Background: Pigment epithelium-derived factor (PEDF) interacts with its receptor PEDF-R to exert cytoprotection.
Results: Alanine scanning of a small fragment (17-mer) of PEDF reveals key interacting residues for binding PEDF-R and alternative retinoprotective peptide versions with higher efficacy.
Conclusion: The 17-mer contains a novel PEDF-R binding region important for retinoprotection.
Significance: Altered PEDF peptides could be exploited pharmacologically to improve protection of photoreceptors from degeneration.
Keywords: cell death, cell surface receptor, molecular modeling, mouse, neuroprotection, peptide interaction, photoreceptor, protein structure, retina, serpin
Abstract
The cytoprotective effects of pigment epithelium-derived factor (PEDF) require interactions between an as of a yet undefined region with a distinct ectodomain on the PEDF receptor (PEDF-R). Here we characterized the area in PEDF that interacts with PEDF-R to promote photoreceptor survival. Molecular docking studies suggested that the ligand binding site of PEDF-R interacts with the neurotrophic region of PEDF (44-mer, positions 78–121). Binding assays demonstrated that PEDF-R bound the 44-mer peptide. Moreover, peptide P1 from the PEDF-R ectodomain had affinity for the 44-mer and a shorter fragment within it, 17-mer (positions 98–114). Single residue substitutions to alanine along the 17-mer sequence were designed and tested for binding and biological activity. Altered 17-mer[R99A] did not bind to the P1 peptide, whereas 17-mer[H105A] had higher affinity than the unmodified 17-mer. Peptides 17-mer, 17-mer[H105A], and 44-mer exhibited cytoprotective effects in cultured retina R28 cells. Intravitreal injections of these peptides and PEDF in the rd1 mouse model of retinal degeneration decreased the numbers of dying photoreceptors, 17-mer[H105A] being most effective. The blocking peptide P1 hindered their protective effects both in retina cells and in vivo. Thus, in addition to demonstrating that the region composed of positions 98–114 of PEDF contains critical residues for PEDF-R interaction that mediates survival effects, the findings reveal distinct small PEDF fragments with neurotrophic effects on photoreceptors.
Introduction
Pathological photoreceptor demise in the eye can perturb the structure and function of the retina and lead to visual loss, as is seen in the retina of patients with retinitis pigmentosa and age-related macular degeneration. Several lines of evidence suggest that vision can be preserved by interfering with photoreceptor cell death (1). Therefore, endogenous neurotrophic factors are likely to play important roles in preventing retinal degeneration. Pigment epithelium-derived factor (PEDF)3 is a naturally occurring neurotrophic factor found in the interphotoreceptor matrix surrounding the photoreceptors. The human retinal pigment epithelium expresses the SERPINF1 gene that encodes the PEDF protein (2, 3). The monolayer of retinal pigment epithelium cells secretes the mature PEDF protein in a directional fashion toward its apical side into the interphotoreceptor matrix (4–6), where it is thought to be responsible for photoreceptor survival and for its avascularity. Several reports have revealed that PEDF levels in eyes of patients and animal models of retinal degeneration are lower than in nondiseased patients or wild type animals, respectively (7–9). Furthermore, administration of PEDF in rodent models of retinitis pigmentosa and age-related macular degeneration results in a delay in photoreceptor degeneration (9–15). PEDF is a potential therapeutic agent for retinal degenerations, and further studies on its mechanism of action are fundamental to develop its therapeutic use.
Although the fundamental mechanisms of action remain unclear, there is accumulated evidence that the modular nature of PEDF helps it to participate in multiple cellular functions (16, 17). As a member of the serine protease inhibitor superfamily of serpins, the PEDF protein shares three-dimensional structural homology with other members. However, it does not have a demonstrable serine protease inhibitory activity and belongs to the subclass of noninhibitory serpins, e.g. ovalbumin and maspin (2, 18). The PEDF protein (50,000-Mr) is a monomeric glycoprotein with a homologous serpin reactive center loop toward its C-terminal end, which is distinct and separated from regions for neurotrophic and antiangiogenic activities and for collagen and glycosaminoglycan binding motifs (2). It is established that PEDF biological effects are independent of its protease inhibitory potential; however, they depend on interactions with receptors on the surface of target cells (16). Structure-function relationship studies have revealed that a region toward the N terminus of the PEDF polypeptide (human amino acid positions 78–121) confers the neurotrophic properties to the molecule. Peptide 44-mer from the neurotrophic region has demonstrable neuronal differentiating activity on retinoblastoma Y-79 cells, as well as survival effects on spinal cord motor neurons, whereas peptide 34-mer from the antiangiogenic region (human position 44–77) does not exhibit neurotrophic activities (19–21).
We have begun to elucidate the molecular mechanisms that govern the cytoprotective effects of PEDF and have identified PEDF-R protein as a receptor for PEDF. This novel protein encoded by PNPLA2 (patatin-like phospholipase domain containing family) is a membrane-linked lipase enzyme, which PEDF binding can stimulate (22). Plasma membranes of retina cells contain PEDF-R, which has affinity for PEDF and is essential for cytoprotective activity of PEDF (23). We have also mapped the ligand-binding domain (LBD) to an extracellular loop of the human PEDF-R (positions 203–232) and shown that peptides derived from this ectodomain act as soluble receptor fragments that block both PEDF/PEDF-R interactions and PEDF-mediating survival action (23). However, the region of PEDF that binds PEDF-R remains to be identified.
Therefore, we hypothesized that the neurotrophic region of PEDF interacts with the LBD of PEDF-R to trigger the necessary downstream molecular cascades leading to cell survival. To test this proposition, we designed experiments to map the region of PEDF that interacts with PEDF-R to exert retinoprotective activity. The biological consequences of these interactions were examined in retina cells in culture and in photoreceptors of a mouse model of retina degeneration in vivo. The data support the pharmacotherapeutic potential of PEDF as a neuroprotectant in retinal degeneration.
Experimental Procedures
Peptides and Protein
Recombinant human PEDF was expressed, purified, and conjugated with fluorescein using fluorescein-5-EX, succinimidyl ester as outlined previously (24). Recombinant PEDF-R with an N-terminal His6/Xpress tag was expressed in the Expressway in vitro transcription/translation system (Life Technologies Corp., Invitrogen), as described previously (22). Human PEDF-R peptide P1 (Thr210–Leu249) and human PEDF peptides 34-mer (Asp44–Asn77) and 44-mer (Val78–Thr121) were synthesized as before (19, 23), and 17-mer (Gln98–Ser115) and the set of 17-mer alanine scan peptides were all chemically synthesized and purified by Biosynthesis Inc. Sequences of peptides are in Table 1. Fluorescently labeled peptides were synthesized with FITC covalently linked to the N terminus via an 6-aminohexylcarbamoyl linker by Biosynthesis Inc. Lyophilized peptides were dissolved in deionized water at 1 mg/ml and stored at −80 °C. The lipase inhibitor atglistatin was purchased from Cayman Chemical.
TABLE 1.
Sequences of peptides used in this study
| Name | Sequence |
|---|---|
| P1 | TSIQFNLRNLYRLSKALFPPEPLVLREMCKQGYRDGLRFL |
| 34-mer | FFVPVNKLAAVSNFGYDLYRVRSSMSPTTN |
| 44-mer | VLLSPLSVATALSALSLGADQRTESIIHRALYYDLISSPDIHGT |
| 17-mer | QRTESIIHRALYYDLIS |
| Q98A | ARTESIIHRALYYDLIS |
| R99A | QATESIIHRALYYDLIS |
| T100A | QRAESIIHRALYYDLIS |
| E101A | QRTASIIHRALYYDLIS |
| S102A | QRTEAIIHRALYYDLIS |
| I103A | QRTESAIHRALYYDLIS |
| I104A | QRTESIAHRALYYDLIS |
| H105A | QRTESIIARALYYDLIS |
| R106A | QRTESIIHAALYYDLIS |
| L108A | QRTESIIHRAAYYDLIS |
| Y109A | QRTESIIHRALAYDLIS |
| Y110A | QRTESIIHRALYADLIS |
| D111A | QRTESIIHRALYYALIS |
| L112A | QRTESIIHRALYYDAIS |
| I113A | QRTESIIHRALYYDLAS |
| S114A | QRTESIIHRALYYDLIA |
We prepared recombinant PEDF with single residue alterations using the full-length human gene SERPINF1 for PEDF protein. Mutant SERPINF1[R99A] and SERPINF1[H105A] were generated by targeting WT SERPINF1 cDNA by site-directed mutagenesis. WT and mutant SERPINF1 cDNA were cloned into p3XFLAG-CMVTM-9 expression vector (Sigma). The resulting expression vectors were transfected into BHK cells using Lipofectamine® 2000. After selection with 0.4 mg/ml neomycin, stably transfected cells were cycled between serum and serum-free media every 24 h. Conditioned serum-free media containing the recombinant proteins were harvested, concentrated by ammonium sulfate precipitation, and purified by a two-step ion-exchange column chromatography as described before (24).
Cell Culture
The retina precursor cell line R28 is widely used and was originally derived from a rat retina at the age of 6 days infected with the psi2 E1A adenovirus and then cloned by limiting dilution (see Kerafast website). R28 cells (Gift from Gail Seigel, University of Buffalo) were cultured in DMEM with 10% FBS and 1% penicillin/streptomycin at 37 °C and 5% CO2 (23). Cell passages 50–58 were used in experiments in this study.
Modeling and Molecular Docking
The P1 peptide folding was modeled using the ab initio fragment assembly protocol of Rosetta (25). One million decoys were generated, and the top 0.5% of models by Rosetta score was clustered using MaxCluster (26). The most representative models from the top five clusters by size were globally docked onto the native PEDF structure (Protein Data Bank code 1IMV, chain A) using both the docking protocol of Rosetta (27) and clusPro (28). For global docking with Rosetta, 10,000 docking models were generated and clustered, and the representative models for each of the top 10 clusters by score were kept, resulting in 50 possible docking models. Each of the 50 possible docking models was subjected to a local refinement docking run, wherein the ligand (P1 peptide) was randomly displaced from its original position and allowed to reposition itself using a least squares search. The hallmark of realistic docking is the so-called “funnel-shaped” plot of score versus distance from the original docking position (25), and clear funnels were seen in only two of the local refinements. For global docking with clusPro, the 5 possible ab initio models gave a total of 60 possible docking models. Each of these docking models was subjected to the same local refinement docking procedure, and only one showed a clear funnel-shaped plot. Of the three docking models, one of the models generated using the all-Rosetta method and the one from clusPro show the same arrangement of P1 bound to the neurotrophic region of PEDF (positions 44–121). The models were visually inspected, and no unusual steric clashes and chemical bonding violations were seen. The modeling data were generated by an individual with no knowledge of the other data in this paper. A file with coordinates of the models is found at the National Institutes of Health Helix Systems website (supplemental data).
