–5470. doi: 10.1093/nar/gki855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung S, Sonntag KC, Andersson T, Bjorklund LM, Park JJ, Kim DW, Kang UJ, Isacson O, Kim KS. Genetic engineering of mouse embryonic stem cells by Nurr1 enhances differentiation and maturation into dopaminergic neurons. Eur J Neurosci. 2002;16:1829–1838. doi: 10.1046/j.1460-9568.2002.02255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conerly ML, MacQuarrie KL, Fong AP, Yao Z, Tapscott SJ. Polycomb-mediated repression during terminal differentiation: what don’t you want to be when you grow up? Genes Dev. 2011;25:997–1003. doi: 10.1101/gad.2054311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway AM, James AB, Zang J, Morris BJ. Regulation of neuronal cdc20 (p55cdc) expression by the plasticity-related transcription factor zif268. Synapse. 2007;61:463–468. doi: 10.1002/syn.20387. [DOI] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Weistead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, Boyer LA, Young RA, Jaenisch R. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci USA. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis MA, Penney EB, Pearson AG, van Roon-Mom WM, Butterworth NJ, Dragunow M, Connor B, Faull RL. Increased cell proliferation and neurogenesis in the adult human Huntington's disease brain. Proc Natl Acad Sci U S A. 2003;100:9023–9027. doi: 10.1073/pnas.1532244100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalgard CL, Zhou Q, Lundell TG, Doughty ML. Altered gene expression in the emerging cerebellar primordium of Neurog1−/− mice. Brain Res. 2011;1388:12–21. doi: 10.1016/j.brainres.2011.02.087. [DOI] [PubMed] [Google Scholar]
- Dasen JS, Tice BC, Brenner-Morton S, Jessell TM. A Hox regulatory network establishes motor neuron pool identity and target-muscle connectivity. Cell. 2005;123:477–491. doi: 10.1016/j.cell.2005.09.009. [DOI] [PubMed] [Google Scholar]
- de Melo J, Qiu X, Du G, Cristante L, Eisenstat DD. Dlx1, Dlx2, Pax6, Brn3b, and Chx10 homeobox gene expression defines the retinal ganglion and inner nuclear layers of the developing and adult mouse retina. J Comp Neurol. 2003;461:187–204. doi: 10.1002/cne.10674. [DOI] [PubMed] [Google Scholar]
- Escurat M, Djabali K, Gumpel M, Gros F, Portier M. Differential expression of two neuronal intermediate-filament proteins, peripherin and the low-molecular-mass neurofilament protein (NF-L), during the development of the rat. J Neurosci. 1990;10:764–784. doi: 10.1523/JNEUROSCI.10-03-00764.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farah MH, Olson JM, Sucic HB, Hume RI, Tapscott SJ, Turner DL. Generation of neurons by transient expression of neural bHLH proteins in mammalian cells. Develop. 2000;127:693–702. doi: 10.1242/dev.127.4.693. [DOI] [PubMed] [Google Scholar]
- Fedtsova N, Turner EE. Signals from the ventral midline and isthmus regulate the development of Brn3.0-expressing neurons in the midbrain. Mech Dev. 2001;105:129–144. doi: 10.1016/s0925-4773(01)00399-9. [DOI] [PubMed] [Google Scholar]
- Fode C, Gradwohl G, Morin X, Dierich A, LeMeur M, Goridis C, Guillemot F. The bHLH protein NEUROGENIN 2 is a determination factor for epibranchial placode-derived sensory neurons. Neuron. 1998;20:483–494. doi: 10.1016/s0896-6273(00)80989-7. [DOI] [PubMed] [Google Scholar]
- Fode C, Ma Q, Casarosa S, Ang SL, Anderson DJ, Guillemot F. A role for neural determination genes in specifying the dorsoventral identity of telencephalic neurons. Genes Dev. 2000;14:67–80. [PMC free article] [PubMed] [Google Scholar]
- Frontelo P, Leader JE, Yoo N, Potocki AC, Crawford M, Kulik M, Lechleider RJ. Suv39h histone methyltransferases interact with Smads and cooperate in BMP-induced repression. Oncogene. 2004;23:5242–5251. doi: 10.1038/sj.onc.1207660. [DOI] [PubMed] [Google Scholar]