Circular Dichroism
To assess the secondary structure of peptide P1 CD was performed. Peptide P1 was diluted in 10 mm sodium phosphate, pH 7.4, 50 mm NaF, and 0.05% Tween 20 to a concentration of 0.1 mg/ml. The CD spectrum was scanned from 190 to 260 nm at 25 °C. Using a water bath to increase the temperature, additional scans were taken at 60, 70, and 80 °C. CD spectra of other peptides used in the study are in Fig. 1D.
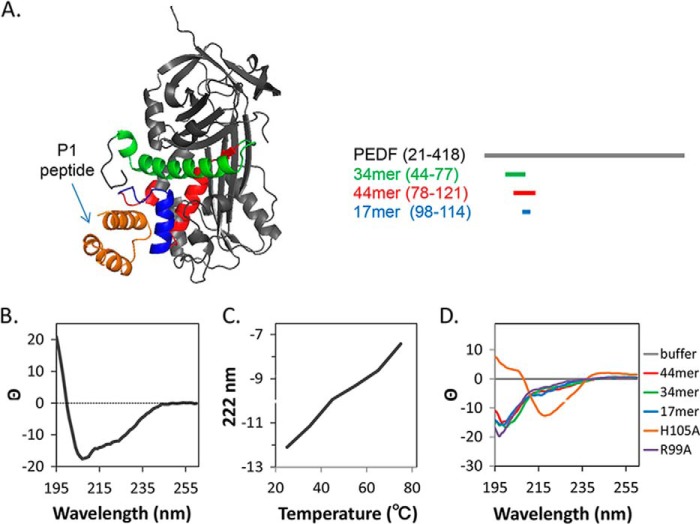
FIGURE 1.
Modeling of binding interaction of LBD of PEDF-R to PEDF. A, the structure of the LBD peptide P1 derived from the ab initio fragment assembly protocol of Rosetta is shown in orange. The resultant P1 peptide structure docked to the PEDF crystal structure (Protein Data Bank code 1IMV) using Rosetta program is shown. P1 peptide docked to a cleft that contained a solvent-exposed region corresponding to α-helix C within the residues 98–114 (17-mer; blue) of the neurotrophic 44 amino acid region (red). The antiangiogenic peptide region (34-mer; green) was not part of the docking region. A linear representation of the PEDF structure is given at right with identical color code as above. B, the CD spectrum of peptide P1 contained the negative ellipticity at 222 and 208 nm and a positive peak at 190 nm that are indicative of α helical structure. C, additionally, the CD spectrum of peptide P1 changed to reflect peptide unfolding as the temperature was increased from 25 to 80 °C, indicating that the P1 fragment is folded at 25 °C and unfolds at higher temperatures. D, CD spectra of peptides used in this study are shown.
Ligand Blot Binding Assay
Binding of fluorescently tagged PEDF or peptides to immobilized P1 peptide was assayed by ligand blot, as reported previously (23). P1 peptide (1 μg) was immobilized on nitrocellulose by vacuum filtration using the convertible filtration manifold system (Invitrogen, Life Technologies). Peptides immobilized in the membrane were stained with Ponceau Red (Sigma-Aldrich) to confirm peptide immobilization and relative amount. Strips of the nitrocellulose membrane containing P1 peptide slots were sliced and incubated in blocking solution (1% BSA in TBS (50 mm Tris, pH 7.5, 150 mm NaCl) plus 0.1% (v/v) Tween 20) for 1 h at 25 °C. Then the strips were incubated in solutions (1 ml) containing each fluorescently tagged PEDF or peptides at concentrations, as indicated, in TBS plus 0.05% Tween 20 for 2 h at room temperature in the dark. The bound material was detected by imaging the fluorescent tag on a laser scanner (TyphoonTM 9410; GE LifeSciences).
Saturation binding experiments were performed from ligand blots with fluorescently tagged ligand at concentrations that varied as indicated. The amount of bound material detected on the scanned images was quantified based on the fluorescence intensity using the analyze gel tool in ImageJ. Each data point was the average of triplicate assays ± standard error. The plots were fit in GraphPad Prism using nonlinear fit with an equation for binding − saturation, one site − specific binding to obtain Kd values.
Pulldown Assay
Binding of full-length PEDF-R and PEDF peptides was determined via pulldown assay. Full-length His6/Xpress-tagged PEDF-R was synthesized in an in vitro translation system (yield of ∼933 ng per 50 μl of reaction) as described before (23). The 34-mer and 44-mer peptides chemically synthesized containing FITC at their N terminus end were used. The in vitro expression reaction was divided into two equal aliquots to ensure equal amounts of recombinant PEDF-R protein (approximate 420 ng) in each of the 34-mer and 44-mer pulldown mixtures, and then FITC labeled 34-mer or 44-mer in Tris buffered saline plus 0.1% Tween 20 (TBS-T) was added to each (final concentration, 300 nm). Mixtures were at an estimated molar ratio of 1:6 for PEDF-R:peptide. After incubation under rotation for 2 h at 4 °C, a suspension of Ni-NTA resin (50 μl of bed volume) (Invitrogen) prewashed in TBS-T, was added to the binding reaction mixture and incubated for 1 h at 4 °C. Following the incubation, the resin mixture was sedimented by centrifugation at 800 × g for 30 s at 4 °C. Then the resin was washed three times with 500 μl of TBS-T to remove unbound material, and the bound proteins were removed from the resin by adding 20 μl of 1× NuPAGE® LDS sample buffer (Life Technologies; 141 mm Tris base, 2% LDS, 10% glycerol, 0.51 mm EDTA, 0.22 mm SERVA Blue G, 0.175 mm Phenol Red, pH 8.5) containing 1.25 mm DTT and heating the resin at 95 °C for 5 min. The bound material was subjected to SDS-PAGE to separate the His6/Xpress-tagged PEDF-R and FITC peptides. The bound fractions were loaded on a NuPAGE® Novex® 4–12% Bis-Tris protein gel (1.5 mm, 10 well; Life Technologies, Inc.) and electrophoresed at 170 V for 40 min in NuPAGE® MES SDS running buffer (Novex®; Life Technologies). Low range standards (20,700–103,000 molecular weight), prestained mixture of six proteins were used (Bio-Rad prestained SDS-PAGE standards 161-0305, catalogue no. 161-0305). The proteins and peptides were transferred to a nitrocellulose membrane. The FITC of peptides were detected with a laser scanner (TyphoonTM 9410; GE LifeSciences). His6/Xpress-tagged PEDF-R was detected via Western blot using anti-Xpress (Invitrogen, diluted at 1:10,000) and HRP-conjugated goat anti-mouse (KPL, diluted at 1:200,000).
Surface Plasmon Resonance (SPR)
Binding of 34-mer and 44-mer peptides to immobilized P1 determined by SPR was performed using a Biacore3000, as described previously (22). Briefly, P1 peptide (5 μg/ml diluted into 10 mm sodium phosphate, pH 7.3) was immobilized on a CM5 sensor chip (Biacore) via amine-coupling chemistry (1500 reference units). A reference surface without peptide was prepared using the same procedure. During binding analysis, the P1 chip was paired with a reference chip, and the binding of 300 μl of PEDF peptide (5 μm in TBS50-T) at a flow rate of 5 ml/min was measured. Subsequently, the chips were washed with TBS50-T, and peptide dissociation was followed for 3 h at the 5 ml/min. The chips were regenerated with regeneration buffer (1% octyl-glucoside, 500 mm NaCl, 10 mm NaOH) at a flow rate of 20 ml/min.
Fluorescence Polarization
The binding assessment of the 17-mer alanine scan peptides was performed using fluorescence polarization. FITC-P1 peptide (20 pm) in TBS, 0.05% Tween 20 was mixed with each of the 17-mer alanine scanning peptides at 0.2 μm, or as indicated, in 50 μl final volume per reaction mixture and incubated at 25 °C for 1 h. The fluorescence polarization was measured with Envision (PerkinElmer) plate reader. The polarization (mP) of each sample was calculated using Equation 1, where S is light detected parallel to the excitation plane, and P is light detected perpendicular to the excitation plane.
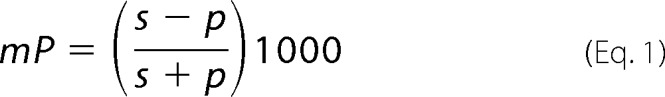 |
The change in polarization (ΔmP) is the polarization for the sample normalized to the polarization of free FITC-P1 peptide.
R28 Cell Survival Assay
The biologic activity of PEDF-derived peptides was determined by the ability to protect retinal cells from serum starvation-mediated cell death, as described (23). Rat retina R28 cells (1.5 × 104/well) were cultured in an 8-well Nunc chamber slide for 16 h in 5% FBS. The cells were then washed with PBS and serum-free media containing effectors was added and incubated at 37 °C for 48 h. For treatment with P1 peptide, PEDF and PEDF-derived peptides were preincubated in 50 μl of medium and rocked at 4 °C for 4 h prior to addition to cells at the indicated final concentration. Antiapoptotic effect was evaluated by performing the TUNEL assay at the end point using an ApopTag fluorescein in situ apoptosis detection kit (Chemicon International, Temecula, CA) following the manufacturer's protocol. The nuclei were counterstained with Hoechst, and images were taken using fluorescence microscopy. Five different fields were imaged per treatment from two independent wells in each experiment. Cells were manually counted using ImageJ 1.42 cell counter plugin. The data are shown as the ratio of TUNEL-positive cells over the total cells per field.