- Fu H, Cai J, Clevers H, Fast E, Gray S, Greenberg R, Jain MK, Ma Q, Qiu M, Rowitch DH, Taylor CM, Stiles CD. A genome-wide screen for spatially restricted expression patterns identifies transcription factors that regulate glial development. J Neurosci. 2009;29:11399–11408. doi: 10.1523/JNEUROSCI.0160-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Dominguez M, Poquet C, Garel S, Charnay P. Ebf gene function is required for coupling neuronal differentiation and cell cycle exit. Develop. 2003;130:6013–6025. doi: 10.1242/dev.00840. [DOI] [PubMed] [Google Scholar]
- Gohlke JM, Armant O, Parham FM, Smith MV, Zimmer C, Castro DS, Nguyen L, Parker JS, Gradwohl G, Portier CJ, Guillemot F. Characterization of the proneural gene regulatory network during mouse telencerphalon development. BMC Biol. 2008;6:15. doi: 10.1186/1741-7007-6-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowan K, Helms AW, Hunsaker TL, Collisson T, Ebert PJ, Odom R, Johnson JE. Cross-inhibitory activities of Neurog1 and Math1 allow specification of distinct dorsal interneurons. Neuron. 2001;31:219–232. doi: 10.1016/s0896-6273(01)00367-1. [DOI] [PubMed] [Google Scholar]
- Gradwohl G, Fode C, Guillemot F. Restricted expression of a novel murine atonal-related bHLH protein in undifferentiated neural precursors. Dev Biol. 1996;180:227–241. doi: 10.1006/dbio.1996.0297. [DOI] [PubMed] [Google Scholar]
- Greber B, Wu G, Bernemann C, Joo JY, Han DW, Ko K, Tapia N, Sabour D, Sterneckert J, Tesar P, Schöler HR. Conserved and divergent roles of FGF signaling in mouse epiblast stem cells and human embryonic stem cells. Cell Stem Cell. 2010;6:215–226. doi: 10.1016/j.stem.2010.01.003. [DOI] [PubMed] [Google Scholar]
- Greber B, Coulon P, Zhang M, Moritz S, Frank S, Müller-Molina AJ, Araúzo-Bravo MJ, Han DW, Pape HC, Schöler HR. FGF signalling inhibits neural induction in human embryonic stem cells. EMBO J. 2011;30:4874–4884. doi: 10.1038/emboj.2011.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillemot F, Lo LC, Johnson JE, Auerbach A, Anderson DJ, Joyner AL. Mammalian achaete-scute homolog 1 is required for the early development of olfactory and autonomic neurons. Cell. 1993;75:463–476. doi: 10.1016/0092-8674(93)90381-y. [DOI] [PubMed] [Google Scholar]
- Hatakeyama J, Bessho Y, Katoh K, Ookawara S, Fujioka M, Guillemot F, Kageyama R. Hes genes regulate size, shape and histogenesis of the nervous system by control of the timing of neural stem cell differentiation. Develop. 2004;131:5539–5550. doi: 10.1242/dev.01436. [DOI] [PubMed] [Google Scholar]
- Helms AW, Battiste J, Henke RM, Nakada Y, Simplicio N, Guillemot F, Johnson JE. Sequential roles for Mash1 and Ngn2 in the generation of dorsal spinal cord interneurons. Develop. 2005;132:2709–2719. doi: 10.1242/dev.01859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helms AW, Johnson JE. Specification of dorsal spinal cord interneurons. Cur Opin Neurobiol. 2003;13:42–49. doi: 10.1016/s0959-4388(03)00010-2. [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Proc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Irioka T, Watanabe K, Mizusawa H, Mizuseki K, Sasai Y. Distinct effects of caudalizing factors on regional specification of embryonic stem cell-derived neural precursors. Brain Res Dev Brain Res. 2005;154:63–70. doi: 10.1016/j.devbrainres.2004.10.004. [DOI] [PubMed] [Google Scholar]