In Vivo Retina Protection
All procedures on mice were conducted at the Centro Servizi Stabulario Interdipartimentale, approved by the Ethical Committee of University of Modena and Reggio Emilia (Protocol number 106 22/11/2012) and by the Italian Ministero della Salute and were in accordance with the Association for Research in Vision and Ophthalmology Statement for the Use of Animals in Ophthalmic and Vision Research. C3H/HeN (rd1) mice were purchased from Harlan Italy (Udine, Italy). Mice were maintained in 12-h light/dark cycles. For intravitreal administration, rd1 mice at the age of 11 days (postnatal day 11; PN11) were anesthetized with an intraperitoneal injection of 2 ml/100 g of body weight of avertin (1.25% (w/v) 2,2,2-tribromoethanol and 2.5% (v/v) 2-methyl-2-butanol; Sigma, Milan, IT). Subsequently, 0.5 μl of PBS containing different concentrations of PEDF or PEDF peptides with or without 10 times molar excess (10×) of P1 peptide were delivered intravitreously via a trans-scleral transchoroidal approach. Control eyes received vehicle or P1 peptide only. Treated mice were sacrificed 16 h postinjection at the age of 12 days (PN12).
Analysis based on counting of dying cells (i.e. TUNEL+) is a reliable estimation of neuroprotection because degeneration is very fast in this model (29). Eyes were enucleated, fixed, embedded in paraffin, and sectioned as previously described (29). DNA fragmentation in dying cells was detected by TUNEL kit (fluorescein; Roche) according to the producer's protocols. Nuclei were then stained with DAPI (Sigma) for 2 min. The slides were mounted with mowiol 4–88 (Sigma) and analyzed at an Axioskop 40 fluorescence microscope (Zeiss). TUNEL-positive cells were counted in sections passing through the optic nerve and expressed as percentages of the total number of photoreceptors (number of DAPI stained in the photoreceptor outer nuclear layer) in each section analyzed. Paired Student's t test analysis compared data derived from at least three different untreated and at least three different treated rd1 retinas or data from at least three different retinas treated with PEDF or PEDF-derived peptides in the presence or absence of the P1 peptide.
Results
Molecular Docking of the LBD of PEDF-R to PEDF
Although the three-dimensional structure of PEDF has been resolved (Protein Data Bank code 1IMV), there are limited experimental and theoretical data on the structure of PEDF-R. Therefore, the structure of P1 peptide (human PEDF-R positions Thr210–Leu249) was modeled using the ab initio fragment assembly protocol of Rosetta. The top five clusters contained 2710, 1200, 249, 235, and 185 members each, respectively. All representative models were dominated by helical structure. The theoretical P1 peptide structure contained three α helices, which was confirmed by circular dichroism (Fig. 1). The modeled P1 peptide structure was docked to the three-dimensional PEDF structure (Protein Data Bank code 1IMV) using two modeling algorithms: Rosetta and ClusPro. The docking models were verified by the strong tendency of the partners to return to the same configuration after random displacement. Both algorithms yielded a model with P1 peptide binding to a cleft on PEDF that contains the 44-mer region (Fig. 1A). The model predicts that the interaction between PEDF and the P1 fragment is centered around Glu101, with a combination of backbone and side chain atoms providing contacts and that likely the first six residues of the 44-mer do not affect binding. The majority of interactions between the 44-mer of PEDF and peptide P1 seem to arise from backbone atoms of PEDF contacting the side chains of the P1 fragment. The results suggest that P1 peptide folds into α helical structure and binds to a cleft in PEDF that contains the neurotrophic 44-mer, with helix C of PEDF being in closer proximity to the P1 peptide region of PEDF-R.
Binding of PEDF Peptide Fragments to PEDF-R
To identify a receptor binding site of PEDF, peptides 34-mer and 44-mer derived from biologically active regions were assayed for ability to interact with PEDF-R protein using a pulldown approach. Binding was performed with recombinant His6/Xpress-tagged PEDF-R protein mixed with a 6-fold molar excess of each peptide labeled at its N terminus with FITC. After separation of unbound, the FITC peptides bound to the recombinant His6/Xpress-PEDF-R in Ni-NTA resin were detected. We found that FITC-44-mer bound to the PEDF-R, whereas no detection of bound FITC-34-mer was observed (Fig. 2A, lanes 1 and 5, respectively). FITC peptides mixed with aliquots of in vitro translation reaction without PEDF-R expression vector (Fig. 2A, lanes 2 and 6), and FITC peptides with Ni-NTA beads only and without in vitro translation reaction mixtures (Fig. 2A, lanes 3 and 7) served as negative controls. Xpress tag immunoreactivity confirmed that the amount of His6/Xpress-tagged PEDF-R was equal in each pulldown mixture (Fig. 2A, bottom blot).
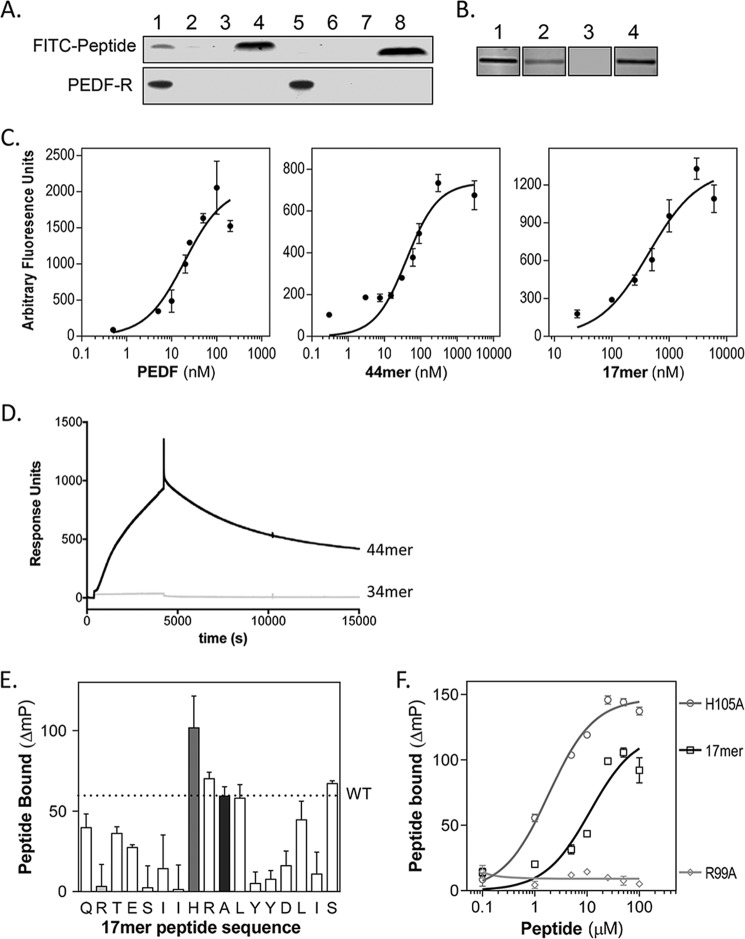
FIGURE 2.
PEDF 44-mer and 17-mer binding to PEDF-R. A, pulldown of FITC-labeled 44-mer and 34-mer peptides bound to recombinant His6/Xpress-tagged PEDF-R with Ni-NTA beads. The top and bottom blots show detection of FITC peptides by fluorescence imaging and PEDF-R detected by immunoreactivity with anti-Xpress, respectively. Lanes 1–4 correspond to 44-mer, and lanes 5–8 to 34-mer, with lanes 1 and 5 for FITC peptide with in vitro expressed His6/Xpress-PEDF-R, lanes 2 and 6 for FITC peptide with in vitro expression reaction without PEDF-R plasmid, and lanes 3 and 7 for FITC peptides incubated without in vitro expression reaction. Total amount of FITC peptides in each binding reaction was loaded in lanes 4 and 8. B, ligand blot of P1 peptide immobilized on nitrocellulose and incubated with solutions of fluorescently labeled ligands. Each condition was performed in triplicate, and a fluorescence image of one representative slice is shown. Concentration of ligands were as follows: lane 1, 50 nm Fl-PEDF; lane 2, 3 μm FITC-44-mer; lane 3, 3 μm FITC-34-mer; lane 4, 3 μm FITC-17-mer. C, saturation binding experiments performed from ligand blots. Concentrations of the fluorescently tagged ligands varied as indicated in the x axis. The y axis corresponds to estimated intensities of fluorescence in scanned ligand blots using ImageJ. Each data point is the average of triplicate assays ± standard error. The plots were fit in GraphPad Prism, and the estimated Kd values for PEDF, 44-mer, and 17-mer were 9.1 ± 1.1, 40.8 ± 5.9, and 412.6 ± 80.1 nm, respectively. D, real time binding to P1 peptide by SPR. Two surfaces were prepared without (reference) and with immobilized P1 peptide on CM5 sensor chips via amine-coupling chemistry. During binding analysis, the P1 chip was paired with a reference chip. The binding of 34-mer or 44-mer peptide (injections of 300 μl at 5 μm) in TBS50-T at a flow rate of 5 ml/min was measured (response units). Subsequently, the chips were washed with TBS50-T, and peptide dissociation was followed for 3 h at 5 ml/min. Sensograms are shown. E and F, alanine scan of 17-mer and binding to peptide P1. E, fluorescence polarization mixtures of each peptide at 10 μm and FITC-P1 peptide at 20 pm. The plot shows the change in FITC-P1 peptide polarization due to 17-mer binding for each peptide. The letters on the x axis show the amino acid in the 17-mer peptide substituted for an alanine in single letter code ordered as in the sequence. The A in the tenth position is for WT 17-mer peptide. Each bar corresponds to binding of each peptide of the alanine scan set. The gray, light gray, and black bars correspond to the WT 17-mer, 17-mer[R99A], and 17-mer[H105A], respectively. F, binding curves of 17-mer peptides. The plot shows the change in FITC-P1 peptide polarization when titrated with peptides 17-mer, 17-mer[H105A], and 17-mer[R99A] at concentrations as indicated with 20 pm FITC-P1. Each point is the average ± standard error of triplicate assays for each condition. The plots were fit in GraphPad Prism, and the estimated half-maximum polarization change (EC50) values were 11.42 ± 3.16 μm for 17-mer[H105A] and 1.83 ± 0.197 μm for 17-mer.
A ligand blot approach was used to examine the binding between PEDF peptides and the PEDF-R ectodomain containing the LBD, peptide P1. Binding was assessed by incubating membrane slices containing immobilized P1 peptide in solutions of increasing concentrations of FITC-34-mer or FITC-44-mer, as ligands (Fig. 2, B and C). Fluorescein-conjugated PEDF (Fl-PEDF) was a positive control. The binding data revealed that only 44-mer bound P1 peptide with an apparent affinity of ∼4.5 times lower than that of the full-length PEDF, whereas binding of the 34-mer was undetectable.