- Jaeger I, Arber C, Risner-Janiczek JR, Kuechler J, Pritzsche D, Chen IC, Naveenan T, Ungless MA, Li M. Temporally controlled modulation of FGF/ERK signaling directs midbrain dopaminergic neural progenitor fate in mouse and human pluripotent stem cells. Development. 2011;138:4363–4374. doi: 10.1242/dev.066746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarman AP, Ahmed I. The specificity of proneural genes in determining Drosophila sense organ identity. Mech Dev. 1998;76:117–125. doi: 10.1016/s0925-4773(98)00116-6. [DOI] [PubMed] [Google Scholar]
- Jessell TM. Neuronal specification in the spinal cord: inductive signals and transcriptional codes. Nat Rev Genet. 2000;1:20–29. doi: 10.1038/35049541. [DOI] [PubMed] [Google Scholar]
- Johnson JE, Birren SJ, Anderson DJ. Two rat homologues of Drosophila achaete-scute specifically expressed in neuronal precursors. Nature. 1990;346:858–861. doi: 10.1038/346858a0. [DOI] [PubMed] [Google Scholar]
- Juan AH, Derfoul A, Feng X, Ryall JG, Dell’Orso S, Pasut A, Zare H, Simone JM, Rudnicki MA, Sartorelli V. Polycomb EZH2 controls self-renewal and safeguards the transcriptional identity of skeletal muscle stem cells. Genes Dev. 2011;25:789–794. doi: 10.1101/gad.2027911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanda S, Tamada Y, Yoshidome A, Hayashi I, Nishiyama T. Over-expression of bHLH genes facilitate neural formation of mouse embryonic stem (ES) cells in vitro. Int J Dev Neurosci. 2004;22:149–156. doi: 10.1016/j.ijdevneu.2004.01.002. [DOI] [PubMed] [Google Scholar]
- Kawaguchi J, Nichols J, Gierl MS, Faisal T, Smith A. Isolation and propagation of enteric neural crest progenitor cells from mouse embryonic stem cells and embryos. Develop. 2010;137:693–704. doi: 10.1242/dev.046896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MH, Gunnersen JM, Tan SS. Mph2 expression in germinal zones of the mouse brain. Dev Dyn. 2005;232:209–215. doi: 10.1002/dvdy.20213. [DOI] [PubMed] [Google Scholar]
- Kim EJ, Hori K, Wyckoff A, Dickel LK, Koundakjian EJ, Goodrich LV, Johnson JE. Spatiotemporal fate map of Neurogenin1 (Neurog1) lineages in the mouse central nervous system. J Comp Neurol. 2011;519:1355–1370. doi: 10.1002/cne.22574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Yoon YS, Kim JW, Jung M, Kim SU, Lee YD, Suh-Kim H. Neurogenin1 is sufficient to induce neuronal differentiation of embryonal carcinoma P19 in the absence of retinoic acid. Cell Mol Neurosci. 2004;24:343–356. doi: 10.1023/B:CEMN.0000022767.74774.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunath T, Saba-El-Leil MK, Almousailleakh M, Wray J, Meloche S, Smith A. FGF stimulation of the Erk 1/2 signalling cascade triggers transition of pluripotent embryonic stem cells from self-renewal to lineage commitment. Develop. 2007;137:2895–2902. doi: 10.1242/dev.02880. [DOI] [PubMed] [Google Scholar]
- Kyba M, Perlingeiro RC, Daley GQ. HoxB4 confers definitive lymphoid-myeloid engraftment potential on embryonic stem cell and yolk sac hematopoietic progenitors. Cell. 2002;109:29–37. doi: 10.1016/s0092-8674(02)00680-3. [DOI] [PubMed] [Google Scholar]
- Lalmansingh AS, Karmakar S, Jin Y, Nagaich AK. Multiple modes of chromatin remodeling by Forkhead box proteins. Biochim Biophys Acta. 2012;1819:707–715. doi: 10.1016/j.bbagrm.2012.02.018. [DOI] [PubMed] [Google Scholar]
- Lanner F, Rossant J. The role of FGF/Erk signaling in pluripotent cells. Develop. 2010;137:3351–3360. doi: 10.1242/dev.050146. [DOI] [PubMed] [Google Scholar]
- Lee JE, Hollenberg SM, Snider L, Turner DL, Lipnick N, Weintraub H. Conversion of Xenopus ectoderm into neurons by NeuroD, a basic helix-loop-helix protein. Science. 1995;268:836–844. doi: 10.1126/science.7754368. [DOI] [PubMed] [Google Scholar]
- Liu A, Li J, Marih-Husstege M, Kageyama R, Fan Y, Gelinas C, Casaccia-Bonnefil P. A molecular insight of Hes5-dependent inhibition of myelin gene expression: old partners and new players. EMBO J. 2006;25:4833–4842. doi: 10.1038/sj.emboj.7601352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo L, Tiveron MC, Anderson DJ. MASH1 activates expression of the paired homeodomain transcription factor Phox2a, and couples pan-neuronal and subtype-specific components of autonomic neuronal identity. Develop. 1998;125:609–620. doi: 10.1242/dev.125.4.609. [DOI] [PubMed] [Google Scholar]
- Lo L, Dormand E, Greenwood A, Anderson DJ. Comparison of the generic neuronal differentiation and neuron subtype specification functions of mammalian achaete-scute and atonal homologs in cultured neural progenitor cells. Develop. 2002;129:1553–1567. doi: 10.1242/dev.129.7.1553. [DOI] [PubMed] [Google Scholar]
- Louvi A, Artavanis-Tsakonas S. Notch signalling in vertebrate neural development. Nat Rev Neurosci. 2006;7:93–102. doi: 10.1038/nrn1847. [DOI] [PubMed] [Google Scholar]
- Ma Q, Anderson DJ, Fritzsch B. Neurogenin 1 null mutant ears develop fewer, morphologically normal hair cells in smaller sensory epithelia devoid of innervation. J Assoc Res Otolaryngol. 2000;1:129–143. doi: 10.1007/s101620010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q, Kintner C, Anderson DJ. Identification of neurogenin, a vertebrate neuronal determination gene. Cell. 1996;87:43–52. doi: 10.1016/s0092-8674(00)81321-5. [DOI] [PubMed] [Google Scholar]
- Ma Q, Sommer L, Cserjesi P, Anderson DJ. Mash1 and neurogenin1 expression patterns define complementary domains of neuroepithelium in the developing CNS and are correlated with regions expressing notch ligands. J Neurosci. 1997;17:3644–3652. doi: 10.1523/JNEUROSCI.17-10-03644.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machold R, Klein C, Fishell G. Genes expressed in Atoh1 neuronal lineages arising from the r1/isthmus rhombic lip. Gene Expr Patterns. 2011;11:349–359. doi: 10.1016/j.gep.2011.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magueron R, Li G, Sarma K, Blais A, Zavadil J, Woodcock CL, Dynlacht BD, Reinberg D. Ezh1 and Ezh2 maintain repressive chromatin through different mechanisms. Mol Cell. 2008;32:503–518. doi: 10.1016/j.molcel.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak SK, Kultz D. Gadd proteins induce G2/M arrest and modulate apoptosis in kidney cells exposed to hyperosmotic stress. J Biol Chem. 2004;297:39075–39084. doi: 10.1074/jbc.M406643200. [DOI] [PubMed] [Google Scholar]
- Mallamaci A, Di Blas E, Briata P, Boncinelli E, Corte G. OTX2 homeoprotein in the developing central nervous system and migratory cells of the olfactory area. Mech Dev. 1996;58:165–178. doi: 10.1016/s0925-4773(96)00571-0. [DOI] [PubMed] [Google Scholar]
- Mangale VS, Hirokawa KE, Stayaki PRV, Gokulchandran N, Chikbire S, Subramanian L, Shetty AS, Martynoga B, Paul J, Mail MV, Li Y, Flanagan LA, Tole SH, Monuki ES. Lhx2 selector activity specifies cortical identity and suppresses hippocampal organizer fate. Science. 2008;319:304–309. doi: 10.1126/science.1151695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin-Husstege M, He Y, Kondo T, Sabilitzky F, Casaccia-Bonnefil P. Multiple roles of Id4 in developmental myelination: predicted outcomes and unexpected findings. Glia. 2006;54:285–296. doi: 10.1002/glia.20385. [DOI] [PubMed] [Google Scholar]
- Mattar P, Britz O, Johannes C, Nieto M, Ma L, Rebeyka A, Klenin N, Polleux F, Guillemot F, Schuumans C. A screen for downstream effectors of neurogenin2 in the embryonic neocortex. Dev Biol. 2004;273:373–389. doi: 10.1016/j.ydbio.2004.06.013. [DOI] [PubMed] [Google Scholar]