Real time binding to P1 peptide was examined by SPR using solutions of 34-mer and 44-mer injected over sensor chips with immobilized P1 peptide (Fig. 2D). Sensograms relative to the reference surface (no P1) showed that only the 44-mer, and not the 34-mer, associated to P1, and that dissociation was observed after injections of the 44-mer solution were stopped. The results indicate that the 44-mer interaction with P1 peptide was reversible.
Given that the first 20 amino acids in the region spanning positions 44–121 are part of a large hydrophobic α helix that is buried in the core of the folded PEDF protein and unlikely to contribute significantly to binding, a fragment of 17 amino acids (residue positions Gln98–Ser114) was synthetized and tested for binding. This region contains the solvent-exposed amino acids and helix C in the folded PEDF protein. Ligand blotting revealed that the FITC-17-mer peptide bound to P1 peptide with an apparent affinity ∼10 and 45 times lower than that for the 44-mer and for PEDF, respectively (Fig. 2D). The estimated Kd values for PEDF, 44-mer, and 17-mer were 9.1 ± 1.1, 40.8 ± 5.9, and 412.6 ± 80.1 nm, respectively. The results imply that the 17-amino acid fragment contains a PEDF-R binding region.
Roles of Arginine 99 and Histidine 105 in PEDF Binding to PEDF-R Peptide P1
To identify the contribution of each side chain in the receptor binding interaction, an alanine scanning methodology was utilized. In total, 17 peptides were designed from the region of PEDF composed of residue positions 98–115: 16 with a single alanine alteration in each consecutive amino acid position and 1 with unmodified sequence given that the 17-mer peptide already contains an alanine at PEDF residue position 107 (Ala107) (Table 1). Binding was assayed by measuring the change in polarization of peptide FITC-P1 in the presence of PEDF peptides (bound) compared with FITC-P1 alone (unbound). The change in FITC-P1 polarization caused by unmodified 17-mer (WT) binding was 59 ΔmP as illustrated by a dotted line in Fig. 2E. Values of peptides with lower affinity for peptide FITC-P1 than the unmodified 17-mer will fall below the line, and those with higher affinity will fall above the line. The alanine scan revealed several amino acid side chains that when altered to those of alanine affected the P1:17-mer interaction. The arginine in the second position in the 17-mer (R99A) and the histidine in the eighth position (H105A) had significant effects on decreasing and increasing P1 peptide affinity, respectively. Fig. 2F shows the change in polarization normalized to free FITC-P1 in the presence of 17-mer, 17-mer[R99A], or 17-mer[H105A] peptides at concentrations between 0 and 100 μm. A titration of the 17-mer[R99A] reveals that even at the highest concentration, there was no detectable binding to the FITC-P1 peptide. The 17-mer[H105A] variant bound to FITC-P1 with a relative affinity 6-fold higher than that of unmodified 17-mer peptide. The data demonstrate the importance of the side chain of Arg99 in PEDF for binding P1 peptide and reveal that an alanine side chain in position 105 would facilitate interactions between ligand and receptor.
Protective Effects of 17-mer Peptides in Retina Precursor Cells
We investigated the cytoprotective effects of PEDF peptides on rat retina R28 cells. The R28 cell line was chosen because PEDF binds the PEDF-R found in their plasma membranes and protects them from death induced by serum starvation (15, 23, 30, 31). Moreover, peptide P1 can hinder the PEDF-mediated protective effects on these cells (23). Fig. 3 (A and B) show that treatments of serum-starved R28 cells with peptides 44-mer, 17-mer, and 17-mer[H105A] for 48 h, each caused a significant decrease of 85, 69, and 68% in the number of TUNEL-positive cells relative to no addition, respectively,like PEDF (85%). However, treatment with 34-mer and 17-mer[R99A] revealed only 30 and 28% decreases in the number of TUNEL-positive cells, which were not significant. We also examined the blocking effects of P1 peptide on the protective activity of PEDF peptides. Serum-starved R28 cells were treated with preincubated mixtures of 44-mer, 17-mer, or 17-mer[H105A], each with 1- and 10-fold molar excess of P1 peptide as soluble receptor fragment. P1 peptide reduced effectively the number of TUNEL-positive nuclei of 44-mer, 17-mer, and 17-mer[H105A], with both P1 peptide concentrations (Fig. 3C). The percentage of TUNEL-positive cells with P1 peptide (35–40%) was at least 2-fold that without P1 peptide (15–20%) and matched those values without PEDF peptides (40%). Peptide P1 by itself did not have significant effect, nor did it change the percentage of TUNEL-positive cells observed for 17-mer[R99A]. The results demonstrate that the PEDF-R ectodomain peptide P1 blocked efficiently the protective effects of 44-mer, 17-mer, and 17-mer[H105A] peptides, as shown before for PEDF, likely by interfering with their binding to cell surface PEDF-R.
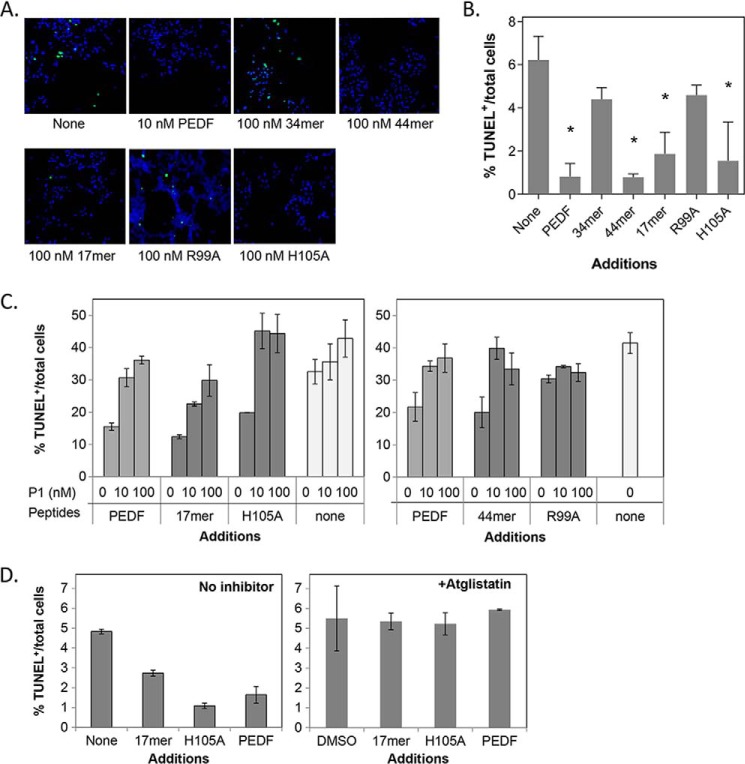
FIGURE 3.
Protective effects of PEDF peptides in cultured cells. R28 cells in serum-free media were treated with PEDF-derived peptides for 48 h. Cytoprotection was assayed by counting TUNEL-positive nuclei (green). A, representative images from each condition show merged TUNEL+ nuclei and Hoechst stained nuclei (blue, total cells). Concentrations of peptides were as indicated. B, quantification of the protective effects by PEDF peptides. Each bar corresponds to the average of the percentage of TUNEL-positive nuclei per total number of cells from triplicate wells ± S.D. (error bars) with treatment as indicated in A. *, p < 0.03. C, blocking of the protective effects of PEDF peptides by P1 peptide. Treatments were with PEDF or peptides at 10 nm mixed with P1 peptide at the indicated concentrations. D, cells were pretreated with atglistatin (3.5 μm) in DMSO or DMSO (vehicle control) for 1 h before a 48-h treatment with or without PEDF peptides and protein at 10 mm (indicated). Cells were fixed and processed for TUNEL staining and counterstained with Hoechst dye for the nucleus. Quantification of TUNEL-positive nuclei is shown. Each bar corresponds to the average of percentage of TUNEL-positive nuclei per total number of cells from duplicate wells per assay ± S.D. Each plot shows assays performed with an individual batch of R28 cells. PEDF and no additions were positive and negative controls, respectively.
We have shown previously that the phospholipase activity of PEDF-R is inhibited by bromoenol lactone and that this inhibitor attenuates the PEDF-mediated cytoprotective activity in R28 cells (22, 23). In this study, we used atglistatin, a selective and competitive inhibitor of the enzymatic activity of PEDF-R/ATGL (32), and investigated its effects on PEDF-mediated cytoprotection. R28 cells were incubated in media with atglistatin before adding PEDF and PEDF peptides. Fig. 3D shows that atglistatin completely abolished the survival activity of PEDF and the peptides 17-mer and H105A, and no effect was seen with vehicle (DMSO) alone. The observations imply that the cell survival effects of PEDF and peptides specifically depend on the enzymatic activity of PEDF-R.
Photoreceptor-protective Effects of Small PEDF Peptides in Vivo
We also assayed the neuroprotective effects of PEDF, 44-mer and 17-mer peptides in a retina degeneration model of retinitis pigmentosa, the rd1 mouse. The rd1 mouse is a well characterized animal model of retinitis pigmentosa that lacks phosphodiesterase activity, which affects phototransduction and photoreceptor viability. The rd1 mutant retina is characterized by a peak of photoreceptor cell death in mice of 11 days of age (PN11) (29, 33). We chose this time during the retinal degeneration process to assess effects of PEDF and of PEDF peptides on photoreceptor cell death. Purified human recombinant PEDF protein was injected into the vitreous of rd1 mutant mice at PN11, and cell death in photoreceptors was evaluated the following day (PN12) by TUNEL. Fig. 4A shows that PEDF at 2, 6, 10, and 20 pmol per eye reduced the number of TUNEL-labeled photoreceptors relative to the vehicle injected contralateral eyes (negative control, 0 in the graph). Doses above 6 pmol per eye plateau to 50% of those without PEDF. The neuroprotective effects of PEDF at 6 pmol per eye assessed by TUNEL staining are shown in Figs. 5 (A, B, A′, and B′). We also tested the neuroprotective activity of PEDF-derived peptides in vivo. Intravitreal injections of peptides 44-mer and 17-mer at doses of 6 pmol per eye and above decreased cell death of rd1 mutant photoreceptors of ∼30 and 50%, respectively (Fig. 4B). Peptide 17-mer[H105A] decreased the number of TUNEL-positive nuclei of rd1 mutant photoreceptors of more than 50% starting at doses of 2 pmol per eye and was more effective compared with PEDF (Fig. 4, A and B). Peptide 17-mer[R99A] was an ineffective peptide also in vivo. The neuroprotective effects of PEDF-derived peptides evaluated by TUNEL labeling are shown in Fig. 5 (D, E, G, H, J, K, M, N, D′, E′, G′, H′, J′, K′, M′, and N′).