- Mizuseki K, Sakamoto T, Watanabe K, Muguruma K, Ikeya M, Nishiyama A, Arakawa A, Suemori H, Nakatsuji N, Kawasaki H, Murakami F, Sasai Y. Generation of neural crest-derived peripheral neurons and floor plate cells from mouse and primate embryonic stem cells. Proc Natl Acad Sci USA. 2003;100:5828–5833. doi: 10.1073/pnas.1037282100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammadi M, McMahon G, Sun L, Tang C, Hirth P, Yeh BK, Hubbard SR, Schlessinger J. Structures of the tyrosine kinase domain of fibroblast growth factor receptor in complex with inhibitors. Science. 1997;276:955–960. doi: 10.1126/science.276.5314.955. [DOI] [PubMed] [Google Scholar]
- Moody SA, Quigg MS, Frankfurter A. Development of the peripheral trigeminal system in the chick revealed by an isotype specific anti-beta-tubulin monoclonal antibody. J Comp Neurol. 1987;279:567–580. doi: 10.1002/cne.902790406. [DOI] [PubMed] [Google Scholar]
- Murray RC, Tapscott SJ, Petersen JW, Calof AL, McCormick MB. A fragment of the Neurogenin1 gene confers regulated expression of a reporter gene in vivo and in vitro. Dev Dyn. 2000;218:189–94. doi: 10.1002/(SICI)1097-0177(200005)218:1<189::AID-DVDY16>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Nadal-Nicolás FM, Jiménez-López M, Sobrado-Calvo P, Nieto-López L, Cánovas-Martínez I, Salinas-Navarro M, Vidal-Sanz M, Agudo M. Brn3a as a marker of retinal ganglion cells: qualitative and quantitative time course studies in naive and optic nerve-injured retinas. Invest. Ophthalmol. Vis Sci. 2009;50:3860–3868. doi: 10.1167/iovs.08-3267. [DOI] [PubMed] [Google Scholar]
- Naka H, Nakamura S, Shimazaki T, Okano H. Requirement for COUP-TFI and II in the temporal specification of neural stem cells in CNS development. Nat Neurosci. 2008;11:10114–10123. doi: 10.1038/nn.2168. [DOI] [PubMed] [Google Scholar]
- Nakada Y, Parab P, Simmons A, Omer-Abdalla A, Johnson JE. Separable enhancer sequences regulate the expression of the neural bHLH transcription factor neurogenin1. Dev Biol. 2004;271:479–487. doi: 10.1016/j.ydbio.2004.04.021. [DOI] [PubMed] [Google Scholar]
- Napoli JL. Quantification of physiological levels of retinoic acid. Methods Enzymol. 1986;123:112–124. doi: 10.1016/s0076-6879(86)23015-3. [DOI] [PubMed] [Google Scholar]
- Orford KW, Scadden DT. Deconstructing stem cell self-renewal: genetic insights into cell-cycle regulation. Nat Rev Genet. 2008;9:115–128. doi: 10.1038/nrg2269. [DOI] [PubMed] [Google Scholar]
- Pang ZP, Yang N, Vierbuchen T, Ostermeier A, Fuentes DR, Yang TQ, Cigtri A, Sebastiano V, Marro S, Sudhof TC, Wernig M. Induction of human neuronal cells by defined transcription factors. Nature. 2011;476:220–223. doi: 10.1038/nature10202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parras CM, Schuurmans C, Scardigli R, Kim J, Anderson DJ, Guillemot F. Divergent functions of the proneural genes Mash1 and Ngn2 in the specification of neuronal subtype identity. Genes Dev. 2002;16:324–338. doi: 10.1101/gad.940902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez SE, Rebelo S, Anderson DJ. Early specification of sensory neuron fate revealed by expression and function of neurogenins in the chick embryo. Develop. 1999;126:1715–1728. doi: 10.1242/dev.126.8.1715. [DOI] [PubMed] [Google Scholar]
- Piao C-S, Li B, Zhang L-J, Zhao L-R. Stem cell factor and granulocyte colony-stimulating factor promote neuronal lineage commitment of neural stem cells. Differentiation. 2012;83:17–25. doi: 10.1016/j.diff.2011.08.006. [DOI] [PubMed] [Google Scholar]
- Potzner MR, Tsarovina K, Binder E, Penzo-Mendez A, Lefebvre V, Rohrer H, Wegner M, Sock E. Sequential requirement of Sox4 and Sox11 during development of the sympathetic nervous system. Develop. 2010;137:775–784. doi: 10.1242/dev.042101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell LM, Jarman AP. Context dependence of proneural bHLH proteins. Curr Opin Genet Dev. 2008;18:411–417. doi: 10.1016/j.gde.2008.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozolli O, Bosetti A, Croci L, Consalez GG, Vetter ML. Xebf3 is a regulator of neuronal differentiation during primary neurogenesis in Xenopus. Dev Biol. 2001;233:495–512. doi: 10.1006/dbio.2001.0230. [DOI] [PubMed] [Google Scholar]
- Quinones HI, Savage TK, Battiste J, Johnson JE. Neurogenin1 (Neurog1) expression in the ventral neural tube is mediated by a distinct enhancer and preferentially marks ventral interneuron lineages. Dev Biol. 2010;30:283–292. doi: 10.1016/j.ydbio.2010.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan XJ, Hassan BA. From skin to nerve: flies, vertebrates and the first helix. Cell Mol Life Sci. 2005;62:2036–2049. doi: 10.1007/s00018-005-5124-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes JH, O’Shea KS, Wys NL, Velkey JM, Prieskorn DM, Wesolowski K, Miller JM, Altschuler RA. Glutamatergic neuronal differentiation of mouse embryonic stem cells after transient expression of neurogenin 1 and treatment with BDNF and GDNF: in vitro and in vivo studies. J Neurosci. 2008;28:12622–12631. doi: 10.1523/JNEUROSCI.0563-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasai Y. Directed differentiation of neural and sensory tissues from embryonic stem cells in vitro. Ernst Schering Res Found Workshop. 2005;54:101–109. doi: 10.1007/3-540-37644-5_7. [DOI] [PubMed] [Google Scholar]
- Satoh J, Obayashi S, Tabunoki H, Wakana T, Kim SU. Stable expression of neurogenin 1 induces LGR5, a novel stem cell marker, in an immortalized human neural stem cell line HB1.F3. Cell Mol Neurobiol. 2010;30:415–426. doi: 10.1007/s10571-009-9466-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo S, Lim J-W, Yellajoshyula D, Chang L-W, Kroll KL. Neurogenin and neuroD direct transcriptional targets and their regulatory enhancers. EMBO J. 2007;26:5093–5108. doi: 10.1038/sj.emboj.7601923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer L, Ma Q, Anderson DJ. Neurogenins, a novel family of atonal-related bHLH transcription factors, are putative mammalian neuronal determination genes that reveal progenitor cell heterogeneity in the developing CNS and PNS. Mol Cell Neurosci. 1996;8:221–241. doi: 10.1006/mcne.1996.0060. [DOI] [PubMed] [Google Scholar]
- Staviridis MP, Collins BJ, Storey KG. Retinoic acid orchestrates fibroblast growth factor signaling to drive embryonic stem cell differentiation. Develop. 2010;137:881–890. doi: 10.1242/dev.043117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streit A, Berliner AJ, Papanayotou C, Sirulnik A, Stern CD. Initiation of neural induction by FGF signaling before gastrulation. Nature. 2000;406:74–78. doi: 10.1038/35017617. [DOI] [PubMed] [Google Scholar]
- Sterneckert J, Stehling M, Bernemann C, Arauzo-Bravo MJ, Greber B, Gentile L, Ortmeier C, Sinn Z, Wu G, Ruau D, Zenke M, Brintrup R, Klein DC, Ko K, Scholer HR. Neural induction intermediates exhibit distinct roles of Fgf signaling. Stem Cells. 2010;28:1772–1781. doi: 10.1002/stem.498. [DOI] [PubMed] [Google Scholar]
- Sun Y, Nadal-Vicens M, Misono S, Lin MZ, Zubiaga A, Hua X, Fan G, Greenberg ME. Neurogenin promotes neurogenesis and inhibits glial differentiation by independent mechanisms. Cell. 2001;104:365–376. doi: 10.1016/s0092-8674(01)00224-0. [DOI] [PubMed] [Google Scholar]
- Tajika M, Yamamoto K, Mekada A, Kani K, Okabe H. Neuronal intermediate filament in the developing rat retina. Acta Histochem Cytochem. 2004;37:95–99. [Google Scholar]
- Takano-Maruyama M, Chen Y, Gaufo GO. Differential contribution of Neurog1 and Neurog2 on the formation of cranial ganglia along the anterior-posterior axis. Dev Dyn. 2012;241:229–241. doi: 10.1002/dvdy.22785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripodi M, Filosa A, Armentano M, Studer M. The Coup-TF nuclear receptors regulate cell migration in the mammalian basal forebrain. Develop. 2004;131:6119–6129. doi: 10.1242/dev.01530. [DOI] [PubMed] [Google Scholar]