FIGURE 4.
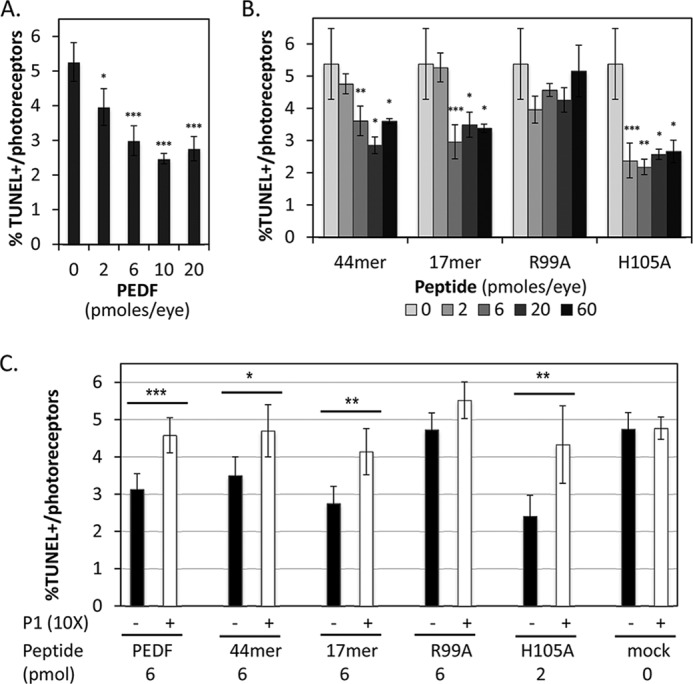
Protective effects of PEDF and PEDF peptides in vivo. PEDF or PEDF peptides were intravitreally injected into rd1 mutant eyes of PN11 mice, and photoreceptor survival was assayed a day later by counting TUNEL-positive nuclei of photoreceptors in retina sections. A, plot of percentages of TUNEL-positive photoreceptor nuclei (TUNEL-positive/total number of photoreceptor nuclei) as a function of amount of PEDF injected. B, plot of percentages of TUNEL-positive photoreceptor nuclei (TUNEL-positive/total number of photoreceptor nuclei) as a function of amount of peptide 44-mer, 17-mer, 17-mer[R99A], and 17-mer[H105A] injected. C, blocking of the neuroprotective effects of PEDF and PEDF derived peptides by P1 peptide. PEDF and peptides were injected at the indicated concentrations mixed (+) or not mixed (−) with 10-fold excess of P1 peptide. Each point corresponds to the average of the percentages of TUNEL-positive photoreceptors (TUNEL-positive/total number of photoreceptor nuclei) from at least three eyes ± S.D. For Student's t test: ***, p ≤ 0.001; **, p ≤ 0.01; *, p ≤ 0.05.
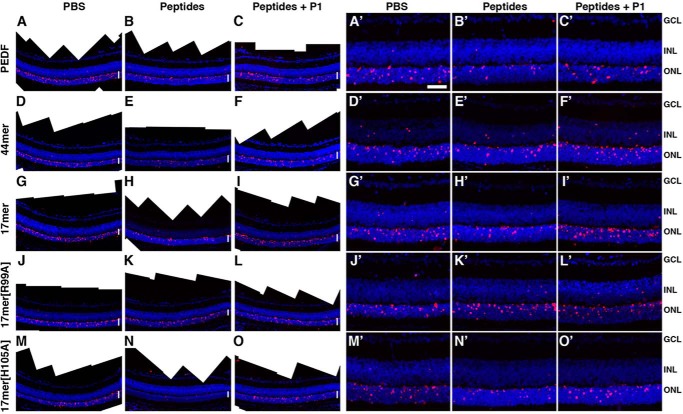
FIGURE 5.
Reduction of dying cells in retinas treated with PEDF and PEDF peptides. A–O, photomerge images of sections visualize a larger area of a representative retina labeled by TUNEL (red). A′–O′, microphotographs of sections of retinas labeled by TUNEL (red) are shown. Nuclei are stained with DAPI (blue). A, A′, D, D′, G, G′, J, J′, M, and M′, mock treated control retinas. B and B′, retina injected with 6 pmol of PEDF. C and C′, contralateral retina injected with 6 pmol of PEDF and 60 pmol of P1 peptide. E and E′, retina injected with 6 pmol of 44-mer. F and F′, contralateral retina injected with 6 pmol of 44-mer and 60 pmol of P1 peptide. H and H′, retina injected with 6 pmol of 17-mer. I and I′, contralateral retina injected with 6 pmol of 17-mer and 60 pmol of P1 peptide. K and K′, retina injected with 6 pmol of 17-mer[R99A]. L and L′, contralateral retina injected with 6 pmol of 17-mer[R99A] and 60 pmol of P1 peptide. N and N′, retina injected with 2 pmol of 17-mer[H105A]. O and O′, contralateral retina injected with 2 pmol of 17-mer[H105A] and 20 pmol of P1 peptide. White vertical lines in A–O indicate the layer of photoreceptor nuclei (outer nuclear layer). Scale bar in A′, 100 μm. GCL, ganglion cell layer; INL, inner nuclear layer; ONL = outer nuclear layer.
To test the contribution of PEDF-R, we injected PEDF or peptides mixed with 10 times molar excess of P1 peptide to block PEDF/PEDF-R interaction. P1 peptide reduced the PEDF-protective effects on the rd1 photoreceptors, implying that PEDF acted via PEDF-R on photoreceptor survival in vivo (Fig. 4C), observations that are in agreement with our results using cells in culture (see above). P1 peptide attenuated the protective effects the 44-mer, 17-mer, and 17-mer[H105A] (Fig. 4C), as was observed with the full-length PEDF (above). Fig. 5 shows the blocking effects of P1 peptide on PEDF, 44-mer, 17-mer, and 17-mer[H105A]. P1 peptide had neither negative nor positive effects on mock injected and 17-mer[R99A] injected rd1 mutant retinas (Fig. 5, L and L′). The results imply that interactions of the fragment of PEDF comprised of residues 98–114 with the P1 peptide region of PEDF-R could trigger downstream effects for protecting the dystrophic retina.
Full-length PEDF with Alterations R99A and H105A
We investigated the effects of R99A and H105A modifications in the full-length PEDF on binding to the receptor and survival activity. Recombinant human PEDF proteins with positions Arg99 or His105 changed to alanine and both with a 3X-FLAG tag at the N terminus were prepared. Binding assays to the PEDF-R peptide P1 showed that a single alteration in Arg99 to alanine resulted in loss of binding, whereas an increase in bound material was observed with the H105A alteration in PEDF (Fig. 6A). Binding of the tagged PEDF versions was lower than that of untagged Fl-PEDF. Effects of the proteins on R28 cell survival with the PEDF[R99A] protein had lower efficacy than that with of PEDF[H105A] (Fig. 6B), but both had lower effects than that of untagged PEDF. Similarly, in rd1 mice in vivo, the H105A was more efficacious in protecting photoreceptors from death than the unmodified PEDF (Fig. 6C), but both were less active than the untagged protein as shown above in Fig. 4. When we analyzed how much PEDF or PEDF[H105A] rescues cell death comparing each injected eye to the contralateral, we noted that PEDF[H105A] always performed better than PEDF (the percentage of rescue of dying cells for 2, 6, and 10 pmol of PEDF was 15, 16, and 19%, whereas for PEDF[H105A] it was 26, 29, and 31%). The results imply that R99A and H105A alterations in the full-length proteins reflected the effects observed with the 17-mer[R99A] and 17-mer[H105A] peptides.
FIGURE 6.
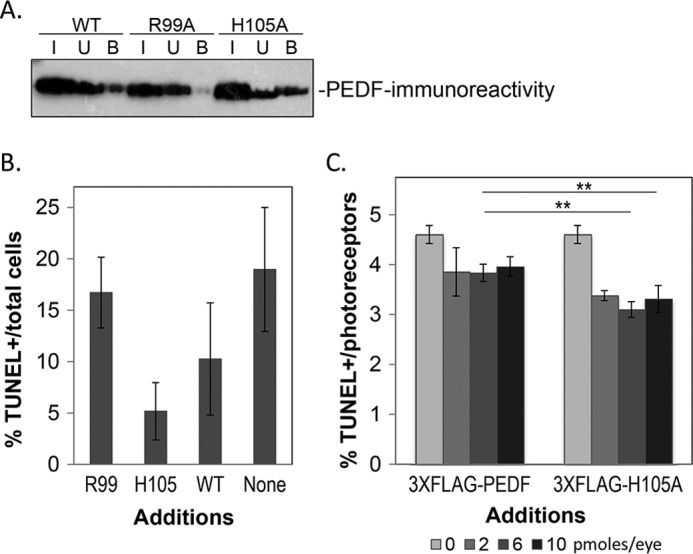
Full-length PEDF with alterations in Arg99 and His105. Full-length PEDF proteins were expressed using an expression cassette for PEDF with a 3X-FLAG tag fused to its N-terminal end. The recombinant proteins were secreted by BHK cells stably transfected with expression vectors containing the particular mutated SERPINF1 cDNA and purified by a two-step ion exchange column chromatography. A, binding assays by peptide P1 affinity chromatography of each recombinant protein was performed, and input (I), unbound (U), and bound (B) material were resolved by SDS-PAGE followed by Western blot using a specific anti-PEDF antibody. Binding reactions and washes were with 50 mm Tris, pH 7.5, 300 mm NaCl, 0.005% Tween 20. Incubations were at 4 °C for 1 h. B, the protective effects of recombinant 3X-FLAG-PEDF proteins assayed in R28 cells. R28 cells in serum-free media were treated with effectors for 48 h. Cytoprotection was assayed by counting TUNEL-positive nuclei relative to total number of cells counted from (Hoechst stained nuclei). Each bar corresponds to the average of percentage of TUNEL-positive nuclei per total number of cells from duplicate wells per assay ± S.D. The recombinant FLAG-fused PEDF proteins are indicated as Arg99, His105, and WT. SFM indicates no treatment. C, protective effects of recombinant 3X-FLAG-PEDF and 3X-FLAG-PEDF[H105A] proteins in vivo. Proteins were intravitreally injected into rd1 mutant eyes of PN11 mice, and photoreceptor survival was assayed a day later as described in Fig. 4. Plot of percentages of TUNEL-positive photoreceptor nuclei (TUNEL-positive/total number of photoreceptor nuclei) as a function of amount of each protein injected. Each point corresponds to the average of the percentages of TUNEL-positive photoreceptors (TUNEL-positive/total number of photoreceptor nuclei) from at least three eyes ± S.D. For Student's t test: **, p ≤ 0.01.