- Tropepe V, Hitoshi S, Sirard C, Mak TW, Rossant J, van der Kooy D. Direct neural fate specification from embryonic stem cells: a primitive mammalian neural stem cell stage acquired through a default mechanism. Neuron. 2001;30:65–78. doi: 10.1016/s0896-6273(01)00263-x. [DOI] [PubMed] [Google Scholar]
- Turner DL, Weintraub H. Expression of achaete-scute homolog 3 in Xenopus embryos converts ectodermal cells to a neural fate. Genes Dev. 1994;8:1434–1447. doi: 10.1101/gad.8.12.1434. [DOI] [PubMed] [Google Scholar]
- White J, Stead E, Faast R, Conn S, Cartwright P, Dalton S. Developmental activation of the Rb-E2F pathway and establishment of cell cycle-regulated cyclin-dependent kinase activity during embryonic stem cell differentiation. Mol Biol Cell. 2005;16:2018–2027. doi: 10.1091/mbc.E04-12-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wichterle H, Lieberam I, Porter JA, Jessell TM. Directed differentiation of embryonic stem cells into motor neurons. Cell. 2002;110:385–397. doi: 10.1016/s0092-8674(02)00835-8. [DOI] [PubMed] [Google Scholar]
- Williams RL, Hilton DJ, Pease S, Wilson TA, Stewart CL, Gearing DP, Wagner EF, Metcalf D, Nicola NA, Gough NM. Myeloid leukemia inhibitory factor maintains the developmental potential of embryonic stem cells. Nature. 1988;336:684–687. doi: 10.1038/336684a0. [DOI] [PubMed] [Google Scholar]
- Wilson L, Maden M. The mechanisms of dorsoventral patterning in the vertebrate neural tube. Dev Biol. 2005;282:1–13. doi: 10.1016/j.ydbio.2005.02.027. [DOI] [PubMed] [Google Scholar]
- Wilson SI, Graziano E, Harland R, Jessell TM, Edlund T. An early requirement for FGF signaling in the acquisition of neural cell fate in the chick embryo. Curr Biol. 2000;10:421–429. doi: 10.1016/s0960-9822(00)00431-0. [DOI] [PubMed] [Google Scholar]
- Wutz A, Rasmussen TP, Jaenisch R. Chromosomal silencing and localization are mediated by different domains of Xist RNA. Nat Genet. 2002;30:167–174. doi: 10.1038/ng820. [DOI] [PubMed] [Google Scholar]
- Ying QL, Nichols J, Chambers I, Smith A. BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with Stat3. Cell. 2003a;115:281–292. doi: 10.1016/s0092-8674(03)00847-x. [DOI] [PubMed] [Google Scholar]
- Ying QL, Stavridis M, Griffiths D, Li M, Smith A. Conversion of embryonic stem cells into neuroectodermal precursors in adherent monoculture. Nat Biotechnol. 2003b;21:183–186. doi: 10.1038/nbt780. [DOI] [PubMed] [Google Scholar]
- Zeng WR, Fabb SR, Haynes JM, Pouton CW. Extended periods of neural induction and propagation of embryonic stem cell-derived neural progenitors with EGF and FGF2 enhances Lmx1a expression and neurogenic potential. Neurochem Int. 2011;59:394–403. doi: 10.1016/j.neuint.2011.04.002. [DOI] [PubMed] [Google Scholar]
- Zhang C, Basta T, Jensen ED, Klymkowsky MW. The beta-catenin/VegT-regulated early zygotic gene Xnr5 is a direct target of SOX3 regulation. Develop. 2003;130:5609–5624. doi: 10.1242/dev.00798. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Functional annotation clustering analysis at 24, 48 and 72h of transgene induction identifies clusters of genes that are mapped to Gene Ontology classifications (GO terms).
KEGG pathway maps at of genes up- and down-regulated at sequential stages of differentiation. n = number of genes, p = significance with Benjamini-Hochberg correction for multiple comparisons.
Forward and reverse primer pairs used in PCR, expected product size (Product), AT = annealing temperature, cycles = RT-PCR cycles employed, Denaturant ± DMSO.