Discussion
We have demonstrated that the small peptide fragment 17-mer from the neurotrophic region of PEDF retains the affinity for PEDF-R and the retinoprotective property of the full-length PEDF. The findings are significant given that PEDF-R is a critical receptor for the PEDF cytoprotective activity (23). The data reveal the region of residues 98–114 of PEDF is retinoprotective and contains a novel site to bind PEDF-R, which in the three-dimensional structure of PEDF maps to a solvent-exposed region with helix C. We provide novel PEDF fragments from this 17-amino acid region with enhanced and attenuated individual affinities for PEDF-R and neuroprotective effects and identify amino acid positions important for the ligand/receptor interactions. The findings led us to conclude that the side chains of amino acids in 17-mer (e.g. positions 99 and 105) are available to interact with the LBD of PEDF-R to likely stimulate downstream effects for protecting photoreceptors from degeneration.
The observation that the neurotrophic 44-mer peptide binds PEDF-R and its LBD (e.g. P1 fragment) agrees with the prediction from molecular docking algorithms and with the neurotrophic nature of PEDF-R. Pulldown, ligand blotting, and real time binding measured by SPR (Fig. 2) corroborated that only the 44-mer and not the antiangiogenic 34-mer associates to P1 peptide. The 17-mer fragment is the shortest known PEDF fragment with PEDF-R affinity and prosurvival activity. Its sequence is identical to previously reported ERT peptide (residues 98–114) (21) except for the first amino acid residue position (human PEDF position 98), which in 17-mer is a glutamine. It was reported that ERT induces neurite outgrowth on human retinoblastoma cells and neuroendocrine differentiation in prostate cancer cells like the full-length PEDF (21). Given that fragments ERT and 17-mer exhibited similar binding to P1 peptide (data not shown), it is expected that they share neurotrophic properties mediated by PEDF-R.
The amino acid positions crucial for receptor binding (from Fig. 2D; Arg99, Ser102, Ile104, Tyr109, and Tyr110) are predicted to be essential for the biological responses mediated by interaction with cell surface PEDF-R. Consequently, a peptide with an alteration R99A with lower affinity for the LBD is less efficacious than unmodified 17-mer, emphasizing the critical arginine at position 99 for survival activity (Figs. 3 and 4). It is possible that the positively charged side chain of Arg99 may attract acidic residues Glu230 and Glu236 of the LBD of PEDF-R that could form the ligand-receptor interface (Fig. 7). However, experiments to test the effects of ionic potential on the 17-mer:P1 peptide binding showed only s minor decrease, if any, in fluorescence polarization with NaCl concentration increases from 0.15 to 2 m (data not shown), suggesting that interactions of other nature, like hydrophobic, dominate over ionic ones. It is worth noting that the arginine residue in position 99 of PEDF is conserved in other species (Table 2; 4 fish, 2 amphibia and 3 aves, 24 mammalian, including human, mouse, rat, monkey, bovine, dog, and horse) in agreement with its requirement for function across species. In addition, serpins maspin and ovalbumin that do not bind PEDF-R (22) have an aspartic and serine in the homologous position to Arg99 of PEDF of their sequences, respectively (Table 3). It is also worth mentioning that residues in P1 peptide region of PEDF-R that are proximal to the 17-mer peptide region (positions 98–114) during docking, Glu230 and Glu236, are conserved among species (Table 4; human, bovine, mouse, rat, dog, Xenopus tropicalis, and Dario rerio). However, we cannot discriminate whether the decrease in apparent affinity of the alanine scan peptides was due to the requirement of such amino acid for interacting with the PEDF-R ectodomain or to effects in peptide conformational change as result of the alteration. In contrast, the 17-mer[H105A] peptide, with a higher observed affinity for LBD than 17-mer, is also more efficacious than its unmodified version in survival of photoreceptors in vivo. The enhanced effect could be explained by the replacement of the basic residue (His) for a neutral one (Ala), which would be in proximity to the hydrophobic Leu222 in the LBD in a more hydrophobic microenvironment to favor affinity increases (Fig. 7).
FIGURE 7.
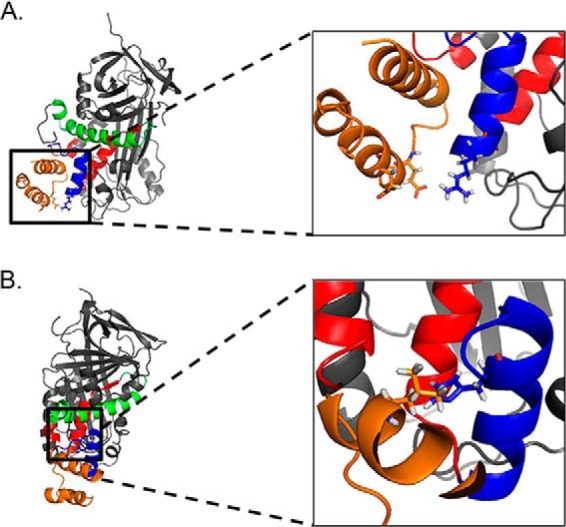
PEDF amino acids that modify binding to the P1 fragment of the PEDF receptor. P1 peptide (orange) interacts with the solvent accessible 17-mer peptide region (blue) within the neuroprotective 44-mer peptide region (red). A, the inset shows the close proximity of the positively charged amino acid Arg99 (blue) of PEDF and two negatively charged amino acids Glu230 and Glu236 (orange) of the LBD of PEDF-R that could be important for binding. B, the molecules are rotated 90° to the right on the y axis and 15° forward on the z axis relative to A. The inset shows the close proximity of the hydrophilic His105 (blue) to the hydrophobic Leu222 (orange) in the P1 fragment. Amino acids Arg99 and His105 of PEDF and Glu230, Glu236, and Leu222 are shown with side chains in sticks.
TABLE 2.
PEDF orthologue alignment
| Accession number | Class | Order | Species | Protein | Starting amino acid | Sequencea | Ending amino acid |
|---|---|---|---|---|---|---|---|
| AAA84914 | Mammalia | Primates | Human | PEDF | 71 | QRTESIIHRALYYDLISSPDIHGTYKELLDTVTAPQKNLKSASR | 141 |
| XP_003816903 | Mammalia | Primates | Bonobo | PEDF | 71 | QRTESIIHRALYYDLISSPDIHGTYKELLDTVTAPQKNLKSASR | 141 |
| XP_001155004 | Mammalia | Primates | Chimpanzee | PEDF | 71 | QRTESIIHRALYYDLISSPDIHGTYKELLDTVTAPQKNLKSASR | 141 |
| XP_002747873 | Mammalia | Primates | Marmoset | PEDF | 71 | QRTESIIHRALYYDLISSPDIHGTYKELLDTVTAPQKNLRSASR | 141 |
| EHH24345 | Mammalia | Primates | Rhesus monkey | PEDF | 71 | QRTESVIHRALYYDLISSPDIHGTYKELLGTVTAPQKNLKSASR | 141 |
| XP_003799139 | Mammalia | Primates | Small eared galago | PEDF | 71 | QQTESVIHRALYYDLINNPDIHGTYKELLATVTDAQKNFKSASR | 141 |
| XP_002826865 | Mammalia | Primates | Sumatran orangutan | PEDF | 71 | QRTESTIHRALYYDLISSPDIHGTYKELLDTVTAPQKNFKSASR | 141 |
| NP_001072130 | Mammalia | Cetartiodactyla | Boar | PEDF | 69 | QRTESSLHRALYYDLISNPDLHGTYKELLAAVTAPQKNLKSASR | 139 |
| XP_003515170 | Mammalia | Rodentia | Chinese hamster | PEDF | 72 | QRTESIIHRALYYDLISNSDIHSTYKELLASVTAPEKSLKSASR | 142 |
| NP_776565 | Mammalia | Artiodactyla | Cow | PEDF | 69 | QRTESNIHRALYYDLISNPDIHGTYKDLLASVTAPQKNLKSASR | 139 |
| NP_001071056 | Mammalia | Carnivora | Dog | PEDF | 49 | QRTESTIHRALYYDLISNPDIHSTYKELLASVTAPEKNFKSASR | 119 |
| XP_003417025 | Mammalia | Proboscidea | Elephant | PEDF | 70 | QRTESTIHRALYYDLISIADIHDTYKELLAAVTAPEKNLKSASR | 140 |
| AES06342 | Mammalia | Carnivora | Ferret | PEDF | 71 | QRTESTIHRALYYDLISNPDIHGTYKELLAAVTAPEKNFKSASR | 141 |
| NP_001166531 | Mammalia | Rodentia | Guinea pig | PEDF | 70 | QRTEATIHRALYYDMISNPDIHSTYKELLATVTAPQKNLKSASR | 140 |
| NP_001137426 | Mammalia | Perissodactyla | Horse | PEDF | 71 | QRTESSIHLALYYDLIKNPDIHGTYKELLASVTAPNKNFKSASR | 141 |
| CAX15544 | Mammalia | Rodentia | Mouse | PEDF | 70 | HRTESVIHRALYYDLITNPDIHSTYKELLASVTAPEKNLKSASR | 140 |
| EHB07111 | Mammalia | Rodentia | Naked mole rat | PEDF | 70 | QRTESIIHRALYYDVISNPDIHGTYKELLATVTGPQKNLKSASR | 140 |
| XP_001371584 | Mammalia | Didelphimorphia | Opossum | PEDF | 69 | PKTQSTIHRALYYDLITNQDIHSTYKELLATVTAAQKNLKSASR | 139 |
| XP_001507178 | Mammalia | Monotremata | Platypus | PEDF | 70 | PRTESLIHRALYYDLIHNPDIHGTYKELLATVTAPQKNLKTASR | 140 |
| EDM05207 | Mammalia | Rodentia | Rat | PEDF | 49 | QRTESVIHRALYYDLINNPDIHSTYKELLASVTAPEKNFKSASR | 119 |
| NP_001132919 | Mammalia | Artiodactyla | Sheep | PEDF | 69 | QRTESSIHRALYYDLISNPDIHGTYKDLLASVTAPQKNLKSASR | 139 |
| XP_003770166 | Mammalia | Dasyuromorphia | Tasmanian devil | PEDF | 72 | QRTESTIHRALYYDLITNPDIHGTYKELLASVTASQKNLKSASR | 142 |
| NP_035470 | Mammalia | Rodentia | Mouse | PEDF | 70 | HRTESVIHRALYYDLITNPDIHSTYKELLASVTAPEKNLKSASR | 140 |
| XP_002918078 | Mammalia | Carnivora | Panda | PEDF | 71 | QRTETTIHRALYYDLITNPDIHGTYKELLASVTAPEKNFKSASR | 141 |
| XP_003226635 | Reptilia | Squama | Carolina anole | PEDF | 263 | ERTEDTIQRALFYDLLDKAQVHEVYKGLLGSLTGPAKAFKTAAR | 333 |
| AAZ22323 | Actinopterygii | Pleuronectiformes | Japanese flounder | PEDF | 60 | ELAERQLFRALRFHTLQDPQLHNTLKDLLASLRSPGKGLSIAAR | 130 |
| XP_003457710 | Actinopterygii | Perciformes | Tilapia | PEDF | 83 | EHAQRQLYRALRYHTLQDPQLHNTLKDMLASVRTAGKGLSTAAR | 153 |
| NP_001004539 | Actinopterygii | Cypriniformes | Zebrafish | PEDF | 61 | ERAEKQIYRALRYHTLQDSQLHDTLRDLLSSLRASAKGFKSAER | 131 |
| NP_001133806 | Actinopterygii | Salmoniformes | Atlantic salmon | PEDF precursor | 61 | DRSERWLYRALRYHTLQDPQLHDTLRDLLASLRAPGKGLSIAAR | 132 |
| NP_001085983 | Amphibia | Anura | African clawed frog | PEDF | 61 | QRTESLIHRSLYYDLLNDPELHATYKELLASLTSHGSGLKSTWR | 131 |
| NP_989086 | Amphibia | Batrachia | Western clawed frog | PEDF | 61 | QRTESLIQRSLYYDLLNDPEVHATYKDLLASFTSQASGLKSTWR | 131 |
| NP_001244218 | Aves | Galliformes | Chicken | PEDF | 69 | ERTEDVISRALFYDLLNKAEVHNTYKDLLASVTGPEKSLKSASR | 139 |
| XP_003211790 | Aves | Galliformes | Turkey | PEDF | 69 | ERTEDVISRALFYDLLNKAEVHNTYKDLLASVTGPEKSLKSASR | 139 |
| XP_002197455 | Aves | Neognathae | Zebra finch | PEDF | 38 | ERTEDVISRALFYDLLNKAEVHDTYKELLSSVTGPEKSMKSASR | 108 |
a Position Arg99 of human PEDF and aligned positions are highlighted.
TABLE 3.
Alignment of the 17-mer sequence of human PEDF among chicken ovalbumin and human maspin
| SERPIN | Sequencea | Ending amino acid | Accession number |
|---|---|---|---|
| PEDF | QRTESIIHRALYYDLIS | 114 | AAA84914 |
| Ovalbumin | DSTRTQINKVVRFDKLP | 64 | AAB59956 |
| Maspin | GDTANEIGQVLHFENVK | 64 | AAA18957 |
a Position Arg99 of human PEDF is highlighted.
TABLE 4.
Alignment of P1 peptide region across species
| Species | Accession number | Sequencea | Ending amino acid |
|---|---|---|---|
| Human | AAH17280.1 | SKALFPPEPLVLREMCK | 239 |
| Mouse | AAH64747.1 | SKALFPPEPMVLREMCK | 239 |
| Mouse | BAC27476.1 | SKALFPPEPMVLREMCK | 239 |
| Rat | XP_341961.1 | SKALFPPEPMVLREMCK | 239 |
| Bovine | XP_878393.1 | SKALFPPEPLVLREMCK | 239 |
| Dog | XP_854164.1 | SKALFPPEPTVLREMCK | 239 |
| X. tropicalis | NP_001015693 | TRALFPPEPTVLGEMCQ | 240 |
| D. rerio | AAH75928.1 | SKALFPPEPKVLAEMCQ | 239 |
a Positions Glu230 and Glu236 of PEDF-R are highlighted.
Because the 44-mer and 17-mer fragments maintain the biological activity of PEDF that is attenuated by the blocking LBD peptide, it seems reasonable to conclude that they are folded somehow similarly as in the overall protein and interact with PEDF-R on cells to trigger the necessary pro-survival signaling events like the full-length PEDF. The data points out that the affinities of P1 peptide for PEDF, 44-mer, and 17-mer decrease as the molecules shorten, which may be a result of differences of the regions when they are in the overall protein and in the peptide. Lack of optimal three-dimensional structure may affect stability and binding affinity of the peptides relative to the native state in the full-length protein. However, the biological efficacy may be insensitive to peptide length as 17-mer, 44-mer, and PEDF exhibited similar potencies in vivo.
This is the first time that PEDF peptides have been tested in an animal model of retina degeneration. PEDF injected into the vitreous of mice diffuses through the retina and retinal pigment epithelium choroid, where it remains biologically active for photoreceptor survival (12) and for inhibiting neovascularization (34). Although the intravitreally injected protein is cleared in less than 24 h in mice (12), the clearance time of PEDF peptides from the eye is unknown. The fact that activity was observed for the peptides suggests that they remain available to bind and stimulate PEDF-R in the retina, which has been identified previously in photoreceptors (22). Blocking the biological effects of the small PEDF fragments with a soluble receptor fragment (LBD peptide P1) implies that the small PEDF fragments act by interacting with PEDF-R. Together with the observations that PEDF cannot bind nor stimulate LBD-truncated PEDF-R (22), the data imply that 17-mer peptides interact with the LBD region of PEDF-R to exert neuroprotective effects on photoreceptors.
Multifunctional proteins combine several independent functions on a single polypeptide chain. To this effect, independence implies that each function is assigned to a different region of the polypeptide chain. As such, PEDF is a multitasking protein that is considered an ocular guardian because it protects the retina from degeneration induced by genetic insults and from pathological neovascularization. Studies of the independent activities of PEDF are challenged by the presence of other properties of the same molecule. The approaches through the use of peptide synthesis, protein chemistry, and recombinant DNA technologies based on the three-dimensional structure of PEDF have been successful to separate and alter them individually (21, 35–38). Our results further emphasize an individual neurotrophic site in PEDF and that its interactions with the ectodomain of PEDF-R are key for the downstream effects leading to the protection of photoreceptors from degeneration. The 17-mer, as derived from the 44-mer, is not expected to confer antiangiogenic or antitumorigenic effects of the 34-mer and PEDF polypeptide (34, 39), as shown before for ERT (21). One advantage for the development of PEDF peptide-based drugs is that small molecules can be more permeable and stable and will facilitate delivery with limited side effects that may be caused by other regions of the entire molecule. Retinitis pigmentosa, a major cause for loss of vision during working age, is associated to mutations in several genes. Identification and characterization of neuroprotective factors targeting common molecular pathways activated in different forms of retinitis pigmentosa may benefit a large group of patients. Thus, we propose that PEDF-based reagents may be used in retina degeneration therapeutics given their retinoprotective properties, which is recapitulated here with the short 17-mer fragments for the rd1 mutant retina.
Author Contributions
J. K. designed the study, performed binding assays, and wrote the first draft of the paper. P. S. performed and analyzed experiments on survival assays in cells. A. C. and V. M. designed and performed experiments with animal models. J. B. expressed and purified full-length proteins and performed binding assays shown in Fig. 6. L. K. performed part of assays of Fig. 2C and survival assays with cells shown in Fig. 3D. F. P. constructed expression vectors for mutated gene. D. H. performed protein modeling. S. P. B. conceived and coordinated the study and wrote the paper. All authors reviewed the results and approved the final version of the manuscript.
Supplementary Material
Acknowledgments
We thank Dr. Gail Siegel for generously providing R28 cells, Dr. Inna Gorshkova for performing SPR experiments, and Dr. Kenneth Jacobson for critical reading of the manuscript. This study utilized the high performance computational capabilities of the Biowulf Linux cluster at the National Institutes of Health.
This work was supported, in whole or in part, by National Eye Institute, National Institutes of Health Intramural Research Program (to S. P. B.). This work was also supported by Fondazione Telethon Grant GGP06096, by E-RARE 2009 (RHORCOD), and by Italian Ministry Grant 20094CZ3M2 (to V. M.). The authors declare that they have no conflicts of interest with the contents of this article.

This article contains supplemental data.
- PEDF
- pigment epithelium-derived factor
- PEDF-R
- pigment epithelium-derived factor receptor
- LBD
- ligand-binding domain
- Ni-NTA
- nickel-nitrilotriacetic acid
- SPR
- surface plasmon resonance
- PNn
- postnatal day n.
References
- 1. Marigo V. (2007) Programmed cell death in retinal degeneration: targeting apoptosis in photoreceptors as potential therapy for retinal degeneration. Cell Cycle 6, 652–655 [DOI] [PubMed] [Google Scholar]
- 2. Becerra S. P. (2006) Focus on Molecules: Pigment epithelium-derived factor (PEDF). Exp. Eye Res. 82, 739–740 [DOI] [PubMed] [Google Scholar]
- 3. Steele F. R., Chader G. J., Johnson L. V., Tombran-Tink J. (1993) Pigment epithelium-derived factor: neurotrophic activity and identification as a member of the serine protease inhibitor gene family. Proc. Natl. Acad. Sci. U.S.A. 90, 1526–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Becerra S. P., Fariss R. N., Wu Y. Q., Montuenga L. M., Wong P., Pfeffer B. A. (2004) Pigment epithelium-derived factor in the monkey retinal pigment epithelium and interphotoreceptor matrix: apical secretion and distribution. Exp. Eye Res. 78, 223–234 [DOI] [PubMed] [Google Scholar]
- 5. Dawson D. W., Volpert O. V., Gillis P., Crawford S. E., Xu H., Benedict W., Bouck N. P. (1999) Pigment epithelium-derived factor: a potent inhibitor of angiogenesis. Science 285, 245–248 [DOI] [PubMed] [Google Scholar]
- 6. Wu Y. Q., Notario V., Chader G. J., Becerra S. P. (1995) Identification of pigment epithelium-derived factor in the interphotoreceptor matrix of bovine eyes. Protein Expr. Purif. 6, 447–456 [DOI] [PubMed] [Google Scholar]
- 7. Holekamp N. M., Bouck N., Volpert O. (2002) Pigment epithelium-derived factor is deficient in the vitreous of patients with choroidal neovascularization due to age-related macular degeneration. Am. J. Ophthalmol. 134, 220–227 [DOI] [PubMed] [Google Scholar]
- 8. Ogata N., Matsuoka M., Imaizumi M., Arichi M., Matsumura M. (2004) Decreased levels of pigment epithelium-derived factor in eyes with neuroretinal dystrophic diseases. Am. J. Ophthalmol. 137, 1129–1130 [DOI] [PubMed] [Google Scholar]
- 9. Wang Y., Subramanian P., Shen D., Tuo J., Becerra S. P., Chan C. C. (2013) Pigment epithelium-derived factor reduces apoptosis and pro-inflammatory cytokine gene expression in a murine model of focal retinal degeneration. ASN Neuro. 5, e00126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Akiyama G., Sakai T., Kuno N., Kimura E., Okano K., Kohno H., Tsuneoka H. (2012) Photoreceptor rescue of pigment epithelium-derived factor-impregnated nanoparticles in Royal College of Surgeons rats. Mol. Vis. 18, 3079–3086 [PMC free article] [PubMed] [Google Scholar]
- 11. Cao W., Tombran-Tink J., Chen W., Mrazek D., Elias R., McGinnis J. F. (1999) Pigment epithelium-derived factor protects cultured retinal neurons against hydrogen peroxide-induced cell death. J. Neurosci. Res. 57, 789–800 [PubMed] [Google Scholar]
- 12. Cayouette M., Smith S. B., Becerra S. P., Gravel C. (1999) Pigment epithelium-derived factor delays the death of photoreceptors in mouse models of inherited retinal degenerations. Neurobiol. Dis. 6, 523–532 [DOI] [PubMed] [Google Scholar]
- 13. Imai D., Yoneya S., Gehlbach P. L., Wei L. L., Mori K. (2005) Intraocular gene transfer of pigment epithelium-derived factor rescues photoreceptors from light-induced cell death. J. Cell. Physiol. 202, 570–578 [DOI] [PubMed] [Google Scholar]
- 14. Miyazaki M., Ikeda Y., Yonemitsu Y., Goto Y., Sakamoto T., Tabata T., Ueda Y., Hasegawa M., Tobimatsu S., Ishibashi T., Sueishi K. (2003) Simian lentiviral vector-mediated retinal gene transfer of pigment epithelium-derived factor protects retinal degeneration and electrical defect in Royal College of Surgeons rats. Gene Ther. 10, 1503–1511 [DOI] [PubMed] [Google Scholar]
- 15. Murakami Y., Ikeda Y., Yonemitsu Y., Onimaru M., Nakagawa K., Kohno R., Miyazaki M., Hisatomi T., Nakamura M., Yabe T., Hasegawa M., Ishibashi T., Sueishi K. (2008) Inhibition of nuclear translocation of apoptosis-inducing factor is an essential mechanism of the neuroprotective activity of pigment epithelium-derived factor in a rat model of retinal degeneration. Am. J. Pathol. 173, 1326–1338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Becerra S. P., Notario V. (2013) The effects of PEDF on cancer biology: mechanisms of action and therapeutic potential. Nat. Rev. Cancer 13, 258–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Craword S. E., Fitchev P., Veliceasa D., Volpert O. V. (2013) The many facets of PEDF in drug discovery and disease: a diamond in the rough or split personality disorder? Expert. Opin. Drug Discov. 8, 769–792 [DOI] [PubMed] [Google Scholar]
- 18. Gettins P. G., Simonovic M., Volz K. (2002) Pigment epithelium-derived factor (PEDF), a serpin with potent anti-angiogenic and neurite outgrowth-promoting properties. Biol. Chem. 383, 1677–1682 [DOI] [PubMed] [Google Scholar]
- 19. Alberdi E., Aymerich M. S., Becerra S. P. (1999) Binding of pigment epithelium-derived factor (PEDF) to retinoblastoma cells and cerebellar granule neurons. Evidence for a PEDF receptor. J. Biol. Chem. 274, 31605–31612 [DOI] [PubMed] [Google Scholar]
- 20. Bilak M. M., Becerra S. P., Vincent A. M., Moss B. H., Aymerich M. S., Kuncl R. W. (2002) Identification of the neuroprotective molecular region of pigment epithelium-derived factor and its binding sites on motor neurons. J. Neurosci. 22, 9378–9386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Filleur S., Volz K., Nelius T., Mirochnik Y., Huang H., Zaichuk T. A., Aymerich M. S., Becerra S. P., Yap R., Veliceasa D., Shroff E. H., Volpert O. V. (2005) Two functional epitopes of pigment epithelial-derived factor block angiogenesis and induce differentiation in prostate cancer. Cancer Res. 65, 5144–5152 [DOI] [PubMed] [Google Scholar]
- 22. Notari L., Baladron V., Aroca-Aguilar J. D., Balko N., Heredia R., Meyer C., Notario P. M., Saravanamuthu S., Nueda M. L., Sanchez-Sanchez F., Escribano J., Laborda J., Becerra S. P. (2006) Identification of a lipase-linked cell membrane receptor for pigment epithelium-derived factor. J. Biol. Chem. 281, 38022–38037 [DOI] [PubMed] [Google Scholar]
- 23. Subramanian P., Locatelli-Hoops S., Kenealey J., DesJardin J., Notari L., Becerra S. P. (2013) Pigment epithelium-derived factor (PEDF) prevents retinal cell death via PEDF receptor (PEDF-R): identification of a functional ligand binding site. J. Biol. Chem. 288, 23928–23942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Aymerich M. S., Alberdi E. M., Martínez A., Becerra S. P. (2001) Evidence for pigment epithelium-derived factor receptors in the neural retina. Invest. Ophthalmol. Vis. Sci. 42, 3287–3293 [PubMed] [Google Scholar]
- 25. Rohl C. A., Strauss C. E., Misura K. M., Baker D. (2004) Protein structure prediction using Rosetta. Methods Enzymol. 383, 66–93 [DOI] [PubMed] [Google Scholar]
- 26. Siew N., Elofsson A., Rychlewski L., Fischer D. (2000) MaxSub: an automated measure for the assessment of protein structure prediction quality. Bioinformatics 16, 776–785 [DOI] [PubMed] [Google Scholar]
- 27. Gray J. J., Moughon S., Wang C., Schueler-Furman O., Kuhlman B., Rohl C. A., Baker D. (2003) Protein-protein docking with simultaneous optimization of rigid-body displacement and side-chain conformations. J. Mol. Biol. 331, 281–299 [DOI] [PubMed] [Google Scholar]
- 28. Comeau S. R., Gatchell D. W., Vajda S., Camacho C. J. (2004) ClusPro: a fully automated algorithm for protein-protein docking. Nucleic Acids Res. 32, W96–W99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sanges D., Comitato A., Tammaro R., Marigo V. (2006) Apoptosis in retinal degeneration involves cross-talk between apoptosis-inducing factor (AIF) and caspase-12 and is blocked by calpain inhibitors. Proc. Natl. Acad. Sci. U.S.A. 103, 17366–17371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Notari L., Miller A., Martínez A., Amaral J., Ju M., Robinson G., Smith L. E., Becerra S. P. (2005) Pigment epithelium-derived factor is a substrate for matrix metalloproteinase type 2 and type 9: implications for downregulation in hypoxia. Invest. Ophthalmol. Vis. Sci. 46, 2736–2747 [DOI] [PubMed] [Google Scholar]
- 31. Subramanian P., Notario P. M., Becerra S. P. (2010) Pigment epithelium-derived factor receptor (PEDF-R): a plasma membrane-linked phospholipase with PEDF binding affinity. Adv. Exp. Med. Biol. 664, 29–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mayer N., Schweiger M., Romauch M., Grabner G. F., Eichmann T. O., Fuchs E., Ivkovic J., Heier C., Mrak I., Lass A., Höfler G., Fledelius C., Zechner R., Zimmermann R., Breinbauer R. (2013) Development of small-molecule inhibitors targeting adipose triglyceride lipase. Nat. Chem. Biol. 9, 785–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hauck S. M., Ekström P. A., Ahuja-Jensen P., Suppmann S., Paquet-Durand F., van Veen T., Ueffing M. (2006) Differential modification of phosducin protein in degenerating rd1 retina is associated with constitutively active Ca2+/calmodulin kinase II in rod outer segments. Mol. Cell. Proteomics 5, 324–336 [DOI] [PubMed] [Google Scholar]
- 34. Amaral J., Becerra S. P. (2010) Effects of human recombinant PEDF protein and PEDF-derived peptide 34-mer on choroidal neovascularization. Invest. Ophthalmol. Vis. Sci. 51, 1318–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Alberdi E., Hyde C. C., Becerra S. P. (1998) Pigment epithelium-derived factor (PEDF) binds to glycosaminoglycans: analysis of the binding site. Biochemistry 37, 10643–10652 [DOI] [PubMed] [Google Scholar]
- 36. Becerra S. P., Perez-Mediavilla L. A., Weldon J. E., Locatelli-Hoops S., Senanayake P., Notari L., Notario V., Hollyfield J. G. (2008) Pigment epithelium-derived factor binds to hyaluronan. Mapping of a hyaluronan binding site. J. Biol. Chem. 283, 33310–33320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Becerra S. P., Sagasti A., Spinella P., Notario V. (1995) Pigment epithelium-derived factor behaves like a noninhibitory serpin. Neurotrophic activity does not require the serpin reactive loop. J. Biol. Chem. 270, 25992–25999 [DOI] [PubMed] [Google Scholar]
- 38. Meyer C., Notari L., Becerra S. P. (2002) Mapping the type I collagen-binding site on pigment epithelium-derived factor. Implications for its antiangiogenic activity. J. Biol. Chem. 277, 45400–45407 [DOI] [PubMed] [Google Scholar]
- 39. Deshpande M., Notari L., Subramanian P., Notario V., Becerra S. P. (2012) Inhibition of tumor cell surface ATP synthesis by pigment epithelium-derived factor: implications for antitumor activity. Int. J. Oncol. 41, 219–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.